Abstract
The wide variety of pathogenic Leptospira serovars and the weak protection offered by the available vaccines encourage the search for protective immunogens against leptospirosis. We found that the secretin GspD of the type II secretion system (T2S) of Leptospira interrogans serovar Canicola was highly conserved amongst pathogenic serovars and was expressed in vivo during infection, as shown by immunohistochemistry. Convalescent sera of hamsters, dogs, and cows showed the presence of IgG antibodies, recognizing a recombinant version of this protein expressed in Escherichia coli (rGspDLC) in Western blot assays. In a pilot vaccination study, a group of eight hamsters was immunized on days zero and 14 with 50 µg of rGspDLC mixed with Freund’s incomplete adjuvant (FIA). On day 28 of the study, 1,000 LD50 (Lethal Dose 50%) of a virulent strain of Leptospira interrogans serovar Canicola (LOCaS46) were inoculated by an intraoral submucosal route (IOSM). Seventy-five percent protection against disease (p = 0.017573, Fisher’s exact test) and 50% protection against infection were observed in this group of vaccinated hamsters. In contrast, 85% of non-vaccinated hamsters died six to nine days after the challenge. These results suggest the potential usefulness of the T2S secretin GspD of Leptospira as a protective recombinant vaccine against leptospirosis.
Keywords: Leptospira, vaccine, recombinant, T2S, GspD, challenge
1. Introduction
Leptospirosis is a neglected and widespread zoonosis caused by the pathogenic species of Leptospira. Humans are accidental hosts who acquire the infection always from an animal source. Cattle, pigs, and dogs are the more commonly affected animal species with leptospirosis, causing abortion, infertility, and general systemic failure. However, rats, mice, and many other feral rodents are commonly infected with minor and unseen signs of disease; however, they are quite important reservoirs that spread, infecting leptospires in the environment through their urine. Rodents are considered as the main source of infection to human populations in rural and suburban areas with poor sanitary conditions [1,2]. More than one million human cases of leptospirosis and more than 50,000 deaths worldwide are reported to occur every year [3]. Human vaccines have been developed and administered to in risk populations only in China, Cuba, France, and Japan [4]. To date, the best way to prevent leptospirosis, besides the improvement of sanitary conditions, is animal vaccination. Currently available vaccines consist of cell suspensions of inactivated leptospires (bacterins), which have proved to be partially protective and limited to specific Leptospira serovars and not eliciting a long-lasting immunity. Immunity obtained with such a kind of vaccine is directed mainly towards leptospiral lipopolysaccharide (LPS), this being the main reason for their limited protection against disease caused by the specific serovars, those included in the bacterins formulation [5,6]. Research has been focused on conserved surface and subsurface leptospiral proteins as candidates for better vaccines against leptospirosis. Some relevant outer membrane proteins (OMPs), such as the porin OmpL1 and the lipoproteins LipL41 [7], LipL32 [8], LipL21 [9], Loa22 [10,11], and other OMPs like the Lig family of proteins A, B, and C [12,13,14], have been tested as vaccines, showing variable and often disappointing results [4]. Reverse vaccinology strategies (RV) have also been applied in the search for suitable protective antigens against leptospirosis without definitive results [15,16]. One functional protein that might be located on the Leptospira’s outer membrane is the secretin GspD, an important component of the type II secretion system (T2S). The OMP GspD is a protein known to be involved in toxin and enzyme secretion in Gram-negative bacteria [17]. In addition, GspD has been found as the twelfth most abundant component of the outer membrane (OM) proteome in Leptospira [18]. In the most complete RV approach so far performed to identify leptospiral vaccine candidates, GspD of Leptospira borgpettersenii serovar Hardjo strain L550 was used in a pool with four other OMPs. Despite a detectable antibody response, the protection was neither observed in this hamster model nor in 47 other protein pools tested in such a study [16]. Despite these results, we considered the GspD outstanding features, such as being a putative metabolically important protein, to be located on the surface of the outer membrane and so exposed to the immune system, and, finally, to be an abundant protein in Leptospira, to study GspD as a potential vaccine against leptospirosis. To determine the protection conferred against leptospirosis, we cloned and expressed the gspD gene of a virulent strain of Leptospira interrogans serovar Canicola (LOCaS46) into the pET-14b vector (Novagen™, Cambridge, MA, USA). The purified recombinant rGspDLC protein expressed in E. coli, when combined with incomplete Freund adjuvant (IFA) and administered twice in a group of eight hamsters, elicited protection against the infection with 1000 LD50 (lethal dose 50%), of a virulent homolog strain. Seventy-five percent of the challenged hamsters in the experimental group survived the infection (p = 0.021, Fisher’s exact test), and even 50% of them showed sterile immunity.
2. Materials and Methods
2.1. Genetic Procedures
2.1.1. Construction of Recombinant Vectors
The gsp locus of the L. interrogans serovar Canicola strain LOCaS46 used in this work was sequenced. The gspD gene (Genbank accession number: MT743253) showed 100% identity with their homologs in several other strains of serovar Canicola available in the databases. Cloning of the gspD gene was performed using the procedure described by Pinne et al. [19], with some modifications. To remove the signal peptide, the 1734 bp PCR amplicon was designed to encode 573 residues of the GspD protein except for the first 23 aa. The DNA fragment was PCR-amplified from genomic DNA of the LOCaS46 strain, using the APM1f (GT GTT ACA TAT GGA CAA ACC AGT CTT T CC) and APM3r (TC ATT TCG GCC GCT CTT TCT CTG ATT TCT CTT TCT TTG) set of primers. These primers were designed with the inclusion of the appropriate NdeI and BamHI restriction sites for cloning into the pET-14b expression vector (Novagen™, Cambridge, MA, USA). The diagram of the pET-14b_gspDLC construct was created with SnapGene™ (Figure 1). The PCR product size was verified by agarose gel electrophoresis, and the product was purified with the QIAquick PCR purification kit (Qiagen™, Redwood City, CA, USA), according to the manufacturer’s instructions. The DNA concentration was measured in a NanoDrop ND-1000 apparatus (Thermo Scientific™, Wilmington, DE, USA) at 260 nm. The ligation mixture was then chemically transformed into the E. coli DH5α cloning strain. The construct was sequenced (Instituto de Biotecnología, UNAM, Chamilpa, Cuernavaca) to verify that the insert was in the frame and subcloned into the BLR(DE3)pLysS E. coli expression strain (Sigma-Aldrich™, San Louis, MO, USA). The fifteen first and last residues of the rGspDLC protein are shown (Figure 1). Recombinant colonies, grown after incubation at 37 °C overnight on LB plates with ampicillin (100 µg/mL), were selected.
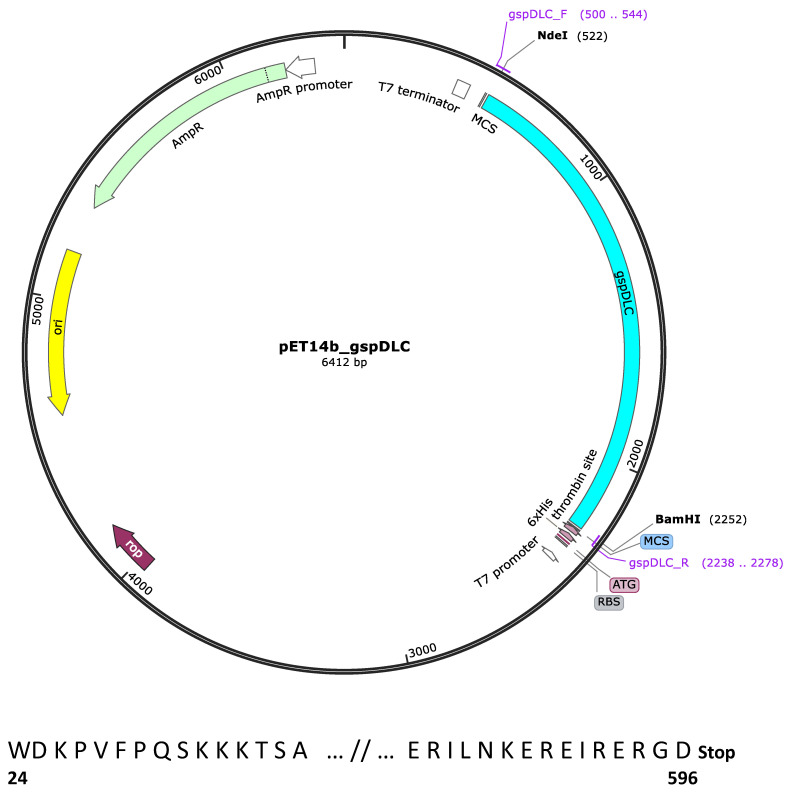
Figure 1.
