Significance
Lipid droplets (LDs) are specialized fat-storage organelles that can be used as a fuel source by many types of cells when nutrients are scarce. One mechanism used by hepatocytes (the functional cells of the liver) for the catabolism of these energy reserves is the lysosome-directed process of autophagy. Traditionally, autophagy necessitates the enclosure of cargo within a double-membrane autophagosome before delivery to the lysosome for degradation. Here, we use live-cell and electron microscopy to demonstrate that stable contacts between LDs and lysosomes in hepatocytes can result in the transfer of both proteins and lipids from LDs directly into the lysosome in the absence of an autophagosomal intermediate. These findings reveal a mechanism used for lipid homeostasis in the liver.
Keywords: lipid droplet, microautophagy, lipolysis, lysosome, hepatocyte
Abstract
Hepatocytes metabolize energy-rich cytoplasmic lipid droplets (LDs) in the lysosome-directed process of autophagy. An organelle-selective form of this process (macrolipophagy) results in the engulfment of LDs within double-membrane delimited structures (autophagosomes) before lysosomal fusion. Whether this is an exclusive autophagic mechanism used by hepatocytes to catabolize LDs is unclear. It is also unknown whether lysosomes alone might be sufficient to mediate LD turnover in the absence of an autophagosomal intermediate. We performed live-cell microscopy of hepatocytes to monitor the dynamic interactions between lysosomes and LDs in real-time. We additionally used a fluorescent variant of the LD-specific protein (PLIN2) that exhibits altered fluorescence in response to LD interactions with the lysosome. We find that mammalian lysosomes and LDs undergo interactions during which proteins and lipids can be transferred from LDs directly into lysosomes. Electron microscopy (EM) of primary hepatocytes or hepatocyte-derived cell lines supports the existence of these interactions. It reveals a dramatic process whereby the lipid contents of the LD can be “extruded” directly into the lysosomal lumen under nutrient-limited conditions. Significantly, these interactions are not affected by perturbations to crucial components of the canonical macroautophagy machinery and can occur in the absence of double-membrane lipoautophagosomes. These findings implicate the existence of an autophagic mechanism used by mammalian cells for the direct transfer of LD components into the lysosome for breakdown. This process further emphasizes the critical role of lysosomes in hepatic LD catabolism and provides insights into the mechanisms underlying lipid homeostasis in the liver.
A hallmark of fatty liver disease and related metabolic disorders is the intracellular accumulation of triacylglycerols and other neutral lipids within organelles called lipid droplets (LDs) (1–3). As a central site of lipid metabolism, the liver usually can tolerate significant fluctuations in the LD content of its parenchymal cells, the hepatocytes (4). Chronic dysregulation in levels of LDs is associated with liver disease and necessitates a better understanding of the mechanics underlying hepatic LD metabolism (5). Delimited by a phospholipid monolayer and decorated with a dynamically changing proteome, a primary function of the LD is to sequester esterified fatty acids away from the aqueous cytoplasm as neutral lipids, thus averting the lipotoxicity associated with high cytoplasmic concentrations of free fatty acids (6–9). In addition to a protective function, LDs also provide a supply of lipids for cellular membrane production and readily oxidizable substrates used to support mitochondrial adenosine triphosphate (ATP) generation (10, 11). While complex, the mechanisms underlying the regulated breakdown of hepatic LDs are primarily thought to require the involvement of two central catabolic pathways: cytosolic lipolysis and autophagy (12).
Hepatic lipolysis is generally regarded as a rapid process promoting the release of free fatty acids from parent triacylglycerols stored within the LD by a cascade of cytoplasmic lipases (13, 14). Autophagy, on the other hand, represents a lysosome-driven process that appears to play an especially important role in the degradation of hepatocellular LDs (15). A selective form of LD-centric autophagy known as lipophagy is thought to involve the recognition of as-of-yet unidentified LD-specific receptors to promote the localized assembly and extension of a sequestering phagophore around the perimeter of the LD (16, 17). How this phagophore is targeted to (and extended around) the LD surface to facilitate lipophagy remains unclear. Once fully enclosed, the double-membrane lipoautophagosome undergoes fusion with the lysosome to form a degradative organelle known as an autolysosome. Lysosomal lipases deposited within the autolysosome are then ultimately responsible for the acid hydrolysis of the LD-stored neutral lipids and subsequent release of free fatty acids (18–20). The two pathways of lipolysis and lipophagy likely work in tandem as coordinated processes (21). Indeed, we recently reported that lipolysis can act to rapidly reduce the size of large LDs to diameters more appropriate in size for engulfment by lipophagic vesicles (22).
To better evaluate the role and consequences of these LD–lysosomal interactions in the context of mammalian lipophagy, we used a combination of live-cell laser-scanning confocal and electron microscopy (EM) to study these contacts further. We identified a subpopulation of these associations that appear to promote the direct exchange of both protein and lipid from LDs that are actively “sampled” by the lysosome. Importantly, this process seems to result in the accumulation of LD-derived protein and lipids in the lysosome in the absence of canonical autophagy proteins. Together, these findings provide evidence for a lysosome-centric, LD catabolic process that is independent of traditional autophagy/lipophagy and plays a meaningful role in hepatocellular lipid homeostasis.
Results
Stable Interactions between Lysosomes and LDs Support LD Protein Transfer in Hepatocytes.
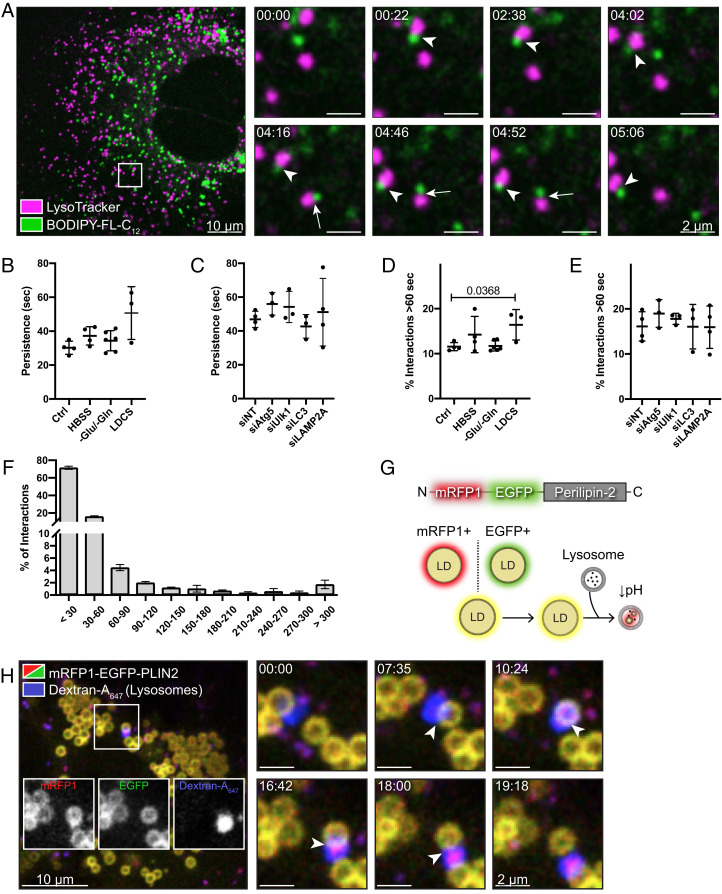
To study the dynamics of LD–lysosome interplay within the hepatocyte, we used AML12 mouse hepatocytes as an initial model imaging system. These cells maintain many characteristics of normal primary hepatocytes (23) and flatten well on collagen-coated glass imaging dishes to support time-lapse imaging of individual organelles in live cells. Labeling of LDs and lysosomes with BODIPY-FL-C12 and LysoTracker Deep Red, respectively, allowed monitoring of dynamic interactions between these two compartments. As shown in Fig. 1A and Movie S1, discrete contacts between the two organelles were observed that were often highly transient. Under basal cell culture conditions (i.e., growth in full-serum medium, 37 °C, 5% CO2), the persistence of thousands of individual interactions were measured and found to average 30.26 s (Fig. 1B, control), a timescale consistent with previously published data (24). Because of the importance of the lysosome to the process of macroautophagy and the central role of the LD as an energy-storage depot, we predicted that culture of hepatocytes under known autophagy-stimulating conditions (i.e., acute culture in Hank’s balanced salt solution [HBSS], amino acid-, or lipoprotein-deficient medium) (25) might significantly accentuate the duration of transient contacts between these organelles when nutrients are scarce. Such an increase in LD–lysosome associations, however, was not observed (Fig. 1B). Similarly, analysis of contacts between LDs and lysosomes in primary rat hepatocytes subjected to 4-h HBSS starvation showed only a slight (P = 0.085) elevation in the mean persistence of interactions between the two organelles (SI Appendix, Fig. S1A).
