Abstract
Globally, approximately 1 in 4 cancers in women are diagnosed as breast cancer (BC). Despite significant advances in the diagnosis and therapy BCs, many patients develop metastases or relapses. Hence, novel therapeutic strategies are required, that can selectively and efficiently kill malignant cells. Direct targeting of the genetic and epigenetic aberrations that occur in BC development is a promising strategy to overcome the limitations of current therapies, which target the tumour phenotype. The clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system, composed of only an easily modifiable single guide RNA (sgRNA) sequence bound to a Cas9 nuclease, has revolutionised genome editing due to its simplicity and efficiency compared to earlier systems. CRISPR/Cas9 and its associated catalytically inactivated dCas9 variants facilitate the knockout of overexpressed genes, correction of mutations in inactivated genes, and reprogramming of the epigenetic landscape to impair BC growth. To achieve efficient genome editing in vivo, a vector is required to deliver the components to target cells. Gold nanomaterials, including gold nanoparticles and nanoclusters, display many advantageous characteristics that have facilitated their widespread use in theranostics, as delivery vehicles, and imaging and photothermal agents. This review highlights the therapeutic applications of CRISPR/Cas9 in treating BCs, and briefly describes gold nanomaterials and their potential in CRISPR/Cas9 delivery.
Keywords: Breast cancer, genome editing, CRISPR/Cas9, gold nanoparticles, gold nanoclusters
Introduction
Advances in the treatment of breast cancers (BCs) have led to significant improvements in the overall survival of patients. Local therapies, including surgery and radiotherapy, in conjunction with adjuvant targeted therapies and chemotherapy are mainstays of BC treatment.1 Based on immunohistochemical staining of hormone receptors (HR), human epidermal growth factor receptor-2 (HER2), and ki67, a marker of cell proliferation, BCs can be divided into four subtypes which respond to different therapies. The majority of BCs are HR+ luminal tumours, which can be further subdivided into luminal A (HR+, HER2-, low ki67) and B (HR+, HER2+/high ki67) subtypes.2 These cancers generally respond well to endocrine therapies targeting the estrogen receptor (ER) and display relatively good prognoses.3 HER2+ BCs, which are HR- and HER2+, are treated using anti-HER2 drugs. Triple negative breast cancers (TNBC) are HR- and HER2-, with chemotherapy and radiotherapy as the options for treatment. The aggressive nature of TNBC coupled with the lack of targeted therapies has led to poor prognosis and high risk of relapse and metastasis compared to other subtypes.3–5
Current treatments such as chemo- and radiotherapy are associated with adverse physical and cognitive side effects, as they are non-specific and also affect healthy cells.6 Many patients show intrinsic or acquired resistance to targeted therapies, chemotherapy, or radiotherapy, causing treatments to fail and leading to metastasis.7,8 Even if tumours display a pathologic complete response, the risk of relapse remains for decades after therapy.9 A major issue with current therapies is their dependence on the tumour phenotype. BCs show great molecular heterogeneity, with mutations in a variety of genes controlling cell growth, epigenetic modification, and transcription ultimately resulting in the malignant phenotype. Thus, genome editing techniques, which can directly target these genetic changes, show great promise in cancer therapy.
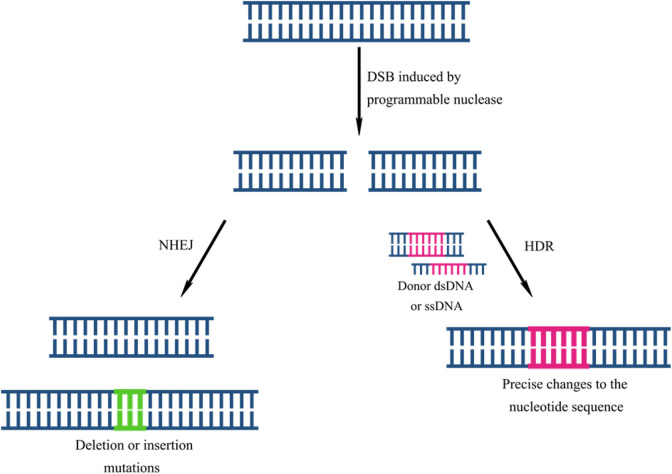
Genome editing involves modification of DNA, through the insertion, removal, or replacement of sequences. Current methods exploit endonucleases that introduce double-stranded breaks (DSB) into the DNA. Following cleavage, the DNA may be repaired by non-homologous end joining (NHEJ) or homology-directed repair (HDR) (Figure 1). HDR uses a donor DNA molecule as a template for repair.10 In this way, precise changes that correct gene function can be introduced into the genome. In contrast, NHEJ is an error-prone process in which the two ends of the DSB are ligated together, often leading to insertion or deletion (indel) mutations that may knock the gene out.10 Since NHEJ targets the gene directly, it allows for more effective silencing than RNA interference (RNAi), which indirectly silences gene expression by targeting mRNA.
Figure 1.
The process of genome editing by exploiting the natural repair mechanism of DSB repair, leading to random insertions or deletions via NHEJ or precise corrections via HDR.
Four genome editing systems with different programmable nucleases have been developed: meganucleases, which have not seen extensive use for genome editing, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system. Despite being the youngest genome editing system, CRISPR/Cas9 has shown the most potential in cancer therapy, and will be discussed in this review.
CRISPR/Cas9
In contrast to earlier genome editing systems, which mediate sequence recognition through protein-DNA interactions, the CRISPR/Cas system uses an RNA molecule to mediate binding. It is derived from an prokaryotic adaptive immune system protecting against invading viruses and plasmids, and is composed of CRISPR loci, comprised of alternating repeat-spacer units, and CRISPR-associated (Cas) proteins.11 Immunisation occurs in three stages: (i) adaptation, in which invading nucleic acids are cleaved by a complex of Cas endonucleases and the resulting fragments, called protospacers, are integrated into CRISPR loci between identical repeats; (ii) expression, in which the locus is transcribed into pre-CRISPR RNA (pre-crRNA) and processed into individual CRISPR RNA (crRNA) molecules; and (iii) interference, where the crRNA directs a single Cas endonuclease or a protein complex to cleave the foreign nucleic acids.12,13
CRISPR/Cas systems are broadly classified into two classes, further divided into six types and numerous subtypes, based on the mechanism by which recognition and cleavage occur.14 Class 1 systems use protein complexes to effect cleavage, while Class 2 systems only utilise one protein, making them more applicable for genome editing.15 All Class 2 systems (types II, V, and VI) have certain targeting constraints. Type VI systems, which utilise Cas13 to cleave RNA, recognise a protospacer flanking sequence (PFS).16 Type II and V systems recognise a conserved 2–5 bp sequence called the protospacer adjacent motif (PAM).17 Type V Cas proteins recognise a PAM directly upstream of the protospacer, such as 5′-TTTN-3′ recognised by the Cas12a, or Cpf1, protein.13,18 In contrast, the Cas9 endonuclease of type II systems recognise a PAM downstream of the protospacer.19
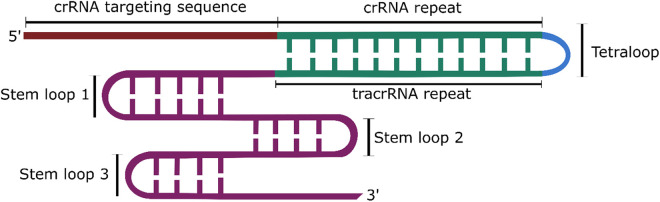
The type II CRISPR/Cas9 system is the best characterised and most commonly used CRISPR system. For cleavage, Cas9 requires an additional RNA molecule called the trans-activating crRNA (tracrRNA), which facilitates crRNA binding and maturation. However, for use in genome editing, the tracrRNA and crRNA can be connected via a linker into one molecule termed the single guide RNA (sgRNA) (Figure 2).
Figure 2.
Structure of the chimeric sgRNA, containing the targeting sequence of the crRNA and the hairpin loops of the tracrRNA. The repeat sequences of the crRNA and tracrRNA sequences are linked via a tetraloop to form one structure.