Map of the pET-14b_gspDLC recombinant expression vector. The 1734 bp gspDLC cloned insert is shown in pale blue color. NdeI and BamHI restriction digestion cloning sites are shown. The map was built with the SnapGene™ 1.1.3 program. The fifteen first and last residues of the cloned peptide are shown, and numbers represent the actual numbered residue of GspD of Leptospira interrogans SV Canicola strain LOCaS46.
2.1.2. GspDLC Expression, Screening, and Purification
Recombinant E. coli colonies were obtained by culture in selective LB-Amp plates (100 µg/mL) at 37 °C overnight and screened by PCR. Recombinants carrying the appropriate insert were sub-cultured in LB-Amp broth (100 µg/mL), shaking at 200 rpm and 18 °C until they reached an OD600 of 0.5. The cultures were then induced with 1 mM IPTG, incubating overnight for the expression of the rGspDLC protein. Cells were centrifuged at 6000× g for 3 min at 4 °C, and the lysis of the pellet was performed with the BugBuster reactive (Novagen™, San Diego, CA, USA), as the manufacturer’s instructions. The cell lysis reactive was mixed with 0.5 mM PMSF (Affimetrix™, Santa Clara, CA, USA) as proteases inhibitor and bovine pancreatic DNAse-I (Sigma™). The rGspDLC-His-tagged protein was detected in the E. coli cells extracts by Western blot assays and later purified by nickel affinity chromatography in Ni-NTA Superflow nickel charged columns (Qiagen™) with sodium phosphate and urea elution buffer, as specified by the manufacturer. The recombinant protein was concentrated by size exclusion using 30 kDa Amicon filter columns (Millipore™, San Louis, MO, USA) and centrifugation at 4000× g.
2.2. Antisera Production
New Zealand rabbits were immunized on days zero, 7, 14, and 21 with either 100 µg of the rGspDLC in PBS mixed with IFA or with a strain LOCaS46 culture and washed and concentrated up to 108 leptospires/mL in PBS mixed with IFA. The rabbits were injected intramuscularly and bled to death on day 28 under ketamine dissociative anesthesia. The anti-rGspDLC polyclonal antiserum was evaluated in Western blot assays, and the anti-LOCaS46 antiserum was evaluated by the microscopic agglutination test (MAT).
2.3. Screening for GspD in Leptospira Serovars
OMP and total protein extracts of Leptospira cultures of serovars Australis, Ballum, Bratislava, Canicola, Hardjobovis, Grippotyphosa, Pomona, and Wolffi were obtained, as per the protocol described by Matsunaga [20]. These protein extracts were screened for the presence of GspD homologs in Western blot assays using the rabbit anti-rGspDLC polyclonal antiserum and a mouse monoclonal anti-rabbit IgG (Jackson ImmunoResearch™, West Grove, PA, USA) as the secondary antibody.
2.4. Leptospira Cultures and LD50 Calculation
An isolate obtained from an asymptomatic carrier dog was used for the challenging assays as well as for preparing the inactivated vaccine for the positive control group C. This isolate was characterized as Leptospira interrogans serovar Canicola by cross agglutination test (CAT), using a collection of serovars and specific rabbit antisera, and its identity was genetically confirmed by Multilocus Sequence typing (MLST) using the Ahmed et al. scheme [21], and such a strain was called LOCaS46. The LD50 of this strain was assessed in groups of eight Syrian hamsters in both intraperitoneal (IP) and intraoral submucosal inoculation (IOSM) [22]. This route of inoculation was attempted to get closer to the natural route of infection. To calculate the concentration of leptospires, 7-day old cultures incubated at 30 °C in Ellinghausen-McCullough-Johnson-Harris (EMJH) liquid medium [23] were ten-fold diluted in the same medium and quantified in triplicate in a Petroff-Hausser counting chamber (Hausser Scientific™). The LD50 was determined by the method of Reed and Müench [24]. Sub-cultures of leptospires were performed in both liquid EMJH and semisolid Fletcher media [25]. Fletcher medium was enriched with 8% of inactivated rabbit serum filter-sterilized at 0.22 µm.
2.5. Antigenicity of GspD in Infected Animals
Sera from five dogs, previously detected as Leptospira carriers by the culture of kidney tissues or urine samples, were used to search for specific anti-rGspDLC antibodies in Western blot assays. The dogs’ sera were previously shown to have a MAT titer equivalent to 1:1600 or higher against the serovars Canicola and/or Pyrogenes. Likewise, sera obtained from eight experimentally infected hamsters used to calculate the LD50 of the virulent strain LOCaS46 were tested in similar assays. The 15 µg of the rGspDLC protein was run in a 12% PAGE and transferred to a nitrocellulose membrane by electrophoresis. A 1:100 dilution of the primary antibody was used to recognize the transferred proteins, and a 1:400 dilution of an HRP-conjugate (as required, dog or hamster anti-IgG) was used as the secondary antibody (Jackson Immuno Research™, West Grove, PA, USA).
2.6. GspD In Situ Expression by Leptospira
To screen for the expression of GspD in tissues of hamsters experimentally infected with the virulent strain LOCaS46, the anti-rGspDLC and anti-LOCas46 rabbit polyclonal antisera were used in immunohistochemistry assays (IHC). Kidney and liver tissues obtained from the hamsters used in the LD50 assays were used for this experiment. A modification of the protocols, suggested by Massone et al. [26] and Haines and Chelack [27], was used as follows. Double serial dilutions of the anti-rGspDLC and anti-LOCaS46 antisera were prepared, from 1:25 as the initial dilution. Sections of each kidney and liver tissue samples were cut 3–5 µm thick using a Leica RM2245 microtome and mounted onto 3% poly-L-lysine-treated glass plates (Sigma™). Sections were dewaxed and washed twice for 30 min in xylol. Tissues were then hydrated 5 min thrice with decreasing concentrations of ethanol diluted in deionized water (96, 80, 70, 50%). To inhibit endogenous peroxidases, tissues were next treated for 30 min at room temperature with a 3.75% dilution of hydrogen peroxide in ethanol. Finally, tissues were washed for 5 min thrice with PBS, then for 20 min with a citrate buffer solution (10 mM sodium citrate, 0.05% Tween 20, pH 6.0), and again for 5 min thrice with PBS. For blocking unspecific reactions, the slides were covered with 60 µL of a 1:10 diluted goat nonimmune serum at 37 °C for 30 min in a humid chamber. After discarding the blocking serum, 60 µL of the rabbit polyclonal anti-rGspDLC or anti-LOCas46 antisera was applied in double serial dilutions from 1:25 up to 1:3200 in PBS pH 7.4 with 0.5% skimmed milk. Slides were incubated for 16 h at 4 °C in a humid chamber. After incubation, 5 min washes with PBS were done thrice, and then the slides were treated with 60 µL of biotinylated anti-rabbit IgG conjugate (Histostain-SP, Invitrogen™, Camarillo, CA, USA) and incubated at 37 °C for 60 min in a humid chamber. After three washes, 5 min each in PBS, 60 µL of the streptavidin-peroxidase conjugate (Histostain-SP, Invitrogen™, Camarillo, CA, USA) was added and incubated at 37 °C for 30 min in a humid chamber. For detection, the chromogen was prepared as follows: 50 µL of aminoethylcarbazol (Histostain-SP, Invitrogen™, Camarillo, CA, USA), 50 µL of the buffer provided by the manufacturer (Histostain-SP, Invitrogen™, Camarillo, CA, USA), and 50 µL of a 0.6% hydrogen peroxide solution in 850 µL deionized water; 60 µL of this chromogen solution was applied to the slides and incubated for 5 min. The reaction was stopped by rinsing in deionized water, and counterstaining was done for 5 min with Harris hematoxylin (Histostain-SP, Invitrogen™, Camarillo, CA, USA), and the slides were rinsed again in water and incubated for 1 min at 37 °C in PBS. Finally, slides were covered with Histomount resin (Histostain-SP, Invitrogen®, Camarillo, CA, USA) and a coverslip. Slides were observed under an AxioImager Z1 microscope (Carl Zeiss MicroImaging, Jena, Germany).
