Abstract
The ability of interferons (IFNs) to inhibit HIV-1 replication in cell culture models has long been recognized, and the therapeutic administration of IFNα to HIV-1-infected patients who are not receiving antiretroviral therapy produces a clear but transient decrease in plasma viral load. Conversely, studies of chronic HIV-1 infection in humans and SIV-infected animal models of AIDS show positive correlations between elevated plasma levels of IFNs, increased expression of IFN-stimulated genes (ISGs), biomarkers of inflammation and disease progression. In this Review, we discuss the evidence that IFNs can control HIV-1 replication in vivo and debate the controversial role of IFNs in promoting the pathological sequelae of chronic HIV-1 infection.
HIV-1 is a retrovirus of the genus Lentivirus that causes persistent infection of humans and arose from cross-species transmissions of SIV of chimpanzees in the first half of the twentieth century1,2. HIV-1 principally infects CD4+ T cells and is the aetiological agent of AIDS, which is characterized by the loss of CD4+ T cells, profound immunodeficiency and susceptibility to fatal opportunistic infections. Acute HIV-1 infection, which often manifests as a flu-like illness in infected patients, is marked by high levels of systemic viral replication and a partial depletion of CD4+ T cells that disproportionally affects some compartments, such as the lymphoid tissue of the gut. After the first few weeks of infection, the development of immune responses against HIV-1, in particular the adaptive cytotoxic CD8+ T cell response, leads to some control of viral replication, the establishment of a stable set-point plasma viral load and significant reconstitution of the CD4+ T cell count. A clinically asymptomatic phase of infection follows, typically lasting 8–10 years and involving persistent HIV-1 replication, systemic immune activation and inflammation, and progressive CD4+ T cell decline, ultimately leading to the development of AIDS.
Viral infections are sensed by components of the innate immune system called pattern recognition receptors (PRRs), which include mainly the membrane-based Toll-like receptors (TLRs) and the cytosolic retinoic acid inducible gene 1 (RIG-I)-like receptors (RLRs)3. PRRs recognize conserved chemical and structural features of pathogens called pathogen-associated molecular patterns (PAMPs), leading to the activation of signalling cascades that culminate in programmes of transcriptional induction and the release of, among other molecules, interferons (IFNs). IFNs, and particularly type I IFNs, are a family of pleiotropic pro-inflammatory and immunomodulatory cytokines that induce an antiviral state through the upregulation of hundreds of genes termed interferon-stimulated genes (ISGs) (FIG. 1).
Figure 1 |. Induction of ISG expression.

Type I interferons (IFNs) bind to type I IFN receptor (IFNAR), which is composed of IFNAR1 and IFNAR2 subunits, leading to tyrosine kinase 2 (TYK2) and Janus kinase 1 (JAK1) activation. These kinases phosphorylate (P) signal transducer and activator of transcription 1 (STAT1) and STAT2 to allow homodimerization (for STAT1) and heterodimerization (STAT1 plus STAT2). These STAT dimers then translocate to the nucleus. STAT1–STAT2 dimers to bind interferon regulatory factor 9 (IRF9) to form the interferon-stimulated gene factor 3 (ISCF3) complex, which engages IFN-stimulated response elements (ISREs), whereas STAT1 homodimers engage gamma-activated seguences (GASs). Binding of the STAT dimers to ISREs and GASs activates transcription of IFN-stimulated genes (ISGs). Other STAT dimers, phosphoinositide 3-kinases and mitogen-activated protein kinases may also be activated downstream of type I IFNs.
The relationship between IFNs and HIV-1 infection has received escalating attention over the past decade owing to a number of important observations: IFNα (a family of type I IFNs) exerts a profound block on HIV-1 replication in cell culture models; the known anti-HIV-1 restriction factors, which are cellular proteins that inhibit viral replication, are themselves encoded by ISGs; acute HIV-1 infection of humans induces a wave of IFNα production in plasma; and IFNs seem to be capable of controlling HIV-1 replication in infected patients, as demonstrated by significant reductions in plasma HIV-1 viral load following therapeutic IFNα administration. Much of the associated research focus has been on identifying ISGs and their products that inhibit viral replication; as a result, a number of anti-HIV-1 ISGs have now been characterized (BOX 1).
Box 1 |. The discovery of HIV and SIV restriction and resistance factors.
APOBEC3G
Viral infectivity factor (Vif) is required for HIV-1 replication in primary cells and some cell lines. APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G) was discovered through correlative RNA expression profiling and functional screening for cDNAs that, when expressed in virus-producing cells, inhibit infection by vif-deficient HIV-1. Subsequent work by many groups revealed that the related proteins APOBEC3D and APOBEC3F, as well as some variants of APOBEC3H, also suppress HIV-1 infection41.
TRIM5α
Analyses of divergent retroviruses in cells from various species, as well as the identification of Fv1 as a post-entry suppressor of infection with Murine leukaemia virus106, pointed to the existence of species-specific inhibitors of HIV-1. Functional screening of rhesus macaque cDNAs for suppressors of HIV-1 infection in target cells revealed rhesus tripartite motif-containing protein 5α (TRIM5α) as an early post-entry inhibitor of HIV-1 (REF. 42).
Tetherin
HIV-1 viral protein unique (Vpu) is essential for efficient virion release from some cell types, a phenotype that is exacerbated by interferon-α (IFNα) treatment in some cell types. Tetherin was identified through correlative RNA expression profiling and functional screening of cDNAs that inhibited the release of vpu-deficient HIV-1 particles from cells45,46.
SAMHD1
The HIV-2 viral protein X (Vpx) promotes infection of myeloid cells, most notably plasmacytoid dendritic cells (pDCs). The finding that Vpx engages an E3 ubiquitin ligase containing cullin 4A (CUL4A), DNA damage-binding protein 1 (DDB1) and DDBl-CUL4-associated factor 1 (DCAF1) suggested that Vpx interacts with a cellular inhibitor of infection to promote the ubiquitylation and proteasomal degradation of this inhibitor. Affinity chromatography using Vpx as the ‘bait’ identified SAMHD1 (SAM and HD domain-containing protein 1) as this interacting factor, and the post-entry suppressor activity of SAMHD1 was revealed in postmitotic target cells43,44.
IFITM
IFNα inhibits the production of infectious HIV-1 from certain cell lines. RNA silencing-based screening of IFN-stimulated genes (ISGs) was carried out on HIV-1-infected cells treated with IFNα, and this approach identified IFN-induced transmembrane protein 1 (IFITM1) as a suppressor of virus infectivity. Follow-up experiments showed that IFITM2 and IFITM3 can also inhibit HIV-1 (REF. 77).
SLFN11
Schlafen (SLFN) proteins are encoded by ISGs, and the differential expression of SLFN11 between 293 cells and 293T cells guided its assignment as a negative regulator of HIV-1 mRNA translation80.
MX2
The inhibitory effects of IFNα on the early stages of HIV-1 infection vary between cell lines and types. Transcriptomics-based screens guided the functional testing of ISG cDNAs for post-entry inhibitors of wild-type HIV-1 infection. The link between myxovirus resistance 2 (MX2) and IFN-mediated suppression was confirmed using MX2-directed RNA silencing approaches70–72.
In parallel, substantial effort has been made to understand the role, if any, of chronic IFN stimulation in the pathogenesis of AIDS. This body of work includes gene expression profiling studies of CD4+ T cells from HIV-1-infected patients, as well as studies of pathogenic SIV infections of non-human primates (NHPs) (such as SIV infection of macaque monkeys), and has revealed that ISGs are among the genes that are abnormally upregulated during chronic pathogenic infections4–6.
In this Review, we discuss recent advances in our understanding of the relationship between HIV-1 and the IFN system, and we try to reconcile the beneficial and detrimental roles of IFNs during natural HIV-1 infection.
