Leray et al. (1) reassuringly conclude that “GenBank is a reliable resource for 21st century biodiversity research” based on an important quantitative assessment of its taxonomic accuracy. However, their insightful analysis focuses only on taxonomic levels above species. GenBank (2) is the key reference database for the growing fields of eDNA and metabarcoding, in which studies frequently report species-level sequence identities (e.g., refs. 3–5). Thus, the reliability of GenBank for species identification should be evaluated before drawing sweeping conclusions about its robustness for biodiversity research.
To assess possible sources of error for species assignment, we created an example dataset comprising all mitochondrial COI sequences available on GenBank for 25 marine species in California’s commercial fisheries and 18 common mid-Atlantic fishery species. Applying filtering criteria used by Leray et al. (1), we restricted sequence length to 100 to 2,000 base pairs and then performed all pairwise comparisons among all sequences for each species using BLASTn (6), excluding duplicates and self-comparisons. We build upon Leray et al.’s (1) analysis by illustrating three categories of intraspecific variation that highlight biological and database constraints for species-level taxonomic assignment.
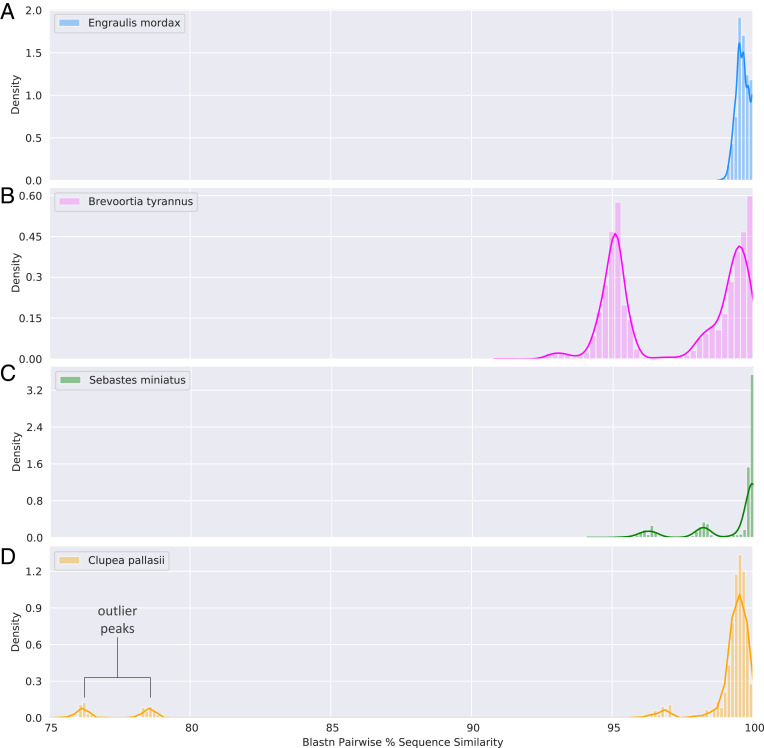
Straightforward species assignment typically occurs when within-species comparisons of sequence similarity form a unimodal distribution (Fig. 1A). However, we found unimodal distributions in only 17 of these 43 species. Multimodal distributions (Fig. 1 B and C) were more common, presenting substantial inferential challenges.
Fig. 1.
Distributions of sequence similarity in intraspecific pairwise comparisons of GenBank mitochondrial COI sequence data for four marine species representing three categories of species-level data. The distribution of Engraulis mordax (A) is unimodal, whereas the distribution of Brevoortia tyrannus (B) is multimodal, likely reflecting hybridization with two congeners (Brevoortia patronus and Brevoortia smithi). For the multimodal distribution of Sebastes miniatus (C), additional modes are likely driven by either misidentification or hybridization including a few individual sequences that identically match two sympatric congeners. Large gaps between modes (≥15%) (D) strongly suggest database errors at higher taxonomic levels that could negatively affect accurate taxonomic identification for incorrectly labeled taxa. In Clupea pallasii (Clupeidae), the two sequences driving the outlier peaks match Agonopsis vulsa (Agonidae) and Limanda punctatissima (Pleuronectidae) at high percent identity (100% and 92.98%, respectively).
