Significance
Intergenerational epigenetic inheritance is usually considered to be of minor significance in mammals because the epigenome is reprogrammed twice between each generation. Yet whether this paradigm holds in nonmammalian organisms is largely unknown. Here, we show that epigenetic marks are transferred across generations in an invertebrate, the honey bee. We found that the epigenome is much more similar between fathers and daughters than between unrelated males and females of different generations within each colony. Epigenetic marks are not only conserved across generations, but also across somatic and germline tissues. Epigenetic reprogramming may thus be a mammalian-specific feature, suggesting a heightened capacity for epigenetic marks to influence evolutionary adaptation across the tree of life.
Keywords: epigenetic inheritance, epigenetic remodeling, DNA methylation, Apis mellifera
Abstract
The evolutionary significance of epigenetic inheritance is controversial. While epigenetic marks such as DNA methylation can affect gene function and change in response to environmental conditions, their role as carriers of heritable information is often considered anecdotal. Indeed, near-complete DNA methylation reprogramming, as occurs during mammalian embryogenesis, is a major hindrance for the transmission of nongenetic information between generations. Yet it remains unclear how general DNA methylation reprogramming is across the tree of life. Here we investigate the existence of epigenetic inheritance in the honey bee. We studied whether fathers can transfer epigenetic information to their daughters through DNA methylation. We performed instrumental inseminations of queens, each with four different males, retaining half of each male’s semen for whole genome bisulfite sequencing. We then compared the methylation profile of each father’s somatic tissue and semen with the methylation profile of his daughters. We found that DNA methylation patterns were highly conserved between tissues and generations. There was a much greater similarity of methylomes within patrilines (i.e., father-daughter subfamilies) than between patrilines in each colony. Indeed, the samples’ methylomes consistently clustered by patriline within colony. Samples from the same patriline had twice as many shared methylated sites and four times fewer differentially methylated regions compared to samples from different patrilines. Our findings indicate that there is no DNA methylation reprogramming in bees and, consequently, that DNA methylation marks are stably transferred between generations. This points to a greater evolutionary potential of the epigenome in invertebrates than there is in mammals.
Epigenetic inheritance refers to a form of information transfer across generations that is not based on the DNA sequence (1). Three main mechanisms are associated with epigenetic processes and are candidates for carrying epigenetic information between generations, namely DNA methylation, histone posttranslational modifications, and small noncoding RNAs (2). DNA methylation is the most thoroughly studied epigenetic mechanism. In eukaryotes, 5-cytosine methylation in CpG dinucleotides is the most common covalent modification to DNA (3). DNA methylation involves DNA methyltransferases (DNMTs) that are responsible for both the maintenance of methylation through cell divisions and the addition of new methylation marks (4). DNA methylation profiles vary widely between eukaryotic groups; yet gene body methylation is evolutionarily conserved (5). Gene body methylation is generally associated with constitutive gene expression, genes with housekeeping functions, and is thought to prevent spurious transcription (6). This points to a homeostatic function of gene body methylation, rather than a role in regulating gene activity during development or in response to environmental change. The function of gene body methylation beyond a homeostatic role remains controversial (7).
Despite generating a lot of attention, the evolutionary significance of epigenetic inheritance remains enigmatic (1, 8, 9). In mammals, almost all DNA methylation marks are erased and reestablished twice during embryogenesis: first after fertilization and second during primordial germ cell formation (10, 11). These two waves of global epigenetic remodeling constitute a significant barrier to transfer of epigenetic information via DNA methylation between generations. As a result, uncontroversial examples of epigenetic inheritance via DNA methylation in mammals are very rare beyond the few genomic regions that are resistant to reprogramming, such as retrotransposons (12) and imprinted genes (13). Whether this pattern is universally found in nonmammalian animals is currently unclear.
In the nonmammalian vertebrate species investigated so far, there is an absence of global DNA methylation remodeling during embryogenesis (14–16). For example, zebrafish inherit the paternal DNA methylome configuration throughout development and only remodel the maternal methylome configuration (14, 16). Transfer of DNA methylation patterns across generations has been shown in zebrafish (17), suggesting that epigenetic inheritance via DNA methylation might be more prevalent in nonmammalian vertebrates (5).
In invertebrates, we know very little about the extent of DNA methylation remodeling during embryogenesis, and whether DNA methylation marks can be inherited across generations (5). Recent investigations in various invertebrate taxa, including honey bees, indicate that DNA methylation levels largely remain constant during development (18–20), suggesting no epigenetic reprogramming. Further, interspecific crosses in Nasonia wasps revealed a stable inheritance of DNA methylation marks in F1 hybrids (21), suggesting the existence of intergenerational inheritance of DNA methylation patterns in insects. While these results are significant, more data are needed to make general conclusions about the general patterns of epigenetic remodeling and inheritance in invertebrates.
An important factor to consider when studying epigenetic inheritance is the existence of genetic variation among samples (8). DNA methylation states are often influenced by the underlying DNA sequence (22, 23). This phenomenon is particularly prevalent in insects (21, 24–27) and can be a serious confounding factor when not controlled by appropriate experimental design (28).
Here, we investigate whether DNA methylation marks are heritable in the honey bee. Due to their colonial organization, their polyandrous mating system, and well-developed technology for artificial insemination, honey bees are a good model to study the functional and evolutionary significance of DNA methylation in invertebrates, especially given the genome-wide depletion of DNA methylation in classical invertebrate models like Drosophila (29, 30). DNA methylation marks are relatively sparse in honey bees and are mainly restricted to gene bodies (3). This feature is common to most invertebrates (5) and greatly facilitates downstream analyses.
A honey bee colony is comprised of thousands of genetically heterogeneous female workers and a single queen who is a mother to all of the workers, all living in a homogeneous environment. The queen mates with many different males early in her life, storing the sperm from each. She uses the sperm of each male to produce worker offspring, each male fathering a distinct worker subfamily (i.e., patriline). Because males are haploid, all their sperm are genetically identical, so workers within a patriline share 100% of their paternal genome (31). This, combined with the availability of instrumental insemination procedures (32), allowed us to design an experiment where the methylomes of fathers and daughter workers (from the same patriline) were compared with the methylomes of unrelated males and workers (from different patrilines) within the same colony. This gave us the unique opportunity to investigate the inheritance of DNA methylation marks in an invertebrate while keeping the effects of genetic and environmental variability to a minimum.
