PURPOSE
In previous analyses of the MURANO study, fixed-duration venetoclax plus rituximab (VenR) resulted in improved progression-free survival (PFS) compared with bendamustine plus rituximab (BR) in patients with relapsed or refractory chronic lymphocytic leukemia (CLL). At the 4-year follow-up, we report long-term outcomes, response to subsequent therapies, and the predictive value of molecular and genetic characteristics.
PATIENTS AND METHODS
Patients with CLL were randomly assigned to 2 years of venetoclax (VenR for the first six cycles) or six cycles of BR. PFS, overall survival (OS), peripheral-blood minimal residual disease (MRD) status, genomic complexity (GC), and gene mutations were assessed.
RESULTS
Of 389 patients, 194 were assigned to VenR and 195 to BR. Four-year PFS and OS rates were higher with VenR than BR, at 57.3% and 4.6% (hazard ratio [HR], 0.19; 95% CI, 0.14 to 0.25), and 85.3% and 66.8% (HR, 0.41; 95% CI, 0.26 to 0.65), respectively. Undetectable MRD (uMRD) at end of combination therapy (EOCT) was associated with superior PFS compared with low MRD positivity (HR, 0.50) and high MRD positivity (HR, 0.15). Patients in the VenR arm who received ibrutinib as their first therapy after progression (n = 12) had a reported response rate of 100% (10 of 10 evaluable patients); patients subsequently treated with a venetoclax-based regimen (n = 14) had a reported response rate of 55% (six of 11 evaluable patients). With VenR, the uMRD rate at end of treatment (EOT) was lower in patients with GC than in those without GC (P = .042); higher GC was associated with shorter PFS. Higher MRD positivity rates were seen with BIRC3 and BRAF mutations at EOCT and with TP53, NOTCH1, XPO1, and BRAF mutations at EOT.
CONCLUSION
Efficacy benefits with fixed-duration VenR are sustained and particularly durable in patients who achieve uMRD. Salvage therapy with ibrutinib after VenR achieved high response rates. Genetic mutations and GC affected MRD rates and PFS.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) remains largely incurable despite recent therapeutic advances. Bendamustine plus rituximab (BR) was previously widely used as salvage therapy based on a high overall response rate and favorable overall survival (OS) data.1 Concurrently, the Bruton tyrosine kinase inhibitor (BTKi) ibrutinib was established as an effective monotherapy option in relapsed or refractory (R/R) CLL.2 However, neither of these regimens are curative, resulting in a patient population enriched for resistance mutations after disease progression (PD). Postprogression therapies include targeted agents such as BTKis or inhibitors of the phosphoinositide 3-kinase (PI3K) pathway. However, these require extended administration until PD or development of unacceptable toxicity.3 Fixed-duration therapies that achieve deep and durable clinical responses are attractive because they may limit the development of resistant subclones and therapy-related toxicity and reduce the financial burden of treatment.3
CONTEXT
Key Objective
This analysis of the phase III MURANO study investigated long-term follow-up, predictive biomarkers, and response to next-line therapy in patients with relapsed or refractory (R/R) chronic lymphocytic leukemia (CLL) treated with fixed-duration venetoclax plus rituximab (VenR) versus bendamustine plus rituximab (BR).
Knowledge Generated
Progression-free and overall survival remained superior with VenR at a median follow-up of 4 years; response rates to novel agents as subsequent therapy were high, including with ibrutinib after venetoclax. The rate of undetectable minimal residual disease at end of treatment was lower in patients with higher genomic complexity and mutations in TP53, NOTCH1, XPO1, and BRAF.
Relevance
These data show a sustained long-term benefit for fixed-duration VenR in R/R CLL and suggest that extended therapy would be unlikely to prevent early progression. Data for next-line therapy indicate the potential of either ibrutinib or venetoclax re-treatment as a treatment option after progression on VenR. The clinical relevance of the biomarker findings requires validation in further studies.
Venetoclax is a highly selective inhibitor of BCL-2, acting independently of TP534 to induce high response rates in patients with R/R CLL and those with traditionally poor prognostic features such as chromosome 17p deletion (del[17p]).5 MURANO (ClinicalTrials.gov identifier: NCT02005471) is an ongoing, global, phase III, open-label, randomized study investigating the efficacy and safety of fixed-duration venetoclax plus rituximab (VenR) therapy compared with BR in patients with R/R CLL.6,7
With chemoimmunotherapy (CIT), minimal residual disease (MRD) status at end of treatment (EOT) predicts clinical outcome; specifically, undetectable MRD (uMRD; < 1 CLL cell per 10,000 leukocytes) is consistently linked to more favorable progression-free survival (PFS) and OS.8 Clinically relevant biomarkers predictive of poor response to CIT include unmutated immunoglobulin heavy chain (IGHV) gene, del(17p), mutated TP53, and cytogenetic or genomic complexity (GC).8-11 Targeted therapies overcome the adverse impact of IGHV status,12,13 but the identification of biomarkers for targeted therapies is required to guide clinical practice. Specific biomarkers that might have predictive value with targeted agents as well as CIT are certain recurrent mutations and GC.10,11,14-20 How to define GC is currently under debate. It was recently found that presence of five or more, but not three or more, chromosomal aberrations is an independent prognostic factor with adverse outcome in CLL after chemotherapy.10,11 Whether this also holds true for targeted agents is currently unknown.
Here, we present a 4-year clinical update of MURANO that describes outcomes after venetoclax cessation and the impact of MRD status, including response to subsequent therapies. Furthermore, we explore the predictive value of baseline molecular characteristics in clinical responses to 2-year fixed-duration therapy with VenR.
PATIENTS AND METHODS
Study Design and Conduct
The study design and eligibility criteria have been published previously.6 The trial was conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonisation of Good Clinical Practice guidelines. Trial protocol approval was obtained from the ethics committee at each participating institution, and written informed consent to participate was provided by all patients. The data cutoff date was May 8, 2019, for PFS, OS, and safety and December 1, 2019, for response to subsequent therapy.
Treatment
Patients were randomly assigned (1:1) to receive either six 28-day cycles of VenR, followed by single-agent venetoclax (400 mg) once daily for a total of 2 years, or six 28-day cycles of standard BR. The 2-year venetoclax treatment period was calculated from day 1 of cycle 1 after venetoclax dose ramp-up. Information on dosing, prophylactic measures, and monitoring has previously been published.6 An optional re-treatment/crossover substudy was added as part of a protocol amendment activated in March 2018. Patients from either study arm with PD after completing treatment, who needed treatment and had not received further anti-CLL therapy, were eligible to receive VenR for an additional fixed duration (approximately 2 years).
Clinical Assessments
The primary efficacy end point was investigator-assessed PFS, defined as the time from randomization to first occurrence of PD, relapse, or death, whichever occurred first. MRD status (peripheral blood [PB] only) was assessed at cycle 4, 2 to 3 months after end of combination therapy (EOCT; secondary end point), and then every 3 to 6 months. Allele-specific oligonucleotide polymerase chain reaction and flow cytometry were used in the central analysis of serial PB MRD samples, as described previously.6,7 uMRD was defined as < 1 CLL cell/10,000 leukocytes (MRD value < 0.0001, or 10−4)8; low MRD positivity was defined as 10−4 to < 10−2, and high MRD positivity was defined as ≥ 10−2.
Other end points included OS, complete response (CR) and partial response (PR) rates (International Workshop on Chronic Lymphocytic Leukemia 2008 criteria21), and safety assessments. Safety data collected for the current analysis period were prespecified adverse events (AEs) of concern, serious AEs (SAEs) related to study drug, and development of a second primary malignancy.
Molecular Assessments
Whole-exome sequencing (WES) and analysis of GC by high-density array comparative genomic hybridization (aCGH) were performed on CD19-enriched baseline samples for 314 of 389 enrolled patients. GC analyses were performed at the Laboratory of Genome Diagnostics of the Amsterdam University Medical Centers, University of Amsterdam (Amsterdam, the Netherlands). WES was performed by EA Genomics using SureSelect (Agilent, Santa Clara, CA) using 100-bp paired-ends reading.
