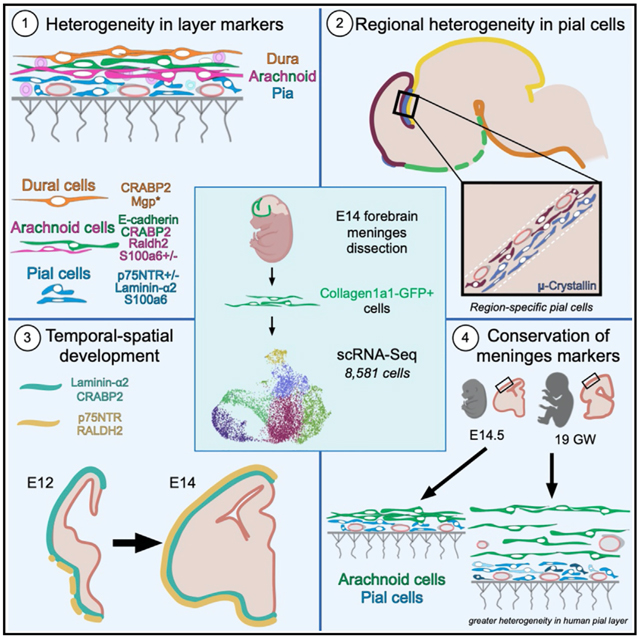
SUMMARY
The meninges are a multilayered structure composed of fibroblasts, blood and lymphatic vessels, and immune cells. Meningeal fibroblasts secrete a variety of factors that control CNS development, yet strikingly little is known about their heterogeneity or development. Using single-cell sequencing, we report distinct transcriptional signatures for fibroblasts in the embryonic dura, arachnoid, and pial. We define new markers for meningeal layers and show conservation in human meninges. We find that embryonic meningeal fibroblasts are transcriptionally distinct between brain regions and identify a regionally localized pial subpopulation marked by the expression of μ-crystallin. Developmental analysis reveals a progressive, ventral-to-dorsal maturation of telencephalic meninges. Our studies have generated an unparalleled view of meningeal fibroblasts, providing molecular profiles of embryonic meningeal fibroblasts by layer and yielding insights into the mechanisms of meninges development and function.
In Brief
DeSisto et al. use scRNA-seq to create a molecular profile of developing mouse meningeal fibroblasts. They show that fibroblasts of the pia, arachnoid, and dura meningeal layers are transcriptionally distinct, some layer markers are conserved in human fetal meninges, and fibroblast subsets demonstrate brain region-localized gene expression.
Graphical Abstract
INTRODUCTION
The meninges, made up of the dura, arachnoid, and pial layers, encases the CNS from its earliest stages of development and persists as a protective covering for the adult CNS. The meninges are composed of fibroblasts but also contain resident immune cells and an extensive network of blood and lymphatic vessels. Meningeal fibroblasts have emerged as critical players in CNS development (reviewed in Dasgupta and Jeong, 2019; Siegenthaler and Pleasure, 2011). They are the major source of basement membrane (BM) proteins that form the glial limitans (Hecht et al., 2010; Halfter et al., 2002; Hartmann et al., 1992). Meningeal production of Cxcl12 controls the migration of Cajal-Retzius cells (Borrell and Marín, 2006), proliferation of cerebellar radial glial progenitor cells (Haldipur et al., 2015, 2014), and proper positioning of granule cell progenitors in the hippocampus and cerebellum (Reiss et al., 2002; Zhu et al., 2002). Bone morphogenic proteins (BMPs) produced by the meninges help control cortical layer formation (Choe and Pleasure, 2018; Choe et al., 2012), while meningeal fibroblast production of retinoic acid directs cortical neurogenesis and cerebrovascular development (Boucherie et al., 2018; Haushalter et al., 2017; Mishra et al., 2016; Siegenthaler et al., 2009). The importance of meningeal fibroblasts in CNS development is underscored by Foxc1 mutant mice, where meningeal populations fail to fully develop and cause severe neocortical and cerebellar defects (Aldinger et al., 2009; Siegenthaler et al., 2009; Zarbalis et al., 2007). FOXC1 mutations underlie human CNS developmental defects, further highlighting the importance of meningeal fibroblasts in human CNS development (Haldipur et al., 2017; Aldinger et al., 2009).
Despite well-documented roles in brain development, characterization of meningeal fibroblasts is still based primarily on histological and electron microscopy (EM) studies. Further, progress on understanding meninges development has not advanced much beyond identifying the probable origins (neural crest and mesoderm [Jiang et al., 2002]) and some work on the timing of their developmental emergence (Siegenthaler et al., 2009). There is a clear need for better molecular characterization of the meningeal fibroblast populations. This will help accelerate the discovery of the origins of these specialized fibroblasts, mechanisms that control their development, and their functions in the CNS. To begin to address this need, we utilized single-cell RNA sequencing (scRNA-seq) analysis to create a molecular profile of E14 mouse forebrain meningeal fibroblasts. We demonstrate heterogeneity among E14 meningeal fibroblasts, showing that fibroblasts of the pia, arachnoid, and dura have distinct transcriptional profiles. We present evidence that E14 meningeal fibroblasts are transcriptionally distinct between brain regions and highlight a unique pial fibroblast subpopulation, marked by μ-crystallin expression, that is regionally localized in mice. These studies greatly advance our understanding of the meninges, providing the first molecular signature of meningeal fibroblasts and generating insights into mechanisms of meninges development and function.
RESULTS
scRNA-seq andClusterAnalysis ofForebrain Embryonic Meningeal Fibroblasts
We carried out scRNA-seq analysis on forebrain meningeal fibroblasts isolated from Collagen-1a1-GFP/+ (Col1a1-GFP/+) E14 embryos (Figures 1A and S1). Col1a1-GFP labels fibroblasts including those in the meninges (Kelly et al., 2016; Soderblom et al., 2013; Lin et al., 2008; Yata et al., 2003), which we confirmed for the E14 meninges (Figures S1A–S1E). To access the E14 brain, the partially ossified calvarium was removed, leaving on the brain surface the leptomeninges (arachnoid, pia) and some areas of the dura that are not removed with the calvarium (Figures S1D and S1E). Therefore this dissection approach effectively isolates leptomeninges but only a small portion of the dura. Forebrain meninges were removed from three litter mate embryos, and GFP+ cells were isolated by flow cytometry (Figure S1F). We captured and sequenced the transcriptomes of 8,581 cells (2,729, 3,278, and 2,574). Clustering analysis using Seurat 3 R-Package segregate captured cells into six principal clusters based on gene expression (Figures 1B and 1C; Table S1). Gene expression in the 6 principal clusters can be queried on our website: https://cuanschutz-devbio.shinyapps.io/Siegenthaler_shiny/. Using published markers of dura (Alpl, Foxc2) (Doro et al., 2019; Cooper et al., 2012; Zarbalis et al., 2007; Angelov, 1990) and arachnoid (Ptgds, Aldh1a2) (Kalamarides et al., 2011; Siegenthaler et al., 2009), we preliminarily classified cells in meninges 4 (M4) as dura fibroblasts (enriched in Fxyd5, Dkk2, and Alpl), and meninges 3 (M3) as arachnoid fibroblasts (enriched in Crabp2, Ogn, Aldh1a2, and Ptgds) (Figure 1C). M4 was the smallest cluster, consistent with only part of the dura being obtained in the dissection. Cells in the meninges 1 (M1) cluster showed enriched expression of S100a6, Col18a1, and Postn (Figure 1C; Table S1). We provisionally classified M1 as pial fibroblasts as Col18a1 is part of the pial BM and localizes to the pial layer in humans and mice (Caglayan et al., 2014). Meninges 2 (M2) had enriched expression of S100a6, Ngfr, and Aldh1a2. This cluster shared enriched genes with M1 and M3, suggesting that it contains a mixture of pial and arachnoid fibroblasts and/or an intermediate cell type.
Figure 1. Transcriptome Clustering Analysis Segregates Embryonic Meningeal Fibroblasts into Four Phenotypic Groups and Two Cycling Groups.
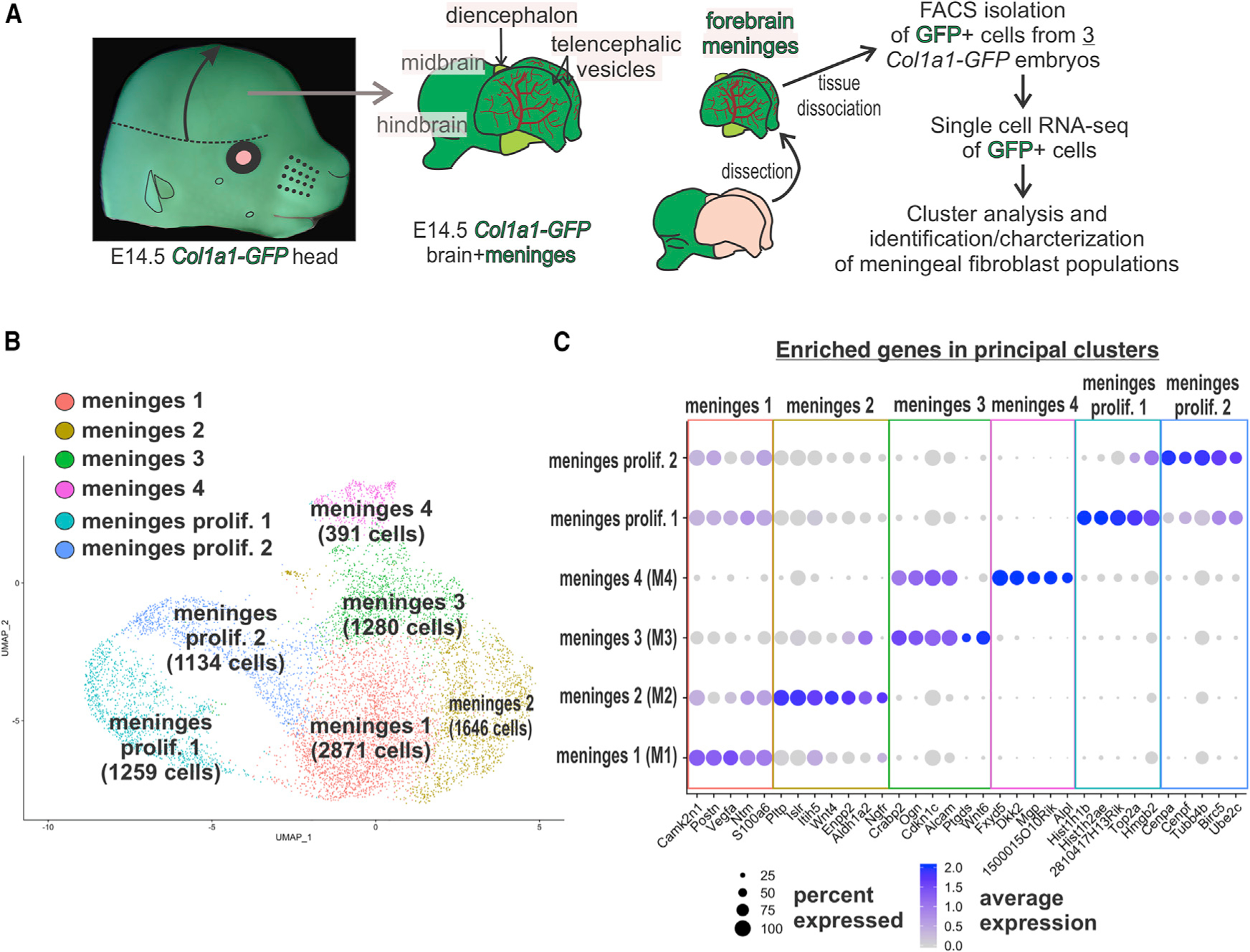
(A) Workflow of brain/meninges dissection, isolation, and single-cell RNA-seq analyses of E14 Col1a1-GFP+ meningeal fibroblast cells.
(B) UMAP clustering analysis resolved four principal phenotypic clusters and two clusters of dividing cells. Cell counts are shown in parentheses.
(C) Dot plot depicting enriched genes, curated from the top 30 based on adjusted p value, in each of the principal cell clusters. Dot color represents average expression level, and size represents percentage of cells expressing each marker.
Meninges proliferation 1 and 2 contained proliferating meningeal fibroblasts enriched in the expression of cell cycle genes (Top2a, Cenpa, and Cenpf) (Figure 1C). We analyzed the expression of S100a6 (M1/2) and Crabp2 (M3/4) to estimate the percentage of M1/M2 cells and M3/M4 cells within the proliferating clusters. Meninges proliferation 1 and 2 contained on average 81% S100a6 (M1/2) and 3.6% Crabp2 (M3/4) cells; 11% of cells had both S100a6/Crabp2 and 4.3% that were low in both genes. Thus, the two proliferating clusters contain a mixture of cells from the other 4 principal clusters with the majority being cells from the M1/M2 clusters that comprise 72% of the cells in the M1–4 clusters.
Embryonic Meningeal Fibroblasts from the Pial, Arachnoid, and Dural Layers Have Distinct Transcriptional Signatures
Immunofluorescence (IF) labeling on E14 Col1a1-GFP/+ embryos was used to determine where cells of the principal clusters localize within meninges layers (pia, arachnoid, and dura). We immunolabeled E14 Col1a1-GFP forebrain sections to detect laminin α2 (Lama2), p75NTR (Ngfr), and S100a6 (S100a6), enriched in M1-M2 clusters (Figure 2A; Table S2). Laminin α2, p75NTR, and S100a6 were observed in the E14 forebrain meninges (Figures 2B, 2C, and 2E) with gene expression for S100a6, Lama1, and Ngfr detected at all levels of the CNS meninges (Figures S2A–S2D). Closer examination of the telencephalic meninges confirmed laminin α2, S100a6, and p75NTR expression in Col1a1-GFP+ cells at the brain surface, consistent with expression in the pia (Figures 2G, 2H, and 2I). p75NTR, whose expression was most enriched in cluster M2, showed stronger expression in the ventral telencephalic and thalamic meninges as compared to the dorsal telencephalon (Figures 2B and 2C). This is consistent with the pia having regionalized expression profiles, closely examined in Figure 4. p75NTR incompletely overlapped with laminin α2+ cells, with some laminin α2+/GFP+ only cells above the laminin α2-p75NTR layer (Figure 2G, open arrows). These data show that cells identified in clusters M1 and M2 localize to the embryonic pial layer and that p75NTR, laminin α1/2, and S100a6 mark cells in the pial layer throughout the developing CNS.
