Abstract
Background:
It is unclear whether microRNAs could be a potential diagnostic biomarker for asthma or not. The objective of this study is to figure out the diagnostic value of microRNAs in asthma.
Methods:
Literature retrieval, screening of publications, specific data extraction, and quality evaluation were conducted according to the standard criteria. Stata 14.0 software was used to analyze the diagnostic value of microRNA for asthma, including the combined sensitivity (Sen), specificity (Spe), the area under the curve (AUC), positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR).
Results:
A total of 72 studies, containing 4143 cases and 2188 controls, were included for this comprehensive analysis. None of the included publications were rated low in quality. We summarized that, compared with controls, more than 100 miRNAs were reported differently expressed in asthma, although the expression trends were inconsistent. Besides, there were five studies among these 72 articles that applied the diagnostic evaluation of microRNAs in asthma. We found that the pooled Sen, Spe, and AUC for the combination of miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p in asthma were 0.87 (95%CI: 0.72–0.95), 0.84 (95%CI: 0.74–0.91), and 0.93 (95%CI: 0.89–0.94) individually, and the PLR, NLR, and DOR were 5.5 (95%CI: 3.1–9.7), 0.15 (95%CI: 0.07–0.36), and 35 (95%CI: 10–127) in asthma, respectively. In terms of subgroup analyses, we found that the Sen for these combination miRNAs from serum was higher than that in plasma, while the Spe in plasma worked better than that in serum. Furthermore, compared with children, the combination of above miRNAs from adults had higher Spe and similar Sen.
Conclusions:
From our analysis, the combination of miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p from peripheral blood could potentially act as a diagnostic biomarker for asthma.
The reviews of this paper are available via the supplemental material section.
Keywords: asthma, diagnostic biomarker, microRNA
Introduction
Asthma is a common disease that affects the health of about 330 million people worldwide.1 As a chronic airway inflammation disease, it is characterized by airway hyperresponsiveness and reversible airway obstruction, resulting in shortness of breath, recurrent wheezing, chest tightness, and coughing.2 Currently, the widely used diagnostic criteria for asthma, such as the Global Initiative for Asthma (GINA),3–6 mainly focus on the medical history of respiratory symptoms and lung function tests, for which there are also some drawbacks. For instance, asthma may be neglected when patients suffer from multiple diseases (heart failure, anemia, etc.) which could cause dyspnea.7 Besides, forced expiratory volume in 1 s, acting as an effective way to detect airway obstruction and an important tool in the diagnosis of asthma,8 cannot be used alone for classification of different types and stages of asthma.9–11 Therefore, a universal and high-specificity biomarker to assist the diagnosis of asthma is required in clinical practice.
MicroRNA (miRNA) is a class of endogenous non-coding small RNAs of approximately 19–22 nucleotides in length, which serve as posttranscriptional regulators of gene expression.12,13 Studies have shown that miRNAs can be involved in the pathogenesis of a variety of allergic diseases, such as asthma, allergic rhinitis,14 eosinophilic esophagitis,15,16 and eczema.17,18 miRNA may play a role in coordinating the phenotypic programming of immune and airway epithelial cells to increase the production of cytokines and other mediators, leading to inflammatory characteristics. Niu et al.19 found that miR-33b can inhibit mast cell degranulation by inhibiting the release of calcium and the inhibitory pathway of antigen- and IgE-dependent aggregation of the high-affinity IgE receptor (FcεRI), and could also play a role in airway inflammation of asthma and mast cell biology. miR-21 was shown to target and inhibit the expression of IL-12p35, induce dendritic cells to produce more IL-12 and CD4+ T cells, and thus reduce the production of interferon-g (IFN-g) and IL-4.20
Recently, miRNA is proposed to be effective in the diagnosis of asthma based on a series of studies.19 Suojalehto et al.21 found that the expression of miR-155 was downregulated in the nasal mucosa of asthmatic patients. Furthermore, the decrease of let-7a-5p in asthma was associated with peripheral blood eosinophil ratio.22 Compared with non-asthmatics and mild-to-moderate asthmatics, plasma miR-155 level was significantly elevated in patients with severe asthma.23 Besides, the increased expression of miR-146 was known to inhibit the nuclear factor-kappa B factor (NF-κB), this limiting to inflammatory responses in plasma from asthma.24 The over-expression of miR-126 in acute asthma was found to be correlated with signs of immune imbalance and could predict the severity of childhood asthma.25
However, the position that miRNAs could be a group of good biomarker candidates for asthma is inconsistent. Tang et al.26 pointed out that the expression of miR-1268 was higher in the bronchoalveolar lavage fluid (BALF) of asthmatic adults, while Levänen et al.27 reported no significant changes of miR-1268 in BALF in asthmatic adult patients compared with the control group. As for the let-7 family, the expression of let-7f was upregulated in bronchial epithelial cells (BECs) from asthmatic patients,28 whereas let-7f was downregulated in CD4+ T cells from peripheral blood mononuclear cells (PBMCs).28,29 Furthermore, the miR-146a was decreased in bronchial biopsies from Australian patients with asthma;30 however, it was increased in plasma from asthma in Egypt.31 All these may partly be explained by different ethnicities, applications for different diagnosis criteria, different sample sources, detection methods, and so on.
To determine whether miRNA could be a good biomarker candidate for the diagnosis of asthma or not, we conducted a comprehensive qualitative and combined quantitative research of the diagnostic value of miRNAs in asthma. We found that more than 100 miRNAs were reported differently expressed in asthma, and the combined miRNAs of miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p from peripheral blood could play a role as diagnostic biomarkers for asthma from our analysis.
Materials and methods
Search strategy
We searched databases including PubMed, Embase, Web of Science, and Cochrane Library through 31 May 2020, with the key terms “asthma,” “miRNA” and “microRNA”. At the same time, the references of relevant literature were manually searched. The whole process of literature searching and screening was done by two independent staff. When there was a dispute, it was discussed with a third party.
Inclusion criteria based on PICOS
The inclusion criteria for the selection of eligible studies were according to the PICOS principle as follows:
(1) Participants (P): the study included asthma patients of different ages, including children and adults. All cases were reported as asthma based on GINA,4–6 American Thoracic Society Guidelines (ATSG),32 National Heart, Lung, and Blood Institute (NHLBI),33 Spanish guidelines of the management of Asthma (GEMA),34 International Study of Asthma and Allergies in Children (ISAAC),35 Chinese guideline for the prevention and management of bronchial asthma (CGBS),36 Guidelines for Diagnosis and Prevention of Bronchial Asthma in Pediatric Group (GDPB),37,38 Guideline of Severe Asthma Research Program (GSARP)39 and asthma patients diagnosed by physician.
(2) Intervention (I): specific designed or commercial arrays, quantitative real-time polymerase chain reaction (qRT-PCR), or next-generation sequencing (NGS) were used to detect the miRNAs expression levels in all participants.
(3) Control (C): all controls were from healthy people or non-asthmatic controls.
(4) Outcomes (O): (1) Qualitative analysis: publications reported the specific miRNA types which were differently expressed between cases and controls. (2) Quantitative analysis: publications reported specific differently expressed miRNAs with data for diagnosis evaluation among all participants, including true positive (TP), false positive (FP), false negative (FN), and true negative (TN).
(5) Studies (S): Case-control design or cohort design.
Exclusion criteria
Studies were excluded if: (1) the literature was not in English; (2) the type of the research was a review, case report, or conference summary; (3) the publication was a small-sized study on the same topic from the same team which also shared overlapped participants with large-sized studies; (4) the data were incomplete for analysis.
Data extraction
The extracted data consisted of two parts: the first part contains basic information such as the first author, the year and region of publication, the numbers and ages of participates, and the details of differently expressed miRNA, including the specific miRNA types, the sources of researched samples, detection methods of reported miRNAs, and expression trend between cases and controls. The second part is the diagnostic data used for quantitative analysis, including TP, FP, FN, and TN for all participants. Two independent researchers extracted the data. Disagreements were discussed and resolved with the third researcher.
Quality assessment for publications
We used the Newcastle–Ottawa Scale (NOS),40 a common tool for quality evaluation of non-randomized study, to estimate all qualitative and quantitative studies included. The total score of the NOS evaluation is 9 points. Five points or more is considered not of low quality. Besides, we also evaluated the papers applied for quantitative analysis by the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2)41 through RevMan5.3 with the levels of “high,” “low” and “unclear.”
Data analysis
In this research, we used the bivariate model for quantitative analysis of the diagnostic value of miRNAs in asthma.42 To test whether the study effect size could be combined or not, the Spearman correlation coefficient was used to determine whether there was a threshold effect in the study. Generally, r = 0.6 is taken as the critical value, and if the Spearman correlation coefficient value is greater than this value, the threshold effect is considered to exist.43 Then, Stata 14.0 statistical software (Stata Corporation, College Station, TX, USA) was used for data analysis and to calculate the pooled sensitivity (Sen), specificity (Spe), positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) the area under curve (AUC) and corresponding 95% credible interval (CI). To show the clinical utility of miRNAs for asthma, Fagan analysis was used to prove the relationship among prior probability, likelihood ratio, and posterior probability under the pre-test probabilities of 25%, 50% and 75% which presented for clinical suspicion of asthma at 25%, 50% and 75%, respectively.
