Abstract
Caenorhabditis elegans survives on ephemeral food sources in the wild, and the species has a variety of adaptive responses to starvation. These features of its life history make the worm a powerful model for studying developmental, behavioral, and metabolic starvation responses. Starvation resistance is fundamental to life in the wild, and it is relevant to aging and common diseases such as cancer and diabetes. Worms respond to acute starvation at different times in the life cycle by arresting development and altering gene expression and metabolism. They also anticipate starvation during early larval development, engaging an alternative developmental program resulting in dauer diapause. By arresting development, these responses postpone growth and reproduction until feeding resumes. A common set of signaling pathways mediates systemic regulation of development in each context but with important distinctions. Several aspects of behavior, including feeding, foraging, taxis, egg laying, sleep, and associative learning, are also affected by starvation. A variety of conserved signaling, gene regulatory, and metabolic mechanisms support adaptation to starvation. Early life starvation can have persistent effects on adults and their descendants. With its short generation time, C. elegans is an ideal model for studying maternal provisioning, transgenerational epigenetic inheritance, and developmental origins of adult health and disease in humans. This review provides a comprehensive overview of starvation responses throughout the C. elegans life cycle.
Keywords: dauer, L1 arrest, starvation, quiescence, WormBook
IN the wild, Caenorhabditis elegans are found in association with decaying plant matter, including rotting fruits, stems, and leaf litter, where they consume a diet of opportunistic microbes (Schulenburg and Félix 2017) .With rapid growth and short generation time, populations can expand rapidly such that food becomes limiting. The worm’s existence is even more perilous given pathogens, predators, and abiotic stressors that likely cooccur with nutrient stress. Deeper understanding of C. elegans natural history has come in recent years, establishing a broader context for experimental research (Frezal and Félix 2015). Nutrient availability can be easily manipulated in culture, and interest in the nutritional dimension of molecular and phenotypic analyses continues to grow. Research in this area has also increased with interest in aging, which is sensitive to nutrient availability and is governed by nutrient-sensing pathways.
Scope of this review
A worm’s response to starvation is a complex and dynamic process. Changes in signaling and behavior can result from changes in sensory perception of food cues and can therefore be near instantaneous. Changes in gene expression and metabolism occur within minutes to hours and continue for days. Starved worms display signs of aging over a period of several days as mortality commences. This review covers the C. elegans starvation responses over all of these time scales and throughout the life cycle. The effects of starvation on development and behavior, including acute and persistent effects as well as molecular and cellular mechanisms, will be covered. These effects are organized by developmental stage and different aspects of behavior. Molecular and metabolic consequences of starvation are reviewed in relation to starvation resistance. Persistent effects of starvation covered include primarily intragenerational and transgenerational effects, reflecting pathological and potentially adaptive responses. Transcriptome- and proteome-wide regulation of the starvation response is also covered.
Effects of Starvation on Development
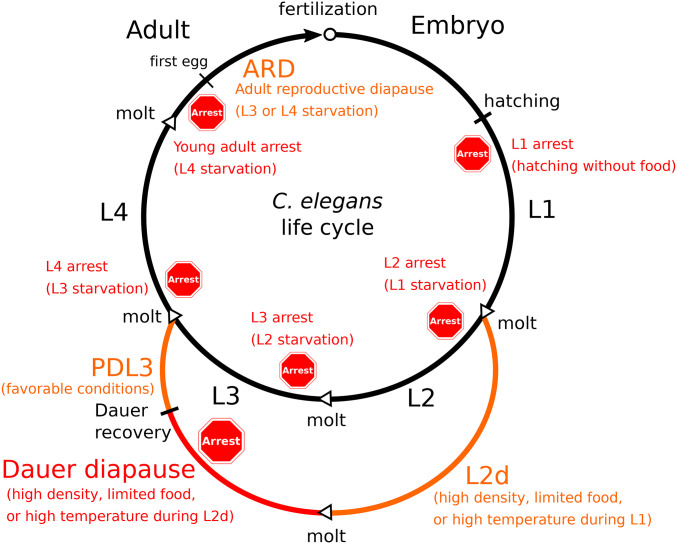
C. elegans displays robust developmental responses to starvation throughout the life cycle (Figure 1). Dauer diapause is the best studied of these and arguably most central to the biology of the organism. However, instantaneous responses to acute starvation at various developmental stages are also critical to environmental adaptation and presumably organismal fitness. Dauer diapause, L1 arrest, late larval arrest, and adult reproductive diapause (ARD) provide an opportunity for comparative analysis of how the animal senses and responds to nutrient availability at different points in its life cycle. Work in this area reveals considerable overlap in regulation of developmental arrest at each stage along with notable distinctions.
Figure 1.
Developmental responses to starvation throughout the life cycle. The progression of developmental stages in well-fed conditions is shown by the black arrow. L2d (predauer) development, PDL3 (postdauer L3) development, and ARD (adult reproductive diapause) are depicted in orange. Dauer diapause, L1 arrest, and other examples of developmental arrest are in red, with a stop sign indicating approximately when they occur relative to stage-specific molts. Specific conditions leading to each developmental response are indicated in parentheses below the response. Molts are depicted by open arrow heads. Hatching, dauer recovery, and onset of reproduction are also indicated with black lines. ARD can occur in response to L3 (Gerisch et al. 2020) or L4 (Angelo and Van Gilst 2009; Seidel and Kimble 2011) starvation, depending on conditions. In contrast, absolute starvation of L3 larvae results in L4 arrest (Schindler et al. 2014). Analogous to L3 and L4 arrest, an L2 arrest potentially results from starving previously fed L1 larvae, although this has not been demonstrated.
Dauer diapause
In contrast to larvae that develop into reproductive adults in replete environments, young larvae that experience high population density, limited nutrient availability, or increased ambient temperature may undergo an alternative developmental trajectory that results in larval arrest in a state of diapause known as dauer (Cassada and Russell 1975; Golden and Riddle 1984b; Riddle and Albert 1997; Hu 2007; Fielenbach and Antebi 2008) (Figure 1). L1 larvae exposed to favorable conditions through the first larval molt develop into L2 larvae and are committed to reproductive development. In contrast, L1 larvae that experience sufficient environmental stress before the first larval molt develop into a predauer stage termed L2d. L2d animals can either molt and undergo dauer arrest if environmental conditions remain unfavorable, or develop into nondauer L3 larvae if ambient conditions improve before the L2d molt (Golden and Riddle 1984b; Schaedel et al. 2012). Dauer larvae are nonfeeding and morphologically and physiologically distinct from reproductively developing larvae in that they have a unique cuticle, a remodeled pharynx, a narrow body, increased lipid stores, and altered metabolism. These dauer-specific adaptations render worms resistant to starvation and other stressors, enabling survival for several months or more in harsh conditions (e.g., desiccation or freezing). Dauers also exhibit a behavior known as nictation that promotes their dispersal (Yang et al. 2020). When favorable environmental conditions ensue, dauers enter a postdauer L3 stage, resuming pharyngeal pumping, feeding, growth, and the implementation of developmental programs characteristic of nondauer L3 larvae, before molting to become L4 larvae and proceeding with reproductive development (Figure 1) (Euling and Ambros 1996a,b; Riddle and Albert 1997; Karp and Ambros 2012). Dauer diapause (Riddle and Albert 1997; Hu 2007; Fielenbach and Antebi 2008) and practical methods for working with dauer larvae (Nika et al. 2016; Karp 2018) have been reviewed in detail elsewhere. For the remainder of this section we will focus on critical background and recent insights, while putting dauer diapause in the context of the life cycle.
Developmental arrest during dauer diapause reflects an ability of C. elegans to anticipate starvation before it occurs, as increased population density is the environmental factor that most strongly promotes dauer arrest. Because dauer development depends on specific environmental cues and involves provisioning in addition to metabolic adaptation, it is a true diapause, in contrast to starvation-induced developmental arrest at other larval stages (Kostál 2006; Baugh 2013). Because L1 and L2 predauer (also known as “L2d”) development must be completed first, larvae require food for dauer development, and larvae that hatch with absolutely no food do not form dauers (Johnson et al. 1984). Larvae cultured at high temperature also enter dauer arrest (Ailion and Thomas 2000), suggesting that dauer development represents a relatively general strategy to survive unfavorable conditions.
The dauer developmental program can be understood as a progression through three phases: perception and integration of environmental information, commitment to the dauer developmental fate, and execution of the dauer program. During the first larval stage after hatching, external cues that provide information about environmental conditions and the near-term availability of nutritional resources are detected (Swanson and Riddle 1981). This information is conveyed systemically through the regulation of conserved DAF-7 TGFβ-like and DAF-2 insulin-like growth factor receptor (IGFR) signaling pathways (Riddle and Albert 1997; Hu 2007; Fielenbach and Antebi 2008). Transcriptional outputs of DAF-7/TGFβ and DAF-2/IGFR pathways converge on a conserved steroid hormone biosynthetic pathway to regulate the biosynthesis of bile-acid-like hormones known as dafachronic acids (DAs), which function as ligands for the nuclear receptor and vitamin D receptor (VDR) homolog DAF-12 (Hu 2007; Fielenbach and Antebi 2008). DAF-12/VDR acts as a switch that controls commitment to either reproductive development or dauer arrest; when DAF-12/VDR is engaged by DA ligands, animals develop reproductively, whereas in the absence of DA ligands, unliganded DAF-12/VDR commits animals to the dauer developmental fate (Schaedel et al. 2012). Upon commitment to dauer arrest, execution of the dauer program culminates in morphological, functional, and behavioral changes characteristic of the dauer larva.
Perception and integration of environmental information:
The initial step in dauer development is induced by external cues that convey information about the relative likelihood that environmental conditions will support development to reproductive adulthood. These cues are perceived by sensory neurons and control the expression of secreted ligands that regulate conserved TGFβ-like and insulin-like signaling pathways throughout the animal. Early events in the perception and integration of environmental dauer-regulatory signals are best understood in the ASI sensory neurons, which promote reproductive development in replete environmental conditions (Bargmann and Horvitz 1991) through the expression of DAF-7/TGFβ and multiple insulin-like peptide (ILP) ligands for DAF-2/IGFR (Ren et al. 1996; Pierce et al. 2001; Li et al. 2003; Cornils et al. 2011).
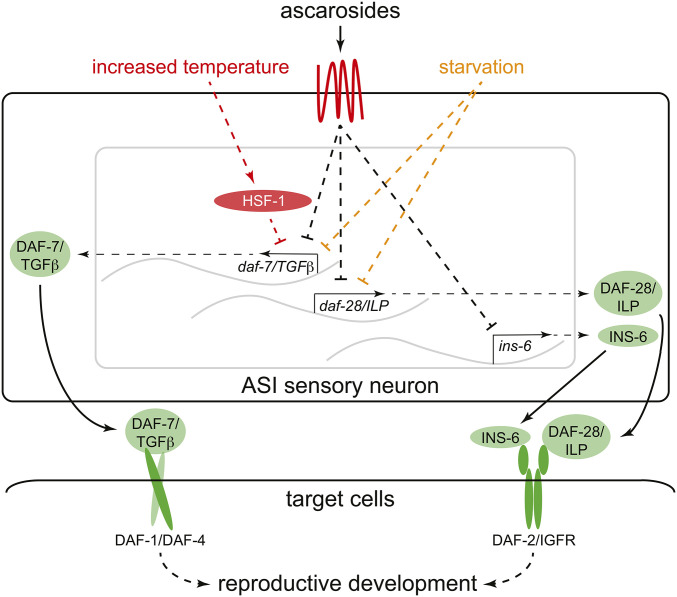
The main environmental cue that promotes dauer arrest is a mixture of secreted ascarosides (collectively known as dauer pheromone) that functions as an indicator of population density (Ludewig and Schroeder 2013). Ascarosides act via specific cell surface receptors expressed in the ASI sensory neurons (McGrath et al. 2011; Park et al. 2012) to induce dauer arrest by reducing the expression of the TGF-β-like peptide DAF-7 (Ren et al. 1996) and the agonist ILPs DAF-28 (Li et al. 2003) and INS-6 (Cornils et al. 2011) (Figure 2). These peptides are secreted from sensory neurons to promote reproductive development in favorable environments (Hu 2007; Fielenbach and Antebi 2008). Additional ILPs, such as the putative DAF-2/IGFR agonists INS-4 and INS-9 and the antagonists INS-1 and INS-18, are also expressed in ASI and may be involved in the initial response to dauer-inducing environmental cues (Pierce et al. 2001; Kao et al. 2007; Matsunaga et al. 2012; Fernandes de Abreu et al. 2014; Hung et al. 2014; Delaney et al. 2017; Li et al. 2019). As dauer induction by pheromone requires the activity of DAF-3/SMAD, DAF-5/SnoN, and DAF-16/ Forkhead box O transcription factor (FoxO) (Ailion and Thomas 2000), which are the major targets of DAF-7/TGFβ and DAF-2/IGFR signaling, respectively, inhibition of DAF-7/TGFβ and DAF-2/IGFR signaling is likely the main mechanism through which ascarosides promote dauer entry.
Figure 2.
Schematic model depicting integration of environmental cues in the ASI sensory neurons. Molecules that promote reproductive development are green, and those that promote dauer arrest are red. A generic transmembrane ascaroside receptor is portrayed as a thick red line. DAF-1 and DAF-4 are homologs of the human type I and type II TGFβ receptor, respectively. See text for details. IGFR, insulin-like growth factor receptor; ILP, insulin-like peptide.
The ability of food to mitigate the induction of dauer arrest by pheromone was recognized some time ago (Golden and Riddle 1984a). A heat-stable, neutral, hydrophilic “food signal” inhibits dauer entry and enhances recovery from dauer arrest (Golden and Riddle 1982, 1984a); reduced levels of this food signal likely account for enhancement of dauer arrest by food scarcity (Golden and Riddle 1984b). Fractionation of Escherichia coli extracts led to the identification of NAD+ as a component of the food signal that can induce dauer recovery (Mylenko et al. 2016). The hydrophilic fraction of bacterial extracts has greater food signal activity than NAD+ alone (Mylenko et al. 2016), and perception of polypeptides also stimulates dauer recovery (Kaplan et al. 2018). In addition, perception of saturated and monounsaturated fatty acids stimulates dauer recovery (Kaul et al. 2014). Thus, diverse molecular components of food may modulate the organismal response to dauer pheromone.
Both starvation and dauer pheromone reduce daf-7/TGFβ and daf-28/ILP expression in sensory neurons (Ren et al. 1996; Li et al. 2003; Neal et al. 2015) (Figure 2). Reduced nutrient availability may promote dauer arrest at least in part by increasing the synthesis of dauer-inducing ascarosides; the dauer-inducing ascaroside icas#9/IC-asc-C5 is detectable in extracts from starved L1 larvae, but not in extracts from well-fed adult or mixed-stage animals (Artyukhin et al. 2013b). However, food scarcity likely also influences dauer arrest independently of ascarosides, as food deprivation inhibits daf-28 expression in both the ASI and ASJ sensory neurons, whereas crude pheromone inhibits daf-28 expression specifically in ASI (Neal et al. 2015).
Temperature also modulates dauer induction by pheromone. Elevations in ambient temperature increase the sensitivity of animals to dauer pheromone (Golden and Riddle 1984b; Ailion and Thomas 2000). This may also occur through the regulation of DAF-7/TGFβ expression, as increased temperature reduces daf-7 expression in ASI (Schackwitz et al. 1996; Ailion and Thomas 2000) in a manner requiring the heat-inducible transcription factor HSF-1 (Barna et al. 2012) (Figure 2). Additionally, increased environmental temperature, which exacerbates organismal defects in the endoplasmic reticulum (ER) unfolded protein response (UPR) (Richardson et al. 2011), could inhibit ILP biogenesis and/or secretion indirectly by increasing ER stress in the ASI sensory neurons (see below) (Kulalert and Kim 2013).
The role of proteostasis in dauer regulation was first hinted at by the discovery that a neomorphic allele of daf-21, which encodes the C. elegans Hsp90 ortholog (Birnby et al. 2000), causes dauer arrest. Detailed analysis of another neomorphic dauer-constitutive allele, daf-28(sa191), has revealed that the physiologic state of the ASI sensory neurons can influence the dauer decision. The DAF-28 R37C mutant protein encoded by daf-28(sa191) induces the ER UPR in the ASI sensory neurons and causes dauer arrest largely due to constitutive phosphorylation of eIF2α at S49 by the C. elegans PERK ortholog PEK-1 (Kulalert and Kim 2013). As the dauer formation-constitutive (Daf-c) phenotype of daf-28(sa191) animals is partially suppressed by loss-of-function mutations in daf-16/FoxO and daf-18/phosphatase and tensin (PTEN), but not daf-3/SMAD or daf-5/SnoN (Malone et al. 1996), translational inhibition due to eIF2α S49 phosphorylation may reduce levels of INS-4, INS-6, INS-9, and/or other agonist ILPs below the threshold needed to promote reproductive development. However, the incomplete suppression of the Daf-c phenotype of daf-28(sa191) by daf-16/FoxO loss-of-function mutations suggests that activation of the ER UPR per se in the ASI sensory neurons may contribute to dauer arrest. Intriguingly, loss-of-function mutations in daf-41, which encodes the C. elegans ortholog of the human HSP90 cochaperone p23/PTGES3, are Daf-c, and their epistatic relationships with dauer formation-defective (Daf-d) mutations overlap partially with those observed for daf-28(sa191) (Horikawa et al. 2015). As daf-41 is expressed in sensory neurons (including ASI) and is required for chemotaxis (Horikawa et al. 2015), DAF-41/p23 may promote reproductive development in part by contributing to proteostasis in the ASI sensory neurons.
Thus, a picture is emerging of the ASI sensory neurons as an initial site where external information about the likelihood that ambient conditions will suffice to sustain reproductive development is integrated with internal information about cellular homeostasis. Inputs that convey information about population density, food availability, and temperature converge to modulate the expression of DAF-7/TGFβ and ILPs. The physiologic state of the ASI sensory neurons may contribute to the dauer decision, both due to, and independent of, its effect on the biogenesis of ILPs. Ultimately, the amount and complement of active DAF-7/TGFβ and ILPs secreted from ASI and other cells act on specific cell surface receptors expressed throughout the animal to regulate the activity of transcription factors that control the molecular switch determining commitment to reproductive or dauer developmental fates (Figure 2).
New signaling components and mechanisms of cross-talk:
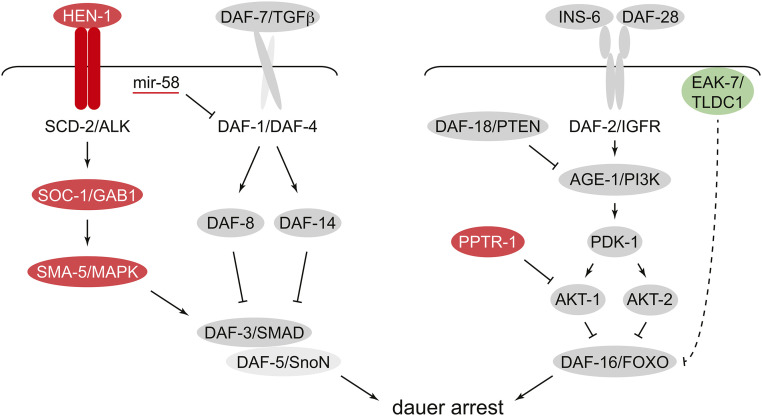
DAF-7/TGFβ and ILPs bind to their cognate cell surface receptors to regulate conserved signaling pathways that control the dauer decision (Hu 2007; Fielenbach and Antebi 2008) (Figure 3). Although early screens for Daf-c and Daf-d mutants revealed most of the major components of the DAF-7/TGFβ and DAF-2/IGFR pathways (Figure 3), more recent work has identified new modulators of both pathways. The suppressor of constitutive dauer (scd) genes emerged from genetic screens for suppressors of the Daf-c phenotype of daf-1/TGFβRI, daf-8/SMAD, and daf-14/SMAD mutants (Inoue and Thomas 2000). A detailed analysis of scd-2, which encodes the C. elegans ortholog of the human anaplastic lymphoma kinase (ALK) receptor tyrosine kinase, led to the elucidation of a conserved signaling module that acts in parallel to the canonical DAF-7/TGFβ pathway to promote DAF-3/SMAD and DAF-5/SnoN activity. The secreted protein HEN-1 is proposed to function as an agonist ligand for SCD-2/ALK that activates DAF-3/SMAD and DAF-5/SnoN through the SOC-1/GAB1 adaptor protein and the MAP kinase family member SMA-5 (Reiner et al. 2008) (Figure 3, left panel). MicroRNAs may also play a modulatory role in DAF-7/TGFβ signaling, as mir-58 family microRNAs can repress daf-1 and daf-4 expression in vivo through their 3′ untranslated regions (de Lucas et al. 2015) (Figure 3).
Figure 3.
Signal transduction pathways regulated by DAF-7/TGFβ (left) and insulin-like peptides (right). Core pathway components are depicted in gray. More recently identified signaling modulators that promote reproductive development or dauer arrest are shown in green and red, respectively. Multiple mechanisms of cross-talk between these pathways are not depicted. See text for details.
A screen for new modulators of DAF-2/IGFR signaling led to the discovery of EAK-7, a conserved plasma-membrane-associated protein that acts in parallel to AKT-1 and AKT-2 to inhibit DAF-16/FoxO activity (Alam et al. 2010) (Figure 3, right panel). Unlike AKT-1 and AKT-2, which inhibit DAF-16/FoxO through phosphorylation and subsequent export from the nucleus to the cytoplasm, EAK-7 inhibits the activity of nuclear DAF-16/FoxO without altering its subcellular localization. The mechanism through which EAK-7 regulates DAF-16/FoxO remains obscure. AKT-1 is negatively regulated by the conserved protein phosphatase 2A (PP2A) regulatory subunit PPTR-1, which activates DAF-16/FoxO by promoting the dephosphorylation of AKT-1 at T350 (Padmanabhan et al. 2009) (Figure 3). T350 on AKT-1 is analogous to T308 on human Akt/Protein Kinase B, the phosphorylation of which by 3-phosphoinositide-dependent protein kinase-1 is required for Akt activation (Alessi et al. 1997; Stokoe et al. 1997).
Although much of the genetic analysis involving Daf-c and Daf-d mutants is consistent with depictions of the DAF-7/TGFβ and DAF-2/IGFR pathways as linear pathways that act in parallel (Riddle and Albert 1997; Hu 2007; Fielenbach and Antebi 2008) (Figure 3), it is clear that channels exist through which these pathways communicate with and reinforce each other in dauer regulation. Dauers that form due to reduced DAF-7/TGFβ signaling require DAF-16/FoxO activity to execute dauer programs in vulval precursor cells (VPCs; Karp and Greenwald 2013) and neurons (Bhattacharya et al. 2019), indicating that DAF-7/TGFβ pathway activity contributes to DAF-16/FoxO inhibition. Conversely, DAF-2/IGFR signaling promotes DAF-7/TGFβ signaling, as daf-7::GFP expression in the ASI sensory neurons is reduced in daf-2/IGFR loss-of-function mutants (Barna et al. 2012). This is likely due, at least in part, to derepression of HSF-1, which is inhibited by DAF-2/IGFR (Chiang et al. 2012) and confers temperature-dependent repression of daf-7 (Barna et al. 2012). Intriguingly, the gene encoding the HEN-1 ligand, which promotes dauer arrest through SCD-2/ALK, DAF-3/SMAD, and DAF-5/SnoN (Reiner et al. 2008) (Figure 3, left panel) is a DAF-16/FoxO target gene that is induced ∼15-fold in daf-2(e1370) mutants (Tepper et al. 2013; Chen et al. 2015). Thus, increasing HEN-1 expression could be a mechanism through which reduction in DAF-2/IGFR signaling reinforces dauer-promoting signals by increasing DAF-3/SMAD and DAF-5/SnoN activity through the HEN-1/SCD-2/SOC-1/SMA-5 pathway (Figure 3). Other genes exhibit complex interactions with dauer regulatory pathways and may function as conduits of signaling cross-talk. These include genes that encode the acid sphingomyelinase homolog ASM-3 (Kim and Sun 2012), the pyruvate dehydrogenase phosphatase homolog PDP-1 (Narasimhan et al. 2011), and the protein kinase C family member PKC-1 (Monje et al. 2011; Kulalert et al. 2017).
Commitment to the dauer developmental fate: the DA-DAF-12/VDR switch:
The nuclear receptor and VDR homolog DAF-12 is the transcriptional switch that commits animals to either reproductive development or dauer arrest. This switch is controlled by the synthesis of DAs (bile-acid-like steroid hormones) that act as DAF-12/VDR ligands (Motola et al. 2006). In favorable environments, hypodermal expression of the cytochrome P450 DAF-9/P450 (Gerisch et al. 2001; Jia et al. 2002), which catalyzes the final step in DA biosynthesis (Motola et al. 2006) (Figure 4), promotes DA biosynthesis and systemic ligand engagement of DAF-12/VDR, committing animals to reproductive development (Gerisch and Antebi 2004; Mak and Ruvkun 2004; Motola et al. 2006; Schaedel et al. 2012). When unfavorable ambient conditions inhibit DAF-7/TGFβ and DAF-2/IGFR signaling (Figure 2), transcriptional programs dependent upon DAF-3/SMAD, DAF-5/SnoN, and DAF-16/FoxO are initiated, and daf-9/P450 expression in the hypodermis is inhibited (Gerisch and Antebi 2004; Mak and Ruvkun 2004). In the absence of hypodermal DA biosynthesis, unliganded DAF-12/VDR promotes dauer arrest (Antebi et al. 2000; Schaedel et al. 2012) through a physical interaction with the SHARP corepressor ortholog DIN-1S (Ludewig et al. 2004).
Figure 4.
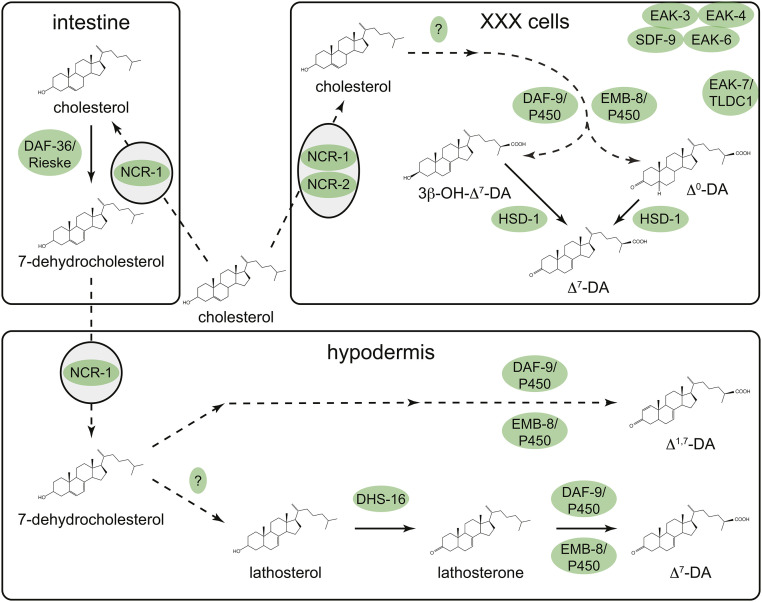
Hypothetical model of pathways involved in dafachronic acid (DA) biosynthesis by enzymes acting in intestine, XXX cells, and hypodermis. Solid arrows denote steps supported by experimental data. See text for details.
Intriguingly, whereas daf-12 is expressed ubiquitously throughout the animal (Antebi et al. 2000), daf-9 is expressed specifically in hypodermis, spermatheca, and the two endocrine XXX cells (Jia et al. 2002; Ohkura et al. 2003; Gerisch and Antebi 2004). A controlled temporal analysis of the dauer decision using crude dauer pheromone and synthetic Δ7-DA supports a model of developmental fate commitment, whereby inputs from DAF-7/TGFβ and DAF-2/IGFR pathways regulate levels of DA synthesis in the XXX cells. DA produced by the XXX cells that exceeds a threshold triggers a positive feedback loop that induces hypodermal DA synthesis, thus locking in the reproductive fate by ensuring systemic engagement of DAF-12/VDR by DA ligands (Schaedel et al. 2012).
Since the initial identification of Δ4- and Δ7-DA as DAF-12/VDR ligands (Motola et al. 2006) and the establishment of the roles of DAF-9/P450 and the Rieske oxygenase DAF-36 in DA biosynthesis (Motola et al. 2006; Rottiers et al. 2006), a number of genetic screens have identified other DA biosynthetic pathway components that act in distinct tissues (Figure 4). With the exception of daf-9/P450, null mutations in which cause nonconditional dauer arrest (Gerisch et al. 2001; Jia et al. 2002) strong loss-of-function mutations in all DA biosynthesis genes identified to date cause weak Daf-c or synthetic dauer-constitutive phenotypes, indicating that they are functionally redundant. In aggregate, the data support a model whereby distinct enzymes acting in multiple tissues contribute to DA biosynthesis in the XXX cells and the hypodermis (Figure 4).
