Significance
The current paradigm for force transmission between cells and the extracellular matrix is the focal adhesion clutch, in which highly dynamic bonds involving the cytoskeletal linker proteins talin and vinculin transmit tension between moving actin filaments and immobile integrins. We found, however, that while dynamic bonds dominate at cell edges where actin flow is rapid, force transfer in adhesions further from the edge that may be linked to actin bundles is transmitted mainly through more stable interactions. Vinculin contributes to stable and not dynamic bonds as previously thought. We identify the protein interactions that mediate these mechanisms and demonstrate modulation by matrix stiffness. These data therefore substantially revise our view of force transmission in cell-matrix adhesions.
Keywords: focal adhesion, talin, stiffness sensing
Abstract
Integrin-dependent adhesions mediate reciprocal exchange of force and information between the cell and the extracellular matrix. These effects are attributed to the “focal adhesion clutch,” in which moving actin filaments transmit force to integrins via dynamic protein interactions. To elucidate these processes, we measured force on talin together with actin flow speed. While force on talin in small lamellipodial adhesions correlated with actin flow, talin tension in large adhesions further from the cell edge was mainly flow-independent. Stiff substrates shifted force transfer toward the flow-independent mechanism. Flow-dependent force transfer required talin’s C-terminal actin binding site, ABS3, but not vinculin. Flow-independent force transfer initially required vinculin and at later times the central actin binding site, ABS2. Force transfer through integrins thus occurs not through a continuous clutch but through a series of discrete states mediated by distinct protein interactions, with their ratio modulated by substrate stiffness.
The mechanical properties of the extracellular matrix (ECM) control the growth, survival, gene expression, and morphogenesis of cells and tissues (1–3). These functions are attributed to the “focal adhesion clutch,” in which moving actin filaments transmit force to integrins via dynamic protein interactions (4–8). This model proposes that F-actin, driven by force from myosin and/or actin polymerization, flows inward from the cell edge over immobilized integrins bound to ECM proteins (7–10). Force transmission between actin and integrins is mediated by dynamic connections that allow variable efficiency of force transmission, rather than through stable protein–protein interactions. Cells sense the mechanical properties of the ECM via cyclic application of contractile forces (11, 12), resulting in changes in loading rate on the molecular components that control both bond lifetime and signaling outputs (6, 13). Talin, which mediates both conformational activation of integrins and linkage to actin (14), is central in these processes (6). Talin-containing adhesions initially form at the leading edge of lamellipodia through a myosin-independent process that requires actin polymerization and retrograde flow (4, 15). These small nascent adhesions are mostly transient, with a small fraction maturing into more stable, larger focal adhesions (FAs). Maturation involves myosin and alpha-actinin cross-linking of actin (15), buildup of force on talin, and recruitment of vinculin, slowing retrograde actin flow as forces from the actin cytoskeleton are transmitted to the substrate. Talin contains an N-terminal head domain that binds integrins and a C-terminal rod domain that binds F-actin via actin binding site (ABS)2 in the center of the rod, ABS3 at the far C terminus, and multiple vinculin binding sites along its length (16). Our previous work, using point mutations of talin’s actin binding sites that reduce affinity by around 70%, showed that mutating ABS2 reduced force on talin in mature FAs, whereas mutating ABS3 had no effect (17). However, ABS3 mutation reduces cell spreading and migration (18).
In its simplest form, the molecular clutch model describes F-actin filaments sliding rearward across a bed of single clutch molecules that rapidly engage and disengage from the actin, with increasing engagement slowing the actin and deforming the substrate (13, 19). In this model, mechanosensitivity is driven by the effect of substrate rigidity on force loading rate. A key prediction of these models is that force transmission peaks at intermediate rigidities. This is because at low rigidities force buildup is too slow for high forces to be reached before clutches unbind, while at high rigidities force buildup is so fast that clutches unbind quickly, resulting in “frictional slippage.” At intermediate rigidity, the rate of force buildup allows for significant force transfer before unbinding but not too fast to overload the individual clutches before more can bind. This behavior was observed in neuronal growth cones and glioma cells (13, 20). Additionally, simultaneous measurements of traction force and actin flow revealed a biphasic relationship between flow and force: In immature adhesions close to the edge force was low and speed was high, while in the maturing adhesions further from the edge force was higher and correlated with actin speed, trending toward zero at low actin speeds (21). These data have been taken to support the clutch model with maximal force transfer at intermediate actin flow rates. These experiments, however, were only performed on the relatively soft substrates (E ∼ 7 kPa) used in traction force microscopy (TFM).
Most cell types, however, display monotonically increasing force and adhesion signaling in the limit of high stiffness. This response is driven by growth and strengthening of FAs under force, mediated in part through unfolding of talin and exposure of additional actin (ABS2) and vinculin binding sites. More recent iterations of the model have therefore included reinforcement parameters that reduce frictional slippage and maintain force transmission at higher stiffness (6). In this model, talin is essential for mechanosensing at high stiffness and increasing stiffness monotonically increases traction forces but decreases actin flow speed, corresponding to most cell types that grow their adhesions in response to stiffness (6). However, these models ignore adhesion maturation and the associated changes in composition and structure, only varying the number of engaged clutches. Further iterations of the clutch model have addressed this through incorporation of multiple structural modules, force-dependent feedback, and two distinct actin networks, allowing for model predictions of growth and traction oscillations (22). Recent work has revised the model to include reversible cross-links within the F-actin network to account for more stable force transfer that is observed for engaged matrix attachments underneath the cell (23). However, these models still emphasize actin flow and dynamic bonds. These questions led us to examine the relationship between actin flow and force transfer to the key clutch molecule talin, using our recently developed tension sensor (17).
Results
Force on Talin Correlates with Actin Speed.
We measured rates of actin flow using quantitative actin speckle microscopy (4) while simultaneously measuring force across talin using a molecular tension sensor in which a fluorescence resonance energy transfer (FRET) pair is separated by an elastic domain, such that tension stretches the spring to decrease FRET efficiency (Fig. 1A) (17, 24). To facilitate actin speckle imaging, we stably expressed SNAP-tagged actin in NIH 3T3 cells. SNAP-actin was expressed at <1% of endogenous actin and did not detectably perturb cytoskeletal organization, cell adhesion, spreading, or migration speed (SI Appendix, Fig. S1 A–D). Addition of cell-permeable SiR647-benzyl guanine (SI Appendix, Fig. S1 D, E, and K) to cells expressing the talin tension sensor (TalinTS) or control sensor (TalinCTS; schematic in Fig. 1A) allowed tracking of actin flow and TalinTS FRET (Fig. 1B) for up to 10 min without significant photobleaching (Movie S1).
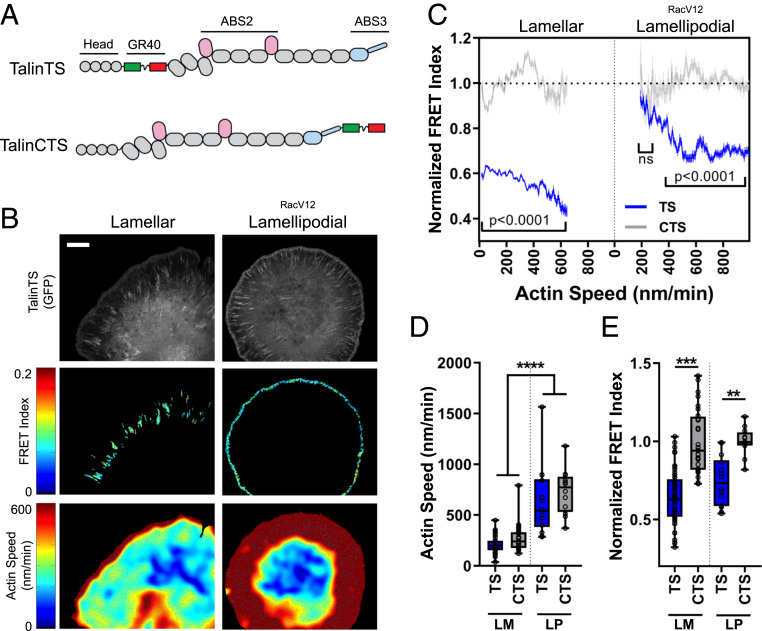
Fig. 1.