3. Vaccines
3.1. rGspDLC Vaccine
The purified rGspDLC protein was washed with PBS and concentrated to 1 µg/µL through a 30 kDa Amicon Ultra-4 filter column (Millipore Sigma™) in a final volume of 200 µL in PBS. Protein concentration was measured with the Micro BCA Protein Assay Kit (ThermoFisher™), according to the manufacturer’s instructions. The recombinant protein was used either alone (group A) or emulsified with IFA (Sigma™) in a 50% v/v suspension (group B) to get 50 µg of rGspDLC in a final volume of 100 µL.
3.2. Killed Cells Vaccine
The strain LOCaS46 of L. interrogans serovar Canicola was cultured in EMJH broth at 30 °C for 10 to 14 days. The cultures were concentrated by centrifugation and diluted in fresh EMJH medium to obtain a concentration of 109 leptospires/mL. The concentration was estimated by counting the cells in a Petroff-Hausser counting chamber (Hausser Scientific™, Horsham, PA, USA). The leptospires suspension was inactivated with 0.05% formalin and emulsified with IFA (50% v/v).
3.3. Pilot Vaccination Study
Five groups of eight weeks old hamsters were treated as follows: group A, eight hamsters, vaccinated twice with 50 µg of the rGspDLC protein alone; group B, eight hamsters, vaccinated twice with 50 µg of the rGspDLC protein mixed 1:1 with IFA; group C, eight hamsters, vaccinated twice with a 109/mL suspension of formalin-inactivated leptospires mixed 1:1 with IFA; group D, thirteen hamsters, injected twice with IFA, and group E, five hamsters, injected twice with sterile saline solution (Table 1). All treatments were freshly prepared in a final volume of 100 µL per hamster and injected subcutaneously on days zero and 14 of the experiment. On day 28, groups A to E were infected with the virulent Leptospira interrogans serovar Canicola strain LOCaS46, using an inoculation route by intraoral submucosal injection (IOSM). A dose of 1000 LD50 (40,000 leptospires) in a volume of 100 µL was applied with a 31G needle under isoflurane anesthesia, as previously described by Asoh et al. [22]. The IOSM route was selected to achieve a closer to the natural route of infection for leptospirosis when compared with the commonly used IP route of infection. An additional group of three non-infected hamsters was used as a control. The hamsters were kept in a 10/14 h light/darkness ratio during the length of the study, and food and clean water were supplied ad libitum. A final point of the experiment was established as the loss of 10% corporal weight to avoid unnecessary suffering of the infected specimens. Hamsters were observed daily and weighed to sense such an established final point. If they were not found dead, when the final point was reached or at the end of the experiment on day 56, the hamsters were killed by an isoflurane anesthesia overdose and necropsy. All animals were observed for 3 min after isoflurane overdose to confirm the death before proceeding to necropsy. During necropsy, kidney, liver, and urine samples were aseptically obtained for culture in EMJH and Fletcher media for PCR and histopathology and sera samples to screen for anti-Leptospira MAT antibodies. A direct macerate of kidney and liver tissues and blood as well as urine samples (when possible) was diluted directly into EMJH medium and observed under dark-field microscopy to assess for the presence of leptospires (X800 magnification).
Table 1.
Treatment protocols applied to hamster groups, protection percentage, infection, and kidney lesion rates.
| Group | Treatment | n | Survivors (Protection %) | Infection Rates a PCR Culture |
NI b | Kidney Lesions Rates | p-Value c | ||
|---|---|---|---|---|---|---|---|---|---|
| A | GspDLC + no adjuvant + challenge | 8 | 5 (65.5) | 8 | 3 | 0 | 4.875 | 0.0555 | NS d |
| B | GspDLC + IFA + challenge | 8 | 6 (75) | 5 | 4 | 3 | 5 | 0.0176 | |
| C | Bacterin + challenge | 8 | 8 (100) | 1 | 1 | 7 | 3.5 | 0.0002 | |
| D | IFA + challenge | 13 | 2 (15.3) | 13 | 8 | 0 | 7.75 | ND e | |
| E | Saline + challenge | 5 | 0 (0) | 5 | 5 | 0 | 8.4 | 1 | NS d |
aLeptospira infection was determined by PCR and culture. b NI, Not infected hamsters. c p-values are two-sided and were obtained through comparison with group D (Fisher’s exact test). d NS, Not significant at p > 0.05. e ND, Not determined.
3.4. Histopathology
A complete kidney and 0.5 cm3 of liver were obtained at necropsy, and the samples were immediately fixed in a 10% buffered formaldehyde solution, routinely processed with paraffin, and consecutively sectioned at 5 µm. For microscopic examination and scoring, some sections were stained with hematoxylin-eosin, and others were processed for immunohistochemistry, as previously described. The histomorphological evaluation of kidney lesions—indicative of Leptospira infection—tubular cortical necrosis, interstitial hemorrhage, tubular epithelium degeneration, intratubular hyaline eosinophilic content (tubular proteinoses), thrombosis, neutrophilic tubulitis, and interstitial inflammation, mainly composed of neutrophils, lymphocytes, and macrophages, was performed. The evaluated hepatic lesions were interstitial hemorrhage, vacuolar degeneration, necrosis, thrombosis, and interstitial inflammation, mainly composed of neutrophils, lymphocytes, and macrophages. The criteria for the histopathology evaluation was rated from zero to 4 according to the severity of the lesions: 0 as normal (0% coverage), 1 as minimal (up to 10% coverage), 2 as mild (up to 25% coverage), 3 as moderate (up to 50% coverage), and 4 as severe (more than 50% coverage). The sum of each lesion score per hamster in a group was finally expressed as an average. Slides were examined under an AxioImager Z1 microscope (Carl Zeiss MicroImaging, Jena, Germany) for histology or under dark-field microscopy to screen for leptospires in tissues.
4. Statistical Analysis
Where appropriate, the data are presented as means and standard deviations of the means. Comparisons of the severity of the renal lesions and the invasion of Leptospira in different tissues among hamster groups A to E were performed using the Student’s t-test and by the one-way ANOVA calculator; comparisons of the survivals in groups A to D were performed using the Fisher’s exact test; a p-value ≤ 0.05 was taken as statistically significant.
Ethical Approval
All animal procedures in this study were performed according to the Mexican Official Standard NOM-062-ZOO-1999 (Technical Specifications for the Production, Care, and Use of Laboratory Animals) and the NOM-033-SAG/ZOO-1995 (Methods to Kill Domestic and Wild Animals) and were approved by the Institutional Committee for Care and Use of Experimental Animals of the School of Veterinary Medicine of the National Autonomous University of Mexico (CICUAE-FMVZ/UNAM), under the registration number: MC2014-27.
5. Results
5.1. Expression of the rGspDLC Protein and Antisera Production
The 1734 bp DNA fragment of the gspD gene of Leptospira interrogans serovar Canicola strain LOCaS46, cloned into the pET-14b (Novagen™ Cambridge, MA, USA) expression vector and chemically transformed into the BL21(DE3) pLysS (Sigma-Aldrich™, San Louis, MO, USA) E. coli cells, expressed the predicted 63 kDa poly-His-tagged protein when induced with IPTG, as described (Figure 2A).
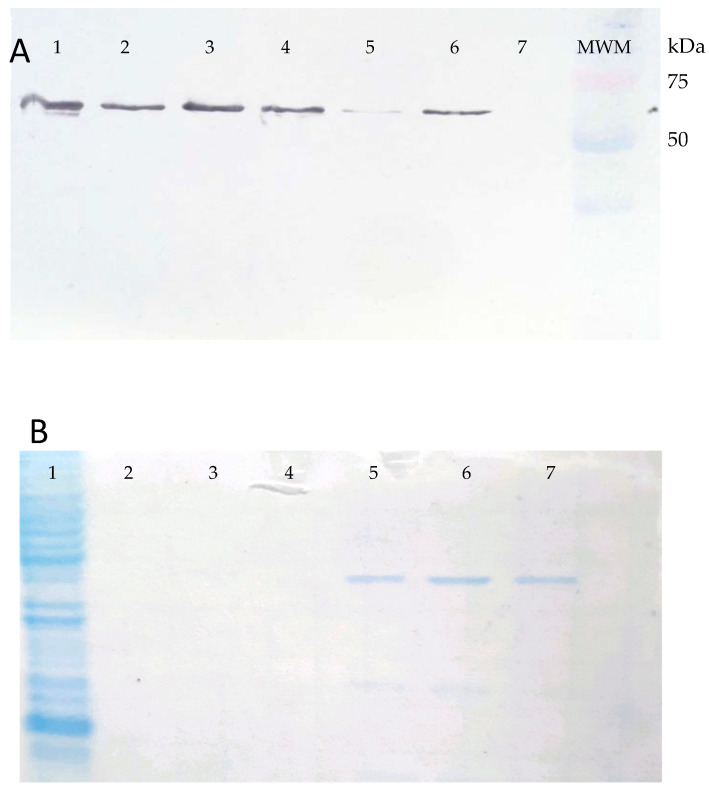
Figure 2.
