Significance
Epigenetic regulations control the accessibility of transcription factors to their target regions. Modulation of chromatin accessibility determines which transcripts to be expressed and therefore, defines cell identity. Chromatin modulation during cell fate determination involves a complex regulatory network, yet the comprehensive view remains to be explored. Here, we provide a global view of chromatin accessibility during muscle stem cell activation. We identified a long noncoding RNA (lncRNA), LncMyoD, which regulates lineage determination and progression through modulating chromatin accessibility. Functional analysis showed that loss of LncMyoD strongly impairs reprogramming of fibroblasts into myogenic lineage and causes defects in muscle stem cell differentiation. Our findings provide an epigenetic mechanism for the regulation of muscle stem cell myogenic lineage progression by an lncRNA.
Keywords: stem cell, lncRNA, chromatin modulation, lineage progression
Abstract
Epigenetics regulation plays a critical role in determining cell identity by controlling the accessibility of lineage-specific regulatory regions. In muscle stem cells, epigenetic mechanisms of how chromatin accessibility is modulated during cell fate determination are not fully understood. Here, we identified a long noncoding RNA, LncMyoD, that functions as a chromatin modulator for myogenic lineage determination and progression. The depletion of LncMyoD in muscle stem cells led to the down-regulation of myogenic genes and defects in myogenic differentiation. LncMyoD exclusively binds with MyoD and not with other myogenic regulatory factors and promotes transactivation of target genes. The mechanistic study revealed that loss of LncMyoD prevents the establishment of a permissive chromatin environment at myogenic E-box–containing regions, therefore restricting the binding of MyoD. Furthermore, the depletion of LncMyoD strongly impairs the reprogramming of fibroblasts into the myogenic lineage. Taken together, our study shows that LncMyoD associates with MyoD and promotes myogenic gene expression through modulating MyoD accessibility to chromatin, thereby regulating myogenic lineage determination and progression.
In eukaryotic chromatin, much of the DNA is buried within nucleosomes, and accessibility of transcription factors (TFs) to their target sequences is sterically hindered by histones and higher-order chromatin structures. The accessibility of chromatin regions determines the transcriptome landscape of the cell, therefore restricting the identity of the cell type (1). Pioneer TFs are capable of de novo binding to their target sites in silent or closed chromatin regions (2). Such initial binding can actively open up the local chromatin for binding by other factors, thereby causing manifest changes in gene regulatory networks crucial for cell fate changes and cellular reprogramming. Cell fate determination by pioneer TFs can be exemplified by the conversion of fibroblasts into induced pluripotent stem cells by the Yamanaka factors (3). However, the core pluripotency pioneer TFs, Oct4, Sox2, and Klf4, have been reported to benefit from the cooperative interaction with other TFs (4, 5). Also, the binding of NRF1 is restricted by the DNA methylation state of its target motif, which is dependent on the binding of surrounding TF motifs (6). All these findings suggest a complex network in how TFs access compact chromatin during lineage commitment, and the detailed mechanism needs to be further investigated.
Exploration of mammalian genomes has revealed a new class of RNA regulator: long noncoding RNAs (lncRNAs) (7, 8). lncRNAs are arbitrarily defined as transcripts longer than 200 nucleotides that have no protein-coding potential (9). This class of RNAs shares similar chromatin properties with messenger RNAs and can be identified with a histone 3 lysine-4 trimethylation (H3K4me3)-marked active promoter and an H3K36me3-marked gene body (8, 10). lncRNAs have been found to play fundamental roles in diverse biological processes, including X chromosome silencing, stem cell maintenance and differentiation, and cell fate programming and reprogramming (11–13). Further mechanistic studies of the functions of lncRNAs indicate that they regulate transcription via chromatin modulation (14). For example, HOTTIP directly binds with WDR5 and displays H3K4me3 marks on its target genes through chromosomal looping to promote transcription (15). On the other hand, HOTAIR physically associates with the Polycomb repressive complex 2 (PRC2) and displays H3K27me3 marks across hundreds of targeting sites (16, 17). Another lncRNA, SRA, associates with p68 to stabilize the interaction of cohesion with CCCTC-binding factor, thus facilitating the establishment of long-range interaction through chromosomal looping (18). Moreover, around 9,000 lncRNAs were identified to associate with PRC2 in mouse embryonic stem cells (19), suggesting an emerging theme of a global lncRNA–chromatin network (20). Still, the detailed mechanism of how lncRNAs modulate chromatin accessibility remains largely elusive.
lncRNAs have emerged to regulate muscle development and differentiation. Dum, LncMyoD, and Linc-YY1 are directly activated by MyoD to promote myogenic differentiation through silencing Dppa2 expression, inhibiting IMP2-mediated mRNA translation, and dissociating YY1/PRC repressive complex from target promoters, respectively (21–23). Core enhancer RNA and Distal regulatory regions RNA promote chromatin remodeling to establish myogenic cell identity (24). Some lncRNAs such as SRA and linc-RAM form a complex with MyoD and other factors to enhance transcription of muscle-specific genes through chromatin modulation (25, 26). Different mechanisms have also been proposed to explain lncRNA functions during myogenic differentiation. Linc-MD1 is a cytoplasmic lncRNA and functions as a sponge of selective microRNAs (27, 28). Munc is produced from the upstream enhancer region of the Myod1 locus and promotes Myod1 and Myog transcription (29). Irm interacts with Mef2D and promotes assembly of MyoD/Mef2D complex to enhance differentiation (30). These findings emphasize the importance of lncRNAs during myogenic differentiation. However, it remains unclear how lncRNAs control myogenic lineage determination in muscle stem cells.
In this study, we showed a dramatic change in chromatin accessibility during muscle stem cell (satellite cell [SC]) activation using Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) analysis. We identified an lncRNA, LncMyoD, in modulating chromatin accessibility for myogenic lineage progression of muscle stem cells. Loss of LncMyoD led to the down-regulation of myogenic-related genes and observable phenotypic defects in SC differentiation. Mechanistically speaking, we observed that LncMyoD facilitates transactivation through direct binding to the MyoD protein and revealed LncMyoD promotes MyoD accessing to myogenic E-box–containing chromatins. Notably, LncMyoD is necessary for the reprogramming of 10T1/2 fibroblasts into myoblasts, indicating its chromatin modulation activity plays a critical role during cell fate determination. Our results collectively suggest that LncMyoD associates with MyoD to promote the establishment of an accessible chromatin environment for myogenic gene activation and is critical for myogenic lineage determination and progression.
Results
Extensive Changes in Chromatin Accessibility during SC Activation Affect the Transcriptome Landscape of SC.