IFN-mediated responses to HIV-1 infection
There are 3 families of IFNs, each of which signal through their respective receptors: type I IFNs, consisting in humans of IFNα (which has 13 subtypes), IFNβ, IFNuω, IFNε and IFNκ; type II IFN (that is, IFNγ); and type III IFNs, consisting of IFNλ1, IFNλ2, IFNλ3 and IFNλ4 (REFS 7,8. Antiviral responses primarily involve type I IFNs, which exhibit broad antiviral effects against multiple viruses in cultured cell models. Type I IFNs act in a paracrine and an autocrine manner to signal through the heterodimeric type I IFN receptor (IFNAR), which is composed of the subunits IFNAR1 and IFNAR2, leading to activation of the receptor-associated protein kinases tyrosine kinase 2 (TYK2) and Janus kinase 1 (JAK1) (FIG. 1). These kinases phosphorylate signal transducer and activator of transcription 1 (STAT1) and STAT2 to allow either homodimerization (for STAT1) or heterodimerization (STAT1–STAT2) and dimer translocation to the nucleus. STAT1–STAT2 dimers bind to IFN regulatory factor 9 (IRF9) to form the ISG factor 3 (ISGF3) complex, which binds to IFN-stimulated response elements (ISREs) in the promoters of ISGs, whereas STAT1 dimers engage gamma-activated sequences (GAS). Binding of STAT dimers to ISREs and GAS activates the transcription of hundreds of ISGs9 (FIG. 1) (for a list of activated ISGs, see Interferome v2.01). Other STAT dimers, phosphoinositide 3-kinases (PI3Ks) and mitogen-activated protein kinases (MAPKs) can also be activated via signalling cascades downstream of IFNAR, leading to additional programmes of gene induction10.
In addition to genes that are clearly related to immunity and host defence, ISGs include genes involved in diverse cellular functions, such as transcription, translation, cytoskeletal organization, DNA damage repair, apoptosis and lipid metabolism11. Although there are many well-characterized ISGs (reviewed in REFS 12,13), the functions of the majority remain largely obscure. In the past few years, high-throughput platforms have been developed for characterizing the antiviral effects of ISG-encoded proteins both individually and in combination14,15. These platforms have identified ISGs displaying broad inhibition against a number of viral families as well as others with more specific antiviral activity14,16. Identification and functional characterization of these genes and their products should lead to a better understanding of both the mechanisms of innate antiviral resistance and the corresponding viral evasion mechanisms, and should provide a clearer picture of the mechanistic underpinnings for the therapeutic application of IFNs.
HIV-1 sensing by components of the IFN system
A number of PRRs are involved in HIV-1 sensing, including the cytoplasmic receptors cyclic GMP–AMP (cGAMP) synthase (cGAS) and IFNγ-inducible protein 16 (IFI16) — both of which recognize viral cDNA — and TLR7, which recognizes viral genomic RNA in endosomes (FIG. 2). Importantly, different cell types seem to have differing capacities to sense infection, with plasmacytoid dendritic cells (pDCs), a cell type that is rich in PRRs, clearly playing a central part in HIV-1 detection and IFNα production.
Figure 2 |. Intracellular sensing of HIV-1 Infection.
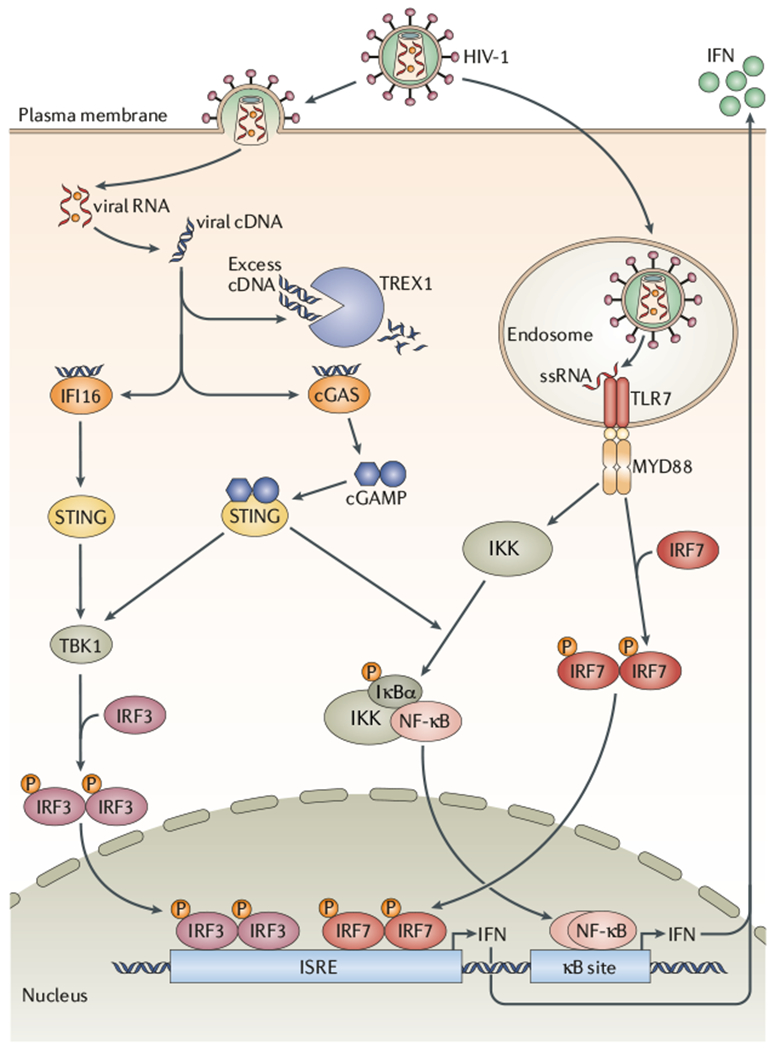
Following HIV-1 entry into the cell, viral RNA is reverse transcribed into cDNA, which is detected by the cytoplasmic receptors cyclic GMP–AMP (cGAMP) synthase (cGAS) and interferon-γ (IFNγ)-inducible protein 16 (IFI16). Following cDNA detection, IFI16 activates stimulator of IFN genes (STING), which leads to the activation of TANK-binding kinase 1 (TBK1) and the subseguent phosphorylation (P) of the IFN regulatory factor 3 (IRF3). Phosphorylated IRF3 can then engage IFN-stimulated response elements (ISREs), thereby inducing the expression of type I IFNs. When cGAS detects viral cDNA, the enzyme produces cGAMP, which leads to the activation of STING. STING then activates the inhibitor of NF-κB (IκB) kinase (IKK) complex and TBK1, leading to the activation of nuclear factor-κB (NF-κB) and IRF3, respectively. These transcription factors induce the expression of genes encoding IFNs and other pro-inflammatory cytokines. The cellular 3′-repair exonuclease 1 (TREX1) helps HIV-1 to evade cytosolic sensing by degrading viral cDNA in the cytoplasm. In addition to sensing cytoplasmic viral cDNA, cells can also sense HIV-1 single-stranded RNA (ssRNA) via Toll-like receptor 7 (TLR7) in endosomes, especially in plasmacytoid dendritic cells (pDCs). TLR7 activation by ssRNA in pDC endosomes results in the activation of myeloid differentiation primary response gene 88 (MYD88) and subseguent induction of IFN via activation of IRF7 and NF-κB.
Several sensors for cytoplasmic DNA have been identified over the past few years (reviewed in REF. 17), and two such sensors, cGAS and IFI16, are capable of detecting HIV-1 cDNA following infection18–22. cGAS has been shown to be responsible for sensing of nascent HIV-1 cDNA in infected monocyte-derived dendritic cells (MDDCs)18,20 as well as in the monocytic THP-1 cell line18. Following activation by DNA binding, cGAS catalyses the synthesis of a cGAMP isomer from ATP and GTP. This cyclic dinucleotide functions as a second messenger that binds to and activates stimulator of IFN genes (STING). STING then activates the inhibitor of NF-κB (IκB) kinase (IKK) and TANK-binding kinase 1 (TBK1), resulting in the induction of IFNs and other cytokines via the activation of the transcription factors nuclear factor-κB (NF-κB) and IRF3 (FIG. 2). Further support for the importance of the signalling pathways initiated by cGAS comes from the analysis of engineered HIV-1 viruses carrying mutations affecting the capsid (CA) region of the Gag protein21. Specifically, mutant viruses with substitutions at residues N74 or P90 induce the production of cGAMP and IFNs following infection of monocyte-derived macrophages (MDMs), whereas wild-type viruses do not21. These mutations prevent CA interactions with host cell proteins such as cyclophilin A (CYPA; also known as PPIA), nucleoporin 358 (NUP358; also known as RANBP2) and cleavage and polyadenylation specific factor 6 (CPSF6)23, whereas it is thought that the recruitment of these host proteins to wild-type viral reverse-transcription complexes (RTCs) shields viral cDNA from sensing by cGAS (and possibly other PRRs)21.