Data processing of WES included mapping to the National Center for Biotechnology Information (NCBI) Build 38 human reference genome using the Genomic Short-Read Nucleotide Alignment Program (GSNAP)22,23 and base quality score recalibration (Genome Analysis Toolkit [GATK] tools BaseRecalibrator and ApplyBQSR).24 Somatic variants were called using LoFreq25 and GATK Mutect2 followed by FilterMutectCalls. Variants were filtered by requiring calls from both LoFreq and Mutect2/FilterMutectCalls, excluding variants present in dbSNP (version b151) and those present in a panel of normals generated from 40 normal samples from patients with solid tumors. Variants were annotated according to Variant Effect Predictor (VEP) version 77.26 Final mutation calls only included predicted deleterious mutations called by Condel score.27 Herein, 47 genes commonly mutated in CLL28 were analyzed in relation to PFS and MRD status at EOCT and EOT (Data Supplement, online only).
aCGH was performed using commercially available 4×180K slides (AMADID 023363; Agilent) according to the manufacturer’s protocol, with NCBI Build 37 as reference genome. Spot intensities were measured with a filter of a minimum of 100 adjacent clones to define an aberration. Array-based GC status was defined by number of genomic aberrations, as follows: noncomplex, zero to two aberrations; low, three to four aberrations; or high, five or more aberrations.11
Statistical Analyses
There is no α spending allocated to the current analysis of end points; therefore, all P values are considered descriptive.7 Kaplan-Meier estimates were used to analyze time-to-event data, including landmark analyses from EOCT and EOT according to MRD status. The log-rank test and Cox proportional hazards regression model were used to compare overall PFS and OS across treatment arms. Fisher’s exact test was performed to compare MRD status at EOCT and EOT and clinical and cytogenetic risk factors in VenR-treated patients with and without PD after EOT. Multivariable analysis was used to determine the impact of gene mutations and GC on MRD and PFS at EOCT and EOT; covariates included IGHV status, Rai stage at baseline, TP53 mutation status and/or del(17p) by aCGH, fludarabine resistance status, and maximum nodal size > 10 cm. Analysis of the impact of TP53 mutation excluded TP53 genomic disruption and del(17p) as covariates.
RESULTS
Patients
In total, 389 patients were enrolled; 194 were assigned to receive VenR, and 195 were assigned to receive BR (Data Supplement). As reported previously, patients’ clinical characteristics were similar between study arms.6 The median duration of follow-up from study enrollment was 48 months.
PFS and OS
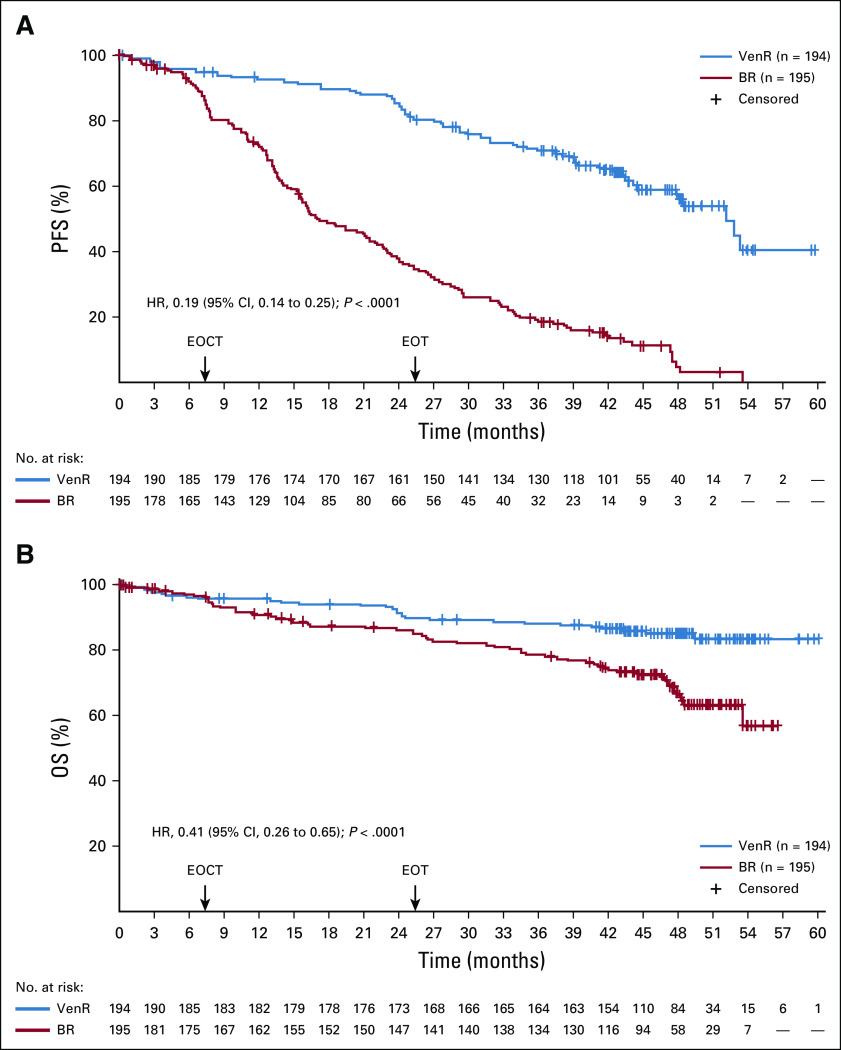
At the 4-year follow-up, the PFS benefit with VenR over BR remained (hazard ratio [HR], 0.19; 95% CI, 0.14 to 0.25; P < .0001; Fig 1). Four-year PFS estimates were 57.3% (95% CI, 49.4% to 65.3%) for VenR and 4.6% (95% CI, 0.1% to 9.2%) for BR. For patients who completed 2 years of venetoclax therapy without developing PD (n = 130; reasons for venetoclax discontinuation [n = 64]: AEs, 45.3% [n = 29]; PD, 34.4% [n = 22]; death, 3.1% [n = 2]; other, 17.2% [n = 11]7), the median follow-up after venetoclax completion was 22 months (range, 1-35 months). The PFS estimates 18 and 24 months after treatment cessation in these VenR patients were 75.5% (95% CI, 67.4% to 83.7%) and 68.0% (95% CI, 57.6% to 78.4%), respectively.
FIG 1.
Kaplan-Meier assessments of (A) progression-free survival (PFS) and (B) overall survival (OS). BR, bendamustine plus rituximab; EOCT, end of combination therapy; EOT, end of treatment; HR, hazard ratio; ITT, intent-to-treat; VenR, venetoclax plus rituximab.
