Abstract
Quantifying fiber diameter is important for characterizing electrospun polymer scaffolds. Many researchers use manual measurement methods, which can be time-consuming and variable. Semi-automated tools exist, but there is room for improvement. The current work used Matlab to develop an image analysis program to quickly and consistently measure fiber diameter in scanning electron micrographs. The new Matlab method, termed “SIMPoly” (Semiautomated Image Measurements of Polymers) was validated by using synthetic images with known fiber size and was found to be accurate. The Matlab method was also applied by three different researchers to scanning electron microscopy (SEM) images of electrospun poly(lactic-co-glycolic acid) (PLGA). Results were compared with the semi-automated DiameterJ method and a manual ImageJ measurement approach, and it was found that the Matlab-based SIMPoly method provided measurements in the expected range and with the least variability between researchers. In conclusion, this work provides and describes SIMPoly, a Matlab-based image analysis method that can simply and accurately measure polymer fiber diameters in SEM images with minimal variation between users.
Impact statement
This article describes Semiautomated Image Measurements of Polymers (SIMPoly), a novel image analysis tool for measuring polymer fiber diameter in scanning electron micrographs of electrospun scaffolds. The Matlab-based SIMPoly method developed is a semi-automated tool that is quick, easy to use, and absent of significant user bias. This tool can be used to accurately measure electrospun polymer fiber diameters, an important characteristic for characterizing tissue-engineered scaffolds.
Keywords: polymer fiber measurement, electrospun scaffolds, Matlab tool, electrospinning
Introduction
Electrospinning is a processing technique used to form micro- and nano-scale fibers of various materials through the application of electrostatic force. The most common implementation of electrospinning in tissue engineering involves the collection of polymer fibers onto a conductive surface after they are drawn out from a solution, or less commonly from a melt.1,2 The solution is expelled from a syringe through a conductive needle charged via a high-voltage power supply and directed toward the grounded conductive surface located some controlled distance from the needle tip. The polymer solution is drawn into a fine fiber as it travels between the needle tip and the grounded surface. The resulting fibrous, porous structure can then be used as a scaffold in a tissue-engineered construct.1–3
Electrospinning has become an increasingly popular fabrication technique in the field of tissue engineering for a variety of reasons, many of which stem from its high degree of tailorability. Fiber and pore characteristics, as well as properties of the bulk scaffold, can be altered by modifying the numerous parameters of the electrospinning process.4 The ability to tailor these properties allows one to match the mechanical properties of native tissue more closely, while still retaining favorable micro- and nano-scale morphology to facilitate cell interaction and proliferation.2,5–7 Further, material selection as part of the electrospinning process can yield a variety of scaffold morphologies and mechanical properties, and it may also be used to tailor scaffold performance to a specific application.2
Controlling fiber size is of particular importance for scaffolds in tissue engineering, as fiber size has been shown to have strong correlations with cell adhesion, growth, and differentiation within a construct.8–14 For example, in vascular applications, total cell count and cell coverage has been observed to increase significantly in scaffolds with sub-micron fiber diameters compared with scaffolds with fiber diameters above 1 μm.8–11 In neural tissue engineering, researchers have shown that cell adhesion, migration, and proliferation are reduced as average fiber diameter increases.12,13 Fiber orientation and alignment within a construct have also been shown to facilitate cell interactions.14 Accurate characterization of fiber morphology on the micro- and nanoscale is, thus, critical in producing desired and consistent tissue scaffolds.15–18
A variety of methods exist to quantify fiber size of an electrospun scaffold. Many laboratories, including ours, use image processing programs such as ImageJ to manually measure the diameter of fibers observed in scaffold images taken with scanning electron microscopy (SEM).19,20 However, manual measurement techniques can be time-consuming and subject to user bias. Potential bias may affect results from a given user across multiple measuring sessions, and affect measurements taken by multiple users, rendering comparison of results between users difficult.20 In addition, the time-consuming nature of manual measurements often means that relatively few images or fibers are actually evaluated.20 Attempts to address these issues include automating some or all of the image analysis process.21–23
One well-known example of semi-automation is the open-source ImageJ plugin called DiameterJ. DiameterJ was established by Hotaling et al. to analyze SEM images of nanofibers to determine fiber diameter within 60 s. DiameterJ is able to identify fibers and take several hundred measurements within a given SEM micrograph, providing a much larger sample size in significantly less time than manual measurement methods. Validation of DiameterJ was performed on digital synthetic images of white lines on a black background and SEM images of monodispersed steel wires. DiameterJ was shown to produce results not statistically different from known values and was found to be significantly faster compared with manual image analysis.21 However, DiameterJ involves a critical user-dependent intermediate step where an appropriately segmented image is selected, which may introduce bias or variation. Further, the initial learning curve for DiameterJ can be steep: Learn DiameterJ, an online training module for the plugin, takes ∼14 h to cover all of the features included in DiameterJ.24
The creation, validation, and impact of DiameterJ supports the need for such tools and suggests that it would be beneficial to establish additional techniques that potentially address the challenges mentioned earlier. It would be ideal to create a semi-automated method that can analyze fiber diameter without any bias or variation; the goal of the current work was to create such a tool. Matlab is a programming platform built for engineers and researchers to quickly perform complex mathematical operations. The applications of Matlab include machine learning, signal processing and communications, image and video processing, and computational biology.25 Matlab was, therefore, chosen for this application as it runs quickly on most computers, contains sophisticated image processing tools, and is familiar to many engineers and researchers.