IFI16 is a pyrin and HIN domain-containing (PYHIN) protein and was identified as a protein bound to cytosolic DNA24. IFI16 induces IFN production via STING and IRF3 (FIG. 2). In cultured quiescent tonsillar CD4+ T cells infected with HIV-1, in which cGAS is not expressed, IFI16 sensed incomplete HIV-1 DNA replication intermediates. In addition to stimulating IFN production, IFI16 activation induces caspase 1 activation and cell death by pyroptosis19,25, in contrast to the caspase 3-mediated apoptosis observed in activated, productively infected T cells. Importantly, in ex vivo human lymphoid aggregate cultures infected with HIV-1, more than 95% of CD4+ T cell death occurs in cells that are in a quiescent state25,26, suggesting that IFI16-mediated pyroptosis may be a major contributing mechanism to the profound decline of CD4+ T cells that characterizes chronic HIV-1 infection.
TLR7 has been shown to be essential for HIV-1 sensing by pDCs and mediates the recognition of viral genomic RNA in endosomes27,28. TLR7 activation causes recruitment of myeloid diffentiation primary response gene 88 (MYD88), which leads to the induction of IFNs and other cytokines through activation of IRF7 and NF-κB29 (FIG. 2). In vitro, pDCs can efficiently sense HIV-1 free particles as well as virus-infected cells, leading to the production of high levels of IFNα27,28. The importance of pDC-mediated viral sensing is underscored by observations from humanized mice, in which depletion of pDCs almost completely abrogates the initial wave of IFN production seen during acute HIV-1 infection30. Additionally, in SIV-infected cynomolgus macaques, the natural reversal of IFN production is strongly associated with pDC exhaustion or death31, and blockade of TLR7-mediated (and TLR9-mediated) virus sensing during acute SIV infection of macaques results in a diminution of IFN production32, further highlighting the importance of TLR sensing in vivo. Interestingly, the production of IFNs in this context is not completely abrogated, consistent with the view that multiple sensors participate in viral detection28,32.
In contrast to the production of IFNs by pDCs following infection, HIV-1 seems to avoid triggering an IFN response in many other cultured cells21,33–35. The host 3′-repair exonuclease 1 (TREX1) plays an important part in this respect by degrading excess HIV-1 cDNA34, which may otherwise be sensed by cGAS or IFI16 (FIG. 2). For instance, in MDMs depleted of TREX1, HIV-1 cDNA induces IFN expression via STING, TBK1 and IRF3 (REF. 34). Therefore, there seems to be competition between recognition of HIV-1 cDNA by cytoplasmic receptors, masking of HIV-1 cDNA by cellular factors and clearance of HIV-1 cDNA by TREX1; the balance between these effects will have an impact on the IFN response and, presumably, pathogenic outcome in the infected host.
Finally, evidence has recently emerged that HIV-1 restriction and sensing can be mediated by the same proteins. For example, two HIV-1 restriction factors, the tripartite motif-containing protein 5α (TRIM5α) and tetherin (also known as BST2), can each detect particulate viral assemblies and initiate intracellular signalling via transforming growth factor-β-activated kinase 1 (TAK1, also known as MP3K7), TNF receptor-associated factor 6 (TRAF6) and NF-κB to enhance the expression of pro-inflammatory genes and cytokines36–38.
The IFN-induced anti-HIV-1 effectors
Efforts to assign single ISG-encoded proteins as effectors of specific inhibitory mechanisms during the HIV-1 life cycle have been somewhat erratic until recently, although the focused screens that led to the identification of several HIV-1 and SIV restriction factors represent exceptions (BOX 1). Interestingly, these two topics are now merging, partly because restriction factors, although frequently expressed constitutively, are also induced to a degree by IFN39,40 and partly because functional screening of cDNAs has become more experimentally mainstream.
HIV-1 restriction factors.
Restriction factors are dominantly acting, cell-intrinsic proteins that can potently suppress HIV and SIV replication. To date, four restriction factors have been unambiguously defined: TRIM5α; sterile α motif domain and histidine aspartic acid (HD) domain 1 (SAMHD1); apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) proteins (specifically, APOBEC3G, APOBEC3D, APOBEC3F and certain allelic forms of APOBEC3H); and tetherin41–46 (FIG. 3).These proteins share a number of features (BOX 2), and their mechanisms of action have been reviewed extensively47,48: TRIM5α, SAMHD1 and APOBEC3 proteins target the early post-entry phases of infection, and tetherin prevents viral release from infected cells (FIG. 3).
Figure 3 |. HIV-1 restriction and resistance factors.

In the absence of viraLLy encoded antagonists (or viral escape), host cell proteins called HIV-1 restriction factors (yellow) inhibit various stages of the replication cycle. The tripartite motif-containing protein 5α (TRIM5α) promotes accelerated fragmentation of viral cores, preventing cDNA synthesis. SAM and HD domain-containing protein 1 (SAMHD1) depletes the cellular levels of 2′-deoxynucleoside 5′-triphosphates (dNTPs), which are reguired for efficient cDNA synthesis. APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3) proteins interfere with the processivity of HIV-1 reverse transcriptase and induce hypermutation of viral cDNA by cytidine deamination. Tetherin prevents the release of budded virions from the infected cell. Several viral proteins (blue) antagonize these cellular restriction factors. Viral infectivity factor (Vif) antagonizes APOBEC3 proteins, viral protein unique (Vpu) antagonizes tetherin, and the HIV-2 viral protein X (Vpx) antagonizes SAMHD1. HIV-1 resistance factors (brown) inhibit other stages of viral replication and are not counteracted by the virus. Myxovirus resistance 2 (MX2) prevents the nuclear import and integration of viral cDNA. Schlafen 11 (SLFN11) suppresses the translation of viral proteins. Interferon-induced transmembrane proteins (IFITMs) inhibit viral entry by interfering with membrane fusion. dsDNA, double-stranded DNA; gRNA, viral genomic RNA; LTR, long terminal repeat; ssDNA, single-stranded DNA.
Box 2 |. Hallmarks of HIV and SIV restriction factors.
Restriction factors that target HIV and SIV are dominantly acting, cell-intrinsic proteins that can potently suppress HIV and SIV replication and that share a number of features.
Germ line encoded
Restriction factors are antiretroviral proteins that are invariant within an individual and are not altered through gene rearrangement or somatic mutation.
Inducible by IFNs
Restriction factors are typically expressed constitutively in many cell types but can also be induced by interferons (IFNs) in some cells, such as macrophages.
Cell autonomous
Restriction factors are sufficient to mediate viral suppression when expressed in single virus-producing cells or viral target cells. Communication with other cells is not required for activity.
Inactive against wild-type viruses replicating in their natural hosts
Owing to effective virally encoded evasion or escape mechanisms, restriction factors are essentially inactive against wild-type viruses in their natural hosts. For example, HIV-1 viral infectivity factor (Vif) preserves viral replication in human cells because it is an efficient antagonist of human APOBEC3 (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3) proteins. Similarly, HIV-1 capsid structures are not recognized by human tripartite motif-containing protein 5α (TRIM5α).
Barriers against cross-species transmission
Despite being essentially inactive against viruses replicating in their natural hosts, restriction factors are potent antiviral effectors against viruses from other host species. For example, HIV-1 Vif fails to antagonize APOBEC3 proteins from African green monkeys, resulting in effective suppression of wild-type HIV-1 in African green monkey cells. Similarly, HIV-1 capsids are efficiently recognized and inhibited by TRIM5α of rhesus macaques. Therefore, restriction factors are thought to be important barriers against cross-species transmission of primate immunodeficiency viruses.
Frequently downregulated or suppressed by viral accessory proteins
Vif antagonizes APOBEC3 proteins, viral protein unique (Vpu) antagonizes tetherin, and HIV-2 viral protein X (Vpx) antagonizes SAM and HD domain-containing protein 1 (SAMHD1). The exception is TRIM5α, the functionality of which is determined not by the interference of an accessory protein but rather by an interaction with the capsid (CA) lattice of post-entry viral cores.
Under Darwinian selection
The coding sequences of restriction factors display the hallmarks of Darwinian selection, as sequence variation in some positions has a propensity to be characterized by a high dN/dS ratio — which is the number of nonsynonymous substitutions per nonsynonymous site (dN) divided by the number of synonymous substitutions per synonymous site (dS). This feature can be indicative of host–pathogen co-evolution, and the sites under this selection may represent sequences encoding protein regions involved in direct pathogen contact, such as the binding site for Vif in APOBEC3G.
