Abstract
Homeobox genes encode transcription factors that have crucial roles in embryogenesis. A recently discovered set of homeobox genes - the Rhox genes - are expressed during both embryogenesis and in adult reproductive tissues. The 33 known mouse Rhox genes are clustered together in a single region on the X chromosome, while likely descendents of the primodial Rhox cluster, Arx and Esx1, have moved to other positions on the X chromosome. Here, we summarize what is known about the regulation and function of Rhox cluster and Rhox-related genes during embryogenesis and gametogenesis. The founding member of the Rhox gene cluster - Rhox5 (previously known as Pem) - has been studied in the most depth and thus is the focus of this review. We also discuss the unusually rapid evolution of the Rhox gene cluster.
Introduction
Homeobox genes encode transcription factors that govern a myriad of processes during development. They were first discovered over 25 years ago in Drosophila melanogaster and have since been identified in a wide variety of eukaryotic species. There are many subfamilies of homeobox transcription factors; all share a structurally conserved 60 amino acid DNA-binding homeodomain motif that recognizes specific DNA sequences (McGinnis & Krumlauf 1992). Homeobox transcription factors have critical roles in specifying spatial positioning of cells, controlling cellular proliferation, and regulating the differentiation of cells during embryonic development. Disruption or improper regulation of homeobox genes can result in drastic changes in developmental programs and elicit alterations in the identity of body segments. A striking example is the discovery that mutations in the fly Antennapedia homeobox gene cause transformation of the head segment into an abdominal segment that has a fully jointed leg in place of the antenna (Postlethwait & Schneiderman 1969). Such ‘homeotic transformations’ are a hallmark of many of the first discovered homeobox genes - the Hox genes - but they are not a universal feature of all homeobox genes. The only unifying feature of all homeobox genes is that they encode a homeodomain, a motif that binds not only to specific DNA sequences, but also to specific proteins, and sometimes even to RNA (Shyu & Wilkinson 2000, Svingen & Tonissen 2006).
This review focuses on the Rhox gene family, a recently discovered set of homeobox genes (MacLean et al. 2005a) clustered on the X chromosome (Fig. 1). As described in more detail in a later section, the mouse Rhox gene cluster harbors at least 33 genes and thus is the largest homeobox gene cluster known. Like most homeobox genes, Rhox genes are expressed during embryonic development (Fig. 2; Hogeveen & Sassone-Corsi 2005, MacLean et al. 2005a, Spitz & Duboule 2005, Zhan et al. 2005, Daggag et al. 2008). However, unlike most homeobox genes, Rhox genes typically remain expressed at high levels after birth, and thus are candidates to control postnatal and adult developmental events, particularly those essential for male and female reproduction (MacLean et al. 2005a). Some Rhox genes are expressed in germ cells, while others are expressed in somatic cells in the reproductive tract, including granulosa, Sertoli, and probably Leydig cells. Individual Rhox genes exhibit different, but sometimes overlapping, expression patterns. This suggests that each Rhox family member has the potential to have both unique and redundant roles in embryonic, postnatal, and adult development, a notion that is only beginning to be examined experimentally.
Figure 1.
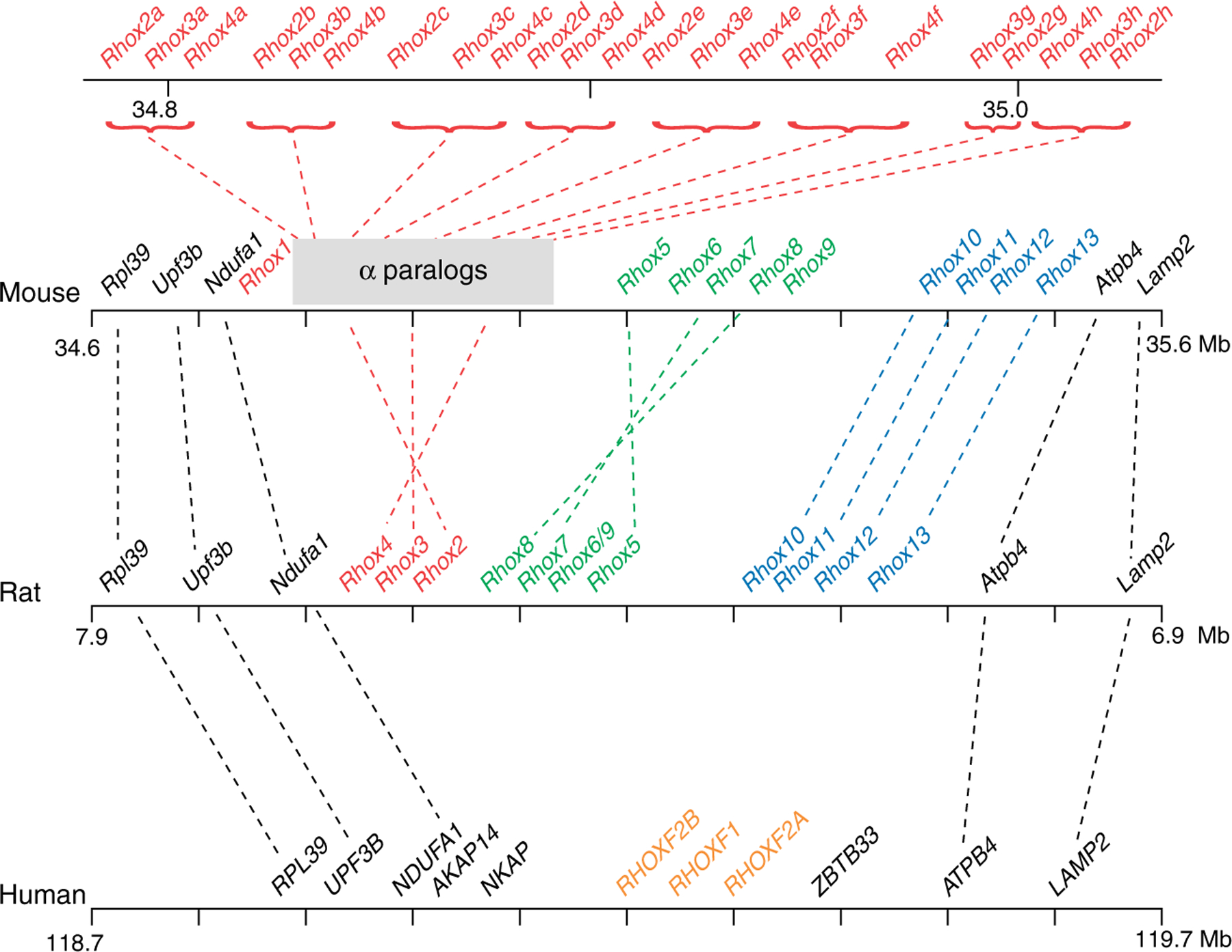
Organization of the Rhox cluster in rodents and humans. The syntenic region of the X chromosome containing the Rhox orthologs and conserved flanking genes is shown. The rodent Rhox gene subclusters are indicated by red (α), green (β), and blue (γ). Established orthologous genes are indicated by dotted lines. The orthologous relationship between human RHOX genes and rodent Rhox genes cannot be clearly assigned because of the rapid evolution of Rhox genes. The map positions shown are according to builds 37.1 (mouse), 3.4 (rat), and 37.1 (human).
Figure 2.
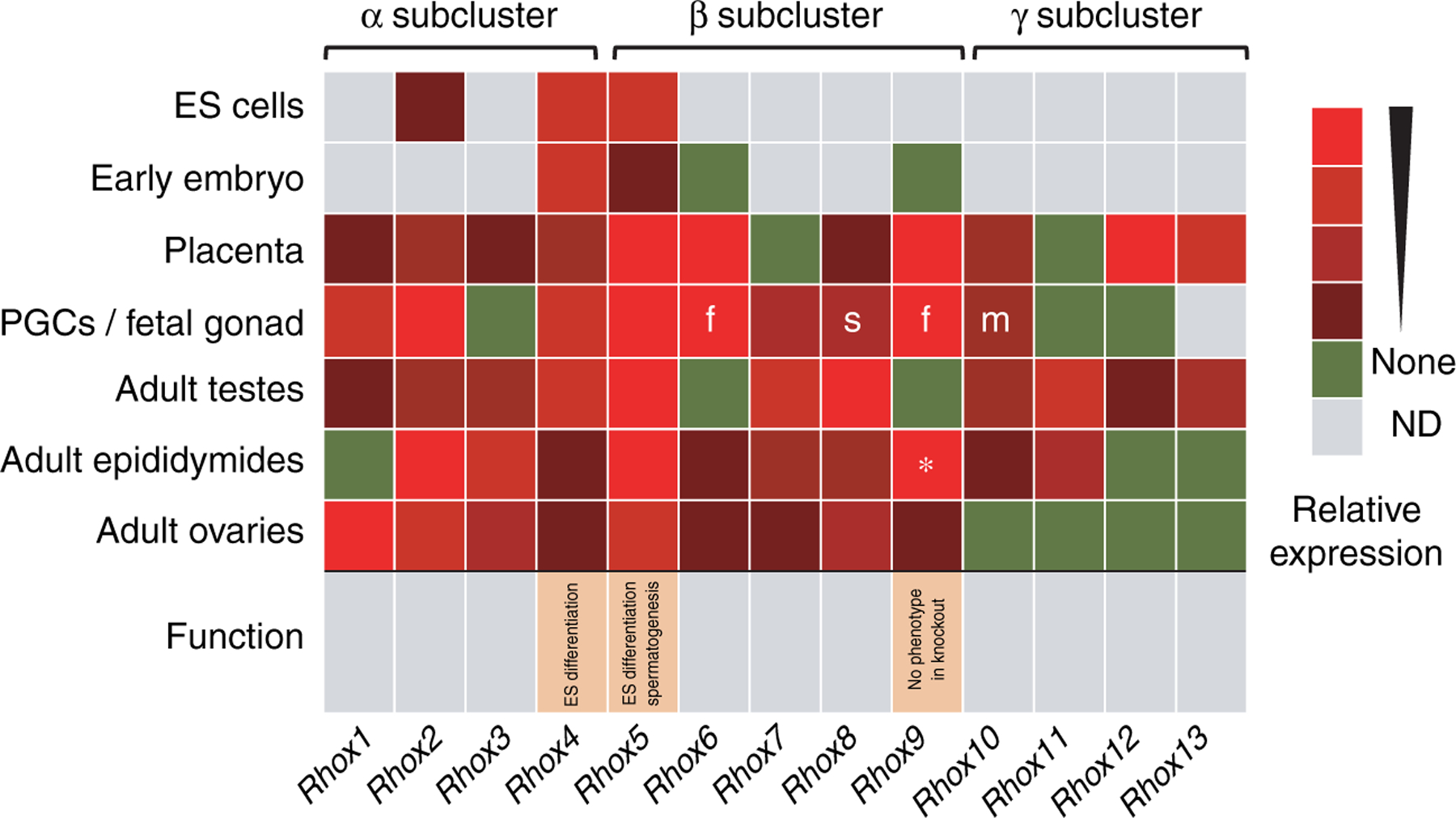
Summary of mouse Rhox cluster gene expression and function. The relative expression of each Rhox gene within the indicated tissue or cell type is shown by the heat map, with highly expressed genes shown in bright red and modestly expressed genes in maroon. The expression of Rhox genes in ES cells was determined by northern blot analysis (Sasaki et al. 1991, Fan et al. 1999), qPCR analysis (Jackson et al. 2002, Fouse et al. 2008), and semi-quantitative PCR analysis (Oda et al. 2006). The expression of Rhox genes in the early embryo (blastocyst to E8.5) was determined by semi-quantitative PCR analysis (Takasaki et al. 2000, Jackson et al. 2002, Chazaud et al. 2006); while Rhox6 and Rhox9 cannot be detected in normal mouse embryos, they can be detected in Dnmt-mutant embryos deficient in DNA methylation by RT-PCR (33 cycles; Oda et al. 2006). The expression of Rhox genes in fetal gonads was determined by qPCR and in situ hybridization analyses (Daggag et al. 2008); these analyses demonstrated that all Rhox genes are predominantly expressed in primordial germ cells (PGCs), except for Rhox8, which was exclusively expressed in gonadal somatic cells (s). ‘m’ and ‘f’ indicate expression specifically in male and female PGCs respectively. The expression of Rhox1 to Rhox12 in placenta and adult testes, epididymides, and ovaries was determined using qPCR analysis and/or ribonuclease protection analysis (MacLean et al. 2005a). The expression of Rhox13 in ovary and testis was determined by northern and semi-quantitative RT-PCR analyses (Geyer & Eddy 2008); its expression in epididymis and placenta was determined by qPCR analysis (JA MacLean & MF Wilkinson 2007, unpublished observations). (*) Rhox9 mRNA was reported as highly expressed in epididymis, but is now known to be an alternatively spliced Rhox9 mRNA that generates an amino domain-truncated protein (JA MacLean & MF Wilkinson 2005, unpublished observations). In vitro and in vivo functional analyses of Rhox5 were performed by Fan et al. (1999) and MacLean et al. (2005a), respectively. In vitro and in vivo functional analyses of Rhox4 and Rhox9 were peformed by Takasaki et al. (2001) and Jackson et al. (2002), respectively.
Here, we review what is known about the regulation, function, and evolution of the Rhox gene cluster, with an emphasis on the role and regulation of its family members in gametogenesis. We also refer readers to other reviews and commentaries covering Rhox genes (Lindsey & Wilkinson 1996a, Hogeveen & Sassone-Corsi 2005, MacLean & Wilkinson 2005, Spitz & Duboule 2005, Hu et al. 2007, Svingen & Koopman 2007, Shanker et al. 2008, Bettegowda & Wilkinson 2010).
Rhox5
Rhox5 (originally called Pem) was the first member of the Rhox gene cluster to be discovered. It was identified in a screen designed to find novel genes functioning in development and cancer. In particular, the screen identified Rhox5 as being differentially expressed between two T-cell lymphoma clones from a common origin that were frozen at different developmental stages and had different malignant properties (Wilkinson et al. 1990). Subsequent work demonstrated that Rhox5 is not only expressed in a subset of T-cell lymphomas, but it is widely expressed in tumors derived from many different cell types and tissue origins (Wilkinson et al. 1990, Maiti et al. 1996b, Wayne et al. 2002a). Its ubiquitous expression in tumors may derive, in part, from its ability to be induced by the Ras proto-oncogene (Rao et al. 2002a, MacLean et al. 2005b). RHOX5 may also have a causal role in tumor formation, as it induces an immune response in mouse tumors (Ono et al. 2000), and it physically interacts with proteins involved in malignancy (Lemmens et al. 2001, Guo et al. 2005).