In summary, there is a common need to quantify fiber diameters of electrospun scaffolds. These scaffolds are often used in the tissue engineering field, and fiber diameter has been shown to impact everything from initial cell adhesion to mechanical properties and performance of the construct. Many researchers use manual image analysis methods to measure fiber diameters, which can be time-consuming and often introduces user bias or variation. Tools such as DiameterJ have been developed and widely utilized, but there is room for new or improved options. The two goals of the current work were to (1) develop a new semi-automated image analysis method using Matlab, and (2) compare the new method with a manual ImageJ analysis technique and the semi-automated DiameterJ approach. This comparison was performed by using image sets from the publication by Hotaling et al.
Methods
To develop and validate the Matlab algorithm, two different image sets were utilized from the Hotaling et al. DiameterJ publication. These included digital synthetic images with known pixel diameters and SEM images of electrospun polymeric nanofibers. These image sets were used during the development of the Matlab method and for the comparison across different measurement methods.
Digital synthetic images
A set of 41 digital synthetic images of white lines with known pixel diameters on a black background was used to initially develop and validate the Matlab algorithm. The white line diameters spanned 10 px to 100 px in diameter, with half of the images being ordered, straight lines and the other half disordered lines. These 41 images were published and provided by Hotaling et al. for the validation of DiameterJ,26 and they were downloaded for use in the current work. This image set was selected, as it was the main validation method for DiameterJ.
Poly(lactic-co-glycolic acid) nanofiber images
An image set of nanofiber poly(lactic-co-glycolic acid) (PLGA) samples was published by Hotaling et al. for the validation of DiameterJ. This image set was also downloaded from the data provided by Hotaling et al.26 Their electrospun polymeric samples were composed of PLGA dissolved in hexafluoroisopropanol, and their images were taken by using a Hitachi S4700 SEM.
Semiautomated Image Measurements of Polymers Matlab algorithm measurement approach
The Matlab algorithm was developed to acquire the SEM micrograph, measure the scale bar, resize the image to remove the SEM information, enhance the image, morphologically reconstruct the image, skeletonize the image, and analyze the fiber diameter. Each of these steps is described later. The resulting algorithm was termed “SIMPoly” (Semiautomated Image Measurements of Polymers), based on its simplicity for users and application to polymer fiber diameters.
Image acquisition, scale bar measurement, and image resize
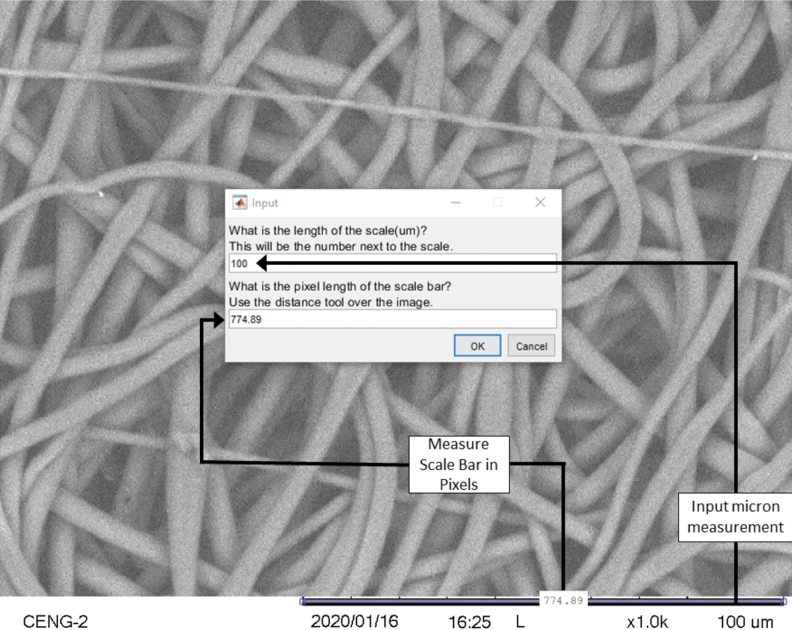
Once the SEM image has been selected, the first step for image analysis is to determine the micrometer to pixel conversion ratio. This information is required to determine the image's fiber diameter in micrometers, rather than pixels. If the conversion ratio (micrometers/pixels) is known, it can be entered manually. If the conversion ratio is unknown, it can be measured within the Matlab algorithm by using the built-in measurement tool. Figure 1 illustrates this built-in measurement tool. If the image has no scale bar, no ratio will be used and the output will be in pixels. Although pixel length should be an integer, the internal Matlab distance measuring tool measures between pixels, the outputted pixel length may contain decimals. After the scale bar has been measured, the image is resized to remove any SEM information.
FIG. 1.