Fig. 1.
Frequent interactions between hepatocellular lysosomes and LDs support LD protein transfer. (A) Live-cell confocal imaging of a single AML12 mouse hepatocyte cultured under basal growth conditions and treated with BODIPY-FL-C12 and LysoTracker Deep Red to label LDs and lysosomes, respectively. Note the dynamic and transient nature of many of these contacts. Panels at Right represent still images from a time series (see corresponding Movie S1). Arrows indicate an example of a single transient interaction lasting <30 s while arrowheads represent a more persistent interaction lasting at least 5 min in length. (B and C) Analysis of the mean persistence (in seconds) of interactions between LDs and lysosomes in AML12 hepatocytes cultured under various nutritional states (B) or following 72-h treatment with multiple siRNAs targeting key components of the macroautophagy or chaperone-mediated autophagy machinery (C). (D and E) Percentage of interactions observed in live-cell imaging experiments in which the two organelles remained associated for >60 s. Values represent calculations of interactions from >5 movies from n ≥ 3 independent experiments. (F) Histogram showing the percentage of interactions occurring for various lengths of time (in seconds). Data collected from AML12 mouse hepatocytes cultured under basal growth conditions (n = 4 independent experiments). (G) Basis for the tandem-fluorescent perilipin reporter. N-terminal fusions of mRFP1 and EGFP to PLIN2 or PLIN5 render the surface of cytoplasmic LDs yellow when individual channels are merged. Upon contact with an acidic organelle (i.e., lysosomes), the EGFP signal is selectively quenched, leaving behind RFP+-only punctae. (H) Live-cell confocal imaging of an AML12 mouse hepatocyte transiently expressing the PLIN2 reporter shows direct contact (arrowhead) of a lysosome (blue) with the surface of an LD and subsequent release from the organelle, “peeling” away a structure that is RFP+ (see corresponding Movie S2).
Lipophagy is known to proceed following the sequestration of LDs within a double-membrane autophagosome (macrolipophagy) and can be regulated by the selective removal of scaffolding proteins (e.g., PLIN2) from the surface of the LD by chaperone-mediated autophagy (CMA) (15, 26). Therefore, we predicted that the knockdown of critical components of these two pathways might meaningfully alter the associations between LDs and lysosomes. To our surprise, siRNA-mediated depletion of the canonical macroautophagy or CMA machinery had little effect on the persistence of these interactions (Fig. 1C and SI Appendix, Fig. S1B), suggesting that these organelle–organelle contacts represent events which occur irrespective of the presence of canonical autophagosomes or molecular chaperones essential to CMA. Further, we also knocked down factors previously shown to play roles in coordinating conventional macrolipophagy (Rab7 and Rab10) as well as proteins recently shown to have lipid transporter functions (Atg2a and Vps13c) (27–31). Genetic depletion of these factors likewise had no significant effect on LD–lysosome contact persistence or frequency (SI Appendix, Fig. S1 C–E). Though the mean interaction persistence for all of these conditions tested was found to be very short (varying in the range of 30 to 50 s) (Fig. 1 B and C and SI Appendix, Fig. S1C), we observed that ∼15% of interactions reproducibly lasted longer than 60 s (Fig. 1 D and E). Binning of the percentages of interactions from AML12 hepatocytes cultured under basal growth conditions further showed that only a small percentage (∼2%) persist for periods longer than 5 min (Fig. 1F). We therefore focused our attention on identifying potential roles for these stable interactions.
As the persistent associations observed could represent a direct lysosomal sampling of proteins from the surface of LDs, cells were transfected to transiently express a dual-fluorescent variant of PLIN2 in AML12 hepatocytes that exhibits differential fluorescence dependent upon the pH of its surrounding environment. We have previously shown that this construct, like wild-type PLIN2, localizes to the surface of LDs and leaves behind small RFP+-only puncta upon interaction with acidic lysosomes (Fig. 1G and SI Appendix, Fig. S1F) (27). Long-term live-cell imaging showed instances of prolonged interactions between lysosomes and reporter-coated LDs that resulted in the eventual formation of RFP+-only puncta that then dissociated from the source LD together with the lysosome, suggestive of a piecemeal removal of protein from the surface of the LD (Fig. 1H and Movie S2). Stable expression of this mRFP1-EGFP-PLIN2 reporter in AML12 hepatocytes revealed significant numbers of RFP+-only puncta spread throughout the cytoplasm, indicating that a considerable quantity of the PLIN2 reporter is continually being sampled by lysosomes (SI Appendix, Fig. S1G), even when cells are cultured in nutrient-rich conditions.
Persistent Interactions between Lysosomes and LDs Support Direct Lipid Transfer.