I2 is applied to measure the heterogeneity, which describes the percentage of variation between analyzed studies.44 If I2 = 25%, it means that slight heterogeneity existed; I2 = 50% meant moderate heterogeneity; high heterogeneity was present when I2 = 75%.45 Next, subgroup analysis and meta-regression analysis were used to find the sources of heterogeneity, which we performed from the aspect of sample sources (serum versus plasma) and ages (adults versus children), respectively. If there was a significant decrease in heterogeneity in either subgroup, it would be considered as the source of heterogeneity. Additionally, meta-regression analysis was carried out by taking age and sample sources as covariables respectively and using the restrictive maximum likelihood method to establish the regression model of effect size to a single covariable. When the tau2, which represented the estimate of between-study variance, decreased significantly in a covariable (sample sources or ages), this covariable would be considered as the source of heterogeneity.45
Sensitivity analysis, which is conducted by excluding one of the included studies in turn, was used to determine whether a single study has an undue influence on the overall result. We used the Funnel plot to evaluate the publication bias. Generally speaking, when the Funnel plot is basically symmetrical, it is considered that there may be no publication bias; otherwise, it may indicate the existence of publication bias.46
Results
Publication selection and quality assessment
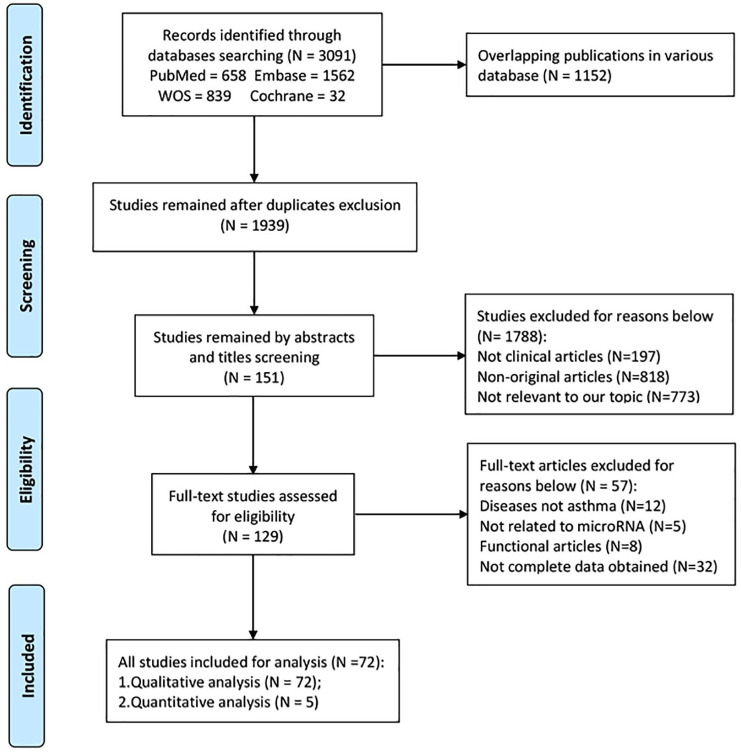
Through the literature retrieval method mentioned above, a total of 3091 studies were initially obtained. According to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) standard,47 the standard literature selection process was conducted as shown in Figure 1. We finally included 72 articles21,22,25–31,48–110 with 4143 patients and 2188 controls based on the inclusion and exclusion criteria. The features of the included research are presented in Table 1. For the quality of all publications included, 36 studies had NOS scores higher than 7, 30 studies had a NOS score of 7, and six studies had a score of 6, indicating that the none of included studies were of low quality.
Figure 1.
Flowchart of study selection based on the inclusion and exclusion criteria.
Table 1.
Characteristics of 72 studies included to explore the diagnostic value of microRNAs in asthma.
| Study ID | Region | No. of cases/controls | Diagnosis criteria | Differentially expressed miRNAs | Specimen sources | Detection method | NOS |
|---|---|---|---|---|---|---|---|
| Children | |||||||
| Ibrahim et al.50 | Egypt | 100/50 | GINA | 196a2 | Serum | qRT-PCR | 8 |
| Bartel et al.63 | Germany | 18/9 | Hospital | 92b, 210, 34a | ASMCs | qRT-PCR | 8 |
| Karam and Abd Elrahman61 | Egypt | 100/100 | GINA | 155, let-7a | Plasma | qRT-PCR | 8 |
| Li et al.60 | China | 23/15 | Hospital | 378 | Peripheral blood, lung tissue | qRT-PCR | 8 |
| Liu et al.58 | China | 180/180 | ISAAC | 155 | Serum | qRT-PCR | 8 |
| Nasser et al.57 | Egypt | 30/20 | GINA | 15a | Serum | qRT-PCR | 8 |
| Dong et al.71 | China | 150/50 | GINA | 27b-3p | BECs | Microarray, qRT-PCR | 8 |
| Hammad et al.30 | Egypt | 27/21 | GINA | 21, 146a | Plasma | qRT-PCR | 8 |
| Tian et al.69 | China | 100/100 | GDPB | 1 | Blood | qRT-PCR | 8 |
| Tian et al.69 | China | 80/80 | CGBS | 126 | Serum | qRT-PCR | 8 |
| Zhang et al.67 | China | 18/15 | Hospital | 192 | CD4+ T cellsa, plasma | qRT-PCR | 8 |
| Zhang et al.64 | China | 52/26 | GINA | 29c | CD4+ T cellsa | Microarray, qRT-PCR | 8 |
| Hu et al.80 | China | 124/382 | Hospital | 149 | PBMCs | qRT-PCR | 8 |
| Dong et al. 93 | China | 62/62 | GINA | 625-5p, 513a-5p, 22-3p | Plasma | Microarray, qRT-PCR | 8 |
| Elbehidy et al. 92 | Egypt | 95/80 | GINA | 21 | Serum | qRT-PCR | 8 |
| Liu et al. 88 | China | 50/50 | GINA | 125b | Sputum | qRT-PCR | 8 |
| Midyat et al. 85 | USA | 95/96 | GINA | let-7e, 98, 497 | Blood | qRT-PCR | 8 |
| Zhang et al. 82 | China | 30/30 | Hospital | 146a | Plasma | qRT-PCR | 8 |
| Sawant et al. 95 | USA | 8/8 | Hospital | 21 | Serum | qRT-PCR | 7 |
| Liu et al. 109 | China | 6/6 | GINA | 483-3p, 221 | Blood | Microarray, qRT-PCR | 8 |
| Adult | |||||||
| ElKashef et al. 51 | Egypt | 30/30 | Hospital | 21, 155 | Serum | qRT-PCR | 8 |
| Liang and Tang 49 | China | 772/441 | GINA | 155 | CD4+ T cellsb | qRT-PCR | 7 |
| Ye et al. 48 | China | 180/90 | GINA | 125a | Plasma | qRT-PCR | 8 |
| Fang et al. 62 | Switzerland | 5/5 | Hospital | 21-5p | ASMCs | qRT-PCR | 6 |
| Huang et al. 22 | China | 70/34 | GINA | let-7a-5p | Peripheral blood | Microarray, qRT-PCR | 8 |
| Liang et al. 59 | China | 50/15 | Hospital | 218-5p | BECs | qRT-PCR | 8 |
| Qiu et al. 75 | China | 52/45 | CGBS | 17 | CD4+ T cellsb | Microarray, qRT-PCR | 8 |
| Rodrigo-Muñoz et al. 55 | Spain | 173/53 | GEMA | 185-5p, 320a, 1246, 144-5p | Serum | NGS, qRT-PCR | 8 |
| Tsai et al. 54 | China | 86/38 | GINA | 10a-5p, 146a-5p | BECs, serum | NGS, qRT-PCR | 7 |
| Wu et al. 53 | China | 53/47 | GINA | 1165-3p | Serum | qRT-PCR | 8 |
| Zhao et al. 52 | China | 80/80 | GINA | 125b | Serum | qRT-PCR | 8 |
| Daniel et al. 72 | USA | 16/28 | NHLBI | 155 | CD4+ T cellsb | Microarray, qRT-PCR | 8 |
| Faiz et al. 31 | Australia | 16/39 | Hospital | 146a | Bronchial biopsy | Microarray, qRT-PCR | 7 |
| Huang et al. 70 | China | 35/15 | GINA | 199a-5p | Plasma, sputum | qRT-PCR | 8 |