Correlating actin speed with force on talin. (A) Schematic of talin tension sensor (TS) and talin C-terminal control sensor (CTS). (B) Heat maps for simultaneous talin tension-sensor FRET index and actin speed in large lamellar adhesions in untransfected cells (Left) and lamellipodial adhesions in RacV12-expressing cells (Right). (Scale bar, 10 µm.) (C) Binned pixelwise correlations of FRET vs. actin speed (mean ± 95% confidence interval, TS n = 49, CTS n = 28, TS RacV12 n = 14, CTS RacV12 n = 12, cells from 10 independent experiments). ns, not statistically significant. (D) Quantification of average actin speed on a per-cell basis and (E) normalized FRET index for cells transfected with talin TS or CTS during spreading on fibronectin-coated glass. FRET index is normalized to the average value of the control sensor. LM: lamellar, LP: lamellipodial (box plots indicate median and quartiles, dots indicate single-cell averages, for data shown in C, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Tukey’s post hoc).
Cells were plated on fibronectin-coated glass coverslips and time-lapse images acquired at 45 to 90 min, when cells have large FAs but are still spreading (Movie S1). Time-lapse videos of talin FRET and actin speckle were recorded for 5 min and quantified for regions of the FAs. The average FRET for 2 × 2-pixel regions was correlated with the average actin velocity. As previously observed (17), the talin tension sensor showed lower FRET compared to the control sensor (Fig. 1 C and E and SI Appendix, Fig. S1F) with similar actin flow speeds (Fig. 1 C and D). We then plotted the average FRET against average actin speed (bin width 5 nm/min) within segmented larger FAs, which formed in the lamellar region 5 to 10 µm back from the cell edges. This analysis revealed that while increasing flow speed correlated with higher force (lower FRET) (Fig. 1 C, Left), the intercept for FRET at 0 actin flow was well below the level for the control sensor. This result indicates residual force on talin and suggests that in mature, lamellar adhesions a large portion of the talin tension is independent of actin flow.
Small, immature adhesions also form at cell edges where actin flow is faster; however, their low signal made imaging difficult. Lamellipodial adhesions are driven by activation of Rac through integrins (25–27); thus, to enhance their formation, we further elevated Rac activity by expression of activated RacV12, which increased actin flow and induced pronounced small lamellipodial adhesions (Fig. 1 B and C, Right). RacV12-induced lamellipodial adhesions had lower tension (higher FRET) than large lamellar adhesions. Talin tension again correlated with actin flow; however, the intercept at zero actin flow matched the control sensor. Thus, the flow-independent component was absent. Estimation of forces in the two adhesion types using a previously described model for tension sensor force (28) indicated that lamellar adhesions have average forces ranging from 1 to 7 pN while lamellipodial adhesions have forces ranging from 1 to 5 pN (SI Appendix, Fig. S1 G–J). Inhibiting myosin II with blebbistatin eliminated large FAs, while in the small lamellipodial adhesions talin tension was partially reduced, indicating that flow-dependent force has both myosin-dependent and -independent components (SI Appendix, Fig. S2 A and B).
Force on Talin in Large Adhesions Is Maintained after Stopping Flow.
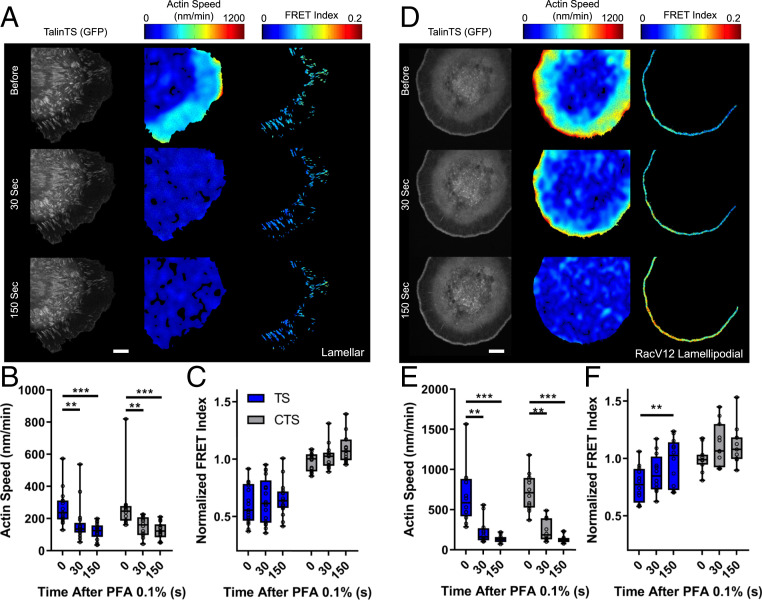
To confirm flow-independent force transmission, we perturbed actin flow and measured effects on talin tension. We used an antibody to fibronectin, which blocks new integrin binding (SI Appendix, Fig. S2 I and J) and signaling to Rac but does not disrupt existing FAs (29, 30). This treatment slowed actin flow as expected (26) but did not change talin tension in large lamellar adhesions (SI Appendix, Fig. S2 C–E). Latrunculin A to sequester G-actin, together with jasplakinolide to stabilize F-actin, also slowed actin flow without affecting talin tension in large FAs (SI Appendix, Fig. S2 C–E). Conversely, blebbistatin decreased tension on talin in large FAs with little change in actin flow (SI Appendix, Fig. S2 F–H), indicating a substantial contribution of myosin to the flow-independent force. While these data support the notion of flow-independent force transmission, we sought a method to completely inhibit actin flow. Addition of low (0.1%) paraformaldehyde (PFA) rapidly and completely stopped actin flow as efficiently as complete fixation (SI Appendix, Fig. S3A). Residual baseline flow in this figure represents noise in the speckle tracking software, as demonstrated by kymography (SI Appendix, Fig. S3B). Stopping actin flow occurred without detectably cross-linking cell protein as assayed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (SI Appendix, Fig. S3C) but did halt adhesion growth and sliding (SI Appendix, Fig. S2K). Stopping flow increased FRET/decreased talin tension in small lamellipodial adhesions in RacV12 cells to nearly the level of the control sensor but had little effect on larger lamellar adhesions (Fig. 2 A–F). We also examined lamellipodial adhesions without expression of RacV12, using two approaches. First, higher expression of the tension sensor improved signal-to-noise sufficiently to determine FRET efficiency in small adhesions. Lamellipodial adhesions in these cells also showed flow-dependent tension on talin (SI Appendix, Fig. S3 D and E). Conversely, large lamellar adhesions in RacV12 cells remained flow-independent (SI Appendix, Fig. S3 G and H). Second, increasing laser power and charge-coupled device camera gain also increased the signal, which showed similar levels of force in wild-type (WT) lamellipodial adhesions (SI Appendix, Fig. S3F). Thus, actin flow is the major determinant of force transmission in small, lamellipodial adhesions but makes only a small contribution in large lamellar adhesions.