Panel (A). Western blot of the BL21(DE3) pLysS (Promega™) E. coli cells transformed with the pET-14b_gspDLC recombinant expression vector and induced with 1 mM IPTG (lanes 1–6). Lane 7, uninduced control. An approximately 63 kDa band was observed corresponding to the rGspDLC protein reactive to an anti-His monoclonal antibody. Panel (B). Coomassie blue-stained acrylamide gel, showing the rGspDLC purified by Ni-affinity chromatography and sodium phosphate/urea elution buffer. Lane 1, recombinant E. coli lysate; lanes 5, 6, and 7 show the purified recombinant protein.
The protein yield of the recombinant GspD secretin (rGspDLC) was approximately 80 to 120 µg/L of culture. The rGspDLC protein was purified by Ni-affinity chromatography and concentrated by size exclusion up to 1 mg/mL using 30 kDa Amicon filter columns (Millipore™), and an estimated 60 to 70% purity of the recombinant protein was estimated based on Coomassie blue-stained acrylamide gels (Figure 2B). The anti-rGspDLC rabbit antiserum reached a titer of 1:32,000 in Western blot assays. On the other hand, antiserum obtained from rabbits immunized four times with a suspension of the heat-inactivated L. interrogans serovar Canicola strain LOCaS46 reached a 1:12,800 antibody titer in the MAT (data not shown).
5.2. GspD in Leptospira Serovars, Antigenicity, and In Situ Expression
A protein of approximately 63 kDa was detected in the extracts of all nine Leptospira serovars tested when the polyclonal rabbit antiserum raised against the rGspDLC protein was used in Western blot assays (Figure 3). An additional band of approximately 45 kDa was also observed in such assays in most of the Leptospira serovars. This finding was maybe due to the presence of an undetermined leptospiral protein with a structural region similar to rGspDLC, which was recognized by the polyclonal rabbit antibody.
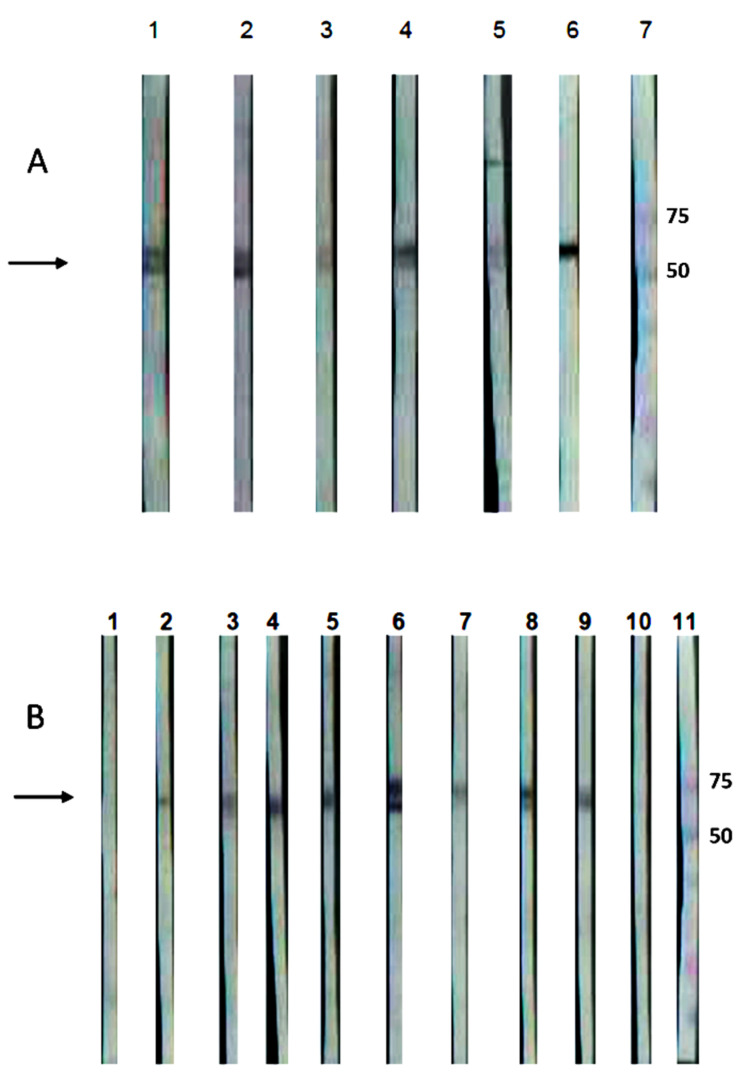
Figure 3.
Western blot of the total protein extracts obtained from cultures of nine Leptospira serovars. An approximately 63 kDa band was shown when a rabbit antiserum against rGspDLC was used in a 1:100 dilution. Aus: Australis; Bal: Ballum; Bra: Bratislava; Can: Canicola; Har: Hardjoprajitno; Gri: Grippotyphosa; Pom: Pomona; Pyr: Pyrogenes; Wol: Wolffi.
Similarly, the sera from five dogs, previously identified as Leptospira interrogans serovar Canicola carriers, and the sera from the experimentally infected hamsters in this work showed IgG antibodies, detecting the 63 kDa band in Western blots of the rGspDLC protein expressed by the E. coli transformants (Figure 4).
Figure 4.
Western blot of the recombinant GspD secretin of Leptospira interrogans serovar Canicola (rGspDLC), expressed by E. coli BL21(DE3)pLysS transformants. Panel (A): Detection of an approximately 63 kDa band (arrow), using five sera from four naturally infected dogs (lanes 1–4) and one convalescent dog with leptospirosis (lane 5). Infection with serovar Canicola was confirmed by culture and MLST. Lane 6, anti-His-HRP positive control; lane 7, molecular weight marker. Panel (B): Detection of an approximately 63 kDa band (arrow), using the sera from eight hamsters experimentally infected with the virulent serovar Canicola strain LOCaS46 (lanes 2–9), and the infection was confirmed by culture. Sera from two non-infected hamsters were used as negative controls (lanes 1 and 10), where a band was not detected; lane 11, molecular weight marker.
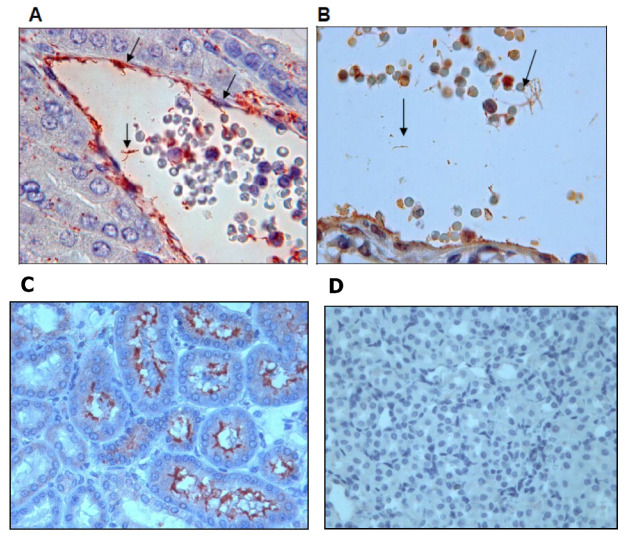
Further IHC assays using a 1:25 dilution of the anti-rGspDLC rabbit polyclonal antiserum detected leptospires in kidney tissue of the experimentally infected hamsters (Figure 5B), and a rabbit polyclonal antiserum against serovar Canicola (anti-LOCaS46) was used as a positive control in such IHC assays (Figure 5A,C). The specificity of both antisera (anti-rGspDLC and anti-LOCaS46) was proved in renal tissue of non-infected hamsters in similar assays that showed no unspecific reactions (Figure 5D).