Figure 2. Genes Enriched in Principal Clusters Encode for Proteins Expressed in Specific Meningeal Layers.
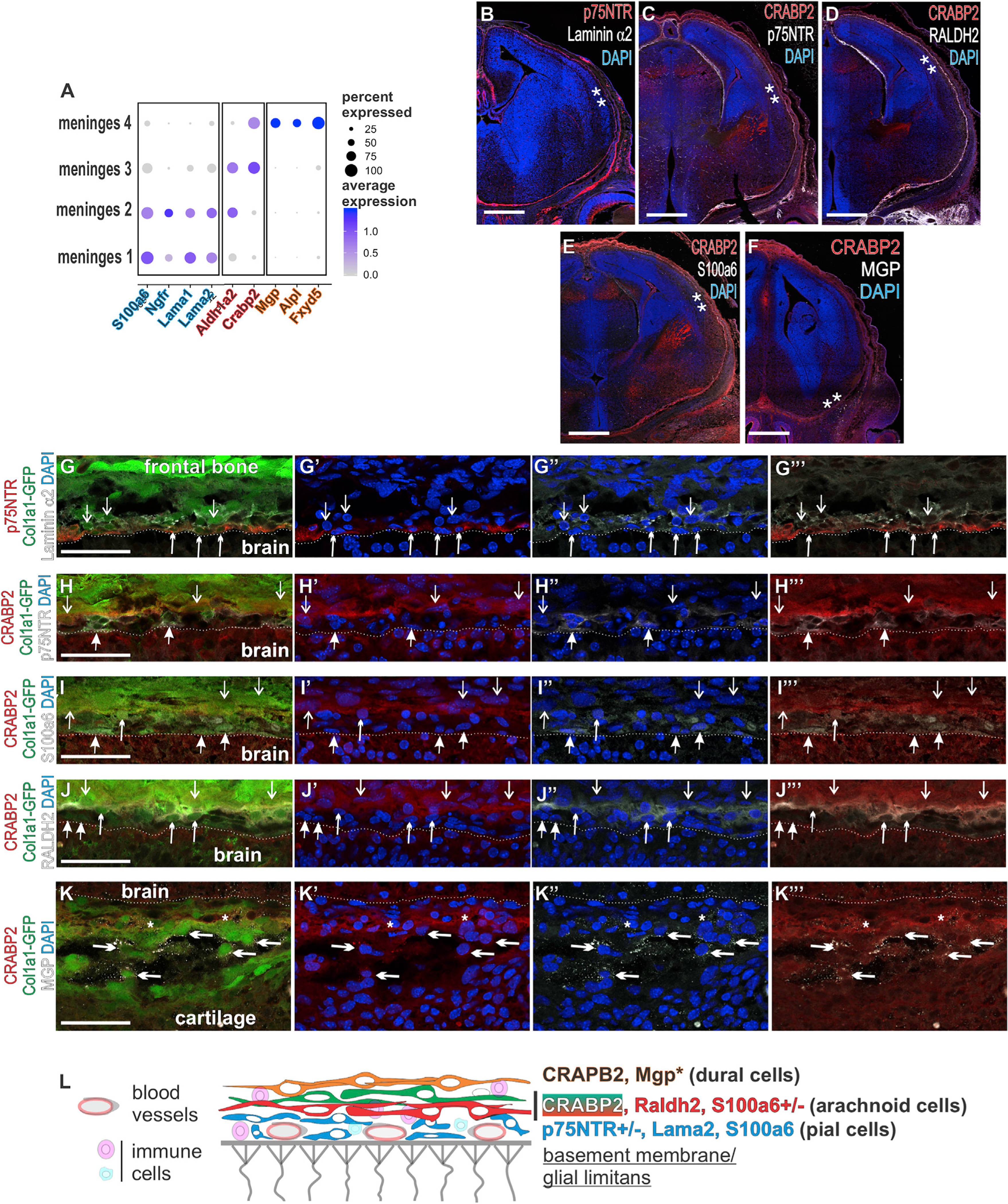
(A) Dot plot depicting select genes enriched in principal clusters and validated by protein expression. Colored type denotes cluster-enriched markers: meninges 1 and 2, blue; meninges 2/3, pink; meninges 4, orange.
(B–F) Coronal sections of the forebrain depicting immunolabeling of meninges enriched genes. ** indicates locations of high-resolution images in (G–K).
(G) Laminin α2+/p75NTR+/GFP+ are indicated by upward short arrowheads and laminin α2+/GFP+ by downward open arrows.
(H) CRABP2+/GFP+/p75NTR cells are indicated by downward open arrows, and p75NTR+/GFP+/ CRABP2– are indicated by upward closed arrows.
(I) S100a6+/GFP+/ CRABP2– are indicated by an upward closed arrow, the S100a6+/GFP+/CRABP2+ cell is indicated by an upward short arrowhead, and CRABP2+/S100a6-/GFP+ layers are indicated by downward open arrows.
(J) Raldh2+/CRABP2+/GFP+ are indicated by upward short arrowheads, Raldh2– / CRABP2– /GFP+ are indicated by upward closed arrows, and CRABP2+/RALDH2– /GFP+ cells are indicated by downward open arrows.
(K) Short arrowheads indicate MGP+/CRABP2+/GFP+ cells immediately adjacent to a CRABP2+/GFP+/MGP– arachnoid layer, though some cells in this layer were MGP+ (open arrow).
(L) Summary diagram showing the expression of meninges layer enriched proteins depicted in (G–K).
MGP* indicates that protein expression was only observed ventrally. Dotted line indicates brain surface in (G–K). Scale bars represent 500 μm in (B–F) and 50 μm in (G–K). Dot color represents average expression level, and size represents percentage of cells expressing each marker.
Figure 4. Subcluster Enriched Genes Localize to Specific Regions of the Embryonic Meninges.
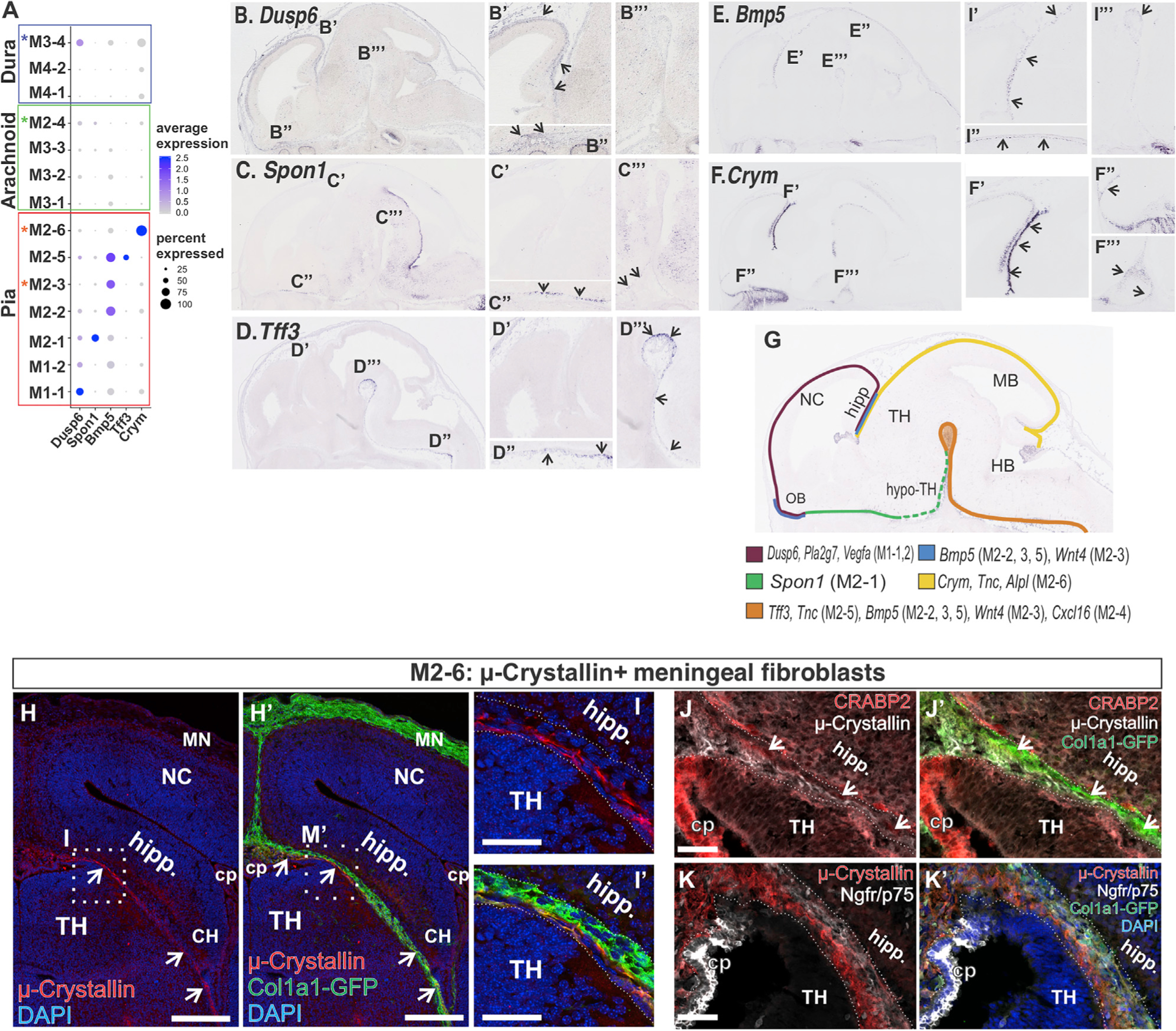
(A) Dot plot depicting genes differentially enriched in M1 and M2 subclusters (B–F) In situ gene expression in E14 sagittal sections depicting the expression of genes identified as enriched in M1 and M2 subclusters. Areas that are magnified are indicated on the low-magnification image in (B–F). Arrows indicate signal detected in meninges.
(G) Summary of meninges expression pattern of subcluster enriched genes based on (F–J) and Figure S4.
(H–M) (L and M) Col1a1-GFP E14 coronal brain section depicts μ-crystallin+/GFP+ cells in the meninges between the future hippocampus and thalamus. Magnified areas in (M and M′) are indicated by a box in (L and L′). (H and I) Col1a1-GFP E14 coronal brain section depicting μ-crystallin+/GFP+ cells in the meninges immediately adjacent to the surface of the thalamus. μ-crystallin+/GFP+ cells are separate from CRABP2+ (arrows in J) or p75NTR+ layers in this region but extend into the choroid plexus (CP) (K). Scale bars represent 200 μm in (H) and 50 μm in (I–K); TH, thalamus; Hipp, hippocampus; MN, meninges; NC, neocortex; CP, choroid plexus. Dot color represents the average expression level, and size represents the percentage of cells expressing each marker.
We next performed IF for CRABP2 and RALDH2 (protein product of Aldh1a2) enriched in the M3 cluster (presumptive arachnoid fibroblasts), while RALDH2 but not CRABP2 was enriched in the M2 cluster (Figure 2A; Table S2). CRABP2 (Figures 2C–2F) and RALDH2 (Figure 2D) were detected in the entire forebrain meninges with those in situ showing expression throughout the CNS meninges (Figures S2E and S2F). RALDH2 and/or CRABP2 was detected in Col1a1-GFP+ cells located directly above the pial layer (Figures 2H–2J), the expected position of arachnoid cells. CRABP2 was detected in the neocortex just below the pial surface, likely representing CRABP2-expressinng radial glial endfeet (Boucherie et al., 2018) and Cajal-Retzius cells (Loo et al., 2019). p75NTR did not overlap with CRABP2 (Figure 2H), and S100a6+/GFP+ were mostly CRABP2; however, we did occasionally observe S100a6+/CRABP2+ cells (Figure 2I). Thus, the RALDH2+/CRABP2+ cells are above the pia, supporting their identity as arachnoid fibroblasts, and arachnoid fibroblasts seem to be present in both the M2 and M3 clusters.
Cells in cluster M4, preliminarily defined as dural fibroblasts, were enriched for Crabp2 and Mgp, with no expression of Aldh1a2 (RALDH2) (enriched in arachnoid), S100a6, Ngfr, or Lama1/2 (enriched in pia) (Figure 2A; Table S2). Layers of CRABP2+/RALDH2– cells are detected above CRABP2+/Raldh2+ arachnoid cells (Figure 2J), and GFP+/MGP+/CRABP2+ were observed in the outer meninges layer (Figure 2K). These are consistent with the anatomic location of the dura. Of note, methyl green pyronin (MGP) staining was seen around a few CRABP2+ cells located closer to the brain surface (Figure 2K″, asterisks), but MGP is a secreted protein; thus, it is not clear if this is co-localization. MGP protein expression did not extend into dorsal telencephalic regions at E14 (Figure 2F), but the Mgp in situ signal extended over the dorsal telencephalon (Figure S2H), indicating that MGP protein detection may lag behind gene detection. In situ gene expression showed Fxyd5, another M4-cluster-enriched gene, appeared in the outermost layer of the meninges and encircled the CNS at all levels but did not extend into the meninges between the thalamus and telencephalon or around the embryonic cerebellum (Figure S2G), a feature of the dural layer. Thus, genes with enriched expression by cells in cluster M4 are detected in the dural layer of the meninges.