Fig. 2.
Evaluating flow dependence. (A) Control cells before and after stopping flow with 0.1% PFA for lamellar adhesions, showing talinTS, actin speed, and talin normalized FRET index. (B) Quantification of actin flow speed before and after addition of 0.1% PFA. (C) Quantified per-cell average talin TS and CTS normalized FRET index in lamellar adhesions normalized to the control sensor. Values for actin flow speed were averaged over 1-min intervals before, 0 to 60 s after (30 s) or 120 to 180 s (150 s) after addition of 0.1% PFA (box plots indicate median and quartiles, dots indicate single-cell averages, n = 14 to 17 cells per group from five independent experiments, #P < 0.05, two-way ANOVA with Tukey’s post hoc test). (D) Representative images of talinTS, actin speed, and talin TS normalized FRET index in RacV12 cells. (E) Average actin flow rates and (F) talin normalized FRET index for RacV12 lamellipodial adhesions (box plots indicate median and quartiles, dots indicate single-cell averages, n = 11 or 12 cells per group from three independent experiments, **P < 0.01, ***P < 0.001, two-way ANOVA with Tukey’s post hoc test). (Scale bars, 10 µm.)
Flow-Independent Force Is Stiffness-Dependent.
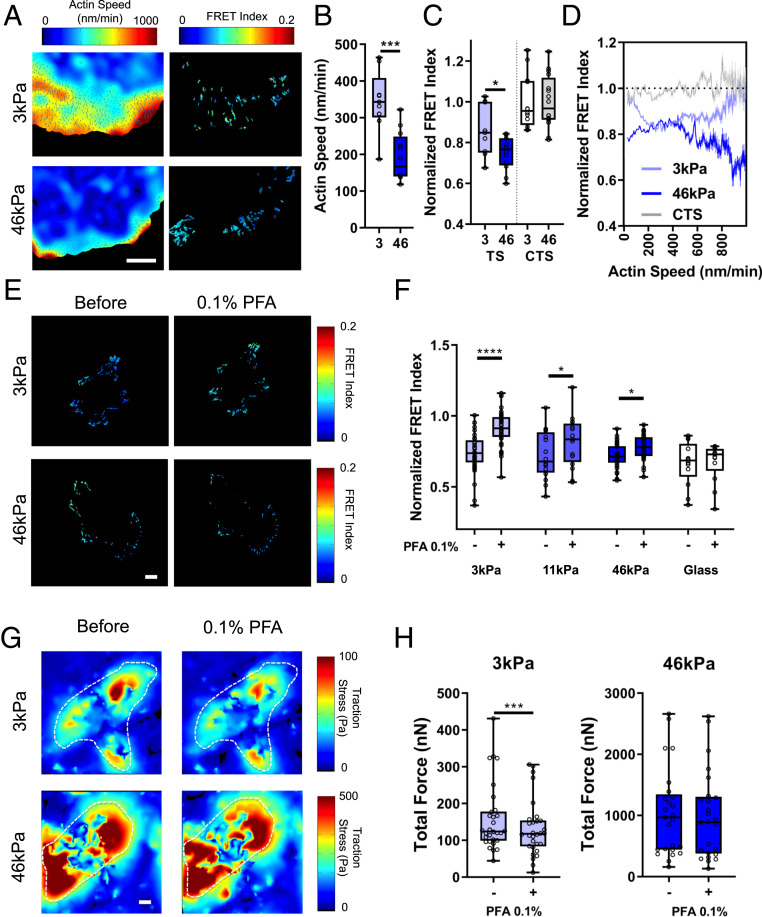
Substrate stiffness is known to regulate both traction forces and actin flow rates, with most cell types displaying higher traction force and slower actin flow on stiff substrates (6). To determine how stiffness affects flow-independent force transfer, we plated cells on fibronectin-coated polyacrylamide substrates of various stiffness. On 46-kPa gels, lamellar adhesions displayed low actin flow rates (Fig. 3 A and B) and high force on talin (low FRET; Fig. 3C) relative to 3-kPa gels. Correlating actin flow rate with talin force in lamellar adhesions on these gels showed substantial tension at 0 actin flow on stiff 46-kPa gels (Fig. 3D). However, on soft 3-kPa gels talin tension trended toward zero at low actin flow rates (Fig. 3D and SI Appendix, Fig. S3L). Talin tension also decreased at the highest actin flow rates on 3-kPa gels, consistent with the dynamic clutch model as the predominant mechanism on soft substrates. This effect was less evident on stiff substrates. This difference may be due to the higher density of adhesion molecules (integrin, talin, and vinculin) within these adhesions that slows actin and prevents slippage, but other mechanisms are also possible. Importantly, stopping flow with 0.1% PFA on gels revealed that talin tension shifted in a dose-dependent manner from flow-dependent to flow-independent with increasing substrate stiffness (Fig. 3 E and F). Traction measurements on soft (3-kPa) and stiff (46-kPa) gels showed that 0.1% PFA (Fig. 3G) decreased total force generation in cells on soft gels but not stiff gels (Fig. 3H and SI Appendix, Fig. S3K). Together, these results indicate that stiffness drives the emergence of flow-independent force transmission.
Fig. 3.
Modulation by substrate stiffness. (A) Heat maps of actin speed (Left) and talin TS normalized FRET index (Right) in cells on soft (3-kPa) or stiff (46-kPa) fibronectin-coated polyacrylamide gels. (Scale bar, 10 µm.) (B) Average per cell actin speed and (C) normalized FRET index (box plots indicate median and quartiles, dots indicate single-cell averages, n = 9 to 16 cells from two independent experiments, *P < 0.01, ***P < 0.001). (D) Binned pixelwise correlations of FRET vs. actin speed (mean ± 95% confidence interval, 3 kPa n = 9, 46 kPa n = 12, CTS 3 kPa n = 9). (E) Heat maps of talin TS normalized FRET index before and after 0.1% PFA for cells on polyacrylamide gels. (Scale bar, 10 µm.) (F) Average talin TS FRET index per cell (normalized to CTS control on same stiffness) before and after 0.1% PFA (box plots indicate median and quartiles, dots indicate single-cell averages, 3 kPa n = 37, 11 kPa n = 16, 46 kPa n = 33, glass n = 13, matched cells per group from five independent experiments. *P < 0.05, ****P < 0.0001, two-way ANOVA with Sidak’s post hoc test). (G) Traction force heat maps for cells on 3-kPa or 46-kPa polyacrylamide gels before and after 0.1% PFA. (Scale bar, 10 µm.) (H) Quantification of total force per cell before (−) and after (+) 0.1% PFA on 3-kPa and 46-kPa gels (box plots indicate median and quartiles, dots indicate single cell averages, 3 kPa n = 30, 46 kPa n = 25, matched cells per group from three independent experiments, ***P < 0.001, two-sided paired t test).
Vinculin Contributes to the Flow-Independent Force.