Figure 5.
Immunohistochemistry assay on renal tissue of experimentally infected hamsters (Mesocricetus auratus). Infection was performed by intraperitoneal inoculation with the virulent strain LOCaS46 of Leptospira interrogans serovar Canicola. Panels (A,C): Detection of leptospires using a 1:200 dilution of rabbit polyclonal hyperimmune antiserum against L. interrogans serovar Canicola. Panel (B): Detection of leptospires using a 1:25 dilution of the rabbit polyclonal antibody against the recombinant GspD secretin of L. interrogans serovar Canicola (rGspDLC). Panel (D): Renal tissue of a non-infected hamster used as a control.
5.3. Assessment of the Immunogenicity of the rGspDLC Protein
Results of the pilot vaccination study are shown in Table 1. The LD50 for Syrian hamsters of the virulent strain LOCaS46 of L. interrogans serovar Canicola was determined to be as low as 4 leptospires in the IP route and 40 leptospires in the IOSM route. Infection rates were determined by PCR and culture. No infection could be determined in seven out of eight hamsters in group C immunized with the homologous bacterin used as a positive control. Encouraging results were observed in the hamsters in group B vaccinated with the rGspDLC-IFA mixture, where a 75% survival was observed, and additionally, three out of the six hamsters that survived were shown to be free of Leptospira infection by culture and PCR (sterile immunity). However, for the hamsters in group A vaccinated with the rGspDLC alone, all of them were shown to be infected regardless of a 65% protection conferred against disease. The negative control group D treated exclusively with IFA showed only a 15% survival (85% lethality; p = 1.000, Fisher’s exact test). In addition, no survivals were observed in the control group E (Saline, n = 5), confirming, in these two control groups, the high virulence of the challenging strain LOCaS46 used in the pilot vaccination study.
5.4. Infection Assessment by Culture and PCR
All hamsters that lost 10% of body weight, established as the final point of the experiment, were either euthanized by an isoflurane overdose or found dead at days 9 or 10 after the challenge. General moderate jaundice was observed in most animals. At necropsy, kidneys and liver showed moderate congestion, and evident hemorrhages were observed in the lungs. The rates of infection assessed by culture and PCR were as high as 100% in the control groups D (IFA) and E (saline) and also in the hamsters of experimental group A (rGspDLC alone), despite the 65% protection against disease observed in this group. In contrast, three out of eight hamsters in group B (rGspDLC + IFA) (37.5%) and seven out of eight hamsters (87.5%) of group C (bacterin) showed no evidence of infection either by culture or PCR (Table 1). Statistically significant lowest infection rates were observed in hamsters of group C (bacterin) (p < 0.0001). Groups A and B also showed statistically significant lower rates of infection when compared with the control group D (IFA); however, there was no statistically significant difference between those two groups (p > 0.5).
5.5. Infection Assessment by Histopathology
Experimental infection with the virulent L. interrogans serovar Canicola strain LOCaS46 produced macroscopic lesions characteristic of active leptospirosis, such as general jaundice, pulmonary hemorrhages (Figure 6A–D), and congestion in kidneys and liver. Microscopic lesions included tubular cortical necrosis, interstitial hemorrhage, tubular epithelium degeneration, intratubular hyaline eosinophilic content, thrombosis, neutrophilic tubulitis, and interstitial inflammation, mainly composed of neutrophils, lymphocytes, and macrophages (Figure 6E–H). The lowest average scores for renal lesions were observed in group C (bacterin) (mean = 3.5; p = < 0.0001). Meanwhile, the experimental groups A (rGspDLC alone) and B (rGspDLC-IFA) showed similar renal lesions scores (group A: mean = 4.875; p = 0.003026) (group B: mean = 5; p = 0.010007). The p-value of the infection assessment was calculated by comparison with the control group D (IFA), which showed the highest score (mean = 7.75). Results were statistically significant in the one-tailed t-test calculator at a significant level of 0.05 (Table 1).
Figure 6.
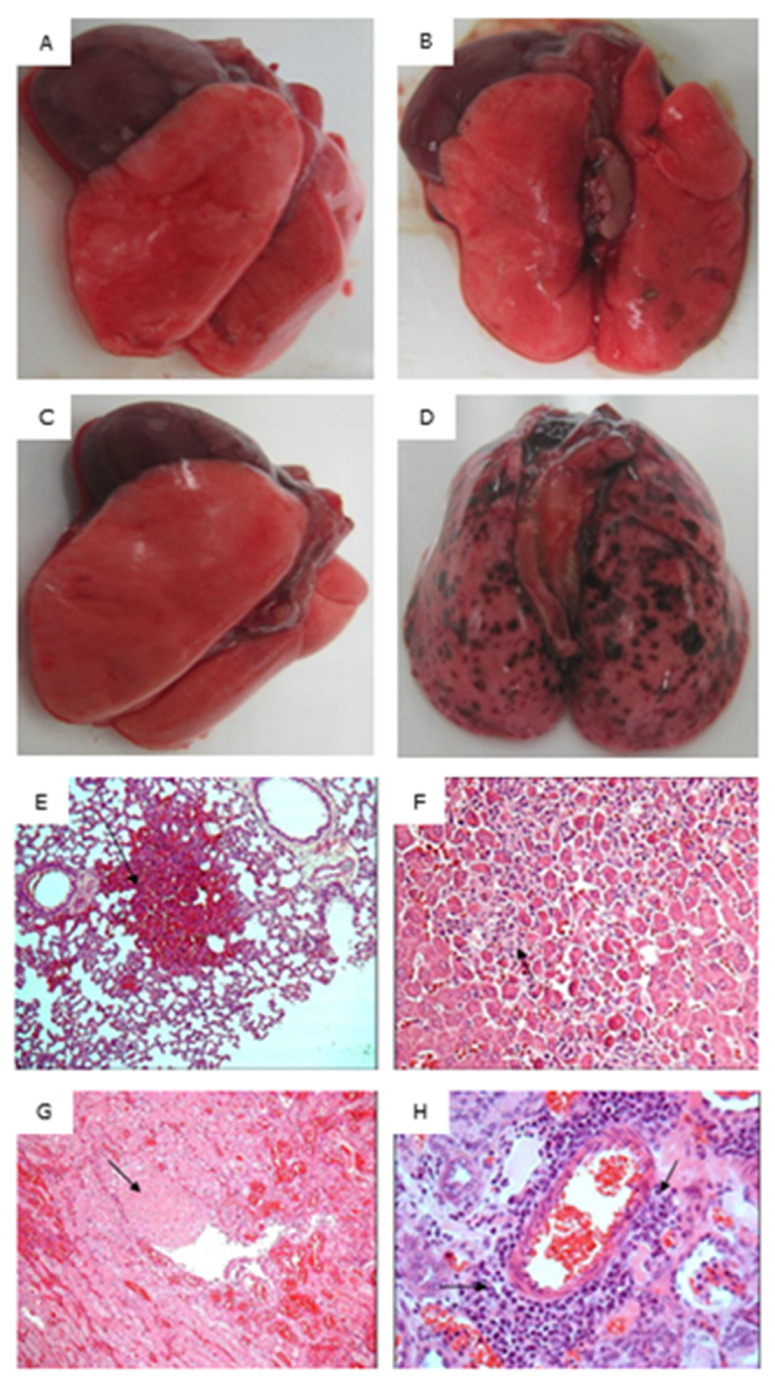
Pathology assessment of lungs from experimentally infected hamsters (Panels (A–D)), and HE 100X histopathology of the lung (Panel (E)), liver (Panel (F)), and kidney (Panels (G,H)) of a hamster in the negative control group D (IFA). Panel (A): Lung of a survival hamster in group A (rGspDLC alone); Panel (B): Lung of a survival hamster in group B (rGspDLC-IFA); Panel (C): Lung of a survival hamster in group C (serovar Canicola bacterin); Panel (D): Lung of a dead hamster in group D (IFA); Panel (E): Pulmonary hemorrhage; Panel F: Hepatocellular degeneration and necrosis and inflammatory infiltration; Panel (G): Renal vascular thrombosis; (H): Renal perivascular inflammatory infiltration, mainly composed of lymphocytes and plasmatic cells.
