Abstract
Background:
Cancer stem cells (CSCs) are considered the main cause of cancer recurrence and metastasis, and DNA methylation is involved in the maintenance of CSCs. However, the methylation profile of esophageal CSCs remains unknown.
Methods:
Side population (SP) cells were isolated from esophageal squamous cell carcinoma (ESCC) cell lines KYSE150 and EC109. Sphere-forming cells were collected from human primary esophageal cancer cells. SP cells and sphere-forming cells were used as substitutes for cancer stem-like cells. We investigated the genome-wide DNA methylation profile in esophageal cancer stem-like cells using reduced representation bisulfite sequencing (RRBS).
Results:
Methylated cytosine (mC) was found mostly in CpG dinucleotides, located mostly in the intronic, intergenic, and exonic regions. Forty intersected differentially methylated regions (DMRs) were identified in these 3 groups of samples. Thirteen differentially methylated genes with the same alteration trend were detected; these included OTX1, SPACA1, CD163L1, ST8SIA2, TECR, CADM3, GRM1, LRRK1, CHSY1, PROKR2, LINC00658, LOC100506688, and NKD2. DMRs covering ST8SIA2 and GRM1 were located in exons. These differentially methylated genes were involved in 10 categories of biological processes and 3 cell signaling pathways.
Conclusions:
When compared to non-CSCs, cancer stem-like cells have a differential methylation status, which provides an important biological base for understanding esophageal CSCs and developing therapeutic targets for esophageal cancer.
Keywords: esophageal cancer, cancer stem cells, methylome, RRBS, DMR
Introduction
Esophageal squamous cell carcinoma (ESCC) is the third most common cancer and the fourth leading cause of cancer-related deaths in China.1,2 Although the treatment of ESCC has improved, the prognosis for esophageal cancer is still very poor owing to the high recurrence and metastasis rates.3 Therefore, development of new therapeutic methods is crucial. Cancer stem cells (CSCs) are a small population of cells in tumors with stem-cell-like properties. CSCs exhibit higher resistance to anti-tumor therapies compared to non-CSCs and they play an important role in tumor recurrence and metastasis.
CSCs were first identified in hematological malignancies,4 but subsequently, they were isolated from many solid tumors, such as lung cancer, breast cancer, and so on.5-9 In our previous studies, we identified that SP cells, sorted using Hoechst 33342 from ESCC cells, overexpressed several stem cell-related genes and shared certain common features of CSCs, such as the ability of self-renewal and tumorigenesis, in vivo.10,11 We also found that sphere-forming cells isolated from human primary ESCC cells overexpressed stem cell-related genes.12 Based on our previous research, we demonstrated that SP and sphere-forming cells were enriched cancer stem-like cells in ESCC cell lines.
DNA methylation also plays an important role in maintaining pluripotency and regulating the differentiation of CSCs. In breast cancer, putative breast CSCs exhibited altered methylation levels in several genes compared to those in the parental tumor cells.13 In hepatocellular carcinoma, SP cells exhibited a differential DNA methylation status when compared to the non-SP cells. The genes regulating embryonic development were hypermethylated in SP cells. The methylation and the consequent silencing of these genes may play an important role in the maintenance of stem-like properties in SP cells.14 However, the methylation profile of esophageal CSCs has not been elucidated.
In this study, we investigated the genome-wide DNA methylation profiles of ESCC-CSCs by RRBS. Our results would advance the knowledge and understanding of the ESCC-CSC methylome.
Methods and Materials
Cell Culture
Human esophageal squamous cell carcinoma cell lines EC9706 and KYSE150 were used in this study. The EC9706 cell line was established and studied by Han et al,15 whereas the KYSE150 cell line was kindly provided by Dr. Shimada (First Department of Surgery, Faculty of Medicine, Kyoto University, Japan). EC9706 and KYSE150 were cultured in RPMI 1640 and DMEM, (BIOROC, Tianjin, China) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA), respectively. S1 primary ESCC cells were isolated from ESCC tissue specimens in our laboratory11 and cultured in K-SFM medium (Gibco, Grand Island, NY, USA). All the cells were routinely incubated at 37°C in a humidified atmosphere of 5% CO2.