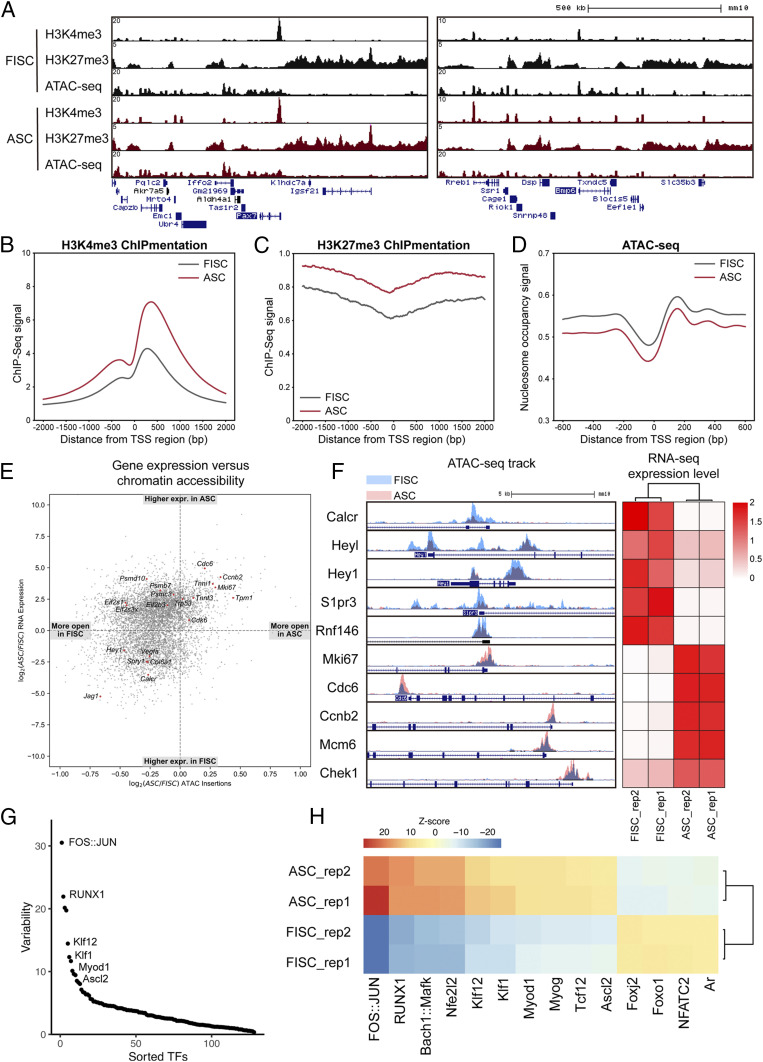
To explore the chromatin structure changes during SC activation, we performed chromatin immunoprecipitation with sequencing library preparation by Tn5 transposase (“tagmentation”) (ChIPmentation) (31) of H3K4me3 and H3K27me3 histone marks and ATAC-seq (32) on freshly isolated satellite cells (FISCs) and activated satellite cells (ASCs) isolated from 2.5-d postinjury hind-limb muscles using fluorescence-activated cell sorting (FACS). The profiling data showed a conventional distribution of histone marks relative to gene features and ATAC-seq signals: genes that are expressed in SCs are occupied by H3K4me3 histone mark at their promoter regions, while nonexpressing genes are strongly marked with H3K27me3 histone marks. As expected, ATAC-seq signals are only enriched at genomic locations where H3K27me3 histone marks are absent (Fig. 1A and SI Appendix, Fig. S1A). Genome-wide analysis of our data revealed that SCs exhibit dramatic chromatin accessibility changes during activation. Consistent with previous reports (33), SCs exhibit increased signal of H3K4me3 and H3K27me3 histone marks during activation (Fig. 1 B and C). Moreover, analysis of nucleosome occupancy using ATAC-seq data suggested that SCs have nucleosome removal events happening around the transcription start site (TSS) regions of genes during activation (Fig. 1D). Genomic annotation of the ATAC-seq data revealed that most of the open regions locate in intronic and distal intergenic regions in both FISCs and ASCs (SI Appendix, Fig. S1B). However, ASCs have a higher percentage of open promoter regions, and the identity of these promoters differs a lot from the those in FISCs (SI Appendix, Fig. S1 B and C). Taken together, our data suggest that SCs undergo dramatic chromatin remodeling upon activation, preparing them for myogenic lineage progression.
Fig. 1.
Chromatin accessibility changes during SC activation. (A) Genome tracks of H3K4me3 ChIP-seq, H3K27me3 ChIP-seq, and ATAC-seq signals in FISCs and ASCs across Pax7 (Left) and Bmp6 loci (Right). (B and C) Distribution of H3K4me3 (B) and H3K27me3 (C) histone marks across all TSS regions in FISCs and ASCs. Normalized tag intensity of 2 kb up- and downstream of the TSSs across the genome is shown. (D) Distribution of nucleosome occupancy signal across all TSS regions in FISCs and ASCs. Normalized tag intensity of 600 bp up- and downstream of the TSSs across the genome is shown. (E) Scatterplot showing fold change of Tn5 insertions (ASC/FISC) vs. fold change of RNA expression (ASC/FISC). Each dot represents a gene identified in RNA-seq. (F) Genome tracks of ATAC-seq signals in FISCs and ASCs across promoters of representative genes and their corresponding RNA-seq expression level. (G) chromVAR analysis of different TF motif accessibility changes between FISCs and ASCs. Graph represents the ranking of the variability of TF motifs accessibility changes. (H) Heat map showing Z scores of top-ranking TFs motif accessibility changes between FISCs and ASCs.
To understand the correlation of chromatin accessibility and gene expression during SC activation, we compared the deep RNA-sequencing (RNA-seq) data on FISCs and ASCs with the corresponding ATAC-seq data. Consistent with previous studies showing that chromatin accessibility is positively correlated with gene expression (34–36), our data showed that genes that are highly expressed in FISCs have higher chromatin accessibility across their promoter regions, and genes that are associated with proliferation and muscle development have higher chromatin accessibility in ASCs (Fig. 1 E and F). Interestingly, stress response genes such as Psmd10, Psmb7, Eif2s1, and Eif2s3x have higher accessibility in FISCs than ASCs, even though their gene expression is higher in ASCs than FISCs (Fig. 1E). This result indicates that SCs undergo environmental stress during the isolation process, and the chromatin accessibility changes are prior to gene expression. Detailed examination of ATAC-seq peaks using Reactome (37) showed that several signaling pathway genes, such as the Notch signaling pathway, are highly accessible in FISCs (SI Appendix, Fig. S1D), while genes associated with cell cycle, muscle contraction, and transcription are highly accessible in ASCs (SI Appendix, Fig. S1E). Functional analysis of differentially expressed genes in FISCs and ASCs also indicates that ASCs are actively involved in proliferation and transcription (SI Appendix, Fig. S1 F and G).
To further determine whether the chromatin structure changes alter TF binding, we analyzed the ATAC-seq data using chromVAR software, which estimates accessibility changes within ATAC-seq peaks sharing the same TF binding motif or annotation (38). We ranked TF motifs according to the variability of their accessibility changes during SC activation (Fig. 1G). Among the top-ranked TF motifs with the highest variability, known key factors that promote proliferation, such as FOS/JUN and RUNX1, were identified to have their binding motifs more accessible in ASCs (Fig. 1G). The binding motifs of myogenic regulatory factors (MRFs; i.e., MyoD and MyoG) were also identified to have their motif accessibility increased during SC activation (Fig. 1H). Taken together, our results suggest that the chromatin structure changes during SC activation affect TFs binding, therefore modulating the transcriptome landscape of SCs for myogenic lineage progression.
Identification of lncRNAs during SC Activation and Differentiation.
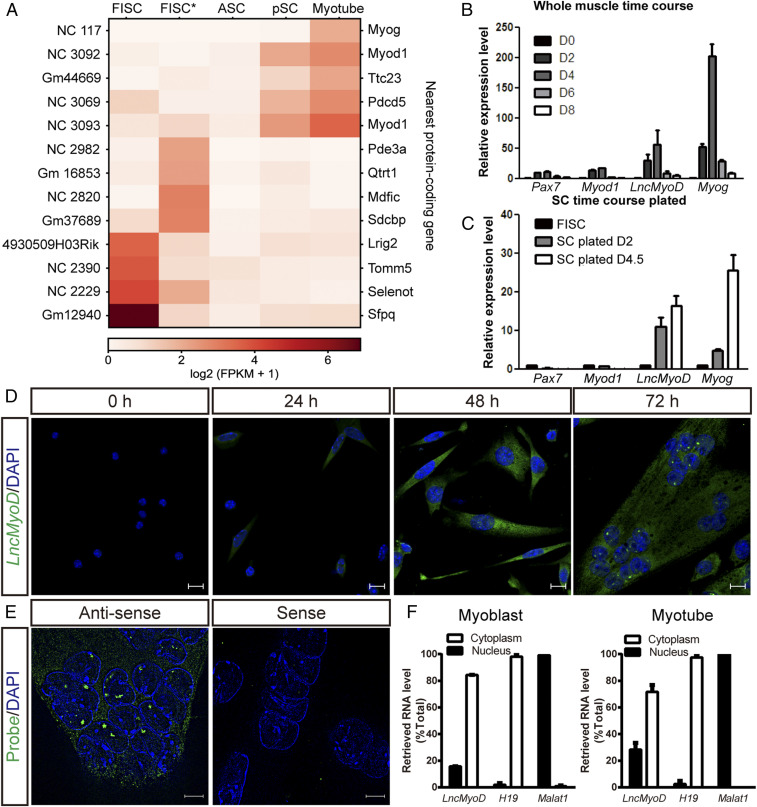
Previous studies have reported lncRNAs as regulators of chromatin remodeling (14). To identify chromatin-modifying lncRNAs that are expressed during SC activation and differentiation, we combined our RNA-seq data on FISCs and ASCs with published RNA-seq data from FISCs*, proliferating satellite cells, and myotubes (39) and performed de novo transcript assembly using the Tuxedo pipeline (40). To ensure identified transcripts are indeed noncoding, we examined their protein-coding potential using the Coding Potential Calculator 2 (CPC2) algorithm (41) and excluded transcripts classified as protein coding. In addition, we eliminated single-exon transcripts and transcripts smaller than 200 nucleotides, which could potentially be primary microRNA transcripts. These efforts yielded 14,947 putative lncRNAs expressed at any stage during SC activation, proliferation, and differentiation. We additionally characterized the expression level of lncRNAs in our RNA-seq datasets with the Gencode M19 annotation (42). We found that among the identified lncRNAs and the annotated lncRNAs, 3,998 lncRNA transcripts were differentially expressed during SC differentiation (Dataset S1). Further examination of their nearby protein-coding genes yielded three lncRNAs that were located near MRFs loci: NC 117, NC 3092, and NC 3093 (Fig. 2A), which matched previously predicted lncRNAs XR_878548 (43), Munc (29), and LncMyoD (22), respectively.
