Significance
Native Americans are neglected in human genetics studies, despite recent interest in the study of ancient DNA of their ancestors. Our findings on Andean and Amazonian populations exemplify how the current pattern of genetic diversity in human populations is influenced by the interaction of history and environment. In the present case, this pattern is influenced by 1) altitudinal and climatic differences among the northern, lower, and fertile Andes versus the southern, higher, and arid Andes and 2) the sharp differences between the Andean highlands and the Amazon lowlands, where natural selection and other evolutionary forces acted for millennia, shaping differences in the frequencies of genetic variants related to immune response, drug response, and cardiovascular and hematological functions.
Keywords: Native Americans, human population genetics, natural selection, gene flow
Abstract
Western South America was one of the worldwide cradles of civilization. The well-known Inca Empire was the tip of the iceberg of an evolutionary process that started 11,000 to 14,000 years ago. Genetic data from 18 Peruvian populations reveal the following: 1) The between-population homogenization of the central southern Andes and its differentiation with respect to Amazonian populations of similar latitudes do not extend northward. Instead, longitudinal gene flow between the northern coast of Peru, Andes, and Amazonia accompanied cultural and socioeconomic interactions revealed by archeology. This pattern recapitulates the environmental and cultural differentiation between the fertile north, where altitudes are lower, and the arid south, where the Andes are higher, acting as a genetic barrier between the sharply different environments of the Andes and Amazonia. 2) The genetic homogenization between the populations of the arid Andes is not only due to migrations during the Inca Empire or the subsequent colonial period. It started at least during the earlier expansion of the Wari Empire (600 to 1,000 years before present). 3) This demographic history allowed for cases of positive natural selection in the high and arid Andes vs. the low Amazon tropical forest: in the Andes, a putative enhancer in HAND2-AS1 (heart and neural crest derivatives expressed 2 antisense RNA1, a noncoding gene related to cardiovascular function) and rs269868-C/Ser1067 in DUOX2 (dual oxidase 2, related to thyroid function and innate immunity) genes and, in the Amazon, the gene encoding for the CD45 protein, essential for antigen recognition by T and B lymphocytes in viral–host interaction.
Living Native Americans, the object of this study, are among the most neglected populations in human genetics studies, despite the increasing interest in the study of ancient DNA (aDNA) of their ancestors (1, 2). Western South America was one of the cradles of civilization in the Americas and the world (3). When the Spanish conqueror Francisco Pizarro arrived in 1532, the pan-Andean Inca Empire ruled in the Andean region and had achieved levels of socioeconomic development and population density unmatched in other parts of South America. The Inca Empire, which lasted for around 200 years before the conquest, with its emblematic architecture such as Machu Picchu and the city of Cuzco, was just the “tip of the iceberg” of a millenary cultural and biological evolutionary process (4, 5). This process started 11,000 to 14,000 years ago (6–8) with the peopling of this region, hereafter called western South America, that involves the entire Andean region and its adjacent and narrow Pacific coast.
Tarazona-Santos et al. (9) proposed in 2001 that cultural exchanges and gene flow along time have led to a current relative genetic, cultural, and linguistic homogeneity between the populations of western South America compared with those of eastern South America (a term that hereafter refers to the region adjacent to the eastern slope of the Andes and eastward, including Amazonia), where populations remained more isolated from each other. For instance, only two languages (Quechua and Aymara) of the Quechumaram linguistic stock predominate in the entire Andean region, whereas in eastern South America natives speak a different and broader spectrum of languages classified into at least four linguistic families (5, 9, 10). This spatial pattern of genetic diversity and its correlation with geography and environmental, linguistic, and cultural diversity was confirmed, enriched, and rediscussed by us and others (2, 4, 5, 9–15).
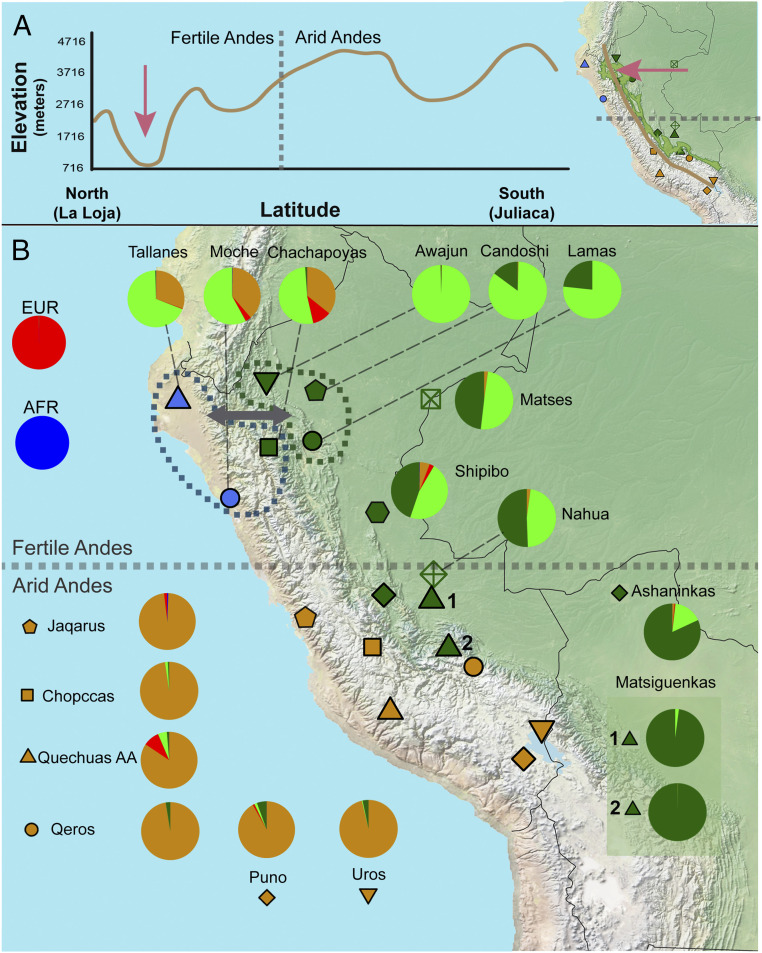
There are, however, pending issues. The first is whether the current dichotomic organization of genetic variation characterized by the between-population homogeneous southern Andes vs. between-population heterogeneous central Amazon extends northward. This is important because scholars from different disciplines emphasize that western South America is not latitudinally homogeneous, differentiating a northern and, in general, lower and wetter fertile Andes and a southern, higher, and more arid Andes (16) (Fig. 1A). These environmental and latitudinal differences are correlated with demography and culture, including different histories and spectra of domesticated plants and animals. Indeed, the development of agriculture, in the first urban centers such as Caral (3) and its associated demographic growth, occurred earlier in the northern fertile Andes (around 5,000 years ago) than in the southern arid Andes (and their associated coast), with products such as cotton, beans, and corn domesticated in the fertile north and the potato, quinoa, and South American camelids domesticated in the arid south (16). In human population genetics studies, the region where the between-population homogeneity was ascertained by Tarazona-Santos et al. (9) was the arid Andes. Consequently, here we test whether the between-population homogenization of western South America and the dichotomy of arid Andes/Amazonia extend to the northward fertile Andes.