Rhox5 expression
While Rhox5 was initially identified as being expressed in a T-lymphoma, it is not normally expressed in either the thymus or normal T cells (Wilkinson et al. 1990). Instead, it exhibits a highly selective and developmentally regulated expression pattern during embryogenesis, postnatal development, and in adult animals. Rhox5 is initially expressed in the unfertilized mouse oocyte, remains expressed at low levels in the blastocyst, and later is mainly confined in its expression to male and female primordial germ cells (PGCs) and extra-embryonic tissues, where it is expressed at very high levels (Wilkinson et al. 1990, Sasaki et al. 1991, Lin et al. 1994, Pitman et al. 1998, Hamatani et al. 2004, Chazaud et al. 2006, Daggag et al. 2008). After birth, its expression is primarily restricted to specific somatic cells in the reproductive tract: i) Sertoli cells in the testes, ii) principal cells in the epididymis, and iii) granulosa cells in the ovary (Wilkinson et al. 1990, Lin et al. 1994, Lindsey & Wilkinson 1996b, Maiti et al. 1996a, Pitman et al. 1998, Rao et al. 2002b). The expression of Rhox5 in Sertoli cells is of interest given that these cells provide a structural framework for spermatogenesis and direct several androgen-dependent steps of spermatogenesis. Indeed, RHOX5 expression in Sertoli cells is induced by androgen and AR and it is specifically expressed in Sertoli cells during the androgen-dependent stages (IV–VIII) of the spermatogenic epithelial cycle, making it a candidate to drive androgen-dependent steps of spermatogenesis (Lindsey & Wilkinson 1996c, Sutton et al. 1998, Rao et al. 2003). In the adult epididymis, Rhox5 expression is restricted to the caput region, where sperm derived from the testis acquire forward motility (Rao & Wilkinson 2002, Rao et al. 2002b). In the adult ovary, RHOX5 protein is exclusively in mural granulosa cells, which line the walls of the developing follicle and respond to the ovulatory LH surge (Pitman et al. 1998). Thus, RHOX5 is expressed in the nurse cells that translate the key hormone signal that promotes final oocyte maturation.
In vivo Rhox5 functions
While analysis of its expression pattern allowed a speculative glimpse into Rhox5’s potential functions, it was not until the generation of Rhox5-null mice (Pitman et al. 1998) that its actual functions came to light. Rhox5-null males have increased germ cell apoptosis, reduced epididymal sperm count, reduced frequency of sperm with normal forward motility, and are subfertile (MacLean et al. 2005a). It is likely that the reduced sperm count results exclusively from the increased germ cell apoptosis, as cell proliferation (measured by bromodeoxyuridine incorporation) is not significantly affected by loss of Rhox5. Apoptosis is a normal event during spermatogenesis; it occurs specifically in a subset of spermatogonia and spermatocytes during stages I–IVand XII, respectively, of the seminiferous epithelial cycle. Rhox5-null males had higher numbers of apoptotic germ cells at these stages of the seminiferous epithelial cycle, indicating that RHOX5 is required for establishing the normal balance of germ cell survival versus death. In addition, Rhox5-null males have apoptotic germ cells in stages of the seminiferous epithelial cycle that normally have none (i.e. stages V–XI). This ‘promiscuous apoptosis’ is intriguing because it suggests that RHOX5 has a role in specifying the stage-specific death cycle that normally occurs in spermatogenesis. RHOX5 is well placed to perform such a role, as it is specifically expressed in Sertoli cells, the central hub of the seminiferous epithelial cycle.
How exactly RHOX5 controls germ cell survival is not known, but it is likely to involve regulation of Sertoli cell genes encoding cell surface and secreted proteins that communicate with germ cells. As described in more detail below, several RHOX5-regulated genes encoding cell surface and secreted proteins that control cell survival and apoptosis have been identified (Hu et al. 2007, 2008; J MacLean II, Z Hu, M Rao, C Wayne & MF Wilkinson 2010, unpublished observations). By regulating such proteins, RHOX5 may be a major mediator of androgen action in the testes. Despite the well-established importance of androgens and testosterone in spermatogenesis, RHOX5 currently stands alone as an androgen-regulated transcription factor whose ablation results in abnormal spermatogenesis (MacLean & Wilkinson 2005, MacLean et al. 2005a, Hu et al. 2007, 2008). Because androgens act through the Sertoli cells, not germ cells, to promote spermatogenesis (De Gendt et al. 2004, Holdcraft & Braun 2004, Chang et al. 2004, MacLean & Wilkinson 2005), this makes RHOX5 a strong candidate to have a role in one or more of the androgen-dependent steps of spermatogenesis.
The defect in sperm motility observed in Rhox5-null males could result from either an epididymal or a testes defect. The most likely site of the defect is the epididymis, as RHOX5 is specifically expressed in the caput region (Rao et al. 2002b), where sperm motility is acquired (Rao & Wilkinson 2002). However, it is possible that the defect is in the testes; in this case, the simplest explanation is that RHOX5 expression in Sertoli cells is necessary for the proper programming of the adjacent developing germ cells so that they are competent to later acquire motility in the epididymis.
The defects that have been detected in Rhox5-null mice are confined to the male reproductive tract. Thus, even though Rhox5 is also expressed in granulosa cells in the ovary, Rhox5-null female mice have normal ovulation, litter frequency, and litter size (Pitman et al. 1998, MacLean et al. 2005a). Mice lacking Rhox5 also have no obvious defects in embryonic development despite Rhox5 normally being expressed in the early embryo, PGCs, and placenta (Pitman et al. 1998). One obvious explanation for this is that other Rhox family members compensate for the loss of Rhox5. Another non-mutually exclusive explanation is that Rhox5-null mice have subtle defects in embryonic development and female reproduction that have so far escaped detection. It is also possible that embryonic and female reproductive defects depend on context and would only be observed in particular environmental or genetic backgrounds. Finally, one cannot rule out that Rhox5 expression in some cell types and developmental stages is ‘noise’ that has no functional significance; i.e. it has not been selected against because it is not deleterious.
While there is no in vivo evidence for an embryonic role of Rhox5, it is worth noting that Fan et al. (1999) showed that overexpression of RHOX5 in embryonic stem (ES) cells results in blockade of their differentiation in vitro. The differentiation block requires the amino terminus of RHOX5, not the homeodomain, suggesting that it might be the result of sequestering RHOX5-interacting factors crucial for ES cell differentiation. It will be interesting in the future to identify the RHOX5-interacting factors that have a role in this differentiation blockade.
Biochemical functions of RHOX5
When Rhox5 was first identified, it was defined as a novel gene with no obvious motifs in common with other genes (Wilkinson et al. 1990). Thus, the biochemical function of its encoded protein was initially unclear. However, subsequent bioinformatic studies revealed that RHOX5 possesses a divergent homeodomain harboring the key amino acids necessary to form the homeodomain structure, but was otherwise substantially different than other homeodomains defined at that time (Rayle 1991, Sasaki et al. 1991). This was an important breakthrough for at least two reasons. First, it indicated that RHOX5 is almost certainly a transcription factor. Second, it suggested that RHOX5 might regulate sets of genes that are different than those regulated by other known homeobox proteins. Indeed, the RHOX5 homeodomain has unique amino acid residues at positions known to make base-specific contacts with DNA (Rayle 1991, Sasaki et al. 1991, MacLean et al. 2005a).
To begin to understand RHOX5’s role as a transcription factor, two main approaches have been taken: i) the identification of genes regulated by RHOX5 and ii) the identification of proteins that interact with RHOX5. With regard to the first approach, Hu et al. (2010) conducted microarray analysis on 15P-1 Sertoli cell clones stably transfected with a Rhox5 expression vector expressing physiological levels of RHOX5. This study identified many genes positively and negatively regulated by RHOX5 in 15P-1 cells; most of these are also regulated by RHOX5 in vivo, based on analysis of testes from Rhox5-null mice. Many of the RHOX5-regulated genes are expressed in Sertoli cells, indicating that they are candidates to be direct targets of RHOX5. A subset of these genes is regulated by androgen receptor (AR), suggesting that these are secondary androgen-response (SAR) genes; i.e. genes mediating testosterone signaling that are indirectly regulated by AR through the action of AR-regulated transcription factors. The best characterized SAR is Unc5c, which RHOX5 negatively regulates at the transcriptional level through the 5′ UTR region of Unc5c (Hu et al. 2007, 2008). Unc5c encodes a netrin receptor that is crucial for axon guidance in the brain and is a tumor suppressor that promotes apoptosis (Arakawa 2004). To determine whether UNC5C also promotes apoptosis in the testes, Hu et al. examined Unc5c-null mice. They found that these UNC5C-deficient mice had reduced germ cell apoptosis (Hu et al. 2010), indicating that indeed UNC5C promotes the death of both germ cells and neurons. This discovery, coupled with the fact that RHOX5 negatively regulates Unc5c transcription (Hu et al. 2010) and promotes germ cell survival (MacLean et al. 2005a), supports a model in which the pro-survival activity of RHOX5 in the testis is mediated, in part, by its ability to repress the expression of the anti-survival gene Unc5c. Other Sertoli cell factors downstream of RHOX5 that may mediate germ cell survival have been identified by in vivo microarray analysis (Hu et al. 2007, J MacLean II, Z Hu, M Rao, C Wayne & MF Wilkinson 2010, unpublished observations). These factors include insulin and other secreted metabolic regulators, as well as key enzymes and transcription factors that control energy metabolism.
Homeodomain transcription factors do not act alone to regulate their target genes, but rather collaborate with other proteins. With this in mind, investigators have identified several proteins that interact with RHOX5, including menin (MEN1), prosaposin (PSAP), inhibitor of MyoD family (I-MFA; also known as MDFIC), and cell division cycle 37 (CDC37; Lemmens et al. 2001, Guo et al. 2005, 2007a, Luo et al. 2006). Interestingly, none of these are known transcriptional co-regulators, and thus their role in RHOX5-mediated transcriptional regulation is not yet clear. The best characterized of these proteins is menin, which is a nuclear protein with tumor suppressor activity that has been shown to interact with other transcription factors, including JunD (Lemmens et al. 2001, Poisson et al. 2003, and references therein). Two groups, Lemmens et al. and Poisson et al., independently demonstrated that menin and RHOX5 directly interact, based on co-immunoprecipitation and GST-pulldown experiments in transfected cells. Lemmens et al. also showed that menin and Rhox5 transcripts co-localize in seminiferous tubules, suggesting that the encoded proteins are normally co-expressed in the testes. However, it remains to be definitively determined whether menin protein is in Sertoli cells, the sole site of RHOX5 expression in the testis. Interestingly, menin has also been shown to interact with the homeobox transcription factor PDX1 in the pancreas to modulate the expression of insulin and insulin-like growth factor-binding protein 2 (La et al. 2006, Shen et al. 2009). This brings up the possibility that a RHOX5-menin interaction might also influence the expression of insulin-2 and other metabolism-related genes regulated by RHOX5 (J MacLean II, Z Hu, M Rao, C Wayne & MF Wilkinson 2010, unpublished observations).
Another protein that directly binds to RHOX5 is CDC37, a cell-cycle regulator that facilitates formation of the CDC28-G1 cyclin complex (Guo et al. 2005). This suggests that RHOX5 and CDC37 might co-operate to promote cell proliferation, which is consistent with the fact that RHOX5 is expressed in highly proliferative cells, including trophoblasts and PGCs. If RHOX5 can only promote proliferation in cells that also contain CDC37, this would provide an explanation for why RHOX5 is also expressed in non-proliferative cells, such as post-mitotic Sertoli and epididymal cells, as these cells may lack CDC37. As part of its role in the cell cycle, RHOX5 may also function as a DNA checkpoint regulator. The evidence for this comes from the finding that transgenic mice overexpressing RHOX5 in Sertoli cells have increased DNA strand breaks in the adjacent germ cells during their maturation into elongated spermatids (Wayne et al. 2002b).
A third protein that has been shown to directly bind to RHOX5 is PSAP, a lysosomal enzyme activator that is necessary for the normal development of the testes and male accessory organs (Guo et al. 2007a). The notion that PSAP and RHOX5 collaborate is consistent with the fact that both have known functions in the testis. However, it remains to be determined whether both normally exist in the same milieu, as RHOX5 is predominantly a nuclear protein, while PSAP is a cytoplasmic and secreted protein (Pitman et al. 1998, Rao et al. 2002b, Guo et al. 2007b). The cytoplasm may be a site where these two proteins interact, as RHOX5 has been shown to be present in the cytoplasm of some cells (Pitman et al. 1998, Rao et al. 2002b, 2003). By sequestering RHOX5 in the cytoplasm, PSAP may serve as a negative regulator of RHOX5’s transcriptional functions in the nucleus.
Rhox5 evolution
As with perhaps the majority of proteins that function in the reproductive system (Swanson & Vacquier 2002), RHOX5 is rapidly evolving. Mouse and rat RHOX5 exhibit only 73% identity; the RHOX5 homeodomain region is even more divergent than the protein as a whole, displaying only 63% identity in mice and rats (Maiti et al. 1996b). This rapid evolution is not likely to be due to random drift; instead, evidence suggests that it is the result of positive selection pressure for alterations in amino acid sequence. The primary evidence for this comes from sequence analysis of a large panel of mouse and rat species, which showed that Rhox5 has an extremely high ratio of non-synonymous-to-synonymous codon substitutions (Sutton & Wilkinson 1997a). Reinforcing the notion that positive selection is the driving force for this divergence was the finding that some Rhox5 exons have undergone more rapid alterations in sequence than some Rhox5 introns. The rate of Rhox5 evolution is comparable with that occurring in the male-determining gene Sry, which encodes another transcription factor important for the male reproductive tract (Swanson & Vacquier 2002).
