Orc6 is a component of the Origin Recognition Complex important for the initiation of DNA replication. In order to study the functions of Orc6 in vivo, Balasov, Akhmetova, and Chesnokov.....
Keywords: DNA replication, Drosophila, Meier–Gorlin syndrome, ORC
Abstract
Meier–Gorlin syndrome (MGS) is a rare, autosomal recessive disorder characterized by microtia, primordial dwarfism, small ears, and skeletal abnormalities. Patients with MGS often carry mutations in genes encoding the subunits of the Origin Recognition Complex (ORC), components of the prereplicative complex and replication machinery. Orc6 is an important component of ORC and has functions in both DNA replication and cytokinesis. A mutation in the conserved C-terminal motif of Orc6 associated with MGS impedes the interaction of Orc6 with core ORC. Recently, a new mutation in Orc6 was also identified; however, it is localized in the N-terminal domain of the protein. To study the functions of Orc6, we used the human gene to rescue the orc6 deletion in Drosophila. Using this “humanized” Orc6-based Drosophila model of MGS, we discovered that unlike the previous Y225S MGS mutation in Orc6, the K23E substitution in the N-terminal TFIIB-like domain of Orc6 disrupts the protein ability to bind DNA. Our studies revealed the importance of evolutionarily conserved and variable domains of Orc6 protein, and allowed the studies of human protein functions and the analysis of the critical amino acids in live animal heterologous system, as well as provided novel insights into the mechanisms underlying MGS pathology.
DNA replication is fundamentally important for tissue development, growth, and homeostasis. Impairments of the DNA replication machinery can have catastrophic consequences for genome stability and cell division. Meier–Gorlin Syndrome (MGS) is an autosomal recessive disorder that is also known as ear, patella, short stature syndrome and/or microtia, absent patella, micrognathia syndrome, traits highlighting the core clinical phenotypes (Gorlin et al. 1975; Bicknell et al. 2011a; de Munnik et al. 2012a,b). The genes affected by MGS mutations include many members of prereplicative complex (pre-RC), such as Origin Recognition Complex (ORC) subunits (Orc1, Orc4, Orc6), Cdc6, Cdt1, CDC45, and MCM5, as well as Geminin (Bicknell et al. 2011a,b; Guernsey et al. 2011; Burrage et al. 2015; Fenwick et al. 2016; Vetro et al. 2017; McDaniel et al. 2020), suggesting that the clinical phenotype is caused by defects in DNA replication initiation. As pre-RC complex is essential for DNA replication, the mutations in its components are expected to impair cell proliferation and to reduce growth.
ORC plays a central role in the initiation of DNA replication but is also involved in nonreplicative functions (Bell and Stillman 1992; Bell 2002; Chesnokov 2007; Sasaki and Gilbert 2007). The smallest subunit of ORC, Orc6 is the most divergent and enigmatic among ORC subunits. Orc6 is important for DNA replication in all species (Lee and Bell 1997; Chesnokov et al. 2001; Chesnokov et al. 2003; Semple et al. 2006; Balasov et al. 2007; Chen et al. 2007; Balasov et al. 2009). It is also essential for cytokinesis in Drosophila and human cells (Prasanth et al. 2002; Chesnokov et al. 2003; Bernal and Venkitaraman 2011). In Drosophila, Orc6 stimulates septin complex GTPase activity and polymerization during filament assembly through protein-protein interactions (Huijbregts et al. 2009; Akhmetova et al. 2015). Metazoan Orc6 proteins consist of two functional domains: a larger N-terminal domain important for binding of DNA and smaller C-terminal domain important for protein-protein interactions (Chesnokov et al. 2003; Balasov et al. 2007; Duncker et al. 2009; Bleichert et al. 2013). It has been shown that in metazoan species, N-terminal domain of Orc6 shares structural homology with the transcription factor TFIIB (Chesnokov et al. 2003; Balasov et al. 2007; Liu et al. 2011). The conserved motif in the C terminus of Orc6 is responsible for interacting with the Orc3 subunit of ORC (Bleichert et al. 2013). A mutation coding for a tyrosine 232 to serine alteration in this region of the protein is linked to the MGS in humans (Bicknell et al. 2011a; de Munnik et al. 2012a). In our earlier studies, we introduced MGS mutation in Drosophila Orc6 (Y225S) and established a fruit fly model of MGS. These mutant flies displayed severe replication and developmental defects due to impaired interaction of mutant Orc6 protein with the rest of the ORC (Balasov et al. 2015).
Recently, a new Orc6 mutation was described (Li et al. 2017) that also resulted in MGS. Unlike previously described MGS mutation (Bicknell et al. 2011a), this amino acid substitution (K23E) localizes in the N-terminal domain of Orc6, which is important for DNA binding. In our study, to investigate the functions of different Orc6 domains in a live animal model, we introduced human, Drosophila, and hybrid human-Drosophila orc6 genes into flies with an orc6 null mutant background. Using these “humanized” fly strains carrying the K23E MGS mutation, we identified a molecular mechanism resulting in a MGS phenotype and compared it with our Y225S MGS fly model. We found that despite having different underlying molecular mechanisms, both MGS mutations resulted in similar phenotypes, deficient pre-RC formation, and reduced DNA replication.
Materials and Methods
Mutagenesis
Conserved lysine at position 23 was replaced with glutamic acid to create the Meier–Gorlin mutation (K23E), following site-directed mutagenesis protocol (Agilent). Drosophila-human and human-Drosophila hybrids were designed by using the PCR technique. Orc6 mutant and hybrid genes were cloned in frame, with GFP into the modified pUAST vector containing UAS promoter and orc6 Drosophila native promoter. All constructs were injected into Drosophila w1118 embryos (Model System Injections, Raleigh, NC) and individual transgenic strains were maintained.
Purification of recombinant Orc6
Wild-type, mutant, or hybrid Orc6 complementary DNAs (cDNAs) were cloned into the pET15b expression vector, and used to transform Escherichia coli strain BL21. Ni-NTA-purified Orc6 proteins were further purified with HiTrap SP HP (GE Life Sciences) and Superdex 75 (GE Life Sciences) columns.
Electrophoretic mobility shift assay
We incubated 1 ng of 32P end-labeled human B2 lamin or Drosophila ori-β fragments with 300 ng of purified protein in reaction buffer (25 mM Tris, pH 8, 60 mM KCl, 5 mM MgCl2, 0.1% NP-40, 0.12 mg/ml BSA, 10% glycerol) at room temperature for 30 min. Then, 10 µl of each reaction was loaded onto a 4% native polyacrylamide gel. Electrophoresis was performed at room temperature using TAE at pH 8.3 for Drosophila Orc6 and pH 9.5 for human Orc6 as a running buffer. The gel was dried on Whatman paper and exposed to X-ray film.
Fly stocks and rescue experiments
We set up the following fly stocks containing endogenous orc635 deletion and GFP-orc6 fused transposon: (1) orc635/Cy; GFP-Orc6-DmWT: wild type Drosophila Orc6; (2) orc635/Cy; GFP-Orc6-DmK23E: Drosophila Orc6 with K23E MGS mutation; (3) orc635/Cy; GFP-Orc6-HsWT: wild type human Orc6; (4) orc635/Cy; GFP-Orc6-HsK23E: human Orc6 with K23E MGS mutation; (5) orc635/Cy; GFP-Orc6-HD: interspecies hybrid; (6) orc635/Cy; GFP-Orc6-DH: interspecies hybrid; and (7) orc635/Cy; GFP-Orc6-HDK23E: interspecies hybrid with K23E MGS mutation.
Bloomington Drosophila Stock Center stock #5138 (y,w; P{w+mC = tubP-GAL4}LL7/TM3,Sb,Ser) expressing GAL4 ubiquitously under the control of the alphaTub84B promoter was used to design orc635/Cy; tub-GAL4/TM3,Sb fly stocks for rescue experiments.
Fly stock orc635/Cy; GFP-Orc6-DmY225S with Y225S MGS mutation (Balasov et al. 2015) was used as a negative control for immunoprecipitation experiments.
In rescue experiments with a native Drosophila promoter, progeny from heterozygous orc635/Cy; GFP-Orc6 were analyzed for the presence of orc635/orc635; GFP-Orc6 adult flies. In rescue experiments with alphaTub84 (tub) promoter, females of the genotype orc635/Cy; GFP-Orc6 were crossed to males orc635/Cy; tub-GAL4/TM3,Sb, and resulting progeny were analyzed for the presence of orc635/orc635;GFP-Orc6/tub-GAL4 adults.
For Orc6 depletion in motor neurons, we used the Orc6 RNA interference fly stock #HMJ22188 (National Institute of Genetics) (NIG-FLY Stock Center, Japan) and the motor-neuron-specific D42-GAL4 driver #8816 (Bloomington Drosophila Stock Center).
BrdU labeling and immunostaining of third instar larval brains
Larval brains were soaked in PBS with 1 µM BrdU for 30 min at 25°. BrdU incorporation was detected by monoclonal antibodies (Becton Dickinson) following the manufacturer’s protocol, as described previously (Sullivan et al. 2000). Images was made with Olympus Fluoview FV300 confocal microscope. BrdU incorporation was calculated as a percentage of BrdU signal from total brain area using cellSens Dimension Desktop software (Olympus).