Results
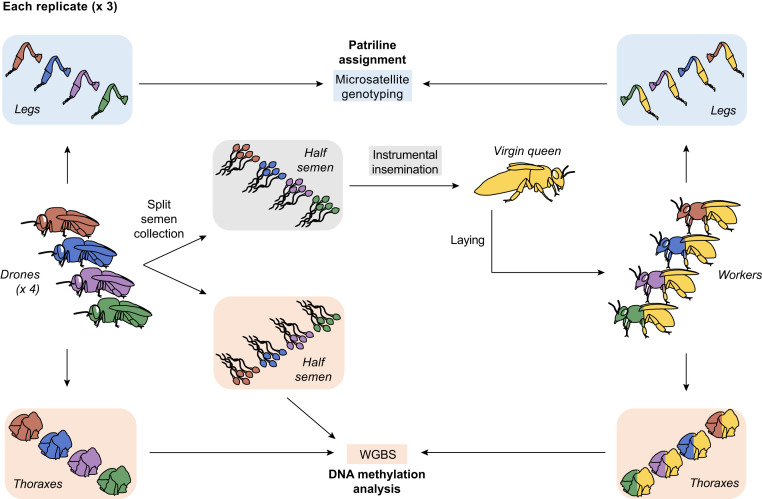
We investigated the transmission of DNA methylation profiles between fathers and daughters by instrumentally inseminating three queens, each with one half of the semen of each of four drones (i.e., males). We then performed whole genome bisulfite sequencing (WGBS) of the remaining half of each drone’s semen (hereafter, “semen”), the thorax of each drone (hereafter, “drone”), and a pool of daughter worker thoraxes from each patriline (hereafter, “worker”; Fig. 1). This allowed us to determine, within each colony, whether DNA methylation profiles were conserved across generations between fathers and daughters by teasing apart the effects of patriline and social environment on DNA methylation states.
Fig. 1.
Experimental design. For each replicate (×3), we collected the semen from four drones and split each semen into two equal parts. We performed instrumental insemination of virgin queens each with half the semen of four drones, before allowing each queen to lay and produce worker offspring. We collected the hind legs of each drone and worker and performed microsatellite genotyping to assign them to their respective patriline. We performed WGBS of each drone’s half semen and thorax, as well as a pool of 20 worker thoraxes from each patriline to analyze their DNA methylation profiles.
The sequencing output yielded a total of 1,263.6 million reads (318.43 Gb) with 27.67 ± 1.48-fold genome coverage across 30 methylomes from three different colonies (SI Appendix, Table S1). There were 6.37 ± 0.17 million CpGs sufficiently covered across all samples, of which 54.80 ± 1.61 thousand sites were significantly methylated (mCpGs; SI Appendix, Table S1). The distribution of mCpGs in various genomic regions was very similar across tissues and colonies. Most mCpGs were located within genes. Most gene body mCpGs were located within exons (SI Appendix, Fig. S1 A–C). These patterns follow what is typically observed in Hymenopteran insects (33).
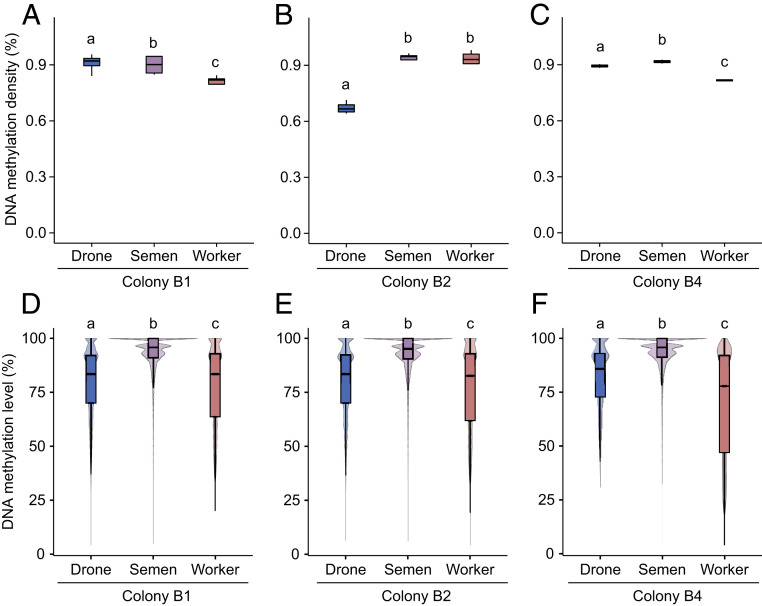
We first compared the methylation density (i.e., the ratio of mCpGs to CpGs across the genome) of drone, semen, and worker samples to test whether there were any global changes to the extent of DNA methylation between tissues and generations. Overall, the methylation density across tissues was just under 1% (Fig. 2 A–C), as is typically observed in honey bees (3, 20). Tissues had significantly different methylation densities; yet this effect was colony dependent (generalized linear mixed effects models [GLMMs], all P < 0.00001; Fig. 2 A–C). Worker samples had a lower methylation density than semen and drone samples. The lower levels of methylation density of drones in colony B2 are likely an artifact of lower coverage (SI Appendix, Table S1).
Fig. 2.
Patterns of DNA methylation in drones, semen, and workers. (A–C) DNA methylation density (% of mCpGs out of all CpGs) across all patrilines for drone, semen, and worker samples in colony B1 (A), B2 (B), and B4 (C). Box plots represent median, interquartile range (IQR), and 1.5 × IQR. (D–F) DNA methylation level [% of C out of (C + T) at each CpG site] of mCpGs across all patrilines for drone, semen, and worker samples in colony B1 (D), B2 (E), and B4 (F). Violin plots represent median, interquartile range (IQR), 1.5 × IQR, and kernel density plot. Different lowercase letters represent significant differences within each colony (GLMMs, all P < 0.0003).
We next compared the methylation level (i.e., ratio of C to [C + T] reads at each CpG) of all samples. Across all colonies, the methylation level of semen samples was the highest, while worker samples had the lowest level of methylation (GLMMs, all P < 0.00001; Fig. 2 D–F). This pattern was similar across exons, introns, and intergenic regions (SI Appendix, Fig. S1 D–F).
Global DNA methylation reprogramming could have occurred transiently during early embryogenesis (i.e., egg or young larval stage), resulting in similar genome-wide DNA methylation densities, but targeting different sites. To rule out this possibility, we directly compared the similarity of DNA methylation patterns between samples both within and between different patrilines in each colony, reasoning that, all else being equal, there should be a greater similarity of methylomes within patrilines than between patrilines if there is intergenerational inheritance of DNA methylation patterns.
