Abstract
Vector-borne plant diseases have significant ecological and economic impacts, affecting farm profitability and forest composition throughout the world. Bacterial vector-borne pathogens have evolved sophisticated strategies to interact with their hemipteran insect vectors and plant hosts. These pathogens reside inplant vascular tissue, and their study represents an excellent opportunity to uncover novel biological mechanisms regulating intracellular pathogenesis and to contribute to the control of some of the world’s most invasive emerging diseases. In this perspective, we highlight recent advances and major unanswered questions in the realm of bacterial vector-borne disease, focusing on liberibacters, phytoplasmas, spiroplasmas, and Xylella fastidiosa.
Keywords: liberibacter, phytoplasma, spiroplasma, Xylella fastidiosa, vector-borne disease, effector, plant immunity
INTRODUCTION
Bacterial vector-borne diseases cause some of the most serious crop losses worldwide. Examples of bacterial vector-borne pathogens include members of the genera Xylella, Candidatus Liberibacter, Spiroplasma, and Ca. Phytoplasma. Whereas X. fastidiosa proliferates in the xylem, liberibacters, spiroplasmas, and phytoplasmas are phloem limited and proliferate in sieve elements (Figure 1). These pathogens are transmitted by different groups of piercing-sucking insects in the order Hemiptera (Jiang et al., 2019) (Figure 2). Citrus, grape, and olive industries have been seriously affected by these bacterial vector-borne pathogens in recent decades. Huanglongbing disease (HLB) caused by Candidatus Liberibacter asiaticus(CLas)isthe mostdamaging disease of citrus, resulting in estimated losses of over $7.8 billion in Florida since 2007 (Court et al., 2018), and the phytoplasma disease lime witches’ broom threatens the lime industry in the Middle