| Tang et al. 26 | China | 49/22 | ATSG | 200b/c, 21, 1268, 663a, let-7a/b, 30a, 99a, 27a, 133a, 155, 24 | BALF | qRT-PCR | 8 |
| Wang et al. 68 | China | 82/80 | CGBS | 21 | Serum | qRT-PCR | 8 |
| Zhang et al. 66 | China | 25/22 | GINA | 221 | BECs | qRT-PCR | 8 |
| Zhang et al. 65 | China | 77/36 | Hospital | 221-3p | BECs, sputum, plasma | qRT-PCR | 8 |
| Chen et al. 81 | China | 20/20 | Hospital | 98 | PBMCs | qRT-PCR | 8 |
| Kärner et al. 79 | Swiss | NA | GINA | 323-3p | PBMCs | qRT-PCR | 8 |
| Lacedonia et al. 78 | Italy | 13/46 | GINA | 145, 338 | Serum, sputum | qRT-PCR | 8 |
| Malmhäll et al. 111 | Sweden | 16/13 | Hospital | 155 | Sputum | qRT-PCR | 8 |
| Milger et al. 76 | Germany | 46/21 | Hospital | 21-5p,15a-5p,27a-3p,29c-3p,223-3p,425-5p, 342-3p | Plasma | qRT-PCR | 8 |
| Qiu et al. 75 | China | 30/25 | Hospital | 371, 138, 544, 145, 214 | CD3+ T cellsb | Microarray, qRT-PCR | 8 |
| Sun et al. 74 | Italy | 9/17 | Hospital | 101, 19a | ASMCs | qRT-PCR | 7 |
| Yin et al. 73 | China | 46/10 | Hospital | 34, 449 | Serum | qRT-PCR | 8 |
| Cai et al. 94 | China | 28/28 | GINA | 194, 375 | Plasma | qRT-PCR | 8 |
| Fan et al. 91 | China | 59/49 | GINA | 145 | CD4+ T cellsb | qRT-PCR | 8 |
| Huo et al. 90 | China | 72/35 | Hospital | 181b-5p, 181a-5p | Bronchial brushing biopsy, plasma | Microarray, qRT-PCR | 8 |
| Li et al. 89 | China | 10/10 | Hospital | 31 | Plasma, BECs | qRT-PCR | 8 |
| Luo et al. 87 | China | 20/25 | Hospital | 98 | PBMCs | qRT-PCR | 8 |
| Maes et al. 86 | Belgium | 76/20 | GINA, ATSG | 629-3p, 223-3p, 142-3p | Sputum | qRT-PCR | 8 |
| Panganiban et al. 84 | USA | 35/44 | Hospital | 16, 223, 148, 146a, 299-5p, 570, 150 | Plasma | qRT-PCR | 8 |
| Pietrusinńska et al. 83 | Poland | 3/6 | GINA | 224, 339-5p, 382 | Plasma | qRT-PCR | 7 |
| Haj-Salem et al. 96 | Canada | 15/9 | ATSG | 19a | BECs | qRT-PCR | 8 |
| Newcomb et al. 29 | USA | 29/9 | GSARP | let-7f | CD4+ T cellsb | qRT-PCR | 8 |
| Martinez-Nunez et al. 102 | UK | 15/13 | Hospital | 18a, 27a, 128, 155 | BECs | Microarray, qRT-PCR | 8 |
| Perry et al. 101 | USA | 18/9 | ATSG | 221 | ASMCs | qRT-PCR | 8 |
| Rijavec et al. 100 | Slovenia | 24/12 | GINA | let-7a | Bronchial biopsy | qRT-PCR | 8 |
| Roff et al. 99 | USA | 10/10 | Hospital | 570-3p | BECs, serum | qRT-PCR | 8 |
| Simpson et al. 98 | USA | 24/8 | Hospital | 19a | CD4+ T cellsc | qRT-PCR | 8 |
| Suojalehto et al. 21 | Finland | 117/33 | GINA | 18a, 126, 155, let-7e, 224, 498, 187, 87, 143, 886-2p | Nasal mucosa | qRT-PCR | 8 |
| Wu et al. 97 | China | 35/12 | GINA | 21, 126 | BECs | qRT-PCR | 8 |
| Levänen et al. 27 | Sweden | 10/10 | Hospital | let-7a, 21, 658, 24,26a, 99a, 200c, 1268 | BALF | Microarray, qRT-PCR | 8 |
| Nicodemus-Johnson et al. 103 | USA | 22/32 | Hospital | 148b | BALF | qRT-PCR | 7 |
| Jardim et al. 28 | USA | 16/16 | ATSG | 487b, 181c, let-7f | BECs | Microarray, qRT-PCR | 7 |
| Panganiban et al. 108 | USA | 10/10 | Hospital | 1248, 26a, let-7a, let-7d | Serum | Microarray, qRT-PCR | 8 |
| Solberg et al. 107 | USA | 35/12 | Hospital | 34c-5p, 34b-5p, 449a, 449b-5p | BECs | Microarray, qRT-PCR | 8 |
| Tsitsiou et al. 106 | UK | 16/8 | ATSG | 146a, 146b, 28a-5p | CD4+ T cellsb, CD8+ T cellsb | Microarray, qRT-PCR | 8 |
| Yamamoto et al. 105 | UK | 7/4 | Hospital | 192 | PBMCs | qRT-PCR | 7 |
| Zhang et al. 104 | China | 50/20 | CGBS(2003) | 155 | CD4+ T cellsb | qRT-PCR | 8 |
| Williams et al. 110 | UK | 8/8 | Hospital | 140, 449, 200c, 30b/c/d, 93, 223, 191, 320, 342,142-3p | Airway biopsies | qRT-PCR | 7 |
From peripheral blood.
From PBMCs.
From BALF.
ASMCs, airway smooth muscle cells; BALF, bronchoalveolar lavage fluid; BECs, bronchial epithelial cells; PBMCs, peripheral blood mononuclear cells.
In particular, there were five publications51,53,55,61,92 among the 72 articles with the data details required for quantitative diagnosis analysis for miRNAs in asthma (Table 2). There is no threshold effect based on the analysis of Spearman (Spearman correlation = 0.5946) which means that the study effect size could be combined. Through the QUADAS-2 quality evaluation of these five studies, we rated that the research quality at the middle and upper grades (Supplemental Figure 1).
Table 2.
Basic diagnostic data extracted from the five articles included in the quantitative analysis.
| Study ID | No. of case/control | miRNA | Level | Sample | Diagnostic power |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | FP | FN | TN | DOR | Sen | Spe | |||||
| Rodrigo-Muñoz et al.55a | 138/39 | miR-320a | ↑ | Serum | 130 | 19 | 8 | 20 | 21.95 | 0.941 | 0.514 |
| Rodrigo-Muñoz et al.55b | 138/39 | miR-185-5p | ↑ | Serum | 123 | 9 | 15 | 30 | 32.5 | 0.89 | 0.8 |
| Rodrigo-Muñoz et al.55c | 138/39 | miR-144-5p | ↓ | Serum | 81 | 9 | 57 | 30 | 24.73 | 0.587 | 0.774 |
| Rodrigo-Muñoz et al.55d | 138/39 | miR-1246 | ↑ | Serum | 68 | 5 | 70 | 34 | 24.14 | 0.496 | 0.865 |
| Karam and Abd Elrahman61a | 100/100 | miR-155 | ↑ | Plasma | 80 | 11 | 20 | 89 | 33.04 | 0.8 | 0.89 |
| Karam and Abd Elrahman61b | 100/100 | Let-7a | ↓ | Plasma | 85 | 10 | 15 | 90 | 85 | 0.85 | 0.9 |
| Elbehidy et al.92 | 95/80 | miR-21 | ↑ | Serum | 88 | 18 | 7 | 62 | 45.55 | 0.93 | 0.78 |
| Wu et al.53 | 53/47 | miR-1165-3p | ↑ | Serum | 44 | 7 | 9 | 15 | 10.83 | 0.83 | 0.69 |
| ElKashef et al.51a | 30/30 | miR-21 | ↑ | Serum | 30 | 2 | 0 | 29 | NA | 1 | 0.95 |
| ElKashef et al.51b | 30/30 | miR-155 | ↑ | Serum | 30 | 0 | 0 | 30 | NA | 1 | 1 |
Independent study from the same article.