Collectively, these results support a model where p75NTR, laminin α2, and S100a6 identify embryonic pial fibroblasts, and RALDH2 and CRABP2 are expressed by arachnoid fibroblasts, with CRABP2 and MGP expressed by fibroblasts in the dura (Figure 2L). These are intermingled with blood vessels of the perineural vascular plexus, located on the brain surface and sprout inward to vascularize the brain (Nakao et al., 1988), and meningeal immune cells present from embryonic stages onward (Utz et al., 2020; Goldmann et al., 2016).
Principle cluster (Figure 1) and IF data (Figure 2) suggest that M1 are pial fibroblasts, M2 contains both pial and arachnoid cells, and M3 cells are arachnoid fibroblasts, while cells in M4 are dural fibroblasts. To confirm these data in an unbiased manner, we performed subclustering analysis on the 4 principle non-proliferating clusters (M1–M4) and then used non-negative matrix (NMF) factorization to obtain unsupervised hierarchical clustering among the subclusters. The subclustering analysis divided the 4 principle clusters into 14 subclusters (2 M1, 6 M2, 4 M3, and 2 M4 subclusters) (Figure 3A, legend includes percentage of cells in each subcluster; Figures S3A–S3D). Gene expression in the 14 subclusters can be queried on our website: https://cuanschutz-devbio.shinyapps.io/Siegenthaler_shiny/.
Figure 3. Unbiased Hierarchal Analyses Confirm Distinct Transcriptional Signatures between Pia, Arachnoid, and Dura Fibroblasts.
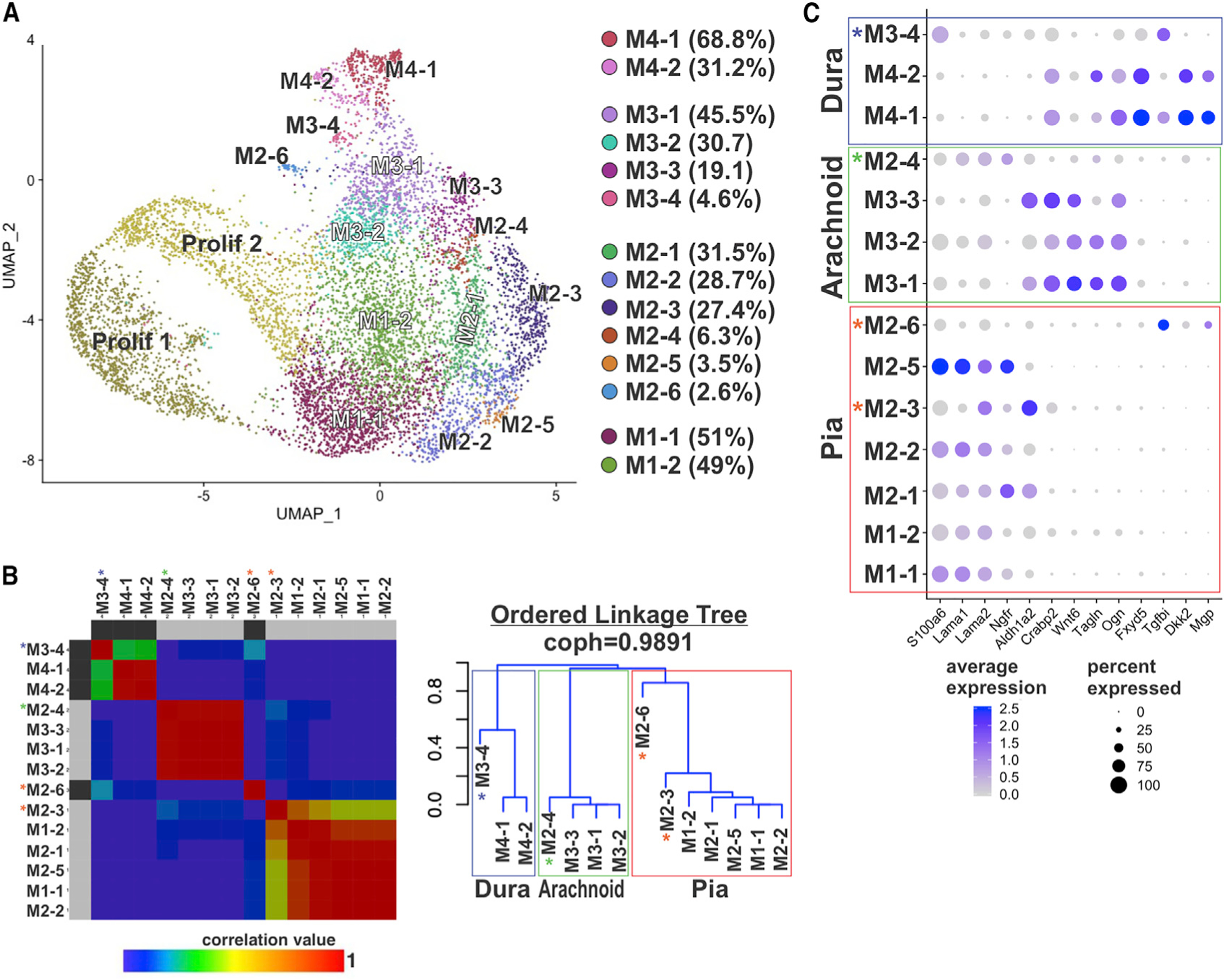
(A) UMAP annotated with 14 subclusters identified by analysis of principal clusters M1–M4. Percentage of cells per subcluster relative to the principal cluster is indicated in the legend.
(B) Non-negative matrix factorization (NMF) matrix and linkage tree or cladogram showing hierarchical relationships among M1–M4 subclusters. Cophenetic correlation is maximized for k = 4. Colors represent correlation values based on consensus clustering that range from 0 (blue, samples are never in the same cluster) to 1 (red, samples are always in the same cluster). Cladogram branches are annotated to indicate “pia” (red), “arachnoid” (green), and “dura” (blue) populations. Asterisks indicate subclusters that are transcriptionally distinct from other subclusters in the “pia” (red asterisk), “arachnoid” (green), and “dura” (blue) clades.
(C) Dot plot depicting genes enriched in pia, arachnoid, and dura subclusters. Dot color represents average expression level, and size represents the percentage of cells expressing each marker.
The hierarchical analysis segregated the meningeal subclusters into three main branches that correspond with our pial, arachnoid, and dura classifications (Figure 3B). A curated list of genes enriched in each subcluster is in Figures S3E and S3F, and enriched genes in M1/2, M3, and M4 subclusters are listed in Table S3. The dura and arachnoid clades each include an outlier subcluster (Figures 3B, M2–4 and M3–4 indicated by green and blue *, respectively), while the pia clade has two transcriptionally distinct outlier subclusters (M2–3 and M2–6) (red* Figure 3B).
Five pia subclusters group closely in the cladogram (M1–1, M1–2, M2–1, M2–2, and M2–5) (Figure 3B). Each of these sub-groups had enriched expression of S100a6 and Lama1/2 with low expression of Crabp2 and Aldh1a2 (Figure 3C), matching what principle cluster and IF analysis support as pial fibroblasts. Of note, the M2–1 and M2–5 subclusters showed enriched expression of Ngfr, while the M2–1 subcluster had higher levels of Aldh1a2 but lacked Crabp2. The outlier pia sub-group M2–3 had high expression of Lama2 but also Aldh1a2 and Crabp2 (arachnoid markers) (Figure 3C; Table S3, Tab 1). Thus, cells in these clusters have both pia and arachnoid transcriptional properties. The other pial outlier subcluster, M2–6, had low expression of S100a6, Lama1/2, and Ngfr but enriched expression of Postn, Crym, and Tgfbi (Figure 3C; Table S3, Tab 1).
The hierarchical analysis classified the M3–1, M3–2, and M3–3 subclusters into the arachnoid clade, with M2–4 as an arachnoid related outlier (Figure 3B). Fibroblasts in the arachnoid clade are enriched for Crabp2 and Aldh1a2, matching principal cluster and IF analysis, along with Wnt6, Tagln, and Ogn (Figure 3C; Table S3, Tab 2). Arachnoid outlier cluster, M2–4, has very low expression of Crabp2 and Aldh1a2 but enriched expression of arachnoid genes Wnt6 and Tagln (Figure 3C; Table S3, Tab 1).
The M4–1 and M4–2 subclusters group as dural fibroblasts in the hierarchical analysis (Figure 3B) and show enriched expression of Crabp2 and Ogn, also in arachnoid clusters, but unique expression of Fxyd5, Dkk2, Tgfbi, and Mgp (Figure 3C). The primary difference is enrichment of cell cycle genes (Birc5, Cenpa, and Mki67) in M4–2 (Figure S3F; Table S3, Tab 3). Dural outlier M3–4 is unique in that it has significantly enriched expression of S100a6 (pia), Fxyd5, and Tgfbi (dura) as compared to other M3 subclusters but lacks expression of Dkk2 or Mgp (Figure 3C; Table S3, Tab 2). Collectively, our hierarchical cluster analysis confirms that pial, arachnoid, and dural fibroblasts have distinct transcriptional signatures.
Subcluster Enriched Genes Localize to Different Regions of the Meninges
We next tested if the unique transcription profiles of the subclusters might correlate with meninges adjacent to specific brain regions. To do this, we analyzed the meninges region expression of genes selectively enriched by a single or small number of subclusters. Using the publicly available gene expression database GenePaint (Visel et al., 2004) of E14 whole embryos, we looked at the in situ pattern for Dusp6, Plα2g7: M1–1, Spon1: M2–1, Bmp5: M2–2, 3, 5, Wnt4: M2–3, Cxcl16, Htra: M2–4, Tff3, Tnc: M2–5, and Crym, Tnc: M2–6 (Figures 4A and S4A). Interestingly, the expression profiles for each of these genes show highly regionalized meninges expression (Figures 4 and S4). For example, Dusp6 (M1–1) was detected in meninges around the dorsal telencephalon and the olfactory bulb (Figure 4B), whereas Spon1 (enriched in M2–1) was only detected in a strip of meninges in the ventral telencephalon and hypothalamus (Figure 4C). A summary expression map of genes analyzed in Figures 4B–4F and S4B–S4H and their respective subcluster enrichment is shown in Figure 4G.
We did not observe the same extent of localized expression patterns for genes enriched in M3 and M4 subclusters (Figures S5A–S5N). For the most part, expression in meninges was observed throughout the rostral-caudal axis with patchy signal in some areas for certain genes, in particular M3–3 enriched genes Ptgds and Fgfbp1 (Figures S5D and S5E). The exceptions were Slc22a2 (high expression in the posterior telencephalon and ventral hindbrain meninges) and Ptgfr (meninges strip around the ventral and posterior telencephalon and at the juncture between the ventral thalamus and hindbrain) (Figures S5F–S5H).
We further characterized expression of μ-crystallin (Crym), which uniquely marked the M2–6 subcluster, a transcriptionally unique population that does not express validated arachnoid or pial markers. Immunostaining for μ-crystallin+ cells in the E14.5 dorsal thalamus meninges showed these cells were Col1a1-GFP+ (Figures 4H and 4I) and, consistent with gene expression analysis (Figure 3C; Table S3 Tab 1), do not co-express p75NTR (pia) or CRABP2 (arachnoid/dura) (Figures 4J and 4K). GFP+/μ-crystallin+ cells were also detected in the 3rd ventricle choroid plexus (Figure 4K). Further, μ-crystallin+ cells do not intermingle with the vasculature of the perineural vascular plexus (Figures S4I and S4J). The expression pattern of μ-crystallin+ in meningeal fibroblasts, immediately adjacent to the dorsal thalamus surface, is consistent with these cells being a region-specific pial layer.
Functional Characterization of Embryonic Meninges Using Single-Cell Data
We used our single-cell meningeal fibroblast data set to provide new knowledge about important known functions of the embryonic meninges, in particular (1) developmental emergence of the arachnoid barrier cells, (2) secretion of factors that regulate brain development, and (3) production of extracellular matrix (ECM) proteins that comprise the pial BM.
Arachnoid barrier (AB) cells are the outer part of the arachnoid layer and are characterized by high expression of E-cadherin (Cdh1) (Ahn et al., 2019; Cha et al., 2014) and Claudin 11 (Cldn11) (Weller et al., 2018). Tight junctions between AB cells form part of the blood-cerebrospinal fluid (CSF) barrier (Balin et al., 1986; Nabeshima et al., 1975); however, AB cells are a poorly characterized cell type. To begin to fill this gap, we identified high Cdh1 expressing cells in our E14 data set: 23 and 21 Cdh1+ cells in the M3 (arachnoid) and M4 (dura) clusters, respectively. Cdh1+ cells were selectively enriched in Ndrg1, Klf5, Thsbd4, Chst2, and Cd55 (Figure 5A; Table S4). The M3 Cdh1+ cells were significantly enriched in junction genes, (Cxadr, Cdh5, and Cldn11), while M4 Cdh1+ were only enriched in one junction gene, Cdh1 (Table S4). Cdh1+ cells had low expression of pia genes (Lama1, Postn, and S100a6) (Figure 5A; Table S4), while the M3 Cdh1+ cells showed enriched expression of Wnt6 and Ptgds, and M4 Cdh1+ cells expressed Fxyd5 and Mgp (Figure 5A; Table S4). IF detection of E-cadherin (Ecad) at E14 showed that E-cad+ cells co-expressed Col1a1-GFP in cells away from the brain surface (Figures 5C and 5D) and were a continuous layer in ventral telencephalon (Figure 5D) but discontinuous dorsally (Figure 5C). E-cad+/GFP+ cells co-labeled with arachnoid/dura marker CRABP2 and were detected between CRABP2+/GFP+/E-cad arachnoid and dural cells (Figure 5E). Dura gene Mgp is selectively enriched in dural Cdh1+ cells, and MGP IF was detected in ventral E-cad+ cells (Figure 5G) but was not apparent in dorsal E-cad+ cells (Figure 5F). Our analyses show that E-cad AB cells are present at E14 and, as in the adult, comprise the outer layer of the arachnoid (Figure S6M) but are a discontinuous and heterogenous population, potentially reflective of ongoing maturation.
Figure 5. Developmental Emergence of Meningeal Fibroblast Arachnoid Barrier Cells and Meningeal Fibroblasts as a Source of Secreted Factors and ECM.