Fig. 2.
LncMyoD is an lncRNA expressed in ASCs. (A) Heat map showing the expression pattern of identified lncRNAs between RNA-seq data from different RNA-seq samples with their nearby protein-coding genes. FPKM, fragments per kilobase of transcript per million mapped reads. (B) Temporal expression of LncMyoD in whole-TA muscle extract and during muscle regeneration (D0, uninjured; D2 to D8, days 2 to 8 after muscle injury). (C) Temporal expression of LncMyoD in FACS-sorted FISCs and SCs plated in vitro under differentiation condition for 2 or 4.5 d after isolation. (D) FISH of endogenous LncMyoD molecules (green) in SCs cultured under differentiation condition for different time. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (Scale bar: 10 μm.) (E) FISH of LncMyoD antisense and sense probes in SCs cultured under differentiation condition for 72 h. The cell nuclei were stained with DAPI (blue). (Scale bar: 5 μm.) (F) qRT-PCR results of LncMyoD expression level in cytoplasm and nucleus in myoblasts and myotubes. H19 is used as a marker for cytoplasmic fraction; Malat1 is used as a marker for nuclear fraction.
To further examine the chromatin structure across these lncRNA loci during SC activation and differentiation, we examined the H3K4me3, H3K27me3 histone marks, and ATAC-seq signal distribution across these lncRNA loci in FISCs and ASCs. Two of the three lncRNAs, XR_878548 and Munc, had little access to their genomic loci in differentiating satellite cells (dSCs) (SI Appendix, Fig. S2 A and B), suggesting that they may function at a time beyond our analysis time points. On the other hand, NC 3092 (LncMyoD) showed an open chromatin environment in ASCs as well as a dramatic gain in chromatin accessibility in dSCs (SI Appendix, Fig. S2C). Although its locus is not marked by H3K4me3 during SC activation, the broad repressive mark, H3K27me3, is decreased (SI Appendix, Fig. S2C), suggesting that this lncRNA is released from a repressed chromatin state during SC activation and is permissive for expression during SC differentiation; 5′ and 3′ rapid amplification of complementary DNA ends (RACE) analysis showed that the full length of this transcript is 622 bp (SI Appendix, Fig. S2D). Gong et al. (22) detected a transcript variant they termed LncMyoD* in C2C12 cells (SI Appendix, Fig. S2E). However, we were unable to detect this isoform in SCs using an RACE assay, possibly due to its low expression level relative to the LncMyoD (SI Appendix, Fig. S2F). Therefore, we focused on LncMyoD for further study.
To examine the expression pattern of LncMyoD, we performed qRT-PCR time-course analysis in whole-muscle extract and cultured SCs (Fig. 2 B and C). LncMyoD was not detected in uninjured tabialis anterior (TA) muscle or FISCs. However, it became detectable in injured TA muscle 2 d postinjury (Fig. 2B). It was also detected in SCs cultured under the differentiating condition (Fig. 2C). To further identify the localization of LncMyoD, we performed fluorescence in situ hybridization (FISH) on FISCs and SCs cultured for different amounts of time (Fig. 2D). Concordant with the qRT-PCR results, LncMyoD was undetectable until 48 h after plating (Fig. 2D). Similar to other regulatory noncoding RNAs, foci of LncMyoD are localized in the nuclei (Fig. 2E). Subcellular fractionation results also confirmed that LncMyoD is expressed in both the cytoplasm and nucleus, but the ratio of the nuclear fraction is increased when cells are differentiating to form myotubes (Fig. 2F), suggesting LncMyoD’s function in regulating SC differentiation.
LncMyoD Is Required for Myogenic Lineage Progression.
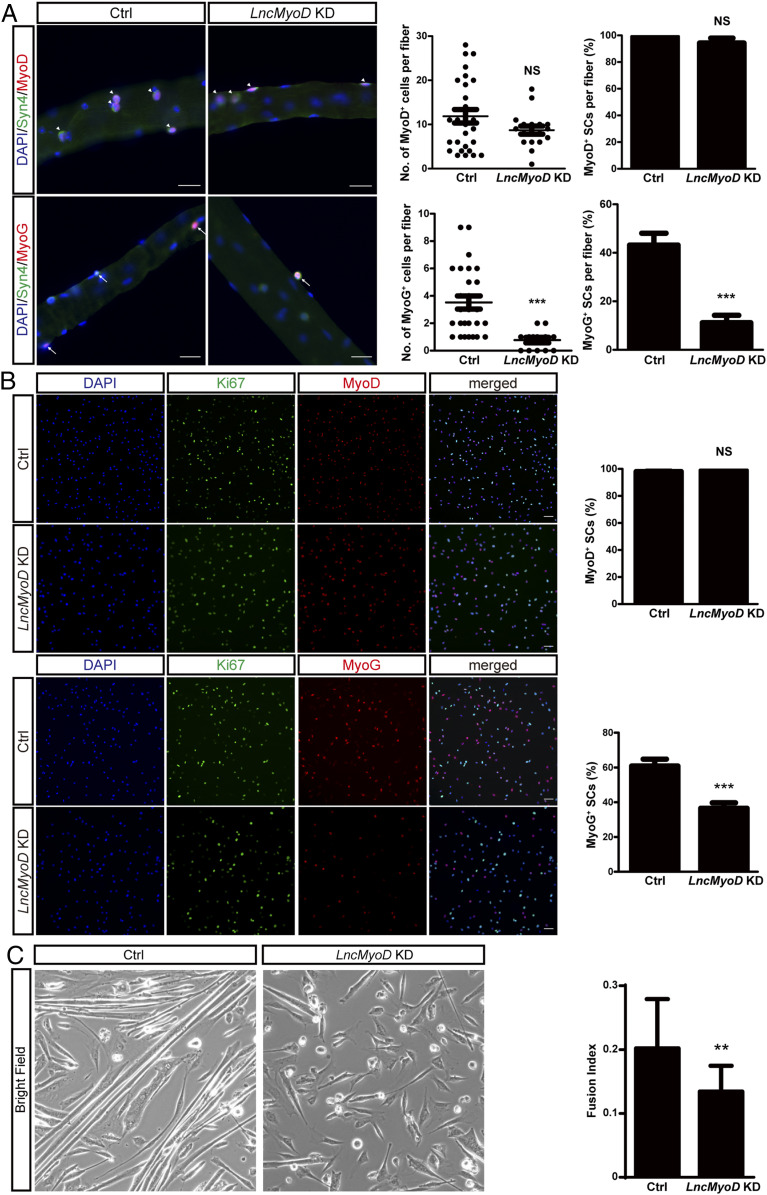
To investigate the role of LncMyoD during myogenic lineage progression, we conducted loss-of-function experiments with small interfering RNA (siRNA) treatment targeting LncMyoD. We confirmed the efficiency of siRNA by qRT-PCR on SCs and SC-derived primary myoblasts (SI Appendix, Fig. S3 A and B). We first conducted loss-of-function experiments on fiber-associated SCs ex vivo using Syndecan 4 (Syn4) as an SC marker on fiber explants (44, 45) and quantified the number of SCs on fibers 3 d after siRNA transfection. Essentially all SCs were MyoD+ in both the control (Ctrl) and LncMyoD knockdown (KD) groups, and there was no difference in the absolute number of MyoD+ SCs per fiber between groups (Fig. 3A). However, the loss of LncMyoD in SCs led to a reduction of MyoG expression (Fig. 3A). These results indicate that the loss of LncMyoD impairs SC differentiation but not SC proliferation.
