Abstract
Objective:
LncRNAs are non-coding RNAs exerting vital roles in the occurrence and development of various cancer types. This study tended to describe the expression pattern of FENDRR in colorectal cancer (CRC), and further investigate the role of FENDRR in CRC cell biological behaviors.
Methods:
Gene expression profile of colon cancer was accessed from the TCGA database, and then processed for differential analysis for identification of differentially expressed lncRNAs and miRNAs. Some in vitro experiments like qRT-PCR, MTT, colony formation assay, wound healing assay and Transwell assay were performed to assess the effect of FENDRR on cell biological behaviors. Dual-luciferase reporter assay was conducted to further validate the targeting relationship between FENDRR and miR-424-5p, and rescue experiments were carried out for determining the mechanism of FENDRR/miR-424-5p underlying the proliferation, migration and invasion of CRC cells.
Results:
Bioinformatics analysis suggested that FENDRR was significantly down-regulated in CRC tissue, and low FENDRR was intimately correlated to poor prognosis. FENDRR overexpression could greatly inhibit cell proliferation, migration and invasion. Besides, there was a negative correlation between FENDRR and miR-424-5p. Dual-luciferase reporter assay indicated that miR-424-5p was a direct target of FENDRR. Rescue experiments discovered that FENDRR exerted its role in cell proliferation, migration and invasion in CRC via targeting miR-424-5p.
Conclusion:
FENDRR is poorly expressed in CRC tissue and cells, and low FENDRR is responsible for the inhibition of cell proliferation, migration and invasion of CRC by means of targeting miR-424-5p.
Keywords: colorectal cancer, FENDRR, miR-424-5p, proliferation, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most common gastrointestinal malignancies, with the morbidity ranking third (10.2%) worldwide and the mortality ranking second (9.2%). According to A Cancer Journal for Clinicians, there were approximately over 1,800,000 newly diagnosed cases in 2018 and 881,000 deaths due to CRC.1 Studies have reported that the inducement for CRC occurrence is intimately associated with genetic factors. For example, various tumor susceptibility genes like K-ras.2,3 and N-ras,4 the mutation of tumor suppressor genes like P535 and PTEN,6 epigenetic modification,7 aberrant regulation of tumor-related signaling pathways8,9 and other factors have been reported to be implicated in CRC occurrence and development. However, much work so far on the mechanism underlying CRC occurrence and development has focused on the mutation and regulatory functions of coding genes, yet the role of non-coding genes in CRC is poorly investigated.
Long non-coding RNAs (lncRNAs) are RNA molecules with a transcript length over 200 nt. LncRNAs have no ability to code proteins, but they are able to regulate gene expression levels in multiple aspects, such as epigenetic regulation, transcriptional and post-transcriptional regulation. Recently, many studies have reported the important role of lncRNAs in the occurrence of human tumors.10-12 Meanwhile, lncRNAs have been verified to regulatory function on CRC occurrence and development. For instance, lncRNA HEIH can antagonize the miR-939 mediated inhibition of Bcl-XL transcription to potentiate the occurrence of CRC.13 LINC00483 can be used as a competing endogenous RNA (ceRNA) for miR-204-3p, thereby promoting cell proliferation and metastasis of CRC via mediating FMNL2.14 Besides, lncRNAs have been reported to have the potential acting as biomarkers for CRC prognosis.15 LncRNA ATB, for example, is significantly elevated in CRC patients with hematogenous migration, and it might be implicated in CRC progression and can be used as a novel biomarker for the poor prognosis of CRC patients.16
LncRNA FENDRR is a non-coding RNA located on 16q24.1. It has been reported that FENDRR directly targets miR-18a-5p to regulate RUNX1, in turn inhibitory functioning on the development of prostate cancer.17 In addition, FENDRR acting as a ceRNA decreases Mdr1 by targeting Hur, thus attenuating the stemness of non-small cell lung cancer cells.18 Moreover, FENDRR has been investigated to play a negative role in CRC progression via reducing Sox4,19 yet the molecular mechanism of FENDRR in CRC remains elusive.
Therefore, it’s crucial to clarify the role of FENDRR in CRC. This study adopted a series of in vitro experiments to exploit the effect of FENDRR on cell biological behaviors in CRC, which helps to extend our knowledge on CRC pathogenesis and gains more insight into future clinical treatment.