East (Donkersley et al., 2018). Moreover, coconut phytoplasmas have destroyed millions of palms for centuries and are spreading locally (Gurr et al., 2016). Grape production has been significantly affected by Pierce’s disease of grapevine (X. fastidiosa), resulting in an annual cost of approximately $100 million in California alone (Tumber et al., 2014). Recently, European olive orchards have been devastated by the emerging disease olive quick decline syndrome caused by X. fastidiosa (Schneider et al., 2020). In the absence of disease control measures, the total economic loss to the olive industry is estimated to reach up to €5.2 billion in Italy and €16.86 billion in Spain over the next 50 years (Schneider et al., 2020).
Figure 1. Bacterial Vector-Borne Diseases That Persist in Plant Vascular Tissues.
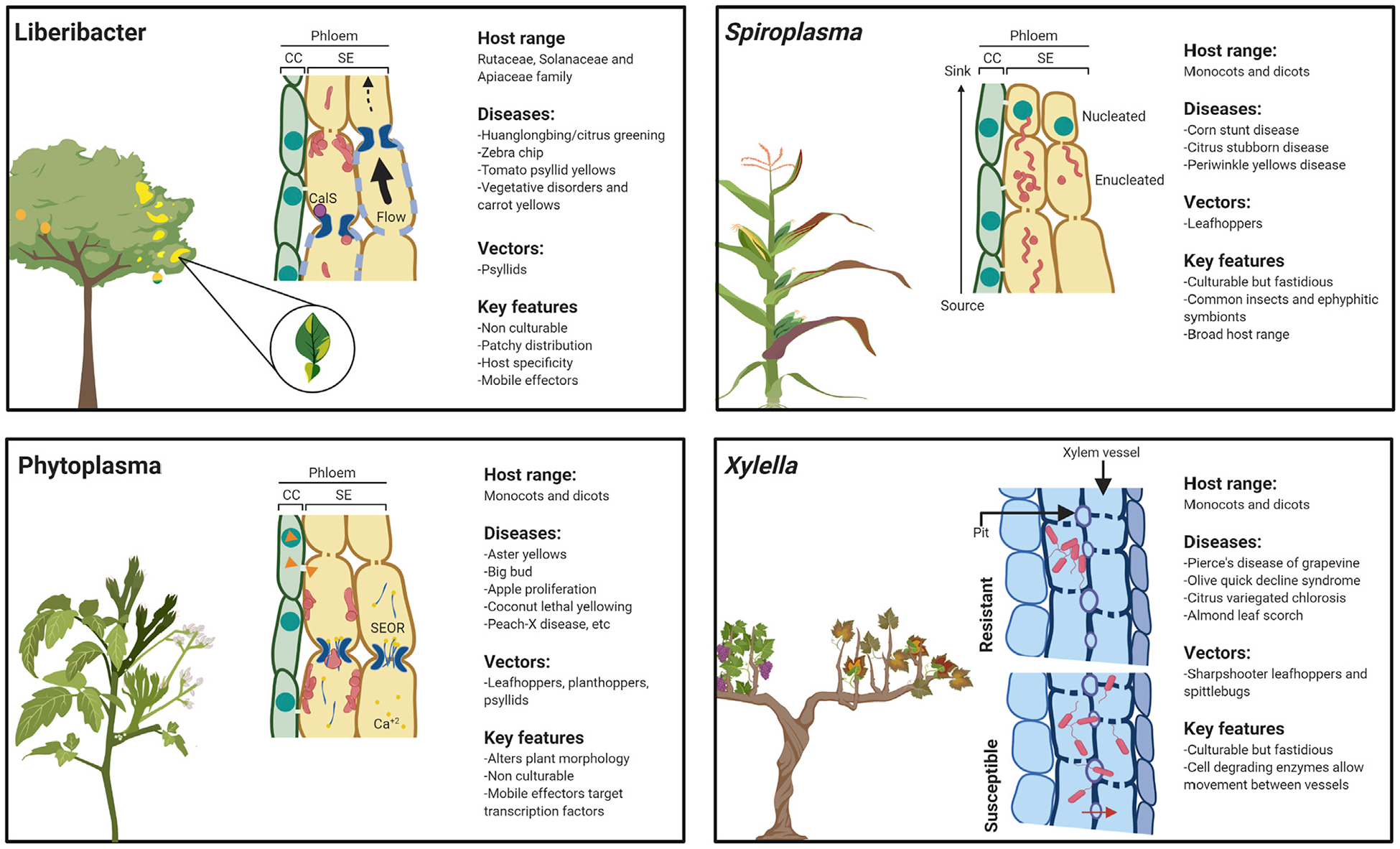
Pathogenic bacteria are delivered directly into the phloem (liberibacter, spiroplasma, and phytoplasma) or xylem (Xylella) during vector feeding. Upon delivery into the phloem, liberibacters and phytoplasmas are confined to sieve elements, move intracellularly through sieve pores, and tend to accumulate in sink tissues. Spiroplasmas preferentially localize near nucleated cells or in phloem parenchyma cells. Phytoplasmas attach to the sieve element membrane. Phloem-limited bacteria are pleomorphic. Vector feeding and bacterial proliferation induce Ca2+ production, accumulation of Sieve Element Occluding Relatives (SEOR) proteins, and callose deposition through callose synthases (CalS). Xylella cells move between xylem vessels through the pit membrane and exhibit differential distribution in resistant hosts.
Figure 2. Circulative Propagative and Non-circulative Propagative Bacterial Localization in Insect Vectors through the Perspective of Phylogenetic Relationships.