Fig. 1.
Genetic and geographic landscape for western South American natives. (A) Elevation vs. latitude plot from a cross-section line in the Andean region from La Loja (Ecuador) to Juliaca (Peru). A vertical line indicates the division between Fertile and Arid Andes (16). The map shows the path used to create the plot elevation and latitude plot and a green area delimiting the area of the Amazon Yunga region. Altitude data were obtained from Google Maps (https://www.google.com.br/maps). (B) Geographic distribution and genetic structure for 18 native populations from the coast, Andes, and Amazon inferred by ADMIXTURE result (K = 5, corresponding to the lowest cross-validation error). Pie charts show the average percentages over individuals for the five ADMIXTURE clusters in each population. Three clusters were related to Native American groups: one Andean (brown) and two Amazonian (green clusters). Two clusters were associated with non-Native continental ancestries (European [red] and African [dark blue]). Blue and green dashed lines delimit the groups that showed a highly significant D statistic value, indicating gene flow (gray arrow, |Z score| > 4, SI Appendix, Figs. S14–S17). Matsiguenkas 1= Matsiguenkas-Sepahua, Matsiguenkas 2= Matsiguenkas-Shimaa. The gray horizontal dashed line in the center of the map shows the approximate division between the fertile Andes and arid Andes (16).
A second open issue is the evolutionary relationship between Andean and Amazonian populations, particularly with the culturally, linguistically, and environmentally different neighboring populations of the Amazon Yunga (the rain forest transitional region between the Andes and Lower Amazonia). Harris et al. (5) inferred that Andean and Amazonian populations diverged around 12,000 years ago. Archaeological findings of recent decades have rejected the traditional view of the Amazonian environment as incompatible with complex pre-Columbian societies and have revealed that the Amazonian basin has produced the earliest ceramics of South America, that endogenous agricultural complex societies developed there, and that population sizes were larger than previously thought (17). Population genetics studies (18) have reported episodes of gene flow in Amazonia which suggest that Amazonian populations were not necessarily isolated groups. Moreover, the ancestors of people living on the Peruvian coast, in the Andes, and in the Amazon Yunga had cultural and commercial interactions during the last millennia, sharing practices such as sweet potato and manioc cultivation, ceramic iconography and styles (e.g., Tutishcanyo, Kotosh, Valdivia, and Corrugate), and traditional coca chewing (19). Therefore, here we address whether gene flow accompanied the cultural and socioeconomic interactions between the ancestors of current Andean and Amazon Yunga populations.
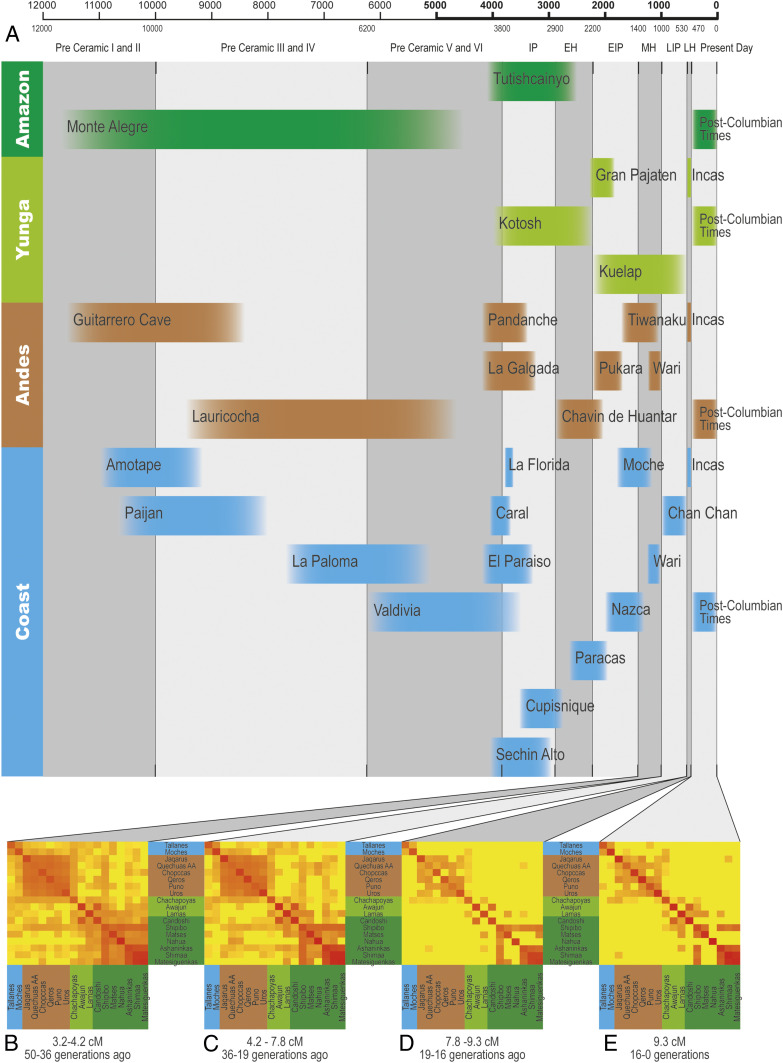
Despite some controversy about definitions and chronology, archeologists identify a unique cultural process in western South America which includes three temporal horizons, Early, Middle, and Late, that correspond to periods of cultural dispersion involving a wide geographic area (20) (Fig. 2). In particular, the Middle and Late Horizons are associated with the expansions of the Wari (∼1,000 to 1,400 years before present [YBP]) and Inca (∼524 to 466 YBP) states, respectively (21–23). The between-population homogeneity currently observed in the arid Andes results from high levels of gene flow in this region, which is commonly associated with the Inca Empire (20). However, Isbell (22) has suggested that the former Wari expansion led to the spread of the Quechua language in the central Andes and that the Wari were pioneers in developing a road system in the Andes called Wari ñam, which was later used by the Incas to develop their network of roads (the Qapaq ñam) (16). A third relevant question is, therefore, when the current between-population genetic homogenization started in the context of the arid Andean chronology (Fig. 2). Particularly, is this a phenomenon restricted to the period of the Inca Empire (Late Horizon), or did it extend backward to the Middle/Wari Horizon?
