Abstract
The genomic proportion that two relatives share identically by descent—their genetic relatedness—can vary depending on the history of recombination and segregation in their pedigree. Previous calculations of the variance of genetic relatedness have defined genetic relatedness as the proportion of total genetic map length (cM) shared by relatives, and have neglected crossover interference and sex differences in recombination. Here, we consider genetic relatedness as the proportion of the total physical genome (bp) shared by relatives, and calculate its variance for general pedigree relationships, making no assumptions about the recombination process. For the relationships of grandparent-grandoffspring and siblings, the variance of genetic relatedness is a simple decreasing function of , the average proportion of locus pairs that recombine in meiosis. For general pedigree relationships, the variance of genetic relatedness is a function of metrics analogous to . Therefore, features of the aggregate recombination process that affect and analogs also affect variance in genetic relatedness. Such features include the number of chromosomes and heterogeneity in their size, the number of crossovers and their spatial organization along chromosomes, and sex differences in recombination. Our calculations help to explain several recent observations about variance in genetic relatedness, including that it is reduced by crossover interference (which is known to increase ). Our methods further allow us to calculate the neutral variance of ancestry among F2s in a hybrid cross, enabling precise statistical inference in F2-based tests for various kinds of selection.
Keywords: Meiosis, recombination, identity by descent, relatedness, heritability, genetic mapping, crossover interference
VARIANCE in the amount of DNA shared by relatives identically by descent (IBD)—variance in genetic relatedness—is an important quantity in genetics (Thompson 2013). It translates to variance in the phenotypic similarity of relatives, and is a vital component of pedigree-based estimates of heritability and the genetic variance of traits (Visscher et al. 2006, 2007; Young et al. 2018). It is also an important consideration when estimating pedigree relationships and the degree of inbreeding from genotype data (Kardos et al. 2015; Wang 2016). Variance in genetic relatedness has also been hypothesized to have important consequences for the evolution of behavior (Barash et al. 1978) and of karyotypes and recombination rates (Sherman 1979; Wilfert et al. 2007). Moreover, as we show elsewhere, variance in genetic relatedness plays a key role in selection against deleterious introgressed DNA following hybridization (Veller et al. 2019a).
For most pedigree relationships, genetic relatedness can vary because of variable patterns of recombination and segregation within the pedigree. For example, it is possible that a mother segregates only crossoverless paternal chromatids to an egg, in which case the resulting offspring inherits one half of its genome from its maternal grandfather and none from its maternal grandmother. On the other hand, if the mother shuffles her maternal and paternal DNA thoroughly into the egg, the offspring will be approximately equally genetically related to its maternal grandparents. Thus, intuitively, a higher degree of genetic shuffling within a pedigree leads to lower variance in genetic relatedness between relatives.
Previous theoretical calculations of the variance of genetic relatedness have largely been restricted to measuring genetic relatedness as the proportion of total genetic map length (in cM) shared IBD by relatives [e.g., Franklin (1977); Hill (1993b); Guo (1996); Visscher et al. (2006); a general treatment is given by Hill and Weir (2011)]. However, measuring genetic relatedness as the proportion of map length shared causes several problems, most notably when the genetic maps of the two sexes differ, as will typically be the case (Lenormand and Dutheil 2005; Sardell and Kirkpatrick 2020). This is easiest to appreciate for the genetic relatedness of an individual to its paternal and maternal grandparents, the values of which are determined in a paternal and a maternal meiosis, respectively. Theoretical calculations of the variance of cM genetic relatedness require the use of genetic map lengths from the relevant meioses, and thus, in these two cases, require different definitions of genetic relatedness: proportion of total male map length for relatedness to paternal grandparents, and proportion of total female map length for relatedness to maternal grandparents. Indeed, in the extreme case where crossing over is absent in one sex—say males, as in Drosophila—cM genetic relatedness to paternal grandparents is undefined in these calculations, because the male map length is 0 cM. Practically speaking, these problems can be sidestepped by defining cM genetic relatedness in terms of a sex-averaged genetic map, but this leads to substantial biases in theoretical calculations of its variance (Caballero et al. 2019).
A natural alternative that avoids such problems is to measure genetic relatedness as the proportion of the physical length of the genome (in bp) shared IBD by relatives. For many purposes, bp genetic relatedness will be the more appropriate measure (White and Hill 2020) and, unlike cM genetic relatedness, bp genetic relatedness is unambiguous when there are sex differences in recombination. Moreover, in the modern genomic era, it will often be the case that a species’ genome has been sequenced before its genetic map has been elucidated, so that only bp genetic relatedness can be assayed.
Translating previous calculations of the variance of cM genetic relatedness to the variance of bp genetic relatedness would be valid only under the assumption of uniform recombination rates along chromosomes. This assumption is unrealistic for most species. For example, crossovers tend to be terminally localized along human chromosomes, especially in males (Holm and Rasmussen 1983; Bojko 1985). White and Hill (2020) have recently developed a procedure to estimate the variance of bp genetic relatedness without the assumption of uniform recombination rates. However, their method still assumes uniform recombination rates in the regions between adjacent markers, making it best applicable to high-density linkage maps (rather than low-density linkage maps or cytological data, which will be more readily available for some species).