SP Cell Sorting by Fluorescence Activated Cell Sorting (FACS)
SP cell sorting was performed as previously described.16 KYSE150 and EC109 cells were incubated with Hoechst 33342 (Sigma-Aldrich, St. Louis, MO, USA) at a final concentration of 5 μg/ml and 7.5 μg/ml, respectively.
Tumor Sphere Culture
Sphere-forming cells were cultured by seeding 2500 cells into 6-well low-adherence plates, culturing with non-FBS DMEM, supplemented with 10 ng/ml basic fibroblast growth factor (PeproTech, Rocky Hill, NJ, USA), 20 ng/ml epidermal growth factor (PeproTech, Rocky Hill, NJ, USA), 5 μg/ml insulin (Sigma-Aldrich, St. Louis, MO, USA), and 20 μl/ml B27 supplement (Invitrogen, Carlsbad, CA, USA). After 14 days of incubation, the spheres were harvested. The sphere-forming cells were dissociated with TrypLE (Gibco, Grand Island, NY, USA) and washed with phosphate buffer saline.
DNA Extraction
1 × 106 cells were used for DNA extraction using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. DNA was quantified using ND-100 NanoDrop and agarose gel electrophoresis.
RRBS Library and Sequencing
The library construction and sequencing services were provided by RiboBio Co., Ltd. (Guangzhou, China) based on previously published RRBS studies.17 40-220 bp fragment sizes were selected to build the RR genome. The genome was separated into 9 categories, which were exonic, intronic, ncRNA (non-coding RNA), splicing, 3’UTR, 5’UTR, upstream (2 kb upstream region of the transcription start site), downstream (2 kb downstream region of the transcription end site), and intergenic regions (the regions excluded from the other categories). CGIs were described as regions >200 bp in length, with a C and G percentage >0.5. The +/− 2 kb regions flanking the CGIs were defined as CGI shores, and the +/− 2 kb regions flanking the CGI shores were defined as CGI shelves. Each samples wasere tsetedtested twice.
RRBS Data Analysis
The methylation level of a cytosine was calculated as the methylated reads of this cytosine divided by the total covered reads. For CpG sites, reads from both strands were combined to calculate the methylation levels. swDMR software was used to identify the differentially methylated regions (DMRs). The DMRs, whose methylation levels varied more than 20% and the P value was smaller than 0.05 (|Δβ|≥20%, P < 0.05), were selected.
The Gene Ontology (GO) project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). The ontology covers 3 domains: biological process, cellular components, and molecular functions. Genes that exhibit at least 1 DMR were subjected to the GO enrichment analysis. Pathway analysis is a functional analysis mapping genes to the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database (http://www.genome.jp/kegg/pathway.html).
Results
Isolation of ESCC-CSCs
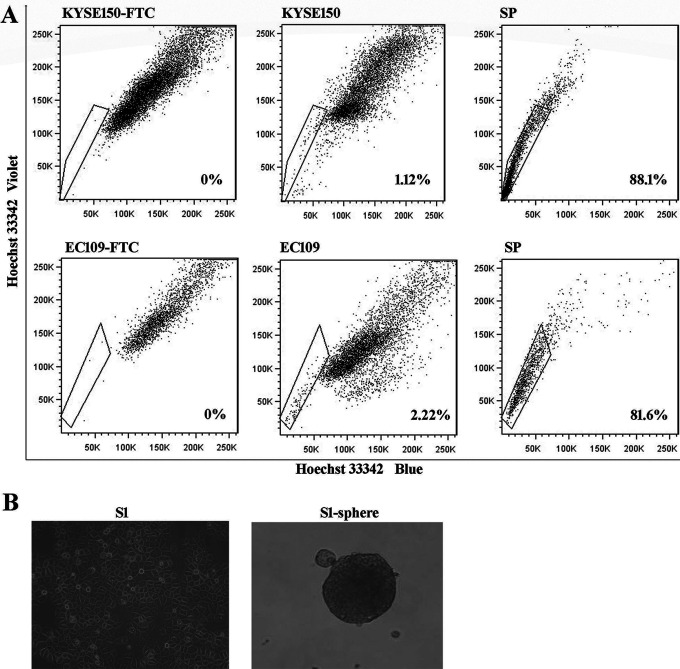
SP cells were isolated using Hoechst 33342 staining, through the FACS technique. The proportion of SP cells was 1.12% in KYSE150 and 2.22% in EC109 ESCC cell lines. The purity of the sorted SP cell population was estimated to be 88.1% for KYSE150 and 81.6% for EC109 (Figure 1A). Tumor sphere culture represents a surrogate in vitro model to study CSCs. We cultured tumor sphere cells(S1-sphere) of primary ESCC cells derived from ESCC patient lesions(S1) using non-adherent conditions (Figure 1B). The sphere formation ratio was nearly 1%. SP cells and sphere-forming cells were enriched for cancer stem-like cells in ESCC cells.