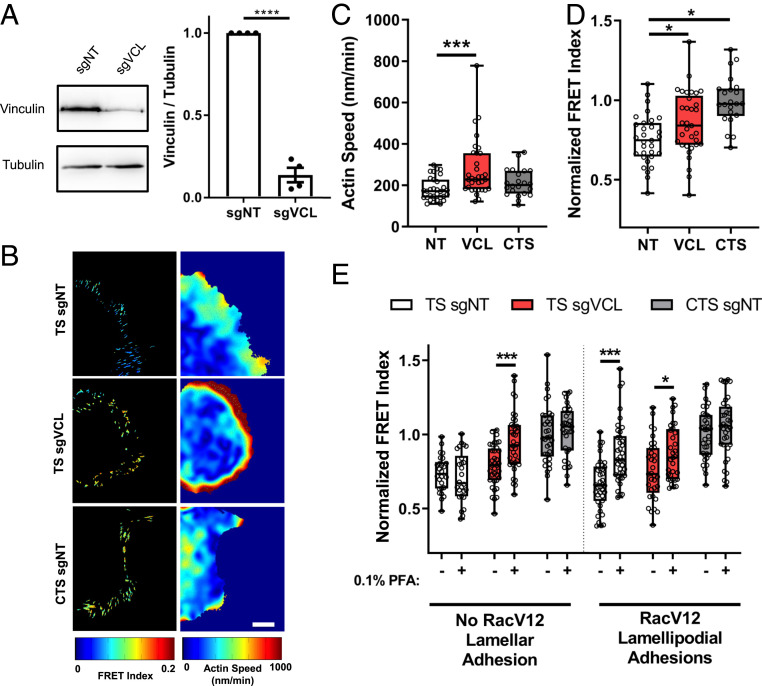
Vinculin deletion accelerates actin flow while reducing cellular traction force, which was interpreted to indicate that vinculin is a component of the FA clutch (31). Vinculin depletion also decreases tension on talin (17) and increases adhesion turnover and dynamics (31). In our system, vinculin depletion using a CRISPR–single-guide RNA vector (Fig. 4A) similarly increased actin speed (Fig. 4 B and C), decreased talin tension (Fig. 4D and SI Appendix, Fig. S4E), and increased adhesion growth rate, sliding velocity, and turnover (SI Appendix, Fig. S4 A–D). However, talin FRET in small lamellipodial adhesions in RacV12 cells showed little change (Fig. 4E). Small lamellipodial adhesions also had lower vinculin levels (SI Appendix, Fig. S5A) consistent with these results. Importantly, vinculin depletion converted large lamellar adhesions from flow-independent to partially flow-dependent (Fig. 4E). There was no significant effect on small lamellipodial adhesions, which remained largely flow-dependent.
Fig. 4.
Vinculin transmits flow-independent force. (A) Western blot and quantification for vinculin after infection with nontargeting (NT) or vinculin guide RNA, with tubulin loading control (n = 4 experiments, ****P < 0.0001, two-sided t test). (B) Representative heat maps of talin FRET index and actin speed. (Scale bar, 10 µm.) (C) Average actin speed and (D) normalized FRET index per cell after vinculin depletion (box plots indicate median and quartiles, dots indicate single-cell averages, NT n = 31, VCL n = 33, CTS n = 22 cells per group from three independent experiments, *P < 0.05, ***P < 0.001, one-way ANOVA with Dunnett’s post hoc test). (E) Normalized FRET index for Talin TS and CTS before and after addition of 0.1% PFA, with vinculin (VCL) or nontargeting control guide RNA (NT) (box plots indicate median and quartiles, dots indicate single-cell averages, TS-NT n = 15, TS-VCL n = 18, CTS-NT n = 17, TS-NT-RacV12 n = 19, TS-VCL-RacV12 n = 15, CTS-NT-RacV12 n = 17, matched cells from two independent experiments, *P < 0.05, ***P < 0.001, two-way ANOVA with Sidak’s post hoc test).
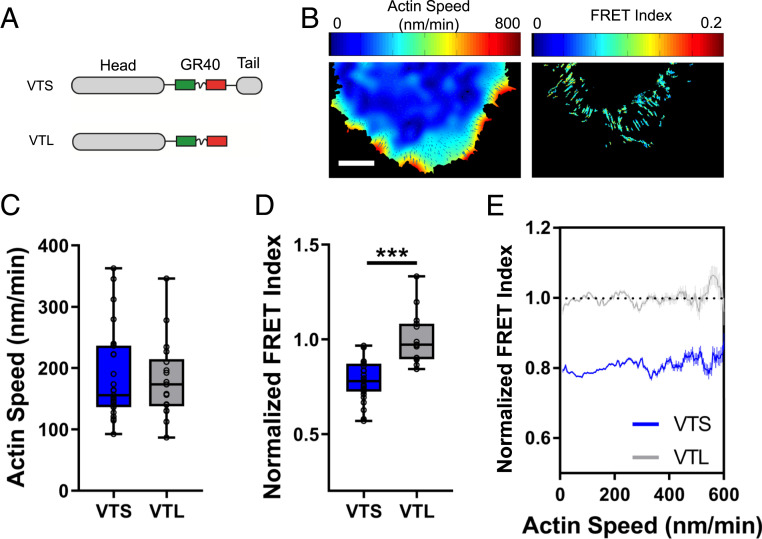
To further examine the mechanism of vinculin force transfer, we used the vinculin tension sensor (Fig. 5A) (17, 24) to correlate actin flow rates with force on vinculin (Fig. 5B). Expression of these constructs did not affect actin flow rates (Fig. 5C). The results showed significant force on vinculin (Fig. 5D), which when plotted according to our pixelwise correlation analysis revealed no correlation between vinculin tension and actin flow rates (Fig. 5E). Further, stopping flow with 0.1% PFA did not change the force on vinculin (SI Appendix, Fig. S4 F and G). Thus, vinculin primarily contributes to actin flow-independent force transfer, rather than flow-dependent force transfer as was previously thought (31).
Fig. 5.
Force on vinculin does not correlate with actin flow. (A) Schematic of the vinculin tension sensor (VTS) and tailless control sensor (VTL). (B) Representative heat maps of actin speed and vinculin FRET index normalized to VTL for lamellar adhesions in WT 3T3 cells. (Scale bar: 10 μm.) (C) Quantification of average per-cell actin speed and (D) vinculin TS and VTL normalized FRET index for lamellar adhesions during spreading on fibronectin-coated glass (box plots indicate median and quartiles, dots indicate single-cell averages, VTS n = 27, VTL n = 16, ***P < 0.001). (E) Binned pixelwise correlations of vinculin sensor FRET vs. actin speed (mean ± 95% confidence interval).
Talin ABS3 Mediates Flow-Dependent Force.