Regulated by protein degradation
The function or downregulation of restriction factors typically involves the cellular ubiquitin–proteasome system, which is involved in protein degradation. For example, Vif binds to and recruits APOBEC3 proteins to an E3 ubiquitin ligase complex containing cullin 5 (CUL5), elongin B (ELOB) and ELOC, and this leads to polyubiquitylation and proteasomal degradation of APOBEC3 proteins; furthermore, TRIM5α is itself a ubiquitin ligase.
TRIM5α is an E3 ubiquitin ligase that binds to the CA lattice that forms the exterior surface of the post-entry viral cores. These viral cores normally mature into RTCs, but the TRIM5α–CA interaction leads to accelerated RTC fragmentation and prevents viral cDNA synthesis. SAMHD1 is a 2′-deoxynucleoside 5′-triphosphate (dNTP) triphosphohydrolase that depletes dNTP levels in non-dividing cells, thereby depriving reverse transcriptase of the substrates required for effective cDNA synthesis49,50. Interestingly, SAMHD1 has also been reported to be a nuclease that targets viral RNA for degradation51. APOBEC3 proteins are cytidine deaminases that are packaged into virions and remain associated with viral RTCs in newly challenged cells, where they suppress viral cDNA synthesis by interfering with the processivity of reverse transcription. In addition, they destructively hypermutate the cDNAs that are made by catalysing excessive C-to-U editing of (mostly) first-strand cDNA (also known as the minus strand). These mutations register as G-to-A transitions in the cDNA strand (also known as the plus strand) and compromise the genetic integrity of the virus. Finally, tetherin is a transmembrane protein that prevents the release of budded virions from the surface of infected cells by forming proteinaceous bridges between viral and cellular membranes (FIG. 3).
HIV-1 evasion of restriction factors.
One of the hallmarks of restriction factors (BOX 2) is that HIVs and SIVs have evolved an array of evasion mechanisms such that these proteins are commonly regarded as ineffective at controlling viral replication in cells of the natural host. For example, human restriction factors have little, or only minor, impact on HIV-1 transmission and replication in the context of human infections. By contrast, restriction factors tend to be very effective at blocking HIV or SIV infection of unnatural hosts, owing to the inability of viral evasion mechanisms to function efficiently in non-cognate species52. As a consequence, restriction factors are viewed as influential barriers to cross-species transmission53.
The viral accessory proteins viral infectivity factor (Vif) and viral protein unique (Vpu) serve as countermeasures for the APOBEC3 proteins and tetherin, respectively. Vif interacts with APOBEC3 proteins and induces the recruitment of an E3 ubiquitin ligase complex containing the scaffold protein cullin 5 (CUL5) and the substrate adaptors elongin B (ELOB; also known as TCEB2) and ELOC (also known as TCEB1), which leads to polyubiquitylation and proteasomal degradation of APOBEC3 proteins54,55. Similarly, Vpu interacts with tetherin, preventing tetherin trafficking to the cell surface and promoting tetherin ubiquitylation and subsequent degradation in endolysosomes56. Human TRIM5α does not suppress HIV-1 because it fails to engage and disrupt the post-entry CA lattice effectively. Escape from SAMHD1 is less well understood and may in fact be a ‘red herring’, as the importance of HIV-1 infection of myeloid cells in vivo remains questionable. Interestingly, the related virus HIV-2 encodes an accessory protein, viral protein X (Vpx), which induces SAMHD1 ubiquitylation and degradation through the recruitment of an E3 ubiquitin ligase complex that contains CUL4A, DNA damage-binding protein 1 (DDB1) and DDB1–CUL4-associated factor 1 (DCAF1; also known as VPRBP). This activity of Vpx suggests that HIV-2 needs to avert SAMHD1 function for replication in vivo. Therefore, the absence of a similar activity in HIV-1 may point to the possible irrelevance of SAMHD1 during natural infection or may indicate that SAMHD1 serves to assist HIV-1 in avoiding sensing in myeloid cells. Furthermore, the reverse transcriptase of HIV-1 can still catalyse DNA synthesis in the presence of low dNTP concentrations, including those found in non-proliferating myeloid cells, indicating that SAMHD1 activity in these cells does not prevent HIV-1 replication57.
Although human restriction factors do not induce pronounced anti-HIV-1 phenotypes in the context of infection with wild-type viruses, there is good evidence that subtle contributions take place during natural infection. In reality, the actions of a restriction factor and its counterbalancing evasion mechanism are likely to be in a state of equilibrium such that neither exhibits complete dominance. Examining the interaction between Vif and APOBEC3 illustrates this equilibrium, as incomplete Vif function (for example, due to allelic variation) or increased APOBEC3 activity (as occurs following IFN induction or treatment58, or on account of intrinsic expression differences) could shift this balance and promote APOBEC3-mediated effects. Indeed, this must happen during natural HIV-1 infection, as G-to-A hypermutated viral sequences are readily detected59, viral sequence evolution takes place at sites of APOBEC3 editing60,61, and higher APOBEC3 expression levels correlate with clinical benefit62. Whether variation in the balance between other restriction factors and their corresponding escape pathways, including the modulatory effects of IFNs63, can affect in vivo HIV-1 infection and disease progression remains to be determined.
HIV-1 resistance factors.
Given evidence from cell culture models that treatment with IFNs can potently suppress HIV and SIV replication64–69, there is an enduring interest in discovering additional anti-HIV and anti-SIV ISGs. Indeed, the observation that IFNα treatment imparts a strong post-entry block to retrovirus infection at the level of viral cDNA accumulation and integration68,69 provoked cDNA screens that led to the identification of myxovirus resistance 2 (MX2) as a significant inhibitor of HIV-1 infection in IFN-treated cultured cells70–72.
MX2 is an IFN-inducible dynamin-like GTPase, although this enzymatic function appears to be dispensable for viral inhibition70,72. Human MX2 localizes to the nuclear envelope, nuclear pore complexes and cytoplasmic puncta72–74, and inhibits divergent HIV-1 strains, but it is less effective against SIVs and inactive against other retroviruses, such as murine leukaemia virus (MLV)70. Viral inhibition occurs after substantial cDNA synthesis, in contrast to the earlier blocks described for the APOBEC3 proteins, TRIM5α and SAMHD1. Although the mechanism of viral inhibition mediated by MX2 is unknown, it may involve direct interactions with the viral CA protein70–72,75,76 and is manifested as a failure of viral cDNA to enter and accumulate in the nucleus70,72 (FIG. 3).
There is growing evidence for the existence of additional IFN-activated mechanisms of HIV-1 control. For example, the IFN-induced transmembrane (IFITM) proteins — specifically, IFITM1, IFITM2 and IFITM3 — are found in various cellular membranes and are incorporated into the viral membrane77,78. In contrast to the quantitative effect of tetherin on viral release, the IFITM proteins interfere with membrane fusion through the combined effects of increasing curvature, decreasing fluidity and altering membrane composition; as a result, IFITM proteins can act in virus particles or in target cells to impede viral entry77–79 (FIG. 3).
A further example is schlafen 11 (SLFN11), which inhibits virion production by suppressing HIV-1 protein synthesis80 (FIG. 3). SFFN11 is a cytoplasmic RNA-binding protein that selectively represses the translation of mRNAs with a codon bias different from that of typical human mRNAs — such as those expressed by HIV-1. This inhibitory mechanism may involve the binding of SLFN11 to tRNAs and their subsequent inactivation or removal.
Much remains to be learnt regarding the molecular details of MX2-, SFLN11- and IFITM-mediated inhibition of HIV-1, and searching for further ISG-encoded proteins that suppress early reverse transcription or other stages of viral replication remain active areas of current investigation. Interestingly, HIV-1-mediated mechanisms for evading MX2-, SLFN11- or IFITM-mediated suppression of viral replication have not been reported, making these ISG-encoded proteins fundamentally different to the restriction factors discussed above, which are either neutralized or avoided by HIV-1 evasion strategies. Accordingly, MX2, IFITM proteins and SLFN11 are more appropriately classified as resistance factors and are good candidates for host proteins that contribute to HIV-1 control during acute infection.