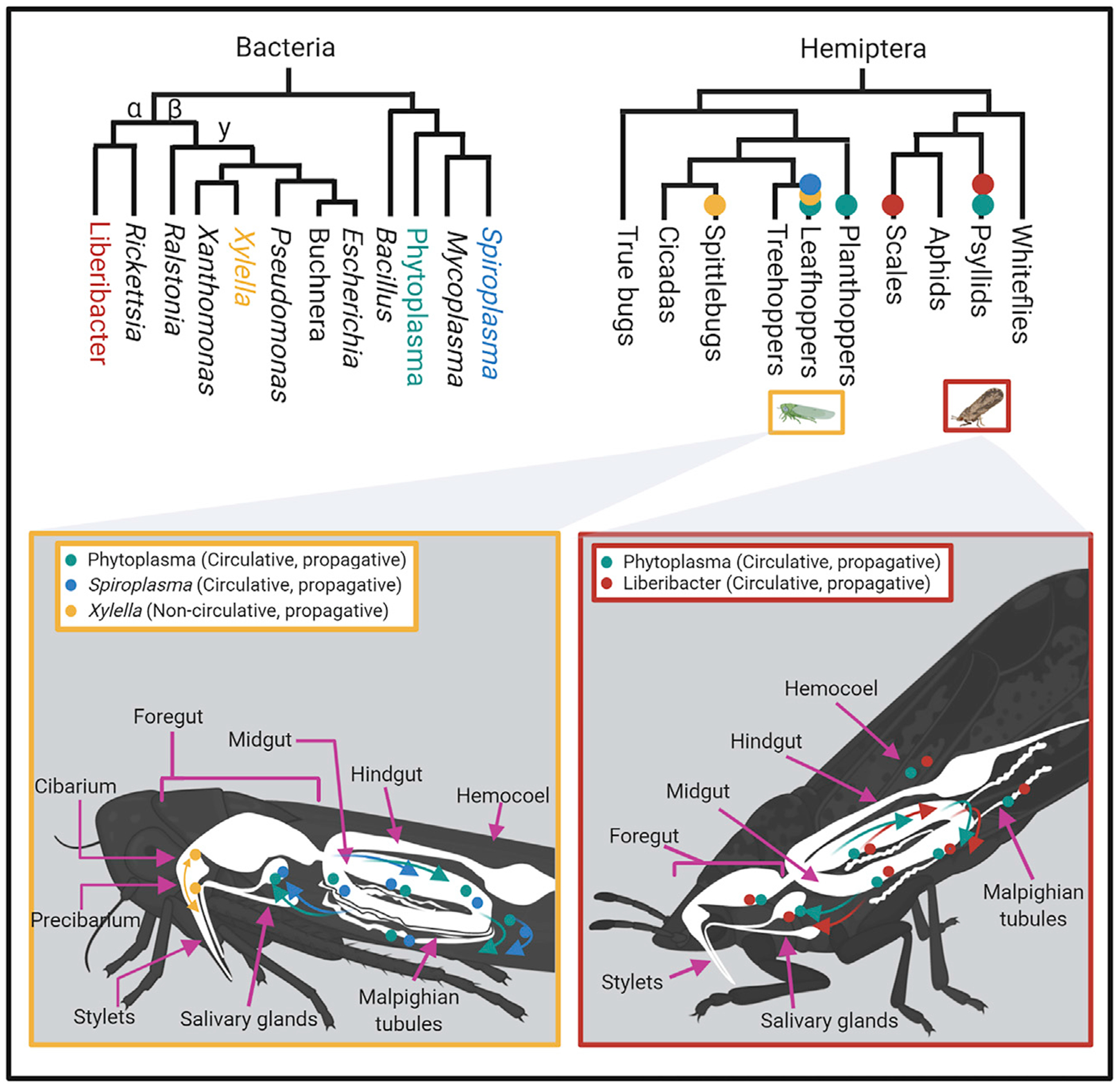
Phylogenetic relationships are simplified and represent relative ancestry. Top left, phylogeny showing hemipteran families and close relatives known to be vectors of each respective pathogen. Top right, phylogeny showing relationships among the four main pathogens discussed in the perspective. Detailed bacterial distribution across main tropic regions in the insect vector body is exemplified using a leafhopper (bottom left) and psyllid (bottom right). Colors are consistent in all four panels. Teal, Ca. Phytoplasma spp.; blue, Spiroplasma spp.; red, Ca. Liberibacter spp.; yellow, Xylella fastidiosa.
Bacterial vector-borne disease research is challenging because of fastidious pathogen growth, the inability to culture multiple species, reliance on an insect vector for transmission, and the lack of model systems (Jiang et al., 2019). However, deciphering the interaction between pathogen, vector, and plants in diverse systems provides new opportunities to discover novel biology that regulates intracellular pathogens and cannot be addressed in more tractable systems. Transmission of bacteria directly into vascular tissues, such as the xylem or phloem, may be an adaptation that allows these pathogens to evade plant immune perception. Their virulence mechanisms are probably different from those of other well-characterized Gram-negative bacteria and provide unique opportunities to uncover novel biological insights (Toruño et al., 2016; Kruse et al., 2019). A greater understanding of pathogen host and vector manipulation, as well as plant responses, is critical for the effective control of vector-borne disease. Here, we highlight major advances and unanswered questions in bacterial vector-borne plant disease and provide a roadmap for future research directions.
WHAT ARE THE MINIMAL REQUIREMENTS FOR LIFE?