Immunostaining of polytene chromosomes
The flies bearing GFP-Orc6 transgene were crossed to Bloomington Drosophila Stock Center fly stock 6870 (w1118; P{Sgs3-GAL4.PD}TP1) expressing GAL4 under Sgs3 promoter. This promoter drives GAL4 in the salivary glands of third instar larvae. Salivary glands expressing different variants of GFP-Orc6 were dissected in PBS with 0.1% NP-40, transferred in fixing solution (2% formaldehyde, 45% acetic acid) for 1 min, squashed in 45% acetic acid, and frozen in liquid nitrogen. The slides were desiccated in 96% alcohol and stored in 70% alcohol at −20°. For immunofluorescence studies, slides were briefly washed in PBS with 0.1% NP-40 and incubated with primary antibodies [mouse anti-GFP (B2), #sc9996; Santa Cruz Biotechnology] diluted in 10% goat antiserum for 2 hr in humid chamber. Secondary goat anti-mouse antibodies (Alexa Fluor 488; Thermo Fisher Scientific) were used to visualize Orc6 on the polytene chromosomes.
Immunoprecipitation from ovaries
Twenty freshly dissected ovaries were crushed in a glass homogenizer with 100 µl of high-salt immunoprecipitation buffer (25 mM HEPES pH 7.6, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 300 mM NaCl, 0.01% Triton X-100) and incubated for 1 hr at 4° with continuous rotation. Extract was centrifuged at 15000 × g for 15 min and supernatant was diluted three times with low-salt immunoprecipitation buffer (25 mM HEPES pH 7.6, 12.5 mM MgCl2, 100 mM KCl, 0.1 mM EDTA, 0.01% Triton X-100). Protein A-Sepharose (#6501-5; BioVision) and rabbit polyclonal antibodies against Drosophila Orc2 subunit were added and incubated with the supernatant overnight. Sepharose beads were washed three times with low-salt immunoprecipitation buffer and diluted in 10 µl of immunoprecipitation buffer. Samples were boiled in loading buffer, separated in 10% SDS-polyacrylamide gel, and transferred on Immobilon-P membrane (#IPVH00010; Millipore, Bedford, MA). The presence of GFP-Orc6 fused protein was detected with a mouse anti-GFP (B2, #sc9996; Santa Cruz Biotechnology) antibody. A mouse anti-Orc5 antibody was used to verify ORC complex immunoprecipitation.
Mitotic chromosome preparation
Preparation of mitotic nuclei was described previously (Lebedeva et al. 2000). Briefly, third instar larval neural ganglia were incubated in 0.075M KCl for 5 min, fixed in methanol with acetic acid (3:1) for 20 min, and then dispersed in a drop of 50% propionic acid on a slide. Then slides were dried and stained with 5% Giemsa’s solution.
Western blot of larval brains
Freshly dissected brains were incubated with 450 mM NaCl, 0.5% NP-40 in PBS buffer for 1 hr, and samples were centrifuged 10 min at 10000 × g. Pellets were washed three times with 100 mM NaCl, 0.5% NP-40 in PBS buffer and used for Western blotting. MCM complex was detected with rabbit polyclonal anti-MCM4 and anti-MCM5 antibodies.
Fluorescence spectroscopy
Purified Orc6 protein (wild type or mutant) was diluted in buffer (50 mM NaH2PO4, pH 7.4, 100 mM NaCl) to 10 ng/µl. Then, 260 µl samples were loaded in quartz cuvette. Fluorescence measurements were performed in the VARIAN CARY Eclipse fluorescence spectrophotometer. The spectral widths of the excitation and the emission bands were 5 and 20 nm, respectively. Excitation was performed at 290 nm wavelength. Emission spectra were recorded from 300 to 400 nm in 1-nm steps. An emission spectrum of a buffer was subtracted from protein spectra.
Microarray analysis
RNA was isolated from 5-day-old females using ZR Tissue and Insect RNA MicroPrep (#R2030; Zymo Research). DNA was removed using TURBO DNase (#AM2238; Invitrogen, Carlsbad, CA) following manufacturer’s recommendations. cDNA was generated from 1 μg of total RNA using ProtoScript II First Strand cDNA Synthesis Kit (#E6560; New England Biolabs).
The microarray experiment was conducted at the Boston University Microarray and Sequencing Resource Core Facility. Drosophila Gene 1.0 ST CEL files were normalized to produce gene-level expression values using the implementation of the robust multiarray average (Irizarry et al. 2003) in the affy package (version 1.48.0) (Gautier et al. 2004) included in the Bioconductor software suite (version 3.2) (Gentleman et al. 2004), and an Entrez Gene-specific probeset mapping (20.0.0) from the Molecular and Behavioral Neuroscience Institute (Brainarray) at the University of Michigan (Dai et al. 2005) (brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF). Array quality was assessed by computing relative log expression and normalized unscaled standard error, using the affyPLM package (version 1.46.0). Principal component analysis was performed using the prcomp R function with expression values that had been normalized across all samples to a mean of zero and a standard deviation of one. Differential expression was assessed using the moderated (empirical Bayesian) t-test implemented in the limma package (version 3.26.9) (i.e., creating simple linear models with lmFit, followed by empirical Bayesian adjustment with eBayes). Correction for multiple hypothesis testing was accomplished using the Benjamini–Hochberg false discovery rate. Human homologs of fly genes were identified using HomoloGene (version 68). All microarray analyses were performed using the R environment for statistical computing (version 3.2.0). Gene ontology terms analysis was conducted using DAVID Bioinformatics Resources 6.8 (david.ncifcrf.gov) (Huang et al. 2009).
Data availability statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.13070288.
Results
Establishing Drosophila strains to study the functions of human Orc6 in vivo
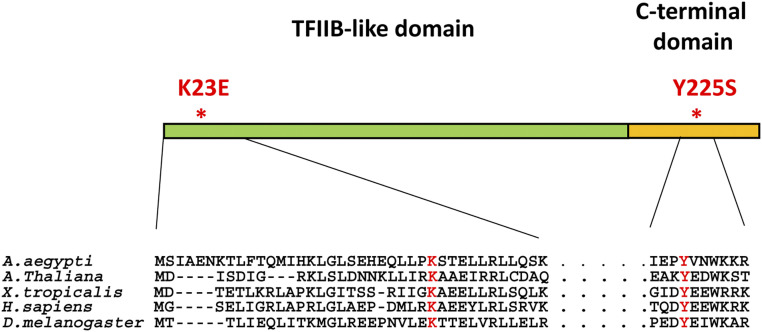
The metazoan Orc6 consists of two domains (Figure 1): a larger N-terminal domain that carries a homology with TFIIB transcription factor and is important for DNA binding (Chesnokov et al. 2003; Balasov et al. 2007; Liu et al. 2011), and a shorter C-terminal domain essential for the interaction of the protein with the core ORC through its Orc3 subunit (Bleichert et al. 2013). The C-terminal domain of metazoan Orc6 is also important for the function of Orc6 in cytokinesis (Prasanth et al. 2002; Chesnokov et al. 2003; Huijbregts et al. 2009; Bernal and Venkitaraman 2011; Akhmetova et al. 2015).
Figure 1.
Domain organization of metazoan Orc6 protein. Orc6 protein consists of the N-terminal TFIIB-like domain important for DNA binding, and C-terminal domain required for the interaction with core ORC. Drosophila and human N-terminal domains share 27% amino acid identity and 48% similarity. C-terminal domains have only 15% amino acid identity and 22% similarity. The resulting amino acid changes from the new MGS mutation, the conservative lysine at position 23 (K23E), as well as the previously described C-terminal MGS mutation (Y225S) are indicated in red.
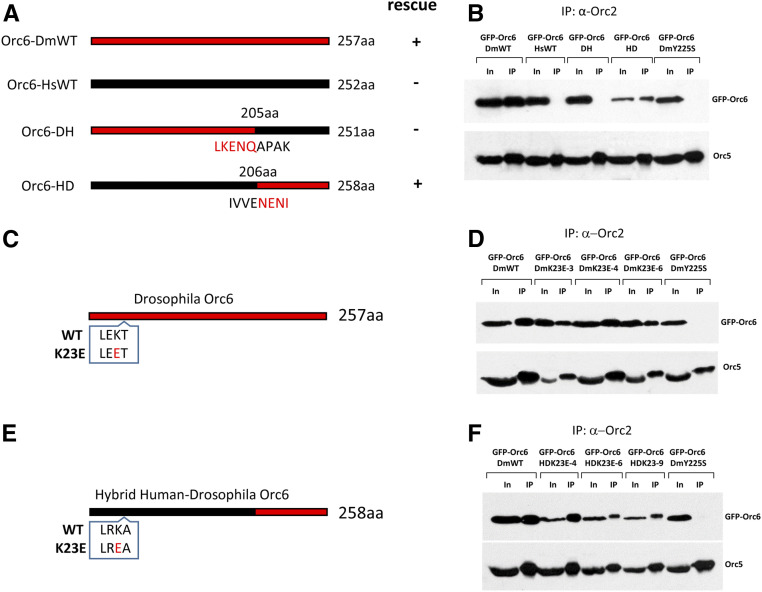
In our previous work we generated a deletion of the orc6 gene in Drosophila allowing the studies of the protein in vivo. We showed that the orc6 deletion resulted in defects in DNA replication as well as abnormal chromosome condensation and segregation (Balasov et al. 2009). Orc6 is the least conserved among ORC subunits, with metazoan Orc6 proteins showing significantly higher homology as compared to budding yeast protein. The full-length human orc6 gene (Orc6-HsWT) under control of native orc6 promoter did not rescue lethality of the Orc6 deficient flies (Balasov et al. 2009). However, human Orc6 protein was able to bind chromosomes when expressed in Drosophila (Liu et al. 2011). We asked if the expression of a hybrid protein designed from the human DNA binding domain of Orc6 and the Drosophila C terminus responsible for the interaction with a core ORC might rescue the lethality associated with the Orc6 deletion. Two constructs were created (Figure 2A). The first consisted of the Drosophila N-terminal TFIIB-like domain and human C terminus (Orc6-DH); the second construct contained human N-terminal domain and Drosophila C terminus (Orc6-HD). Both constructs were tested for their ability to rescue orc6 deletion. The expression of Orc6-DH did not rescue the lethality; however, the expression of Orc6-HD resulted in a full rescue and obtained flies were undistinguishable from the flies rescued with wild-type Drosophila Orc6 (Orc6-DmWT) (Figure 2A).
