Significance
The Agaricomycetes is a conspicuous and successful group of Fungi, containing ∼36,000 described species. The group presents striking diversity in fruiting bodies, including those with a pileus (cap) and stipe (stalk), puffballs, coral fungi, crust-like forms, etc. Agaricomycetes also comprise ecologically diverse species, including decayers, mycorrhizal symbionts, and pathogens. We assembled a “megaphylogeny” with 8,400 species that represent ∼23% of the known diversity of Agaricomycetes and used it to investigate the relative impact of fruiting body forms and nutritional modes on diversification rates. Across all Agaricomycetes, a pileate-stipitate fruiting body is associated with increased diversification compared to other forms. No such relationship was found for nutritional modes, including mycorrhizal symbiosis. We conclude that morphological innovation has driven diversification in Agaricomycetes.
Keywords: Agaricomycetes, diversification, ectomycorrhizal fungi, gasteroid forms, megaphylogeny
Abstract
With ∼36,000 described species, Agaricomycetes are among the most successful groups of Fungi. Agaricomycetes display great diversity in fruiting body forms and nutritional modes. Most have pileate-stipitate fruiting bodies (with a cap and stalk), but the group also contains crust-like resupinate fungi, polypores, coral fungi, and gasteroid forms (e.g., puffballs and stinkhorns). Some Agaricomycetes enter into ectomycorrhizal symbioses with plants, while others are decayers (saprotrophs) or pathogens. We constructed a megaphylogeny of 8,400 species and used it to test the following five hypotheses regarding the evolution of morphological and ecological traits in Agaricomycetes and their impact on diversification: 1) resupinate forms are plesiomorphic, 2) pileate-stipitate forms promote diversification, 3) the evolution of gasteroid forms is irreversible, 4) the ectomycorrhizal (ECM) symbiosis promotes diversification, and 5) the evolution of ECM symbiosis is irreversible. The ancestor of Agaricomycetes was a saprotroph with a resupinate fruiting body. There have been 462 transitions in the examined morphologies, including 123 origins of gasteroid forms. Reversals of gasteroid forms are highly unlikely but cannot be rejected. Pileate-stipitate forms are correlated with elevated diversification rates, suggesting that this morphological trait is a key to the success of Agaricomycetes. ECM symbioses have evolved 36 times in Agaricomycetes, with several transformations to parasitism. Across the entire 8,400-species phylogeny, diversification rates of ectomycorrhizal lineages are no greater than those of saprotrophic lineages. However, some ECM lineages have elevated diversification rates compared to their non-ECM sister clades, suggesting that the evolution of symbioses may act as a key innovation at local phylogenetic scales.
The class Agaricomycetes is one of the largest and most conspicuous groups of Fungi. It contains ∼36,000 described species (1) that display striking diversity in both morphology of fruiting bodies (reproductive structures) and nutritional modes. We assembled the largest phylogeny of Agaricomycetes to date and used it to reconstruct historical patterns of morphological and ecological evolution in Agaricomycetes. We also sought to address whether form or nutritional mode is the primary driver of diversification in Agaricomycetes.
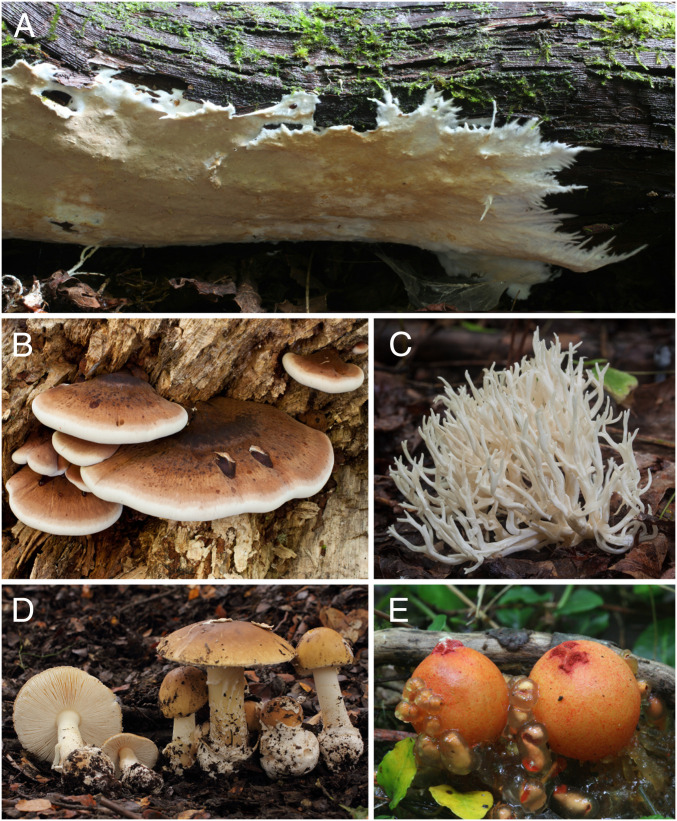
Fruiting bodies of Agaricomycetes vary from relatively simple resupinate or crust-like forms to more complex forms, such as clavarioid-coralloid (club- or candelabra-shaped), pileate-stipitate (with cap and stalk), pileate-sessile (with lateral cap and no stalk), or gasteroid forms. The latter produce spores internally and include forms such as puffballs, bird nest fungi, stinkhorns, and false truffles (Fig. 1). Morphological evolution in Agaricomycetes is marked by extensive convergence (2–9). Previous studies suggested that the resupinate morphology is plesiomorphic and that there is a directional trend toward the evolution of pileate-stipitate forms (8, 10).
Fig. 1.