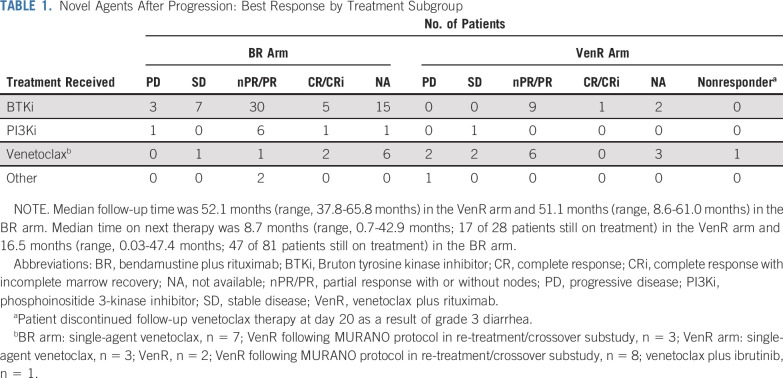
OS benefit with VenR versus BR remained (HR, 0.41; 95% CI, 0.26 to 0.65; P < .0001; Fig 1). Four-year OS rates were 85.3% with VenR and 66.8% with BR. This benefit with VenR was seen despite a high proportion of patients in the BR arm receiving novel targeted agents after PD (81 of 103 patients; 79%), including BTKis (n = 60), PI3K inhibitors (PI3Kis; n = 9), venetoclax (n = 10), or other investigational medicinal products (IMPs; n = 2). Twenty-eight (67%) of 42 patients in the VenR arm with subsequent therapy after PD received novel agents: BTKis (n = 12), PI3Kis (n = 1), reintroduction of venetoclax (n = 14), or IMPs (n = 1). Among patients treated with ibrutinib after venetoclax (n = 12), the response rate was 100% in evaluable patients (10 of 10 patients; all PRs). At a follow-up of 6.2-42.9 months, eight patients were still receiving ibrutinib therapy, and three patients had PD (two of three patients died as a result of PD; Table 1; Data Supplement). Among patients treated with a venetoclax-based regimen after venetoclax therapy (n = 14), the response rate was 55% (six of 11 evaluable patients; all PRs); two patients achieved stable disease, one was considered a nonresponder, and three had PD (Table 1).
TABLE 1.
Novel Agents After Progression: Best Response by Treatment Subgroup
MRD
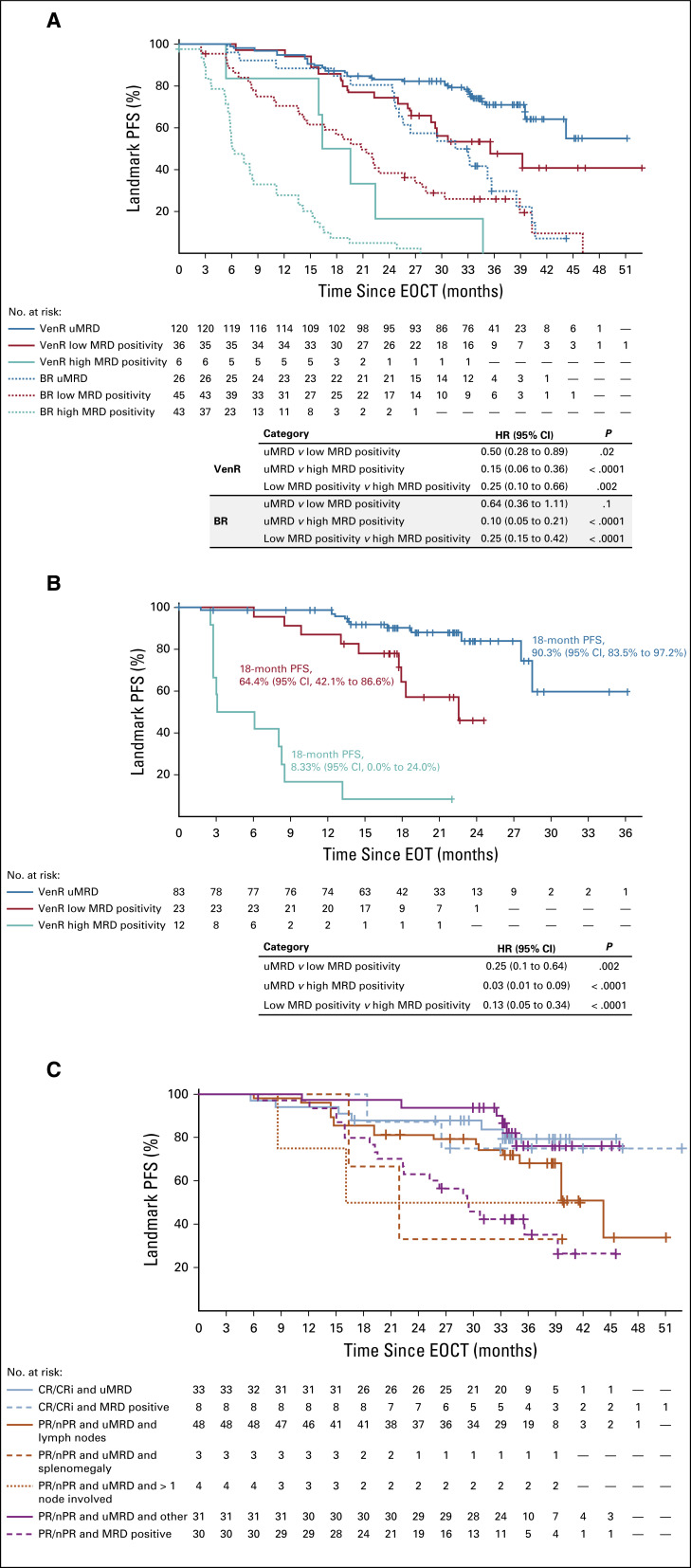
Patients achieving uMRD at EOCT had the most favorable outcome in both treatment arms, with HRs for PFS in the VenR arm of 0.50 (95% CI, 0.28 to 0.89) versus low MRD positivity and 0.15 (95% CI, 0.06 to 0.36) versus high MRD positivity (Fig 2A). During post-venetoclax follow-up in the VenR arm, with a median of 22 months off therapy, 73.1% of patients (95 of 130 patients) who completed 2 years of venetoclax without PD remained progression free (Data Supplement). PFS rates at 18 months after EOT were 90.3% (95% CI, 83.5% to 97.2%) in patients with uMRD, 64.4% (95% CI, 42.1% to 86.6%) in patients with low MRD positivity, and 8.3% (95% CI, 0.0% to 24.0%) in patients with high MRD positivity (MRD status at EOT; Fig 2B). Few patients with high MRD positivity at EOT ever achieved uMRD (three of 14 patients); most patients in this group demonstrated increasing PB MRD before venetoclax cessation (13 of 14 patients; Data Supplement).
FIG 2.
Landmark Kaplan-Meier analyses. (A) Progression-free survival (PFS) from end of combination therapy (EOCT) in both study arms based on minimal residual disease (MRD) status at EOCT. (B) PFS from end of treatment (EOT) in patients in the venetoclax plus rituximab (VenR) arm who completed 2 years of venetoclax, based on MRD status at EOT (excludes two patients who completed venetoclax but experienced disease progression before MRD measurement). (C) PFS from EOCT in the VenR arm based on MRD status at EOCT and involvement of lymph nodes. BR, bendamustine plus rituximab; CR, complete response; CRi, complete response with incomplete marrow recovery; nPR, nodular partial response; PR, partial response; uMRD, undetectable minimal residual disease.
VenR-treated patients who achieved a best response of PR solely as a result of residual lymph node enlargement but who achieved uMRD in PB at EOCT had PFS similar to that of patients attaining CR or CR with incomplete marrow recovery (CRi; Fig 2C). With this duration of follow-up, MRD status did not appear to affect PFS in patients achieving CR/CRi; all had either uMRD (n = 33) or low MRD positivity (n = 8).
Molecular Biomarkers and Clinical Outcomes
Baseline characteristics, uMRD status at EOCT, and efficacy outcomes in the subset of 314 patients in whom molecular assessments were performed were similar to those in the intent-to-treat population (Data Supplement). The number of events after VenR treatment cessation remained low; because MRD status proved to be a strong predictor of PD after treatment cessation, MRD status was used as a surrogate for PFS.