Figure 1.
Isolation of esophageal cancer stem cells. A. Side population (SP) analysis in KYSE150 and EC109 cell lines. B. Tumor spheres (S1-sphere) formed by primary ESCC cells (S1).
Methylation of the Captured CpGs
Each sample generated more than 34.73 million paired reads. After quality control of the raw data, each library produced approximately 3 Gb of clean data for further analysis.
Methylated cytosine (mC) was mainly located in CpG dinucleotides, followed by CHH and CHG. Approximately, more than 70% of the cytosine residues in CpG dinucleotides were methylated in all samples, and 3-10% of the cytosines were methylated in CHG and CHH. In KYSE150 cell lines, the methylation level of CG in SP cells was lower than that in NSP cells (76.22% vs 79.22%). However, in EC109 cell lines, the methylation level of CG in SP cells was higher than that in NSP cells (77.96% vs 71.76%). The methylation level of sphere-forming cells was lower than that of the parental primary S1 cells (67.53 vs 84.39%). The methylation levels of CGIs and promoters were similar to those of the RR genome (Table 1).
Table 1.
Methylation Levels of the RR Genome, CGIs, and Promoters.
| Samples | Pattern | CG (%) | CHG (%) | CHH (%) |
|---|---|---|---|---|
| KYSE150-NSP | RR genome methylation level | 79.22 | 5.15 | 4.84 |
| CGI methylation level | 70.66 | 4.55 | 4.90 | |
| Promoter methylation level | 56.30 | 4.60 | 4.51 | |
| KYSE150-SP | RR genome methylation level | 76.22 | 4.92 | 4.62 |
| CGI methylation level | 67.90 | 4.12 | 4.48 | |
| Promoter methylation level | 53.35 | 4.16 | 4.04 | |
| EC109-NSP | RR genome methylation level | 71.76 | 3.76 | 3.81 |
| CGI methylation level | 59.39 | 3.07 | 3.17 | |
| Promoter methylation level | 42.04 | 3.12 | 3.30 | |
| EC109-SP | RR genome methylation level | 77.96 | 3.88 | 3.92 |
| CGI methylation level | 66.66 | 3.26 | 3.35 | |
| Promoter methylation level | 50.97 | 3.34 | 3.49 | |
| S1 | RR genome methylation level | 84.39 | 32.16 | 39.45 |
| CGI methylation level | 72.73 | 31.83 | 29.07 | |
| Promoter methylation level | 56.79 | 35.42 | 41.06 | |
| S1-sphere | RR genome methylation level | 67.53 | 7.84 | 8.34 |
| CGI methylation level | 56.58 | 7.11 | 6.77 | |
| Promoter methylation level | 35.44 | 7.01 | 8.11 |
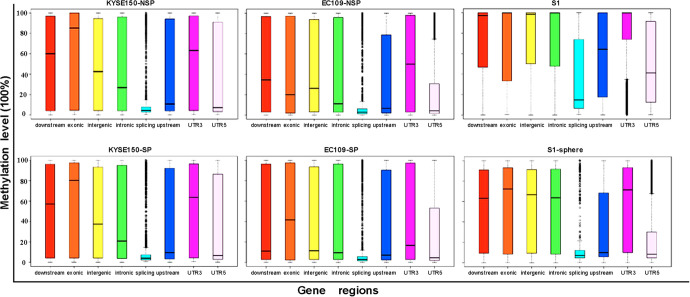
We studied the location of mC by ANNOVAR software. In both cell lines as well as the primary ESCC S1 cells, mC was mainly located in intronic, intergenic, and exonic regions (Figure 2).