Examples of the basic fruiting body forms in Agaricomycetes. (A) Resupinate (Phanerochaete sp.). (B) Pileate-sessile (Ischnoderma resinosum). (C) Clavarioid-coralloid (Ramariopsis kunzei). (B and C) Image credit: Mike Wood (photographer). (D) Pileate-stipitate (Amanita diemii). (E) Gasteroid (Calostoma cinnabarinum).
The evolution of gasteroid forms is correlated with the loss of a mechanism of forcible spore discharge, called ballistospory, and the evolution of enclosed spore-bearing structures. It has been hypothesized that the loss of ballistospory is irreversible because it involves a complex suite of anatomical features generating a “surface tension catapult” (8, 11). The effect of gasteroid fruiting body forms on diversification rates has been assessed in Sclerodermatineae, Boletales, Phallomycetidae, and Lycoperdaceae, where it was found that lineages with this type of morphology have diversified at higher rates than nongasteroid lineages (12). However, these four selected clades represent some of the most species-rich, highly modified groups of gasteroid forms; many other clades of gasteromycetes are species poor and often have fruiting bodies that are intermediate in morphology between the most derived gasteroid forms and their putative nongasteroid ancestors (which are defined by possession of exposed spore-bearing structures and ballistospory).
The Agaricomycetes is also ecologically diverse; they are saprotrophs in a wide variety of substrates, symbionts with plants, animals, algae and cyanobacteria, and pathogens. Agaricomycetes contain the major concentrations of wood-decay fungi, including white rot and brown rot fungi, and ectomycorrhizal (ECM) symbionts. The impact of ECM symbiosis on diversification has been of particular interest. It has been hypothesized that this symbiosis facilitates the use of unexploited niches and consequently promotes diversification (13, 14). In the genus Laccaria (Agaricales), the ECM mode has been suggested to be a driver of diversification (i.e., a key innovation) (15). However, studies in other groups of Agaricales did not support the ECM mode as a key innovation (13, 16) and suggested that the transition to ECM lifestyle promotes diversification only in some lineages (16). So far, there have been no analyses of the impact of ECM associations on diversification across all Agaricomycetes.
Varga et al. (17) studied the impact of fruiting body morphologies on diversification rates using a 5,284-species phylogeny of Agaricomycetes, finding that the pileate lineages have higher diversification rates than nonpileate lineages. However, their study did not focus on nutritional modes. We assembled and analyzed a five-gene dataset for 8,472 species of Agaricomycetes using a pipeline for assembly of supermatrices and tested the following five hypotheses on the evolution of morphology and ecological lifestyles in Agaricomycetes: 1) resupinate forms are plesiomorphic, 2) pileate-stipitate forms promote diversification, 3) the evolution of gasteroid forms is irreversible, 4) the ECM symbiosis promotes diversification, and 5) the evolution of ECM symbiosis is irreversible.
Results and Discussion
Megaphylogeny Recovers Expected Agaricomycete Topology.
Using the pipeline PifCoSm (Pipeline for Construction of Supermatrices, available through https://github.com/RybergGroup/PifCoSm), we created an alignment of 8,472 nuclear ribosomal large subunit (nLSU) sequences, 1,872 nuclear ribosomal small subunit (nSSU) sequences, 784 largest subunit of RNA polymerase II (rpb1) sequences, 1,683 second-largest subunit of RNA polymerase II (rpb2) sequences, and 856 translation elongation factor 1-alpha (tef-1-α) sequences (SI Appendix, Table S1). The resulting phylogeny recovered the 21 monophyletic orders accepted in the Agaricomycetes, with a topology that is in general agreement with previous phylogenies (18–20). The Russulales was placed as a sister group of a large clade containing the Polyporales, Thelephorales, Gloeophyllales, Jaapiales, and Agaricomycetidae, as shown in previous studies (19, 20), although in other studies, it has also been recovered as sister to Jaapiales and Gloeophyllales (17). The close relationship between Phallomycetidae, Stereopsidales, and Trechisporales was previously recovered (19, 21). Within the Agaricomycetidae, we recovered a grouping that included Atheliales, Lepidostromatales, and Boletales as a sister to Agaricales and Amylocorticiales. The placement of Lepidostromatales within the Agaricomycetidae is uncertain; it was previously suggested as a sister to the Boletales, Amylocorticiales, and Agaricales but with low support (22).
Our time-calibrated phylogeny suggests that some clades are older than previously estimated, but most of our age estimates are within the CIs of prior studies (17, 23–26). Most orders were inferred to have a Jurassic origin (Agaricales, Amylocorticiales, Atheliales, Boletales, Corticiales, Gloeophyllales, Hymenochaetales, Jaapiales, Lepidostromatales, Polyporales, and Thelephorales), in agreement with a previous megaphylogeny (17, 25), in some cases with a subsequent diversification in the Cretaceous (SI Appendix, Figs. S1–S3). We estimated the ages of the stem and crown nodes of the Agaricomycetidae at 217 Mya and 189 Mya, respectively, which approximately coincides with the diversification of Pinaceae (25, 27) and the later diversification of angiosperms (25, 28).
Morphological Evolution.
The ancestor of agaricomycetes had a resupinate fruiting body.