What selective forces have driven the rapid evolution of Rhox5? While the answer to this question is not known, it is worth first noting that even though the RHOX5 homeodomain as a whole has undergone a very high rate of sequence change, the sequences encoding two classes of amino acids within the RHOX5 homeodomain region have remained relatively intact. One class is the amino acids that are crucial for homeodomain structure and the other is the amino acids in the third helix, the ‘recognition helix’ that makes all the base-specific contacts with DNA (Sutton & Wilkinson 1997a). This suggests that purifying selection has fixed sequences crucial for maintaining both the RHOX5 homeodomain structure and its DNA targets. Why are the other amino acids in the RHOX5 homeodomain undergoing rapid change? Analysis of a large number of different homeobox transcription factors from different species has revealed that a common feature of their homeodomain region is that they bind not only DNA but also specific proteins (Moens & Selleri 2006). This leads to the hypothesis that the rapid evolution of the RHOX5 homeodomain has been driven by the proteins with which it interacts. If true, it will be important to determine whether these are already identified RHOX5-interacting proteins or novel ones. It will also be intriguing to know whether the RHOX5 homeodomain has co-evolved with its binding partners over time or instead is undergoing selection to bind to new protein partners in different species.
Rhox5 regulation
The Rhox5 gene is regulated by many different stimuli, including differentiation signals, oncogenic signals, hormones, and DNA methylation. Sasaki et al. (1991) was the first study to demonstrate that Rhox5 is regulated by differentiation signals. These authors discovered that Rhox5 mRNA levels are dramatically upregulated when ES cells are induced to differentiate into embyroid bodies, peaking 9 days after induction of differentiation. While they did not determine which specific cell types in differentiating ES cells express elevated levels of Rhox5, examination of the F9 embryocarcinoma cell line provided some insight into this issue. F9 cells are malignant but have stem-like qualities and thus they can be induced to specifically differentiate into visceral or parietal endoderm. Sasaki et al. (1991) found that F9 cells strongly upregulated Rhox5 mRNA when they are induced to differentiate into either visceral endoderm or parietal endoderm. This recapitulates what occurs in vivo, as Rhox5 is highly and specifically expressed in both the visceral and parietal endoderm components of the yolk sac (Wilkinson et al. 1990, Lin et al. 1994).
Rhox5 is regulated by DNA methylation
A regulatory mechanism that mounting evidence suggests is crucial for controlling Rhox5 expression is DNA methylation. The first evidence for this was the finding by Sasaki et al. (1991) that treatment of mouse 10T1/2 mesenchymal stem cells with the DNA methyltransferase (DNMT) inhibitor 5-aza-2′-deoxycytidine (5AzaC) led to a dramatic (O50-fold) increase in Rhox5 mRNA levels. Rhox5 mRNA was specifically induced in 10T1/2 derivatives that committed to the muscle cell lineage (myoblasts), not to cells that committed to the cartilage (chondrocyte) or fat cell (adipocyte) lineages. This induction may be physio-logically relevant, as Rhox5 transcripts, including a muscle-specific alternatively spliced form, are expressed at modest levels in rat skeletal muscle (Maiti et al. 1996a). However, Rhox5 mRNA is not detectably expressed in mouse skeletal muscle (MacLean et al. 2005a), indicating that if indeed Rhox5 does have a role in muscle differentiation, it is not a conserved role.
Another interpretation of the observation that Rhox5 is dramatically induced by chemical inhibition of DNA methylation in muscle cells is that this reflects Rhox5 being transcriptionally repressed by DNA methylation in inappropriate cell lineages. Two subsequent studies - both of which employed genetic manipulation techniques to knockout genes encoding DNMTs - obtained evidence that indeed this is the case. Jackson-Grusby et al. used microarray analysis to identify genes upregulated in cultured fibroblasts that lack DNMT1, the ‘maintenance DNMT’ essential for maintaining heritable DNA methylation patterns in dividing cells. Rhox5 was one of the most upregulated genes (by ~14-fold) identified in these DNA methylation-deficient cells (Jackson-Grusby et al. 2001). Given that Rhox5 is not normally expressed in fibroblasts, this provided evidence that DNA methylation is sufficient to prevent its expression in this cell type. However, it did not rule out that DNA demethylation triggers other events, such as changes in histone modifications, which contributed to the derepression of Rhox5 transcription.
Oda et al. (2006) examined whether DNA methylation is responsible for preventing Rhox5 expression in mid-gestation embryos. Consistent with this, they observed that the Rhox5 promoter shifts from being in an unmethylated state in eight-cell embryos and blastocysts - both of which express Rhox5 - to a methylated state in mid-gestation (e8.5) embryos, which lack detectable Rhox5 expression (Lin et al. 1994, Hamatani et al. 2004). It should be noted, however, that Oda et al. examined the methylation status of the Rhox5 alternative promoter not appreciably expressed in early embryos (Rao et al. 2003; see next section for a description of the alternative Rhox5 promoters). To obtain direct evidence for a role of DNA methylation, Oda et al. examined the effect of loss of DNMT3a and DNMT3b, the ‘de novo synthesis DNMTs’ that are responsible for new DNA methylation events in ES cells, trophoblast tissues, and PGCs (Okano et al. 1999). Mice lacking both of these enzymes fail to produce viable offspring, exhibit abnormal growth rate and development, and die prior to e11.5 (Okano et al. 1999). They found that Dnmt3a−/−Dnmt3b−/− e8.5–9.5 embryos harbor a hypomethylated Rhox5 gene promoter that is dere-pressed, allowing high levels of Rhox5 transcripts to be expressed. The repression of Rhox5 expression in early embryos can be mediated by either of the de novo synthesis DNMTs, as single-mutant (Dnmt3a−/− or Dnmt3b−/−) embryos did not exhibit a detectable increase in Rhox5 mRNA expression. Surprisingly, single-mutant Dnmt3b−/− embryos did exhibit reduced Rhox5 promoter DNA methylation. This suggests that either i) DNA demethylation is not sufficient for Rhox5 transcription, ii) a threshold of reduced DNA methylation is required to activate Rhox5 transcription, or iii) DNA demethylation triggers only a modest increase in Rhox5 transcription that was not detected by the investigators.
In contrast to the embryo proper, extra-embryonic tissues constitutively express high levels of Rhox5 (Wilkinson et al. 1990), which was not significantly further increased by loss of DNMT3a and DNMT3b (Oda et al. 2006). Together with an earlier report that found that Rhox5 is selectively hypomethylated in placenta (Schutt et al. 2003), these data collectively indicate that DNA methylation defines the tissue-specific expression pattern of Rhox5 by virtue of its ability to prevent Rhox5 expression in mid-gestation embryos. In the future, it will be interesting to determine whether the induction of Rhox5 expression in PGCs in later-stage embryos (Pitman et al. 1998, Daggag et al. 2008) is the result of cell type-specific DNA demethylation.
Rhox5 is an imprinted gene
DNA methylation controls genomic imprinting, an epigenetic process that results in the selective repression of one of the two alleles of a given gene. Paternally imprinted genes are methylated and transcriptionally silenced on the paternal allele, whereas maternally imprinted genes are methylated and transcriptionally silenced on the maternal allele. Genomic imprinting acts only on a selected subset of genes (<100 are known in humans and mice), and its evolutionary origin and functional importance - while under intense investigation - remain unclear (Moore 2001, Laprise 2009). Imprinting would seem to put organisms at a disadvantage since it causes genes to be functionally haploid, and thus it increases the risk that mutations in them will lead to phenotypic defects. Genomic imprinting may have evolved as a result of selection pressure arising from opposing needs of the two parental genomes; the paternal genome selects for features that promote placental and prenatal growth, while the maternal genome selects for features that conserve resources and thereby reduce growth.
Recently, Rhox5 has been demonstrated to be a gene imprinted in the early embryo. This was uncovered by Kobayashi et al. (2006) whose initial goal was to identify genes differentially expressed in male and female blastocysts. Using offspring from male mice that harbored a green fluorescent protein (GFP)-expressing transgene on their X chromosome, these authors sorted GFP-expressing female and non-expressing male blastocysts and found - using expression profiling - that ~600 genes were differentially expressed by twofold or more, with roughly half being expressed at higher levels in female blastocysts and the other half being preferentially expressed in male blastocysts. Of those preferentially expressed in female blastocysts, only two genes were found to be exclusively in female blastocysts: Xist and Rhox5. The finding that Xist was exclusively expressed in female blastocysts was expected, as it is a non-coding transcript specifically expressed in female cells that is crucial for X-chromosome inactivation and dosage compensation in female cells (Chow & Heard 2009). The discovery that Rhox5 exhibits a female-specific expression pattern was unexpected; subsequent investigation revealed the reason: Rhox5 is exclusively expressed from the paternal X chromosome (Kobayashi et al. 2006), which is not present in male blastocysts. The authors went on to show that the paternal-specific expression of Rhox5 is established by the eight-cell stage and is maintained until e6.5.
It remains for future investigations to understand the mechanism and functional significance of Rhox5 imprinting in the early embryo. As described earlier, Rhox5 controls the expression of several genes regulating energy metabolism, including those encoding secreted proteins and master transcriptional regulators that are likely to influence large batteries of downstream energy metabolism genes (Hu et al. 2007). This suggests the possibility that Rhox5 is subject to imprinting because of competition between male and female early embryos for limited energy resources. Because Rhox5 is on the X chromosome, it may also be subject to another form of imprinting. Most X-linked genes are preferentially expressed from the maternal allele in extra-embryonic tissue as a result of a chromosome-wide paternal imprinting mechanism (Reik & Lewis 2005). While Kobayashi et al. (2006) did not address whether Rhox5 is subject to this mechanism, they did obtain evidence that e7.5 embryos express higher levels of Rhox5 from the maternal allele than from the paternal allele, consistent with this possibility. Furthermore, two X-linked Rhox5-related genes, Rhox6 and Esx1, have been shown to be paternally imprinted in extraembryonic tissue (Li & Behringer 1998, Oda et al. 2006). To directly address this issue, we examined placental tissue from crosses of two related species with recognizably different Rhox5 alleles (Mus musculus musculus and Mus musculus molossinus) and found that Rhox5 was preferentially expressed from the maternal allele, regardless of its species of origin (J MacLean II, Z Hu, M Rao, C Wayne & MF Wilkinson 2010, unpublished observations). Coupled with the work of Kobayashi et al. (2006), this demonstrates that Rhox5 is an unusual gene that is paternally imprinted in extra-embryonic tissues and maternally imprinted in the early embryo. Few other mammalian genes are known to be both paternally and maternally imprinted; it will be interesting to know the functional consequences of this unique expression pattern. It will also be important to determine whether Rhox5 undergoes a switch in imprinting (from paternal to maternal) in some cell types or instead undergoes different kinds of imprinting (maternal or paternal) in different cell types.
Because Rhox5 has roles in spermatogenesis (MacLean et al. 2005a), it will also be important to know whether Rhox5 imprinting pattern has consequences for male fertility. Indeed, abnormalities in both maternal and paternal imprinting have been associated with low sperm quality and hypofertility in humans (Kobayashi et al. 2007, Marques et al. 2010). Furthermore, human sperm harboring genes that are abnormally imprinted have been shown to be less likely to be competent for IVF and ICSI (Kobayashi et al. 2007). Thus, it will be interesting to determine whether human RHOX genes are subject to imprinting and, if so, whether RHOX imprinting abnormalities have a negative impact on human fertility.
Rhox5 has two promoters
Rhox5 mRNA is transcribed from two promoters that are independently regulated but give rise to the same protein. The proximal promoter (Pp) is of interest because it is directly regulated by the nuclear hormone receptor AR, which along with other transcription factors restricts its expression to specific somatic cell types in the testes and epididymis. The distal promoter (Pd) is also expressed in a tissue- and cell type-specific manner, but its primary sites of expression are in the early embryo and somatic cells in adult female reproductive tissues. The Pd is also expressed in a wide variety of tumors. In this section, we describe the expression pattern of these two promoters in more detail, and then review studies that have examined the cis elements and trans-acting factors that control their unique expression patterns.
The Pp
The Pp gives rise to full-length mRNA specifically in the testis and epididymis in both mice and rats (Maiti et al. 1996a, 1996b, Sutton et al. 1998, Rao et al. 2003). In mice testes, where Pp-derived transcripts are dramatically induced between P8 and P9, Pp expression is restricted to Sertoli cells (Lindsey & Wilkinson 1996c, Sutton et al. 1998, Rao et al. 2003). Pp expression is further restricted to Sertoli cells in stages IV–VII of the seminiferous epithelial cycle in adult mouse testes, as Rhox5 mRNA and RHOX5 protein are only observable in these stages (Lindsey & Wilkinson 1996c, Sutton et al. 1998, Rao et al. 2003). In the mouse epididymis, the Pp is specifically expressed in somatic cells (primarily in principal cells) in segments 2–4 of the caput region (Rao et al. 2002b). Together, these data indicate that the Pp is an exquisitely regulated promoter that is expressed in a cell type-, region-, stage-, and tissue-specific manner.
Pp transcription in both the testes and epididymis depends on AR and testosterone. This conclusion comes from several lines of evidence, including: i) Rhox5 mRNA levels (measured with either Rhox5 or Pp-specific probes) are dramatically reduced in animals deficient in the ability to produce LH (the peptide hormone responsible for inducing testosterone production in Leydig cells) as a result of hypophysectomy, chemical treatment, or genetic mutations (Lindsey & Wilkinson 1996b, 1996c, Maiti et al. 1996a, Sutton et al. 1998); ii) testosterone reverses the defect in Rhox5 expression caused by loss of LH (Lindsey & Wilkinson 1996b); iii) androgens induce Rhox5 and/or Pp-driven reporter expression in primary Sertoli cell cultures and Sertoli cell lines (Rao et al. 2003, Sneddon et al. 2005, Zhou et al. 2005); iv) AR-deficient mice have reduced testicular Rhox5 expression (De Gendt et al. 2004, Holdcraft & Braun 2004, Tan et al. 2005, O’Shaughnessy et al. 2007, Schauwaers et al. 2007); v) Rhox5 expression is drastically reduced in testes lacking AR specifically in Sertoli cells (De Gendt et al. 2004, Holdcraft & Braun 2004, Denolet et al. 2006); vi) Rhox5, as well as some other androgen-dependent genes, exhibits modestly reduced expression in mice lacking AR in peritubular myoid cells and smooth muscle cells, suggesting that AR can also exert effects on Sertoli cells indirectly; vii) Rhox5 has been shown to be positively regulated by AR and/or androgen in several independent expression profiling (microarray) studies conducted in different model systems in both mice and cell lines (Sadate-Ngatchou et al. 2004, Zhou et al. 2005, Denolet et al. 2006, Eakin & Hadjantonakis 2006, Kobayashi et al. 2006, Eacker et al. 2007, O’Shaughnessy et al. 2007, Prante et al. 2008, De Gendt et al. 2009); viii) RHOX5 is localized to Sertoli cells specifically during the androgen-dependent stages of spermatogenesis (Lindsey & Wilkinson 1996b, Sutton et al. 1998); and ix) the Pp possesses four androgen-response elements (AREs), each of which is required for its AR- and androgen-dependent expression in transfected cell lines (Barbulescu et al. 2001, Geserick et al. 2003, Bhardwaj et al. 2008, Faus & Haendler 2008). Together, these studies strongly suggest that the Rhox5 Pp is a direct target of AR in Sertoli cells.