Figure 2.
Methylation level at different gene locations.
Selection of DMRs
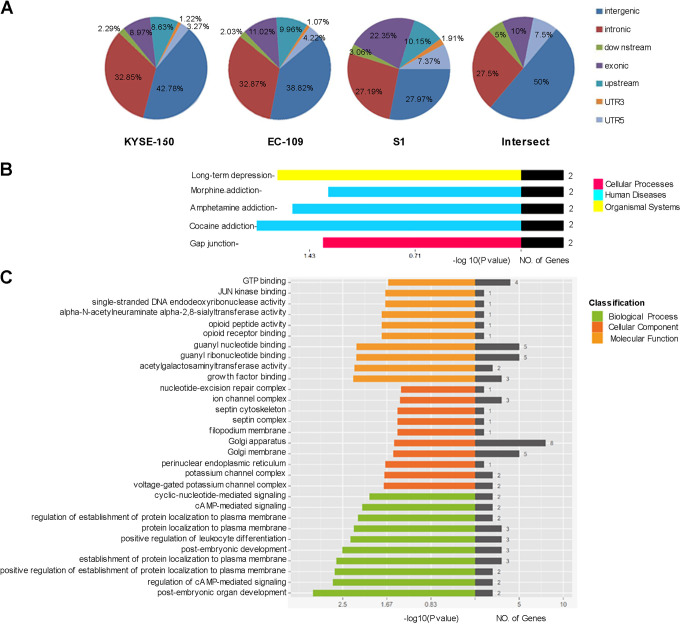
Compared to the respective NSP cells, KYSE150 SP cells had 13,465 hypermethylated DMRs and 43,190 hypomethylated DMRs, while EC109 SP cells had 8,581 hypermethylated DMRs and 7,892 hypomethylated DMRs. Compared with parental primary S1 cells, S1-sphere cells had 16,642 hypermethylated DMRs and 48,893 hypomethylated DMRs. The DMRs occurred predominantly in intergenic (42.78%, 32.32%, 27.97%) and intronic regions (32.85%, 32.87%, 27.19%) and less frequently in the 3’UTR (1.22%, 1.07%, 1.19%). Furthermore, 40 overlapping DMRs were detected among these 3 groups, frequently located in intergenic (50%), intronic (27.5%), exonic (10%), 5’UTR (7.5%) and downstream (5%) regions (Figure 3A).
Figure 3.
Differentially methylated regions (DMRs) analysis. A. The distribution of DMRs. B. The GO analysis of intersect DMRs in these 3 groups of specimens. C. The KEGG analysis of intersected DMRs in these 3 groups.
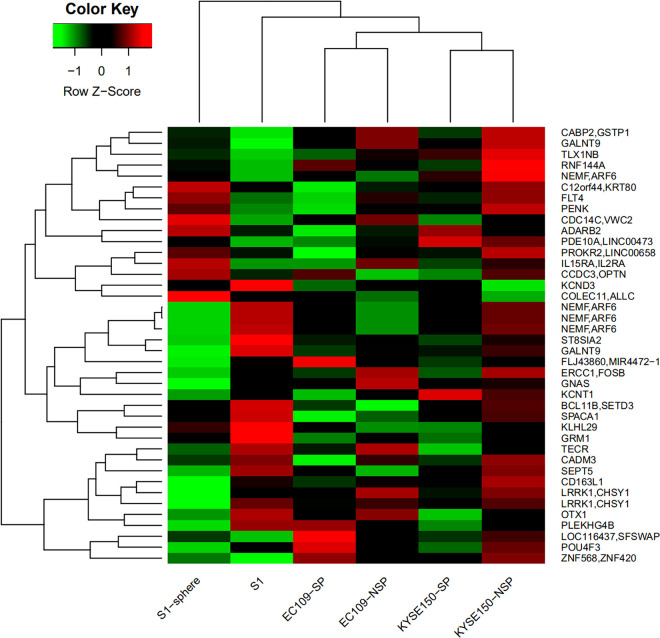
These 40 overlapping DMRs covered 53 genes (Figure 4). Among these overlapping DMRs, 11 DMRs exhibited the same alteration trend, which were hypomethylated in SP cells and sphere cells represented as cancer stem-like cells. These 11 DMRs covered 13 genes, including OTX1, SPACA1, CD163L1, ST8SIA2, TECR, CADM3, GRM1, LRRK1, CHSY1, PROKR2, LINC00658, LOC100506688, and NKD2. DMRs covering genes ST8SIA2 and GRM1 were located in the exonic region.