The SIMPoly algorithm includes an initial built-in step that uses the SEM micrograph scale bar to convert pixels to micrometers. This allows fiber diameter measurements to be output in microns. If the micrometer/pixel ratio is already known, it can be entered without measurement of the scale bar, or if it is unknown but there is no scale bar, the output will be in pixels. SEM, scanning electron microscopy; SIMPoly, Semiautomated Image Measurements of Polymers.
Image enhancement, morphological reconstruction, and skeletonization
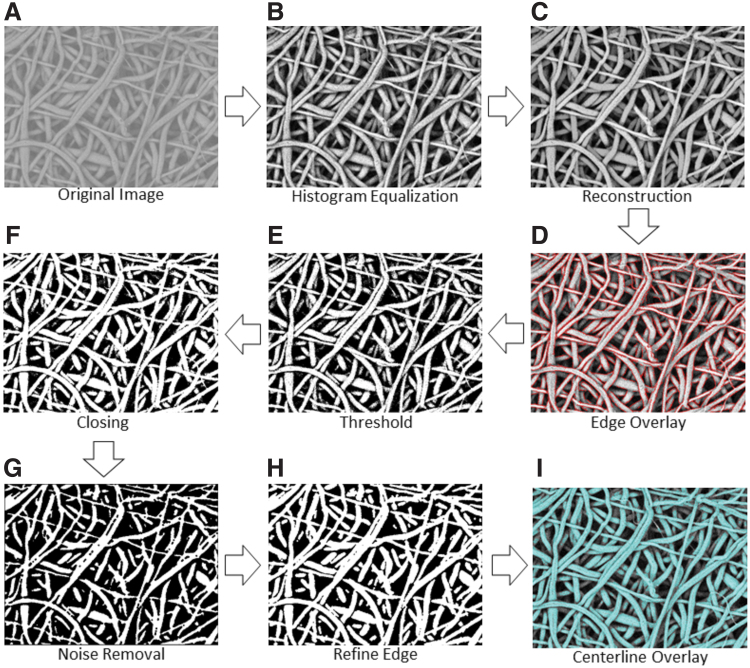
The next step for fiber diameter analysis is to enhance, reconstruct, and skeletonize the selected SEM micrograph. Figure 2 displays the workflow for these steps in the algorithm. The image's contrast is enhanced by using locally adaptive histogram equalization (Fig. 2B). Next, the image is morphologically reconstructed to minimize noise for improved thresholding results in the subsequent steps (Fig. 2C). The edge of each fiber is found by using a Canny detector and overlaid in red for later background noise removal (Fig. 2D). Next, the image is binarized with a global threshold by using Otsu's method, which minimizes the intraclass variance of the thresholded black and white pixels. The binary image is created with a filter that removes any background noise below a certain value (Fig. 2E). The binary image then undergoes several morphological operations to remove noise pixels and minimize analysis error. Morphological closing is used to mitigate noise and imperfections from thresholding by filling holes and sharpening edges within the fibers (Fig. 2F).
FIG. 2.
Diagram of the overall workflow in the SIMPoly fiber analysis algorithm. Starting with an original image (A), image processing steps include contrast enhancement through histogram equalization (B), reconstruction to brighten desired fibers (C), edge overlay for later background noise removal (D), thresholding for initial removal of background noise (E), image dilation and erosion for “closing” to correct any gaps created from previous filtering step (F), median filter application to smooth fiber edges and remove any leftover noise (G), dilation of fibers to brighten edges to normal size without presence of background noise (H), and skeletonization of image and overlay of skeleton onto the binary image (I). Color images are available online.
Multiple iterations of median filtering are performed to further clean the binary image and smooth fiber edges (Fig. 2G). Fibers that are not clearly represented are removed, and the fibers that remain are cleaned up. The image is then dilated to thicken the remaining fibers, as subsequent cleaning will have thinned all fibers, leading to an underestimation of the true diameters (Fig. 2H). In an additional noise removal step, the previous edge overlay information (Fig. 2D) is used to remove any noise a certain distance away from any edge reading. The fiber centerline is estimated by using an axial thinning algorithm that skeletonizes the image by reducing all fiber midlines to 1-pixel wide lines. The skeletonized image is overlaid over the binary image (Fig. 2I). This centerline overlaid image (Fig. 2I) contains the histogram equalized image (Fig. 2B) overlaid with the skeletonized centerline in red and the segmented image in blue. The distance between each pixel on the midline of each fiber and the nearest non-zero pixel is multiplied by 2 to cover the entire diameter.
Fiber diameter analysis, histogram, and color map
The measured diameters are plotted on a histogram, and a Gaussian model is fit over the data. The centroid location is used to determine the average diameter, and the peak width is used to determine standard deviation. The algorithm was designed to output several image steps (including the raw image, segmented image, and a centerline overlaid image) (Fig. 2I), as well as quantitative results of image analysis, a histogram with Gaussian fit, and a color map. The raw and segmented images can be used for side-by-side comparison to allow the user to confirm the accuracy of the segmentation if desired. Further, the centerline overlaid image can be used to directly compare the accuracy of the segmentation. If the edge of the segmented blue fiber is continuous with the original fiber, then the diameter analysis is likely accurate. The quantitative results include the average fiber diameter and standard deviation in pixels and microns. If there is no scale bar in the image, only pixel measurements are output. The histogram displays the frequency of each fiber diameter throughout the image. The color map displays the distribution of fiber diameters based on standard deviation overlaid onto the image. The color map is a visual representation of the range of diameters present in the image.