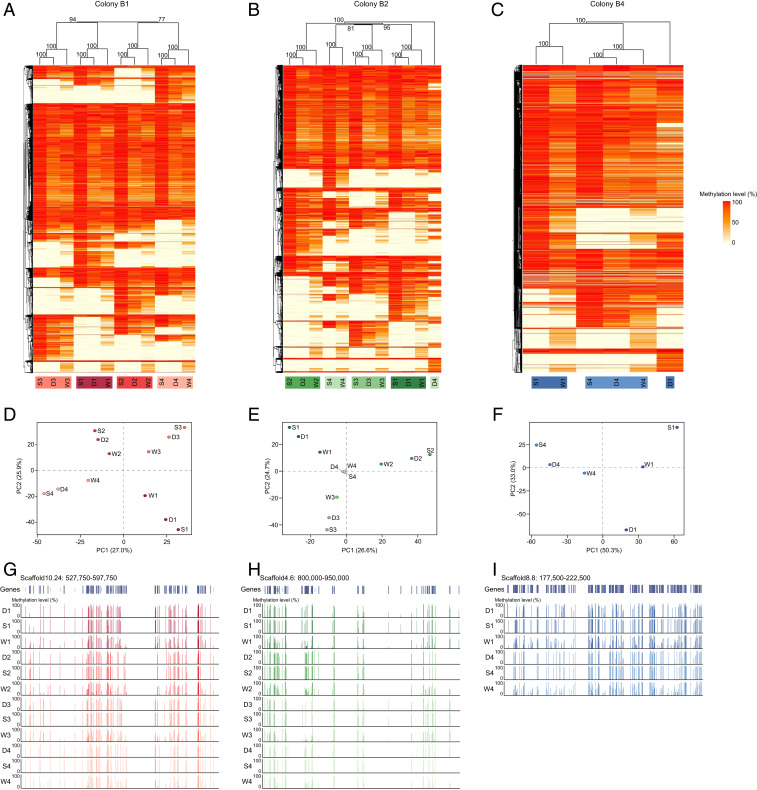
We performed a hierarchical clustering based on the methylation levels of all shared mCpGs across all semen, drone, and worker samples within each colony to test whether their methylomes clustered by patriline, as opposed to by caste, tissue, or randomly. Across all three colonies, there was a very clear clustering of methylation patterns by patriline, which was strongly supported statistically (Fig. 3). As expected, drone and semen samples of each patriline clustered in most cases, as these samples originated from the same individual. Yet, drone, semen, and daughter worker samples of each patriline also clustered in 8 of 10 patrilines (Fig. 3 A–C). Further, in the other 2 patrilines, there was still a clustering of semen and worker samples by patriline (Fig. 3 B and C). Principal component analyses (PCAs) also showed that samples from each patriline were well separated from all other samples along the first two principal components (Fig. 3 D–F). Visual inspection of DNA methylation patterns revealed clear patriline-specific gene body methylation in each colony (Fig. 3 G–I).
Fig. 3.
Clustering of samples according to their methylation status in each colony. (A–C) Hierarchical clustering (average agglomerative method on correlation distances) of methylation status of all shared mCpGs across all patrilines for drone (D), semen (S), and worker (W) samples in colony B1 (A), B2 (B), and B4 (C). Bootstrap support values (%) are depicted above each node. Heatmaps show the level of DNA methylation for each site (ranging from 0%, yellow to 100%, red). (D–F) PCA of DNA methylation level of all shared mCpGs across all patrilines for drone (D), semen (S), and worker (W) samples in colony B1 (D), B2 (E), and B4 (F). Percentage of variance explained is depicted on each axis. (G–I) Exemplar genome browser snapshots showing gene body methylation patterns in each sample in colony B1 (G), B2 (H), and B4 (I). Vertical lines show the level of DNA methylation for each CpG. Different colors represent different patrilines.
We repeated the analysis using the samples from all three colonies together. There was again a very clear clustering by patriline, with semen and worker samples of each patriline clustering for all 10 patrilines, and drone, semen, and worker samples of each patriline clustering for 8 of the 10 patrilines (SI Appendix, Fig. S2). These results thus show that patriline has a much stronger effect on methylomes than tissue, caste, or colony. DNA methylation patterns were positively correlated between all samples (all Pearson’s r > 0.52, all P < 0.00001; SI Appendix, Fig. S3), with a higher correlation within patrilines (r = 0.85 ± 0.02) than between patrilines (r = 0.63 ± 0.01) and between colonies (r = 0.64 ± 0.01).
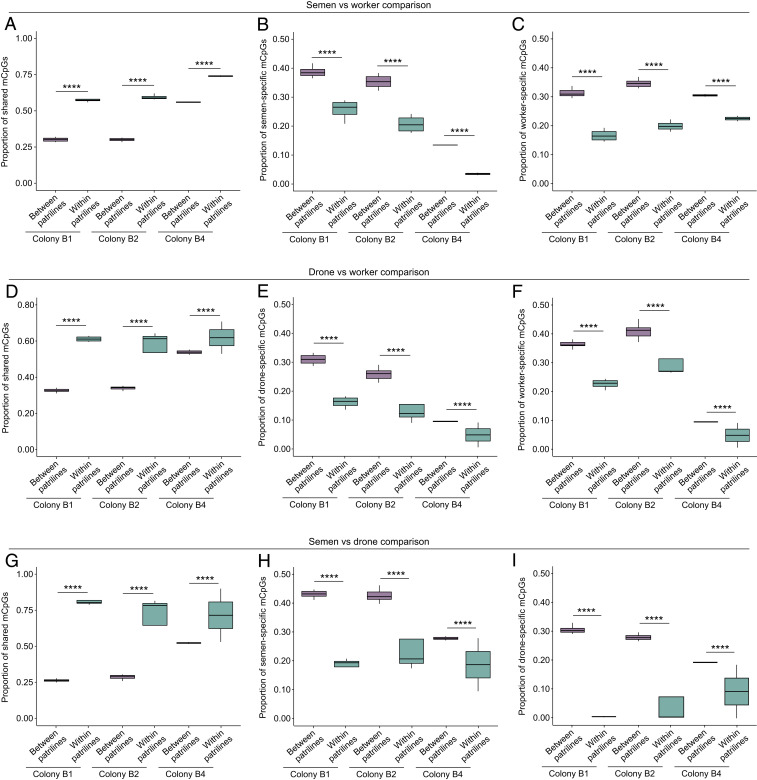
To confirm these findings, we compared the similarity of methylomes within and between patrilines. The proportion of shared mCpGs out of the total number of shared CpGs was significantly higher within patrilines than between patrilines for every pairwise comparison (i.e., semen vs. workers; drones vs. workers; and semen vs. drones) in each colony (GLMMs, all P < 0.00001; Fig. 4 A–C). By contrast, the proportion of sample-specific mCpGs was significantly lower within patrilines than between patrilines for each sample pairwise comparison in each colony (all P < 0.00001; Fig. 4 D–I). The same pattern held true at the gene level. The proportion of shared methylated genes (MGs) out of the total number of shared genes was significantly higher within patrilines than between patrilines for each pairwise sample comparison in each colony (GLMMs, all P < 0.0026; SI Appendix, Fig. S4 A–C). The proportion of sample-specific MGs was lower within patrilines than between patrilines for each pairwise sample comparison in each colony, although this effect was not always statistically significant (SI Appendix, Fig. S4 D–I). Considering all shared mCpGs, the proportion of sites with an identical methylation status in both fathers and daughters was 81.0 ± 0.4% in colony B1, 81.4 ± 0.3% in colony B2, and 81.7 ± 0.3% in colony B4. We estimate that the proportion of mCpGs being specifically inherited from fathers to daughters was 27.5 ± 1.3% in colony B1, 29.4 ± 1.1% in colony B2, and 18.0 ± 0.1% in colony B4.