Figure 4.
The genes were covered by 40 over-lapped DMRs in these 3 groups. The red represents hypermethylated, and the green represents hypomethylated.
Functional and Signaling Pathway Analysis of Differentially Methylated Genes
According to the GO analysis, the differentially methylated genes were involved in 10 categories of biological processes, including post-embryonic organ development, regulation of cAMP-mediated signaling, positive regulation of establishment of protein localization to plasma membrane, and establishment of protein localization to plasma membrane; 6 categories in the domain of cellular components, including Golgi apparatus, Golgi membrane, and ion channel complex; 5 categories in the domain of molecular functions, such as guanyl nucleotide binding (Figure 3B). KEGG analysis showed that the overlapping differentially methylated genes were mainly involved in long-term depression gap junction and drug addiction such as cocaine addiction (Figure 3C).
Analysis of the Hypomethylated Genes using the Gene Expression Profiling Interactive Analysis (GEPIA) Database
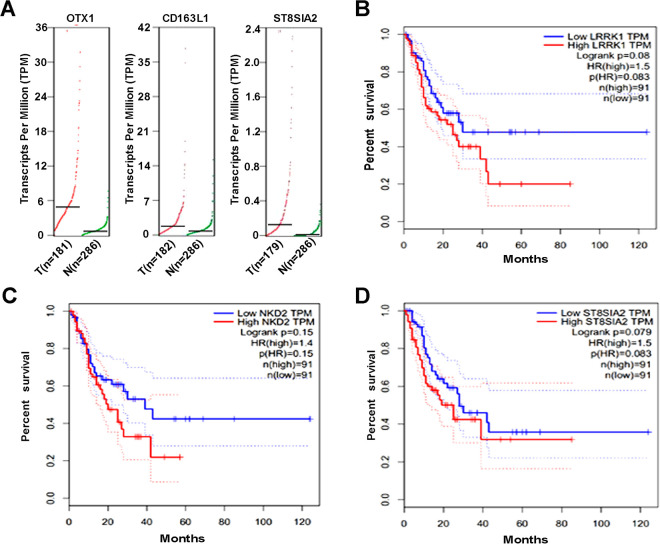
We investigated the mRNA expression of the hypomethylated genes using the online tools at GEPIA. mRNA expression levels of OTX1, CD163L1, and ST8SIA2 were higher in esophageal cancer tissues than in normal tissues (Figure 5A). Esophageal carcinoma patients with high levels of LRRK1, NKD2, or ST8SIA2 exhibited a shorter disease-free survival, but there was no statistically significant difference (Figure 5B-5C). None of these 13 genes was associated with overall survival .
Figure 5.
Analysis of the hypomethylated genes using GEPIA. A. The mRNA expression of OTX1, CD163L1, and ST8SIA2 in esophageal cancer. B. Expression of LRRK1, NKD2, or ST8SIA2 is correlated with poor diseasefree survival in ESCC.
Discussion
Cancer is most likely a disease of stem cells. More and more studies suggest that CSCs play an important role in the initiation and progression of tumors.18,19 Therefore, CSCs might be the ultimate target in cancer therapy. CSCs have many stem cell properties, such as self-renewal and multi-differentiation abilities. However, the mechanism underlying the maintenance of these properties remains unclear.
DNA methylation is one of the most important epigenetic mechanisms that play a significant role in regulating the differentiation of CSCs.20,21 Recent studies found that CSCs had a methylome different from that of non-CSCs. In breast cancer, putative breast CSCs showed altered methylation levels of several genes compared to parental tumor cells.13 Zhai also revealed that hepatocellular carcinoma SP cells possessed a differential DNA methylation status in comparison to the NSP cells.14 However, to date, the DNA methylation features of esophageal CSCs have not been documented.