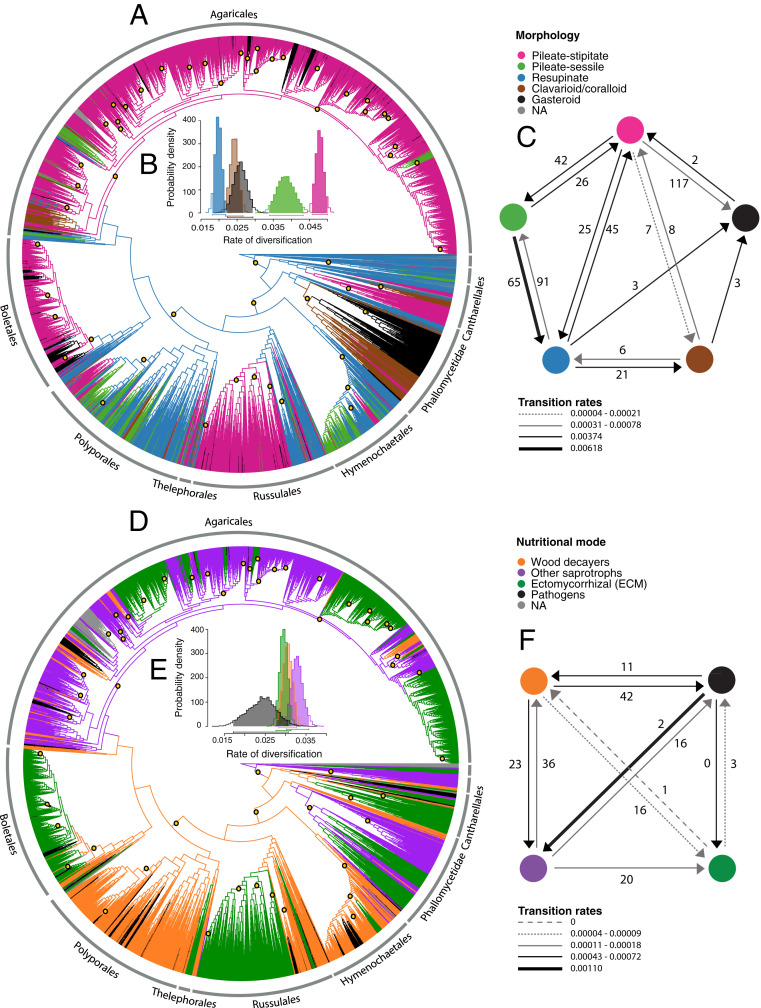
Ancestral state reconstruction suggests that the ancestor of Agaricomycetes had a resupinate fruiting body, with a posterior probability of 0.99 according to the BayesTraits Markov chain Monte Carlo (MCMC) analyses and a posterior probability of 1 according to stochastic mapping. Based on the average number of transitions from stochastic mapping, we inferred 160 transitions from resupinate forms to other morphologies and 96 reversals to the resupinate form. The clavarioid fruiting body morphology is the least common in Agaricomycetes. Only 3% of the taxa included here present this form, which was inferred to have originated 28 times and lost 17 times according to our analyses. Pileate-sessile forms were inferred to have evolved 133 times and transitioned to other morphologies 91 times. The pileate-stipitate form is the dominant fruiting body morphology, which was inferred to have evolved 85 times and to have given rise to other morphologies 191 times (Fig. 2C and SI Appendix, Fig. S4).
Fig. 2.
(A–C) Macroevolutionary dynamics of fruiting body forms in Agaricomycetes. (D–F) Macroevolutionary dynamics of nutritional modes in Agaricomyctes. (A and D) Stochastic mapping of five fruiting body forms and four nutritional modes, respectively. The yellow dots indicate shifts in diversification rates according to BAMM. Only major orders of Agaricomycetes are labeled. (B and E) The posterior probability distributions of net diversification rates (speciation minus extinction) from the MuSSE model. The bar underneath each distribution represents the 95% credible interval obtained from the posterior distribution. (C and F) The number of average transitions between character states estimated from a set of 100 stochastic maps by SIMMAP and transition rates estimated by MuSSE (NA = not applicable). The absence of arrows between states indicates zero transitions as well as transition rates.
Pileate-stipitate forms promote diversification across agaricomycetes.
Multi-State Speciation and Extinction (MuSSE) analyses, in which we evaluated the effects of five different fruiting body morphologies on diversification rates, suggested that clades with pileate-stipitate forms have the highest diversification rates, followed by clades with pileate-sessile forms. Clades with resupinate fruiting bodies were inferred to have the lowest diversification rates (likelihood ratio [LR] test, P < 0.001) (Fig. 2B and SI Appendix, Tables S2 and S3). To test the impact of alternative coding regimes, we repeated the analyses using Binary State Speciation and Extinction (BiSSE), which yielded results that were, in general, consistent with those from MuSSE (SI Appendix, Fig. S5 and Table S4). In all cases, the unconstrained model, in which all speciation, extinction, and transformation rates were allowed to vary, was significantly better than any of the constrained models (LR test, P < 0.001) (SI Appendix, Table S5).
Gasteroid forms have evolved more than 100 times.
We resolved 123 unique origins of gasteroid forms. Six transitions to gasteroid forms were inferred to have occurred via clavarioid/coralloid or resupinate ancestors (three each), but the vast majority, 117 transitions, were inferred from pileate-stipitate ancestors (Fig. 2 A and C). Most of the gasteroid lineages are young (4–50 Ma), include fewer than five species (SI Appendix, Fig. S6 and Table S6), and are nested within lineages of nongasteroid fungi such as Agaricus, Amanita, Cortinarius, Entoloma, Lactarius, and Russula (SI Appendix, Fig. S7). Some of these genera have been redelimited to include gasteroid species (29–32). The oldest gasteroid lineages are in the subclass Phallomycetidae and correspond to the orders Hysterangiales, Phallales (216 Ma), and Geastrales (211 Ma) (SI Appendix, Fig. S6), which include false truffles, stinkhorns, and earthstars and cannonball-fungi (respectively). These types of fruiting bodies (particularly those of Phallales and Geastrales) involve a high degree of differentiation and developmental organization and are among the most complex forms in fungi.
Trait-dependent diversification analysis with BiSSE suggested that diversification rates of gasteroid lineages are lower than those of nongasteroid lineages (LR test, P < 0.001), whereas MuSSE analyses suggested that diversification rates of gasteroid fungi are similar to that of clavarioid/coralloid forms and greater than resupinate forms but below that of pileate-stipitate or pileate-sessile forms (LR test, P < 0.001) (Fig. 2B and SI Appendix, Tables S2 and S4). The BiSSE model that prohibited reversals from a gasteroid to a nongasteroid state was significantly worse than the unconstrained model (LR test, P < 0.01) (SI Appendix, Table S5).