Fig. 4.
Overlap of mCpGs within and between patrilines in colonies B1, B2, and B4. (A–C) Semen vs. worker comparison showing the proportion of (A) shared mCpGs, (B) semen-specific mCpGs, and (C) worker-specific mCpGs. (D–F) Drone vs. worker comparison showing the proportion of (D) shared mCpGs, (E) drone-specific mCpGs, and (F) worker-specific mCpGs. (G–I) Semen vs. drone comparison showing the proportion of (G) shared mCpGs, (H) semen-specific mCpGs, and (I) drone-specific mCpGs. Box plots represent median, IQR, and 1.5 × IQR. GLMMs: ****P < 0.0001.
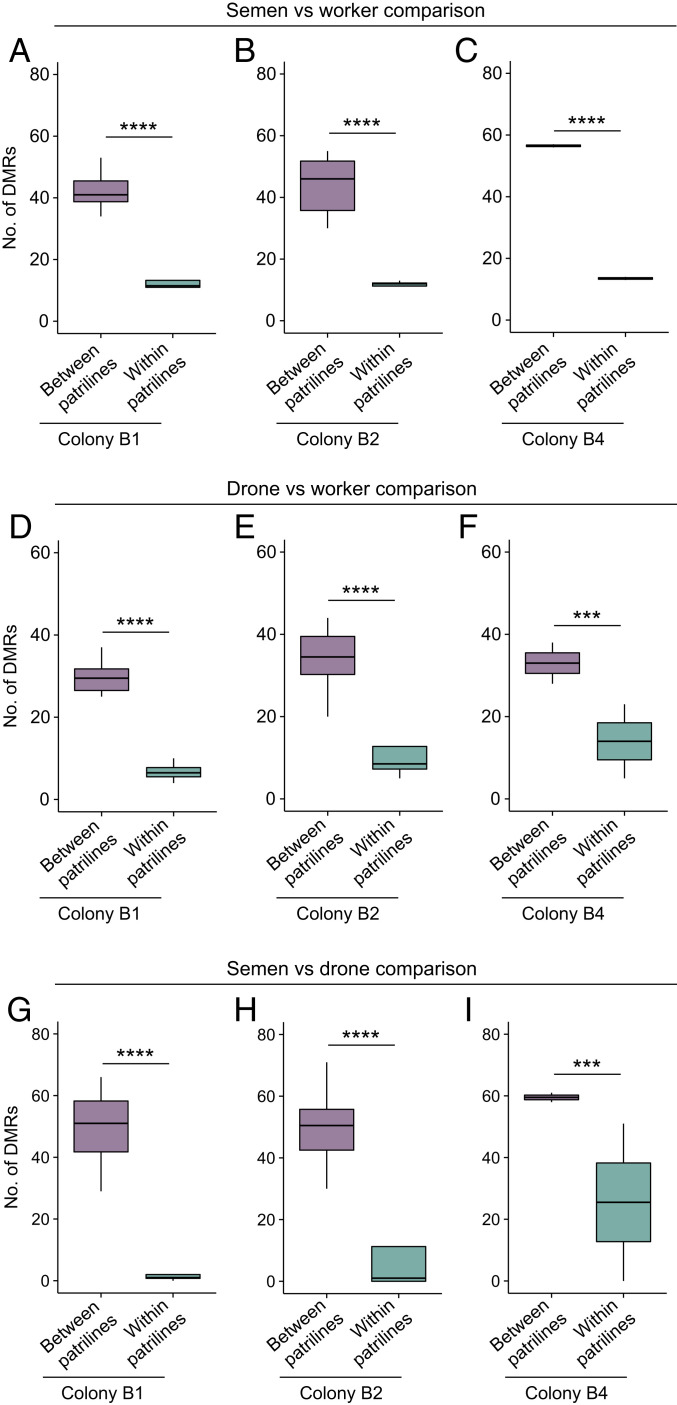
Finally, we compared the extent of differential methylation within and between patrilines. There was a significantly lower number of differentially methylated regions (DMRs) within patrilines than between patrilines for each sample pairwise comparison in each colony (GLMMs, all P < 0.00078; Fig. 5 and SI Appendix, Figs. S5–S13). Likewise, the number of differentially methylated genes (DMGs) was significantly lower within patrilines than between patrilines for each sample pairwise comparison in each colony (GLMMs, all P < 0.0018; SI Appendix, Fig. S14). Thus, the similarity of methylomes was consistently greater within patrilines than between patrilines.
Fig. 5.
DMRs within and between patrilines. (A–C) Number of DMRs between semen and workers from different patrilines and from the same patriline in colony B1 (A), B2 (B), and B4 (C). (D–F) Number of DMRs between drones and workers from different patrilines and from the same patriline in colony B1 (D), B2 (E), and B4 (F). (G–I) Number of DMRs between semen and drones from different patrilines and from the same patriline in colony B1 (G), B2 (H), and B4 (I). Box plots represent median, IQR, and 1.5 × IQR. ***P < 0.001; ****P < 0.0001.
Out of a total of 345 DMGs found across all sample pairwise comparisons, only 79 DMGs (22.9%) were present in every colony, whereas 175 DMGs (50.7%) were colony specific. Within each colony, most DMGs differed specifically between patrilines (respectively 80.3%, 66.7%, and 47.8% in colonies B1, B2, and B4), while almost none differed specifically within patrilines (respectively 0.4%, 0.4%, and 6.0%). A relatively minor subset of DMGs showed differential methylation patterns both within and between patrilines (respectively 19.2%, 32.9%, and 46.3%). Thus, DMGs had patriline-specific methylation patterns overall. Gene ontology analyses did not reveal any significant enrichment for any particular molecular function or biological process across all sets of DMGs.
Discussion
In this study we set out to investigate whether DNA methylation profiles are transferred between generations in honey bees. After controlling for genetic and environmental effects, we found that the similarity of methylomes is much greater within patrilines than it is between patrilines, that is, between fathers and daughters as opposed to between unrelated males and workers from the same colony. There were no major differences in the density of DNA methylation between tissues and generations. Samples from the same patriline, particularly semen and worker samples, consistently clustered with respect to their DNA methylation patterns. These samples shared twice as many methylated sites and had four times fewer differentially methylated regions compared to samples from different patrilines. Our results thus confirm that there is no DNA methylation reprogramming during embryogenesis in the honey bee and clearly show that there is an intergenerational transfer of DNA methylation marks between fathers and daughters in this invertebrate.