In addition, previous theoretical calculations of the variance of genetic relatedness (including those for bp genetic relatedness) have assumed that crossover interference is absent. However, it has recently been shown, by computer simulation of various forms of crossover patterning along chromosomes, that crossover interference tends to decrease variances of genetic relatedness (Caballero et al. 2019). Since crossover interference is a nearly ubiquitous feature of meiosis (Hillers 2004; Otto and Payseur 2019), its neglect in previous calculations of the variance of genetic relatedness further limits their generality.
In this paper, we derive a general, assumption-free formulation for the variance of bp genetic relatedness. We show that the variance of genetic relatedness is a simple, decreasing function of certain newly developed metrics of genome-wide genetic shuffling: and analogs (Veller et al. 2019b). These metrics, in a natural and intuitive way, take into account features of the aggregate recombination process, such as the number of chromosomes and heterogeneity in their size, the number of crossovers and their location along the chromosomes, the spatial relations of crossovers with respect to each other (e.g., crossover interference), and sex differences in recombination.
Our formulation of the variance of genetic relatedness in terms of and analogs allows the effects that the above meiotic features have on the variance of genetic relatedness to be reinterpreted—often with greater intuition—in terms of their effects on aggregate genetic shuffling. For example, the fact that crossover interference decreases the variance of genetic relatedness (Caballero et al. 2019) can be explained by the intuitive fact that crossover interference, by spreading crossovers out evenly along chromosomes, increases the amount of genetic shuffling that they cause (Gorlov and Gorlova 2001; Veller et al. 2019b).
In the calculations below, the number of loci in the genome is assumed to be very large. Loci i and j are recombinant in a random gamete with probability rij (e.g., if i and j are on different chromosomes). Sex-specific recombination rates, and , are distinguished where necessary. We assume that there is no inbreeding; for a treatment of the variance of cM genetic relatedness in finite populations, in which a degree of inbreeding is inevitable, see Carmi et al. (2013). “Genetic relatedness” refers to bp genetic relatedness, unless specified otherwise.
Relationships of Direct Descent
Pedigree relationships of direct descent (or “lineal” relationships) involve a single lineage, from an ancestor to one of its descendants. We will focus here on the specific example of grandparent-grandoffspring; calculations of the variance of genetic relatedness for general relationships of direct descent are given in Supplemental Material, File S1, Section S1.
Grandparent-grandoffpsring
Let the random variable IBDgrand be the proportion of a grandoffspring’s genome inherited from a specified grandparent. Consider the gamete produced by the grandoffspring’s parent, and let be the fraction of this gamete’s genome that derives from the focal grandparent (so that, by Mendelian segregation, ). We first wish to calculate . To do so, we use an approach very similar to that of Hill (1993a) and Visscher et al. (2006), but we define genetic relatedness in terms of bp shared rather than cM shared, and make no assumptions about the recombination process (in File S1, Section S3, we discuss technical differences between our calculations of the variance of bp genetic relatedness and previous calculations of the variance of cM genetic relatedness). We calculate (details in File S1, Section S1) that
| (1) |
where is the probability that a randomly chosen locus pair recombines in meiosis (Veller et al. 2019b). Because half of the grandoffspring’s genome comes from this gamete, , so that is the coefficient of relationship, and
| (2) |
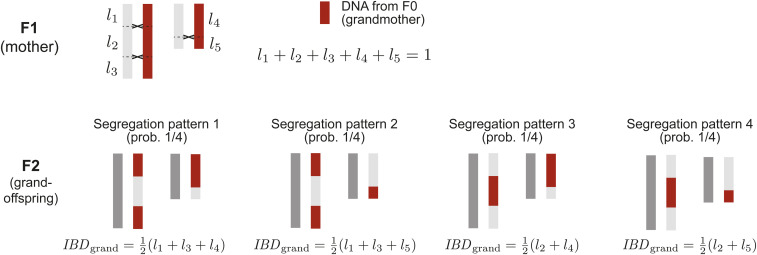
A graphical demonstration of Equation 2, based on the possible segregation patterns of a given parental meiosis, is shown in Figure 1.
Figure 1.
The variance of genetic relatedness between grandoffspring and grandparent, calculated from the possible segregation patterns of a single parental meiosis. In the figure, the positions of crossovers in a maternal meiosis (and the chromatids involved) are specified, but the segregation pattern in the resulting egg (and therefore offspring) is not. Averaging across the four segregation patterns, we find , and, from Equation 1 in Veller et al. (2019b), . Across the four possible segregation patterns, and which is Equation 2.
Note that the formulation in Equation 2 and other such formulations in this paper apply to the whole genome, or a single chromosome, or any specific genomic region. In the latter cases, is the probability that a randomly chosen pair of loci within the region of interest recombine in meiosis. In addition, because the recombination process often differs between the sexes, the value of can differ between spermatogenesis and oogenesis. In calculating the variance of genetic relatedness between a grandoffspring and one of its maternal grandparents, the value for oogenesis, , would be used; the value for spermatogenesis, , would be used for paternal grandparents.
can be estimated from various kinds of data, including cytological data of crossover positions at meiosis I, sequence data from gametes, and linkage maps (Veller et al. 2019b). We used cytological data from Lian et al. (2008) to calculate chromosome-specific and genome-wide values of in human male, and the linkage map of Kong et al. (2010) to calculate analogous values in human female (translating map distances to recombination rates using Kosambi’s map function, which incorporates a model of crossover interference). Substituting these values of into Equation 2 yields the variance of genetic relatedness to paternal and maternal grandparents in humans, for each chromosome and genome-wide. Table 1 displays the standard deviations, together with the corresponding standard deviations of cM genetic relatedness, calculated by substituting the sex-specific chromosome map lengths reported by Kong et al. (2010) into the relevant formula of Hill and Weir (2011).