A synthetic dauer-constitutive screen performed in a ncr-1 mutant background led to the identification of hsd-1, which encodes a putative 3-β-hydroxysteroid dehydrogenase that is expressed specifically in the XXX cells (Patel et al. 2008). Although HSD-1 is predicted to catalyze the conversion of cholesterol to the Δ4-DA precursor 4-cholesten-3-one, animals with a hsd-1 null mutation have wild-type levels of 4-cholesten-3-one (Wollam et al. 2012). Intriguingly, the identities of metabolites detected in hsd-1 null mutants, but not wild type, suggest that HSD-1 may act downstream of DAF-9/P450 in the XXX cells to synthesize Δ7-DA, possibly as a 3-β-hydroxysteroid dehydrogenase on 3β-OH-Δ7-DA and/or as a 7-desaturase on a novel DA, Δ0-DA (Mahanti et al. 2014) (Figure 4). hsd-1 mutations also emerged from a screen for enhancers of the akt-1 (eak) dauer-constitutive phenotype (Dumas et al. 2010); the fact that other EAK proteins are expressed specifically in the XXX cells (Ohkura et al. 2003; Hu et al. 2006), and likely act in the same pathway as HSD-1 (Hu et al. 2006; Alam et al. 2010; Dumas et al. 2010), suggests that they may also play a role in DA biosynthesis in the XXX cells (Figure 4). This is supported by a recent report describing a natural variant in the cis-regulatory region of eak-3 in a wild C. elegans isolate that reduces eak-3 expression, sensitizes animals to dauer-inducing cues, and retards development in favorable environments. The dauer sensitization and delayed development phenotypes are both rescued by exogenous Δ7-DA, suggesting that reduction of eak-3 activity impairs DA biosynthesis (Billard et al. 2020). A daf-36 enhancer screen revealed the identities of two other DA biosynthetic enzymes; DHS-16 is a 3-β-hydroxysteroid dehydrogenase that converts lathosterol to the Δ7-DA precursor lathosterone (Motola et al. 2006; Wollam et al. 2012), and EMB-8 is a cytochrome P450 oxidoreductase that may act as a cofactor for DAF-9/P450 in DA biosynthesis (Wollam et al. 2012) (Figure 4).
As commitment to the reproductive developmental fate occurs when DA concentrations exceed a threshold and trigger feed-forward DA synthesis in the hypodermis (Schaedel et al. 2012), it can be regulated through modulation of either DA production or the systemic threshold beyond which animals commit to reproductive development. A feature common to many of these regulatory mechanisms is the incorporation of inputs reflecting the internal metabolic state of the developing organism into the dauer commitment decision. Starvation directly inhibits DA biosynthesis, as C. elegans cannot synthesize the DA precursor cholesterol de novo and must obtain it through food consumption. Therefore, in addition to its effects on DAF-7/TGFβ and ILP production in ASI and other cells (Figure 2), starvation enhances dauer arrest by limiting the availability of cholesterol and other precursors for DA biosynthesis. Moreover, under conditions of nutrient deprivation (i.e., exogenous cholesterol limitation), endogenous metabolites such as phosphorylated glycosphingolipids and endocannabinoids influence commitment by promoting mobilization of sterols from internal stores (Boland et al. 2017; Galles et al. 2018). The levels and activity of DA biosynthetic enzymes are also subject to regulation. The nuclear receptor NHR-8 enhances Δ7-DA biosynthesis by promoting the intestinal expression of daf-36 (Magner et al. 2013). Recently, it has been reported that organismal NADPH levels are correlated with commitment to reproductive development (Penkov et al. 2015); this may occur through the regulation of DAF-9/P450 activity, as NADPH is an obligate cofactor for DAF-9/P450 (Motola et al. 2006). Enzymes such as the sterol methyltransferase STRM-1 enhance commitment to dauer arrest through covalent modification and subsequent shunting of cholesterol and DA precursors (Hannich et al. 2009; Mahanti et al. 2014). Finally, ascaroside pheromones, in addition to reducing expression of DAF-7/TGFβ and ILPs in sensory neurons (Figure 2), also promote dauer arrest by raising the threshold of DA needed to commit animals to the reproductive developmental fate (Schaedel et al. 2012). The mechanistic basis for this observation is not understood.
A number of questions pertaining to DA biosynthesis linger. Enzymes that catalyze established steps in DA biosynthesis (e.g., the reductase that catalyzes the conversion of 7-dehydrocholesterol to lathosterol; Figure 4) remain to be identified. In addition, the molecular nature of biosynthetic intermediates that link cholesterol to the putative Δ7-DA precursors 3β-OH-Δ7-DA and Δ0-DA, and 7-dehydrocholesterol to the novel DAF-12/VDR ligand Δ1,7-DA (Mahanti et al. 2014), is not known (Figure 4). Intriguingly, the lack of anatomic overlap in the expression of DAF-36/Rieske and DAF-9/P450 (Gerisch et al. 2001; Jia et al. 2002; Rottiers et al. 2006) implies that 7-dehydrocholesterol is transported from tissues and cells expressing DAF-36/Rieske to the hypodermis. The mechanisms underlying intercellular transport of DAs and their precursors are not fully understood, but may involve the Niemann-Pick C1 homologs NCR-1 and NCR-2 (Sym et al. 2000; Li et al. 2004) (Figure 4) and/or other transmembrane transporters required for the secretion and uptake of Drosophila steroid hormones (Yamanaka et al. 2015; Okamoto et al. 2018). Finally, pathways beyond current models for DA biosynthesis (Figure 4) remain to be discovered. For example, dhs-16;hsd-1 double null mutants are deficient in lathosterone but have wild-type levels of Δ7-DA (Wollam et al. 2012), indicating that lathosterone-independent routes to Δ7-DA biosynthesis exist.
Execution of the dauer developmental program:
Although the existence of dauer-specific morphological features, such as pharyngeal and cuticular remodeling, has been known for decades, the mechanistic basis for dauer execution is poorly understood. Recent work has revealed that traversal of the dauer developmental fate induces unexpected changes in gene expression, epigenetic regulation, behavior, morphology, metabolism, and genetic wiring, many of which require the activity of at least one of the terminal transcription factors regulated by DAF-7/TGFβ, ILPs, and DAs (Figure 5). Further, although DAs and DAF-12/VDR act downstream of DAF-3/SMAD, DAF-5/SnoN, and DAF-16/FoxO as a dauer commitment switch, these transcription factors appear to have additional, independent functions in the execution phase.
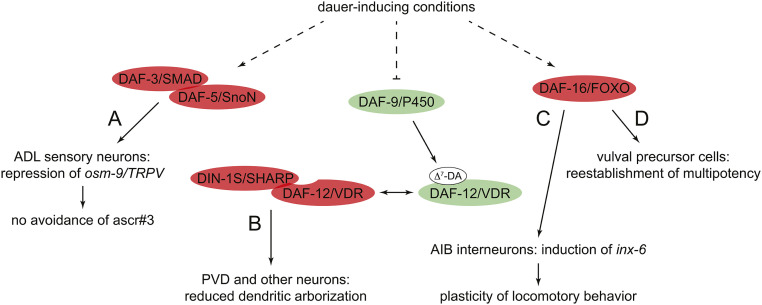
Figure 5.
Examples of dauer execution programs (A–D). Molecules depicted in green and red promote reproductive development and dauer arrest, respectively. See text for details.
Some dauer execution programs result in changes in neuronal gene expression and/or morphology that underlie dauer-specific behaviors. For example, DAF-16/FoxO-dependent expression of the innexin inx-6 in the AIB interneurons of dauers underlies dauer-specific plasticity in locomotory behavior (Bhattacharya et al. 2019) (Figure 5C). Nictation, which is a dauer-specific behavior that enhances dispersal (Yang et al. 2020), may be influenced by the induction of dendritic arborization of a subset of IL2 sensory neurons during dauer traversal (Schroeder et al. 2013). In contrast, dauer arrest results in DAF-12/VDR-dependent inhibition of dendritic arborization of the PVD neuron (Richardson et al. 2019) (Figure 5B); the functional consequences of dauer-induced changes in PVD morphology are not known.
Other dauer-specific changes ensure the fidelity of reproductive development after dauer exit. For example, DAF-16/FoxO ensures the reestablishment of multipotency in VPCs during dauer arrest (Karp and Greenwald 2013) (Figure 5D). Passage through dauer also induces rewiring of genetic networks that control developmental timing, as exemplified by dauer-induced changes in the regulatory machinery responsible for stage-specific repression of the Hunchback-like transcription factor gene hbl-1 (Ilbay and Ambros 2019).
Dauer execution also involves metabolic remodeling. Fat storage increases in dauers, and this requires autophagy (Meléndez et al. 2003). The observation that defects in autophagy result in the formation of abnormal dauers rather than suppression of dauer arrest supports the contention that increase in fat storage is part of a postcommitment dauer execution program. Intriguingly, studies on the autophagy gene atg-18 reveal that autophagy in sensory neurons promotes systemic fat storage nonautonomously. Dauer-specific lipids may also contribute to aspects of dauer morphogenesis; a novel class of glycolipids termed maradolipids that are only detected after the dauer commitment time point may contribute to dauer-specific specialization of the intestinal lumen (Penkov et al. 2010).
Intriguingly, traversal through dauer can cause long-term postdauer phenotypic changes, suggesting that a “memory” of adverse environmental conditions during larval development generates phenotypic plasticity in genetically identical adult animals with distinct life histories (Hall et al. 2010) (see Effects of dauer diapause in later life). In one specific instance of dauer-induced “memory,” passage through dauer induces DAF-3/SMAD- and DAF-5/SnoN-dependent repression of the TRPV channel gene osm-9 in the ADL chemosensory neurons of postdauer adults, resulting in abrogation of avoidance behavior toward the ascaroside ascr#3 (Sims et al. 2016) (Figure 5A).
Epigenetic regulation of the dauer decision:
As the implementation of specific transcriptional programs is required for the dauer decision, factors involved in epigenetic control of gene expression have also been implicated in dauer regulation. Most of these factors enhance dauer arrest by controlling aspects of sensory perception, signal transduction, and transcription factor activity upstream of the DA-DAF-12/VDR commitment switch. NURF-1, a conserved component of the NURF chromatin remodeling complex, may enhance the expression of molecules involved in the response to specific ascarosides, as it is required for the induction of dauer arrest by ascr#2 and ascr#3, but is dispensable for ascr#5-induced dauer formation (Large et al. 2016). Components of the RNA interference (RNAi) machinery act upstream of DAF-7/TGFβ and DAF-2/IGFR in sensory neurons to promote dauer arrest in response to pheromones (Bharadwaj and Hall 2017), and may also control the expression of genes encoding components of signaling pathways directly activated by ascarosides. The dosage compensation proteins DPY-21 and SET-4, which repress X-chromosome gene expression by controlling the methylation state of histone H4 lysine 20 (Brejc et al. 2017; Delaney et al. 2017), promote dauer arrest in hermaphrodites by reinforcing a feedback loop that activates DAF-16/FoxO through repression of the X-linked genes akt-2 and ins-9 (Delaney et al. 2017). The SWI/SNF chromatin remodeling complex also potentiates DAF-16/FoxO activity, doing so through physical interactions with DAF-16/FoxO at target gene promoters (Riedel et al. 2013). The heterochromatin protein family member HPL-2, which acts as a general repressor of gene expression (Couteau et al. 2002), may antagonize DAF-16/FoxO- and DAF-3/DAF-5 SMAD/SnoN-dependent transcriptional programs, as hpl-2 loss-of-function mutations enhance dauer arrest in daf-2/IGFR and daf-7/TGFβ mutants (Meister et al. 2011).
Developmental timing regulators and dauer arrest:
In addition to its role as a dauer commitment switch, DAF-12/VDR also has a general function in developmental timing (Antebi et al. 1998). Heterochronic genes that interact with daf-12 can also influence the dauer decision. For example, lin-42, which encodes the C. elegans ortholog of Period circadian proteins (Jeon et al. 1999), acts at the level of daf-12/VDR to inhibit dauer arrest (Tennessen et al. 2010). The heterochronic gene hbl-1, which encodes a Hunchback-like transcription factor (Abrahante et al. 2003; Lin et al. 2003), also influences dauer arrest through complex interactions with DAF-2/IGFR, DAF-7/TGFβ, and DA pathways (Karp and Ambros 2011). The transcriptional repressor BLMP-1, a major target of the E3 ubiquitin ligase and heterochronic protein DRE-1 (Horn et al. 2014), is required for dauer arrest in daf-2/IGFR, daf-7/TGFβ, and daf-9/P450 mutants, and may act at the level of dauer commitment as well as in the execution phase to promote dauer-specific epidermal remodeling (Horn et al. 2014; Hyun et al. 2016).
L1 arrest
In contrast to dauer diapause, larvae that hatch in the complete absence of food arrest development without morphological modification, in a state known as L1 arrest (also referred to as L1 diapause) (Baugh 2013) (Figure 1). Larvae can survive L1 arrest for weeks, and they resume development upon feeding. Greenwald and Horvitz first reported that wild-type larvae arrest development in the L1 stage after hatching in the absence of bacteria (Greenwald and Horvitz 1982). Johnson et al. (1984) reported survival and movement of L1 larvae with no growth for up to 12 days of starvation, indicating that larvae arrested by absolute starvation upon hatching do not form dauer larvae. Arrested L1 larvae could also be recovered by feeding with grossly normal subsequent development. Protocols for preparation of larvae in L1 arrest became routine for sterilization, synchronization, and freezing of strains (Lewis and Fleming 1995; Stiernagle 2006). L1 arrest reflects active regulation as opposed to a passive consequence of limited nutrition (Baugh and Sternberg 2006; Fukuyama et al. 2006), and it has become an important model for nutritional control of development. L1 arrest is reviewed elsewhere (Baugh 2013; Fukuyama 2018). Here, we will focus on critical background and recent progress on signaling and developmental regulation during L1 arrest.
Assaying L1 developmental progression:
L1 development can be tracked by monitoring a variety of cell types (Sulston and Horvitz 1977). Q neuroblasts migrate and divide early during L1 development to form six neurons. Lateral epidermal seam cells of the V, H, and T lineages undergo a stereotyped progression of asymmetric division, fusion, and elongation, providing greater developmental resolution than cell division alone. P neuroblasts migrate ventrally and divide during L1 development, and some of their descendants differentiate into motor neurons near the L1 molt. There is a single M cell at hatching, and it undergoes a series of divisions to produce 18 cells by the end of the L1 stage providing facile quantification late L1 development. In addition, primordial germ cells (PGCs) Z2 and Z3 divide during L1 development. A number of reporter genes that facilitate analysis of these developmental events are available (Kaplan et al. 2015; Roy et al. 2018; Zheng et al. 2018b). Size can also be assessed with image-based analysis (Moore et al. 2013), and cuticular alae can be examined to determine when the L1 molt has occurred (Page and Johnstone 2007).
Insulin/insulin-like growth factor signaling in regulation of L1 arrest:
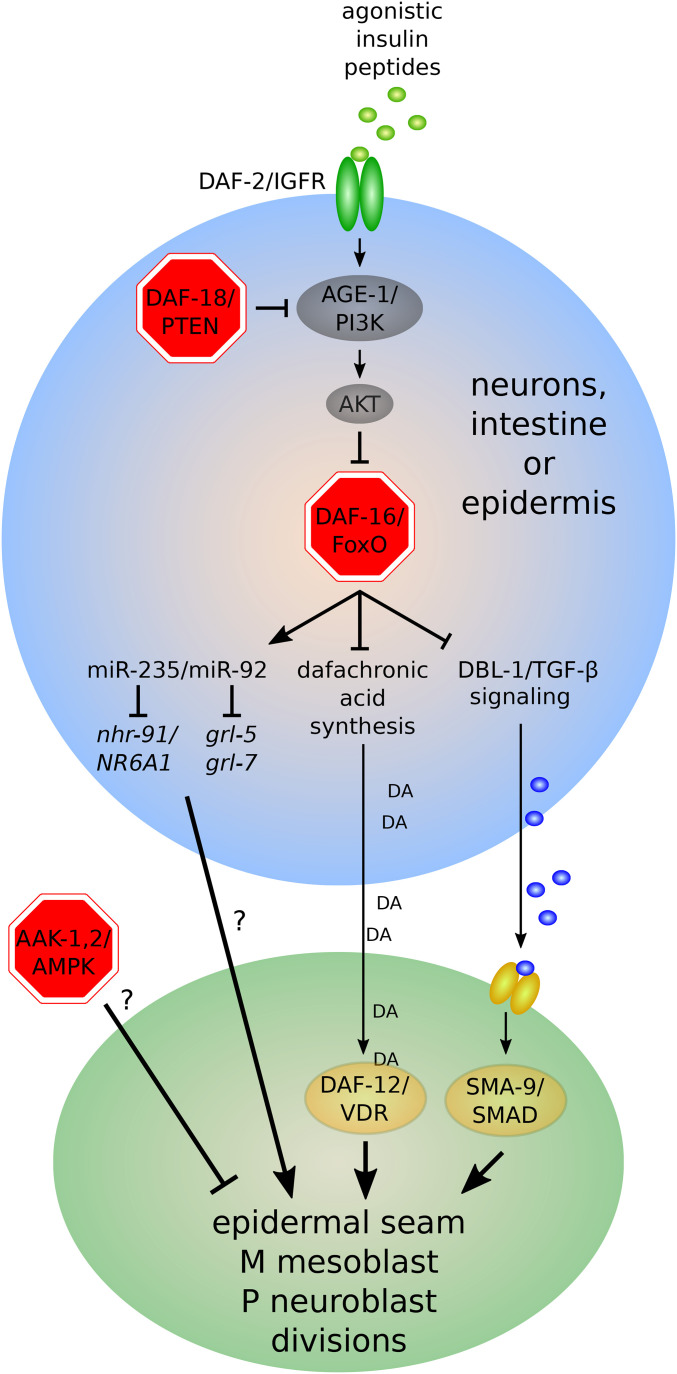
Insulin/insulin-like growth factor signaling (IIS) governs L1 development (Figures 6–8). Mutation of the only known IGFR, daf-2/IGFR, causes L1-stage developmental arrest in fed larvae at high temperature (Gems et al. 1998). However, arrest is reversible at low temperature, consistent with physiological regulation as opposed to impaired development (Baugh and Sternberg 2006). Disruption of ILP secretion also causes constitutive L1 arrest, confirming physiological regulation of development (Kao et al. 2007). daf-2/IGFR mutants are also resistant to L1 starvation (see Energy homeostasis regulators) (Munoz and Riddle 2003; Baugh and Sternberg 2006). Disruption of the chromatin remodeler let-418/CHD4 causes an L1 arrest phenotype at high temperature (Erdelyi et al. 2017), and genetic interactions with daf-2/IGFR suggest a regulatory relationship between IIS and chromatin modification (Saudenova and Wicky 2018). DAF-2/IGFR signaling acts via the phosphoinositide 3-kinase (PI3K) pathway to antagonize activity of the FoxO DAF-16 (Figure 6) (Lin et al. 1997; Ogg et al. 1997). PI3K signaling results in AKT-mediated phosphorylation of DAF-16/FoxO, causing localization of DAF-16/FoxO to the cytoplasm. IIS is reduced during starvation, resulting in nuclear localization of DAF-16/FoxO (Henderson and Johnson 2001). Nuclear DAF-16/FoxO activity promotes stress resistance during starvation (Henderson et al. 2006). daf-16/FoxO mutants fail to arrest somatic development in starved L1 larvae (Baugh and Sternberg 2006), and they are sensitive to starvation and die rapidly (Munoz and Riddle 2003). daf-16/FoxO is epistatic to daf-2/IGFR for both phenotypes.
Figure 6.
Regulation of lateral epidermal seam cell, M mesoblast, and P neuroblast divisions during L1 arrest. Factors required to arrest cell divisions are shown in red. See text for details.
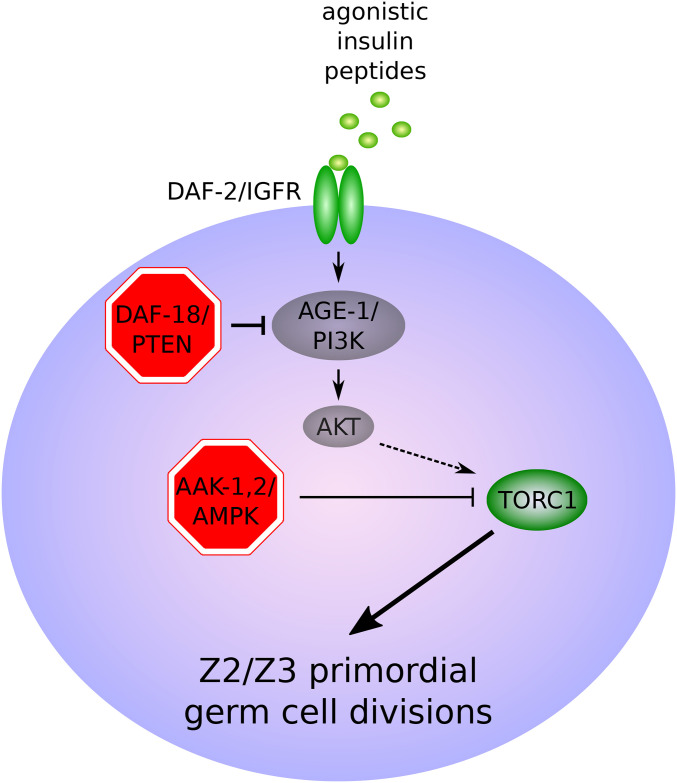
Figure 8.
Regulation of Z2/Z3 primordial germ cell divisions during L1 arrest. IIS is activated in response to feeding, presumably by unknown agonistic ILPs, which is hypothesized to result in activation of TORC1 and Z2/Z3 divisions (Fukuyama et al. 2012). Factors required to arrest cell divisions are shown in red. See text for details.
daf-16/FoxO regulates expression of a variety of genes directly or indirectly to promote developmental arrest. daf-16/FoxO inhibits signaling pathways that promote L1 development including seam, P, and M cell divisions (Figure 6; see below) (Kaplan et al. 2015). daf-16/FoxO promotes expression of the microRNA miR-235 (ortholog of mammalian miR-92) in starved L1 larvae, and mir-235 is also required for arrest of P and M cells. Upregulation of the miR-235 target nhr-91/NR6A1 contributes to the arrest-defective phenotype of the mir-235 mutant (Kasuga et al. 2013). The cyclin-dependent kinase inhibitor cki-1/p27 is required to arrest seam cell divisions during L1 starvation (Hong et al. 1998), and cki-1/p27 is not appropriately expressed in the seam cells of starved L1 daf-16/FoxO mutants, suggesting a mechanism of cell cycle regulation (Baugh and Sternberg 2006). daf-16/FoxO also represses expression of the microRNA lin-4 (Baugh and Sternberg 2006), which promotes L2 identity and progression of developmental time (Feinbaum and Ambros 1999; Olsen and Ambros 1999). How DAF-16/FoxO inhibits other aspects of development, including cell migration and fusion, is unknown.
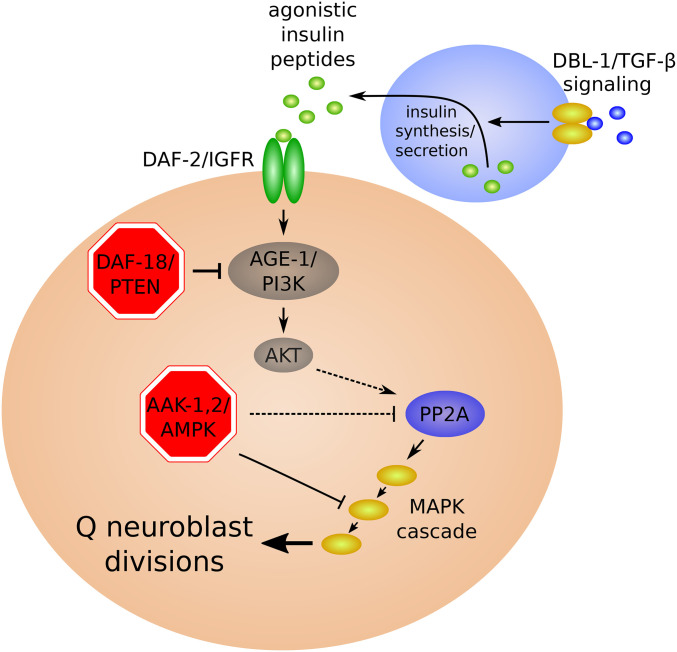
In contrast to the epidermal seam cells, P neuroblasts, and M mesoblasts (Baugh and Sternberg 2006; Kaplan et al. 2015), the germ cells and Q neuroblasts remain arrested in starved L1 daf-16/FoxO mutants (Fukuyama et al. 2006; Fukuyama 2018; Zheng et al. 2018b) (Figures 6–8). These observations suggest the existence of an effector of DAF-2/IGFR signaling distinct from DAF-16/FoxO in the germ cells and Q neuroblasts. The PTEN ortholog DAF-18, a negative regulator of DAF-2/IGFR signaling, is required for somatic arrest during L1 starvation (Fukuyama et al. 2015; Zheng et al. 2018b), as is the AMP-activated protein kinase (AMPK) (Baugh and Sternberg 2006; Fukuyama et al. 2012; Zheng et al. 2018b). Both are also required for L1 starvation survival (see Energy homeostasis regulators) (Baugh and Sternberg 2006; Fukuyama et al. 2012). Critically, daf-18/PTEN and AMPK are each required to arrest germ cell and Q neuroblast divisions during L1 starvation, but daf-16/FoxO is not (Fukuyama et al. 2006; Fukuyama et al. 2012; Fukuyama et al. 2015; Zheng et al. 2018b). In germ cells, daf-18/PTEN and AMPK function in parallel and converge on inhibition of TOR complex 1 (TORC1) (Fukuyama et al. 2012). AMPK functions downstream of, or in parallel to, daf-18/PTEN in regulation of Q cell divisions, and AMPK inhibits PP2A to maintain Q cell quiescence (Zheng et al. 2018b). In daf-18/PTEN and AMPK mutants, PP2A is abnormally activated during L1 starvation, leading to activation of the MAP kinase MPK-1 via LIN-45/RAF to promote Q cell divisions (Zheng et al. 2018b). In mammals, FoxO transcription factors, PTEN, and AMPK function as tumor suppressors (Paik et al. 2007; Chalhoub and Baker 2009; Zadra et al. 2015), indicating conserved roles in regulation of cell proliferation.
Cell-autonomous and -nonautonomous function of IIS:
The IIS pathway regulates development cell-autonomously and -nonautonomously during L1 arrest and recovery (Figures 6–8). Genetic mosaic analysis as well as tissue-specific transgenic rescue of insulin/insulin-like growth factor (IGF) pathway components revealed nonautonomous function in regulation of aging (Apfeld and Kenyon 1998; Wolkow et al. 2000; Libina et al. 2003). Tissue-specific transgenic rescue of daf-16/FoxO null mutants was performed in a daf-2/IGFR mutant background (Libina et al. 2003). This study suggested that the constitutive developmental-arrest phenotype of daf-2 mutants (L1 and dauer arrest) can result from daf-16 activity in specific somatic tissues. Similarly, transgenic expression of daf-16 exclusively in the intestine, epidermis, or nervous system of a daf-16 null mutant is sufficient to rescue developmental arrest of the M mesoblast, epidermal seam cells, and VB neurons during L1 starvation (Kaplan et al. 2015). Likewise, tissue-specific expression of daf-18/PTEN, a positive regulator of daf-16/FoxO, in the intestine, epidermis or nervous system is sufficient to rescue arrest of the P neuroblasts (Fukuyama et al. 2015). These sites of cell-nonautonomous action have also been reported for daf-16 in regulation of aging (Libina et al. 2003; Zhang et al. 2013). These observations suggest that DAF-16/FoxO regulates signaling from multiple tissues to promote developmental arrest. Tissue-specific transgenic expression of a gain-of-function akt-1 allele revealed cell-nonautonomous regulation of P neuroblast and M mesoblast divisions, although in this case the epidermis was found to be the salient site of action (Fukuyama et al. 2015). However, gain-of-function akt-1 can also activate M mesoblast divisions cell-autonomously (Fukuyama et al. 2015), and daf-18/PTEN regulates Q neuroblast divisions cell-autonomously (Zheng et al. 2018b). These cell-type-specific distinctions presumably reflect intricacies of the IIS pathway in an organismal context, although technical limitations of the transgenic approaches used may also be a factor.