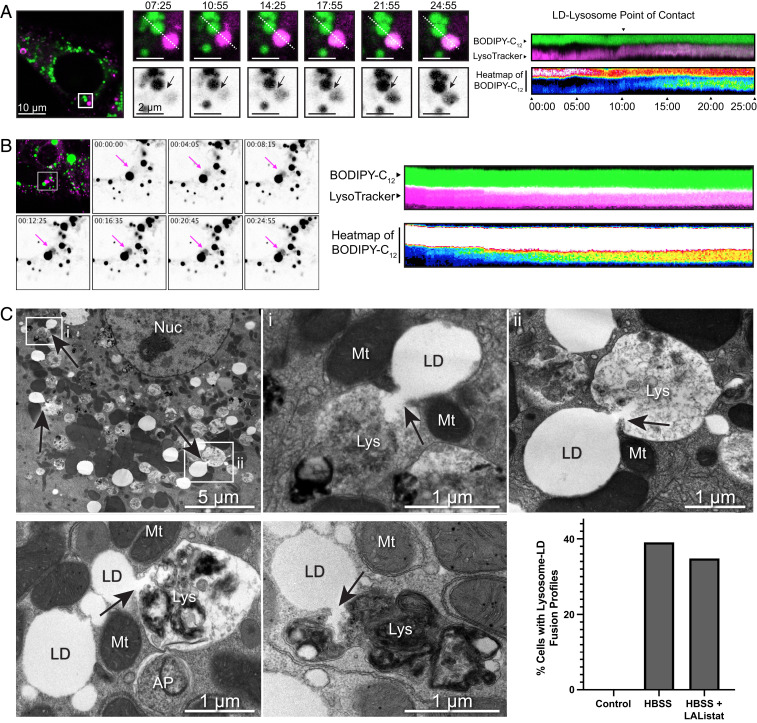
Aside from proteins residing on the LD surface, another cargo that could be exchanged as a result of direct contacts between the two organelles is lipid itself. Live-cell imaging of AML12 hepatocytes cultured under nutrient-replete conditions was therefore performed to determine whether direct lipid transfer might occur between LDs and lysosomes. As shown in Figs. 2 A and B and Movie S3, stable contacts between lysosomes and LDs were observed during which fluorescently (BODIPY-558/568) labeled C12 fatty acids began to accumulate in adjacent lysosomes (labeled with LysoTracker Deep Red) over time. Heatmap kymographs of the BODIPY fluorophore in both compartments (Fig. 2 A and B, Rightmost panels) showed a concomitant increase in lysosomal BODIPY-558/568 fluorescence upon sustained contact of the lysosome with the LD. As with light microscopy-based evidence of protein transfer (Fig. 1H), these putative lipid-transfer events were also infrequent and challenging to observe routinely, leading us to examine higher-resolution approaches for addressing the nature of LD–lysosome contacts. Accordingly, we performed electron microscopy on both primary rat hepatocytes and AML12 hepatocytes incubated in 150 μM bovine serum albumin (BSA)-complexed oleate (BSA:oleate) overnight (to promote LD accumulation) followed by an acute 4-h starvation in HBSS (to facilitate the catabolism of these LDs). Strikingly, cells subjected to this acute nutrient starvation revealed numerous instances of apparent lipid transfer between the LD and adjacent degradative structures that possess morphologies consistent with late endosomes, lysosomes, or autolysosomes (Fig. 2C and SI Appendix, Fig. S2 A and B). Interestingly, this transfer resembles a direct “injection” of hydrophobic lipid into the aqueous interior of the lysosome (Fig. 2C, arrows).
Fig. 2.
Persistent interactions between lysosomes and LDs facilitate direct lipid transfer between compartments. (A and B) Live-cell confocal imaging examples of AML12 mouse hepatocytes cultured under basal growth conditions showing the appearance of BODIPY-positive signal within a lysosome throughout a persistent contact lasting >18 min (see corresponding Movie S3). To the Right of each panel is a kymograph indicating that the transfer appears to be initiated upon contact of the organelles. (C) Electron micrographs of primary rat hepatocytes subjected to culture in media containing 150 μM oleic acid overnight (to stimulate LD formation) and serum-starved in HBSS (to promote LD catabolism) for 4 h before fixation and processing for TEM. Arrows indicate regions of contact between lysosomal and LD compartments. Note the appearance of clear tension on individual LDs with lipid appearing to be directly transferred into adjacent degradative compartments. Interestingly, mitochondria were frequently observed to be adjacent to these sites of contact, perhaps hinting at the possibility of nodes for transfer of fatty acids (FAs) liberated from LDs for oxidation in nearby mitochondria. (Nuc, nucleus; Mt, mitochondrion; Lys, lysosome; AP, autophagosome; LD, lipid droplet). Quantification reflects the percentage of primary rat hepatocytes (n = at least 23 cells for each condition) from single 100-nm sections that contained profiles similar to those displayed in this figure.
It is important to note three critical observations concerning this exchange process. First, quantification of numerous EM fields revealed that this interorganelle lipid transfer was infrequently observed in cells cultured under basal growth conditions (by EM), suggesting that this event is a regulated process that is stimulated by culture in nutrient-depleted (i.e., HBSS) treatment conditions. Second, the recipient compartments contain intralumenal vesicles or dark protease-containing granules found in degradative compartments, suggesting that these structures represent late endosomes/lysosomes. To confirm this premise, cells were cultured in horseradish peroxidase (HRP) for 60 min before a 150-min chase in label-free medium ahead of processing for EM. HRP is internalized by endocytosis and ultimately accumulates in late endocytic compartments to produce an electron-dense reaction product upon subsequent incubation with 3,3-diaminobenzidine (DAB). As shown in SI Appendix, Fig. S2B, the LD-interactive organelles stain darkly with HRP in primary hepatocytes, consistent with a late endosomal/lysosomal compartment. Finally, the recipient lysosomes appear to have only a single limiting unit membrane bilayer, and the LDs with which they are interacting are not observed to be surrounded by a double-membrane characteristic of canonical autophagosomes. We were able to resolve double-membrane macroautophagic profiles easily in these nutrient-deprived cells, which often contain other organelles such as mitochondria and endoplasmic reticulum (ER) (SI Appendix, Fig. S3); however, LDs did not appear to be sequestered within identifiable autophagosomes (SI Appendix, Fig. S3).
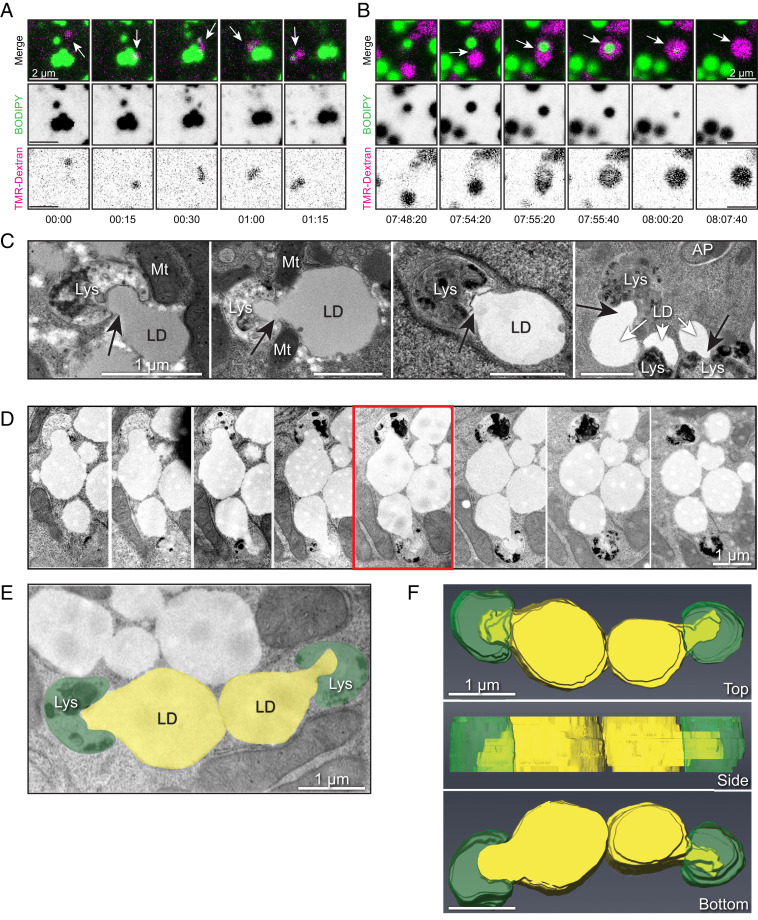
In addition to the perceived injection of lipid from LD to lysosome seen at the light and EM levels, other apparent mechanisms of transfer were also observed. Additional live-cell imaging of HBSS-starved primary hepatocytes showed occasional instances of lysosomes compressing and physically remodeling the LD, resulting in the removal and ingestion of either a peripheral portion of each LD (Fig. 3A) or engulfment of the entire droplet (Fig. 3B and Movie S4). These processes appeared to be remarkably rapid and could occur within just 1 to 2 min. Corresponding EMs of HBSS-starved primary hepatocytes (Fig. 3C) show further evidence for early stages in the deformation of LDs, with multiple examples of lysosomes consuming nearby LDs. In some instances, several lysosomes are observed to deform a single LD simultaneously. To gain a better topological appreciation of this process, we performed serial sections through two LDs undergoing lipid transfer for three-dimensional (3D) volume reconstruction using Amira software (Thermo Scientific). This perspective (Fig. 3 D–F) further suggests that lysosomes are exerting deliberate force on the perimeter of LDs in possible preparation for their direct piecemeal degradation, again in the absence of any recognizable autophagosomal intermediate.