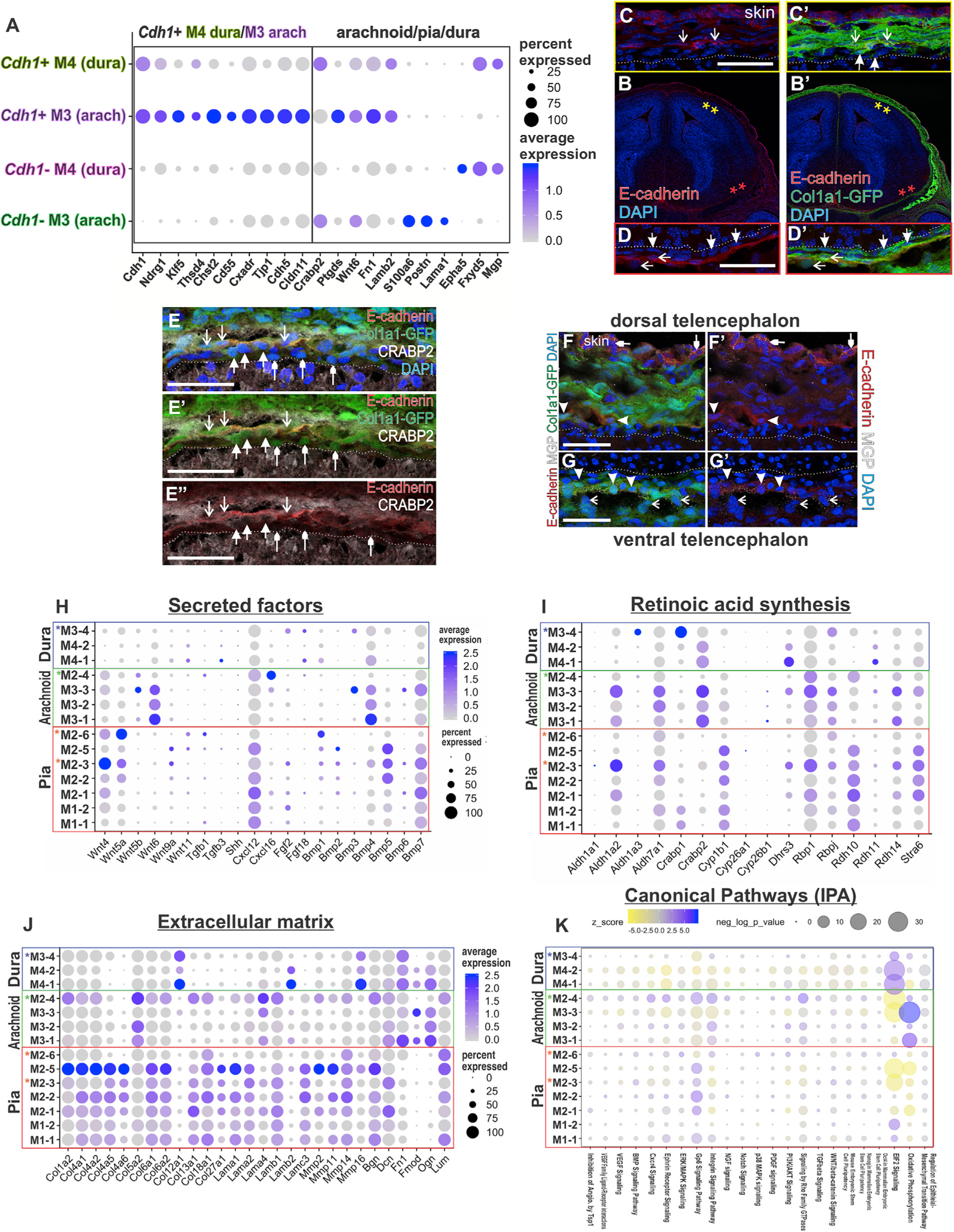
(A) Dot plot depicting enriched genes in Cdh1+ in M3 and M4 clusters curated from the top 15 enriched genes ranked on adjusted p value and genes enriched in principal clusters.
(B–D) E14 Col1a1-GFP coronal section at the level of the telencephalon with E-cadherin antibody labeling. Magnified view of meninges in dorsal (C) and ventral telencephalon (D). Closed arrows indicate E-cadherin-/GFP+ cells, and open arrows indicate E-cadherin+/GFP+ cells.
(E) Col1a1-GFP dorsal telencephalon meninges with antibody labeling for E-cadherin and CRABP2. Downward facing open arrows indicate CRABP2+/E-cadherin+/GFP+ cells, upward facing closed arrows indicate CRABP2+/E-cadherin-/GFP+ cells, and upward facing small arrowheads indicate CRABP2–/E-cadherin-/GFP+ cells.
(F and G) Col1a1-GFP dorsal and ventral telencephalon meninges with antibody labeling for E-cadherin and MGP. Closed arrowheads in (F and G) indicate E-cadherin+/GFP+ cells; these only co-express MGP in ventral areas (G). Open arrows in (G) indicate MGP+/E-cadherin-/GFP+ cells. Small closed arrowheads indicate MGP+/E-cadherin+/GFP–epithelial cells in the skin. Scale bars, 50 μm (C–G).
(H–J) Dot plots depicting secreted factors (H), retinoic acid synthesis and transport pathway genes (I), and extracellular matrix forming and remodeling enzymes
(J). Dot color represents the average expression level, and size represents the percentage of cells expressing each marker.
(K) Ingenuity pathway analysis (IPA) canonical pathway results showing pathway enrichment (purple hues) and depletion (yellow hues) by subcluster. Dot color represents the z score, and size represents the Benjamini Hochberg multiple comparison corrected p value.
Secreted morphogens showed differential gene expression across meningeal subclusters. For example, Wnt4 and Wnt5a are enriched in pial M2 subclusters, while Wnt6 showed high expression within the three arachnoid M3 subclusters (Figure 5H; Table S4). Meningeal fibroblast expression of Cxcl12 is enriched in the pial M1/2 subclusters (Figure 5H; Table S4). Bmp4, Bmp5, and Bmp7 are the predominantly expressed BMPs, with the arachnoid M3 clusters showing enriched Bmp4 expression and select pial M2 subclusters expressing Bmp5 (Figure 5H; Table S4). Genes for Tgfb and Fgf ligands and Shh were very low or not detected, suggesting the E14 meningeal fibroblasts are not a significant source of these factors.
Meningeal production of retinoic acid is critical for brain development. We identified enrichment of retinol transporter Stra6 and retinal synthesis gene Rdh10, both essential for the first steps in retinoic acid synthesis, in the pial M1/2 subclusters (Figure 5I; Table S4). Enrichment of Aldh1a2, which encodes for RALDH2 the enzyme responsible for the second step of retinoic acid synthesis, was observed in several arachnoid M2/3 sub-clusters (Figure 5I; Table S4). Crabp2, whose gene encodes a retinoic acid binding protein that controls intracellular transport, also showed expression in arachnoid subclusters as well as the dura (Figure 5I; Table S4).
We assessed the expression of a suite of ECM-related genes among our meningeal fibroblasts subclusters. The pial M1/2 subclusters had the highest expression of ECM-related genes, including collagens, laminins, matrix metalloproteinases, and proteoglycans (Figure 5J; Table S4, Tab 5). In contrast, M3/4 arachnoid and dural subclusters show uniquely enriched expression of Col5a2, Col12a1, Ogn, and Fn1 (Figure 5J; Table S4).
To identify potential signaling or pathway significance of gene expression patterns observed in the meningeal fibroblasts, we performed ingenuity pathway analysis (IPA). We plotted the results by subcluster, showing relative depletion and enrichment z scores and p values for the canonical pathways (Figure 5K; Table S4). The greatest levels of differential expression were in EIF2 signaling (enriched in dura subclusters and depleted in arachnoid and pia subclusters) and oxidative phosphorylation (enriched in arachnoid subclusters and depleted in pia subclusters). The GP6 signaling pathway, a collagen signaling receptor, and integrin pathways were upregulated in the pial subclusters, with a pattern similar to that observed in collagen gene enrichment (Figures 5J and 5K). Several signaling pathways that we anticipated might show differential enrichment, including VEGF, BMP, Ephrin, and NGF, showed minimal differential enrichment among the meningeal fibroblast populations.
Ventral-to-Dorsal Maturation of Telencephalic Meningeal Layers
The timing of meningeal layer differentiation has not previously been studied as layer specific markers were not known. To determine when and where the meningeal layer markers identified in Figures 1, 2, and 3 (p75NTR [pia], Lama2 [pia], CRABP2 [arachnoid/dura], RALDH2 [arachnoid]) are first expressed in the telencephalon, we analyzed E12 and E13 Col1a1-GFP embryos. We selected E12 and E13 because by E14 our meningeal layer markers are seen over the entire telencephalon (Figure 2), and we previously showed that RALDH2+ meningeal cells only partially cover the telencephalon at E12.5 (Siegenthaler et al., 2009). At E12, pia markers p75NTR and laminin α2 were detected in the inner layer of Col1a1-GFP+ fibroblasts, directly adjacent to the brain and consistent with pial identity (Figures 6A, 6C, S6B, and S6C). Laminin α2+/GFP+ cells were observed both ventrally and dorsally (Figures S6B and S6C), whereas p75NTR+/GFP+ cells were observed ventrally (Figure 6C) but not dorsally in the telencephalon (Figure 6D). However, by E13 p75NTR+/GFP+ cells were observed in both dorsal and ventral telencephalic meninges (Figures 6B and 6E).
Figure 6. Meningeal Layer Markers Appear in a Temporally Restricted Manner, and Diverse Meningeal Fibroblasts Populations Are Labeled by Tbx18-CreErt2.
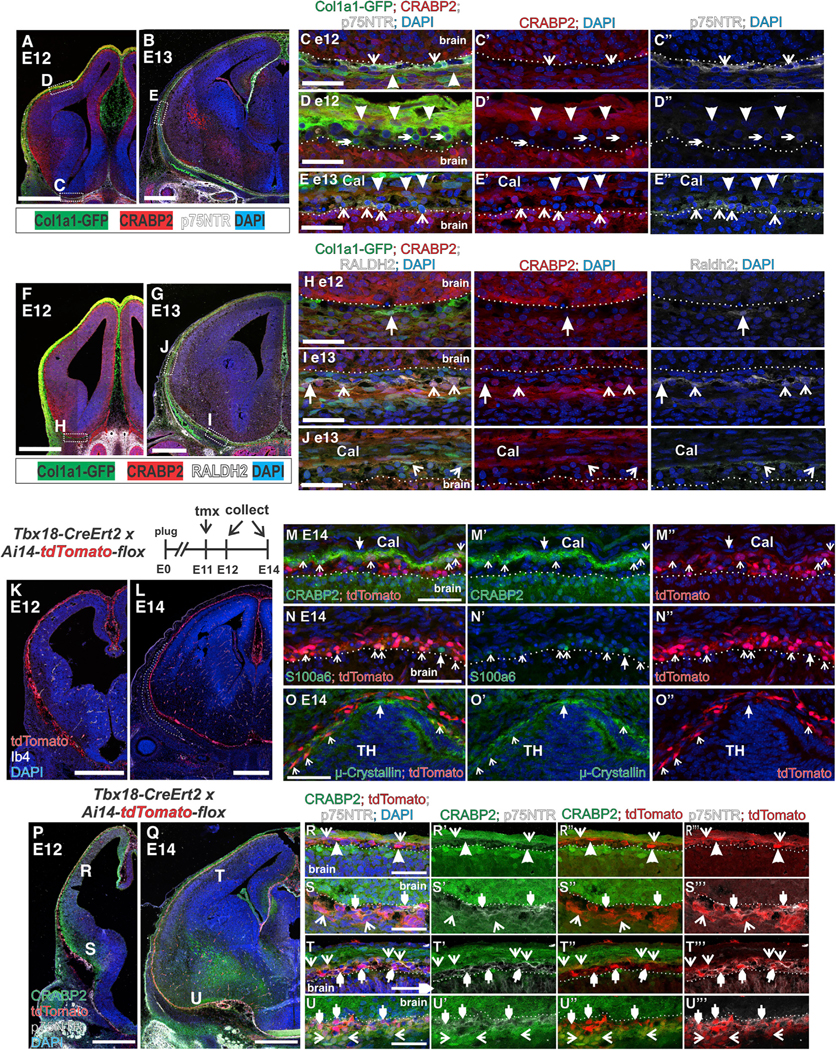
(A and B) Low-magnification images of Col1a1-GFP, CRABP2, and p75NTR expression in (A) E12 and (B) E13 telencephalon. Boxes in (A and B) indicate the region of the magnified image in (C, D, and E).
(C–E) CRABP2+/p75NTR–/GFP+ and p75NTR+/CRABP2–/GFP+ at E12 (C and D) and E13 (E) are indicated by closed arrowheads and open arrowheads, respectively. p75NTR–/CRABP2–/GFP+ at E12 in (D) are indicated by small arrowheads.
(F and G) Low-magnification images of Col1a1-GFP, CRABP2, and RALDH2 expression in (F) E12 and (G) E13 telencephalon. Boxes in (F and G) indicate region of magnified image in (H, I, and J).
(H–J) GFP+/Raldh2+/CRABP cells in ventral E12/E13 meninges indicated by closed arrowheads in (H and I). GFP+/Raldh2+/CRABP2+ population indicated by open arrowheads in (I and J).
(K and L) Low-magnification images of Tbx18-CreErt2/+; Ai14-flox/+ depicting tdTomato, Ib4 (blood vessels), and DAPI at (K) E12 and (L) E14. Outlined area in L indicates the frontal bone.
(M–O) tdTomato+ cells co-expressing CRAPB2 (M), S100a6 (N), and μ-crystallin in the thalamic meninges (O) are indicated by open arrows. TdTomato–cells that are CRABP2+, S100a6+ or μ-crystallin+ are indicated by closed arrows.