An insulin/IGF, dbl-1/TGF-β, daf-12/VDR, and hedgehog-like signaling network:
Nonautonomous function of insulin/IGF pathway components suggests that daf-16/FoxO directly or indirectly regulates activity of one or more additional signaling pathways in controlling L1 development. Dauer development is regulated by the daf-7/TGF-β pathway and daf-12/VDR steroid hormone signaling in addition to IIS and daf-16/FoxO (Hu 2007; Fielenbach and Antebi 2008). The dbl-1/TGF-β pathway is largely distinct from the daf-7/TGF-β pathway, and it regulates adult body size and male tail development (Savage-Dunn 2005). Genetic epistasis and gene expression analyses suggest that daf-16/FoxO inhibits dbl-1/TGF-β and daf-12/VDR signaling to promote arrest of M mesoblast and epidermal seam cell divisions during L1 starvation (Kaplan et al. 2015) (Figure 6). Furthermore, disruption of dbl-1/TGF-β or daf-12/VDR signaling reduces the rate of L1 development in fed larvae, showing that these pathways support L1 development (Kaplan et al. 2015). Together, these results suggest that daf-16/FoxO causes developmental arrest in starved L1 larvae by inhibiting pathways that otherwise promote development.
mir-235/miR-92, which is activated by daf-16/FoxO, cell-nonautonomously regulates P neuroblast divisions from the epidermis (Kasuga et al. 2013; Fukuyama et al. 2015) (Figure 6). miR-235 represses expression of the hedgehog-related genes grl-5 and grl-7 during L1 arrest, and their expression is upregulated in the epidermis in response to feeding as miR-235 levels decrease (Kume et al. 2019). Forced expression of grl-5 or grl-7 in the epidermis of starved larvae activates P neuroblast divisions, and grl-5 and grl-7 are required for the arrest-defective phenotype of the mir-235 mutant (Kume et al. 2019). How miR-235 or its targets nhr-91, grl-5, and grl-7 regulate P cell divisions cell-nonautonomously is not understood.
daf-18/PTEN regulates Q neuroblast divisions in cell-autonomous fashion during L1 arrest (Zheng et al. 2018b) (Figure 7). Mutations affecting the dbl-1/TGF-β pathway suppress Q cell divisions in starved daf-18/PTEN mutants. In contrast to regulation of M mesoblast and epidermal seam cell divisions (Kaplan et al. 2015), dbl-1 signaling appears to function upstream of the insulin/IGF pathway to regulate Q cell divisions (Zheng et al. 2018b). Disruption of dbl-1 signaling reduces expression of the agonist ILP genes ins-3 and ins-4, reducing DAF-2/IGFR pathway activity. Overexpression of these peptides is also sufficient to activate Q cell divisions in starved dbl-1 mutant larvae, consistent with ins-3 and ins-4 functioning downstream of dbl-1 (Zheng et al. 2018b). Together with Kaplan et al. (2015), these results suggest that dbl-1/TGF-β signaling is potentially involved upstream or downstream of IIS depending on the cell type in question (Zheng et al. 2018b).
Figure 7.
Regulation of Q neuroblast divisions during L1 arrest. ins-3, ins-4, and presumably other agonistic ILPs secreted from chemosensory neurons and possibly other tissues in response to food activate IIS, leading to the activation of PP2A and the RAF-MEK-ERK MAP kinase cascade and Q neuroblast divisions (Zheng et al. 2018b). Factors required to arrest cell divisions are shown in red. See text for details.
ILPs governing L1 arrest and development:
The C. elegans genome encodes 40 ILPs that function broadly as either putative agonists or antagonists of DAF-2/IGFR based on phenotypic analysis (Pierce et al. 2001; Fernandes de Abreu et al. 2014). ILP sequences and predicted structure enabled classification of each as alpha, beta, or gamma (Pierce et al. 2001). One study found that beta classification is a good predictor of agonist function (Zheng et al. 2018a). Although hindered by genetic redundancy, loss-of-function phenotypic analysis is the most reliable way to determine whether individual ILPs activate or inhibit IIS in their native context. Such analysis of nearly all ILPs has been completed for a variety of IIS-regulated phenotypes with the exception of L1 arrest and development (Fernandes de Abreu et al. 2014).
Identification of IIS as a critical regulator of L1 arrest begged the question of which ILPs are affected by nutrient availability to govern L1 development. Expression analysis revealed complex expression patterns in time and space, with a large proportion of insulin-like genes expressed in chemosensory neurons and the intestine (Pierce et al. 2001; Baugh et al. 2011; Ritter et al. 2013). Time-series analysis of messenger RNA (mRNA) expression for all 40 peptides in fed and starved L1 larvae identified 13 candidate agonists and 8 candidate antagonists based on whether they are positively or negatively regulated by food, respectively (Chen and Baugh 2014). Ethanol and/or amino acids are sufficient to induce expression of several of these putative agonists in otherwise starved L1 larvae (Fukuyama et al. 2015), and perception of food is sufficient to induce expression of daf-28 and ins-6 (Kaul et al. 2014; Kaplan et al. 2018). Expression-based classification agreed remarkably well with published genetic analyses of a variety of phenotypes, and it successfully identified ins-4, ins-6, and daf-28 as functional agonists in L1 larvae (Chen and Baugh 2014). As a complementary approach to classify ILPs as putative agonists or antagonists, peptides were overexpressed pan-neuronally and L1 starvation survival, Q cell divisions, dauer formation, and fat accumulation were measured (Zheng et al. 2018a). L1 starvation survival and Q cell divisions are the only L1 arrest-specific phenotypes. This analysis identified 16 putative agonists, 8 antagonists, and 11 with function contingent on the observed phenotype. Caveats apply to both of these classification approaches (i.e., expression does not necessarily correlate with function, and function may be context-dependent), and although there is substantial overlap in the resulting classifications, there are also discrepancies.
Genetic analysis suggests that at least three putative agonist ILPs, daf-28, ins-4, and ins-6, function redundantly in L1 larvae. Disruption of any one of these three alone has no detectable phenotypic consequence, but simultaneous disruption of daf-28 and ins-4 increases starvation resistance, phenocopying reduction of daf-2/IGFR function (Chen and Baugh 2014). These putative agonists are expressed in starved L1 larvae, albeit at relatively low levels compared to fed larvae, suggesting basal levels of IIS reduce starvation resistance. Overexpression of daf-28, ins-4, or ins-6 with their own promoters is sufficient to promote M cell division in starved L1 larvae (Chen and Baugh 2014), and pan-neuronal overexpression of daf-28, ins-4, or ins-6, as well as 10 other ILPs, is sufficient to promote Q cell divisions (Zheng et al. 2018a). Simultaneous disruption of five candidate agonists (daf-28, ins-4, ins-5, ins-6, and ins-7) had no detectable effect of the rate of M or seam cell divisions in fed larvae, although disruption of daf-2/IGFR clearly did (Chen and Baugh 2014), consistent with extensive functional redundancy among ILPs (Pierce et al. 2001). Together, these results suggest that ins-4, ins-6, daf-28 and possibly other agonists redundantly promote L1 development in response to feeding.
Putative antagonists presumably counteract function of agonists, promoting developmental arrest and starvation resistance. Function of putative antagonists during L1 arrest has not been investigated by loss-of-function analysis, but pan-neuronal overexpression of eight different ILPs increased L1 starvation survival in a wild-type background (Zheng et al. 2018a). In addition, overexpression of the same eight genes suppressed Q cell divisions in a starved L1 arrest-defective daf-18/PTEN mutant (Zheng et al. 2018a).
A common set of ILPs appears to regulate L1 arrest and dauer diapause. In addition to regulating L1 arrest, daf-28, ins-4, and ins-6 function as agonists in regulation of dauer development, promoting dauer bypass or recovery (Li et al. 2003; Kao et al. 2007; Cornils et al. 2011; Fernandes de Abreu et al. 2014; Hung et al. 2014; Zheng et al. 2018a). Reporter gene analysis of daf-28, ins-4, and ins-6 in L1 larvae revealed expression in chemosensory neurons and intestine (Chen and Baugh 2014). This expression pattern is consistent with other developmental stages, including dauer entry and exit (Li et al. 2003; Cornils et al. 2011; Ritter et al. 2013; Hung et al. 2014). Common signals and signaling centers suggest that regulation of L1 arrest and dauer formation is distinguished in part by the degree of IIS, with L1 arrest resulting from very low signaling activity and dauer arrest resulting from relatively low to moderate activity, although the timing of signaling could also be a factor. The penetrance of constitutive arrest as L1 or dauer larvae is correlated across allelic series of daf-2/IGFR mutants, with the penetrance of L1 arrest consistently lower (Gems et al. 1998), although biochemical or other evidence that signaling activity correlates with phenotype would strengthen the model. Nonetheless, additional signaling pathways contribute to distinct developmental outcomes as well.
Nutrient sensing and feedback regulation of IIS:
Feedback regulation of IIS is widespread and likely contributes to homeostasis. daf-16/FoxO represses transcription of the putative DAF-2/IGFR agonist ins-7, producing positive feedback and FoxO-to-FoxO signaling (Murphy et al. 2003; Murphy et al. 2007). Likewise, daf-16/FoxO activates transcription of the putative antagonist ins-18, again producing positive feedback (Murphy et al. 2003; Matsunaga et al. 2012). Disruption of insulin gene function and measurement of insulin gene expression revealed several instances of insulin-to-insulin signaling, as predicted for feedback regulation (Fernandes de Abreu et al. 2014). Expression analysis of insulin-like mRNAs over time during recovery from L1 arrest in daf-2/IGFR and daf-16/FoxO mutants revealed extensive positive and negative feedback, with the majority of insulin peptides contributing (Kaplan et al. 2019). Feedback regulation presumably couples signaling activity across tissues in support of organismal homeostasis. Indeed, regulation of dauer development involves insulin-mediated signaling between the intestine and nervous system (Hung et al. 2014). Chemosensory neurons likely respond to external conditions, and the intestine likely responds to internal conditions, with feedback integrating these inputs by propagating signaling throughout the animal to achieve a coherent physiological state and coordination of postembryonic development (Kaplan and Baugh 2016). Consistent with this model, sensory perception of food, or even just polypeptide, without feeding is sufficient to activate daf-28-mediated IIS in starved L1 larvae, affecting gene expression and lipid metabolism (Kaplan et al. 2018). However, specific contributions of feedback regulation to developmental dynamics and homeostasis have not been determined.
Lipid-TORC1 signaling and neuronal differentiation:
Somatic cells in C. elegans generally do not divide, migrate, or fuse during stringent L1 arrest (suspended in buffer), based on observation of lateral epidermal seam cells, P neuroblasts, Q neuroblasts, and the M mesoblast (Baugh and Sternberg 2006; Fukuyama et al. 2015; Zheng et al. 2018b). The germ cells Z2 and Z3 also do not divide (Fukuyama et al. 2006). These events represent most but not all of the developmental events that occur during the L1 stage (Sulston and Horvitz 1977). The AWC sensory neurons sense volatile components of food and direct chemotaxis (Bargmann et al. 1993), and they complete their differentiation during postembryonic development (Troemel et al. 1999). Unlike other L1 developmental events, AWC differentiation does not depend on feeding (Kniazeva et al. 2015). Rather, AWC neurons differentiate in L1 larvae whether fed or starved, although differentiation is slower in starved larvae (Kniazeva et al. 2015). Monomethyl branched-chain fatty acids, which are synthesized by the worm, are essential to postembryonic growth and development (Kniazeva et al. 2008). The TORC1 kinase mediates regulation of growth and development by a critical monomethyl branched-chain fatty acid-derived sphingolipid (Zhu et al. 2013). This lipid-TORC1 pathway functions in the intestine of fed and starved L1 larvae to cell-nonautonomously promote maturation of AWC sensory neurons, supporting foraging behavior (Kniazeva et al. 2015). Such function of TORC1 is surprising in that it is usually inactive during starvation (TOR signaling is reviewed elsewhere; Blackwell et al. 2019). Foraging during starvation is clearly important to the animal, illustrating an effect of cellular energy homeostasis pathways on animal behavior.
Larval starvation and vulval induction
Larval starvation influences vulval development in mutants that exhibit defects in vulval induction. A specialized somatic gonad cell, the anchor cell, secretes LIN-3/EGF during the L3 stage to activate RAS-MAPK signaling in the adjacent VPC, thereby specifying the 1° vulval cell fate. EGF-RAS-MAPK activation causes lateral inhibition of the 1° fate and induction of the 2° fate in the neighboring VPCs via activation of the Delta-Notch pathway (Sternberg 2005). It was originally reported that dauer formation or larval starvation without dauer formation suppressed the incompletely penetrant vulvaless (Vul) phenotype of several mutants (lin-2, lin-3, lin-7, lin-24, lin-33, and let-23) affecting the EGF-RAS-MAPK pathway, suggesting that starvation increases inductive signaling (Ferguson and Horvitz 1985). Braendle and Félix systematically characterized vulval development in six different ecologically relevant conditions, including 48-hr L2 starvation period and dauer-forming conditions (before being returned to standard culture conditions with food, allowed to develop, and assayed) (Braendle and Félix 2008). They found that the vulval phenotype of 26 out of 41 mutants examined was modified by environmental conditions, with starvation and dauer formation producing the most frequent and consistent effects. Consistent with Ferguson et al., they found that L2 starvation or dauer formation suppressed the Vul phenotype of loss-of-function lin-3/EGF and let-23/EGFR mutants. However, another study subjecting L2 larvae to a 36-hr starvation period reported suppression of the multivulva phenotype caused by a let-60/RAS gain-of-function mutant, suggesting that starvation actually inhibits inductive signaling (Battu et al. 2003). To resolve this paradox, it has been proposed that L2 starvation elicits antagonistic effects on vulval induction, with a positive effect of starvation emanating from internal nutrient status and a smaller negative effect from external conditions (Grimbert et al. 2018). Notably, L2 starvation has no appreciable effect on wild-type vulval development despite modifying mutant phenotypes (Braendle and Félix 2008). It is intriguing to speculate that the effects of starvation or dauer formation revealed by mutants reflect compensatory mechanisms that confer developmental robustness to environmental variation.
Late larval arrest
L1 larvae are not unique in their ability to arrest development as an acute starvation response (Figure 1). In the initial characterization of L1 arrest, it was noted that there is no growth in L2 or L4 larvae subjected to starvation (for 24 and 48 hr after isolation of eggs, respectively), but that they continued moving (Johnson et al. 1984). Thirty years later, it was reported that worms that are starved as L2 or L3 larvae complete a molt and arrest development at the beginning of the next larval stage, revealing a developmental checkpoint just before initiation of the molting cycle in L3 and L4 larvae (Schindler et al. 2014). Arrested L3 and L4 larvae have no morphological modification, similar to L1 arrest, and larvae in L3 arrest do not display dauer-specific features (Schindler et al. 2014). Survival of late larval starvation is also more similar to L1 than dauer arrest, lasting for weeks rather than months, although L3 larvae survive starvation longer than newly hatched (unfed) L1 larvae (Hibshman et al. 2018). There are three VPCs that undergo three rounds of cell division during L3 development to generate the 22 cells that comprise the mature vulva (Sulston and Horvitz 1977). Given the discrete nature of the L3 and L4 developmental checkpoints, larvae starved near the beginning of the L3 stage arrest with either three VPCs or 22 vulva cells (Schindler et al. 2014). The fact that intermediate points of developmental progression were not observed indicates that if an individual larva bypasses a given checkpoint it completes the larval stage despite being starved and arrests at the next checkpoint. Notably, TORC1 activity is necessary for developmental progression through each larval stage, suggesting TORC1 signaling licenses development in fed larvae (Duong et al. 2020).
IIS regulates late larval arrest:
Consistent with L1 and dauer arrest, IIS regulates late larval arrest (Schindler et al. 2014) (Figure 6). daf-16/FoxO null mutants are arrest-defective, and many of the larvae starved during the L2 or L3 stage bypass the L3 or L4 checkpoint, respectively. Shifting temperature-sensitive daf-2(e1370) mutants from the permissive to the restrictive temperature at the mid-L2 stage (which is beyond the dauer decision time point) resulted in transient developmental delay at the L3 and L4 checkpoints in fed larvae (Schindler et al. 2014). As is the case for the daf-2/IGFR constitutive L1 arrest phenotype, these transient delays are daf-16/FoxO-dependent. Likewise, daf-16/FoxO functions cell-nonautonomously to promote L3 and L4 arrest (Schindler et al. 2014). Epidermal expression of a daf-16/FoxO transgene with a tissue-specific promoter had the strongest effect of the tissues examined, but this effect was weak compared to expression of DAF-16/FoxO with its own promoter (Schindler et al. 2014), suggesting it may function in other tissues as well, as in L1 arrest (see Cell-autonomous and -nonautonomous function of IIS) (Kaplan et al. 2015).
daf-9/P450 promotes late larval development:
IIS likely regulates steroid hormone signaling during late larval development. The cytochrome P450-encoding gene daf-9/P450 operates in a steroid hormone pathway including daf-12/VDR that regulates dauer development (Gerisch and Antebi 2004; Mak and Ruvkun 2004). Disruption of daf-9/P450 function suppressed the late larval arrest-defective phenotype of a daf-16/FoxO mutant (Schindler et al. 2014), revealing an additional similarity to regulation of L1 arrest and dauer formation. In addition, overexpression of daf-9/P450 caused bypass of both L3 and L4 developmental checkpoints during starvation (Schindler et al. 2014). Overexpression of daf-9/P450 also suppressed developmental arrest caused by loss of TORC1 activity (Duong et al. 2020). Curiously, daf-12/VDR does not appear to regulate progression through L3 and L4 checkpoints. Daf-d mutants with the daf-12(rh61rh411) null allele did not bypass the L3 checkpoint during starvation, and this mutant did not suppress the arrest-defective phenotype of a daf-16/FoxO null mutant or daf-9/P450 overexpression (Schindler et al. 2014). These results suggest that daf-16/FoxO inhibits daf-9/P450 activity to promote L3 and L4 arrest during starvation, like L1 and dauer arrest, but that daf-9/P450 acts through an effector other than daf-12/VDR, unlike L1 and dauer arrest. daf-9/P450 functions in the biosynthetic pathway for DA, a DAF-12/VDR ligand involved in regulation of dauer development (Motola et al. 2006). However, the genome encodes 284 nuclear hormone receptors (NHRs), mostly uncharacterized (Antebi 2006, 2015). It is intriguing to speculate that daf-9/P450 participates in an unidentified steroid hormone pathway to regulate nutrient-dependent progression through late larval development.
ARD
Starved L3 and L4 larvae can continue development and arrest as reproductive adults in the ARD (Figure 1) (Angelo and Van Gilst 2009). ARD was recently reviewed elsewhere (Carranza-García and Navarro 2020), but a systematic characterization of the physiology and genetic requirements of ARD was subsequently published (Gerisch et al. 2020). The germline is dramatically reduced to a small number of quiescent stem cells during ARD (Angelo and Van Gilst 2009; Seidel and Kimble 2011), and the soma is also morphologically modified, resulting in “mini-adults” (Gerisch et al. 2020). Worms in ARD are behaviorally quiescent, appearing flaccid and not moving unless provoked. Remarkably, a substantial portion of the population can recover from ARD within a day of feeding, with resumption of germline proliferation, reproduction, and movement (Angelo and Van Gilst 2009; Seidel and Kimble 2011; Gerisch et al. 2020). Sperm lose viability during ARD, as self-fertility upon recovery decreases with longer periods of starvation, while mating preserves fertility (Angelo and Van Gilst 2009). Nonetheless, ARD enables mature worms to adapt to starvation by postponing reproduction.
Understanding ARD has been hampered by conflicting reports on its induction and properties. It was reported that embryogenesis is arrested in utero during ARD (Angelo and Van Gilst 2009), but this conclusion was challenged (Seidel and Kimble 2011). Population density was originally reported to affect induction (Angelo and Van Gilst 2009), but subsequent work suggests this is not the case (Seidel and Kimble 2011; Gerisch et al. 2020). ARD can involve exceptional longevity (nearly threefold at 20° and over fivefold at 15°) (Gerisch et al. 2020), but this depends on culture conditions. Exceptional longevity apparently requires initiation of starvation specifically in mid-L3 larvae with subsequent culture on nematode growth medium (NGM) plates (Gerisch et al. 2020). However, absolute starvation (salt buffer or plates without peptone) of L4 larvae can also cause developmental arrest as adults, although without such exceptional longevity (Seidel and Kimble 2011; Schindler et al. 2014). It is possible that nutrition provided from the peptone in NGM medium is necessary for exceptional longevity (Kaplan et al. 2018) and possibly other reported ARD properties (see Commentary on starvation conditions). In any case, ARD survival requires that animals avoid “bagging” (internal hatching of embryos; see Egg laying) (Angelo and Van Gilst 2009; Seidel and Kimble 2011).
Regulation of ARD is not as well understood as other developmental responses to starvation. It is clear that the genetic requirements for ARD are distinct from dauer diapause (Gerisch et al. 2020). Factors required for survival of L1 starvation and dauer arrest such as daf-16/FoxO and AMPK are required to survive ARD, but other pathways critical to dauer formation are dispensable for ARD. Moreover, hlh-30/TFEB is a master regulator of ARD (Gerisch et al. 2020). The NHR nhr-49 is not required for ARD (Gerisch et al. 2020), although it was originally reported to be (Angelo and Van Gilst 2009). Gonad shrinkage was reportedly due to apoptosis (Angelo and Van Gilst 2009), but it has been reported that shrinkage occurs independent of apoptosis, suggesting that shrinkage is due to ongoing ovulation (Seidel and Kimble 2011; Carranza-García and Navarro 2019). Phosphorylation of the MAP kinase MPK-1 promotes meiotic progression and oocyte maturation (Lee et al. 2007). In fed worms, DAF-2/IGFR stimulates and DAF-18/PTEN inhibits phosphorylation of MPK-1 in daf-16/FoxO-independent fashion, and phosphorylation is lost within hours of starvation, causing oogenesis to stall (Lopez et al. 2013). GLP-1/Notch signaling is required for germline stem cell maintenance in fed worms, but it is dispensable during starvation-induced cell cycle quiescence (Seidel and Kimble 2015). The rate of germ cell proliferation during recovery from adult starvation is comparable to the rate in fed L3 larvae, but the number of proliferative germ cell nuclei plateaus with ∼30% fewer than in adults that were continuously fed (Roy et al. 2016). Delayed reproduction in response to starvation depends, in part, on mgl-1/GRM3 and neuropeptide signaling in AIY neurons, consistent with neuroendocrine regulation (Jeong and Paik 2017). “Heritable stress” in prg-1/Piwi mutants, which results in a transgenerational mortal-germline phenotype, promotes an ARD-like state, suggesting that stressors other than starvation can cause reproductive quiescence (Heestand et al. 2018).
Germline development
C. elegans larvae hatch with two PGCs, Z2 and Z3. The PGCs begin proliferation during the L1 larval stage and continue dividing throughout larval development. Meiosis begins during the L4 larval stage, and approximately 150 sperm are produced by each of the two hermaphrodite gonad arms in well-fed animals. Worms irreversibly switch from spermatogenesis to oogenesis around the L4 molt, and germline proliferation continues throughout adulthood. Germline development is reviewed in depth elsewhere (Hubbard and Greenstein 2005; Albert Hubbard and Schedl 2019). All stages of germline development are sensitive to nutrient availability [reviewed in Fukuyama (2018)], and the number of germ cells in the mature germline is determined by the relative rates of proliferation and apoptosis.
Germline proliferation:
Starvation presumably causes germ cell proliferation to stop at any larval stage, since starvation causes developmental arrest (Johnson et al. 1984; Schindler et al. 2014). Regulation of germ cell proliferation during arrest has only been investigated in L1 larvae, dauers and adults. Dietary restriction and amino acid deprivation during larval development reduce adult germ cell number by affecting proliferation via cell-autonomous alterations to insulin/IGF and TOR signaling (Michaelson et al. 2010; Korta et al. 2012; Hibshman et al. 2016). Starvation causes germ cell proliferation to halt in adults, and proliferation resumes with feeding (see ARD).
Starvation of newly hatched L1 larvae arrests development by inducing cell cycle arrest in somatic and germline precursor cells (see L1 arrest). Unlike somatic cells, which arrest in the G0/G1 phase of the cell cycle during L1 arrest (Hong et al. 1998), Z2 and Z3 have condensed chromosomes and duplicated centrosomes in L1 arrest, and they are capable of dividing in the presence of the DNA synthesis inhibitor hydroxyurea (Fukuyama et al. 2006). Thus, Z2 and Z3 are likely arrested in G2. Z2 and Z3 quiescence requires daf-18/PTEN and AMPK (Fukuyama et al. 2006; Fukuyama et al. 2012) (Figure 8). Proliferation of Z2 and Z3 in daf-18/PTEN mutants requires age-1/PI3K and akt-1, indicating that DAF-18/PTEN maintains G2 arrest in Z2 and Z3 by restraining AGE-1/PI3K and AKT-1. However, Z2 and Z3 remain quiescent in starved daf-16/FoxO mutant L1 larvae, indicating that AGE-1/PI3K and AKT-1 promote Z2 and Z3 cell division by regulating an AKT-1 substrate distinct from DAF-16/FoxO (Fukuyama et al. 2006). daf-18/PTEN and AMPK function in parallel and converge on inhibition of TORC1 to maintain Z2/Z3 quiescence (Fukuyama et al. 2012).
Germ cells also require daf-18/PTEN and AMPK for quiescence during dauer arrest (Narbonne and Roy 2006). As in L1 arrest (Fukuyama et al. 2006), quiescence does not depend on daf-16/FoxO (Tenen and Greenwald 2019). AMPK functions cell-nonautonomously in somatic cells to maintain germ cell quiescence and integrity, likely by regulating an endogenous small RNA pathway (Kadekar and Roy 2019). Although daf-18/PTEN regulates nongonadal somatic cell divisions cell-nonautonomously in L1 and dauer arrest (Fukuyama et al. 2015; Tenen and Greenwald 2019), daf-18/PTEN functions in the somatic gonad to govern somatic gonadal blast cell and germ cell divisions in dauer larvae (Tenen and Greenwald 2019). Sophisticated genetic analysis addressing necessity and sufficiency of function in different sites suggests that DAF-18/PTEN activity leads to the production of an unidentified “pro-quiescence” signal from multiple sites within the somatic gonad, with the effect of this signal somehow being restricted to the gonad (Tenen and Greenwald 2019). Given similarities to L1 arrest, this work raises the question of whether daf-18/PTEN functions in Z1 and Z4 (L1 somatic gonad precursors) to promote quiescence of both somatic and germline precursors via a similar signaling mechanism.
Germline apoptosis:
During normal development, hundreds of cells in the hermaphrodite germline undergo programmed cell death in a process known as physiological germline apoptosis (PGA) (Gumienny et al. 1999; reviewed in Gartner et al. (2008)]. PGA occurs in the syncytial region of the germline and is thought to enable provisioning of adequate cytoplasmic contents to maturing oocytes (Gumienny et al. 1999). The regulation of PGA and somatic cell apoptosis differs, as PGA occurs independently of the BH3 family proteins EGL-1 and CED-9, which control somatic cell apoptosis (Gumienny et al. 1999). Starvation of animals for 6 hr increases germline apoptosis by greater than twofold (Salinas et al. 2006). Similar to PGA, stress-induced germline apoptosis is EGL-1- and CED-9-independent. It also occurs independently of the p53 homolog CEP-1 and components of the DNA damage response that are required for germline apoptosis induced by genotoxic insults [reviewed in Gartner et al. (2008)]. The upstream pathways that govern induction of germ cell death by starvation differ from those that regulate apoptosis induced by other stresses; unlike apoptosis induced by oxidative, hyperosmolar, or heat stress, starvation-induced apoptosis requires neither the MAPKK family members MEK-1 and SEK-1 nor the nonreceptor tyrosine kinase ABL-1. Apoptosis induction by all four stresses requires the RNA binding protein TIAR-1 (Silva-Garcia and Estela Navarro 2013).
Effects of Starvation on Behavior
Starvation influences a variety of behaviors in C. elegans, including feeding and foraging, as well as sleep (Table 1). Worms are also capable of learning to associate starvation with specific features of the environment, affecting their preferences for environmental conditions. However, the effect of starvation on behavior has generally received less attention than developmental or molecular consequences of starvation.
Table 1. Effects of starvation on behavior.
| Behavior | Effect of starvation | References |
|---|---|---|
| Locomotion | Dispersal, enhanced slowing when fed | (Sawin et al. 2000; Gray et al. 2005; Artyukhin et al. 2013b) |
| Pharyngeal pumping | Initial decrease followed by recovery, greater increase when fed | (Avery and Horvitz 1990; Dwyer and Aamodt 2013) |
| Egg laying | Inhibited, “bagging” | (Chen and Caswell-Chen 2004; Schafer 2005) |
| Sleep | Induced | (Skora et al. 2018; Wu et al. 2018b) |
| Carbon dioxide avoidance | Attraction rather than repulsion | (Bretscher et al. 2008; Rengarajan et al. 2019) |
| Associative learning | Modifies learning in multiple paradigms | (Saeki et al. 2001; Tomioka et al. 2006; Lin et al. 2010; Russell et al. 2014) |
Starvation has complex, time-dependent effects on a variety of behaviors.
Locomotion
A worm’s locomotory response to food depends on its feeding state. Well-fed animals move more slowly on a lawn of E. coli than they do in its absence, illustrating a “basal slowing response” to food (Sawin et al. 2000). Removal from food provokes local search behavior comprised of reversals and omega-shaped turns (Gray et al. 2005). Reversals and omegas are subsequently suppressed as behavior shifts toward dispersal over the following 30 min. AAK-2/AMPK acts in AIY and AIB interneurons to promote the transition from local to distal exploration during starvation (Ahmadi and Roy 2016). In addition, animals that have been deprived of food for 30 min decrease their rate of locomotion even more than well-fed animals when they encounter a lawn, illustrating an “enhanced slowing response” (Sawin et al. 2000). Bacteria are sensed mechanically, and the basal and enhanced slowing responses rely on dopaminergic and serotonergic neural signaling, respectively. Neuronal AMPK inhibition mediates the effect of serotonin signaling on locomotion (Cunningham et al. 2014). These behavioral responses to food and feeding history presumably reflect adaptive responses by increasing the amount of time an individual spends in the presence of food during foraging.