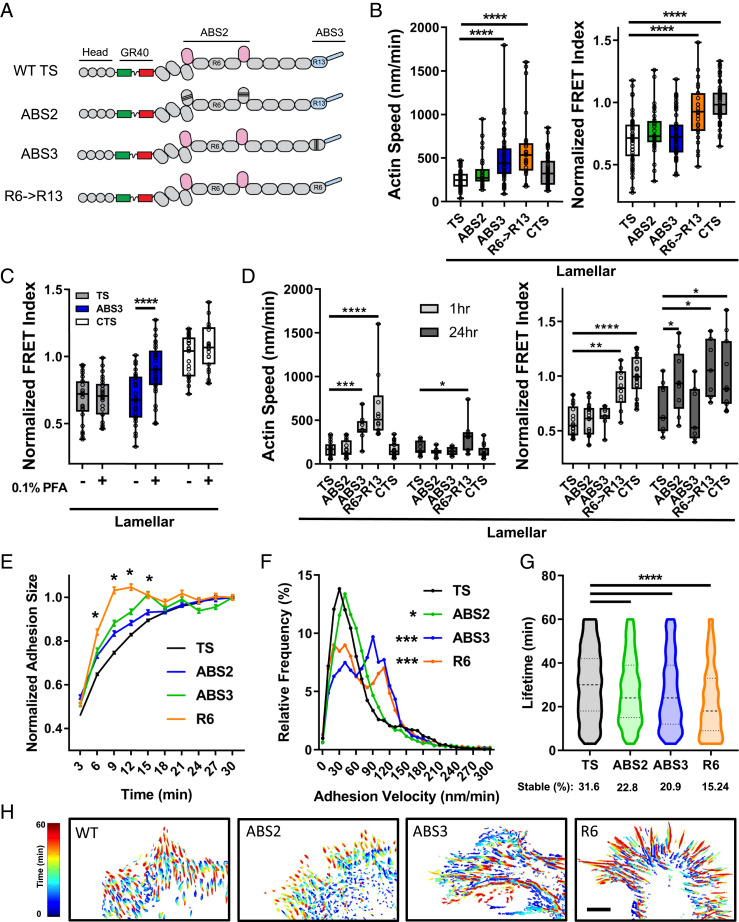
To address the function of the actin binding sites in force transfer, we replaced endogenous talin1 with the ABS2 or ABS3 mutants (32) via a CRISPR-Cas9 mutation/reexpression strategy (Fig. 6A and SI Appendix, Fig. S5 D–J). In RacV12-induced lamellipodial adhesions, neither mutation affected talin tension (SI Appendix, Fig. S6A). However, the ABS3 mutation dramatically increased actin speed (SI Appendix, Fig. S6A and Movies S2 and S3). The ABS3 mutation also increased actin speed in large lamellar (flow-independent) adhesions without affecting talin tension (Fig. 6B). Stopping actin flow in cells with ABS3 mutant talin decreased talin tension (increased FRET) in lamellar adhesions (Fig. 6C) similar to normal lamellipodial adhesions (Fig. 2F). Thus, reduced ABS3 affinity shifts large adhesions from mainly flow-independent to flow-dependent force transfer. In addition, these adhesions become more elongated, with increased turnover and growth rate and decreased lifetime (Fig. 6 E–H and SI Appendix, Figs. S5I and S6E). The ABS3 mutation also decreased the vinculin/talin ratio in lamellar adhesions (SI Appendix, Fig. S5C), consistent with conversion to flow-dependent force transfer (SI Appendix, Fig. S5A).
Fig. 6.
Actin binding site mutations in talin. (A) Schematic of the ABS2 (six point mutations), ABS3 (three point mutations), and R6->R13 swap constructs in the talin tension sensor. (B) Average per-cell actin speed and normalized FRET index for lamellar adhesions in cells with talin mutants (box plots indicate median and quartiles, dots indicate single cell averages, n = 25 t 31 cells per group from three independent experiments, ****P < 0.0001, one-way ANOVA with Tukey’s post hoc test). (C) Talin normalized FRET index in lamellar adhesions of cells with WT or ABS3 mutant talin before (−) and after (+) 0.1% PFA (box plots indicate median and quartiles, dots indicate single-cell averages, TS: n = 25, ABS3 n = 28, CTS n = 23, matched cells from five independent experiments, ****P < 0.0001). (D) Actin speed and talin normalized FRET index for lamellar adhesions in cells at 1 vs. 24 h (box plots indicate median and quartiles, dots indicate single-cell averages, n = 7 to 22 cells per group, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA with Sidak’s post hoc test). Adhesion dynamics in talin-depleted cells reconstituted with WT (TS) or mutant (ABS2, ABS3, R6) talinTS. (E) Average adhesion size normalized to their maximal size (mean ± SEM, WT n = 20,732 adhesions from 49 cells, ABS2 n = 6,643 adhesions from 18 cells, ABS3 n = 3,571 adhesions from 18 cells, R6 n = 3,895 adhesions from 18 cells, *P < 0.01 vs. TS). (F) Adhesion velocity and adhesion lifetime (G) distributions for talin actin binding site mutants (violin plot with median and quartiles, cells imaged for 1 h at one frame per 3 min. *P < 0.05, ***P < 0.001, one-way ANOVA with Dunnett’s post hoc test). (H) Representative heat maps of adhesion dynamics over 1 h. (Scale bar, 10 µm.)
These effects suggested that faster flow might compensate for the ∼70% lower affinity of the ABS3 mutation. To test this idea, we replaced the ABS3-containing R13 domain with R6, which completely eliminates actin binding (18) but does not disrupt talin dimerization (which occurs if R13 is deleted). R6→R13 talin (Fig. 6A) further increased retrograde actin flow (Fig. 6B and Movie S4) but, unlike the ABS3 point mutant, abolished talin tension even in lamellar adhesions (Fig. 6B). Quantification of traction forces using TFM confirmed significant loss of force with this R6 mutation (SI Appendix, Fig. S6 C and D). Combined, these data indicate that ABS3 mediates force transmission via the flow-dependent mechanism, which occurs mainly in small lamellipodial adhesions. ABS3-dependent slowing of actin flow is also required for the subsequent arrest and flow-independent force transfer in large lamellar adhesions.
We next examined the ABS2 mutant, which we previously found reduced talin tension in large FAs at 24 h after plating (17), confirmed in Fig. 6D. However, at ∼1 h after plating, the ABS2 mutation had only a weak effect (Fig. 6D), with no effect on actin flow. By contrast, vinculin depletion had a large impact on talin force and actin flow in lamellar adhesions of 1-h spreading cells but only a small effect in mature, 24-h cells (SI Appendix, Fig. S6B). Thus, vinculin mediates the initial arrest of flowing actin and flow-independent force transmission during cell spreading, whereas ABS2 mediates force transfer in fully mature adhesions at later times.
All three of the mutations altered adhesion dynamics. The ABS2 mutation moderately increased adhesion growth rate, velocity, and turnover (Fig. 6 E–H) similar to effects of vinculin depletion (SI Appendix, Fig. S4 A–D) (31). The ABS3 mutation and the R6 mutation induced larger changes in adhesion growth rate, sliding velocity, and turnover (Fig. 6 E–H) likely due to the combination of reduced vinculin recruitment (SI Appendix, Fig. S5C) and reduced actin binding. However, FAs were readily evident in all cases.
Discussion
Together, these studies identify three distinct mechanisms of force transfer operating at different locations and conditions. In immature adhesions in lamellipodia at cell edges, actin flow speed is high and flow-dependent force transfer through ABS3 predominates. For cells on stiff substrates, lamellar adhesions further from the edge have slower actin flow and force transfer is mainly flow-independent, with only a small contribution from flow-dependent force transfer. This mechanism is mediated by vinculin at early times after plating and by ABS2 at later times when cells are quiescent and lack lamellipodia. These results substantially reinterpret vinculin’s role in the FA clutch, which was previously proposed to mediate flow-dependent force transfer (31). Importantly, on soft substrates, actin flow in both lamellipodial and lamellar adhesions is faster and flow-dependent force transfer makes a larger contribution. These quantitative shifts in the relative contributions of flow-dependent vs. independent are thus mechanosensitive.
The traditional view of the “molecular clutch” involves constant slippage of actin during force transmission (7). Recent modifications add adhesion strengthening to increase force transfer at low actin flow rates but force transfer still varies in a continuous manner and flow remains the defining feature. The present results lead to a model in which force transfer occurs via a series of discrete states, only one of which is flow-dependent. In this model, ABS3 transfers force via a dynamic mechanism with high on and off rates in small adhesions at the cell edge, thereby slowing actin flow. When stiffness is high, actin decelerates and vinculin binds to form stable attachments, analogous to a fully engaged clutch. ABS2 mediates an additional flow-independent force transfer mechanism that requires further adhesion maturation. This view is consistent with the multivalent nature of the talin–vinculin complex and the catch bond behavior of the vinculin–actin bond (33), which are difficult to reconcile with the rapid kinetics required of flow-dependent force transmission. Actin stress fibers linked to FAs very likely represent one type of connection with high force and low actin flow, though such regions without clear actin bundles are also prevalent, suggesting that a high degree of actin bundling is not a requirement.