(P–Q) Tbx18-CreErt2/+; Ai14/+ depicting tdTomato, CRABP2, and p75NTR labeling at (P) E12 and (Q) E14. Letter annotation in (P and Q) indicate the region of the magnified image in (R–U).
(R–U) tdTomato+/CRABP2+/p75NTR– are indicated with open arrows, tdTomato+/p75NTR–/ CRABP2– cells (R only) are indicated by closed arrowheads, tdTomato+/p75NTR / CRABP2– cells are indicated by small closed arrowheads. Scale bars, 500 μm (A, B, F, G, K, L, P, and Q) and 50 μm (C–E, H–J, M–O, and R–U), Cal, calvarium; TH, thalamus.
For development of the arachnoid fibroblasts, we analyzed CRABP2, RALDH2, and E-cad expression (Figures 6F, 6G, and S6D). At E12 and E13, CRABP2+/GFP+ cells were detected adjacent to the GFP+ pial layer both ventrally and dorsally (Figures 6H–6J). In contrast, RALDH2 protein expression followed a temporally restricted expression pattern in the E12-E13 telencephalon. At E12, a rare RALDH2+/ CRABP2– cell was observed ventrally (Figure 6H), and no RALDH2+ cells were observed in the dorsal telencephalon (data not shown). By E13, RALDH2+/GFP+ cells were present in ventral and dorsal telencephalic meninges, most were CRABP2+, but some in the ventral region were CRABP2 (Figures 6I and 6J). A similar ventral-to-dorsal expansion of E-cad+/GFP+ cells was detected from E13 to E14. E-cad+/GFP+ cells were readily seen in the ventral telencephalon at E13 but were absent dorsally (Figures S6D–S6F). By E14, E-cad labeling is apparent throughout the entire telencephalic meninges, though incomplete dorsally (Figures 5C–5E). Collectively, these data show that expression of pial marker p75NTR, arachnoid markers RALDH2, and E-cad appear in a staggered, ventral-to-dorsal pattern from E12 to E14 during development.
Inducible Cre driver mouse lines that recombine embryonic meningeal fibroblasts have so far not been identified. Therefore, we used our embryonic meningeal fibroblast markers to test the suitability of the Tbx18-CreErt2, documented to recombine fibroblasts in the adult meninges along with pericytes and vSMCs throughout the body (Guimarães-Camboa et al., 2017), to label embryonic meningeal fibroblasts pre- and post-marker expression. Tamoxifen was given to pregnant females at E11, and Tbx18-CreErt2; Ai14-tdTomato embryos were collected at E12 and E14. TdTomato+ cells were detected in the meninges and brain mural cells at both E12 (Figures 6K and S6G–S6K) and E14 (Figures 6L–6O) but not in adjacent cranial mesenchyme structures like the frontal bone (Figure 6L, white outline). At E14, tdTomato was detected in pia (S100a6), arachnoid/dura (CRABP2), and μ-crystallin pial populations (Figures 6M–6O), indicating that Tbx18-CreErt recombines transcriptionally diverse meningeal fibroblast populations. We next tested if Tbx18-CreErt2 recombines meningeal fibroblasts early in their maturation process, focusing on p75NTR that appears in pial cells in a ventral-to-dorsal pattern. Tbx18-CreErt2-recombined cells were detected at the pial surface both dorsally and ventrally at E12, but p75NTR expression was limited to TdTomato+ cells ventrally (Figures 6P, 6R, and 6S). CRABP2, in contrast, was expressed in TdTomato+ cells both ventrally and dorsally at E12 (Figures 6P, 6R, and 6S). By E14, p75NTR+/tdTomato+ pial and CRABP2/tdTomato+ arachnoid/dura populations are observed both dorsally and ventrally (Figures 6Q, 6T, and 6U).
Dorsally, we detected a CRABP2+/tdTomato– layer of cells above the CRABP2+/tdTomato+ layer (Figure 6T); possibly, this is unossified calvarial mesenchyme that will eventually contribute to the frontal bone. Laminin α2 labels pial fibroblasts dorsally at E12, prior to expression of p75NTR in this region, and we detected laminin α2 +/tdTomato+ cells in dorsal regions at E12 (Figure S6L), consistent with laminin α2 being an early marker of pial fibroblasts. Collectively, these data support Tbx18-CreErt2 as a useful tool for studying meningeal fibroblast development and show that despite the large transcriptional diversity of meningeal fibroblasts that they can all be labeled with a Tbx18-CreErt2.
Conservation of Mouse Meningeal Fibroblast Subtype Markers in the Human Fetal Meninges
To determine whether meningeal layer specific markers are conserved between mouse and human fetal meninges, expression of S100a6 and p75NTR (pia in mouse), μ-crystallin (pial subcluster in mouse), and CRABP2 (arachnoid/dura in mouse) were analyzed in the neocortical meninges human fetal brain (19 gestational weeks or GW) (Figures 7A–7D). S100a6 and p75NTR labeled cells immediately adjacent to the brain (Figures 7B and 7D), showing that S100a6 and p75NTR are expressed by pial meningeal fibroblasts in the fetal human brain. Interestingly, μ-crystallin+ cells, which label a regionally specific subset of meningeal fibroblasts in embryonic mouse meninges, could be seen in the pial layer throughout the fetal human neocortex (Figures 7C and S7B). CRABP2 did not overlap with μ-crystallin, p75NTR, or S100a6 and was expressed by two morphologically distinct meningeal cell layers, a loose network of CRABP2+ cells (closed arrows in 7C and 7D), and a compact, outer layer that seemed to contain only CRABP2+ cells (small arrowheads in 7C and 7D).
Figure 7. Meningeal Fibroblast Layer Markers in Human Fetal Meninges and Postnatal/Adult Mouse Meninges.
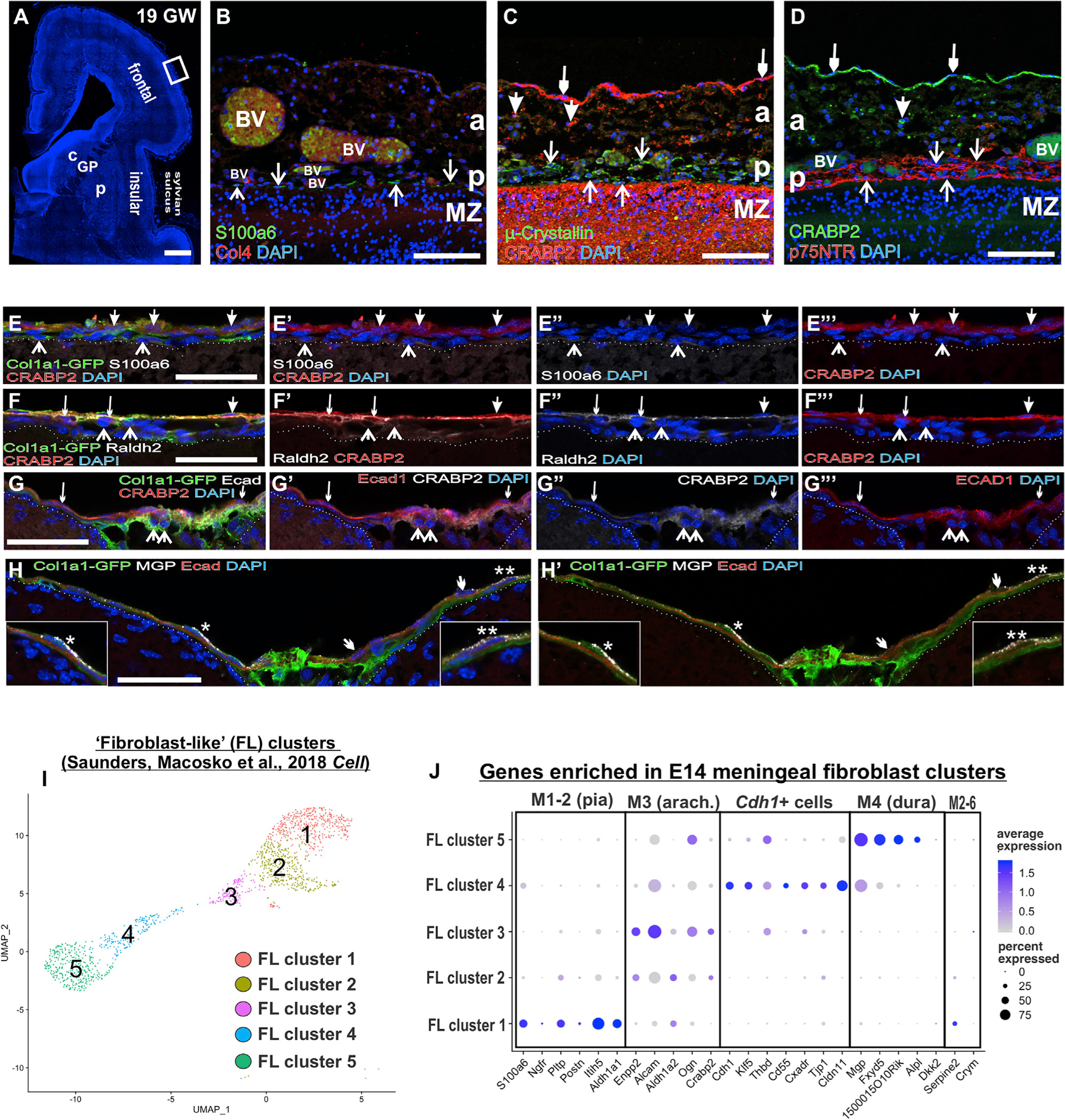
(A) Coronal section of a 19 gestational week (GW) human telencephalon. The box indicates magnified regions in (B–D).
(B) Arrows indicate S100a6+ immediately adjacent to the brain, consistent with being in the pia next to Col4+ blood vessels (BV).
(C) CRABP2 labels loosely (big closed arrows) and tightly packed cells (small closed arrows) in the meninges distal from the brain, whereas μ-crystallin+ cells (open arrows) were immediately adjacent to the brain surface and slightly distal (open arrows).
(D) p75NTR+ cells (open arrows) had a pattern similar to that of μ-crystallin in the pia and were spatially separate from CRABP2+ cells in the arachnoid (closed arrows).
(E) Postnatal day (P) 15 Col1a1-GFP meninges overlaying the cortex; the closed arrows indicate GFP+/CRABP2+ cells, and open arrows indicate GFP+/ S100a6+ cells.
(F) Closed arrow indicates GFP+/CRABP2+/Raldh2 cell, small closed arrows indicate GFP+/CRABP2+/Raldh2+ cells, and open arrows indicate GFP+/–CRABP2 /Raldh2+ cells.
(G) P15 Col1a1-GFP meninges overlaying cerebellum; closed small arrows indicate E-cadherin+/CRABP2+ cells, and open arrows indicate GFP+/E-cad–/CRABP2+ cells.
(H) Closed small arrow indicates E-cadherin+/MGP+; *, inset is GFP+/E-cad–/MGP+ cells above an E-cad+ cell; **, inset is E-cad+/MGP+ cell.
(I) UMAP of cells derived from “fibroblast-like” clusters within frontal and posterior cortex datasets first published in Saunders et al. (2018). Five principal clusters (FL 1–5) are identified.
(J) Dot plot depicting genes identified as enriched in embryonic pia, arachnoid, arachnoid barrier cells (Cdh1+), and dura clusters. C, caudate; GP, globus pallidus; P, putamen (A); P, pia (B–D); A, arachnoid. Scale bars represent 1 mm in (A), 100 μm in (B–D), and 50 μm in (E–H). Dot color represents average expression level, and size represents the percentage of cells expressing each marker.
Meninges overlaying the Sylvian sulcus displayed a similar staining pattern for CRABP2, S100a6, μ-crystallin+, and p75NTR (Figures S7A–S7D), but the meninges was thicker in this area, and the CRABP2+ layer of loosely packed cells was expanded (Figures S7B and S7D). Immediately below this was a cell sparse region containing long strips of tissue, some of which contained CRABP2+ cells (Figures S7B–S7D). These are characteristic of arachnoid trabeculae that traverse the subarachnoid space that are most prominent in higher vertebrate and primate meninges (Mortazavi et al., 2018). Of note, in the Sylvian sulcus meninges, μ-crystallin+ cells at the pial surface also expressed S100a6; however, some μ-crystallin+ cells were also detected above the outer pia layer, and these cells were S100a6 negative (Figures S7E and S7F).
This analysis demonstrates that arachnoid and pial markers are conserved between mouse and human fetal meninges. Further, we find that μ-crystallin labels a more widespread pial population in humans than in mice.
Persistence of Meningeal Layer Markers in the Postnatal and Adult Mouse Meninges
To determine if markers of fetal meningeal fibroblasts persist in the postnatal and adult brain, we performed IF on postnatal day 15 (P15) Col1a1-GFP brains for S100a6 (pia), RALDH2 (arachnoid), E-cad (arachnoid barrier), CRABP2 (arachnoid/dura), MGP (dura and subset of the arachnoid barrier), and μ-crystallin (pial subcluster). CRABP2+/GFP+ cells occupied the outer layer, separated from the brain surface by a CRABP2/–GFP+ cell layer (Figure 7E). Some of the CRABP2– inner layer cells were S100a6+/GFP+ and lay immediately adjacent to the brain surface (Figure 7E). Co-staining of CRABP2 and Raldh2 revealed an outer population of CRABP+/Raldh2– cells immediately opposed to inner CRABP2+/Raldh2+/GFP+ cells (Figure 7F). CRABP–/Raldh2+/GFP+ cells were also detected (Figure 7F) closer to the brain surface. CRABP2 co-staining with E-cad, a marker of AB cells, showed an outer population of E-cad+/CRABP2+ immediately above CRABP2+/E-cad– cells (Figure 7G). Of note, the E-cad+ cells comprise the outer detectable continuous layer in sections because most of the dura adheres to the underside of the calvarium, which is removed to permit brain sectioning. Staining for MGP, which is enriched in dura and a subset of E-cad+ in the fetal meninges, consistently labeled E-cad+ cells (Figures 7H and 7H′. right inset). and we also noted occasional E-cad–/MGP+ in the outer layer (Figures 7H and 7H′, left inset). Therefore, some dural cells remain adhered to the leptomeninges following calvarium removal. These are likely dural border cells that are very closely associated with the arachnoid barrier layer (Nabeshima et al., 1975). μ-crystallin, which marks a regionalized pial subset of E14 meninges, was not detected in the P15 meninges in any region but was observed in a subset of Col1a1-GFP+ stroma of the choroid plexus (Figures S7G and S7H).