Once the Matlab algorithm, SIMPoly, was created with the tools and approach outlined earlier, it was used to analyze the digital synthetic and polymer nanofiber image sets (see SIMPolyMatlabCode.m for final code). One researcher measured each of the digital synthetic images. Due to the digital synthetic images having no scale bar, the image set had no point of measurement bias or variation. As a result, a single researcher was able to take the synthetic image measurements. In addition, three researchers individually measured every image from the PLGA image set by using the new Matlab-based SIMPoly tool. All results were recorded and used for analysis and comparison against the manual and DiameterJ measurement approaches.
Manual measurement approach
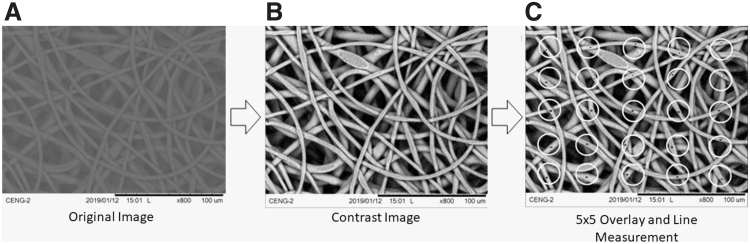
To compare outputs from the Matlab-based SIMPoly method, fiber diameters for the PLGA nanofiber image set were measured manually by using the Line tool in ImageJ. Figure 3 illustrates the workflow for manual measurement of SEM micrographs. First, the image was selected and opened in ImageJ (Fig. 3A). Using the line tool, the scale bar for the SEM micrograph was measured in pixels. Using this measurement, the micrometer-to-pixel ratio was inputted into the scale function of ImageJ to convert all future line measurements into micrometers. The image was enhanced by increasing the number of saturated pixels to 15% of those present in the image (Fig. 3B).
FIG. 3.
Workflow for manual fiber diameter measurements using ImageJ. Original SEM images were selected and opened in ImageJ (A) and enhanced (B). Then, a 5 × 5 circle template was overlaid onto each image and 25 fiber diameter measurements were made by using the line tool to manually measure the fiber closest to the middle of each circle (C).
Next, a 5 × 5 circle template was overlaid onto each image (circle diameter ∼125 pixels) and 25 fiber diameter measurements were made per image by selecting the fiber closest to the middle of each circle that was fully visible (Fig. 3C). Three researchers experienced in this practice performed 25 fiber diameter measurements for each micrograph in the PLGA nanofiber image set. The 25 values taken by each researcher were averaged for each image to determine the researcher's average fiber diameter. The averages from each researcher were used for the comparison to the SIMPoly and DiameterJ measurement methods. In the same way that DiameterJ was validated, manual measurement was not seen as the “source of truth” for the images; however, the comparison was made, as manual measurements are the most common source of measurements in literature for SEM images.20 These measurements were also used to compare the variation between researchers for the three measurement methods.
DiameterJ measurement approach
Using the ImageJ plugin DiameterJ, the PLGA nanofiber image sets were analyzed by three researchers using the fiber diameter Histogram method. The Histogram method was chosen in DiameterJ, over the Super Pixel method, due to Hotaling et al.'s recommendation to use the Histogram algorithm with peak fitting for most cases.21 Once each SEM micrograph was opened in ImageJ, the scale bar was measured by using the line tool in ImageJ to determine the micrometer-to-pixel ratio for the SEM micrograph. Next, the DiameterJ plugin was opened within ImageJ and the folder containing the designated micrographs was selected for segmentation. The segmented image folder was opened, and each researcher used their best discretion to select the “best” segmented image based on its similarity to the original in terms of features displayed. Once the “best” segmented image was selected, the DiameterJ 1-018 Histogram analysis was run, and the median and standard deviation fiber diameter information was recorded. Three researchers measured each SEM micrograph in the PLGA nanofiber image set using DiameterJ. The diameter measurements were averaged between researchers for each image, and the data were used for analysis and comparison to the SIMPoly and manual methods. These measurements were also used to compare the variation between researchers for the three measurement methods. For more information on the specifics of the DiameterJ measurement approach, see “DiameterJ: A Validated Open Source Nanofiber Diameter Measurement Tool” by Hotaling et al.21
Data analysis
For the synthetic images, percent error and uncertainties were calculated by comparing the measured fiber diameters with the known values. For the PLGA images, where no “true” value was known, the percent difference was calculated between each researcher for each image and these values were averaged, resulting in a single percent difference for each image. These six values were averaged for each measurement method to obtain an average percent difference between users for each measurement method.