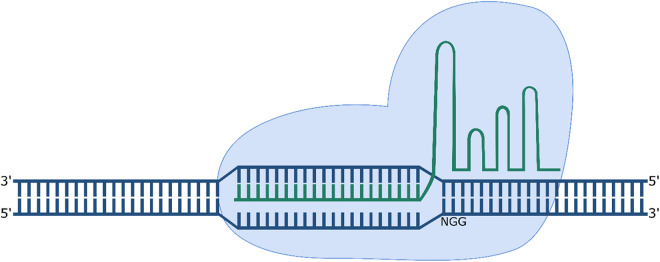
Cas9 undergoes a conformational change following binding of the gRNA, allowing it to search for the PAM sequence and cleave the target strand 3 bp upstream of the PAM (Figure 3).20 Cleavage is dependent not only on the presence of the PAM, but also on the complementarity of the “seed” sequence, the 10–12 nt of the target sequence adjacent to the PAM.21 The Streptococcus pyogenes Cas9 (spCas9) is the most popular Cas9 used for genome editing. It is a large 1368 amino acid protein; however, it recognises a short 5′-NGG-3′ PAM sequence compared to orthologues from other bacterial species.19
Figure 3.
CRISPR/Cas9 mediated cleavage of DNA. The sgRNA mediates binding to the 20 nt directly upstream of the 5′-NGG-3′ PAM sequence on the target strand.
The simplicity of CRISPR/Cas9 has made it the most used genome editing system. Compared to earlier systems, it is cheaper, easier to design, and can be retargeted without the need for protein engineering.22 The PAM requirement limits the choice of target sequence; however this may be overcome by using different Cas proteins, or spCas9 variants that have been modified to recognise different PAMs.23,24 The CRISPR system is also highly versatile and catalytically inactivated Cas9 s (dCas9), with mutated nuclease domains, have been conjugated to a variety of proteins and enzymes to perform different functions. CRISPR interference (CRISPRi) and activation (CRISPRa) techniques fuse dCas9 s with transcription repressors, such as the Krüppel associated box (KRAB) domain, or activators, to inhibit or promote transcription, respectively.25,26 Fusions with DNA methyltransferases (DNMT) and DNA deaminases can be used to modify methylation patterns and specific bases. These variants greatly broaden the scope of CRISPR applications in treating diseases such as cancer, which result from a variety of mutations.
Therapeutic potential of CRISPR in BC
CRISPR/Cas9 is a powerful tool for the study and treatment of various cancers, including BCs. Genes involved in cancer development are often categorised into two broad groups: proto-oncogenes, which promote cell growth and proliferation and, when mutated or activated drive tumour development; and tumour suppressor genes (TSGs), which are involved in DNA repair and control of cell growth, which when mutated or inactivated, lead to genomic instability and uncontrolled proliferation. Many different types of mutations, at the nucleotide, transcriptional, and epigenetic levels, may lead to their aberrant or inhibited expression. CRISPR/Cas9 and its variants can target mutations at each level and thus have great potential in treating BCs.
Targeting oncogenes and TSGs
Genes encoding growth factors and their receptors, transcription factors (TFs), signalling transducers, and chromatin remodelling proteins have oncogenic potential.27 CRISPR/Cas9 can be used to target these oncogenes directly, knocking them out and inhibiting cancer growth through various mechanisms. The technique has been successfully applied to knocking out both cellular and viral oncogenes in diverse cancer models, including leukaemia,28 cervical cancer,29 endometrial cancer,30 and prostate cancer.31 The PI3KCA, HER2/ErbB2, and MYC oncogenes have been implicated in BC.32 Knockout of HER2 has been observed to reduce the viability of HER2+ BT-474 and SKBR-3 cells.33 Notably, targeting of HER2 exon 12 produced a truncated protein with a dominant negative function rather than knocking out protein expression, suggesting that knockout of all copies of an oncogene in cancer cells may not always be necessary to exert a therapeutic effect. In vitro CRISPR/Cas9-mediated knockout of the Lipocalin 2 (Lcn2) oncogene, implicated in BC growth and metastasis in TNBC cells, did not show reduced cell proliferation, but instead inhibited cell migration by suppressing epithelial to mesenchymal transition, while in vivo treatment led to significantly reduced tumour growth.34
Alternatively, oncogenes can be targeted indirectly, by inhibiting aberrant transcriptional programmes that lead to overexpression. The upregulation of the MYC oncogene, overexpressed in 30–50% of high grade BCs, is often mediated by super enhancers, regions surrounding the gene bound by enhancer elements that bind TFs.35–37 TF binding can be impaired through CRISPR/Cas9-mediated mutagenesis or dCas9-DNMT-mediated methylation of the binding site, both of which have been observed to reduce MYC expression and cell proliferation in vitro.36 CRISPRi strategies have also shown potential in suppressing oncogene expression in squamous cell carcinoma cells and may be applied to BC therapy.38
Mutations in multiple TSGs such as PTEN, BRCA1 and BRCA2 have been identified in BCs. These TSGs play central roles in maintaining genome integrity by directing repair of DSBs through HR and NHEJ, and by ensuring progression of replication forks and restarting stalled forks.39–41 Restoring TSG function is more difficult than knocking out an oncogene.42 TSG expression, which may be repressed by dysregulated TFs or hypermethylated promoters, can be promoted using CRISPR variants. Expression of the PTEN TSG, whose loss is associated with more aggressive BC, has been activated in TNBC SUM159 cells using a CRISPRa approach, fusing dCas9 with the VPR domain consisting of the transcriptional activators VP64, p65, and Rta.43,44 Hypermethylation of TSGs, including PTEN and BRCA1, represses their expression.45–47 Removal of methylation can be facilitated by fusion with ten-eleven translocation (TET) dioxygenases. These enzymes convert 5′-mC to 5-hydroxymethylcytosine (5-hmc), which is corrected to C during DNA replication, removing methylation.48,49 dCas9-TET fusions have been used to demethylate the BRCA1 gene in vitro in MCF-7 and HeLa cells, upregulating BRCA1 expression and enhancing the cytotoxic effect of the chemotherapeutic Mitomycin-C.49 Fusion of dCas9 with an R2-stemloop, a short RNA sequence that recruits the DNMT1 enzyme and inhibits its activity, has also shown potential as a demethylation strategy.50
CRISPR/Cas9-mediated HDR can be used to correct small mutations, such as single nucleotide polymorphisms (SNPs) or indels, that knock out TSGs. The TP53 gene is estimated to be mutated in 30–35% of BCs and 80% of TNBCs.51 While it is not a “pure” TSG, as it may undergo gain-of-function mutations that drive oncogenesis, TP53 remains a prominent target for therapy.52 Correction of the TP53 414delC null mutation in PC-3 prostate cancer cells, leading to increased protein expression and apoptosis, has been facilitated using CRISPR, highlighting its potential for correcting mutations in BC.53 Base editing techniques provide a more precise method of correcting SNPs than HDR, as they facilitate corrections without inducing a DSB, reducing the chance of NHEJ-mediated repair. These techniques conjugate dCas9 to cytidine or adenosine deaminases. Correction of the TP53 Tyr163Cys mutation in HCC1954 BC cells has been achieved by fusing a Cas9 nickase with cytidine deaminase and uracil DNA glycosylase inhibitor (UGI) proteins, facilitating conversion of the mutant C-G pair to T-A.54 Adenosine deaminases convert A to inosine, which bonds C, ultimately leading to replacement of an A-T bp with C-G.55
Targeting DNA repair pathways
In addition to TSGs such as BRCA1/2 and PTEN, genome integrity is also maintained by poly-(ADP-ribose) polymerase (PARP) enzymes, involved in single-stranded break (SSB) repair and restarting stalled replication forks.56,57 Their inhibition impedes repair and replication, and has synthetic lethality with HR deficiency in BRCA1/2-deficient cancers. However, PARP knockout in these cancers may not be as effective as treatment with inhibitors, as the cytotoxic effect results from PARP trapping at the SSBs rather than from PARP downregulation.58–60 Instead, PARP gene knockout may be used in conjunction with platinum-based chemotherapeutics that induce DNA damage. CRISPR targeting of the PARP1 gene has been shown to enhance the cytotoxicity of cisplatin in ovarian cancer cells.61 The gene also presents a therapeutic target in BC patients receiving concurrent chemotherapy, as co-treatment with PARP inhibitors and the platinum-based drugs has shown improved survival in metastatic TNBC patients.62
Targeting the kinome
The kinome refers to kinase proteins involved in the phosphorylation of proteins and lipids.63 Their dysregulation is a common feature of many cancers, including BC. The tyrosine kinase family, which phosphorylate tyrosine residues, includes transmembrane receptor and cytoplasmic kinases.64 Well-studied oncogenes such as HER2, PI3KCA, and FGFR that are susceptible to knockout via CRISPR/Cas9, are members of this family.65
Cyclin-dependent kinases (CDKs) are serine/threonine kinases. They bind cyclins and regulate cell cycle progression through phosphorylation of the retinoblastoma (Rb) TSG, and transcription and RNA splicing through phosphorylation of RNA polymerase II and TFs.66 CDK4/6 inhibitors have been approved for combination therapy for HR+ BCs and THZ inhibitors have been developed for transcriptional CDKs (tCDKs); however, the development of drug resistance remains a potential issue.67,68 CRISPR/Cas9 knockout of the cell cycle CDK2 has been observed to induce cell cycle arrest in vitro in cutaneous melanoma cells.69 This CDK is a potential target for TNBC treatment, as it has been observed to promote tumourogenesis in vivo, with inhibition inducing ER expression in TNBC MDA-MB-231 cells, sensitising them to ER-targeted therapies.70 Overactivity of tCDKs can lead to transcriptional addiction, where cancers become “addicted” to transcription of genes that drove the initial stages of tumour formation, but remain necessary for cancer cell survival after tumourogenesis.71 Their knockout may thus induce tumour death. Knockout of tCDKs has identified CDK7 and CDK9 to be required for TNBC growth.72 CDK7 dependence was observed to be TNBC-specific, with knockout leading to reduced proliferation in vitro and reduced tumour growth in vivo.