Fig. 3.
LncMyoD is required for myogenic lineage progression. (A, Left) Single fibers were isolated and cultured for 3 d under differentiation condition after siRNA KD and stained for Syn4 and MyoD/MyoG. Syn4, MyoD double-positive cells are highlighted by arrowheads. Syn4, MyoG double-positive cells are highlighted by arrows. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (Scale bar: 50 μm.) (A, Right) Quantification of MyoD+/MyoG+ cells per fiber and percentage of Syn4+ cells that are MyoD+/MyoG+ in Ctrl and LncMyoD KD fibers. NS, not significant. ***P < 0.001. (B, Left) FACS-sorted SCs were cultured for 3 d under differentiation condition after siRNA KD and stained for Ki67 and MyoD/MyoG. The cell nuclei were stained with DAPI (blue). (Scale bar: 100 μm.) (B, Right) Quantification of the percentage of MyoD+ and MyoG+ cells in Ctrl and LncMyoD KD SCs. NS, not significant. ***P < 0.001. (C, Left) Morphology of FACS-sorted SCs cultured 4 d under differentiation condition after siRNA treatment. (C, Right) Fusion index calculated from FACS-sorted SCs cultured 4 d under differentiation condition after siRNA treatment. **P < 0.01.
To examine the effect of LncMyoD KD on cultured SCs in vitro, we performed immunostaining for myogenic markers on SCs with siRNA treatment targeting LncMyoD. We cultured FACS-sorted SCs in differentiation medium for 48 h after LncMyoD KD and quantified various myogenic markers. Similar to our previous results, there was no difference in MyoD expression between the Ctrl and LncMyoD KD groups (Fig. 3B). However, the LncMyoD KD group exhibited a significant decrease in MyoG expression (Fig. 3B). Furthermore, when SCs were induced to differentiate for a more extended period, loss of LncMyoD led to defects in myotube formation (Fig. 3C). The fusion index also indicated a reduction in fusing potential in LncMyoD KD SCs (Fig. 3C). These results suggest that the expression of LncMyoD is required prior to myogenic differentiation.
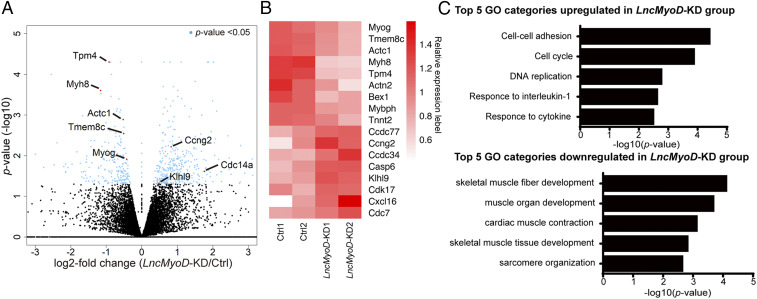
To further analyze how LncMyoD regulates SC transcriptome, we treated FACS-sorted SCs with siRNA targeting LncMyoD, followed by RNA-seq. There was a significant number of differentially expressed genes between the Ctrl and LncMyoD KD groups (Fig. 4A). Typical genes known to regulate myogenesis, such as Myog, Actc1, and Tmem8c (Myomaker), exhibited decreased expression in the LncMyoD KD group compared with the Ctrl group (Fig. 4 A and B). Gene ontology analysis of biological processes using DAVID (46) demonstrated that muscle formation-related genes were highly down-regulated after LncMyoD KD. In contrast, genes associated with cell proliferation and cell-state transition were highly enriched in the LncMyoD KD group (Fig. 4C). Taken together, these results indicate that loss of LncMyoD leads to a marked deficiency in the myogenic differentiation program.
Fig. 4.
Loss of LncMyoD leads to deficiencies in myogenic lineage progression. (A) Volcano plot showing gene expression-level changes between the Ctrl and LncMyoD KD groups. Blue dots represent genes with P values <0.05. (B) Heat map plot showing relative expression level of representative genes between the Ctrl and LncMyoD KD samples. (C) Gene ontology (GO) analysis of differentially expressed genes in the LncMyoD KD group.
LncMyoD Directly Binds with MyoD to Promote Gene Transcription.
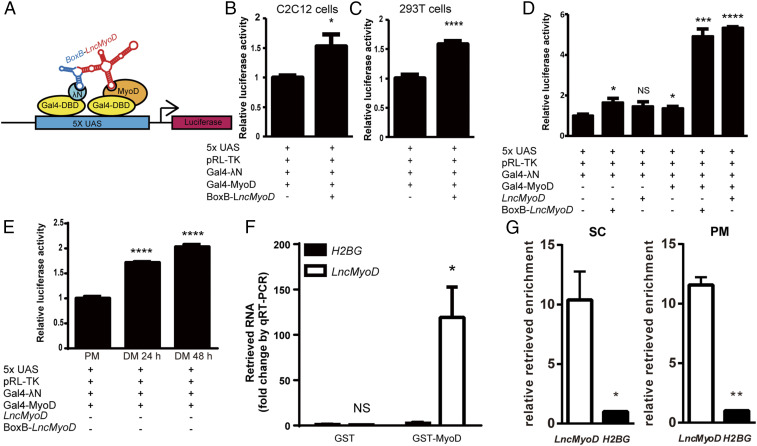
To explore the mechanism of the LncMyoD function, we first examined its potential binding partners using the CatRAPID algorithm (47). CatRAPID computationally predicts the binding propensity of protein–RNA pairs by calculating secondary structure, hydrogen bonding, and van der Waals contributions. Since the RNA-seq analysis points to the notion that LncMyoD regulates SC differentiation, we focused our search among the essential myogenic regulators. After predicting the binding potential of Pax7 and four MRFs with LncMyoD, MyoD stood out as a potential target (SI Appendix, Fig. S4A). Further comparison of LncMyoD and MyoD sequences revealed potential binding hot spots (SI Appendix, Fig. S4B). To understand the interaction between LncMyoD and MyoD protein, we utilized the BoxB tethering assay system (15, 48) (Fig. 5A). In this system, an RNA (i.e., LncMyoD) is tethered to the BoxB RNA, which interacts with λN protein. Both the λN and MRFs are fused with Gal4-DNA binding domain (Gal4-DBD) so that the interaction of LncMyoD and individual MRF can be dissected through examining the signal of luciferase activity.
Fig. 5.
LncMyoD facilitates transactivation via binding with MyoD. (A) Illustration of the BoxB tethering assay system. UAS, upstream activating sequence. (B and C) Luciferase activity after cotransfection of the indicated constructs in C2C12 cells (B) and 293T cells (C). *P < 0.05; ****P < 0.0001. (D) Luciferase activity after cotransfection of the indicated constructs in C2C12 cells. NS, not significant. *P < 0.05; ***P < 0.001; ****P < 0.0001. (E) Luciferase activity after cotransfection of the indicated construct in C2C12 cells cultured in differentiation medium (DM) for 24 or 48 h. ****P < 0.0001. (F) qRT-PCR results of retrieved LncMyoD level in GST binding assay. *P < 0.05. (G) qRT-PCR results of retrieved LncMyoD level with MyoD RIP on cultured SCs and SC-derived primary myoblasts (PMs). *P < 0.05; **P < 0.01.