The beneficial roles of IFNs
Studies in different animal models have demonstrated the importance of the IFN system in controlling HIV and SIV infections.
IFNs in the control of acute infection.
IFNAR-deficient mice are highly susceptible to infection with a range of different viruses (reviewed in REF. 81). IFNs are also crucial for the control of lentiviral infections of NHPs82. For example, in a pathogenic SIV rhesus macaque model using intrarectal challenge, administration of an IFNAR antagonist led to higher viral loads during acute infection than in untreated controls, and treated animals progressed to AIDS and death within 30 weeks post-infection, whereas all untreated control animals survived through the 44-week follow-up82. Using the same model, pegylated-IFNα2a administration before and during viral challenge and for the subsequent 4 weeks after infection reduced the frequency of viral transmission82. Therefore, an intact IFN response during acute lentiviral infection seems to be crucial for viral control and the amelioration of subsequent disease and, when stimulated, can help to suppress initial viral transmission.
Importantly, this state of relative SIV suppression in IFN-treated macaques was not durable, as ISGs were downregulated within the first few weeks of IFNα treatment, and viral loads became higher than in untreated controls once the animals were infected. Downregulation of ISGs was associated with upregulation of the gene encoding forkhead box O3a (FOXO3a), which is a negative regulator of IFN signalling, suggesting that desensitization to the IFN response was a result of endogenous homeostatic control. Furthermore, corroborating the findings of these NHP studies, which indicate a beneficial role of IFNs during the early stages of infection, depletion of pDCs in a humanized mouse model of HIV-1 infection leads to reduced production of IFNα and to increased HIV-1 replication in the acute phase30. However, in contrast to the marked T cell decline observed in primates, the subsequent loss of T cells in these humanized mice was much less severe30.
As noted above, primary HIV-1 infection of humans is followed by an intense cytokine storm involving IFNα (from a median baseline of 4.6 pg ml−1 to a median peak of 37.5 pg ml−1) and many other pro-inflammatory and immunomodulatory cytokines83 that collectively precede the establishment of the set-point viral load. In addition, transmitted/founder viruses (T/F viruses) are commonly less sensitive to inhibition by IFNα in cultured cell models than the corresponding viruses that are present during chronic infection, suggesting that resisting the inhibitory effects of IFNα may provide a selective advantage during transmission and acute infection84,85. Taken together, these observations demonstrate that a robust type I IFN response helps to control initial HIV and SIV infection.
Clinical effects of IFN treatment on HIV-1 infection.
The virological benefits of administering IFNα to patients infected with HIV-1 have been recognized for some years. In the pre-highly active antiretroviral therapy (HAART) era, a randomized controlled trial of 12 weeks of IFNα2b treatment was conducted in asymptomatic patients infected with HIV-1; this trial showed that treatment led to a decrease in the frequency of viral isolation by culture and fewer patients developing AIDS during follow-up relative to patients receiving placebo86. However, interest in the use of IFNs for the control of HIV-1 infection declined with the development and introduction of HAART in the later 1990s. Nonetheless, the capacity of IFNα to reduce the plasma HIV-1 viral load was recognized and characterized in the context of its administration for other conditions such as hepatitis C virus (HCV) co-infection and Kaposi sarcoma87,88.
A more recent study investigated the virological and immunological effects of 12 weeks of pegylated-IFNα treatment in patients who were infected with HIV-1 alone and who were immunologically stable in the absence of HAART89. Weekly virological analysis showed that IFNα treatment induced a rapid decline in viral load, which reached a nadir at 2 weeks with a median reduction of 1.3 log10 copies ml−1 from baseline. This rapid initial decline in viral load following treatment with IFNα was followed by a partial rebound in HIV-1 viraemia prior to treatment discontinuation (FIG. 4). Follow-up work has examined the expression levels of a set of canonical ISGs, including MX2, in the peripheral blood mononuclear cells (PBMCs) of these patients and found correlations between sustained ISG upregulation and viral-load reduction90.
Figure 4 |. The effect of IFNα treatment on plasma HIV-1 viral load.

The graph shows plasma HIV-1 viral load (PVL) responses in patients who are infected with HIV-1 and who have not received antiretroviral therapy, during 12 weeks of treatment with pegylated-interferon-α (IFNα). The thick dashed line indicates the median PVL. IFNα treatment induces a rapid decline in PVL in most patients, whereas a minority fail to respond. The PVL reaches a nadir at 2 weeks (median reduction of 1.3 log10 copies ml−1 from baseline), followed by partial reversal of the response. Adapted from Asmuth, D. M. et al., Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon alfa-2a in HIV-l-monoinfected participants: a phase II clinical trial, J. Infect. Dis., 2010, 201, 11, 1686–1196, by permission of Oxford University Press.
Interestingly, a minority of patients did not exhibit a significant reduction in viral load, and this lack of response was associated with high baseline expression of ISGs in PBMCs and poor upregulation of ISGs following treatment. Interestingly, similarly poor ISG responses have been noted in hepatocytes of patients who are infected with HCV and fail to respond to IFN-based therapy91. Notably, although IFNα treatment induces sustained ISG induction in human PBMCs, this is not always observed in NHPs. For example, a reversal of ISG induction was seen in rhesus macaques after the first week of treatment (with or without acute SIV infection)82 and between weeks 3 and 12 in studies of SIV-infected sooty mangabeys92. The basis for this difference between humans and NHPs is not understood, but probably reflects important points of variation in the biology of NHP models of SIV infection and human HIV-1 infections.
The maintenance of ISG expression throughout the 12 weeks of IFNα therapy in humans raises the question of what events underpin the partial reversal of the viral-load decline seen early in therapy. One possibility is that HIV-1 evolves to escape the activity of IFN-induced effectors. Indeed, there is some evidence that Vpu evolution during IFNα treatment leads to the selection of alleles that downregulate tetherin more effectively58. Similarly, the IFN-induced upregulation of APOBEC3 proteins may contribute to increased rates of HIV-1 evolution61. Nevertheless, despite the limitations of IFNα monotherapy described above, another study has shown that treatment with IFNα alone appears to be capable of maintaining virological suppression (defined as fewer than 400 HIV-1 copies ml−1) for at least 12 weeks after HAART discontinuation in 45% of patients with prior virological control on HAART93. Although this subject is not the focus of this Review, it is possible that IFNα also enhances the antiviral activity of many different immune cells, and this would potentially contribute to improved viral control (BOX 3).
Box 3 |. Regulation of immune cells by type I IFNs.
Although there is convincing evidence that upregulation of interferon (IFN)-stimulated gene (ISG) expression contributes to cell-autonomous resistance to infection and is an important component of the prophylactic and therapeutic effects of IFNα, there is debate about the role of other immune effector mechanisms in mediating the beneficial effects observed during these responses. For example, both direct and indirect effects of type I IFNs have been described for almost all immune cells and, for many cells types, type I IFNs can stimulate or inhibit effector functions depending on the timing and context of the exposure (reviewed in REF. 98).
Natural killer (NK) cells and CD8+ T cells have a recognized role in the clearance of virally infected cells. Type I IFNs are important regulators of NK cell function and promote their activation and proliferation through the induction of interleukin-15 (IL-15)107–109. IL-15 production occurs directly in NK cells, through signalling via type I IFN receptor (IFNAR) and also from IFN-activated conventional dendritic cells (DCs)109. Type I IFNs are also important for the survival of antiviral CD8+ effector T cells, and removal of IFN signalling during CD8+ T cell activation limits their proliferation100,110. Type I IFNs induce the apoptosis of memory CD4+ and CD8+ T cells111 and of regulatory T cells during acute viral infection112, and this may facilitate the development of an optimal functional cytolytic T cell response100.
Studies in non-human primate (NHP) models of SIV infection point towards the importance of NK cells, as opposed to T cells, in mediating protective type I IFN responses. IFNα2a treatment of macaques leads to resistance to SIV infection via rectal challenge, and this resistance is associated with ISG upregulation in various cell types, and also with increased frequencies of CD56+ NK cells in the blood and of CD16+ NK cells in the rectum82. SIV-specific CD4+ and CD8+ T cell responses are not affected by IFNα2a treatment in this model, suggesting that these cell types do not play a crucial part in the observed IFNα-induced protection. Similarly, treatment with a type I IFN antagonist before SIV infection suppressed ISG induction and NK cell numbers at >12 weeks post-infection, but had no observable effect on CD4+ or CD8+ T cell responses82. Supporting these findings, treatment of chronically SIV-infected sooty mangabeys with a recombinant type I IFN agonist decreased the SIV viral load — similar to the effect observed for IFNα treatment in humans infected with HIV-1 — but had no effect on SIV-specific CD8+ T cells responses92.