Fig. 3.
Direct interactions between LDs and lysosomes drive piecemeal lipid transfer. (A) Live-cell confocal imaging of primary rat hepatocyte incubated with fluorescent dextran (magenta) to label lysosomes and BODIPY-FL-C12 (green) to label LDs. Panels show time series stills of an example of a lysosome contacting an individual LD and pulling away a portion of fluorescent lipid signal, suggestive of direct lipid transfer between compartments. (B) Live-cell confocal imaging of a primary rat hepatocyte labeled as in A, showing an example of a lysosome directly enveloping and quickly degrading the internalized LD within less than 1 min. See corresponding Movie S4. (C) Additional electron micrographs of primary rat hepatocytes subjected to culture in media containing 150 μM oleic acid overnight (to stimulate LD formation) and serum-starved in HBSS (to promote LD catabolism) for 4 h before fixation and processing for TEM. Note the appearance of profiles of piecemeal degradation of the lipid droplet that appear to be directly transferred into adjacent degradative compartments. (All scale bars, 1 μm.) (D) Serial section electron microscopy through an instance of putative lipid transfer structures in primary rat hepatocytes loaded overnight with 150 μM oleate and subjected to 4-h HBSS starvation. Sections are ∼100 nm apart in successive panels. (E and F) The 3D representation of the serial sections used in D, generated using Amira Software (Thermo Scientific). Lipid droplets are pseudocolored yellow, while degradative structures are pseudocolored green (E). Note the apparent protrusion of lipid content into the adjacent degradative structures from a 3D reconstruction of a series of sections taken through the midplane of two LDs (F).
Inhibition of Macroautophagy or Chaperone-Mediated Autophagy Does Not Affect Lipid/Protein Transfer.
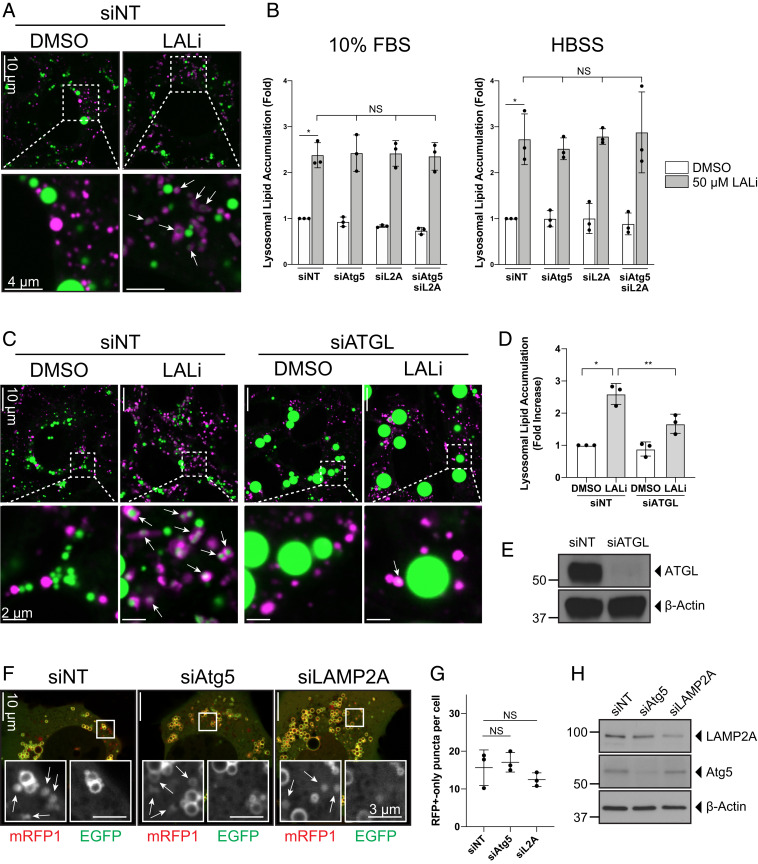
As direct, real-time visualization of the functional consequences of these interactions is difficult from a quantitative standpoint, we chose to use endpoint readouts of incorporation of lipid or protein incorporation into the lysosome in response to a genetic perturbation to various components of the macroautophagy or CMA machinery. As lipid is catabolized rapidly within the lysosome, the lysosomal acid lipase (LAL) inhibitor lalistat was utilized to slow lysosomal LD catabolism and preserve the number of lysosomes exhibiting ingested LDs following culture under control conditions or after knockdown of components of the autophagic machinery. AML12 hepatocytes were subjected to 72-h knockdowns of Atg5 or LAMP2A (individually, or in combination), both in full-serum (10% fetal bovine serum [FBS]) or starvation (4 h HBSS) conditions, in the presence or absence of 50 μM lalistat (Fig. 4 A and B and SI Appendix, Fig. S4A). Live cells were then imaged to determine the intensity of LipidTOX staining within LysoTracker-labeled lysosomes. As expected, the addition of lalistat resulted in a significant accumulation of lysosomal structures containing lipid and reinforces the importance of this organelle in LD catabolism (Fig. 4 A and B), in agreement with previous results (22). Notably, the combination of lalistat with a knockdown of key components of the CMA (LAMP2A) or macroautophagy (Atg5) machinery did not result in a significant decrease in the accumulation of neutral lipid within the lysosomal compartment (Fig. 4B). A complementary study in fixed cells demonstrated the same effect, showing that Atg5 or LAMP2A were not required for lipid accumulation in LAMP1-positive lysosomes (SI Appendix, Fig. S4 B–F). siRNA-mediated silencing of either protein, however, did show a significant increase in total cellular LDs (SI Appendix, Fig. S4 D–F), confirming that the knockdowns are effective and that these perturbations alter cellular lipid content. These findings are consistent with the biochemical analysis of triglyceride levels in these cells (SI Appendix, Fig. S4G) as well as previously published reports (15, 26).
Fig. 4.
Knockdown of macroautophagy/CMA machinery does not affect lipid/protein transfer between lysosomes and LDs. AML12 hepatocytes were treated for 72 h with a nontargeting control siRNA (siNT), siAtg5, siLAMP2A, or a combination of both siAtg5 and siLAMP2A, to examine possible compensation between these pathways. Cells were treated for 24 h ± 50 μM lalistat and cultured for 4 h in full-serum (10% FBS) or HBSS starvation conditions. (A) Confocal live-cell imaging stills of siNT-treated cells cultured under full-serum conditions ±50 μM lalistat. Lysosomes are stained in magenta with LysoTracker Deep Red, and LDs are stained with LipidTOX green. Note the accumulation of lipid within lysosomes upon 24 h LAL inhibitor (LALi) treatment (arrows). (B) Quantification of lysosomal lipid accumulation from live cells. LALi treatment results in significant retention of lipids within the lysosome. Knockdowns of macroautophagy or CMA components do not perturb this lipid accumulation. Summary of n = 3 independent experiments in which >5 fields of ∼40 cells were quantified. *P < 0.05 as measured by paired t test. Error bars reflect SD. NS, not significant. (C and D) siRNA-mediated knockdown of ATGL negatively affects lysosomal lipid accumulation. Confocal images (C) using the same uptake assay conditions as in A, reveals a ∼40% reduction (D) in lysosomal lipid uptake upon ATGL knockdown. Summary of n = 3 independent experiments in which >5 fields of ∼40 cells were quantified. *P < 0.05, **P < 0.01 as measured by paired t test. Error bars reflect SD. (E) Immunoblotting to demonstrate the knockdown efficiency of siATGL treatment in the experiments used in C and D. (F) Confocal images of AML12 mouse hepatocytes transiently expressing the PLIN2-mRFP1-EGFP reporter and subjected to 72-h treatment with a nontargeting control siRNA (siNT), siAtg5, or siLAMP2A shows the presence of RFP+-only puncta in all three cases. Insets show individual RFP+ and EGFP channels with arrows indicating RFP+-only puncta. (G) Quantification of the number of RFP+-only puncta from >10 cells from n = 3 independent experiments revealed no significant difference in the ability of the reporter to be internalized into the lysosome when macroautophagy or CMA machinery was compromised by 72-h treatment with indicated siRNAs. NS, not significant. (H) Immunoblotting to demonstrate the knockdown efficiency of siAtg5 or siLAMP2A treatment in the experiments used in F and G.
