Significance
We use a single-run, data-independent acquisition–based mass spectrometry approach to generate and compare dozens of yeast proteomes in less than a day, and provide a comprehensive resource detailing changes to the yeast proteome following commonly used stress treatments in yeast. Our systems biology approach identifies and validates regulatory targets of an E3 ubiquitin ligase during a metabolic switch, providing insights into the interplay of metabolic pathways. The speed, simplicity, and scalability of this workflow makes it particularly well-suited for screens in any cellular system to investigate specific effects of deletions or mutants or other perturbations to obtain the response of biological system on a global level.
Keywords: yeast systems biology, mass spectrometry, proteomics, stress, GID E3 ligase
Abstract
The yeast Saccharomyces cerevisiae is a powerful model system for systems-wide biology screens and large-scale proteomics methods. Nearly complete proteomics coverage has been achieved owing to advances in mass spectrometry. However, it remains challenging to scale this technology for rapid and high-throughput analysis of the yeast proteome to investigate biological pathways on a global scale. Here we describe a systems biology workflow employing plate-based sample preparation and rapid, single-run, data-independent mass spectrometry analysis (DIA). Our approach is straightforward, easy to implement, and enables quantitative profiling and comparisons of hundreds of nearly complete yeast proteomes in only a few days. We evaluate its capability by characterizing changes in the yeast proteome in response to environmental perturbations, identifying distinct responses to each of them and providing a comprehensive resource of these responses. Apart from rapidly recapitulating previously observed responses, we characterized carbon source-dependent regulation of the GID E3 ligase, an important regulator of cellular metabolism during the switch between gluconeogenic and glycolytic growth conditions. This unveiled regulatory targets of the GID ligase during a metabolic switch. Our comprehensive yeast system readout pinpointed effects of a single deletion or point mutation in the GID complex on the global proteome, allowing the identification and validation of targets of the GID E3 ligase. Moreover, this approach allowed the identification of targets from multiple cellular pathways that display distinct patterns of regulation. Although developed in yeast, rapid whole-proteome–based readouts can serve as comprehensive systems-level assays in all cellular systems.
Proteome remodeling has repeatedly proven to be a vital cellular mechanism in response to stress, changes in environmental conditions, and toxins or pathogens. Cells must both synthesize proteins which enable them to adapt to the new environmental condition and inactivate or degrade proteins which are detrimental or no longer needed. For each environmental perturbation, the proteome must be precisely and distinctly remodeled to ensure healthy and viable cells (1). Indeed, decreases in proteome integrity are hallmarks of many human diseases, including cancer, Alzheimer’s disease, muscular dystrophies, and cystic fibrosis (2–4). Despite the importance of cellular stress responses, our understanding of how cellular pathways interact during adaptation remains incomplete. Therefore, knowing precisely how the proteome changes at a global level in response to environmental cues is crucial for identifying the underlying molecular mechanisms that facilitate cellular adaptation.
The yeast Saccharomyces cerevisiae is a powerful model system that is widely used to probe biological pathways, due to its ease of manipulation and rapid growth compared to mammalian models. In addition, the availability of extensive genetic resources in yeast, including deletion libraries (5, 6), green fluorescent protein–tagged libraries (7, 8), overexpression libraries (9), and the recently developed SWAp-tag library (10–12), has made yeast a premier model system for conducting transcriptomics, proteomics, interactomics, or metabolomics screens (13–19). Indeed, systems-wide biology screens and large-scale proteomics were both pioneered in the yeast model. Furthermore, the cellular interaction networks and molecular mechanisms ascertained in yeast can be readily applied to other systems (20–22).
Early genome-wide studies showed that over 4,000 proteins are expressed during log-phase growth in yeast and this organism was the first whose entire proteome was mapped by mass spectrometry (MS)-based proteomics (23). Subsequently, yeast has served as a model of choice for the development of ever-more-sensitive and faster proteomics workflows (23–34). Remarkably, the optimized sample preparation coupled with MS analysis performed on the Orbitrap hybrid mass spectrometer allowed identification of around 4,000 yeast proteins over a 70-min liquid chromatography (LC)-MS/MS run (24, 30). However, the necessity of technological expertise and lengthy analysis times for high-quality, in-depth yeast proteome measurements has so far precluded the widespread adoption of cutting-edge proteomics workflows in the yeast research community. With further advances in technology and new acquisition modes, such as data-independent acquisition (DIA) (35, 36), we hypothesized that it would now be possible to obtain accurate and high yeast proteome coverage by a straightforward and rapid single-run approach, enabling researchers to easily study biological processes on a global scale. Such a system could then serve as a template for more complex proteomes, including the human proteome.
One mechanism of maintaining proteome integrity is the marking and degradation of proteins that are damaged or no longer needed with ubiquitin. The conjugation of ubiquitin to its targets is catalyzed by E3 ubiquitin ligases, a diverse group of enzymes that recognize and bind target proteins and facilitate ubiquitin transfer together with an E2, ubiquitin-conjugating enzyme. Ubiquitination relies on a variety of cellular signals to direct E3 ligases to their target proteins, and tight regulation of this process is crucial for cellular viability (37). For instance, during carbon starvation, yeast cells induce expression of the inactive GID (glucose-induced degradation) E3 ligase, which is subsequently activated upon glucose replenishment. Following its activation, the GID E3 ligase targets gluconeogenic enzymes, leading to their degradation and sparing the yeast from energetically costly metabolic pathways that are unnecessary in fermentable carbon sources (38–40). In addition, ubiquitin ligases also serve as crucial regulators in response to oxidative, heavy metal, and protein folding stresses (41–43). Despite the importance of ubiquitination during cellular adaptation, our knowledge of the E3-dependent responses to cellular perturbation remains incomplete.
Here, we describe a systems biology approach employing rapid, single-run, data-independent (DIA) mass spectrometric analysis, which we use to comprehensively map changes to the yeast proteome in response to a variety of yeast stresses. We investigate growth conditions commonly used in yeast research, including growth media, heat shock, osmotic shock, amino acid starvation, and nitrogen starvation. Our DIA-based approach is sufficiently sensitive and robust to detect quantitative proteome remodeling in response to all these stresses. We then apply this methodology to probe a specific biological question to identify novel regulation by the GID E3 ligase during a metabolic switch. We use a combination of a core subunit deletion and a structure-based catalytic mutant to identify all of the known substrates of the GID E3 ligase and discover two previously unknown targets which display distinct patterns of regulation.
Results
Streamlined and Scalable Yeast Proteome Analysis Employing DIA.
