Significance
Ischemic stroke triggers peripheral immunosuppression, increasing the susceptibility to poststroke pneumonia that is linked with poor survival. The poststroke brain initiates intensive communication with the immune system, and acetylcholine contributes to these messages; but the responsible molecules are yet unknown. We discovered a “changing of the guards,” where microRNA levels decreased but small transfer RNA fragments increased in poststroke blood. This molecular switch may rebalance acetylcholine signaling in CD14+ monocytes by regulating their gene expression and modulating poststroke immunity. Our observations point to transfer RNA fragments as molecular regulators of poststroke immune responses that may be potential therapeutic targets.
Keywords: acetylcholine, immunology, ischemic stroke, microRNA, transfer RNA fragment
Abstract
Stroke is a leading cause of death and disability. Recovery depends on a delicate balance between inflammatory responses and immune suppression, tipping the scale between brain protection and susceptibility to infection. Peripheral cholinergic blockade of immune reactions fine-tunes this immune response, but its molecular regulators are unknown. Here, we report a regulatory shift in small RNA types in patient blood sequenced 2 d after ischemic stroke, comprising massive decreases of microRNA levels and concomitant increases of transfer RNA fragments (tRFs) targeting cholinergic transcripts. Electrophoresis-based size-selection followed by qRT-PCR validated the top six up-regulated tRFs in a separate cohort of stroke patients, and independent datasets of small and long RNA sequencing pinpointed immune cell subsets pivotal to these responses, implicating CD14+ monocytes in the cholinergic inflammatory reflex. In-depth small RNA targeting analyses revealed the most-perturbed pathways following stroke and implied a structural dichotomy between microRNA and tRF target sets. Furthermore, lipopolysaccharide stimulation of murine RAW 264.7 cells and human CD14+ monocytes up-regulated the top six stroke-perturbed tRFs, and overexpression of stroke-inducible tRF-22-WE8SPOX52 using a single-stranded RNA mimic induced down-regulation of immune regulator Z-DNA binding protein 1. In summary, we identified a “changing of the guards” between small RNA types that may systemically affect homeostasis in poststroke immune responses, and pinpointed multiple affected pathways, which opens new venues for establishing therapeutics and biomarkers at the protein and RNA level.
Stroke is a global burden of growing dimensions, accounting for ∼5.5 million deaths annually, and leaving most of the surviving patients permanently disabled (1). The immune system is one of the main players in the pathophysiology of stroke. Brain injury dampens immune functions in the periphery, which limits the inflammatory response and infiltration of immune cells into the CNS and may pose a neuroprotective mechanism in stroke patients. However, this systemic immunosuppression simultaneously increases the risk of infectious complications (2), for example, by inducing lymphocyte apoptosis and decreasing the production of proinflammatory cytokines such as lymphocytic IFN-γ and monocytic TNF-α (3). Therefore, poststroke recovery largely depends on a delicate balance between inflammation, which exacerbates the severity of symptoms, and the poststroke suppression of immune functions, which increases the susceptibility to infections (3). This involves incompletely understood molecular regulators of cholinergic and sympathetic signaling and the hypothalamus–pituitary–adrenal gland axis (HPA). Among other processes, brain injury leads to activation of the vagus nerve, which mediates antiinflammatory signaling through the cholinergic efferent fibers and the noradrenergic splenic nerve (4). Binding of acetylcholine (ACh) to the nicotinic α7 receptors on monocytes/macrophages decreases the production of proinflammatory cytokines (4, 5) in a manner susceptible to suppression by microRNA (miR) regulators of cholinergic signaling, such as miR-132 (6). We hypothesized that this and other small RNA fine-tuners of innate immune responses, including miRs and the recently rediscovered transfer RNA (tRNA) fragments (tRFs), may contribute to regulation of poststroke processes.
Both miRs and tRFs may control entire biological pathways, such that their balanced orchestration could modulate brain-induced systemic immune functioning. miRs are small noncoding RNAs whose expression requires transcription yet can be rapidly induced, enabling degradation and translational suppression of target genes carrying a complementary motif. One miR may suppress the expression of many targets involved in the same biological pathway, and many miRs may cotarget the same transcripts, enabling cooperative suppression. Hence, miR regulators of ACh signaling may regulate the role of ACh in both cognitive function and systemic inflammation (6, 7).
Recent reports highlight tRNA as another major source of small-noncoding RNA (8), including tRNA halves (tiRNAs) and smaller tRNA fragments, tRFs. tiRNAs are created by angiogenin cleavage at the anticodon loop (9), raising the possibility that the poststroke angiogenin increase might change their levels (10). Among other functions (11, 12), smaller fragments derived from the 3′- or 5′-end of tRNA (3′-tRF/5′-tRF) or internal tRNA parts (i-tRF) may incorporate into Argonaute (Ago) protein complexes and act like miRs to suppress their targets (13). Differential expression of tRFs was reported under hypoxia, oxidative stress, ischemic reperfusion (9, 14), and in epilepsy (15), which are all involved in ischemic stroke complications. tRFs may be generated via enzymatic degradation of tRNA, independent of de novo transcription, which implies that tRF levels may be modulated more rapidly than miR levels. However, whether brain–body communication and immune suppression after ischemic stroke in human patients involves blood tRF changes has not yet been studied.
Taking into consideration that the cholinergic system is one of the controllers of immune functions, we investigated changes in the levels of miR- and tRF-regulators, with a specific focus on those that may control the ACh-mediated suppression of poststroke immune functions. We performed small and long RNA sequencing (RNA-seq) of whole-blood samples collected from ischemic stroke patients 2 d after stroke onset, mined RNA-seq datasets of blood cell transcripts, and sought potential links between perturbed miRs and tRFs, poststroke immune responses, and the cholinergic antiinflammatory pathway.
Results
Stroke-Perturbed Small RNAs Display a Cholinergic-Associated Shift from miRs to tRFs.
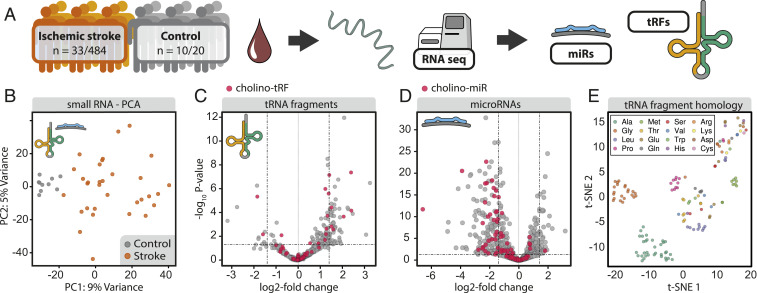
To seek poststroke small RNA regulators of body–brain communication, we first performed small RNA-seq of whole-blood samples collected on day 2 after ischemic stroke from 33 male patients of the PREDICT cohort (484 participants) (16) and 10 age- and sex-matched controls (Fig. 1A; see demographic data in Dataset S1). Principal component analysis (PCA) of the differentially expressed (DE) small RNAs completely segregated the stroke and control groups (Fig. 1B). The respective direction of change among the two small RNA classes involved a statistically significant decline in miRs and a parallel increase in tRFs, indicating a “changing of the guards” from miRs to tRFs. Specifically, 87% of the 143 DE tRFs were up-regulated, whereas 63% of the 420 DE miRs were down-regulated (Benjamini–Hochberg corrected P < 0.05) (Fig. 1 C and D). Of the 143 DE tRFs, 87 were 3′-tRFs and 30 of those (all up-regulated) were derived from alanine binding tRNA (SI Appendix, Fig. S1), indicating nonarbitrary fragment generation.
Fig. 1.
Poststroke differential expression of small RNA species and tRF homology clustering. (A) Whole-blood total RNA samples were collected on day 2 poststroke from patients of the PREDICT cohort (NCT01079728) (16) and age-matched controls. (B) PCA of DE tRFs/miRs in patients’ blood separated stroke and control samples. (C) Volcano plot of DE tRFs from stroke patients and controls (horizontal line at adjusted P = 0.05) showing up-regulation of most DE tRFs. (D) Volcano plot of DE miRs shows predominant down-regulation in stroke patients compared with controls (horizontal line at adjusted P = 0.05). Red dots in C and D reflect Cholino-tRFs and Cholino-miRs, respectively. (E) t-SNE visualization of tRF homology based on pairwise alignment scores of sequences of all detected tRFs shows grouped tRFs of several specific amino acid origins.