Multimodal distributions of sequence similarity can indicate a variety of processes that could hinder species assignments. Our dataset includes one example (Fig. 1B) where documented hybridization between congeners leads to shared mitochondrial DNA haplotypes (7) that prevent species assignment without insights from nuclear DNA. Multimodal distributions can also arise from misidentification of congeners (Fig. 1C), whereby identical or highly similar reference sequences are attributed to two closely related species in GenBank.
Large gaps between modes (Fig. 1D) may be driven by misattributions at the genus level or above. This scenario arose twice in our exploration: Sequences with low similarity were attributed to the same species, but querying them against the full GenBank database revealed comparably strong matches to multiple disparate taxa. Barring errors, this result would indicate low differentiation at a locus. Such sequences are included in the error rates Leray et al. report and could be avoided if researchers performed a BLAST search on sequences prior to submission, as previously noted (1).
Depending on the taxon, it may be infeasible to obtain robust species-level identification from barcoding genes. Many studies instead report genus- or higher-level assignments (8, 9), the reliability of which Leray et al. (1) have rigorously demonstrated. Regardless of taxonomic scale, GenBank’s reliability depends upon researchers verifying their contributions, adding “environmental” tags for eDNA and metabarcoding data, and using alternative repositories (i.e., Sequence Read Archive) when taxonomic identity cannot be verified directly (e.g., gut contents) (10). GenBank is an unmatched resource, yet biodiversity researchers using eDNA and metabarcoding must be cognizant of how both biological processes and human errors can complicate inferences of species identity.
Acknowledgments
This work was supported by a gift from the David R. and Patricia D. Atkinson Foundation to the Cornell Atkinson Center for Sustainability and Environmental Defense Fund.
Footnotes
The authors declare no competing interest.
Data Availability.
The data and code used to produce Fig. 1 have been deposited on GitHub and are available at https://github.com/mistergroot/genbankspecieslevel/.
References
- 1.Leray M., Knowlton N., Ho S.-L., Nguyen B. N., Machida R. J., GenBank is a reliable resource for 21st century biodiversity research. Proc. Natl. Acad. Sci. U.S.A. 116, 22651–22656 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D. A., Karsch-Mizrachi I., Lipman D. J., Ostell J., Wheeler D. L., GenBank. Nucleic Acids Res. 36, D25–D30 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drinkwater R., et al. , Using metabarcoding to compare the suitability of two blood-feeding leech species for sampling mammalian diversity in North Borneo. Mol. Ecol. Resour. 19, 105–117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimmerling N., et al. , Quantitative species-level ecology of reef fish larvae via metabarcoding. Nat. Ecol. Evol. 2, 306–316 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Brown E. A., Chain F. J. J., Zhan A., MacIsaac H. J., Cristescu M. E., Early detection of aquatic invaders using metabarcoding reveals a high number of non-indigenous species in Canadian ports. Divers. Distrib. 22, 1045–1059 (2016). [Google Scholar]
- 6.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson J. D., Karel W. J., Genetic evidence for asymmetric hybridization between menhadens (Brevoortia spp.) from peninsular Florida. J. Fish Biol. 71, 235–249 (2007). [Google Scholar]
- 8.Djurhuus A., et al. , Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 11, 254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leray M., Knowlton N., DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc. Natl. Acad. Sci. U.S.A. 112, 2076–2081 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belcaid M., Poisson G., Detecting anomalies in the Cytochrome C Oxidase I amplicon sequences using minimum scoring segments. Appl. Comput. Rev. 17, 6–14 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and code used to produce Fig. 1 have been deposited on GitHub and are available at https://github.com/mistergroot/genbankspecieslevel/.