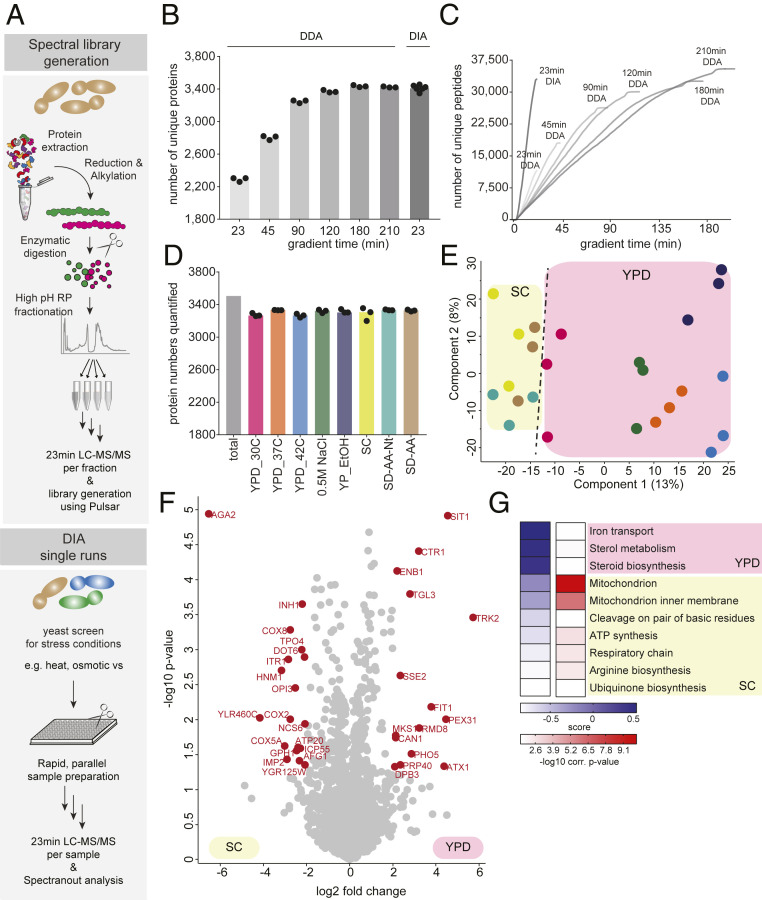
In order to establish a fast and scalable single-run analysis approach for yeast proteome profiling, we explored a DIA strategy on an Orbitrap mass spectrometer. Unlike data-dependent acquisition (DDA), a DIA method isolates coeluting peptide ions together in predefined mass windows, fragmenting and analyzing all ions simultaneously (36). This strategy overcomes the limited sequencing speed of sequential DDA, enabling fast and scalable single-shot analysis workflows. On Orbitrap-based mass analyzers, it yields substantially higher number of identified proteins with unprecedented quantitative accuracy (44). To generate a yeast-specific and comprehensive spectral library that is generally used for this approach, we cultured yeast under various growth and stress conditions. After extraction and digestion of proteins, we separated peptides obtained from each condition by basic reversed-phase (RP) chromatography into eight fractions. The resulting 64 fractions (8 fractions × 8 conditions) were measured using a DDA method with a 23-min LC gradient and analyzed with the Spectronaut software (Fig. 1A). Together with LC overhead time this took about half an hour, allowing for the analysis of 45 samples per day—almost half a 96-well plate. Our library comprised more than 74,103 precursors which mapped into 4,712 unique proteins, covering 87% of the expressed yeast proteome according to a previous report that computationally aggregated 21 different large-scale datasets (45). The median sequence coverage was 27% and on average 12 peptides were detected per protein.
Fig. 1.
Fast and scalable yeast proteome analysis using DIA. (A) Experimental workflow for yeast spectral library construction (Top) and fast, single-run DIA-based analysis of yeast proteomes (Bottom). (B) Number of identified proteins using DDA with varying LC gradient lengths compared to 23-min DIA. (C) Cumulative number of identified unique yeast peptides over time using DDA with varying LC gradient lengths and the 23-min DIA method. (D) Number of quantified proteins in growth and stress conditions. (E) PCA of conditions along with their biological replicates based on their proteomic expression profiles. (F) Volcano plot of the (−log10) P values vs. the log2 protein abundance differences between yeast grown in YPD vs. SC. The proteins marked in red change significantly (P < 0.05 and at least fourfold change in both directions). (G) GO-term enrichment in the YPD vs. SC fold change dimension (one-dimensional annotation enrichment, FDR <5%). Terms with positive enrichment scores are enriched in YPD over SC and vice versa.
Combined with our own comprehensive spectral library, the 23-min DIA method on average identified 33,909 peptides and 3,413 distinct proteins in single measurements of six replicates (Q-value less than 1% at protein and precursor levels; Fig. 1 B and C). This implies that ∼73% of proteins in the deep yeast spectral library were matched into the single runs. Note that the single runs represent only yeast grown in rich media (yeast extract peptone dextrose [YPD]), whereas the library combines the proteomes of yeast grown under several growth conditions and therefore contains proteins which are not expressed during growth in YPD. Therefore, the degree of proteome completeness is likely much higher than 73%. Measurements were highly reproducible with Pearson coefficients greater than 0.92 between replicates (SI Appendix, Fig. S1A) and coefficients of variation <20% for 68% of all common proteins between the six replicates. In comparison, a single-run, data-dependent acquisition strategy with the same LC gradient quantified only 11,883 peptides and 2,289 distinct proteins on average (Fig. 1 B and C). To more directly compare the performance of the 23-min DIA method to the DDA method we analyzed the same sample with increasing gradient lengths. We could only reach the same depth using the DDA method with at least 180-min-long LC gradients (33,425 peptides and 3,435 proteins) (Fig. 1 B and C). Thus, the DIA method allows us to obtain coverage comparable to DDA in a high-throughput and in-depth fashion while taking considerably less MS time.
Large-Scale and Quantitative Analysis of Yeast Stress Response in Half a Day.
Using this DIA-based systems biology approach, we next comprehensively and quantitatively analyzed proteome changes in response to various stresses in yeast. Each condition was processed in three biological replicates and—after tryptic digestion—the peptides were analyzed in single runs using the rapid DIA method. We quantified 3,506 distinct proteins in total (Fig. 1D and Dataset S1). Reproducibility was high, with Pearson correlations >0.93 between the three biological replicates (SI Appendix, Fig. S1B). Strikingly, over 90% of all detected proteins were consistently quantified at varying levels across all conditions (Dataset S1). Principal component analysis (PCA) demonstrated that the first component accounted for 13% of the variability and segregated with the different conditions and growth media as the major effectors (Fig. 1E).
We first looked more closely at the differences in protein expression during growth in YPD (rich media) and SC (synthetic complete media), the two most common growth media used in yeast research. YPD and SC media differ in their nutrient composition as well as their pH. During growth in YPD, the three most significantly up-regulated proteins (Sit1, Ctr1, and Enb1) are regulators of copper and iron transport (Fig. 1 F, Right and G), consistent with the fact that copper and iron are limiting factors for the growth of yeast at more alkaline pH (46). Conversely, during growth in SC, many mitochondrial proteins were up-regulated compared to YPD (Fig. 1 F, Left and G), including the cytochrome c oxidase subunits Cox8, Cox2, and Cox5a, the mitochondrial adenosine 5′-triphosphate (ATP) synthase Atp20, and the mitochondrial aminopeptidase Icp55. Yeast mitochondria reproduce through fission and must be inherited by daughter cells during cell division (47). The up-regulation of many mitochondrial proteins is thus consistent with the faster growth rate of our yeast strains in SC compared to YPD. Because the choice of media is often considered crucial in experimental design, these data on differentially regulated proteins in pathways of interest provide an important resource for yeast biologists.