Materials and Methods
Cell Culture and Sample Collection
Human normal colon cell line FHC (ATCC®CRL-1831) and CRC cell lines HCT116 (ATCC® CCL-247EMT), SNU-C2B (ATCC® CCL-250), NCI-H498 (ATCC® CCL-254) and HCT-15 (ATCC® CCL-225) were all ordered from Genetimes ExCell Technology, Inc. (Shanghai, China). All cells were cultured in the Roswell Park Memorial Institute-1640 medium (RPMI-1640; 11875093; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; 10099141C; Gibco, Grand Island, NY, USA), and maintained in 5% CO2 at 37 °C.
A panel of CRC tissue samples (n = 20) and corresponding adjacent normal tissue samples (n = 20) were collected from the Lishui Municipal Central Hospital. All samples were preserved immediately in liquid nitrogen at −80 °C after being resected. This study had been approved by the Ethics Committee of Lishui Municipal Central Hospital and informed consent had been obtained from all subjects.
Bioinformatics Analysis
MiRNA expression profile of CRC was accessed from TCGA database, and then processed for differential analysis using the “edgeR” package (|logFC|>2, padj < 0.05). Afterward, starBase database was applied to screen interacted differentially expressed lncRNAs and miRNAs (DElncRNAs and DEmiRNAs). Survival analysis was performed on the target genes in the TCGA-COAD dataset with the “survival” package of R language.
Cell Transfection
miR-424-5p mimic and mimic NC were purchased from GenePharma (Shanghai, China). Small interfering RNAs (siRNAs) against FENDRR were synthesized by Sangon, Co., Ltd (Shanghai, China). pEGFP1-FENDRR recombinant plasmids were constructed using pEGFP1 overexpression vectors. Then, the mimics, siRNAs, recombinant plasmids and corresponding controls were transfected into CRC cells by applying Lipofectamine®3000 (Invitrogen, USA) following the standard process, respectively.
Real-Time Quantitative PCR (qRT-PCR)
Trizol (Invitrogen, USA) was used to isolate total RNA according to the manufacturer’s instructions. Inverse transcription assay kit (Invitrogen, USA) was applied for cDNA synthesis. qRT-PCR was run on the ABI 7900HT instrument (Applied Biosystems, USA) using the miScript SYBR Green PCR Kit (Qiagen, Germany) under the follow-up thermal cycling conditions: initial denaturation at 95 °C for 10 min, 95 °C for 2 min, 40 cycles of 95 °C for 5 s and 60 °C for 30 s. GAPDH and U6 were applied as endogenous regulators of FENDRR and miR-424-5p, respectively. The results were normalized using the 2-ΔΔCt method. Primers were listed in Supplementary Table 1.
MTT
HCT116 cells (5 × 103) were planted into 96-well plates with 3 repeated wells for each treatment. At 1, 2, 3, 4 and 5 days, sterile MTT reagent (Beyotime) was added into each well per the manufacturer’s protocol. Enzyme-labeled instruments (Molecular Devices, Sunnyvale, CA, USA) were employed to read the absorbance at 490 nm.
Colony Formation Assay
6 repeated wells were set for each treatment (103 cells/well). After 14 days of incubation, cells were fixed with 30% formaldehyde (Thermo Fisher Scientific, USA) for 15 min, and then stained in 0.1% crystal violet (Thermo Fisher Scientific, USA). An optimal microscope was used to observe the colonies (Colonies with over 50 cells were qualified).
Wound Healing Assay
6-well plates were used to culture cells. When cells were grown to 80% in confluence, the tip of a 200 ul pipette was used to make a scratch on the monolayer. Medium was applied to wash off the separated cells, and the remained cells were nurtured for further 24 h with fresh mediums. Cell migration at 0 and 24 h were monitored under a microscope and then photographed.
Transwell
24-well Transwell inserts (8 µm in aperture, BD Biosciences) were used for cell invasion assessment. An estimate of 2 × 104 cells suspended by serum-free mediums were seeded into Matrigel matrix-coated upper chambers, and the Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (Thermo Fisher Scientific, USA) was added into lower chambers. After 48 h of incubation at room temperature, cotton swabs were employed to softly remove the uninvaded cells, and cells invaded to the lower chambers were stained in 0.5% crystal violet. The number of invaded cells was calculated under a microscope.