Alarm pheromone:
Starved L1 larvae produce an alarm pheromone. Larvae in L1 arrest secrete relatively large amounts of the octopamine succinyl ascaroside osas#9. Worms of all developmental stages avoid this synthetic pheromone, or buffer conditioned with larvae in L1 arrest, unless food is present (Artyukhin et al. 2013b). This work shows that in addition to functioning as a neurotransmitter and neurohormone during starvation, octopamine decorates a pheromone, reflecting a central role in the starvation response (Figure 9).
Figure 9.
Functions of octopamine during starvation. Octopamine functions as a neurotransmitter to mediate several of the effects of starvation on behavior, functions as a neurohormone to alter lipid metabolism in support of starvation survival, and is used to decorate an ascaroside-based pheromone in starved larvae to produce an alarm pheromone. References: 1 (Horvitz et al. 1982), 2 (Bayer and Hobert 2018), 3 (Rengarajan et al. 2019), 4 (Artyukhin et al. 2013b), and 5 (Tao et al. 2016).
Pharyngeal pumping
Starvation has complex effects on pharyngeal pumping [reviewed in Avery and You (2012)]. Well-fed worms exhibit a low pumping rate when initially deprived of food and increased pumping in the presence of bacteria (Avery and Horvitz 1990; Dwyer and Aamodt 2013). Exogenous serotonin stimulates pumping (Horvitz et al. 1982), suggesting serotonin signaling mediates the effect of food. Longer than 4 hr without food results in a heightened increase in pumping induced by bacteria (Avery and Horvitz 1990). Within 24 hr of starvation, the rate of pumping increases to a rate comparable to that in fed worms (Dwyer and Aamodt 2013). Sustained pumping during starvation requires autophagy (Kang et al. 2007). Genetic and pharmacologic approaches implicate a signal transduction pathway in the pharynx that is activated by starvation and required for starvation-induced changes in pumping. Starvation acts through the muscarinic acetylcholine receptor GAR-3, the Gq alpha subunit EGL-30, and the protein kinase C family member TPA-1 to activate the MAP kinase MPK-1 (You et al. 2006). The critical MPK-1 substrates that control starvation-induced changes in pharyngeal pumping are not known. Octopamine likely contributes to the effects of starvation on pumping (Figure 9).
Egg laying
Starvation inhibits egg laying [reviewed in Schafer (2005)]. Serotonin promotes and octopamine inhibits egg laying (Horvitz et al. 1982). Because starved worms hold their eggs, the eggs hatch inside the mother to produce a “bag of worms,” ultimately causing her to die from bagging. Larvae that hatch inside their mother are nourished by eating her, and they are more likely to arrest in dauer diapause (Chen and Caswell-Chen 2004). Such matricide by internal hatching is therefore likely to be an adaptive starvation response.
Sleep
Starvation induces sleep in C. elegans. Sleep is reviewed in depth elsewhere (Flavell et al. in press). An hour of food deprivation increased behavioral responsiveness to abrupt decreases in ambient O2 concentrations, while 16 hr of food deprivation attenuated responsiveness to decreased O2 levels (Skora et al. 2018). In particular, worms starved for 16 hr displayed sustained slowing of locomotory activity and transient bouts of behavioral quiescence in response to decreased O2 levels. In ambient conditions, starvation for 16 hr or more in L1 larvae, dauers, and adults increased the frequency of bouts of behavioral quiescence and depolarization of RIS neurons, interpreted as sleep (Skora et al. 2018; Wu et al. 2018b). Surprisingly, global neural activity was not decreased in worms starved for 16 hr while awake (Skora et al. 2018), despite reduced fat reserves and presumably energy availability (Witham et al. 2016). However, global neural activity did decrease during sleep, suggesting that the increased frequency of sleep episodes during long-term starvation helps maintain energy homeostasis (Skora et al. 2018). Indeed, sleep during L1 starvation promotes survival, as demonstrated by ablation of RIS neurons (Wu et al. 2018b).
Energy homeostasis regulators govern starvation-induced sleep. DAF-16/FoxO, DAF-18/PTEN, and AMPK promote starvation resistance (see Energy homeostasis regulators). daf-2/IGFR mutants display excess sleep when well-fed, and daf-16/FoxO, daf-18/PTEN, AMPK, and sir-2.1/SIRT1 mutants are starvation-induced sleep-defective, with daf-16/FoxO and AMPK functioning in parallel to promote sleep during long-term starvation (24 hr) (Wu et al. 2018b). Paradoxically, daf-2/IGFR acts through daf-16/FoxO to promote behavioral responsiveness to reduced O2 levels in worms starved for 1 hr (Skora et al. 2018).
UV radiation and heat shock also induce sleep in a phenomenon known as stress-induced sleep (Hill et al. 2014). However, 24 hr of starvation in young adults suppresses stress-induced sleep (Goetting et al. 2018). Conditions used to evoke stress-induced sleep were optimized for ALA neuron-dependent effects (Goetting et al. 2018), whereas starvation-induced sleep relies on RIS and RMG neurons (Skora et al. 2018; Wu et al. 2018b). Stress-induced sleep was also not affected by mutation of daf-2/IGFR (Goetting et al. 2018), revealing an additional distinction from starvation-induced sleep (Skora et al. 2018; Wu et al. 2018b). Thus, distinct signaling pathways and neural circuits appear to underlie stress-induced and starvation-induced sleep, consistent with different effects of nutrient availability on these types of sleep.
Carbon dioxide attraction
The response to CO2 depends on feeding state. CO2 is an ambiguous signal to C. elegans in that it can be associated with a variety of sources, both favorable and unfavorable. Well-fed worms raised at ambient CO2 levels are repelled by it, and well-fed worms raised at high CO2 levels are attracted to it (Guillermin et al. 2017). Furthermore, starved animals that were raised at ambient CO2 levels are attracted to it (Bretscher et al. 2008; Rengarajan et al. 2019). The reversal from repulsion to attraction happens within hours of starvation, with dopamine promoting repulsion in well-fed animals and octopamine promoting attraction during starvation (Figure 9).
Associative learning paradigms
Starvation has been used to establish simple paradigms of behavioral plasticity that have permitted the elucidation of the mechanistic basis for associative learning. Well-fed animals respond to defined environmental stimuli in a characteristic manner but alter these responses if they are conditioned to these stimuli in the context of food deprivation.
Salt chemotaxis learning:
Worms are normally attracted to sodium chloride (NaCl). However, if they are starved in the presence of NaCl, their chemotaxis toward NaCl and other water-soluble attractants decreases substantially (Saeki et al. 2001). This behavior has been referred to as “salt chemotaxis learning” (Tomioka et al. 2006).
The DAF-2 IIS pathway is required for salt chemotaxis learning. Loss-of-function mutations in daf-2/IGFR, age-1/PI3K, pdk-1, and akt-1 all reduce or abolish salt chemotaxis learning (Tomioka et al. 2006; Vellai et al. 2006), indicating that IIS promotes associative learning. Intriguingly, the impairment in associative learning caused by reduction of IIS requires daf-18/PTEN but is independent of daf-16/FoxO (Tomioka et al. 2006; Vellai et al. 2006), suggesting that undiscovered molecular targets of AKT-1 govern salt chemotaxis learning. This is reminiscent of the daf-2/IGFR-dependent, daf-16/FoxO-independent, L1 starvation-induced arrest of primordial germ and Q cells (see L1 arrest). Cell-specific rescue experiments support a model in which the release of the ILP INS-1 from the AIA interneuron promotes learning by activating the DAF-2/IGFR pathway in the ASER sensory neuron (Tomioka et al. 2006). Starvation may also modulate the response to NaCl by inhibiting an EGL-30/Gq–diacylglycerol–TTX-4/nPKC pathway in the ASER neuron (Adachi et al. 2010).
Benzaldehyde-starvation associative plasticity:
Animals that are normally attracted to the odorant benzaldehyde in replete conditions develop aversion to benzaldehyde if preconditioned for 1 hr with benzaldehyde in the absence of food (Lin et al. 2010). Similar to salt chemotaxis learning, ins-1, daf-2/IGFR, and age-1/PI3K mutants are defective in benzaldehyde-aversion learning. However, the DAF-2/IGFR pathway acts in distinct cells to promote salt chemotaxis and benzaldehyde-aversion learning. The benzaldehyde learning defect in ins-1 mutants is partially rescued by expression of ins-1 in AIA (as is the case in salt chemotaxis learning; Tomioka et al. 2006) or ASI and fully rescued by expression in both cells. In contrast, whereas the salt chemotaxis phenotype of age-1/PI3K mutants is rescued by expression of age-1/PI3K in the ASER sensory neuron (Tomioka et al. 2006), the benzaldehyde phenotype in age-1/PI3K mutants is rescued by expression of age-1/PI3K in the AWC neuron, but not the ASER neuron (Lin et al. 2010). As is the case in salt chemotaxis learning, INS-1 acts as a DAF-2/IGFR agonist to promote benzaldehyde-aversion learning. These data support a model of associative learning whereby starvation indirectly alters the response of specific sensory neurons to defined environmental stimuli by inducing INS-1 biosynthesis in and/or release from AIA interneurons (and possibly other neurons). INS-1 then alters the response of sensory neurons to specific stimuli by activating DAF-2/IGFR signaling in distinct neurons, the identity of which is dependent upon the nature of the environmental stimulus. How different stimuli act in conjunction with starvation to control the anatomical specificity of DAF-2/IGFR pathway activation in response to INS-1 secretion is not known. Moreover, the molecular basis for the context-dependence of INS-1 action on DAF-2/IGFR signaling (i.e., functioning as an agonist ligand in associative learning and as a DAF-2/IGFR antagonist in dauer regulation; Pierce et al. 2001; Hung et al. 2014) remains obscure.
Humidity preference (hygrosensation):
Under replete conditions, animals do not exhibit preference for specific ambient humidity levels. However, if they are exposed to low- or high-humidity environments in the context of food deprivation, they exhibit an aversion for the ambient humidity level at which they experienced starvation (Russell et al. 2014). This recognition and aversion of ambient humidity levels associated with starvation, known as hygrosensation, requires components of both mechanosensory and thermosensory neuronal pathways. mec-6, mec-10, and asic-1, which encode components of a putative ion channel complex required for mechanosensation (Goodman 2006), act in FLP neurons, and the TAX-4 cGMP-dependent cation channel acts in the thermosensory AFD neurons to promote hygrosensation. Whether the DAF-2/IGFR pathway plays a role in hygrosensation associated with food deprivation is not known.
Persistent Effects of Starvation
In addition to reversible developmental responses, starvation can have persistent effects on phenotype throughout the life cycle and across generations (Table 2). For example, larval starvation can subsequently affect neuronal and reproductive development, enabling use of C. elegans as a model to study developmental origins of adult health and disease. In addition, cellular rejuvenation mechanisms are activated during recovery from starvation, mitigating pathological consequences. Larval starvation can also have lasting effects on fertility, including heritable effects (see also Transgenerational epigenetic inheritance of starvation resistance).
Table 2. Persistent effects of starvation.
| Starvation regimen | Phenotypic consequence | References |
|---|---|---|
| Brief L1, L3, or dauer arrest in wild type | Altered sex-specific axon pruning and adult behavior | (Bayer and Hobert 2018) |
| Extended L1 arrest in wild type | Persistent protein aggregates despite rejuvenation during recovery | (Roux et al. 2016) |
| Extended L1 arrest in wild type | Decreased growth rate and increased variation | (Lee et al. 2012; Jobson et al. 2015) |
| Extended L1 or dauer arrest in wild type | Gonadal abnormalities and reduced fertility | (Kim and Paik 2008; Lee et al. 2012; Wolf et al. 2014; Ow et al. 2018; Webster et al. 2018; Jordan et al. 2019) |
| Brief L1 starvation in daf-18/PTEN or AMPK mutants | Gonadal abnormalities and reduced fertility | (Wolf et al. 2014; Demoinet et al. 2017) |
| Extended L1 or dauer arrest in wild type | Transgenerational epigenetic inheritance of increased starvation survival and life span | (Rechavi et al. 2014; Jobson et al. 2015; Webster et al. 2018) |
| Brief L1 starvation in AMPK mutants | Transgenerational epigenetic inheritance of reduced fertility | (Demoinet et al. 2017) |
The starvation regimen, including genotype, stage, and approximate duration, is included along with the phenotypic consequences and reference. Extended L1 starvation is a week or more, extended dauer arrest is several weeks, and brief L1 starvation is ∼1 day, although experimental details vary by study.
Neuronal development
Larval starvation has lasting effects on the connectivity and function of the nervous system in male worms. Sex-specific patterns of synaptic connectivity develop in the L4 stage through synaptic pruning of “sex-hybrid” juvenile connectivity (Jarrell et al. 2012; Oren-Suissa et al. 2016). Passage through dauer arrest or 24 hr of starvation during the L1 or L3 stage inhibits sex-specific synaptic pruning in males but not hermaphrodites (Bayer and Hobert 2018), revealing that sexually dimorphic wiring is sensitive to early life starvation. Changes in wiring have functional consequences, as poststarvation males retain juvenile sensory acuity that is lost with pruning in well-fed males, but they are less efficient at mating (Bayer and Hobert 2018). Extrasynaptic serotonin signaling promotes pruning in well-fed L4 males, and L1 starvation causes an octopamine-dependent decrease in serotonin signaling that persists to the L4 stage (Bayer and Hobert 2018) (Figure 9). The molecular mechanism for the long-term effect on serotonin synthesis is unknown. Notably, octopamine activates the cAMP-response element binding protein CRH-1/CREB in SIA neurons, affecting CREB-dependent gene expression (Suo et al. 2006). This mechanism may be involved in acute behavioral responses to starvation mediated by octopamine (Horvitz et al. 1982). In addition, crh-1/CREB is implicated in long-term associative memory (Kauffman et al. 2010), suggesting a possible mechanism for persistent effects of starvation on serotonin signaling. Early life stress has lasting effects on the adult nervous system and behavior in vertebrates, which also involves changes in serotonin signaling (Lajud and Torner 2015; Houwing et al. 2017), suggesting that work in C. elegans will contribute to elucidation of molecular mechanisms by which early life experience affects adult behavior.
Effects of dauer diapause in later life
The conditions that drive dauer formation influence the phenotypes displayed later in life after recovery (see also Larval starvation reduces reproductive success). Dauer formation can be driven primarily by nutrient limitation or population density (Hu 2007; Fielenbach and Antebi 2008). With population density as the driver, postdauer worms have increased brood size and life span as well as altered olfactory behavior (Hall et al. 2010; Hall et al. 2013; Sims et al. 2016). With nutrient limitation as the driver, they have decreased brood size (Ow et al. 2018). These dauer-forming conditions also have reciprocal effects on gene expression, with a large number of genes up- or downregulated in postdauer adults depending on dauer-forming conditions (Ow et al. 2018). Genetic and genomic analyses suggest that histone modification and endogenous RNAi pathways mediate these effects of early life experience on adult gene expression and life history traits (Hall et al. 2010; Hall et al. 2013; Sims et al. 2016; Ow et al. 2018). That different dauer-forming conditions elicit different effects in postdauer worms suggests that these effects reflect potentially adaptive plastic responses to environmental conditions, although it is also possible that different conditions impose different developmental constraints.
Rejuvenation during recovery from starvation
Seminal studies on dauer arrest, L1 arrest, and starved adults each found that time spent in these developmentally quiescent states does not affect adult life span upon feeding (Klass and Hirsh 1976; Johnson et al. 1984; Angelo and Van Gilst 2009). Each of these studies concluded that aging was somehow suspended during developmental arrest. However, recent work suggests that aging does occur during developmental arrest, and that feeding triggers a rejuvenation process. Various hallmarks of aging increase over time during L1 arrest, including mitochondrial fission, production of reactive oxygen species (ROS), protein aggregation, sensitivity to proteotoxic stress, decreased mobility, and increased mortality (Roux et al. 2016). Remarkably, all of these signs of aging, with the exception of protein aggregates, are reversed within hours of feeding (Roux et al. 2016). These results suggest that normal adult life span following prolonged developmental arrest is not due to suspension of aging during arrest but instead physiological reversal of aging during recovery from arrest. Not all surviving larvae resume development when fed, and age-related phenotypes are better predictors than fat content of the ability of individual worms to recover from arrest (Roux et al. 2016). These observations suggest that starved larvae die at least in part due to aging rather than simply depletion of nutrient stores. Furthermore, the ER UPR sensor IRE-1/ERN-1 is required for reversal of age-related phenotypes during recovery (Roux et al. 2016). Mutation of ire-1/ERN-1 does not affect the duration of L1 starvation survival (scored by movement), but it greatly reduces the ability of arrested larvae to develop upon feeding. The transcriptional effector of IRE-1/ERN-1 signaling, XBP-1, is only partially required for recovery potential, suggesting an additional effector mechanism. The MAP kinase kgb-1/MAPK10 is also required for complete recovery potential, and KGB-1/MAPK10 phosphorylation is reduced in an ire-1/ERN-1 mutant, suggesting that IRE-1/ERN-1 maintains KGB-1/MAPK10 phosphorylation during L1 arrest to promote recovery potential (Roux et al. 2016). It is intriguing to imagine that deeper insights into the molecular basis of reversal of aging-related phenotypes during recovery from larval starvation could lead to the development of interventions that reverse aging in humans.
Characterization of developmental dynamics during recovery from L1 arrest reveals an initial lag phase followed by a developmental rate that is comparable to continuously fed larvae (Olmedo et al. 2019). The length of this lag phase increases in daf-16/FoxO mutants and as a function of the duration of L1 starvation (Olmedo et al. 2019). These observations are consistent with increased aging requiring more time for reversal before development commences.
Recovery from adult starvation also appears to involve physiological rejuvenation. Starved adults display gross signs of tissue and cellular aging, including atrophy of the intestine and somatic gonad, as well as shrinkage of the germline (Angelo and Van Gilst 2009) (see ARD). Remarkably, recovery from starvation is marked by drastic reversal of these phenotypes and normal adult life span (Angelo and Van Gilst 2009). Somatic rejuvenation does not require germline signaling or replication of nuclear or mitochondrial DNA (Burnaevskiy et al. 2018). Rejuvenation is accompanied by expansion of nucleolar size, and it requires expansion of the somatic RNA pool and processing of ribosomal RNA (rRNA) (Burnaevskiy et al. 2018). Likewise, ribosomal profiling revealed that translation of ribosomal proteins is dramatically upregulated immediately upon feeding arrested L1 larvae (Stadler and Fire 2013), and translation is required to initiate postembryonic growth and development (see Translation and the proteome) (Cenik et al. 2019). These observations suggest that translational capacity is reduced during starvation, and that it must be restored to reverse age-related phenotypes that occur over time during starvation.
Reproductive development and fertility
Although developmental arrest enables larvae to postpone reproduction and endure starvation, there are nonetheless reproductive costs associated with larval starvation.
Larval starvation reduces reproductive success:
Extended L1 starvation delays growth and reduces fertility upon recovery (Lee et al. 2012; Jobson et al. 2015). Morphological abnormalities in the gonad, including but not limited to germ cell tumors and uterine masses, are visible in early adulthood of wild-type worms and are associated with dramatically reduced fertility (Lee et al. 2012; Wolf et al. 2014; Jordan et al. 2019). Likewise, extended dauer arrest subsequently delays development and reduces fecundity, with developmental abnormalities evident in the gonad (Kim and Paik 2008). Reduced fecundity following dauer arrest has been reported in two additional studies using different dauer-inducing conditions (Ow et al. 2018; Webster et al. 2018). However, brood size actually increases following relatively short-term dauer arrest induced by high population density rather than nutrient limitation, suggesting that limited nutrient availability during arrest, rather than dauer development itself, leads to decreased fertility (Hall et al. 2010; Ow et al. 2018) (see Effects of dauer diapause in later life).
Genes required to survive L1 starvation also protect larvae from developing gonadal abnormalities. daf-16/FoxO, daf-18/PTEN, and AMPK mutants are each starvation-sensitive and die rapidly during L1 arrest (Munoz and Riddle 2003; Baugh and Sternberg 2006; Fukuyama et al. 2012; Demoinet et al. 2017) (see Energy homeostasis regulators). Each of these genes is also required to arrest cell division during L1 starvation (Baugh and Sternberg 2006; Fukuyama et al. 2006; Fukuyama et al. 2015) (see L1 arrest). In addition, daf-18/PTEN and AMPK mutants enhance L1 starvation-induced gonad abnormalities and loss of fertility (Fukuyama et al. 2012; Wolf et al. 2014; Demoinet et al. 2017). Notably, these abnormalities are not observed at appreciable frequencies when the mutants are not starved. Development of gonadal abnormalities is not simply due to failure to arrest PGC divisions during L1 starvation (Demoinet et al. 2017), although PGC hyperplasia during L1 starvation may contribute. There is pharmacological evidence that unchecked mitochondrial activity leads to gonadal abnormalities in daf-18/PTEN mutants (Wolf et al. 2014), and that aberrant histone modification and transcriptional elongation lead to abnormalities in AMPK mutants (Demoinet et al. 2017). Rapid activation of transcription in PGCs during recovery from L1 arrest is mutagenic (Butuči et al. 2015), suggesting that incomplete DNA repair could potentially contribute to developmental abnormalities. Reducing IIS with RNAi during development following extended L1 starvation in wild-type worms reduces penetrance of gonadal abnormalities in daf-16/FoxO and skn-1/Nrf-dependent fashion, suggesting that molecular and cellular damage incurred during starvation is at least partially reversible (Jordan et al. 2019). These results demonstrate that the signaling pathways that preserve viability during starvation also impinge on developmental fidelity and reproductive success following starvation.
Heritable effects of starvation on reproductive success:
Apparent fitness costs of extended L1 arrest persist in the next generation. F1 progeny of worms subjected to extended L1 starvation are themselves relatively small, grow slower, include an increased frequency of males, and can have reduced fertility (Jobson et al. 2015). AMPK is not only required to preserve reproductive fitness in worms subjected to L1 starvation, but progeny of AMPK mutants starved as L1 larvae have a marked decrease in fertility (Demoinet et al. 2017). AMPK regulates the COMPASS complex, which methylates histone 3 lysine 4 (H3K4) during L1 arrest (see AMPK and histone modification). Remarkably, increased levels of H3K4 trimethylation that occur during L1 starvation in AMPK mutants are transmitted through the germline to descendants. These and other observations suggest that AMPK prevents trimethylation of H3K4 during L1 arrest, and that aberrant accumulation of this epigenetic mark contributes to epigenetic inheritance of decreased fertility (Demoinet et al. 2017). Molecular mechanisms responsible for heritable effects of starvation in AMPK mutants may also contribute to heritable effects in wild-type worms, with AMPK mutants sensitized such that these effects are evident with shorter periods of starvation. The heritable sterility of AMPK mutants following L1 starvation is reminiscent of the mortal germline (transgenerational sterility) phenotype seen in mutants affecting telomerase, some histone modification enzymes, nuclear RNAi genes, and prg-1/Piwi (Smelick and Ahmed 2005; Katz et al. 2009; Buckley et al. 2012; Simon et al. 2014). These observations suggest that extended starvation of larvae can result in a heritable form of aberrant germline gene regulation.
Starvation Resistance
As a trait, starvation resistance is likely critical to fitness of C. elegans in the wild, and it is relevant to metabolic disease risk and aging in humans. Remarkably, adult worms survive longer on plates without food than with food (given prevention of reproduction and therefore death due to bagging) (Kaeberlein et al. 2006; Lee et al. 2006; Gerisch et al. 2020). Furthermore, time spent in starvation does not subsequently affect life span after feeding, leading to an increase in total life span. Klass and Hirsh originally showed that larvae can be arrested as dauers for as much as 60 days with no effect on postdauer life span (Klass and Hirsh 1976). Johnson et al. subsequently showed that L1 arrest also has no effect on adult life span (Johnson et al. 1984), and starvation during adulthood does not affect life span after feeding (Angelo and Van Gilst 2009). If aging results from accumulation of molecular or cellular damage, then prolonged arrest should lead to increased accumulation of damage and decreased life span. Instead, it was suggested that developmental arrest is an “ageless” or “nonaging” state (Klass and Hirsh 1976; Johnson et al. 1984) (see Rejuvenation during recovery from starvation for an alternative perspective), revealing an intimate relationship between development and aging. Understanding how worms adapt to starvation and endure developmental arrest is therefore fundamental to understanding aging.
Energy homeostasis regulators
IIS and AMPK, both critical regulators of energy homeostasis, mediate metabolic adaptation to starvation with profound effects on starvation resistance (Figure 10 and Table 3). Mutation of daf-2/IGFR or age-1/PI3K increases L1 starvation survival in a daf-16/FoxO-dependent fashion, and daf-16/FoxO single mutants survive starvation approximately half as long as wild type (Munoz and Riddle 2003; Baugh and Sternberg 2006). Furthermore, mutation of unc-31/CAPS, a calcium-activated regulator of neural dense-core vesicle release, also increases L1 starvation survival in a daf-16-dependent fashion (Lee and Ashrafi 2008), consistent with physiological control of starvation resistance through regulation of ILP secretion. Although daf-18/PTEN inhibits IIS mediated through the PI3K pathway, daf-18/PTEN null mutants are even more sensitive to L1 starvation than daf-16/FoxO null mutants (Fukuyama et al. 2012; Cui et al. 2013). This result indicates that daf-18/PTEN promotes starvation resistance by doing more than just activating daf-16/FoxO, consistent with genetic analyses of L1 starvation-induced germline and Q cell quiescence (Fukuyama et al. 2006; Zheng et al. 2018b), and salt chemotaxis learning (Tomioka et al. 2006; Vellai et al. 2006). Disruption of AMPK reduces L1 starvation survival (Baugh and Sternberg 2006; Lee et al. 2012) and dauer survival (Narbonne and Roy 2009). AMPK inhibition in ASI sensory neurons mediates effects of serotonin signaling on neuroendocrine secretion (e.g., DAF-7/TGF-β and DAF-28/ILP) and peripheral fat metabolism (Cunningham et al. 2014), suggesting neuronal AMPK promotes starvation resistance. Simultaneous disruption of aak-1 and aak-2, two α-catalytic subunits of AMPK, has a dramatic effect on starvation survival, comparable to loss of daf-18/PTEN (Fukuyama et al. 2012). Not only does this double mutant reveal redundant function of aak-1 and aak-2, but it also shows that AMPK, like daf-18/PTEN, is more important to starvation survival than daf-16/FoxO. In addition, an aak-1; aak-2; daf-18/PTEN triple mutant is apparently more starvation-sensitive than the AMPK double mutant or daf-18/PTEN alone, suggesting that AMPK and daf-18/PTEN act in parallel to promote starvation resistance (Fukuyama et al. 2012).
Figure 10.
Factors affecting starvation resistance. Genes, molecules, and aspects of life history that impinge on starvation resistance are depicted on the outside, and processes that mediate effects on resistance are on the inside. Regulatory interactions between processes are also indicated with arrows. All known factors are not included. See also Tables 3 and 4.