The highly androgen/AR-dependent expression of Rhox5 has lead to Rhox5 being widely used as a marker to examine perturbations disturbing androgen signaling (Sutton et al. 1998, Barbulescu et al. 2001, Geserick et al. 2003, De Gendt et al. 2004, MacLean & Wilkinson 2005, Terada et al. 2005, Bhardwaj et al. 2008, Prante et al. 2008). Because the Rhox5 gene encodes a transcription factor, another implication of its androgen/AR-dependent expression is that it is a candidate to regulate the transcription of genes that mediate androgen action during spermatogenesis (MacLean & Wilkinson 2005). Indeed, as described earlier, RHOX5 controls the expression of several AR-regulated genes (Hu et al. 2010). To date, no other androgen- and AR-inducible transcription factors have been identified in Sertoli cells.
Analysis of transgenic mice harboring different lengths of Pp 5′ upstream sequences by Rao et al. (2002b, 2003) revealed that all of the elements necessary for androgen-dependent and cell type-specific expression of Rhox5 are present within the first ~600 nt upstream of the Pp transcription start site. This sequence is divided into two ~300 nt regulatory domains: regions I and II. Region I contains one or more repressor elements that dictate, at least in part, the specificity of Pp expression. In the testis, regionI helpsconfer the stage-specific expression of Rhox5 during seminiferous epithelial cycle (Lindsey & Wilkinson 1996b, 1996c, Sutton et al. 1998) by inhibiting Pp expression in other stages (Rao et al. 2003). In the epididymis, region I inhibits inappropriate expression of the Pp in epididymal regions other than the caput (Rao et al. 2002b).
Region II confers the other properties of the Pp, including its i) androgen-dependent expression, ii) tissue-specific expression in the testis and epididymis, and iii) Sertoli cell-specific expression in the testes (Rao et al. 2002b, 2003). Androgen-dependent expression is conferred by the four AREs in region II (Barbulescu et al. 2001, Geserick et al. 2003, Bhardwaj et al. 2008). These AREs also contribute to the tissue- and cell type-specific expression pattern of the Pp, as recently shown by Bhardwaj et al. (2008). Consensus AREs have also been identified in the Rhox5-related human RHOXF2A gene (Wayne et al. 2002a), but to date, AR-dependent expression has only been demonstrated for the RHOXF1 gene (Geserick et al. 2002).
Bhardwaj et al. (2008) also identified other cis elements in region II that collaborate with AREs to drive cell type-specific expression. Most importantly, they identified several GATA transcription factor-binding sites crucial for the expression of the Pp in Sertoli cells, at least one of which is crucial for Pp transcription in vivo. This conclusion is based on several lines of evidence, including: i) mutation of the GATA consensus sites in region II reduces Pp transcription in transiently transfected Sertoli cells, ii) mutation of the GATA site most crucial for Pp expression in transfected cells also strongly reduces Pp expression in transgenic mice, iii) forced expression of GATA factors, in combination with AR, induces Rhox5 expression in Sertoli cell lines that constitutively express low levels of the GATA factors, iv) this ability of GATA factors to increase Pp expression is abrogated by mutation of the crucial GATA site important in vivo, and v) several GATA factors are recruited to the Pp in adult testes and the MSC1 Sertoli cell line, as shown by chromatin immunoprecipitation (ChIP) analysis.
The induction of Pp expression by GATA transcription factors is likely to depend on their ability to co-operate with AR (Bhardwaj et al. 2008). An indirect line of evidence for this is that the GATA site crucial for Pp transcription in vivo is <25 nt from two AREs in region II. More compelling evidence is that GATA factors and AR form a complex in vitro. Depletion of either GATA-binding site factors or ARE-binding factors ablates the formation of this complex. Furthermore, forced expression of GATA transcription factors rescues expression from Pp reporter constructs harboring mutations in any of the four AREs in region II. Finally, the level of GATA factors bound to the Pp (as measured by ChIP) in MSC1 cells is increased in response to the addition of AR and androgen.
It remains for future studies to determine which of the six known GATA transcription factors are responsible for co-operating with AR to induce Pp expression. GATA1 is one candidate, as Gata1 mRNA levels increase concomitantly with Ar and Pp mRNA levels during postnatal testis development (Bhardwaj et al. 2008). Other candidates are GATA4 and GATA6, as they are expressed at high levels in Sertoli cells (Viger et al. 2008). However, Gata4 and Gata6 mRNAs are downregulated during postnatal testes development (Bhardwaj et al. 2008), suggesting that their role in regulating Pp might primarily be in the early portion of the first wave of spermatogenesis. Of the other three GATA factors, only GATA2 is significantly expressed in Sertoli cells (Bhardwaj et al. 2008), making GATA2 another candidate to regulate Pp transcription. This notion is supported by the finding that GATA2 is bound to the Pp in both the adult testes and the MSC1 Sertoli cell line (Bhardwaj et al. 2008).
Restriction of Pp expression to Sertoli cells is conferred not only by transcription-promoting factors but also by negative regulatory influences. While not well defined, region II appears to have a repressor element that inhibits Rhox5 expression in non-Sertoli cells (Bhardwaj et al. 2008). In addition, as described above, a regulatory region resides upstream of this element - within region I - that represses Pp expression in Sertoli cells in stages of the seminiferous epithelial cycle that normally do not express RHOX5 (Rao et al. 2003). The putative inhibitor factors that confer negative regulation by binding to these inhibitory elements in regions I and II have not yet been identified.
The Pd
The Pd is expressed in the ovary and placenta, and is also co-expressed with the Pp in the testes (Maiti et al. 1996a, 2001, MacLean et al. 2005b). The likely cellular site of Pd expression in the ovary is mural granulosa cells, as that is where RHOX5 protein has been shown to be specifically expressed (Pitman et al. 1998). The Pd is broadly expressed in the placenta, based on the finding that RHOX5 protein is present in spongiotrophoblasts, labyrinthine trophoblasts, and trophoblast giant cells (Lin et al. 1994, Pitman et al. 1998). The Pd is also responsible for expression of Rhox5 in the embryo proper and ES cells (Rao et al. 2003; A Bhardwaj & MF Wilkinson 2009, unpublished observations). In contrast to its highly specific expression in normal tissues, the Pd is widely expressed in tumors, regardless of cell lineage or tissue of origin (Wilkinson et al. 1990, Rao et al. 2002a).
A minimal element required for Pd transcription has been defined (~100 nt upstream of its transcription start site) that is only 22 nt in length (Rao et al. 2002a, MacLean et al. 2005b). This 22-nt element is sufficient to activate the transcription of an inactive heterologous promoter in primary granulosa cells and tumor cells. It consists of two ETS family-binding sites and a single SP1 family-binding site. One ETS family member, GABP, and two SP1 family members, SP1 and SP3, have been shown to act through these binding sites to activate Pd transcription, based on the following lines of evidence: i) overexpression of any of these three transcription factors elicits a dose-dependent increase in Pd expression in primary mouse or rat granulosa cells; ii) dominant-negative forms of these transcription factors decrease Pd transcriptional activity; iii) all three of these transcription factors bind to the Pd, as assessed by EMSA analyses, and iv) mutation of any one of the binding sites for these three factors reduces Pd transcription. In the future, it will be important to determine whether other members of the large ETS family besides GABP control Pd transcription. The ETS family member ELF1 binds to the Pd, but its functional role in Pd transcription is not known (Rao et al. 2002a). Interestingly, ELF1 upregulation has been associated with gynecological cancers of the cervix, uterus, and breast (Andrews et al. 2008, Nicol et al. 2008), but whether it has a role in ovarian tumors has not been determined. It will also be important to determine whether the Pd minimal element drives Rhox5 expression in cell types besides primary granulosa cells and tumor cells.
Because the processes of ovulation and tumorigenesis are both induced and propagated by related receptor-mediated signal transduction events via growth factors and inflammatory cytokines, it is perhaps not surprising to find that the Pd is transcriptionally activated by such signaling events. A key signal transduction event required for Pd transcripion is initiated by the G-protein RAS (Rao et al. 2002a, MacLean et al. 2005b). Gain-of-function evidence for this is that transient transfection of a constitutive form of RAS induces Pd expression in primary granulosa cells, mesenchymal stem cells, and tumor cells. Conversely, a dominant-negative form of RAS inhibits Pd transcription in both primary granulosa cells and tumor cells. The ability of RAS-mediated signaling to induce the Pd in granulosa cells is interesting in light of the fact that the LH surge that triggers ovulation activates K-RAS in granulosa cells (Fan et al. 2008). K-RAS signaling is also required for the phosphorylation and activation of MAPK3/1 (ERK1/2), which, in turn, is required for granulosa cells to promote LH-induced resumption of germ cell meiosis, ovulation, and luteinization (Fan et al. 2009). Gonadotropin treatment induces peak MAPK3/1 activity in rodent granulosa cells at a similar time point as that for maximal Pd mRNA levels (MacLean et al. 2005b, Fan et al. 2009), suggesting the possibility that Pd transcription also requires MAPK3/1. While this has not been tested, it is known that other components of the MAP kinase pathway - namely the stress-activated protein kinase/c-Jun amino-terminal kinases - are required for Pd transcription (Rao et al. 2002a, MacLean et al. 2005b).
The finding that RAS signaling activates the Pd strongly suggests that RAS transformation is responsible for the expression of Rhox5 in a broad array of tumor cells. Furthermore, because RAS is known to stimulate the activity of SP1 and ETS factors (Kivinen et al. 1999, Yordy & Muise-Helmericks 2000), this provides an obvious mechanism for how RAS induces Pd transcription. While RAS activation typically occurs as a result of mutation in tumor cells, it remains unclear what activates RAS in normal cells. The identification of upstream factors, including extracellular factors, which activate RAS in granulosa cells and other cell types, will be an important future goal for the field.
Further insights into the regulation of Pd transcription have come from a recent study by Li et al. (2009) who used ‘epigenetic’ inhibitors to study the role of DNA methylation and histone modifications in regulating the Pd. The drugs used by the investigators were the DNMT inhibitor 5AzaC, the histone deacetylase inhibitor MS-275, and the chemotherapeutic drug arsenic trioxide (ATO), all of which trigger both DNA demethylation and histone modifications favorable for transcription. They found that all three drugs induced Pd-derived transcripts in several different cell lines. ATO and MS-275 induced only Pd-derived transcripts, not Pp-derived transcripts, consistent with other studies showing that these two promoters are independently regulated. Interestingly, even though these two inhibitors decreased global DNMT levels, they did not lead to significant demethylation of the Pd, suggesting that these inhibitors induced the Pd by a DNA methylation-independent mechanism. Indeed, both inhibitors elicited transcription-promoting histone modifications at the Pd. 5AzaC was unique in its ability to induce both Pd- and Pp-derived transcripts. It probably acted by directly demethylating these promoters, as both the Pd and Pp underwent a dramatic decrease in methylation in response to 5AzaC. This is probably also the mechanism responsible for the dramatic induction of Rhox5 transcripts in 10T1/2 mesenchymal stem cells reported earlier (Sasaki et al. 1991). However, 5AzaC treatment also increased the levels of transcription-promoting histone modifications at the Pd, complicating the interpretation of its mechanism of action (Li et al. 2009). In the future, it will be important to untangle the independent contributions of DNA methylation and histone modifications, a challenging task given that these two events are intimately linked at many levels (Berger 2007).
Rhox5 is not alone on the X chromosome
Rhox5 is not an orphan homeobox gene on the X chromosome, as the original studies on Rhox5 had suggested (Lin et al. 1994, Maiti et al. 1996b, Sutton & Wilkinson 1997b), but instead rests in the middle of a large homeobox gene cluster (MacLean et al. 2005a; Fig. 1). The best-characterized Rhox gene cluster is in mice, but Rhox gene clusters also exist at the syntenic position of the X chromosome in other mammals, including rats and humans (Lindsey & Wilkinson 1996b, Sutton et al. 1998, Geserick et al. 2002, Wayne et al. 2002a, MacLean et al. 2005a, Zhan et al. 2005, Morris et al. 2006). All the genes in this cluster were probably derived from a common ancestral homeobox gene that was duplicated multiple times by unequal crossing over (Jackson et al. 2006, MacLean et al. 2006, Morris et al. 2006, Wang & Zhang 2006, Geyer & Eddy 2008). Evidence suggests that this ancestral homeobox gene was also the precursor gene that gave rise to the single-copy aristaless homeobox gene in D. melanogaster (Maiti et al. 1996b, Sutton & Wilkinson 1997a, MacLean et al. 2005a). The common origin of the Rhox genes is strongly suggested by both their close physical proximity and the fact that all Rhox genes have two introns at positions within the homeobox region that are unique to this subfamily of homeobox genes (MacLean et al. 2005a, 2006). In addition, Rhox genes encode homeodomains more similar to each other than those encoded by other homeobox genes (Maiti et al. 1996b, MacLean et al. 2005a). Finally, some Rhox genes encode related motifs in the amino-terminal region (MacLean et al. 2005a, 2006, Jackson et al. 2006).