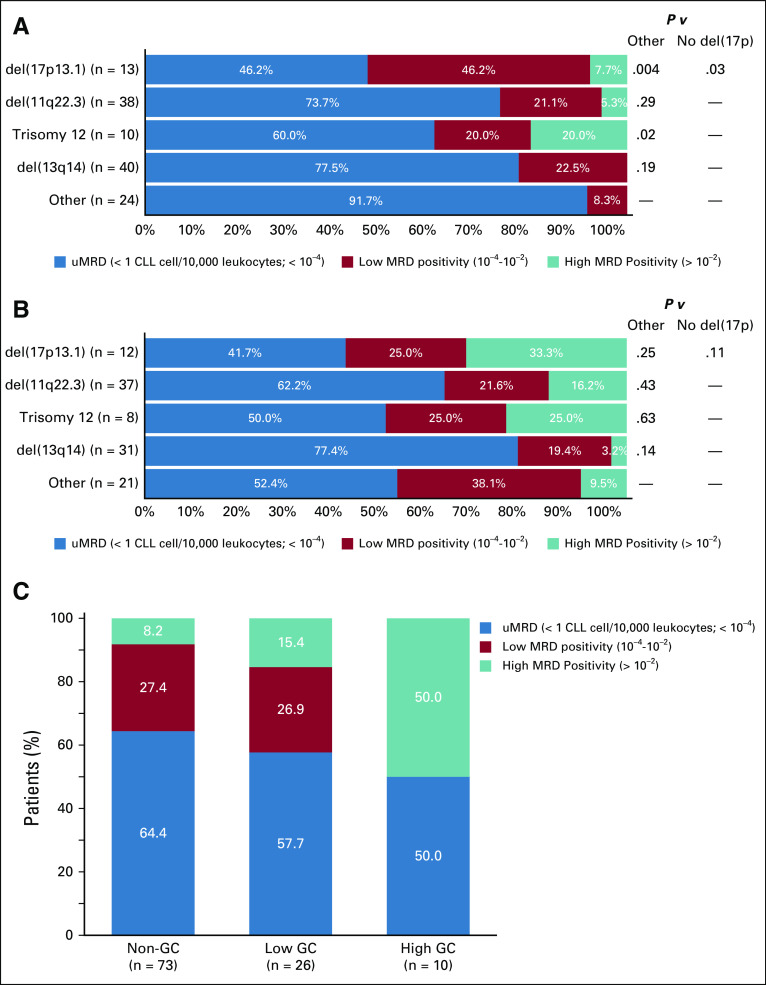
In this study, there was 83.5% concordance between detection of del(17p) by clinically applied aCGH (≥ 100 adjacent clones) and the fluorescence in situ hybridization (FISH) assay by central laboratory (Vysis CLL FISH Probe Kit, Abbott Laboratories, Abbott Park, IL) using a detection limit of 7% (P < .0001; Cohen’s κ = 0.502), with most of the discordance being a result of the detection of low-clone del(17p) by FISH that was not detected by aCGH (Data Supplement). Analysis revealed that patients with low-clone del(17p) by FISH had similar PFS to patients with normal 17p (Data Supplement); therefore, we used aCGH as the determinant of 17p status in this analysis. At baseline, major genomic aberrations were detected by aCGH in the VenR arm at the following rates: 44.4% del(13q14) monoallelic, 31.7% del(11q22.3), 16.9% del(13q14) biallelic, 12.0% del(17p13.1), and 9.9% trisomy 12 (Data Supplement). Most VenR patients (79 of 121 patients; 65.3%) had multiple abnormalities, whereas 42 (34.7%) of 121 patients harbored one aberration. In the VenR arm, MRD positivity (low or high MRD positivity) at EOCT was significantly more frequently observed using the Döhner hierarchical classification system29 in patients with del(17p) versus those without del(17p) (P = .031) or without any of the four major alterations (P = .004; Fig 3A). There was also an association between MRD-positive status at EOCT and trisomy 12 (P = .024), although this was based on data for only 10 patients. In contrast, no association with MRD status was observed at EOT for any of the four major cytogenetic alterations (biomarker-evaluable population; Fig 3B).
FIG 3.
Impact of genomic alterations on minimal residual disease (MRD) response in patients treated with venetoclax plus rituximab (VenR; biomarker-evaluable population [BEP]). MRD status at (A) end of combination therapy and (B) end of treatment (EOT) according to major cytogenetic alterations, using Döhner hierarchical classification. Missing values resulted from disease progression (n = 12), death (n = 11), missing visit (n = 8), or MRD technical issues (n = 2). Other group includes all patients not harboring one of the four named abnormalities; del13q14 group includes both mono- and biallelic deletions. P values were calculated using Fisher’s exact test. (C) MRD status at EOT according to genomic complexity (GC) status; samples with missing MRD values were not included in the BEP for this analysis. CLL, chronic lymphocytic leukemia.
Of 288 patients with GC data, 194 (67.3%) had noncomplex status (zero to two aberrations), 63 (21.9%) had low GC (three to four aberrations), and 31 (10.8%) had high GC (five or more aberrations). High- and low-GC status correlated with an increased frequency of high MRD positivity at EOT (P = .042; Fig 3C).
We assessed the association between baseline mutations in 47 commonly mutated genes28 (Fig 4A details mutations seen in ≥ 5% of patients in the VenR arm) and MRD status at EOCT and at EOT. The number of patients in the biomarker-evaluable population with high MRD positivity harboring mutations was low, especially at EOCT, because of the high response rates to VenR in this cohort, resulting in low statistical power for all analyses. Numerically lower uMRD rates were seen at EOCT in VenR patients with BRAF or BIRC3 mutations compared with wild-type patients (Data Supplement). As expected, a larger number of patients with high MRD positivity were present at the EOT time point, increasing the power of analyses at that time. Numerically lower uMRD rates at EOT were reported in VenR patients with the following mutated genes: TP53, NOTCH1, XPO1, and BRAF (Data Supplement). Overall, a reduced uMRD rate was observed for patients harboring one or more driver mutations in both the BR and VenR arms at EOCT, but this was not statistically significant (Data Supplement).
FIG 4.
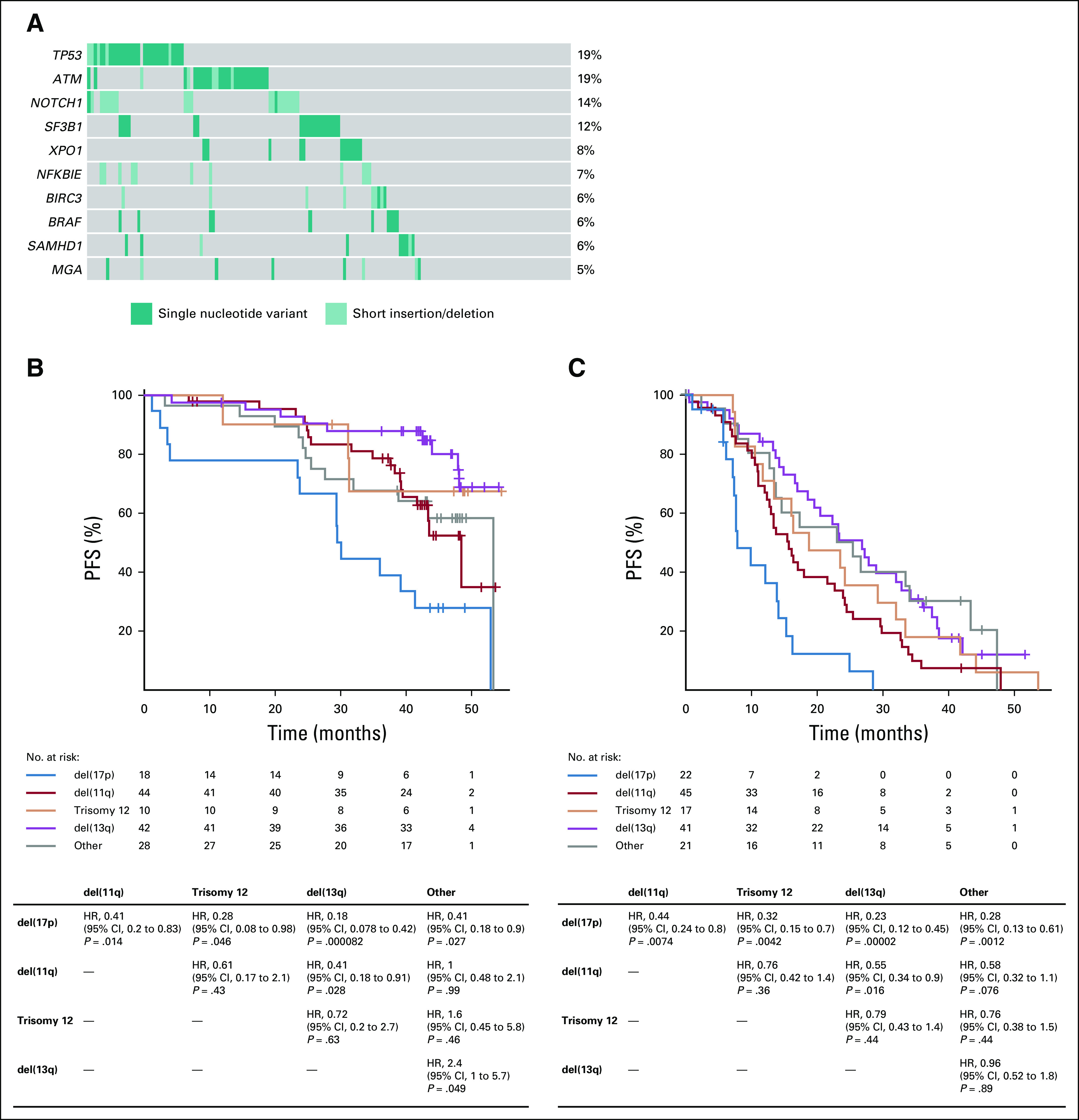
(A) Oncoprint of most frequently mutated genes (≥ 5%) in the venetoclax plus rituximab (VenR) arm. Progression-free survival (PFS) in patients with cytogenetic alterations, using Döhner hierarchical classification, in the (B) VenR arm and (C) bendamustine plus rituximab (BR) arm. HR, hazard ratio.