Future work will be required to elucidate the regulatory mechanisms that control utilization of these distinct mechanisms, the biophysical properties of the binding sites that mediate these distinct types of force transfer and to provide a more complete understanding of their functions in different biological systems.
Methods
Cell Culture, Transfection, and SNAP Labeling.
SNAP tag was cloned onto the N terminus of human beta-actin in a modified pEGFP-NT vector (Clontech) and transfected into NIH 3T3 cells. Cells were sorted to obtain a homogeneous population of low-level SNAP-actin–expressing cells and passaged for no more than 14 passages under G418 selection after sorting. Relative expression of SNAP-actin vs. endogenous actin was determined by RT-PCR with primers for native mouse actin (forward [fw]: 5′-cgagcgtggctacagcttc-3′; reverse [rv]: 5′-gccatctcctgctcgaagtc-3′), human SNAP-actin (fw: 5′-ggtcgccgtgaaagagtgg-3′; rv: 5′-gacgagcgcggcgatatca-3′), and mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (fw: 5′-accacagtccatgccatcac-3′; rv: 5′-tccaccaccctgttgctgta-3′). Messenger RNA was isolated using the mRNeasy kit (Qiagen) and complementary DNA (cDNA) was synthesized using iScript cDNA synthesis kit (Bio-Rad). PCR was run for 40 cycles on a Bio-Rad CFX96 RT-PCR machine using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). SNAP-actin 3T3 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) with sodium pyruvate (Gibco), 10% fetal bovine serum (FBS; Sigma), 1% penicillin/streptomycin (Gibco), and 250 µg/mL Geneticin (G418 sulfate; Gibco) at 37 °C and passaged 1:20 two times per week. SNAP labeling was performed using SNAP-Cell SiR647 (New England Biolabs). After titration to optimize signal-to-noise and speckle tracking (SI Appendix, Fig. S1 D and E), cells were labeled with 3 µM dye for 15 min at 37 °C in subsequent experiments.
Talin or vinculin tension and control sensors were transiently transfected into SNAP-actin NIH 3T3 cells. All plasmid expression constructs were prepped from Stbl3 cells using a Nucleobond Xtra Midi kit (Machery-Nagel). SNAP-actin 3T3 cells were plated at ∼50% confluence in six-well plates 1 d prior to transfection and transfected with Lipofectamine 2000 according to the manufacturer’s specifications. Briefly, two tubes per transfection were prepared, one with 10 µL of Lipofectamine 2000 in 250 µL of OPTI-MEM without serum (Gibco) and a second with 4 µg of the expression construct in 250 µL of OPTI-MEM. After 5-min incubation at room temperature the two tubes were mixed dropwise and incubated 20 min at room temperature prior to adding all 500 µL to a well of the six-well plate containing cells and 2 mL of OPTI-MEM with 4% FBS. Cells were incubated for 14 to 16 h and media replaced with normal culture media. Cells were used 38 to 50 h posttransfection.
The day of an experiment, cells were trypsinized and 8 × 105 cells per group were suspended in 200 µL of phenol-free DMEM with 0.1% bovine serum albumin (BSA) and 3 µM SNAP-Cell SiR647 and then incubated at 37 °C for 15 min. Cells were then spun at 300 × g for 5 min and washed three times with 400 µL of phenol-free DMEM with 0.1% BSA; 4 × 105 cells suspended in 200 µL phenol-free DMEM with Hepes and 0.1% BSA were held in suspension on BSA-coated 96-wells at 37 °C for up to 4 h prior to seeding (allowing for multiple staggered seedings). Cells were examined at 45 to 90 min after seeding on coverglass-bottomed dishes (#1.5 coverslip, 22-mm coverslip, 35-mm dish; MatTek) coated with fibronectin (10 µg/mL overnight at 4 °C in PBS). Standard medium for imaging was phenol-free DMEM with Hepes and 2% FBS. For experiments with fibronectin-blocking antibody, serum-free medium was supplemented with 10 µM lysophosphatidic acid (Sigma), 1× Insulin-Transferrin-Selenium (Gibco), and 0.1% BSA.
FRET and Speckle Image Acquisition.
Time-lapse images were acquired using a PerkinElmer UltraVIEW VoX spinning-disk confocal attached to a Nikon Eclipse Ti microscope with a 100× oil objective (numerical aperture 1.45) equipped with a temperature- and CO2-controlled environmental chamber. Fourteen-bit images were acquired for four channels (each with 0.5-s exposure) on an electron-multiplying charged-coupled device camera (C9100-50; Hamamatsu Photonics) using Volocity software (PerkinElmer). Speckle channel (siR647) was acquired at 6 to 10 frames per minute and FRET channels (eGFP, tagRFP, and FRET) were acquired at one frame per minute. Images were acquired with the following filter combinations: donor (GFP) channel with a 488-nm (excitation) and 527/55 filter (emission), acceptor (tagRFP) with a 561-nm laser (excitation) and 615/79 filter (emission), and FRET channel with a 488-nm laser (excitation ) and 615/70 filter (emission). For all FRET constructs, only cells with an average acceptor intensity within a prespecified range (800 to 5,000) were analyzed (to provide adequate signal-to-noise). Cells that failed to spread were excluded.
Quantification of Speckle Time-Lapse Images.
Quantification of speckle time-lapse images was performed using a previously developed open-source software in MATLAB (MathWorks) (4, 34). Analysis was performed on 21 to 61 frames with default settings for noise model calibration, thresholding, and mask refinement and speckle detection (for the given frame rate [6 to 10 frames per min], wavelength [720 nm], resolution [71.8 nm per pixel], and numerical aperture of the lens [1.4]). Flow tracking was performed and averaged over 1-min intervals then matched with FRET data from images taken in the middle of that interval.
Quantification of Three-Cube FRET Imaging and Calculation of FRET Efficiency and Force.
Quantification of FRET data was performed as previously described using custom software in MATLAB (MathWorks) (17). All three FRET images (eGFP [enhanced green fluorescent protein], TagRFP [red fluorescent protein], and FRET) were corrected for illumination gradient, pixel shift, and background subtraction followed by three-point smoothening. Bleed-through and cross-excitation coefficients were calculated for the laser power by imaging cells transfected with eGFP-talin or TagRFP-talin (SI Appendix, Fig. S1F). The slope of the pixelwise donor or acceptor channel intensity vs. FRET channel intensity gives donor leakage () and acceptor leakage () fractions, respectively. Heat maps of FRET and pixelwise FRET index were calculated using the following equation:
where , , and are the shade, shift, and background-corrected pixel intensities for each of the respective channels (fret, donor, and acceptor) and is the corrected FRET intensity. Since FRET index and leakage coefficients are dependent on the exact laser power and microscope settings (SI Appendix, Fig. S1F) but the percent difference relative to control sensor is not, FRET index was then normalized to control sensor to give normalized FRET index:
Pixelwise correlations of normalized FRET and actin flow rates were performed for 2- × 2-pixel areas, with flow data calculated and averaged over 30 s before and after the matched FRET data. Additionally, average normalized FRET per cell was calculated for each experiment and used for statistical comparisons between groups.
To estimate force on talin, FRET efficiency was calculated and converted to estimated forces using a previously developed method for three-cube FRET efficiency calculation (35) and a worm-like chain model for estimated forces (28). FRET efficiency () was calculated using the equation
where is the corrected FRET intensity (from above) and G is a proportionality constant that relates the increase in acceptor intensity to the decrease in donor intensity (due to sensitized emission and quenching). G was calculated using GFP–RFP sensors with two different-sized linkers (40 and 80 amino acids, 8 or 16 GPGGA repeats) and diffuse cytoplasmic signal was estimated from cells expressing similar levels of unlinked eGFP and TagRFP.