Furthermore, as with a lack of observable effect on LD–lysosome interactions, knockdown of macrolipophagy factors (Rab7, Rab10) or lipid transporters (Atg2a, Vps13c) also did not affect the lalistat-induced accumulation of lipid within lysosomes (SI Appendix, Fig. S5 A and B). Consistent with previous data showing that lipophagy acts primarily on those LDs that have been reduced in size by lipolysis (22), we found that lysosomal lipid content in AML12 hepatocytes treated with siRNA targeting adipose triglyceride lipase (ATGL) had a blunted capacity for uptake of lipid into the lysosome (Fig. 4 C–E). This lysosomal lipid accumulation appears not to be due to the secretion and reuptake of fluorescent lipoprotein particles via the endocytic pathway, as cells treated with conditioned media following a BODIPY-C12 chase period exhibited little detectable accumulation of fluorescent LDs within lysosomes (SI Appendix, Fig. S5 C and D).
Finally, in agreement with data on lipid uptake into the lysosome, we also measured the accumulation of the same LD-associated PLIN2 reporter used in Fig. 1 following siRNA-mediated knockdown of both LAMP2A and Atg5 (Fig. 4 F–H) and again found that these perturbations had no significant effect on the ability of the lysosome to internalize this LD-specific protein. Taken together, these findings are consistent with the premise that direct LD ingestion by lysosomes contributes in a meaningful way to lipid catabolism in the hepatocyte and occurs by a lysosome-directed process that is distinct from canonical autophagic mechanisms.
Discussion
In this study, we provide insights into a mechanism by which the hepatocyte catabolizes its LD stores. Laser-scanning confocal microscopy of LD–lysosome interactions in both primary hepatocytes and a nonneoplastic hepatocyte cell line (AML12) revealed a subpopulation (∼15%) of persistent contacts between these organelles lasting longer than 60 s in cells cultured under nutrient-limited conditions (HBSS), suggesting a functional contribution of these interactions to LD catabolism (Fig. 1). Live-cell imaging provided further evidence for the transfer of both LD-localized proteins as well as neutral lipids directly from the LD to the lysosomal compartment (Figs. 1 and 2). Subsequent ultrastructural examination of oleate-loaded, HBBS-starved hepatocytes by electron microscopy confirmed the existence of intimate associations between the two organelles as well as a direct exchange or injection of neutral lipid from LDs into the lysosomal/autolysosomal lumen in the absence of a defined autophagosomal intermediate (Figs. 2 and 3). Importantly, this transfer of lipid and protein between organelles persists in hepatocytes in which core canonical macroautophagy (Atg5) or CMA (LAMP2A) components are reduced by siRNA (Fig. 4). Together, these data imply the existence of an additional mechanism of lysosome-mediated catabolism that can be used for the turnover of LDs in hepatocytes.
An Additional Autophagic Mechanism for Lysosome-Mediated LD Turnover.
Our findings suggest that a previously unrecognized mechanism used by the hepatocyte for the catabolism of LDs occurs not via the assembly of a traditional sequestering phagophore using membranes derived from other organelles nor localized de novo biogenesis (32–34), but rather the efficient docking of a degradative organelle directly onto the targeted LD. This study does not eliminate the role of macroautophagy or CMA in hepatocellular LD turnover but does identify a functionally relevant lipid catabolic process that also occurs in these cells. Indeed, it is likely that these related (but distinct) degradative mechanisms each contributes to the vital process of LD catabolism for use as an essential energy source to ensure cell survival.
Several observations differentiate the process described here from canonical macroautophagy. First, the initial steps in macroautophagy involve the recruitment of a core lipid synthetic machinery and the Atg5-dependent lipidation of LC3. These events promote the construction and extension of a phagophore around the organelle for encapsulation within a double-membrane autophagosome (19, 35). Our findings suggest that protein and lipids can be transferred directly from the LD to the lysosome effectively, even in the absence of Atg5. Consistent with this observation are reports of Atg5-independent mechanisms of autophagy, suggesting that there is some inherent flexibility in the process of cargo capture for delivery to the lysosome (36, 37). We were able to easily identify conventional double-membrane-delimited autophagosomes and mitoautophagosomes by EM in the cell models examined in this study (SI Appendix, Fig. S3). Though intact LDs could be observed inside other organelles, the morphologies of these compartments are most consistent with degradative organelles such as lysosomes or autolysosomes and not autophagosomes; indeed, no double-membrane structures were observed that could be unambiguously classified as true “lipoautophagosomes.” This could potentially reflect a lower frequency of autophagosome-based macrolipophagy relative to processes such as bulk macroautophagy or mitophagy.
Furthermore, the findings that LAMP2A knockdown did not perturb the transfer of LD protein or lipid import into the lysosome (Fig. 4) suggests that the process we observe can proceed in a CMA-independent manner. As CMA works primarily at the level of individual peptides, it is thought to play more of a regulatory function in orchestrating the downstream lipolytic and lipophagic degradation of the LD (17, 38–40). A recent study revealed that the LD-specific protein PLIN2 harbors a CMA motif, promoting its recognition by the cytosolic Hsc70 chaperone and delivery to the lysosome for degradation in a LAMP2A-dependent manner (26). CMA-mediated turnover of PLIN2 was found to be enhanced by nutrient starvation and shown to precede the recruitment of lipolytic or macroautophagy machinery to the LD, suggesting that CMA regulates access of different catabolic systems to the LD via the removal of physical barriers from the LD surface (26). The results from this previous study showed significant LD accumulation in cultured mouse embryonic fibroblasts, as well as hepatic fat accumulation following LAMP2A knockout and are consistent with our observation that siRNA-mediated LAMP2A knockdown in hepatocytes also results in significant LD accumulation (SI Appendix, Fig. S4 D–F). However, our findings also show that despite the resulting increase in cellular LD content, LAMP2A knockdown does not affect the internalization of labeled fluorescent lipids into lysosomes, suggesting the existence of an additional means of lipid/protein ingress into the lysosome that is independent of CMA.