Fig. 2.
Changes in IBD sharing over time between the Pacific coast, central Andes, Amazon Yunga, and Amazon and its relationship with the archaeological chronology of the Andes. (A) Key historical events (cultures and archeological sites) of Peruvian history in four Peruvian longitudinal regions, coast, Andes, Amazon Yunga, and Amazonia. This is a simplified chronology of Peruvian archaeological history based on different dating records. To account for temporal uncertainties, we depicted the events in the chronology plot without clearly defined chronological borders. The timeline on the top and bottom is represented in years before present. IP: initial period, EH: Early Horizon, EIP: early intermediate period, MH: Middle Horizon, LIP: late intermediate period, LH: Late Horizon. Adapted from ref. 4, which is licensed under CC BY 4.0. (B–E) Heat maps of the average pairwise relatedness (85) among Native Americans of the Natives 1.9M dataset. Each heat map represents an interval of IBD segment lengths, which correspond to interval times (36).
Finally, Native Americans had to adapt to different and contrasting environments and stresses. The high and arid Andes are characterized by high ultraviolet radiation, cold, dryness, and hypoxia (a stress that does not allow for cultural adaptations and requires biological changes) (24, 25). The Amazon has a low incidence of light, a warm and humid climate typical of the rain forest, and high biodiversity, including pathogens (26). Here we infer episodes of genetic adaptation to the arid Andes and the Amazonian tropical forest.
Results and Discussion
We used data from Harris et al. (5) for 74 indigenous individuals and additional data from 289 unpublished individuals from 18 Peruvian Native populations, genotyped for ∼2.5 million single nucleotide polymorphisms (SNPs) (Fig. 1B and Dataset S1). For population genetics analyses, we created three datasets with different SNP densities and populations (27–30) (SI Appendix, Fig. S1 and section 1.3, and Datasets S2 and S3). The institutional review boards of participants’ institutions approved this research. The study was led by Peruvian institutions and investigators who have a long record of community engagement activities as an intrinsic component of their research protocols. Bioinformatics pipelines are described in (31).
The Between-Population Homogenization of Western South America and the Dichotomy of Arid Andes/Amazonia do not Extend to the Northward Fertile Andes.
By applying ADMIXTURE (32) and principal component analyses (Fig. 1B and SI Appendix, Figs. S2–S7), as well as haplotype-based methods (33, 34) (SI Appendix, Figs. S8–S13 and sections 2.1.1 and 2.1.2), we confirmed that populations in the arid Andes are genetically homogeneous, appearing as an almost panmictic unit, with an ancestry pattern differentiated with respect to Amazonian populations (Fig. 1B). Conversely, populations of the northern coast (Moches and Tallanes) and in the northern Amazon Yunga (i.e., Chachapoyas) share the same ancestry profile between them (Fig. 1B and SI Appendix, Figs. S8–S13), which is different from the populations from the arid Andes. Thus, the between-population homogenization of the arid Andes and its differentiation with respect to Amazonian populations of similar latitudes do not extend northward and are not characteristic of all western South America. Instead, the genetic structure of western South Amerindian populations recapitulates the environmental and cultural differentiation between the northern fertile Andes and the southern arid Andes. Nakatsuka et al. (2) (their figure 2), studying aDNA from 86 pre-Columbian individuals, showed that some level of north–south population structure predates the arrival of Spaniards to Peru in 1532. They claim that there was a strong pre-Columbian north–south population structure in the western Andes in pre-Columbian times. However, their claim partly depends on removing from the results of their figure 2 sixteen out of the 86 studied pre-Columbian individuals whom they call “outliers” (18% of their aDNA dataset). The inclusion of these so-called outliers [see SI Appendix, figure S4 of Nakatsuka et al. (2)] shows that the north–south pre-Columbian population structure was not as strong as they claimed.
Longitudinal Gene Flow between the North Coast, Andes, and Amazonia Accompanied the Well-Documented Cultural and Socioeconomic Interactions.
Haplotype-based inferences (ChromoPainter/Globetrotter methods) (33, 34) (Fig. 1B and SI Appendix, Figs. S11–S13 and section 2.1.3), statistical tests of treeness (35) (Fig. 1B and SI Appendix, Figs. S14 and S15 and section 3.2.1), and admixture graphs (35) (SI Appendix, Figs. S16–S19 and section 3.2.2) reveal genetic signatures of gene flow between coastal/Andean and Amazon Yunga populations in latitudes of the northern fertile Andes but not in the southern arid Andes. Thus, longitudinal gene flow between the north coast, Andes, and Amazonia accompanied cultural and socioeconomic interactions documented by archeology, which include ceramic styles and crops, as well as the critical role that Chachapoyas may have played (see Introduction and SI Appendix, section 3.1). This pattern of gene flow recapitulates the differentiation between the fertile north, where altitudes are lower, and the arid south, where the Andes altitudes are higher (Fig. 1A) and may have acted as a barrier to gene flow, imposing a sharper environmental differentiation between the Andes and the Amazon Yunga. Formal comparison of admixture graphs (35) (SI Appendix, Figs. S16–S19) representing different scenarios shows that gene flow was more intense from the north coast to the Amazon than in the opposite direction and that in latitudes of the fertile north, gene flow included important ethnic groups such as the current Chachapoyas of the Amazon Yunga, as well as eastward Lower Amazonian populations such as those of the Jivaro linguistic family (Awajun and Candoshi) and Lamas (Fig. 1B and SI Appendix, Figs. S16–S19). These results are consistent with those of Nakatsuka et al. (2) based on current and pre-Hispanic individuals.
The Homogenization of the Central Arid Andes Started at least during the Wari Expansion (1,400 to 1,000 YBP).