Figure 2.
Design of hybrid Orc6 and Orc6 mutants. (A) Diagrams of transgenic constructs. Black represents human Orc6 and red represents Drosophila Orc6. Orc6-DmWT and Orc6-HsWT indicate wild-type Drosophila and human proteins, respectively. Orc6-DH is the hybrid protein containing first 205 amino acids of Drosophila and last 46 amino acids of human Orc6. Orc6-HD is the hybrid protein containing first 206 amino acids of human and last 52 amino acids of Drosophila Orc6. (B) Immunoprecipitation of the ORC complex with anti-Orc2 antibodies from ovaries expressing GFP-tagged Orc6 shown in A. (C) The position of the K23E substitution in Drosophila Orc6 (red). (D) Immunoprecipitation of ORC complex using Orc2 antibodies from Drosophila Orc6-K23E mutant fly ovaries. Immunoprecipitations from three independent transgenic fly stocks are shown: DmK23E-3, DmK23E-4, and DmK23E-6. (E) The position of the K23E in hybrid human-Drosophila Orc6. The substitution is colored in red. (F) Immunoprecipitation of ORC complex using Orc2 antibodies from hybrid human-Drosophila K23E mutant fly ovaries. Immunoprecipitations from three independent transgenic fly stocks are shown: HDK23E-4, HDK23E-6, and HDK23E-9. Orc6-DmY225S was used as a negative control.
Orc6 is a subunit of the ORC complex. Therefore, we analyzed the ability of Orc6 proteins described above to associate with Drosophila ORC complex. We employed anti-Orc2 antibodies for immunoprecipitation of ORC complex, and antibodies against Orc5 subunit were used as a rigorous control for the presence of the whole ORC complex in immunoprecipitated material. We found that Orc6-HD but not Orc6-DH or wild-type human Orc6-HsWT immunoprecipitated with the rest of the ORC complex (Figure 2B).
Furthermore, the MCM helicase association with the chromatin was diminished in Orc6-DH as compared to the Orc6-HD strain (Supplemental Material, Figure S1A), indicating that the pre-RC formation is impaired in the presence of Orc6-DH form of the protein.
Since Orc6-HD compensated for the orc6 deletion similar to the wild-type Drosophila Orc6, this transgene can be used for studying mutations in the human large N-terminal TFIIB-like domain portion of Orc6 protein in the live animal model.
The analysis of flies carrying MGS mutation localized in the N-terminal domain of Orc6
In our previous analysis of Orc6 functions in vivo, we showed that the mutation of the conserved tyrosine to serine (Y225S) in the C-terminal domain of Drosophila Orc6 resulted in third instar lethality associated with defects in DNA replication and chromosome abnormalities (Balasov et al. 2015). In humans, the corresponding mutation (Y232S) is associated with MGS. The molecular analyses of the mutation in this Drosophila model of MGS revealed that it disrupted the interaction between Orc6 and the rest of the ORC through the Orc3 subunit, thereby impairing pre-RC formation and MCM recruitment, which is essential for DNA replication (Bleichert et al. 2013; Balasov et al. 2015).
Recently, the first diagnosed MGS case in China was described (Li et al. 2017). The patient had a short stature, microtia, small patella, and craniofacial abnormalities characteristic for MGS. It was reported that the patient carried a novel homozygous mutation in the orc6 gene, resulting in a change of A to G at position 67, which corresponds to the conversion of lysine at position 23 to glutamic acid (K23E). Interestingly, this mutation is localized within N-terminal TFIIB-like domain of the protein, unlike the earlier described Y232S MGS mutation (Bicknell et al. 2011a; de Munnik et al. 2012a), which effects the C-terminal domain of the protein (Figure 1).
We then set about determining the molecular consequences of the new MGS mutation. Since only wild-type Drosophila Orc6 and hybrid Orc6-HD resulted in viable adults, we incorporated K23E mutation into Drosophila Orc6 gene and human N-terminal domain of the Orc6-HD hybrid construct (Figure 2, C and E). The resulting constructs, Orc6–DmK23E and Orc6-HDK23E, were introduced into flies carrying the homozygous deletion of the orc6 gene. Three independent transgenic fly stocks were set up for each mutant construct (Figure S2).
To analyze the ability of mutant proteins to interact with the rest of the complex, we immunoprecipitated ORC from obtained transgenic flies using Orc2 antibodies. Drosophila Orc6 carrying Y225S mutation (Orc6-DmY225S) was used as a negative control for complex association since this mutation abolished the interaction of Orc6 protein with ORC complex (Balasov et al. 2015). As expected, Orc6-DmY225S protein did not immunoprecipitate with ORC (Figure 2, D and F). However, both Drosophila and hybrid Human-Drosophila Orc6 proteins carrying the K23E substitution were present in the immunoprecipitated material indicating that the interaction with core ORC was not affected by the mutation (Figure 2, D and F). It was previously shown that metazoan Orc6 associated with ORC via its Orc3 subunit (Bleichert et al. 2013). Therefore, we tested whether K23E mutation would disrupt a direct interaction between Orc6 and Orc3 subunits. As shown in Figure S3, mutant Orc6 protein carrying the K23E mutation was able to pull down Orc3 in the in vitro transcription/translation reaction as well as the wild-type Drosophila Orc6. We conclude that K23E mutation in Orc6 does not affect its association with the core ORC.
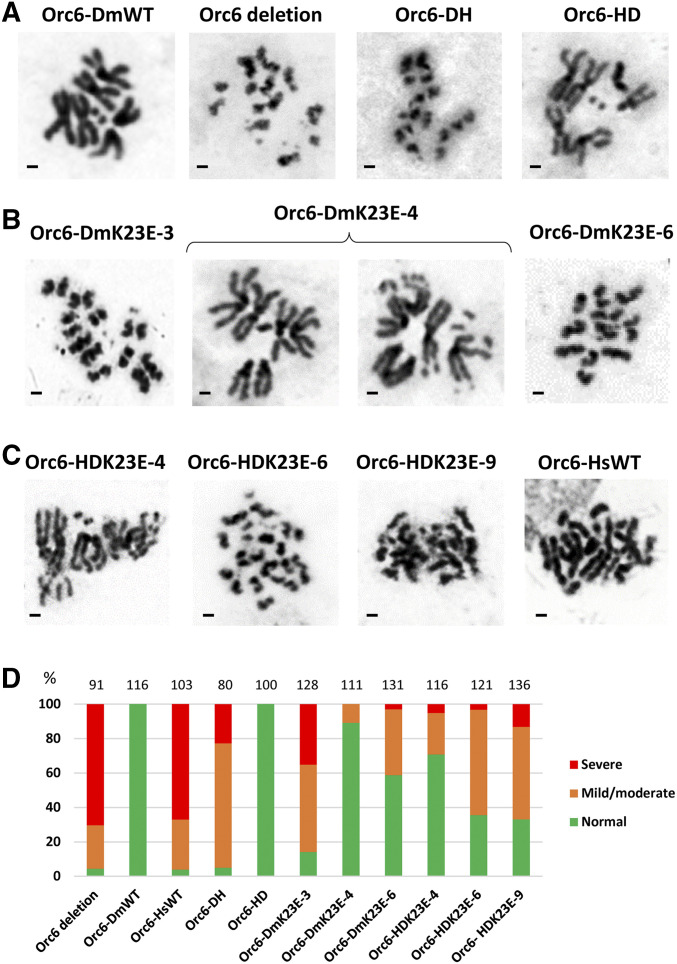
Replication defects often result in a loss of chromosome integrity. Therefore, we investigated mitotic chromosomes in developing larval brains, since this tissue is known to provide the best mitotic figures. The normal Drosophila melanogaster karyotype consists of four pairs of chromosomes (Figure 3A, Orc6-DmWT). In larvae carrying orc6 deletion (Figure 3A, Orc6 deletion) or expressing Orc6-DH (Figure 3A, Orc6-DH) or Orc6-HsWT (Figure 3C, Orc6-HsWT) constructs on orc6 deletion background, chromosomes lost parts of their arms, appeared aberrantly condensed, and fragmented. On the other hand, Orc6-HD transgene expression restored mitotic chromosome structure (Figure 3A, Orc6-HD). The incorporation of the K23E mutation into Orc6 resulted in chromosome defects and fragmentations (Figure 3, B and C). To quantify chromosome integrity, we counted percentages of severely affected mitoses (as in Orc6 deletion mutant), mildly/moderately affected (as in Orc6-HDK23E-4), and normal mitoses (as in Orc6-DmWT) (Figure 3D). As expected, two transgenic stocks, Orc6-DmWT and Orc6-HD, restored all karyotypes (100%) to the normal pattern. Orc6 deletion, Orc6-DH, and Orc6-HsWT contained >95% of defective mitoses. Orc6-DmK23E and Orc6-HDK23E mutants all contained some defective mitoses with different degree of severity (Figure 3D). The minor chromosome defects were also reported for the human patient carrying the K23E mutation (Li et al. 2017).