Type I IFNs have also been shown to enhance B cell-mediated immune responses113 and probably facilitate other adaptive immune responses indirectly. For example, type I IFN-mediated activation and maturation of DCs and other antigen-presenting cells leads to increased antigen presentation, the secretion of cytokines and chemokines, and the presence of co-stimulatory signals114.
There has been some interest in the potential use of IFNs in HIV-1 eradication efforts. It has been reported that for patients who are co-infected with HIV and HCV and who are receiving IFNα-based therapy combined with HAART, there is a reduction in total and integrated (proviral) HIV-1 DNA levels in PBMCs94. Similar results were seen in patients who maintained virological suppression when treated with IFNα monotherapy after HAART discontinuation (see above)93. Although the overall effects were modest and effects between patients were highly variable, it is important to ascertain the underlying mechanism, particularly in the context of patient stratification based on general IFN responsiveness (that is, the extent of ISG upregulation from baseline levels). This report and others may hint at a potential role for IFNα in decreasing the size of the HIV-1-infected cellular reservoir, a highly desirable step with direct relevance for HIV-1 cure efforts95.
The detrimental roles of IFNs
In contrast to the studies reporting the beneficial roles of IFNs during acute SIV infection and the virological effects of IFN treatment during HIV-1 infection, several studies associate IFN signalling and stimulation of ISGs with progressive HIV or SIV-related disease. In the chronic phase of HIV-1 infection, a positive correlation between the plasma levels of IFNα and the plasma HIV-1 viral load has been reported, and there is an inverse correlation between the plasma levels of IFNα and CD4+ T cell counts96.
Numerous studies have characterized the gene expression profiles of CD4+ T cells isolated from patients who are infected with HIV-1 and who exhibit different viral loads and different rates of disease progression4–6. In general, patients with higher viral loads, increased rates of disease progression and later-stage HIV-1 infection have increasingly abnormal CD4+ T cell gene expression profiles. In addition to a pronounced increase in the expression of genes involved in the cell cycle, these studies show a relative overexpression of ISGs in patients with higher viral loads and faster rates of disease progression. These observations led to the hypothesis that chronic IFN signalling and generalized inflammation may be partially responsible for CD4+ T cell dysfunction and loss during chronic HIV-1 infection. According to this model, IFN signalling and other drivers of CD4+ T cell activation, such as viral antigens and microbial translocation, increase the available number of activated CD4+ T cells, which are the permissive substrates for productive viral infection, and thereby facilitate HIV-1 replication. This promotes the establishment of a detrimental and perpetual cycle of immune activation and viral replication, as well as cell death through various mechanisms, among which may be IFN-dependent apoptosis mediated by tumournecrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)97. Furthermore, IFNα can have inhibitory effects on cell-mediated immunity through the production of immunosuppressive cytokines such as interleukin-10 (IL-10) and the upregulation of ligands for inhibitory receptors such as programmed death ligand 1 (PDL1), and these effects may contribute further to the loss of HIV-1 immune control10,98–100. A cautionary interpretation of such studies that relate disease markers to both plasma IFNα levels and ISG expression is that activation of the IFN system in individuals with more advanced disease might simply be a consequence of increased viral replication, and that similar correlations could be made for many other pro-inflammatory or antiviral cytokines.
Observations from different NHP SIV models have been influential in suggesting a role for IFNα in the pathogenesis of HIV and SIV infection, particularly through the comparison of pathogenic models — those in which the SIV-infected animals generally succumb to the development of AIDS, such as SIVmac infection of macaques — with non-pathogenic models — those in which an SIV infection of a particular NHP host does not appear to cause disease, such as SIVagm infection of African green monkeys. As with HIV-1 infection of humans, IFNα levels correlate positively with viral load and negatively with CD4+ T cell count in pathogenic NHP models101. Furthermore, immune responses during the acute phase of SIV infection have been extensively investigated in pathogenic and non-pathogenic NHP models in order to identify immune signatures that could provide an explanation for the differences in subsequent disease outcomes102–104. Gene expression profiling of CD4+ T cells from blood and lymph nodes have reproducibly demonstrated the upregulation of ISGs in the acute phase of SIV infection in both pathogenic and non-pathogenic models. However, in the non-pathogenic models, the upregulation of ISGs and the expression of immune activation markers are reversed during the first few weeks post-infection, and subsequent viral replication occurs in the absence of further IFN stimulation or immune hyperactivation102–104. By contrast, in the pathogenic models, the upregulation of ISGs is maintained over time. Although the basis for this dichotomy remains poorly understood, these data suggest that sustained IFN-mediated stimulation could contribute to disease pathogenesis. Interestingly, administration of high-dose IFNαto African green monkeys (a non-pathogenic model) from days 9 to 24 post-infection does not impede the resolution of immune activation and the reversal of ISG induction seen during the primary infection105, suggesting that other inflammatory mediators may be responsible for the continuing ISG elevation and immune activation that are a hallmark of pathogenic infections.
In summary, there are reproducible associations between levels of IFNα in plasma, expression of ISGs in CD4+ T cells and disease progression in HIV-1 or SIV infection. In addition, the downregulation of ISG expression after the acute SIV infection appears to distinguish non-pathogenic from pathogenic infections of NHPs. Taken together, these findings point towards dysregulation of IFN signalling pathways in chronic persistent HIV or SIV-related disease, although a more causal role for type I IFNs in pathogenesis remains to be proved.
Conclusion and outlook
IFNα is capable of suppressing HIV-1 replication in infected patients, and an intact IFN response seems to be crucial for the control of acute lentiviral infection of primates and for control of subsequent disease. IFNs are thought to control HIV-1 replication principally through the upregulation of ISGs in the target cells for infection. In addition to the IFN-mediated increase in the expression of HIV-1 restriction factors, which may assist in overcoming viral evasion and escape mechanisms, the recently identified HIV-1 resistance factors MX2, SLFN11 and IFITMs may also be important for this response. However, several aspects of the role of IFNs during HIV-1 infection require further investigation, and future studies should aim to: identify pathways of HIV-1 detection in all infected cell types, including activated CD4+ T cells; determine the relevance of HIV-1 sensing in non-productively infected cells for pathogenesis; and define additional HIV-1-suppressive ISGs, such as those responsible for the early post-entry block to HIV-1 infection.
A more complete understanding of the in vivo mechanism (or mechanisms) of HIV-1 viral-load reduction during IFNα treatment is a central research focus, and there are several key questions relating to this that should be addressed. Which ISGs are important in vivo? Do other IFN-stimulated branches of the immune system, such as natural killer cells, B cells and T cells, contribute to these mechanisms? Does HIV-1 escape from these effector mechanisms, or is the host response limited by reversal of the type I IFN responses at the immunological level? Are type I IFNs capable of reducing HIV-1 DNA loads in CD4+ T cells, and if so, what is the underpinning biology? In addition, findings in NHP models indicate that pre-exposure prophylactic strategies that rely on the use of IFNs might be beneficial for controlling many viral infections, but future studies are needed to determine the optimal timing for such interventions.
In summary, there is a complex relationship between HIV-1 sensing by the innate immune system, the evocation of antiviral states, the pathways of viral evasion, and the induction and perpetuation of pathogenic cellular processes. Deciphering this relationship will require incisive molecular and translational research. We envisage that further elucidating the interplay between HIV-1 and the IFN system will yield broadly applicable principles for understanding many viral infections and will help to guide future intervention and therapeutic strategies.
Acknowledgements
The authors are funded by grants and fellowships from the UK Medical Research Council, the European Commission, the Wellcome Trust, the US National Institutes of Health, and the Department of Health via a UK National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s and St. Thomas’ National Health Service (NHS) Foundation Trust in partnership with King’s College London and King’s College Hospital NHS Foundation Trust.
Glossary
- Retrovirus
A positive-sense single-stranded RNA virus of tire family Retroviridae. These viruses replicate via a DNA intermediate that is synthesized by the reverse transcriptase enzyme. Retroviruses integrate their DNA into the host cell chromosome.