We analyzed the distribution of identity-by-descent (IBD) segment lengths between individuals of different arid Andean populations, which is informative about the dynamics of past gene flow (5, 36). We observed a signature of gene flow in the interval between 1,400 and 1,000 YBP, within the Wari expansion in the Middle Horizon (Fig. 2). Thus, the homogenization of the central arid Andes is not only due to migrations during the Inca Empire or later during the Spanish Viceroyalty of Peru, when migrations (often forced) occurred (37). The Wari expansion (1,400 to 1,000 YBP) was also accompanied by intensive gene flow whose signature is still present in the between-population genetic homogeneity of the arid central Andes region. We also observed that during the Wari/Middle Horizon the effective population size (Ne) was rising in the arid Andes (SI Appendix, Fig. S22), a trend that stopped with the European contact, when Ne started to decline, consistent with demographic records (38) and with genetic studies by Lindo et al. (39). Because IBD analysis on current individuals does not allow for inferences of gene flow that occurred more than 75 generations ago (36), ancient DNA analysis at the population level will be necessary to infer whether the between-population homogenization of the Andes started even earlier.
Episodes of Genetic Adaptation Occurred in the Arid Andes and the Amazonian Tropical Forest.
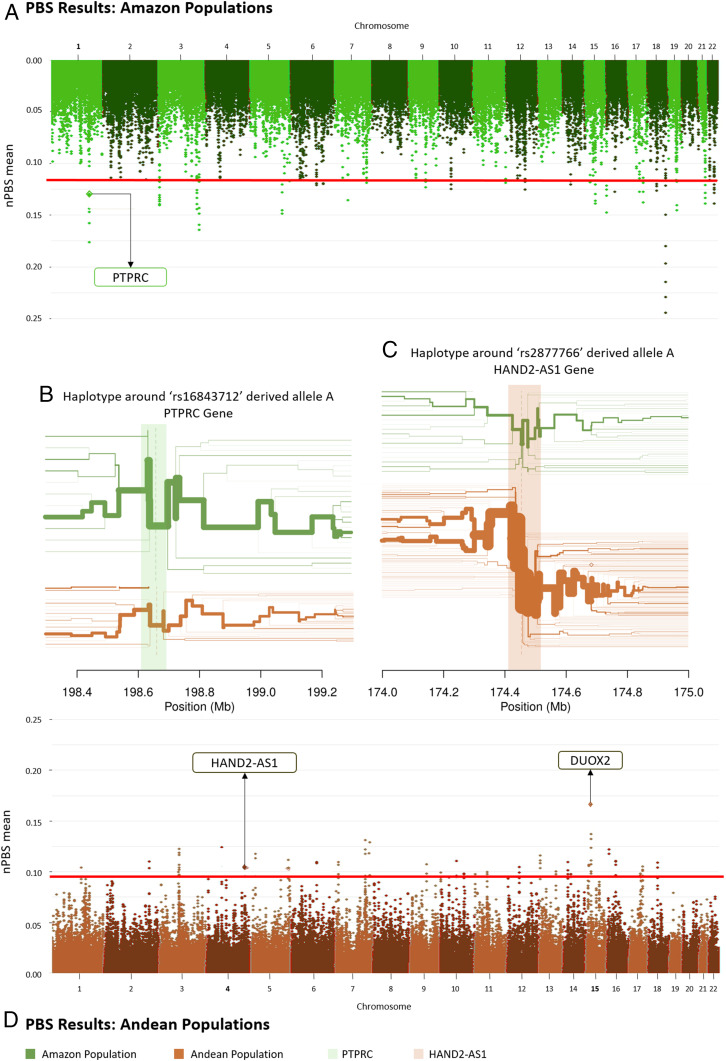
Populations from the high and arid Andes and those from the Amazon (Fig. 1B) settled in these contrasting environments more than 5,000 years ago (40) and show little evidence of gene flow between them (i.e., that would homogenize allele frequencies, potentially concealing the effect of diversifying natural selection). We performed genome-wide scans in these two groups of populations using two tests of positive natural selection: 1) population branch statistics (PBSn) comparing arid Andeans (Chopccas, Quechuas_AA, Qeros, Puno, Jaqarus, and Uros; n = 102) vs. Amazonian populations (Ashaninkas, Matsiguenkas, Matses, and Nahua; n = 75) with a Chinese population (Dai in Xishuangbanna, China; n = 100) from 1000 Genomes as an out-group (41) (SI Appendix, section 5.2.1) and 2) long-range haplotypes (xpEHH) (42) estimated for the two groups of populations (Fig. 3 and SI Appendix, Figs. S24–S27 and section 5.2.2). The complete lists of SNPs with high PBSn and xpEHH statistics for Andean and Amazonian populations are in Datasets S4–S7.
Fig. 3.
Sandwich-like representation of natural selection signatures in the arid Andes and the Amazon tropical forest. Manhattan plots (the bread of the sandwich) correspond to the PBSn estimated from the sliding window in Amazon (A) and Andean (D) populations. The horizontal red line shows the 99.95th percentile of PBSn values. The filling of the sandwich is the long-range haplotype representations. (B) Long-range haplotypes flanking the rs16843712 derived allele A (frequencies: Amazon, 0.811; Andean, 0.324) in the PTPRC gene (light green vertical shading). (C) Haplotype flanking the rs2877766 derived allele A (frequencies: Amazon, 0.453; Andean, 0.880) in the HAND2-AS1 gene (light brown vertical shading). Green plots refer to the Amazon populations; brown plots refer to the Andean populations.