Our ancestral state reconstruction identified two putative reversals from gasteroid to nongasteroid forms, both in the Boletales. The first was in the Sclerodermatineae, where the nongasteroid genus Gyroporus was nested among gasteroid lineages (SI Appendix, Fig. S7). However, a recent phylogenetic study of Boletales using 87 genes suggested that the gasteroid taxa of Sclerodermatineae form a monophyletic sister group to Gyroporus (33). The second putative reversal was in the Suillineae, where we recovered the genus Rhizopogon, a group of hypogeous false truffles, as paraphyletic to the nongasteroid genera Suillus, Gomphidius, and Chroogomphus. Taxa representing the putative reversals all have typical ballistosporic basidia, with characteristic curved sterigmata and asymmetrical spores with a hilar appendage, which are the anatomical features that facilitate the forcible spore discharge.
To examine the sensitivity of our conclusions to the topology of Boletales, we tested the same models but pruned the taxa in which the putative reversals occurred. Using the pruned tree, the model that prohibited reversals from a gasteroid to a nongasteroid state was not significantly different from the unconstrained model (LR test, P = 1) (SI Appendix, Table S7). Thus, while BiSSE analyses of the entire dataset suggested that reversibility of gasteroid forms could not be rejected, the pruned phylogeny indicated that irreversibility of gasteroid forms could not be rejected. Reversals from gasteroid to nongasteroid forms seem unlikely based on the anatomical complexity of ballistospory.
Evolution of Nutritional Modes.
Across Agaricomycetes, ECM lineages do not have higher net diversification rates than saprotrophic lineages.
For nutritional modes, we explored four different coding regimes (I to IV), which included two- to five-character states. Coding regime I included two-character states: non-ECM and ECM. The unconstrained model obtained under this coding suggested that ECM lineages diversified at a higher rate than non-ECM clades (SI Appendix, Fig. S8 and Table S8). However, a model that constrained net diversification rates to be equal could not be rejected (LR test, P = 0.5) (SI Appendix, Table S9). Coding regime II separated nutritional modes into three categories: saprotrophs, ECM, and pathogens. Analyses under this coding suggested that diversification rates of saprotrophs are higher than those of ECM and that pathogens have the lowest rate (SI Appendix, Fig. S9 and Table S10). Once again, the model that constrained saprotrophs and ECM lineages to have equal diversification rates could not be rejected (LR test, P = 0.3) (SI Appendix, Table S11). Coding regime III included four-character states: wood decayers, other saprotrophs (i.e., on soil, litter, and dung), ECM, and pathogens. Under this coding, fungi that degrade soil, litter, and dung have the highest diversification rate, followed by wood decayers, ECM, and pathogens (LR test, P < 0.001) (Fig. 2D and SI Appendix, Tables S12 and S13). Finally, coding regime IV included five-character states: wood decayers that produce a brown rot, wood decayers that produce a white rot, other saprotrophs, ECM, and pathogens. Analyses under this coding suggest that fungi that produce a white rot diversify faster than brown rot fungi (LR test, P < 0.001) (SI Appendix, Fig. S10 and Tables S14 and S15).
None of our analyses, under any coding regime, suggest that ECM lineages have greater diversification rates than saprotrophic lineages. These results were consistent under alternative coding regimes that consider saprotrophs with pathogenic capabilities as saprotrophs (SI Appendix, Tables S8–S17).
Evolution of ECM may promote diversification at local phylogenetic scales.
When examining overall diversification rates across the entire Agaricomycetes, we did not find an increase in diversification rates in ECM lineages compared to other nutritional modes (Fig. 2E and SI Appendix, Tables S10, S12, S14, and S16). Nevertheless, several ECM lineages are particularly species rich (34), and this ecological lifestyle has been linked to an increase in diversification rates in some clades (13, 15, 16, 33). Therefore, we evaluated individual ECM clades and compared their diversification rates with their non-ECM sister clades (SI Appendix, Table S18). Results of these comparisons suggest that the ECM lineages Cortinarius, Tricholoma, Hygrophorus, Boletaceae, and Russulaceae have experienced increases in diversification rates compared to their closest relatives. Thus, the ECM nutritional mode is not correlated with an elevated rate of diversification across all Agaricomycetes, but it may act as a key innovation at local phylogenetic scales (or its impact on diversification is contingent on other evolutionary innovations). We also identified several non-ECM clades that have higher diversification rates than their ECM sister clades, such as Psathyrellaceae, a family of deliquescent mushrooms, and the clade containing Bolbitius and Conocybe, many of which also detected by Varga et al. (17).
Increases in diversification rates in ECM lineages have been previously inferred in Laccaria (15), Tricholoma, the Rhodopolioid clade of Entoloma (16), and Boletaceae (33). On the other hand, some ECM lineages in the suborder Tricholomatineae and in the order Boletales did not show diversification rate increases, and it has been suggested that other factors such as historical climate change and host availability can create the necessary conditions for a rapid diversification (16, 33). For example, in the family Inocybaceae, which is uniformly ECM, the highest rates of diversification occurred well after the origin of the crown group, long after the origin of ECM lifestyle in this lineage (35). Further hypotheses about the factors that drive diversification in ECM lineages should be tested, such as host specificity, geographic origin and distribution, or combinations of biotic and abiotic factors.
Many gains, and few losses, of ECM symbiosis.