Dual-Luciferase Reporter Assay
To validate the combination of miR-424-5p and FENDRR 3′-UTR, psiCHECK luciferase vectors (Sangon, Co., Ltd, Shanghai, China) with wild type (WT) and mutant (MUT) FENDRR 3′-UTR were generated, called FENDRR-WT and FENDRR-MUT. After being cultured in 48-well plates for 24 h, FENDRR-WT and FENDRR-MUT vectors were co-transfected with miR-424-5p mimic or mimic NC into HCT116 cells. Dual-luciferase assay kit (Promega, Fitchburg, WI, USA) was applied for determination of luciferase activity in each transfection group.
Statistical Analysis
Measurement data were presented as mean ± standard deviation. Student’s t test and one-way analysis of variance (ANOVA) were applied to analyze the comparisons between 2 groups and among the multiple groups under the SPSS 22.0 software, respectively. All experiments were repeated at least 3 times. P < 0.05 was considered statistically significant.
Results
FENDRR Is Down-Regulated in CRC Tissues and Cells
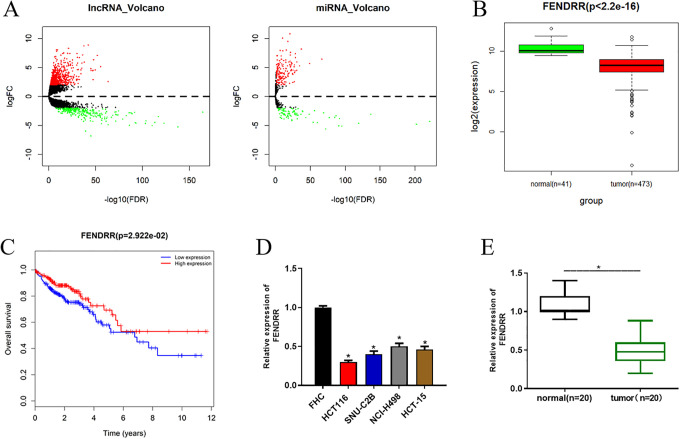
In all, 1,135 DElncRNAs and 271 DEmiRNAs were screened from the TCGA-COAD dataset (Figure 1A). Expression and survival analysis were performed and it was found that the lncRNA FENDRR was significantly decreased in tumor tissue (Figure 1B), and patients with low FENDRR had a much shorter survival time than those with high expression (Figure 1C). It has been reported that FENDRR is greatly down-regulated in various cancer tissues, which induces the malignant progression and leads to poor prognosis of cancer20,21. Hence, FENDRR was selected as our research object. To determine the level of FENDRR in cancer cases, qRT-PCR was conducted to test the level of FENDRR in CRC cells and clinical tissue samples. As shown in Figure 1D and 1E, FENDRR was remarkably reduced in CRC cells and tumor tissue relative to the normal colon cell and matched adjacent normal tissue. The results above collectively demonstrated that lncRNA FENDRR was aberrantly poorly expressed in CRC tissue and cells.
Figure 1.
FENDRR is significantly down-regulated in CRC tissue and cells. (A) Differential analysis was performed to identify DElncRNAs and DEmiRNAs in the TCGA-COAD dataset using the “edgeR” package. Among the DElncRNAs, (B) FENDRR was observed to be down-regulated in tumor tissue in the TCGA-COAD dataset and (C) survival analysis was conducted with red representing high expression and blue representing low expression. qRT-PCR was carried out to determine the level of FENDRR in (D) CRC cell lines and normal colon cell line, as well as (E) in clinical cancer tissue samples and paired adjacent normal tissue samples (* p < 0.05).