The gene with the consensually strongest signal of adaptation (both from PBSn and xpEHH statistics: PBSn = 0.205, P value = 0.003; xpEHH = 4.481, P value < 0.00001) to the Andean environment (Fig. 3 and Dataset S4) is a long noncoding RNA gene called HAND2-AS1 (heart and neural crest derivatives expressed 2 RNA antisense 1, chromosome 4), that modulates cardiogenesis by regulating the expression of the nearby HAND2 gene (43, 44). This result is consistent with 1) the natural selection genome-wide scan by Crawford et al. (41), who identified three genes related to the cardiovascular system in Andeans, including TBX5, which works together with HAND2 in reprogramming fibroblasts to cardiac-like myocytes (45, 46), and 2) a pattern of adaptation of Andean populations preferentially mediated by the cardiovascular system. The derived allele rs2877766-A (frequencies: Amazonians, 0.453; Andeans, 0.880) is the core of the extended haplotype. HAND2-AS1 is located in the antisense 5′ region of HAND2, and the positively selected six SNPs core haplotype is ∼18-kilobase and encompasses a putative human enhancer (GeneHancer identifier GH04J173536, SI Appendix, Fig. S29). Considering the limitation of our data that come from genotyping arrays, we further recovered from the sequencing data by Harris et al. (5) all nearby SNPs in linkage disequilibrium in Andean populations (r2 > 0.80) with the core SNP rs2877766. We found that the positively selected haplotype includes the SNP rs3775587, mapped within the putative enhancer GH04J173536. Altogether, these results suggest (but do not demonstrate) that the HAND2-AS1 signature of natural selection is related to regulation of gene expression by an enhancer and reflects cardiovascular adaptations. Andeans have cardiovascular adaptations to high altitude that differ from those of lowlanders exposed to hypoxia and from those of other highlanders, showing higher pulmonary vasoconstrictor response to hypoxia, lower resting middle cerebral flow velocity than Tibetans, and higher uterine artery blood flow than Europeans and lowlanders raised in high altitude (47).
DUOX2 (dual oxidase 2, chromosome 15) is the gene with the highest signal of adaptation to the Andean environment by PBSn analysis (PBSn = 0.22, P value = 0.002) (Fig. 3 and SI Appendix, Fig. S24). It has already been reported as a natural selection target in the Andes (48, 49). DUOX2 encodes a transmembrane component of an NADPH oxidase, which produces hydrogen peroxide (H2O2), and is essential for the synthesis of the thyroid hormone and for the production of the microbicidal hypothiocyanite anion (OSCN−) during mucosal innate immunity response against bacterial and viral infections in the airways and intestines (50, 51). Mutations in DUOX2 produce inherited hypothyroidism (52). Here we report the following: 1) The PBSn signal for DUOX2 comprises several SNPs, including two missense mutations (rs269868: C > T: Ser1067Leu, C allele frequencies: Amazon, 0.01, Andean, 0.53; rs57659670: T > C: His678Arg, C allele frequencies: Amazon, 0.01, Andean, 0.53); 2) bioinformatics analysis reveals that rs269868 is located in an A-loop, 1064-1078 amino acids, which is a region of interaction of DUOX2 with its coactivator DUOXA2. Mutations in this region of the protein can affect the stability and maturation of the dimer and, consequently, the conversion of the intermediate product O2 to the final product H2O2 and their released proportions (53). If the natural selection signal is related to this effect, then the standing ancestral allele has been positively selected in the Andes. It is not clear whether the DUOX2 natural selection signal is related to thyroid function or innate immunity. Before the introduction of the public health program of supplementing manufactured salt with iodine, one of the environmental stresses of the Andes for human populations was iodine deficiency, which impairs thyroid hormone synthesis, increasing the risk of developing hypothyroidism, goiter, obstetric complications, and cognitive impairment (54, 55).
Natural selection studies in Amazon populations are scarce. Studies targeting rain forest populations in Africa and Asia have found natural selection signals in genes related to height and immune response (56). In the Amazon region, the strongest natural selection PBSn signal (PBSn = 0.302, P value = 0.002) is in a long noncoding RNA gene on chromosome 18 with unknown function (Dataset S5 and SI Appendix, Fig. S25). The second-highest signal (which also shows a significant long-range haplotype signal: PBSn = 0.265, P value = 0.004; xpEHH = −4.222, P value = 0.0003) corresponds to the gene PTPRC (Fig. 3), which encodes the protein CD45, essential in antigen recognition by T and B lymphocytes in pathogen–host interaction, in particular for viruses such as human adenovirus type 19 (57), HIV-1-induced cell apoptosis (58, 59), hepatitis C (60, 61), and herpes simplex virus 1 (62), even if we cannot exclude a role for unknown viruses endemic in the Amazon region. The core haplotype flanks the rs16843712 derived allele A (frequencies: Amazonia, 0.811; Andes, 0.324), within the putative human enhancer GH01J198660 (sensu GeneHancer; SI Appendix, Fig. S30), and includes the A (Thr193) allele of the nonsynonymous SNP rs4915154 (A > G: Thr193Ala) in exon 6 that affects alternative splicing and alters a potential O- and N-linked glycosylation site. The positively selected allele A (Thr193) has been associated (63) with a lower proportion of CD45R0+ T memory cells and an increased amount of naive phenotype T cells expressing A (exon 4), B (exon 5), and C (exon 6) isoforms. This result is consistent with the hypothesis of CD45 evolution driven by a host–virus arms race model (64).
In addition to the natural selection PBSn and xpEHH signals, we used the bioinformatics platform MASSA (Multi-Agent System for SNP Annotations) (65) to annotate the 1,985 (0.1%) most differentiated SNPs (FCT > 0.318) between the same Andean and Amazonian groups that we tested for natural selection. Notably, we found three TMPRSS6 (transmembrane serine protease 6) variants, rs855791-T (2246T > C Val727Ala: Andean = 0.60, Amazon = 0.92), rs4820268-G (Andean = 0.59 Amazon = 0,98), and rs2413450-T (Andean = 0.60, Amazon = 0.98; Dataset S8), more common in the Amazon region and associated with a broad spectrum of hematological phenotypes such as lower hemoglobin, iron, ferritin, and glycated hemoglobin and higher hepcidin/ferritin ratio (a hormone that decreases iron absorption and distribution) levels in blood, as well as mean corpuscular volume (sensu Genome-Wide Association Study [GWAS] Catalog, that includes GWASs with Latin American admixed individuals) (66–68).
We use DANCE [Disease Ancestry Network (69)] to present the allele frequencies of our total Native American samples for 30,270 GWAS hits and its associated complex phenotypes (sensu GWAS Catalog, https://www.ebi.ac.uk/gwas/), in comparison with African, European, and Asian allele frequencies from the 1000 Genome Project. While this information is relevant, we recall that the allelic architecture of the complex diseases presented in the GWAS Catalog is biased by the underrepresentation of individuals with non-European ancestry in genetic studies.