Altering the epigenome
Epigenetic modifications are heritable changes to the DNA that do not involve changes to the nucleotide sequence. These modifications work together to regulate gene expression, and their dysregulation is thus a feature of tumourogenesis. The major mechanisms are DNA methylation, histone modification, and non-coding RNAs (ncRNA).
DNA methylation commonly occurs on the C of CpG repeats in promoter regions, called CpG islands, preventing TF binding and thus inhibiting gene expression.73 The process is carried out by DNMT1, DNMT3A, and DNMT3B enzymes, which convert C to 5′-methylcytosine (5′-mC) and ensure methylation is maintained during cell division.74 In addition to TSG hypermethylation, hypomethylation at the promotor level, facilitating expression of genes promoting tumour growth; and at the genome level, potentially destabilising chromosomes, have been observed in BCs.75–77 These abnormal methylation patterns can be altered using dCas9 variants, as described above, or by inhibiting mutated or overexpressed DNMT and TET enzymes.78–80 Current DNMT inhibitors are associated with adverse side effects, and their non-specific mechanism of action can induce global hypomethylation or demethylation of oncogenes, potentially promoting cancer growth.81,82 RNAi-mediated knockdown of DNMT1 has been observed to inhibit transformation without inducing excessive demethylation.81 DNMT1 knockout by CRISPR/Cas9 has shown significant anti-tumour activity in in vivo ovarian cancer models, and can thus potentially facilitate knockout of specific DNMT genes in BCs, inhibiting cancer growth with fewer side effects than DNMT inhibitors.83 TET1 has also been observed to be overexpressed in TNBC cells, inducing hypomethylation, with CRISPR knockout reducing cell migration and proliferation.84
Histone post-translational modifications in transcription start sites similarly influence DNA accessibility to TFs, by controlling whether the chromosomes are tightly bound, forming heterochromatin with repressed TF binding; or loosely bound, forming euchromatin open for transcription.85 Histone acetylation is associated with weakened DNA binding, promoting transcription, while methylation may increase or inhibit transcription.86 Various aberrant modifications have been identified in BC that alter the chromatin state and gene expression patterns, resulting from dysregulation of histone-modifying enzymes, such as lysine-specific demethylase 1 (LSD1).87–92 RNAi knockout of LSD1 has been observed to inhibit cellular proliferation in BC cell lines.90 Histone-modifying enzymes also modify non-histone proteins, altering their activity and potentially promoting tumour growth.93–95 Thus, overexpressed enzymes present a therapeutic target for CRISPR/Cas9 knockout. Alternatively, fusions of dCas9 with acetyltransferases,96 deacetyltransferases,97 methyltransferases,98,99 and demethylases can potentially be used to correct abnormal modifications, repressing oncogenes and activating TSGs.
Most non-coding DNA is transcribed as ncRNA, some of which conduct essential housekeeping and regulatory functions. The most widely studied ncRNA are microRNA (miRNA) and long non-coding RNA (lncRNA). miRNA (∼22 nt) regulate gene expression by binding complementary mRNA, leading to its degradation or repressed translation100; while lncRNA (>200 nt) alter chromatin states and TF binding, and influence mRNA and miRNA activity.101 Their dysregulated activity has been associated with chemoresistance and metastasis in BCs.102–106 However, their knockdown by RNAi is inhibited by the short length of miRNAs, and the nuclear localisation of some lncRNAs.107,108 CRISPR/Cas9 knockout of lncRNA and miRNA has been observed to impair growth and invasion in bladder, ovarian, and hepatocellular carcinoma models, and can potentially be applied to knockout in BC.107,109,110 Moreover, CRISPR has been observed to induce long-term knockouts of miRNA in vitro in colon cancer cells.108 However, NHEJ may not efficiently knockout ncRNAs, as non-coding sequences may tolerate indels.111 This may be overcome by using multiple sgRNA to delete the gene, a strategy which has shown potential for lncRNA knockout.112,113 CRISPRi has also shown potential for inhibition of ncRNA transcription in a study which identified the PVT1 promoter for the PVT1 lncRNA to act as a tumour suppressor, and can thus potentially be used to inhibit oncogenic ncRNA.114 Type VI Cas13 systems may also be exploited to target and cleave ncRNA.
Reversing drug resistance
BCs may develop resistance to chemotherapeutics through a variety of mechanisms. Resistance in HR+ and HER+ BCs may result from the loss of ERα and HER2 expression, often resulting from epigenetic changes that silence these genes.115,116 Multiple SNPs associated with resistance, such as ERα Tyr537Ser and Asp538Gly, which drive constitutive expression by promoting interactions with coactivators, and HER2 mutations Lys753Glu and Leu755Ser, have also been associated with resistance to lapatinib and trastuzumab.117–120 Resistance to PARP inhibitors may result from BRCA reactivation or PARP1 point mutations.121,122 These mutations can be altered using CRISPR/Cas9 HDR, or the previously described Cas9 variants, conjugated to epigenetic modulators or base editors, to re-sensitise tumours to therapy.
Multidrug resistance is often mediated by overexpression of the ATP-binding cassette (ABC) transporters that remove drugs from cancer cells before therapeutically active concentrations can accumulate. The P-glycoprotein (P-gp), Multidrug Resistance-Associated Protein 1 (MRP1), and Breast Cancer Resistance Protein (BCRP) transporters have been recognised as playing a major role in resistance.123 Knockout of these efflux pumps facilitates re-sensitisation to existing drugs and avoids the need to develop new therapies.124 CRISPR-mediated knockout of P-gp has been observed to increase chemosensitivity of A2780/ADR ovarian cancer cells, and increase the intracellular doxorubicin concentrations in resistant MCF-7/ADR cells, leading to increased cell death following treatment compared to unedited cells.124,125
Immunotherapy
Immunotherapy involves boosting the immune response to tumour cells. Cancers may avoid immune responses through overexpression of immune checkpoint proteins, normally responsible for preventing autoimmune responses.126 These proteins bind receptors on the surface of immune cells, averting immune attack. The expression of various checkpoint proteins has been observed in BCs and particularly in TNBC, including CD155 and programmed cell death-ligand 1 (PD-L1), which recognises the programmed cell death 1 (PD-1) receptor on immune cells.127–130 CRISPR/Cas9 knockout of either PD-L1 or its receptor may trigger an immune response against the tumour.131,132 CRISPR knockout of CDK5 has also been shown to downregulate PD-L1 expression, inhibiting tumour growth in vitro and in vivo.133,134 shRNA-mediated knockdown of CD155 has also been observed to inhibit the growth of in vitro and in vivo BC models, highlighting its potential in BC therapy.135
Knockdown of checkpoint proteins may be used to enhance the efficacy of chimeric antigen receptor (CAR) T-cells, which express CARs recognising tumour-associated antigens (TAAs).136 Multiple TAAs, such as HER2, mucin1, and TEM8 have shown potential as targets for BC CAR T-cell therapy.137–139 CRISPR/Cas9 knockout of PD-1 has been shown to enhance the anti-cancer activity of CAR T-cells targeting mesothelin, overexpressed in TNBC BT-459 cells.140 However, T-cells must be isolated from the patient and edited ex vivo in a laborious and lengthy process. Universal T-cells eliminate the need for isolation; however, class I human leucocyte antigens (HLA) and T-cell receptors (TCR) present on the surface of donor T-cells must be removed to prevent graft-vs-host disease.141 CRISPR/Cas9-mediated HDR allows for simultaneous knockout of TCRs and/or HLAs and knockin of CAR-encoding genes. Introduction of the anti-CD19 CAR into the TCR locus has been shown using CRISPR, leading to effective CAR expression while avoiding T-cell exhaustion.142 Multiplex strategies, allowing simultaneous knock out of TCRs, beta-2 microglobulin (B2 M), a subunit of HLA-I, and other proteins including PD-1 and CTLA-4, have also been used to generate allogeneic CAR T-cells with enhanced anti-cancer activity.141,143,144
Targeting cellular adhesion