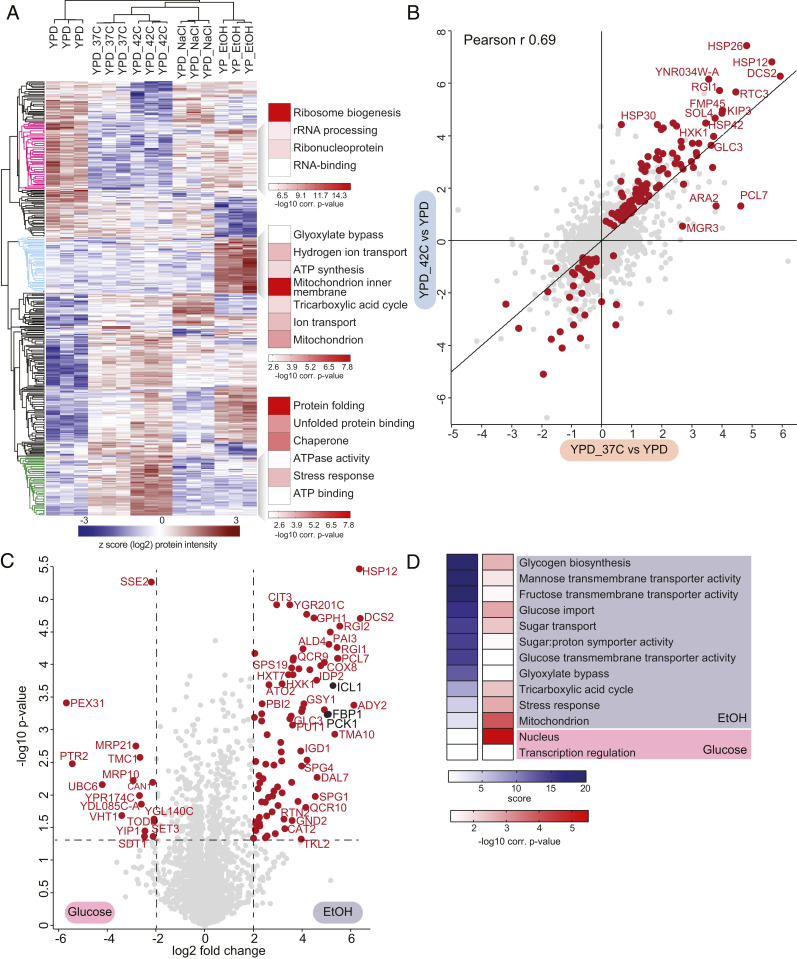
Next, we investigated proteome changes in yeast grown under various stress conditions. Here, we focused on those commonly utilized in yeast research: heat shock, osmotic shock, carbon starvation, amino acid starvation, and nitrogen starvation. Each produced a discrete stress response, resulting in synthesis or degradation of a distinct set of proteins (Fig. 2A). For example, yeast cells grown under heat shock induce expression of chaperones and stress-response proteins, a well-characterized response that allows the cell to quickly recover from global heat-induced protein misfolding (48–50). Importantly, our data also revealed that the heat-shock response is dose-dependent, with higher induction of the stress response at 42 °C compared to 37 °C (Fig. 2 A, green cluster and B). Yeast experiencing osmotic shock, on the other hand, induced distinct proteome changes, with the most enriched Gene Ontology (GO) term under this condition being actin-cortical patch (SI Appendix, Fig. S2 A and B). This is consistent with the fact that yeast cells rapidly disassemble and remodel the actin cytoskeleton during osmotic stress and favor the formation of actin patches over filaments, a mechanism that lowers the turgor pressure and allows continued growth of yeast under high osmolarity (51, 52). In addition, one of the most up-regulated proteins during osmotic stress is Ena1 (SI Appendix, Fig. S2A), a sodium efflux pump that plays a crucial role in allowing salt tolerance (53). Growth during amino acid or nitrogen starvation primarily resulted in the induction of amino acid biosynthetic pathways, with arginine and cysteine synthesis being particularly up-regulated (SI Appendix, Fig. S2 C–F).
Fig. 2.
Large-scale and quantitative analysis of yeast proteomes under different stresses. (A) Heat map of z-scored protein abundances (log2 DIA intensities) of the differentially expressed proteins (ANOVA, FDR <0.01) after hierarchical clustering of stress conditions performed in YPD and YPE. Fisher exact test was performed to identify significantly enriched GO terms in the most prominent profiles (FDR <5%). (B) Correlation of log2 fold-changes of all the quantified proteins during heat shock. The proteins that change significantly in either 37 °C or 42 °C compared to 30 °C YPD control are colored in red (t test, FDR <5%). (C) Volcano plot of the (−log10) P values vs. the log2 protein abundance differences between glucose starvation (ethanol) vs. YPD. Red dots indicate significantly different proteins, determined based on P < 0.05 and at least fourfold change in both directions. (D) GO-term enrichment in the ethanol vs. YPD fold change dimension (one-dimensional annotation enrichment, FDR <5%). Terms with positive enrichment scores are enriched in stress condition over glucose (YPD) control and vice versa.
In addition to temperature and nutrient availability, carbon source is a crucial determinant of yeast growth. We compared the proteomes of yeast grown in the aerobic carbon source, glucose, with the nonfermentable carbon source, ethanol. Yeast will preferentially metabolize aerobic carbon sources, such as glucose, when they are present in the media. When only nonfermentable carbon sources, such as ethanol, are present, yeast cells will instead metabolize them through several pathways, including gluconeogenesis to generate glucose and conversion of ethanol into pyruvate to allow for ATP generation in the mitochondria via the tricarboxylic acid cycle (54, 55). Consistent with this, we observe a general up-regulation of mitochondrial proteins and those involved in the tricarboxylic acid cycle during growth in ethanol (Fig. 2 A, light blue cluster, C, and D). In addition, many proteins involved in carbon metabolism are differentially regulated in glucose and ethanol-containing media. For example, we see a greater than 16-fold up-regulation of the gluconeogenic enzymes Fbp1, Pck1, and Icl1 (Fig. 2C). In the absence of glucose, both Hxt7, a glucose transporter, and Hxk1, a hexokinase, are up-regulated (Fig. 2C), allowing the cell to quickly import and metabolize any glucose in the environment. These results are consistent with the idea that yeast have “anticipatory” programming, which not only allows them to adapt to the current stressor but also facilitates a rapid response to shifts in environmental conditions (40, 56). Moreover, apart from identifying proteins that have altered levels in response to a shift in environmental conditions, we also accurately determined their fold changes, giving valuable insight into the protein content under different stress and growth conditions that is indispensable for systems-level modeling.
Taken together, our results indicate that the fast and robust DIA-based approach described here can reliably and quantitatively retrieve the known differences and even reveal new and biologically meaningful regulation of protein expression, thereby providing a near-comprehensive resource for yeast researchers and a valuable platform to support future studies in quantitative biology.
Global Regulation of the Yeast Proteome during Glucose Starvation and Recovery.
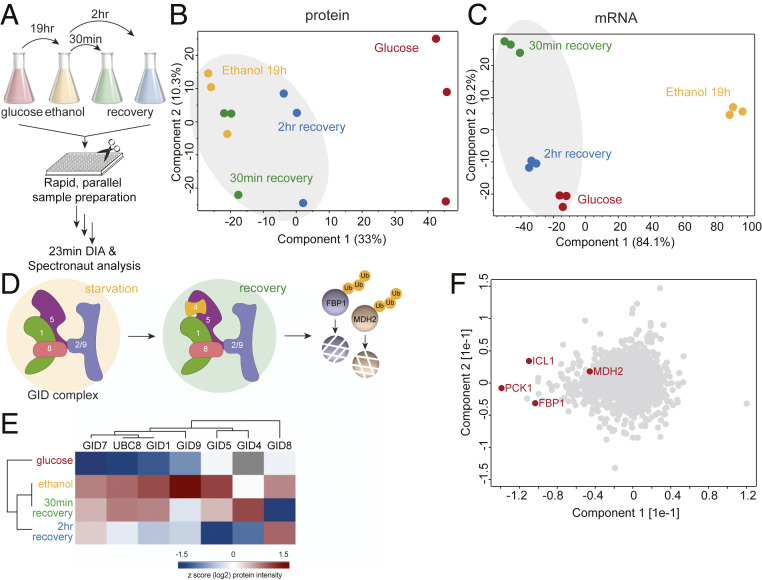
To gain better insights into how yeast regulate metabolism in response to a change in carbon source, we next expanded our analysis to investigate glucose starvation and glucose recovery. Yeast cultures were first grown to logarithmic phase in glucose then switched to media containing ethanol as a nonfermentable carbon source. Following 19 hours of growth in ethanol, glucose was replenished and the yeast were allowed to recover for 30 minutes or 2 hours (Fig. 3A). In these growth conditions, we quantified 3,602 distinct proteins in total (Dataset S2). The first PCA component segregated the growth conditions, with glucose being largely separated from the ethanol and recovery conditions (Fig. 3B and SI Appendix, Fig. S3A). To further investigate the regulation of metabolism in alternate carbon sources, we compared the proteome changes with those of the transcriptome. PCA analysis of the transcriptome also showed that the first component separated the growth conditions. Interestingly, in this case cells grown in ethanol were largely separated from the glucose (never starved) and glucose recovery conditions (Fig. 3C and SI Appendix, Fig. S3B), suggesting that during this metabolic shift yeast cells remodel their gene expression first through rapid changes in transcription, which facilitates production of new proteins, and then remove proteins that are no longer required.