Notably, the 420 DE miRs included several miRs known to be perturbed in stroke: hsa-miR-532-5p (log2 fold-change [log2FC] = −2.27, P = 1.81e-33) (17), hsa-miR-148a-3p (log2FC = −2.30, P = 9.61e-19), and hsa-let-7i-3p (log2FC = −1.07, P = 4.31e-04) (18). To test the potential involvement of miRs and tRFs in regulating the cholinergic antiinflammatory pathway after stroke, we performed targeting analysis of DE miRs and tRFs toward cholinergic transcripts (SI Appendix, Expanded Methods; for a complete list of cholinergic genes, see Dataset S2) via an in-house integrative database (miRNeo) (19) containing comprehensive transcription factor (TF)- and miR-targeting data, complemented by de novo prediction of tRF-targeting using TargetScan (20). A restrictive approach identified 131 miRs and 64 tRFs containing complementary motifs to at least five cholinergic-associated transcripts each (further termed “Cholino-miRs” and “Cholino-tRFs”) (Fig. 1 C and D and SI Appendix, Fig. S2; for full lists see Datasets S3 and S4). Permutation targeting analysis showed an enrichment of cholinergic targets for both DE miRs (100,000 permutations, P = 0.0036) and DE tRFs (100,000 permutations, P = 2e-05). Further indicating nonrandom generation of these fragments, the tRFs identified in our dataset clustered into oligonucleotide families with high sequence homology via t-distributed stochastic neighbor embedding (t-SNE) (Fig. 1E), including families known to associate with Ago and suppress growth and proliferation via posttranscriptional down-regulation in lymphocytes (e.g., tRF-22-WE8SPOX52 from tRNAGly) (21) and metastatic cancer cells (tRF-18-HR0VX6D2 from tRNALeu) (22). This supported our prediction that the concomitant elevation of tRF levels and decline of miR levels in poststroke blood could contribute to the poststroke changes in cholinergic signaling pathways.
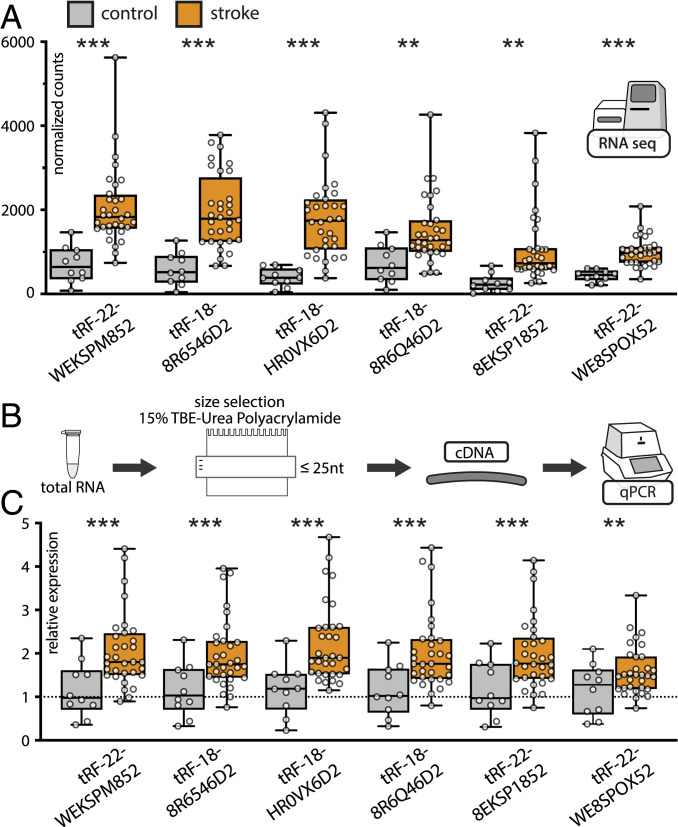
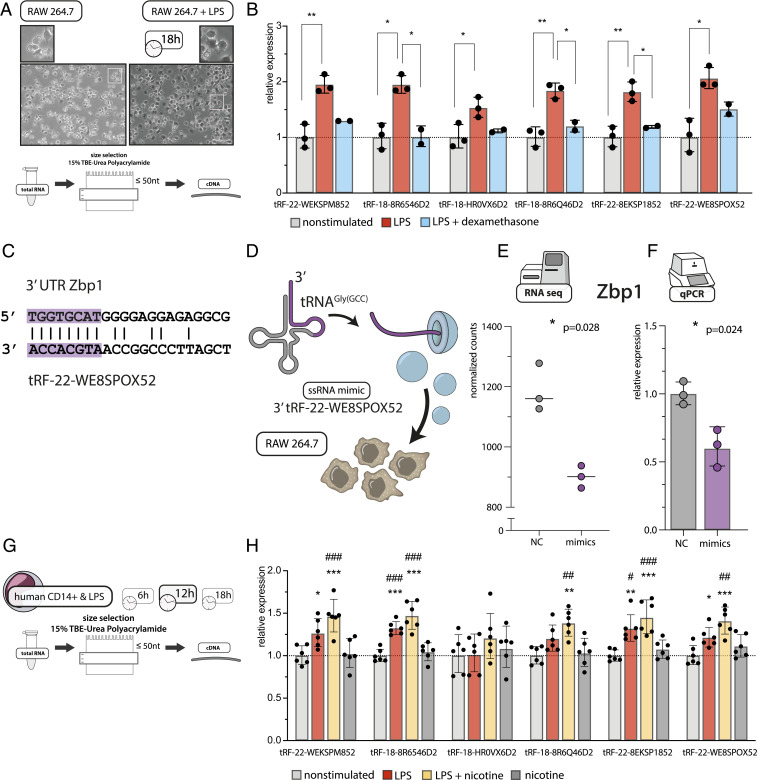
To further challenge our findings, we validated the expression levels of prominently DE tRFs identified by RNA-seq in a separate cohort of PREDICT patients (16). Standard qPCR techniques cannot distinguish between the full-length tRNA molecules and their 3′-tRF cleavage products. Therefore, to experimentally validate tRF changes (Fig. 2A), we implemented an electrophoresis size selection-based strategy followed by cDNA synthesis from the selected small RNAs and qRT-PCR (maximum 25 nt) (Fig. 2B). This procedure validated the top six up-regulated tRFs identified in RNA-seq data (tRF-22-WEKSPM852, tRF-18-8R6546D2, tRF-18-HR0VX6D2, tRF-18-8R6Q46D2, tRF-22-8EKSP1852, and tRF-22-WE8SPOX52, according to count-change) (Fig. 2 B and C and SI Appendix, Expanded Methods) and demonstrated significant increases accompanied by higher variability in the blood levels of these tRFs in poststroke patients compared to controls.
Fig. 2.
qRT-PCR validation of the top six up-regulated tRFs in PREDICT stroke patients following size selection for small RNA. (A) RNA-seq counts normalized to the size of the library [using DESeq2 (23)] of the top six up-regulated tRFs (from left to right). Asterisks indicate adjusted P values of Wald test via DESeq2, **P < 0.01, ***P < 0.001; shown are box-plots with whiskers minimum to maximum. (B) Size-selection workflow for validations in a separate subgroup of PREDICT stroke patients (n = 32) using the same control group (n = 10). (C) qRT-PCR validations using normalized expression (hsa-miR-30d-5p, hsa-let7d-5p, hsa-miR-106b-3p, and hsa-miR-3615 served as housekeeping transcripts) (SI Appendix, Expanded Methods), relative to the control group (line at mean normalized expression for the control group = 1) confirmed up-regulation of top six DE tRFs identified in RNA-seq, one-way ANOVA, **P < 0.01, ***P < 0.001, box-plots with whiskers minimum to maximum.
Stroke-Perturbed Whole-Blood tRFs Are Biased toward Cellular Blood Compartments.