Experiment
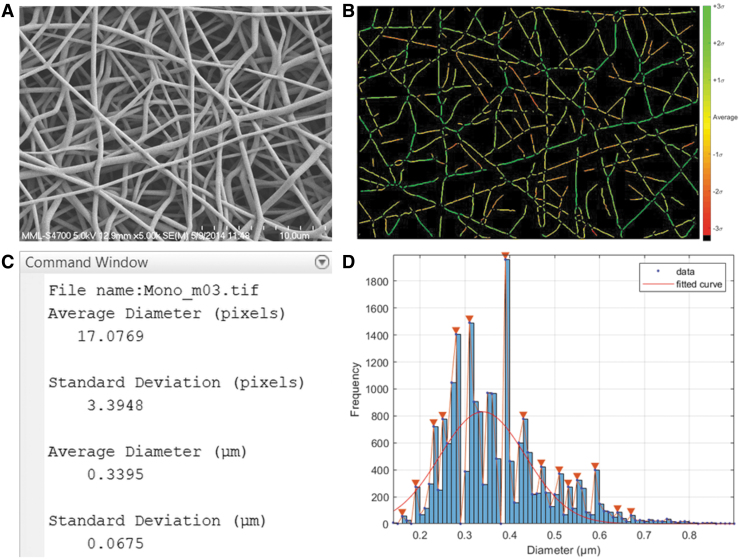
SIMPoly, the new Matlab-based method, was successfully developed such that it could accurately analyze fiber diameter with minimal bias or variation. The program outputs several image steps, as well as the quantitative fiber diameter results, a histogram, and a standard deviation color map. Figure 4 displays an example output of the SIMPoly algorithm from one of the PLGA images, including the original SEM image for comparison (Fig. 4A), a color map displaying distribution of fibers based on standard deviation (Fig. 4B), average fiber diameter measurements and standard deviation (Fig. 4C), and a histogram of all measured fibers (Fig. 4D).
FIG. 4.
Example output of the SIMPoly algorithm after analyzing a PLGA nanofiber image. (A) Original SEM Image. (B) Color map displaying distribution of fibers based on standard deviation. (C) Command Window displaying average diameter and standard deviation of fiber diameter measurements in pixels and microns. (D) Histogram of measured fibers with Gaussian curve fit over data. PLGA, poly(lactic-co-glycolic acid). Color images are available online.
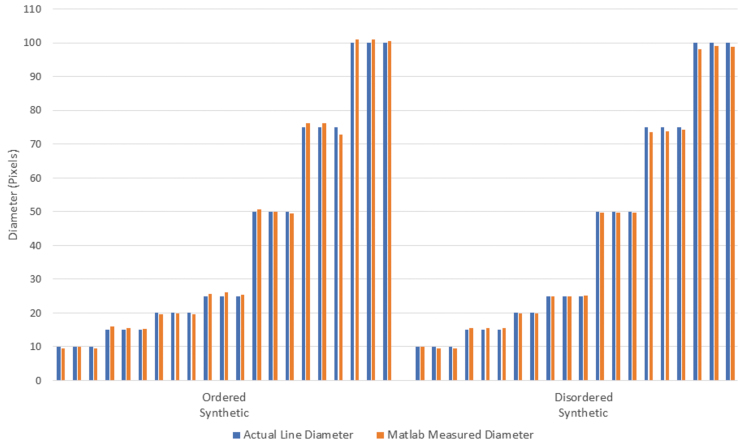
To validate the accuracy of the Matlab-based SIMPoly method, fiber diameter measurements were obtained from the synthetic image sets and the measured diameters were compared with the known diameters. The measured diameters, displayed in Figure 5, all fell within 10% of the known diameters. As seen in Table 1, the average percent error between the measured and known diameters was 2.1% ± 1.7% for the ordered synthetic images and 1.6% ± 1.5% for the disordered synthetic images, where results are given as percent error plus or minus the uncertainty. For reference, the average percent error between measured and known diameters published in the DiameterJ paper was 2.5% ± 1.9% for the ordered synthetic images and 4.7% ± 1.4% for the disordered synthetic images.21
FIG. 5.
Bar graph displaying the actual diameter versus the diameter measured by using the SIMPoly method for each synthetic image. The average percent error between the measured and known diameters was 2.1% for the synthetic ordered images, and 1.6% for the synthetic disordered images. Color images are available online.
Table 1.
Percent Error of Measured Diameters Compared with Known Diameters for Ordered and Disordered Synthetic Images
| SIMPoly average percent error | Published diameterJ average percent error21 | |
|---|---|---|
| Ordered synthetic | 2.1% ± 1.7% | 2.5% ± 1.9% |
| Disordered synthetic | 1.6% ± 1.5% | 4.7% ± 1.4% |
Measured SIMPoly percent error and published DiameterJ percent errors with uncertainties are included.
SIMPoly, Semiautomated Image Measurements of Polymers.