PFS was superior with VenR compared with BR in all molecular subsets (Figs 4B, 4C, and 5; Data Supplement). Within the VenR cohort, despite the observed differences in uMRD rates, a trend for inferior PFS was observed only in patients with del(17p13.1); however, the difference did not reach statistical significance in multivariable adjustment (Figs 4B and 4C). GC had a major impact on clinical outcome, although VenR showed superiority in each GC category (Fig 5A). Within the VenR cohort, patients with noncomplex GC showed better PFS than those with either high-GC or low-GC status (HR, 2.9; 95% CI, 1.1 to 3.6; P = .0057; and HR, 2.0; 95% CI, 1.4 to 6.3; P = .025, respectively). Patients with high-GC status showed a trend toward inferior PFS versus those with low-GC status (HR, 1.5; 95% CI, 0.7 to 3.4; P = .29). Similar outcome patterns according to GC were observed in the BR arm (Fig 5A). In the VenR arm, a significant impact of del(17p) on PFS (P < .01) was seen in patients with high-GC status but not patients with low-GC status or noncomplex GC. In contrast, del(17p) had most effect on PFS in non-GC patients in the BR arm (Data Supplement). A significant impact of mutation burden on PFS by multivariable analysis was observed only in the BR arm (HR, 2.2; 95% CI, 1.1 to 4.7; P = .033) when one or more driver mutations were present (Data Supplement), but no effect was seen in either arm when two or more recurrently mutated genes had mutations (Fig 5B).
FIG 5.
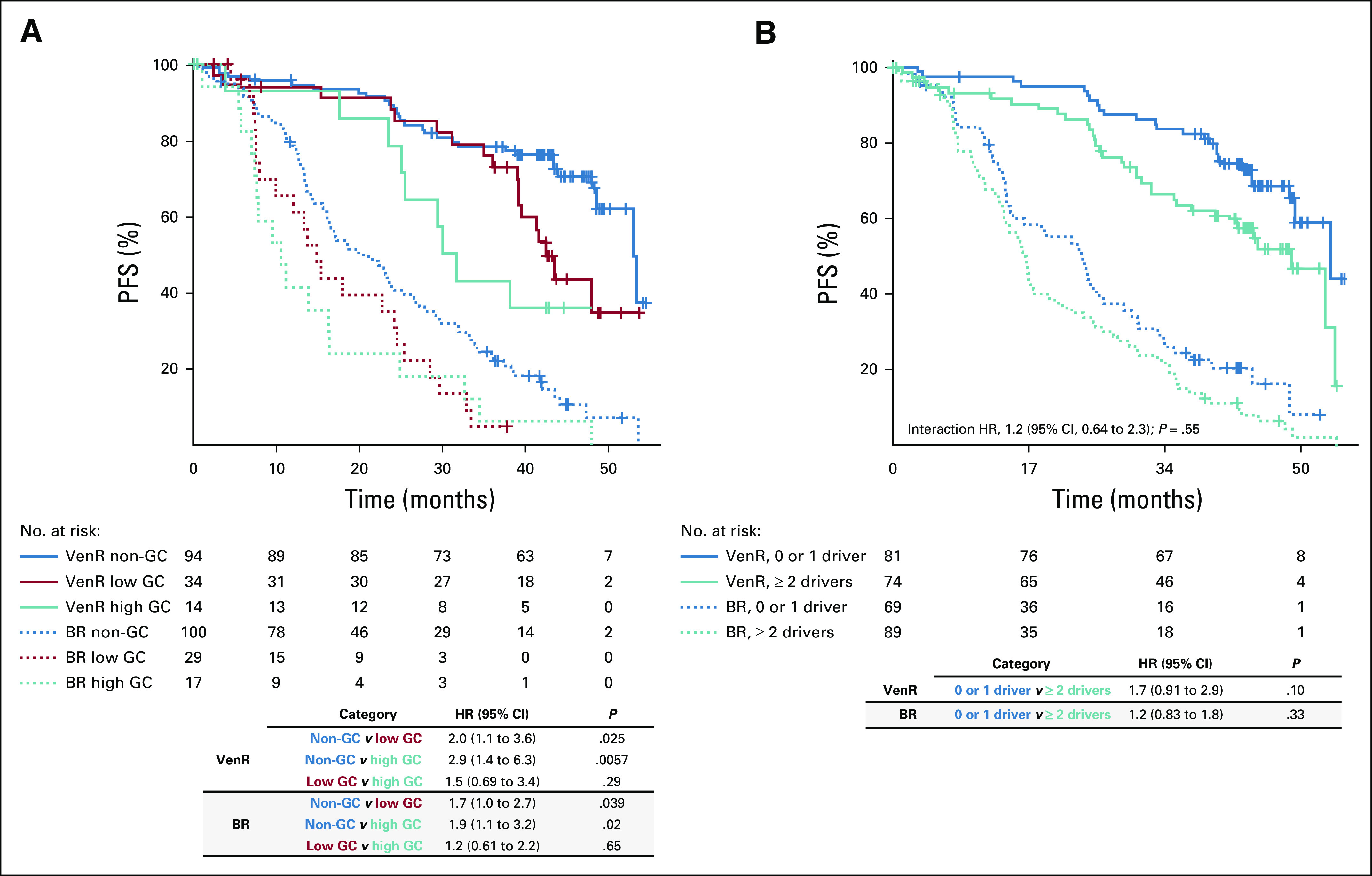
Multivariable analysis of progression-free survival (PFS) according to (A) genomic complexity (GC) status and (B) driver mutation burden (number of recurrently mutated genes with mutations). Covariates were IGHV status, Rai stage at baseline, TP53 mutation status and/or del(17p) by array comparative genomic hybridization, fludarabine resistance status, and maximum nodal size > 10 cm. BR, bendamustine plus rituximab; HR, hazard ratio; VenR, venetoclax plus rituximab.
Safety
The current analysis showed no new SAEs considered related to the study drug. Excluding nonmelanoma skin malignancies, three additional second primary malignancies were detected since the previous analysis (BR, n = 1 [melanoma]; VenR, n = 2 [melanoma and breast cancer]). There were no new reports of Richter transformation after an additional 12-month follow-up (VenR, n = 7; BR, n = 6; Data Supplement).