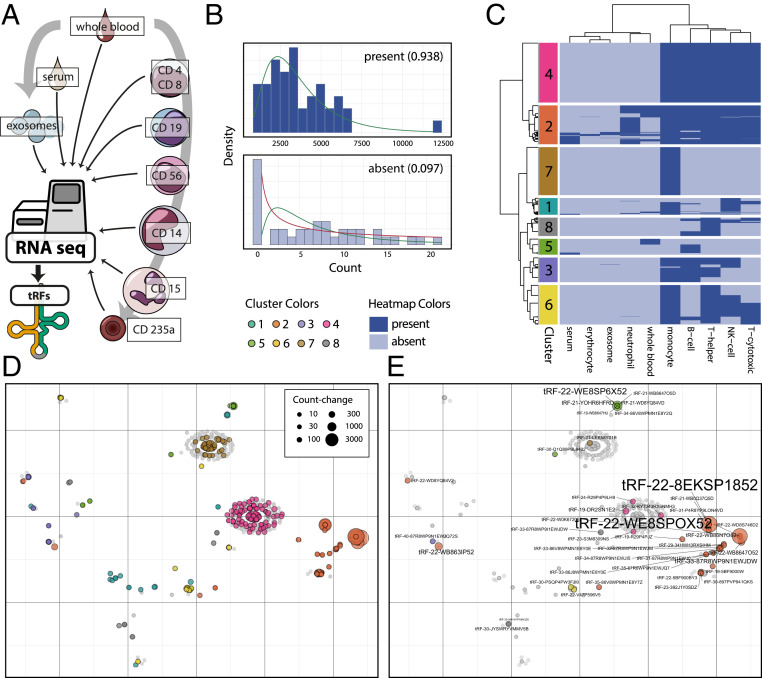
To clarify the distribution of stroke-perturbed tRFs among the immunologically relevant blood cell types, we mined an RNA-seq dataset comprising sorted cell populations collected from healthy volunteers: CD4+ T helper cells, CD8+ T cytotoxic cells, CD56+ NK cells, CD19+ B cells, CD14+ monocytes, CD15+ neutrophils, CD235a+ erythrocytes, serum, exosomes, and whole blood (450 samples in total) (24) (Fig. 3A). Predicting that log-normal distribution of the counts in different samples would point toward biological significance, we categorized all tRFs found in this dataset into present/absent in a specific blood compartment (without introducing a limit for counts) (Materials and Methods and Fig. 3B). Two main clusters of specific blood compartments could be identified based on their specific tRF profile: 1) monocytes, B-, T-, and NK cells; 2) neutrophils, whole blood, serum, exosomes, and erythrocytes (SI Appendix, Fig. S3). Furthermore, we distinguished eight tRF subclusters, based on the presence/absence of specific tRFs in blood compartments (Fig. 3C), with cluster four comprising molecules expressed specifically in monocytes, B-, T-, and NK cells, and cluster seven consisting of tRFs expressed only in monocytes.
Fig. 3.
Immune cell tRF expression clustering and cell type-specific analysis. (A) Analysis of RNA-seq datasets from T lymphocytes (CD4+ T helper cells and CD8+ T cytotoxic cells), B lymphocytes (CD19+), NK cells (CD56+), monocytes (CD14+), neutrophils (CD15+), erythrocytes (CD235a+), serum, exosomes, and whole blood (24) yielded a blood tRF profile. (B) Definition of presence/absence of small RNAs in these blood compartments via statistical assertion of log-normal count distribution (values between 0 and 1, closer to 1: present). (C) Detailed analysis of identified tRFs found eight subclusters based on cell types expressing specific molecules. (D) t-SNE of all found tRFs represented by gray dots, DE tRFs identified in the PREDICT study are marked with cluster-specific color. (E) t-SNE of all tRFs found, Cholino-tRFs identified in the PREDICT study are marked with cluster-specific color.
Based on this methodology, we conducted a census of small RNA species found intra- vs. extracellularly: We detected 1,624 distinct intracellular tRFs but only 93 extracellular tRFs; 149 in whole blood, but 1,417 in CD14+ monocytes alone. Similarly, we detected 559 distinct intracellular miRs but 145 extracellular miRs, 475 in monocytes alone, and 331 in whole blood. Using the presence/absence measure for analyzing the poststroke DE tRFs (Fig. 3D and SI Appendix, Fig. S4 for the top 20 stroke DE tRFs), we detected 77 DE tRFs from the PREDICT dataset as expressed in immune cells (Fig. 3E; for a detailed list of tRFs and affiliated clusters, see Dataset S5), including 10 Cholino-tRFs. Notably, tRFs previously shown to function posttranscriptionally in a miR-like manner [e.g., tRF-22-WE8SPOX52 from tRNAGly (21) and tRF-18-HR0VX6D2 from tRNALeu, alias hsa-miR-1280 (22)] segregated into whole blood, monocyte, T-, B-, and NK-cell compartments rather than into erythrocyte, serum, or exosome compartments. Thus, the poststroke-modified tRFs may be functionally involved in regulating the leukocytic poststroke response.
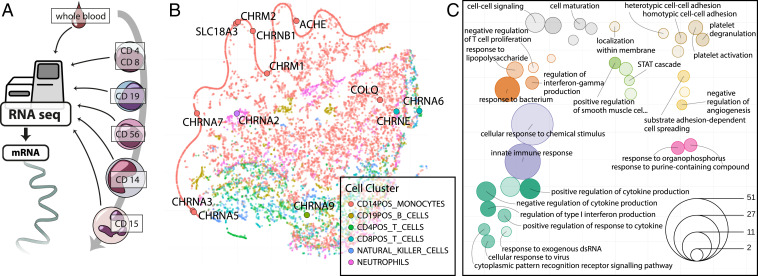
CD14+ Monocytes Show Highest Cholinergic-Related Transcriptional Repertoire.
The enrichment of DE Cholino-miRs and Cholino-tRFs identified in the PREDICT dataset and the contribution of the cholinergic antiinflammatory pathway to peripheral immunosuppression called for pinpointing the immune compartments in which these small RNAs might affect poststroke immune suppression. Analysis of long RNA regulatory circuits (25) specific to blood-borne leukocytes (Fig. 4A) identified CD14+ monocytes as the main cell type expressing cholinergic core and receptor genes (Fig. 4B and Dataset S2). To confirm the relevance of this effect, we performed long RNA-seq in blood samples from 20 stroke patients from the PREDICT study and 4 controls. This showed 204 up-regulated and 490 down-regulated long RNA transcripts. Gene ontology (GO) enrichment analyses of the most implicated genes yielded highly specific terms relevant to innate immunity, vascular processes, and cholinergic links (Fig. 4C; for a list of all significant terms, see SI Appendix, Table S1). More specifically, terms linked to innate immune processes in poststroke blood involved responses to lipopolysaccharide (LPS) mediated by interferons and other cytokines (Fig. 4C, Left); vascular processes comprised platelet activation and degranulation, control of cell–cell adhesion, and regulation of angiogenesis (Fig. 4C, Right). Intriguingly, differentially regulated genes also showed involvement in response to organophosphorus agents, which are known acetylcholinesterase (AChE) inhibitors, supporting a cholinergic participation.
Fig. 4.
Immune cell gene-expression clustering and long RNA pathways perturbed in stroke blood. (A) Published cell type-specific long RNA profiles (25) were used to visualize transcriptomes of T lymphocytes (CD4+ T helper cells and CD8+ T cytotoxic cells), B lymphocytes (CD19+), NK cells (CD56+), monocytes (CD14+), and neutrophils (CD15+). (B) t-SNE visualization of 15,032 genes on the basis of their expression in blood-borne immune cells extrapolated from transcriptional activities in regulatory circuits (25). Genes are colored by the cell type in which their expression was highest. Cholinergic core and receptor genes were mainly found in the CD14+ monocytic compartment. (C) Enrichment of poststroke DE genes (log2FC > 1.4) in circulation- and immunity-related pathways, presented as t-SNE of GO terms by their shared genes (SI Appendix, Expanded Methods); color denotes t-SNE cluster, size denotes number of significant genes in term; deeper color indicates lower enrichment P value (all P < 0.05). Distance between terms indicates the number of shared genes between the GO terms, closer meaning more shared genes.
Stroke Leads to Perturbation of miRNA Regulatory Networks.