DNA methylation states are often directly influenced by the underlying DNA sequence in honey bees (26, 27) and other insects (21, 25). A clear prediction from these studies is that DNA methylation marks should be conserved across generations, yet direct evidence for this has been missing. We corroborate these findings by showing that DNA methylation patterns are much more strongly influenced by genotype than by tissue, caste, or colony. Previous studies used artificial interspecific crosses to compare cis-mediated methylation states between parental and hybrid offspring (21). Here we directly compared genome-wide parental, gamete, and offspring methylation states to provide the missing piece to the puzzle by showing unequivocally that DNA methylation marks are indeed transferred across generations in the honey bee.
Our results also corroborate recent studies reporting no remodeling of DNA methylation during honey bee embryogenesis (18, 20). It must be pointed out that these studies did not fully control for environmental and genetic effects, but rather looked at patterns of DNA methylation across different developmental stages. By contrast, our experimental design compared samples within patrilines and between patrilines in several colonies, allowing us to tease apart any sample-, genetic-, or condition-specific effects and to specifically identify the transfer of DNA methylation marks across generations.
Absence of DNA methylation remodeling during embryogenesis has been observed in all invertebrates investigated thus far: sponges, ctenophores, cnidarians, insects, sea urchins, and sea squirts (18–20). Nonmammalian vertebrates, as exemplified by fish and frogs (14–16), also appear to lack any global DNA methylation reprogramming (5). By contrast, the near-complete erasure and remodeling of DNA methylation marks during embryogenesis is seen across mammals (10, 11). This inventory is clearly incomplete, and more data are needed across metazoans before drawing final conclusions (5, 34). Yet, the emerging picture is that mammals are the exception rather than the rule when it comes to resetting of DNA methylation patterns during embryogenesis. Several potential mechanisms have been suggested to explain the existence of DNA methylation reprogramming in mammals (35). For example, DNA methylation plays a key role in genomic imprinting (13). Genomic imprinting in animals is thought to be an evolutionary innovation of mammals, which could have driven the evolution of DNA methylation reprogramming in this clade (36). Likewise, X-chromosome inactivation, another mammal-specific feature that is dependent on DNA methylation (37), could also explain why DNA methylation reprogramming is required in mammals.
The methylation level was higher in semen samples than in somatic tissues, indicating a greater fidelity in the maintenance of DNA methylation patterns in the germline, in accordance with previous findings (20, 26). Yet, the high similarity of methylomes between semen and drone samples from the same genetic background (i.e., originating from the same individual) suggests a passive, rather than an active (i.e., involving TET enzymes) (38) demethylation in the soma, resulting from loss of methylation by dilution after millions of cell divisions. It is also possible that some changes in DNA methylation patterns can accumulate as a consequence of a drone’s ontogeny. The greater heterogeneity of DNA methylation patterns observed in worker samples likely results from their greater genetic heterogeneity. Each worker sample was a pool of 20 diploid individuals, whereas each semen and drone sample was a single haploid individual. Further, worker samples also probably inherited DNA methylation marks from their mother queen, some with a different methylation status than the ones inherited from the workers’ father. Indeed, there is no reason to expect any more erasure of DNA methylation marks from the female germline than the male germline. Honey bee eggs carry DNA methylation marks (27, 39). Moreover, DNA methylation patterns from both parents were found in the F1 generation of crosses between two species of Nasonia wasps (21). Thus we suggest that the inheritance of DNA methylation marks from both parents is a general phenomenon in honey bees.
Another consequence of the stability of methylomes through both meiosis and mitosis is the similarity of DNA methylation patterns between different tissues, exemplified here by semen and thorax. This appears to be true across invertebrates (18, 40, 41), arguing against a significant role for DNA methylation in generating cell and tissue identity during development in this lineage. Invertebrates therefore differ strongly from mammals, where methylation plays a fundamental role in tissue differentiation and development (42).
What are the evolutionary consequences of our findings? We have demonstrated the existence of intergenerational (i.e., parent to offspring) epigenetic inheritance in the honey bee. Given that there is no apparent remodeling of DNA methylation marks during embryogenesis (18, 20), it is highly likely that DNA methylation patterns in honey bees are transmitted across several generations, and therefore represent a case of true transgenerational epigenetic inheritance (1, 9). Transgenerational transfer of epigenetic signals has been very clearly demonstrated in other clades, particularly in plants (43) and nematodes (44). Across species, there should be a direct negative correlation between the extent of epigenetic reprogramming during embryogenesis and the propensity to faithfully retain epigenetic marks across several generations (8, 9). We thus suspect that examples of transgenerational epigenetic inheritance in invertebrates and possibly nonmammalian vertebrates will accumulate with future studies. Yet, whether such processes are adaptive or able to influence macroevolution remains unclear (1, 8, 9, 45). Demonstrating the transfer of epigenetic marks across generations is a first necessary step to understanding the role of heritable methylation patterns in the life history and macroevolution of honey bees. For example, there is the potential for epialleles, i.e., genes that have been modified by methylation, to become targets of selection in the same way as changes to sequence. The capacity of epialleles to generate phenotypic variation in complex traits is well established in plants (46), but as far as we are aware, is unknown in insects. It is thus more than ever necessary to uncover the molecular consequences of DNA methylation if we are to understand its evolutionary implications.
Material and Methods
Instrumental Insemination Procedure.
Apis mellifera drones were collected at a natural mating lek (drone congregation area) (47) located at the University of Sydney in September 2017. Sexually mature drones were lured inside a Williams drone trap (48), a net containing baits impregnated with artificial queen pheromone (E)-9-oxodec-2-enoic acid (9-ODA) and suspended from a helium balloon. Drones were immediately transferred to the laboratory to be used in artificial insemination. Following eversion of the endophallus, we collected the semen from each drone’s ejaculate into a glass insemination tip (32). Each ejaculate was then split into two equal parts (∼0.5 μL each). The first half to be used for insemination was transferred to an Eppendorf tube containing saline diluent to prevent desiccation (32), while the other half was immediately stored at −80 °C for later sequencing. Each drone’s body was also stored at −80 °C for later microsatellite genotyping.
We then pooled the semen from the four drones into a single insemination tip, gently mixing the ejaculates as they were drawn into the tip in order to maximize equal representation of each drone in the worker offspring. The pooled semen was used to inseminate a virgin queen of standard Australian commercial stock (mainly Apis mellifera ligustica) (32). We marked the inseminated queen with a numbered tag (Opalith Plättchen) and introduced her into a new colony, standardized for strength to four frames of brood and workers. This protocol was repeated for three different queens (Fig. 1). Visual inspections confirmed that each queen had started to lay 1 wk later.