Table 1. Standard deviations of genetic relatedness to a paternal and maternal grandparent, and to a sibling, in humans, for both bp and cM measures of genetic relatedness.
| Chromosome | Grandparent | Sibling | ||||
|---|---|---|---|---|---|---|
| Paternal | Maternal | |||||
| bpa | cMb | bpc | cMb | bpd | cMb | |
| 1 | 0.145 | 0.156 | 0.115 | 0.126 | 0.146 | 0.147 |
| 2 | 0.152 | 0.161 | 0.117 | 0.130 | 0.152 | 0.152 |
| 3 | 0.164 | 0.168 | 0.126 | 0.138 | 0.165 | 0.162 |
| 4 | 0.171 | 0.174 | 0.127 | 0.139 | 0.170 | 0.166 |
| 5 | 0.173 | 0.176 | 0.128 | 0.142 | 0.173 | 0.169 |
| 6 | 0.183 | 0.181 | 0.133 | 0.145 | 0.184 | 0.175 |
| 7 | 0.179 | 0.178 | 0.135 | 0.148 | 0.181 | 0.176 |
| 8 | 0.187 | 0.184 | 0.139 | 0.152 | 0.192 | 0.183 |
| 9 | 0.186 | 0.186 | 0.149 | 0.157 | 0.199 | 0.188 |
| 10 | 0.184 | 0.182 | 0.139 | 0.152 | 0.188 | 0.181 |
| 11 | 0.187 | 0.189 | 0.142 | 0.157 | 0.193 | 0.190 |
| 12 | 0.179 | 0.182 | 0.140 | 0.154 | 0.183 | 0.183 |
| 13 | 0.177 | 0.192 | 0.152 | 0.170 | 0.192 | 0.204 |
| 14 | 0.175 | 0.195 | 0.160 | 0.178 | 0.195 | 0.214 |
| 15 | 0.180 | 0.197 | 0.154 | 0.172 | 0.196 | 0.209 |
| 16 | 0.190 | 0.194 | 0.155 | 0.169 | 0.207 | 0.204 |
| 17 | 0.193 | 0.195 | 0.150 | 0.168 | 0.204 | 0.204 |
| 18 | 0.198 | 0.201 | 0.158 | 0.173 | 0.213 | 0.213 |
| 19 | 0.199 | 0.203 | 0.178 | 0.182 | 0.226 | 0.223 |
| 20 | 0.195 | 0.211 | 0.167 | 0.184 | 0.213 | 0.231 |
| 21 | 0.198 | 0.219 | 0.189 | 0.205 | 0.235 | 0.260 |
| 22 | 0.201 | 0.218 | 0.188 | 0.205 | 0.236 | 0.259 |
| Genome | 0.040 | 0.040 | 0.031 | 0.034 | 0.041 | 0.040 |
Calculated from cytological data of Lian et al. (2008).
Calculated from formulas in Hill and Weir (2011), using chromosome map lengths of Kong et al. (2010). Does not take into account crossover interference. cM relatedness to paternal and maternal grandparents defined, respectively, in terms of the male and female map; cM relatedness of siblings defined in terms of the sex-averaged map.
Calculated from linkage maps of Kong et al. (2010) using Kosambi’s map function.
Calculated from cytological data of Lian et al. (2008) (male meiosis) and linkage maps of Kong et al. (2010) using Kosambi’s map function (female meiosis).
Several observations emerge from Table 1. First, the variance of genetic relatedness for each individual chromosome is substantially larger than the genome-wide variance. This is because the majority of genetic shuffling in humans is due to independent assortment of chromosomes, rather than crossing over (Crow 1988; Veller et al. 2019b). Second, the variance of genetic relatedness to a paternal grandparent is greater than to a maternal grandparent, for each chromosome and genome-wide. This is because male meiosis involves less genetic shuffling than female meiosis (lower ), owing to fewer crossovers and their more terminal localization along the chromosomes in males (Veller et al. 2019b).
In comparing the variances of bp and cM genetic relatedness, three meiotic features are relevant. Per-chromosome comparisons are affected by the location of crossovers along chromosomes (crossover distribution) and with respect to each other (crossover interference). The genome-wide comparisons are additionally influenced by independent assortment of chromosomes. We discuss the effects of these features in turn.
First, proterminal localization of crossovers in humans (especially males) reduces relative to a uniform distribution of crossovers (Veller et al. 2019b), increasing the variance of bp vs. cM genetic relatedness (since crossovers are uniformly distributed along the genetic map, by definition). To isolate this effect of nonuniform recombination rates, we artificially eliminate crossover interference in the calculation of by using linkage maps and Haldane’s map function (which, unlike Kosambi’s map function, assumes no crossover interference). Calculating in this way, we find that the chromosome-specific variances of bp genetic relatedness are typically larger than their corresponding cM values (File S1, Section S4), more so in males because of their more terminal distribution of crossovers.