- Lentivirus
A particular genus of retroviruses that are primarily characterized by infections with long clinical incubation periods, often years to decades. Lentiviruses infect primate and non-primate hosts.
- Set-point plasma viral load
The semi-stable plasma level of HIV-1 RNA that is readied after the period of acute HIV-1 infection in most patients, in the absence of antiretroviral therapy
- Pattern recognition receptors
(PRRs). Germ line-encoded receptors that recognize the pathogen-associated molecular patterns which characterize pathogenic microorganisms.
- Pathogen-associated molecular patterns
(PAMPs). Biomolecules of diverse nature (ranging from lipopolysaccharides to forms of nucleic acids) that are characteristic of pathogenic microorganisms.
- Apoptosis
A mode of programmed cell death that leads to the elimination of the cell without the release of inflammatory mediators.
- Plasmacytoid dendritic cells
(pDCs). DCs that are specialized in the detection of microbial pathogens and the production of interferon-α (IFNα). pDCs are thought to be particularly important for HIV-1 sensing.
- Monocyte-derived dendritic cells
(MDDCs). DCs that have been derived by inducing their differentiation from primary monocytes in vitro.
- Monocyte-derived macrophages
(MDMs). Macrophages that have been derived by inducing their differentiation from primary monocytes in vitro.
- Reverse-transcription complexes
(RTCs). Complexes of viral nucleic acid, viral proteins (for example, reverse transcriptase) and cellular proteins that mediate viral DNA synthesis. RTCs are derived from viral capsids following virus entry into the cytoplasm during infection.
- Pyroptosis
A mode of programmed cell death that leads to the release of mediators of inflammation and that is often triggered by recognition of pathogenic microorganisms.
- Lymphoid aggregate cultures
Cultures composed of small blocks of lymphoid tissue usually derived from the tonsils or the spleen. This experimental system is used in an attempt to replicate the spatial organization and cytokine milieu of in vivo lymphoid tissue.
- Humanized mice
Mice that congenitally lack T cells, B cells and natural killer cells, and that are transplanted with human haematopoietic stem cells, leading to the reconstitution of a human-derived immune system.
- Nuclear pore complexes
Large protein complexes that form the channels in the nuclear envelope which allow the transport of molecules between the nucleus and the cytoplasm.
- Pegylated
Covalently conjugated to polyethylene glycol (PEG). This alters the pharmacokinetic behaviour of a drug, allowing the dosing frequency to be reduced in the case of interferon-α (IFNα).
- Transmitted/founder viruses
(T/F viruses). Viruses that are responsible for the establishment of initial HIV-1 infection and from which the viral population seen in later infection is thought to be derived.
- Highly active antiretroviral therapy
(HAART). A combination of antiretroviral drugs used to suppress HIV replication.
- Kaposi sarcoma
A common tumour associated with advanced HIV-1 infection. The tumour is caused by human herpesvirus 8 (HHV8) and presents as a purplish-brown vascular lesion either on the skin or in internal organs.
- HIV-1 eradication
Clearance of replication-competent HIV-1 from the body of an infected person. Achieving this goal is generally thought to require the inhibition of any ongoing HIV-1 replication during conventional antiretroviral therapy and the elimination of infected cells harbouring latent (or transcriptionally inactive) but replication-competent HIV-1. The term reservoir is generally used to denote the pool of latently infected cells.
- Microbial translocation
The emergence of microorganisms and microbial products, into the portal and systemic circulation from the gut, owing to compromise of the host gastrointestinal immune system.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Faria NR et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science 346, 56–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Arc M et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc. Natl Acad. Sci. USA 112, E1343–E1352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeuchi O & Akira S Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Rotger M et al. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6, e1000781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study characterizes the gene expression profile abnormalities of the CD4+ T cells of patients with HIV-1 infection according to plasma viral load.
- 5.Hyrcza MD et al. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 81, 3477–3486 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedaghat AR et al. Chronic CD4+ T-cell activation and depletion in human immunodeficiency virus type 1 infection: type I interferon-mediated disruption of T-cell dynamics. J. Virol. 82, 1870–1883 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelbi-Alix MK & Wietzerbin J Interferon, a growing cytokine family: 50 years of interferon research. Biochimie 89, 713–718 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Prokunina-Olsson L et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature Genet. 45, 164–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takaoka A & Yanai H Interferon signalling network in innate defence. Cell. Microbiol. 8, 907–922 (2006). [DOI] [PubMed] [Google Scholar]
- 10.McNab F, Mayer-Barber K, Sher A, Wack A & O’Garra A Type I interferons in infectious disease. Nature Rev. Immunol. 15, 87–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Veer MJ et al. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69, 912–920 (2001). [PubMed] [Google Scholar]
- 12.MacMicking JD Interferon-inducible effector mechanisms in cell-autonomous immunity. Nature Rev. Immunol. 12, 367–382 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoggins JW Interferon-stimulated genes: roles in viral pathogenesis. Curr. Opin. Virol. 6, 40–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoggins JW et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu SY, Sanchez DJ, Aliyari R, Lu S & Cheng G Systematic identification of type I and type II interferon-induced antiviral factors. Proc. Natl Acad. Sci. USA 109, 4239–4244 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoggins JW et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goubau D, Deddouche S & Reis e Sousa C Cytosolic sensing of viruses. Immunity 38, 855–869 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao D et al. Cyclic GMP–AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 341,903–906 (2013).This study identifies cGAS as an important sensor of HIV.
- 19.Monroe KM et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahaye X et al. The capsids of HIV-1 and HIV-2 determine immune detection of the viral cDNA by the innate sensor cGAS in dendritic cells. Immunity 39, 1132–1142 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Rasaiyaah J et al. HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jakobsen MR et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl Acad. Sci. USA 110, E4571–E4580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matreyek KA & Engelman A Viral and cellular requirements for the nuclear entry of retroviral preintegration nucleoprotein complexes. Viruses 5, 2483–2511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unterholzner L et al. IFI16 is an innate immune sensor for intracellular DNA. Nature Immunol. 11, 997–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doitsh G et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505, 509–514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doitsh G et al. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143, 789–801 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports that the majority of CD4+ T cells depleted from HIV-1-infected tissues are abortively infected resting cells that die via IFI16-dependent pyroptosis.
- 27.Beignon AS et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor–viral RNA interactions. J. Clin. Invest. 115, 3265–3275 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lepelley A et al. Innate sensing of HIV-infected cells. PLoS Pathog. 7, e1001284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blasius AL & Beutler B Intracellular Toll-like receptors. Immunity 32, 305–315 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Li G et al. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog. 10, e1004291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruel T et al. Plasmacytoid dendritic cell dynamics tune interferon-alfa production in SIV-infected cynomolgus macaques. PLoS Pathog. 10, e1003915 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kader M et al. Blocking TLR7- and TLR9-mediated IFN-α production by plasmacytoid dendritic cells does not diminish immune activation in early SIV infection. PLoS Pathog. 9, e1003530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsang J et al. HIV-1 infection of macrophages is dependent on evasion of innate immune cellular activation. AIDS 23, 2255–2263 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA & Lieberman J The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nature Immunol. 11, 1005–1013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manel N & Littman DR Hiding in plain sight: how HIV evades innate immune responses. Cell 147, 271–274 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertel T et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galao RP, Le Tortorec A, Pickering S, Kueck T & Neil SJ Innate sensing of HIV-1 assembly by tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe 12, 633–644 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galao RP, Pickering S, Curnock R & Neil SJ Retroviral retention activates a Syk-dependent HemITAM in human tetherin. Cell Host Microbe 16, 291–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neil S & Bieniasz P Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29, 569–580 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan N & Chen ZJ Intrinsic antiviral immunity. Nature Immunol. 13, 214–222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehy AM, Gaddis NC, Choi JD & Malim MH Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Stremlau M et al. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Laguette N et al. SAMHD1 is the dendritic-and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hrecka K et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neil SJ, Zang T & Bieniasz PD Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Van Damme N et al. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malim MH & Bieniasz PD HIV restriction factors and mechanisms of evasion. Cold Spring Harb.Perspect. Med. 2, a006940 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides an overview of HIV restriction factors.