We first checked the tethering system by transfecting Gal4-MyoD alone into C2C12 cells. As expected, the luciferase activity increased with the addition of MyoD in a dosage-dependent manner (SI Appendix, Fig. S4D). We subsequently transfected both BoxB-LncMyoD and Gal4-MyoD into the system and observed a significant increase in luciferase activity in both the myogenic cell line (Fig. 5B) and nonmyogenic cell line (Fig. 5C). This observation indicates that LncMyoD and MyoD synergistically promote luciferase expression. Furthermore, an increase in the amount of BoxB-LncMyoD in the system dramatically boosted the luciferase signal (SI Appendix, Fig. S4E), suggesting that LncMyoD functions in a dosage-dependent manner. However, when recruiting BoxB-LncMyoD to other MRFs fused with Gal4-DBD, no induction of luciferase activity was observed (SI Appendix, Fig. S4C), indicating that the interaction between LncMyoD and MyoD is unique and specific among MRFs.
The MyoD-LncMyoD interaction described above is based on the specific binding ability of the BoxB sequence to the λN protein. To determine whether LncMyoD can be recruited to MyoD protein without external aid, we performed a luciferase assay with LncMyoD instead of BoxB-LncMyoD. The addition of LncMyoD also significantly increased luciferase activity (Fig. 5D). In addition, when C2C12 cells were induced to differentiate, we also observed an increase in luciferase activity (Fig. 5E). This observation suggests that endogenous LncMyoD expressed during differentiation can be recruited to the tethering system by MyoD.
We further investigated whether MyoD and LncMyoD directly interact with each other using a glutathione S-transferase (GST) binding assay system (15). Purified GST and GST-fused MyoD protein were incubated with in vitro-transcribed LncMyoD to examine their binding potential (SI Appendix, Fig. S4F). Because of the binding specificity between GST protein and glutathione beads, only the RNAs that are directly bound to the fusion protein can be retrieved. LncMyoD was only detected in the presence of MyoD protein, indicating the existence of a physical binding between MyoD protein and LncMyoD RNA (Fig. 5F). To validate the LncMyoD–MyoD interaction in vivo, we performed MyoD RNA immunoprecipitation (RIP) on cultured SCs and SC-derived primary myoblasts. The results showed that LncMyoD can be pulled down by MyoD in both SCs and primary myoblasts (Fig. 5G), suggesting that endogenous LncMyoD and MyoD interact together in vivo.
To further identify the LncMyoD-interacting domain of MyoD, we generated the MyoD truncation proteins through combinations of its three functional domains (i.e., N terminal, basic helix–loop–helix domain [bHLH], and C terminal). Gal4-DBD fused MyoD truncations were cotransfected with LncMyoD-containing plasmid into 293T cells, and RNA was retrieved through immunoprecipitation of Gal4-DBD. The results show that all MyoD truncations can pull down LncMyoD compared with Gal4DBD (SI Appendix, Fig. S4G). However, none of the truncations interact with LncMyoD as efficient as the full-length MyoD. Among the truncations we generated, the truncations containing the bHLH domain pulled down the most amount of LncMyoD compared with others (SI Appendix, Fig. S4G), suggesting that the bHLH domain could be the major LncMyoD-interacting domain. Since the bHLH domain recognizes E-box–containing DNA, this result agrees with our conclusion that LncMyoD facilitates MyoD binding to its target regions. Taken together, our results suggest that LncMyoD forms a complex with MyoD protein to promote gene transcription.
LncMyoD Functions by Modulating Chromatin Accessibility for Myogenic Lineage Progression.
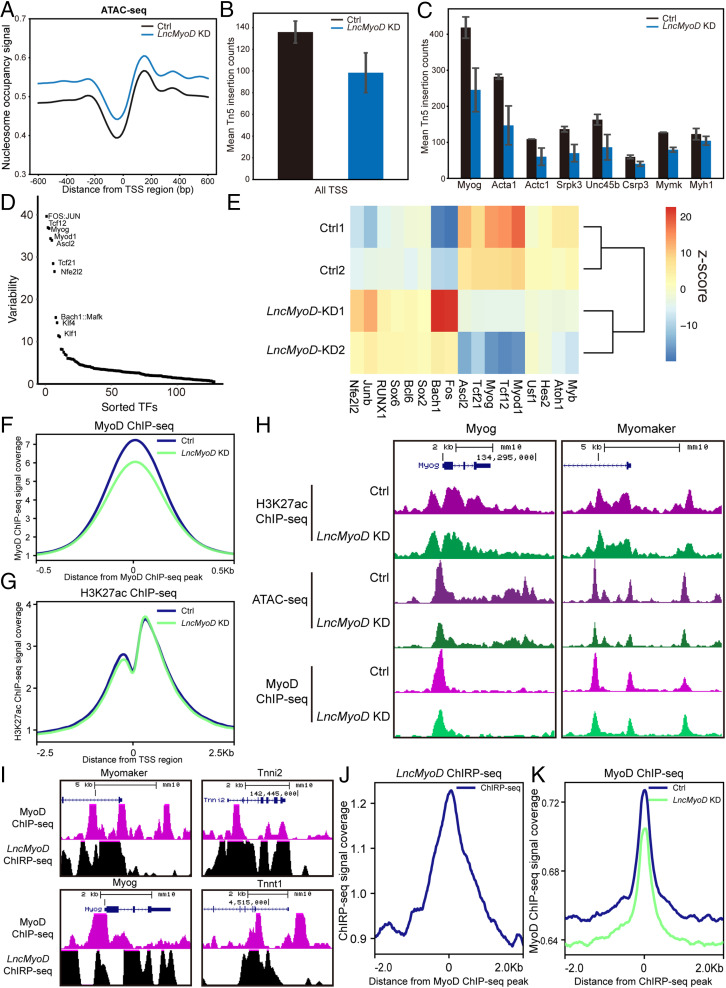
To understand whether LncMyoD influences chromatin accessibility during SC activation and differentiation, we performed ATAC-seq on FACS-sorted SCs with siRNA-induced LncMyoD KD (SI Appendix, Fig. S5A). LncMyoD KD led to an increased nucleosome occupancy signal and a decreased Tn5 insertion count around all TSSs, suggesting that global genome accessibility has changed (Fig. 6 A and B). Additionally, LncMyoD KD samples contain a much higher relative proportion of single-nucleosome spanning reads compared with the Ctrl samples, suggesting chromatin structure is changed upon loss of LncMyoD (SI Appendix, Fig. S5B). To further examine the effect of LncMyoD KD on the chromatin environment, we utilized published chromatin immunoprecipitation followed by sequencing (ChIP-seq) data of H3K4me3 in C2C12 myoblasts and myotubes from the Encyclopedia of DNA Elements (ENCODE) portal (49). We retrospectively extracted regions that were enriched for H3K4me3 histone mark in C2C12 myoblasts and myotubes and assessed their accessibility after LncMyoD KD using ATAC-seq reads. After LncMyoD KD, genome accessibility was reduced globally as indicated by decreased signals across all H3K4me3 peaks (SI Appendix, Fig. S5C). Of note, the decrease was more dramatic across H3K4me3 peaks from myotubes, suggesting that LncMyoD has a more pronounced effect during myogenic differentiation. Detailed examination of TSS accessibility confirmed the dramatic loss of chromatin accessibility across the genomic loci of typical myogenic genes (Fig. 6C).
Fig. 6.