We used a published single-cell RNA-seq data set of adult mouse brain (Saunders et al., 2018) to examine the transcriptional profile of adult leptomeninges. We performed principal component, uniform manifold approximation and projection (UMAP), and cluster analyses on cells isolated from the frontal and posterior cortex classified as “fibroblast-like” (FL) clusters based on the expression of Col1a1 and Col1a2 (Saunders et al., 2018). We analyzed FL cells from these two brain regions because (1) they collectively contained the largest FL clusters by cell number, and (2) these regions had attached leptomeninges versus internal brain regions. The 5 clusters (FL cluster 1–5) had unique transcriptional profiles that included many genes identified and validated as expressed in embryonic meningeal fibroblasts (Figures 7I, 7J, and S7G; Table S5).
FL-1 was notable for expression of S100a6, Pltp, and Itih5 (Figure 7J; Table S5), which are significantly enriched in M1 and/or M2 embryonic fibroblast clusters that contain pial fibroblasts (Figure 1C). Ngfr and Postn, expressed in embryonic pial subclusters, are absent from adult clusters. Interestingly, FL-1 also showed enriched expression of Aldh1a1, a retinoic acid synthesis protein in the same family as Aldh1a2, that was not seen in our fetal single-cell transcriptomes. This is consistent with reports that RALDH1 (encoded by Aldh1a1) is detected in the meninges but only in postnatal and adult animals (Niederreither et al., 2002).
FL 2 and 3 were enriched in genes identified as expressed by M3 arachnoid embryonic clusters, including Aldh1a2, Crabp2, Ogn, and Alcam (Figure 7J; Table S5). Enpp2 is also selectively enriched in FL 2 and 3; in fetal meninges, Enpp2 is enriched in the M2–3 subcluster that has characteristics of arachnoid cells, in particular high Crabp2 and Aldh1a2 (Table S3). The FL-4 cluster had enriched expression of genes we identified as uniquely expressed by fetal Cdh1+ arachnoid barrier cells, including tight junctions genes Cldn11, Tjp1, and Cxadr as well as Klf5, Cd55, and Fn1 (Figure 7J; Table S5). FL-5 was enriched in genes we identified as dura, including Fxyd5, Nov, Mgp, 1500015O10Rik, and Alpl. Dkk2 is not expressed; this is different from the enrichment observed in embryonic dura subclusters. This cluster likely contains dural border cells that adhere to the leptomeninges following calvarium removal. Mgp is also enriched in putative arachnoid barrier cells within FL-4 (Figure 7J; Table S5), consistent with MGP protein expression in postnatal E-cad+ arachnoid barrier cells. Expression of some subcluster enriched genes with regionalized expression embryonically persisted in adult meningeal fibroblast clusters, including Ptgfr (M3–4) and Slc22a2 (M3–3) (Figure S7G). Other genes were not detected, including Crym and Serpine2 that are enriched in M2–6, a regionalized pial population in the embryonic meninges.
Collectively, this analysis demonstrates that some genes enriched in the embryonic meningeal fibroblast clusters persist in the adult and highlights potentially important differences among embryonic and adult meningeal fibroblasts.
DISCUSSION
Here, we present the most comprehensive analyses to date of developing meningeal fibroblasts. Our data are the first to show that distinct molecular signatures for pial, arachnoid, and dural fibroblasts are present by E14 and that subclusters exist within layer groups, reflecting meninges from different regions around the brain. Heterogeneity so early in development suggests unique developmental functions among subpopulations. One particularly unique subpopulation M2–6, a pial outlier, showed high expression of Crym (crystallin-μ) and Serpine2 (PN-1) and had a distinct regionalization above the dorsal thalamus and into the 3rd ventricle choroid plexus. Possibly, this population has region-specific functions related to CNS development or acts locally to regulate the development of meninges-located cell types and structures (immune cells or blood vasculature) contributing to specialization of these areas of the meninges. For example, PN-1 (Serpine2) is a serine protease inhibitor that can inhibit angiogenesis (Selbonne et al., 2012), and Serpine2−/− mice display hypervascularity in the developing retina and muscle (Selbonne et al., 2015).
What could account for this regionalized meninges gene expression? It may reflect differences in tissue germ layer origin; anterior meninges arise from the neural crest, whereas posterior meninges originate from the mesoderm (Jiang et al., 2000). However, the exact border between neural crest and mesoderm derived meninges has not been rigorously mapped. Possibly, meninges are regionally patterned by signals from the adjacent brain and calvarium/skull. There is considerable evidence that the meninges produce factors that help control brain and calvarium development (reviewed in Dasgupta and Jeong, 2019); therefore, signal(s) traveling in the opposite direction to locally control meningeal fibroblast gene expression is plausible. Importantly, these regional differences may reflect ongoing differentiation; this needs to be resolved with more single-cell studies at earlier and later time points. More studies are needed to identify root causes of regional differences in E14 meninges gene expression and whether meninges regionalization, if it persists, is important for meninges function in the developing and adult brain.
Meningeal fibroblast production of ECM is critical to CNS development and our data highlight differential production of ECM components among meningeal fibroblast populations. Pial fibroblasts help form the pial BM that serves as an essential boundary between the brain parenchyma and the meninges (Halfter et al., 2002). Pial subclusters located throughout the forebrain were enriched in expression of collagens, laminins, proteoglycans, and MMPs. This is consistent with functional specialization to produce the pial BM. Interestingly, dura and arachnoid subclusters had a different ECM production profile: Lamb2, Col12a1, proteoglycans Ogn and Fmod, and fibronectin (Fn1). Ogn, which encodes the secreted proteoglycan osteoglycin, has been implicated in the regulation of bone formation (Lee et al., 2018). Ogn is enriched in arachnoid and dura that are adjacent to the calvarial mesenchyme that is actively undergoing inter-membranous ossification at E14. The meninges, in particular the dura, have previously been shown to play a role in regulating calvarial development (Dasgupta and Jeong, 2019; Greenwald et al., 2000; Rice et al., 2000; Mehrara et al., 1999); however, which meningeal fibroblast subtypes and the signals involved are incompletely understood.
Meninges production of signaling molecules, like Cxcl12, BMPs, and retinoic acid, are mechanisms through which the meninges control brain development. Our profiling yielded insight into localized meninges production of certain factors.
Wnts and BMP genes are the clearest example of region selective expression at E14. Wnt4 was selectively enriched in the M2–3 pial outlier subcluster with a posterior forebrain expression pattern. Wnt6, by contrast, is enriched in the three major arachnoid subclusters suggestive of broad distribution across the CNS. Wnt4 and Wn6 expression were also identified in a recent bulk RNA-seq study of E13 cranial mesenchyme that included meninges (Dasgupta et al., 2019). Expression of these Wnt ligands by the embryonic meninges has not been previously demonstrated, and it could be acting locally to control meninges, vascular, or immune cell development. WNT-β-catenin signalling in neural crest cells is required for meninges development (Choe et al., 2014), and our IPA shows significant WNT-β-catenin signaling in certain meningeal subclusters. That said, a study abrogating meninges WNT secretion did not overtly effect meninges or brain development (DiNuoscio and Atit, 2019).
Therefore, more studies are needed to test the function of meninges-derived WNTs. BMP genes also showed layer and apparent region-specific expression at E14. Bmp7 is broadly expressed by several subclusters, whereas Bmp5 is enriched in posterior located meninges, and Bmp4 was selectively enriched in M3/4 arachnoid and dura clusters. Bmp7 and Bmp4 are the best studied of the meninges-derived Bmps, implicated in the development of the corpus collosum (Bmp7) and corticogenesis (Bmp4 and Bmp7) (Choe and Pleasure, 2018; Choe et al., 2012; Segklia et al., 2012); no studies thus far have identified Bmp5 as a meninges-derived factor. Interestingly, retinoic acid synthesis genes Rdh10 and Aldh1a2 showed differential gene enrichment in pial versus arachnoid clusters, respectively. This expression pattern suggests that pial and arachnoid fibroblasts work in conjunction to make retinoic acid, with each layer responsible for a different synthesis step. Our analysis revealed potential alternative retinoic acid synthesis pathways, including enrichment in non-canonical retinoic acid synthesis gene Cyb1b1 (Chambers et al., 2007) in pial populations. Meningeal retinoic acid synthesis is likely more complicated than previously appreciated, involving all layers and proteins other than RDH10 and RALDHs; more studies are needed to test this possibility.
Our developmental analysis of layer markers in the telencephalon reveals a progressive ventral-to-dorsal protein expression of some markers. The ventral-to dorsal progression between E12 and E14 parallels events in the adjacent neocortex, in particular migration and early accumulation of projection neurons in the cortical plate and the start of subpial migration of interneurons (Greig et al., 2013; Anderson et al., 1997). E-cad, p75NTR, and RALDH2 all exhibit this ventral-to-dorsal appearance; however, the timing is different, potentially reflecting distinct differentiation mechanisms. Our data indicate that meningeal cells, labeled by Tbx18-CreErt2 mouse line and expressing CRABP2 and laminin α2, are present around the telencephalon before other markers are detected. Consistent with this, Cxcl12, which guides Cajal-Retzius cell migration, is detected as early as E10 in the telencephalic meninges (Borrell and Marín, 2006). Our studies support a model of progressive maturation of existing meningeal fibroblasts as opposed to migration of meningeal cells expressing layer specific markers around the telencephalon, though more studies are needed to distinguish between these possibilities.
Our studies of human fetal meninges show expression of certain markers are conserved between mice and humans (pia, S100a6 and p75NTR; arachnoid, CRABP2). This indicates that meningeal fibroblast development is potentially similar between mice and humans, with important functions like secretion of signals that act on progenitors and migrating neurons potentially conserved (Haldipur et al., 2017; Aldinger et al., 2009). Widespread μ-crystallin+ cells in the human fetal pial layer overlying the neocortex was different from the restricted expression in mice. Our data also indicate greater cellular diversity in human fetal pial fibroblasts. We observe μ-crystallin+ meningeal fibroblasts expressing S100a6 and negative for S100a6 overlying the cortex, implying that there are potentially two cellular subtypes in the developing human pia. More work is needed to test this idea; however, it is tempting to speculate that meningeal fibroblast diversity evolved in parallel with the CNS to support the unique needs of forming a larger and more complex brain.
We show that some layer specific genes identified at E14 persist in postnatal and adult meninges. That said, several genes with enriched cluster expression at E14, such as Postn, Ngfr, and Dkk2, are no longer detected in the adult. Possibly, the genes are downregulated, the populations disappear, or this is a limitation of the adult data set. Of note, the leptomeninges (pia and arachnoid) in the adult are considerably thinner than the E14 leptomeninges (compare panels in Figure 2 with Figure 7). We predict that as the brain grows the meningeal layers may thin out to cover the brain surface, though more studies are needed to test this idea. Fetal and adult E-cad-expressing arachnoid barrier cells are quite similar in their gene expression, with enriched expression in genes required for tight and adherens junctions formation and genes such as Klf5, previously studied for its role in intestinal epithelial cell proliferation (McConnell et al., 2011).
Three limitations of our single-cell analysis are (1) the under-representation of the dura layer in the embryonic and adult data sets, (2) the possible presence of fibroblasts in the meninges that do not express Col1a1-GFP+ and therefore were not captured, and (3) the presence of perivascular Col1a1 expressing fibroblasts from the brain parenchyma in the adult single-cell data set. With regard to the dura, it is very likely that fibroblast complexity exists in the dural layer (e.g., suture versus non-suture dura). The relatively small population of dural cells captured in our data set and the small number detected in the adult fibroblast-like clusters means our analysis is not suitable for comprehensive evaluation of dural fibroblast heterogeneity or function. Studies capturing the entire embryonic and adult leptomeninges and dura are needed to address this and to identify fibroblast populations that do not express collagen-1. The lineage relationship between perivascular and meningeal fibroblasts has not been rigorously explored, though we and others have previously shown that collagen 1-expressing perivascular fibroblasts and meningeal fibroblasts express RALDH1, RALDH2, and PDGFRα (Vanlandewijck et al., 2018; Kelly et al., 2016). These genes are high in the adult Fl-1 cluster that is also enriched in pial genes, suggesting that FL-1 also contains collagen-1-expressing perivascular fibroblasts. This raises the possibility that pial and perivascular fibroblasts are transcriptionally very similar, if not identical, though more careful studies comparing these adult populations are required.
In summary, our analysis provides much needed characterization of fibroblasts that make up the embryonic meninges. This data set will serve as a resource for researchers studying meninges-related structure, process, malformation, and disease during development and in the adult. Further, it will serve as an important tool to identify the mechanisms that direct fibroblast diversity and specialization in the meninges and help accelerate the discovery of new meningeal fibroblast subtype functions.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Julie Siegenthaler (julie.siegenthaler@cuanschutz.edu).
Materials Availability
This study did not generate new unique materials.
Data and Code Availability
The embryonic meninges single cell data generated during this study are available at NCBI Gene Expression Omnibus (GEO), accession number: GSE150219.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All mice were housed in specific-pathogen-free facilities approved by AALAC and were handled in accordance with protocols approved by the IACUC committee on animal research at the University of Colorado, Anschutz Medical Campus. The following mouse lines was used in this study: Col1a1-GFP (Yata et al., 2003), Tbx18-CreErt2 (Guimarães-Camboa et al., 2017) generously provided by Sylvia Evans, UCSD also available from Jackson Laboratories (stock#: 031520), and Ai14-tdTomato-flox (Madisen et al., 2010) B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J Jackson Laboratories Stock#: 007914). All mouse lines were maintained on a C57/BLK6J background. Tamoxifen (100ul of 20mg/ml) was administered in the morning of E11 to timed pregnant Ai14-tdTo-mato-fl/fl mice crossed with Tbx18-CreErt2-Cre/+ male. GFP+ and tdTomato+ embryos were identified by fluorescence detection using a stereoscope equipped with a fluorescent light source.