Figure 3.
Mitoses in larval neuroblasts. Brains of third instar larvae expressing specified transgenes on orc6 deletion background were analyzed for the karyotype. (A) Orc6 deletion mutant and hybrid Orc6 (Orc6-DH, Orc6-HD) compared to Drosophila wild type (Orc6-DmWT). (B) Three independent DmOrc6 transgenics bearing the K23E mutation (Orc6-DmK23E-3, Orc6-DmK23E-4, Orc6-DmK23E-6). (C) Three independent hybrid Orc6-HD transgenic lines bearing the mutation in human portion of Orc6 (Orc6-HDK23E-4, Orc6-HDK23E-6, Orc6-HDK23E-9) compared to human wild-type Orc6 (Orc6-HsWT). Bars represents 1 µm. (D) Percentages of defective (orange and red) and normal (green) karyotypes. Total number of mitoses analyzed is shown above the chart.
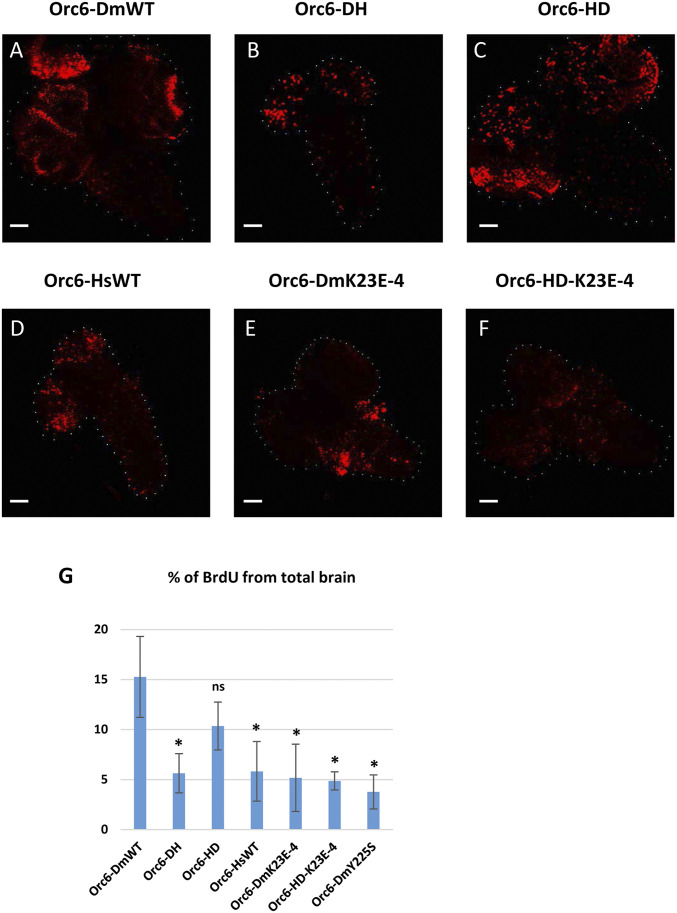
To investigate replication defects more rigorously, we analyzed BrdU incorporation in larval brains of the fly stocks expressing hybrid or mutant proteins. Brains were underdeveloped and BrdU incorporation was reduced in flies carrying the Orc6-HsWT or Orc6-DH constructs (Figure 4, B and4D). No difference was observed between the Orc6-DmWT and Orc6-HD hybrid constructs (Figure 4, A and C). The larval brains of the fly stocks expressing mutant proteins Orc6-DmK23E (Figure 4E) or Orc6-HDK23E (Figure 4F) were also underdeveloped and showed reduced BrdU labeling. Quantifications confirmed that Orc6-HD was not statistically different from Orc6-DmWT, whereas all other analyzed mutants were defective (Figure 4G).
Figure 4.
BrdU incorporation in larval brains. Brains of third instar larvae expressing specified transgenes on orc6 deletion background were labeled with BrdU. Transgenes are as follows: (A) wild-type Drosophila Orc6, (B) Orc6-DH hybrid, (C) Orc6-HD hybrid, (D) wild-type human Orc6, (E) Drosophila Orc6-K23E mutant, and (F) Orc6-HD hybrid with the K23E mutation. Bars represents 50 µm. (G) Percentages of BrdU-labeled area relative to total brain area (n = 5). * indicates a statistically significant difference compared to Orc6-DmWT; ns indicates no significant difference.
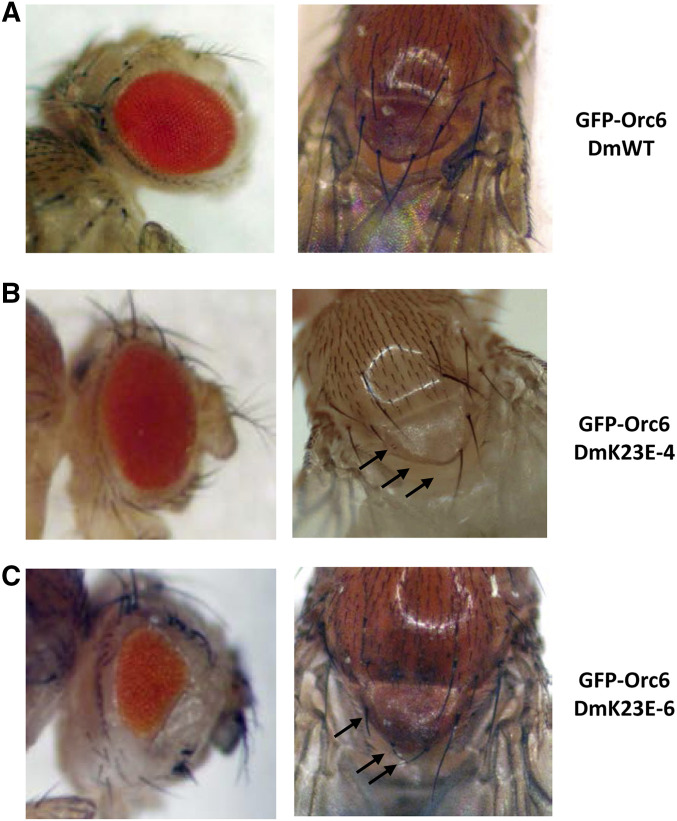
Next, we calculated the percentage of homozygous flies rescued with different transgenes driven by native Orc6 promoter (Table 1). We found that stock Orc6-DmK23E-4 resulted in 28% of adult viability. The majority of rescued flies (82%) were indistinguishable from wild type (Figure 5A). Minor defects (missing scutellar bristles) appeared in 18% of flies (Figure 5B). It should be noted that stock Orc6-DmK23E-4 had the highest expression level of Orc6-DmK23E protein compared to others, and promoted flies to the adult stage (Figure S2 and Table 1). Another stock Orc6-DmK23E-6 produced very few (3%) homozygous adults (Table 1). All of the surviving flies had defective scutellar bristles and reduced eye size (Figure 5C). The eye of Drosophila is a well-established model to study proliferation defects. The compound eye of Drosophila consists of uniform regular facets, and the final number of them is determined during several mitotic divisions in third instar larvae. Any delay or disturbance during the course of divisions result in reduced size and abnormal shape of the adult eye. This phenotype was consistent with a disrupted Orc6 function in replication and cell cycle progression. Another interesting observed phenotype was the inability of mutant flies to fly. Rescued adult flies carrying Orc6-DmK23E could not fly but were able to jump and walk. Previously we reported that flies with C-terminal Orc6 MGS mutation (Y225S) also missed scutellar bristles and were flightless (Balasov et al. 2015). The observed bristle defect and flightless phenotype could be a common MGS feature in Drosophila.