Worker Collection.
Around 2 mo postinsemination, we removed combs of emerging worker brood from each colony and placed them in separate cages in a 35 °C incubator overnight to control the age of the workers. We marked 353.7 ± 108.5 (mean ± SE) workers, <24 h old, from each colony in 1- to 3-age cohorts with color paint marks (Posca Paint Pens, Mitsubishi Pencil Co.) that uniquely identified their age and colony of origin. All marked workers from each donor colony were then introduced into a different host colony (colonies B1, B2, and B4). Each host colony consisted of a naturally mated queen and four frames of brood and workers. Fourteen days later, we collected as many marked workers as possible, ensuring that they had the correct color (to avoid collecting drifted bees). Waiting 14 d allowed us to investigate long-lasting similarities in DNA methylation profiles between fathers and daughters. We collected 176.7 ± 58.1 marked workers from each colony on dry ice and stored them at −80 °C for later genotyping and sequencing (Fig. 1).
Genotyping.
To assign each worker into her respective patriline, we extracted DNA from one hind leg of all drones used for inseminations and all marked workers using the Chelex method (49). DNA was amplified at five polymorphic microsatellite loci, A8, A24, A29, A79, and B124 (50), using standard PCR conditions (51). PCR products were analyzed on a 3130XL Genetic Analyzer (Applied Biosystems) and fragment length was scored using GeneMapper software 3.7 (Applied Biosystems). We grouped workers sharing the same paternal alleles as a fathering drone into the same patriline (Dataset S1).
Methylation states can be influenced by the local DNA sequence (allele-specific methylation) (26). We grouped workers by patriline to reduce the effect of cryptic genetic variability to a minimum. Within each worker patriline, there was a combination of three alleles, one coming from the father and common to all workers and two coming from the mother, each present in 50%, on average of the workers. To further minimize biases arising from unequal representation of maternal alleles in the worker offspring, we only used patrilines that contained at least 20 workers to be used for next-generation sequencing. All four worker patrilines could be used in colonies B1 and B2, whereas only two patrilines (1, 4) could be used in colony B4 (Dataset S1).
Whole Genome Bisulfite Sequencing.
To evaluate the conservation of DNA methylation patterns between fathers and daughters, we sequenced each drone’s thorax and half semen, as well as a pool of 20 worker thoraxes from each patriline. We chose thoraxes from both fathers and daughters to control for possible tissue differences.
We isolated DNA using a standard phenol/chloroform/isoamyl alcohol extraction protocol (52). We measured the concentration of DNA on a Qubit fluorometer (Life Technologies) and added unmethylated lambda phage DNA (0.1% wt/wt, Promega) as a spike-in control to assess bisulfite conversion efficiency. Library preparation was performed using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England BioLabs) following the manufacturer’s instructions. Briefly, we sheared DNA to 250-bp target fragments on a Covaris E220 sonicator (20% duty factor, 50 cycles/bursts, 18-W peak incident power, 80-s duration). DNA fragments were then end repaired, A tailed, and ligated with NEBNext methylated adapter oligos for Illumina (New England BioLabs). Bisulfite conversion was performed using the EZ DNA Methylation Direct Kit (Zymo Research) according to the manufacturer’s instructions. We amplified converted DNA fragments for seven cycles with KAPA HiFi HotStart Uracil+ Readymix (Roche). Libraries were cleaned with Agencourt AMPure XP beads (Beckman Coulter). We controlled the size distribution and molarity of the libraries on a TapeStation (Agilent Technologies) and quantified library concentration on a Qubit fluorometer (Life Technologies). A total of 30 methylomes (i.e., 4 × drone’s thorax + 4 × drone’s semen + 4 × workers’ thorax pool for colonies B1 and B2 [total 12 methylomes per colony]; 2 × drone’s thorax + 2 × drone’s semen + 2 × workers’ thorax pool for colony B4 [6 methylomes]) were sequenced at the Australian Genome Research Facility (Melbourne, Australia) on HiSeq2500 system (Illumina) using eight lanes of 125-bp paired-end sequencing. We have deposited WGBS data for the 10 drones’ thorax samples, the 10 drones’ semen samples, and the 10 workers’ thorax pool samples to the National Center for Biotechnology Information (NCBI) Sequence Read Archive under accession no. PRJNA623232 (53).
Methylation Analysis.
We used FastQC 0.11.15 (54) to check the quality of the reads. We trimmed adapter sequences and removed low-quality reads (Phred score <30) and short reads (length <36 bp) with TrimGalore 0.4.1 (55). The remaining reads were mapped to the honey bee reference genome assembly Amel_4.5 (56) using Bismark 0.16.1 (57) with Bowtie2 2.2.9 (58). We excluded reads that fell within unplaced and nonnuclear regions. Duplicated reads were removed with Bismark and the methylation status of each cytosine was determined using BWASP (59). Bisulfite treatment selectively converts nonmethylated cytosines into thymines, but leaves methylated cytosines unaltered. The methylation level at each site was thus calculated as the proportion of C to (C + T) reads (60). Strands were merged for each CpG site (61). Sites were deemed sufficiently covered for subsequent analyses if they had at least 10 reads (26). We determined the methylation status for each site using a binomial test with the bisulfite conversion rate for each sample as the probability of success using BWASP (62). Sites were deemed significantly methylated (hereafter mCpG) if they had a Bonferroni-corrected P value <0.01. We determined the methylation density for each sample as the proportion of mCpGs relative to the total number of sufficiently covered CpG sites. We annotated genomic features with HOMER 4.9.1 (63) using the A. mellifera official gene set amel_OGSv3.2 (64). We calculated the methylation level for each gene as the average methylation level for all sufficiently covered CpG sites across that particular gene. We only considered genes with at least 10 sufficiently covered sites. Genes having a methylation level of at least 5% were deemed as being methylated (26, 61).
We investigated the inheritance of DNA methylation patterns by comparing the methylomes of drones, semen, and workers using four complementary analyses. First, we compared the methylation density (mCpGs/CpGs across the genome) and the methylation level [C/(C + T) at each CpG site] for drones, semen, and workers across all patrilines for each colony. This analysis was aimed at identifying global changes in DNA methylation between tissues and generations. We used GLMMs with a binomial error distribution and a logit-link function using the package lme4 (65) in R 3.3.3 (66). We included patriline as a random factor in the models and corrected P values for multiple comparisons following the Benjamini and Hochberg procedure (67).
