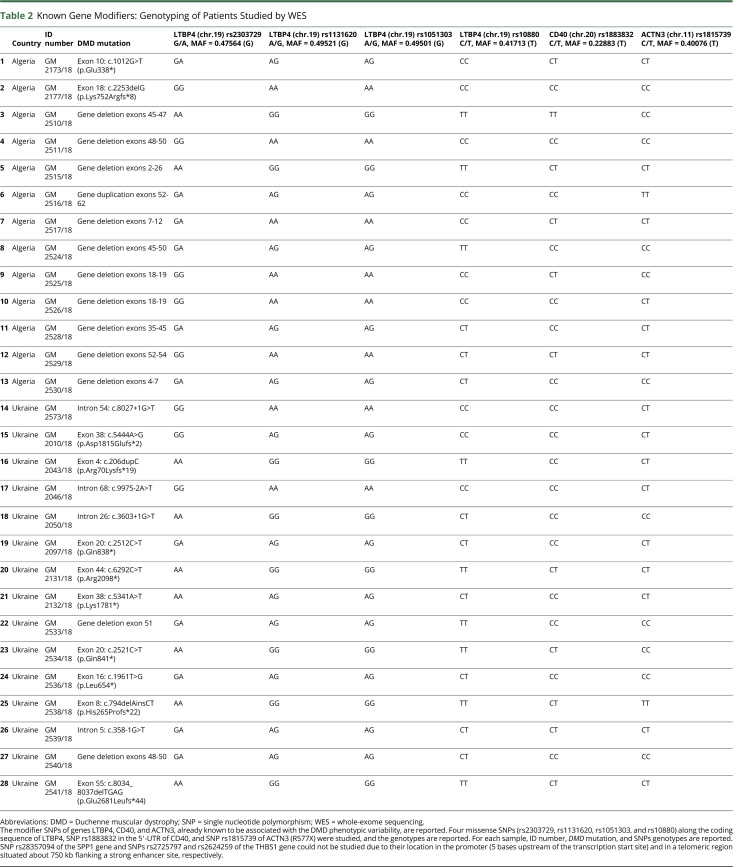
Rita Selvatici
Rita Selvatici, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Rachele Rossi
Rachele Rossi, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Fernanda Fortunato
Fernanda Fortunato, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Cecilia Trabanelli
Cecilia Trabanelli, MSc
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Yamina Sifi
Yamina Sifi, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Alice Margutti
Alice Margutti, MSc
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Marcella Neri
Marcella Neri, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Francesca Gualandi
Francesca Gualandi, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Lena Szabò
Lena Szabò, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Balint Fekete
Balint Fekete, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Lyudmilla Angelova
Lyudmilla Angelova, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Ivan Litvinenko
Ivan Litvinenko, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Ivan Ivanov
Ivan Ivanov, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Yurtsever Vildan
Yurtsever Vildan, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Oana Alexandra Iuhas
Oana Alexandra Iuhas, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Mihaela Vintan
Mihaela Vintan, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Carmen Burloiu
Carmen Burloiu, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Butnariu Lacramioara
Butnariu Lacramioara, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Gabriela Visa
Gabriela Visa, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Diana Epure
Diana Epure, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Cristina Rusu
Cristina Rusu, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Daniela Vasile
Daniela Vasile, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Magdalena Sandu
Magdalena Sandu, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Dmitry Vlodavets
Dmitry Vlodavets, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Monica Mager
Monica Mager, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Theodore Kyriakides
Theodore Kyriakides, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Sanja Delin
Sanja Delin, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Ivan Lehman
Ivan Lehman, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Jadranka Sekelj Fureš
Jadranka Sekelj Fureš, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Veneta Bojinova
Veneta Bojinova, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Mariela Militaru
Mariela Militaru, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Velina Guergueltcheva
Velina Guergueltcheva, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Birute Burnyte
Birute Burnyte, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Maria Judith Molnar
Maria Judith Molnar, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Niculina Butoianu
Niculina Butoianu, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Selma Dounia Bensemmane
Selma Dounia Bensemmane, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Samira Makri-Mokrane
Samira Makri-Mokrane, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Agnes Herczegfalvi
Agnes Herczegfalvi, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Monica Panzaru
Monica Panzaru, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Adela Chirita Emandi
Adela Chirita Emandi, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Anna Lusakowska
Anna Lusakowska, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Anna Potulska-Chromik
Anna Potulska-Chromik, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Anna Kostera-Pruszczyk