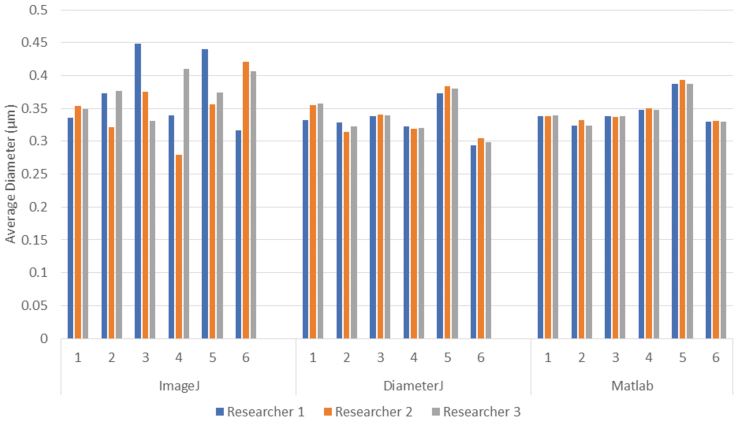
The fiber diameters for PLGA images were measured by using the manual ImageJ method, DiameterJ, and the SIMPoly method. Plotting the data for each image and each approach revealed that the SIMPoly method resulted in fiber diameter measurements in the expected range, as compared with the measurements from the manual ImageJ and DiameterJ methods (Fig. 6). These data also revealed more consistency between researchers for each image by using the SIMPoly method. The average percent difference between users was 15.39% for the manual ImageJ method, 2.23% for the DiameterJ method, and 0.73% for the SIMPoly method, illustrating that user variability was highest for the manual ImageJ method and lowest for the SIMPoly method.
FIG. 6.
Bar graph displaying the average fiber diameter measurements made by each researcher using each of the three analysis methods (manual ImageJ, DiameterJ, and Matlab-based SIMPoly) for the PLGA nanofiber images. Although there is no known “true” fiber size for these samples, the new Matlab-based SIMPoly method provided appropriate fiber diameter measurements, as compared with the more well-established manual and DiameterJ approaches, and the SIMPoly method had the least variation between researchers. Color images are available online.
Discussion
The aim of this work was to develop and validate a semi-automated image analysis method with Matlab that is quick, easy to use, and absent of significant user bias. The Matlab method was developed and validated by using image sets from the Hotaling et al. publication, including a set of synthetic images of known fiber size and a set of SEM images of electrospun PLGA. The new Matlab-based method was termed SIMPoly.
The SIMPoly algorithm was first validated by using the ordered and disordered synthetic images with known pixel diameters. It was shown that the SIMPoly method could accurately measure the pixel diameter within a 10% error between the known diameter for all synthetic images. The SIMPoly method was also compared against manual ImageJ and DiameterJ methods for quantifying fiber diameters of PLGA nanofibers, and results illustrated that the SIMPoly output was in the expected range and had less variability between researchers.
The new Matlab tool, SIMPoly, possesses several significant advantages, including its simplicity, speed, and lack of user bias. The SIMPoly method requires only two steps to quantify an image's fiber diameter: selecting the image and, if appropriate, measuring the scale bar. In contrast, the DiameterJ program has ∼10 steps required per image. An image can be fully quantified by the SIMPoly method in around 15 s, with the generation of around 4500 to 7000 fiber diameter counts per image. In comparison, the manual ImageJ method has been shown to take ∼100 s to yield 10s of measurements and the DiameterJ program takes ∼60 s to yield thousands of measurements.21
With regard to the outputted measurements, the SIMPoly and DiameterJ methods performed similarly in the current work. However, it is worth noting that, as seen in Figure 6, there was an ∼20 nm difference for image 4 and an ∼25 nm difference for image 6 between the SIMPoly and DiameterJ methods. Image 4 has a number of small background fibers that the SIMPoly algorithm removed during segmentation, thus slightly increasing the diameter. Image 6 is at a slightly higher magnification than the other images, which could have resulted in the higher variation due to a smaller amount of fibers to measure. However, the measurements for these images were still all within the expected diameter range determined by using the manual method and were more consistent between researchers.
In terms of consistency and repeatability between researchers, the SIMPoly method has a single point of potential user bias, which is the measurement of the scale bar. However, this point of user bias is very limited, which is why the variation between users was lowest for the SIMPoly method at 0.73%, compared with 15.39% and 2.23% for the manual ImageJ and DiameterJ methods, respectively. The DiameterJ program requires the user to select the best segmented image for further analysis, whereas the SIMPoly method automates this step. Further, the SIMPoly method offers the ability for the scale bar to be measured directly within the program (Fig. 1) and displays a color map (Fig. 4B) that gives a visual representation of the standard deviation of the fibers.
The SIMPoly method was only validated to analyze nonconductive polymer samples. Conductive materials, such as metal, imaged under SEM were not included in this work. Therefore, researchers should perform their own validation of the SIMPoly method before measuring any conductive material images. It should be noted that small specks of dust or debris seen on very high-quality SEM images of conductive materials could interfere with the measurement of the image. However, given the prevalence of nonconductive polymers for electrospun scaffolds, and the commonality of SEM images that have similar resolutions to the PLGA samples utilized for this work, there are a wide range of research groups and scaffold types that could benefit from this new tool. Further, the SIMPoly method has the potential to measure natural matrices, such as collagen, elastin, or other extracellular matrices.