Integrins are transmembrane proteins that function in cell adhesion to the extracellular matrix (ECM).145 When dysregulated, they play important roles in both tumour development and metastasis, by promoting cell motility, growth and survival, modifying the ECM to promote growth, and facilitating survival of circulating cancer cells and metastatic colonisation.145 CRISPR knockout of these integrins may thus reduce the tumour-forming and metastatic potential of BC cells, as shown by knockout of integrin α5 (ITGA5), an integrin that promotes tumour cell migration and metastasis to the lymph nodes and lungs.146,147
Delivery of CRISPR/Cas9
Therapeutic efficacy of CRISPR/Cas9 is dependent on its ability to reach target cells with minimal biodegradation. The Cas9 and sgRNA may be delivered in the form of the ribonucleoprotein (RNP), a plasmid, or as a combination of Cas9 mRNA and sgRNA. Plasmids are stable and easily synthesised, and can simultaneously encode and deliver all required components. However, their large size, with the ∼4.2 kbp spCas9 gene, and strong negative charge hinder delivery.148 Moreover, plasmids must first undergo transcription and translation, with expression continuing for prolonged periods after transfection, leading to delayed editing and increased off-targets compared to RNP delivery.149 In contrast, delivery of the RNP produces rapid therapeutic effects, and its relatively short expression time before protease degradation reduces the chance of off-targets. However, the large protein size and the net negative charge of the complex may interfere with cellular uptake.150 Delivery of the mRNA and sgRNA similarly avoids the need for nuclear localisation and leads to transient Cas9 expression with reduced off-target effects.151 However, the instability of RNA and the long Cas9 mRNA length of ∼4500 nt complicates delivery.152
Delivery methods can be broadly categorised as physical, viral or non-viral. Physical methods include electroporation, hydrodynamic injection, and microinjection. However, these techniques are often difficult to apply in vivo and may damage cells.153 Viral vectors have seen widespread use due to their high transfection efficiencies; however, issues such as their limited packaging size, difficult synthesis, and immunogenic and carcinogenic risks have resulted in a shift to non-viral vectors, and nanoparticles (NP) in particular.154,155 NPs, ranging from 1–100 nm in size, have shown great potential as gene and drug delivery vehicles due to their lower immunogenicity, tunable synthesis, and large loading capacity. Their large surface area-to-volume ratio allows for coating with polymers that facilitate CRISPR/Cas9 binding and protection, and functionalisation with compounds that promote targeting and increased circulation times.156 Inorganic NPs, composed of metals, magnetic compounds, selenium and silica, have been widely investigated as delivery vehicles for cancer therapy. Gold nanomaterials are among the most popular and have the potential as carriers for CRISPR/Cas9-mediated genome editing.
Gold nanomaterials
Gold nanomaterials display many unique optical and physiochemical properties that facilitate their use as imaging agents, biosensors, and vectors. Among their attractive properties are their small size, and facile synthesis and functionalisation. Their highly tunable synthesis allows for modification of NP size and shape to optimise characteristics for therapy. Moreover, the ability of gold nanomaterials to convert absorbed light energy into heat following irradiation with near infrared light, leading to thermal ablation of surrounding tumour tissue, permits their use as photothermal therapy agents.157
Spherical gold nanoparticles (AuNPs) are among the most popular NPs for gene and drug delivery. Their unique optical properties include their localised surface plasmon resonance (LSPR), the phenomenon where free electrons on the NP surface oscillate in response to light exposure. The electrons absorb and scatter light energy, facilitating the use of AuNP as imaging agents.158 AuNP are most commonly synthesised using the citrate reduction method, in which chloroauric acid (HAuCl4) is reduced by trisodium citrate, to produce citrate-capped AuNPs 10–20 nm in diameter and with a net negative charge.159,160 This method can be easily modified by varying the ratios of reagents to produce AuNPs of 15–150 nm in diameter.161
Gold nanoclusters (AuNCs) have more recently been investigated as carriers, as their advantageous qualities facilitate their use as simultaneous delivery and imaging agents. These ultrasmall (<2 nm) NPs consist of only a few to tens of Au atoms, imparting them with characteristics unique from conventional AuNPs, such as the absence of SPR and size-dependent fluorescence.162 They display a large Stokes shift, good photostability, resistance to photobleaching, and low toxicity compared to conventional fluorophores such as dyes and quantum dots.163,164 These optical properties have led to AuNCs being widely studied as sensors and cellular imaging agents.165–168 AuNCs are commonly synthesised through reduction of HAuCl4 with biomolecules such as peptides,169,170 proteins,168,171 and polymers.164,172,173 Glutathione (GSH) is often used as a capping agent in a simple and eco-friendly synthesis process requiring no additional reducing agents, to produce biocompatible AuNCs with a relatively high quantum yield.174,175
For use as delivery agents, inorganic NPs are often coated with polymers and conjugated to various ligands to enhance stability, biocompatibility, and transfection. As-synthesised capped AuNPs and AuNCs can easily bond with cationic polymers, such as chitosan and polyethyleneimine (PEI), facilitating interactions with anionic cell membrane components and anionic groups of other ligands. Functionalisation can alternatively exploit the semi-covalent interactions between sulphur and gold to bind thiol groups, allowing conjugation with peptides, proteins, and thiol-modified nucleic acids. The hydrophilic polymer polyethylene glycol (PEG) is widely used to inhibit interactions with plasma proteins, thus preventing uptake by the reticuloendothelial system and increasing circulation times and bioavailability. Specific tumour accumulation may be achieved through functionalisation with targeting ligands that bind receptors on the surface of the target cells, facilitating uptake by receptor-mediated endocytosis. In addition to HRs and HER2, BCs have been shown to overexpress folate and transferrin receptors, and their ligands are commonly attached to NPs for targeting.176–178 The difficulties associated with nuclear localisation can be overcome through conjugation with nuclear localisation signals (NLS), peptides that promote interactions with the nuclear pores.179 The cationic HIV-1 trans-activator of transcription (TAT) peptide is the most popular NLS, and also acts as a cell-penetrating peptide (CPP) promoting transport across the cell membrane.180
Gold nanomaterials for CRISPR delivery
AuNPs and AuNCs are relatively novel vehicles for CRISPR delivery and have not been exploited, compared to viral vectors and organic lipid and polymeric NPs. While they have not been used to mediate genome editing of BC, they have shown their potential in studies delivering CRISPR components to various malignant cells in vivo and in vitro, achieving editing efficiencies comparable to other delivery systems.
Arginine-coated AuNPs have been shown capable of delivering RNPs, and facilitating AASV1 and PTEN knockdown in vitro.181,182 AuNPs were able to enter cells via a membrane fusion process, avoiding potential lysosomal degradation, leading to editing efficiencies of ∼20–30%. RNP binding has been achieved by binding a thiol-modified crRNA to the AuNP, and reacting the resulting AuNP-crRNA with Cas9 to form the RNP.183 Delivery of the large CRISPR/Cas9 plasmid targeting the Plk-1 gene has been described using lipid-encapsulated TAT-coated AuNPs in in vitro and in vivo melanoma tumours.184 Controlled irradiation with 514 nm light following transfection was used to promote plasmid release without inducing cell death, producing a synergistic effect in tumour ablation.
The delivery of an HDR template can be achieved in multiple ways using AuNPs, and may be performed simultaneously with the Cas9 and sgRNA due to AuNPs’ large loading capacity. A layer-by-layer approach has been used, in which the ssDNA template was complexed with a PEI layer coating the AuNP-RNP.183 Another study utilised AuNPs complexed with thiol-linked ssDNA molecules that bound the donor DNA and RNP, and coated with the polymer PAsp(DET).185 These AuNPs achieved an in vitro HDR frequency of 3–4%, significantly higher than lipofectamine transfection, and facilitated in vivo correction of the dystrophin gene.