Anna Kostera-Pruszczyk, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Andriy Shatillo
Andriy Shatillo, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Djawed Bouchenak Khelladi
Djawed Bouchenak Khelladi, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Oussama Dendane
Oussama Dendane, MD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Mingyan Fang
Mingyan Fang, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Zhiyuan Lu
Zhiyuan Lu, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,
Alessandra Ferlini
Alessandra Ferlini, MD, PhD
1From the Medical Genetics Unit (R.S., R.R., F.F., C.T., A.M., M.N., F.G., A.F.), Department of Medical Sciences, University of Ferrara, Italy; Neurologie (Y.S.), CHU de Benbadis, Constantine, Algérie; 2nd Department of Paediatrics Clinic (L.S., A.H.), Semmelweis University; Institute of Genomic Medicine and Rare Disorders (B.F., M.J.M.), Semmelweis University, Budapest, Hungary; Department of Medical Genetics (L.A.), Medical University, Varna, Bulgaria; Department of Pediatrics (I. Litvinenko), Medical University Sofia; Department of Child Neurology (I. Litvinenko), University Pediatric Hospital “Prof. Ivan Mitev”, Sofia; Department Pediatrics (I.I.), St. George University Hospital, Medical University Plovdiv, Bulgaria; Pediatric Neurology Department (Y.V.), Pediatric Neurologist County Clinical Emergency Hospital of Constanta; G. Curteanu Municipal Clinical Hospital Oradea (O.A.I.); Department of Neuroscience (M.V., M. Militaru), Iuliu Hatieganu University of Medicine and Pharmacy, Cluj Napoca; Pediatric Neurology Department (C.B., N.B.), “Alexandru Obregia” Clinical Psychiatry Hospital, Bucharest; “Grigore T Popa” University of Medicine and Pharmacy (B.L., C.R., M.P.); “Sfanta Maria” Children's Hospital (B.L., C.R., M.P.); Pediatric Clinical Hospital Sibiu (G.V.); “Dr. Victor Gomoiu” Children's Hospital (D.E., D. Vasile, M.S.); “Carol Davila” University of Medicine and Pharmacy (M.S., N.B.), Bucharest, Romania; Russian Children Neuromuscular Center (D. Vlodavets), Veltischev Clinical Pediatric Research Institute of Pirogov Russian National Research Medical University, Moscow, Russia; Department of Neuroscience Neurology and Pediatric Neurology “Iuliu Hatieganu” University of Medicine and Pharmacy (M. Mager), Faculty of Medicine; Pediatric Neurology Department (M. Mager), Emergency Clinical Hospital for Children, Cluj-Napoca, Romania; Department of Basic and Clinical Sciences (T.K.), University of Nicosia, Cyprus; Department of Pediatrics (S.D.), General Hospital Zadar, Zadar, Croatia; Department of Paediatrics (I. Lehman, J.S.F.); University Hospital Centre Zagreb, ; Faculty of Medicine University of Osijek (J.S.F.), Croatia; University Hospital of Neurology and Psychiatry Sveti Naum (V.B.); Clinic of Neurology (V.G.), University Hospital Sofiamed; Sofia University “St. Kliment Ohridski” (V.G.)Bulgaria; Institute of Biomedical Sciences (B.B.), Faculty of Medicine, Vilnius University, Lithuania; Ali Ait Idir Hospital (S.D.B., S.M.-M.), Algiers, Algeria; University of Algiers I. Algeria (S.D.B., S.M.-M.); Center of Genomic Medicine (A.C.E.), University of Medicine and Pharmacy Victor Babes Timisoara; Regional Center of Medical Genetics Timis (A.C.E.), Clinical Emergency Hospital for Children Louis Turcanu Timisoara, Romania; Department of Neurology (A.L., A.P., A.K.-P.), Medical University of Warsaw, Poland; Institute of Neurology (A.S.), Psychiatry and Narkology National Academy of Medical Science of Ukraine; Neurologie (D.B.K., O.D.), CHU Tidjani Damerdji, Tlemcen, Algerie; and BGI-Shenzhen (M.F., Z.L.), Shenzhen, China.
1,✉