A limitation of the SIMPoly method is that it does not replicate all the potential capabilities of DiameterJ. DiameterJ provides mesh hole analysis as well as fiber orientation analysis,21 whereas our new SIMPoly method was solely developed for fiber diameter measurement. DiameterJ was also validated for samples with unimodal or multimodal distributions, whereas the SIMPoly method has only been validated for samples with unimodal distribution. Future work can include testing and potentially expanding the code to accommodate bimodal images.
An additional limitation of this method is the cost of the required Matlab software. DiameterJ and ImageJ are free programs, whereas Matlab must be purchased to run the SIMPoly algorithm. Although many universities, research institutions, and companies have access to Matlab, if a researcher did not already have access to the program, it could cost as much as $2000 to purchase a license. For researchers that do not have access to Matlab, or for those unfamiliar with the Matlab software, DiameterJ may still be a preferred tool. However, many undergraduate engineering programs are increasingly incorporating Matlab coursework into their curriculum. As a result, not only do many researchers have access, but also labs with engineering backgrounds and/or experience with Matlab may prefer the Matlab method over DiameterJ due to its speed, lack of a user selection step, and ease of use.
Researchers who choose to implement the SIMPoly Matlab-based tool should note that it only uses a Gaussian fit to determine fiber diameter. Although a Gaussian fit is a good choice for the majority of polymer fiber images, it is not the best approach for every single image. Future work or project-specific implementation could include expanding the program to include additional measurement approaches. In addition, the SIMPoly method is limited to fibers with at least 10 px and less than 100 px should be effectively used. For example, if the smallest fiber in the image is 1 μm, SEM imaging should be conducted at a magnification of at least 1500 × . DiameterJ is limited to the same specifications for image analysis.21 However, despite these specifications, the SIMPoly method does provide the primary function of accurately and efficiently measuring fiber diameters and can be applied to a large range of image magnifications.
As labs or researchers consider implementing the SIMPoly method, it will be helpful that Matlab is an open-source program that provides the opportunity for researchers to modify it to fit their needs. Although the current work clearly provides and describes the new SIMPoly tool and demonstrates its accuracy and consistency with the previously published datasets, future work can look at applying the tool to new images. These images could include micrographs taken from a variety of different SEMs as well as polymers of varying types and sizes. In our own research lab, we use the SIMPoly method to characterize electrospun PCL scaffolds (Supplementary Figs. S1 and S2). When researchers first apply the SIMPoly method to their images, it will be important to validate it by using known values or outputs from their existing methods. It is recommended that researchers initially validate the code by either performing their existing internal “gold standard” approach for comparison or using the manual measurement approach described in Methods using the 5 × 5 ImageJ grid. A researcher should apply the known or manual method on at least three images within their image set. Once they determine the SIMPoly method is measuring the correct diameter, they can move forward with using the method on the rest of their images.
Conclusion
The new Matlab-based method, SIMPoly, presented in this article is a simple, modifiable, image analysis tool for measuring polymer fiber diameter in scanning electron micrographs. The method was validated in this article by using synthetic fiber images and was applied by researchers to evaluate electrospun PLGA polymer fibers. The method was shown to be quick and accurate, without significant bias or variation between users.
Supplementary Material
Authors' Contributions
R.M.: Conceptualization, Methodology, Software, Investigation, Writing—Original Draft, Writing—Review and Editing, Visualization; A.T.: Formal analysis, Investigation, Validation, Data Curation, Writing—Review and Editing, Visualization; L.B.: Software, Methodology, Writing—Review and Editing; E.D.: Conceptualization, Writing—Original Draft, Writing—Review and Editing; B.G.: Investigation, Writing—Review and Editing; K.O'H.C.: Conceptualization, Supervision, Project administration, Writing—Original Draft, Writing—Review and Editing.
Data Availability
The raw and processed data required to reproduce these findings are available to download from
Disclosure Statement
No competing financial interests exist.