Our ability to capture large numbers of these cargo transfer events in live cells may be hampered by a lack of sufficient spatiotemporal resolution. As shown in Fig. 1, most interactions between the compartments are fleeting. Less than 2% of interactions persist for longer than 5 min. Furthermore, many of these interactions are occurring between small organelles less than 500 nm in diameter (see below), further complicating their direct observation by light microscopy. It was only through the use of EM that we were able to observe numerous instances of lipid transfer between organelles reliably; at this resolution, we could identify multiple events co-occurring in the same cell (Fig. 2C). It is therefore possible that many of the transient contacts between the organelles also result in some small degree of lipid transfer but that this exchange is only visible at the EM and not light level. By EM, we found lipid transfer profiles in ∼40% of cells examined; however, the number seen in each 100-nm section was variable, suggesting that this process is infrequent enough to make live-cell capture difficult but ubiquitous enough to result in the catabolism of substantial numbers of LDs over a long period. This process may therefore represent a physiologically meaningful mechanism for LD turnover in cells that is more rapid and efficient than macrolipophagy.
Direct Sampling and Engulfment of LDs by the Lysosome: Microlipophagy in Hepatocytes.
Direct ingestion of various cellular organelles by the lysosome has been previously reported in both mammalian cells and yeast (41–45). In yeast, LDs are known to be directly catabolized by the vacuole in a specialized process referred to as microlipophagy (46). Following nutritional deprivation, liquid-ordered microdomains form on the yeast vacuolar membrane that allow for the direct uptake of LDs. The formation and stabilization of these sterol-enriched domains appear to be mediated by ATG proteins, including ATG14p, ATG6p, ATG21, and ATG32 (47, 48); however, it seems that this process in yeast occurs independently of canonical macroautophagy, as core components such as ATG7 do not appear to be required (49). Following acute culture in nutrient-limiting conditions, our EM observations in the hepatocyte are most consistent with a yeast microlipophagy-like mechanism for hepatic LD turnover in that lipid from these LDs appear to 1) be directly injected into or 2) “fuse” with adjacent lysosomal structures. This premise is further supported by evidence from electron micrographs found in the recent literature of a microlipophagy-like phenomenon resembling the observations presented here (50, 51). The biomechanics whereby an organelle with a phospholipid bilayer (lysosome) might fuse with an organelle bounded by a phospholipid monolayer (lipid droplet) are not clear. Previous studies of LD interactions with other double-membrane organelles such as peroxisomes proposed the existence of hemifusion events such that the inner leaflet of the peroxisomal membrane “invades” the lipid core of the droplet following fusion of the outer leaflet of the peroxisome with the LD monolayer. These structures, referred to as “pexopodia,” may serve to permit luminal peroxisomal enzymes access to TAG or LD-associated proteins (3, 52). Similar events may be occurring between the LD and lysosome, as we describe here. Additional work will be necessary to understand better the nature of the dynamics at these contact sites.
The LD itself possesses several properties that make it a unique substrate for the process of mammalian microautophagy. The first is its size relative to the lysosome: the mammalian lysosome is often much smaller than its yeast counterpart, the vacuole. Mammalian lysosomes can have diameters as little as 100 nm, depending on the cell type and nutrient availability (53). The yeast vacuole, on the other hand, can be more than an order of magnitude larger in diameter (54). For this reason, there must exist clear limitations on the sizes of those hepatocellular LDs susceptible to direct microlipophagic engulfment by the mammalian lysosome. Along these lines, knockdown of ATGL, the rate-limiting enzyme of lipolysis, was found to negatively affect lysosomal lipid uptake (Fig. 4C). This result is consistent with our previously published data suggesting a critical role for this enzyme in reducing the size of cytoplasmic LDs to a diameter compatible with endolysosomes (22). The impairment of lysosomal lipid uptake in siATGL-treated cells may therefore indicate that direct lysosomal catabolism can occur only on very small LDs. Consequently, a “piecemeal microautophagy” mechanism of LD degradation may therefore be required for the turnover of the largest LDs by small mammalian lysosomes, similar to piecemeal degradation of the nucleus by the yeast vacuole (55) or lysosomal degradation of mitochondrial-derived vesicles in mammalian cells (56, 57). Further work will be required to elucidate the regulatory role of ATGL in the context of microlipophagy.
The spherical shape of the LD presents another biophysical challenge to the process of microlipophagy. Our EM images provide evidence for the existence of significant force-generating membrane dynamics on the LD surface by surrounding lysosomes. The forces required to cause such deformations would likely be substantial and require considerable input from the actomyosin cytoskeletal machinery as well as dedicated membrane-deforming proteins such as members of the BAR or ESCRT families of proteins (58). Furthermore, the Laplace pressure differential across the LD monolayer is proposed to be higher in small LDs as compared to large LDs (59), possibly influencing the degree of lysosome-mediated deformation that can occur at contact sites with LDs of different sizes. The machinery responsible for these dynamics at the LD–lysosome interface remains unknown. However, there is evidence for the formation of small Arf1/COPI-mediated budding events in adipocytes to produce “nano-LDs” with 60-nm diameters that might represent attractive targets to the smaller lysosomes of mammalian cells (60). Further analysis represents an essential next step in understanding this process in the hepatocyte.
Implications for the Existence of Mammalian Microlipophagy and Future Directions.
This study raises several questions and areas of unmet need. First, it will be critical to identify the molecular tethers or regulatory proteins that promote bridging of the lysosome and LD to sustain the persistent contacts occurring during microlipophagy. Interactions of LDs with other organelles have been described to occur with the aid of multiple tethers. For example, the yeast ER integral membrane protein Mdm1 and its eukaryotic homolog, Snx14, have recently been demonstrated to help coordinate ER–LD interactions (61–63). Also in yeast, the proteins Ldo16 and Ldo45 were identified to mediate ER–LD contacts and to additionally play a role in the uptake of LDs into the vacuole (64). The identity of proteins that bridge mammalian LDs and lysosomes are currently unknown. However, the numerous Rab GTPases and membrane trafficking proteins residing on the LD may help regulate such interorganelle interactions, as has been previously suggested (27, 28). Secondly, we find by EM that lipid transfer profiles are more prevalent in HBSS-starved cells, consistent with the findings in yeast that nutritional limitation results in changes to the vacuolar membrane to support microlipophagy (47, 48); hepatic microlipophagy may therefore represent a critical mechanism that can be rapidly activated for cellular energy production during periods of nutrient deprivation.
As a consequence, it will be essential to understand how microlipophagy works in parallel with other means of hepatocellular LD utilization (macroautophagy, CMA, and lipolysis) moving forward. Finally, it will be crucial to place this process within the context of advanced liver disease; for example, understanding how chronic exposure to a high-fat diet or hepatotoxins such as alcohol or acetaminophen (known to disrupt conventional lipophagy and mitophagy) (65–67) might affect microlipophagy will be important areas of future study. Also key will be an understanding of whether genetic factors known to be linked to fatty liver disease (i.e., the pathogenic PNPLA3 I148M mutant) interfere with hepatic microlipophagy in any way. Ultimately, this information will help guide our understanding of how this autophagic process might be manipulated during the progression of fatty liver disease.
Materials and Methods
Cell Culture, Plasmids, and Reagents.