FENDRR Inhibits Cell Proliferation and Colony Formation Ability in CRC
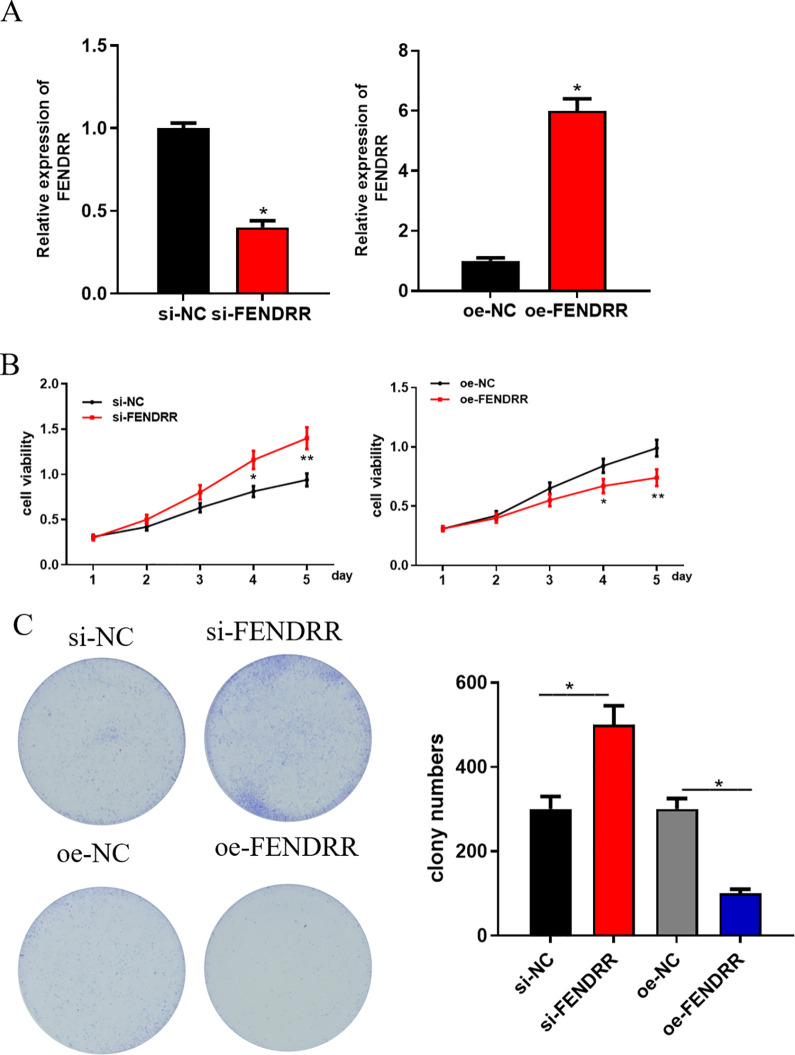
To investigate the role of FENDRR in cell proliferation and colony formation ability in CRC, si-NC, si-FENDRR, oe-NC and oe-FENDRR were transfected into HCT116 cells. qRT-PCR was conducted to assess the transfection efficiency, finding that FENDRR was significantly decreased in si-FENDRR transfected cells, but increased in oe-FENDRR transfected cells (Figure 2A). Afterward, MTT and colony formation assay revealed that silencing FENDRR could obviously enhance cell proliferation and colony formation ability, while opposite effect was detected when FENDRR was overexpressed (Figure 2B and C). Taken together, FENDRR could inhibitory function on cell proliferation and colony formation ability in CRC.
Figure 2.
FENDRR inhibits CRC cell proliferation and colony formation ability si-NC, si-FENDRR, oe-NC and oe-FENDRR were transfected into HCT116 cells. (A) qRT-PCR was performed to detect the transfection efficiency. Then, the cells were collected for (B) MTT and (C) colony formation assay to assess the cell proliferation and colony formation ability of CRC (**p < 0.05).
FENDRR Suppresses Cell Migration and Invasion in CRC
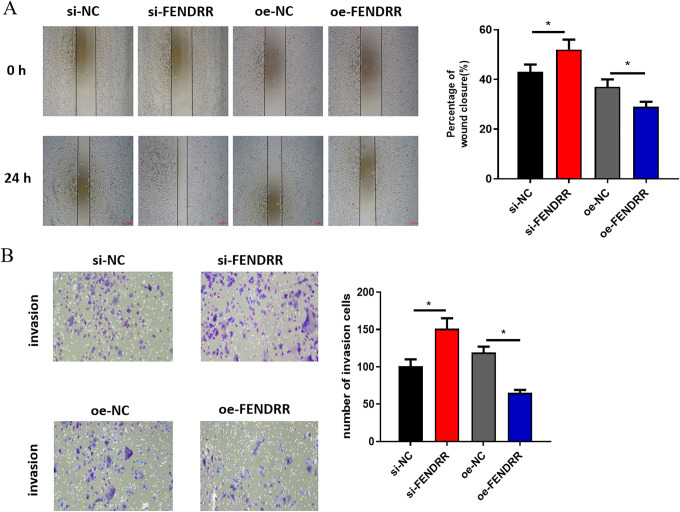
The role of FENDRR in cell migration and invasion in CRC was further explored. Wound healing assay and Transwell assay were performed and it was discovered that silencing FENDRR could considerably strengthen cell migration and invasion, whereas overexpressing FENDRR had the opposite effects (Figure 3A and B). Thus, it could be concluded that FENDRR inhibited cell migration and invasion of CRC.