Funding Information
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Supplementary Material
References
- 1. Bhardwaj N., and Kundu S.C.. Electrospinning: a fascinating fiber fabrication technique. Biotechnol Adv 28, 325, 2010 [DOI] [PubMed] [Google Scholar]
- 2. Awad N.K., Niu H., Ali U., Morsi Y.S., and Lin T.. Electrospun fibrous scaffolds for small-diameter blood vessels: a review. Membranes (Basel) 8, 15, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Khorshidi S., Solouk A., Mirzadeh H., et al. A review of key challenges of electrospun scaffolds for tissue-engineering applications. J Tissue Eng Regen Med 10, 715, 2016 [DOI] [PubMed] [Google Scholar]
- 4. Li Z., and Wang C.. One Dimensional Nanostructures Electrospinning Technique and Unique Nanofibers. One-Dimensional Nanostructures. Berlin, Heidelberg: Springer, 2013, p. 1 [Google Scholar]
- 5. Pektok E., Nottelet B., Tille J.C., et al. Degradation and healing characteristics of small-diameter poly(ɛ-caprolactone) vascular grafts in the rat systemic arterial circulation. Circulation 118, 2563, 2008 [DOI] [PubMed] [Google Scholar]
- 6. Bergmeister, H., Grasl, C., Walter, I., et al. Electrospun small-diameter polyurethane vascular grafts: ingrowth and differentiation of vascular-specific host cells, Artif Organs 36, 54, 2012 [DOI] [PubMed]
- 7. Mrówczyński W., Mugnai D., De Valence S., et al. Porcine carotid artery replacement with biodegradable electrospun poly-e-caprolactone vascular prosthesis. J Vasc Surg 59, 210, 2014 [DOI] [PubMed] [Google Scholar]
- 8. Whited B.M., and Rylander M.N.. The influence of electrospun scaffold topography on endothelial cell morphology, alignment, and adhesion in response to fluid flow. Biotechnol Bioeng 111, 184, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keun Kwon I., Kidoaki S., and Matsuda T.. Electrospun nano- to microfiber fabrics made of biodegradable copolyesters: structural characteristics, mechanical properties and cell adhesion potential. Biomaterials 26, 3929, 2005 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Ji Y., Ghosh K., Clark R.A.F., Huang L., and Rafailovich M.H.. Effects of fiber orientation and diameter on the behavior of human dermal fibroblasts on electrospun PMMA scaffolds. J Biomed Mater Res 90, 1092, 2009 [DOI] [PubMed] [Google Scholar]
- 11. Xu C.Y., Inai R., Kotaki M., and Ramakrishna S.. Aligned biodegradable nanofibrous structure: a potential scaffold for blood vessel engineering. Biomaterials 25, 877, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Yoshimoto H., Shin Y.M., Terai H., and Vacanti J.P.. A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials 24, 2077, 2003 [DOI] [PubMed] [Google Scholar]
- 13. Christopherson G.T., Song H., and Mao H.Q.. The influence of fiber diameter of electrospun substrates on neural stem cell differentiation and proliferation. Biomaterials 30, 556, 2009 [DOI] [PubMed] [Google Scholar]
- 14. Wu H., Fan J., Chu C.C., and Wu J.. Electrospinning of small diameter 3-D nanofibrous tubular scaffolds with controllable nanofiber orientations for vascular grafts. J Mater Sci Mater Med 21, 3207, 2010 [DOI] [PubMed] [Google Scholar]
- 15. Noriega S.E. Hasanova, G.I., Schneider, M.J., Larsen, G.F., and Subramanian, A. Effect of fiber diameter on the spreading, proliferation and differentiation of chondrocytes on electrospun chitosan matrices. Cells Tissues Organs 195, 207, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Badami A.S., Kreke M.R., Thompson M.S., Riffle J.S., and Goldstein A.S.. Effect of fiber diameter on spreading, proliferation, and differentiation of osteoblastic cells on electrospun poly(lactic acid) substrates. Biomaterials 27, 596, 2006 [DOI] [PubMed] [Google Scholar]
- 17. Lowery J.L., Datta N., and Rutledge G.C.. Effect of fiber diameter, pore size and seeding method on growth of human dermal fibroblasts in electrospun poly(ɛ-caprolactone) fibrous mats. Biomaterials 31, 491, 2010 [DOI] [PubMed] [Google Scholar]
- 18. Cui W., Li X., Zhu X., Yu G., Zhou S., and Weng J.. Investigation of drug release and matrix degradation of electrospun poly(DL-lactide) fibers with paracetanol inoculation. Biomacromolecules 7, 1623, 2006 [DOI] [PubMed] [Google Scholar]
- 19. Herting S., DiBartolomeo A., Pipes T., et al. Human umbilical versus coronary cell sources for tissue-engineered blood vessel mimics. Appl Vitr Toxicol 2, 175, 2016 [Google Scholar]
- 20. Narayanan G., Tekbudak M.Y., Caydamli Y., Dong J., and Krause W.E.. Accuracy of electrospun fiber diameters: the importance of sampling and person-to-person variation. Polym Test 61, 240, 2017 [Google Scholar]
- 21. Hotaling N.A., Bharti K., Kriel H., and Simon C.G.. DiameterJ: a validated open source nanofiber diameter measurement tool. Biomaterials 61, 327, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziabari M., Mottaghitalab V., and Haghi A.K.. Application of direct tracking method for measuring electrospun nanofiber diameter. Brazilian J Chem Eng 26, 53, 2009 [Google Scholar]
- 23. Tomba E., Facco P., Roso M., Modesti M., Bezzo F., and Barolo M.. Artificial vision system for the automatic measurement of interfiber pore characteristics and fiber diameter distribution in nanofiber assemblies. Ind Eng Chem Res 49, 2957, 2010 [Google Scholar]
- 24. Learn DiameterJ, (n.d.). https://sites.google.com/site/diameterj/ (accessed September3, 2020)
- 25. MATLAB—MathWorks—MATLAB & Simulink, (n.d.). https://www.mathworks.com/products/matlab.html (accessed September3, 2020)
- 26. Hotaling N.A., Bharti K., Kriel H., and Simon C.G.. Dataset for the validation and use of DiameterJ an open source nanofiber diameter measurement tool. Data Br 5, 13, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data required to reproduce these findings are available to download from