Second, we tested whether the methylomes of all samples clustered by patriline as opposed to by caste, by tissue, or randomly. We performed a hierarchical clustering of DNA methylation levels of all shared mCpGs across all semen, drone, and worker samples using the average agglomerative method based on correlation distances (26, 61) and computed heatmaps using the R package ComplexHeatmap (68). Bootstrap resampling probability was used to estimate the statistical support of the heatmap clustering (69) using the R package pvclust (70). We restricted the analysis to those CpGs that were methylated in at least one sample while being sufficiently covered across all samples. These results were also visualized using PCAs with the R package FactoMineR (71). This analysis was conducted separately for each colony and for all three colonies combined. We calculated Pearson’s correlation of DNA methylation levels of all shared mCpGs between all pairwise sample comparisons and computed correlograms using the R package corrplot (72).
Third, we compared the overlap of mCpGs and MGs within and between patrilines for each colony between each sample pairwise comparison (i.e., semen vs. workers; drones vs. workers, and semen vs. drones), as well as the extent of sample-specific mCpGs and MGs within and between patrilines for each colony. For example, in colony B1, the methylome of semen from patriline 1 was compared against the methylome of workers from patriline 1 (within-patriline comparison), and then against the methylome of workers from patrilines 2, 3, and 4 (between-patriline comparisons). This analysis aimed to test whether there was a greater similarity of methylomes (i.e., more shared mCpGs and MGs and less sample-specific mCpGs and MGs) within patrilines than between patrilines. To control for the likely influence of the queens’ alleles on the workers’ methylomes, we performed pairwise comparisons within each colony separately. We restricted the analysis to all CpGs and genes that were methylated in at least one sample while being sufficiently covered across all samples. We further removed from the analysis all CpGs (genes) that were consistently methylated across all samples, as these sites (genes) are not informative. We then compared the proportion of shared and sample-specific mCpGs (MGs) out of the total amount of shared CpGs (genes) within and between patrilines using binomial GLMMs in R as described above.
We used the above lists of shared mCpGs within and between patrilines to estimate the intergenerational inheritance of mCpGs between semen and worker samples. For each semen sample, we calculated the average proportion of shared mCpGs across all between-patriline semen-worker pairwise comparisons. This value represents the average proportion of mCpGs that a particular semen sample is expected to share with workers regardless of their relatedness. We then subtracted this value from the proportion of shared mCpGs between that same semen sample and the worker sample from its own patriline to calculate an estimate of how heritable mCpGs are between fathers and daughters.
Fourth, we investigated the extent of differential methylation (i.e., the number of DMRs and DMGs) within and between patrilines for each colony between each pairwise sample comparison (i.e., semen vs. workers, drones vs. workers, and semen vs. drones). This analysis aimed to determine whether there was greater similarity of methylomes (i.e., fewer DMRs and DMGs) within patrilines than between patrilines. We used the R package methylKit (73) to determine DMRs between each pairwise sample comparison within and between each patriline. We used a sliding window approach (200-bp windows, 100-bp step size). DMRs were defined as each window with a methylation difference of 15% or greater between the two samples, and a q-value (Fisher’s exact test corrected P value) (74) of 0.01 or less (27). DMGs were defined as any gene intersecting with at least one DMR. We compared the number of DMRs and DMGs found within patrilines and between patrilines using GLMMs with a Poisson error distribution and a log-link function using the R package lme4. We included patriline as a random factor in the models. We performed gene ontology analyses using DAVID (75).
Supplementary Material
Acknowledgments
We thank Joey Lai for his assistance with library preparation. This work was supported by the Hermon Slade Foundation (HSF1801 to B.P.O., B.Y., and E.J.R.) and the Australian Research Council (DP150100151 to B.P.O. and Alyson Ashe, and DP1801011696 to B.P.O. and Amro Zayed). Library preparation for WGBS was performed at the Westmead Scientific Platforms (Sydney, Australia), which are supported by the Westmead Research Hub, the Westmead Institute for Medical Research, the Cancer Institute New South Wales, the National Health and Medical Research Council, and the Ian Potter Foundation. Three anonymous reviewers provided helpful comments on the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017094117/-/DCSupplemental.
Data Availability.
Whole genome bisulfite sequencing data have been deposited in NCBI Sequence Read Archive (PRJNA623232) (53).
References
- 1.Perez M. F., Lehner B., Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 21, 143–151 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Skvortsova K., Iovino N., Bogdanović O., Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 19, 774–790 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Zemach A., McDaniel I. E., Silva P., Zilberman D., Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Lyko F., The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81–92 (2018). [DOI] [PubMed] [Google Scholar]
- 5.de Mendoza A., Lister R., Bogdanovic O., Evolution of DNA methylome diversity in eukaryotes. J. Mol. Biol. 432, 1687–1705 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Neri F., et al. , Intragenic DNA methylation prevents spurious transcription initiation. Nature 543, 72–77 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Zilberman D., An evolutionary case for functional gene body methylation in plants and animals. Genome Biol. 18, 87 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heard E., Martienssen R. A., Transgenerational epigenetic inheritance: Myths and mechanisms. Cell 157, 95–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radford E. J., Exploring the extent and scope of epigenetic inheritance. Nat. Rev. Endocrinol. 14, 345–355 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Morgan H. D., Santos F., Green K., Dean W., Reik W., Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14, R47–R58 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Lee H. J., Hore T. A., Reik W., Reprogramming the methylome: Erasing memory and creating diversity. Cell Stem Cell 14, 710–719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lane N., et al. , Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Reik W., Walter J., Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2, 21–32 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Skvortsova K., et al. , Retention of paternal DNA methylome in the developing zebrafish germline. Nat. Commun. 10, 3054 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogdanovic O., et al. , Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Res. 21, 1313–1327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ortega-Recalde O., Day R. C., Gemmell N. J., Hore T. A., Zebrafish preserve global germline DNA methylation while sex-linked rDNA is amplified and demethylated during feminisation. Nat. Commun. 