TP, true positive; FP, false positive; FN, false negative; TN, true negative; DOR, diagnostic odds ratio; Sen, sensitivity; Spe, specificity
Profile of miRNAs in asthma
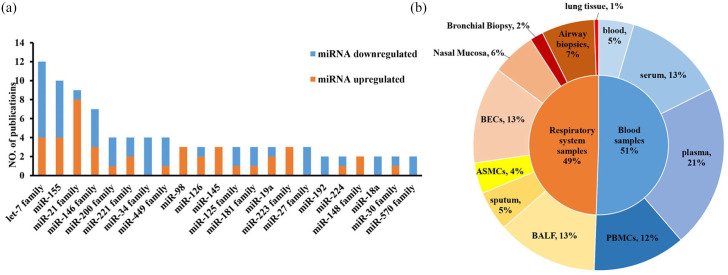
Here, we conducted a qualitative analysis from all included studies. Firstly, we found that there were more than 100 miRNAs differently expressed between asthma and controls in all included publications (Table 1). Then we created a category for miRNAs that were studied in more than two articles. We summed up that there were three miRNAs families (miR-34 family, miR-27 family, and miR-570 family) and two miRNAs (miR-192 and miR-18a) consistently downregulated. Besides, there were two miRNAs (miR-98 and miR-145) and two miRNA families (miR-148 family and miR-223 family) upregulated in asthma. However, the expression trends were different in the other four miRNAs (miR-155, miR-126, miR-19a, and miR-224) and nine miRNA families (let-7 family, miR-21 family, miR-146 family, miR-200 family, miR-221 family, miR-449 family, miR-125 family, miR-181 family, miR-30 family) (Figure 2a).
Figure 2.
Profile of miRNAs in asthma. (a) differentially expressed miRNAs between asthma patients and controls reported in more than two studies. The orange and blue individually represent upregulated and downregulated miRNA in asthma patients compared with controls; (b) distribution of the specimen for all differently expressed miRNA between asthma patients and controls, and each color stands for a specific sample source. All these data were summarized from 72 included publications according to our listed criteria.
Then, we analyzed the distribution characteristics for the sources of all specimens. Since asthma is a kind of respiratory system disease, upper or lower airway-derived sample sources containing BECs (13%), airway biopsies (7%), nasal mucosa (6%), sputum (5%), airway smooth muscle cells (ASMCs) (4%), BALF (13%), bronchial biopsy (2%), and lung tissue (1%), could directly reflect the pathologic change of the disease and have a good representative role (Figure 2b). We also found that blood or specific part separated from plasma (21%), peripheral blood including serum (13%), and PBMCs (12%) are possible good options for candidate biomarker sources as they are easy and non-invasive to access (Figure 2b).
From Figure 2a, we discovered that the most widely reported differently expressed miRNAs in asthma were the let-7 family (let-7a, let-7d, let-7e, and let-7f), miR-155, and miR-21 family (miR-21 and miR-21-5p). When we tried to explore the relationships between these differently expressed miRNAs and their specimen sources, we found that the expression levels of miR-155 and let-7 family were differently expressed in sputum, BECs, BALF, plasma, serum, and PBMCs of asthma patients, which reflects the specificity of these miRNAs in different sample sources.
The diagnostic value of combined different miRNAs for asthma
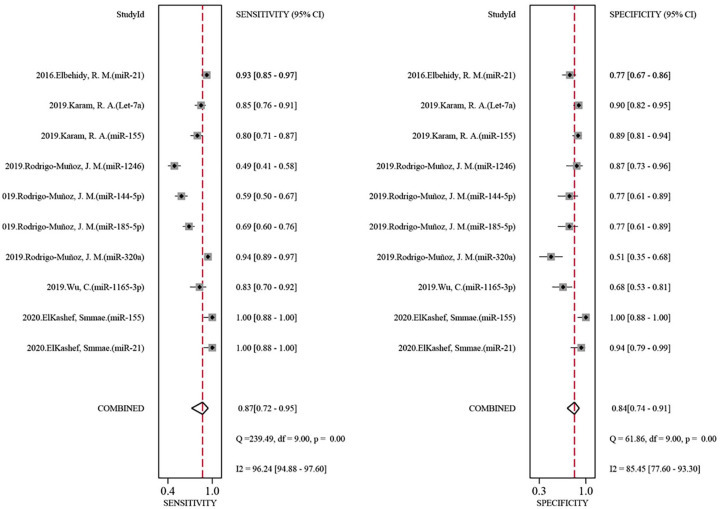
Based on the listed criteria for the diagnostic data, we evaluated the diagnostic value of a combination of eight miRNAs (miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p) for asthma (Table 2) from all reported differently expressed miRNA in five articles included for quantitative analysis. We analyzed that the pooled sensitivity and specificity of the above miRNAs are 0.87 (95%CI: 0.72–0.95) and 0.84 (95%CI: 0.74–0.91) respectively (Figure 3). The pooled AUC is 0.93 (95%CI: 0.89–0.94) (Figure 4). Additionally, we calculated that the pooled PLR, NLR, and DOR of the eight miRNAs for asthma were 5.5 (95%CI: 3.1–9.7), 0.15 (95%CI: 0.07–0.36), and 35 (95%CI: 10–127) individually, which indicates that the probability of asthma increased by 5.5 times when the miRNA test was positive, while the incidence of asthma only increased by 0.15 times when miRNA was negative. This information supports that the combination of these miRNAs has a relatively good biomarker role in asthma.
Figure 3.
Forest plots for the diagnostic value of the combination of eight miRNAs in asthma. Left is the pooled sensitivity analysis. Right is the pooled specificity analysis. The combined miRNAs contain miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p.
Figure 4.
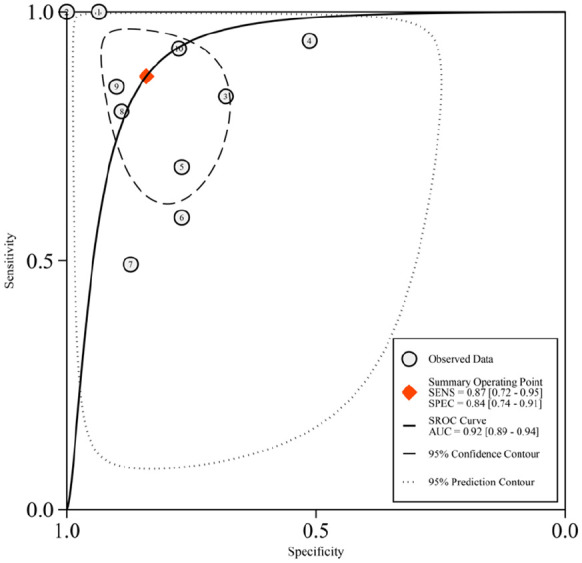
The summary receiver operating characteristic curve for the diagnosis value of the combination of the eight miRNAs in asthma. The combined miRNAs contain miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p. The red diamond represents the pooled analysis of all included studies. The number of each black circle represents each separate study as follows: ① 2020, ElKashef, Smmae.(miR-21); ② 2020, ElKashef, Smmae.(miR-155); ③ 2019, Wu, C. (miR-1165-3p); ④ 2019, Rodrigo-Muñoz, J. M. (miR-320a); ⑤ 2019, Rodrigo-Muñoz, J. M. (miR-185-5p); ⑥ 2019, Rodrigo-Muñoz, J. M. (miR-144-5p); ⑦ 2019, Rodrigo-Muñoz, J. M. (miR-1246); ⑧ 2019, Karam, R. A. (miR-155); ⑨ 2019, Karam, R. A. (Let-7a); ⑩ 2016, Elbehidy, R. M. (miR-21).
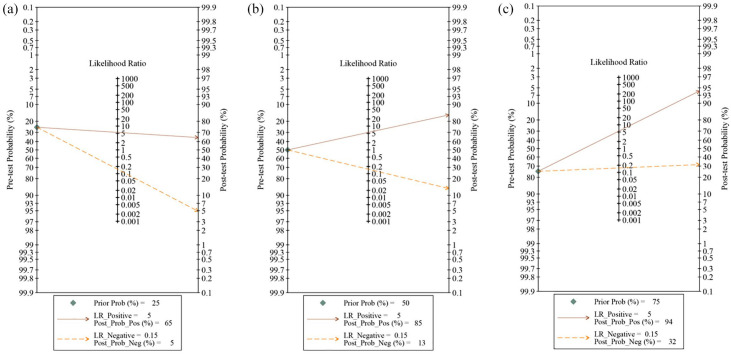
When testing the possibility of application, we deduced that given a pre-test probability (Prob) of 25% (low clinical suspicion of asthma: 25%), the posterior probability positive and negative were 65% and 5%, respectively (Figure 5a). Similarly, the combination of these miRNAs could increase the diagnosis of asthma to 85% (Figure 5b) and 94% (Figure 5c) separately when setting the pre-test prob at 50% (clinical suspicion of asthma: 50%) or 75% (high clinical suspicion of asthma: 75%). These results indicate that the combination of these miRNAs could increase the diagnosis rates of asthma, and imply the potential application value of these miRNAs in the future.
Figure 5.
Fagan plots of the combined of eight miRNAs for asthma. (a–c) were performed under the prior probability value of 25%, 50%, and 75%, respectively. The combined miRNAs contain miR-185-5p, miR-155, Let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p.