METHOD DETAILS
Meninges Sample Preparation
Meningeal fibroblasts were derived from 3 littermate E14 Col1a1-GFPGFP/+animals. To access the meninges, we removed the skin and calvarium, exposing GFP+ meningeal fibroblasts on the brain surface. The GFP+ meninges were then peeled off the forebrain (telencephalon and diencephalon, composed of thalamus/hypothalamus) using a dissecting scope and dissociated. Complete removal of the GFP+ forebrain meninges was confirmed by sectioning and imaging brains post-removal of skin/calvarium (Figure S1D) and post-removal of forebrain meninges (Figure S1E). We did note on the dorsal brain surface only post-removal of skin/calvarium, there were fewer GFP+ cells above the presumptive RALDH2+ arachnoid layer (Figure S1D). This indicates that a limitation of our dissection is that dorsal dura and possibly arachnoid cells are removed along with the calvarium in our dissection process, but these are present in ventral meninges. GFP+ cells were isolated from the meningeal tissue by first incubating the tissue in 1.5 mL of digestion solution consisting of HBSS with Cα2+/Mg2+ (Invitrogen), 2% w/v bovine serum albumin (Calbiochem), 1% w/v glucose, 5 mg/mL Type II collagenase (Worthington) and 2% DNase, for 15 minutes at 37° C. The tissue was then triturated until well dispersed in the digestion solution. Liberation of the cells from the tissue into a single cell suspension was confirmed by viewing samples of the solution under a microscope, with additional incubation time at 37° C if needed to completely disaggregate the cells. Fluorescently-activated cell sorting (MoFlo XDP cell sorter; Beckman Coulter) was then performed using the Col1a1-GFP+/GFP marker to isolate GFP+ fibroblasts from other cell types within the meninges, with gating optimized for selection of singlets and GFP+ cell recovery (see Figure S1F for representative scatter plots). The percent recovery of GFP+ cells for each sample was 26.01% (18,517 events), 24.73% (13,379 events), and 27.66% (15,506 events).
Single Cell Capture, Library Preparation and Sequencing
To capture, label, and generate transcriptome libraries of individual cells, we used the 10X genomics Chromium Single Cell 3’ Library and Gel Bead Kit v3 following the manufacture’s protocols. Briefly, the single cell suspension, RT PCR master mix, gel beads and partitioning oil were loaded into a Single Cell A Chip 10 genomics chip, placed into the Chromium controller, and the Chromium single cell A program was run to generate GEMs (Gel Bead-In-EMulsion) that contain RT-PCR enzymes, cell lysates and primers for sequencing, barcoding, and poly-DT sequences. GEMs are then transferred to PCR tubes and the RT-PCR reaction is run to generate barcoded single cell identified cDNA. Barcoded cDNA is used to make sequencing libraries for sequencing analysis. Sequencing was performed on an Illumina NovaSeq 6000 using paired end 150 cycle 2×150 reads. Cell capture, library prep and sequencing were performed by the Genomics and Microarray core at the University of Colorado, Anschutz Medical Campus.
Analysis of Single Cell RNA-Seq Data
Cellranger (3.1.0) count module was used for alignment, filtering, barcode counting and UMI counting of the single cell FASTQs and Seurat (3.1.1) (Stuart et al., 2019; Butler et al., 2018) was used for cluster analysis. The scRNAseq data from the 3 replicates were merged, filtered to remove cells with <500 genes, >8000 genes or percent mitochondrial genes >20% and clusters found using Find-Neighbors(reduction=‘pca’, dims=1:40) and FindClusters(resolution=0.2).
Non-fibroblast clusters (GFP negative cells captured inadvertently) were identified by gene expression as containing endothelial cells/mural cells (84 cells total; Pecam, Cldn5, Kdr, Tagln, Abcc9, Cspg6), interneurons (18 cells total, Gabra2, Vash1, Lhfpl4), other neural cells (10 cells total, Tubb3) and monocytes (19 cells total; CD68). These cells were removed from further analysis and represented 2% of cells captured. One cluster had low overall gene expression per cell and low number of individual genes expressed per cell indicative of low quality (415 cells total, 5% of cells captured), these were removed from any further analysis. Following removal of non-fibroblast clusters and low quality cells, data were reanalyzed as described above.
Subclustering of the M1-M4 principal clusters was done by creating a subset of the desired cluster of cells and using FindNeighbors(reduction=‘pca’, dims=1:40) and FindClusters(resolution=0.3) on the subset. Cdh1+ M3 and Cdh1+ M4 cells were found and labeled by subsetting either M3 (arachnoid) or dura M4 cluster cells expressing Cdh1 using subset(subset=Cdh1>1). All dot plots were generated setting col.min = 0. Counts for sample specific and total subcluster cell number is provided in Table S6. The expression of Crabp2 and S100a6 in cells in the two clusters of proliferating cells was used to characterize these proliferating cells as M3/4 (Crabp2 >1 and S100a6 <1; proliferating 1: 41 cells, proliferating 2: 46 cells), M1/2 (S100a6 >1 and Crabp2<1; proliferating 1: 1033 cells, proliferating 2: 907 cells), both high (Crabp2>1 and S100a6>1; proliferating 1: 127 cells, proliferating 2:136 cells) or both low (Crabp2<1 and S100a6<1; proliferating 1: 58 cells, proliferating 2: 45 cells).
Adult single cell RNAseq data were downloaded from dropviz.org. scRNAseq data files F_GRCm38.81.P60Cortex_noRep5_FRONTALonly.raw.dge.txt.gz and F_GRCm38.81.P60Cortex_noRep5_POSTERIORonly.raw.dge.txt.gz were used along with cluster data in F_GRCm38.81.P60Cortex_noRep5_FRONTALonly.cluster.assign.RDS and F_GRCm38.81.P60Cortex_noRep5_POS-TERIORonly.cluster.assign.RDS and analyzed with Seurat. The frontal and posterior single cell RNAseq data were merged, cluster 14, the Fibroblast-like (FL) cells, were subset out and this subset was used to calculate UMAP coordinates and cluster analysis of just the adult FL cells. When these clusters were examined for enriched genes, 3 clusters of cells were identified as non-fibroblasts (endothelial, glial, and neuronal) and removed from further analyses. The remaining five FL clusters were analyzed using Seurat as described above.
In order to further characterize gene expression among the meningeal fibroblast subclusters identified through the UMAP clustering analysis, we performed unsupervised hierarchical clustering using non-negative matrix factorization (NMF) (Brunet et al., 2004; Lee and Seung, 1999). For the NMF analysis, we calculated the arithmetic mean of gene expression values for the cells comprising each subcluster. We conducted pathway analyses using Ingenuity Pathway Analysis (IPA, Qiagen). For the input data, we calculated the arithmetic mean of gene expression values for the cells comprising each subcluster and filtered to remove data with p values exceeding 0.01. To display the data, we exported Z-scores and p values (corrected for multiple comparisons using the Benjamini Hochberg procedure) for each subcluster. Dot plots were prepared in R using ggplot2. Dot plots for Canonical Pathways are in Figure 5K and all data for all Canonical Pathways is provided in Table S4.
Immunofluorescence and Imaging
Embryos were collected and whole heads fixed overnight with 4% paraformaldehyde followed by 20% sucrose and frozen in OCT compound (Tissue-Tek). Tissue was cryosectioned in 12μm increments and tissue-mounted slides were subjected to antigen retrieval by immersing the slides in solution of 0.01M citric acid, 0.05% Tween, pH 6, and heating in a pressure cooker (Cuisinart Model CPC-600) for 6 min. No antigen retrieval was performed when staining for Laminin α2 or E-cadherin. The tissue was permeabilized by incubating for 10 min at room temperature in PBS with 0.1% Triton-X (Sigma), blocked in 2% lamb serum/0.05% Triton-X solution for 40 min at room temperature and primary. and secondary solution was. 0.05% Triton-X in PBS. The exception to this was staining with antibody to laminin α2, for this blocking solution was 3% bovine serum albumin in 0.25% Triton-X for 1 h, primary and secondary antibodies were 3% BSA. Incubation in the following primary antibodies was conducted overnight at 4° C in appropriate solution: rabbit anti-S100A6 (1:100; Novus NBP2–44492), rat anti-Laminin α2 (1:200, Sigma-Aldrich L0663), mouse anti-μ-crystallin (1:250; Invitrogen; PIMA525192), rabbit anti-CRABP2 (1:100; Proteintech 10225), mouse anti-CRABP2 (1:100 Millipore; MAB5488), rabbit anti-μ-crystallin (1:100; Proteintech 12495), rabbit anti-Raldh2 (1:100, Sigma-Aldrich HPA010022), chicken anti-GFP (1:500, Invitrogen A10262), mouse anti-E-cadherin (1:100, BD Transduction Laboratories 610181), rabbit anti-p75NTR (1:200, Cell Signaling Technology D4B3 8238), chicken anti-RFP (1:500, Rockland 600–901-379). Following incubation with primary antibodies, tissue sections were incubated for 60 min with appropriate Alexafluor-conjugated secondary antibodies (Invitrogen), Alexafluor 647-conjugated isolectin-B4 (1:100; Invitrogen I324550) and DAPI (1:1000; Invitrogen).
Confocal images were obtained using a Zeiss 780 Laser Scanning Microscope with associated Zeiss Zen Black software. Images were processed using Adobe Photoshop. To draw lines indicating the border between the brain and meninges, we use the Col1a1-GFP+ cells as a guide. Laminin α2 is enriched in the pial basement membrane, the well accepted outer surface of the brain. We consistently find that Col1a1-GFP+ cells of the meninges stop at the laminin α2 labeled basement membrane, supporting that Col1a1-GFP+ cells can be used to demarcate the border between the meninges and brain.
Human Fetal Tissue Collection and Immunostaining
Human fetal tissue collection was obtained from a spontaneous abortion and after parents’ informed consent given to the Foetopathologie Unit at Hôpital Robert Debré and in accordance with French legislation. The gestational age (19 weeks) was based on first-trimester sonography crown-rump length measurement and confirmed at autopsy by the evaluation of fetal biometry and organ and skeletal maturations. Brains were removed and fixed in 4% buffered formalin added with 3 g/L of ZnSO4 for approximately 2 weeks before to be processed for paraffin embedding and sectioned at 6μm increments. Following de-parafinization, tissue immunofluorescent detection and imaging was performed as described for mouse tissue.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-S100A6 | Novus Biologicals | Cat#: NBP1–89388 |
| Rat monoclonal anti-Laminin-a2 | Sigma-Aldrich | Cat#: L0663 |
| Mouse monoclonal anti-μ-crystallin | Invitrogen | Cat#: PIMA525192 |
| Rabbit polyclonal anti-CRABP2 | Proteintech | Cat#: 10225–1-AP |
| Mouse monoclonal anti-CRABP2 | Millipore | Cat#: MAB5488 |
| Rabbit polyclonal anti-μ-crystallin | Proteintech | Cat#: 12495–1-AP |
| Rabbit polyclonal anti-ALDH1A2 | Sigma-Aldrich | Cat#: HPA010022 |
| Chicken polyclonal anti-GFP | Invitrogen | Cat#: A10262 |
| Mouse monoclonal anti-E-Cadherin | BD Transduction Laboratories | Cat#: 610181 |
| Rabbit monoclonal anti-p75NTR | Cell Signaling Technology | Cat#: D4B3 Product#: 8238 |
| Chicken polyclonal anti-RFP | Rockland Immunochemicals | Cat#: 600–901-379 |
| Isolectin GS-IB4 Alexa Fluor 647 Conjugate | Invitrogen | Cat#: I32450 |
| Biological Samples | ||
| Human fetal brain (paraffin embedded sections) | Foetopathologie Unit at Hôpital Robert Debré | http://robertdebre.aphp.fr/equipes-cliniques/pole-biologie/foetopathologie/ |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ GEM, Library & Gel Bead Kit v3 | 10x Genomics | PN-1000075 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE150219 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Tbx18tm3.1(cre/ERT2)Sev/J | The Jackson Laboratory | Stock No: 031520 |
| Mouse: Tg(Col1a1-EGFP)#Dab | Yata et al., 2003 | MGI: 4458034 |
| Mouse: B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | Stock No: 007914 |
| Software and Algorithms | ||
| Cellranger (3.1.0) | 10x Genomics | http://software.10xgenomics.com/single-cell/overview/welcome |
| R (3.6.3), R(3.6.1) | R Foundation for Statistical Computing | https://cran.r-project.org/ |
| Seurat (3.1.1) | Butler et al., 2018; Stuart et al., 2019, | http://satijalab.org/seurat/ |
| Ingenuity Pathway Analysis (IPA) (September 2019 Release) | QIAGEN | Cat#: 830018 |
| Non-negative matrix factorization (NMF) Consensus Clustering (version 5) | Brunet et al., 2004; Lee and Seung, 1999 | GenePattern |
| ggplot2 | R Studio | https://ggplot2.tidyverse.org/ |
| Other | ||
| DropViz Single-cell RNAseq data | Saunders et al., 2018 | http://www.dropviz.org |
| E14 mouse embryo RNA in situ expression data | GenePaint | http://www.gp3.mpg.de |
| Web browser for searching and visualizing gene expression in E14 meningeal fibroblast single cell RNAseq data set | This paper | https://cuanschutz-devbio.shinyapps.io/Siegenthaler_shiny/ |
Highlights.