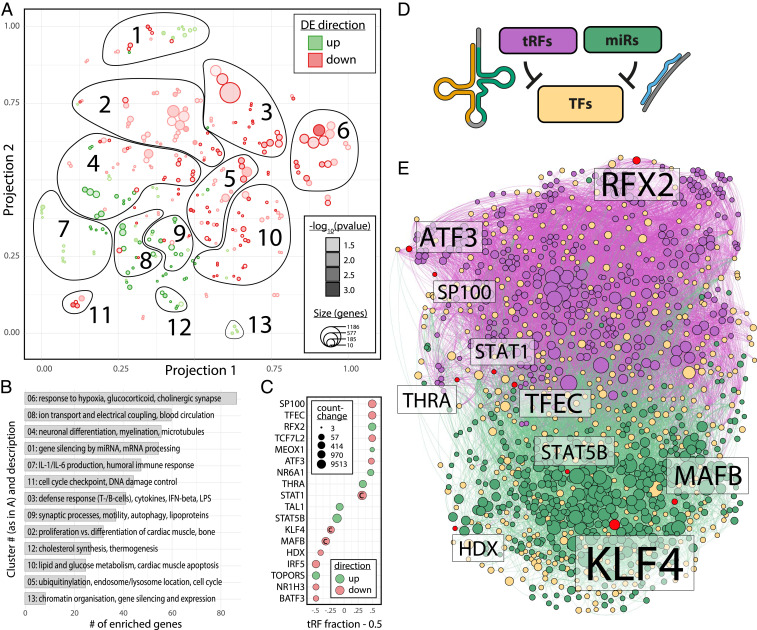
Notably, miRs up-regulated after stroke both appear in smaller numbers compared to down-regulated miRs and possess significantly fewer gene targets per individual miR [via miRNeo, mean (up vs. down) 463 vs. 804, median 266 vs. 717, one-way ANOVA P = 2.1e-07, F(1, 352) = 28.06]. To unravel target genes of these two miR populations, we performed gene target enrichment via permutation inside miRNeo (10,000 permutations). Significantly enriched gene targets (permutation P < 0.05) were subjected to GO analyses and visualized in a t-SNE projection, yielding 13 clusters of related terms, indicating most-affected pathways (Fig. 5A and SI Appendix, Fig. S5). Within each cluster, we determined the most relevant genes via hypergeometric enrichment (Fisher’s exact test). Ranking of the clusters by the absolute number of enriched genes (Benjamini–Hochberg adjusted P < 0.05) revealed the putative biological processes that were most influenced by the miR perturbation following ischemic stroke in our patients (Fig. 5B).
Fig. 5.
GO enrichment of miR targets and perturbed pathways; divergent influence of miRs and tRFs in CD14+ TF regulatory circuits. (A) t-SNE visualization of GO terms enriched in the targets of miRs perturbed by stroke, performed separately for positively (green) and negatively (red) perturbed miRs, segregated into 13 functional clusters. Size of circles represents the number of genes in the respective GO term; depth of color represents enrichment P value (all P < 0.05). (B) Bar graph of clusters identified in A ordered by the number of enriched genes (Fisher’s exact test, Benjamini–Hochberg adjusted P < 0.05) shows most pertinent processes with miR involvement. (C) The top 18 DE TFs in stroke patients’ blood present a gradient of targeting by miRs and/or tRFs (left = 100% miR targeting, right = 100% tRF targeting; value shown as “tRF fraction – 0.5” to center on 50/50 regulation by miRs and tRFs). Size of points and color denote absolute count-change and direction of differential regulation, respectively. “C” marks TFs targeting cholinergic core or receptor genes. (D) Small RNA targeting of TFs active in CD14+ cells was analyzed using miRNeo (19). (E) Force-directed network of all TFs active in CD14+ monocytes self-segregates to form largely distinct TF clusters targeted by DE tRFs and miRs in stroke patients’ blood. Yellow = TF, red = TF DE in stroke patients’ blood, green = miR, purple = tRF. Size of node denotes activity toward targets.
The significantly derepressed cluster no. 6 (84 enriched genes) pointed toward perturbation of pathways involved in responses to hypoxia (GO:0036293, P = 0.008) and drugs (GO:0042493, P = 0.035), including antibiotics (GO:0071236, P = 0.013), glucocorticoids (GO:0051384, P = 0.007), and the cholinesterase-blocking organophosphorus agents (GO:0046683, P = 0.019), which reinforced the notion of cholinergic participation. Cross-check of enriched genes via DAVID (26) confirmed a role of cluster no. 6 in hypoxia (GO:0071456, P = 8.3e-13), drug response (GO:0042493, 5.4e-10), and the cholinergic synapse (Kyoto Encyclopedia of Genes and Genomes pathway 04725, P = 0.006). Cluster no. 8 (with second-most 58 enriched genes) highlighted perturbed transmembrane ion conductivity, particularly in regulation of cardiac muscle cell action (GO:0098901, P = 0.044) and negative regulation of blood circulation (GO:1903523, P = 0.016). DAVID analysis confirmed involvement in “regulation of cardiac muscle contraction by the release of sequestered calcium ion” (GO:0010881, P = 2.1e-15) and regulation of heart rate (GO:0002027, P = 1.2e-07). The subsequent clusters indicate further involvement in nerve cell regulatory processes (cluster no. 4), regulation of gene silencing by miRNA (cluster no. 1), and humoral immunity via IL-1 and IL-6 (cluster no. 7) (SI Appendix, Fig. S5). The entire list of clusters and gene enrichments is available in Dataset S6.
tRFs May Suppress Inflammation and Cholinergic-Associated TFs Alone or in Cooperation with miRs.
Cellular responses to different stimuli are coordinated by cell type-specific transcriptional regulatory circuits. To facilitate understanding of the role of miRs and tRFs in regulating the transcriptional state of CD14+ monocytes after stroke, we generated a monocyte-specific transcriptional interaction network of small RNAs targeting TFs (via miRNeo) (19), combined with differential expression of long and small RNAs from the PREDICT cohort. The gradually divergent targeting of these TFs by miRs and tRFs implied largely separate domains of regulation by these small RNA species (Fig. 5C). This notion was topologically strengthened by the fact that the force-directed network of all TFs active in CD14+ monocytes self-segregated to form two distinct clusters of TFs, which were primarily targeted either by miRs or tRFs, including numerous TFs DE in stroke patient blood (Fig. 5 D and E). Among the implied TFs are proteins known for their influence on cholinergic genes, as well as their involvement in inflammation, such as STAT1 or KLF4 (27, 28). Notably, eight DE TFs were not predicted to be targeted by any miR or tRF present in CD14+ monocytes.
Stroke-Induced tRFs Show Evolutionarily Conserved Participation in Macrophage/Monocyte Responses to Inflammatory Stimuli.
To test if the stroke-induced tRFs are involved in the inflammatory response of monocytes and macrophages, we subjected murine RAW 264.7 cells to LPS stimulation with or without dexamethasone suppression of their inflammatory reactions (Fig. 6A). By 18 h after LPS stimulation, qRT-PCR analysis after size-selection (for ≤50-nt fragments) detected pronounced up-regulation of the top six poststroke up-regulated tRFs (Fig. 6B). Moreover, dexamethasone suppression of the LPS response led to down-regulation of those tRFs, along with a diminished inflammatory response (SI Appendix, Fig. S6). Predicted targets of these molecules comprise members of mitogen-activated protein kinases (MAPK) and TNF receptor-associated factors (Dataset S7), further pointing toward their regulatory role in response to inflammatory stimuli. Notably, one of the top six stroke-perturbed tRFs, tRF-22-WE8SPOX52, is complementary to the 3′UTR sequence of murine Z-DNA binding protein Zbp1 and therefore a predicted regulator of the Zbp1 transcript and its immune system activity (Fig. 6C). To challenge the functional activity of tRF-22-WE8SPOX52 in murine RAW 264.7 macrophages, we overexpressed this tRF using single-stranded RNA (ssRNA) mimics (Fig. 6D and SI Appendix, Fig. S7), which significantly reduced the expression of its Zbp1 target compared to negative control, as quantified by long RNA-seq (Fig. 6E) and validated by qRT-PCR in an independent experiment (Fig. 6F).