In conclusion, in western South America, there is an environmental and cultural differentiation between the fertile north of the Andes, where altitudes are lower, and the arid south of the Andes, where these mountains are higher, defining sharp environmental differences between the Andes and Amazonia. This has influenced the genetic structure of western South Amerindian populations. Indeed, the between-population homogenization of the central southern Andes and its differentiation with respect to Amazonian populations of similar latitudes do not extend northward. Gene flow between the northern coast of Peru, the Andes, and Amazonia accompanied cultural and socioeconomic interactions revealed by archeology, but in the central southern Andes, these mountains have acted as a genetic barrier to gene flow (70). We provide insights on the dynamics of the genetic homogenization between the populations of the arid Andes which is not only due to migrations during the Inca Empire or the subsequent colonial period but started at least during the earlier expansion of the pre-Inca Wari Empire (600 to 1,000 YBP). Nakatsuka et al. (2), comparing ancient with modern individuals from western South America, make the general claim that the genetic structure of current populations “strongly echoed” and “are most closely related to the ancient individuals from their region” (i.e., 500 to 2,000 years ago). However, this general statement is not supported by their own results (see their SI Appendix, figure S7). From nine ancient (500 to 2,000 years ago) vs. current comparisons of populations from the same region, this statement is true only for the five cases of the Southern Highlands of Peru and for Chile (their SI Appendix, figure S7 J and K) and not for the four comparisons from the Peruvian coast and north of Peru (their SI Appendix, figure S7 F–I). Thus, Nakatsuka et al.’s (2) results emphasize and add a temporal perspective to the dichotomy observed by us between the current northern fertile Andes (more associated with trans-Andean gene flow) and the southern arid Andes (more homogeneous between populations and differentiated from the Amazonia). The evolutionary journey of western South Amerindians was accompanied by episodes of adaptive natural selection to the high and arid Andes vs. the low Amazon tropical forest: the noncoding gene HAND2-AS1 (related to cardiovascular function and with the positively selected haplotype encompassing a putative human enhancer) and DUOX2 (related to thyroid function and innate immunity) in the Andes. In the Amazon forest, the gene encoding for the protein CD45, essential for antigen recognition by T and B lymphocytes and viral–host interactions, shows a signature of positive natural selection, consistent with the host–virus arms race hypothesis. Our results and other studies (70) continue to show how Andean highlanders and Amazonian dwellers provide examples of how the interplay between geography and culture influences the genetic structure and adaptation of human populations.
Materials and Methods
The protocol for the Peruvian Genome Diversity Project was approved by the Research and Ethics Committee (OI003-11 and OI-087-13) of the Peruvian National Institute of Health, and all participants who had samples collected in this project provided informed consent. We genotyped 289 present-day Native Americans from Peru using the Human Omni array of Illumina for 2.5 million SNPs as part of the Peruvian Genome Diversity Project. Quality control was performed using PLINK (71) and Laboratório de Diversidade Genética Humana bioinformatics protocols and scripts (31). We merged our individuals with public datasets (1, 28–30) and Kaqchikel individuals from M.D. lab from National Cancer Institute. For D statistics and admixture graph analyses, we generate masked data, after phasing our datasets with SHAPEIT2 (72) and inferring the non-Native DNA segments with RFMix (73). To infer population structure, we used two approaches: 1) principal component analysis in Eigenstrat (74) and genetic clustering on ADMIXTURE software (32) using a linkage disequilibrium pruned dataset and 2) fineSTRUCTURE (33), MIXTURE MODEL (34, 75), and SOURCEFIND (76) for haplotype-based analyses, after phase inference. Historical relationships were inferred using D statistics (77) and Admixture Graphs (35). IBD was inferred using refinedIBD (78) and IBDNe (79). For the genetic differentiation analyses, the pairwise genetic distances (F statistics) between Native South American groups (FST) and between populations within groups (FSC) were calculated for multilocus and individual loci using 4P software (80) and the hierfstat R package (81), respectively. The linkage disequilibrium was inferred by the software Haploview (82). Natural selection scans were performed using population branch statistics (41, 83) and xpEHH from the package Selscan (42, 84).
Supplementary Material
Acknowledgments
We thank the Peruvian populations for their participation. We thank the members of the Laboratório de Diversidad Genética Humana, Mateus Gouveia, Kelly Nunes, Garrett Hellenthal, Mark Lipson, Marcia Beltrame, Fabrício Santos, Claudio Struchiner, Ricardo Santos, Luis Guillermo Lumbreras, Sandra Romero-Hidalgo, Víctor Acuña-Alonzo, Miguel Ortega, and Juliana Lacerda, for discussions or technical assistance; Harrison Montejo, Silvia Capristano, Juana Choque, and Marco Galarza from Laboratorio de Biotecnologia y Biologia Molecular of Instituto Nacional de Salud (Peru) for collaborating with the Peruvian Genome Project and conducting the genotyping; and Rafael Tou, Lucas Faria, Livia Metzker, and Alex Teixeira for their final reading of SI Appendix. This work was supported by the Peruvian National Institute of Health (INS), the Brazilian Conselho Nacional de Desenvolvimento Científico e Tecnológico, Pró-Reitoria de pesquisa at the Universidade Federal de Minas Gerais (UFMG), Fundação de Amparo à Pesquisa de Minas Gerais (FAPEMIG, grant number RED00314‐16), and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) programs: the Programa de Excelência Acadêmica (PROEX) and the Programa Institucional de Internacionalização (PRINT). V.B. was a CAPES/Programa de Estudantes-Convênio de Pós-Graduação (PEC-PG) fellow (grant number 88882.195664/2018-01). P.E.R. was funded by the Fondo Nacional de Desarrollo Científico, Tecnológico y de Innovación Tecnológica (Fondecyt - Perú) (grant number 34-2019, “Proyecto de Mejoramiento y Ampliación de los Servicios del Sistema Nacional de Ciencia, Tecnología e Innovación Tecnológica”). Datasets were processed in the Sagarana HPC cluster at the Centro de Laboratórios Multiusuários at Instituto de Ciências Biológicas-UFMG. This work is a product of the collaboration between investigators from the Peruvian Genome Project at the INS and the Genomics and Bioinformatics group of the Project Proproject Epidemiologia Genômica de Coortes Brasileiras de base populagional (EPIGEN-Brazil, https://epigen.grude.ufmg.br/), funded by the Departamento de Ciência e Tecnologia/Ministério de Saúde (DECIT-MS, Brazil).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2013773117/-/DCSupplemental.
Data Availability.