The murine AML12 hepatocyte cell line was obtained from ATCC (CRL-2254). The mRFP-EGFP-PLIN2 LD reporter construct for lysosome–LD interactions was as used previously (27). LysoTracker Deep Red (L12492), BODIPY-FL-C12 (D3822), BODIPY-558/568-C12 (D3835), dextran-tetramethylrhodamine 10,000 MW (D1817), dextran-Alexa 647 10,000 MW (D22914), and Lipofectamine 3000 transfection reagent (L3000008) were obtained from Thermo Fisher Scientific. The β-actin antibody (A2066), lipoprotein-depleted serum (S5394), BSA:oleate (O1008), Oil Red O (O0625), and chloroquine hydrochloride (C6628) were obtained from Sigma-Aldrich. The PLIN2 antibody (B3121) was obtained from LS Biosciences. The ATGL antibody (2439) was from Cell Signaling. The Atg5 antibody (D5F5U) was from Cell Signaling. The Atg2a antibody (PD041) was from MBL International Corp. The LAMP2A antibody (ab18528) was from AbCam. The Ulk1 antibody (8054) was from Cell Signaling. The Rab7 antibody (9367) was from Cell Signaling. The Rab10 antibody (GTX82800) was from GeneTex. The LAMP1 antibody (1D4B) was from the Developmental Studies Hybridoma Bank (University of Iowa). The LC3 antibody (NB600-1384) was obtained from Novus. The VPS13c antibody was generously provided by James Granneman, Wayne State University, Detroit, MI. The LAL inhibitor lalistat-2 (25347) and Bafilomycin A1 (11038) were obtained from Cayman Chemical. The monodansylpentane (MDH) was from Abgent (SM1000a). Hank’s balanced salt solution (1× with calcium and magnesium) used for nutrient deprivation experiments (23-21-020-CV) was from Corning CellGro.
Primary Hepatocyte Isolation.
Primary rat hepatocytes were isolated from Sprague–Dawley rats (Envigo) as described previously (22, 68) and in accordance with an approved protocol by the Institutional Animal Care and Use Committee at Mayo Clinic. Briefly, collagenase digestion of liver tissue was performed with portal vein perfusion and physical dissociation of digested tissue. Isolation of viable hepatocytes was facilitated by multiple centrifugation steps, including a Percoll (Sigma-Aldrich) gradient for separation of viable cells that were subsequently plated onto collagen-coated coverslips or imaging dishes.
Live-Cell Imaging.
Cells were cultured on 35-mm Petri dishes with an 18-mm well and no. 1.5 coverglass (Cell E&G, GBD00004-200) that had been coated overnight with collagen (type I collagen, rat tail, Corning, 354236). Imaging was performed on a Zeiss LSM780 confocal microscope using a 40× oil 1.4 numerical aperture (NA) objective lens with frames acquired approximately every 2 s for a minimum of 5 min. Raw .czi files were imported into Fiji v.1.52 (69) for downstream analysis. After applying a Gaussian blur to remove background noise, interaction persistence was calculated using manually thresholded lysosomal and LD channels from each movie, subsequent segmentation of regions of pixel overlap between the two channels, and use of the 3D Objects Counter analysis plugin to identify object persistence across multiple frames. For these laser scanning confocal microscopy-based live-cell analyses, a “contact” was defined as an overlap of thresholded signals >5 adjacent pixels in area and representing an xy resolution-limited overlap of ∼250 nm (70). To measure lysosomal lipid accumulation, we first applied a Gaussian blur to remove background noise from the image. The lysosomal channel was thresholded using the Bernsen method (radius = 18). Intensity of the LipidTox channel within the thresholded lysosomal area was then calculated.
siRNA Knockdown.
Cells were transfected using Lipofectamine RNAiMAX (13778-150) from Thermo Fisher according to the manufacturer’s recommended protocol. siGENOME pools (Dharmacon/Horizon Discovery) targeting murine Atg2a (M-056681-01), Rab7 (M-040859-01), Rab10 (M-040862-01), Vps13c (M-053177-01), ATGL (M-040220-01), Ulk1 (M-040155-00), Atg5 (M-064838-02), Map1LC3B (M-040989-01), and LAMP2 (M-059036-01), or a nontargeting control siRNA (D-001210-01), were used for knockdowns of 72 h.
Transmission Electron Microscopy.
For standard transmission electron microscopy (TEM), cells on carbon-coated coverslips were rinsed in 37 °C HBSS and fixed with 37 °C primary fixative (100 mM cacodylate, pH 7.4, 60 mM sucrose, 2.5% glutaraldehyde) for 1 h at room temperature (RT), rinsed three times with washing buffer (100 mM cacodylate, pH 7.4, 200 mM sucrose) then fixed in the secondary fixative (50 mM cacodylate, pH 7.4, 100 mM sucrose, 1% OsO4) for 1 h at room temperature, rinsed three times in water, and fixed in 1% uranyl acetate in water for 1 h at room temperature. Samples were then dehydrated in a graded ethanol series, embedded in Quetol 651 (Ted Pella) and polymerized in a 65 °C oven overnight. After removal from the oven, the coverslip was removed from the bottom of the sample, the block trimmed down to a trapezoid 1 mm wide at the base, and 100-nm thin sections were cut and viewed on a Jeol 1200 transmission electron microscope (Jeol Ltd).
BODIPY-C12 Chase and Lysosome Retention.
AML12 cells were labeled with 7.5 µM BODIPY-(558/568)-C12 lipid for 2 h, washed with two exchanges of HBSS, and placed into regular culture media for 24 h with or without 50 µM lalistat. Following this 24 h “chase” period, cells were fixed and processed for immunofluorescence using a LAMP1 antibody (Cat. no. 1D4B, Developmental Studies Hybridoma Bank [DSHB], University of Iowa) to detect remaining fluorescent lipid in the lysosome.
BODIPY-C12 Conditioned Medium Experiment.
AML12 cells were plated to confluency on smaller 10-mm coverslips in a six-well dish. Control cells were loaded for 2 h with 150 μM BSA:oleate + 7.5 μM BODIPY-FL-C12, then washed four times in HBSS and chased 24 h in regular growth medium containing 50 μM lalistat as seen in Fig. 4A and SI Appendix, Fig. S4A. For conditioned medium experiments, “donor” AML12 cells were created from a confluent monolayer of a single well of a six-well plate by loading for 2 h with oleic acid (OA) + BODIPY-FL-C12. In an adjacent well, “acceptor” AML12 cells were plated to 10-mm coverslips and also loaded 2 h with 150 μM OA, but with no BODIPY-FL-C12. Both donor and acceptor cells were washed four times in HBSS; a coverslip of acceptor cells was then placed into the donor cell dish for 24 h in regular medium containing 50 μM lalistat. After 24 h, coverslips were fixed in paraformaldehyde and immunolabeled with a LAMP1 antibody obtained from the DSHB (Cat. no. 1D4B).
Triglyceride Measurement.
Lipid was extracted from AML12 cells using 1 mL chloroform/methanol (2:1) and vigorous shaking for 1 h at RT. A total of 200 μL double-distilled H2O was added, and samples were vortexed and centrifuged at 3,000 × g for 5 min. The lower lipid phase was collected (∼700 μL), dried overnight, and resuspended in a mixture containing tert-butanol (471712; Sigma-Aldrich), Triton X-114 (X114; Sigma-Aldrich), and methanol (9:4:2). TAG quantification was done in AML12 cells using a colorimetric kit from Pointe Scientific (T7532, T7531-STD) according to manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank all members of the M.A.M. laboratory for helpful discussions. We are incredibly grateful for expert technical assistance from Bridget Mehall. This work was supported by NIH grants R01AA020735 (M.A.M. and C.A.C.), R01DK044650 (M.A.M.), T32DK007352 (R.J.S. and M.B.S.), K99AA026877 (M.B.S.), and the Optical Microscopy Core of the Mayo Clinic Center for Cell Signaling in Gastroenterology (P30DK084567); a Gilead Sciences Research Scholars in Liver Disease award (R.J.S.); and the US Department of Veterans Affairs (C.A.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. T.C.W. is a guest editor invited by the Editorial Board.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2011442117/-/DCSupplemental.
Data Availability.
All study data are included in the article and supporting information.