Estimated forces were calculated based on a previously described worm-like chain model for the mechanical sensitivity of tension sensor modules (28). Using this model and parameters for our sensor [eGFP-(GPGGA)8-TagRFP, Ro = 5.7 nm, Rfp = 1.15 nm, Lp = 0.5 nm, nAA = 40] allows estimation of a FRET Efficiency vs. force relationship (SI Appendix, Fig. S1H) which can then be used to convert FRET efficiency to an estimated force in piconewtons (SI Appendix, Fig. S1 I and J).
Knockdown and Western Blot.
Knockdown of talin 1 and vinculin was performed using lentivirally delivered small-guide RNAs coexpressed with Cas9 (sgNT 5′-GCGAGGTATTCGGCTCCGCG-3′; sgVCL 5′-GCCGTCAGCAACCTCGTCC-3′; sgTLN 5′-GATGTTAGACGGAACGGTGA-3′). Lentiviruses were generated by transfecting Lenti-X 293T cells (Clontech) (10-cm dishes) with packaging vectors (2.5 µg VSV; 5 µg pxPAX2; Addgene) and the pLentiCRISPR DNA vector (7.5 µg) using Lipofectamine 2000 (Invitrogen). Virus-containing supernatant was collected 36 and 60 h posttransfection. SNAP-actin 3T3 cells were infected with 1 mL of viral supernatant mixed with 4 mL of fresh media and 8 µM polybrene (Sigma) and incubated overnight. Medium was changed 12 to 16 h postinfection and puromycin (7.5 µM) was added 3 d after infection. After 4 d of selection, cells were transfected with tension sensors and used for experiments. On the day of the experiment, protein samples were taken and knockdown efficiency was assayed by Western blot.
For Western blots, cells were lysed in 2× protein sample buffer (80 mM Tris, pH 6.8, 4% SDS, 10% glycerol, and 0.0006% bromophenol blue) with protease and phosphatase inhibitor mixture (Halt 100×; Thermo) and scraped from plates on ice. Samples were sonicated 30 times at 30% power, 1 Hz, 50% duty cycle on ice; 2.5% beta-mercaptoethanol was added and samples heated for 5 min at 95 °C. Samples were spun at 5,000 rpm for 30 s in a microfuge then 20 µL was loaded onto polyacrylamide gels and run 2 h at 110 V in Tris-glycine-SDS running buffer. Transfer was performed using a Bio-Rad Trans-Blot Turbo with 30-min transfer protocol and Tris-glycine/methanol buffer on Immun-Blot poly(vinylidene difluoride) membrane (Bio-Rad). Membranes were blocked with 5% milk (Omni-Block; AmericanBio) in Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 1 h and incubated with rabbit anti-Talin 1 (1:2,500; Abcam), mouse anti-Vinculin (1:2,500; Sigma), or rabbit anti-GAPDH (1:3,000; Cell Signaling) in TBS-T at 4 °C. After washing three times for 5 min each, membranes were incubated with anti-rabbit horseradish peroxidase (HRP) or anti-mouse HRP (1:4,000; Vector Labs) for 1 h at room temperature in TBS-T. After washing, blots were developed with Super Signal West Pico chemiluminescent substrate (Thermo) using a SYNGENE G-Box imager. For analysis of total protein cross-linking (SI Appendix, Fig. S3B), gels were washed with water three times for 5 min then stained with Imperial Protein Stain (1×; Thermo Scientific) for 1 h and destained in water overnight.
Immunostaining and Antibodies.
Cells on fibronectin-coated glass or polyacrylamide gels were fixed with 4% PFA (Electron Microscopy Sciences) in PBS for 20 min at room temperature and permeabilized with 0.05% Triton X-100 in PBS supplemented with 320 mM sucrose and 6 mM MgCl2. Cells were washed three times with PBS and blocked for 30 min with 1% BSA in PBS. Cells were incubated overnight at 4 °C with primary antibodies (anti-YAP 1:200, sc-101199; Santa Cruz Biotechnology and anti-vinculin 1:200, V9131; Sigma-Aldrich), diluted in 1% BSA in PBS. Cells were washed three times with PBS and incubated at room temperature for 1 h with secondary antibodies (Alexa-647 anti-mouse, 1:1,000; Molecular Probes) or Alexa-647 conjugated phalloidin (1:1,000; Molecular Probes). Cells were washed 3× with PBS and mounted in Fluoromout-G containing DAPI (SouthernBiotech). Cell areas were quantified from phalloidin images using ImageJ by subtracting the background, thresholding to generate cell masks, and determining area using the Analyze Particles function. Ratiometric images of talin and vinculin were analyzed by masking adhesion areas of interest in imageJ and then ratioing the background subtracted, channel-shade, and channel-shift corrected images in MATLAB (MathWorks).
Perturbing Actin Flow.
To slow actin flow, a combination of Latrunculin A (600 nM; Sigma) and Jasplikinolide (1 µM; Tocris Biosciences) was added to cells. This treatment slowed flow within 90 s of addition and gave interpretable results for ∼120 s, before disruption of the actin network. For blocking new adhesion formation, cells in serum-free media with 0.1% BSA were treated with 25 µg/mL of 16G3 anti-fibronectin blocking antibody for 10 min (Hybridoma generated at University of Virginia Antibody Engineering and Technology Core). To completely arrest actin flow, cells were perfused with 0.1% PFA in normal imaging media. For these experiments, actin flow was measured continuously and averages were calculated for the 1 min before PFA (before), the interval centered at 30 s (0 to 60 s after PFA), and the interval centered at 150 s (120 to 180 s after PFA).
Polyacrylamide Substrate Preparation.
Polyacrylamide TFM and speckle microscopy substrates were fabricated as described previously (36). Briefly, 20-mm coverslip-bottomed dishes (#0 coverslip; Mattek) were silanized with a 2% solution of 3-aminopropyltrimethoxysilane in isopropanol for 10 min at room temperature. After washing with double-distilled H2O (ddH2O) and drying, coverslips were incubated with 1% glutaraldehyde solution in ddH2O for 30 min and then washed three times. Polyacrylamide gels were cast onto the silanized surface by preparing acrylamide/bis-acrylamide solutions (Bio-Rad) of various ratios (SI Appendix, Table S1) and polymerizing with ammonium persulfate (American Bio) and TEMED (Sigma). For TFM experiments, fluorescently labeled beads were mixed. Gels were cast between the silanized surface and a 12-mm uncoated glass coverslips with a volume of 8 μL. After casting, gels were treated with fresh sulfo-SANPAH (Sigma) in ddH2O (2 mg/mL) and exposed to ultraviolet (UV) light for 3 min (8 W, 254-nm wavelength at a distance of 2 to 3 in). After UV, gels were washed with ddH2O and then covered with fibronectin (200 μg/mL in PBS at pH 7.4) overnight at 4 °C. Prior to seeding, gels were washed three times with PBS and preincubated with media for 1 h.
TFM.