Fig. 6.
tRF changes upon LPS stimulation of murine RAW 264.7 macrophages and human CD14+ monocytes and tRF-mimic transfection. (A) LPS stimulation of RAW 264.7 murine macrophage cells induced clear morphologic changes within 18 h. Extracted RNA was subjected to size selection and cDNA synthesized from the ≤50-nt fraction alone. (Scale bar, 100 μm, magnification in the upper panel 2.5×) (B) LPS-stimulated RAW 264.7 cells show dexamethasone-suppressible elevated levels of poststroke induced tRFs. Normalized qRT-PCR values (using mmu-miR-30d-5p, mmu-let7d-5p as housekeeping transcripts) (SI Appendix, Expanded Methods), compared to unstimulated controls. Each dot represents two to four technical replicates, ANOVA with Tukey post hoc, *P < 0.05, **P < 0.01, bar graphs ±SD(lg). (C) Murine Zbp1 sequence carries an 8-nt-long fragment in the 3′UTR complementary to tRF-22-WE8SPOX52. (D) To test the miR-like mechanism of action, RAW 264.7 cells were transfected with ssRNA tRF-22-WE8SPOX mimics or negative control ssRNA, and RNA was extracted 24 h after transfection and subjected to polyA-selected RNA-seq (E) and qRT-PCR (F). (E) Long RNA-seq of cells transfected with ssRNA tRF-22-WE8SPOX52 mimics revealed significantly down-regulated expression of Z-DNA binding protein (Zbp1) as compared to negative control (NC). *P < 0.05, shown is adjusted P value of Wald test via DESeq2, bar graph ±SD. (F) qRT-PCR from an independent cell culture experiment confirmed the down-regulation of Zbp1 expression after ssRNA tRF-22-WE8SPOX52 mimic transfection (relative normalized expression using Gapdh as a housekeeping gene), *P < 0.05 one-way ANOVA, each dot represents a technical cell culture replicate, bar graph ±SD(lg). (G) MACS-sorted CD14+ cells from healthy human donors were stimulated with LPS with or without addition of nicotine and collected 6, 12, and 18 h thereafter. Extracted RNA was subjected to size selection and cDNA synthesized from the ≤50-nt fraction. Timepoints 6 h and 18 h are shown in SI Appendix, Fig. S8. (H) At 12 h after LPS stimulation, human monocytes exhibited up-regulation of poststroke induced tRFs as compared to unstimulated controls or cells treated with nicotine alone. This reaction was boosted by the addition of nicotine. Shown is relative expression (hsa-miR-30d and hsa-let7d-5p were used as housekeeping transcripts) (SI Appendix, Expanded Methods) normalized to the nonstimulated group. Each dot represents one donor. ANOVA with Tukey post hoc, *P < 0.05, **P < 0.01, ***P < 0.001 vs. nonstimulated cells; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. cells upon addition of nicotine, bar graphs ±SD(lg).
Finally, we aimed to validate the functional implications of these findings in primary human cells. Therefore, we performed LPS stimulation experiments in magnetic-activated cell-sorted (MACS) CD14+ monocytes from healthy volunteers and collected the cells at 6, 12, and 18 h after LPS addition (Fig. 6 G and H and SI Appendix, Fig. S8 A and B for 6- and 18-h timepoints). To further challenge the cholinergic link, we used nicotine as an immunosuppressive agent (SI Appendix, Fig. S8C) (29). LPS-stimulated primary CD14+ cells presented significant up-regulation of four of the six stroke-induced tRFs at 12 h, an effect that was augmented by the addition of nicotine. Interestingly, at 18 h the tRF levels in the LPS + nicotine group were comparable to those of nonstimulated cells (SI Appendix, Fig. S8B). Together, these findings demonstrate evolutionarily conserved and cholinergic-regulated increases of stroke-induced tRFs under proinflammatory insults.
Discussion
To date, few studies have simultaneously assessed the joint impact of blood miR and tRF changes in human disease. Here we have discovered a stroke-induced decline of miRs and concomitant elevation of tRFs in whole blood, and demonstrated that this shift may be associated with the poststroke cholinergic blockade of immune function. To validate our RNA-seq findings of tRFs in a way that circumvents the ambiguous detection of full-length tRNA, we developed and used a size selection-based qRT-PCR test in an independent cohort of patients. Mining transcriptomic datasets identified CD14+ monocytes as likely pivotal in the cholinergic control of immunity, demonstrated that the stroke-induced tRFs may target specific monocytic TFs, and showed that at least some of those tRFs may actively control processes linked to inflammatory responses. Moreover, several of the stroke-induced tRFs were also induced in LPS-exposed murine macrophages and in human CD14+ primary cells and showed time-dependent nicotine- and dexamethasone-induced up-regulation/suppression, supporting the notion that the elevation of tRFs is an evolutionarily conserved response mechanism. Overexpression of tRF-22-WE8SPOX52 using ssRNA mimics led to the down-regulation of its Zbp1 target, which is involved in regulating inflammatory responses. This concept of integrated fine-tuning of poststroke immune responses opens new venues for stroke diagnostics and therapeutics.
The cholinergic antiinflammatory reflex plays a substantial role in regulating peripheral immune responses after CNS injury, along with the HPA axis and sympathetic signaling (3). Excessive cholinergic responses suppress pulmonary innate immunity, including macrophage and alveolar epithelial cell responses; this may facilitate the development of pneumonia (30), a major factor of nonrecovery (31). However, while reduced AChE activities in poststroke patients’ serum associate with poor survival (32), stroke-induced immunosuppression may be brain-protective (2), calling for caution when considering therapeutic boosting of immune reaction in the periphery to limit infections. Therefore, an in-depth understanding and characterization of the molecular regulators of immune responses and the cholinergic pathway after CNS-injury is of utmost importance at both the system and mechanism levels.
System-Level Perturbations Associated with Ischemic Stroke.
We identified CD14+ monocytes to be the most likely immune cell subpopulation for a transcriptional cholinergic response. Monocytes play established roles in responses to stroke, including prolonged monocytosis, deactivation, and functional impairment of circulating monocytes/macrophages observed in experimental models (33) and human patients (34). Moreover, stroke leads to overproduction of CD14++/CD16− (classical) and CD14++/CD16+ (intermediate) monocytes with simultaneous decrease in CD14+/CD16++ (nonclassical) monocytes, which correlates with stroke-associated infection (35). Relatedly, immune cells in general, and monocytes in particular appear to be enriched in specific small RNA species (compare Fig. 3C and ref. 36), and several of the most highly perturbed small RNAs are abundantly expressed in monocytes (compare SI Appendix, Fig. S4 and ref. 37).
Our in-depth analysis of the pathways targeted by perturbed miRs supports a tie between stroke-induced changes and a cholinergic response. Both the identified clusters as well as the genes enriched in each cluster may be further investigated for identifying the diverse mechanisms involved. A recent whole-blood microarray survey identified 15 miRs, 11 of which were replicated in our study, to be suppressed within less than 72 h in intracerebral hemorrhage patients compared to controls (38). Those miRs pointed toward the same processes we found, including inflammation and humoral immunity via JAK/STAT-activating cytokines, vascular integrity, and the cellular immune response (38).
Stroke is a sudden incident with rapid onset and drastic systemic changes within a short time frame. In the response to such immunologic emergencies, translational control is an important tool (39). That tRFs may be rapidly produced by regulated nuclease cleavage of preexisting tRNAs in a “burst-like” fashion makes them particularly appropriate for handling acute situations. Recent reports demonstrate production of 3′-tRFs by specific nucleases, and 3′- and 5′-tRNALeu fragments were shown to regulate T cell activation (40). Furthermore, tRFs can perform different molecular roles, including Ago-mediated suppression of target genes carrying complementary sequence motifs (13). At least two of the stroke-induced tRFs up-regulated after LPS stimulation show miR-like function: tRF-22-WE8SPOX52 regulates B cell growth via suppressing the expression of Replication Protein A1 (RPA1) (21) and a 17-nt-long analog of tRF-18-HR0VX6D2 limits cancer cell proliferation by impacting the cholinergic-regulating Notch signaling pathway (22). Interestingly, hsa-miR-1260b, identified in our study and by others as perturbed poststroke (41), differs from tRF-18-HR0VX6D2 by only 1 nt at position 9 and an additional nucleotide at the 3′-end (SI Appendix, Fig. S9), which indicates that hsa-miR-1260b may actually be a tRF (42), and calls for further investigation.
Our study identified two main factors that may lead to an overall decrease in transcript regulation by miRs after stroke. First, the majority of miRs perturbed in our patient collective were down-regulated, and second, the down-regulated miRs possessed significantly more targets than the up-regulated ones. For many processes regulated by these miRs, the resultant effect will hence be de-repression of targeted genes (Fig. 5A). Additionally, we have identified a stark dichotomy between the target sets of miRs and tRFs, indicating much complementarity and only little cooperative overlap of affected transcripts between those two small RNA species (Fig. 5E). However, these changes may still lead to a homeostatic functional cooperation. In summary, the poststroke “changing of the guards” in the small RNA response may lead to preferential de-repression of miR targets and concomitant repression of tRF targets, and the de-repression of miR targets may be as pivotal for regulating the initial inflammatory response and subsequent peripheral immunosuppression as the tRF elevation we identified.