The potential of AuNCs for CRISPR delivery has been highlighted in several studies. Lipid-encapsulated TAT-coated AuNCs carrying both the Cas9 protein and Plk1 sgRNA encoded as a plasmid produced editing efficiencies of 26.2% in vitro, and significantly inhibited tumour growth in vivo in melanoma models.186 A proof-of-concept study showed that GSH-AuNCs can self-assemble with Cas9 proteins at physiological pH, producing spCas9-AuNC complexes which dissociate under acidic pH.187 Transfection with these complexes and sgRNA targeting the viral E6 oncogene in HeLa cells produced editing efficiencies of 34% and significantly reduced protein expression. These Cas9-AuNCs could facilitate genome editing in systems where the components (Cas9, sgRNA, template for HDR) are delivered separately.
While AuNCs have not been exploited to deliver CRISPR nucleic acids, they have shown potential as gene delivery agents. PEI-coated AuNCs have facilitated improved delivery of the EGFP gene compared to PEI in a proof-of-concept study.172 AuNCs capped with the positive tetrapeptide K4 have also been shown capable of assembling with DNA and RNA.188
Conclusion
The use of CRISPR/Cas9 has exploded since it was first adapted for use in genome editing in 2013.189,190 Its simplicity, ease of use, and versatility have led to its widespread use in all aspects of cancer research. CRISPR/Cas9 has the ability to target any oncogenic mutation through simple redesign of an RNA sequence, and provides a means of exploiting the molecular heterogeneity of BCs to tailor therapies to specific individuals, thus avoiding treatment with ineffective or cytotoxic drugs. Personalised treatments for BC, targeting the hormone and HER2 receptors, have significantly improved the outcomes of many patients. However, CRISPR now allows for extension of these tailored treatments to individuals suffering from TNBC.
However, before the progression of CRISPR therapies into clinical settings, their safety must be thoroughly evaluated to avoid adverse side effects. Off-target cleavage is a concern, as mismatches outside the PAM and seed sequences in the sgRNA are tolerated.191 The use of bioinformatics tools that evaluate sgRNA and identify possible off-targets and spCas9 variants with improved fidelity reduces non-specific activity.192–194 Studies have also observed innate and adaptive immunity against the spCas9 protein since it originates from pathogenic bacteria.195,196 Cas9-induced immune responses may also lead to responses being raised against the edited cells. Further studies are required to assess the likelihood of adverse immune responses as, thus far, the majority of CRISPR clinical trials (as listed on U.S. National Library of Medicine’s site, https://ClinicalTrials.gov) focus on ex vivo editing of T-cells. However, an ongoing trial (NCT03872479) is assessing the safety of adenoviral-packaged CRISPR/Cas9 correcting the CEP290 gene, in the first trial to attempt editing in humans.197,198
Efficient and selective delivery of therapeutics is a universal issue faced in gene and drug therapy. However, the development of CRISPR delivery systems will benefit from the extensive research that has already been conducted. AuNPs have proven themselves as delivery agents for gene therapy, and show great promise for the delivery of CRISPR/Cas9 systems. Their capacity for multi-functionalisation allows for efficient delivery of all CRISPR/Cas9 formats. Before their clinical translation, the biodistribution and fate of various sizes and shapes of AuNPs and AuNCs must be clearly determined, as gold is non-biodegradable.
Overall, the CRISPR/Cas9 system can be applied to the treatment of BCs to develop highly effective precision medicines. With further optimisation, these systems may produce treatments that overcome the limitations faced by current therapies and significantly improve the survival of patients suffering from BC.
Footnotes
Declaration of Conflicting Interests: The author(s) declare no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors acknowledge the National Research Foundation of South Africa, grant numbers 113850/81289, for supporting the research of the authors.
ORCID iD: Moganavelli Singh  https://orcid.org/0000-0002-9985-6567
https://orcid.org/0000-0002-9985-6567
References
- 1. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA 2019; 321: 288–300. [DOI] [PubMed] [Google Scholar]
- 2. McCubrey JA, Davis NM, Abrams SL, et al. Targeting breast cancer initiating cells: advances in breast cancer research and therapy. Adv Biol Regul 2014; 56: 81–107. [DOI] [PubMed] [Google Scholar]
- 3. Darbeheshti F, Izadi P, Razavi ANE, et al. Comparison of BRCA1 expression between triple-negative and luminal breast tumors. Iran Biomed J 2018; 22: 210–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costa RLB, Gradishar WJ. Triple-negative breast cancer: current practice and future directions. J Oncol Pract 2017; 13: 301–303. [DOI] [PubMed] [Google Scholar]
- 5. Wu X, Baig A, Kasymjanova G, et al. Pattern of local recurrence and distant metastasis in breast cancer by molecular subtype. Cureus 2016; 8: e924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chang L, Weiner LS, Hartman SJ, et al. Breast cancer treatment and its effects on aging. J Geriatr Oncol 2019; 10: 346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vu T, Claret FX. Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol 2012; 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haque MM, Desai K V. Pathways to endocrine therapy resistance in breast cancer. Front Endocrinol (Lausanne) 2019; 10: 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ignatov A, Eggemann H, Burger E, et al. Patterns of breast cancer relapse in accordance to biological subtype. J Cancer Res Clin Oncol 2018; 144: 1347–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhan T, Rindtorff N, Betge J, et al. CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol 2019; 55: 106–119. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Peng N. Endogenous CRISPR-Cas system-based genome editing and antimicrobials: review and prospects. Front Microbiol 2019; 10: 2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc B Biol Sci 2016; 371: 20150496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015; 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pyenson NC, Gayvert K, Varble A, et al. Broad targeting specificity during bacterial type III CRISPR-Cas immunity constrains viral escape. Cell Host Microbe 2017; 22: 343–353.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mir A, Edraki A, Lee J, et al. Type II-C CRISPR-Cas9 biology, mechanism, and application. ACS Chem Biol 2018; 13: 357–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burmistrz M, Krakowski K, Krawczyk-Balska A. RNA-targeting CRISPR–Cas systems and their applications. Int J Mol Sci 2020; 21: 1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gleditzsch D, Pausch P, Müller-Esparza H, et al. PAM identification by CRISPR-Cas effector complexes: diversified mechanisms and structures. RNA Biol 2019; 16: 504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Z, Edwards H, Xu P. CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab Eng Commun 2020; 10: e00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leenay RT, Beisel CL. Deciphering, communicating, and engineering the CRISPR PAM. J Mol Biol 2017; 429: 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Rhun A, Escalera-Maurer A, Bratovič M, et al. CRISPR-Cas in Streptococcus pyogenes. RNA Biol 2019; 16: 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ribeiro LF, Ribeiro LFC, Barreto MQ, et al. Protein engineering strategies to expand CRISPR-Cas9 applications, Platero RA. (ed.). Int J Genomics 2018; 2018: 1652567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lone BA, Karna SKL, Ahmad F, et al. CRISPR/Cas9 system: a bacterial tailor for genomic engineering. Genet Res Int 2018; 2018: 3797214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018; 556: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nishimasu H, Shi X, Ishiguro S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018; 361: 1259–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015; 12: 326–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tavassoli M, Pezzella F. Oncogenesis and tumour suppression In: Pezzella F, Tavassoli M, Kerr DJ. (eds) Oxford Textbook of Cancer Biology, 1st ed Oxford: Oxford University Press, 2019. pp. 136–154. [Google Scholar]
- 28. Narimani M, Sharifi M, Jalili A. Knockout of BIRC5 gene by CRISPR/Cas9 induces apoptosis and inhibits cell proliferation in leukemic cell lines, HL60 and KG1. Blood Lymphat Cancer Targets Ther 2019; 9: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu Z, Yu L, Zhu D, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int 2014; 2014: 612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cai J, Huang S, Yi Y, et al. Ultrasound microbubble-mediated CRISPR/Cas9 knockout of C-erbB-2 in HEC-1A cells. J Int Med Res 2019; 47: 2199–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawamura N, Nimura K, Nagano H, et al. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget 2015; 6: 22361–22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pranavathiyani G, Thanmalagan RR, Leimarembi Devi N, et al. Integrated transcriptome interactome study of oncogenes and tumor suppressor genes in breast cancer. Genes Dis 2019; 6: 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang H, Sun W. CRISPR-mediated targeting of HER2 inhibits cell proliferation through a dominant negative mutation. Cancer Lett 2017; 385: 137–143. [DOI] [PubMed] [Google Scholar]