Table 3. Genes that affect starvation resistance.
| Gene | Effect on starvation resistance | Function | References |
|---|---|---|---|
| ocr-2/TRPV | Decreases | Chemosensation | (Lee and Ashrafi 2008) |
| osm-1/IFT172 | Decreases | Chemosensation | (Lee and Ashrafi 2008) |
| osm-3/KIF17 | Decreases | Chemosensation | (Lee and Ashrafi 2008) |
| osm-6/IFT52 | Decreases | Chemosensation | (Lee and Ashrafi 2008) |
| che-11/IFT140 | Decreases | Chemosensation | (Lee and Ashrafi 2008) |
| unc-31/CAPS | Decreases | ILP secretion | (Lee and Ashrafi 2008; Cui et al. 2013; Cui et al. 2017) |
| ins-1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 13, 16, 18, 19, 20, 25, 29, 30, 31, 32, and daf-28 | Decreases | ILPs | (Chen and Baugh 2014; Zheng et al. 2018a) |
| ins-12, 14, 17, 22, 28, 34, 37, and 39 | Increases | ILPs | (Zheng et al. 2018a) |
| daf-2/IGFR | Decreases | Insulin/IGF signaling | (Munoz and Riddle 2003; Baugh and Sternberg 2006) |
| age-1/PI3K | Decreases | Insulin/IGF signaling | (Munoz and Riddle 2003; Cui et al. 2013; Cui et al. 2017) |
| daf-18/PTEN | Increases | Insulin/IGF signaling | (Fukuyama et al. 2012; Cui et al. 2013) |
| daf-16/FoxO | Increases | Insulin/IGF signaling and transcriptional regulation | (Munoz and Riddle 2003; Baugh and Sternberg 2006; Cui et al. 2013; Cui et al. 2017) |
| mir-71 | Increases | Targets insulin/IGF signaling | (Zhang et al. 2011) |
| ain-1 | Increases | miRISC component | (Zhang et al. 2011) |
| aak-2/AMPK | Increases | Energy homeostasis | (Baugh and Sternberg 2006; Fukuyama et al. 2012; Lee et al. 2012; Webster et al. 2017) |
| pha-4/FoxA | Increases | Transcriptional regulation | (Zhong et al. 2010; Wu et al. 2018a) |
| skn-1/Nrf | Ambiguous | Transcriptional regulation | (Paek et al. 2012) |
| lin-35/Rb | Increases | Transcriptional regulation | (Cui et al. 2013; Cui et al. 2017) |
| daf-12/VDR | Increases | Transcriptional regulation | (Tao et al. 2017) |
| din-1/SPEN | Increases | Transcriptional regulation | (Tao et al. 2017) |
| nhr-49/NHR | Increases | Transcriptional regulation of lipid metabolism | (Goh et al. 2018) |
| hlh-30/TFEB | Increases | Transcriptional regulation of lipid metabolism | (O’Rourke and Ruvkun 2013; Settembre et al. 2013) |
| lipl-4 | Increases | Lipase | (O’Rourke et al. 2013) |
| lipl-5 | Decreases | Lipase | (Buis et al. 2019) |
| sptl-2/SPTLC2 | Increases | Ceramide biosynthesis | (Cui et al. 2017) |
| hyl-1/CERS5 | Increases | Ceramide biosynthesis | (Cui et al. 2017) |
| hyl-2/CERS5 | Increases | Ceramide biosynthesis | (Cui et al. 2017) |
| sphk-1/SPHK2 | Increases | Ceramide biosynthesis | (Cui et al. 2017) |
| acs-20/SLC27A1 | Increases | Ceramide biosynthesis | (Cui et al. 2017) |
| tbc-2/TBC1D2 | Increases | Yolk provisioning | (Chotard et al. 2010) |
| rme-1/EHD1 | Increases | Yolk provisioning | (Chotard et al. 2010) |
| rme-6/GAPVD1 | Increases | Yolk provisioning | (Chotard et al. 2010) |
| rme-2/LDL receptor | Increases | Yolk provisioning | (Perez et al. 2017; Jordan et al. 2019; Olmedo et al. 2019) |
| ceh-60/PBX1 | Increases | Yolk provisioning | (Van Rompay et al. 2015) |
| vrp-1 | Increases | Yolk provisioning | (Van Rompay et al. 2015) |
| vit-1/yolk protein | Increases | Yolk provisioning | (Jordan et al. 2019) |
| vit-5/yolk protein | Increases | Yolk provisioning | (Perez et al. 2017; Jordan et al. 2019) |
| pks-1 | Increases | Polyketide synthase | (Shou et al. 2016) |
| nrps-1 | Increases | Nonribosomal peptide synthetase | (Shou et al. 2016) |
| gpb-2/Gβ5 | Increases | Autophagy | (Kang et al. 2007) |
| bec-1/atg-6/beclin1 | Increases | Autophagy | (Kang et al. 2007) |
| atg-18/WIPI1 | Increases | Autophagy | (Hibshman et al. 2018) |
| unc-51/atg-1/ULK1 | Increases | Autophagy | (Hibshman et al. 2018) |
| pink-1/PINK1 | Decreases | Autophagy | (Hibshman et al. 2018) |
| cep-1/p53 | Increases | DNA repair | (Derry et al. 2001) |
| pcm-1/PCMT1 | Increases | Damaged protein repair | (Gomez et al. 2007) |
| icl-1 | Increases | Glyoxylate cycle | (Hibshman et al. 2017) |
| pck-1/PCK1 | Increases | Gluconeogenesis | (Hibshman et al. 2017) |
| tps-1 | Increases | Trehalose synthesis | (Hibshman et al. 2017) |
| alh-6 | Increases | Proline catabolism | (Pang et al. 2014) |
| eat-2 | Increases | Pharyngeal pumping | (Lee and Ashrafi 2008) |
| isp-1/ISP | Increases | Mitochondrial respiration | (Lee and Ashrafi 2008) |
| clk-1/COQ7/CAT5 | Increases | Mitochondrial respiration | (Lee and Ashrafi 2008) |
| tbh-1/DBH | Increases | Octopamine biosynthesis | (Tao et al. 2016) |
| hsf-1/HSF | Increases | Transcriptional regulation of heat shock response | (Baugh and Sternberg 2006) |
| rpn-6 | Increases | Proteostasis | (Webster et al. 2017) |
| hsp-4/HSP70 | Increases | ER unfolded protein response | (Jo et al. 2009) |
| ire-1/ERN-1 | Increases | ER unfolded protein response | (Jo et al. 2009; Roux et al. 2016) |
| xbp-1/XBP1 | Increases | ER unfolded protein response | (Roux et al. 2016) |
| kgb-1/MAPK10 | Increases | ER unfolded protein response | (Roux et al. 2016) |
| ife-2/eIF4E | Decreases | Translation initiation | (Lee et al. 2012) |
| ifg-1/eIF4G | Decreases | Translation initiation | (Pan et al. 2007) |
| rsks-1/RPS6KB1 | Decreases | Translational activator | (Pan et al. 2007; Wu et al. 2018a) |
| efk-1I/eEF2K | Increases | Translational repressor | (Leprivier et al. 2013) |
| rrp-8/RRP8 | Decreases | Ribosomal RNA processing | (Wu et al. 2018a) |
| kars-1 | Decreases | Aminoacyl tRNA synthetase | (Webster et al. 2017) |
| wars-1 | Decreases | Aminoacyl tRNA synthetase | (Webster et al. 2017) |
| vars-2 | Decreases | Aminoacyl tRNA synthetase | (Webster et al. 2017) |
The effect of the gene’s function on starvation resistance is indicated along with the general function of the gene product and the citation. The effect on starvation resistance was mostly inferred from loss of function, but in relatively rare cases gain-of-function or overexpression alleles were analyzed. L1 starvation survival was most often assayed, but in many cases starvation survival at other stages (typically L4/young adult) was assayed, or other aspects of starvation resistance were assayed (e.g., growth rate during recovery).
Despite the importance of daf-16/FoxO, daf-18/PTEN, and AMPK in promoting starvation resistance, relatively few effector mechanisms have been identified. Mutations affecting dbl-1/TGF-β or daf-12/VDR steroid hormone signaling suppress the L1 arrest-defective phenotype of daf-16/FoxO mutants, but not the starvation-sensitive phenotype (Kaplan et al. 2015) (see L1 arrest). This observation reveals that daf-16/FoxO mutants do not die rapidly during starvation due to inappropriate development. Likewise, the mir-235/miR-92 mutant also has an L1 arrest-defective phenotype, but is not starvation sensitive (Kasuga et al. 2013). Thus, DAF-16/FoxO promotes developmental arrest and starvation resistance through regulation of different target genes. DAF-16/FoxO promotes starvation resistance in part by orchestrating a metabolic shift toward gluconeogenesis and trehalose synthesis (see Central carbon metabolism). In addition, DAF-16/FoxO and AMPK promote sleep during starvation, which supports survival (see Sleep). Nonetheless, additional effector mechanisms remain to be discovered.
Nemamides A and B and IIS
Nematodes are exceptional as metazoans for having polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) genes. pks-1 and nrps-1 are required for synthesis of the hybrid polyketide-nonribosomal peptides Nemamide A and Nemamide B (Shou et al. 2016). Nemamide A and B levels are elevated during L1 starvation, and pks-1 and nrps-1 mutants are sensitive to L1 starvation, with reduced survival and developmental rate upon recovery. pks-1 and nrps-1 mutants do not have an L1 arrest defective-phenotype, but the putative daf-2/IGFR agonists daf-28, ins-4 and ins-5 are upregulated during L1 arrest in the mutants (Shou et al. 2016). daf-28 and ins-4 promote L1 development and limit starvation survival (Chen and Baugh 2014) (see ILPs governing L1 arrest and development), suggesting that Nemamides A and B reduce IIS activity to promote L1 starvation resistance (Shou et al. 2016). Investigation into nutrient-dependent regulation of Nemamide A and B synthesis, as well as their mechanism of action, will shed new light on the starvation response and regulation of IIS.
Autophagy
Autophagy promotes starvation resistance [reviewed in Meléndez and Levine (2009)]. Genetic or pharmacological disruption of autophagy reduces L1 starvation survival (Kang et al. 2007; Hibshman et al. 2018). Autophagy maintains pharyngeal pumping during starvation (Kang et al. 2007), and it functions in other tissues as well (Chapin et al. 2015). Mitochondrial content and function decline during larval starvation, as measured from whole animals, and genetic analysis suggests that nonselective autophagy contributes to degradation of mitochondria during starvation (Hibshman et al. 2018). Expression of the lipase lipl-4 is induced within hours of starvation in young adults, and overexpression of lipl-4 increases levels of ω-6 polyunsaturated fatty acids, which activate autophagy, promoting starvation survival (O’Rourke et al. 2013). It is presumed that autophagy supports starvation resistance by recycling cellular material and liberating energy, but this has not been directly demonstrated in C. elegans.
Stress-response pathways
A variety of stress-response pathways promote starvation resistance. Disruption of the heat shock factor hsf-1 reduces L1 starvation survival (Baugh and Sternberg 2006). Proteasome function and the ER UPR also support starvation survival and recovery (Jo et al. 2009; Roux et al. 2016; Webster et al. 2017). Protein homeostasis is reviewed in depth elsewhere (Hoppe and Cohen 2020). Furthermore, levels of the disaccharide trehalose increase during starvation and a nonhydrolyzable form of trehalose increases starvation survival (Hibshman et al. 2017). Trehalose is known to preserve integrity of macromolecules (Erkut et al. 2011), suggesting it contributes to proteostasis and membrane integrity during starvation. In addition, DNA and protein repair pathways support starvation survival (Derry et al. 2001; Gomez et al. 2007), and recovery from L1 arrest is mutagenic to the germline (Butuči et al. 2015). These observations suggest that a variety of molecular insults are actively resisted in support of starvation survival.
Central carbon metabolism
Glyoxylate cycle, gluconeogenesis, and trehalose synthesis:
Carbohydrate metabolism is modified in response to starvation [reviewed in Braeckman et al. (2009)]. During embryogenesis and early larval development, metabolism is shifted away from the tricarboxylic acid (TCA) cycle toward the carbon-conserving glyoxylate cycle. The glyoxylate cycle converts two acetyl-CoA molecules into succinate and malate, and in C. elegans it depends on the bifunctional isocitrate lyase/malate synthase enzyme ICL-1 (Liu et al. 1995). ICL-1 activity and expression peak at hatching in fed larvae and decrease during larval development, but they are upregulated in response to starvation as L1 larvae or in later stages (Khan and McFadden 1982; Liu et al. 1997). Genome-wide expression analyses of starved L1 and dauer larvae suggest that increased reliance on the glyoxylate cycle during starvation is accompanied by upregulation of gluconeogenesis and trehalose (a disaccharide of glucose) synthesis as well as downregulation of the TCA cycle and oxidative phosphorylation (Wang and Kim 2003; McElwee et al. 2006; Hibshman et al. 2017). Genetic and pharmacological analyses suggest that the shift from glycolysis toward gluconeogenesis and trehalose synthesis supports starvation survival (Hibshman et al. 2017). Trehalose contributes to starvation resistance both by protecting macromolecules and functioning as a transportable metabolic energy source (Erkut et al. 2011; Hibshman et al. 2017), although the trafficking of trehalose in nematodes is not understood.
ATP synthesis:
Levels of the TCA cycle intermediate α-ketoglutarate are increased in starved worms (Chin et al. 2014), as in bacteria, yeast, and vertebrate liver (Kaminsky et al. 1982; Brauer et al. 2006). α-ketoglutarate inhibits ATP synthase, causing reduced ATP content and oxygen consumption, increased autophagy, and increased life span (Chin et al. 2014). An effect on starvation survival has not been investigated, but these observations suggest that α-ketoglutarate may promote starvation resistance.
Fat metabolism
Fat metabolism in C. elegans is thoroughly reviewed elsewhere (Ashrafi 2007; Watts and Ristow 2017); here, we focus on regulation of fat metabolism in response to starvation. Fatty acid oxidation increases during starvation (Elle et al. 2012). However, starvation effects fat metabolism and the lipid profile in complex ways (Van Gilst et al. 2005b; Macedo et al. 2020).
nhr-49/NHR:
In mammals, NHRs of the Peroxisome Proliferator-Activated Receptor (PPAR) family are critical regulators of lipid metabolism and energy homeostasis (Chawla et al. 2001; Evans et al. 2004). Despite the large number of NHR genes in C. elegans, none display significant sequence homology to the PPAR family (Sluder and Maina 2001; Gissendanner et al. 2004). Disruption of the NHR nhr-49 (a homolog of HNF4α) causes a high-fat, short-lived phenotype (Van Gilst et al. 2005a), and nhr-49 mutants have reduced L1 starvation survival (Goh et al. 2018). nhr-49 and its coactivator, the mediator subunit mdt-15, are responsible for many of the changes in fat metabolism during starvation, contributing to fat utilization (Van Gilst et al. 2005a; Taubert et al. 2006). Despite homology to HNF4α, nhr-49 function is therefore more similar to mammalian PPARα and its coactivator PGC-1 (Ashrafi 2007).
hlh-30/TFEB and lipid mobilization:
Loss of hlh-30/TFEB function renders worms extremely sensitive to L1 starvation (O’Rourke and Ruvkun 2013; Settembre et al. 2013). hlh-30/TFEB activates expression of multiple lipase genes within hours of starvation, promoting lipid mobilization and starvation survival (O’Rourke and Ruvkun 2013; Settembre et al. 2013). In fed worms, mTOR represses hlh-30/TFEB and mxl-3/MAX represses lipase expression, providing conditional control of lipase activity (O’Rourke and Ruvkun 2013). hlh-30/TFEB is also required to activate autophagy during starvation (O’Rourke et al. 2013; O’Rourke and Ruvkun 2013). Regulation of autophagy by hlh-30/TFEB may be due to transcriptional regulation of autophagy genes, coordinating lipid metabolism and autophagy (O’Rourke and Ruvkun 2013). However, accumulation of ω-6 polyunsaturated fatty acids due to hlh-30/TFEB activation of the lipase lipl-4 activates autophagy on its own, indicating that changes in lipid metabolism influence autophagy (O’Rourke et al. 2013). Effects of vitellogenin lipoproteins on aging and starvation resistance suggest that hlh-30/TFEB-mediated lipid mobilization affects physiology systemically through lipid transport (Seah et al. 2016; Harvald et al. 2017).
In addition to displaying increased mortality during L1 starvation, hlh-30/TFEB mutants are defective at recovery from L1 starvation (Murphy et al. 2019). Lysosomal acidification is deficient in hlh-30/TFEB mutants, and TOR activation is impaired in response to feeding E. coli OP50. The hlh-30/TFEB target lipl-2 has a similar recovery-defective phenotype, implicating lipid metabolism in recovery from starvation. However, recovering starved hlh-30/TFEB mutants with a completely defined medium (CeMM), or specifically glucose and linoleic acid, restores lysosomal acidification as well as TOR activation and recovery upon subsequent feeding with OP50 (Murphy et al. 2019). This work reveals a critical role of hlh-30/TFEB regulation of lysosomal function during recovery from starvation, and it shows that supplementation with glucose and linoleic acid can bypass this requirement.
The lipase lipl-5 is also induced by starvation in adults, promoting increased fat catabolism (Buis et al. 2019). In contrast to lipl-4 in starved L1 larvae (O’Rourke et al. 2013), lipl-5 function decreases starvation resistance in adults (Buis et al. 2019). lipl-5 appears to function in coelomocytes (Buis et al. 2019), systemically affecting mitochondrial membrane composition and function (Macedo et al. 2020).
Translation:
As a major anabolic process, translation is energetically costly. Disruption of the translation initiation factors ifg-1/eIF4G or ife-2/eIF4E or ribosomal S6 kinase rsks-1/RPS6KB1, all positive regulators of translation, limits growth rate and fecundity while increasing life span, starvation survival, and resistance to other forms of stress (Pan et al. 2007; Lee et al. 2012; Wu et al. 2018a). Conversely, mutation of translation elongation factor 2 kinase efk-1/eEF2K, a negative regulator of translation, causes mild reduction in life span but relatively severe reduction in L1 starvation survival (Leprivier et al. 2013). ifg-1/eIF4G mRNA expression is downregulated in dauer larvae and upregulated during recovery from L1 starvation (Pan et al. 2007; Baugh et al. 2009), and efk-1/eEF2K mRNA is upregulated during L1 arrest (Baugh et al. 2009; Leprivier et al. 2013), suggesting nutrient-dependent transcriptional regulation of translational capacity. In addition, AMPK activates and mTORC1 inhibits eEF2K, and mTORC1 activates ribosomal S6 kinase, all in response to nutrient availability in mammalian cells (Proud 2007), suggesting additional, post-translational mechanisms of nutrient-dependent regulation.
Translation and proteostasis are linked to lipid metabolism. Disruption of ribosomal DNA transcription or rRNA processing causes nucleolar stress, causing lipid accumulation and increased starvation resistance (Wu et al. 2018a). PHA-4/FoxA expression is upregulated by nucleolar stress, and PHA-4/FoxA promotes expression of lipogenic enzymes (Wu et al. 2018a). AMPK activity is also regulated by translational capacity, suggesting feedback regulation. Total RNA (tRNA) synthetase deficiency activates AMPK to promote starvation resistance in a proteasome-dependent fashion (Webster et al. 2017). Consequently, reducing translation by disruption of aminoacyl tRNA synthetase function increases fat mass and starvation resistance, while disruption of proteasomal function does the opposite (Webster et al. 2017). Whether pha-4/FoxA is required for disruption of tRNA charging to increase lipid accumulation and starvation resistance is unknown. Together, these observations suggest that PHA-4/FoxA and AMPK contributes to homeostasis during fluctuations in nutrient availability by coordinating translational capacity and lipid metabolism.
ER UPR:
Surprisingly, the ER UPR affects lipid metabolism. ER UPR genes ire-1/ERN-1 and hsp-4/BiP are required for upregulation of lipases and hydrolysis of fat stores within hours of starvation, and ire-1 and hsp-4 mutants have reduced starvation survival (Jo et al. 2009). These results implicate ER-resident proteins in nutrient sensing and lipid metabolism. Notably, mutations affecting the nhr-49/NHR coactivator mdt-15, which regulates expression of fatty acid desaturases (Van Gilst et al. 2005a; Van Gilst et al. 2005b; Taubert et al. 2006), cause constitutive ER UPR activation in the absence of protein aggregation (Hou et al. 2014). Instead, activation of the ER UPR is due to changes in ER membrane composition and fluidity (Hou et al. 2014). Together, these results suggest that membrane disequilibrium, in addition to proteostasis, is monitored to maintain ER homeostasis.
skn-1/Nrf:
skn-1/Nrf links proline catabolism with lipid metabolism during starvation. skn-1/Nrf overexpression is reported to promote starvation resistance, but skn-1/Nrf gain-of-function mutants gave inconsistent results, and data are limiting (Paek et al. 2012). Mutation of alh-6/ALDH4A1, a mitochondrial enzyme involved in proline catabolism, accelerates lipid mobilization within hours of starvation, suggesting a connection between amino acid and lipid metabolism (Pang et al. 2014). Expression of fasting-induced lipase fil-1, as well as several fatty acid oxidation genes, is upregulated by mutation of alh-6 during starvation, and L4 starvation survival is reduced. In addition, skn-1/Nrf is activated in alh-6 mutants, and skn-1/Nrf is required for upregulation of most fatty acid oxidation genes as well as accelerated lipid mobilization in alh-6 mutants (Pang et al. 2014). SKN-1/Nrf physically interacts with the mediator subunit MDT-15 (Goh et al. 2014), an nhr-49/NHR coactivator (Taubert et al. 2006), and mdt-15 is required for skn-1/Nrf regulation of fatty acid oxidation genes (Pang et al. 2014). SKN-1/Nrf also physically interacts with MXL-3/MAX (Paek et al. 2012), which represses lipase expression in fed worms (O’Rourke and Ruvkun 2013). Together these observations suggest SKN-1/Nrf is at a nexus of regulatory pathways coordinating lipid metabolism with fluctuations in nutrient availability and other forms of stress.
Octopamine:
The effects of exogenous octopamine on adult behavior are similar to the effects of starvation (see Effects of Starvation on Behavior) (Horvitz et al. 1982), reflecting a role of octopamine in the starvation response (Figure 9). In addition to functioning as a neurotransmitter, octopamine functions as a neurohormone to mediate indirect, systemic effects of DAF-12/VDR on lipid metabolism. DAF-12/VDR activates expression of the tyramine beta-hydroxylase gene tbh-1/DBH, which is critical to biosynthesis of octopamine, in the RIC neurons within hours of starvation, leading to an increase in systemic octopamine levels (Tao et al. 2016). tbh-1 mutant adults are starvation sensitive, and resistance is rescued with exogenous octopamine (Tao et al. 2016). Systemic octopamine induces expression of the lipase lips-6 in the intestine via its receptor SER-3, resulting in lipid mobilization (Tao et al. 2016). Whether octopamine induction of lips-6 expression involves HLH-30/TFEB is unknown.
Ceramides:
Synthesis of ceramides, waxy lipids containing sphingosine and fatty acid moieties, supports starvation survival. Although understudied in the context of starvation, ceramides play signaling roles in a number of cell biological contexts (Hannun and Obeid 2008). There are three ceramide synthase genes in C. elegans: hyl-1, hyl-2, and lagr-1. hyl-1 is required for synthesis of ceramides and sphingolipids with particularly long acyl chains (≥C24), and hyl-2 is required for synthesis with shorter acyl chains (≤C22) (Menuz et al. 2009). Genetic disruption of ceramide biosynthesis dramatically reduces L1 starvation survival (Cui et al. 2017). Mutation of hyl-1 has a stronger effect than hyl-2, suggesting that ceramides with particularly long acyl chains are more important to survival, similar to what has been observed for life span (Mosbech et al. 2013). Gene expression analysis suggests that ceramides promote starvation resistance at least in part by affecting nutrient-responsive gene expression, possibly interacting with IIS and LIN-35/Rb (Cui et al. 2017) (see Transcriptional regulators), but specific mechanisms by which ceramides influence starvation resistance remain to be determined.
ROS
ROS can limit starvation survival. ROS increase during L1 starvation (Roux et al. 2016), and DAF-16/FoxO activates the superoxide dismutase gene sod-3 to protect starved larvae from oxidative stress (Henderson et al. 2006). Mammalian FoxO3a activates expression of manganese superoxide dismutase to protect glucose-starved, quiescent cells from oxidative stress, suggesting conserved function (Kops et al. 2002). daf-16/FoxO promotes starvation resistance (Munoz and Riddle 2003; Baugh and Sternberg 2006), and these observations suggest upregulation of antioxidant enzymes as an effector mechanism. Ligand-free DAF-12/VDR together with its coactivator DIN-1/SPEN also activates expression of antioxidant enzyme genes (gst-4 and gst-10) during starvation in young adults (Tao et al. 2017). daf-12/VDR and din-1/SPEN null mutants display systemic necrosis during adult starvation and do not survive as long as wild-type worms (Tao et al. 2017). Notably, survival of daf-12/VDR and din-1/SPEN null mutants as well as a daf-12/VDR gain-of-function (Daf-c) mutant is indistinguishable from wild-type animals during L1 arrest (Lee and Ashrafi 2008; Kaplan et al. 2015). Tao et al. starved adult worms, as opposed to L1-stage larvae, but they also starved them on NGM plates as opposed to a simple buffer as used by Kaplan et al. and Lee et al. (Tao et al. 2017; Kaplan et al. 2015; Lee and Ashrafi 2008). The discrepant results for daf-12/VDR and starvation survival could therefore be due to developmental stage or starvation conditions (see Commentary on starvation conditions).
Environmental factors affecting starvation resistance
A variety of environmental factors affect starvation resistance (Table 4). Availability of a carbon source to otherwise starved L1 larvae increases survival. As little as 1 mM ethanol increases L1 starvation survival (Castro et al. 2012). Supplementation with n-propanol or n-butanol also extends survival, but methanol and isopropanol do not, suggesting that ethanol is converted to acetate and used as a carbon source (Castro et al. 2012). Consistent with this hypothesis, supplementation with sodium acetate also increases survival (L.R.B., unpublished results), and the hydrogen atoms from ethanol are incorporated into fatty acids and amino acids (Castro et al. 2012). Ethanol also affects behavior of starved worms (Artyukhin et al. 2015). Notably, when cholesterol is added to S-basal, the ethanol solvent results in a final ethanol concentration of 20 mM (0.1%), which is sufficient to double L1 starvation survival (Castro et al. 2012). In contrast, M9 does not contain ethanol. It is important that researchers are cognizant of whether the buffer they are using contains ethanol. For example, the L1 arrest-defective phenotype of daf-16/FoxO mutants, is only displayed in the presence of ethanol (or other appropriate carbon source) (Baugh and Sternberg 2006). Supplementation with glucose, trehalose, or other sugars can also approximately double starvation survival (Hibshman et al. 2017). Strictly speaking, worms supplemented with a carbon source are not completely starved, but none of these carbon sources is sufficient to promote L1 development beyond limited divisions of the epidermal seam cells in wild-type worms (Baugh and Sternberg 2006; Castro et al. 2012; Fukuyama et al. 2015; Hibshman et al. 2017).
Table 4. Environmental factors that affect starvation resistance.
| Environmental factor | References |
|---|---|
| Temperature | (Lee et al. 2012) |
| Ethanol, n-propanol, and n-butanol | (Castro et al. 2012) |
| Sodium acetate | L.R.B., unpublished results |
| Trehalose, glucose, or maltose | (Hibshman et al. 2017) |
| Actinomycin D | (Wu et al. 2018a) |
| Population density | (Artyukhin et al. 2013a) |
| Perception of food | (Kaplan et al. 2018) |
| Test tube closures | L. Avery and A. B. Artyukhin, personal communicationa |
Results are based on analysis of L1 starvation survival or recovery from L1 starvation. Drugs and toxins are not included as environmental factors.
See the WormBreeder’s Gazette article “Artifacts in L1 Starvation Assay.”, http://wbg.wormbook.org/2012/06/25/artifacts-in-l1-starvation-assay/ Ambient light may also affect starvation survival since it affects life span (De Magalhaes Filho et al. 2018).
A variety of additional environmental factors are known to influence L1 starvation survival, illustrating the need for carefully considering and controlling experimental conditions. For example, larvae survive L1 starvation longer at 15° and shorter at 25° compared to 20° (Lee et al. 2012). Larvae survive L1 starvation longer at higher population density, and this effect does not require daf-22/Scp2 (Artyukhin et al. 2013a). daf-22 is required for ascaroside biosynthesis (Ludewig and Schroeder 2013), suggesting that the molecule(s) responsible for population-density-dependent L1 starvation survival is either a novel pheromone or some other excreted metabolite that is synthesized in a DAF-22/Scp2-independent fashion. The disaccharide trehalose, which is produced during starvation (Hibshman et al. 2017) (see Central carbon metabolism), is detectable at low micromolar concentrations in the buffer of starved L1s, and such concentrations are sufficient to mimic the effect of high density in low-density cultures (A. Mata-Cabana and M. Olmedo, personal communication). The type of test tube or closure used can affect L1 starvation survival (L. Avery and A. B. Artyukhin, personal communication; see WormBreeder’s Gazette article “Artifacts in L1 Starvation Assay”, http://wbg.wormbook.org/2012/06/25/artifacts-in-l1-starvation-assay/), highlighting potential effects of seemingly trivial experimental factors. It is also likely that ambient light reduces starvation survival, since it has dramatic effects on adult life span (De Magalhaes Filho et al. 2018). The perception of food through the chemical senses during starvation has the curious effect of rendering L1 arrest irreversible (Kaplan et al. 2018). The perception of food does not affect survival per se, as mortality is not affected, but it does affect starvation resistance in that they are incapable of growth and reproduction thereafter.
Nongenetic inheritance of starvation resistance
Potentially adaptive responses to nutrient stress appear to extend across generations. Although it is clear that C. elegans encounters starvation frequently in the wild, the dynamics of fluctuations in food availability are unclear. Nonetheless, it is conceivable that anticipation of starvation based on parental or ancestral conditions could be beneficial in certain circumstances. Evidence now exists for maternal (intergenerational) and transgenerational epigenetic effects on starvation resistance (Figure 11).
Figure 11.
Nongenetic inheritance of starvation responses. Multigenerational effects of environmental conditions are classified by the number of generations they are inherited. Phenotypic effects that persist for only one or two generations (F1 or F2 progeny) are “intergenerational.” In contrast, effects that persist for three or more generations (F3 and beyond) indicate “transgenerational epigenetic inheritance.” This distinction is made since direct effects on germ cells can theoretically persist for two generations, whereas epigenetic inheritance requires germline mechanisms that actively maintain a regulatory state. Worm images were borrowed with permission from WormAtlas (Altun and Hall 2009).