DISCUSSION
The benefits of fixed-duration VenR versus BR were sustained after a median follow-up of 48 months, confirming this fixed-duration treatment strategy as broadly effective. There has been some doubt as to whether drug cessation is the optimal approach for patients with high MRD positivity at 24 months, because of high rates of early progression in this population. However, the data presented here show that nearly all of these patients already had increasing MRD while on venetoclax therapy. Although this indicates emerging disease resistance and a probable lack of benefit from ongoing single-agent venetoclax, it is possible that treatment slowed the MRD increase, a hypothesis currently under investigation.
The enduring PFS benefit and progressively greater OS benefit of VenR, despite subsequent high rates of novel agent use after progression in the BR arm, support the early use of targeted therapies such as VenR in patients with R/R CLL and indicate that treatment benefits are durable. Better outcomes with earlier use have also been reported with ibrutinib.30 Importantly, this study also presents evidence that salvage therapy with ibrutinib or re-treatment with a venetoclax-based regimen is effective in patients who develop PD after VenR. Overall, 10 of 10 patients achieved a response with ibrutinib as their first postprogression therapy, and six of 11 patients achieved a response with a venetoclax-based regimen, most of whom received VenR. These data align with previously reported high response rates to salvage therapy with ibrutinib after venetoclax.16,31,32 However, longer-term follow-up and a larger patient cohort will be required to confirm these findings.
Here, aCGH was used to assess genomic aberrations in preference over FISH; there was moderate agreement between aCGH and FISH detection of del(17p). Previously, del(17p) was considered overrepresented and clinically overperformed in the BR arm, in comparison with other published data sets,33-35 when detected by FISH with the standard cutoff value of 7%. Increasing the cutoff value to 20% rectified these anomalous findings, which were attributable to a high proportion of patients with low-clone del(17p), a subgroup who had similar outcomes to patients with normal 17p. Similar outcomes were seen in the aCGH del(17p) and FISH high-clone del(17p) populations; we consider aCGH to be the more robust and clinically informative determinant of 17p status in this cohort.
This analysis of MURANO study data establishes novel associations between candidate biomarkers and clinical outcomes (MRD and PFS or OS) with VenR therapy. We expected to find the association between del(17p) and reduced MRD response, as shown here in the hierarchical analysis, but trisomy 12 CLL has previously been considered to be associated with an intermediate prognosis with CIT. However, it is recognized that trisomy 12 CLL is associated with unmutated IGHV and NOTCH1 mutations,36 and further studies of larger cohorts are needed to determine the independent contribution of these biologic factors. In contrast, del(11q) is an established negative prognostic marker in CLL,8 but the impact found here on PFS and MRD response appeared greater in the BR arm than the VenR arm. This aligns with a recent report from the CLL14 study, in which del(11q) was identified as an independent prognostic factor for PFS with chlorambucil plus obinutuzumab but not venetoclax plus obinutuzumab.37
Prognosis was superior in patients with noncomplex or low-GC status; high-GC status (five or more aberrations) was associated with inferior PFS. Use of chromosome banding analysis (CBA) for the detection of GC recently determined that the presence of five or more aberrations is more clinically relevant than the previously used cutoff of three or more abnormalities.10 The use of genomic arrays for GC detection has been shown to perform at least as well as CBA analysis and reinforced the cutoff of five or more aberrations11; as a result of their high resolution, array-based methods are capable of better defining small aberrant regions.
The likelihood of achieving uMRD status at EOCT in the VenR arm was better in patients without mutations in BIRC3 or BRAF; BRAF mutations have previously been shown to arise in patients developing venetoclax resistance, whereas BIRC3 mutations are associated with CIT resistance.38,39 Although BRAF mutations were also associated with lower uMRD at EOT, this was not the case for BIRC3 mutations, suggesting that a longer duration of venetoclax therapy is effective in overcoming the negative impact of this mutation. Lower uMRD rates at EOT were also seen with NOTCH1 and TP53 mutations. NOTCH1 mutations promote CLL cell proliferation and are associated with inferior outcomes with CIT, single-agent venetoclax or ibrutinib, and higher risk of transformation.40-43 The current analysis of VenR therapy did not detect an adverse impact on PFS from mutations in NOTCH1, despite the lower uMRD rate.
The MURANO study has already shown that the safety profile of venetoclax therapy is favorable.6,7 The lack of long-term drug-related SAEs in the present analysis is encouraging. The fixed duration of venetoclax therapy may be advantageous over continuous therapies because it limits the period during which patients are likely to experience AEs.
Strengths of this study include the long duration of follow-up, the large study population, and detailed molecular characterization of the enrolled patient population. Limitations are that the numbers of patients in specific biomarker subsets are modest, necessitating further studies for validation of these results, and also that these patients, who were enrolled in 2014-2015, had not been exposed to targeted agents such as BTKis, which is a major difference from current frontline management approaches and limits generalizability. Another aspect requiring further investigation is the optimal management of patients who develop PD after venetoclax therapy.
In this analysis, the benefits in PFS and OS with VenR versus BR in patients with R/R CLL were sustained over a median follow-up time of 48 months. With VenR, genetic mutations affected MRD response, and differences in outcomes were observed according to BRAF mutations and GC status. The PFS rate 24 months after treatment cessation was 68.0% in patients completing 2 years of venetoclax, with patients attaining PB uMRD showing particularly durable responses. These data provide further support for the application of fixed-duration VenR in R/R CLL.
ACKNOWLEDGMENT
We thank the patients and their families and all MURANO study team members and investigators. Venetoclax is being developed in a collaboration between Genentech and AbbVie. Genentech and AbbVie participated in the design and conduct of the study, analysis and interpretation of data, and writing, review, and approval of the article.
PRIOR PRESENTATION
Presented, in part, at the 61st Annual Meeting and Exposition of the American Society of Hematology, Orlando, FL, December 7-10, 2019.
SUPPORT
Supported by Genentech and AbbVie. Third-party medical writing assistance under the direction of A.P.K. and J.F.S. was provided by Stephanie Cumberworth and Kate Rijnen of Gardiner-Caldwell Communications and was funded by F. Hoffmann-La Roche.
CLINICAL TRIAL INFORMATION
NCT02005471 (MURANO)
EQUAL CONTRIBUTION
A.P.K. and J.Q.W. contributed equally to article development and share first authorship.