- 34. Guo P, Yang J, Huang J, et al. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc Natl Acad Sci U S A 2019; 116: 18295–18303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen D, Zhao Z, Huang Z, et al. Super enhancer inhibitors suppress MYC driven transcriptional amplification and tumor progression in osteosarcoma. Bone Res 2018; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schuijers J, Manteiga JC, Weintraub AS, et al. Transcriptional dysregulation of MYC reveals common enhancer-docking mechanism. Cell Rep 2018; 23: 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fallah Y, Brundage J, Allegakoen P, et al. MYC-driven pathways in breast cancer subtypes. Biomolecules 2017; 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshida M, Yokota E, Sakuma T, et al. Development of an integrated CRISPRi targeting △Np63 for treatment of squamous cell carcinoma. Oncotarget 2018; 9: 29220–29232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou S-Q, Ouyang M, Brandmaier A, et al. PTEN in the maintenance of genome integrity: from DNA replication to chromosome segregation. Bioessays 2017; 39 DOI: 10.1002/bies.201700082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gorodetska I, Kozeretska I, Dubrovska A. BRCA genes: the role in genome stability, cancer stemness and therapy resistance. J Cancer 2019; 10: 2109–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sulkowski PL, Scanlon SE, Oeck S, et al. PTEN regulates nonhomologous end joining by epigenetic induction of NHEJ1/XLF. Mol Cancer Res 2018; 16: 1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Morris LGT, Chan TA. Therapeutic targeting of tumor suppressor genes. Cancer 2015; 121: 1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moses C, Nugent F, Waryah CB, et al. Activating PTEN tumor suppressor expression with the CRISPR/dCas9 system. Mol Ther – Nucleic Acids 2019; 14: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li S, Shen Y, Wang M, et al. Loss of PTEN expression in breast cancer: association with clinicopathological characteristics and prognosis. Oncotarget 2017; 8: 32043–32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 2000; 92: 564–569. [DOI] [PubMed] [Google Scholar]
- 46. Zhu X, Shan L, Wang F, et al. Hypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast Cancer Res Treat 2015; 150: 479–486. [DOI] [PubMed] [Google Scholar]
- 47. Luo S, Chen J, Mo X. The association of PTEN hypermethylation and breast cancer: a meta-analysis. Onco Targets Ther 2016; 9: 5643–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu XS, Wu H, Ji X, et al. Editing DNA methylation in the mammalian genome. Cell 2016; 167: 233–247.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choudhury SR, Cui Y, Lubecka K, et al. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016; 7: 46545–46556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu A, Wang J, Sun W, et al. Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation. Cell Discov 2019; 5: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Duffy MJ, Synnott NC, Crown J. Mutant p53 in breast cancer: potential as a therapeutic target and biomarker. Breast Cancer Res Treat 2018; 170: 213–219. [DOI] [PubMed] [Google Scholar]
- 52. Soussi T, Wiman KG. TP53: an oncogene in disguise. Cell Death Differ 2015; 22: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Batır MB, Şahin E, Çam FS. Evaluation of the CRISPR/Cas9 directed mutant TP53 gene repairing effect in human prostate cancer cell line PC-3. Mol Biol Rep 2019; 46: 6471–6484. [DOI] [PubMed] [Google Scholar]
- 54. Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016; 533: 420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017; 551: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bryant HE, Petermann E, Schultz N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 2009; 28: 2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ström CE, Johansson F, Uhlén M, et al. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 2011; 39: 3166–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434: 913–917. [DOI] [PubMed] [Google Scholar]
- 59. Fong PC, Boss DS, Yap TA, et al. Inhibition of poly(ADP-Ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361: 123–134. [DOI] [PubMed] [Google Scholar]
- 60. Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Molecul Oncol 2011; 5: 387–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim SM, Yang Y, Oh SJ, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release 2017; 266: 8–16. [DOI] [PubMed] [Google Scholar]
- 62. O’Shaughnessy J, Osborne C, Pippen JE, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 2011; 364: 205–214. [DOI] [PubMed] [Google Scholar]
- 63. Duong-Ly KC, Peterson JR. The human kinome and kinase inhibition. Curr Protoc Pharmacol 2013; 60: 2.9.1–2.9.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paul MK, Mukhopadhyay AK. Tyrosine kinase – role and significance in cancer. Int J Med Sci 2012; 1: 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hynes NE. Tyrosine kinase signalling in breast cancer. Breast Cancer Res 2000; 2: 154–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sánchez-Martínez C, Lallena MJ, Sanfeliciano SG, et al. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs: recent advances (2015–2019). Bioorganic Med Chem Lett 2019; 29: 126637. [DOI] [PubMed] [Google Scholar]
- 67. McCartney A, Migliaccio I, Bonechi M, et al. Mechanisms of resistance to CDK4/6 inhibitors: potential implications and biomarkers for clinical practice. Front Oncol 2019; 9: 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao Y, Zhang T, Terai H, et al. Overcoming resistance to the THZ series of covalent transcriptional CDK Inhibitors. Cell Chem Biol 2018; 25: 135–142.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu H, Li Z, Huo S, et al. Induction of G0/G1 phase arrest and apoptosis by CRISPR/Cas9-mediated knockout of CDK2 in A375 melanocytes. Mol Clin Oncol 2020; 12: 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nie L, Wei Y, Zhang F, et al. CDK2-mediated site-specific phosphorylation of EZH2 drives and maintains triple-negative breast cancer. Nat Commun 2019; 10: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell 2017; 168: 629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Y, Zhang T, Kwiatkowski N, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 2015; 163: 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kang JG, Park JS, Ko JH, et al. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci Rep 2019; 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 2018; 19: 81–92. [DOI] [PubMed] [Google Scholar]
- 75. Roll JD, Rivenbark AG, Sandhu R, et al. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol 2013; 95: 276–287. [DOI] [PubMed] [Google Scholar]
- 76. Sharma G, Mirza S, Parshad R, et al. CpG hypomethylation of MDR1 gene in tumor and serum of invasive ductal breast carcinoma patients. Clin Biochem 2010; 43: 373–379. [DOI] [PubMed] [Google Scholar]
- 77. Kim SJ, Kang H-S, Chang HL, et al. Promoter hypomethylation of the N-acetyltransferase 1 gene in breast cancer. Oncol Rep 2008; 19: 663–668. [PubMed] [Google Scholar]
- 78. Shin E, Lee YK, Koo JS. Differential expression of the epigenetic methylation-related protein DNMT1 by breast cancer molecular subtype and stromal histology. J Transl Med 2016; 14: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kullmann K, Deryal M, Ong MF, et al. DNMT1 genetic polymorphisms affect breast cancer risk in the central European Caucasian population. Clin Epigenetics 2013; 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Girault I, Tozlu S, Lidereau R, et al. Expression analysis of DNA methyltransferases 1, 3A, and 3B in sporadic breast carcinomas. Clin Cancer Res 2003; 9: 4415–4422. [PubMed] [Google Scholar]
- 81. Chik F, Szyf M. Effects of specific DNMT gene depletion on cancer cell transformation and breast cancer cell invasion; toward selective DNMT inhibitors. Carcinogenesis 2011; 32: 224–232. [DOI] [PubMed] [Google Scholar]
- 82. Ateeq B, Unterberger A, Szyf M, et al. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia 2008; 10: 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. He ZY, Zhang YG, Yang YH, et al. In vivo ovarian cancer gene therapy using CRISPR-Cas9. Hum Gene Ther 2018; 29: 223–233. [DOI] [PubMed] [Google Scholar]
- 84. Good CR, Panjarian S, Kelly AD, et al. TET1-Mediated hypomethylation activates oncogenic signaling in triple-Negative breast cancer. Cancer Res 2018; 78: 4126–4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Marcum JA. The cancer epigenome: a review. J Biotechnol Biomed 2019; 2: 067–083. [Google Scholar]
- 86. Yen CY, Huang HW, Shu CW, et al. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett 2016; 373: 185–192. [DOI] [PubMed] [Google Scholar]
- 87. Jin W, Li QZ, Liu Y, et al. Effect of the key histone modifications on the expression of genes related to breast cancer. Genomics 2020; 112: 853–858. [DOI] [PubMed] [Google Scholar]
- 88. Xi Y, Shi J, Li W, et al. Histone modification profiling in breast cancer cell lines highlights commonalities and differences among subtypes. BMC Genomics 2018; 19: 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Karsli-Ceppioglu S, Dagdemir A, Judes G, et al. The epigenetic landscape of promoter genome-wide analysis in breast cancer. Sci Rep 2017; 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lim S, Janzer A, Becker A, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 2010; 31: 512–520. [DOI] [PubMed] [Google Scholar]