Kinetically, stroke is characterized by an initial inflammatory response followed by immunosuppression facilitated by, among others, the cholinergic antiinflammatory reflex (3). Therefore, the time-dependent elevation of tRFs and Cholino-tRFs in particular may offer new mechanisms of homeostatic fine-tuning in response to cerebral ischemia. Furthermore, not only the peripheral but also the central immune response at the site of the injury is of great importance for stroke prognosis. Brain injury triggers activation of microglia and infiltration of peripheral immune cells, including monocyte-derived macrophages, which accumulate at the lesion site 3 to 7 d after stroke (35). Experimental evidence highlights essential roles of these cells in CNS-repair processes and neuronal protection (43, 44), and our own studies indicate small RNA-regulated cross-talk between neuronal and immunological regulation by JAK/STAT-related mechanisms (19).
Our present study presents tRFs as potential players in regulating the poststroke inflammatory responses. For example, KLF4, identified as down-regulated in our sequencing dataset, is involved in controlling the macrophage response to LPS (28) and the differentiation of monocytes toward an inflammatory phenotype (45). Therefore, a decrease in miRs targeting this TF may contribute to proinflammatory monocytic response observed in the initial phase of stroke. Similarly, MAFB is essential in facilitating the clearance of damage-associated molecular patterns (DAMPs) in the ischemic brain and, consequently, limiting the inflammatory response while supporting recovery (46). MAFB de-repression in peripheral immune cells may be a mechanism supporting monocyte infiltration of the brain. Conversely, STAT1 and ATF3 may be preferentially repressed due to their targeting by tRFs. STAT1 is essential in IFN- and IL-6–mediated inflammatory response, and ATF3 is similarly induced by IFNs and contributes to STAT activity via inhibition of STAT-dephosphorylating phosphatases (47, 48). Additionally, ATF3 down-regulates AChE expression during stress (49). Whether these processes contribute to body homeostasis after the damaging event, or rather to pathologic derailment of immune function, requires detailed kinetic studies of circulating monocytes and brain-infiltrating monocyte-derived macrophages, with simultaneous profiling of short and long transcripts.
Mechanistic Implications of tRF Regulation after Ischemic Stroke.
To gain new insight into the regulation of inflammation by stroke-induced tRFs, we quantified the top six perturbed tRFs in RAW 264.7 murine macrophages and primary LPS-stimulated CD14+ human monocytes. Both cell types responded to LPS by up-regulation of these tRFs within 12 to 18 h. Interestingly, dexamethasone prevented or subsequently down-regulated the increased expression of tRFs in the RAW 264.7 cells. To further seek cholinergic links of these stroke-regulated tRFs, we used nicotine as an immunosuppressive stimulus in LPS-stimulated human CD14+ cells. Monocytes and macrophages express the cholinergic nicotinic α7 receptor, which after binding of ACh down-regulates the production of inflammatory cytokines (e.g., TNF-α) (29). In human CD14+ cells, the levels of the top six stroke-induced tRFs were transiently elevated by the addition of nicotine at the 12-h timepoint (back to baseline by 18-h poststimulation). Thus, the elevated levels of blood tRFs 2 d after ischemic stroke may reflect potentiated cholinergic signaling, which remains to be investigated in the clinical setting.
In human patients, the stroke response in blood is cell type- and time-specific. For example, day 2 postischemia features a transient increase in STAT3 phosphorylation of monocyte subsets, which is also detected in patients after major surgery (50). Conversely, STAT3 signaling causes immune stimulation in monocytes but is linked to immunosuppression in monocytic myeloid-derived suppressor cells (50). Therefore, the biological activities of stroke-induced tRFs are very likely also cell type- and context-specific. Our ssRNA tRF-22-WE8SPOX52 mimic experiments further support tRF involvement in the posttranscriptional regulation of genes implicated in inflammatory responses. Zbp1, which was significantly down-regulated under tRF-22-WE8SPOX52 overexpression, is a DAMP-sensor that induces IFN responses, programmed cell death, and NLRP3 inflammasome formation (51). The ZBP1 protein has often been linked to the response to viral infections, but some studies point to its role in the reaction to bacterial pathogens, where it may be involved in the induction of necroptosis (51). Incidentally, the ZBP1 transcript is also down-regulated in our long RNA-seq of patient blood (log2FC = −1.7, adjusted P = 0.001); the exact nature of the interaction of tRF-22-WE8SPOX52 and ZBP1 should be subject of future studies.
Limitations.
We hypothesized that the tRFs identified in our study are of cellular origin and therefore reanalyzed the small RNA-seq data provided by Juzenas et al. (24) (GSE100467). While some studies identified an enrichment of tRFs in exosomes (40), our reanalysis found the tRFs mostly in the cellular blood compartments. Although the main immune populations are included in the analyzed dataset, it should be noted that the tRFs identified in our study may also originate from immune cells not sorted/sequenced by Juzenas et al. (24). Additionally, considering the different roles of specific immune subpopulations, identifying the specific source of tRFs and their roles in immune function should be the goal of further investigations. For example, monocyte subsets are differentially regulated after stroke, and since these cells all express the CD14 marker (35), a higher cellular resolution is called for. Furthermore, tRFs are induced upon cellular stress (52), such that the sorting procedure may affect their expression, and they may reside in extracellular compartments (15, 53), calling for testing the impact of sample processing, RNA isolation, and sequencing techniques on the detectability of tRFs. Additionally, a method for the direct comparison of miR and tRF levels in the same sequencing experiment will be important for discerning the true difference in detected counts after alignment, possibly via spike-in procedures. The currently maturing technology of single-cell sequencing is an obvious candidate for achieving the goals of higher cellular resolution along with avoiding stressors associated with sample preparation, but its shallow sequencing depth is still an issue.
Given the sex-related differences in cholinergic responses (19, 54), the molecular regulators of cholinergic signaling and immunity should be investigated in detail in both males and females. However, to increase the consistency of our results, we only included male stroke patients, which is a further limitation of this study. Interestingly, the overall impact of stroke is greater in women, as their higher life expectancy is linked to increased stroke incidence in older adults and they face worse recovery prospects (55). Last, but not least, tRFs may have functions other than their miR-like activities; for example, tRNALeu-CAG fragments facilitate translation and ribosome biogenesis (12), whereas tRNAGly-, tRNAGlu-, tRNAAsp-, and tRNATyr-derived tRFs displace RNA-binding proteins leading to mRNA destabilization (11). Therefore, potential functions of stroke-perturbed tRFs other than Ago-mediated suppression of translation should be further examined.
Conclusion
While the specific roles of tRFs in regulating local neuroinflammatory responses and functional modulation of specific peripheral monocyte subsets remain to be elucidated, our findings point toward tRFs/miRs as homeostatic regulators of poststroke immune responses and potential biomarkers for increased infection risk in these patients. The cumulative role of tRFs and miRs as general postdamage mediators of CNS-immune communication thus calls for seeking small RNAs, and tRFs in particular, as involved in other traumatic pathologies, such as spinal cord injury, traumatic brain injury, concussion, as well as neuroinflammatory brain diseases.
Materials and Methods
Expanded methods can be found in SI Appendix.
Clinical Cohort.
PREDICT was a prospective multicenter study with sites in Germany and Spain (https://www.clinicaltrials.gov/, NCT01079728) (16) that analyzed 484 acute ischemic stroke patients. Patients underwent daily screenings for stroke-associated pneumonia, dysphagia, and inflammation markers and their clinical outcome was recorded 3 mo poststroke. To exclude very severe cases of stroke, we only considered for sequencing samples from patients with modified Rankin Scale (mRS) values of 3 and below at discharge from the hospital, leaving n = 240 relevant cases. Blood was collected into RNA stabilizing tubes (Tempus Blood RNA tubes, Applied Biosystems) on each day of hospitalization. Blood samples collected on the second day were subjected to small and long RNA-seq, with time from stroke occurrence to blood withdrawal varying between 0.94 and 2.63 d (average: 1.98 d). Blood samples from age- and ethnicity-matched healthy controls were obtained at matched circadian time from donors (ZenBio). These samples were collected from an FDA Regulated Blood Bank. All procedures, polices, forms and consents used in the donor screening, collection and manufacturing process were submitted to the FDA for review and approval through the Prior Approval Supplement process described in 21 CFR 601.12(b) (ZenBio).