Second, crossover interference increases by spreading crossovers out more evenly along chromosomes (Veller et al. 2019b), thus decreasing the variances of bp genetic relatedness relative to the corresponding variances of cM genetic relatedness (the calculations of which do not take into account crossover interference). Thus, in spite of the tendency of nonuniform recombination rates to increase the per-chromosome variances of bp genetic relatedness, these variances are nevertheless smaller than the corresponding variances of cM genetic relatedness when crossover interference is taken into account (Table 1). The negative effect of crossover interference on the variance of genetic relatedness was previously identified by Caballero et al. (2019). Interestingly, in human male, the per-chromosome variances of genetic relatedness calculated from raw (cytological) crossover data are smaller than those calculated from linkage maps using Kosambi’s map function (File S1, Section S4), suggesting that Kosambi’s map function does not capture the full influence of crossover interference on genetic shuffling in human male.
Finally, in humans, chromosome lengths are more variable when measured in bp than in cM (File S1, Section S3). This causes the contribution of independent assortment of chromosomes to to be smaller than if the bp lengths of the chromosomes were only as variable as the cM lengths (Veller et al. 2019b), which, in turn, increases the genome-wide variance of bp vs. cM genetic relatedness to grandparents (the mathematical details of this effect are explained in File S1, Section S3). Because of this effect, although the chromosome-specific variances of bp genetic relatedness to grandparents are substantially smaller than their cM counterparts, the genome-wide variances of bp and cM genetic relatedness are more similar (Table 1).
Indirect Relationships
Indirect relationships involve two descendants of at least one individual in the pedigree. In the case of multiancestor pedigrees, we restrict our attention to two-ancestor pedigrees where the two ancestors were a mating pair (so that the focal descendants are, for example, full siblings, or aunt-nephew, etc.). We focus here on half-siblings and full-siblings; the calculations for general indirect relationships of this kind are given in File S1, Section S2.
Half-siblings
Let the random variable IBDh-sib be the proportion of two half-siblings’ genomes that they share IBD, if they have the same father but unrelated mothers. Then is the coefficient of relationship, and
| (3) |
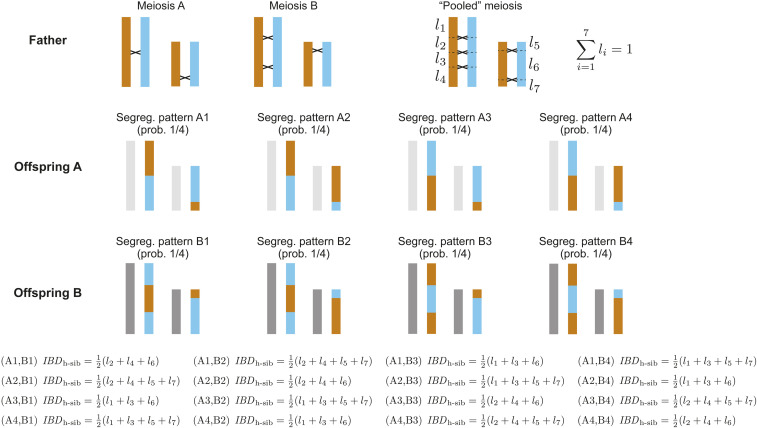
where is the probability that a randomly chosen locus pair recombines when the crossovers of two of the father’s meioses are pooled into one hypothetical meiosis (see Figure 2 for an example of a pooled meiosis). If the common parent were instead the mother, would replace . A graphical demonstration of Equation 3, based on the possible segregation patterns of two meioses in the parent, is given in Figure 2.
Figure 2.
The variance of genetic relatedness between half-siblings, calculated from the possible segregation patterns of two meioses of their common father. The positions of crossovers in the two paternal meioses (and the chromatids involved) are specified, but the segregation patterns in the resulting sperm cells (and therefore the two offspring) are not. Applying Equation 1 in Veller et al. (2019b) to the “pooled meiosis” in which the crossovers from the two actual meioses have been combined, we find Across the 16 possible segregation patterns , and which is Equation 3.
Siblings:
Let the random variable IBDsib be the proportion of two full-siblings’ genomes that they share IBD, assuming their mother and father to be unrelated. Then is the coefficient of relationship, and
| (4) |
Like , can be estimated from various kinds of data, including cytological data of crossover positions at meiosis I, sequence data from gametes, and linkage maps. Table 1 lists the chromosome-specific and genome-wide standard deviations of bp genetic relatedness of human siblings, calculated using cytological data from Lian et al. (2008) for male meiosis and the linkage map of Kong et al. (2010) for female meiosis (with map distances converted to recombination rates using Kosambi’s map function). Also shown are the corresponding standard deviations of cM genetic relatedness of siblings, defined as the proportion of the sex-averaged genetic map that they share IBD.