Table 1. Rescue of the orc6 deletion with the expression of different Orc6 transgenes under control of native or tubulin promoter.
| Orc6-Dm WT | Orc6-Dm K23E-3 | Orc6-Dm K23E-4 | Orc6-Dm K23E-6 | Orc6-DH | Orc6-HD | Orc6-HD K23E-4 | Orc6-HD K23E-6 | Orc6-HD K23E-9 | Orc6-Hs WT | Orc6-Hs K23E | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Native promoter | + | – | + | +/- | – | + | – | – | – | – | – |
| 80% | 0% | 28% | 3% | 0% | 65% | 0% | 0% | 0% | 0% | 0% | |
| (495) | (411) | (422) | (276) | (678) | (683) | (393) | (301) | (296) | (351) | (413) | |
| Tubulin promoter | + | + | + | + | + | + | + | + | + | + | – |
| 91% | 39% | 54% | 48% | 20% | 80% | 74% | 79% | 33% | 27% | 0% | |
| (596) | (622) | (427) | (496) | (790) | (495) | (591) | (569) | (670) | (774) | (315) |
Percentages of rescued flies were calculated based on expected segregation of phenotypes. Total progeny analyzed is shown in parentheses. (+), the expression of transgene restores viability of orc6‐deleted flies to adult stage; (−), no rescue, (±) very low rescue percentage.
Figure 5.
Phenotypes of orc6 deletion flies rescued with Orc6-DmK23E. (A) Flies expressing wild-type Drosophila Orc6. (B) Orc6DmK23E-4 mutant flies have mild phenotype: eye size and facets number are normal; however, 18% of flies have missing or defective scutellar bristles (arrows). (C) All surviving flies carrying Orc6DmK23E-6 showed reduced eye facets number and multiple defects of scutellar bristles (arrows).
In our previous study describing our fly model of MGS based on Orc6 Y225S mutation (Balasov et al. 2015), we showed that the elevated expression of Orc6 Y225S transgene rescued the orc6 deletion phenotype. We investigated whether it would be the case for K23E mutation. Similar to the Y225S Orc6 mutant, all K23E orc6 transgenes have an UAS promoter upstream of native orc6 promoter. The UAS promoter allows increased Orc6 transgene expression by crossing to flies bearing the tubulin promoter-driven GAL4 (see Materials and Methods). As summarized in Table 1, elevated expression of both Drosophila and human-Drosophila hybrid transgenes carrying the K23E mutation promoted flies to adult stage. Remarkably, these rescued flies also had defects of the scutellar bristles and were not able to fly, similar to the phenotypes observed in the fly model of MGS based on the Orc6 Y225S mutation (Balasov et al. 2015).
Overall, we concluded that both MGS mutations resulted in reduced DNA replication and chromosome defects, manifesting in similar phenotypes; however, the molecular mechanisms driving this phenotype might be different. The Y225S substitution impedes Orc6 binding to the ORC complex (Bleichert et al. 2013; Balasov et al. 2015). The K23E substitution resides in the N-terminal domain of Orc6 that is important for DNA binding. Therefore, we investigated the ability of Drosophila, human, and hybrid Orc6 proteins carrying the K23E mutation to bind DNA.
K23E mutations disrupt DNA binding ability of Orc6 protein
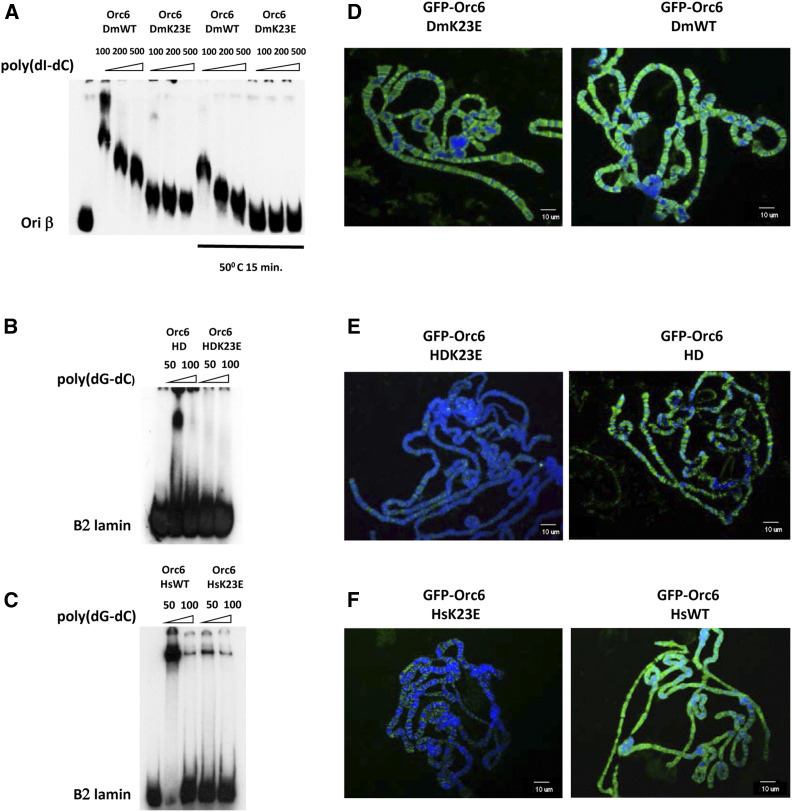
Six recombinant proteins were purified using an E. coli expression system: Drosophila (Orc6-DmWT and Orc6-DmK23E), hybrid human/Drosophila (Orc6-HD and Orc6-HDK23E), and human (Orc6-HsWT and Orc6-HsK23E) (Figure S4A). As shown in Figure 6A, both Drosophila Orc6 wild-type protein and Orc6 carrying the K23E mutation were able to bind in vitro with the ori-β DNA fragment derived from the origin of DNA replication associated with a chorion gene cluster. Binding was very tight, with the majority of the labeled probe bound by the protein, as we observed before (Balasov et al. 2007), suggesting the presence of multiple binding sites for Orc6 within the ori-β fragments. However, the mobility of DNA-protein complex changed significantly when Orc6-K23E was used in the reaction. The mutant complex migrated faster compared to the wild-type protein, probably due to a reduced template occupancy resulting from the K23E mutation. Increasing the concentration of the mutant protein in electrophoretic mobility shift assay (EMSA) reactions slightly improved the DNA binding of Orc6-K23E protein (Figure S4B).
Figure 6.
DNA and chromosome binding ability of hybrid and mutant Orc6. Electrophoretic mobility shift assay (EMSA) and chromosome binding of (A and D) Drosophila Orc6 wild type and K23E mutant; (B and E) hybrid Orc6 HD wild type and Orc6 HD-K23E mutant proteins; and (C and F) human Orc6 wild type and K23E mutant. “50°C 15 min” indicates that protein probe was heat-treated before adding to the reaction. The amount of competitor poly(dI-dC) and poly(dG-dC) is shown in nanograms. Polytene chromosomes isolated from salivary glands expressing GFP-tagged Orc6 were immunostained with anti-GFP antibodies, and DNA was stained with DAPI.
When the K23E substitution was made in the human Orc6 protein or in a hybrid protein Orc6-HD containing human N-terminal TFIIB-like domain, the overall DNA binding ability of the resulting mutant proteins was significantly diminished (Figure 6, B and C). We would like to point out that the DNA binding pattern for human Orc6 is different from Drosophila protein, with the majority of the DNA-protein complex migrating at the top of the EMSA gel. This difference between fly and human Orc6 binding to DNA was reported previously (Balasov et al. 2007; Liu et al. 2011).
To visualize the Orc6 mutants binding in vivo, we used GFP-Orc6 bearing fly stocks described in Materials and Methods. The UAS promoter in the construct allows for GAL4-induced expression using the GAL4/UAS binary system (Brand and Perrimon 1993; Duffy 2002). GFP-Orc6 expression was induced in salivary glands of third instar larvae to test chromosome binding of Orc6 mutants. Nuclei of Drosophila salivary glands contain polytene chromosomes that can be easily visualized with microscopy. Both Gal4-induced Orc6-DmWT and Orc6-DmK23E (Figure 6D) were found to be associated with polytene chromosomes. In contrast, over-expressed Orc6-HDK23E and Orc6-HsK23E mutants showed significantly reduced DNA binding and association with chromosomes (Figure 6, E and F), in agreement with the EMSA DNA binding experiments shown in Figure 6, B and C. Together, our data on DNA binding suggest that K23E mutation has a debilitating effect on DNA binding ability of Orc6.