Figure 3.
FENDRR inhibits cell migration and invasion of CRC si-FENDRR, oe-FENDRR and their negative controls were transfected into HCT116 cells. (A) Wound healing assay and (B) Transwell assay were conducted to assess cell migration and invasion (* p < 0.05).
miR-424-5p Is a Direct Target of FENDRR
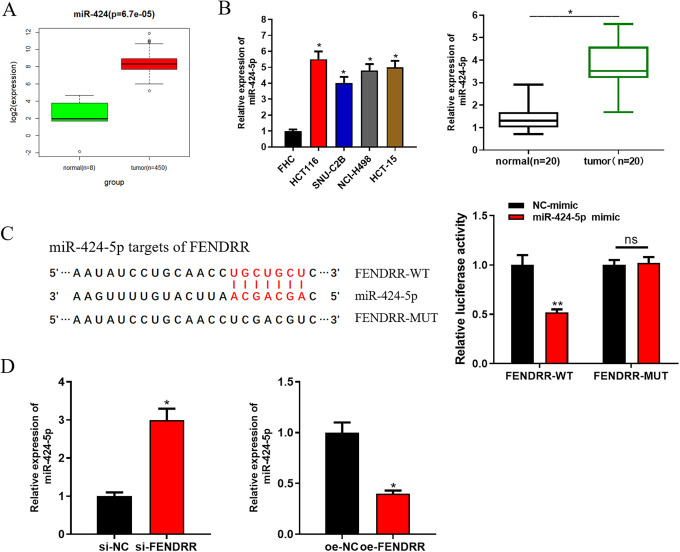
Recently, many studies have revealed that lncRNAs can serve as ceRNAs to targeted bind with miRNAs, in turn exerting their regulatory roles in tumorigenesis and development.22,23 In view of this, we reasoned that FENDRR might function in CRC via sponging targeted miRNAs. To validate such speculation, starBase database was used to predict the potential targets of FENDRR. miR-424-5p was predicted to harbor the binding sites with FENDRR and observed to be significantly up-regulated in tumor tissue in the TCGA-COAD dataset (Figure 4A). In addition, qRT-PCR was performed and suggested the remarkable high expression of miR-424-5p in both CRC cells and clinical cancer tissue samples relative to the normal controls (Figure 4B). Moreover, dual-luciferase reporter assay was conducted for further validation of their targeting relationship. As indicated in Figure 4C, the luciferase activity in cells transfected with miR-424-5p mimic and FENDRR-WT was greatly decreased, whereas there was no obvious difference in FENDRR-MUT transfected cells. Furthermore, we detected miR-424-5p expression in cells with si-FENDRR or oe-FENDRR, finding that miR-424-5p was up-regulated when FENDRR was silenced, but down-regulated when FENDRR was overexpressed (Figure 4D). Overall, these findings elucidated that FENDRR could directly target miR-424-5p.
Figure 4.
FENDRR directly targets miR-424-5p. Database starBase was used and it was found that miR-424-5p bore the targeted binding sites with FENDRR and (A) miR-424-5p was observed to be up-regulated in tumor tissue in the TCGA-COAD dataset. (B) qRT-PCR was performed to detect miR-424-5p in CRC cells and clinical tissue samples. (C) Dual-luciferase reporter assay was conducted to verify the targeting relationship between FENDRR and miR-424-5p. Afterward, si-FENDRR, oe-FENDRR and their negative controls were transfected into CRC cells. (D) qRT-PCR was carried out to assess miR-424-5p expression in each transfection group (* p < 0.05; ** p < 0.01).