- 48.Schaller T, Goujon C & Malim MH AIDS/HIV. HIV interplay with SAMHD1. Science 335, 1313–1314 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Goldstone DC et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480, 379–382 (2011). [DOI] [PubMed] [Google Scholar]
- 50.Lahouassa H et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nature Immunol. 13, 223–228 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ryoo J et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nature Med. 20, 936–941 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirchhoff F Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67 (2010). [DOI] [PubMed] [Google Scholar]
- 53.Sharp PM & Hahn BH Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu X et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science 302, 1056–1060 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Guo Y et al. Structural basis for hijacking CBF-β and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505, 229–233 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Neil SJ The antiviral activities of tetherin. Curr. Top. Microbiol. Immunol. 371, 67–104 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Diamond TL et al. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J. Biol. Chem. 279, 51545–51553 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pillai SK et al. Role of retroviral restriction factors in the interferon-α-mediated suppression of HIV-1 in vivo. Proc. Natl Acad. Sci. USA 109, 3035–3040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armitage AE et al. Conserved footprints of APOBEC3G on hypermutated human immunodeficiency virus type 1 and human endogenous retrovirus HERV-K(HML2) sequences. J. Virol. 82, 8743–8761 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wood N et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 5, e1000414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim EY et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 10, e1004281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albin JS & Harris RS Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev Mol. Med. 12, e4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neil SJ, Sandrin V, Sundquist WI & Bieniasz PD An interferon-α-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2, 193–203 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho DD et al. Recombinant human interferon alfa-A suppresses HTLV-III replication in vitro. Lancet 326, 602–604 (1985). [DOI] [PubMed] [Google Scholar]
- 65.Kornbluth RS, Oh PS, Munis JR, Cleveland PH & Richman DD Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J. Exp. Med. 169, 1137–1151 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shirazi Y & Pitha PM Alpha interferon inhibits early stages of the human immunodeficiency virus type 1 replication cycle. J. Virol. 66, 1321–1328 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meylan PR, Guatelli JC, Munis JR, Richman DD & Kornbluth RS Mechanisms for the inhibition of HIV replication by interferons-α, -β, and -γ in primary human macrophages. Virology 193, 138–148 (1993). [DOI] [PubMed] [Google Scholar]
- 68.Goujon C & Malim MH Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J. Virol. 84, 9254–9266 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheney KM & McKnight A Interferon-α mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS ONE 5, e13521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goujon C et al. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z et al. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410 (2013). [DOI] [PubMed] [Google Scholar]
- 72.Kane M et al. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 70–72 describe the anti-HIV-1 activities of MX2.
- 73.Melen K et al. Human MxB protein, an interferon-α-inducible GTPase, contains a nuclear targeting signal and is localized in the heterochromatin region beneath the nuclear envelope. J. Biol. Chem. 271, 23478–23486 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Goujon C et al. Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J. Virol. 88, 9017–9026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fricke T et al. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11, 68 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fribourgh JL et al. Structural insight into HIV-1 restriction by MxB. Cell Host Microbe 16, 627–638 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Compton AA et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 16, 736–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tartour K et al. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 11, 103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu J et al. The IFITM proteins inhibit HIV-1 infection. J. Virol. 85, 2126–2137 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li M et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491 , 125–128 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Alsharifi M, Mullbacher A & Regner M Interferon type I responses in primary and secondary infections. Immunol. Cell Biol. 86, 239–245 (2008). [DOI] [PubMed] [Google Scholar]
- 82.Sandler NG et al. Type I interferon responses in rhesus macaques prevent SIV infection and slow disease progression. Nature 511, 601–605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper highlights the importance of type I interferons in the control of acute SIV infection.
- 83.Stacey AR et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J. Virol. 83, 3719–3733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parrish NF et al. Phenotypic properties of transmitted founder HIV-1. Proc. Natl Acad. Sci. USA 110, 6626–6633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fenton-May AE et al. Relative resistance of HIV-1 founder viruses to control by interferon-a. Retrovirology 10, 146 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lane HC et al. Interferon-a in patients with asymptomatic human immunodeficiency virus (HIV) infection: a randomized, placebo-controlled trial. Ann. Intern. Med. 112, 805–811 (1990). [DOI] [PubMed] [Google Scholar]
- 87.Torriani FJ et al. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection in HIV-infected patients. N. Engl. J. Med. 351, 438–450 (2004). [DOI] [PubMed] [Google Scholar]
- 88.Neumann A et al. Differential antiviral effect of PEG-interferon-α-2b on HIV and HCV in the treatment of HIV/HCV co-infected patients. AIDS 21, 1855–1865 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Asmuth DM et al. Safety, tolerability, and mechanisms of antiretroviral activity of pegylated interferon alfa-2a in HIV-1-monoinfected participants: a Phase II clinical trial. J. Infect. Dis. 201, 1686–1696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study clearly characterizes the effect of IFNα therapy on HIV-1 plasma viral load in patients infected with HIV-1.
- 90.Hubbard JJ et al. Host gene expression changes correlating with anti-HIV-1 effects in human subjects after treatment with peginterferon alfa-2a. J. Infect. Dis. 205, 1443–1447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sarasin-Filipowicz M et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl Acad. Sci. USA 105, 7034–7039 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vanderford TH et al. Treatment of SIV-infected sooty mangabeys with a type-I IFN agonist results in decreased virus replication without inducing hyperimmune activation. Blood 119, 5750–5757 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Azzoni L et al. Pegylated interferon alfa-2a monotherapy results in suppression of HIV type 1 replication and decreased cell-associated HIV DNA integration. J. Infect. Dis. 207, 213–222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun H et al. Hepatitis C therapy with interferon-α and ribavirin reduces CD4 T-cell-associated HIV-1 DNA in HIV-1/hepatitis C virus-coinfected patients. J. Infect. Dis. 209, 1315–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barouch DH & Deeks SG Immunologic strategies for HIV-1 remission and eradication. Science 345, 169–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hardy GA et al. Interferon-α is the primary plasma type-I IFN in HIV-1 infection and correlates with immune activation and disease markers. PLoS ONE 8, e56527 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Herbeuval JP et al. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood 106, 3524–3531 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Douek DC, Roederer M & Koup RA Emerging concepts in the immunopathogenesis of AIDS. Annu. Rev. Med. 60, 471–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marshall HD, Urban SL & Welsh RM Virus-induced transient immune suppression and the inhibition of T cell proliferation by type I interferon. J. Virol. 85, 5929–5939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marshall HD & Kaech SM Regulating the diverse outcomes of interferon’s interference. Trends Immunol. 35, 353–354 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fraietta JA et al. Type I interferon upregulates Bak and contributes to T cell loss during human immunodeficiency virus (HIV) infection. PLoS Pathog. 9, e1003658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bosinger SE et al. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 119, 3556–3572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jacquelin B et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 119, 3544–3555 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harris LD et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J. Virol. 84, 7886–7891 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 102–104 identify the differences in the type I IFN responses of pathogenic and non-pathogenic NHP models following SIV infection.
- 105.Jacquelin B et al. Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-α during primary SIVagm infection. PLoS Pathog. 10, e1004241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Best S, Le Tissier P, Towers G & Stoye JP Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382, 826–829 (1996). [DOI] [PubMed] [Google Scholar]
- 107.Paolini R, Bernardini G, Molfetta R & Santoni A NK cells and interferons. Cytokine Growth Factor Rev. 10.1016/j.cytogfr.2014.11.003 (2014). [DOI] [PubMed] [Google Scholar]
- 108.Swann JB et al. Type I IFN contributes to NK cell homeostasis, activation, and antitumor function. J. Immunol. 178, 7540–7549 (2007). [DOI] [PubMed] [Google Scholar]
- 109.Zanoni I et al. IL-15 cis presentation is required for optimal NK cell activation in lipopolysaccharide-mediated inflammatory conditions. Cell Rep. 4, 1235–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Kolumam GA, Thomas S, Thompson LJ, Sprent J & Murali-Krishna K Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 202, 637–650 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bahl K, Huebner A, Davis RJ & Welsh RM Analysis of apoptosis of memory T cells and dendritic cells during the early stages of viral infection or exposure to Toll-like receptor agonists. J. Virol. 84, 4866–4877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Srivastava S, Koch MA, Pepper M & Campbell DJ Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J. Exp. Med. 211, 961–974 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Le Bon A et al. Type I interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 14, 461–470 (2001). [DOI] [PubMed] [Google Scholar]
- 114.Hervas-Stubbs S et al. Direct effects of type I interferons on cells of the immune system. Clin. Cancer Res. 17, 2619–2627 (2011). [DOI] [PubMed] [Google Scholar]