K23E mutation results in the instability of the Drosophila, but not human, Orc6
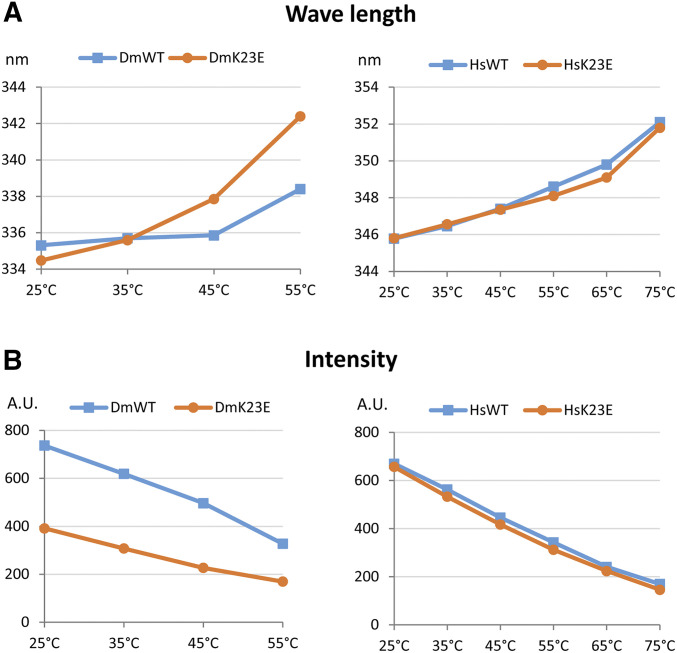
Often mutations lead to a protein unfolding that may affect its functions, such as DNA binding. Therefore, we tested the effect of the K23E substitution on protein stability. In this experiment the unfolding of the protein was studied by measuring the intrinsic fluorescence intensity at a wavelength of excitation corresponding to tryptophan. Tryptophan fluorescence is very sensitive to protein conformational changes and thus provides information about changes in secondary and/or tertiary structure. A decrease in fluorescence intensity and shift in the maximal emission wavelength reflect protein unfolding (Royer 2006). We measured the tryptophan fluorescence of Drosophila and human Orc6 proteins at the increasing temperatures and compared them with mutants carrying the K23E mutation. As presented in Figure 7A, the K23E mutation in human Orc6 did not change a stability of the mutant protein relative to wild type at all tested temperatures; however, the Drosophila Orc6 carrying the K23E mutation was more susceptible to the heat and began unfolding at temperatures over 35°, suggesting that the amino acid substitution causes an instability in fly protein. Importantly, fluorescence intensity of the Drosophila mutant relative to wild type was significantly lower even at 25° (Figure 7B). This indicates that some part of the mutant protein is already unfolded at temperatures optimal for Drosophila development. We hypothesized that elevated temperature may increase protein unfolding and further affect the DNA binding ability of Drosophila Orc6. To test this hypothesis we repeated EMSA with wild-type and K23E mutant protein probes heat shocked at 50° for 15 min. Mutant Orc6 did not show any DNA binding under these conditions, whereas the wild-type Orc6 bound DNA but migrated faster compared to the protein not subjected to heat treatment (Figure 6A and Figure S4B). We suggest that the instability of Orc6-DmK23E protein contributes to the loss of DNA binding ability and ultimately to the MGS phenotype in Drosophila.
Figure 7.
Tryptophan fluorescence of Drosophila and human Orc6 proteins. (A) Emission maximum wavelengths at gradually increasing temperatures are shown. Shift in the maximal emission wavelength reflects protein unfolding. (B) Absolute intensity at each temperature point. DmWT and DmK23E indicate Drosophila Orc6 wild type and Orc6-K23E mutant, respectively, and HsWT and HsK23E indicate human Orc6 wild type and Orc6 HsK23E mutant, respectively.
Elevated expression of the wild-type human Orc6 in Drosophila rescues a lethality associated with deletion of the orc6 gene
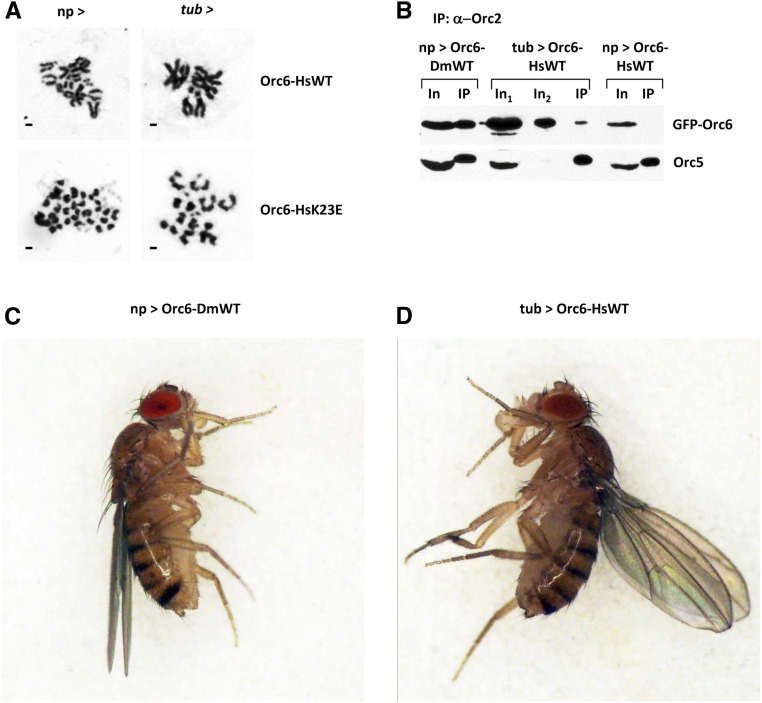
In our previous work (Balasov et al. 2015), we found that Drosophila C-terminal MGS mutation Y225S was lethal in flies despite the mild MGS clinical appearance in humans. Importantly, human Orc6 is loosely associated with the core ORC (Vashee et al. 2001; Vashee et al. 2003; Ranjan and Gossen 2006), whereas Drosophila Orc6 associates with core ORC much more tightly (Gossen et al. 1995; Chesnokov et al. 1999; Chesnokov et al. 2001). This might explain why the Y225S MGS mutation in Orc6 has a more dramatic effect on survival in Drosophila compared to the corresponding Y232S mutation in humans. We also found that elevated expression of mutant protein carrying Y225S substitution rescued lethality, restored normal karyotype, and allowed detection of Orc6 with the rest of ORC complex (Balasov et al. 2015). The expression of the wild-type human Orc6 by the native orc6 promoter could not rescue a lethality associated with Drosophila orc6 gene deletion (Table 1). We asked if overexpression of the human Orc6 would rescue orc6 deficiency in Drosophila. Using a GAL4/UAS binary system, we boosted the expression of human wild type and K23E mutant in flies. We found that the elevated expression of the wild-type human Orc6 was able to rescue flies to viability (Table 1) with restoration of normal karyotype (Figure 8A). We also observed human Orc6 with other ORC subunits after immunoprecipitation with anti-Orc2 antibodies (Figure 8B). Remarkably, rescued adult flies displayed an upheld wing phenotype (Figure 8, C and D) and were not able to fly (File S1, movie file), similar to the Drosophila C-terminal MGS mutation Y225S flies (Balasov et al. 2015). However, when K23E mutation was introduced into the human Orc6, the resulting mutant Orc6-HsK23E transgene was not able to rescue orc6 deficient flies to viability even under overexpressed conditions (Table 1), and no improvements were observed during development of the flies (Figure 8A, lower row). The loose association of human Orc6 with the core complex somewhat mimics the disruptive effect of Y225S substitution in Drosophila Orc6 for interaction with ORC. As a result, the Orc6-HsK23E mutant appears to be defective in both DNA binding (Figure 6, C and F) and in interacting with the core ORC, therefore it was not possible to rescue a lethality regardless of the protein expression level (Table 1).
Figure 8.
Rescue of orc6 deletion flies with human Orc6. (A) Mitoses in neuroblasts of orc6 deletion mutant rescued with human Orc6 wild type or with the K23E MGS mutation. Expression of transgenes was driven with either native orc6 promoter (np >) or with strong constitutive tubulin promoter (tub >). (B) Immunoprecipitation of the ORC complex with anti-Orc2 antibodies from ovaries expressing GFP-tagged Orc6 transgenes. In1 and In2 indicate 1/10 and 1/50 of total immunoprecipitation reaction, respectively. (C) Adult flies rescued with wild-type Drosophila Orc6. (D) Adult flies rescued with tubulin promoter driven overexpression of wild-type human Orc6.
Microarray analysis of the MGS mutation in Drosophila
In our previous study, we found that flies carrying Y225S mutation had rough spots in the eye, irregular hair patterns, and missing bristles—features possibly associated with minor cell polarity defects (Balasov et al. 2015). These phenotypes could indicate potential role of Orc6 in other processes or pathways apart from replication. To investigate this possibility further, we conducted a microarray analysis of gene expression to identify the genes/pathways that are affected by the MGS mutations. Orc6 carrying Drosophila Y225S MGS mutation rescued adult flies only when overexpressed, therefore orc635/orc635; Orc6-DmY225S/tubP-GAL4 adults were subjected to the microarray analysis against orc635/orc635; Orc6-DmWT/tubP-GAL4 adults. From the total analyzed 13557 genes, 1446 genes were downregulated more than two times and 1363 genes were upregulated more than two times. Gene ontology analysis revealed downregulated DNA replication, DNA repair, and cell proliferation processes in the top 10 scores (Table S1). One of the Orc6-DmK23E MGS mutant transgenes rescued flies to the viability without overexpression; therefore, orc635/orc635; Orc6-DmK23E-4/Orc6-DmK23E-4 were analyzed and compared to orc635/orc635; Orc6-DmWT/Orc6-DmWT. In this case, only 33 genes were downregulated more than two times and 25 genes were upregulated more than two times. This result is consistent with a relatively mild phenotype associated with K23E mutation compared to Y225S mutation. Next, we filtered up- and downregulated genes in both mutants for common genes. The end results consisted of two short lists of 9 downregulated genes and 14 upregulated genes (Table S1). We analyzed the expression patterns of genes based on modeENCODE RNA-seq data (flybase.org) and found that all nine downregulated genes had maximum expression level in ovaries. The ovary is the organ where the majority of the replication events happens in adult flies, and so they are more sensitive to replication defects. The 14 upregulated genes showed high expression in head and digestive system. Most of them encoded products with endopeptidase and ubiquitin-transferase activities. We hypothesize that the elevated expression of these genes could reflect degradation/utilization processes in tissues where proliferation defects resulted in cell damage. Overall microarray analysis did not reveal any additional genes/pathways affected by MGS mutations with the exception of replication and proliferation. It should be noted that RNA was isolated from the whole body of the adult flies, which might obscure the difference between specific tissue and developmental stages. For a more rigorous conclusion, tissue-/stage-specific microarrays should be performed.