RRBS is a cost-effective method to generate genome-wide DNA methylation profiles at single-nucleotide resolution.22 We found CSCs and non-CSCs have the same methylation distribution in ESCC cell lines using the RRBS analysis. mC was mainly located in CpG dinucleotides, and more than 70% of the cytosines in CpG dinucleotides were methylated. The distribution of mC is mainly in the intronic, intergenic, and exonic regions, in both CSCs and non-CSCs. CpG sites played important roles in regulating gene expression and in mammalian evolution.23 The methylation of CpG islands located in promoters is associated with transcriptional silencing.24 In addition, CGIs in the body of genes can act as alternative promoters or enhancers and display tissue-specific methylation.25,26 In all ESCC samples, more than 50% of cytosines in CGIs were methylated, which indicates that methylation plays an important role in ESCC.
CSCs and non-CSCs have different methylation levels. Many DMRs were detected in our 3 experimental groups. Among these 3 groups, 40 DMRs were overlapping. Eleven DMRs covering 13 genes were hypomethylated in CSCs, indicating that these genes may be highly expressed in CSCs.
Among the 13 differentially methylated genes, ST8SIA2 attracted our attention. The protein encoded by this gene is a type II membrane protein, which functions in the attachment of polysialic acid (PSA) to the neural cell adhesion molecule (NCAM).27,28 Re-expression of PSA-NCAM and its transferases in tumors is closely related to tumor progression and poor prognosis of patients.29,30 Overexpression of ST8SIA2 in small cell lung cancer (SCLC) cells enhances the invasion and migration abilities of SCLC cells.31 In our study, we found that ST8SIA2 is hypomethylated in ESCC-CSCs. GEPIA analysis revealed that the expression of ST8SIA2 in esophageal cancer tissues was higher than that in normal tissues and that it is correlated with poor disease-free survival. However, the expression and function of ST8SIA2 in CSCs of ESCC need further analysis.
Our study revealed that the cancer stem-like cells of ESCC possessed a differential DNA methylation pattern compared to non-CSCs. These differentially methylated genes may contribute to maintenance of stem-like properties in cancer stem-like cells. To our knowledge, this is the first study to analyze the genome-wide DNA methylome of ESCC-CSCs. Our findings provide valuable clues for further investigations of epigenetic regulation in sustaining ESCC-CSCs.
Abbreviations
- CSCs
cancer stem cells
- ESCC
esophageal squamous cell carcinoma
- 3′UTR
3′untranslated region
- 5′UTR
5′untranslated region
- CDS
coding sequence
- CGIs
CpG islands
- RRBS
reduced representation bisulfite sequencing
- DMR
differentially methylated region
Footnotes
Authors’ Note: Xiying Yu and Ying Teng contributed equally to this work both as the first authors. The data sets used or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contribution: Wei Jiang was involved in the conception and design of the study. Xiying Yu and Ying Teng performed the cellular and molecular experiments. Xingran Jiang and Hui Yuan assisted with the statistical analysis. Xiying Yu and Ying Teng were major contributors in drafting the initial manuscript.
Ethical Statement: Our study did not require an ethical board approval because it did not involve human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article: This study was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (Grant no. 2016-I2M-1-001), National Natural Science Foundation of China (81972572, 81602138).