Maternal provisioning and intergenerational adaptation to nutrient stress:
Maternal age and diet affect oocyte provisioning and starvation resistance in L1 larvae. The C. elegans genome contains six vitellogenin genes, which encode the protein subunits of the low-density lipoprotein particle commonly referred to as yolk (Sharrock 1983; Blumenthal et al. 1984). Vitellogenin is synthesized in the intestine, secreted into the body cavity, and taken up by developing oocytes in the gonad via receptor-mediated endocytosis (Greenstein 2005). Vitellogenin expression is regulated by a variety of pathways such that it is tissue- and stage-specific, sexually dimorphic, and subject to physiological regulation (DePina et al. 2011; Balklava et al. 2016; Goszczynski et al. 2016). Mutations disrupting synthesis, endocytosis, or embryonic storage of vitellogenin reduce L1 starvation survival (Chotard et al. 2010; Van Rompay et al. 2015). Dietary restriction causes worms to produce fewer but larger eggs, and such progeny retain greater reproductive success after extended L1 starvation (see Larval starvation reduces reproductive success) (Harvey and Orbidans 2011; Hibshman et al. 2016). Maternal reduction of IIS also increases progeny size and L1 starvation resistance, as measured by growth and fecundity following extended L1 starvation (Hibshman et al. 2016). In addition, maternal age affects progeny size and L1 starvation resistance due to differences in vitellogenin provisioning to oocytes (Perez et al. 2017). Likewise, maternal dietary restriction and reduced IIS increase vitellogenin provisioning to oocytes, which reduces IIS in progeny, protecting them from pathological consequences of extended L1 starvation (Jordan et al. 2019). These observations suggest that IIS mediates adaptation to nutrient stress across generations. Together these results show differential oocyte provisioning of vitellogenin, and possibly other materials, underlies phenotypic variation and intergenerational phenotypic plasticity.
Transgenerational epigenetic inheritance of starvation resistance:
Because altered gamete provisioning or other aspects of the maternal environment can potentially affect offspring for up to two generations, demonstration of germline epigenetic inheritance generally requires transgenerational effects in F3 descendants (Figure 11). In one study, extended L1 arrest increased L1 starvation resistance and heat resistance in F3 progeny (Jobson et al. 2015). In another study, it increased F3 life span and was reported to affect small RNA expression, but a role of small RNAs in regulation of gene expression or life span was not addressed (Rechavi et al. 2014). Long-term dauer arrest also increases L1 starvation resistance and life span in F3 progeny, with changes in mRNA expression of very small magnitude affecting a very large number of nutrient-responsive genes (Webster et al. 2018). Notably, long-term dauer arrest reduces L1 size and starvation resistance in F1 progeny, suggesting epigenetic effects are initially obscured by pathological maternal effects (Webster et al. 2018). A similar pattern in F1 and F3 progeny was seen with extended L1 arrest (Jobson et al. 2015). This dauer study corroborated prior reports that extended L1 arrest caused increased starvation resistance and life span three generations later (Rechavi et al. 2014; Jobson et al. 2015). Despite the relatively small effect size (∼10% increase in L1 starvation survival), starvation resistance as a heritable consequence of extended starvation is consistent with epigenetic memory contributing to transgenerational environmental adaptation (Webster et al. 2018). In addition, 24 hr starvation of L4 larvae is reported to increase life span and oxidative stress resistance in F1 progeny, and these effects could be transmitted through males, suggesting germline epigenetic inheritance (Kishimoto et al. 2017). Germline transmission mechanisms and specificity of transgenerational responses to starvation are unclear. Nuclear RNAi and histone modifications are implicated in epigenetic inheritance of gene regulation, but whether these mechanisms mediate adaptive organismal responses to starvation or other environmental conditions has not been sufficiently addressed (Perez and Lehner 2019). In addition, it is unclear if starvation or other stressors provoke epigenetic inheritance of a general stress response or if epigenetic memories are specific to their stimuli.
Global Regulation of Gene Expression During Starvation
Nutrient availability is of paramount significance in gene regulation (Figure 12). Effects of starvation on expression of numerous individual genes has been reported in C. elegans. In this section we will review genome-wide analyses of gene expression that broadly characterize the starvation response and define regulatory mechanisms that contribute to it.
Figure 12.
Global regulation of gene expression in response to nutrient availability. AMPK restrains H3K4 methylation activity of the COMPASS complex during starvation, limiting transcription (Demoinet et al. 2017). RNA Polymerase II is recruited to promoters of housekeeping genes during starvation, but initiation and elongation are inhibited without feeding (Maxwell et al. 2014). Alternative mRNA isoforms are expressed in fed and starved larvae, in particular those encoding splicing factors themselves (Maxwell et al. 2012). Translation is repressed during starvation but dramatically upregulated in response to feeding, with ribosomal proteins being synthesized immediately (Stadler and Fire 2013).
Transcription
Time-series analyses of mRNA expression reveal a profound effect of starvation on gene expression (Baugh et al. 2009; Harvald et al. 2017). In contrast, ∼5% of known microRNA genes are differentially expressed in fed and starved L4 larvae (Garcia-Segura et al. 2015). The starvation response occurs rapidly within hours of hatching in the absence of food, and expression changes relatively slowly after that (Baugh et al. 2009). Nutrient availability also affects mRNA isoform expression in L1 larvae, affecting genes involved in mRNA splicing, translation, and signaling, revealing post-transcriptional effects on gene expression (Maxwell et al. 2012).
Transcriptional regulators:
Given such pervasive effects of nutrient availability on transcription, there must be a variety of transcription regulators that contribute to the starvation response. Mutation of the tumor suppressor lin-35/Rb causes sensitivity to L1 starvation (Cui et al. 2013). Microarray analysis of these mutants in fed and starved L1 larvae revealed relatively little overlap in genes affected in each condition, suggesting conditional specificity of LIN-35/Rb function (Cui et al. 2013). By comparing the effects of lin-35/Rb in starved L1 larvae to the effects of being fed or starved (Baugh et al. 2009), it was determined that lin-35/Rb contributes to aspects of the starvation response, accounting for starvation sensitivity of the mutant (Cui et al. 2013). daf-16/FoxO mutants are also starvation sensitive (Munoz and Riddle 2003; Baugh and Sternberg 2006), but daf-16/FoxO function is not required for the bulk of the early L1 starvation response (Hibshman et al. 2017). daf-16/FoxO affects more genes later in L1 starvation (∼12 hr compared to 2–4 hr) (Kaplan et al. 2015), suggesting indirect effects or possibly late targets. hsf-1/HSF is required for L1 starvation survival (Baugh and Sternberg 2006), suggesting transcriptional regulation of proteostasis. Overexpression of skn-1/Nrf or pha-4/FoxA increases L1 starvation survival (Zhong et al. 2010; Paek et al. 2012). Chromatin immunoprecipitation reveals thousands of binding sites for PHA-4/FoxA during L1 arrest, and binding correlates with gene expression, suggesting that pha-4/FoxA also contributes to the starvation response (Zhong et al. 2010). Although daf-16/FoxO, lin-35/Rb, hsf-1/HSF, skn-1/Nrf, and pha-4/FoxA, as well as nhr-49/NHR, mdt-15, and hlh-30/TFEB, all contribute to the starvation response, other transcriptional regulators are likely to also be involved.
Postrecruitment regulation of RNA Polymerase II:
Genes associated with growth are poised for transcriptional activation during L1 arrest. The transcriptional response to feeding occurs rapidly during recovery from L1 arrest, with many genes upregulated in 1 hr or less (Maxwell et al. 2012), suggesting a mechanism for global activation of the feeding response in starved larvae. RNA Polymerase II (Pol II) accumulates at the 5′ end of many genes during L1 arrest, suggesting postrecruitment regulation of transcription (Baugh et al. 2009). Analysis of Pol II elongation in conjunction with binding and mRNA expression revealed that Pol II is paused during early elongation at actively transcribed stress-response genes during L1 arrest (Kruesi et al. 2013; Maxwell et al. 2014). However, during L1 arrest Pol II also accumulates immediately upstream of genes associated with housekeeping and growth, but in these locations Pol II has not initiated and begun elongation (Maxwell et al. 2014). This observation suggests postrecruitment regulation of transcription initiation, as if to poise growth genes for rapid activation upon feeding. Indeed, 5′ accumulation of uninitiated Pol II (“docked” Pol II) decreases and mRNA abundance increases rapidly in response to feeding, consistent with poising of Pol II at growth genes during starvation (Maxwell et al. 2014). This work suggests that gene expression is coordinated with nutrient availability through postrecruitment regulation of growth and stress genes during starvation at initiation and elongation, respectively.
AMPK and histone modification:
AMPK contributes to repression of transcriptional elongation during L1 starvation. H3K4me3 is associated with active transcriptional elongation, and H3K4me3 levels are abnormally high in the PGCs of AMPK mutants during L1 arrest. Remarkably, high levels of H3K4me3 persist in L4 larvae following L1 arrest and in the PGCs of fed L1 larvae for several generations following starvation (Demoinet et al. 2017). H3K4 trimethylation is catalyzed by the COMPASS complex, and all but one of the COMPASS complex subunits is a predicted phosphorylation target of AMPK (Demoinet et al. 2017). Multiple lines of evidence support a model in which AMPK inhibits COMPASS activity during L1 arrest, maintaining a chromatin state that is not permissive to transcriptional elongation. Transgenerational effects of extended starvation in wild-type worms has been reported (see Transgenerational epigenetic inheritance of starvation resistance) (Rechavi et al. 2014; Jobson et al. 2015; Webster et al. 2018), and it remains to be determined if H3K4me3 contributes to germline transmission of those traits as well.
Translation and the proteome
Nutrient availability has a profound effect on translation. Analysis of mRNA expression together with ribosome profiling during L1 arrest and 3 hr after feeding allowed nutrient-dependent differences in steady-state mRNA levels and translation efficiency to be deconvolved (Stadler and Fire 2013). This analysis confirmed pervasive effects on transcription (Baugh et al. 2009; Maxwell et al. 2012), revealed that effects on translation efficiency are comparable in magnitude, and showed that effects on transcription and translation are generally concordant. Comparative analysis in C. briggsae, C. remanei, and C. brenneri shows that regulation of translation is more conserved than transcription (Stadler and Fire 2013). Critically, this work discovered that mRNAs for ribosomal proteins are translationally repressed during starvation, and that they are among the most translationally upregulated transcripts upon feeding (Stadler and Fire 2013). Transcription and mRNA splicing of ribosomal proteins are also regulated during transition from arrest to growth in L1 larvae (Maxwell et al. 2012), highlighting the importance of regulating translational capacity. Likewise, normal recovery from adult starvation requires expansion of the somatic RNA pool and processing of rRNA (see Rejuvenation during recovery from starvation) (Burnaevskiy et al. 2018). In addition, ribosomal protein mRNAs are translationally repressed in yeast during glucose starvation (Arribere et al. 2011), suggesting a conserved gene regulatory mechanism controlling quiescence and growth.
There is no growth during embryogenesis in C. elegans, and maternal ribosomes are sufficient to complete embryogenesis (Cenik et al. 2019). Larvae lacking ability to synthesize new ribosomes arrest as L1 larvae upon hatching with food. Notably, developmental arrest in this context is independent of daf-16/FoxO and daf-18/PTEN (Cenik et al. 2019), components of the insulin/IGF pathway that are required for starvation-induced L1 arrest (see L1 arrest). Nonetheless, ribosome insufficiency-induced arrest reflects active regulation as opposed to a passive consequence of insufficient translational capacity. Arrested larvae are capable of translation with maternally provided ribosomes. Furthermore, genetic mosaic larvae arrest when only about half of their cells are incapable of synthesizing ribosomes (Cenik et al. 2019). These observations reveal the action of a novel, systemic mechanism by which worms sense and respond to their translational capacity.
Nutrient-dependent regulation of translation in adult males may differ from larval stages or adult hermaphrodites. That is, young adult males utilize fat to actually increase translation as an initial (1–2 days) response to starvation (Tan et al. 2011). A speculative interpretation of this seemingly paradoxical observation is that it is in the evolutionary interest of adult males to mate, and so they invest resources in translation and spermatogenesis at the cost of long-term survival. Transient (2-day) starvation in young adult males actually increases meiosis and sperm number (Chou et al. 2019), consistent with this model. It should be emphasized that this is a speculative model and further investigation is warranted.
Proteomic analyses of the starvation response reveal widespread effects on the proteome and metabolic enzymes in particular. Analysis of L4/young adult worms found that changes to the proteome plateaued within ∼8–16 hr of starvation, with prominent effects on lipid metabolic enzymes and histone variants (Larance et al. 2015). The latter suggests global alteration of chromatin state during starvation. A 16-hr starvation time series analyzing the transcriptome and proteome in parallel in mid-L4 larvae revealed a relatively strong positive correlation between mRNA and protein levels, and it confirmed the importance of hlh-30/TFEB on regulation of lipid metabolism (Harvald et al. 2017) (see Fat metabolism).
Perspective
Commentary on starvation resistance
In an evolutionary or ecological sense, starvation resistance is defined by the effect of starvation on fitness, but fitness is difficult to measure. Whether a worm dies from starvation relates to fitness but is incomplete. Starvation survival can be measured “directly,” by scoring each worm in a starved population as live or dead based on movement or necrosis, or it can be measured “indirectly,” by feeding a starved population and scoring worms as live or dead based on their ability to recover from starvation after some period of time (typically 2 days). Some mutants and starvation conditions do not result in mortality but render larvae incapable of development thereafter, reflecting reduced starvation resistance (Roux et al. 2016; Kaplan et al. 2018). In addition, in certain conditions survival is not detectably affected whether scored directly or indirectly, but growth rate and fecundity upon recovery are affected (Hibshman et al. 2016). In summary, the trait starvation resistance is best thought of as an integral of effects on mortality, growth rate and reproduction.
Experimental evaluation of starvation resistance poses practical challenges. Direct and indirect scoring of survival are both subjective. For direct scoring, there is typically not a discrete transition between life and death, since movement of starving worms gradually becomes more and more sporadic, and since necrosis can develop in parts of the body (often the posterior) while other parts remain capable of movement (often the head or pharynx). For indirect scoring, the longer larvae are starved the slower they grow and develop upon recovery, and with greater variation (Lee et al. 2012; Jobson et al. 2015; Webster et al. 2018). In some cases, the investigator may simply require that the worm be still alive after some recovery period, or they may require that they develop to a particular stage after the recovery period. An alternative is to measure size after a defined recovery period, producing a quantitative rather than binary result. Like growth rate, reproductive success is also decreased following starvation, and brood size can be measured following recovery from starvation. Scoring eggs laid after a defined recovery period (rather than total brood size) integrates effects of starvation on developmental rate and fertility, arguably providing the best proxy for fitness since population size increases faster with a shorter effective generation time (Hodgkin and Barnes 1991).
Commentary on starvation conditions
Experimental starvation conditions vary across studies, potentially confounding results. Worms are sometimes starved on NGM plates, or they are starved in buffer (typically M9 or S-basal). NGM contains peptone, an enzymatic digest of protein, but buffers do not. Feeding worms an enzymatic digest of protein dramatically increases L1 starvation survival without promoting development (Kaplan et al. 2018). In addition, starvation on NGM plates without food may actually result in extremely slow development, although typically described as developmental arrest. In addition, cholesterol is typically added to S-basal but not M9 buffer, resulting in 0.1% ethanol final concentration (cholesterol is dissolved in ethanol). Ethanol provides a carbon source that can double L1 starvation survival without appreciably promoting development in wild-type worms (see Environmental factors affecting starvation resistance) (Castro et al. 2012; Fukuyama et al. 2015). The use of antibiotics and other drugs could also affect results in unintended ways.
Differences in starvation conditions can potentially account for discrepant results reported in the literature. For example, starvation of L3 larvae in buffer results in L4 arrest while starvation on NGM plates induces ARD (see Late larval arrest and ARD). In addition, daf-12/VDR promotes starvation resistance in adult worms starved on plates (Tao et al. 2017), but it does not affect L1 starvation survival in buffer (with or without ethanol) (Lee and Ashrafi 2008; Kaplan et al. 2015) (see ROS).
Investigation of starvation responses in C. elegans would benefit from more nuanced starvation conditions. In studying starvation, the worm field generally compares conditions with and without a bacterial food source. In other systems, distinct responses to starvation of specific elements (e.g., C, N, or P) or macronutrients (e.g., protein, carbohydrates, or fats) have been characterized, but experimental systems to address such forms of starvation are not available for C. elegans. The effects of starvation may also be confounded with abrupt transition to feeding in rich conditions. For example, developmental abnormalities resulting from extended L1 starvation do not occur when starved larvae are recovered in dauer-forming or diet-restriction conditions (Jordan et al. 2019) (see Reproductive development and fertility). This observation suggests that pathology can result from severe mismatch of conditions in addition to starvation itself.
Conclusions and future directions
Much has been learned about C. elegans starvation responses in recent years, but much remains unknown. For example, a number of genes that are critical to starvation resistance and developmental arrest are not essential in standard laboratory conditions. Unbiased genetic approaches are needed to identify the complete set of genes that are essential for during starvation. In addition, the signaling pathways underlying various starvation responses comprise an organismal signaling network of staggering complexity. Sophisticated ways to perturb gene function in time and space as well as approaches that allow real-time imaging of molecular events while manipulating nutrient availability are necessary to dissect this network. Furthermore, male starvation responses are largely uncharacterized, and few sex-specific differences have been identified (see Neuronal development and Translation) although more are likely present. Nonetheless, C. elegans has proven to be a powerful integrative organismal system to investigate how animals adapt to starvation. Discoveries made in C. elegans will continue providing insights in other animals, elucidating environmental adaptation and the molecular basis of disease.
Acknowledgments
We would like to thank members of the Baugh laboratory, past and present, for critical feedback; Dennis Kim and Frank Schroeder for discussions pertaining to the respective roles of proteostasis and dafachronic acid biosynthetic intermediates in dauer regulation; and Iva Greenwald for input on details pertaining to dauer recovery. Work in the Baugh laboratory is supported by the National Institutes of Health (grant R01-GM-117408).
Footnotes
Communicating editor: A. Antebi
Literature Cited
- Abrahante J. E., Daul A. L., Li M., Volk M. L., Tennessen J. M. et al. , 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. 10.1016/S1534-5807(03)00127-8 [DOI] [PubMed] [Google Scholar]
- Adachi T., Kunitomo H., Tomioka M., Ohno H., Okochi Y. et al. , 2010. Reversal of salt preference is directed by the insulin/PI3K and Gq/PKC signaling in Caenorhabditis elegans. Genetics 186: 1309–1319. 10.1534/genetics.110.119768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi M., and Roy R., 2016. AMPK acts as a molecular trigger to coordinate glutamatergic signals and adaptive behaviours during acute starvation. eLife 5: e16349 10.7554/eLife.16349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailion M., and Thomas J. H., 2000. Dauer formation induced by high temperatures in Caenorhabditis elegans. Genetics 156: 1047–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam H., Williams T. W., Dumas K. J., Guo C., Yoshina S. et al. , 2010. EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. 12: 30–41. 10.1016/j.cmet.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert Hubbard E. J., and Schedl T., 2019. Biology of the Caenorhabditis elegans germline stem cell system. Genetics 213: 1145–1188. 10.1534/genetics.119.300238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi D. R., James S. R., Downes C. P., Holmes A. B., Gaffney P. R. et al. , 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr. Biol. 7: 261–269. 10.1016/S0960-9822(06)00122-9 [DOI] [PubMed] [Google Scholar]
- Altun Z. F., and Hall D. H., 2009. Introduction in, WormAtlas; doi:10.3908/wormatlas.1.1. [Google Scholar]
- Angelo G., and Van Gilst M. R., 2009. Starvation protects germline stem cells and extends reproductive longevity in C. elegans. Science 326: 954–958. 10.1126/science.1178343 [DOI] [PubMed] [Google Scholar]
- Antebi A., 2006. Nuclear hormone receptors in C. elegans (January 3, 2006), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.64.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A., 2015. Nuclear receptor signal transduction in C. elegans. (June 9, 2015), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.64.2, http://www.wormbook.org. 10.1895/wormbook.1.64.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A., Culotti J. G., and Hedgecock E. M., 1998. daf-12 regulates developmental age and the dauer alternative in Caenorhabditis elegans. Development 125: 1191–1205. [DOI] [PubMed] [Google Scholar]
- Antebi A., Yeh W. H., Tait D., Hedgecock E. M., and Riddle D. L., 2000. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 14: 1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Apfeld J., and Kenyon C., 1998. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell 95: 199–210. 10.1016/S0092-8674(00)81751-1 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Doudna J. A., and Gilbert W. V., 2011. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol. Cell 44: 745–758. 10.1016/j.molcel.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyukhin A. B., Schroeder F. C., and Avery L., 2013a Density dependence in Caenorhabditis larval starvation. Sci. Rep. 3: 2777 10.1038/srep02777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyukhin A. B., Yim J. J., Srinivasan J., Izrayelit Y., Bose N. et al. , 2013b Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J. Biol. Chem. 288: 18778–18783. 10.1074/jbc.C113.477000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artyukhin A. B., Yim J. J., Cheong Cheong M., and Avery L., 2015. Starvation-induced collective behavior in C. elegans. Sci. Rep. 5: 10647 10.1038/srep10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K., 2007. Obesity and the regulation of fat metabolism (March 9, 2007), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.130.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L., and Horvitz H. R., 1990. Effects of starvation and neuroactive drugs on feeding in Caenorhabditis elegans. J. Exp. Zool. 253: 263–270. 10.1002/jez.1402530305 [DOI] [PubMed] [Google Scholar]
- Avery L., and You Y. J., 2012. C. elegans feeding (May 21, 2012), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.150.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balklava Z., Rathnakumar N. D., Vashist S., Schweinsberg P. J., and Grant B. D., 2016. Linking gene expression in the intestine to production of gametes through the phosphate transporter PITR-1 in Caenorhabditis elegans. Genetics 204: 153–162. 10.1534/genetics.116.188532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann C. I., and Horvitz H. R., 1991. Control of larval development by chemosensory neurons in Caenorhabditis elegans. Science 251: 1243–1246. 10.1126/science.2006412 [DOI] [PubMed] [Google Scholar]
- Bargmann C. I., Hartwieg E., and Horvitz H. R., 1993. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515–527. 10.1016/0092-8674(93)80053-H [DOI] [PubMed] [Google Scholar]
- Barna J., Princz A., Kosztelnik M., Hargitai B., Takacs-Vellai K. et al. , 2012. Heat shock factor-1 intertwines insulin/IGF-1, TGF-beta and cGMP signaling to control development and aging. BMC Dev. Biol. 12: 32 10.1186/1471-213X-12-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battu G., Hoier E. F., and Hajnal A., 2003. The C. elegans G-protein-coupled receptor SRA-13 inhibits RAS/MAPK signalling during olfaction and vulval development. Development 130: 2567–2577. 10.1242/dev.00497 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., 2013. To grow or not to grow: nutritional control of development during Caenorhabditis elegans L1 arrest. Genetics 194: 539–555. 10.1534/genetics.113.150847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L. R., and Sternberg P. W., 2006. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr. Biol. 16: 780–785. 10.1016/j.cub.2006.03.021 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., Demodena J., and Sternberg P. W., 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94. 10.1126/science.1169628 [DOI] [PubMed] [Google Scholar]
- Baugh L. R., Kurhanewicz N., and Sternberg P. W., 2011. Sensitive and precise quantification of insulin-like mRNA expression in Caenorhabditis elegans. PLoS One 6: e18086 10.1371/journal.pone.0018086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E. A., and Hobert O., 2018. Past experience shapes sexually dimorphic neuronal wiring through monoaminergic signalling. Nature 561: 117–121. 10.1038/s41586-018-0452-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharadwaj P. S., and Hall S. E., 2017. Endogenous RNAi pathways are required in neurons for dauer formation in Caenorhabditis elegans. Genetics 205: 1503–1516. 10.1534/genetics.116.195438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A., Aghayeva U., Berghoff E. G. and Hobert O., 2019. Plasticity of the Electrical Connectome of C. elegans. Cell 176: 1174–1189.e16. 10.1016/j.cell.2018.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billard B., Vigne P., and Braendle C., 2020. A natural mutational event uncovers a life history trade-off via hormonal pleiotropy. Curr. Biol. S0960-9822(20)31158-1. 10.1016/j.cub.2020.08.004 [DOI] [PubMed] [Google Scholar]
- Birnby D. A., Link E. M., Vowels J. J., Tian H., Colacurcio P. L. et al. , 2000. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans. Genetics 155: 85–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell T. K., Sewell A. K., Wu Z., and Han M., 2019. TOR signaling in Caenorhabditis elegans development, metabolism, and aging. Genetics 213: 329–360. 10.1534/genetics.119.302504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T., Squire M., Kirtland S., Cane J., Donegan M. et al. , 1984. Cloning of a yolk protein gene family from Caenorhabditis elegans. J. Mol. Biol. 174: 1–18. 10.1016/0022-2836(84)90361-9 [DOI] [PubMed] [Google Scholar]
- Boland S., Schmidt U., Zagoriy V., Sampaio J. L., Fritsche R. F. et al. , 2017. Phosphorylated glycosphingolipids essential for cholesterol mobilization in Caenorhabditis elegans. Nat. Chem. Biol. 13: 647–654. 10.1038/nchembio.2347 [DOI] [PubMed] [Google Scholar]
- Braeckman B. P., Houthoofd K., and Vanfleteren J. R., 2009. Intermediary metabolism (February 16, 2009), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.146.1, http://www.wormbook.org. 10.1895/wormbook.1.146.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braendle C., and Félix M. A., 2008. Plasticity and errors of a robust developmental system in different environments. Dev. Cell 15: 714–724. 10.1016/j.devcel.2008.09.011 [DOI] [PubMed] [Google Scholar]
- Brauer M. J., Yuan J., Bennett B. D., Lu W., Kimball E. et al. , 2006. Conservation of the metabolomic response to starvation across two divergent microbes. Proc. Natl. Acad. Sci. USA 103: 19302–19307. 10.1073/pnas.0609508103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brejc K., Bian Q., Uzawa S., Wheeler B. S., Anderson E. C. et al. , 2017. Dynamic control of X chromosome conformation and repression by a histone H4K20 demethylase. Cell 171: 85–102.e23. 10.1016/j.cell.2017.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. J., Busch K. E., and de Bono M., 2008. A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 105: 8044–8049. 10.1073/pnas.0707607105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A. et al. , 2012. A nuclear argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis A., Bellemin S., Goudeau J., Monnier L., Loiseau N. et al. , 2019. Coelomocytes regulate starvation-induced fat catabolism and lifespan extension through the lipase LIPL-5 in Caenorhabditis elegans. Cell Rep. 28: 1041–1049.e4. 10.1016/j.celrep.2019.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N., Chen S., Mailig M., Reynolds A., Karanth S. et al. , 2018. Reactivation of RNA metabolism underlies somatic restoration after adult reproductive diapause in C. elegans. eLife 7: e36194 10.7554/eLife.36194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butuči M., Williams A. B., Wong M. M., Kramer B., and Michael W. M., 2015. Zygotic genome activation triggers chromosome damage and checkpoint signaling in C. elegans primordial germ cells. Dev. Cell 34: 85–95. 10.1016/j.devcel.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Carranza-García E., and Navarro R. E., 2019. Apoptosis contributes to protect germ cells from the oogenic germline starvation response but is not essential for the gonad shrinking or recovery observed during adult reproductive diapause in C. elegans. PLoS One 14: e0218265 10.1371/journal.pone.0218265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranza-García E., and Navarro R. E., 2020. Insights into the hypometabolic stage caused by prolonged starvation in L4-adult Caenorhabditis elegans hermaphrodites. Front. Cell Dev. Biol. 8: 124 10.3389/fcell.2020.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada R. C., and Russell R. L., 1975. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 46: 326–342. 10.1016/0012-1606(75)90109-8 [DOI] [PubMed] [Google Scholar]