FENDRR Inhibits CRC cell Proliferation, Migration and Invasion Via Targeting miR-424-5p
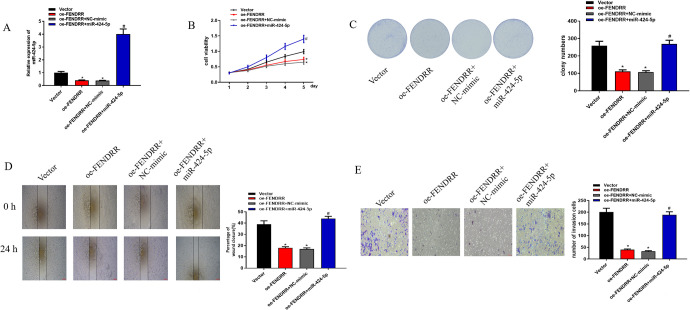
As aforementioned, FENDRR could targeted bind with miR-424-5p. To gain more insight into the mechanism of FENDRR in CRC occurrence and development through targeting miR-424-5p, cells were divided into 4 groups: vector, oe-FENDRR, oe-FENDRR+NC-mimic and oe-FENDRR+miR-424-5p mimic. As revealed in Figure 5A, FENDRR overexpression-induced miR-424-5p reduction could be elevated after miR-424-5p was overexpressed. In addition, cell biological behaviors were assayed. As shown in Figure 5B-E, overexpressing FENDRR could suppress cell proliferation, colony formation ability, migration and invasion, but such inhibitory effects could be reversed after miR-424-5p was overexpressed simultaneously. These findings demonstrated that FENDRR could inhibit cell proliferation, migration and invasion in CRC via targeting miR-424-5p.
Figure 5.
FENDRR inhibits CRC cell proliferation, migration and invasion via targeting miR-424-5p. All cells were divided into 4 groups: vector, oe-FENDRR, oe-FENDRR+NC-mimic and oe-FENDRR+miR-424-5p mimic. (A) qRT-PCR was performed to test miR-424-5p in each transfection group. Then, the cells were harvested for (B) MTT, (C) colony formation assay, (D) wound healing assay and (E) Transwell assay to determine the proliferation, colony formation ability, cell migration and invasion of CRC cells (* means relative to the vector group, p < 0.05; # means relative to the oe-FENDRR+NC-mimic group, p < 0.05).
Discussion
Increasing evidence has discovered that lncRNAs play important roles in tumorigenesis and development, which makes the molecular function of lncRNAs in tumor the present research focus. In this study, we analyzed the miRNA expression data from the TCGA-COAD dataset, and found that FENDRR was significantly decreased in CRC and showed a close correlation with poor prognosis. It has been reported that FENDRR can suppress cell proliferation of breast cancer, and high FENDRR is related to good prognosis.21 Besides, in gastric cancer, low FENDRR can predict poor prognosis, and FENDRR is able to mediate cell metastasis via targeting fibronectin 1.24 Moreover, down-regulation of FENDRR has been discovered to be implicated with the poor prognosis of prostate cancer.25 In view of these, we believed that FENDRR could function in CRC by acting as a tumor suppressor.
To confirm our speculation, we collected clinical tissue samples (tumor samples: n = 20; adjacent normal samples: n = 20), 4 CRC cells lines and 1 normal colon cell line, finding that FENDRR was significantly decreased in both CRC tissue and cells relative to the normal counterparts. Thereafter, the role of FENDRR in cell biological behaviors of CRC was exploited. FENDRR siRNA and pEGFP1-FENDRR were transfected into cells for construction of FENDRR silencing and overexpression. A series of in vitro experiments revealed that FENDRR overexpression could obviously attenuate cell proliferation, migration and invasion, while FENDRR silencing played a promotive role. These results indicate that FENDRR can be used as a tumor suppressor to decrease cell proliferation, migration and invasion of CRC.
Nowadays, numerous studies have revealed that lncRNAs can act as ceRNAs to target miRNAs, thereby regulating mRNAs and consequently mediating tumorigenesis and development.26,27 Hence, we speculated that FENDRR might play a regulatory role during CRC occurrence and development by serving as a ceRNA. We examined its potential targeted miRNAs by searching the starBase database, which were then intersected with the DEmiRNAs screened from the TCGA-COAD dataset. Eventually, we found that FENDRR could target miR-424-5p. To further clarify the underlying mechanism, bioinformatics analysis and qRT-PCR were performed and it was suggested that there was a negative correlation between FENDRR and miR-424-5p in CRC tissue and cells. Dual-luciferase reporter assay validated the targeting relationship between miR-424-5p and FENDRR, while rescue experiments indicated that the inhibitory effect of FENDRR on cell proliferation, migration and invasion of CRC could be reversed by miR-424-5p overexpression.