Analysis of the flightless phenotype
The flightless phenotype is a characteristic of all analyzed Orc6-based MGS mutants in Drosophila, including previously described Y225S (Balasov et al. 2015) and a new K23E mutations. In our current study, GAL4 > UAS overexpression of Drosophila K23E mutant rescued the lethality and bristle defects but did not restore the ability to fly. We assayed flight performance by dropping flies individually into a 90-cm cylinder. Flies expressing wild-type Drosophila Orc6 immediately escaped the cylinder or landed on the wall and flew away in seconds. In contrast, mutants landed on the bottom of the cylinder, they did not try to escape and were easy to collect. Interestingly, the flies rescued with human Orc6 were not able to fly either (File S1, movie file).
To understand the cellular mechanism of flightless phenotype, we performed histological analyses of the major thoracic muscles. In Drosophila, large indirect flight muscles (IFMs) mediate flight by contraction and expansion of the thoracic cuticle. IFMs are composed of six dorsal longitudinal muscles and seven dorsoventral muscles (Bernard et al. 2003). Therefore, we analyzed histological transverse sections of IFMs for signs of degeneration or atrophy. Light microscopy revealed that all large muscle fiber groups were present in MGS mutants (Figure S5). An inability to fly might also result from defects in IFM innervation. To test this, we induced RNA interference of orc6 specifically in motor neurons. Remarkably, reduction of orc6 expression in motor neurons also caused an inability to fly (data not shown). This experiment suggests that Orc6 might participate in motor neuron development during metamorphosis.
Discussion
MGS is a rare human disease associated with microcephaly, short stature, and multiple developmental defects (Bicknell et al. 2011a; de Munnik et al. 2012a,b; Kerzendorfer et al. 2013). Mutations in a number of factors involved in DNA replication have been found to be causative for this disease (Bicknell et al. 2011a,b; Guernsey et al. 2011; Burrage et al. 2015; Fenwick et al. 2016; Vetro et al. 2017; McDaniel et al. 2020). Several mutations causing MGS were found in ORC subunits, including Orc6 (Bicknell et al. 2011a,b; Guernsey et al. 2011). In humans, Y232S substitution in Orc6 corresponds to a mild clinical appearance of MGS (Bicknell et al. 2011a; de Munnik et al. 2012a); however, the corresponding mutation in Drosophila (Y225S) is lethal and molecular and cell analysis revealed significantly reduced DNA replication and chromosome fragmentation in cells and tissues (Balasov et al. 2015). Importantly, flies carrying the Orc6-Y225S mutation can be rescued by the elevated expression of the transgene (Balasov et al. 2015).
The second known mutation in human Orc6 related to MGS is a homozygous deleterious mutation within the gene. Small deletion c.602-605delAGAA generated stop codon after 202 codons (Shalev et al. 2015). The severity of abnormal embryological development in humans coincided with a lethal embryonic phenotype in Drosophila for Orc6 having only first 200 amino acids (Balasov et al. 2009). The elevated expression of this mutant protein did not rescue a lethal phenotype of flies (Balasov et al. 2015).
A third known mutation in human Orc6 related to MGS was recently reported as the mutation responsible for the K23E substitution (Li et al. 2017). This mutation caused a phenotype in Drosophila that varies in severity from lethality to relatively normal adults. This newly described K23E mutation localizes in the DNA binding N terminus of the protein. Not surprisingly, biochemical and cytological analysis revealed that K23E mutation in both Drosophila and human Orc6 reduced DNA binding in gel-shift experiments. In vivo, this mutation leads to the inability of Orc6-HsK23E or Orc6-HDK23E to bind with chromosomes, but the chromosome association of Orc6-DmK23E is not significantly affected. This observation coincided with the survival rates as flies carrying Orc6-DmK23E often were able to progress to the adulthood. Interestingly, K23E mutation leads to the instability of the fly protein, with a difference already noticeable at 37°. However, in the case of human protein, the same mutation did not change protein conformation compared to the wild type, even at the higher temperatures. Normal body temperature for humans, 37°, is lethal for Drosophila, suggesting that evolutionary changes occurred in warm-blooded animals to withstand thermal challenges otherwise lethal for insects. In summary, the two known MGS substitution mutations in Orc6 affect different functional domains of the protein and result in either impaired DNA binding (K23E) by Orc6 or a loss of the protein association with the core ORC (Y232S). The consequences of both mutations include the reduced amounts of hexameric ORC on DNA, an impaired pre-RC formation, and fewer origin firings. This in turn leads to the replication, proliferation, and development defects manifesting in similar clinical features in both cases.
Drosophila and human Orc6 proteins have only 28% of sequence identity but are structurally similar and critical for the initiation of DNA replication (Duncker et al. 2009). The metazoan Orc6 consists of two domains. The larger N-terminal domain (∼200 amino acids) carries a homology with the transcription factor TFIIB and is important for DNA binding (Chesnokov et al. 2003; Balasov et al. 2007; Liu et al. 2011). The shorter C-terminal domain is important for the function of Orc6 in cytokinesis (Prasanth et al. 2002; Chesnokov et al. 2003; Huijbregts et al. 2009; Bernal and Venkitaraman 2011; Akhmetova et al. 2015), as well as for the interaction of Orc6 with core ORC (Bleichert et al. 2013). Full-length human Orc6 could not rescue a lethality associated with the loss of the fly gene when expressed under native orc6 promoter. Therefore, we created the hybrid Orc6 protein containing human N-terminal domain and Drosophila C terminus. This transgene rescued orc6 deletion flies to viability and they were indistinguishable from the wild-type animals. This indicates that the N-terminal TFIIB-like domain of both proteins involved in the same conserved functions between organisms. On the other hand, the C terminus of both Drosophila and human Orc6 contains conserved motifs responsible for association with ORC complex. Both flies and humans with C-terminal truncations do not survive to the adult stage (Balasov et al. 2009; Shalev et al. 2015). However, the point mutation (Y232S) in human Orc6 manifests in a mild postnatal phenotype (Bicknell et al. 2011a; de Munnik et al. 2012a), while the corresponding Drosophila Y225S MGS mutant shows no difference from the lethal orc6 deletion. It is known that human Orc6 is loosely associated with the core ORC (Vashee et al. 2001; Vashee et al. 2003; Ranjan and Gossen 2006), whereas Drosophila Orc6 associates with other ORC subunits significantly more tightly (Gossen et al. 1995; Chesnokov et al. 1999; Chesnokov et al. 2001). This might explain why the Y225S MGS mutation has a more dramatic effect on survival in Drosophila than the corresponding MGS (Y232S) mutation in humans.
In our earlier study (Balasov et al. 2015) we found that an elevated expression of the fly protein carrying the Y225S mutation rescued third instar lethality, restored normal karyotype, and the mutant protein was detected on DNA with the rest of ORC complex. Similarly, here we found that the elevated expression of human Orc6 allowed the formation of the functional six-subunit ORC on DNA and rescued flies carrying orc6 deletion. In some way human Orc6 in fly system mimics the effect of the C-terminal MGS mutation in Drosophila Orc6. In both cases the interaction of the proteins with core ORC is impaired resulting in similar phenotypes between flies carrying either Drosophila Orc6 with MGS mutation or the human protein as a sole source of Orc6.
In this study, we showed that hybrid protein Orc6-HD containing intact human N-terminal TFIIB-like domain (∼80% of the protein length) and Drosophila C terminus tightly associated with ORC and rescued Orc6-deficient flies to viable adults phenotypically undistinguishable from wild-type animals. This hybrid approach revealed the importance of evolutionary conserved and variable domains of Orc6 protein, and allowed our studies of human protein functions and the analysis of the critical amino acids in a live animal system. We believe that hybrid approach not only opens a broad avenue to study new Orc6 mutations for medical and general science purposes, but also might be useful in other humanized models. In summary, the humanized fly model presented in our studies has the unique advantage of being able to differentially test both fly, human, and chimeric Orc6 proteins to reveal conserved and divergent features of the protein and its functions in the cells of metazoan organisms. The fly strains generated during this work will be useful in analyzing the functions of human protein in a convenient heterologous system. Specifically, our studies revealed novel insights into molecular mechanisms underlying MGS pathology, and provide important clues about disease origin and development.
Acknowledgments
We thank Adam Gower from the Boston University Microarray and Sequencing Resource Core Facility. We also thank Dr. Peter Detloff for the critical reading of the manuscript. All research materials and data from our studies will be freely available to other investigators. This work was supported by a grant from the National Institute of General Medical Sciences (GM121449 to I.C.).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.13070288.