Data have been deposited in the European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega/home (accession nos. EGAD00010001958, EGAD00010001990, EGAD00010001991, EGAD00010001992).
References
- 1.Posth C., et al. , Reconstructing the deep population history of central and South America. Cell 175, 1185–1197.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakatsuka N., et al. , A paleogenomic reconstruction of the deep population history of the Andes. Cell 181, 1131–1145.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solis R. S., Haas J., Creamer W., Dating Caral, a preceramic site in the Supe Valley on the central coast of Peru. Science 292, 723–726 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Scliar M. O., et al. , Bayesian inferences suggest that Amazon Yunga natives diverged from Andeans less than 5000 ybp: Implications for South American prehistory. BMC Evol. Biol. 14, 174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris D. N., et al. , Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire. Proc. Natl. Acad. Sci. U.S.A. 115, E6526–E6535 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahaye C., et al. , New insights into a late-Pleistocene human occupation in America: The Vale da Pedra Furada complete chronological study. Quat. Geochronol. 30, 445–451 (2015). [Google Scholar]
- 7.Dillehay T. D., et al. , Monte Verde: Seaweed, food, medicine, and the peopling of South America. Science 320, 784–786 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Dillehay T. D., et al. , New archaeological evidence for an early human presence at Monte Verde, Chile. PLoS One 10, e0141923 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarazona-Santos E., et al. , Genetic differentiation in South Amerindians is related to environmental and cultural diversity: Evidence from the Y chromosome. Am. J. Hum. Genet. 68, 1485–1496 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell L., American Indian Languages: The Historical Linguistics of Native America (Oxford University Press, 2000). [Google Scholar]
- 11.Fuselli S., et al. , Mitochondrial DNA diversity in South America and the genetic history of Andean highlanders. Mol. Biol. Evol. 20, 1682–1691 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Lewis C. M. Jr., Long J. C., Native South American genetic structure and prehistory inferred from hierarchical modeling of mtDNA. Mol. Biol. Evol. 25, 478–486 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Wang S., et al. , Genetic variation and population structure in native Americans. PLoS Genet. 3, e185 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandoval J. R. et al.; Genographic Project Consortium , The genetic history of indigenous populations of the Peruvian and Bolivian Altiplano: The legacy of the Uros. PLoS One 8, e73006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gnecchi-Ruscone G. A., et al. , Dissecting the pre-Columbian genomic ancestry of Native Americans along the Andes-Amazonia divide. Mol. Biol. Evol. 36, 1254–1269 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lumbreras L. G., Los orígenes de la civilización en el Perú; (Instituto Andino de Estudios Arqueológico-Sociales, 2015). [Google Scholar]
- 17.Roosevelt A., “The maritime, highland, forest dynamic and the origins of complex culture” in The Cambridge History of the Native Peoples of the Americas, F. Salomon, S. B. Schwartz, Eds. (Cambridge University Press, 1999), pp. 264–349. [Google Scholar]
- 18.Barbieri C., et al. , The current genomic landscape of western South America: Andes, Amazonia and Pacific coast. Mol. Biol. Evol. 36, 2698–2713 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman H., Isbell W., Eds., The Handbook of South American Archaeology (Springer, New York, 2008). [Google Scholar]
- 20.Haas J., Pozorski S., Pozorski T., The Origins and Development of the Andean State (Cambridge University Press, 1987). [Google Scholar]
- 21.Lanning E. P., Peru before the Incas (Prentice-Hall, 1967). [Google Scholar]
- 22.Isbell W. H., “Wari and Tiwanaku: International identities in the central Andean Middle Horizon” in The Handbook of South American Archaeology, Silverman H., Isbell W. H., Eds. (Springer, New York, 2008), pp. 731–759. [Google Scholar]
- 23.Valverde G., et al. , Ancient DNA analysis suggests negligible impact of the Wari Empire expansion in Peru’s central coast during the Middle Horizon. PLoS One 11, e0155508 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarazona-Santos E., Lavine M., Pastor S., Fiori G., Pettener D., Hematological and pulmonary responses to high altitude in Quechuas: A multivariate approach. Am. J. Phys. Anthropol. 111, 165–176 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Moore L. G., Measuring high-altitude adaptation. J. Appl. Physiol. (1985) 123, 1371–1385 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amorim C. E. G., Daub J. T., Salzano F. M., Foll M., Excoffier L., Detection of convergent genome-wide signals of adaptation to tropical forests in humans. PLoS One 10, e0121557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis G. R. et al.; 1000 Genomes Project Consortium , An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reich D., et al. , Reconstructing Native American population history. Nature 488, 370–374 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raghavan M., et al. , Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349, aab3884 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallick S., et al. , The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magalhães W. C. S. et al.; Brazilian EPIGEN Consortium , EPIGEN-Brazil initiative resources: A Latin American imputation panel and the scientific workflow. Genome Res. 28, 1090–1095 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lawson D. J., Hellenthal G., Myers S., Falush D., Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hellenthal G., et al. , A genetic atlas of human admixture history. Science 343, 747–751 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patterson N., et al. , Ancient admixture in human history. Genetics 192, 1065–1093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palamara P. F., Lencz T., Darvasi A., Pe’er I., Length distributions of identity by descent reveal fine-scale demographic history. Am. J. Hum. Genet. 91, 809–822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cook N. D., “Migration in colonial Peru: An overview” in Migration in Colonial Spanish America, Robinson D. J., Ed. (Cambridge University Press, 1990), pp. 41–61. [Google Scholar]
- 38.Sanchez-Albornoz N., The Population of Latin America: A History (University of California Press, Berkeley, 1974). [Google Scholar]
- 39.Lindo J., et al. , The genetic prehistory of the Andean highlands 7000 years BP though European contact. Sci. Adv. 4, eaau4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eriksen L., Nature and Culture in Prehistoric Amazonia: Using G.I.S. to Reconstruct Ancient Ethnogenetic Processes from Archeology, Linguistics, Geography, and Ethnohistory (Department of Human Geography, Human Ecology Division, Lund University, 2011). [Google Scholar]