Prior to seeding, TFM gels were washed three times with PBS. Traction force experiments were performed in phenol-free, high-glucose DMEM (Gibco) with 2% FBS (Sigma). The day of the experiment, cells were trypsinized and seeded on TFM substrates at low density (∼3,000 cells per cm2). Cells and fluorescent beads were imaged on a spinning-disk confocal microscope (UltraVIEW VoX; PerkinElmer) attached to a Nikon A-1 microscope equipped with a temperature- and CO2-controlled incubation chamber and 60× 1.4 numerical aperture lens. Fluorescent images of Alexa Fluor 647 beads and differential interference contrast images of cells were acquired before, after 3-min treatment with 0.1% PFA, and after cell lysis with 0.2% SDS. Images were drift corrected and bead displacements were quantified using a previously developed open-source TFM software in MATLAB 2015a (37). Force fields and traction stresses were calculated using FTTC force reconstruction with regularization parameter set at 3 kPa = 0.01, 46 kPa = 0.0001. Total force per cell was calculated as the average traction stress under the cell multiplied by the cell area.
Adhesion Dynamics.
Adhesion dynamics were quantified from 60× 1-h time-lapse images in fully spread cells acquired at one frame every 3 min on a spinning-disk confocal using the RFP channel of the talin tension sensor. Analysis was performed using the FA analysis server (Gomez laboratory, University of North Carolina at Chapel Hill) (38). For FAAS the threshold was set to 2, minimum adhesion size was set to 20 pixels, minimum aspect ratio of 3, and a minimum phase length of 10. Postprocessing of tracked FA data was performed using custom scripts in MATLAB. For adhesion growth, only adhesions that achieved a size of larger than 1.5 μm2 that grew by more than 30% and were tracked for a minimum of 10 frames (30 min) were analyzed. Adhesion size was normalized to the maximum size achieved and then plotted as the normalized adhesion size. For adhesion velocity, only adhesions that achieved a size of larger than 1.5 um2 and were tracked for a minimum of 10 frames were analyzed. For lifetime measurements, only adhesions that were present at the start of the video were considered and tracked until their disappearance. Adhesions that lasted for the entire duration of the 1-h time course were reported as the percent stable adhesions.
Statistics.
All statistical analysis was performed using Prism version 7.01 (GraphPad). Details regarding the specific statistical tests used, the sample size, P values, and number of independent experiments are all included in the figure legends.
Supplementary Material
Acknowledgments
We thank Gaudenz Danuser and Sangyoon Han for their help with the QFSM software and discussions about the data. This work was supported by NIH R01GM047214 (M.A.S.) and NIH F32GM119286 (T.P.D.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.K.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2010292117/-/DCSupplemental.
Data Availability.
Quantitative fluorescence speckle microscopy (QFSM) software (34) and TFM software (37) are open source and available on GitHub (https://github.com/DanuserLab). MATLAB code and code used to prepare movie database files (for QFSM), analyze FRET, and correlate FRET with actin flow are available at https://github.com/TristanDriscoll/FRET-Speckle (39).
References
- 1.Dupont S., et al. , Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E., Geometric control of cell life and death. Science 276, 1425–1428 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Ingber D. E., Mechanical control of tissue growth: Function follows form. Proc. Natl. Acad. Sci. U.S.A. 102, 11571–11572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ponti A., Machacek M., Gupton S. L., Waterman-Storer C. M., Danuser G., Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Hu K., Ji L., Applegate K. T., Danuser G., Waterman-Storer C. M., Differential transmission of actin motion within focal adhesions. Science 315, 111–115 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Elosegui-Artola A., et al. , Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Elosegui-Artola A., Trepat X., Roca-Cusachs P., Control of mechanotransduction by molecular clutch dynamics. Trends Cell Biol. 28, 356–367 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Case L. B., Waterman C. M., Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchison T., Kirschner M., Cytoskeletal dynamics and nerve growth. Neuron 1, 761–772 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Suter D. M., Forscher P., An emerging link between cytoskeletal dynamics and cell adhesion molecules in growth cone guidance. Curr. Opin. Neurobiol. 8, 106–116 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Plotnikov S. V., Pasapera A. M., Sabass B., Waterman C. M., Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannone G., et al. , Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 128, 561–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan C. E., Odde D. J., Traction dynamics of filopodia on compliant substrates. Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Klapholz B., Brown N. H., Talin–The master of integrin adhesions. J. Cell Sci. 130, 2435–2446 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Choi C. K., et al. , Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goult B. T., Yan J., Schwartz M. A., Talin as a mechanosensitive signaling hub. J. Cell Biol. 217, 3776–3784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar A., et al. , Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J. Cell Biol. 213, 371–383 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atherton P., et al. , Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 6, 10038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bangasser B. L., Rosenfeld S. S., Odde D. J., Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment. Biophys. J. 105, 581–592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bangasser B. L., et al. , Shifting the optimal stiffness for cell migration. Nat. Commun. 8, 15313 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardel M. L., et al. , Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183, 999–1005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z., Plotnikov S. V., Moalim A. Y., Waterman C. M., Liu J., Two distinct actin networks mediate traction oscillations to confer focal adhesion mechanosensing. Biophys. J. 112, 780–794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan S. J., et al. , Regulation and dynamics of force transmission at individual cell-matrix adhesion bonds. Sci. Adv. 6, eaax0317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grashoff C., et al. , Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nobes C. D., Hall A., Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Price L. S., Leng J., Schwartz M. A., Bokoch G. M., Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9, 1863–1871 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.del Pozo M. A., Price L. S., Alderson N. B., Ren X. D., Schwartz M. A., Adhesion to the extracellular matrix regulates the coupling of the small GTPase Rac to its effector PAK. EMBO J. 19, 2008–2014 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaCroix A. S., Lynch A. D., Berginski M. E., Hoffman B. D., Tunable molecular tension sensors reveal extension-based control of vinculin loading. eLife 7, e33927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzima E., et al. , Activation of Rac1 by shear stress in endothelial cells mediates both cytoskeletal reorganization and effects on gene expression. EMBO J. 21, 6791–6800 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzima E., del Pozo M. A., Shattil S. J., Chien S., Schwartz M. A., Activation of integrins in endothelial cells by fluid shear stress mediates Rho-dependent cytoskeletal alignment. EMBO J. 20, 4639–4647 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thievessen I., et al. , Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calderwood D. A., Campbell I. D., Critchley D. R., Talins and kindlins: Partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D. L., Bax N. A., Buckley C. D., Weis W. I., Dunn A. R., Vinculin forms a directionally asymmetric catch bond with F-actin. Science 357, 703–706 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza M. C., Besson S., Danuser G., Quantitative fluorescent speckle microscopy (QFSM) to measure actin dynamics. Curr. Protoc. Cytom. 62, 2.18.1–2.18.26 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H., Puhl H. L. 3rd, Koushik S. V., Vogel S. S., Ikeda S. R., Measurement of FRET efficiency and ratio of donor to acceptor concentration in living cells. Biophys. J. 91, L39–L41 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aratyn-Schaus Y., Oakes P. W., Stricker J., Winter S. P., Gardel M. L., Preparation of complaint matrices for quantifying cellular contraction. J. Vis. Exp. 46, e2173 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han S. J., Oak Y., Groisman A., Danuser G., Traction microscopy to identify force modulation in subresolution adhesions. Nat. Methods 12, 653–656 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berginski M. E., Gomez S. M., The focal adhesion analysis server: A web tool for analyzing focal adhesion dynamics. F1000 Res. 2, 68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driscoll T., FRET-Speckle. GitHub. https://github.com/TristanDriscoll/FRET-Speckle. Accessed 20 November 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Quantitative fluorescence speckle microscopy (QFSM) software (34) and TFM software (37) are open source and available on GitHub (https://github.com/DanuserLab). MATLAB code and code used to prepare movie database files (for QFSM), analyze FRET, and correlate FRET with actin flow are available at https://github.com/TristanDriscoll/FRET-Speckle (39).