LncMyoD functions as a chromatin modifier to promote the transcription of myogenesis-related genes. (A) Distribution of nucleosome occupancy signal across all TSS regions (B and C) Calculation of Tn5 insertion counts of all TSS regions (B) and TSS regions of typical myogenesis-related genes (C). (D) chromVAR analysis of different TF motif accessibility changes between the Ctrl and LncMyoD KD groups. Graph represents the ranking of the variability of TF motifs accessibility changes. (E) Heat map showing Z score of TF motif accessibility changes between the Ctrl and LncMyoD KD groups. (F) Distribution of MyoD ChIP-seq signal across all MyoD binding sites. (G) Distribution of H3K27ac ChIP-seq signal across all MyoD binding sites. (H) Genome tracks of the Myog and Myomaker loci showing ATAC-seq, H3K27ac ChIP-seq, and MyoD ChIP-seq across their promoter regions in the Ctrl and LncMyoD KD groups. (I) Genome tracks of the Myog, Myomaker, Tnni2, and Tnnt1 loci showing LncMyoD ChIRP-seq and MyoD ChIP-seq across their promoter regions. (J) Distribution of LncMyoD ChIRP-seq signal across all MyoD binding sites. (K) Distribution of MyoD ChIP-seq signal across all LncMyoD binding sites.
To further determine if LncMyoD-mediated chromatin structure changes alter TF binding, we analyzed our ATAC-seq data using chromVAR. Interestingly, this analysis reveals that binding motifs of E-proteins exhibited the largest decrease in accessibility after LncMyoD KD (Fig. 6 D and E), suggesting that LncMyoD potentially has a role in rendering E-box regions accessible. To validate the chromatin changes across TF binding sites, we utilized published ChIP-seq data of different TFs on C2C12 myoblasts and myotubes from the ENCODE portal (49) and extracted binding sites of these TFs to assess the accessibility changes using our ATAC-seq data. The examined TF binding sites all exhibited a decrease in accessibility after LncMyoD KD (SI Appendix, Fig. S5D). However, when we calculated the extent of accessibility changes, only MRFs (i.e., MyoD and MyoG) exhibited larger reductions in binding site access in myotubes than in myoblasts (SI Appendix, Fig. S5E). Taken together, these analyses indicate that loss of LncMyoD reduces the permissiveness of the chromatin environment at myogenic E-box regions, thereby preventing the binding of MRFs during myogenic differentiation.
To directly assess the impact of LncMyoD on chromatin accessibility, we performed MyoD ChIPmentation on cultured SCs with siRNA treatment targeting LncMyoD. The results showed that MyoD ChIP-seq signals decreased dramatically after the loss of LncMyoD (Fig. 6F). Typical myogenic genes such as Myog and Myomaker showed a decrease of ATAC-seq signals as well as a decrease of MyoD ChIP-seq signals across their promoter regions (Fig. 6H and SI Appendix, Fig. S5F), suggesting that the loss of LncMyoD reduces the chromatin accessibility of MyoD binding sites, thereby preventing the binding of MyoD and inhibiting myogenic differentiation. To investigate whether the decrease of myogenic gene expression after the loss of LncMyoD is related to altered promoter/enhancer activities, we examined the H3K27ac level with LncMyoD KD. No decrease in H3K27ac marks across all TSS regions was observed after LncMyoD KD (Fig. 6 G and H and SI Appendix, Fig. S5F), suggesting that LncMyoD does not affect the promoter/enhancer activities.
To further explore the genomic binding sites of LncMyoD, we performed the Chromatin Isolation by RNA Purification followed by sequencing (ChIRP-seq) (50) using biotin-labeled DNA probes targeting LncMyoD (Fig. 6I and SI Appendix, Fig. S5G). We found a dramatic enrichment of LncMyoD ChIRP-seq signal across MyoD binding sites (Fig. 6J), suggesting the coexistence of MyoD and LncMyoD at specific genomic loci. Furthermore, MyoD ChIP-seq signal decreased dramatically across LncMyoD binding sites after LncMyoD KD (Fig. 6K), indicating the involvement of LncMyoD in MyoD binding to the genome. Detailed examination of myogenic genes loci showed that MyoD and LncMyoD co-occupy myogenic gene promoter regions (Fig. 6I and SI Appendix, Fig. S5H). Taken together, our results show that LncMyoD facilitates MyoD binding to myogenic gene promoter regions, thereby promoting myogenic differentiation.
LncMyoD Is Required to Mediate Transdifferentiation of Fibroblasts to Myoblasts.
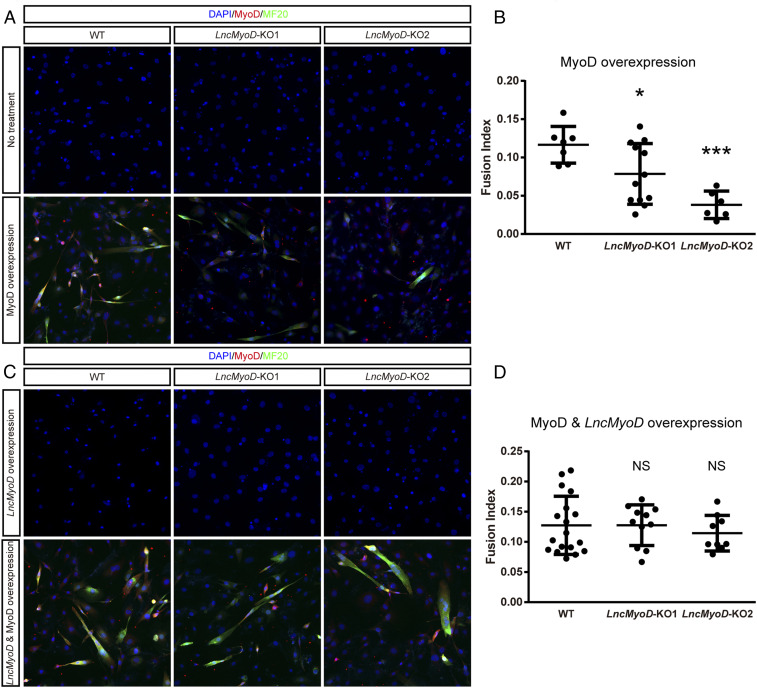
MyoD is considered a master regulator for myogenic lineage reprogramming (51) in that ectopic expression of MyoD in fibroblasts can result in transdifferentiation into myoblast lineage cells (52, 53). Mechanistic studies have revealed the function of MyoD in transdifferentiation: it can access repressive chromatin and recruit histone acetyltransferases (HATs) and switch/sucrose-nonfermentable (SWI/SNF) complexes for chromatin remodeling (54–57). Considering that LncMyoD has a strong physical binding affinity to MyoD protein and has chromatin remodeling function, we next tested whether LncMyoD is involved during the lineage reprogramming process. To this end, we generated LncMyoD knockout (KO) 10T1/2 fibroblast cell lines using the CRISPR-Cas9 system (58). We confirmed the deletion of LncMyoD exon 2 by PCR and sequencing (SI Appendix, Fig. S6 A and B and Dataset S2). To induce MyoD overexpression, we infected 10T1/2 fibroblasts with red fluorescent protein (RFP)-labeled MyoD adenovirus and cultured them in the differentiation medium. Without any treatment, neither wild-type (WT) clones nor LncMyoD KO clones transdifferentiated into myoblasts; only WT clones differentiated and fused into myotubes after MyoD overexpression (Fig. 7A). LncMyoD KO clones underwent cell fusion due to the potent effect of MyoD but could not form large myotubes. Furthermore, the fusion index showed that WT clones had greater fusing potential than LncMyoD KO clones (Fig. 7B). Taken together, these data suggest that LncMyoD is required for the MyoD-induced transdifferentiation of fibroblasts into myoblasts.
Fig. 7.
LncMyoD is essential for the establishment of the myogenic program in 10T1/2 fibroblasts. (A) WT and LncMyoD KO 10T1/2 fibroblast clones were induced to differentiate for 8 d after transfection of the indicated MyoD adenovirus. Cells were then fixed to visualize myotube formation. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (B) Fusion index calculation of WT and LncMyoD KO clones for MyoD adenovirus treatment. *P < 0.05, ***P < 0.001. (C) WT and LncMyoD KO 10T1/2 fibroblast clones were induced to differentiate for 8 d after transfection of the indicated LncMyoD plasmid and MyoD adenovirus. Cells were then fixed to visualize myotube formation. The cell nuclei were stained with DAPI (blue). (D) Fusion index calculation of WT and LncMyoD KO clones for MyoD adenovirus and LncMyoD-containing plasmid treatment. NS, not significant.