RNA Extraction, Quality Control, and Sequencing.
RNA was extracted from 3 mL of whole blood of 484 PREDICT patients using a Tempus Spin RNA isolation kit (Invitrogen, Thermo Fisher Scientific). Presequencing Bioanalyzer 6000 (Agilent) tests showed high RNA quality (RIN values 7.9 to 9.9, median 8.8). Libraries constructed from 600 ng total RNA of 43 samples were subjected to small RNA-seq (NEBNext Multiplex Small RNA library prep set for Illumina, New England Biolabs), and 24 of the small RNA-seq samples served for PolyA-selected long mRNA sequencing (1,000 ng total RNA per sample, TruSeq RNA library preparation kit;Illumina). Sequencing (24 or 12 samples per flow cell for small and long RNAs, respectively) was performed on the Illumina NextSeq 500 platform at the Hebrew University’s Center for Genomic Technologies.
Alignment and Count Table Generation of RNA-Seq Reads.
Quality control was performed using FastQC, v0.11.2 (56); more details can be found in SI Appendix, Expanded Methods. Flexbar (with parameters “-q TAIL -qf Sanger -qw 4 -min-read-length 16”) (57) served for adapter trimming and quality-based filtering of all raw reads. Long RNA was aligned to the human reference transcriptome (ENSEMBL GRCh38 release 79) using salmon (58) with default parameters. Small RNA was aligned to miRBase v21 using miRExpress 2.1.4 (59) with default parameters but skipping adapter trimming for miR expression, and to the tRNA transcriptome using the MINTmap pipeline (60) with default parameters for tRF expression (using only reads mapping exclusively to the tRNA space). Raw gene-expression data of small and long RNA-seq and technical covariates of all experiments are available via the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE158312) (61).
Size Selection for tRF Quantification.
Standard qRT-PCR methods do not allow to distinguish between full-length tRNA molecules and 3′-tRFs. To exclude longer RNA species in the qRT-PCR quantifications, we performed RNA size selection on 15% TBE-Urea-Polyacrylamide gels, selecting only RNA molecules ≤25 nt for validations in the clinical cohort and ≤50 nt for the assessment of tRF expression in LPS-stimulated RAW 264.7 cells and human CD14+ monocytes. A detailed description can be found in SI Appendix, Expanded Methods.
Analysis of the Presence/Absence of Specific tRFs in Blood Compartments.
In descriptive analysis of small RNA expression, a threshold (e.g., at least five counts in at least 80% of samples) is often used to define presence or absence of small RNAs. However, since this definition relies heavily on sequencing depth, and depth can vary widely even in methodically robust sequencing experiments depending on a large number of variables, we defined our own test for descriptive analysis of the presence or absence of lowly expressed small RNAs in each of the sample types. Briefly, this definition comprises estimation of a log-normal distribution on the expression profile of the small RNA across all samples in the individual cell types, and a statistical test to refute the null hypothesis that the distribution is in fact log-normal. For each small RNA, the distribution mean and SD of the expression values per cell type were estimated using the fitdist function of the R/fitdistrplus package (62). The count distribution was then tested against a log-normal distribution with the estimated mean and SD via the R implementation of the Kolmogorov–Smirnov test, with a cutoff of 0.1. The small RNA was defined as present if the test failed to reject the null hypothesis (see SI Appendix, Fig. S4 for examples). The code implementation is available at https://github.com/slobentanzer/stroke-trf.
LPS Stimulation of Murine Macrophages.
Murine RAW 264.7 cells (ATCC TIB-71) cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal calf serum, 1% penicillin-streptomycin-amphotericin B, and 1% l-glutamine (all reagents from Biological Industries) were collected using a cell scraper and stimulated with LPS (from Escherichia coli 0127:B8, Sigma Aldrich) following a modified protocol (63). Briefly, 2e05 cells were stimulated with 100 ng/mL LPS ± 0.5 μM dexamethasone per well (Sigma Aldrich) in 12-well cell culture plates. Cells were collected in Tri-Reagent (Sigma Aldrich) 18 h after LPS stimulation and RNA was isolated using miRNeasy kit (Qiagen). For the size selection, 1 μg of total RNA was used and cDNA was synthesized from 500 pg of size-selected RNA using qScript microRNA cDNA Synthesis Kit (Quanta Biosciences) and following standard protocol (for further details, see SI Appendix, Expanded Methods). Data presented in the report is derived from three independent experiments (two of them with dexamethasone treatment) with two to four technical replicates in each group.
Transfection Experiments with tRF-22-WE8SPOX52 Mimics, RNA-Seq, and qRT-PCR.
Transfections were performed using HiPerFect transfection agent (Qiagen) following a standard protocol for transfecting RAW 264.7 macrophages. Briefly, 2e05 RAW 264.7 cells per well were seeded in 24-well plates and transfected using 6 μL HiPerFect reagent per well and 50 nM final ssRNA tRF-mimics: /5PHOS/rArU rCrCrC rArCrC rGrCrU rGrCrC rAmCmC rA, using NC5 /5PHOS/rGrC rGrArC rUrArU rArCrG rCrGrC rArArU mAmUrG (both from IDT) as negative control. Cells were collected after 24 h following the transfection in Tri-Reagent (Sigma Aldrich) and total RNA isolated using the miRNeasy kit (Qiagen). Two separate experiments were performed: 1) 180 ng RNA was subjected to long RNA-seq (KAPA stranded mRNA-seq kit, Roche) and 2) Zbp1 levels were quantified by qRT-PCR (relative expression normalized to Gapdh) after cDNA synthesis (qScript Kit; Quanta Biosciences) from 100 ng RNA.
Isolation and Ex Vivo Stimulation of Human Monocytes.
This study was approved by the ethics committees of the Charité-Universitätsmedizin Berlin (MG Cohort: EA1/281/10). Peripheral blood mononuclear cells were separated from whole-blood anticoagulated with heparin by density gradient centrifugation over Ficoll (Biocoll separating solution, Biochrom). Untouched monocytes were isolated by using a commercially available Pan Monocyte Isolation Kit (Miltenyi Biotec). Cells (2e06 cells/mL) were cultured in RPMI medium 1640 (VWR), supplemented with 1% penicillin-streptomycin (Biochrom), 2 mM l-glutamine (Biochrom), and 10% autologous serum and stimulated with LPS (1 ng/mL, 0127:B8; Sigma) in the presence or absence of nicotine (300 µM, Sigma) for 6, 12, and 18 h at 37 °C. Unstimulated monocytes and monocytes stimulated with nicotine served as controls. TNF-α concentration was measured in cell culture supernatant by using a commercially available DuoSet ELISA kit (R&D Systems). Cells were collected in Tri-Reagent (Sigma Aldrich) and RNA was isolated using miRNeasy kit (Qiagen). For the size selection, 600 ng of total RNA (or maximum loading volume of 20 μL) were used and cDNA was synthesized from 500 pg of size-selected RNA using qScript microRNA cDNA Synthesis Kit (Quanta Biosciences) and following standard protocol (for further details, see SI Appendix, Expanded Methods).
Statistical Analysis.
Data analysis was performed using R (v4.0.2), the code is available at https://github.com/slobentanzer/stroke-trf (including code for analyses of qRT-PCR data) (64). False-discovery rate correction was applied whenever applicable. qRT-PCR data were analyzed using Bio-Rad CFX Maestro software (Bio-Rad, v4.1.2433.1219) and plotted in GraphPad Prism 8.0 (GraphPad Prism Software).
Supplementary Material
Acknowledgments
We thank Dr. Simonas Juzenas (Saarbrücken) and Prof. Andreas Keller (Saarbrücken/Stanford) for their support concerning the blood compartments RNA-sequencing dataset, and Dr. Iftach Shaked (San Diego) for fruitful discussions. This study was supported by the European Research Council Advanced Award 321501; the Israel Science Foundation Grant 1016/18; the Israeli Ministry of Science, Technology and Space Grant No. 53140 (to H.S.); the German Research Foundation (Exc257, TR84, SFB/TRR167) (to A.M. and C.M.); the Leducq Foundation (19CVD01); and the Einstein Foundation, Berlin (A-2017-406) (to A.M. and H.S.). Further support was provided by a NeuroCure visiting fellowship (to H.S.), as well as by Edmond and Lily Safra Center of Brain Science (ELSC) postdoctoral fellowships (to S.S.-T and K.W.). K.W. is a Shimon Peres Post-doctoral Fellow at the ELSC and S.L. received an ELSC fellowship for visiting PhD students.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013542117/-/DCSupplemental.