- 91. Yokoyama Y, Matsumoto A, Hieda M, et al. Loss of histone H4K20 trimethylation predicts poor prognosis in breast cancer and is associated with invasive activity. Breast Cancer Res 2014; 16: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J Cancer 2008; 122: 1207–1213. [DOI] [PubMed] [Google Scholar]
- 93. Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene 2007; 26: 5420–5432. [DOI] [PubMed] [Google Scholar]
- 94. Patel JH, Du Y, Ard PG, et al. The c-MYC oncoprotein is a substrate of the acetyltransferases hGCN5/PCAF and TIP60. Mol Cell Biol 2004; 24: 10826–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Carlson SM, Gozani O. Nonhistone lysine methylation in the regulation of cancer pathways. Cold Spring Harb Perspect Med 2016; 6: a026435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hilton IB, D’Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 2015; 33: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Xiao B, Yin S, Hu Y, et al. Epigenetic editing by CRISPR/dCas9 in Plasmodium falciparum. Proc Natl Acad Sci USA 2019; 116: 255–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fukushima HS, Takeda H, Nakamura R. Targeted in vivo epigenome editing of H3K27me3. Epigenetics Chromatin 2019; 12: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Cano-Rodriguez D, Gjaltema RAF, Jilderda LJ, et al. Writing of H3K4Me3 overcomes epigenetic silencing in a sustained but context-dependent manner. Nat Commun 2016; 7: 12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Loh HY, Norman BP, Lai KS, et al. The regulatory role of microRNAs in breast cancer. Int J Mol Sci 2019; 20: 4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Dai X, Kaushik AC, Zhang J. The emerging role of major regulatory RNAs in cancer control. Front Oncol 2019; 9: 920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Shi SJ, Wang LJ, Yu B, et al. LncRNA-ATB promotes trastuzumab resistance and invasion metastasis cascade in breast cancer. Oncotarget 2015; 6: 11652–11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xue X, Yang YA, Zhang A, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016; 35: 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kastl L, Brown I, Schofield AC. MiRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat 2012; 131: 445–454. [DOI] [PubMed] [Google Scholar]
- 105. Zhu S, Wu H, Wu F, et al. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 2008; 18: 350–359. [DOI] [PubMed] [Google Scholar]
- 106. Mansoori B, Mohammadi A, Ghasabi M, et al. miR-142-3p as tumor suppressor miRNA in the regulation of tumorigenicity, invasion and migration of human breast cancer by targeting Bach-1 expression. J Cell Physiol 2019; 234: 9816–9825. [DOI] [PubMed] [Google Scholar]
- 107. Zhen S, Hua L, Liu YH, et al. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 2017; 8: 9634–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chang H, Yi B, Ma R, et al. CRISPR/Cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep 2016; 6: 22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou S-J, Deng YL, Liang HF, et al. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and notch signaling in hepatocellular carcinoma. Cell Death Differ 2017; 24: 1577–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Huo W, Zhao G, Yin J, et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J Cancer 2017; 8: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yang J, Meng X, Pan J, et al. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol 2018; 15: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Ho T-T, Zhou N, Huang J, et al. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res 2015; 43: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Han J, Zhang J, Chen L, et al. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol 2014; 11: 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Cho SW, Xu J, Sun R, et al. Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 2018; 173: 1398–1412.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang X, Phillips DL, Ferguson AT, et al. Synergistic activation of functional estrogen receptor (ER)-α by DNA methyltransferase and histone deacetylase inhibition in human ER-α-negative breast cancer cells. Cancer Res 2001; 61: 7025–7029. [PubMed] [Google Scholar]
- 116. Ottaviano YL, Issa JP, Pari FF, et al. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res 1994; 54: 2552–2555. [PubMed] [Google Scholar]
- 117. Fiorillo M, Sanchez-Alvarez R, Sotgia F, et al. The ER-alpha mutation Y537S confers Tamoxifen-resistance via enhanced mitochondrial metabolism, glycolysis and Rho-GDI/PTEN signaling: implicating TIGAR in somatic resistance to endocrine therapy. Aging (Albany NY) 2018; 10: 4000–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Jia S, Miedel MT, Ngo M, et al. Clinically observed estrogen receptor alpha mutations within the ligand-binding domain confer distinguishable phenotypes. Oncol 2018; 94: 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Toy W, Shen Y, Won H, et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 2013; 45: 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zuo WJ, Jiang YZ, Wang YJ, et al. Dual characteristics of novel HER2 kinase domain mutations in response to HER2-targeted therapies in human breast cancer. Clin Cancer Res 2016; 22: 4859–4869. [DOI] [PubMed] [Google Scholar]
- 121. Francica P, Rottenberg S. Mechanisms of PARP inhibitor resistance in cancer and insights into the DNA damage response. Genome Med 2018; 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Pettitt SJ, Krastev DB, Brandsma I, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 2018; 9: 1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mao Q, Unadkat JD. Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport – an update. AAPS J 2015; 17: 65–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ha JS, Byun J, Ahn DR. Overcoming doxorubicin resistance of cancer cells by Cas9-mediated gene disruption. Sci Rep 2016; 6: 22847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Norouzi-Barough L, Sarookhani M, Salehi R, et al. CRISPR/Cas9, a new approach to successful knockdown of ABCB1/P-glycoprotein and reversal of chemosensitivity in human epithelial ovarian cancer cell line. Iran J Basic Med Sci 2018; 21: 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015; 27: 450–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sobral-Leite M, Van de Vijver K, Michaut M, et al. Assessment of PD-L1 expression across breast cancer molecular subtypes, in relation to mutation rate, BRCA1-like status, tumor-infiltrating immune cells and survival. Oncoimmunology 2018; 7: e1509820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Yuan C, Liu Z, Yu Q, et al. Expression of PD-1/PD-L1 in primary breast tumours and metastatic axillary lymph nodes and its correlation with clinicopathological parameters. Sci Rep 2019; 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang M, Sun H, Zhao S, et al. Expression of PD-L1 and prognosis in breast cancer: a meta-analysis. Oncotarget 2017; 8: 31347–31354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li Y-C, Zhou Q, Song Q-K, et al. Overexpression of an immune checkpoint (CD155) in breast cancer associated with prognostic significance and exhausted tumor-infiltrating lymphocytes: a cohort study. J Immunol Res 2020; 2020: 3948928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Yahata T, Mizoguchi M, Kimura A, et al. Programmed cell death ligand 1 disruption by clustered regularly interspaced short palindromic repeats/Cas9-genome editing promotes antitumor immunity and suppresses ovarian cancer progression. Cancer Sci 2019; 110: 1279–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Zhao Z, Shi L, Zhang W, et al. CRISPR knock out of programmed cell death protein 1 enhances anti-tumor activity of cytotoxic T lymphocytes. Oncotarget 2018; 9: 5208–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Deng H, Tan S, Gao X, et al. Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm Sin B 2020; 10: 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Tu K, Deng H, Kong L, et al. Reshaping tumor immune microenvironment through acidity-responsive nanoparticles featured with CRISPR/Cas9-mediated PD-L1 attenuation and chemotherapeutics-induced immunogenic cell death. ACS Appl Mater Interfaces 2020; 12(14): 16018–16030. [DOI] [PubMed] [Google Scholar]
- 135. Gao J, Zheng Q, Shao Y, et al. CD155 downregulation synergizes with adriamycin to induce breast cancer cell apoptosis. Apoptosis 2018; 23: 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Li C, Mei H, Hu Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Brief Funct Genomics. 2020; 19: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bajgain P, Tawinwung S, D’Elia L, et al. CAR T cell therapy for breast cancer: harnessing the tumor milieu to drive T cell activation. J Immunother Cancer 2018; 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Byrd TT, Fousek K, Pignata A, et al. TEM8/ANTXR1-specific CAR T cells as a targeted therapy for triple-negative breast cancer. Cancer Res 2018; 78: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Criscitiello C. Tumor-associated antigens in breast cancer. Breast Care 2012; 7: 262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Hu W, Zi Z, Jin Y, et al. CRISPR/Cas9-mediated PD-1 disruption enhances human mesothelin-targeted CAR T cell effector functions. Cancer Immunol Immunother 2019; 68: 365–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ren J, Liu X, Fang C, et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res 2017; 23: 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017; 543: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Liu X, Zhang Y, Cheng C, et al. CRISPR-Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res 2017; 27: 154–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ren J, Zhang X, Liu X, et al. A versatile system for rapid multiplex genome-edited CAR T cell generation. Oncotarget 2017; 8: 17002–17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Hamidi H, Ivaska J. Every step of the way: integrins in cancer progression and metastasis. Nat Rev Cancer 2018; 18: 533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Ju JA, Godet I, Ye IC, et al. Hypoxia selectively enhances integrin α5β1 receptor expression in breast cancer to promote metastasis. Mol Cancer Res 2017; 15: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xiao Y, Li Y, Tao H, et al. Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett 2018; 433: 199–209. [DOI] [PubMed] [Google Scholar]