Fig. 3.
Global proteome changes of yeast under glucose starvation and recovery. (A) Rapid yeast proteome profiling under glucose starvation and recovery. (B and C) PCA plot of growth conditions along with their biological replicates based on their protein expression (B) and mRNA abundance (C) profiles. (D) The GID E3 ubiquitin ligase is a key regulator of the switch from gluconeogenic to glycolytic growth as it degrades the gluconeogenic enzymes, including Fbp1 and Mdh2. (E) Heat map of z-scored protein abundances (log2) of the GID complex subunits under glucose starvation and recovery in wild-type yeast cells. (F) PCA plot of proteins during glucose starvation and recovery. Proteins marked in red represent the known GID complex substrates.
Several regulatory mechanisms contribute to carbohydrate metabolism, including allosteric regulation, reversible enzyme inactivation through covalent modifications, and irreversible loss of enzyme activity through proteolysis (reviewed in ref. 57). Importantly, we observed that protein turnover during glucose recovery occurs rapidly and in less than one cell division, suggesting an active mechanism of protein degradation. One such mechanism that has been well-characterized by our group and others is the ubiquitination and degradation of gluconeogenic enzymes by the GID E3 ubiquitin ligase. GID E3 ligase subunits are present at low levels in all growth conditions. However, during growth in ethanol, most of the GID subunits are induced, leading to the formation of a yet-inactive anticipatory complex: GIDAnt. Following glucose replenishment, the substrate receptor, Gid4, is rapidly induced and joins the complex, allowing the recognition and subsequent degradation of the gluconeogenic proteins Fbp1, Mdh2, Icl1, and Pck1 via the Pro/N-degron pathway (Fig. 3D) (38–40, 58–61). Indeed, our analysis confirmed that most components of the GID E3 ligase are up-regulated around fourfold during growth in ethanol, with the exception of Gid4, which is rapidly and transiently up-regulated within 30 min of glucose replenishment (Fig. 3E).
Intriguingly, PCA analysis of individual proteins revealed that the known substrates of the GID E3 ubiquitin ligase, Fbp1, Pck1, Icl1 and, to a lesser extent, Mdh2 are the major contributors to the segregation based on growth condition (Fig. 3F). While the GID E3 ligase is known to be an important contributor to the regulation of yeast metabolism during the switch from gluconeogenic to glycolytic conditions, and is thought to have additional substrates, the lack of an obvious phenotype in GID mutants has made the identification of further substrates challenging. Thus, we applied a DIA-based workflow to search for novel regulatory targets of the GID E3 ligase.
Identifying GID Ligase-Dependent Regulation during Recovery from Carbon Starvation.
The structure and molecular mechanism of the GID E3 ligase are known, but the pathways it regulates are only beginning to be elucidated (38, 40, 61). While the role of the GID ligase in the regulation of gluconeogenesis is well-characterized, the conservation of this multiprotein complex throughout eukaryotes suggests that it likely regulates additional pathways. For example, the GID/CTLH complex has a role in erythropoiesis and spermatogenesis in human cells and in embryogenesis in Drosophila (62–65). Thus, we set out to uncover additional pathways regulated by the GID E3 ligase in yeast by utilizing a combination of mutants. First, we used a deletion of the substrate receptor, Gid4, which targets proteins with either an N-terminal proline or a proline at position 2 via the Pro/N-degron pathway (38, 59, 60, 66). Deletion of Gid4 therefore should prevent substrate binding to the GID complex and thereby inhibit degradation. However, Gid4, while conserved in human cells, is not conserved throughout all eukaryotes. For example, the GID complex in Drosophila lacks an identifiable Gid4 homolog (65), suggesting an alternate mode of recognition. In addition, in yeast, the protein Gid10 has been identified as an alternate substrate receptor of the GID complex (40, 67), although no Gid10-dependent cellular substrates have been identified to date. To identify pathways regulated by the GID complex by an alternative recognition pathway, we used a structure-based point mutant in the RING-domain-containing subunit, Gid2K365A, which eliminates catalytic activity without altering folding or complex assembly (40).
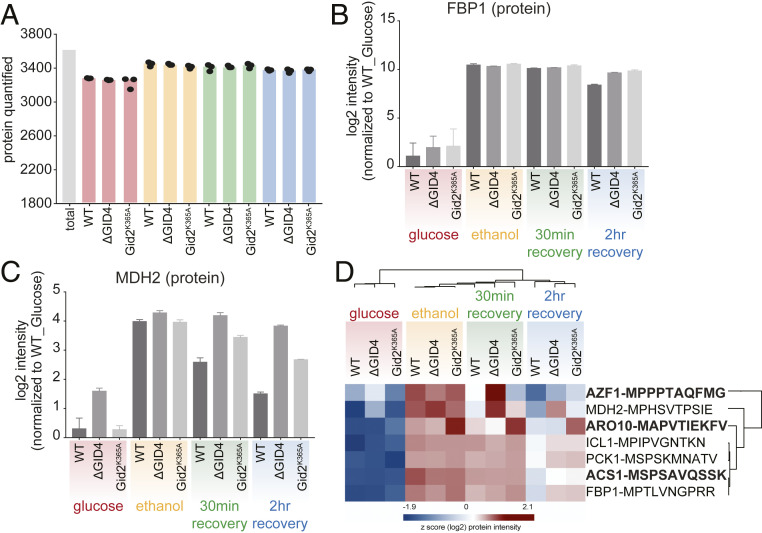
We compared the transcriptomes and proteomes of wild-type yeast to yeast containing either a Gid4 deletion or a Gid2 mutant (Gid2K365A) grown under the glucose starvation and recovery conditions described previously. Each condition was measured in triplicate using the rapid DIA method (Fig. 4A). Importantly, there were no GID-dependent differences in messenger RNA (mRNA) levels following glucose replenishment (SI Appendix, Fig. S4A), demonstrating that the GID E3 ligase does not regulate protein synthesis but rather the fate of existing proteins. To confirm that a DIA-based approach would be able to recognize bona fide GID substrates, we first examined the expression patterns of the well-characterized substrates Fbp1 and Mdh2. Indeed, in wild-type cells, Fbp1 and Mdh2 protein levels are induced during growth in ethanol and then turned over within 2 hours of glucose recovery, with Fbp1 and Mdh2 protein levels reduced by around eightfold and 5.7-fold, respectively. As expected, both proteins are also stabilized in the GID4-deleted and gid2-mutant cells (Fig. 4 B and C), confirming that we can robustly identify changes in expression of known substrates.
Fig. 4.
Rapid and robust DIA-based approach identifies GID substrates during recovery after glucose starvation. (A) Number of quantified proteins in wild type (WT), ΔGID4, and Gid2K365A yeast cells during glucose starvation and recovery. (B and C) Bar graphs showing abundances (log2) of Fbp1 (B) and Mdh2 (C) proteins that are normalized to WT glucose (never starved) condition in WT, ΔGID4, and Gid2K365A yeast cells during glucose starvation and recovery. (D) Heat map of z-scored protein abundances (log2) of the proteins which have the criteria of GID substrates.