Data Availability.
Code data have been deposited in GitHub (https://github.com/slobentanzer/stroke-trf) (64). RNA sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database, (accession no. GSE158312) (61).
References
- 1.Johnson C. O. et al.; GBD 2016 Stroke Collaborators , Global, regional, and national burden of stroke, 1990-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 18, 439–458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dirnagl U., et al. , Stroke-induced immunodepression: Experimental evidence and clinical relevance. Stroke 38 (suppl.2), 770–773 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Meisel C., Schwab J. M., Prass K., Meisel A., Dirnagl U., Central nervous system injury-induced immune deficiency syndrome. Nat. Rev. Neurosci. 6, 775–786 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Rosas-Ballina M., et al. , Acetylcholine-synthesizing T cells relay neural signals in a vagus nerve circuit. Science 334, 98–101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borovikova L. V., et al. , Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature 405, 458–462 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Shaked I., et al. , MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Soreq H., Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci. 38, 448–458 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Torres A. G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L., Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U.S.A. 116, 8451–8456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki S., Ivanov P., Hu G. F., Anderson P., Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L., Guo H., Cheng M., Zhao Y., Jin X., The kinetic change of the serum angiogenin level in patients with acute cerebral infarction. Eur. Neurol. 58, 224–227 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Goodarzi H., et al. , Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161, 790–802 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H. K., et al. , A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552, 57–62 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar P., Anaya J., Mudunuri S. B., Dutta A., Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 12, 78 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q., et al. , tRNA-derived small non-coding RNAs in response to ischemia inhibit angiogenesis. Sci. Rep. 6, 20850 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogg M. C., et al. , Elevation in plasma tRNA fragments precede seizures in human epilepsy. J. Clin. Invest. 129, 2946–2951 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann S. et al.; PREDICT Investigators , Stroke-induced immunodepression and dysphagia independently predict stroke-associated pneumonia—The PREDICT study. J. Cereb. Blood Flow Metab. 37, 3671–3682 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li P., et al. , Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell. Mol. Neurobiol. 35, 433–447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jickling G. C., et al. , microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS One 9, e99283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobentanzer S., Hanin G., Klein J., Soreq H., Integrative transcriptomics reveals sexually dimorphic control of the cholinergic/neurokine interface in schizophrenia and bipolar disorder. Cell Rep. 29, 764–777.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal V., Bell G. W., Nam J. W., Bartel D. P., Predicting effective microRNA target sites in mammalian mRNAs. eLife 4, e05005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maute R. L., et al. , tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. U.S.A. 110, 1404–1409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang B., et al. , tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 77, 3194–3206 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juzenas S., et al. , A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 45, 9290–9301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marbach D., et al. , Tissue-specific regulatory circuits reveal variable modular perturbations across complex diseases. Nat. Methods 13, 366–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Shuai K., et al. , Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76, 821–828 (1994). [DOI] [PubMed] [Google Scholar]
- 28.Feinberg M. W., et al. , Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J. Biol. Chem. 280, 38247–38258 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Fujii T., et al. , Expression and function of the cholinergic system in immune cells. Front. Immunol. 8, 1085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engel O., et al. , Cholinergic pathway suppresses pulmonary innate immunity facilitating pneumonia after stroke. Stroke 46, 3232–3240 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Finlayson O. et al.; Canadian Stroke Network; Stroke Outcome Research Canada (SORCan) Working Group , Risk factors, inpatient care, and outcomes of pneumonia after ischemic stroke. Neurology 77, 1338–1345 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Ben Assayag E., et al. , Serum cholinesterase activities distinguish between stroke patients and controls and predict 12-month mortality. Mol. Med. 16, 278–286 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCulloch L., Alfieri A., McColl B. W., Experimental stroke differentially affects discrete subpopulations of splenic macrophages. Front. Immunol. 9, 1108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urra X., et al. , Monocyte subtypes predict clinical course and prognosis in human stroke. J. Cereb. Blood Flow Metab. 29, 994–1002 (2009). [DOI] [PubMed] [Google Scholar]
- 35.ElAli A., Jean LeBlanc N., The role of monocytes in ischemic stroke pathobiology: New avenues to explore. Front. Aging Neurosci. 8, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leidinger P., Backes C., Meder B., Meese E., Keller A., The human miRNA repertoire of different blood compounds. BMC Genomics 15, 474 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lobentanzer S., “Small RNA dynamics in cholinergic systems,” PhD dissertation, Goethe University, Frankfurt, Germany: (2020). [Google Scholar]
- 38.Cheng X., et al. , MicroRNA and their target mRNAs change expression in whole blood of patients after intracerebral hemorrhage. J. Cereb. Blood Flow Metab. 40, 775–786 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piccirillo C. A., Bjur E., Topisirovic I., Sonenberg N., Larsson O., Translational control of immune responses: From transcripts to translatomes. Nat. Immunol. 15, 503–511 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Chiou N.-T., Kageyama R., Ansel K. M., Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 25, 3356–3370.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan J. R., et al. , Blood microRNAs in low or no risk ischemic stroke patients. Int. J. Mol. Sci. 14, 2072–2084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schopman N. C. T., Heynen S., Haasnoot J., Berkhout B., A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 7, 573–576 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Wattananit S., et al. , Monocyte-derived macrophages contribute to spontaneous long-term functional recovery after stroke in mice. J. Neurosci. 36, 4182–4195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cserép C., et al. , Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science 367, 528–537 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Feinberg M. W., et al. , The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 26, 4138–4148 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shichita T., et al. , MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat. Med. 23, 723–732 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Labzin L. I., et al. , ATF3 is a key regulator of macrophage IFN responses. J. Immunol. 195, 4446–4455 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Glal D., et al. , ATF3 sustains IL-22-induced STAT3 phosphorylation to maintain mucosal immunity through inhibiting phosphatases. Front. Immunol. 9, 2522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinrich R., et al. , ATF3 regulates the expression of AChE during stress. Front. Mol. Neurosci. 11, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai A. S., et al. , A year-long immune profile of the systemic response in acute stroke survivors. Brain 142, 978–991 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuriakose T., Kanneganti T. D., ZBP1: Innate sensor regulating cell death and inflammation. Trends Immunol. 39, 123–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson D. M., Parker R., Stressing out over tRNA cleavage. Cell 138, 215–219 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Godoy P. M., et al. , Large differences in small RNA composition between human biofluids. Cell Rep. 25, 1346–1358 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shenhar-Tsarfaty S., et al. , Weakened cholinergic blockade of inflammation associates with diabetes-related depression. Mol. Med. 22, 156–161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reeves M. J., et al. , Sex differences in stroke: Epidemiology, clinical presentation, medical care, and outcomes. Lancet Neurol. 7, 915–926 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andrews S., Fast QC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 57.Roehr J. T., Dieterich C., Reinert K., Flexbar 3.0–SIMD and multicore parallelization. Bioinformatics 33, 2941–2942 (2020). [DOI] [PubMed] [Google Scholar]
- 58.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W. C., et al. , miRExpress: analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinformatics 10, 328 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loher P., Telonis A. G., Rigoutsos I., MINTmap: Fast and exhaustive profiling of nuclear and mitochondrial tRNA fragments from short RNA-seq data. Sci. Rep. 7, 41184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Winek K., Lobentanzer S., Soreq H., GEO submission. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE158314. Deposited 21 September 2020.
- 62.Delignette-Muller M. L., Dutang C., fitdistrplus: An R package for fitting distributions. J. Stat. Softw. 64, 1–34 (2015). [Google Scholar]
- 63.Bartosh T. J., Ylostalo J. H., Macrophage inflammatory assay. Bio Protoc. 4, e1180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobentanzer S., GitHub repository. https://github.com/slobentanzer/stroke-trf. Deposited 17 April 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code data have been deposited in GitHub (https://github.com/slobentanzer/stroke-trf) (64). RNA sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database, (accession no. GSE158312) (61).