scRNA-seq shows transcriptional profiles corresponding to E14 meningeal layers
Regional heterogeneity in E14 meningeal fibroblast gene expression
Temporal-spatial maturation of telencephalic meningeal layers
Conservation of meningeal layer enriched markers in human fetal meninges
ACKNOWLEDGMENTS
We thank all members of the Siegenthaler lab for comments on the manuscript and Katrina Diener, Okyong Cho, and Brian Woessner in the Genomics Core at the University of Colorado Anschutz Medical Campus for essential advice and technical work on single-cell experiments. J.A.S. would like to acknowledge R.M. and Pamela Siegenthaler for their valuable support throughout her life and scientific career. This work was supported by the National Institutes of Health/National Institute of Neurological Disorders and Stroke R03 NS104566 and R01 NS098273 to J.A.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.devcel.2020.06.009.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ahn JH, Cho H, Kim JH, Kim SH, Ham JS, Park I, Suh SH, Hong SP, Song JH, Hong YK, et al. (2019). Meningeal lymphatic vessels at the skull base drain cerebrospinal fluid. Nature 572, 62–66. [DOI] [PubMed] [Google Scholar]
- Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, and Millen KJ (2009). FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat. Genet. 41, 1037–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, and Rubenstein JL (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476. [DOI] [PubMed] [Google Scholar]
- Angelov DN (1990). Distribution of activity of alkaline phosphatase and Mg-dependent adenosine triphosphatase in the cranial dura mater-arachnoid interface zone of the rat. Cell Tissue Res 260, 595–600. [DOI] [PubMed] [Google Scholar]
- Balin BJ, Broadwell RD, Salcman M, and el-Kalliny M. (1986). Avenues for entry of peripherally administered protein to the central nervous system in mouse, rat, and squirrel monkey. J. Comp. Neurol. 251, 260–280. [DOI] [PubMed] [Google Scholar]
- Borrell V, and Marín O. (2006). Meninges control tangential migration of hem-derived Cajal-Retzius cells via CXCL12/CXCR4 signaling. Nat. Neurosci. 9, 1284–1293. [DOI] [PubMed] [Google Scholar]
- Boucherie C, Boutin C, Jossin Y, Schakman O, Goffinet AM, Ris L, Gailly P, and Tissir F. (2018). Neural progenitor fate decision defects, cortical hypoplasia and behavioral impairment in Celsr1-deficient mice. Mol. Psychiatry 23, 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, and Mesirov JP (2004). Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA 101, 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caglayan AO, Baranoski JF, Aktar F, Han W, Tuysuz B, Guzel A, Guclu B, Kaymakcalan H, Aktekin B, Akgumus GT, et al. (2014). Brain malformations associated with Knobloch syndrome – review of literature, expanding clinical spectrum, and identification of novel mutations. Pediatr. Neurol. 51, 806–813.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JH, Wee HJ, Seo JH, Ahn BJ, Park JH, Yang JM, Lee SW, Kim EH, Lee OH, Heo JH, et al. (2014). AKAP12 mediates barrier functions of fibrotic scars during CNS repair. PLoS One 9, e94695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers D, Wilson L, Maden M, and Lumsden A. (2007). RALDH-independent generation of retinoic acid during vertebrate embryogenesis by CYP1B1. Development 134, 1369–1383. [DOI] [PubMed] [Google Scholar]
- Choe Y, and Pleasure S. (2018). Meningeal Bmps regulate cortical layer formation. Brain plasticity 4, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y, Siegenthaler JA, and Pleasure SJ (2012). A cascade of morphogenic signaling initiated by the meninges controls corpus callosum formation. Neuron 73, 698–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe Y, Zarbalis KS, and Pleasure SJ (2014). Neural crest-derived mesenchymal cells require Wnt signaling for their development and drive invagination of the telencephalic midline. PLoS One 9, e86025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, Durham EL, Cray JJ Jr., Siegel MI, Losee JE, and Mooney MP (2012). Tissue interactions between craniosynostotic dura mater and bone. J. Craniofac. Surg. 23, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta K, Chung JU, Asam K, and Jeong J. (2019). Molecular patterning of the embryonic cranial mesenchyme revealed by genome-wide transcriptional profiling. Dev. Biol. 455, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta K, and Jeong J. (2019). Developmental biology of the meninges. Genesis 57, e23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNuoscio G, and Atit RP (2019). Wnt/beta-catenin signaling in the mouse embryonic cranial mesenchyme is required to sustain the emerging differentiated meningeal layers. Genesis 57, e23279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doro D, Liu A, Grigoriadis AE, and Liu KJ (2019). The osteogenic potential of the neural crest lineage may contribute to craniosynostosis. Mol. Syndromol. 10, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Jordão MJ, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, et al. (2016). Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 17, 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald JA, Mehrara BJ, Spector JA, Chin GS, Steinbrech DS, Saadeh PB, Luchs JS, Paccione MF, Gittes GK, and Longaker MT (2000). Biomolecular mechanisms of calvarial bone induction: immature versus mature dura mater. Plast. Reconstr. Surg. 105, 1382–1392. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, and Macklis JD (2013). Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães-Camboa N, Cattaneo P, Sun Y, Moore-Morris T, Gu Y, Dalton ND, Rockenstein E, Masliah E, Peterson KL, Stallcup WB, et al. (2017). Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo. Cell Stem Cell 20, 345–359.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldipur P, Dang D, Aldinger KA, Janson OK, Guimiot F, Adle-Biasette H, Dobyns WB, Siebert JR, Russo R, and Millen KJ (2017). Phenotypic outcomes in mouse and human Foxc1 dependent Dandy-Walker cerebellar malformation suggest shared mechanisms. eLife 6, e20898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldipur P, Gillies G, Janson OK, Chizhikov VV, and Millen KJ (2015). ISDN2014_0119: Mesenchymal Foxc1 non-autonomously controls cerebellar development through SDF1α-CXCR4 maintenance of radial glial cells. Int. J. Dev. Neurosci. 47, 34. [Google Scholar]
- Haldipur P, Gillies GS, Janson OK, Chizhikov VV, Mithal DS, Miller RJ, and Millen KJ (2014). Foxc1 dependent mesenchymal signalling drives embryonic cerebellar growth. eLife 3, e03962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfter W, Dong S, Yip YP, Willem M, and Mayer U. (2002). A critical function of the pial basement membrane in cortical histogenesis. J. Neurosci. 22, 6029–6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D, Sievers J, Pehlemann FW, and Berry M. (1992). Destruction of meningeal cells over the medial cerebral hemisphere of newborn hamsters prevents the formation of the infrapyramidal blade of the dentate gyrus. J. Comp. Neurol. 320, 33–61. [DOI] [PubMed] [Google Scholar]
- Haushalter C, Schuhbaur B, Dollé P, and Rhinn M. (2017). Meningeal retinoic acid contributes to neocortical lamination and radial migration during mouse brain development. Biol. Open 6, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht JH, Siegenthaler JA, Patterson KP, and Pleasure SJ (2010). Primary cellular meningeal defects cause neocortical dysplasia and dyslamination. Ann. Neurol. 68, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, and Morriss-Kay GM (2002). Tissue origins and interactions in the mammalian skull vault. Dev. Biol. 241, 106–116. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, and Sucov HM (2000). Fate of the mammalian cardiac neural crest. Development 127, 1607–1616. [DOI] [PubMed] [Google Scholar]
- Kalamarides M, Stemmer-Rachamimov AO, Niwa-Kawakita M, Chareyre F, Taranchon E, Han ZY, Martinelli C, Lusis EA, Hegedus B, Gutmann DH, and Giovannini M. (2011). Identification of a progenitor cell of origin capable of generating diverse meningioma histological subtypes. Oncogene 30, 2333–2344. [DOI] [PubMed] [Google Scholar]
- Kelly KK, MacPherson AM, Grewal H, Strnad F, Jones JW, Yu J, Pierzchalski K, Kane MA, Herson PS, and Siegenthaler JA (2016). Col1a1+ perivascular cells in the brain are a source of retinoic acid following stroke. BMC Neurosci. 17, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DD, and Seung HS (1999). Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791. [DOI] [PubMed] [Google Scholar]
- Lee NJ, Ali N, Zhang L, Qi Y, Clarke I, Enriquez RF, Brzozowska M, Lee IC, Rogers MJ, Laybutt DR, et al. (2018). Osteoglycin, a novel coordinator of bone and glucose homeostasis. Mol. Metab. 13, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SL, Kisseleva T, Brenner DA, and Duffield JS (2008). Pericytes and perivascular fibroblasts are the primary source of collagen-producing cells in obstructive fibrosis of the kidney. Am. J. Pathol. 173, 1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo L, Simon JM, Xing L, McCoy ES, Niehaus JK, Guo J, Anton ES, and Zylka MJ (2019). Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Kim SS, Yu K, Ghaleb AM, Takeda N, Manabe I, Nusrat A, Nagai R, and Yang VW (2011). Kruppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology 141, 1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrara BJ, Most D, Chang J, Bresnick S, Turk A, Schendel SA, Gittes GK, and Longaker MT (1999). Basic fibroblast growth factor and transforming growth factor beta-1 expression in the developing dura mater correlates with calvarial bone formation. Plast. Reconstr. Surg. 104, 435–444. [DOI] [PubMed] [Google Scholar]
- Mishra S, Choe Y, Pleasure SJ, and Siegenthaler JA (2016). Cerebrovascular defects in Foxc1 mutants correlate with aberrant WNT and VEGF-A pathways downstream of retinoic acid from the meninges. Dev. Biol. 420, 148–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi MM, Quadri SA, Khan MA, Gustin A, Suriya SS, Hassanzadeh T, Fahimdanesh KM, Adl FH, Fard SA, Taqi MA, et al. (2018). Subarachnoid trabeculae: a comprehensive review of their embryology, histology, morphology, and surgical significance. World Neurosurg 111, 279–290. [DOI] [PubMed] [Google Scholar]
- Nabeshima S, Reese TS, Landis DM, and Brightman MW (1975). Junctions in the meninges and marginal glia. J. Comp. Neurol. 164, 127–169. [DOI] [PubMed] [Google Scholar]
- Nakao T, Ishizawa A, and Ogawa R. (1988). Observations of vascularization in the spinal cord of mouse embryos, with special reference to development of boundary membranes and perivascular spaces. Anat. Rec. 221, 663–677. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Fraulob V, Garnier JM, Chambon P, and Dollé P. (2002). Differential expression of retinoic acid-synthesizing (RALDH) enzymes during fetal development and organ differentiation in the mouse. Mech. Dev. 110, 165–171. [DOI] [PubMed] [Google Scholar]
- Reiss K, Mentlein R, Sievers J, and Hartmann D. (2002). Stromal cell-derived factor 1 is secreted by meningeal cells and acts as chemotactic factor on neuronal stem cells of the cerebellar external granular layer. Neuroscience 115, 295–305. [DOI] [PubMed] [Google Scholar]
- Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, and Thesleff I. (2000). Integration of FGF and TWIST in calvarial bone and suture development. Development 127, 1845–1855. [DOI] [PubMed] [Google Scholar]
- Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, Bien E, Baum M, Bortolin L, Wang S, et al. (2018). Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segklia A, Seuntjens E, Elkouris M, Tsalavos S, Stappers E, Mitsiadis TA, Huylebroeck D, Remboutsika E, and Graf D. (2012). Bmp7 regulates the survival, proliferation, and neurogenic properties of neural progenitor cells during corticogenesis in the mouse. PLoS One 7, e34088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbonne S., Azibani F., Iatmanen S., Boulaftali Y., Richard B., Jandrot-Perrus M., Bouton MC., and Arocas V. (2012). In vitro and in vivo antiangiogenic properties of the serpin protease nexin-1. Mol. Cell. Biol. 32, 1496–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbonne S, Francois D, Raoul W, Boulaftali Y, Sennlaub F, Jandrot-Perrus M, Bouton MC, and Arocas V. (2015). Protease nexin-1 regulates retinal vascular development. Cell. Mol. Life Sci. 72, 3999–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, et al. (2009). Retinoic acid from the meninges regulates cortical neuron generation. Cell 139, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegenthaler JA, and Pleasure SJ (2011). We have got you ‘covered’: how the meninges control brain development. Curr. Opin. Genet. Dev. 21, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderblom C, Luo X, Blumenthal E, Bray E, Lyapichev K, Ramos J, Krishnan V, Lai-Hsu C, Park KK, Tsoulfas P, and Lee JK (2013). Perivascular fibroblasts form the fibrotic scar after contusive spinal cord injury. J. Neurosci. 33, 13882–13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R. (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utz SG, See P, Mildenberger W, Thion MS, Silvin A, Lutz M, Ingelfinger F, Rayan NA, Lelios I, Buttgereit A, et al. (2020). Early fate defines microglia and non-parenchymal brain macrophage development. Cell 181, 557–573.e18. [DOI] [PubMed] [Google Scholar]
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, et al. (2018). A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480. [DOI] [PubMed] [Google Scholar]
- Visel A, Thaller C, and Eichele G. (2004). GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller RO, Sharp MM, Christodoulides M, Carare RO, and Møllgård K. (2018). The meninges as barriers and facilitators for the movement of fluid, cells and pathogens related to the rodent and human CNS. Acta Neuropathol. 135, 363–385. [DOI] [PubMed] [Google Scholar]
- Yata Y, Scanga A, Gillan A, Yang L, Reif S, Breindl M, Brenner DA, and Rippe RA (2003). DNase I-hypersensitive sites enhance alpha1(I) collagen gene expression in hepatic stellate cells. Hepatology 37, 267–276. [DOI] [PubMed] [Google Scholar]
- Zarbalis K, Siegenthaler JA, Choe Y, May SR, Peterson AS, and Pleasure SJ (2007). Cortical dysplasia and skull defects in mice with a Foxc1 allele reveal the role of meningeal differentiation in regulating cortical development. Proc. Natl. Acad. Sci. USA 104, 14002–14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Yu T, Zhang XC, Nagasawa T, Wu JY, and Rao Y. (2002). Role of the chemokine SDF-1 as the meningeal attractant for embryonic cerebellar neurons. Nat. Neurosci. 5, 719–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The embryonic meninges single cell data generated during this study are available at NCBI Gene Expression Omnibus (GEO), accession number: GSE150219.