To identify novel targets, we searched for proteins with an expression profile similar to the known substrates based on the following criteria: 1) the protein should be expressed more highly in ethanol than glucose, 2) its levels should decrease during glucose replenishment, and 3) after 2 h of glucose replenishment it should have a higher expression level in the GID4-deleted and/or gid2-mutant cells, compared to wild type (SI Appendix, Fig. S4B). This provided a list of 31 proteins, including all four known GID substrates (Fbp1, Mdh2, Pck1, and Icl1) (SI Appendix, Fig. S4C). To further prioritize candidates, we limited our search to proteins with an N-terminal proline or a proline in the second position, a genetic and structural requirement of all known cellular substrates (38, 40, 60). The resulting list of seven proteins consisted of the four known substrates, the transcription factor Azf1, and the metabolic enzymes Aro10 and Acs1 (Fig. 4D). Interestingly, Azf1 has already been implicated in regulation of GID4 transcription (68), suggesting its up-regulation in the GID-deficient cells may be a cellular compensation mechanism. However, because we did not observe any GID-dependent mRNA expression changes (SI Appendix, Fig. S4A), we eliminated Azf1 from further analysis. Acs1 was significantly stabilized in both the GID4-deleted and gid2-mutant cells, whereas Aro10 was only significantly stabilized in the gid2 mutant.
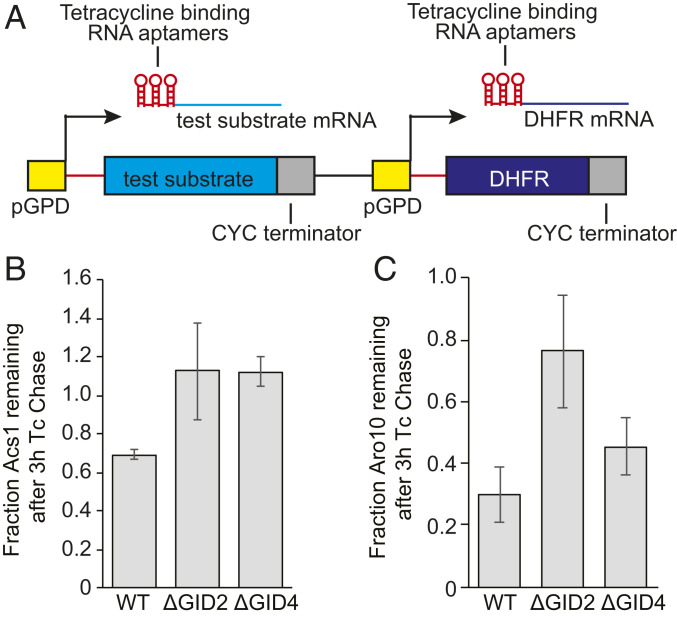
In order to validate Aro10 and Acs1 as GID targets in vivo, we used the promoter reference technique (38, 69), a transcription-independent method to examine protein turnover. In this method, yeast cells are transformed with a plasmid expressing the test substrate and the control protein DHFR from identical promoters (Fig. 5A). The transcribed products carry tetracycline-binding RNA aptamers which inhibit protein expression at the level of translation following the addition of tetracycline to the media, allowing the fate of the existing protein to be monitored. Importantly, this method selectively terminates synthesis of our test proteins, and thus the induction of Gid4 and activation of the GID complex is not impaired. In agreement with our proteomic findings, the Acs1 protein is completely stabilized in both GID2-and GID4-deleted cells (Fig. 5B), while the Aro10 protein is stabilized in GID2-deleted but not GID4-deleted cells (Fig. 5C), indicating a potential Gid4-independent regulation. Thus, Acs1 and Aro10 are confirmed to be regulatory targets of the GID E3 ligase during the switch from gluconeogenic to glycolytic conditions.
Fig. 5.
In vivo validation of GID targets. (A) Schematic of constructs used in the promoter reference technique. (B and C) Quantification of Acs1 (B) and Aro10 (C) degradation, based on at least four independent replicates. Bars represent mean values, and error bars represent SD.
Discussion
Here, we described a straightforward, streamlined, and reproducible systems biology approach for yeast proteome profiling using DIA to analyze biological pathways much faster and with greater depth. The minimalistic workflow employs plate-based preparation of digested yeast cell lysate and requires only a few micrograms of yeast as input and no labeling or special equipment, making it especially amenable for application in nonspecialized research groups. Despite its simplicity, it robustly and quantitatively profiles hundreds of largely covered yeast proteomes [80% of the expressed proteome at normal growth conditions (7)] within an unprecedented throughput (100 samples in ∼2.2 days).
The ability of cells to adapt to stress or changes in environmental conditions relies on extensive proteome remodeling (70–75). Understanding these changes provides broad insight into the molecular mechanisms underlying many processes including heat stress, adaptation to nutrient availability, and regulation of cell division. Applying the DIA-based approach to profile protein levels during response to several stress and growth conditions demonstrated its systems-wide robustness and specificity. In addition, our work provides an in-depth resource on stress mediators regulated at the protein level, which will complement the widely available yeast transcriptome data and further allow yeast researchers to probe numerous biological pathways of interest, including stress response pathways, autophagy, and nutrient signaling pathways.
In addition to identifying proteome changes during stress, we used the DIA-based systems biology approach to identify proteins that are regulated by the GID E3 ubiquitin ligase, a key regulator in the switch from gluconeogenic to glycolytic conditions (54, 61, 76). Despite the importance of the GID complex in metabolic regulation, identification of additional substrates has been hindered by the lack of an obvious phenotype, variable kinetics of protein degradation, and the necessity for a sensitive readout. Our generic and unbiased approach, however, robustly identified two protein regulatory targets of the GID complex, Acs1 and Aro10, further highlighting the importance and need for quantitative proteome datasets to provide a basis for functional studies.
Interestingly, both Acs1 and Aro10, while not considered gluconeogenic enzymes, are important regulators of metabolism and cellular respiration during anaerobic growth. Acs1 encodes one of two isoforms of yeast acetyl-CoA synthetase, which catalyzes the formation of acetyl-CoA from acetate and CoA. Acs1 has a much higher affinity for acetate than its isoform Acs2, making it more desirable for acetyl-CoA production when acetate is limiting, as is the case during growth on nonfermentable carbon sources (77). During glycolytic growth, however, the main energy flux does not require Acs1/2 function, Acs1 expression is suppressed, and existing Acs1 protein must be degraded. Aro10 encodes a phenylpyruvate decarboxylase that catalyzes an irreversible step in the Ehrlich pathway, which provides a more energetically favorable means of NADH (reduced nicotinamide-adenine dinucleotide) regeneration during anaerobic growth. Following glucose replenishment, NADH is regenerated through glycolysis, and thus Aro10 function is no longer required (78, 79).
Here, we show that both Acs1 and Aro10 turnover are dependent on the catalytic activity of the GID complex, via its RING-containing subunit, Gid2. Intriguingly, only Acs1 turnover is dependent on the well-characterized substrate receptor, Gid4, suggesting an alternate mode of recognition for Aro10. Indeed, an additional substrate receptor, Gid10, has recently been identified (40, 67), raising the possibility that Aro10 may be the first substrate identified in this recognition pathway. Alternatively, Aro10 recognition may be facilitated by a yet-to-be identified substrate receptor or an alternative mechanism. In either case, the regulation of Aro10 suggests that the GID E3 ligase may function with separable catalytic and substrate recognition elements, a mechanism previously described for SCF (Skp1-Cullin-Fbox) E3 ligases (80, 81) that provides a flexible means for linking a single E3 to a greater number of substrates. Intriguingly, expression of the GID substrate receptors is induced during several other cellular stresses, including osmotic shock, heat shock, and nitrogen starvation (40, 67, 71), suggesting that the GID complex may play an important role in rewiring metabolic pathways during adaptation to a wide variety of stress conditions.