Bacterial Vector-Borne Pathogenicity Has Evolved at Least Four Times
Considering their small genome size, limited metabolic capacity, and life cycle that involves two hosts, vector-borne bacteria have an important role to play as models for study in research fields as varied as synthetic biology, evolution, phytopathology, and biochemistry. Genome reductions are predominantly caused by loss of metabolic genes, making these bacteria dependent on the acquisition of diverse metabolites from their hosts via various transporters, predominantly ABC transporters (Kube et al., 2012; Oshima et al., 2004). Ca. Liberibacter and X. fastidiosa are both Gram-negative bacteria but belong to different orders, i.e., Rhizobiales (class Alphaproteobacteria) and Xanthomonadales (class Gammaproteobacteria), respectively (Figure 2). Spiroplasmas and phytoplasmas both belong to the class Mollicutes. However, spiroplasmas and phytoplasmas have different codon usages, and spiroplasmas share a common ancestry with mycoplasma and phytoplasmas with the saprotrophic acholeplasmas (Gundersen et al., 1994; Gupta et al., 2019) (Figure 2). Spiroplasmas are common commensals, pathogens, or obligate symbionts of diverse arthropods, and only three Spiroplasma sp. (S. citri, S. kunkelii, and S. phoeniceum) are vector-borne plant pathogens. By contrast, all phytoplasmas are insect-borne plant pathogens. The origin of the phytoplasma lineage dates back to ~641 million years ago (Ma) and predates land plants and hemipteran insects, and the crown group of phytoplasmas began to diversify ~316 Ma, roughly coinciding with the origin of these two hosts (Cao et al., 2020). The evolutionary history of these bacteria indicates that the requirement for insect transmission evolved independently multiple times. In all four cases, their evolution involved genome reductions and the retention and acquisition of virulence genes that enabled them to colonize animal and plant hosts.
Bacterial Vector-Borne Pathogens Have Distinct Metabolic Requirements
The four taxa of vector-borne plant pathogenic bacteria exhibit signatures of differential genome reduction rates, ranging from 2.5 Mb for X. fastidiosa to ±1.7–1.2 Mb for spiroplasma and liberibacters to <1 Mb for phytoplasmas. X. fastidiosa and S. citri are fastidious but can be cultured and genetically modified (Bové, 1997; Bové and Garnier, 2003; Perilla-Henao and Casteel, 2016). By contrast, Ca. Liberibacter sp. and phytoplasmas have largely resisted transformation (Ha et al., 2019; Naranjo et al., 2019). Thus, it is likely that each pathogen relies on its own set of genes for growth, and culturability is not necessarily associated with genome size but rather with genes that were lost and are required for life in vitro. For example, mycoplasmas with genome sizes similar to those of phytoplasmas were cultured in rich artificial medium (Tully et al., 1977). Phytoplasmas lack different metabolic genes compared with other bacteria in the class Mollicutes (Chen et al., 2012). For instance, compared with mycoplasmas and S. citri, phytoplasmas lack a phosphotransferase system for the import of sugars and seem to import disaccharides by means of ABC transporter systems, import malate instead of lactate, and lack genes for generating their own ATP (which must be sequestered from insect and plant hosts) (Oshima et al., 2004; Bai et al., 2006; Kube et al., 2014).
How Can Current Knowledge Inform Minimal Requirements for Life?
Current data indicate that the minimal requirements for life and pathogenesis differ among vector-borne bacteria. They involve a minimal genome for basic metabolism that is adapted for living in the xylem and insect foregut for X. fastidiosa and in the phloem and various insect organs for Ca. Liberibacter, Spiroplasma spp., and Ca. Phytoplasma. Comparisons with closely related culturable bacteria will aid in the synthesis of genomes to generate minimal cells that grow independently in defined media. Genes essential for minimal life have been a major research axis in the “top–down” synthetic biology field, which aims to design novel systems based on known biology (Roberts et al., 2013). Identifying the minimal requirements for life in vitro is important for gaining insight into fundamental biological processes, both for synthetic biology that aims to construct artificial cell parts and for efforts to define the origin of life.
Technological achievements that enable whole small genome transplantation (Lartigue et al., 2007) could be used to engineer the phytoplasma genome. Adding metabolic and transporter genes for transplantation into a recipient cell could facilitate biochemical advances and growth under axenic conditions. However, genome transplantation requires a culturable recipient cell that is phylogenetically close to the donor cell (Labroussaa et al., 2016). Genome transplantation has not yet been achieved in acholeplasma, but the panel of transplantable species is increasing. Because S. citri and phytoplasmas share many physiological and ecological traits, S. citri, which is culturable, can serve as a model for a better understanding of phytoplasma biology (Renaudin et al., 2015). These studies will require expanding the toolbox available for S. citri genetic manipulation.