Subgroup analysis and meta-regression analysis
To explore the sources of heterogeneity for combined effect size (I2 = 79.3%), we performed a subgroup analysis firstly from the aspect of sample sources. We found that the combination of the above eight miRNAs from serum performed better in Sen than plasma (0.89, 95%CI: 0.77–1.00 versus 0.83, 95%CI: 0.54–0.96), and plasma performed better in Spe than serum (0.90, 95%CI: 0.79–1.00 versus 0.81, 95%CI: 0.72–0.91). The I2 of the serum group was 0.0%; however, the I2 value in the plasma group was still high (I2 = 77.3%) (Supplemental Figure 2a), which indicated the sample sources might be one of the sources of heterogeneity and needed to be further confirmed by regression analysis. Then, in terms of ages, we found that children with asthma had higher Spe (0.87, 95%CI: 0.75–0.98 versus 0.86, 95%CI: 0.74–0.98) than adults with asthma. Adults and children have same Sen (0.87, 95%CI: 0.69–1.00 versus 0.87, 95%CI: 0.87–1.00). Similarly, the I2 of the children group was 0.0%, but the I2 in the adult group was still high (I2 = 81.6%) (Supplemental Figure 2b), suggesting age might be one of the sources of heterogeneity and needed further exploration.
Next, meta-regression analysis was used to further verify the source of heterogeneity and explore the heterogeneity contribution of ages and sample sources. Sample sources (serum and plasma) and ages (adults and children) were taken as covariables, respectively. Through the regression analysis of age, it was found that the tau2 (represent for variance) decreased from 0.9775 (p = 0.000) to 0.04453 (p = 0.035), a decrease of 95.44%, indicating that age could explain the 95.44% of heterogeneity source. However, from the aspect of sample types, there was no significant change in tau2 (0.9775 versus 0.9055), suggesting that the specimen types may not be a main source of heterogeneity (Supplemental Figure 3).
Sensitivity analysis and publication bias
Through sensitivity analysis, we found that there is no significant variation of the compiled value when excluding one study in turn for all included data, indicating that the results for the overall studies were not overly dependent on one study (Supplemental Table 1). Moreover, the Funnel plot was symmetrical and showed no significant publication bias in the present analysis (Supplemental Figure 4).
Discussion
We conducted a qualitative and combined quantitative study to evaluate the diagnostic value of miRNA in asthma. To sum up, there were more than 100 miRNAs from all included studies differently expressed in asthma. Though the expression levels and trends were different depending on different publications, we found that the pooled miRNAs (miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p) had a high diagnostic value (Sen = 0.87, 95%CI: 0.72–0.95; Spe = 0.84, 95%CI: 0.74–0.91; AUC = 0.93, 95%CI: 0.89–0.94). As far as we know, this is the first comprehensive analysis of the diagnostic value of miRNA for asthma.
It is reported in many studies that microRNA could act as a new potential biomarker for a series of respiratory diseases. The role of miRNAs in lung cancer has been confirmed. In 2018, Pan et al.112 found that miR-33a-5p and miR-128-3p had high sensitivity and specificity in the early diagnosis of lung cancer, either in combination or alone. The diagnostic value of miR-339-5p and miR-21 in lung adenocarcinoma, especially in early stage, has also been demonstrated in the study of Zhang et al.113 Additionally, Wang et al.114 proposed that miR-210 was significantly decreased in peripheral blood of patients with chronic obstructive pulmonary disease, and had a high diagnostic value of Sen (85.6%), Spe (72.6%), and AUC (0.821). Similar situations were reported in pulmonary tuberculosis (PTB). According to the research of Zhang et al.,113 six kinds of serum-derived miRNAs (miR-378, miR-483-5p, miR-22, miR-29c, miR-101, and miR-320b) were significantly different in PTB compared with healthy people, and other respiratory diseases had a high Sen, Spe, and AUC of 95.0%, 91.8%, and 95%, respectively. Furthermore, by comparing acute lung injury (ALI) and healthy patients, the application of miR-140 from peripheral blood had a high AUC of 93.5% in ALI.115 To some extent, the evidence supports the potential and possibility of miRNA in the application of diagnosis for asthma.
Related function and mechanism studies also support the pathogenesis role for miRNAs, such as miR-185-5p, miR-155, let-7a, miR-21, and miR-1165-3p, as biomarkers for asthma. Studies have shown that miR-155 could inhibit interleukin 13 (IL-13)-induced bronchial smooth muscle cell proliferation and migration by targeting TAB2 and affecting asthma signaling pathways, and also participates in allergic airway inflammation by regulating the transcription factor PU1.111,116 miR-21 could stimulate fibroblasts through TGF-1, affecting relevant targets of the Wnt pathway, and thereby affect lung disease, especially lung fibroblast function in asthma according to the study of Ong et al.117 Besides, Kumar et al.118 reported that Let-7a could regulate the imbalance of T-lymphocyte subsets in asthma and changes the expression of Th2 and Th17-related cytokines IL-13 and IL-17. Furthermore, the upregulation of miR-1165-3p could lead to a decrease in airway hyperreactivity and airway inflammation through directly targeting IL-13.119 As for miR-185-5p, it participates in calcium signaling by targeting NFAT and CaMKII proteins in cardiomyocytes, and may play a role in muscle cell hypertrophy, proliferation, and cell contraction in asthma.120,121 These results suggest that these biomarker candidates play a role in the pathogenesis of asthma.
According to our results, the combined miRNAs (miR-185-5p, miR-155, let-7a, miR-21, miR-320a, miR-1246, miR-144-5p, and miR-1165-3p) could be a potential biomarker for the diagnosis of asthma based on a relatively high Sen of 0.87, Spe of 0.84, and high AUC of 0.93. Sensitivity is the ability to correctly find an individual with a specific disease, while specificity is to correctly classify the person as disease-free. When faced with inconsistent trends of Sen and Spe, we need to consider the diseases themselves in the clinical setting. For instance, for diseases with high mortality like cancers, biomarkers with high sensitivity are essential for early screening. Meanwhile, in the course of disease diagnosis and treatment, especially for those with obvious side effects, a tool with high specificity is required. Besides, the value of AUC can directly reflect the diagnostic effect. AUC between 0.5 and 0.7 indicates a low diagnostic value, a value between 0.7 and 0.9 is good, and above 0.9 is considered very good.
However, there was some heterogeneity in the pooled quantitative statistics for the combination of the above eight miRNAs. From the results of subgroup analysis and meta-regression analysis, we found that age may be one of the main causes of heterogeneity, which suggested that it would be better to perform analyses in different age groups to explore the diagnosis role of miRNAs for asthma in future research. Besides, the qualitative analysis also provided some evidence for understanding the origin of heterogeneity to some extent. For instance, the expression level for miR-155 could go in a different direction when compared with different control groups.49,72 Moreover, depending on the cut-off of expression value of miRNAs, the Sen and Spe for miR-155 could be as high as 100%51 or go down to 80% and 89%, respectively.61 Similar situations of samples derived from different regions or different cut-off also affect the results of miR-21 for asthma.26,27,51,92 These indicate that further research could be optimized from the above aspects to increase the homogeneity.
There are still some limitations to this analysis. Firstly, there were only five studies available for quantitative analysis, resulting in a small sample size of patients and controls included in the study, which may weaken the diagnostic value of miRNAs in asthma. Besides, due to the limited amount of available data, we were unable to explore the diagnostic value of individual miRNAs for asthma. Nor can we compare the diagnostic power between a single miRNA and the combination of specific miRNAs in asthma. Additionally, we did not explore the relationship between miRNAs and related clinical phenotypes. Finally, although the bivariate model is widely used in diagnostic meta-analysis,122–125 we were not able to verify the results in an independent data set due to the limitations of the included data.
In conclusion, this study suggests that the combination of miR-185-5p, miR-155, let-7a, miR-320a, miR-1246, miR-144-5p, miR-21, and miR-1165-3p may have a potential diagnostic value for asthma.
Supplemental Material
Supplemental material, sj-docx-10-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-11-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-6-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-7-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-8-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-9-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-1-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-2-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-3-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-4-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-5-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This research was supported by the National Natural Science Foundation of China (No. 82001357, No. 31500999), the Hunan Provincial Natural Science Foundation of China (No. 2020JJ5951), the Youth Science Foundation of Xiangya Hospital (No. 2019Q17), the International Postdoctoral Exchange Fellowship Program (No. 2020118), the Degree & Postgraduate Education Reform Project of Central South University (No. 2020JGB125), the Undergraduate Education Reform Project of Central South University (No. 2020jy146), and the Fundamental Research Funds for the Central Universities of Central South University (No. 2020zzts432).
ORCID iD: Yuan Zhang  https://orcid.org/0000-0003-0577-7129
https://orcid.org/0000-0003-0577-7129
Supplemental material: The reviews of this paper are available via the supplemental material section.
Contributor Information
Li Xu, Department of Respiratory Medicine, Xiangya Hospital, Central South University, Changsha, Hunan, China; School of Life Sciences, Central South University, Changsha, Hunan, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan, China.
Minhan Yi, School of Life Sciences, Central South University, Changsha, Hunan, China.