- 148. Ran FA, Cong L, Yan WX, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015; 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Kim S, Kim D, Cho SW, et al. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 2014; 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 2015; 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Liang X, Potter J, Kumar S, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 2015; 208: 44–53. [DOI] [PubMed] [Google Scholar]
- 152. Miller JB, Zhang S, Kos P, et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chemie – Int Ed 2017; 56: 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Glass Z, Lee M, Li Y, et al. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol 2018; 36: 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Li L, Hu S, Chen X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials 2018; 171: 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003; 302: 415–419. [DOI] [PubMed] [Google Scholar]
- 156. Mendes R, Fernandes A, Baptista P. Gold Nanoparticle approach to the selective delivery of gene silencing in cancer – The case for combined delivery? Genes (Basel) 2017; 8: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Moon HJ, Ku M, Lee H, et al. Implantable photothermal agents based on gold nanorods-encapsulated microcube. Sci Rep 2018; 8: 13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Singh P, Pandit S, Mokkapati VRSS, et al. Gold nanoparticles in diagnostics and therapeutics for human cancer. Int J Mol Sci 2018; 19: 1979–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Turkevich J, Stevenson PC, Hillier J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss Faraday Soc 1951; 11: 55. [Google Scholar]
- 160. Elahi N, Kamali M, Baghersad MH. Recent biomedical applications of gold nanoparticles: a review. Talanta 2018; 184: 537–556. [DOI] [PubMed] [Google Scholar]
- 161. Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold suspensions. Nat Phys Sci 1973; 241: 20–22. [Google Scholar]
- 162. Li D, Chen Z, Mei X. Fluorescence enhancement for noble metal nanoclusters. Adv Colloids Interface Sci 2017; 250: 25–39. [DOI] [PubMed] [Google Scholar]
- 163. Lien NTH, Hoang ND, Nghia NT, et al. Synthesis and characterization of fluorescent gold nanoclusters. Vietnam J Chem 2018; 56: 460–465. [Google Scholar]
- 164. Duan Y, Duan R, Liu R, et al. Chitosan-stabilized self-assembled fluorescent gold nanoclusters for cell imaging and biodistribution in vivo. ACS Biomater Sci Eng 2018; 4: 1055–1063. [DOI] [PubMed] [Google Scholar]
- 165. Liu Y, Ai K, Cheng X, et al. Gold-nanocluster-based fluorescent sensors for highly sensitive and selective detection of cyanide in water. Adv Funct Mater 2010; 20: 951–956. [Google Scholar]
- 166. Zhang H, Liu Q, Wang T, et al. Facile preparation of glutathione-stabilized gold nanoclusters for selective determination of chromium (III) and chromium (VI) in environmental water samples. Anal Chim Acta 2013; 770: 140–146. [DOI] [PubMed] [Google Scholar]
- 167. Zhang C, Li C, Liu Y, et al. Gold nanoclusters-based nanoprobes for simultaneous fluorescence imaging and targeted photodynamic therapy with superior penetration and retention behavior in tumors. Adv Funct Mater 2015; 25: 1314–1325. [Google Scholar]
- 168. Selvaprakash K, Chen YC. Using protein-encapsulated gold nanoclusters as photoluminescent sensing probes for biomolecules. Biosens Bioelectron 2014; 61: 88–94. [DOI] [PubMed] [Google Scholar]
- 169. Liang G, Jin X, Zhang S, et al. RGD peptide-modified fluorescent gold nanoclusters as highly efficient tumor-targeted radiotherapy sensitizers. Biomaterials 2017; 144: 95–104. [DOI] [PubMed] [Google Scholar]
- 170. Li Y, Yuan M, Khan AJ, et al. Peptide-gold nanocluster synthesis and intracellular Hg2+ sensing. Colloids Surfaces A Physicochem Eng Asp 2019; 579: 123666. [Google Scholar]
- 171. Le Guével X, Daum N, Schneider M. Synthesis and characterization of human transferrin-stabilized gold nanoclusters. Nanotechnology 2011; 22: 275103. [DOI] [PubMed] [Google Scholar]
- 172. Tao Y, Li Z, Ju E, et al. Polycations-functionalized water-soluble gold nanoclusters: a potential platform for simultaneous enhanced gene delivery and cell imaging. Nanoscale 2013; 5: 6154–6160. [DOI] [PubMed] [Google Scholar]
- 173. Gröhn F, Bauer BJ, Akpalu YA, et al. Dendrimer templates for the formation of gold nanoclusters. Macromolecules 2000; 33: 6042–6050. [Google Scholar]
- 174. Luo Z, Yuan X, Yu Y, et al. From Aggregation-induced emission of au(I)–thiolate complexes to ultrabright Au(0)@Au(I)–thiolate core–shell nanoclusters. J Am Chem Soc 2012; 134: 16662–16670. [DOI] [PubMed] [Google Scholar]
- 175. Halawa MI, Lai J, Xu G. Gold nanoclusters: synthetic strategies and recent advances in fluorescent sensing. Mater Today Nano 2018; 3: 9–27. [Google Scholar]
- 176. O’Shannessy DJ, Somers EB, Maltzman J, et al. Folate receptor alpha (FRA) expression in breast cancer: Identification of a new molecular subtype and association with triple negative disease. Springerplus 2012; 1: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Cheung A, Bax HJ, Josephs DH, et al. Targeting folate receptor alpha for cancer treatment. Oncotarget 2016; 7: 52553–52574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Habashy HO, Powe DG, Staka CM, et al. Transferrin receptor (CD71) is a marker of poor prognosis in breast cancer and can predict response to tamoxifen. Breast Cancer Res Treat 2010; 119: 283–293. [DOI] [PubMed] [Google Scholar]
- 179. Pan L, Liu J, Shi J. Cancer cell nucleus-targeting nanocomposites for advanced tumor therapeutics. Chem Soc Rev 2018; 47: 6930–6946. [DOI] [PubMed] [Google Scholar]
- 180. Rizzuti M, Nizzardo M, Zanetta C, et al. Therapeutic applications of the cell-penetrating HIV-1 Tat peptide. Drug Discov Today 2015; 20: 76–85. [DOI] [PubMed] [Google Scholar]
- 181. Mout R, Rotello V. Cytosolic and nuclear delivery of CRISPR/Cas9-ribonucleoprotein for gene editing using arginine functionalized gold nanoparticles. Bio-Protocol 2017; 7: e2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Mout R, Ray M, Tonga GY, et al. Efficient gene editing through direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein. ACS Nano 2017; 11: 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Shahbazi R, Sghia-Hughes G, Reid JL, et al. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat Mater 2019; 18: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Wang P, Zhang L, Zheng W, et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew Chemie – Int Ed 2018; 57: 1491–1496. [DOI] [PubMed] [Google Scholar]
- 185. Lee K, Conboy M, Park HM, et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng 2017; 1: 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wang P, Zhang L, Xie Y, et al. Genome editing for cancer therapy: delivery of cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core-shell nanocarrier. Adv Sci 2017; 4: 1700175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Ju E, Li T, Ramos Da Silva S, et al. Gold nanocluster-mediated efficient delivery of cas9 protein through pH-induced assembly-disassembly for inactivation of virus oncogenes. ACS Appl Mater Interfaces 2019; 11: 34717–34724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Wang Y, Wang X, Ma X, et al. Coassembly of gold nanoclusters with nucleic acids: sensing, bioimaging, and gene transfection. Part Part Syst Charact 2019; 36: 1900281. [Google Scholar]
- 189. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (80-) 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science (80-) 2013; 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys 2017; 46: 505–529. [DOI] [PubMed] [Google Scholar]
- 192. Slaymaker IM, Gao L, Zetsche B, et al. Rationally engineered Cas9 nucleases with improved specificity. Science (80-) 2016; 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Chen JS, Dagdas YS, Kleinstiver BP, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 2017; 550: 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zischewski J, Fischer R, Bortesi L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol Adv 2017; 35: 95–104. [DOI] [PubMed] [Google Scholar]
- 195. Wang D, Mou H, Li S, et al. Adenovirus-mediated somatic genome editing of Pten by CRISPR/Cas9 in mouse liver in spite of Cas9-specific immune responses. Hum Gene Ther 2015; 26: 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Simhadri VL, McGill J, McMahon S, et al. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol Ther – Methods Clin Dev 2018; 10: 105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Li B, Niu Y, Ji W, et al. Strategies for the CRISPR-based therapeutics. Trends Pharmacol Sci 2020; 41: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Li H, Yang Y, Hong W, et al. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Signal Transduct Target Ther 2020; 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]