We subsequently conducted a rescue study by restoring LncMyoD expression during the transdifferentiation process. We examined the transcriptome of both WT and KO clones to validate the rescue effect (SI Appendix, Fig. S6 C–E). Overexpression of LncMyoD alone was not able to induce Myog expression or mediate the transition from fibroblasts into myoblasts in WT or LncMyoD KO clones (Fig. 7C and SI Appendix, Fig. S6 C–E). However, the fusion capacity was rescued in LncMyoD KO clones with LncMyoD and MyoD overexpression, while little difference was observed in WT clones (Fig. 7C). We also observed an increase in Myog expression when LncMyoD is cotransfected with MyoD in KO clones (SI Appendix, Fig. S6 C–E). Moreover, the fusion index of rescued LncMyoD KO clones was restored to the same level as that of WT clones (Fig. 7D). These results collectively suggest that LncMyoD associates with MyoD and promote myogenic lineage determination.
Although MyoD is considered the master regulator of myogenic lineage determination, several cell lines such as HeLa cells and embryonic cell lines have been reported to be “refractory” to myogenic conversion (59–61). To investigate whether LncMyoD can release this resistance, we performed MyoD-induced transdifferentiation on HeLa cells (SI Appendix, Fig. S6F). However, no transdifferentiation phenotype (i.e., Myotube formation) was observed after MyoD induction with or without LncMyoD overexpression, suggesting that LncMyoD is insufficient to release resistance to myogenic conversion. Nevertheless, our result indicates that LncMyoD is essential for promoting myogenic lineage determination.
Discussion
Modulation of chromatin accessibility is critical for cell identity determination. lncRNAs have recently emerged to play important roles in various gene regulatory networks through chromatin modulation (14). However, in contrast to its rapid identifications, functional characterization of lncRNAs remains largely elusive. In this study, we gained insights into the biological functions of a previously reported lncRNA, LncMyoD, using a loss-of-function approach combined with multiple next-generation sequencing techniques. We demonstrated that LncMyoD forms chromatin regulator complex with MyoD and renders E-box regions more accessible for TF binding, thereby promoting myogenic lineage determination and progression. Thus, our study provides another perspective on lncRNA function in cell fate determination.
It has been reported that FISCs can be identified with distinct transcriptome by RNA-seq (62, 63). Here, we extended the finding by analyzing the chromatin structure changes during SC activation using ATAC-seq. Our data suggest that genes associated with the Notch signaling pathway, which is known to regulate quiescence maintenance of SCs (64), are highly accessible in FISCs. In ASCs, genes related to programmed cell death and cell cycle are highly accessible. Interestingly, stress response genes such as Psmd10, Psmb7, Eif2s1, and Eif2s3x are more accessible in FISCs than in ASCs (Fig. 1E), suggesting SCs experience environmental stress during the isolation process. This result also indicates that chromatin accessibility changes prior to gene expression.
In our study, one of the outstanding findings is that LncMyoD participates in the cell fate reprogramming process (Fig. 7). It has been reported that enhancer RNAs promote chromatin accessibility at distinct regulatory regions during myogenic differentiation (24). Similarly, our mechanistic analysis revealed that LncMyoD modulates the accessibility to E-box regions of MRFs (i.e., MyoD and MyoG) (Fig. 6 D and E and SI Appendix, Fig. S5 D and E). Previous ChIP-seq analyses of MyoD and MyoG in C2C12 myotubes showed that 70% of the MyoG binding sites are shared with MyoD (24); thus, MyoG can still benefit from the remodeling events happening at the overlapping binding sequences without direct binding to LncMyoD. In this way, LncMyoD establishes a permissive chromatin environment across myogenic E-box regions and facilitates MyoD binding to its target sites. After the myogenic lineage is determined, the E-box regions remain accessible for the subsequent binding of other TFs. Concordant with this notion, when we induced differentiation in LncMyoD KO primary myoblast cells, the differentiation process was not as largely affected as that in SCs or nonmyogenic cells (SI Appendix, Fig. S6G). Given that myogenic cell lines are already committed to the myogenic lineage, their chromatin environment is fully established for differentiation. Therefore, loss of LncMyoD does not disturb further progression of myogenesis in myogenic cell lines. Taken together, our data suggest that LncMyoD associates with MyoD and establishes permissive chromatin to allow subsequent bindings of other myogenic factors.
Although multiple cell lines can transdifferentiate into myoblasts by MyoD (59), a variety of refractory cell lines such as HeLa cells or embryonic stem cells resists myogenic conversion (60, 61, 65). Subsequent studies showed that chromatin modifiers such as Baf60c could release this resistance (60), and the recruitment of MyoD to a silent Myog promoter requires a prebound homeodomain Pbx as an accessory factor to establish the myogenic-specific lineage (66). Considering that numerous lncRNAs have been reported to recruit regulatory factors and alter genomic architecture (67), other pioneer TFs might also access their target DNA sequences via assistance from chromatin-remodeling lncRNAs. Although LncMyoD fails in releasing myogenic resistance in HeLa cells, our results still provide mechanistic insights into how pioneer TFs may work together with lncRNAs to regulate compact chromatin.
In our study, we explored the genome-wide occupancy of LncMyoD using the ChIRP-seq approach. Around 28% of total LncMyoD was retrieved from the biotin-labeled DNA probes, while only 0.02% GAPDH was retrieved (SI Appendix, Fig. S5G), suggesting the successful pull down of LncMyoD. We also observed enrichment of the ChIRP-seq signal across LncMyoD locus (SI Appendix, Fig. S5H), indicating the active transcription of LncMyoD during differentiation. Combining MyoD ChIP-seq and LncMyoD ChIRP-seq data, we observed enrichment of LncMyoD ChIRP-seq signal across MyoD binding sites (Fig. 6I), suggesting the co-occupancy of LncMyoD and MyoD across the genome. This co-occupancy is disrupted after the loss of LncMyoD (Fig. 6J), indicating the essential role of LncMyoD in maintaining the accessible chromatin states. Taken together, our ChIRP-seq data provide a comprehensive view of LncMyoD binding to the genome and highlight the role of LncMyoD in regulating chromatin accessibility to facilitate MyoD accessing myogenic promoter regions during differentiation.
LncMyoD is reported to bind to IMP2 protein to block IMP2-mediated translation (22). Here, we found that MyoD is another binding partner of LncMyoD and operates to regulate myogenic lineage progression by modulating chromatin accessibility. The RIP assay indicates the interaction between MyoD and LncMyoD in vivo, and the GST pull-down assay validated the direct binding of MyoD and LncMyoD in vitro. Further investigation using the cross-linking and immunoprecipitation of RNA–protein complexes technique will definitively characterize the direct interaction between MyoD and LncMyoD in vivo. The results that LncMyoD interacts with multiple proteins indicate that LncMyoD has multifaceted functions and raise the possibility that LncMyoD functions through cooperating with other RNA binding proteins. Furthermore, in our luciferase assay, LncMyoD alone promoted luciferase activity (Fig. 5D), indicating that other potential TFs are involved in the process of transcription activation. The recruitment potential of LncMyoD is noteworthy in this process, demonstrating the primary role of LncMyoD during transcriptional regulation. Nevertheless, the identification of other associated TFs will advance our understanding of LncMyoD-mediated regulation during SC myogenic lineage progression.
In conclusion, we demonstrated the roles of a previously identified lncRNA, LncMyoD, in regulating myogenic lineage determination and progression. LncMyoD binds directly with MyoD to expose compact E-box–containing chromatin and promotes MRFs to access their target regulatory regions. Furthermore, loss of LncMyoD leads to defects in SC differentiation and strongly impairs the reprogramming of nonmyogenic cells into myogenic cell lineage. Thus, our results provide insights into how lncRNAs modulate chromatin accessibility to regulate cell lineage determination and progression.