In conclusion, this study clarifies the remarkably decreased level of FENDRR in CRC tissue and cells. Meanwhile, there is a negative correlation between FENDRR and miR-424-5p levels, and FENDRR can directly target miR-424-5p to suppress CRC cell proliferation, migration and invasion. Our study uncovers the molecular function of FENDRR in CRC that FENDRR as a tumor suppressor has the potential serving as a diagnostic biomarker or a novel therapeutic target toward CRC treatment.
Supplemental Material
Supplemental Material, Supplemental_file for FENDRR Sponges miR-424-5p to Inhibit Cell Proliferation, Migration and Invasion in Colorectal Cancer by Chuan Cheng, Huixia Li, Jiujian Zheng, Jie Xu, Peng Gao and Jianping Wang in Technology in Cancer Research & Treatment
Footnotes
Authors’ Note: The data and materials in the current study are available from the corresponding author on reasonable request.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by the funds from Zhejiang Lishui Science & Technology Bureau (2016zdxk06). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ORCID iD: Jianping Wang  https://orcid.org/0000-0002-1193-4568
https://orcid.org/0000-0002-1193-4568
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551 (2019) [DOI] [PubMed] [Google Scholar]
- 2. Fang Y, Sun B, Li Z, Chen Z, Xiang J. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2016;55(9):1369–1377. doi:10.1002/mc.22380 (2016) [DOI] [PubMed] [Google Scholar]
- 3. Cardenas-Ramos SG, Alcázar-González G, Reyes-Cortés LM, et al. The frequency and type of K-RAS mutations in Mexican patients with colorectal cancer: a national study. Am J Clin Oncol. 2017;40(3):274–276. doi:10.1097/COC.0000000000000143 (2017) [DOI] [PubMed] [Google Scholar]
- 4. Wang Y, Velho S, Vakiani E, et al. Mutant N-RAS protects colorectal cancer cells from stress-induced apoptosis and contributes to cancer development and progression. Cancer Discov. 2013;3(3):294–307. doi:10.1158/2159-8290.CD-12-0198 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cheng J, Dwyer M, Okolotowicz KJ, Mercola M, Cashman JR. A novel inhibitor targets both Wnt signaling and ATM/p53 in colorectal cancer. Cancer Res. 2018;78(17):5072–5083. doi:10.1158/0008-5472.CAN-17-2642 (2018) [DOI] [PubMed] [Google Scholar]
- 6. Ran H, Zhu Y, Deng R, et al. Stearoyl-CoA desaturase-1 promotes colorectal cancer metastasis in response to glucose by suppressing PTEN. J Exp Clin Cancer Res. 2018;37(1):54 doi:10.1186/s13046-018-0711-9 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143 doi:10.1186/s12943-019-1079-y (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jiang L, Zhao XH, Mao YL, Wang JF, Zheng HJ, You QS. Long non-coding RNA RP11-468E2.5 curtails colorectal cancer cell proliferation and stimulates apoptosis via the JAK/STAT signaling pathway by targeting STAT5 and STAT6. J Exp Clin Cancer Res. 2019;38(1):465 doi:10.1186/s13046-019-1428-0 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou H, Xiong Y, Peng L, Wang R, Zhang H, Fu Z. LncRNA-cCSC1 modulates cancer stem cell properties in colorectal cancer via activation of the Hedgehog signaling pathway. J Cell Biochem. 2020;121(3):2510–2524. doi:10.1002/jcb.29473 (2020) [DOI] [PubMed] [Google Scholar]
- 10. Sporn MB, Roberts AB, Roche NS, Kagechika H, Shudo K. Mechanism of action of retinoids. J Am Acad Dermatol. 1986;15(4 pt 2):756–764. doi:10.1016/s0190-9622(86)70231-4 (1986) [DOI] [PubMed] [Google Scholar]