Of the 21 orders of Agaricomycetes, only the Hysterangiales is resolved as plesiomorphically ECM; the rest have saprotrophic ancestors (SI Appendix, Figs. S11 and S12). We inferred 36 independent origins of the ECM lifestyle (Fig. 2 D and F and SI Appendix, Fig. S13 and Table S18) and four losses. Three of the losses of ECM correspond to transitions to putative pathogens or parasites in Entoloma (E. clypeatum group), Boletaceae (Pseudoboletus parasiticus), and the suborder Suillineae (Chroogomphus and Gomphidius). The parasitic nature of Pseudoboletus, Chroogomphus, and Gomphidius is not fully established; some evidence points to the ability of these fungi to form ECM associations (36–38), while other lines of evidence suggest that they can parasitize other ECM fungi such as Scleroderma, Suillus, and Rhizopogon (39–42). The E. clypeatum group is considered a weak, obligate parasite due to the formation of an abnormal ECM structure that damages the root cap region of some species of Rosaceae (43). Because of the uncertainty in their ecological lifestyle, these losses are tentative and may not be complete (i.e., mixed ECM/parasitic lifestyles).
The fourth loss of ECM was inferred in Thelephorales, which contains putative white rot saprotrophs in the genera Lenzitopsis and Amaurodon. Our analysis suggests that Lenzitopsis and Amaurodon form a clade that is derived from ECM ancestors. However, it has been suggested that the ECM mode evolved several times in this order, and based on previous works (44, 45) and our phylogeny, the phylogenetic position of these non-ECM genera within the Thelephorales is not well resolved.
Comparative genomic evidence of multiple lineages within Agaricomycetes indicates that transitions from saprotrophy to ECM are correlated with massive losses of genes encoding plant cell wall–degrading enzymes (PCWDEs) (26). Thus, reversals from ECM to saprotrophy are unlikely. It is possible that parasitic fungi, particularly mycoparasites, do not require large repertoires of PCWDE, making ECM-to-parasite transitions plausible. Unfortunately, there are no genomes available for the lineages of the putative parasites and saprotrophs that we resolve as potentially derived from ECM ancestors.
The oldest ECM lineages belong to the Sebacinales and Cantharellales, with stem ages of 260 to 289 Mya, which overlaps with the estimated interval between the stem and crown nodes of Pinaceae (25, 27). Most of the ECM lineages had a more recent origin in the Cretaceous and Paleogene (SI Appendix, Fig. S14), which coincides with the diversification of angiosperm hosts, such as oaks, beeches, dipterocarps, and eucalypts (25). An explanation for species richness disparity is that older clades are species rich because they had more time to accumulate diversity (46). Therefore, we explored the correlation between species richness and age. In this case, young ECM lineages have a species richness comparable to that of the oldest lineages, so species richness is not correlated with clade age (Pearson correlation test P = 0.83).
Conclusions
The 36,000 species of Agaricomycetes comprise the vast majority of conspicuous mushrooms and the major concentrations of both wood decayers and ECM symbionts. Collectively, they represent about a quarter of the known species of Fungi (47). We assembled the largest phylogeny of Agaricomycetes to date and asked whether morphological or ecological traits are primarily responsible for their evolutionary success.
Across Agaricomycetes, morphological transitions are the primary drivers of diversification, not nutritional modes. Specifically, lineages with pileate-stipitate forms have significantly elevated diversification rates across all clades and ecological habits. These findings highlight the importance of spore production and dispersal for evolutionary success in Fungi. However, the functional bases for the success of pileate-stipitate forms is not entirely clear. While there has been much research on ballistospory (48, 49), the biomechanics of whole fruiting bodies has received little attention. The cap may protect the spore-bearing structures from rain and other environmental insults, and evaporation off the cap may create convective airflows that disperse spores more efficiently, allowing them to reach longer distances than spores of nonpileate forms (50). Comparative functional studies on the performance of different fruiting body types are needed to address why pileate-stipitate forms have been so successful.
Some studies in Agaricomycetes have found that the ECM habit promotes diversification, while others have failed to show such a relationship (12, 15, 16). Our results suggest that the ECM nutritional mode is not linked to an overall increase in diversification at the level of the entire Agaricomycetes. Nevertheless, our clade-by-clade analyses suggest that some ECM lineages diversify faster than their non-ECM sister clades. The converse is also true; some saprotrophic lineages have higher diversification rates than their ECM sister clades. Elaboration of saprotrophic mechanisms, including the vast array of PCWDEs (23, 24), may have contributed to the evolutionary success of individual saprotrophic lineages.
Although there is a general trend toward reduced complements of PCWDEs in ECM lineages, the specific gene families that are retained (or expanded) vary considerably from group to group (26). Thus, the nonhomologous instances of ECM are not all functionally equivalent, and they may have unique diversification dynamics. Further sampling of genomes from phylogenetically and ecologically pivotal groups, particularly parasites and saprotrophs that may be derived from ECM ancestors, could shed light on reversibility of ECM and diversification in Agaricomycetes.
Materials and Methods
Agaricomycetes Phylogeny.
We mined GenBank for all available sequences of Agaricomycetes of the following gene regions: nLSU, nSSU, rpb1, rpb2, and tef-1-α. To process the data and produce a concatenated alignment, we used the pipeline PifCoSm, a Perl software that uses a SQLite database to organize and parse data from a file with data in full GenBank format. This software is divided in modules that allow the user to manually clean and check the accuracy of the metadata associated with each of the entries. This ensures that when the sequences of different gene regions are concatenated, the user can be confident that they belong to the same species and more likely to the same specimen. It uses hidden Markov Models to identify what genes the sequences belong to and sorts them into different tables. One of the modules identifies individuals that are used to link sequences of different gene regions, which will ultimately be concatenated.