Materials and Methods
Animals and Treatments.
Mice used in this study were around 2 to 3 mo old, and they were housed and maintained in the Animal and Plant Care Facility (APCF) of The Hong Kong University of Science and Technology (HKUST). Mice used in each experiment were age and gender matched. C57BL/6 mice were obtained from the APCF of HKUST. For muscle injury, mice were anesthetized using 2,2,2-Tribromoethanol (250 mg/kg; Sigma-Aldrich) through intraperitoneal injection. Muscle injury was induced by injecting 50 μL of 1.2% barium chloride (Sigma-Aldrich) into the lower hind-limb muscles. All animal experiments were approved by the HKUST Animal Ethics Committee.
Supplementary Material
Acknowledgments
We thank Dawn Cornelison from the University of Missouri for providing the Syn4 antibody and Mingjie Zhang at the HKUST for providing the pGEX-4T-1 plasmid. We also thank the ENCODE Consortium and the laboratory of Barbara Wold for generating C2C12 RNA-seq and ChIP-seq data. This work was supported by Hong Kong Research Grant Council Research Grants 26100114, AoE/M-604/16, T13-607/12R, and T13-605/18-W; National Key Research and Development Program of China Research Grant 2018YFE0203600; Guangdong Provincial Key Science and Technology Program Research Grant 2018B030336001; Lee Hysan Foundation Research Grant LHF17SC01; the Hong Kong Epigenome Project (the Lo Ka Chung Charitable Foundation); and Croucher Innovation Award CIA14SC04 from the Croucher Foundation. This study was supported in part by Innovation and Technology Commission Grant ITCPD/17-9. S.L. is a recipient of the Hong Kong PhD Fellowship Scheme. T.H.C. is the S. H. Ho Associate Professor of Life Science at HKUST.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2005868117/-/DCSupplemental.
Data Availability.
Raw and processed data for all ChIP-seq, ATAC-seq, ChIRP-seq, and RNA-seq datasets have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession nos. GSE129768, GSE113631, GSE108040, and GSE159131–GSE159133. The RNA-seq datasets for FISCs and ASCs have been previously deposited (accession no. GSE113631) (68).
References
- 1.Perino M., Veenstra G. J. C., Chromatin control of developmental dynamics and plasticity. Dev. Cell 38, 610–620 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zaret K. S., Carroll J. S., Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi K., Yamanaka S., Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Oldfield A. J., et al. , Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 55, 708–722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soufi A., et al. , Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell 161, 555–568 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domcke S., et al. , Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Carninci P., et al. , Molecular biology: The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Derrien T., et al. , The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guttman M., Russell P., Ingolia N. T., Weissman J. S., Lander E. S., Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guttman M., et al. , Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockdorff N., et al. , Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351, 329–331 (1991). [DOI] [PubMed] [Google Scholar]
- 12.Flynn R. A., Chang H. Y., Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 14, 752–761 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S., Shan G., LncRNAs in stem cells. Stem Cells Int. 2016, 2681925 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geisler S., Coller J., RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 14, 699–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K. C., et al. , A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn J. L., et al. , Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai M. C., et al. , Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao H., et al. , Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 24, 2543–2555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao J., et al. , Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinn J. L., Chang H. Y., Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L., et al. , LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25, 335–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong C., et al. , A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Develop. Cell 34, 181–191 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Zhou L., et al. , Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6, 10026 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Mousavi K., et al. , eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caretti G., et al. , The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 11, 547–560 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Yu X., et al. , Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8, 14016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cesana M., et al. , A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I., A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell 53, 506–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller A. C., et al. , MUNC, a long noncoding RNA that facilitates the function of MyoD in skeletal myogenesis. Mol. Cell Biol. 35, 498–513 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sui Y., Han Y., Zhao X., Li D., Li G., Long non-coding RNA irm enhances myogenic differentiation by interacting with MEF2D. Cell Death Dis. 10, 181 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidl C., Rendeiro A. F., Sheffield N. C., Bock C., ChIPmentation: Fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods 12, 963–965 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu L., et al. , Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastor W. A., et al. , MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 5, 5795 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lara-Astiaso D., et al. , Chromatin state dynamics during blood formation HHS public access. Science 345, 943–949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ampuja M., et al. , Integrated RNA-seq and DNase-seq analyses identify phenotype-specific BMP4 signaling in breast cancer. BMC Genomics 18, 68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fabregat A., et al. , Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinformatics 18, 142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schep A. N., Wu B., Buenrostro J. D., Greenleaf W. J., chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wüst S., et al. , Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 27, 1026–1039.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Pertea M., Kim D., Pertea G. M., Leek J. T., Salzberg S. L., Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang Y. J., et al. , CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frankish A., et al. , GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sayers E. W., et al. , Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 48, D9–D16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka K. K., et al. , Syndecan-4-expressing muscle progenitor cells in the SP engraft as satellite cells during muscle regeneration. Cell Stem Cell 4, 217–225 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olguin H. C., Olwin B. B., Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev. Biol. 275, 375–388 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Agostini F., et al. , catRAPID omics: A web server for large-scale prediction of protein-RNA interactions. Bioinformatics 29, 2928–2930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baron-Benhamou J., Gehring N. H., Kulozik A. E., Hentze M. W., Using the lambdaN peptide to tether proteins to RNAs. Methods Molecular Biol. (2004). [DOI] [PubMed] [Google Scholar]
- 49.Davis C. A., et al. , The encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu C., Quinn J., Chang H. Y., Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp., 3912 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tapscott S. J., The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Pinney D. F., Pearson-White S. H., Konieczny S. F., Latham K. E., Emerson C. P. Jr, Myogenic lineage determination and differentiation: Evidence for a regulatory gene pathway. Cell 53, 781–793 (1988). [DOI] [PubMed] [Google Scholar]
- 53.Lassar A. B., Paterson B. M., Weintraub H., Transfection of a DNA locus that mediates the conversion of 10T1/2 fibroblasts to myoblasts. Cell 47, 649–656 (1986). [DOI] [PubMed] [Google Scholar]
- 54.Gerber A. N., Klesert T. R., Bergstrom D. A., Tapscott S. J., Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes Dev. 11, 436–450 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Puri P. L., et al. , Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1, 35–45 (1997). [DOI] [PubMed] [Google Scholar]
- 56.Sartorelli V., Huang J., Hamamori Y., Kedes L., Molecular mechanisms of myogenic coactivation by p300: Direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Molecular Cell. Biol. 17, 1010–1026 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sartorelli V., et al. , Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4, 725–734 (1999). [DOI] [PubMed] [Google Scholar]
- 58.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis R. L., Weintraub H., Lassar A. B., Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 60.Albini S., et al. , Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 3, 661–670 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dekel I., Magal Y., Pearson-White S., Emerson C. P., Shani M., Conditional conversion of ES cells to skeletal muscle by an exogenous MyoD1 gene. New Biol. 4, 217–224 (1992). [PubMed] [Google Scholar]
- 62.Machado L., et al. , In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 63.van Velthoven C. T. J., de Morree A., Egner I. M., Brett J. O., Rando T. A., Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bjornson C. R. R., et al. , Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232–242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weintraub H., et al. , Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. USA 86, 5434–5438 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berkes C. A., et al. , Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell 14, 465–477 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Engreitz J. M., Ollikainen N., Guttman M., Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Yue L., Wan R., Luan S., Zeng W., Cheung T. H., Dek modulates global intron retention during muscle stem cells quiescence exit. Dev. Cell. 53, 661–676.e6 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed data for all ChIP-seq, ATAC-seq, ChIRP-seq, and RNA-seq datasets have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession nos. GSE129768, GSE113631, GSE108040, and GSE159131–GSE159133. The RNA-seq datasets for FISCs and ASCs have been previously deposited (accession no. GSE113631) (68).