The Rhox gene cluster
The mouse Rhox gene cluster contains three subclusters: α, β, and γ (Fig. 1). The α subcluster was originally defined as having four genes (MacLean et al. 2005a), but following the refinement of the X chromosome genomic map and sequence analysis, four groups simultaneously reported multiple tandem duplications of three of the α subcluster genes, Rhox2, Rhox3, and Rhox4, making a total of 24 genes in the α subcluster (Jackson et al. 2006, MacLean et al. 2006, Morris et al. 2006, Wang & Zhang 2006). The duplicated copies (paralogs) of each of these three genes are almost identical in sequence (~95–100%). While it is not clear what selective forces are responsible for the recent amplification of these three genes in mice, it is clear that most of these paralogs are expressed and all but one (Rhox3d) encode full-length proteins and thus are presumably functional (Jackson et al. 2006, MacLean et al. 2006, Morris et al. 2006, Wang & Zhang 2006). The β subcluster has five genes, including the founding Rhox family member - Rhox5 - and three other genes that had previously been given other names: Rhox6 (Psx1), Rhox8 (Tox), and Rhox9 (Psx2 and Gpbox) (Chun et al. 1999, Takasaki et al. 2000, Kang et al. 2004). The γ subcluster was originally defined as having three genes (MacLean et al. 2005a), but recently, a fourth member has been identified: Rhox13 (Geyer & Eddy 2008). In total, 33 homeobox genes have so far been identified in the mouse Rhox gene cluster.
The Rhox gene cluster is rapidly evolving. One dramatic demonstration of this came from the discovery that the rat RHOX gene cluster is smaller than its mouse counterpart (Fig. 1), primarily because it has only single copies of the α subcluster paralogs that in mice each have seven to eight copies (MacLean et al. 2005a, 2006, Jackson et al. 2006, Morris et al. 2006). This, coupled with the nearly identical sequence of the members of each paralog family, indicates that the expansion of the α subcluster paralogs occurred very recently, after the split of mice and rats in the rodent lineage. Also, unlike the mouse Rhox gene cluster, which contains two similar genes in the β subcluster (Rhox6 and Rhox9), the rat Rhox gene cluster contains only one (MacLean et al. 2005a, 2006, Morris et al. 2006). The order of the genes in the mouse and rat Rhox gene clusters also appears to differ to some extent (Waterston et al. 2002 (build 37.1), Gibbs et al. 2004 (RGSC v3.4)), indicating the likelihood of gene rearrangements since the mouse/rat split (Fig. 1). However, this remains uncertain because the precise organization of both mouse and rat clusters has not yet been fully refined.
The human RHOX gene cluster is even smaller than the rat Rhox cluster. Only two human RHOX genes have been characterized at the syntenic portion of the X chromosomes (Geserick et al. 2002, Wayne et al. 2002a). One of them is RHOXF1 (originally called OTEX and hPEPP1), which is androgen regulated and selectively expressed in human testes and thus is likely to have a role in spermatogenesis (Geserick et al. 2002, Wayne et al. 2002a). The other human RHOX gene is RHOXF2A (originally called hPEPP2), which, like RHOXF1, is selectively expressed in the testis (Geserick et al. 2002, Wayne et al. 2002a). Interestingly, RHOXF2A encodes a protein that has been shown to interact with both itself and a wide variety of other proteins, including nuclear transcription factors and cytoplasmic RNA decay-promoting proteins (Lehner & Sanderson 2004, Lim et al. 2006, Prasad et al. 2009). RHOXF2A has been classified as a member of the growing ‘cancer/testes’ gene family, a large group of genes that are not necessarily similar in sequence, but instead share the characteristic of being selectively expressed in the testis and a wide variety of tumors (Hofmann et al. 2008). Both human RHOXF1 and RHOXF2A are capable of regulating Rhox5 gene targets in mouse Sertoli cells (Hu et al. 2010), but it is not known what human genes they might regulate. At least one other RHOX gene exists in the human RHOX cluster - RHOXF2B - but neither it nor exonic remnants of other RHOX genes in the RHOX gene cluster have been characterized in terms of expression or function (Lander et al. 2001 (build 37.1)).
The rapid evolution of the Rhox gene cluster makes it a good candidate to have a role in speciation, a process that evidence suggests is largely driven by rapidly evolving X-linked genes (Turner & Hoekstra 2008). In addition to alterations in gene number and gene cluster organization in different species, rapid alterations in the sequence of individual Rhox genes over evolutionary time may have contributed to speciation. As described earlier, the founding member of the Rhox family, Rhox5, has probably been subject to strong positive selection to undergo amino acid changing alterations (Sutton & Wilkinson 1997a); it remains to be determined whether the same is true for other Rhox family members. It would not be surprising if this was the case, as positive selection for changes in amino acid identity is common in large gene families, particularly those involved in reproduction (Chakrabarty et al. 2006, Soares et al. 2007, Rawn & Cross 2008). A definitive assessment of RHOX gene cluster evolution will require complete analysis and annotation of X chromosomes from a full set of mammals. At this point in time, even the mouse X chromosome remains to be fully analyzed; for example, a portion of the mouse Rhox cluster (between Rhox2g and Rhox5) has neither been cloned nor sequenced (as determined by manually evaluating BAC sequences available from NCBI and Ensembl).
Expression and functional role of Rhox cluster genes
All Rhox genes are selectively expressed in reproductive organs, and thus it is probable that they regulate events involved in reproduction (MacLean & Wilkinson 2005, MacLean et al. 2005b, Morris et al. 2006). In the mouse, analysis of a panel of 18 adult tissues demonstrated that all Rhox genes are selectively expressed in the ovary, testis, epididymis, and placenta (MacLean et al. 2005a; Fig. 2). Only Rhox4, Rhox7, and Rhox8 were also expressed in non-reproductive organs; in thymus, stomach, and intestine respectively. Within the placenta, Rhox4 is expressed in the chorionic trophoblast cells in the labyrinth and spongiotrophoblast layers of the placenta, suggesting that it may serve several different functions in this tissue (Jackson et al. 2003). The selective expression pattern of Rhox genes in reproductive organs is conserved, as the rat Rhox genes are also selectively expressed in the ovary, testis, epididymis, and placenta (Lindsey & Wilkinson 1996b, Maiti et al. 1996a, MacLean et al. 2005b, Morris et al. 2006; JA MacLean, unpublished observations), and the two best-characterized human RHOX genes appear to be specifically expressed in the human testis (Geserick et al. 2002, Wayne et al. 2002a). While Rhox genes normally exhibit highly specific and regulated expression patterns, some Rhox family members are ubiquitously expressed in tumors, including mouse Rhox5, Rhox6, and Rhox9, as well as human RHOXF1 (Wilkinson et al. 1990, Rao et al. 2002a, Wayne et al. 2002a, Li et al. 2009).
Little is known about which cell types in adult reproductive organs express individual Rhox genes. Most progress on this front has been conducted on Rhox5, which, as described earlier, is expressed in mural granulosa cells in the ovary, principal cells in the caput epididymis, and Sertoli cells in the testis (Lindsey & Wilkinson 1996b, 1996c, Pitman et al. 1998, Sutton et al. 1998, Rao et al. 2002b). Many other Rhox genes may be expressed in Sertoli cells, based on their expression in an enriched mouse Sertoli cell fraction (MacLean et al. 2005a), but confirmation by immunohistochemistry has not been done on most RHOX proteins. Rhox4 is expressed in enriched adult interstitial cell fraction, suggesting that Rhox4 is expressed in Leydig cells (MacLean et al. 2005a), but this also requires confirmation by immunohistochemistry. Recently, a careful analysis performed by Geyer & Eddy (2008) has demonstrated that Rhox13 mRNA and RHOX13 protein are expressed in differentiating spermatogonia and preleptotene spermatocytes, indicating that Rhox13 is a good candidate to regulate early germ cell differentiation events and/or the transition from mitosis to meiosis. Rhox3 and Rhox11 are expressed in male germ cells at later stages of germ cell development than Rhox13 (A Bettegowda & MF Wilkinson, unpublished observations), indicating that these two Rhox genes are candidates to regulate post-meiotic events in germ cells. Interestingly, Mueller et al. (2008) recently demonstrated that the mouse X chromosome is enriched for multi-copy genes that are expressed in post-meiotic male germ cells, suggesting that the Rhox cluster may be part of a large contingent of X-linked genes devoted to functions in the late stages of spermatogenesis.
While the cell types that express Rhox genes in adult reproductive organs are only beginning to be explored, a recent study conducted by Daggag et al. (2008) has given us a detailed view of the expression of Rhox genes in specific cell types within mouse embryonic gonads. Their study, which used both whole mount in situ hybridization and qPCR analyses, demonstrated that most Rhox genes are expressed in mouse embryonic gonads (Fig. 2). All but one of the Rhox genes are expressed in PGCs; the one exception, Rhox8, was expressed in the somatic cells neighboring the PGCs. Their study not only confirmed previous reports that Rhox5 and Rhox9 are expressed in PGCs (Pitman et al. 1998, Takasaki et al. 2001), but it also greatly extended the number of genes involved, particularly since they showed that PGCs express the Rhox2 and Rhox4 paralogs, which together consist of 15 genes. Interestingly, a recent report has found that Rhox8 is dramatically upregulated in embryonic gonads from mice that have undergone sex reversal in response to loss of the ‘pro-female’ transcription factor FOXL2 (Uhlenhaut et al. 2009). This suggests that Rhox8 might be part of the male-promoting program that converts the ‘indifferent’ early gonad into a male gonad. While most of the Rhox genes are expressed in both male and female PGCs, Daggag et al. found that three exhibit sex-specific expression. Rhox10 is specifically expressed in male PGCs, while Rhox6 and Rhox9, which are highly related in sequence, are expressed predominantly in female PGCs. Not only did some Rhox genes exhibit sex-specific expression, but many of them also exhibited regulated temporal expression in PGCs during gonad development. Together, these data indicate that Rhox genes are good candidates to have roles in developmentally regulated and sex-specific events that occur in embryonic gonads.
In postnatal and adult mice, the expression patterns of the individual genes in the Rhox cluster are quite complex; many exhibit tissue-specific and developmentally regulated gene expression in both male and female reproductive tissues (Lindsey & Wilkinson 1996c, Pitman et al. 1998, MacLean et al. 2005a, 2005b). Their expression patterns suggest that Rhox genes may have both specific and broad roles regulating gametogenesis in both females and males. To date, the in vivo roles of only Rhox5 and Rhox9 (Pitman et al. 1998, Takasaki et al. 2000) have been reported (Fig. 2). As described earlier, Rhox5-null males have increased germ cell apoptosis and sperm motility defects, and they are subfertile (MacLean et al. 2005a). By contrast, Rhox9-null mice were not found to have any observable defects (Takasaki et al. 2001). One explanation is that the functions of Rhox9 are compensated for by Rhox6, as these two genes are highly similar in sequence and expression pattern (Chun et al. 1999, Takasaki et al. 2000). But if Rhox6 does compensate for Rhox9, it does so without being upregulated, as Rhox6 mRNA is not increased in level in trophoblast cells, the testis, or ovary of Rhox9-null mice (Takasaki et al. 2001).
Most of the other genes in the mouse Rhox gene cluster have both unique and overlapping developmental expression patterns, so it is reasonable to suppose that they possess both unique and partially redundant functions (MacLean et al. 2005a). Circumstantial evidence supporting the notion that many RHOX transcription factors have distinct biological functions is that most possess homeodomains differing in amino acid residues at one or more of the four positions in the third helix known to make base-specific contacts with DNA (MacLean et al. 2005a). For example, RHOX5, RHOX6, RHOX9, and RHOX12, all of which are co-expressed in the placenta, have different amino acids at two or more of these four positions. Thus, it seems likely that these four RHOX transcription factors regulate, at least in part, distinct downstream targets (of note, however, is that the two different ‘DNA-recognition’ amino acids in RHOX6 and RHOX9 are conservative/similar substitutions). To ultimately discern the unique and redundant roles of the Rhox cluster genes, it will probably require a combination of approaches that inactivate/deplete individual Rhox genes, combinations of Rhox genes, and even the entire Rhox gene cluster.
In vitro studies will no doubt also contribute to our understanding of the biological roles of Rhox cluster genes (Fig. 2). In this regard, Jackson et al. (2002) showed that Rhox4b (Ehox) is expressed in ES cells and that antisense Rhox4b RNA inhibited ES cell differentiation in vitro. Conversely, ES cell differentiation was promoted in response to Rhox4b overexpression. These experiments, along with assays to measure self-renewal capacity, provided evidence that Rhox4b has two functions in ES cells: it promotes their differentiation and inhibits their self-renewal. It will be interesting to determine whether other Rhox4 paralogs share this property, as the antisense RNA used by Jackson et al. probably targeted all of the other six Rhox4 gene paralogs. A role for Rhox4 genes in early development is consistent with in situ hybridization analyses showing that Rhox4 transcripts exhibit a developmentally regulated pattern of expression in the early embryo; they are initially restricted to extraembryonic endoderm (in e6.5 embryos), and later (e8.5–10.5) they are expressed in the embryo proper, first in the anterior foregut endoderm and then in the pharyngeal pouches (Jackson et al. 2003).
Regulation of Rhox cluster transcription
The field is only just beginning to explore the mechanisms that regulate most of the members of the Rhox gene cluster. One notable exception is the founding member, Rhox5, whose regulation has been examined in detail, as described earlier in this review. Here, we summarize what is known about the regulation of other Rhox genes, including two studies that have uncovered putative global regulatory mechanisms controlling several genes in the Rhox gene cluster.
To ensure proper development, the well-known Hox homeobox gene clusters have evolved specialized regulatory mechanisms to control the time and place that the transcription factors encoded by the genes within these clusters are expressed. Perhaps the most striking discovery that has been made regarding Hox gene clusters is that they exhibit a co-linear pattern of expression, such that their physical order on a chromosome correlates with their expression pattern. In particular, the timing, position, and level of expression of individual Hox genes during embryonic development follow their position within a given Hox gene cluster (Spitz et al. 2003). Similarly, MacLean et al. (2005a) found that testis-expressed Rhox genes exhibit a temporal and quantitative co-linear expression pattern in which the order of these genes within each subcluster on the X chromosome correlates with their timing and relative strength of expression during postnatal testes development. It will be interesting to determine the functional relevance of this co-linear expression pattern, as well as the underlying mechanism responsible for it. With regard to the latter, one could envisage it being directed by global transcriptional enhancer elements. By analogy, a global enhancer has been identified in the Hoxd cluster that drives the co-linear expression pattern of its genes during embryonic development (Spitz et al. 2003).