Yun Tan, School of Life Sciences, Central South University, Changsha, Hunan, China.
Zixun Yi, School of Life Sciences, Central South University, Changsha, Hunan, China.
Yuan Zhang, Department of Respiratory Medicine, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China; National Clinical Research Center for Geriatric Disorders, Xiangya Hospital, Central South University, Changsha, Hunan 410008, China.
References
- 1. Papi A, Brightling C, Pedersen SE, et al. Asthma. Lancet 2018; 391: 783–800. [DOI] [PubMed] [Google Scholar]
- 2. Boulet LP, O’Byrne PM. Asthma and exercise-induced bronchoconstriction in athletes. N Engl J Med 2015; 372: 641–648. [DOI] [PubMed] [Google Scholar]
- 3. GINA Report. Global Strategy for Asthma Management and Prevention, https://ginasthma.org/gina-reports (2020, accessed 31 May 2020).
- 4. Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, http://www.ginasthma.org/ (2016, accessed 31 May 2020).
- 5. Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, http://www.ginaasthma.org/ (2015, accessed 31 May 2020).
- 6. Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, http://www.ginasthma.org/ (2018, accessed 31 May 2020).
- 7. Gibson PG, McDonald VM, Marks GB. Asthma in older adults. Lancet 2010; 376: 803–813. [DOI] [PubMed] [Google Scholar]
- 8. Brigham EP, West NE. Diagnosis of asthma: diagnostic testing. Int Forum Allergy Rhinol 2015; 5 Suppl 1: S27–S30. [DOI] [PubMed] [Google Scholar]
- 9. Heijkenskjöld Rentzhog C, Janson C, Berglund L, et al. Overall and peripheral lung function assessment by spirometry and forced oscillation technique in relation to asthma diagnosis and control. Clin Exp Allergy 2017; 47: 1546–1554. [DOI] [PubMed] [Google Scholar]
- 10. Aaron SD, Vandemheen KL, FitzGerald JM, et al. Reevaluation of diagnosis in adults with physician-diagnosed asthma. JAMA 2017; 317: 269–279. [DOI] [PubMed] [Google Scholar]
- 11. Lommatzsch M, Virchow JC. Severe asthma: definition, diagnosis and treatment. Dtsch Arztebl Int 2014; 111: 847–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ambros V. The functions of animal microRNAs. Nature 2004; 431: 350–355. [DOI] [PubMed] [Google Scholar]
- 13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 14. Zeng Q, Liu W, Luo R, et al. MicroRNA-181a and microRNA-155 are involved in the regulation of the differentiation and function of regulatory T cells in allergic rhinitis children. Pediatr Allergy Immunol 2019; 30: 434–442. [DOI] [PubMed] [Google Scholar]
- 15. Lu LF, Boldin MP, Chaudhry A, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 2010; 142: 914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lu TX, Lim EJ, Wen T, et al. MiR-375 is downregulated in epithelial cells after IL-13 stimulation and regulates an IL-13-induced epithelial transcriptome. Mucosal Immunol 2012; 5: 388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol 2013; 132: 3–13; quiz 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Chen Y, Yuan W, et al. MicroRNA-155-5p is a key regulator of allergic inflammation, modulating the epithelial barrier by targeting PKIα. Cell Death Dis 2019; 10: 884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Niu R, Xiao X, Liu B, et al. Inhibition of airway inflammation in a cockroach allergen model of asthma by agonists of miRNA-33b. Sci Rep 2017; 7: 7409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20. Liu Y, Li H, Xiao T, et al. Epigenetics in immune-mediated pulmonary diseases. Clin Rev Allergy Immunol 2013; 45: 314–330. [DOI] [PubMed] [Google Scholar]
- 21. Suojalehto H, Lindstrom I, Majuri ML, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol 2014; 163: 168–178. [DOI] [PubMed] [Google Scholar]
- 22. Huang Z, Cao Y, Zhou M, et al. Hsa_circ_0005519 increases IL-13/IL-6 by regulating hsa-let-7a-5p in CD4(+) T cells to affect asthma. Clin Exp Allergy 2019; 49: 1116–1127. [DOI] [PubMed] [Google Scholar]
- 23. Qiu L, Zhang Y, Do DC, et al. miR-155 Modulates cockroach allergen- and oxidative stress-induced cyclooxygenase-2 in asthma. J Immunol 2018; 201: 916–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taganov KD, Boldin MP, Chang KJ, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 2006; 103: 12481–12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian M, Ji Y, Wang T, et al. Changes in circulating microRNA-126 levels are associated with immune imbalance in children with acute asthma. Int J Immunopathol Pharmacol 2018; 32: 2058738418779243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang X, Wu F, Fan J, et al. Posttranscriptional regulation of interleukin-33 expression by microRNA-200 in bronchial asthma. Mol Ther 2018; 26: 1808–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levänen B, Bhakta NR, Torregrosa Paredes P, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol 2013; 131: 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jardim MJ, Dailey L, Silbajoris R, et al. Distinct microRNA expression in human airway cells of asthmatic donors identifies a novel asthma-associated gene. Am J Respir Cell Mol Biol 2012; 47: 536–542. [DOI] [PubMed] [Google Scholar]
- 29. Newcomb DC, Cephus JY, Boswell MG, et al. Estrogen and progesterone decrease Let-7F microRNA expression and increase IL-23/IL-23 receptor signaling and IL-17A production in patients with severe asthma. J Allergy Clin Immunol 2015; 136: 1025–1034.e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hammad Mahmoud Hammad R, Hamed D, Eldosoky M, et al. Plasma microRNA-21, microRNA-146a and IL-13 expression in asthmatic children. Innate Immun 2018; 24: 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faiz A, Weckmann M, Tasena H, et al. Profiling of healthy and asthmatic airway smooth muscle cells following interleukin-1beta treatment: a novel role for CCL20 in chronic mucus hypersecretion. Eur Respir J 2018; 52: 1800310. [DOI] [PubMed] [Google Scholar]
- 32. American Thoracic Society. https://www.thoracic.org/ (accessed 31 May 2020).
- 33. Guidelines for the Diagnosis and Management of Asthma (EPR-3). National Heart, Lung, and Blood Institute (NHLBI). 2018. [Google Scholar]
- 34. Plaza Moral V. GEMA(4.0). Guidelines for asthma management. Archivos de bronconeumologia 2015; 51 Suppl 1: 2–54. [DOI] [PubMed] [Google Scholar]
- 35. Park YM, Lee SY, Kim WK, et al. Risk factors of atopic dermatitis in Korean schoolchildren: 2010 international study of asthma and allergies in childhood. Asian Pac J Allergy Immunol 2016; 34: 65–72. [DOI] [PubMed] [Google Scholar]
- 36. Asthma Workgroup, Society of Respiratory Diseases, Society of General Practitioners, Chinese Medical Association (CMA). Chinese guideline for the prevention and management of bronchial asthma (Primary Health Care Version). J Thorac Dis 2013; 5: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guidelines for diagnosis and prevention of bronchial asthma in pediatric group (GDPB) (2016). Chin J Med Inform 2016; 54: 167–181. [Google Scholar]
- 38. Guidelines for diagnosis and prevention of bronchial asthma in pediatric group (GDPB) (revised in 2008). Chin J Med Inform 2009: 20–21. [Google Scholar]
- 39. Moore WC, Bleecker ER, Curran-Everett D, et al. Characterization of the severe asthma phenotype by the National Heart, Lung, and Blood Institute’s Severe Asthma Research Program. J Allergy Clin Immunol 2007; 119: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newcastle Ottawa Scale (NOS). http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp (accessed 31 May 2020).