As for the case of genetic relatedness to grandparents, several meiotic features affect the comparison of the variances of bp and cM genetic relatedness of siblings. First, the bp variances are increased by the proterminal distribution of crossovers along chromosomes in humans, which tends to decrease . Thus, when the variance of bp genetic relatedness of siblings is calculated using linkage maps and Haldane’s map function (to eliminate the effect of crossover interference), the chromosome-specific and genome-wide estimates are substantially larger than the corresponding cM variances (File S1, Section S4). However, crossover interference, by increasing genetic shuffling, increases , thus decreasing the bp variance. These opposing effects of proterminal localization of crossovers and crossover interference roughly cancel in this case, so that our estimates of the variance of bp and cM genetic relatedness of siblings are coincidentally similar, at both the chromosome-specific and genome-wide levels (Table 1).
Within- vs. cross-pedigree variance
The calculations above and in Appendices S1 and S2 are for the variance of genetic relatedness in a given instance of a specified pedigree relationship. This variance derives from the randomness of recombination and segregation in the meiotic processes of the individuals involved in that particular pedigree. For some applications, however, we are interested in the variance of genetic relatedness across instances of a specified pedigree relationship [e.g., using variation in the genetic relatedness of different sibling pairs to estimate the heritability of some trait (Visscher et al. 2006)]. To calculate this ‘population variance’ of genetic relatedness, variation across individuals in their recombination processes must be taken into account. Applying the law of total variance (details in File S1, Section S5), we find that the variance of genetic relatedness across instances of a specified pedigree relationship is equal to the average within-pedigree variance. We have shown that within-pedigree variances are functions of metrics of aggregate recombination such as and ; to calculate the cross-pedigree variance, these metrics must simply be averaged across pedigrees.
A complication arises when using pooled recombination data (such as linkage maps) to estimate the cross-pedigree variance of genetic relatedness, because for all such metrics of aggregate recombination except , calculation of the metric from averaged recombination data does not return the average of the metric across pedigrees (File S1, Section S5). It is therefore technically invalid, in such cases, to use pooled recombination data to calculate the cross-pedigree variance of genetic relatedness (although it is valid in the case of grandoffspring-grandparent).
To get a sense for how large an error the use of pooled recombination data can cause, we focus on the case of paternal half-siblings. Using crossover data generated by Bell et al. (2020) by single-cell sequencing of large numbers of sperm from 20 human male donors, we calculated values of for each individual donor, from which we calculated a value of averaged across individuals. We also calculated a value of from recombination rates that were averaged across individuals. The values of from both individual and pooled recombination rates were calculated genome-wide and per chromosome. Using the two estimates of , we calculated the variance of genetic relatedness of paternal half-siblings according to Equation 3. We found that the values based on pooled recombination fractions differed only slightly from the correctly calculated values: the genome-wide variances differed by about 0.25%, and the chromosome-specific variances differed by comparable amounts. Details of these calculations are given in File S1, Section S5.
Therefore, the bias introduced by using linkage maps to calculate the population variance of genetic relatedness is likely to be small. Nevertheless, when available, disaggregated data in which crossover positions are inferred for individual nuclei (e.g., cytological data for individual meiocytes or sequencing data for individual gametes) are preferable for calculating cross-pedigree variances.
Application: Ancestry Variance and Selection Among F2s
A common experimental design involves mating individuals from two lines, populations, or species (A and B) to form a hybrid “F1” generation, and then mating F1s to produce an F2 generation. Every F1 carries exactly half of its DNA from each species, but there is ancestry variance among F2s because of recombination and segregation in the F1s’ meioses (Hill 1993a).
Each F2 derives from an F1 mother’s egg and an F1 father’s sperm. Let the random variables and be the respective proportions of species-A DNA in the egg and sperm (measured in bp), and let P be the proportion of species-A DNA in an F2’s genome. Then , and, from Equation 1, and . Finally, because and are independent, the ancestry variance among F2s is
| (5) |
[If the F2s instead derived from a backcross of F1s to one of the parental species, the ancestry variance among F2s would be , with calculated for the sex of the F1s involved. The corresponding variance for cM-based ancestry of backcross F2s, and those in later backcross generations, have been calculated by Hill (1993a), assuming no crossover interference.]
The calculation above assumes that there is no systematic selection among F2s in favor of alleles from one of the two species, and no meiotic drive in F1s, both of which would shift the distribution of ancestry among F2s toward one of the two species. For example, Matute et al. (2020) generated two crosses, each between one widely distributed species of Drosophila and one closely related island endemic. In the resulting admixed populations, island ancestry was replicably selected against over time. If viability selection plays a role in this effect, then an ancestry skew toward the widespread species would be expected among adult F2s in these crosses.
An F2-based test for selection of this kind would involve comparing the observed average ancestry among F2s against the neutral null expectation of 1/2. In this case, Equation 5 gives the appropriate null variance for the purpose of statistical inference; the standard error of the test is
| (6) |
where n is the sample size of F2s for which ancestry proportions have been assayed.