A different line of evidence for a global Rhox cluster-regulatory mechanism was obtained by Oda et al. (2006). As described earlier, these authors demonstrated that Rhox5 is selectively expressed in extra-embryonic tissues, not the embryo proper, as a result of DNA methylation blocking Rhox5 expression in the latter. Extending their analysis to other Rhox genes, they found that, like the Rhox5 Pp, the promoter regions of the Rhox2, Rhox6, and Rhox9 genes switch from a hypomethylated state in eight-cell embryos and blastocysts to a hypermethylated state in mid-gestation (e8.5–e9.5) embryos. They found that methylation of these promoters occurs selectively in the embryo proper, not extra-embryonic tissue (trophoblasts and yolk sac), consistent with a repression mechanism specifically preventing the expression of Rhox genes in the embryo proper. Causal evidence that this is indeed the case was the authors’ finding that mouse embryos lacking both de novo DNMTs - DNMT3a and DNMT3b - had greatly elevated expression of most Rhox genes. In contrast, most other genes on the X chromosome - including the Rhox-related Esx1 gene, which is far distant from the Rhox gene cluster - were not upregulated in the double-mutant embryos. Repression was mediated specifically in the embryo proper, as trophoblasts from double-mutant embryos did not exhibit significantly upregulated Rhox cluster gene expression. This was consistent with the fact that Rhox gene cluster promoters are normally already hypomethylated in extra-embryonic tissues (Schutt et al. 2003, Oda et al. 2006).
DNMTs can regulate transcription either directly or indirectly. Oda et al. addressed which might be the case for Rhox cluster genes by comparing the methylation status of their promoters in double-mutant embryos versus control mouse embryos. They found that all the Rhox gene promoters that they tested were less methylated in double-mutant embryos than control embryos, providing evidence that DNA methylation directly regulates their transcription. Methylation of the Rhox6 and Rhox9 promoters required both DNMT3b and DNMT1, but not DNMT3a, based on analysis of single-mutant mice for these three factors. Likewise, the transcriptional repression of Rhox6 and Rhox9 required DNMT3b, not DNMT3a (DNMT1 was not tested). In contrast, Rhox2 promoter methylation could be mediated by either DNMT3a or DNMT3b, as only double-mutant (Dnmt3a−/− Dnmt3b−/−) embryos, not compound heterozygous (e.g. Dnmt3a−/+Dnmt3b−/−) mouse embryos, had a hypomethylated Rhox2 promoter. Together, these data supported the notion that the Rhox gene cluster is controlled by a global DNA methylation-mediated mechanism that confines the expression of most Rhox genes to extra-embryonic tissues in mid-gestation embryos.
Oda et al. obtained evidence that this global regulatory mechanism regulates not only the Rhox gene cluster, but also a short region upstream and a ~0.3 Mb region downstream, which together with the Rhox gene cluster form a domain on the X chromosome that is R1 Mb in length. The evidence that this ‘methylation-targeted domain’ extends a short region upstream of the Rhox gene cluster came from their identification of a gene just ~2 kb upstream of Rhox1, called Gm9, whose promoter region was demethylated in double-mutant mouse embryos lacking DNMT3a and DNMT3b. Accompanying this demethylation event was the ectopic expression of Gm9 in double-mutant embryos. In contrast, the genes upstream of Gm9 (including one <20 kb upstream) did not exhibit altered methylation or expression, implying that the upstream boundary of the methylation-targeted domain is just upstream of Gm9. The evidence that the methylation-targeted domain extends downstream of the Rhox gene cluster was the finding that two CpG island-containing gene promoters downstream of the Rhox gene cluster become demethylated in double-knockout mice embryos. However, there is doubt as to the relevance of this demethylation event, as their respective genes did not appear to be expressed at higher levels in double-knockout mice than in control mice. Furthermore, a Rhox gene at the 3′ end of the Rhox cluster, Rhox12, did not exhibit upregulated expression in double-knockout embryos. Given that Rhox12 is in the γ subcluster, this suggests the possibility that the downstream boundary of the methylation-targeted domain is between the β and γ subclusters. This predicts that all the genes in the α and β subclusters should be upregulated in double-knockout mice. Indeed, all the α and β subclusters that were tested (Rhox1, Rhox2, Rhox3, Rhox4, Rhox5, Rhox6, and Rhox9) exhibited this regulation. In conclusion, the available evidence from Oda et al. suggests the existence of a methylation-targeted domain in the X chromosome that is comprised of the 5′ end of the Rhox gene cluster (the α and β subclusters) and a short region upstream. Whether this domain also includes the 3′ end of the Rhox cluster (the γ subcluster) and genes downstream of that remains to be definitively determined.
The role of DNA methylation in regulating Rhox gene cluster transcription has also been studied in cultured cell lines. Li et al. (2009) demonstrated that the mouse Rhox5, Rhox6, and Rhox9 genes are induced by the DNA methylation inhibitor 5AzaC in mouse cell lines, implying that several Rhox genes are repressed in cell lines by DNA methylation. Likewise, they showed that two best-characterized human RHOX genes, RHOXF1 and RHOXF2B, are induced by 5AzaC in human cell lines. Further evidence that Rhox genes are repressed by DNA methylation was obtained by Oda et al. (2006) who found that the Rhox2, Rhox5, Rhox6, and Rhox9 gene promoters are hypermethylated in ES cells. Analysis of ES cells lacking different combinations of DNMTs demonstrated that the methylation of these Rhox promoters can be mediated by either DNMT3a or DNMT3b. In contrast, the requirement for DNMT1 depended on the Rhox gene promoter tested; Rhox6 and Rhox9 promoter methylation was dramatically inhibited in Dnmt1-mutant ES cells, whereas Rhox2 and Rhox5 promoter methylation was little affected. While these findings suggest that Rhox genes are transcriptionally repressed by DNA methylation in ES cells, this was not directly tested, as the authors did not examine the expression levels of Rhox genes in control and Dnmt-mutant ES cells. However, it seems likely to be the case, as a recent study has demonstrated that a surprisingly large number of X-linked genes, including Rhox2, are upregulated in DNMT-deficient ES cells (Fouse et al. 2008). Given the utility of using cell lines for in-depth molecular studies, it will be important to continue conducting in vitro studies in combination with in vivo studies to obtain an ‘integratedview’ of Rhox cluster gene regulation.
The only Rhox gene promoter that has been studied in depth besides Rhox5’s alternative promoters is the Rhox4b (Ehox) promoter (Lee et al. 2006). Lee et al. (2006) found that the Rhox4b promoter contains a critical CCAAT box within the first ~100 bp of 5′ flanking sequence that is necessary for Rhox4b transcription (as defined by a transfected reporter) and binds to the transcription factor NFY. Downstream of this NFY-binding element is an SP1 transcription factor family consensus-binding site that is necessary for maximal Rhox4b promoter activity. This is of interest given that SP1 family members have been shown to bind and regulate the expression of the Rhox5Pd in primary granulosa cells and cell lines (Rao et al. 2002a, MacLean et al. 2005b). Furthermore, preliminary characterization of the Rhox8 promoter has also identified a key SP1 consensus site required for Rhox8 transcription (J A MacLean, unpublished observations). Thus, regulation by SP1 family members may be a common feature of Rhox genes. Like most Rhox cluster genes, Rhox4b is expressed in ES cells and the placenta, but it also has the unique property of being expressed in the thymus (Jackson et al. 2002, 2003). It will be interesting to determine what peculiar aspects of the Rhox4b promoter allow it to be expressed in the thymus. Given the wealth of knowledge in the field of immunology (and T cells in particular), it may be instructive to learn about genes regulated by Rhox4b in the thymus as a means to gain insight into Rhox4b’s function in embryonic development and the placenta. Rhox4b is one of the seven gene paralogs almost identical in sequence; most encode identical homedomain regions, and the entire deduced amino acid sequence of RHOX4B only differs from that of RHOX4F and RHOX4G by two amino acids (MacLean et al. 2006). It will be interesting to know whether all seven Rhox4 paralogs are regulated in an identical manner and what selection pressures might cause them to undergo divergent regulation over future evolutionary time.
Rhox cluster-derived genes outside of the Rhox cluster
As described above, the Rhox gene cluster has undergone several relatively recent alterations, including its expansion in rodents, particularly in mice, which have tandem duplications of most of the α subcluster genes. Conversely, ancient events have probably reduced the size of the Rhox gene cluster. In particular, two genes were almost certainly relocated from the Rhox gene cluster to distant sites on the X chromosome as a result of ancient chromosomal inversion events (Svingen & Koopman 2007). These two ‘vagabond genes’ are Esx1 (also called Spx1 in mice; ESX1 in humans) and Arx (ARX in humans; Fig. 1). The evidence for their close relationship with the Rhox gene family is threefold. First, the Esx1 and Arx genes are both on the X chromosome, like the Rhox cluster. Second, the ESX1 and ARX homeodomains are similar in sequence to homedomains in RHOX family members. Third, both the Esx1 and Arx genes have the hallmark exon-intron structure specifically characteristic of Rhox genes (i.e. introns located at two particular sites within the homeodomain-encoding region).
Esx1/ESX1
Esx1 is selectively expressed in testes and placenta in both humans and mice (Branford et al. 1997, Li & Behringer 1998, Fohn & Behringer 2001, Yeh et al. 2005). In the mouse testis, in situ hybridization analyses indicated that Esx1 transcripts are specifically expressed in spermatogonia, spermatocytes, and round spermatids in stages IV–VII of the seminiferous epithelial cycle (Branford et al. 1997). Although Esx1 transcripts are first detectable in pre-meiotic germ cells, ESX1 protein is only detectable in post-meiotic round spermatids, suggesting that ESX1 is subject to translation control (Yeh et al. 2005). However, despite its expression in round spermatids, ESX1 does not appear to have a non-redundant role in spermatogenesis, as Esx1-null male mice are fertile and do not exhibit obvious testicular defects (Li & Behringer 1998). Instead, these mice exhibit defective placental development involving the maternal-fetal interface, leading to reduced embryo size and increased placental size. Also consistent with a placental-specific defect, Esx1-null mice are ~20% smaller than normal at birth, but later reach normal size. The growth defect was only observed when the mutant Esx1 gene was inherited from the mother, consistent with the maternal-specific (i.e. imprinted) expression of mouse X-chromosomal genes in placenta. The growth-promoting ability of Esx1 during embryonic development contrasts with its ability to inhibit the growth of tumor cells. One study showed that a naturally produced C-terminal ESX1 fragment inhibits the degradation of cyclins and thereby causes cell cycle arrest (Ozawa et al. 2004), while another study showed that ESX1 slows tumor growth by homeodomain-dependent inhibition of Kras transcription and amino-terminal domain-dependent reduction in tumorgenicity (Nakajima et al. 2008). Another intriguing aspect of Esx1 is that, like Rhox5, it has undergone rapid evolution in mammals (Wang & Zhang 2007). Evidence suggests that positive selection for rapid changes has primarily occurred in the C-terminal region, which is the region that regulates cyclin degradation and cell division. It will be interesting to discover the selective forces that might be responsible for this, and what consequences this has on ESX1’s role in reproduction.
Arx/ARX
The Arx gene exhibits a different expression pattern than Esx1. Arx is specifically in developing male gonads as well as several regions of the developing and adult brain (Kitamura et al. 2002, Gecz et al. 2006). Most studies on Arx have focused on its role in the brain, as it is commonly mutated in mental retardation patients, and studies in Arx-mutant mice have shown that ARX is crucial for development of the forebrain (Kitamura et al. 2002, Gecz et al. 2006). Indeed, because of the dramatic brain defects that result from its loss, Arx-null mice typically undergo epileptic seizures and perinatal death, precluding analysis of ARX’s role in fertility (Kitamura et al. 2002, Marsh et al. 2009). However, this has not prevented an analysis of the role of ARX in the developing male gonad in embryos. Analysis of Arx-mutant male embryos revealed that their testes had a dysplastic interstitium accompanied by reduced expression of a Leydig cell marker, which together suggested that these mice had a defect in fetal Leydig cell differentiation (Kitamura et al. 2002). This defect in Leydig cells is probably not a primary event, as ARX protein is barely detectable in Leydig cells; instead, it is expressed at high levels in fibroblast-like cells adjacent to the Leydig cells in the interstitium. A role for ARX in Leydig cell function is likely to be conserved, as some humans with mutations in the ARX gene have reduced numbers of Leydig cells and reduced testerone levels (Ogata et al. 2000). It will be important in the future to clarify the role of ARX in fetal Leydig cell function, including their ability to produce androgens.
Perspective.
The discovery of the Rhox gene cluster has opened up a new frontier in reproductive biology. The 33 homeobox genes in this cluster are selectively expressed in the male and female reproductive tract, suggesting that Rhox genes probably encode transcription factors devoted to regulating and promoting male and female fertility. In the future, it will be intriguing to dissect the redundant and non-redundant functions of the Rhox genes in mice. To understand the molecular networks that drive these biological functions, the direct gene targets of RHOX transcription factors in different reproductive cell types will need to be identified. Some progress has already been made on this front with regard to androgen/AR-regulated gene networks in somatic cells (i.e. via the androgen/AR-regulated Rhox5 gene), but it will be important to also identify the Rhox gene-regulated networks in developing germ cells. While the study of Rhox gene function in mice has the potential to illuminate the role of Rhox genes in human fertility, there is clearly also a need to directly study the expression and function of human RHOX genes. For example, it will be crucial to assess the function of RHOX genes in human ES cells, including how their development into gametes is regulated by RHOX genes. There is also a need to obtain a deeper understanding of how Rhox genes are transcriptionally regulated. Rhox genes are one of the few genes whose cell type-specific expression has been shown to be regulated by DNA methylation. This makes them a model system to elucidate the underpinings and physiological importance of this key ‘epigenetic’ regulatory mechanism. Finally, the unusual evolution of the Rhox gene cluster and its individual family members is worth studying in more depth, particularly since the rapid evolution of Rhox genes may have contributed to speciation and the development of different reproductive strategies in different organisms.