We defined individuals based on strain, isolate, and voucher with species name as the individual condition. This means that sequences were only linked if they have the same species name and the same voucher, isolate, or strain identifier. Sometimes sequences belonging to the same species and even the same specimen are deposited in GenBank under different names (synonyms); this is particularly problematic since the species name is indicated as the individual condition for sequences to be linked. Therefore, we used a custom Perl script that downloads the current name according to Index Fungorum (1), and we replaced the synonyms; this also resulted in a lot of taxa having the same name but likely representing different species. In order to avoid species redundancy, the individuals were clustered based on similarity using CD-hit version 4.6 (51) and 0.99 as the cutoff value for the nLSU region. This cutoff value was chosen because it has been shown that nLSU can separate fungal strains at the species level with a threshold of 99.81% (52). Then, we linked the individuals and aligned the sequences using MAFFT version 7.402 (53), specifying nLSU as an anchor gene, which means that an individual will be included in the alignment only if a sequence of nLSU is present. The concatenated alignment that resulted from this last step was exported for further manual editing and trimmed using trimAL version 1.2 with the option –gt 0.1 (54). We performed several rounds of filtering to remove misidentified sequences; this was done by manually inspecting maximum likelihood (ML) trees built with RAxML version 8.2.4 (55). Taxa that were placed outside the expected phylogenetic position (i.e., genus, family, or order) were removed from the alignment.
We estimated an ML phylogeny on a gene partitioned dataset using RAxML (55) with 1,000 bootstrap replicates. To obtain a time-calibrated phylogeny, we used the chronos function from the R package ape (56) and incorporated information (minimum and maximum age) from six fungal fossils (SI Appendix, Table S19). We used Dacrymycetales and Tremellales as outgroups, which were removed for the subsequent diversification analysis.
Character Coding.
The fruiting body forms were divided in five different morphotypes: 1) pileate-stipitate (5,256 taxa), 2) pileate-sessile (745 taxa), 3) resupinate (1,291 taxa), 4) clavarioid/coralloid (392 taxa), and 5) gasteroid (691 taxa). Pileate stipitate forms included those that have a well-defined cap and a stipe. Forms that have a cap but lack a stipe or present a rudimentary or reduced stipe were considered pileate-sessile. Resupinate forms were defined as those that lay flat on the substrate, such as crust-like, effused-reflexed, and cyphelloid forms. Club-shaped and branched forms were considered as coralloid/clavarioid. Gasteroid forms included those with enclosed fruiting bodies that produce spores internally and, in most cases, do not forcibly discharge spores. Taxa with no fruiting bodies or with fruiting body forms that could not be assigned to any of the previous categories were coded as not applicable (NA; 71 taxa). We analyzed these five morphotypes as multistate and binary characters.
For nutritional modes, we explored four different binary and multistate coding regimes: I) 0 = non-ECM (4,716 taxa) and 1 = ECM (3,731 taxa); II) 1 = saprotrophs (4,122 taxa), 2 = ECM (3,731 taxa), and 3 = pathogens (392 taxa); III) 1 = wood-decayers (1,787 taxa), 2 = saprotrophs on soil, litter, and dung (2,335 taxa), 3 = ECM (3,731 taxa), and 4 = pathogens (392 taxa); and IV) 1 = brown rot decayers (231 taxa), 2 = white rot decayers (1,556 taxa), 3 = saprotrophs on soil, litter, dung, and wood decayers of uncertain rot type (2,335 taxa), 4 = ECM (3,731 taxa), and 5 = pathogens (392 taxa). Saprotrophic fungi that are also pathogens were included in the category of pathogens, but alternative coding regimes in which those taxa were coded as saprotrophs were also explored. Taxa with uncertain nutritional mode or that could not be assigned to any of the categories proposed were coded as NA. ECM taxa were coded following Rinaldi et al. (57), Tedersoo et al. (44), and Tedersoo and Smith (58). Other nutritional modes were coded based on information available in FUNGuild (59) and a variety of literature sources.
Ancestral State Reconstruction.
We inferred the character histories using the function make.simmap of the R package phytools (60), which implements a stochastic character mapping accommodating uncertainty in the phylogenetic history. This approach assumes that the character state change follows a continuous-time Markov process and estimates the place along the branches where a change has occurred (61). We used a transition matrix with all rates different estimated from our data and default values for the rest of the parameters. We simulated the character histories 100 times and summarized the set of stochastic maps. Additionally, we performed ML and Bayesian ancestral state reconstructions using BayesTraits V3 (62). For the ML analyses, we executed 100 attempts of maximizing the likelihood. The Bayesian analysis was run for 50 million generations, sampling every 5,000 generations with a burn-in of 5 million generations.
Trait-Dependent Diversification.
To assess the effect of different fruiting body forms and nutritional modes on diversification rates, we used the MuSSE and the BiSSE implemented in the diversitree package in R (63, 64), accounting for random and incomplete phylogenies (65) by assigning specific sampling fractions to the character states. We estimated the total number of species missing for each genus according to data from Index Fungorum (1). The MuSSE and BiSSE models use a maximum likelihood optimization to estimate absolute rates of speciation (λ), extinction (µ), and transition (q) of each character state, by maximizing the likelihood of these parameters for a given topology with branch lengths (63). We fitted different models and evaluated them based on log-likelihoods (SI Appendix, Tables S2–S5 and S7–S17). We performed a likelihood ratio test and computed Akaike information criterion (AIC) weights to identify the best models, and we conducted MCMC estimations of those models for 10 to 15 thousand generations.
Trait-Independent Diversification.
Bayesian Analysis of Macroevolutionary Mixtures (BAMM) version 2.5 (66) was used to estimate diversification rates independently of character states. This approach assumes that different diversification regimes occurred across the branches of a phylogeny and accounts for rate variation through time and among lineages. It provides estimates of the number of diversification-rate shifts across the branches of a phylogenetic tree and estimates diversification-rate parameters. We ran four reversible jump MCMCs for 100 million generations with a sampling frequency of 10,000 and a burn-in of 30%. We used the package BAMMtools (67) in R to estimate speciation and extinction priors and to evaluate the outputs. We assumed incomplete sampling and assigned clade-specific sampling probabilities based on the estimated number of species for each of the genera represented in our phylogeny according to Index Fungorum (1).