AUTHOR CONTRIBUTIONS
Conception and design: Arnon P. Kater, Jenny Qun Wu, Thomas Kipps, Peter Hillmen, Su Young Kim, Elizabeth Punnoose, Yanwen Jiang, Marcus Lefebure, Michelle Boyer, Kathryn Humphrey, John F. Seymour
Financial support: Su Young Kim
Administrative support: Thomas Kipps
Provision of study materials or patients: Arnon P. Kater, Peter Hillmen, James D’Rozario, Sarit Assouline, Carolyn Owen, Tadeusz Robak, Ulrich Jaeger, Guillaume Cartron, Anne-Marie Van Der Kevie-Kersemaekers, John F. Seymour
Collection and assembly of data: Arnon P. Kater, Jenny Qun Wu, Thomas Kipps, James D’Rozario, Sarit Assouline, Tadeusz Robak, Ulrich Jaeger, Guillaume Cartron, Julie Dubois, Clemens Mellink, Anne-Marie Van Der Kevie-Kersemaekers, Elizabeth Punnoose, Yanwen Jiang, Marcus Lefebure, Michelle Boyer, Kathryn Humphrey, John F. Seymour
Data analysis and interpretation: Arnon P. Kater, Jenny Qun Wu, Thomas Kipps, Barbara Eichhorst, Peter Hillmen, Carolyn Owen, Javier de la Serna, Guillaume Cartron, Marco Montillo, Eric Eldering, Clemens Mellink, Anne-Marie Van Der Kevie-Kersemaekers, Brenda Chyla, Elizabeth Punnoose, Christopher R. Bolen, Zoe June Assaf, Yanwen Jiang, Jue Wang, Marcus Lefebure, Michelle Boyer, Kathryn Humphrey, John F. Seymour
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Venetoclax Plus Rituximab in Relapsed Chronic Lymphocytic Leukemia: 4-Year Results and Evaluation of Impact of Genomic Complexity and Gene Mutations From the MURANO Phase III Study
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Arnon P. Kater
Consulting or Advisory Role: AbbVie
Research Funding: AbbVie/Genentech, Janssen Roche
Travel, Accommodations, Expenses: Roche
Honoraria: AbbVie
Jenny Qun Wu
Employment: Genentech
Stock and Other Ownership Interests: Genentech
Travel, Accommodations, Expenses: Genentech
Thomas Kipps
Employment: UC San Diego Health, Moores Cancer Center
Stock and Other Ownership Interests: Oncternal Therapeutics
Honoraria: Pharmacyclics, AbbVie, Janssen, Celgene, Genentech, Roche, Gilead Sciences, DAVAOncology
Consulting or Advisory Role: AbbVie, Pharmacyclics, Celgene, Genentech, Roche, Janssen, DAVAOncology, Gilead Sciences, Ascerta/AstraZeneca, Genentech/Roche, Gilead, Janssen, Loxo Oncology, Octernal Therapeutics, Pharmacyclics/AbbVie, TG Therapeutics, VelosBio, Verastem
Speakers’ Bureau: Verastem/Pharmacyclics, Pharmacyclics/Janssen, AbbVie/Genentech, Gilead Sciences, DAVA Pharmaceuticals
Research Funding: Pharmacyclics/Janssen (Inst), Breast Cancer Research Foundation (Inst), The University of Texas MD Anderson Cancer Center (Inst), Oncternal Therapeutics (Inst), Leukemia and Lymphoma Society (Inst), California Institute for Regenerative Medicine (Inst), National Cancer Institute (Inst), National Institutes of Health (Inst), Velos (Inst), Velos (Inst), Celgene (Inst) Patents, Royalties, Other Intellectual Property: Cirmtuzumab was developed by Thomas J. Kipps in the Thomas J. Kipps laboratory and licensed by the University of California to Oncternal Therapeutics, which provided stock options and research funding to the Thomas J. Kipps laboratory (Inst), Ascerta/AstraZeneca, Genentech/Roche, Gilead, Janssen, Loxo Oncology, Pharmacyclics/AbbVie, TG Therapeutics, VelosBio, Verastem
Travel, Accommodations, Expenses: AbbVie/Pharmacyclics, Genentech, Janssen, Gilead Sciences, National Cancer Institute, Celgene, Indy Heme Review, University of Nebraska Medical Center, Society of Hematologic Oncology, Shenzhen Cancer Center, European Research Initiative on CLL (ERIC), DAVAOncology, Patient Power, Breast Cancer Research Foundation, German CLL Study Group, International Workshop on Non-Hodgkin Lymphoma, National Comprehensive Cancer Network, TG Therapeutics, Verastem, Bionest Partner, OncLive
Barbara Eichhorst
Consulting or Advisory Role: Gilead Sciences, Janssen, Roche, AbbVie, Novartis, Celgene, ArQule
Speakers’ Bureau: Janssen, Gilead Sciences, Celgene, AbbVie, Novartis, Roche
Research Funding: Roche, AbbVie, Gilead Sciences, Janssen, BeiGene
Peter Hillmen
Honoraria: Janssen, AbbVie
Research Funding: Janssen (Inst), Pharmacyclics (Inst), Roche (Inst), Gilead Sciences (Inst), AbbVie (Inst), Apellis
James D'Rozario
Leadership: Roche
Honoraria: Roche Australia, Alexion Pharmaceuticals
Consulting or Advisory Role: AbbVie, Alexion, Celgene
Travel, Accommodations, Expenses: Roche Australia
Carolyn Owen
Honoraria: Janssen, Teva, AstraZeneca, Roche Canada, AbbVie, Gilead, Merk
Consulting or Advisory Role: AbbVie, AstraZeneca
Research Funding: Roche Canada(Inst), Acerta Pharma/AstraZeneca (Inst), Gilead Sciences (Inst), Celgene (Inst), Janssen (Inst), AbbVie (Inst)
Tadeusz Robak
Honoraria: AbbVie, Janssen
Consulting or Advisory Role: AbbVie, Janssen, Takeda, Gilead, BeiGene, Amgen, and Roche
Research Funding: AbbVie/Genentech (Inst), Jansen, Takeda, UCB, Acerta, Morphosys AG, Roche, Gilead, Beigene
Travel, Accommodations, Expenses: Roche, AbbVie, Amgen, Janssen
Javier de la Serna
Leadership: Roche, AbbVie, Gilead, Janssen, Novartis
Consulting or Advisory Role: AbbVie, Janssen, Roche, Gilead, Novartis
Speakers’ Bureau: AbbVie, Janssen, Roche, Gilead
Ulrich Jager
Honoraria: Amgen, AbbVie, Roche, Novartis, Gilead Sciences, Janssen Takeda, Bristol Myers Squibb, Eisai, MSD, Sanofi, Miltenyi
Consulting or Advisory Role: Roche, Novartis, Sandoz and MSD
Research Funding: Novartis (Inst), Gilead Sciences (Inst), Roche (Inst), AbbVie (Inst), Celgene (Inst), Takeda (Inst)
Guillaume Cartron
Honoraria: Sanofi, Gilead Sciences, Jansen, Celgene, Roche
Consulting or Advisory Role: Roche, Celgene
Marco Montillo
Honoraria: AbbVie, Janssen, Acerta, Verastem, Gilead
Consulting or Advisory Role: AbbVie, Janssen, Acerta, Gilead, Verastem, Roche
Speakers’ Bureau: AbbVie, Janssen and Gilead
Julie Dubois
Research Funding: Genentech
Eric Eldering
Research Funding: Genentech (Inst), Roche and AbbVie
Su Young Kim
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie
Brenda Chyla
Employment: AbbVie
Stock and Other Ownership Interests: AbbVie
Elizabeth Punnoose
Employment: Genentech
Stock and Other Ownership Interests: Roche
Christopher R. Bolen
Employment: Genentech
Stock and Other Ownership Interests: F Hoffmann La Roche
Zoe June Assaf
Employment: Genentech
Stock and Other Ownership Interests: Genentech, Natera
Yanwen Jiang
Employment: Genentech
Stock and Other Ownership Interests: Genentech, Roche/Genentech
Jue Wang
Employment: Genentech
Stock and Other Ownership Interests: Roche
Marcus Lefebure
Employment: Roche
Stock and Other Ownership Interests: Roche
Michelle Boyer
Employment: Roche
Kathryn Humphrey
Employment: Roche
John F. Seymour
Honoraria: AbbVie, Janssen, Roche, Sunesis Pharmaceuticals, Takeda, BMS, Celgene, Gilead, Mei Pharma
Consulting or Advisory Role: AbbVie, Janssen, Roche, Sunesis Pharmaceuticals, Takeda, BMS, Celgene, Mei Pharma
Speakers’ Bureau: AbbVie, Roche
Research Funding: AbbVie, Celgene, Janssen, Roche
Expert Testimony: Roche
Travel, Accommodations, Expenses: AbbVie, Roche, BMS and Janssen
No other potential conflicts of interest were reported.