Acknowledgements
The authors would like to thank Dr JoAnne Richards (Baylor College of Medicine) and fellows from the Wilkinson laboratory (Marcy Beildeck, Madhuvanthi Ramaiah, Anilkumar Bettegowda, and Hye-Won Song) for their helpful comments during the preparation of this review.
Funding
The authors are funded by the National Institute of Health, grants HD45595, HD53808, and HD55268, which supported work on the Rhox gene cluster.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Andrews PG, Kennedy MW, Popadiuk CM & Kao KR 2008. Oncogenic activation of the human Pygopus2 promoter by E74-like factor-1. Molecular Cancer Research 6 259–266. [DOI] [PubMed] [Google Scholar]
- Arakawa H 2004. Netrin-1 and its receptors in tumorigenesis. Nature Reviews. Cancer 4 978–987. [DOI] [PubMed] [Google Scholar]
- Barbulescu K, Geserick C, Schuttke I, Schleuning WD & Haendler B 2001. New androgen response elements in the murine pem promoter mediate selective transactivation. Molecular Endocrinology 15 1803–1816. [DOI] [PubMed] [Google Scholar]
- Berger SL 2007. The complex language of chromatin regulation during transcription. Nature 447 407–412. [DOI] [PubMed] [Google Scholar]
- Bettegowda A & Wilkinson MF 2010. Transcription and post-transcriptional regulation of spermatogenesis. Philosophical Transactions of the Royal Society of London. Series B 365 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Rao MK, Kaur R, Buttigieg MR & Wilkinson MF 2008. GATA factors and androgen receptor collaborate to transcriptionally activate the Rhox5 homeobox gene in Sertoli cells. Molecular and Cellular Biology 28 2138–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford WW, Zhao GQ, Valerius MT, Weinstein M, Birkenmeier EH, Rowe LB & Potter SS 1997. Spx1, a novel X-linked homeobox gene expressed during spermatogenesis. Mechanisms of Development 65 87–98. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A, MacLean JA II, Hughes AL, Roberts RM & Green JA 2006. Rapid evolution of the trophoblast kunitz domain proteins (TKDPs) - a multigene family in ruminant ungulates. Journal of Molecular Evolution 63 274–282. [DOI] [PubMed] [Google Scholar]
- Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H & Yeh S 2004. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. PNAS 101 6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Yamanaka Y, Pawson T & Rossant J 2006. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Developmental Cell 10 615–624. [DOI] [PubMed] [Google Scholar]
- Chow J & Heard E 2009. X inactivation and the complexities of silencing a sex chromosome. Current Opinion in Cell Biology 21 359–366. [DOI] [PubMed] [Google Scholar]
- Chun JY, Han YJ & Ahn KY 1999. Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Developmental Dynamics 216 257–266. [DOI] [PubMed] [Google Scholar]
- Daggag H, Svingen T, Western PS, van den Bergen JA, McClive PJ, Harley VR, Koopman P & Sinclair AH 2008. The rhox homeobox gene family shows sexually dimorphic and dynamic expression during mouse embryonic gonad development. Biology of Reproduction 79 468–474. [DOI] [PubMed] [Google Scholar]
- Denolet E, De Gendt K, Allemeersch J, Engelen K, Marchal K, Van Hummelen P, Tan KA, Sharpe RM, Saunders PT, Swinnen JV et al. 2006. The effect of a Sertoli cell-selective knockout of the androgen receptor on testicular gene expression in prepubertal mice. Molecular Endocrinology 20 321–334. [DOI] [PubMed] [Google Scholar]
- Eacker SM, Shima JE, Connolly CM, Sharma M, Holdcraft RW, Griswold MD & Braun RE 2007. Transcriptional profiling of androgen receptor (AR) mutants suggests instructive and permissive roles of AR signaling in germ cell development. Molecular Endocrinology 21 895–907. [DOI] [PubMed] [Google Scholar]
- Eakin GS & Hadjantonakis AK 2006. Sex-specific gene expression in preimplantation mouse embryos. Genome Biology 7 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Melhem MF & Chaillet JR 1999. Forced expression of the homeobox-containing gene Pem blocks differentiation of embryonic stem cells. Developmental Biology 210 481–496. [DOI] [PubMed] [Google Scholar]
- Fan HY, Shimada M, Liu Z, Cahill N, Noma N, Wu Y, Gossen J & Richards JS 2008. Selective expression of KrasG12D in granulosa cells of the mouse ovary causes defects in follicle development and ovulation. Development 135 2127–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HY, Liu Z, Shimada M, Sterneck E, Johnson PF, Hedrick SM & Richards JS 2009. MAPK3/1(ERK1/2) in ovarian granulosa cells are essential for female fertility. Science 324 938–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faus H & Haendler B 2008. Androgen receptor acetylation sites differentially regulate gene control. Journal of Cellular Biochemistry 104 511–524. [DOI] [PubMed] [Google Scholar]
- Fohn LE & Behringer RR 2001. ESX1L, a novel X chromosome-linked human homeobox gene expressed in the placenta and testis. Genomics 74 105–108. [DOI] [PubMed] [Google Scholar]
- Fouse SD, Shen Y, Pellegrini M, Cole S, Meissner A, Van Neste L, Jaenisch R & Fan G 2008. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gecz J, Cloosterman D & Partington M 2006. ARX: a gene for all seasons. Current Opinion in Genetics & Development 16 308–316. [DOI] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C et al. 2004. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. PNAS 101 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K, McKinnell C, Willems A, Saunders PT, Sharpe RM, Atanassova N, Swinnen JV & Verhoeven G 2009. Organotypic cultures of prepubertal mouse testes: a method to study androgen action in Sertoli cells while preserving their natural environment. Biology of Reproduction 81 1083–1092. [DOI] [PubMed] [Google Scholar]
- Geserick C, Weiss B, Schleuning WD & Haendler B 2002. OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochemical Journal 366 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geserick C, Meyer HA, Barbulescu K & Haendler B 2003. Differential modulation of androgen receptor action by deoxyribonucleic acid response elements. Molecular Endocrinology 17 1738–1750. [DOI] [PubMed] [Google Scholar]
- Geyer CB & Eddy EM 2008. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 423 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428 493–521. [DOI] [PubMed] [Google Scholar]
- Guo F, Li YQ, Li SQ, Luo ZW, Zhang X, Tang DS & Zhou TH 2005. Interaction of mouse Pem protein and cell division cycle 37 homolog. Acta Biochimica et Biophysica Sinica 37 784–787. [DOI] [PubMed] [Google Scholar]
- Guo F, Huang X, Li S, Sun L, Li Y, Li H, Zhou Y, Chu Y & Zhou T 2007a. Identification of prosaposin as a novel interaction partner for Rhox5. Journal of Genetics and Genomics 34 392–399. [DOI] [PubMed] [Google Scholar]
- Guo F, Li SQ, Chu YH, Huang XF, Sun LM, Li YQ, Li HJ & Zhou TH 2007b. High-level expression, polyclonal antibody preparation and sub-cellular localization analysis of mouse Rhox5 protein. Protein Expression and Purification 54 247–252. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Daikoku T, Wang H, Matsumoto H, Carter MG, Ko MS & Dey SK 2004. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. PNAS 101 10326–10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A et al. 2008. Genome-wide analysis of cancer/testis gene expression. PNAS 105 20422–20427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogeveen KN & Sassone-Corsi P 2005. Homeobox galore: when reproduction goes RHOX and roll. Cell 120 287–288. [DOI] [PubMed] [Google Scholar]
- Holdcraft RW & Braun RE 2004. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 131 459–467. [DOI] [PubMed] [Google Scholar]
- Hu Z, MacLean JA, Bhardwaj A & Wilkinson MF 2007. Regulation and function of the Rhox5 homeobox gene. Annals of the New York Academy of Sciences 1120 72–83. [DOI] [PubMed] [Google Scholar]
- Hu Z, Shanker S, MacLean JA II, Ackerman SL & Wilkinson MF 2008. The RHOX5 homeodomain protein mediates transcriptional repression of the netrin-1 receptor gene Unc5c. Journal of Biological Chemistry 283 3866–3876. [DOI] [PubMed] [Google Scholar]
- Hu Z, Dandekar D, O’Shaughnessy PJ, De Gendt K, Verhoeven G & Wilkinson MF 2010. Androgen-induced Rhox homeobox genes modulate the expression of AR-regulated genes. Molecular Endocrinology 24 60–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Baird JW, Cambray N, Ansell JD, Forrester LM & Graham GJ 2002. Cloning and characterization of Ehox, a novel homeobox gene essential for embryonic stem cell differentiation. Journal of Biological Chemistry 277 38683–38692. [DOI] [PubMed] [Google Scholar]
- Jackson M, Baird JW, Nichols J, Wilkie R, Ansell JD, Graham G & Forrester LM 2003. Expression of a novel homeobox gene Ehox in trophoblast stem cells and pharyngeal pouch endoderm. Developmental Dynamics 228 740–744. [DOI] [PubMed] [Google Scholar]
- Jackson M, Watt AJ, Gautier P, Gilchrist D, Driehaus J, Graham GJ, Keebler J, Prugnolle F, Awadalla P & Forrester LM 2006. A murine specific expansion of the Rhox cluster involved in embryonic stem cell biology is under natural selection. BMC Genomics 7 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E et al. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nature Genetics 27 31–39. [DOI] [PubMed] [Google Scholar]
- Kang YL, Li H, Chen WH, Tzeng YS, Lai YL & Hsieh-Li HM 2004. A novel PEPP homeobox gene, TOX, is highly glutamic acid rich and specifically expressed in murine testis and ovary. Biology of Reproduction 70 828–836. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K et al. 2002. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nature Genetics 32 359–369. [DOI] [PubMed] [Google Scholar]
- Kivinen L, Tsubari M, Haapajarvi T, Datto MB, Wang XF & Laiho M 1999. Ras induces p21Cip1/Waf1 cyclin kinase inhibitor transcriptionally through Sp1-binding sites. Oncogene 18 6252–6261. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Isotani A, Mise N, Yamamoto M, Fujihara Y, Kaseda K, Nakanishi T, Ikawa M, Hamada H, Abe K et al. 2006. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Current Biology 16 166–172. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N & Arima T 2007. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Human Molecular Genetics 16 2542–2551. [DOI] [PubMed] [Google Scholar]
- La P, Desmond A, Hou Z, Silva AC, Schnepp RW & Hua X 2006. Tumor suppressor menin: the essential role of nuclear localization signal domains in coordinating gene expression. Oncogene 25 3537–3546. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al. 2001. Initial sequencing and analysis of the human genome. Nature 409 860–921. [DOI] [PubMed] [Google Scholar]
- Laprise SL 2009. Implications of epigenetics and genomic imprinting in assisted reproductive technologies. Molecular Reproduction and Development 76 1006–1018. [DOI] [PubMed] [Google Scholar]
- Lee WK, Kim YM, Malik N, Ma C & Westphal H 2006. Cloning and characterization of the 5′-flanking region of the Ehox gene. Biochemical and Biophysical Research Communications 341 225–231. [DOI] [PubMed] [Google Scholar]
- Lehner B & Sanderson CM 2004. A protein interaction framework for human mRNA degradation. Genome Research 14 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens IH, Forsberg L, Pannett AA, Meyen E, Piehl F, Turner JJ, Van de Ven WJ, Thakker RV, Larsson C & Kas K 2001. Menin interacts directly with the homeobox-containing protein Pem. Biochemical and Biophysical Research Communications 286 426–431. [DOI] [PubMed] [Google Scholar]
- Li Y & Behringer RR 1998. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nature Genetics 20 309–311. [DOI] [PubMed] [Google Scholar]
- Li Q, Bartlett DL, Gorry MC, O’Malley ME & Guo ZS 2009. Three epigenetic drugs up-regulate homeobox gene Rhox5 in cancer cells through overlapping and distinct molecular mechanisms. Molecular Pharmacology 76 1072–1081. [DOI] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE et al. 2006. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125 801–814. [DOI] [PubMed] [Google Scholar]
- Lin TP, Labosky PA, Grabel LB, Kozak CA, Pitman JL, Kleeman J & MacLeod CL 1994. The Pem homeobox gene is X-linked and exclusively expressed in extraembryonic tissues during early murine development. Developmental Biology 166 170–179. [DOI] [PubMed] [Google Scholar]
- Lindsey S & Wilkinson MF 1996a. Homeobox genes and male reproductive development. Journal of Assisted Reproduction and Genetics 13 182–192. [DOI] [PubMed] [Google Scholar]
- Lindsey JS & Wilkinson MF 1996b. An androgen-regulated homeobox gene expressed in rat testis and epididymis. Biology of Reproduction 55 975–983. [DOI] [PubMed] [Google Scholar]
- Lindsey JS & Wilkinson MF 1996c. Pem: a testosterone- and LH-regulated homeobox gene expressed in mouse Sertoli cells and epididymis. Developmental Biology 179 471–484. [DOI] [PubMed] [Google Scholar]
- Luo ZW, Guo F, Li YQ, Li SQ, Zhang X, Li HJ & Zhou TH 2006. Screening and detecting of proteins interacting with mPem. Sheng Wu Gong Cheng Xue Bao 22 125–130. [PubMed] [Google Scholar]
- MacLean JA II & Wilkinson MF 2005. Gene regulation in spermatogenesis. Current Topics in Developmental Biology 71 131–197. [DOI] [PubMed] [Google Scholar]
- MacLean JA II, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C & Wilkinson MF 2005a. Rhox: a new homeobox gene cluster. Cell 120 369–382. [DOI] [PubMed] [Google Scholar]
- MacLean JA II, Rao MK, Doyle KM, Richards JS & Wilkinson MF 2005b. Regulation of the Rhox5 homeobox gene in primary granulosa cells: preovulatory expression and dependence on SP1/SP3 and GABP. Biology of Reproduction 73 1126–1134. [DOI] [PubMed] [Google Scholar]
- MacLean JA II, Lorenzetti D, Hu Z, Salerno WJ, Miller J & Wilkinson MF 2006. Rhox homeobox gene cluster: recent duplication of three family members. Genesis 44 122–129. [DOI] [PubMed] [Google Scholar]