Taken together, the GID-dependent regulation of Acs1 and Aro10, along with the previously known substrates, suggests that the GID complex is a multifunctional metabolic regulator that influences multiple cellular pathways simultaneously to allow for an efficient switch from gluconeogenic to glycolytic conditions. Moreover, our findings demonstrate that the DIA-based systems biology approach is capable of simultaneously identifying changes to multiple cellular pathways which are integrated to maintain cellular homeostasis. While we here identified specific targets of an E3 ligase, this workflow can be readily adopted by the community to probe numerous cellular pathways, including kinase signaling pathways or cell-cycle-dependent changes. Furthermore, its speed allows the analyzing of at least 15 conditions, in triplicate, per day, making it particularly well-suited for screens. For example, the effect of each of the ∼80 yeast E3 ligases on the global proteome could be ascertained in just 5 days, or each of the ∼117 yeast kinases in about 1 week. In addition, the DIA-based workflow can be easily adapted to identify changes in posttranslational modifications including phosphorylation, ubiquitination, and acetylation, when coupled with an enrichment step (44, 82–84).
Thus, the speed and reproducibility of the DIA-based approach presented here allows researchers to probe complex biological pathways and identify novel regulatory mechanisms. We are currently integrating an HPLC system into our approach as it eliminates the overhead time between sample pickup and start of MS measurement by using preformed gradients (85). Simplified workflows like the one described here could be extended to other organisms, generating high-quality quantitative proteome datasets which are required to explain biological processes on a system-wide level (86, 87). Furthermore, we believe that library-free approaches using prediction tools will further increase the speed of DIA-based proteome profiling workflows like the one presented here. Given that the expressed human proteome (around 15,479 proteins, https://www.proteomicsdb.org) is only around three times larger than the expressed yeast proteome [5,391 proteins, (45)], with only three fold increase in proteomic depth, we anticipate fast single run DIA approaches will also be suitable for rapid generation of human proteomes.
Materials and Methods
Yeast Strains and Growth Conditions.
All yeast strains used in this study are derivatives of BY4741 and are listed in Table 1. For rich conditions, yeast cultures were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) or SC (0.67% yeast nitrogen base without amino acids, 2% glucose, containing 87.5 mg/L alanine, arginine, asparagine, aspartic acid, cysteine, glutamine, glutamic acid, glycine, leucine, lysine, methionine, myo-inositol, isoleucine, phenylalanine, proline, serine, threonine, tyrosine and valine, 43.7 mg/L histidine, tryptophan and uracil, 22.5 mg/L adenine, and 8.7 mg/L para-aminobenzoic acid) media. Unless otherwise specified, yeast cultures were grown at 30 °C. For heat-shock conditions, yeast cultures were grown in YPD to an optical density at 600 nm (OD600) of 1.0 and then shifted to the indicated temperature for 1 h. For osmotic shock conditions, yeast cells were grown in YPD to and OD600 of 1.0, pelleted at 3,000 rpm for 3 min, and resuspended at an OD600 of 1.0 in prewarmed YPD + 0.5 M NaCl. For glucose starvation, yeast cells were grown in YPD to an OD600 of 1.0 to 2.0, pelleted at 3,000 rpm for 3 min, washed once with YPE (1% yeast extract, 2% peptone, and 2% ethanol), resuspended in prewarmed YPE at an OD600 of 1.0, and grown at 30 °C for 19 h. For glucose recovery, yeast cells were pelleted after 19 h of growth in YPE, resuspended to an OD600 of 1.0 in YPD, and allowed to grow at 30 °C for 30 min or 2 h. For amino acid starvation, yeast cells were grown in SC to an OD600 of 1.0 to 2.0, pelleted at 3,000 rpm for 3 min, washed once with SD-AA (0.67% yeast nitrogen base without amino acids, 2% glucose, and 20 mg/L uracil), resuspended in SD-AA to an OD600 of 1.0, and allowed to grow for 1 h. For nitrogen depletion, yeast cells were grown in SC to an OD600 of 1.0 to 2.0, pelleted at 3,000 rpm for 3 min, washed once with SD-N (0.17% yeast nitrogen base without amino acids or ammonium sulfate and 2% glucose), resuspended in SD-N to an OD600 of 1.0, and allowed to grow for 1 h. For proteomics analysis, 50 ODs of cells were pelleted at 3,000 rpm for 3 min, flash-frozen in liquid nitrogen, and stored at −80 °C until lysis. For transcriptomes analysis, 10 ODs of yeast were pelleted, flash-frozen in liquid nitrogen, and stored at −80 °C.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| CRLY12 | BY4741 GID4::KANMX | This study |
| CRLY30 | BY4741 GID2::KANMX | This study |
| CRLY131 | BY4741 gid2::3xFLAG-GID2K365A | Ref. 40 |
Protein Degradation Assays (Promoter Reference Technique).
Protein degradation assays using the promoter reference technique were done as previously described (69). Plasmids used are listed in Table 2. Cells were transformed with plasmid expressing a test substrate and DHFR from identical promoters containing tetracycline-repressible RNA-binding elements. Yeast cells were then grown in SC media lacking histidine, starved in SE (2% ethanol) media lacking histidine for 19 h, and then allowed to recover for the indicated times in SC media lacking histidine. At each time point, 1.0 ODs of yeast cells were pelleted, flash-frozen in liquid nitrogen, and stored at −80 °C until lysis.
Table 2.
Plasmids used in this study
| Plasmid | Source | |
| CRLP47 | pRS313-PTDH3(modified)-Aro103xFlag-CYC-pTDH3(modified)-flagDHFRha-CYC | This study |
| CRLP48 | pRS313-PTDH3(modified)-Acs13xFlag-CYC-pTDH3(modified)-flagDHFRha-CYC | This study |
For lysis, yeast cells were resuspended in 0.8 mL of 0.2 M NaOH, followed by incubation on ice for 20 min, and then pelleted at 11,200 × g for 1 min. The supernatant was removed and the pellet resuspended in 50 μL HU buffer and incubated at 70 °C for 10 min. The lysate was precleared by centrifugation at 11,200 × g for 5 min and then loaded onto a 12% sodium dodecyl sulfate polyacrylamide gel. Protein samples were transferred to a nitrocellulose membrane and then visualized by Western blot using αFLAG (F1804; Sigma) and α-hemagglutinin (H6908; Sigma) primary antibodies and Dylight 633 goat anti-Mouse (35512; Invitrogen) and Dylight 488 goat anti-rabbit (35552; Invitrogen) secondary antibodies. Membranes were imaged on a typhoon scanner (Amersham). Bands were quantified with ImageStudio software (LI-COR).
mRNA Sequencing.
Harvested and frozen cells were sent to Novogene Co., Ltd. (Hong Kong) for RNA extraction, library preparation, mapping, and bioinformatics analysis. Briefly, 3 μg of RNA was used for library generation using NEB Next Ultra RNA Library Prep Kit for Illumina (NEB). The library preparations were sequences on an Illumina Hi-Seq platform and 125-base pair (bp)/150-bp paired-end reads were generated. Reads were indexed using Bowtie v2.2.3 and paired-end clean reads were aligned to the reference genome using TopHat v2.0.12. HTSeq v0.6.1 was used to count the read numbers mapped to each gene, and then FPKM (expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced) of each gene was calculated based on the length of the gene and read counts mapped to the gene. The transcriptome data analysis was performed as explained in Data Processing and Bioinformatics Analysis.
Sample Preparation for MS Analysis.