- 41. Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155: 529–536. [DOI] [PubMed] [Google Scholar]
- 42. Reitsma JB, Glas AS, Rutjes AWS, et al. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol 2005; 58: 982–990. [DOI] [PubMed] [Google Scholar]
- 43. Sedgwick P. Spearman’s rank correlation coefficient. BMJ 2018; 362: k4131. [DOI] [PubMed] [Google Scholar]
- 44. Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods 2006; 11: 193–206. [DOI] [PubMed] [Google Scholar]
- 45. Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003; 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Biljana M, Jelena M, Branislav J, et al. Bias in meta-analysis and funnel plot asymmetry. Stud Health Technol Inform 1999; 68: 323–328. [PubMed] [Google Scholar]
- 47. Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 2009; 339: b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ye S, Zhu S, Feng L. LncRNA ANRIL/miR-125a axis exhibits potential as a biomarker for disease exacerbation, severity, and inflammation in bronchial asthma. J Clin Lab Anal 2020; 34: e23092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liang Z, Tang F. The potency of lncRNA MALAT1/miR-155/CTLA4 axis in altering Th1/Th2 balance of asthma. Biosci Rep 2020; 40. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50. Ibrahim AA, Ramadan A, Wahby AA, et al. Evaluation of miR-196a2 expression and Annexin A1 level in children with bronchial asthma. Allergol Immunopathol 2020; 48: 458–464. [DOI] [PubMed] [Google Scholar]
- 51. ElKashef S, Ahmad SE, Soliman YMA, et al. Role of microRNA-21 and microRNA-155 as biomarkers for bronchial asthma. Innate Immun 2020: 1753425920901563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao M, Juanjuan L, Weijia F, et al. Expression levels of microRNA-125b in serum exosomes of patients with asthma of different severity and its diagnostic significance. Curr Drug Metab 2019; 20: 781–784. [DOI] [PubMed] [Google Scholar]
- 53. Wu C, Xu K, Wang Z, et al. A novel microRNA miR-1165-3p as a potential diagnostic biomarker for allergic asthma. Biomarkers 2019; 24: 56–63. [DOI] [PubMed] [Google Scholar]
- 54. Tsai M-J, Tsai Y-C, Chang W-A, et al. Deducting microRNA-mediated changes common in bronchial epithelial cells of asthma and chronic obstructive pulmonary disease. A next-generation sequencing-guided bioinformatic approach. Int J Mol Sci 2019; 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodrigo-Munoz JM, Canas JA, Sastre B, et al. Asthma diagnosis using integrated analysis of eosinophil microRNAs. Allergy 2019; 74: 507–517. [DOI] [PubMed] [Google Scholar]
- 56. Qiu YY, Wu Y, Lin MJ, et al. LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/ RORgammat. Biomed Pharmacother 2019; 111: 386–394. [DOI] [PubMed] [Google Scholar]
- 57. Nasser M, Fahmey S, Geogry D, et al. Expression of serum microRNAs 221, 222, 15a and level of VEGF-A in children with bronchial asthma. Egypt J Immunol 2019; 26: 133–144. [PubMed] [Google Scholar]
- 58. Liu Q, Wang W, Jing W. Indoor air pollution aggravates asthma in Chinese children and induces the changes in serum level of miR-155. Int J Environ Health Res 2019; 29: 22–30. [DOI] [PubMed] [Google Scholar]
- 59. Liang Y, Feng Y, Wu W, et al. MicroRNA-218-5p plays a protective role in eosinophilic airway inflammation via targeting delta-catenin, a novel catenin in asthma. Clin Exp Allergy. Epub ahead of print 13 September 2019. DOI: 10.1111/cea.13498. [DOI] [PubMed] [Google Scholar]
- 60. Li P, Lang X, Xia S. Elevated expression of microRNA-378 in children with asthma aggravates airway remodeling by promoting the proliferation and apoptosis resistance of airway smooth muscle cells. Exp Ther Med 2019; 17: 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Karam RA, Abd Elrahman DM. Differential expression of miR-155 and Let-7a in the plasma of childhood asthma: potential biomarkers for diagnosis and severity. Clin Biochem 2019; 68: 30–36. [DOI] [PubMed] [Google Scholar]
- 62. Fang L, Wang X, Sun Q, et al. IgE downregulates PTEN through MicroRNA-21-5p and stimulates airway smooth muscle cell remodeling. Int J Mol Sci 2019; 20: 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bartel S, La Grutta S, Cilluffo G, et al. Human airway epithelial extracellular vesicle miRNA signature is altered upon asthma development. Allergy 2019; 75: 346–356 [DOI] [PubMed] [Google Scholar]
- 64. Zhang X, Zhao X, Sun H, et al. The role of miR-29c/B7-H3 axis in children with allergic asthma. J Transl Med 2018; 16: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang K, Liang Y, Feng Y, et al. Decreased epithelial and sputum miR-221-3p associates with airway eosinophilic inflammation and CXCL17 expression in asthma. Am J Physiol Lung Cell Mol Physiol 2018; 315: L253–l264. [DOI] [PubMed] [Google Scholar]
- 66. Zhang H, Sun Y, Rong W, et al. miR-221 participates in the airway epithelial cells injury in asthma via targeting SIRT1. Exp Lung Res 2018; 44: 272–279. [DOI] [PubMed] [Google Scholar]
- 67. Zhang D, Wu Y, Sun G. miR-192 suppresses T follicular helper cell differentiation by targeting CXCR5 in childhood asthma. Scand J Clin Lab Invest 2018; 78: 236–242. [DOI] [PubMed] [Google Scholar]
- 68. Wang XY, Chen Q, Sun YH, et al. Expression of microRNA-21 in serum exosomes of patients with different severities of asthma and its diagnostic value. Acad J Second Mil Med Univ 2018; 39: 740–744. [Google Scholar]
- 69. Tian M, Zhou Y, Jia H, et al. The clinical significance of changes in the expression levels of microRNA-1 and inflammatory factors in the peripheral blood of children with acute-stage asthma. Biomed Res Int 2018; 2018: 7632487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang Y, Zhang S, Fang X, et al. Plasma miR-199a-5p is increased in neutrophilic phenotype asthma patients and negatively correlated with pulmonary function. PLoS One 2018; 13: e0193502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dong X, Zhong N, Fang Y, et al. MicroRNA 27b-3p modulates syk in pediatric asthma induced by dust mites. Front Pediatr 2018; 6: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Daniel E, Roff A, Hsu MH, et al. Effects of allergic stimulation and glucocorticoids on miR-155 in CD4(+) T-cells. Am J Clin Exp Immunol 2018; 7: 57–66. [PMC free article] [PubMed] [Google Scholar]
- 73. Yin H, Zhang S, Sun Y, et al. MicroRNA-34/449 targets IGFBP-3 and attenuates airway remodeling by suppressing Nur77-mediated autophagy. Cell Death Dis 2017; 8: e2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun Q, Liu L, Wang H, et al. Constitutive high expression of protein arginine methyltransferase 1 in asthmatic airway smooth muscle cells is caused by reduced microRNA-19a expression and leads to enhanced remodeling. J Allergy Clin Immunol 2017; 140: 510–524.e513. [DOI] [PubMed] [Google Scholar]
- 75. Qiu YY, Zhang YW, Qian XF, et al. miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am J Transl Res 2017; 9: 3184–3199. [PMC free article] [PubMed] [Google Scholar]
- 76. Milger K, Gotschke J, Krause L, et al. Identification of a plasma miRNA biomarker signature for allergic asthma: a translational approach. Allergy 2017; 72: 1962–1971. [DOI] [PubMed] [Google Scholar]
- 77. Malmhall C, Johansson K, Winkler C, et al. Altered miR-155 expression in allergic asthmatic airways. Scand J Immunol 2017; 85: 300–307. [DOI] [PubMed] [Google Scholar]
- 78. Lacedonia D, Palladino GP, Foschino-Barbaro MP, et al. Expression profiling of miRNA-145 and miRNA-338 in serum and sputum of patients with COPD, asthma, and asthma–COPD overlap syndrome phenotype. Int J COPD 2017; 12: 1811–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kärner J, Wawrzyniak M, Tankov S, et al. Increased microRNA-323-3p in IL-22/IL-17-producing T cells and asthma: a role in the regulation of the TGF-β pathway and IL-22 production. Allergy 2017; 72: 55–65. [DOI] [PubMed] [Google Scholar]
- 80. Hu D, Zhang Z, Ke X, et al. A functional variant of miRNA-149 confers risk for allergic rhinitis and comorbid asthma in Chinese children. Int J Immunogenet 2017; 44: 62–70. [DOI] [PubMed] [Google Scholar]
- 81. Chen L, Xu J, Chu X, et al. MicroRNA-98 interferes with thrombospondin 1 expression in peripheral B cells of patients with asthma. Biosci Rep 2017; 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang Y, Xue Y, Liu Y, et al. MicroRNA-146a expression inhibits the proliferation and promotes the apoptosis of bronchial smooth muscle cells in asthma by directly targeting the epidermal growth factor receptor. Exp Ther Med 2016; 12: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pietrusińska M, Pajak A, Gorski P, et al. Preliminary studies: differences in microRNA expression in asthma and chronic obstructive pulmonary disease. Postepy Dermatol Alergol 2016; 33: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol 2016; 137: 1423–1432. [DOI] [PubMed] [Google Scholar]
- 85. Midyat L, Gulen F, Karaca E, et al. MicroRNA expression profiling in children with different asthma phenotypes. Pediatr Pulmonol 2016; 51: 582–587. [DOI] [PubMed] [Google Scholar]
- 86. Maes T, Cobos FA, Schleich F, et al. Asthma inflammatory phenotypes show differential microRNA expression in sputum. J Allergy Clin Immunol 2016; 137: 1433–1446. [DOI] [PubMed] [Google Scholar]