- Maiti S, Doskow J, Li S, Nhim RP, Lindsey JS & Wilkinson MF 1996a. The Pem homeobox gene. Androgen-dependent and -independent promoters and tissue-specific alternative RNA splicing. Journal of Biological Chemistry 271 17536–17546. [DOI] [PubMed] [Google Scholar]
- Maiti S, Doskow J, Sutton K, Nhim RP, Lawlor DA, Levan K, Lindsey JS & Wilkinson MF 1996b. The Pem homeobox gene: rapid evolution of the homeodomain, X chromosomal localization, and expression in reproductive tissue. Genomics 34 304–316. [DOI] [PubMed] [Google Scholar]
- Maiti S, Meistrich ML, Wilson G, Shetty G, Marcelli M, McPhaul MJ, Morris PL & Wilkinson MF 2001. Irradiation selectively inhibits expression from the androgen-dependent Pem homeobox gene promoter in Sertoli cells. Endocrinology 142 1567–1577. [DOI] [PubMed] [Google Scholar]
- Marques CJ, Francisco T, Sousa S, Carvalho F, Barros A & Sousa M 2010. Methylation defects of imprinted genes in human testicular spermatozoa. Fertility and Sterility 94 585–594. [DOI] [PubMed] [Google Scholar]
- Marsh E, Fulp C, Gomez E, Nasrallah I, Minarcik J, Sudi J, Christian SL, Mancini G, Labosky P, Dobyns W et al. 2009. Targeted loss of Arx results in a developmental epilepsy mouse model and recapitulates the human phenotype in heterozygous females. Brain 132 1563–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W & Krumlauf R 1992. Homeobox genes and axial patterning. Cell 68 283–302. [DOI] [PubMed] [Google Scholar]
- Moens CB & Selleri L 2006. Hox cofactors in vertebrate development. Developmental Biology 291 193–206. [DOI] [PubMed] [Google Scholar]
- Moore T 2001. Genetic conflict, genomic imprinting and establishment of the epigenotype in relation to growth. Reproduction 122 185–193. [DOI] [PubMed] [Google Scholar]
- Morris L, Gordon J & Blackburn CC 2006. Identification of a tandem duplicated array in the Rhox alpha locus on mouse chromosome X. Mammalian Genome 17 178–187. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC & Turner JM 2008. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nature Genetics 40 794–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima J, Ishikawa S, Hamada J, Yanagihara M, Koike T & Hatakeyama M 2008. Anti-tumor activity of ESX1 on cancer cells harboring oncogenic K-ras mutation. Biochemical and Biophysical Research Communications 370 189–194. [DOI] [PubMed] [Google Scholar]
- Nicol AF, Pires AR, de Souza SR, Nuovo GJ, Grinsztejn B, Tristao A, Russomano FB, Velasque L, Lapa e Silva JR & Pirmez C 2008. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA). BMC Cancer 8 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E & Okano M 2006. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes and Development 20 3382–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata T, Matsuo N, Hiraoka N & Hata JI 2000. X-linked lissencephaly with ambiguous genitalia: delineation of further case. American Journal of Medical Genetics 94 174–176. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA & Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99 247–257. [DOI] [PubMed] [Google Scholar]
- Ono T, Sato S, Kimura N, Tanaka M, Shibuya A, Old LJ & Nakayama E 2000. Serological analysis of BALB/C methylcholanthrene sarcoma Meth A by SEREX: identification of a cancer/testis antigen. International Journal of Cancer 88 845–851. [DOI] [PubMed] [Google Scholar]
- O’Shaughnessy PJ, Abel M, Charlton HM, Hu B, Johnston H & Baker PJ 2007. Altered expression of genes involved in regulation of vitamin A metabolism, solute transportation, and cytoskeletal function in the androgen-insensitive tfm mouse testis. Endocrinology 148 2914–2924. [DOI] [PubMed] [Google Scholar]
- Ozawa H, Ashizawa S, Naito M, Yanagihara M, Ohnishi N, Maeda T, Matsuda Y, Jo Y, Higashi H, Kakita A et al. 2004. Paired-like homeodomain protein ESXR1 possesses a cleavable C-terminal region that inhibits cyclin degradation. Oncogene 23 6590–6602. [DOI] [PubMed] [Google Scholar]
- Pitman JL, Lin TP, Kleeman JE, Erickson GF & MacLeod CL 1998. Normal reproductive and macrophage function in Pem homeobox gene-deficient mice. Developmental Biology 202 196–214. [DOI] [PubMed] [Google Scholar]
- Poisson A, Zablewska B & Gaudray P 2003. Menin interacting proteins as clues toward the understanding of multiple endocrine neoplasia type 1. Cancer Letters 189 1–10. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH & Schneiderman HA 1969. A clonal analysis of determination in antennapedia a homoeotic mutant of Drosophila melanogaster. PNAS 64 176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prante BC, Garman KL, Sims BN & Lindsey JS 2008. Matrix-coated transwell-cultured TM4 Sertoli cell testosterone-regulated gene expression mimics in vivo expression. In Vitro Cellular & Developmental Biology. Animal 44 434–443. [DOI] [PubMed] [Google Scholar]
- Prasad TS, Kandasamy K & Pandey A 2009. Human protein reference database and human proteinpedia as discovery tools for systems biology. Methods in Molecular Biology 577 67–79. [DOI] [PubMed] [Google Scholar]
- Rao M & Wilkinson MF 2002. Homeobox genes and the male reproductive tract In The Epididymis, pp 269–283. Eds Robaire B, Hinton BT & Orgebin-Crist M-C. New York: Kluwer Academic/Plenum Publishers. [Google Scholar]
- Rao MK, Maiti S, Ananthaswamy HN & Wilkinson MF 2002a. A highly active homeobox gene promoter regulated by Ets and Sp1 family members in normal granulosa cells and diverse tumor cell types. Journal of Biological Chemistry 277 26036–26045. [DOI] [PubMed] [Google Scholar]
- Rao MK, Wayne CM & Wilkinson MF 2002b. Pem homeobox gene regulatory sequences that direct androgen-dependent developmentally regulated gene expression in different subregions of the epididymis. Journal of Biological Chemistry 277 48771–48778. [DOI] [PubMed] [Google Scholar]
- Rao MK, Wayne CM, Meistrich ML & Wilkinson MF 2003. Pem homeobox gene promoter sequences that direct transcription in a Sertoli cell-specific, stage-specific, and androgen-dependent manner in the testis in vivo. Molecular Endocrinology 17 223–233. [DOI] [PubMed] [Google Scholar]
- Rawn SM & Cross JC 2008. The evolution, regulation, and function of placenta-specific genes. Annual Review of Cell and Developmental Biology 24 159–181. [DOI] [PubMed] [Google Scholar]
- Rayle RE 1991. The oncofetal gene Pem specifies a divergent paired class homeodomain. Developmental Biology 146 255–257. [DOI] [PubMed] [Google Scholar]
- Reik W & Lewis A 2005. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nature Reviews. Genetics 6 403–410. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Pouchnik DJ & Griswold MD 2004. Identification of testosterone-regulated genes in testes of hypogonadal mice using oligonucleotide microarray. Molecular Endocrinology 18 422–433. [DOI] [PubMed] [Google Scholar]
- Sasaki AW, Doskow J, MacLeod CL, Rogers MB, Gudas LJ & Wilkinson MF 1991. The oncofetal gene Pem encodes a homeodomain and is regulated in primordial and pre-muscle stem cells. Mechanisms of Development 34 155–164. [DOI] [PubMed] [Google Scholar]
- Schauwaers K, De Gendt K, Saunders PT, Atanassova N, Haelens A, Callewaert L, Moehren U, Swinnen JV, Verhoeven G, Verrijdt G et al. 2007. Loss of androgen receptor binding to selective androgen response elements causes a reproductive phenotype in a knockin mouse model. PNAS 104 4961–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt S, Florl AR, Shi W, Hemberger M, Orth A, Otto S, Schulz WA & Fundele RH 2003. DNA methylation in placentas of interspecies mouse hybrids. Genetics 165 223–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanker S, Hu Z & Wilkinson MF 2008. Epigenetic regulation and downstream targets of the Rhox5 homeobox gene. International Journal of Andrology 31 462–470. [DOI] [PubMed] [Google Scholar]
- Shen HC, He M, Powell A, Adem A, Lorang D, Heller C, Grover AC, Ylaya K, Hewitt SM & Marx SJ 2009. Recapitulation of pancreatic neuroendocrine tumors in human multiple endocrine neoplasia type I syndrome via Pdx1-directed inactivation of Men1. Cancer Research 69 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB & Wilkinson MF 2000. The double lives of shuttling mRNA binding proteins. Cell 102 135–138. [DOI] [PubMed] [Google Scholar]
- Sneddon SF, Walther N & Saunders PT 2005. Expression of androgen and estrogen receptors in Sertoli cells: studies using the mouse SK11 cell line. Endocrinology 146 5304–5312. [DOI] [PubMed] [Google Scholar]
- Soares MJ, Konno T & Alam SM 2007. The prolactin family: effectors of pregnancy-dependent adaptations. Trends in Endocrinology and Metabolism 18 114–121. [DOI] [PubMed] [Google Scholar]
- Spitz F & Duboule D 2005. Developmental biology: reproduction in clusters. Nature 434 715–716. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F & Duboule D 2003. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113 405–417. [DOI] [PubMed] [Google Scholar]
- Sutton KA & Wilkinson MF 1997a. Rapid evolution of a homeodomain: evidence for positive selection. Journal of Molecular Evolution 45 579–588. [DOI] [PubMed] [Google Scholar]
- Sutton KA & Wilkinson MF 1997b. The rapidly evolving Pem homeobox gene and Agtr2, Ant2, and Lamp2 are closely linked in the proximal region of the mouse X chromosome. Genomics 45 447–450. [DOI] [PubMed] [Google Scholar]
- Sutton KA, Maiti S, Tribley WA, Lindsey JS, Meistrich ML, Bucana CD, Sanborn BM, Joseph DR, Griswold MD, Cornwall GA et al. 1998. Androgen regulation of the Pem homeodomain gene in mice and rat Sertoli and epididymal cells. Journal of Andrology 19 21–30. [PubMed] [Google Scholar]
- Svingen T & Koopman P 2007. Involvement of homeobox genes in mammalian sexual development. Sexual Development 1 12–23. [DOI] [PubMed] [Google Scholar]
- Svingen T & Tonissen KF 2006. Hox transcription factors and their elusive mammalian gene targets. Heredity 97 88–96. [DOI] [PubMed] [Google Scholar]
- Swanson WJ & Vacquier VD 2002. The rapid evolution of reproductive proteins. Nature Reviews. Genetics 3 137–144. [DOI] [PubMed] [Google Scholar]
- Takasaki N, McIsaac R & Dean J 2000. Gpbox (Psx2), a homeobox gene preferentially expressed in female germ cells at the onset of sexual dimorphism in mice. Developmental Biology 223 181–193. [DOI] [PubMed] [Google Scholar]
- Takasaki N, Rankin T & Dean J 2001. Normal gonadal development in mice lacking GPBOX, a homeobox protein expressed in germ cells at the onset of sexual dimorphism. Molecular and Cellular Biology 21 8197–8202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KA, De Gendt K, Atanassova N, Walker M, Sharpe RM, Saunders PT, Denolet E & Verhoeven G 2005. The role of androgens in Sertoli cell proliferation and functional maturation: studies in mice with total or Sertoli cell-selective ablation of the androgen receptor. Endocrinology 146 2674–2683. [DOI] [PubMed] [Google Scholar]
- Terada K, Yomogida K, Imai T, Kiyonari H, Takeda N, Kadomatsu T, Yano M, Aizawa S & Mori M 2005. A type I DnaJ homolog, DjA1, regulates androgen receptor signaling and spermatogenesis. EMBO Journal 24 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM & Hoekstra HE 2008. Causes and consequences of the evolution of reproductive proteins. International Journal of Developmental Biology 52 769–780. [DOI] [PubMed] [Google Scholar]
- Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, Kress J, Treier AC, Klugmann C, Klasen C, Holter NI et al. 2009. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139 1130–1142. [DOI] [PubMed] [Google Scholar]
- Viger RS, Guittot SM, Anttonen M, Wilson DB & Heikinheimo M 2008. Role of the GATA family of transcription factors in endocrine development, function, and disease. Molecular Endocrinology 22 781–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X & Zhang J 2006. Remarkable expansions of an X-linked reproductive homeobox gene cluster in rodent evolution. Genomics 88 34–43. [DOI] [PubMed] [Google Scholar]
- Wang X & Zhang J 2007. Rapid evolution of primate ESX1, an X-linked placenta- and testis-expressed homeobox gene. Human Molecular Genetics 16 2053–2060. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420 520–562. [DOI] [PubMed] [Google Scholar]
- Wayne CM, MacLean JA, Cornwall G & Wilkinson MF 2002a. Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene 301 1–11. [DOI] [PubMed] [Google Scholar]
- Wayne CM, Sutton K & Wilkinson MF 2002b. Expression of the pem homeobox gene in Sertoli cells increases the frequency of adjacent germ cells with deoxyribonucleic acid strand breaks. Endocrinology 143 4875–4885. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Kleeman J, Richards J & MacLeod CL 1990. A novel oncofetal gene is expressed in a stage-specific manner in murine embryonic development. Developmental Biology 141 451–455. [DOI] [PubMed] [Google Scholar]
- Yeh YC, Yang VC, Huang SC & Lo NW 2005. Stage-dependent expression of extra-embryonic tissue-spermatogenesis-homeobox gene 1 (ESX1) protein, a candidate marker for X chromosome-bearing sperm. Reproduction, Fertility, and Development 17 447–455. [DOI] [PubMed] [Google Scholar]
- Yordy JS & Muise-Helmericks RC 2000. Signal transduction and the Ets family of transcription factors. Oncogene 19 6503–6513. [DOI] [PubMed] [Google Scholar]
- Zhan M, Miura T, Xu X & Rao MS 2005. Conservation and variation of gene regulation in embryonic stem cells assessed by comparative genomics. Cell Biochemistry and Biophysics 43 379–405. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Shima JE, Nie R, Friel PJ & Griswold MD 2005. Androgen-regulated transcripts in the neonatal mouse testis as determined through microarray analysis. Biology of Reproduction 72 1010–1019. [DOI] [PubMed] [Google Scholar]