6. Discussion
We report here that a recombinant form of the GspD protein of Leptospira interrogans serovar Canicola (rGspDLC) expressed in E. coli offered a 75% protection against disease in hamsters challenged with 1000 LD50 of a virulent homolog strain (p = 0.021, Fisher’s exact test). Although the minimal 80% protection established by the World Organization for Animal Health (OIE) was not achieved, the results are encouraging in the light that in addition to protection against disease, the experimental group B (rGspDLC-IFA) showed that three out of the six hamsters that survived had no evidence of infection either by culture or PCR from urine, liver, or kidney samples (Table 1). Protection was also inferred by the lesions observed at necropsy. Rates of the lesions observed in kidneys and liver in the experimental group B (mean = 5) were statistically significantly lower than the rates in control group D (IFA) (mean = 7.75) (p < 0.02). Besides, despite that group C (bacterin) showed the lowest rate of lesions, there was no statistically significant difference in the lesions rate between this and the two experimental groups A and B, as was analyzed by the one-way ANOVA calculator (p > 0.05) (Table 1).
The IP route of infection has been widely used for testing the efficiency of vaccines against leptospirosis, despite not being the natural route of infection [4,28]. In our experimental vaccination assay, we tested the previously described IOSM route of infection [22] applied under isoflurane anesthesia; the aim of the method was to mimic a closer to the natural route of infection. The LD50 for hamsters of the challenging strain L. interrogans serovar Canicola LOCaS46, using the IOSM route, was equivalent to 40 leptospires; this dose was four times higher than the lethal dose observed for serovar Losbanos strain K37, using the same infection route [22]. On the other hand, the IOSM LD50 of strain LOCaS46 was ten times higher than the IP route of infection for the same strain (data not shown) but still was very suitable for the experimental infection assays. Coutinho et al. tested the subcutaneous (SC) and intradermal (ID) routes of infection in a study to compare the kinetics of leptospiral infection. A dose of 2 × 106 of L. interrogans serovar Copenhageni strain Fiocruz L1-130 was used, and only a slight difference in the burden of infection in the kidney was found among both routes of infection, reported at days 3 to 4 for the ID route and at days 4 to 5 for the SC route. The IP route was not tested in such a study [28].
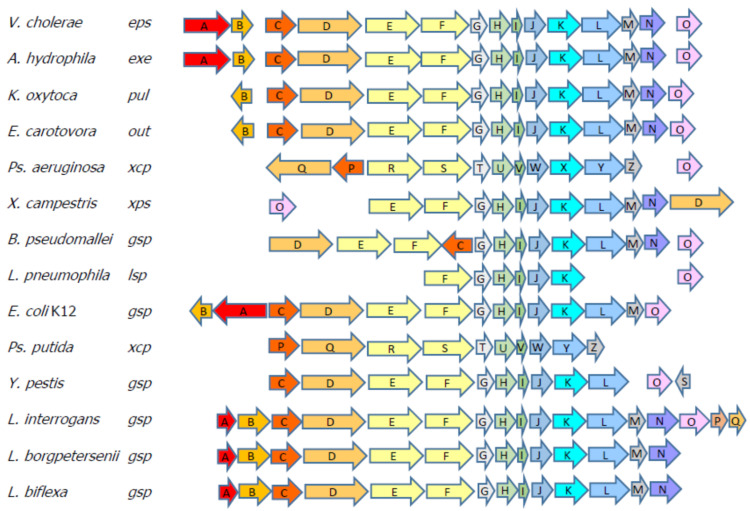
Surface proteins act as potential protective antigens due to their direct interaction with the cells of the host, and so both pathogenesis and immune response develop in parallel [29]. In Leptospira, surface and subsurface proteins, such as LipL32, LipL21, LipL41 [7,9,30], and others like OmpL1, LemA, LigA, LigB [13,31,32], have been shown to be localized in the Leptospira surfaceome and to be conserved among pathogenic serovars; for these reasons, they have been considered as candidates to develop wide spectrum vaccines that may offer heterologous immunity [20,33,34,35]. The secretin subcomplex of the type II secretion system (T2S) has been shown to be built as a pentadecamer of the GspD protein, and that arrangement provides a pore through to the outer membrane in the Proteobacteria [17]. Sequencing of Leptospira genomes has revealed a T1S and T2S but no other secretion systems, with complete Sec and TAT translocases systems [36]. We have shown that the GspD protein of the T2S is expressed by every Leptospira serovar that was screened in this work, as shown in the Western blot using an anti-rGspDLC polyclonal antibody (Figure 3). The detection of an additional 45 kDa band in most of the serovars tested in the assay (all of them but Ballum) might be the result of an undetermined immunoreactive leptospiral protein with a similar structural region that was detected by the polyclonal rabbit antisera. On the other hand, we also showed that GspDLC might well be a surface protein, as shown by the immunohistochemistry assays (Figure 5). Bioinformatic comparison of Gram-negative genomes revealed that some of the T2S genes are common but not universally conserved. Bacteria, such as Klebsiella, Pseudomonas, Vibrio, and Aeromonas, have been used as models for the study of the T2S components and the understanding of their secretion mechanisms [37,38]. While highly prevalent in γ– and β–proteobacteria, the T2S is also recognized in members of the δ and α classes [17,39]. About 12 to 15 different components have been determined as part of the T2S in the Gram-negative cell envelope, spanning from the cytoplasmic membrane to the outer membrane [40]. The first insight of the presence of a T2S in Leptospira has been revealed by MALDI-TOF mass spectrometry of Triton X-114 outer membrane extracts of L. interrogans serovar Lai grown at 20, 30, and 37 C. The analysis has identified, amongst others, the presence of pL18, an 18 kDa protein that is expressed under all tested conditions [33]. The in silico analysis of the predicted encoding sequence of 14 residues of the pL18 protein has shown significant similarity with GspG, a pseudopilin of the T2S in E. coli, Pseudomonas, and other Gram-negative bacteria [41,42,43,44]. Further chromosome walking sequencing has found a series of 14 ORFs corresponding to homologs of the T2S components in the serovar Hardjobovis L550 genome [45]. In Leptospira, 17 genes encoding Gsp components are found in serovars Lai and Copenhageni [46,47], but only 14 homologs are present in serovar Hardjobovis and in the nonpathogenic serovar Patoc [45,48]. Despite that Leptospira biflexa also harbors an encoded T2S locus, their genes show significant divergence when compared with those of the pathogenic L. interrogans or L. borgpetersenii serovars, with a Clustal W general identity of 59% amongst their gspD genes. The gsp loci in Leptospira show a similar order of genes when compared with Gram-negative bacteria [38] (Figure 7).
Figure 7.
Schematic of the type II secretion system (T2S) loci in different bacteria. L. interrogans shows 17 ORFs in the gsp locus; meanwhile, L. borpetersenii and L. biflexa both show only 14 ORFs in their loci. (Modified from Sandkvist, 2001 [38]).
The T2S in Gram-negative bacteria is considered the main secretion system involved in delivering a variety of toxins and enzymes to the outer surface or the extracellular space. Such secreted products include acyltransferases, amylases, kinases, celluloses, pectinases, ADP-ribosylation enzymes, proteases, lipases, and phosphatases, which break down complex biochemical components, thus conferring survival advantages to both pathogenic as well as environmental bacteria [17,49,50]. It has been shown that Leptospira synthetizes diverse hydrolytic enzymes, such as hyaluronidase [51], catalase [52], lipases [53], putative cytochrome oxidases [54,55], and others like the virulence modifying proteins (VMPs) [56] and the putative Quorum sensing (QS) proteins [57], which might well be secreted by the leptospiral T2S. The presence of a T2S in Leptospira implies its role in metabolism, but it also might well be related to pathogenesis, as it has been reported for other Gram-negatives, such as the Vibrio cholerae CLT toxin [58], the E. coli LT toxin [59], the Pseudomonas aeruginosa elastase, exotoxin A, phospholipase C, alkaline phosphatase, and lipases [41,60], as well as other proteins in plant pathogens [17], which are all secreted through their T2S. Some pathogenicity processes facilitated by the T2S include tissue damage, cytotoxicity, adherence, and dissemination [38], and such processes have also been reported in pathogenic Leptospira serovars [56]. The role in metabolism and pathogenesis of the T2S of Leptospira still waits to be explored.
Some reverse vaccinology strategies have been applied to search for potential immunogenic antigens in Leptospira [15]; however, only one study has considered the GspD secretin as a candidate for vaccine trials [16]. In an extensive study of 238 predicted outer membrane or secreted proteins, the authors found GspD as a seroreactive protein. In such a study, GspD was used in a pool of five proteins, including three putative lipoproteins and a TolC-like protein. Despite that all these five proteins were recognized by the sera of vaccinated hamsters, the authors found no protection against leptospirosis in the hamster model [16]. Those findings are in contrast with our results, where the rGspDLC protein mixed with IFA protected up to 75% of hamsters against lethal infection.