10, 3053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamstra J. H., Sales L. B., Aleström P., Legler J., Differential DNA methylation at conserved non-genic elements and evidence for transgenerational inheritance following developmental exposure to mono(2-ethylhexyl) phthalate and 5-azacytidine in zebrafish. Epigenetics Chromatin 10, 20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu X., et al. , Evolutionary transition between invertebrates and vertebrates via methylation reprogramming in embryogenesis. Natl. Sci. Rev. 6, 993–1003 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Mendoza A., et al. , Convergent evolution of a vertebrate-like methylome in a marine sponge. Nat. Ecol. Evol. 3, 1464–1473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris K. D., Lloyd J. P. B., Domb K., Zilberman D., Zemach A., DNA methylation is maintained with high fidelity in the honey bee germline and exhibits global non-functional fluctuations during somatic development. Epigenetics Chromatin 12, 62 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Werren J. H., Clark A. G., Allele-specific transcriptome and methylome analysis reveals stable inheritance and cis-regulation of DNA methylation in Nasonia. PLoS Biol. 14, e1002500 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie W., et al. , Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148, 816–831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shoemaker R., Deng J., Wang W., Zhang K., Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 20, 883–889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X., et al. , Lineage and parent-of-origin effects in DNA methylation of honey bees (Apis mellifera) revealed by reciprocal crosses and whole-genome bisulfite sequencing. Genome Biol. Evol. 12, 1482–1492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall H., Jones A. R. C., Lonsdale Z. N., Mallon E. B., Bumblebee workers show differences in allele-specific DNA methylation and allele-specific expression. Genome Biol. Evol. 12, 1471–1481 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yagound B., Smith N. M. A., Buchmann G., Oldroyd B. P., Remnant E. J., Unique DNA methylation profiles are associated with cis-variation in honey bees. Genome Biol. Evol. 11, 2517–2530 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Remnant E. J., et al. , Parent-of-origin effects on genome-wide DNA methylation in the Cape honey bee (Apis mellifera capensis) may be confounded by allele-specific methylation. BMC Genomics 17, 226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Libbrecht R., Oxley P. R., Keller L., Kronauer D. J., Robust DNA methylation in the clonal raider ant brain. Curr. Biol. 26, 391–395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takayama S., et al. , Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 24, 821–830 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simpson V. J., Johnson T. E., Hammen R. F., Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 14, 6711–6719 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page R. E., Laidlaw H. H., Full sisters and super sisters: A terminological paradigm. Anim. Behav. 36, 944–945 (1988). [Google Scholar]
- 32.Harbo J., “Propagation and instrumental insemination” in Bee Genetics and Breeding, Rinderer T., Ed. (Academic Press, Orlando, FL, 1986), pp. 361–389. [Google Scholar]
- 33.Glastad K. M., Hunt B. G., Goodisman M. A. D., Epigenetics in insects: Genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 64, 185–203 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Schmitz R. J., Lewis Z. A., Goll M. G., DNA methylation: Shared and divergent features across eukaryotes. Trends Genet. 35, 818–827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortega-Recalde O., Hore T. A., DNA methylation in the vertebrate germline: Balancing memory and erasure. Essays Biochem. 63, 649–661 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Kelsey G., Feil R., New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heard E., Clerc P., Avner P., X-chromosome inactivation in mammals. Annu. Rev. Genet. 31, 571–610 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Kohli R. M., Zhang Y., TET enzymes, TDG and the dynamics of DNA demethylation. Nature 502, 472–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drewell R. A., et al. , The dynamic DNA methylation cycle from egg to sperm in the honey bee Apis mellifera. Development 141, 2702–2711 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gatzmann F., et al. , The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenetics Chromatin 11, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki M. M., et al. , Identical sets of methylated and nonmethylated genes in Ciona intestinalis sperm and muscle cells. Epigenetics Chromatin 6, 38 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith Z. D., Meissner A., DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Quadrana L., Colot V., Plant transgenerational epigenetics. Annu. Rev. Genet. 50, 467–491 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Rechavi O., Lev I., Principles of transgenerational small RNA inheritance in Caenorhabditis elegans. Curr. Biol. 27, R720–R730 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Bošković A., Rando O. J., Transgenerational epigenetic inheritance. Annu. Rev. Genet. 52, 21–41 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Johannes F., et al. , Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 5, e1000530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koeniger G., Koeniger N., Ellis J., Connor L., Mating Biology of Honey Bees (Apis mellifera) (Wicwas Press, Kalamazzo, MI, 2014). [Google Scholar]
- 48.Williams J. L., Wind-directed pheromone trap for drone honey bees (Hymenoptera: Apidae). J. Econ. Entomol. 80, 532–536 (1987). [Google Scholar]
- 49.Walsh P. S., Metzger D. A., Higuchi R., Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513 (1991). [PubMed] [Google Scholar]
- 50.Solignac M., et al. , Five hundred and fifty microsatellite markers for the study of the honeybee (Apis mellifera L.) genome. Mol. Ecol. Notes 3, 307–311 (2003). [Google Scholar]
- 51.Estoup A., Solignac M., Cornuet J.-M., Precise assessment of the number of patrilines and of genetic relatedness in honeybee colonies. Proc. Biol. Sci. 258, 1–7 (1994). [Google Scholar]
- 52.Sambrook J., Russell D. W., Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc. 2006, pdb-prot4455 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Yagound B., Remnant E. J., Buchmann G., Oldroyd B. P., Intergenerational transfer of epigenetic marks in the honey bee. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/PRJNA623232. Deposited 11 April 2020. [DOI] [PMC free article] [PubMed]
- 54.Bioinformatics Babraham, FastQC, Version 0.11.15. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 13 November 2020.
- 55.Bioinformatics Babraham, TrimGalore, Version 0.4.1. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore. Accessed 13 November 2020.
- 56.Honeybee Genome Sequencing Consortium , Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Toth A. L., Ozturk M., Sankaranarayanan S., Brendel V. P., BWASP. https://github.com/BrendelGroup/BWASP. Accessed 13 November 2020.
- 60.Glastad K. M., et al. , Variation in DNA methylation is not consistently reflected by sociality in Hymenoptera. Genome Biol. Evol. 9, 1687–1698 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arsenault S. V., Hunt B. G., Rehan S. M., The effect of maternal care on gene expression and DNA methylation in a subsocial bee. Nat. Commun. 9, 3468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Standage D. S., et al. , Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol. Ecol. 25, 1769–1784 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elsik C. G. et al.; HGSC production teams; Honey Bee Genome Sequencing Consortium , Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics 15, 86 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bates D., Machler M., Bolker B. M., Walker S. C., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 66.R Core Team , R: A Language and Environment for Statistical Computing (Version 3.3.3, R Foundation for Statistical Computing, Vienna, Austria, 2017). [Google Scholar]
- 67.Benjamini Y., Hochberg Y., Controlling the false discovery rate–A practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995). [Google Scholar]
- 68.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Rehan S. M., Berens A. J., Toth A. L., At the brink of eusociality: Transcriptomic correlates of worker behaviour in a small carpenter bee. BMC Evol. Biol. 14, 260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki R., Shimodaira H., Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Lê S., Josse J., Husson F., FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008). [Google Scholar]
- 72.R Core Team , R package corrplot. https://github.com/taiyun/corrplot. Accessed 13 November 2020.
- 73.Akalin A., et al. , methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13, R87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang H.-Q., Tuominen L. K., Tsai C.-J., SLIM: A sliding linear model for estimating the proportion of true null hypotheses in datasets with dependence structures. Bioinformatics 27, 225–231 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Huang W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome bisulfite sequencing data have been deposited in NCBI Sequence Read Archive (PRJNA623232) (53).