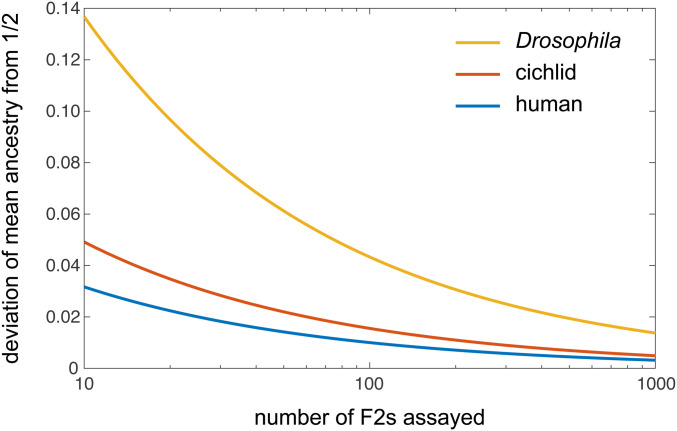
Substituting known values of into Equation 6 then shows, for a given sample of F2s, how much their average ancestry proportion must deviate from 1/2 for us to reject the null hypothesis of neutrality. For example, using a linkage map generated from a cross of two closely related cichlid fish species (Feulner et al. 2018), together with Kosambi’s map function, we calculate a sex-averaged value of . If ancestry fractions were measured for 10 F2s from this cross, then Equation 6 tells us that a 4.9% or greater deviation of the average ancestry from the null expectation of 50% would be statistically distinguishable at the 5% significance level; if 100 F2s were assayed, the threshold detectable deviation would be 1.6%. Threshold deviations for a range of sample sizes are shown in Figure 3 for the recombination processes of cichlids, humans, and Drosophila melanogaster. It is clearly seen that, because of D. melanogaster’s low sex-averaged value of (see Discussion), much greater ancestry deviations among F2s are required for the null hypothesis of neutrality to be rejected, compared to cichlids and humans.
Figure 3.
The minimum deviation of mean ancestry among F2s that can be statistically distinguished from the null expectation of 1/2 at the 5% significance level.
Note that the test described above can also be carried out for a specific genomic region of interest, by using region-specific values of . Alternatively, in a genome-wide scan, regions of the genome where ancestry deviations are particularly large can be statistically identified using region-specific values of , correcting for multiple hypothesis testing.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at figshare: https://doi.org/10.25386/genetics.13138577.
Discussion
Relatives of a given pedigree relationship vary in how much of their DNA they share IBD, because of variable patterns of recombination and chromosome segregation in their pedigrees. Previous theoretical calculations of the variance of genetic relatedness have been limited to measuring the genetic relatedness of two individuals as the proportion of the total genetic map (in cM) that they share IBD. Such calculations have failed to accommodate crossover interference and sex differences in recombination, both of which are near-universal features of meiosis that are known to substantially affect variance in genetic relatedness (Caballero et al. 2019).
Here, we have shown that when genetic relatedness is instead measured as the proportion of the total physical genome (in bp) shared IBD by relatives, the variance of genetic relatedness is determined by aggregate recombination, as quantified by and analogous metrics. These metrics take into account all features of the aggregate recombination process, including the number of chromosomes and heterogeneity in their size, the number of crossovers and their spatial organization along the chromosomes, and sex differences in recombination (Veller et al. 2019b). In addition to incorporating the above meiotic features into theoretical calculations of the variance of genetic relatedness, our treatment further allows these features’ effects on genetic relatedness to be understood intuitively in terms of their effects on aggregate genetic shuffling. Several examples are discussed below.
Sex differences in recombination
In many species, male and female meiosis differ both in the number and location of crossovers [reviewed by Lenormand and Dutheil (2005); Sardell and Kirkpatrick (2020)]. In human male, there are fewer crossovers and they are more terminally localized along the chromosomes than in human female. Both factors decrease the total amount of genetic shuffling in male meiosis (Veller et al. 2019b), providing an intuitive explanation of the observation of Caballero et al. (2019) that, in humans, relatives who are related predominantly via males have a higher variance of genetic relatedness than relatives related predominantly via females. In our calculations, for example, the standard deviation of genetic relatedness to a paternal grandparent is about 30% greater than that to a maternal grandparent (Table 1).
Such effects will be especially pronounced in species with no crossing over in one sex (e.g., male Drosophila and female Lepidoptera). Using chromosome lengths from Release 6 of the D. melanogaster reference genome (Hoskins et al. 2015) and the female linkage map produced by Comeron et al. (2012) (together with Kosambi’s map function), we calculate autosomal values of and . Substituting these values into Equation 2, we find that the standard deviations of (autosomal) genetic relatedness to paternal and maternal grandparents are 0.175 and 0.125, respectively—a difference of 40%.
Chromosome number and size
Because most genetic shuffling in meiosis is due to independent assortment of chromosomes rather than crossing over (Crow 1988; Veller et al. 2019b), the most important contributor to cross-species differences in the variance of genetic relatedness is karyotypic differences. Two karyotypic features affect genetic shuffling, and therefore the variance of genetic relatedness.
First, the greater the number of chromosomes, the greater the genetic shuffling associated with their independent assortment. For example, humans have 22 autosomes while D. melanogaster has only two major autosomes. Therefore, the contribution of independent assortment to [and, equivalently, to ] is large in humans (0.473) but small in D. melanogaster (0.253). Thus, were there no crossing over in either species, the SD of the genetic relatedness of full siblings would be about 8% in humans and 25% in D. melanogaster (Equation 4).
Second, the more homogenously sized the chromosomes are, the greater the genetic shuffling associated with their independent assortment. We have used this fact to explain why independent assortment of chromosomes in humans is more effective at decreasing the variance of cM vs. bp genetic relatedness, because the chromosomes are more homogenously sized when measured in cM.