Our findings showing that the rGspDLC protein conferred up to 75% protection against disease in the hamster model, the fact that GspD is conserved, abundant [18], and a putative functionally important protein in pathogenic Leptospira species, suggest that the T2S GspD secretin of Leptospira might well be considered as a potential immunogen to be used for protection against leptospirosis. Further efforts could be directed to demonstrate that GspD as a vaccine protects against other Leptospira serovars and species as well as conferring protection in other animal species against leptospirosis, as well as to define the immunogenic region or epitopes of the GspD protein and finally search for the optimization of the expression of such peptides into more efficient hosts other than E. coli.
Acknowledgments
We thank David A. Haake for providing technical support and expertise to fulfill part of the molecular biology of this work at his laboratory.
Author Contributions
Conceptualization, A.d.l.P.-M.; methodology, J.M., D.A.H., E.A.R.R., C.A.C.G., A.d.l.P.-M.; validation, J.M., D.A.H., I.E.C.A.; formal analysis, A.d.l.P.-M.; investigation, S.P.L.S., L.O.C.S., G.C.M., E.A.R.R., L.O.L., R.M.B., L.E.A.S., M.G.N.C., R.O.J.M., C.A.C.G., I.E.C.A.; resources, A.d.l.P.-M.; writing—original draft preparation, A.d.l.P.-M.; writing—review and editing, A.C.G., I.E.C.A., A.d.l.P.-M.; visualization, A.d.l.P.-M.; supervision, D.A.H., J.M., A.d.l.P.-M.; project administration, E.A.R.R., C.A.C.G., L.O.C.S., A.d.l.P.-M.; funding acquisition, A.d.l.P.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants of the PAPIIT research program of the National Autonomous University of Mexico, UNAM (IN222806, IN222913, IN216316).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Adler B., de la Peña-Moctezuma A. Leptospira and leptospirosis. Vet. Microbiol. 2010;140:287–296. doi: 10.1016/j.vetmic.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Ko A.I., Goarant C., Picardeau M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009;7:736–747. doi: 10.1038/nrmicro2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa F., Hagan J.E., Calcagno J., Kane M., Torgerson P., Martinez-Silveira M.S., Stein C., Abela-Ridder B., Ko A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop Dis. 2015;9:e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler B. Vaccines against leptospirosis. Curr. Top. Microbiol. Immunol. 2015;387:251–272. doi: 10.1007/978-3-662-45059-8_10. [DOI] [PubMed] [Google Scholar]
- 5.Bulach D.M., Kalambaheti T., de la Peña-Moctezuma A., Adler B. Lipopolysaccharide biosynthesis in Leptospira. J. Mol. Microbiol. Biotechnol. 2000;2:375–380. [PubMed] [Google Scholar]
- 6.Vinh T., Faine S., Handley C.J., Adler B. Immunochemical studies of opsonic epitopes of the lipopolysaccharide of Leptospira interrogans serovar hardjo. FEMS Immunol. Med. Microbiol. 1994;8:99–107. doi: 10.1111/j.1574-695X.1994.tb00431.x. [DOI] [PubMed] [Google Scholar]
- 7.Haake D.A., Mazel M.K., McCoy A.M., Milward F., Chao G., Matsunaga J., Wagar E.A. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 1999;67:6572–6582. doi: 10.1128/IAI.67.12.6572-6582.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seixas F.K., Fernandes C.H., Hartwig D.D., Conceicao F.R., Aleixo J.A., Dellagostin O.A. Evaluation of different ways of presenting LipL32 to the immune system with the aim of developing a recombinant vaccine against leptospirosis. Can. J. Microbiol. 2007;53:472–479. doi: 10.1139/w06-138. [DOI] [PubMed] [Google Scholar]
- 9.He H.J., Wang W.Y., Wu Z.D., Lv Z.Y., Li J., Tan L.Z. Protection of guinea pigs against Leptospira interrogans serovar Lai by LipL21 DNA vaccine. Cell Mol. Immunol. 2008;5:385–391. doi: 10.1038/cmi.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koizumi N., Watanabe H. Molecular cloning and characterization of a novel leptospiral lipoprotein with OmpA domain. FEMS Microbiol. Lett. 2003;226:215–219. doi: 10.1016/S0378-1097(03)00619-0. [DOI] [PubMed] [Google Scholar]
- 11.Umthong S., Buaklin A., Jacquet A., Sangjun N., Kerdkaew R., Patarakul K., Palaga T. Immunogenicity of a DNA and recombinant protein vaccine combining LipL32 and Loa22 for leptospirosis using chitosan as a delivery system. J. Microbiol. Biotechnol. 2015;25:526–536. doi: 10.4014/jmb.1408.08007. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho M.L., Choy H.A., Kelley M.M., Matsunaga J., Babbitt J.T., Lewis M.S., Aleixo J.A., Haake D.A. A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl. Trop. Dis. 2011;5:e1422. doi: 10.1371/journal.pntd.0001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koizumi N., Watanabe H. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine. 2004;22:1545–1552. doi: 10.1016/j.vaccine.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Yan W., Faisal S.M., McDonough S.P., Divers T.J., Barr S.C., Chang C.F., Pan M.J., Chang Y.F. Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect. 2009;11:230–237. doi: 10.1016/j.micinf.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Dellagostin O.A., Grassmann A.A., Rizzi C., Schuch R.A., Jorge S., Oliveira T.L., McBride A.J., Hartwig D.D. Reverse Vaccinology: An approach for identifying leptospiral vaccine candidates. Int. J. Mol. Sci. 2017;18:158. doi: 10.3390/ijms18010158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray G.L., Lo M., Bulach D.M., Srikram A., Seemann T., Quinsey N.S., Sermswan R.W., Allen A., Adler B. Evaluation of 238 antigens of Leptospira borgpetersenii serovar Hardjo for protection against kidney colonisation. Vaccine. 2013;31:495–499. doi: 10.1016/j.vaccine.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 17.Cianciotto N.P., White R.C. Expanding role of Type II Secretion in bacterial pathogenesis and beyond. Infect. Immun. 2017;85:e00014-17. doi: 10.1128/IAI.00014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malmstrom J., Beck M., Schmidt A., Lange V., Deutsch E.W., Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira Interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinne M., Haake D.A. A comprehensive approach to identification of surface-exposed, outer membrane-spanning proteins of Leptospira Interrogans. PLoS ONE. 2009;4:e6071. doi: 10.1371/journal.pone.0006071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsunaga J., Barocchi M.A., Croda J., Young T.A., Sanchez Y., Siqueira I., Bolin C.A., Reis M.G., Riley L.W., Haake D.A., et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 2003;49:929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed N., Devi S.M., Valverde Mde L., Vijayachari P., Machang’u R.S., Ellis W.A., Hartskeerl R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006;5:28. doi: 10.1186/1476-0711-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asoh T., Saito M., Villanueva S.Y., Kanemaru T., Gloriani N., Yoshida S. Natural defense by saliva and mucosa against oral infection by Leptospira. Can. J. Microbiol. 2014;60:383–389. doi: 10.1139/cjm-2014-0016. [DOI] [PubMed] [Google Scholar]
- 23.Ellinghausen H.C., Jr., McCullough W.G. Nutrition of Leptospira Pomona and growth of 13 other serotypes: A serum-free medium employing oleic albumin complex. Am. J. Vet. Res. 1965;26:39–44. [PubMed] [Google Scholar]
- 24.Reed L.J., Muench H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;23:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 25.Fletcher W. Recent work on leptospirosis, Tsutsugamushi disease and typhus in the Federated Malay States. Trans. Roy. Soc. Trop Med. Hyg. 1928;21:265–288. doi: 10.1016/S0035-9203(28)90019-X. [DOI] [Google Scholar]
- 26.Massone A.R., Martin A.A., Ibargoyen G.S., Gimeno E.J. Immunohistochemical methods for the visualization of Mycobacterium paratuberculosis in bovine tissues. Zent. Vet. B. 1990;37:251–253. doi: 10.1111/j.1439-0450.1990.tb01054.x. [DOI] [PubMed] [Google Scholar]
- 27.Haines D.M., Chelack B.J. Technical considerations for developing enzyme immunohistochemical staining procedures on formalin-fixed paraffin-embedded tissues for diagnostic pathology. J. Vet. Diagn Investig. 1991;3:101–112. doi: 10.1177/104063879100300128. [DOI] [PubMed] [Google Scholar]