- 41.Crawford J. E., et al. , Natural selection on genes related to cardiovascular health in high-altitude adapted Andeans. Am. J. Hum. Genet. 101, 752–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabeti P. C. et al.; International HapMap Consortium , Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson K. M., et al. , Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng X., Jiang H., Long non-coding RNA HAND2-AS1 downregulation predicts poor survival of patients with end-stage dilated cardiomyopathy. J. Int. Med. Res. 47, 3690–3698 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashimoto H., et al. , Cardiac reprogramming factors synergistically activate genome-wide cardiogenic stage-specific enhancers. Cell Stem Cell 25, 69–86.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez-Perez A., et al. , Hand2 selectively reorganizes chromatin accessibility to induce pacemaker-like transcriptional reprogramming. Cell Rep. 27, 2354–2369.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julian C. G., Moore L. G., Human genetic adaptation to high altitude: Evidence from the Andes. Genes (Basel) 10, 150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou D., et al. , Whole-genome sequencing uncovers the genetic basis of chronic mountain sickness in Andean highlanders. Am. J. Hum. Genet. 93, 452–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacovas V. C., et al. , Selection scan reveals three new loci related to high altitude adaptation in Native Andeans. Sci. Rep. 8, 12733 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Vliet A., Danyal K., Heppner D. E., Dual oxidase: A novel therapeutic target in allergic disease. Br. J. Pharmacol. 175, 1401–1418 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Deken X., Corvilain B., Dumont J. E., Miot F., Roles of DUOX-mediated hydrogen peroxide in metabolism, host defense, and signaling. Antioxid. Redox Signal. 20, 2776–2793 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Maruo Y., et al. , Natural course of congenital hypothyroidism by dual oxidase 2 mutations from the neonatal period through puberty. Eur. J. Endocrinol. 174, 453–463 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Ueyama T., et al. , The extracellular A-loop of dual oxidases affects the specificity of reactive oxygen species release. J. Biol. Chem. 290, 6495–6506 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pretell E. A., et al. , Elimination of iodine deficiency disorders from the Americas: A public health triumph. Lancet Diabetes Endocrinol. 5, 412–414 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Pan L., Fu Z., Yin P., Chen D., Pre-existing medical disorders as risk factors for preeclampsia: An exploratory case-control study. Hypertens. Pregnancy 38, 245–251 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Fan S., Hansen M. E. B., Lo Y., Tishkoff S. A., Going global by adapting local: A review of recent human adaptation. Science 354, 54–59 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Windheim M., et al. , A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proc. Natl. Acad. Sci. U.S.A. 110, E4884–E4893 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anand A. R., Ganju R. K., HIV-1 gp120-mediated apoptosis of T cells is regulated by the membrane tyrosine phosphatase CD45. J. Biol. Chem. 281, 12289–12299 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Meer S., Perner Y., McAlpine E. D., Willem P., Extraoral plasmablastic lymphomas in a high human immunodeficiency virus endemic area. Histopathology 76, 212–221 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Dawes R., et al. , Altered CD45 expression in C77G carriers influences immune function and outcome of hepatitis C infection. J. Med. Genet. 43, 678–684 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsiao J.-L., Ko W.-S., Shih C.-J., Chiou Y.-L., The changed proportion of CD45RA+/CD45RO+ T cells in chronic hepatitis C patients during pegylated Interferon-α with ribavirin therapy. J. Interferon Cytokine Res. 37, 303–309 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Caignard G., et al. , Genome-wide mouse mutagenesis reveals CD45-mediated T cell function as critical in protective immunity to HSV-1. PLoS Pathog. 9, e1003637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stanton T., et al. , A high-frequency polymorphism in exon 6 of the CD45 tyrosine phosphatase gene (PTPRC) resulting in altered isoform expression. Proc. Natl. Acad. Sci. U.S.A. 100, 5997–6002 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thiel N., Zischke J., Elbasani E., Kay-Fedorov P., Messerle M., Viral interference with functions of the cellular receptor tyrosine phosphatase CD45. Viruses 7, 1540–1557 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soares-Souza G., Novas Abordagens para Integração de Bancos de Dados e Desenvolvimento de Ferramentas Bioinformáticas para Estudos de Genética de Populações (PhD Thesis, Universidade Federal de Minas Gerais, Belo Horizonte, MG, 2014). [Google Scholar]
- 66.Hodonsky C. J., et al. , Genome-wide association study of red blood cell traits in Hispanics/Latinos: The Hispanic community health study/study of Latinos. PLoS Genet. 13, e1006760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kowalski M. H. et al.; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Hematology & Hemostasis Working Group , Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 15, e1008500 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raffield L. M., et al. , Genome-wide association study of iron traits and relation to diabetes in the Hispanic community health study/study of Latinos (HCHS/SOL): Potential genomic intersection of iron and glucose regulation? Hum. Mol. Genet. 26, 1966–1978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Araújo G. S., et al. , Integrating, summarizing and visualizing GWAS-hits and human diversity with DANCE (Disease-ANCEstry networks). Bioinformatics 32, 1247–1249 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Mendes M., Alvim I., Borda V., Tarazona-Santos E., The history behind the mosaic of the Americas. Curr. Opin. Genet. Dev. 62, 72–77 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delaneau O., Marchini J., Zagury J.-F., A linear complexity phasing method for thousands of genomes. Nat. Methods 9, 179–181 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Maples B. K., Gravel S., Kenny E. E., Bustamante C. D., RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patterson N., Price A. L., Reich D., Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leslie S. et al.; Wellcome Trust Case Control Consortium 2; International Multiple Sclerosis Genetics Consortium , The fine-scale genetic structure of the British population. Nature 519, 309–314 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chacón-Duque J.-C., et al. , Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat. Commun. 9, 5388 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Green R. E., et al. , A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Browning B. L., Browning S. R., Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics 194, 459–471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Browning S. R., Browning B. L., Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am. J. Hum. Genet. 97, 404–418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Benazzo A., Panziera A., Bertorelle G., 4P: Fast computing of population genetics statistics from large DNA polymorphism panels. Ecol. Evol. 5, 172–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goudet J., Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Resour. 5, 184–186 (2005). [Google Scholar]
- 82.Barrett J. C., Fry B., Maller J., Daly M. J., Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Yi X., et al. , Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Szpiech Z. A., Hernandez R. D., selscan: An efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31, 2824–2827 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baharian S., et al. , The Great Migration and African-American Genomic Diversity. PLoS Genetics 12, e1006059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data have been deposited in the European Genome-phenome Archive (EGA), https://www.ebi.ac.uk/ega/home (accession nos. EGAD00010001958, EGAD00010001990, EGAD00010001991, EGAD00010001992).