- 11. Huo X, Han S, Wu G, et al. Dysregulated long noncoding RNAs (lncRNAs) in hepatocellular carcinoma: implications for tumorigenesis, disease progression, and liver cancer stem cells. Mol Cancer. 2017;16(1):165 doi:10.1186/s12943-017-0734-4 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang Y, He Y, Zhang P, et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17(1):77 doi:10.1186/s12943-018-0825-x (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939 mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50(3):992–1008. doi:10.4143/crt.2017.226 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yan Y, Wang Z, Qin B. A novel long noncoding RNA, LINC00483 promotes proliferation and metastasis via modulating of FMNL2 in CRC. Biochem Biophys Res Commun. 2019;509(2):441–447. doi:10.1016/j.bbrc.2018.12.090 (2019) [DOI] [PubMed] [Google Scholar]
- 15. Shademan M, Salanghuch AN, Zare K, et al. Expression profile analysis of two antisense lncRNAs to improve prognosis prediction of colorectal adenocarcinoma. Cancer Cell Int. 2019;19(1):278 doi:10.1186/s12935-019-1000-1 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iguchi T, Uchi R, Nambara S, et al. A long noncoding RNA, lncRNA-ATB, is involved in the progression and prognosis of colorectal cancer. Anticancer Res. 2015;35(3):1385–1388. [PubMed] [Google Scholar]
- 17. Zhang G, Han G, Zhang X, et al. Long non-coding RNA FENDRR reduces prostate cancer malignancy by competitively binding miR-18a-5p with RUNX1. Biomarkers. 2018;23(5):435–445. doi:10.1080/1354750X.2018.1443509 (2018) [DOI] [PubMed] [Google Scholar]
- 18. Gong F, Dong D, Zhang T, Xu W. Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR. Eur J Pharmacol. 2019;853:345–352. doi:10.1016/j.ejphar.2019.04.022 (2019) [DOI] [PubMed] [Google Scholar]
- 19. Liu J, Du W. LncRNA FENDRR attenuates colon cancer progression by repression of SOX4 protein. Onco Targets Ther. 2019;12:4287–4295. doi:10.2147/OTT.S195853 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo T, Zhao J, Lu Z, et al. Characterization of long non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in diffuse gastric cancer. Transl Oncol. 2018;11(5):1080–1089. doi:10.1016/j.tranon.2018.06.007 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Y, Zhang W, Liu P, et al. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. Onco Targets Ther. 2018;11:1403–1412. doi:10.2147/OTT.S149511 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng L, Xiang C, Li X, et al. STARD13-correlated ceRNA network-directed inhibition on YAP/TAZ activity suppresses stemness of breast cancer via co-regulating Hippo and Rho-GTPase/F-actin signaling. J Hematol Oncol. 2018;11(1):72 doi:10.1186/s13045-018-0613-5 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhan Y, Chen Z, Li Y, et al. Long non-coding RNA DANCR promotes malignant phenotypes of bladder cancer cells by modulating the miR-149/MSI2 axis as a ceRNA. J Exp Clin Cancer Res. 2018;37(1):273 doi:10.1186/s13046-018-0921-1 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu TP, Huang MD, Xia R, et al. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63 doi:10.1186/s13045-014-0063-7 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y, Yang B, Su Y, Xiang Q, Li Q. Downregulation of long noncoding RNA LINC00683 associated with unfavorable prognosis in prostate cancer based on TCGA. J Cell Biochem. 2019;120(8):14165–14174. doi:10.1002/jcb.28691 (2019) [DOI] [PubMed] [Google Scholar]
- 26. Lyu L, Xiang W, Zhu JY, et al. Integrative analysis of the lncRNA-associated ceRNA network reveals lncRNAs as potential prognostic biomarkers in human muscle-invasive bladder cancer. Cancer Manag Res. 2019;11:6061–6077. doi:10.2147/CMAR.S207336 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bai Y, Long J, Liu Z, et al. Comprehensive analysis of a ceRNA network reveals potential prognostic cytoplasmic lncRNAs involved in HCC progression. J Cell Physiol. 2019;234(10):18837–18848. doi:10.1002/jcp.28522 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, Supplemental_file for FENDRR Sponges miR-424-5p to Inhibit Cell Proliferation, Migration and Invasion in Colorectal Cancer by Chuan Cheng, Huixia Li, Jiujian Zheng, Jie Xu, Peng Gao and Jianping Wang in Technology in Cancer Research & Treatment