- 28.Coutinho M.L., Matsunaga J., Wang L.C., de la Peña-Moctezuma A., Lewis M.S., Babbitt J.T., Aleixo J.A., Haake D.A. Kinetics of Leptospira interrogans infection in hamsters after intradermal and subcutaneous challenge. PLoS Negl. Trop Dis. 2014;8:e3307. doi: 10.1371/journal.pntd.0003307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cash P. Investigating pathogen biology at the level of the proteome. Proteomics. 2011;11:3190–3202. doi: 10.1002/pmic.201100029. [DOI] [PubMed] [Google Scholar]
- 30.Seixas F.K., da Silva E.F., Hartwig D.D., Cerqueira G.M., Amaral M., Fagundes M.Q., Dossa R.G., Dellagostin O.A. Recombinant Mycobacterium bovis BCG expressing the LipL32 antigen of Leptospira interrogans protects hamsters from challenge. Vaccine. 2007;26:88–95. doi: 10.1016/j.vaccine.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 31.Hartwig D.D., Forster K.M., Oliveira T.L., Amaral M., McBride A.J., Dellagostin O.A. A Prime-boost strategy using the novel vaccine candidate, LemA, protects hamsters against leptospirosis. Clin. Vaccine Immunol. 2013;20:747–752. doi: 10.1128/CVI.00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lourdault K., Wang L.C., Vieira A., Matsunaga J., Melo R., Lewis M.S., Haake D.A., Gomes-Solecki M. Oral immunization with Escherichia coli expressing a lipidated form of LigA protects hamsters against challenge with Leptospira interrogans serovar Copenhageni. Infect. Immun. 2014;82:893–902. doi: 10.1128/IAI.01533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cullen P.A., Cordwell S.J., Bulach D.M., Haake D.A., Adler B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infect. Immun. 2002;70:2311–2318. doi: 10.1128/IAI.70.5.2311-2318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haake D.A., Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect. Immun. 2002;70:4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsunaga J., Sanchez Y., Xu X., Haake D.A. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 2005;73:70–78. doi: 10.1128/IAI.73.1.70-78.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng L., Zhang Y., Zhu Y., Yin H., Zhuang X., Zhu W., Guo X., Qin J. Extracellular proteome analysis of Leptospira interrogans serovar Lai. OMICS. 2013;17:527–535. doi: 10.1089/omi.2013.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Filloux A. The underlying mechanisms of type II protein secretion. Biochim. Biophys. Acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 38.Sandkvist M. Type II secretion and pathogenesis. Infect. Immun. 2001;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abby S.S., Rocha E.P.C. Bacterial Protein Secretion Systems. Volume 1615. Humana Press; New York, NY, USA: 2017. Identification of protein secretion systems in bacterial genomes using MacSyFinder; pp. 1–21. [DOI] [PubMed] [Google Scholar]
- 40.Korotkov K.V., Sandkvist M. Architecture, function, and substrates of the Type II Secretion System. Ecosal. Plus. 2019;8:227–244. doi: 10.1128/ecosalplus.ESP-0034-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ball G., Durand E., Lazdunski A., Filloux A. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 2002;43:475–485. doi: 10.1046/j.1365-2958.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 42.Douzi B., Durand E., Bernard C., Alphonse S., Cambillau C., Filloux A., Tegoni M., Voulhoux R. The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. J. Biol. Chem. 2009;284:34580–34589. doi: 10.1074/jbc.M109.042366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Francetic O., Pugsley A.P. The cryptic general secretory pathway (gsp) operon of Escherichia coli K-12 encodes functional proteins. J. Bacteriol. 1996;178:3544–3549. doi: 10.1128/JB.178.12.3544-3549.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu N.T., Leu W.M., Lee M.S., Chen A., Chen S.C., Song Y.L., Chen L.Y. XpsG, the major pseudopilin in Xanthomonas campestris pv. campestris, forms a pilus-like structure between cytoplasmic and outer membranes. Biochem. J. 2002;365:205–211. doi: 10.1042/bj20020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulach D.M., Zuerner R.L., Wilson P., Seemann T., McGrath A., Cullen P.A., Davis J., Johnson M., Kuczek E., Alt D.P., et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc. Natl. Acad. Sci. USA. 2006;103:14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nascimento A.L., Verjovski-Almeida S., Van Sluys M.A., Monteiro-Vitorello C.B., Camargo L.E., Digiampietri L.A., Harstkeerl R.A., Ho P.L., Marques M.V., Oliveira M.C., et al. Genome features of Leptospira interrogans serovar Copenhageni. Braz J. Med. Biol. Res. 2004;37:459–477. doi: 10.1590/S0100-879X2004000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ren S.X., Fu G., Jiang X.G., Zeng R., Miao Y.G., Xu H., Zhang Y.X., Xiong H., Lu G., Lu L.F., et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422:888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 48.Picardeau M., Bulach D.M., Bouchier C., Zuerner R.L., Zidane N., Wilson P.J., Creno S., Kuczek E.S., Bommezzadri S., Davis J.C., et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS ONE. 2008;3:e1607. doi: 10.1371/journal.pone.0001607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korotkov K.V., Sandkvist M., Hol W.G. The type II secretion system: Biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 2012;10:336–351. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikora A.E. Proteins secreted via the type II secretion system: Smart strategies of Vibrio cholerae to maintain fitness in different ecological niches. PLoS Pathog. 2013;9:e1003126. doi: 10.1371/journal.ppat.1003126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller N.G., Froehling R.C., White R.J. Activity of leptospires and their products on L cell monolayers. Am. J. Vet. Res. 1970;31:371–377. [PubMed] [Google Scholar]
- 52.Corin R.E., Cox C.D. Characterization of leptospiral catalase and peroxidase. Can. J. Microbiol. 1980;26:121–129. doi: 10.1139/m80-018. [DOI] [PubMed] [Google Scholar]
- 53.Yanagihara Y., Taniyama T., Misaki H., Suzuki Y., Matsumoto M., Mifuchi I. Phospholipases of Leptospira. I. Presence of phospholipase A1 and lysophospholipase in Leptospira biflexa. Microbiol. Immunol. 1984;28:747–756. doi: 10.1111/j.1348-0421.1984.tb00730.x. [DOI] [PubMed] [Google Scholar]
- 54.Baseman J.B., Cox C.D. Terminal electron transport in Leptospira. J. Bacteriol. 1969;97:1001–1004. doi: 10.1128/JB.97.3.1001-1004.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Catalano-Dupuy D.L., Musumeci M.A., Lopez-Rivero A., Ceccarelli E.A. A highly stable plastidic-type ferredoxin-NADP(H) reductase in the pathogenic bacterium Leptospira interrogans. PLoS ONE. 2011;6:e26736. doi: 10.1371/journal.pone.0026736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fouts D.E., Matthias M.A., Adhikarla H., Adler B., Amorim-Santos L., Berg D.E., Bulach D., Buschiazzo A., Chang Y.F., Galloway R.L., et al. What makes a bacterial species pathogenic? Comparative genomic analysis of the genus Leptospira. PLoS Negl. Trop. Dis. 2016;10:e0004403.:e0004403. doi: 10.1371/journal.pntd.0004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao G., Kong L., Che R., Yi Y., Zhang Q., Yan J., Lin X. Identification and characterization of c-di-GMP metabolic enzymes of Leptospira interrogans and c-di-GMP fluctuations after thermal shift and infection. Front. Microbiol. 2018;20:764. doi: 10.3389/fmicb.2018.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandkvist M., Michel L.O., Hough L.P., Morales V.M., Bagdasarian M., Koomey M., DiRita V.J., Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J. Bacteriol. 1997;179:6994–7003. doi: 10.1128/JB.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tauschek M., Gorrell R.J., Strugnell R.A., Robins-Browne R.M. Identification of a protein secretory pathway for the secretion of heat-labile enterotoxin by an enterotoxigenic strain of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:7066–7071. doi: 10.1073/pnas.092152899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Filloux A., Michel G., Bally M. GSP-dependent protein secretion in gram-negative bacteria: The Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]