Crossover positions
White and Hill (2020) have shown that terminal placement of crossovers tends to increase the variance of genetic relatedness, relative to more central placement of crossovers. This can be explained by the intuitive fact that a crossover near the tip of a chromosome causes less genetic shuffling than a crossover in the middle (Veller et al. 2019b). We have used this fact to explain why, after controlling for the effects of crossover interference, per-chromosome variances of genetic relatedness are smaller for bp than for cM genetic relatedness in humans, because crossovers are proterminally distributed along the physical chromosome maps but, by definition, are uniformly distributed along the genetic maps.
Crossover interference
It has recently been shown, by computer simulation of various forms of crossover patterning along chromosomes, that crossover interference tends to decrease the variance of genetic relatedness between relatives (Caballero et al. 2019). Veller et al. (2019b) demonstrated that interference among crossovers increases the amount of genetic shuffling that they cause (increasing and analogs). The intuition is that, when two crossovers occur very close to each other along a bivalent chromosome at meiosis I (the stage at which crossover interference operates), they cancel each other’s effect on genetic shuffling, together behaving more like a single crossover. Such “stepping on toes” is prevented by crossover interference, thus increasing genetic shuffling. This provides an intuitive explanation of the result of Caballero et al. (2019).
Using a simulation method employed by Mancera et al. (2008) and Wang et al. (2012) to resample empirically observed crossovers in an interference-less way, Veller et al. (2019b) calculated that, in human male, interference among crossovers increases their contribution to by about 15%. By this measure, crossover interference in human male meiosis decreases the genome-wide standard deviation of genetic relatedness to a paternal grandparent from 0.043 to 0.040 (Equation 2), a decrease of about 7%.
Crossover covariation
It has recently been shown for diverse eukaryotes that the number of crossovers per chromosome covaries positively across chromosomes within individual meiotic nuclei (Wang et al. 2019). This “crossover covariation” substantially increases the variance of crossover number per gamete, which will clearly affect the distribution of genetic relatedness among relatives. However, because crossover covariation does not change the (unconditional) probability that a given pair of loci are recombinant in a gamete, it does not alter or analogs (since these are averages of functions of individual pairwise recombination rates—see Appendices S1, S2, and S5). Therefore, crossover covariation does not affect the variance of genetic relatedness among relatives (but it will affect higher-order moments).
Conclusion
We have shown that the variance of genetic relatedness is a function of and analogous metrics. Since these metrics can readily be estimated from modern cytological and sequencing data [see Veller et al. (2019b) and above], our results make it possible to calculate the variance of genetic relatedness in a precise, general, and unambiguous way.
Acknowledgments
We are grateful to Nicolas Bierne, Erin Calfee, Shai Carmi, Graham Coop, Jim Mallet, Molly Schumer, Sivan Yair, and the anonymous referees for helpful discussions and comments. This work was funded in part by the Office of Naval Research (grant N00014-16-1-2914 to M.A.N.), the Defense Advanced Research Projects Agency (grant W911NF-18-2-0265 to M.A.N.), and the John Templeton Foundation (grant 55832 to M.A.N.).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.13138577.
Communicating editor: S. Browning
Literature Cited
- Barash D. P., Holmes W. G., and Greene P. J., 1978. Exact vs. probabilistic coefficients of relationship: some implications for sociobiology. Am. Nat. 112: 355–363. 10.1086/283278 [DOI] [Google Scholar]
- Bell A. D., Mello C. J., Nemesh J., Brumbaugh S. A., Wysoker A. et al. , 2020. Insights into variation in meiosis from 31,228 human sperm genomes. Nature 583: 259–264. 10.1038/s41586-020-2347-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojko M., 1985. Human meiosis IX. Crossing over and chiasma formation in oocytes. Carlsberg Res. Commun. 50: 43–72. 10.1007/BF02907137 [DOI] [Google Scholar]
- Caballero M., Seidman D. N., Qiao Y., Sannerud J., Dyer T. D. et al. , 2019. Crossover interference and sex-specific genetic maps shape identical by descent sharing in close relatives. PLoS Genet. 15: e1007979 10.1371/journal.pgen.1007979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi S., Palamara P. F., Vacic V., Lencz T., Darvasi A. et al. , 2013. The variance of identity-by-descent sharing in the Wright–Fisher model. Genetics 193: 911–928. 10.1534/genetics.112.147215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., and Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905 10.1371/journal.pgen.1002905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow J. F., 1988. The importance of recombination, pp. 56–73 in The Evolution of Sex: An Examination of Current Ideas, edited by Michod R. E., and Levin B. R.. Sinauer, Sunderland. [Google Scholar]
- Feulner P. G. D., Schwarzer J., Haesler M. P., Meier J. I., and Seehausen O., 2018. A dense linkage map of Lake Victoria cichlids improved the Pundamilia genome assembly and revealed a major QTL for sex-determination. G3 (Bethesda) 8: 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin I. R., 1977. The distribution of the proportion of the genome which is homozygous by descent in inbred individuals. Theor. Popul. Biol. 11: 60–80. 10.1016/0040-5809(77)90007-7 [DOI] [PubMed] [Google Scholar]