- 87. Luo X-Q, Yang S-B, Qiu S-Q, et al. Post-transcriptional regulation of interleukin-10 in peripheral B cells of airway allergy patients. Am J Transl Res 2016; 8: 5766–5772. [PMC free article] [PubMed] [Google Scholar]
- 88. Liu Z, Chen X, Wu Q, et al. miR-125b inhibits goblet cell differentiation in allergic airway inflammation by targeting SPDEF. Eur J Pharmacol 2016; 782: 14–20. [DOI] [PubMed] [Google Scholar]
- 89. Li L, Hui Y, Xing C, et al. miroRNA-31 affects the expression of asthma-related cytokines via regulation of CD44. Int J Clin Exp Med 2016; 9: 21506–21513. [Google Scholar]
- 90. Huo X, Zhang K, Yi L, et al. Decreased epithelial and plasma miR-181b-5p expression associates with airway eosinophilic inflammation in asthma. Clin Exp Allergy 2016; 46: 1281–1290. [DOI] [PubMed] [Google Scholar]
- 91. Fan L, Wang X, Fan L, et al. MicroRNA-145 influences the balance of Th1/Th2 via regulating RUNX3 in asthma patients. Exp Lung Res 2016; 42: 417–424. [DOI] [PubMed] [Google Scholar]
- 92. Elbehidy RM, Youssef DM, El-Shal AS, et al. MicroRNA-21 as a novel biomarker in diagnosis and response to therapy in asthmatic children. Mol Immunol 2016; 71: 107–114. [DOI] [PubMed] [Google Scholar]
- 93. Dong X, Xu M, Ren Z, et al. Regulation of CBL and ESR1 expression by microRNA-223p, 513a-5p and 625-5p may impact the pathogenesis of dust mite-induced pediatric asthma. Int J Mol Med 2016; 38: 446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cai K, Ke J, Wang Y, et al. MIR-194 is related to the pathogenesis of asthma by regulating TLR4 expression. Int J Clin Exp Pathol 2016; 9: 1327–1334. [Google Scholar]
- 95. Sawant DV, Yao W, Wright Z, et al. Serum microRNA-21 as a biomarker for allergic inflammatory disease in children. Microrna 2015; 4: 36–40. [DOI] [PubMed] [Google Scholar]
- 96. Haj-Salem I, Fakhfakh R, Bérubé JC, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFβR2 gene in severe asthma. Allergy 2015; 70: 212–219. [DOI] [PubMed] [Google Scholar]
- 97. Wu XB, Wang MY, Zhu HY, et al. Overexpression of microRNA-21 and microRNA-126 in the patients of bronchial asthma. Int J Clin Exp Med 2014; 7: 1307–1312. [PMC free article] [PubMed] [Google Scholar]
- 98. Simpson LJ, Patel S, Bhakta NR, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nat Immunol 2014; 15: 1162–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Roff AN, Craig TJ, August A, et al. MicroRNA-570-3p regulates HuR and cytokine expression in airway epithelial cells. Am J Clin Exp Immunol 2014; 3: 68–83. [PMC free article] [PubMed] [Google Scholar]
- 100. Rijavec M, Korosec P, Zavbi M, et al. Let-7a is differentially expressed in bronchial biopsies of patients with severe asthma. Sci Rep 2014; 4: 6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Perry MM, Baker JE, Gibeon DS, et al. Airway smooth muscle hyperproliferation is regulated by microRNA-221 in severe asthma. Am J Respir Cell Mol Biol 2014; 50: 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Martinez-Nunez RT, Bondanese VP, Louafi F, et al. A microRNA network dysregulated in asthma controls IL-6 production in bronchial epithelial cells. PLoS One 2014; 9: e111659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Nicodemus-Johnson J, Laxman B, Stern RK, et al. Maternal asthma and microRNA regulation of soluble HLA-G in the airway. J Allergy Clin Immunol 2013; 131: 1496–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang YY, Zhong M, Zhang MY, et al. Expression and clinical significance of miR-155 in peripheral blood CD4(+);T cells of patients with allergic asthma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 2012; 28: 540–543. [PubMed] [Google Scholar]
- 105. Yamamoto M, Singh A, Ruan J, et al. Decreased miR-192 expression in peripheral blood of asthmatic individuals undergoing an allergen inhalation challenge. BMC Genomics 2012; 13: 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Tsitsiou E, Williams AE, Moschos SA, et al. Transcriptome analysis shows activation of circulating CD8+ T cells in patients with severe asthma. J Allergy Clin Immunol 2012; 129: 95–103. [DOI] [PubMed] [Google Scholar]
- 107. Solberg OD, Ostrin EJ, Love MI, et al. Airway epithelial miRNA expression is altered in asthma. Am J Respir Crit Care Med 2012; 186: 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Panganiban RP, Pinkerton MH, Maru SY, et al. Differential microRNA expression in asthma and the role of miR-1248 in regulation of IL-5. Am J Clin Exp Immunol 2012; 1: 154–165. [PMC free article] [PubMed] [Google Scholar]
- 109. Liu F, Qin HB, Xu B, et al. Profiling of miRNAs in pediatric asthma: upregulation of miRNA-221 and miRNA-485-3p. Mol Med Rep 2012; 6: 1178–1182. [DOI] [PubMed] [Google Scholar]
- 110. Williams AE, Larner-Svensson H, Perry MM, et al. MicroRNA expression profiling in mild asthmatic human airways and effect of corticosteroid therapy. PLoS One 2009; 4: e5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Malmhäll C, Alawieh S, Lu Y, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. J Allergy Clin Immunol 2014; 133: 1429–1438, 1438.e1421-1427. [DOI] [PubMed] [Google Scholar]
- 112. Pan J, Zhou C, Zhao X, et al. A two-miRNA signature (miR-33a-5p and miR-128-3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep 2018; 8: 16699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhang X, Guo J, Fan S, et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS One 2013; 8: e81076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wang LQ, Wang CL, Xu LN, et al. The expression research of miR-210 in the elderly patients with COPD combined with ischemic stroke. Eur Rev Med Pharmacol Sci 2016; 20: 4756–4760. [PubMed] [Google Scholar]
- 115. Li X, Wang J, Wu H, et al. Reduced peripheral blood mir-140 may be a biomarker for acute lung injury by targeting toll-like receptor 4 (Tlr4). Exp Ther Med 2018; 16: 3632–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shi Y, Fu X, Cao Q, et al. Overexpression of miR-155-5p inhibits the proliferation and migration of IL-13-induced human bronchial smooth muscle cells by suppressing TGF-β-activated kinase 1/MAP3K7-binding protein 2. Allergy Asthma Immunol Res 2018; 10: 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Ong J, Timens W, Rajendran V, et al. Identification of transforming growth factor-beta-regulated microRNAs and the microRNA-targetomes in primary lung fibroblasts. PLoS One 2017; 12: e0183815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kumar M, Ahmad T, Sharma A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol 2011; 128: 1077–1085.e1071-1010. [DOI] [PubMed] [Google Scholar]
- 119. Wang Z, Ji N, Chen Z, et al. MiR-1165-3p suppresses Th2 differentiation via targeting IL-13 and PPM1A in a mouse model of allergic airway inflammation. Allergy Asthma Immunol Res 2020; 12: 859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim JO, Song DW, Kwon EJ, et al. miR-185 plays an anti-hypertrophic role in the heart via multiple targets in the calcium-signaling pathways. PLoS One 2015; 10: e0122509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tang H, Wang Z, Liu X, et al. LRRC4 inhibits glioma cell growth and invasion through a miR-185-dependent pathway. Curr Cancer Drug Targets 2012; 12: 1032–1042. [DOI] [PubMed] [Google Scholar]
- 122. Xiao G, Zhu S, Xiao X, et al. Comparison of laboratory tests, ultrasound, or magnetic resonance elastography to detect fibrosis in patients with nonalcoholic fatty liver disease: a meta-analysis. Hepatology 2017; 66: 1486–1501. [DOI] [PubMed] [Google Scholar]
- 123. Reiman MP, Goode AP, Cook CE, et al. Diagnostic accuracy of clinical tests for the diagnosis of hip femoroacetabular impingement/labral tear: a systematic review with meta-analysis. Br J Sports Med 2015; 49: 811. [DOI] [PubMed] [Google Scholar]
- 124. Fuccio L, Hassan C, Laterza L, et al. The role of K-ras gene mutation analysis in EUS-guided FNA cytology specimens for the differential diagnosis of pancreatic solid masses: a meta-analysis of prospective studies. Gastrointest Endosc 2013; 78: 596–608. [DOI] [PubMed] [Google Scholar]
- 125. Hegedus EJ, Goode AP, Cook CE, et al. Which physical examination tests provide clinicians with the most value when examining the shoulder? Update of a systematic review with meta-analysis of individual tests. Br J Sports Med 2012; 46: 964–978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-10-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-11-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-6-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-7-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-8-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-docx-9-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-1-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-2-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-3-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-4-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease
Supplemental material, sj-pdf-5-tar-10.1177_1753466620981863 for A comprehensive analysis of microRNAs as diagnostic biomarkers for asthma by Li Xu, Minhan Yi, Yun Tan, Zixun Yi and Yuan Zhang in Therapeutic Advances in Respiratory Disease