- Castro P. V., Khare S., Young B. D., and Clarke S. G., 2012. Caenorhabditis elegans battling starvation stress: low levels of ethanol prolong lifespan in L1 larvae. PLoS One 7: e29984 10.1371/journal.pone.0029984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik E. S., Meng X., Tang N. H., Hall R. N., Arribere J. A. et al. , 2019. Maternal ribosomes are sufficient for tissue diversification during embryonic development in C. elegans. Dev. Cell 48: 811–826.e6. 10.1016/j.devcel.2019.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N., and Baker S. J., 2009. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 4: 127–150. 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin H. C., Okada M., Merz A. J., and Miller D. L., 2015. Tissue-specific autophagy responses to aging and stress in C. elegans. Aging (Albany NY) 7: 419–434. 10.18632/aging.100765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A., Repa J. J., Evans R. M., and Mangelsdorf D. J., 2001. Nuclear receptors and lipid physiology: opening the X-files. Science 294: 1866–1870. 10.1126/science.294.5548.1866 [DOI] [PubMed] [Google Scholar]
- Chen J., and Caswell-Chen E. P., 2004. Facultative vivipary is a life-history trait in Caenorhabditis elegans. J. Nematol. 36: 107–113. [PMC free article] [PubMed] [Google Scholar]
- Chen Y., and Baugh L. R., 2014. Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev. Biol. 394: 314–326. 10.1016/j.ydbio.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Chen A. T., Guo C., Itani O. A., Budaitis B. G., Williams T. W. et al. , 2015. Longevity genes revealed by integrative analysis of isoform-specific daf-16/FoxO mutants of Caenorhabditis elegans. Genetics 201: 613–629. 10.1534/genetics.115.177998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W. C., Ching T. T., Lee H. C., Mousigian C., and Hsu A. L., 2012. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148: 322–334. 10.1016/j.cell.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin R. M., Fu X., Pai M. Y., Vergnes L., Hwang H. et al. , 2014. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature 510: 397–401. 10.1038/nature13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotard L., Skorobogata O., Sylvain M. A., Shrivastava S., and Rocheleau C. E., 2010. TBC-2 is required for embryonic yolk protein storage and larval survival during L1 diapause in Caenorhabditis elegans. PLoS One 5: e15662 10.1371/journal.pone.0015662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou W. Y., Lin Y. C., and Lee Y. H., 2019. Short-term starvation stress at young adult stages enhances meiotic activity of germ cells to maintain spermatogenesis in aged male Caenorhabditis elegans. Aging Cell 18: e12930 10.1111/acel.12930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils A., Gloeck M., Chen Z., Zhang Y., and Alcedo J., 2011. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development 138: 1183–1193. 10.1242/dev.060905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Guerry F., Muller F., and Palladino F., 2002. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3: 235–241. 10.1093/embo-reports/kvf051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Cohen M. L., Teng C., and Han M., 2013. The tumor suppressor Rb critically regulates starvation-induced stress response in C. elegans. Curr. Biol. 23: 975–980. 10.1016/j.cub.2013.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Wang Y., Cavaleri J., Kelson T., Teng Y. et al. , 2017. Starvation-induced stress response is critically impacted by ceramide levels in Caenorhabditis elegans. Genetics 205: 775–785. 10.1534/genetics.116.194282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K. A., Bouagnon A. D., Barros A. G., Lin L., Malard L. et al. , 2014. Loss of a neural AMP-activated kinase mimics the effects of elevated serotonin on fat, movement, and hormonal secretions. PLoS Genet. 10: e1004394 10.1371/journal.pgen.1004394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M. P., Saez A. G., and Lozano E., 2015. miR-58 family and TGF-beta pathways regulate each other in Caenorhabditis elegans. Nucleic Acids Res. 43: 9978–9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Magalhaes Filho C. D., Henriquez B., Seah N. E., Evans R. M., Lapierre L. R. et al. , 2018. Visible light reduces C. elegans longevity. Nat. Commun. 9: 927 10.1038/s41467-018-02934-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney C. E., Chen A. T., Graniel J. V., Dumas K. J., and Hu P. J., 2017. A histone H4 lysine 20 methyltransferase couples environmental cues to sensory neuron control of developmental plasticity. Development 144: 1273–1282. 10.1242/dev.145722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoinet E., Li S., and Roy R., 2017. AMPK blocks starvation-inducible transgenerational defects in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 114: E2689–E2698. 10.1073/pnas.1616171114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina A. S., Iser W. B., Park S. S., Maudsley S., Wilson M. A. et al. , 2011. Regulation of Caenorhabditis elegans vitellogenesis by DAF-2/IIS through separable transcriptional and posttranscriptional mechanisms. BMC Physiol. 11: 11 10.1186/1472-6793-11-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derry W. B., Putzke A. P., and Rothman J. H., 2001. Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance. Science 294: 591–595. 10.1126/science.1065486 [DOI] [PubMed] [Google Scholar]
- Dumas K. J., Guo C., Wang X., Burkhart K. B., Adams E. J. et al. , 2010. Functional divergence of dafachronic acid pathways in the control of C. elegans development and lifespan. Dev. Biol. 340: 605–612. 10.1016/j.ydbio.2010.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T., Rasmussen N. R., Ballato E., Mote F. S., and Reiner D. J., 2020. The Rheb-TORC1 signaling axis functions as a developmental checkpoint. Development 147: dev181727 10.1242/dev.181727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer D. S., and Aamodt E. J., 2013. Insulin/IGF-1 signaling, including class II/III PI3Ks, beta-arrestin and SGK-1, is required in C. elegans to maintain pharyngeal muscle performance during starvation. PLoS One 8: e63851 10.1371/journal.pone.0063851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elle I. C., Rodkaer S. V., Fredens J., and Faergeman N. J., 2012. A method for measuring fatty acid oxidation in C. elegans. Worm 1: 26–30. 10.4161/worm.19564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi P., Wang X., Suleski M., and Wicky C., 2017. A network of chromatin factors is regulating the transition to postembryonic development in Caenorhabditis elegans. G3 (Bethesda) 7: 343–353. 10.1534/g3.116.037747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkut C., Penkov S., Khesbak H., Vorkel D., Verbavatz J. M. et al. , 2011. Trehalose renders the dauer larva of Caenorhabditis elegans resistant to extreme desiccation. Curr. Biol. 21: 1331–1336. 10.1016/j.cub.2011.06.064 [DOI] [PubMed] [Google Scholar]
- Euling S., and Ambros V., 1996a Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell 84: 667–676. 10.1016/S0092-8674(00)81045-4 [DOI] [PubMed] [Google Scholar]
- Euling S., and Ambros V., 1996b Reversal of cell fate determination in Caenorhabditis elegans vulval development. Development 122: 2507–2515. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Barish G. D., and Wang Y. X., 2004. PPARs and the complex journey to obesity. Nat. Med. 10: 355–361. 10.1038/nm1025 [DOI] [PubMed] [Google Scholar]
- Feinbaum R., and Ambros V., 1999. The timing of lin-4 RNA accumulation controls the timing of postembryonic developmental events in Caenorhabditis elegans. Dev. Biol. 210: 87–95. 10.1006/dbio.1999.9272 [DOI] [PubMed] [Google Scholar]
- Ferguson E. L., and Horvitz H. R., 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics 110: 17–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes de Abreu D. A., Caballero A., Fardel P., Stroustrup N., Chen Z. et al. , 2014. An insulin-to-insulin regulatory network orchestrates phenotypic specificity in development and physiology. PLoS Genet. 10: e1004225 10.1371/journal.pgen.1004225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N., and Antebi A., 2008. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 22: 2149–2165. 10.1101/gad.1701508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell, S. W., D. M. Raizen and Y. J. You, 2020 Behavioral states. Genetics 216: 2 315-332. 10.1534/genetics.120.303539. [DOI] [PMC free article] [PubMed]
- Frezal L., and Félix M. A., 2015. C. elegans outside the petri dish. eLife 4: e05849 10.7554/eLife.05849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama, M., 2018 Nutritional control of the germline development in Caenorhabditis elegans, pp. 69–101, in Reproductive and Developmental Strategies, edited by K. Kobayashi, T. Kitano, Y. Iwao, and M. Kondo. Springer, Tokyo. [Google Scholar]
- Fukuyama M., Rougvie A. E., and Rothman J. H., 2006. C. elegans DAF-18/PTEN mediates nutrient-dependent arrest of cell cycle and growth in the germline. Curr. Biol. 16: 773–779. 10.1016/j.cub.2006.02.073 [DOI] [PubMed] [Google Scholar]
- Fukuyama M., Sakuma K., Park R., Kasuga H., Nagaya R. et al. , 2012. C. elegans AMPKs promote survival and arrest germline development during nutrient stress. Biol. Open 1: 929–936. 10.1242/bio.2012836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuyama M., Kontani K., Katada T., and Rougvie A. E., 2015. The C. elegans hypodermis couples progenitor cell quiescence to the dietary state. Curr. Biol. 25: 1241–1248. 10.1016/j.cub.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Galles C., Prez G. M., Penkov S., Boland S., Porta E. O. J. et al. , 2018. Endocannabinoids in Caenorhabditis elegans are essential for the mobilization of cholesterol from internal reserves. Sci. Rep. 8: 6398 10.1038/s41598-018-24925-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Segura L., Abreu-Goodger C., Hernandez-Mendoza A., Dimitrova Dinkova T. D., Padilla-Noriega L. et al. , 2015. High-throughput profiling of Caenorhabditis elegans starvation-responsive microRNAs. PLoS One 10: e0142262 10.1371/journal.pone.0142262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Boag P. R., and Blackwell T. K., 2008. Germline survival and apoptosis (September 4, 2008), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.145.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D., Sutton A. J., Sundermeyer M. L., Albert P. S., King K. V. et al. , 1998. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150: 129–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B., and Antebi A., 2004. Hormonal signals produced by DAF-9/cytochrome P450 regulate C. elegans dauer diapause in response to environmental cues. Development 131: 1765–1776. 10.1242/dev.01068 [DOI] [PubMed] [Google Scholar]
- Gerisch B., Weitzel C., Kober-Eisermann C., Rottiers V., and Antebi A., 2001. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1: 841–851. 10.1016/S1534-5807(01)00085-5 [DOI] [PubMed] [Google Scholar]
- Gerisch B., Tharyan R. G., Mak J., Denzel S. I., Popkes-van Oepen T. et al. , 2020. HLH-30/TFEB is a master regulator of reproductive quiescence. Dev Cell. 53: 316–329.e5. 10.1016/j.devcel.2020.03.014 [DOI] [PubMed] [Google Scholar]
- Gissendanner C. R., Crossgrove K., Kraus K. A., Maina C. V., and Sluder A. E., 2004. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev. Biol. 266: 399–416. 10.1016/j.ydbio.2003.10.014 [DOI] [PubMed] [Google Scholar]
- Goetting D. L., Soto R., and Van Buskirk C., 2018. Food-dependent plasticity in Caenorhabditis elegans stress-induced sleep is mediated by TOR-FOXA and TGF-beta signaling. Genetics 209: 1183–1195. 10.1534/genetics.118.301204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y., Martelli K. L., Parhar K. S., Kwong A. W., Wong M. A. et al. , 2014. The conserved mediator subunit MDT-15 is required for oxidative stress responses in Caenorhabditis elegans. Aging Cell 13: 70–79. 10.1111/acel.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G. Y. S., Winter J. J., Bhanshali F., Doering K. R. S., Lai R. et al. , 2018. NHR-49/HNF4 integrates regulation of fatty acid metabolism with a protective transcriptional response to oxidative stress and fasting. Aging Cell 17: e12743 10.1111/acel.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J. W., and Riddle D. L., 1982. A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218: 578–580. 10.1126/science.6896933 [DOI] [PubMed] [Google Scholar]
- Golden J. W., and Riddle D. L., 1984a A Caenorhabditis elegans dauer-inducing pheromone and an antagonistic component of the food supply. J. Chem. Ecol. 10: 1265–1280. 10.1007/BF00988553 [DOI] [PubMed] [Google Scholar]
- Golden J. W., and Riddle D. L., 1984b The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev. Biol. 102: 368–378. 10.1016/0012-1606(84)90201-X [DOI] [PubMed] [Google Scholar]
- Gomez T. A., Banfield K. L., Trogler D. M., and Clarke S. G., 2007. The L-isoaspartyl-O-methyltransferase in Caenorhabditis elegans larval longevity and autophagy. Dev. Biol. 303: 493–500. 10.1016/j.ydbio.2006.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman, M. B., 2006 Mechanosensation (January 6, 2006), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.62.1, http://www.wormbook.org.
- Goszczynski B., Captan V. V., Danielson A. M., Lancaster B. R., and McGhee J. D., 2016. A 44 bp intestine-specific hermaphrodite-specific enhancer from the C. elegans vit-2 vitellogenin gene is directly regulated by ELT-2, MAB-3, FKH-9 and DAF-16 and indirectly regulated by the germline, by daf-2/insulin signaling and by the TGF-beta/Sma/Mab pathway. Dev. Biol. 413: 112–127. 10.1016/j.ydbio.2016.02.031 [DOI] [PubMed] [Google Scholar]
- Gray J. M., Hill J. J., and Bargmann C. I., 2005. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 102: 3184–3191. 10.1073/pnas.0409009101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein D., 2005. Control of oocyte meiotic maturation and fertilization (December 28, 2005), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.53.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., and Horvitz H. R., 1982. Dominant suppressors of a muscle mutant define an essential gene of Caenorhabditis elegans. Genetics 101: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimbert S., Vargas Velazquez A. M., and Braendle C., 2018. Physiological starvation promotes Caenorhabditis elegans vulval induction. G3 (Bethesda) 8: 3069–3081. 10.1534/g3.118.200449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillermin M. L., Carrillo M. A. and Hallem E. A., 2017. A single set of interneurons drives opposite behaviors in C. elegans. Curr. Biol. 27: 2630–2639.e6. 10.1016/j.cub.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny T. L., Lambie E., Hartwieg E., Horvitz H. R., and Hengartner M. O., 1999. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Hall S. E., Beverly M., Russ C., Nusbaum C., and Sengupta P., 2010. A cellular memory of developmental history generates phenotypic diversity in C. elegans. Curr. Biol. 20: 149–155. 10.1016/j.cub.2009.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall S. E., Chirn G. W., Lau N. C., and Sengupta P., 2013. RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans. RNA 19: 306–319. 10.1261/rna.036418.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich J. T., Entchev E. V., Mende F., Boytchev H., Martin R. et al. , 2009. Methylation of the sterol nucleus by STRM-1 regulates dauer larva formation in Caenorhabditis elegans. Dev. Cell 16: 833–843. 10.1016/j.devcel.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Hannun Y. A., and Obeid L. M., 2008. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9: 139–150. 10.1038/nrm2329 [DOI] [PubMed] [Google Scholar]
- Harvald E. B., Sprenger R. R., Dall K. B., Ejsing C. S., Nielsen R. et al. , 2017. Multi-omics analyses of starvation responses reveal a central role for lipoprotein metabolism in acute starvation survival in C. elegans. Cell Syst. 5: 38–52.e4. 10.1016/j.cels.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Harvey S. C., and Orbidans H. E., 2011. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS One 6: e25840 10.1371/journal.pone.0025840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heestand B., Simon M., Frenk S., Titov D., and Ahmed S., 2018. Transgenerational sterility of piwi mutants represents a dynamic form of adult reproductive diapause. Cell Rep. 23: 156–171. 10.1016/j.celrep.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S. T., and Johnson T. E., 2001. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 11: 1975–1980. 10.1016/S0960-9822(01)00594-2 [DOI] [PubMed] [Google Scholar]
- Henderson S. T., Bonafe M., and Johnson T. E., 2006. daf-16 protects the nematode Caenorhabditis elegans during food deprivation. J. Gerontol. A Biol. Sci. Med. Sci. 61: 444–460. 10.1093/gerona/61.5.444 [DOI] [PubMed] [Google Scholar]
- Hibshman J. D., Hung A., and Baugh L. R., 2016. Maternal diet and insulin-like signaling control intergenerational plasticity of progeny size and starvation resistance. PLoS Genet. 12: e1006396 [corrigenda: PLoS Genet. 148: e1007639 (2008)]. 10.1371/journal.pgen.1006396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibshman J. D., Doan A. E., Moore B. T., Kaplan R. E., Hung A. et al. , 2017. daf-16/FoxO promotes gluconeogenesis and trehalose synthesis during starvation to support survival. eLife 6: e30057 10.7554/eLife.30057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibshman J. D., Leuthner T. C., Shoben C., Mello D. F., Sherwood D. R. et al. , 2018. Nonselective autophagy reduces mitochondrial content during starvation in Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 315: C781–C792. 10.1152/ajpcell.00109.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A. J., Mansfield R., Lopez J. M., Raizen D. M., and Van Buskirk C., 2014. Cellular stress induces a protective sleep-like state in C. elegans. Curr. Biol. 24: 2399–2405. 10.1016/j.cub.2014.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J., and Barnes T. M., 1991. More is not better: brood size and population growth in a self-fertilizing nematode. Proc. Biol. Sci. 246: 19–24. 10.1098/rspb.1991.0119 [DOI] [PubMed] [Google Scholar]
- Hong Y., Roy R., and Ambros V., 1998. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development 125: 3585–3597. [DOI] [PubMed] [Google Scholar]
- Hoppe T., and Cohen E., 2020. Organismal protein homeostasis mechanisms. Genetics 215: 889–901. 10.1534/genetics.120.301283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa M., Sural S., Hsu A. L., and Antebi A., 2015. Co-chaperone p23 regulates C. elegans lifespan in response to temperature. PLoS Genet. 11: e1005023 10.1371/journal.pgen.1005023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn M., Geisen C., Cermak L., Becker B., Nakamura S. et al. , 2014. DRE-1/FBXO11-dependent degradation of BLMP-1/BLIMP-1 governs C. elegans developmental timing and maturation. Dev. Cell 28: 697–710. 10.1016/j.devcel.2014.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H. R., Chalfie M., Trent C., Sulston J. E., and Evans P. D., 1982. Serotonin and octopamine in the nematode Caenorhabditis elegans. Science 216: 1012–1014. 10.1126/science.6805073 [DOI] [PubMed] [Google Scholar]
- Hou N. S., Gutschmidt A., Choi D. Y., Pather K., Shi X. et al. , 2014. Activation of the endoplasmic reticulum unfolded protein response by lipid disequilibrium without disturbed proteostasis in vivo. Proc. Natl. Acad. Sci. USA 111: E2271–E2280. 10.1073/pnas.1318262111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing D. J., Buwalda B., van der Zee E. A., de Boer S. F., and Olivier J. D. A., 2017. The serotonin transporter and early life stress: translational perspectives. Front. Cell. Neurosci. 11: 117 10.3389/fncel.2017.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E. J., and Greenstein D., 2005. Introduction to the germ line (September 1, 2005), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.18.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, P. J., 2007 Dauer (August 8, 2007), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.144.1, http://www.wormbook.org.
- Hu P. J., Xu J., and Ruvkun G., 2006. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2: e99 10.1371/journal.pgen.0020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W. L., Wang Y., Chitturi J., and Zhen M., 2014. A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development 141: 1767–1779. 10.1242/dev.103846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun M., Kim J., Dumur C., Schroeder F. C., and You Y. J., 2016. BLIMP-1/BLMP-1 and metastasis-associated protein regulate stress resistant development in Caenorhabditis elegans. Genetics 203: 1721–1732. 10.1534/genetics.116.190793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbay O., and Ambros V., 2019. Pheromones and nutritional signals regulate the developmental reliance on let-7 family microRNAs in C. elegans. Curr. Biol. 29: 1735–1745.e4. 10.1016/j.cub.2019.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., and Thomas J. H., 2000. Suppressors of transforming growth factor-beta pathway mutants in the Caenorhabditis elegans dauer formation pathway. Genetics 156: 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell T. A., Wang Y., Bloniarz A. E., Brittin C. A., Xu M. et al. , 2012. The connectome of a decision-making neural network. Science 337: 437–444. 10.1126/science.1221762 [DOI] [PubMed] [Google Scholar]
- Jeon M., Gardner H. F., Miller E. A., Deshler J., and Rougvie A. E., 1999. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286: 1141–1146. 10.1126/science.286.5442.1141 [DOI] [PubMed] [Google Scholar]
- Jeong H., and Paik Y. K., 2017. MGL-1 on AIY neurons translates starvation to reproductive plasticity via neuropeptide signaling in Caenorhabditis elegans. Dev. Biol. 430: 80–89. 10.1016/j.ydbio.2017.08.014 [DOI] [PubMed] [Google Scholar]
- Jia K., Albert P. S., and Riddle D. L., 2002. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development 129: 221–231. [DOI] [PubMed] [Google Scholar]
- Jobson M. A., Jordan J. M., Sandrof M. A., Hibshman J. D., Lennox A. L. et al. , 2015. Transgenerational effects of early life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics 201: 201–212. 10.1534/genetics.115.178699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. E., Mitchell D. H., Kline S., Kemal R., and Foy J., 1984. Arresting development arrests aging in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 28: 23–40. 10.1016/0047-6374(84)90150-7 [DOI] [PubMed] [Google Scholar]
- Jordan J. M., Hibshman J. D., Webster A. K., Kaplan R. E. W., Leinroth A. et al. , 2019. Insulin/IGF signaling and vitellogenin provisioning mediate intergenerational adaptation to nutrient Stress. Curr. Biol. 29: 2380–2388.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo H., Shim J., Lee J. H., Lee J., and Kim J. B., 2009. IRE-1 and HSP-4 contribute to energy homeostasis via fasting-induced lipases in C. elegans. Cell Metab. 9: 440–448. 10.1016/j.cmet.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Kadekar P., and Roy R., 2019. AMPK regulates germline stem cell quiescence and integrity through an endogenous small RNA pathway. PLoS Biol. 17: e3000309 10.1371/journal.pbio.3000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein T. L., Smith E. D., Tsuchiya M., Welton K. L., Thomas J. H. et al. , 2006. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell 5: 487–494. 10.1111/j.1474-9726.2006.00238.x [DOI] [PubMed] [Google Scholar]
- Kaminsky Y. G., Kosenko E. A., and Kondrashova M. N., 1982. Metabolites of citric acid cycle, carbohydrate and phosphorus metabolism, and related reactions, redox and phosphorylating states of hepatic tissue, liver mitochondria and cytosol of the pigeon, under normal feeding and natural nocturnal fasting conditions. Comp. Biochem. Physiol. B 73: 957–963. 10.1016/0305-0491(82)90343-1 [DOI] [PubMed] [Google Scholar]
- Kang C., You Y. J., and Avery L., 2007. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 21: 2161–2171. 10.1101/gad.1573107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao G., Nordenson C., Still M., Ronnlund A., Tuck S. et al. , 2007. ASNA-1 positively regulates insulin secretion in C. elegans and mammalian cells. Cell 128: 577–587. 10.1016/j.cell.2006.12.031 [DOI] [PubMed] [Google Scholar]
- Kaplan R. E., and Baugh L. R., 2016. L1 arrest, daf-16/FoxO and nonautonomous control of post-embryonic development. Worm 5: e1175196 10.1080/21624054.2016.1175196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R. E., Chen Y., Moore B. T., Jordan J. M., Maxwell C. S. et al. , 2015. dbl-1/TGF-beta and daf-12/NHR signaling mediate cell-nonautonomous effects of daf-16/FOXO on starvation-induced developmental arrest. PLoS Genet. 11: e1005731 10.1371/journal.pgen.1005731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R. E. W., Webster A. K., Chitrakar R., Dent J. A., and Baugh L. R., 2018. Food perception without ingestion leads to metabolic changes and irreversible developmental arrest in C. elegans. BMC Biol. 16: 112 10.1186/s12915-018-0579-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan R. E. W., Maxwell C. S., Codd N. K., and Baugh L. R., 2019. Pervasive positive and negative feedback regulation of insulin-like signaling in Caenorhabditis elegans. Genetics 211: 349–361. 10.1534/genetics.118.301702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., 2018. Working with dauer larvae (August 9, 2018), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.180.1, http://www.wormbook.org. 10.1895/wormbook.1.180.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., and Ambros V., 2011. The developmental timing regulator HBL-1 modulates the dauer formation decision in Caenorhabditis elegans. Genetics 187: 345–353. 10.1534/genetics.110.123992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., and Ambros V., 2012. Dauer larva quiescence alters the circuitry of microRNA pathways regulating cell fate progression in C. elegans. Development 139: 2177–2186. 10.1242/dev.075986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp X., and Greenwald I., 2013. Control of cell-fate plasticity and maintenance of multipotency by DAF-16/FoxO in quiescent Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 110: 2181–2186. 10.1073/pnas.1222377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga H., Fukuyama M., Kitazawa A., Kontani K., and Katada T., 2013. The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497: 503–506. 10.1038/nature12117 [DOI] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V., and Kelly W. G., 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A. L., Ashraf J. M., Corces-Zimmerman M. R., Landis J. N., and Murphy C. T., 2010. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 8: e1000372 10.1371/journal.pbio.1000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul T. K., Reis Rodrigues P., Ogungbe I. V., Kapahi P., and Gill M. S., 2014. Bacterial fatty acids enhance recovery from the dauer larva in Caenorhabditis elegans. PLoS One 9: e86979 10.1371/journal.pone.0086979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan F. R., and McFadden B. A., 1982. Caenorhabditis elegans: decay of isocitrate lyase during larval development. Exp. Parasitol. 54: 47–54. 10.1016/0014-4894(82)90109-6 [DOI] [PubMed] [Google Scholar]
- Kim S., and Paik Y. K., 2008. Developmental and reproductive consequences of prolonged non-aging dauer in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 368: 588–592. 10.1016/j.bbrc.2008.01.131 [DOI] [PubMed] [Google Scholar]
- Kim Y., and Sun H., 2012. ASM-3 acid sphingomyelinase functions as a positive regulator of the DAF-2/AGE-1 signaling pathway and serves as a novel anti-aging target. PLoS One 7: e45890 10.1371/journal.pone.0045890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto S., Uno M., Okabe E., Nono M., and Nishida E., 2017. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat. Commun. 8: 14031 10.1038/ncomms14031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass M., and Hirsh D., 1976. Non-ageing developmental variant of Caenorhabditis elegans. Nature 260: 523–525. 10.1038/260523a0 [DOI] [PubMed] [Google Scholar]
- Kniazeva M., Euler T., and Han M., 2008. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 22: 2102–2110. 10.1101/gad.1692008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniazeva M., Zhu H., Sewell A. K., and Han M., 2015. A lipid-TORC1 pathway promotes neuronal development and foraging behavior under both fed and fasted conditions in C. elegans. Dev. Cell 33: 260–271. 10.1016/j.devcel.2015.02.015 [DOI] [PubMed] [Google Scholar]
- Kops G. J., Dansen T. B., Polderman P. E., Saarloos I., Wirtz K. W. et al. , 2002. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419: 316–321. 10.1038/nature01036 [DOI] [PubMed] [Google Scholar]
- Korta D. Z., Tuck S., and Hubbard E. J., 2012. S6K links cell fate, cell cycle and nutrient response in C. elegans germline stem/progenitor cells. Development 139: 859–870. 10.1242/dev.074047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostál V., 2006. Eco-physiological phases of insect diapause. J. Insect Physiol. 52: 113–127. 10.1016/j.jinsphys.2005.09.008 [DOI] [PubMed] [Google Scholar]
- Kruesi W. S., Core L. J., Waters C. T., Lis J. T., and Meyer B. J., 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. eLife 2: e00808 10.7554/eLife.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulalert W., and Kim D. H., 2013. The unfolded protein response in a pair of sensory neurons promotes entry of C. elegans into dauer diapause. Curr. Biol. 23: 2540–2545. 10.1016/j.cub.2013.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulalert W., Sadeeshkumar H., Zhang Y. K., Schroeder F. C., and Kim D. H., 2017. Molecular determinants of the regulation of development and metabolism by neuronal eIF2alpha phosphorylation in Caenorhabditis elegans. Genetics 206: 251–263. 10.1534/genetics.117.200568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume M., Chiyoda H., Kontani K., Katada T., and Fukuyama M., 2019. Hedgehog-related genes regulate reactivation of quiescent neural progenitors in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 520: 532–537. 10.1016/j.bbrc.2019.10.045 [DOI] [PubMed] [Google Scholar]
- Lajud N., and Torner L., 2015. Early life stress and hippocampal neurogenesis in the neonate: sexual dimorphism, long term consequences and possible mediators. Front. Mol. Neurosci. 8: 3 10.3389/fnmol.2015.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larance M., Pourkarimi E., Wang B., Brenes Murillo A., Kent R. et al. , 2015. Global proteomics analysis of the response to starvation in C. elegans. Mol. Cell. Proteomics 14: 1989–2001. 10.1074/mcp.M114.044289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Large E. E., Xu W., Zhao Y., Brady S. C., Long L. et al. , 2016. Selection on a subunit of the NURF chromatin remodeler modifies life history traits in a domesticated strain of Caenorhabditis elegans. PLoS Genet. 12: e1006219 10.1371/journal.pgen.1006219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. H., and Ashrafi K., 2008. A TRPV channel modulates C. elegans neurosecretion, larval starvation survival, and adult lifespan. PLoS Genet. 4: e1000213 10.1371/journal.pgen.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R. et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G. D., Wilson M. A., Zhu M., Wolkow C. A., de Cabo R. et al. , 2006. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell 5: 515–524. 10.1111/j.1474-9726.2006.00241.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Hendrix A., Kim J., Yoshimoto J., and You Y. J., 2012. Metabolic rate regulates L1 longevity in C. elegans. PLoS One 7: e44720 10.1371/journal.pone.0044720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G., Remke M., Rotblat B., Dubuc A., Mateo A. R. et al. , 2013. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153: 1064–1079. 10.1016/j.cell.2013.04.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., and Fleming J. T., 1995. Basic culture methods, pp. 4–27 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by Epstein H. F., and Shakes D. C., Academic Press, San Diego. [Google Scholar]