Sodium deoxycholate (SDC) lysis buffer (1% SDC and 100 mM Tris, pH 8.4) were added to the frozen cell pellets to achieve a protein concentration of ∼2 to 3 mg per ml. Lysates were immediately heat-treated for 5 min at 95 °C to facilitate lysis and to inactivate endogenous proteases and transferred to a 96-well plate. Lysates were next homogenized with sonication. Protein concentrations were estimated by tryptophan assay (27) and then all samples were diluted to equal protein concentrations in a 96-well plate. To reduce and alkylate proteins, samples were incubated for 5 min at 45 °C with CAA and TCEP, final concentrations of 40 mM and 10 mM, respectively. Samples were digested overnight at 37 °C using trypsin (1:100 wt/wt; Sigma-Aldrich) and LysC (1/100 wt/wt; Wako). The following day, peptide material was desalted using SDB-RPS StageTips (Empore) (27). Briefly, samples were diluted with 1% trifluoroacetic acid (TFA) in isopropanol to a final volume of 200 µL and loaded onto StageTips and subsequently washed with 200 µL of 1% TFA in isopropanol and 200 µL 0.2% TFA/2% ACN (acetonitrile). Peptides were eluted with 80 µl of 1.25% Ammonium hydroxide (NH4OH)/80% ACN and dried using a SpeedVac centrifuge (Concentrator Plus; Eppendorf). Samples were resuspended in buffer A* (0.2% TFA/2% ACN) prior to LC-MS/MS analysis. Peptide concentrations were measured optically at 280 nm (Nanodrop 2000; Thermo Scientific) and subsequently equalized using buffer A*. Three hundred nanograms of peptide was subjected to LC-MS/MS analysis.
To generate the spectral library for DIA measurements cells were lysed in SDC buffer, followed by sonication, protein quantification, reduction, and alkylation and desalting using SDB-RPS StageTips (discussed above). Around 8 or 30 µg of peptides were fractionated into 8 or 24 fractions, respectively, by high-pH reversed-phase chromatography as described earlier (88). Fractions were concatenated automatically by shifting the collection tube during the gradient and subsequently dried in a vacuum centrifuge, and resuspended in buffer A*.
LC-MS/MS Measurements.
Samples were loaded onto a 20-cm reversed-phase column (75-μm inner diameter, packed in-house with ReproSil-Pur C18-AQ 1.9 μm resin [Dr. Maisch GmbH]). The column temperature was maintained at 60 °C using a homemade column oven. A binary buffer system, consisting of buffer A (0.1% formic acid [FA]) and buffer B (80% ACN plus 0.1% FA), was used for peptide separation, at a flow rate of 450 nL/min. An EASY-nLC 1200 system (Thermo Fisher Scientific), directly coupled online with the mass spectrometer (Q Exactive HF-X, Thermo Fisher Scientific) via a nano-electrospray source, was employed for nano-flow liquid chromatography. We used a gradient starting at 5% buffer B, increased to 35% in 18.5 min, 95% in a minute, and stayed at 95% for 3.5 min. The mass spectrometer was operated in Top10 data-dependent mode (DDA) with a full scan range of 300 to 1,650 m/z at 60,000 resolution with an automatic gain control (AGC) target of 3e6 and a maximum fill time of 20 ms. Precursor ions were isolated with a width of 1.4 m/z and fragmented by higher-energy collisional dissociation (HCD) (normalized collision energy [NCE] 27%). Fragment scans were performed at a resolution of 15,000, an AGC of 1e5, and a maximum injection time of 60 ms. Dynamic exclusion was enabled and set to 30 s. For DIA measurements full MS resolution was set to 120,000 with a full scan range of 300 to 1,650 m/z, a maximum fill time of 60 ms, and an AGC target of 3e6. One full scan was followed by 12 windows with a resolution of 30,000 in profile mode. Precursor ions were fragmented by stepped HCD (NCE 25.5, 27, and 30%).
Data Processing and Bioinformatics Analysis.
Spectronaut version 13 (Biognosys) was used to generate the spectral libraries from DDA runs by combining files of respective fractionations using the yeast FASTA file (UniProt, 2018). For the generation of the proteome library default settings were left unchanged. DIA files were analyzed using the proteome library with default settings and enabled cross-run normalization. The Perseus software package versions 1.6.0.7 and 1.6.0.9 and GraphPad Prism version 7.03 were used for the data analysis (89). Protein intensities and mRNA abundances were log2-transformed for further analysis. The datasets were filtered to make sure that identified proteins and mRNAs showed expression or intensity in all biological triplicates of at least one condition and the missing values were subsequently replaced by random numbers that were drawn from a normal distribution (width = 0.3 and downshift = 1.8). PCA analysis of stress and growth conditions and biological replicates was performed as previously described in ref. 90. Multisample test (ANOVA) for determining if any of the means of stress and growth conditions were significantly different from each other was applied to both mRNA and protein datasets. For truncation, we used permutation-based false discovery rate (FDR) which was set to 0.05 in conjunction with an S0-parameter of 0.1. For hierarchical clustering of significant genes and proteins, median protein or transcript abundances of biological replicates were z-scored and clustered using Euclidean as a distance measure for row clustering. GO annotations were matched to the proteome data based on UniProt identifiers. Annotation term enrichment was performed with either Fisher exact test or the 1D tool in Perseus. Annotation terms were filtered for 5% FDR after Benjamini–Hochberg correction.
Supplementary Material
Acknowledgments
This work was supported by the Max Planck Society for the Advancement of Science. We thank Arno Alpi, Florian Meier, Igor Paron, Christian Deiml, Johannes B. Mueller, Viola Beier, Kirby Swatek, Laura A. Hehl, and all the members of the departments of Proteomics and Signal Transduction and Molecular Machines and Signaling at Max Planck Institute of Biochemistry for their assistance and helpful discussions. B.A.S. is funded from the Deutsche Forschungsgemeinschaft (SCHU 3196/1-1). B.A.S. and M.M. are supported by the Max Planck Society.
Footnotes
The authors declare no competing interest.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020197117/-/DCSupplemental.
Data Availability.
All MS proteomics data have been deposited on ProteomeXchange via the PRIDE database with the dataset identifier PXD021559. All other data supporting findings of this study are available within this paper, SI Appendix, and Datasets S1 and S2.
References
- 1.Sui X., et al. , Widespread remodeling of proteome solubility in response to different protein homeostasis stresses. Proc. Natl. Acad. Sci. U.S.A. 117, 2422–2431 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berner N., Reutter K. R., Wolf D. H., Protein quality control of the endoplasmic reticulum and ubiquitin-proteasome-triggered degradation of aberrant proteins: Yeast pioneers the path. Annu. Rev. Biochem. 87, 751–782 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Hanna J., Guerra-Moreno A., Ang J., Micoogullari Y., Protein degradation and the pathologic basis of disease. Am. J. Pathol. 189, 94–103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hipp M. S., Park S. H., Hartl F. U., Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Steinmetz L. M., et al. , Systematic screen for human disease genes in yeast. Nat. Genet. 31, 400–404 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Giaever G., et al. , Functional profiling of the Saccharomyces cerevisiae genome. Nature 418, 387–391 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Ghaemmaghami S., et al. , Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Huh W. K., et al. , Global analysis of protein localization in budding yeast. Nature 425, 686–691 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Jones G. M., et al. , A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat. Methods 5, 239–241 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Weill U., et al. , Genome-wide SWAp-Tag yeast libraries for proteome exploration. Nat. Methods 15, 617–622 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yofe I., et al. , One library to make them all: Streamlining the creation of yeast libraries via a SWAp-tag strategy. Nat. Methods 13, 371–378 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meurer M., et al. , Genome-wide C-SWAT library for high-throughput yeast genome tagging. Nat. Methods 15, 598–600 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Costanzo M., et al. , A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gavin A. C., et al. , Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Ho Y., et al. , Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 (2002). [DOI] [PubMed] [Google Scholar]
- 16.DeRisi J. L., Iyer V. R., Brown P. O., Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Kumar A., et al. , Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lashkari D. A., et al. , Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc. Natl. Acad. Sci. U.S.A. 94, 13057–13062 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagalakshmi U., et al. , The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costanzo M., et al. , Global genetic networks and the genotype-to-phenotype relationship. Cell 177, 85–100 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janssens G. E., et al. , Protein biogenesis machinery is a driver of replicative aging in yeast. eLife 4, e08527 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petranovic D., Nielsen J., Can yeast systems biology contribute to the understanding of human disease? Trends Biotechnol. 26, 584–590 (2008). [DOI] [PubMed] [Google Scholar]