ECM sister clade comparison.
In order to assess the effect of the ECM mode on diversification rates in each ECM clade separately, we calculated the diversification rate of each ECM lineage and compared it to the diversification rate of its non-ECM sister clade using the function getCladeRates from the R package BAMMtools. This approach allowed us to compare two clades while eliminating the bias because of differences in clade age.
Supplementary Material
Acknowledgments
This research was supported by the NSF (award DEB-1456588 to D.S.H.), the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 758161 to L.G.N.), and the Hungarian National Research, Development and Innovation Office (contract no. Ginop-2.3.2-15-00001 and Ginop-2.3.2-15-00052 to L.G.N.). We thank Mike Wood for providing two of the photographs presented in Fig. 1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1922539117/-/DCSupplemental.
Data Availability.
The data (concatenated alignment, phylogenetic tree, character coding of morphology, and nutritional modes) are available via the Open Science Framework and can be accessed at https://osf.io/y2vns/.
References
- 1.Index Fungorum. Index Fungorum Partnership; Kew R. B. G., Mycology, landcare research-NZ: Mycology and chinese academy or science: Institute of microbiology. www.indexfungorum.org. Accessed 12 December 2017.
- 2.Birkebak J. M., Adamčík S., Looney B. P., Matheny P. B., Multilocus phylogenetic reconstruction of the Clavariaceae (Agaricales) reveals polyphyly of agaricoid members. Mycologia 108, 860–868 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Binder M., Larsson K.-H., Matheny P. B., Hibbett D. S., Amylocorticiales ord. nov. and Jaapiales ord. nov.: Early diverging clades of agaricomycetidae dominated by corticioid forms. Mycologia 102, 865–880 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Hosaka K., et al. , Molecular phylogenetics of the gomphoid-phalloid fungi with an establishment of the new subclass Phallomycetidae and two new orders. Mycologia 98, 949–959 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Binder M., Hibbett D. S., Molecular systematics and biological diversification of Boletales. Mycologia 98, 971–981 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Larsson K.-H., et al. , Hymenochaetales: A molecular phylogeny for the hymenochaetoid clade. Mycologia 98, 926–936 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Miller S. L., Larsson E., Larsson K.-H., Verbeken A., Nuytinck J., Perspectives in the new Russulales. Mycologia 98, 960–970 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Hibbett D. S., Pine E. M., Langer E., Langer G., Donoghue M. J., Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. Proc. Natl. Acad. Sci. U.S.A. 94, 12002–12006 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binder M., et al. , The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst. Biodivers. 3, 113–157 (2005). [Google Scholar]
- 10.Hibbett D., Trends in morphological evolution in homobasidiomycetes inferred using maximum likelihood: A comparison of binary and multistate approaches. Syst. Biol. 53, 889–903 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Thiers H. D., The secotioid syndrome. Mycologia 76, 1–8 (1984). [Google Scholar]
- 12.Wilson A. W., Binder M., Hibbett D. S., Effects of gasteroid fruiting body morphology on diversification rates in three independent clades of fungi estimated using binary state speciation and extinction analysis. Evolution 65, 1305–1322 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Ryberg M., Matheny P. B., Asynchronous origins of ectomycorrhizal clades of Agaricales. Proc. Biol. Sci. 279, 2003–2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brundrett M. C., Mycorrhizal associations and other means of nutrition of vascular plants: Understanding the global diversity of host plants by resolving conflicting information and developing reliable means of diagnosis. Plant Soil 320, 37–77 (2009). [Google Scholar]
- 15.Wilson A. W., Hosaka K., Mueller G. M., Evolution of ectomycorrhizas as a driver of diversification and biogeographic patterns in the model mycorrhizal mushroom genus Laccaria. New Phytol. 213, 1862–1873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sánchez-García M., Matheny P. B., Is the switch to an ectomycorrhizal state an evolutionary key innovation in mushroom-forming fungi? A case study in the Tricholomatineae (Agaricales). Evolution 71, 51–65 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Varga T., et al. , Megaphylogeny resolves global patterns of mushroom evolution. Nat. Ecol. Evol. 3, 668–678 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasanna A. N., et al. , Model choice, missing data, and taxon sampling impact phylogenomic inference of deep basidiomycota relationships. Syst. Biol. 69, 17–37 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao R. L., et al. , A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 84, 43–74 (2017). [Google Scholar]
- 20.Nagy L. G., et al. , Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol. Biol. Evol. 33, 959–970 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Sjökvist E., Pfeil B. E., Larsson E., Larsson K. H., Stereopsidales–A new order of mushroom-forming fungi. PLoS One 9, e95227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodkinson B. P., Moncada B., Lücking R., Lepidostromatales, a new order of lichenized fungi (Basidiomycota, Agaricomycetes), with two new genera, Ertzia and Sulzbacheromyces, and one new species, Lepidostroma winklerianum. Fungal Divers. 64, 165–179 (2014). [Google Scholar]
- 23.Eastwood D. C., et al. , The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 333, 762–765 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Floudas D., et al. , The paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Lutzoni F., et al. , Contemporaneous radiations of fungi and plants linked to symbiosis. Nat. Commun. 9, 5451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohler A. et al.; Mycorrhizal Genomics Initiative Consortium , Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Lu Y., Ran J. H., Guo D. M., Yang Z. Y., Wang X. Q., Phylogeny and divergence times of gymnosperms inferred from single-copy nuclear genes. PLoS One 9, e107679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wikström N., Savolainen V., Chase M. W., Evolution of the angiosperms: Calibrating the family tree. Proc. Biol. Sci. 268, 2211–2220 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller S. L., McClean T. M., Walker J. F., Buyck B., A molecular phylogeny of the Russulales including agaricoid, gasteroid and pleurotoid taxa. Mycologia 93, 344–354 (2001). [Google Scholar]