- Libina N., Berman J. R., and Kenyon C., 2003. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. 10.1016/S0092-8674(03)00889-4 [DOI] [PubMed] [Google Scholar]
- Li J., Brown G., Ailion M., Lee S., and Thomas J. H., 2004. NCR-1 and NCR-2, the C. elegans homologs of the human Niemann-Pick type C1 disease protein, function upstream of DAF-9 in the dauer formation pathways. Development 131: 5741–5752. 10.1242/dev.01408 [DOI] [PubMed] [Google Scholar]
- Li W., Kennedy S. G., and Ruvkun G., 2003. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17: 844–858. 10.1101/gad.1066503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Itani O. A., Haataja L., Dumas K. J., Yang J. et al. , 2019. Requirement for translocon-associated protein (TRAP) alpha in insulin biogenesis. Sci Adv 5: eaax0292 10.1126/sciadv.aax0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., and Kenyon C., 1997. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278: 1319–1322. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Johnson S. M., Abraham M., Vella M. C., Pasquinelli A. et al. , 2003. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell 4: 639–650. 10.1016/S1534-5807(03)00124-2 [DOI] [PubMed] [Google Scholar]
- Lin C. H., Tomioka M., Pereira S., Sellings L., Iino Y. et al. , 2010. Insulin signaling plays a dual role in Caenorhabditis elegans memory acquisition and memory retrieval. J. Neurosci. 30: 8001–8011. 10.1523/JNEUROSCI.4636-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Thatcher J. D., Barral J. M., and Epstein H. F., 1995. Bifunctional glyoxylate cycle protein of Caenorhabditis elegans: a developmentally regulated protein of intestine and muscle. Dev. Biol. 169: 399–414. 10.1006/dbio.1995.1156 [DOI] [PubMed] [Google Scholar]
- Liu F., Thatcher J. D., and Epstein H. F., 1997. Induction of glyoxylate cycle expression in Caenorhabditis elegans: a fasting response throughout larval development. Biochemistry 36: 255–260. 10.1021/bi9623800 [DOI] [PubMed] [Google Scholar]
- Lopez A. L. III, Chen J., Joo H. J., Drake M., Shidate M. et al. , 2013. DAF-2 and ERK couple nutrient availability to meiotic progression during Caenorhabditis elegans oogenesis. Dev. Cell 27: 227–240. 10.1016/j.devcel.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig A. H., Kober-Eisermann C., Weitzel C., Bethke A., Neubert K. et al. , 2004. A novel nuclear receptor/coregulator complex controls C. elegans lipid metabolism, larval development, and aging. Genes Dev. 18: 2120–2133. 10.1101/gad.312604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig A. H., and Schroeder F. C., 2013. Ascaroside signaling in C. elegans (January 18, 2013), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.155.1, http://www.wormbook.org. 10.1895/wormbook.1.155.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macedo F., Martins G. L., Luevano-Martinez L. A., Viana G. M., Riske K. A. et al. , 2020. Lipase-like 5 enzyme controls mitochondrial activity in response to starvation in Caenorhabditis elegans. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865: 158539 10.1016/j.bbalip.2019.158539 [DOI] [PubMed] [Google Scholar]
- Magner D. B., Wollam J., Shen Y., Hoppe C., Li D. et al. , 2013. The NHR-8 nuclear receptor regulates cholesterol and bile acid homeostasis in C. elegans. Cell Metab. 18: 212–224. 10.1016/j.cmet.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanti P., Bose N., Bethke A., Judkins J. C., Wollam J. et al. , 2014. Comparative metabolomics reveals endogenous ligands of DAF-12, a nuclear hormone receptor, regulating C. elegans development and lifespan. Cell Metab. 19: 73–83. 10.1016/j.cmet.2013.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak H. Y., and Ruvkun G., 2004. Intercellular signaling of reproductive development by the C. elegans DAF-9 cytochrome P450. Development 131: 1777–1786. 10.1242/dev.01069 [DOI] [PubMed] [Google Scholar]
- Malone E. A., Inoue T., and Thomas J. H., 1996. Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics 143: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga Y., Gengyo-Ando K., Mitani S., Iwasaki T., and Kawano T., 2012. Physiological function, expression pattern, and transcriptional regulation of a Caenorhabditis elegans insulin-like peptide, INS-18. Biochem. Biophys. Res. Commun. 423: 478–483. 10.1016/j.bbrc.2012.05.145 [DOI] [PubMed] [Google Scholar]
- Maxwell C. S., Antoshechkin I., Kurhanewicz N., Belsky J. A., and Baugh L. R., 2012. Nutritional control of mRNA isoform expression during developmental arrest and recovery in C. elegans. Genome Res. 22: 1920–1929. 10.1101/gr.133587.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell C. S., Kruesi W. S., Core L. J., Kurhanewicz N., Waters C. T. et al. , 2014. Pol II docking and pausing at growth and stress genes in C. elegans. Cell Rep. 6: 455–466. 10.1016/j.celrep.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee J. J., Schuster E., Blanc E., Thornton J., and Gems D., 2006. Diapause-associated metabolic traits reiterated in long-lived daf-2 mutants in the nematode Caenorhabditis elegans. Mech. Ageing Dev. 127: 458–472. 10.1016/j.mad.2006.01.006 [DOI] [PubMed] [Google Scholar]
- McGrath P. T., Xu Y., Ailion M., Garrison J. L., Butcher R. A. et al. , 2011. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature 477: 321–325. 10.1038/nature10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P., Schott S., Bedet C., Xiao Y., Rohner S. et al. , 2011. Caenorhabditis elegans Heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol. 12: R123 10.1186/gb-2011-12-12-r123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez A., and Levine B., 2009. Autophagy in C. elegans (August 24, 2009), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.147.1, http://www.wormbook.org. 10.1895/wormbook.1.147.1 [DOI] [Google Scholar]
- Meléndez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H. et al. , 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 301: 1387–1391. 10.1126/science.1087782 [DOI] [PubMed] [Google Scholar]
- Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I. et al. , 2009. Protection of C. elegans from anoxia by HYL-2 ceramide synthase. Science 324: 381–384. 10.1126/science.1168532 [DOI] [PubMed] [Google Scholar]
- Michaelson D., Korta D. Z., Capua Y., and Hubbard E. J., 2010. Insulin signaling promotes germline proliferation in C. elegans. Development 137: 671–680. 10.1242/dev.042523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje J. M., Brokate-Llanos A. M., Perez-Jimenez M. M., Fidalgo M. A., and Munoz M. J., 2011. pkc-1 regulates daf-2 insulin/IGF signalling-dependent control of dauer formation in Caenorhabditis elegans. Aging Cell 10: 1021–1031. 10.1111/j.1474-9726.2011.00747.x [DOI] [PubMed] [Google Scholar]
- Moore B. T., Jordan J. M., and Baugh L. R., 2013. WormSizer: high-throughput analysis of nematode size and shape. PLoS One 8: e57142 10.1371/journal.pone.0057142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech M. B., Kruse R., Harvald E. B., Olsen A. S., Gallego S. F. et al. , 2013. Functional loss of two ceramide synthases elicits autophagy-dependent lifespan extension in C. elegans. PLoS One 8: e70087 10.1371/journal.pone.0070087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola D. L., Cummins C. L., Rottiers V., Sharma K. K., Li T. et al. , 2006. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell 124: 1209–1223. 10.1016/j.cell.2006.01.037 [DOI] [PubMed] [Google Scholar]
- Munoz M. J., and Riddle D. L., 2003. Positive selection of Caenorhabditis elegans mutants with increased stress resistance and longevity. Genetics 163: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C. T., McCarroll S. A., Bargmann C. I., Fraser A., Kamath R. S. et al. , 2003. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283. 10.1038/nature01789 [DOI] [PubMed] [Google Scholar]
- Murphy C. T., Lee S. J., and Kenyon C., 2007. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 19046–19050. 10.1073/pnas.0709613104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. T., Liu H., Ma X., Shaver A., Egan B. M. et al. , 2019. Simple nutrients bypass the requirement for HLH-30 in coupling lysosomal nutrient sensing to survival. PLoS Biol. 17: e3000245 10.1371/journal.pbio.3000245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylenko M., Boland S., Penkov S., Sampaio J. L., Lombardot B. et al. , 2016. NAD+ is a food component that promotes exit from dauer diapause in Caenorhabditis elegans. PLoS One 11: e0167208 10.1371/journal.pone.0167208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan S. D., Yen K., Bansal A., Kwon E. S., Padmanabhan S. et al. , 2011. PDP-1 links the TGF-beta and IIS pathways to regulate longevity, development, and metabolism. PLoS Genet. 7: e1001377 10.1371/journal.pgen.1001377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narbonne P., and Roy R., 2006. Inhibition of germline proliferation during C. elegans dauer development requires PTEN, LKB1 and AMPK signalling. Development 133: 611–619. 10.1242/dev.02232 [DOI] [PubMed] [Google Scholar]
- Narbonne P., and Roy R., 2009. Caenorhabditis elegans dauers need LKB1/AMPK to ration lipid reserves and ensure long-term survival. Nature 457: 210–214. 10.1038/nature07536 [DOI] [PubMed] [Google Scholar]
- Neal S. J., Takeishi A., O’Donnell M. P., Park J., Hong M. et al. , 2015. Feeding state-dependent regulation of developmental plasticity via CaMKI and neuroendocrine signaling. eLife 4: e10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nika L., Gibson T., Konkus R., and Karp X., 2016. Fluorescent beads are a versatile tool for staging Caenorhabditis elegans in different life histories. G3 (Bethesda) 6: 1923–1933. 10.1534/g3.116.030163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L. et al. , 1997. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999. 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- Ohkura K., Suzuki N., Ishihara T., and Katsura I., 2003. SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development 130: 3237–3248. 10.1242/dev.00540 [DOI] [PubMed] [Google Scholar]
- Okamoto N., Viswanatha R., Bittar R., Li Z., Haga-Yamanaka S. et al. , 2018. A membrane transporter is required for steroid hormone uptake in Drosophila. Dev Cell 47: 294–305.e7. 10.1016/j.devcel.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo M., Mata-Cabana A., Jesus Rodriguez-Palero M., Garcia-Sanchez S., Fernandez-Yanez A. et al. , 2019. Prolonged quiescence delays somatic stem cell-like divisions in Caenorhabditis elegans and is controlled by insulin signaling. Aging Cell 19: e13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen P. H., and Ambros V., 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. 10.1006/dbio.1999.9523 [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M., Bayer E. A., and Hobert O., 2016. Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533: 206–211. 10.1038/nature17977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M. C., Borziak K., Nichitean A. M., Dorus S., and Hall S. E., 2018. Early experiences mediate distinct adult gene expression and reproductive programs in Caenorhabditis elegans. PLoS Genet. 14: e1007219 10.1371/journal.pgen.1007219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E. J., Kuballa P., Xavier R., and Ruvkun G., 2013. Omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 27: 429–440. 10.1101/gad.205294.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke E. J., and Ruvkun G., 2013. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat. Cell Biol. 15: 668–676. 10.1038/ncb2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S., Mukhopadhyay A., Narasimhan S. D., Tesz G., Czech M. P. et al. , 2009. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell 136: 939–951. 10.1016/j.cell.2009.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J., Lo J. Y., Narasimhan S. D., Nguyen T. N., Glover-Cutter K. et al. , 2012. Mitochondrial SKN-1/Nrf mediates a conserved starvation response. Cell Metab. 16: 526–537. 10.1016/j.cmet.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A. P., and Johnstone I. L., 2007. The cuticle (March 19, 2007), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.138.1, http://www.wormbook.org. [Google Scholar]
- Paik J. H., Kollipara R., Chu G., Ji H., Xiao Y. et al. , 2007. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323. 10.1016/j.cell.2006.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S., Lynn D. A., Lo J. Y., Paek J., and Curran S. P., 2014. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat. Commun. 5: 5048 10.1038/ncomms6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K. Z., Palter J. E., Rogers A. N., Olsen A., Chen D. et al. , 2007. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell 6: 111–119. 10.1111/j.1474-9726.2006.00266.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., O’Doherty I., Somvanshi R. K., Bethke A., Schroeder F. C. et al. , 2012. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 109: 9917–9922. 10.1073/pnas.1202216109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D. S., Fang L. L., Svy D. K., Ruvkun G., and Li W., 2008. Genetic identification of HSD-1, a conserved steroidogenic enzyme that directs larval development in Caenorhabditis elegans. Development 135: 2239–2249. 10.1242/dev.016972 [DOI] [PubMed] [Google Scholar]
- Penkov S., Mende F., Zagoriy V., Erkut C., Martin R. et al. , 2010. Maradolipids: diacyltrehalose glycolipids specific to dauer larva in Caenorhabditis elegans. Angew. Chem. Int. Ed. Engl. 49: 9430–9435. 10.1002/anie.201004466 [DOI] [PubMed] [Google Scholar]
- Penkov S., Kaptan D., Erkut C., Sarov M., Mende F. et al. , 2015. Integration of carbohydrate metabolism and redox state controls dauer larva formation in Caenorhabditis elegans. Nat. Commun. 6: 8060 10.1038/ncomms9060 [DOI] [PubMed] [Google Scholar]
- Perez M. F., and Lehner B., 2019. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21: 143–151. 10.1038/s41556-018-0242-9 [DOI] [PubMed] [Google Scholar]
- Perez M. F., Francesconi M., Hidalgo-Carcedo C., and Lehner B., 2017. Maternal age generates phenotypic variation in Caenorhabditis elegans. Nature 552: 106–109. 10.1038/nature25012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S. B., Costa M., Wisotzkey R., Devadhar S., Homburger S. A. et al. , 2001. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15: 672–686. 10.1101/gad.867301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C. G., 2007. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem. J. 403: 217–234. 10.1042/BJ20070024 [DOI] [PubMed] [Google Scholar]
- Rechavi O., Houri-Ze’evi L., Anava S., Goh W. S. S., Kerk S. Y. et al. , 2014. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell 158: 277–287. 10.1016/j.cell.2014.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner D. J., Ailion M., Thomas J. H., and Meyer B. J., 2008. C. elegans anaplastic lymphoma kinase ortholog SCD-2 controls dauer formation by modulating TGF-beta signaling. Curr. Biol. 18: 1101–1109. 10.1016/j.cub.2008.06.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan S., Yankura K. A., Guillermin M. L., Fung W., and Hallem E. A., 2019. Feeding state sculpts a circuit for sensory valence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 116: 1776–1781. 10.1073/pnas.1807454116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P., Lim C. S., Johnsen R., Albert P. S., Pilgrim D. et al. , 1996. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274: 1389–1391. 10.1126/science.274.5291.1389 [DOI] [PubMed] [Google Scholar]
- Richardson C. E., Kinkel S., and Kim D. H., 2011. Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity. PLoS Genet. 7: e1002391 10.1371/journal.pgen.1002391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C. E., Yee C., and Shen K., 2019. A hormone receptor pathway cell-autonomously delays neuron morphological aging by suppressing endocytosis. PLoS Biol. 17: e3000452 10.1371/journal.pbio.3000452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle, D. L., and P. S. Albert, 1997 Genetic and Environmental Regulation of Dauer Larva Development in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Riedel C. G., Dowen R. H., Lourenco G. F., Kirienko N. V., Heimbucher T. et al. , 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat. Cell Biol. 15: 491–501. 10.1038/ncb2720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A. D., Shen Y., Fuxman Bass J., Jeyaraj S., Deplancke B. et al. , 2013. Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. 23: 954–965. 10.1101/gr.150466.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V., Motola D. L., Gerisch B., Cummins C. L., Nishiwaki K. et al. , 2006. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev. Cell 10: 473–482. 10.1016/j.devcel.2006.02.008 [DOI] [PubMed] [Google Scholar]
- Roux A. E., Langhans K., Huynh W., and Kenyon C., 2016. Reversible age-related phenotypes induced during larval quiescence in C. elegans. Cell Metab. 23: 1113–1126. 10.1016/j.cmet.2016.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Michaelson D., Hochman T., Santella A., Bao Z. et al. , 2016. Cell cycle features of C. elegans germline stem/progenitor cells vary temporally and spatially. Dev. Biol. 409: 261–271. 10.1016/j.ydbio.2015.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Kahler D. J., Yun C., and Hubbard E. J. A., 2018. Functional interactions between rsks-1/S6K, glp-1/notch, and regulators of Caenorhabditis elegans fertility and germline stem cell maintenance. G3 (Bethesda) 8: 3293–3309. 10.1534/g3.118.200511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J., Vidal-Gadea A. G., Makay A., Lanam C., and Pierce-Shimomura J. T., 2014. Humidity sensation requires both mechanosensory and thermosensory pathways in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 111: 8269–8274. 10.1073/pnas.1322512111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki S., Yamamoto M., and Iino Y., 2001. Plasticity of chemotaxis revealed by paired presentation of a chemoattractant and starvation in the nematode Caenorhabditis elegans. J. Exp. Biol. 204: 1757–1764. [DOI] [PubMed] [Google Scholar]
- Salinas L. S., Maldonado E., and Navarro R. E., 2006. Stress-induced germ cell apoptosis by a p53 independent pathway in Caenorhabditis elegans. Cell Death Differ. 13: 2129–2139. 10.1038/sj.cdd.4401976 [DOI] [PubMed] [Google Scholar]
- Saudenova M., and Wicky C., 2018. The chromatin remodeler LET-418/mi2 is required cell non-autonomously for the post-embryonic development of Caenorhabditis elegans. J. Dev. Biol. 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C., 2005. TGF-beta signaling (September 9, 2005), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.22.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin E. R., Ranganathan R., and Horvitz H. R., 2000. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron 26: 619–631. 10.1016/S0896-6273(00)81199-X [DOI] [PubMed] [Google Scholar]
- Schackwitz W. S., Inoue T., and Thomas J. H., 1996. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron 17: 719–728. 10.1016/S0896-6273(00)80203-2 [DOI] [PubMed] [Google Scholar]
- Schaedel O. N., Gerisch B., Antebi A., and Sternberg P. W., 2012. Hormonal signal amplification mediates environmental conditions during development and controls an irreversible commitment to adulthood. PLoS Biol. 10: e1001306 10.1371/journal.pbio.1001306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, W. R., 2005 Egg-laying (December 14, 2005), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.38.1, http://www.wormbook.org.
- Schindler A. J., Baugh L. R., and Sherwood D. R., 2014. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 10: e1004426 10.1371/journal.pgen.1004426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder N. E., Androwski R. J., Rashid A., Lee H., Lee J. et al. , 2013. Dauer-specific dendrite arborization in C. elegans is regulated by KPC-1/Furin. Curr. Biol. 23: 1527–1535. 10.1016/j.cub.2013.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenburg H., and Félix M. A., 2017. The natural biotic environment of Caenorhabditis elegans. Genetics 206: 55–86. 10.1534/genetics.116.195511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seah N. E., de Magalhaes Filho C. D., Petrashen A. P., Henderson H. R., Laguer J. et al. , 2016. Autophagy-mediated longevity is modulated by lipoprotein biogenesis. Autophagy 12: 261–272. 10.1080/15548627.2015.1127464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., and Kimble J., 2011. The oogenic germline starvation response in C. elegans. PLoS One 6: e28074 10.1371/journal.pone.0028074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel H. S., and Kimble J., 2015. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. eLife 4: e10832 10.7554/eLife.10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F. et al. , 2013. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15: 647–658. 10.1038/ncb2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock W. J., 1983. Yolk proteins of Caenorhabditis elegans. Dev. Biol. 96: 182–188. 10.1016/0012-1606(83)90321-4 [DOI] [PubMed] [Google Scholar]
- Shou Q., Feng L., Long Y., Han J., Nunnery J. K. et al. , 2016. A hybrid polyketide-nonribosomal peptide in nematodes that promotes larval survival. Nat. Chem. Biol. 12: 770–772. 10.1038/nchembio.2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Garcia C. G., and Estela Navarro R., 2013. The C. elegans TIA-1/TIAR homolog TIAR-1 is required to induce germ cell apoptosis. Genesis 51: 690–707. 10.1002/dvg.22418 [DOI] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A. L., Goldstein L. D. et al. , 2014. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7: 762–773. 10.1016/j.celrep.2014.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J. R., Ow M. C., Nishiguchi M. A., Kim K., Sengupta P. et al. , 2016. Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. eLife 5: e11642 10.7554/eLife.11642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora S., Mende F., and Zimmer M., 2018. Energy scarcity promotes a brain-wide sleep state modulated by insulin signaling in C. elegans. Cell Rep. 22: 953–966. 10.1016/j.celrep.2017.12.091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder A. E., and Maina C. V., 2001. Nuclear receptors in nematodes: themes and variations. Trends Genet. 17: 206–213. 10.1016/S0168-9525(01)02242-9 [DOI] [PubMed] [Google Scholar]
- Smelick C., and Ahmed S., 2005. Achieving immortality in the C. elegans germline. Ageing Res. Rev. 4: 67–82. 10.1016/j.arr.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Stadler M., and Fire A., 2013. Conserved translatome remodeling in nematode species executing a shared developmental transition. PLoS Genet. 9: e1003739 10.1371/journal.pgen.1003739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P. W., 2005. Vulval development (June 25, 2005), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.6.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community WormBook, doi/10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe D., Stephens L. R., Copeland T., Gaffney P. R., Reese C. B. et al. , 1997. Dual role of phosphatidylinositol-3,4,5-trisphosphate in the activation of protein kinase B. Science 277: 567–570. 10.1126/science.277.5325.567 [DOI] [PubMed] [Google Scholar]
- Sulston J. E., and Horvitz H. R., 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Suo S., Kimura Y., and Van Tol H. H., 2006. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J. Neurosci. 26: 10082–10090. 10.1523/JNEUROSCI.0819-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson M. M., and Riddle D. L., 1981. Critical periods in the development of the Caenorhabditis elegans dauer larva. Dev. Biol. 84: 27–40. 10.1016/0012-1606(81)90367-5 [DOI] [PubMed] [Google Scholar]
- Sym M., Basson M., and Johnson C., 2000. A model for Niemann-pick type C disease in the nematode Caenorhabditis elegans. Curr. Biol. 10: 527–530. 10.1016/S0960-9822(00)00468-1 [DOI] [PubMed] [Google Scholar]
- Tan K. T., Luo S. C., Ho W. Z., and Lee Y. H., 2011. Insulin/IGF-1 receptor signaling enhances biosynthetic activity and fat mobilization in the initial phase of starvation in adult male C. elegans. Cell Metab. 14: 390–402. 10.1016/j.cmet.2011.06.019 [DOI] [PubMed] [Google Scholar]
- Tao J., Ma Y. C., Yang Z. S., Zou C. G., and Zhang K. Q., 2016. Octopamine connects nutrient cues to lipid metabolism upon nutrient deprivation. Sci. Adv. 2: e1501372 10.1126/sciadv.1501372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Wu Q. Y., Ma Y. C., Chen Y. L., and Zou C. G., 2017. Antioxidant response is a protective mechanism against nutrient deprivation in C. elegans. Sci. Rep. 7: 43547 10.1038/srep43547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubert S., Van Gilst M. R., Hansen M., and Yamamoto K. R., 2006. A mediator subunit, MDT-15, integrates regulation of fatty acid metabolism by NHR-49-dependent and -independent pathways in C. elegans. Genes Dev. 20: 1137–1149. 10.1101/gad.1395406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen C. C., and Greenwald I., 2019. Cell non-autonomous function of daf-18/PTEN in the somatic gonad coordinates somatic gonad and germline development in C. elegans dauer larvae. Curr. Biol. 29: 1064–1072.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Opperman K. J., and Rougvie A. E., 2010. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development 137: 3501–3511. 10.1242/dev.048850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper R. G., Ashraf J., Kaletsky R., Kleemann G., Murphy C. T. et al. , 2013. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154: 676–690. 10.1016/j.cell.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M., Adachi T., Suzuki H., Kunitomo H., Schafer W. R. et al. , 2006. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron 51: 613–625. 10.1016/j.neuron.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Troemel E. R., Sagasti A., and Bargmann C. I., 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398. 10.1016/S0092-8674(00)81525-1 [DOI] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., Jolly A., and Yamamoto K. R., 2005a Nuclear hormone receptor NHR-49 controls fat consumption and fatty acid composition in C. elegans. PLoS Biol. 3: e53 10.1371/journal.pbio.0030053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gilst M. R., Hadjivassiliou H., and Yamamoto K. R., 2005b A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. USA 102: 13496–13501. 10.1073/pnas.0506234102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rompay L., Borghgraef C., Beets I., Caers J., and Temmerman L., 2015. New genetic regulators question relevance of abundant yolk protein production in C. elegans. Sci. Rep. 5: 16381 10.1038/srep16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T., McCulloch D., Gems D., and Kovacs A. L., 2006. Effects of sex and insulin/insulin-like growth factor-1 signaling on performance in an associative learning paradigm in Caenorhabditis elegans. Genetics 174: 309–316. 10.1534/genetics.106.061499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., and Kim S. K., 2003. Global analysis of dauer gene expression in Caenorhabditis elegans. Development 130: 1621–1634. 10.1242/dev.00363 [DOI] [PubMed] [Google Scholar]
- Watts J. L., and Ristow M., 2017. Lipid and carbohydrate metabolism in Caenorhabditis elegans. Genetics 207: 413–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster C. M., Pino E. C., Carr C. E., Wu L., Zhou B. et al. , 2017. Genome-wide RNAi screen for fat regulatory genes in C. elegans identifies a proteostasis-AMPK Axis critical for starvation survival. Cell Rep. 20: 627–640. 10.1016/j.celrep.2017.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster A. K., Jordan J. M., Hibshman J. D., Chitrakar R., and Baugh L. R., 2018. Transgenerational effects of extended dauer diapause on starvation survival and gene expression plasticity in Caenorhabditis elegans. Genetics 210: 263–274. 10.1534/genetics.118.301250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witham E., Comunian C., Ratanpal H., Skora S., Zimmer M. et al. , 2016. C. elegans body cavity neurons are homeostatic sensors that integrate fluctuations in oxygen availability and internal nutrient reserves. Cell Rep. 14: 1641–1654. 10.1016/j.celrep.2016.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf T., Qi W., Schindler V., Runkel E. D., and Baumeister R., 2014. Doxycyclin ameliorates a starvation-induced germline tumor in C. elegans daf-18/PTEN mutant background. Exp. Gerontol. 56: 114–122. 10.1016/j.exger.2014.04.002 [DOI] [PubMed] [Google Scholar]
- Wolkow C. A., Kimura K. D., Lee M. S., and Ruvkun G., 2000. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290: 147–150. 10.1126/science.290.5489.147 [DOI] [PubMed] [Google Scholar]
- Wollam J., Magner D. B., Magomedova L., Rass E., Shen Y. et al. , 2012. A novel 3-hydroxysteroid dehydrogenase that regulates reproductive development and longevity. PLoS Biol. 10: e1001305 10.1371/journal.pbio.1001305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Jiang X., Li Y., Zhu T., Zhang J. et al. , 2018a PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 9: 1195 10.1038/s41467-018-03531-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Masurat F., Preis J. and Bringmann H., 2018b Sleep counteracts aging phenotypes to survive starvation-induced developmental arrest in C. elegans. Curr. Biol. 28: 3610–3624.e8. 10.1016/j.cub.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Marques G., and O’Connor M. B., 2015. Vesicle-mediated steroid hormone secretion in Drosophila melanogaster. Cell 163: 907–919. 10.1016/j.cell.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lee B. Y., Yim H., and Lee J., 2020. Neurogenetics of nictation, a dispersal strategy in nematodes. J. Neurogenet. 10.1080/01677063.2020.1788552 [DOI] [PubMed] [Google Scholar]
- You Y. J., Kim J., Cobb M., and Avery L., 2006. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 3: 237–245. 10.1016/j.cmet.2006.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadra G., Batista J. L., and Loda M., 2015. Dissecting the dual role of AMPK in cancer: from experimental to human studies. Mol. Cancer Res. 13: 1059–1072. 10.1158/1541-7786.MCR-15-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zabinsky R., Teng Y., Cui M., and Han M., 2011. microRNAs play critical roles in the survival and recovery of Caenorhabditis elegans from starvation-induced L1 diapause. Proc. Natl. Acad. Sci. USA 108: 17997–18002. 10.1073/pnas.1105982108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Judy M., Lee S. J., and Kenyon C., 2013. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 17: 85–100. 10.1016/j.cmet.2012.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Chiu H., Boudreau J., Papanicolaou T., Bendena W. et al. , 2018a A functional study of all 40 Caenorhabditis elegans insulin-like peptides. J. Biol. Chem. 293: 16912–16922. 10.1074/jbc.RA118.004542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Qu Z., Zanetti M., Lam B., and Chin-Sang I., 2018b C. elegans PTEN and AMPK block neuroblast divisions by inhibiting a BMP-insulin-PP2A-MAPK pathway. Development 145: dev166876. [DOI] [PubMed] [Google Scholar]
- Zhong M., Niu W., Lu Z. J., Sarov M., Murray J. I. et al. , 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet. 6: e1000848 10.1371/journal.pgen.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Shen H., Sewell A. K., Kniazeva M., and Han M., 2013. A novel sphingolipid-TORC1 pathway critically promotes postembryonic development in Caenorhabditis elegans. eLife 2: e00429 10.7554/eLife.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]