Communicating editor: B. Calvi
Literature Cited
- Akhmetova K., Balasov M., Huijbregts R. P., and Chesnokov I., 2015. Functional insight into the role of Orc6 in septin complex filament formation in Drosophila. Mol. Biol. Cell 26: 15–28. 10.1091/mbc.e14-02-0734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Huijbregts R. P., and Chesnokov I., 2007. Role of the Orc6 protein in origin recognition complex-dependent DNA binding and replication in Drosophila melanogaster. Mol. Cell. Biol. 27: 3143–3153 (erratum: Mol. Cell. Biol. 27: 4206). 10.1128/MCB.02382-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Huijbregts R. P., and Chesnokov I., 2009. Functional analysis of an Orc6 mutant in Drosophila. Proc. Natl. Acad. Sci. USA 106: 10672–10677. 10.1073/pnas.0902670106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasov M., Akhmetova K., and Chesnokov I., 2015. Drosophila model of Meier-Gorlin syndrome based on the mutation in a conserved C-terminal domain of Orc6. Am. J. Med. Genet. A. 167A: 2533–2540. 10.1002/ajmg.a.37214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16: 659–672. 10.1101/gad.969602 [DOI] [PubMed] [Google Scholar]
- Bell S. P., and Stillman B., 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134. 10.1038/357128a0 [DOI] [PubMed] [Google Scholar]
- Bernal J. A., and Venkitaraman A. R., 2011. A vertebrate N-end rule degron reveals that Orc6 is required in mitosis for daughter cell abscission. J. Cell Biol. 192: 969–978. 10.1083/jcb.201008125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard F., Lalouette A., Gullaud M., Jeantet A. Y., Cossard R. et al. , 2003. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev. Biol. 260: 391–403. 10.1016/S0012-1606(03)00255-0 [DOI] [PubMed] [Google Scholar]
- Bicknell L. S., Bongers E. M., Leitch A., Brown S., Schoots J. et al. , 2011a Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 43: 356–359. 10.1038/ng.775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell L. S., Walker S., Klingseisen A., Stiff T., Leitch A. et al. , 2011b Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 43: 350–355. 10.1038/ng.776 [DOI] [PubMed] [Google Scholar]
- Bleichert F., Balasov M., Chesnokov I., Nogales E., Botchan M. R. et al. , 2013. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife 2: e00882 10.7554/eLife.00882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., and Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Burrage L. C., Charng W. L., Eldomery M. K., Willer J. R., Davis E. E. et al. , 2015. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier-Gorlin syndrome. Am. J. Hum. Genet. 97: 904–913. 10.1016/j.ajhg.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., de Vries M. A., and Bell S. P., 2007. Orc6 is required for dynamic recruitment of Cdt1 during repeated Mcm2–7 loading. Genes Dev. 21: 2897–2907. 10.1101/gad.1596807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I. N., 2007. Multiple functions of the origin recognition complex. Int. Rev. Cytol. 256: 69–109. 10.1016/S0074-7696(07)56003-1 [DOI] [PubMed] [Google Scholar]
- Chesnokov I., Gossen M., Remus D., and Botchan M., 1999. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes Dev. 13: 1289–1296. 10.1101/gad.13.10.1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I., Remus D., and Botchan M., 2001. Functional analysis of mutant and wild-type Drosophila origin recognition complex. Proc. Natl. Acad. Sci. USA 98: 11997–12002. 10.1073/pnas.211342798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokov I. N., Chesnokova O. N., and Botchan M., 2003. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc. Natl. Acad. Sci. USA 100: 9150–9155. 10.1073/pnas.1633580100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M., Wang P., Boyd A. D., Kostov G., Athey B. et al. , 2005. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 33: e175 10.1093/nar/gni179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik S. A., Bicknell L. S., Aftimos S., Al-Aama J. Y., van Bever Y. et al. , 2012a Meier-Gorlin syndrome genotype-phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur. J. Hum. Genet. 20: 598–606. 10.1038/ejhg.2011.269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Munnik S. A., Otten B. J., Schoots J., Bicknell L. S., Aftimos S. et al. , 2012b Meier-Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. Am. J. Med. Genet. A. 158A: 2733–2742. 10.1002/ajmg.a.35681 [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. 10.1002/gene.10150 [DOI] [PubMed] [Google Scholar]
- Duncker B. P., Chesnokov I. N., and McConkey B. J., 2009. The origin recognition complex protein family. Genome Biol. 10: 214 10.1186/gb-2009-10-3-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick A. L., Kliszczak M., Cooper F., Murray J., Sanchez-Pulido L. et al. , 2016. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier-Gorlin syndrome and craniosynostosis. Am. J. Hum. Genet. 99: 125–138. 10.1016/j.ajhg.2016.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., and Irizarry R. A., 2004. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315. 10.1093/bioinformatics/btg405 [DOI] [PubMed] [Google Scholar]
- Gentleman R. C., Carey V. J., Bates D. M., Bolstad B., Dettling M. et al. , 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80 10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlin R. J., Cervenka J., Moller K., Horrobin M., and Witkop C. J. Jr, 1975. Malformation syndromes. A selected miscellany. Birth Defects Orig. Artic. Ser. 11: 39–50. [PubMed] [Google Scholar]
- Gossen M., Pak D. T., Hansen S. K., Acharya J. K., and Botchan M. R., 1995. A Drosophila homolog of the yeast origin recognition complex. Science 270: 1674–1677. 10.1126/science.270.5242.1674 [DOI] [PubMed] [Google Scholar]
- Guernsey D. L., Matsuoka M., Jiang H., Evans S., Macgillivray C. et al. , 2011. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 43: 360–364. 10.1038/ng.777 [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., and Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huijbregts R. P., Svitin A., Stinnett M. W., Renfrow M. B., and Chesnokov I., 2009. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Mol. Biol. Cell 20: 270–281. 10.1091/mbc.e08-07-0754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J. et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C., Colnaghi R., Abramowicz I., Carpenter G., and O’Driscoll M., 2013. Meier-Gorlin syndrome and Wolf-Hirschhorn syndrome: two developmental disorders highlighting the importance of efficient DNA replication for normal development and neurogenesis. DNA Repair (Amst.) 12: 637–644. 10.1016/j.dnarep.2013.04.016 [DOI] [PubMed] [Google Scholar]
- Lebedeva L. I., Trunova S. A., and Omel’ianchuk L. V., 2000 [Genetic control of mitosis. Adaptive modifications of v158 mutation expression]. Genetika 36: 1348–1354. [PubMed] [Google Scholar]
- Lee D. G., and Bell S. P., 1997. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell. Biol. 17: 7159–7168. 10.1128/MCB.17.12.7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ding Y., Chang G., Cheng Q., Li X. et al. , 2017 [A boy with Meier-Gorlin syndrome carrying a novel ORC6 mutation and uniparental disomy of chromosome 16]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 34: 68–72. [DOI] [PubMed] [Google Scholar]
- Liu S., Balasov M., Wang H., Wu L., Chesnokov I. N. et al. , 2011. Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB. Proc. Natl. Acad. Sci. USA 108: 7373–7378. 10.1073/pnas.1013676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel S. L., Hollatz A. J., Branstad A. M., Gaskill M. M., Fox C. A. et al. , 2020. Tissue-specific DNA replication defects in Drosophila melanogaster caused by a Meier-Gorlin syndrome mutation in Orc4. Genetics 214: 355–367. 10.1534/genetics.119.302938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., and Stillman B., 2002. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science 297: 1026–1031. 10.1126/science.1072802 [DOI] [PubMed] [Google Scholar]
- Ranjan A., and Gossen M., 2006. A structural role for ATP in the formation and stability of the human origin recognition complex. Proc. Natl. Acad. Sci. USA 103: 4864–4869. 10.1073/pnas.0510305103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer C. A., 2006. Probing protein folding and conformational transitions with fluorescence. Chem. Rev. 106: 1769–1784. 10.1021/cr0404390 [DOI] [PubMed] [Google Scholar]
- Sasaki T., and Gilbert D. M., 2007. The many faces of the origin recognition complex. Curr. Opin. Cell Biol. 19: 337–343. 10.1016/j.ceb.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Semple J. W., Da-Silva L. F., Jervis E. J., Ah-Kee J., Al-Attar H. et al. , 2006. An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes. EMBO J. 25: 5150–5158. 10.1038/sj.emboj.7601391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev S. A., Khayat M., Etty D. S., and Elpeleg O., 2015. Further insight into the phenotype associated with a mutation in the ORC6 gene, causing Meier-Gorlin syndrome 3. Am. J. Med. Genet. A. 167A: 607–611. 10.1002/ajmg.a.36906 [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., and Hawley R. S., 2000. Drosophila Protocols, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Vashee S., Simancek P., Challberg M. D., and Kelly T. J., 2001. Assembly of the human origin recognition complex. J. Biol. Chem. 276: 26666–26673. 10.1074/jbc.M102493200 [DOI] [PubMed] [Google Scholar]
- Vashee S., Cvetic C., Lu W., Simancek P., Kelly T. J. et al. , 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17: 1894–1908. 10.1101/gad.1084203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetro A., Savasta S., Russo Raucci A., Cerqua C., Sartori G. et al. , 2017. MCM5: a new actor in the link between DNA replication and Meier-Gorlin syndrome. Eur. J. Hum. Genet. 25: 646–650. 10.1038/ejhg.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental material available at figshare: https://doi.org/10.25386/genetics.13070288.