- 30.Co-David D., Langeveld D., Noordeloos M. E., Molecular phylogeny and spore evolution of Entolomataceae. Persoonia 23, 147–176 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Justo A., Morgenstern I., Hallen-Adams H. E., Hibbett D. S., Convergent evolution of sequestrate forms in Amanita under Mediterranean climate conditions. Mycologia 102, 675–688 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Peintner U., et al. , Multiple origins of sequestrate fungi related to Cortinarius (Cortinariaceae). Am. J. Bot. 88, 2168–2179 (2001). [PubMed] [Google Scholar]
- 33.Sato H., Toju H., Timing of evolutionary innovation: Scenarios of evolutionary diversification in a species-rich fungal clade, Boletales. New Phytol. 222, 1924–1935 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Kirk P. M., Cannon P. F., Minter D. W., Stalpers J. A., Dictionary of the Fungi (CAB International, Wallingford, ed. 10, 2008). [Google Scholar]
- 35.Matheny P. B., Fordyce J. A., Not all ectomycorrhizal fungal lineages are equal. New Phytol. 222, 1670–1672 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Richter D., Bruhn J., Pinus resinosa ectomycorrhizae: Seven host-fungus combinations synthesized in pure culture. Symbiosis 7, 211–228 (1989). [Google Scholar]
- 37.Agerer R., Beenken L., Christan J., Gomphus clavatus (Pers.:Fr.) S. F. Gray. + Picea abies (L.) Karst. Descr. Ectomycorrhizae 3, 25–29 (1998). [Google Scholar]
- 38.Agerer R., Fungal relationships and structural identity of their ectomycorrhizae. Mycol. Prog. 5, 67–107 (2006). [Google Scholar]
- 39.Agerer R., Studies on Ectomycorrhizae XXIV. Ectomycorrhizae of Chroogomphus helveticus and C. rutilus (Gomphidiaceae, Basidiomycetes) and their relationship to those of Suillus and Rhizopogon. Nova Hedwigia 50, 1–63 (1990). [Google Scholar]
- 40.Agerer R., Studies on ectomycorrhizae XXIV. Mycorrhizae of Gomphidius glutinosis and of G. roseus with some remarks on Gomphidiaceae (Basidiomycetes). Nova Hedwigia 53, 127–170 (1991). [Google Scholar]
- 41.Olsson P. A., Münzenberger B., Mahmood S., Erland S., Molecular and anatomical evidence for a three-way association between Pinus sylvestris and the ectomycorrhizal fungi Suillus bovinus and Gomphidius roseus. Mycol. Res. 104, 1372–1378 (2000). [Google Scholar]
- 42.Miller O. K., Monograph of Chroogomphus (Gomphidiaceae). Mycologia 56, 526–549 (1964). [Google Scholar]
- 43.Agerer R., Waller K., Mycorrhizae of Entoloma saepium: Parasitism or symbiosis? Mycorrhiza 3, 145–154 (1993). [Google Scholar]
- 44.Tedersoo L., May T. W., Smith M. E., Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 20, 217–263 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Zhou L. W., Kõlljalg U., A new species of Lenzitopsis (Thelephorales, Basidiomycota) and its phylogenetic placement. Mycoscience 54, 87–92 (2013). [Google Scholar]
- 46.McPeek M. A., Brown J. M., Clade age and not diversification rate explains species richness among animal taxa. Am. Nat. 169, E97–E106 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Hawksworth D. L., Lücking R., Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 5, 79–95 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Pringle A., Patek S. N., Fischer M., Stolze J., Money N. P., The captured launch of a ballistospore. Mycologia 97, 866–871 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Liu F., et al. , Asymmetric drop coalescence launches fungal ballistospores with directionality. J. R. Soc. Interface 14, 20170083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dressaire E., Yamada L., Song B., Roper M., Mushrooms use convectively created airflows to disperse their spores. Proc. Natl. Acad. Sci. U.S.A. 113, 2833–2838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Vu D., et al. , Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Rinaldi A. C., Comandini O., Kuyper T. W., Ectomycorrhizal fungal diversity: Separating the wheat from the chaff. Fungal Divers. 33, 1–45 (2008). [Google Scholar]
- 58.Tedersoo L., Smith M. E., Biogeography of Mycorrhizal Symbiosis (2017). [Google Scholar]
- 59.Nguyen N. H., et al. , FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016). [Google Scholar]
- 60.Revell L. J., phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 61.Huelsenbeck J. P., Nielsen R., Bollback J. P., Stochastic mapping of morphological characters. Syst. Biol. 52, 131–158 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Pagel M., Meade A., Barker D., Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673–684 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Maddison W. P., Midford P. E., Otto S. P., Estimating a binary character’s effect on speciation and extinction. Syst. Biol. 56, 701–710 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Fitzjohn R. G., Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol. Evol. 3, 1084–1092 (2012). [Google Scholar]
- 65.FitzJohn R. G., Maddison W. P., Otto S. P., Estimating trait-dependent speciation and extinction rates from incompletely resolved phylogenies. Syst. Biol. 58, 595–611 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Rabosky D. L., Automatic detection of key innovations, rate shifts, and diversity-dependence on phylogenetic trees. PLoS One 9, e89543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabosky D. L., et al. , BAMMtools: An R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5, 701–707 (2014). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data (concatenated alignment, phylogenetic tree, character coding of morphology, and nutritional modes) are available via the Open Science Framework and can be accessed at https://osf.io/y2vns/.