ORCID iD: Wei Jiang  https://orcid.org/0000-0002-1527-2856
https://orcid.org/0000-0002-1527-2856
References
- 1. Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349(23):2241–2252. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 3. Gamliel Z, Krasna MJ. Multimodality treatment of esophageal cancer. Surg Clin North Am. 2005;85(3):621–630. [DOI] [PubMed] [Google Scholar]
- 4. Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. [DOI] [PubMed] [Google Scholar]
- 5. Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121(6):823–835. [DOI] [PubMed] [Google Scholar]
- 6. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23(43):7274–7282. [DOI] [PubMed] [Google Scholar]
- 8. Woodward WA, Chen MS, Behbod F, Rosen JM. On mammary stem cells. J Cell Sci. 2005;118(Pt 16):3585–3594. [DOI] [PubMed] [Google Scholar]
- 9. Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311(5769):1880–1885. [DOI] [PubMed] [Google Scholar]
- 10. Huang D, Gao Q, Guo L, et al. Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev. 2009;18(3):465–473. [DOI] [PubMed] [Google Scholar]
- 11. Li H, Gao Q, Guo L, Lu SH. The PTEN/PI3K/Akt pathway regulates stem-like cells in primary esophageal carcinoma cells. Cancer Biol Ther. 2011;11(11):950–958. [DOI] [PubMed] [Google Scholar]
- 12. Harris RA, Wang T, Coarfa C, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balic M, Schwarzenbacher D, Stanzer S, et al. Genetic and epigenetic analysis of putative breast cancer stem cell models. BMC Cancer. 2013;13:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhai JM, Yin XY, Hou X, et al. Analysis of the genome-wide DNA methylation profile of side population cells in hepatocellular carcinoma, Dig Dis Sci. 2013;58(7):1934–1947. [DOI] [PubMed] [Google Scholar]
- 15. Han Y, Wei F, Xu X, et al. [Establishment and comparative genomic hybridization analysis of human esophageal carcinomas cell line EC9706]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19(6):455–457. [PubMed] [Google Scholar]
- 16. Yu X, Jiang X, Li H, Guo L, Jiang W, Lu SH. miR-203 inhibits the proliferation and self-renewal of esophageal cancer stem-like cells by suppressing stem renewal factor Bmi-1. Stem Cells Dev. 2014;23(6):576–585. [DOI] [PubMed] [Google Scholar]
- 17. Yuan XL, Gao N, Xing Y, et al. Profiling the genome-wide DNA methylation pattern of porcine ovaries using reduced representation bisulfite sequencing. Sci Rep. 2016;6:22138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Spillane JB, Henderson MA. Cancer stem cells: a review. ANZ J Surg. 2007;77(6):464–468. [DOI] [PubMed] [Google Scholar]
- 19. Ishii H, Iwatsuki M, Ieta K, et al. Cancer stem cells and chemoradiation resistance. Cancer Sci. 2008;99(10):1871–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Broske AM, Vockentanz L, Kharazi S, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41(11):1207–1215. [DOI] [PubMed] [Google Scholar]
- 21. Trowbridge JJ, Orkin SH. Dnmt3a silences hematopoietic stem cell self-renewal. Nat Genet. 2011;44(1):13–14. [DOI] [PubMed] [Google Scholar]
- 22. Chatterjee A, Rodger EJ, Stockwell PA, Weeks RJ, Morison IM. Technical considerations for reduced representation bisulfite sequencing with multiplexed libraries. J Biomed Biotechnol. 2012:741542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103(5):1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lou S, Lee HM, Qin H, et al. Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol. 2014;15(7):408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SM, Lee YG, Bae JB, et al. HBx induces hypomethylation of distal intragenic CpG islands required for active expression of developmental regulators. Proc Natl Acad Sci U S A. 2014;111(26):9555–9560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janas T. Membrane oligo- and polysialic acids. Biochim Biophys Acta. 2011;1808(12):2923–2932. [DOI] [PubMed] [Google Scholar]
- 28. Foley DA, Swartzentruber KG, Colley KJ. Identification of sequences in the polysialyltransferases ST8Sia II and ST8Sia IV that are required for the protein-specific polysialylation of the neural cell adhesion molecule. NCAM. J Biol Chem. 2009;284(23):15505–15516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Falconer RA, Errington RJ, Shnyder SD, Smith PJ, Patterson LH. Polysialyltransferase: a new target in metastatic cancer. Curr Cancer Drug Targets. 2012;12(8):925–939. [DOI] [PubMed] [Google Scholar]
- 30. Al-Saraireh YM, Sutherland M, Springett BR, et al. Pharmacological inhibition of polysialyltransferase ST8SiaII modulates tumour cell migration. PLoS One. 2013;8(8):e73366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gong L, Zhou X, Yang J, Jiang Y, Yang H. Effects of the regulation of polysialyltransferase ST8SiaII on the invasiveness and metastasis of small cell lung cancer cells. Oncol Rep. 2017;37(1):131–138. [DOI] [PubMed] [Google Scholar]