- Gorlov I. P., and Gorlova O. Y., 2001. Cost–benefit analysis of recombination and its application for understanding of chiasma interference. J. Theor. Biol. 213: 1–8. 10.1006/jtbi.2001.2397 [DOI] [PubMed] [Google Scholar]
- Guo S.-W., 1996. Variation in genetic identity among relatives. Hum. Hered. 46: 61–70. 10.1159/000154328 [DOI] [PubMed] [Google Scholar]
- Hillers K. J., 2004. Crossover interference. Curr. Biol. 14: R1036–R1037. 10.1016/j.cub.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Hill W. G., 1993a Variation in genetic composition in backcrossing programs. J. Hered. 84: 212–213. 10.1093/oxfordjournals.jhered.a111319 [DOI] [Google Scholar]
- Hill W. G., 1993b Variation in genetic identity within kinships. Heredity 71: 652–653. 10.1038/hdy.1993.190 [DOI] [PubMed] [Google Scholar]
- Hill W. G., and Weir B. S., 2011. Variation in actual relationship as a consequence of Mendelian sampling and linkage. Genet. Res. 93: 47–64. 10.1017/S0016672310000480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm P. B., and Rasmussen S. W., 1983. Human meiosis VI. Crossing over in human spermatocytes. Carlsberg Res. Commun. 48: 385–413. 10.1007/BF02911918 [DOI] [Google Scholar]
- Hoskins R. A., Carlson J. W., Wan K. H., Park S., Mendez I. et al. , 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. 10.1101/gr.185579.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardos M., Luikart G., and Allendorf F. W., 2015. Measuring individual inbreeding in the age of genomics: marker-based measures are better than pedigrees. Heredity 115: 63–72. 10.1038/hdy.2015.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A. et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. 10.1038/nature09525 [DOI] [PubMed] [Google Scholar]
- Lenormand T., and Dutheil J., 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3: e63 10.1371/journal.pbio.0030063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J., Yin Y., Oliver-Bonet M., Liehr T., Ko E. et al. , 2008. Variation in crossover interference levels on individual chromosomes from human males. Hum. Mol. Genet. 17: 2583–2594. 10.1093/hmg/ddn158 [DOI] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., and Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. 10.1038/nature07135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute D. R., Comeault A. A., Earley E., Serrato-Capuchina A., Peede D. et al. , 2020. Rapid and predictable evolution of admixed populations between two Drosophila species pairs. Genetics 214: 211–230. 10.1534/genetics.119.302685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto S. P., and Payseur B. A., 2019. Crossover interference: shedding light on the evolution of recombination. Annu. Rev. Genet. 53: 19–44. 10.1146/annurev-genet-040119-093957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardell J. M., and Kirkpatrick M., 2020. Sex differences in the recombination landscape. Am. Nat. 195: 361–379. 10.1086/704943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman P. W., 1979. Insect chromosome numbers and eusociality. Am. Nat. 113: 925–935. 10.1086/283445 [DOI] [Google Scholar]
- Thompson E. A., 2013. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics 194: 301–326. 10.1534/genetics.112.148825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veller, C., N. B. Edelman, P. Muralidhar, and M. A. Nowak, 2019a Recombination, variance in genetic relatedness, and selection against introgressed DNA. BioRxiv doi: 10.1101/846147 (Preprint posted November 18, 2019). [Google Scholar]
- Veller C., Kleckner N., and Nowak M. A., 2019b A rigorous measure of genome-wide genetic shuffling that takes into account crossover positions and Mendel’s second law. Proc. Natl. Acad. Sci. USA 116: 1659–1668. 10.1073/pnas.1817482116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Medland S. E., Ferreira M. A. R., Morley K. I., Zhu G. et al. , 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLoS Genet. 2: e41 10.1371/journal.pgen.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher P. M., Macgregor S., Benyamin B., Zhu G., Gordon S. et al. , 2007. Genome partitioning of genetic variation for height from 11,214 sibling pairs. Am. J. Hum. Genet. 81: 1104–1110. 10.1086/522934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Fan H. C., Behr B., and Quake S. R., 2012. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150: 402–412. 10.1016/j.cell.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Veller C., Sun F., Ruiz-Herrera A., Shang Y. et al. , 2019. Per-nucleus crossover covariation and implications for evolution. Cell 177: 326–338.e16. 10.1016/j.cell.2019.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., 2016. Pedigrees or markers: which are better in estimating relatedness and inbreeding coefficient? Theor. Popul. Biol. 107: 4–13. 10.1016/j.tpb.2015.08.006 [DOI] [PubMed] [Google Scholar]
- White I. M. S., and Hill W. G., 2020. Effect of heterogeneity in recombination rate on variation in realised relationship. Heredity 124: 28–36. 10.1038/s41437-019-0241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilfert L., Gadau J., and Schmid-Hempel P., 2007. Variation in genomic recombination rates among animal taxa and the case of social insects. Heredity 98: 189–197. 10.1038/sj.hdy.6800950 [DOI] [PubMed] [Google Scholar]
- Young A. I., Frigge M. L., Gudbjartsson D. F., Thorleifsson G., Bjornsdottir G. et al. , 2018. Relatedness disequilibrium regression estimates heritability without environmental bias. Nat. Genet. 50: 1304–1310. 10.1038/s41588-018-0178-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Supplemental material available at figshare: https://doi.org/10.25386/genetics.13138577.