- 23.de Godoy L. M., et al. , Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Hebert A. S., et al. , The one hour yeast proteome. Mol. Cell. Proteomics 13, 339–347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marguerat S., et al. , Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller J. B., et al. , The proteome landscape of the kingdoms of life. Nature 582, 592–596 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M., Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Nagaraj N., et al. , System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteomics 11, M111.013722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R., Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards A. L., et al. , One-hour proteome analysis in yeast. Nat. Protoc. 10, 701–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soufi B., et al. , Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol. Biosyst. 5, 1337–1346 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Thakur S. S., et al. , Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webb K. J., Xu T., Park S. K., Yates J. R. 3rd, Modified MuDPIT separation identified 4488 proteins in a system-wide analysis of quiescence in yeast. J. Proteome Res. 12, 2177–2184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu R., et al. , Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol. Cell. Proteomics 10, M111.009654 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillet L. C., et al. , Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ludwig C., et al. , Data-independent acquisition-based SWATH-MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 14, e8126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varshavsky A., The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Chen S. J., Wu X., Wadas B., Oh J. H., Varshavsky A., An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 355, eaal3655 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hämmerle M., et al. , Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 25000–25005 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Qiao S., et al. , Interconversion between anticipatory and active GID E3 Ubiquitin ligase conformations via metabolically driven substrate receptor assembly. Mol. Cell 77, 150–163.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Kaiser P., Su N. Y., Yen J. L., Ouni I., Flick K., The yeast ubiquitin ligase SCFMet30: Connecting environmental and intracellular conditions to cell division. Cell Div. 1, 16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi A., et al. , Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol. Cell. Biol. 26, 221–229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang D. D., Hannink M., Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23, 8137–8151 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bekker-Jensen D. B., et al. , Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries. Nat. Commun. 11, 787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho B., Baryshnikova A., Brown G. W., Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192–205.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Serrano R., Bernal D., Simón E., Ariño J., Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279, 19698–19704 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Bereiter-Hahn J., Behavior of mitochondria in the living cell. Int. Rev. Cytol. 122, 1–63 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Pastor R., Burchfiel E. T., Thiele D. J., Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartl F. U., Molecular chaperones in cellular protein folding. Nature 381, 571–579 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Vabulas R. M., Raychaudhuri S., Hayer-Hartl M., Hartl F. U., Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2, a004390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blomberg A., Adler L., Physiology of osmotolerance in fungi. Adv. Microb. Physiol. 33, 145–212 (1992). [DOI] [PubMed] [Google Scholar]
- 52.Chowdhury S., Smith K. W., Gustin M. C., Osmotic stress and the yeast cytoskeleton: Phenotype-specific suppression of an actin mutation. J. Cell Biol. 118, 561–571 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Platara M., et al. , The transcriptional response of the yeast Na(+)-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J. Biol. Chem. 281, 36632–36642 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Gancedo J. M., Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schüller H. J., Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 43, 139–160 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Tagkopoulos I., Liu Y. C., Tavazoie S., Predictive behavior within microbial genetic networks. Science 320, 1313–1317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barnett J. A., Entian K. D., A history of research on yeasts 9: Regulation of sugar metabolism. Yeast 22, 835–894 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Chen S. J., Melnykov A., Varshavsky A., Evolution of substrates and components of the Pro/N-degron pathway. Biochemistry 59, 582–593 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20, 1298–1345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong C., et al. , Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway. Nat. Chem. Biol. 14, 466–473 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Santt O., et al. , The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol. Biol. Cell 19, 3323–3333 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao W. X., et al. , Precise temporal regulation of post-transcriptional repressors is required for an orderly Drosophila maternal-to-zygotic transition. Cell Rep. 31, 107783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Puverel S., Barrick C., Dolci S., Coppola V., Tessarollo L., RanBPM is essential for mouse spermatogenesis and oogenesis. Development 138, 2511–2521 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soni S., Bala S., Hanspal M., Requirement for erythroblast-macrophage protein (Emp) in definitive erythropoiesis. Blood Cells Mol. Dis. 41, 141–147 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Zavortink M., et al. , The E2 Marie Kondo and the CTLH E3 ligase clear deposited RNA binding proteins during the maternal-to-zygotic transition. eLife 9, e53889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tasaki T., Sriram S. M., Park K. S., Kwon Y. T., The N-end rule pathway. Annu. Rev. Biochem. 81, 261–289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Melnykov A., Chen S. J., Varshavsky A., Gid10 as an alternative N-recognin of the Pro/N-degron pathway. Proc. Natl. Acad. Sci. U.S.A. 116, 15914–15923 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slattery M. G., Liko D., Heideman W., The function and properties of the Azf1 transcriptional regulator change with growth conditions in Saccharomyces cerevisiae. Eukaryot. Cell 5, 313–320 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oh J. H., Chen S. J., Varshavsky A., A reference-based protein degradation assay without global translation inhibitors. J. Biol. Chem. 292, 21457–21465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berry D. B., Gasch A. P., Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol. Biol. Cell 19, 4580–4587 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gasch A. P., et al. , Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hahn J. S., Hu Z., Thiele D. J., Iyer V. R., Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 24, 5249–5256 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lindquist S., The heat-shock response. Annu. Rev. Biochem. 55, 1151–1191 (1986). [DOI] [PubMed] [Google Scholar]
- 74.Morano K. A., Grant C. M., Moye-Rowley W. S., The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mühlhofer M., et al. , The heat shock response in yeast maintains protein homeostasis by chaperoning and replenishing proteins. Cell Rep. 29, 4593–4607.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Regelmann J., et al. , Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: A genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol. Biol. Cell 14, 1652–1663 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van den Berg M. A., et al. , The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 271, 28953–28959 (1996). [DOI] [PubMed] [Google Scholar]
- 78.Hazelwood L. A., Daran J. M., van Maris A. J., Pronk J. T., Dickinson J. R., The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74, 2259–2266 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pires E. J., Teixeira J. A., Brányik T., Vicente A. A., Yeast: The soul of beer’s aroma–A review of flavour-active esters and higher alcohols produced by the brewing yeast. Appl. Microbiol. Biotechnol. 98, 1937–1949 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Bai C., et al. , SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274 (1996). [DOI] [PubMed] [Google Scholar]
- 81.Skowyra D., Craig K. L., Tyers M., Elledge S. J., Harper J. W., F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219 (1997). [DOI] [PubMed] [Google Scholar]
- 82.Hansen F. M., et al. , Data-independent acquisition method for ubiquitinome analysis reveals regulation of circadian biology. bioRxiv: 10.1101/2020.07.24.219055 (25 July 2020). [DOI] [PMC free article] [PubMed]
- 83.Stukalov A., et al. , Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. biorxiv:10.1101/2020.06.17.156455 (17 June 2020).
- 84.Baeza J., et al. , Revealing dynamic protein acetylation across subcellular compartments. J. Proteome Res. 19, 2404–2418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bache N., et al. , A novel LC system embeds analytes in pre-formed gradients for rapid, ultra-robust proteomics. Mol. Cell. Proteomics 17, 2284–2296 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahmad Y., Lamond A. I., A perspective on proteomics in cell biology. Trends Cell Biol. 24, 257–264 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Larance M., Lamond A. I., Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16, 269–280 (2015). [DOI] [PubMed] [Google Scholar]
- 88.Kulak N. A., Geyer P. E., Mann M., Loss-less nano-fractionator for high sensitivity, high coverage proteomics. Mol. Cell. Proteomics 16, 694–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tyanova S., et al. , The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Deeb S. J., et al. , Machine learning-based classification of diffuse large B-cell lymphoma patients by their protein expression profiles. Mol. Cell. Proteomics 14, 2947–2960 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All MS proteomics data have been deposited on ProteomeXchange via the PRIDE database with the dataset identifier PXD021559. All other data supporting findings of this study are available within this paper, SI Appendix, and Datasets S1 and S2.