REFERENCES
- 1.Fischer K Cramer P Busch R, et al. : Bendamustine combined with rituximab in patients with relapsed and/or refractory chronic lymphocytic leukemia: A multicenter phase II trial of the German Chronic Lymphocytic Leukemia Study Group. J Clin Oncol 29:3559-3566, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Byrd JC Hillmen P O’Brien S, et al. : Long-term follow-up of the RESONATE phase 3 trial of ibrutinib vs ofatumumab. Blood 133:2031-2042, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuneo A, Foà R: Relapsed/refractory chronic lymphocytic leukemia: Chemoimmunotherapy, treatment until progression with mechanism-driven agents or finite-duration therapy? Mediterr J Hematol Infect Dis 11:e2019024, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson MA Deng J Seymour JF, et al. : The BCL2 selective inhibitor venetoclax induces rapid onset apoptosis of CLL cells in patients via a TP53-independent mechanism. Blood 127:3215-3224, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stilgenbauer S Eichhorst B Schetelig J, et al. : Venetoclax for patients with chronic lymphocytic leukemia with 17p deletion: Results from the full population of a phase II pivotal trial. J Clin Oncol 36:1973-1980, 2018 [DOI] [PubMed] [Google Scholar]
- 6.Seymour JF Kipps TJ Eichhorst B, et al. : Venetoclax-rituximab in relapsed or refractory chronic lymphocytic leukemia. N Engl J Med 378:1107-1120, 2018 [DOI] [PubMed] [Google Scholar]
- 7.Kater AP Seymour JF Hillmen P, et al. : Fixed duration of venetoclax-rituximab in relapsed/refractory chronic lymphocytic leukemia eradicates minimal residual disease and prolongs survival: Post-treatment follow-up of the MURANO phase III study. J Clin Oncol 37:269-277, 2019 [DOI] [PubMed] [Google Scholar]
- 8.Hallek M Cheson BD Catovsky D, et al. : iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 131:2745-2760, 2018 [DOI] [PubMed] [Google Scholar]
- 9.Boddu P, Ferrajoli A: Prognostic factors in the era of targeted therapies in CLL. Curr Hematol Malig Rep 13:78-90, 2018 [DOI] [PubMed] [Google Scholar]
- 10.Baliakas P Jeromin S Iskas M, et al. : Cytogenetic complexity in chronic lymphocytic leukemia: Definitions, associations, and clinical impact. Blood 133:1205-1216, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leeksma AC Baliakas P Moysiadis T, et al. : Genomic arrays identify high-risk chronic lymphocytic leukemia with genomic complexity: A multi-center study. Haematologica 10.3324/haematol.2019.239947 [epub ahead of print on January 23, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shanafelt TD Wang XV Kay NE, et al. : Ibrutinib-rituximab or chemoimmunotherapy for chronic lymphocytic leukemia. N Engl J Med 381:432-443, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woyach JA Ruppert AS Heerema NA, et al. : Ibrutinib regimens versus chemoimmunotherapy in older patients with untreated CLL. N Engl J Med 379:2517-2528, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herling CD Klaumünzer M Rocha CK, et al. : Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood 128:395-404, 2016 [DOI] [PubMed] [Google Scholar]
- 15.Thompson PA O’Brien SM Wierda WG, et al. : Complex karyotype is a stronger predictor than del(17p) for an inferior outcome in relapsed or refractory chronic lymphocytic leukemia patients treated with ibrutinib-based regimens. Cancer 121:3612-3621, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson MA Tam C Lew TE, et al. : Clinicopathological features and outcomes of progression of CLL on the BCL2 inhibitor venetoclax. Blood 129:3362-3370, 2017 [DOI] [PubMed] [Google Scholar]
- 17.Burger JA Landau DA Taylor-Weiner A, et al. : Clonal evolution in patients with chronic lymphocytic leukaemia developing resistance to BTK inhibition. Nat Commun 7:11589, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landau DA Sun C Rosebrock D, et al. : The evolutionary landscape of chronic lymphocytic leukemia treated with ibrutinib targeted therapy. Nat Commun 8:2185, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leeksma AC Taylor J Wu B, et al. : Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia 33:390-402, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Sawaf O Lilienweiss E Bahlo J, et al. : High efficacy of venetoclax plus obinutuzumab in patients with complex karyotype and chronic lymphocytic leukemia. Blood 135:866-870, 2020 [DOI] [PubMed] [Google Scholar]
- 21.Hallek M Cheson BD Catovsky D, et al. : Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 111:5446-5456, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo MA Banks E Poplin R, et al. : A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491-498, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu TD, Nacu S: Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873-881, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A Hanna M Banks E, et al. : The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297-1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilm A Aw PP Bertrand D, et al. : LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res 40:11189-11201, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLaren W Gil L Hunt SE, et al. : The Ensembl variant effect predictor. Genome Biol 17:122, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.González-Pérez A, López-Bigas N: Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 88:440-449, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landau DA Tausch E Taylor-Weiner AN, et al. : Mutations driving CLL and their evolution in progression and relapse. Nature 526:525-530, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Döhner H Stilgenbauer S Benner A, et al. : Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med 343:1910-1916, 2000 [DOI] [PubMed] [Google Scholar]
- 30.O’Brien SM Byrd JC Hillmen P, et al. : Outcomes with ibrutinib by line of therapy and post-ibrutinib discontinuation in patients with chronic lymphocytic leukemia: Phase 3 analysis. Am J Hematol 94:554-562, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mato AR Roeker LE Jacobs R, et al. : Assessment of the efficacy of therapies following Venetoclax discontinuation in CLL reveals BTK inhibition as an effective strategy. Clin Cancer Res 26:3589-3596, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin VS Lew TE Handunnetti SM, et al. : BTK inhibitor therapy is effective in patients with CLL resistant to venetoclax. Blood 135:2266-2270, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zelenetz AD Barrientos JC Brown JR, et al. : Idelalisib or placebo in combination with bendamustine and rituximab in patients with relapsed or refractory chronic lymphocytic leukaemia: Interim results from a phase 3, randomised, double-blind, placebo-controlled trial. Lancet Oncol 18:297-311, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuneo A Follows G Rigolin GM, et al. : Efficacy of bendamustine and rituximab as first salvage treatment in chronic lymphocytic leukemia and indirect comparison with ibrutinib: A GIMEMA, ERIC and UK CLL FORUM study. Haematologica 103:1209-1217, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser G Cramer P Demirkan F, et al. : Updated results from the phase 3 HELIOS study of ibrutinib, bendamustine, and rituximab in relapsed chronic lymphocytic leukemia/small lymphocytic lymphoma. Leukemia 33:969-980, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abruzzo LV Herling CD Calin GA, et al. : Trisomy 12 chronic lymphocytic leukemia expresses a unique set of activated and targetable pathways. Haematologica 103:2069-2078, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tausch E Schneider C Robrecht S, et al. : Prognostic and predictive impact of genetic markers in patients with CLL treated with obinutuzumab and venetoclax. Blood 135:2402-2412, 2020 [DOI] [PubMed] [Google Scholar]
- 38.Herling CD Abedpour N Weiss J, et al. : Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat Commun 9:727, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diop F Moia R Favini C, et al. : Biological and clinical implications of BIRC3 mutations in chronic lymphocytic leukemia. Haematologica 105:448-456, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts AW Ma S Kipps TJ, et al. : Efficacy of venetoclax in relapsed chronic lymphocytic leukemia is influenced by disease and response variables. Blood 134:111-122, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villamor N Conde L Martínez-Trillos A, et al. : NOTCH1 mutations identify a genetic subgroup of chronic lymphocytic leukemia patients with high risk of transformation and poor outcome. Leukemia 27:1100-1106, 2013 [DOI] [PubMed] [Google Scholar]
- 42.Stilgenbauer S Schnaiter A Paschka P, et al. : Gene mutations and treatment outcome in chronic lymphocytic leukemia: Results from the CLL8 trial. Blood 123:3247-3254, 2014 [DOI] [PubMed] [Google Scholar]
- 43.Del Papa B Baldoni S Dorillo E, et al. : Decreased NOTCH1 activation correlates with response to ibrutinib in chronic lymphocytic leukemia. Clin Cancer Res 25:7540-7553, 2019 [DOI] [PubMed] [Google Scholar]