References
- 1.Walther T. C., Farese R. V. Jr, Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Welte M. A., Expanding roles for lipid droplets. Curr. Biol. 25, R470–R481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olzmann J. A., Carvalho P., Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20, 137–155 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulze R. J., Schott M. B., Casey C. A., Tuma P. L., McNiven M. A., The cell biology of the hepatocyte: A membrane trafficking machine. J. Cell Biol. 218, 2096–2112 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gluchowski N. L., Becuwe M., Walther T. C., Farese R. V. Jr, Lipid droplets and liver disease: From basic biology to clinical implications. Nat. Rev. Gastroenterol. Hepatol. 14, 343–355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen T. B., et al. , DGAT1-dependent lipid droplet biogenesis protects mitochondrial function during starvation-induced autophagy. Dev. Cell 42, 9–21 e25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piccolis M., et al. , Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Mol. Cell 74, 32–44.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Listenberger L. L., et al. , Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walther T. C., Chung J., Farese R. V. Jr, Lipid droplet biogenesis. Annu. Rev. Cell Dev. Biol. 33, 491–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rambold A. S., Cohen S., Lippincott-Schwartz J., Fatty acid trafficking in starved cells: Regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell 32, 678–692 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henne W. M., Reese M. L., Goodman J. M., The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37, e98947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zechner R., Madeo F., Kratky D., Cytosolic lipolysis and lipophagy: Two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18, 671–684 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Sathyanarayan A., Mashek M. T., Mashek D. G., ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 19, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schott M. B., et al. , β-Adrenergic induction of lipolysis in hepatocytes is inhibited by ethanol exposure. J. Biol. Chem. 292, 11815–11828 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh R., et al. , Autophagy regulates lipid metabolism. Nature 458, 1131–1135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R., Cuervo A. M., Lipophagy: Connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012, 282041 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulze R. J., Drižytė K., Casey C. A., McNiven M. A., Hepatic lipophagy: New insights into autophagic catabolism of lipid droplets in the liver. Hepatol. Commun. 1, 359–369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu K., Czaja M. J., Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 20, 3–11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatica D., Lahiri V., Klionsky D. J., Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warner T. G., Dambach L. M., Shin J. H., O’Brien J. S., Purification of the lysosomal acid lipase from human liver and its role in lysosomal lipid hydrolysis. J. Biol. Chem. 256, 2952–2957 (1981). [PubMed] [Google Scholar]
- 21.Martinez-Lopez N., et al. , Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the Brown adipose tissue and liver. Cell Metab. 23, 113–127 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schott M. B., et al. , Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 218, 3320–3335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J. C., Merlino G., Fausto N., Establishment and characterization of differentiated, nontransformed hepatocyte cell lines derived from mice transgenic for transforming growth factor alpha. Proc. Natl. Acad. Sci. U.S.A. 91, 674–678 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valm A. M., et al. , Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik S., Cuervo A. M., Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schroeder B., et al. , The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology 61, 1896–1907 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Z., et al. , A novel Rab10-EHBP1-EHD2 complex essential for the autophagic engulfment of lipid droplets. Sci. Adv. 2, e1601470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar N., et al. , VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valverde D. P., et al. , ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramseyer V. D., Kimler V. A., Granneman J. G., Vacuolar protein sorting 13C is a novel lipid droplet protein that inhibits lipolysis in brown adipocytes. Mol. Metab. 7, 57–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schütter M., Giavalisco P., Brodesser S., Graef M., Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell 180, 135–149.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Hailey D. W., et al. , Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 141, 656–667 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puri C., Renna M., Bento C. F., Moreau K., Rubinsztein D. C., Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 154, 1285–1299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakatogawa H., Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Honda S., et al. , Association between atg5-independent alternative autophagy and neurodegenerative diseases. J. Mol. Biol. 432, 2622–2632 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Nishida Y., et al. , Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461, 654–658 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Schulze R. J., Sathyanarayan A., Mashek D. G., Breaking fat: The regulation and mechanisms of lipophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1862, 1178–1187 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drizyte-Miller K., Schott M. B., McNiven M. A., Lipid droplet contacts with autophagosomes, lysosomes, and other degradative vesicles. Thousand Oaks 3, 1–13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaushik S., Cuervo A. M., The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Duve C., Wattiaux R., Functions of lysosomes. Annu. Rev. Physiol. 28, 435–492 (1966). [DOI] [PubMed] [Google Scholar]
- 42.Kissová I., et al. , Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy 3, 329–336 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Sakai Y., Koller A., Rangell L. K., Keller G. A., Subramani S., Peroxisome degradation by microautophagy in Pichia pastoris: Identification of specific steps and morphological intermediates. J. Cell Biol. 141, 625–636 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuck S., Gallagher C. M., Walter P., ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J. Cell Sci. 127, 4078–4088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuttle D. L., Dunn W. A. Jr, Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25–35 (1995). [DOI] [PubMed] [Google Scholar]
- 46.van Zutphen T., et al. , Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C. W., Miao Y. H., Chang Y. S., A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 206, 357–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seo A. Y., et al. , AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. eLife 6, e21690 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vevea J. D., et al. , Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosch M., Parton R. G., Pol A., Lipid droplets, bioenergetic fluxes, and metabolic flexibility. Semin. Cell Dev. Biol., 10.1016/j.semcdb.2020.02.010 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Li Y., et al. , Impaired fasting-induced adaptive lipid droplet biogenesis in liver-specific Atg5-deficient mouse liver is mediated by persistent nuclear factor-like 2 activation. Am. J. Pathol. 188, 1833–1846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Binns D., et al. , An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu H., Ren D., Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Klionsky D. J., Eskelinen E. L., The vacuole versus the lysosome: When size matters. Autophagy 10, 185–187 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krick R., et al. , Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 19, 4492–4505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soubannier V., et al. , A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22, 135–141 (2012). [DOI] [PubMed] [Google Scholar]
- 57.McLelland G. L., Lee S. A., McBride H. M., Fon E. A., Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214, 275–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ebrahim S., Liu J., Weigert R., The actomyosin cytoskeleton drives micron-scale membrane remodeling in vivo via the generation of mechanical forces to balance membrane tension gradients. BioEssays 40, e1800032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiam A. R., Farese R. V. Jr, Walther T. C., The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14, 775–786 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilfling F., et al. , Arf1/COPI machinery acts directly on lipid droplets and enables their connection to the ER for protein targeting. eLife 3, e01607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hariri H., et al. , Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J. Cell Biol. 218, 1319–1334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Datta S., Liu Y., Hariri H., Bowerman J., Henne W. M., Cerebellar ataxia disease-associated Snx14 promotes lipid droplet growth at ER-droplet contacts. J. Cell Biol. 218, 1335–1351 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen T. B., Olzmann J. A., Getting a handle on lipid droplets: Insights into ER-lipid droplet tethering. J. Cell Biol. 218, 1089–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teixeira V., et al. , Regulation of lipid droplets by metabolically controlled Ldo isoforms. J. Cell Biol. 217, 127–138 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams J. A., Ni H. M., Ding Y., Ding W. X., Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 309, G324–G340 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chao X., et al. , Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology 155, 865–879 e12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schulze R. J., et al. , Ethanol exposure inhibits hepatocyte lipophagy by inactivating the small guanosine triphosphatase Rab7. Hepatol. Commun. 1, 140–152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen L., Hillebrand A., Wang D. Q., Liu M., Isolation and primary culture of rat hepatic cells. J. Vis. Exp. 3917 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scorrano L., et al. , Coming together to define membrane contact sites. Nat. Commun. 10, 1287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.