IMPORTANCE OF TISSUE TROPISM FOR PLANT COLONIZATION
Tropism refers to the ability of a given organism to move and colonize a specific tissue. Research in vector-borne disease presents unique opportunities to investigate vascular patterning, vascular defense, and novel componentsthat regulate pathogen movement. Some insectvector-borne plant viruses encode an active strategy in the viral genome to maintain phloem localization, and this strategy may facilitate dispersal by phloem-feeding insect vectors and/or evasion of plant host immune responses (Peter et al., 2009).
Pathogen localization in a host is probably a combination of vascular flow, host genotype, and possibly bacterial preference for a tropic region (Figure 1). Accumulation in sink tissues, including roots and growing tissues, is often observed in phloem-limited pathogens; however, their distribution within the plant is variable (Figure 1). This distribution extends to individual sieve tubes in the phloem where, for example, CLas massively proliferates in some tubes while barely colonizing others (Achor et al., 2020; Hartung et al., 2010), and spiroplasmas accumulate near undifferentiated phloem cells (Bové et al., 2003). Recovery of apple trees from apple proliferation phytoplasma is associated with increased callose depositions that prevent recolonization of the apple tree crowns during the spring and with enhanced overall defense responses of recovered apple trees (Musetti et al., 2010). Characterization of CLas and X. fastidiosa localization in citrus and related genotypes indicates that replication and establishment are correlated with the host’s ability to resist infection, leading to lower pathogen populations in genotypes that exhibit some resistance (Coletta-Filho et al., 2020; Ramadugu et al., 2016). X. fastidiosa exhibits differential tropism in the xylem of susceptible and resistant citrus genotypes; it is able to move to secondary xylem in susceptible genotypes but is confined to the primary xylem in resistant genotypes (Figure 1) (Niza et al., 2015). We lack a mechanistic understanding of how host genetic variability affects pathogen colonization and tropism. However, studies have correlated, for example, pit membrane carbohydrate composition with the ability of X. fastidiosa to colonize different grapevine varieties (Ingel et al., 2019). It is evident that individuals in a population respond differently to infection and that disease pressure and the environment play a role in mediating host responses. The composition of microbial communities can also influence disease. For example, the composition and diversity of citrus leaf and root microbiota were associated with HLB symptom severity (Blaustein et al., 2017).
To understand how the host regulates and responds to pathogen colonization, tools must be developed to investigate vascular biology; these may include inducible expression systems, cell-specific promoters, and robust silencing and gene-editing platforms. Recent advancements in phloem-limited viral vectors that enable expression of small genes and RNAi, such as Citrus tristeza viral vectors, hold promise for testing the importance of specific loci (Dawson et al., 2015). The development of advanced microscopy techniques will enable researchers to investigate bacterial movement during the course of infection and colonization across different pathogen strains and host genotypes (Newman et al., 2003). These tools will enable the identification of key loci important for vascular responses, including those that affect vascular patterning, limit pathogen distribution, and affect tropism. A greater understanding of how pathogens are distributed holds great promise for disease control by facilitating early detection of diseased plants, a task that is complicated due to uneven pathogen distribution. Early and efficient detection of bacterial pathogens that infect perennial woody plants, such as CLas, X. fastidiosa, S. citri, and Ca. Phytoplasma palmicola, will enable the removal of infected plants that serve as a reservoir for pathogen spread. A detailed understanding of vascular biology is also needed to develop delivery strategies for therapeutic molecules to control bacterial infection in plants.
Pathogen Movement and Host Colonization
Phloem-limited bacteria must rely on intracellular communication to move and colonize different sections of the phloem. The late systemic spread in sieve elements colonized by CLas and phytoplasmas coupled with the slow seasonal migration of Ca. Phytoplasma mali (apple proliferation phytoplasma) from roots to new apple flush suggest that these pathogens can also move against the phloem flow, albeit at a slow rate (Hilf and Luo, 2020; Schaper and Seemüller, 1984; Wei et al., 2004). Phytoplasmas lack genes coding for cytoskeletal elements or flagella (Music et al., 2019; Oshima et al., 2004). CLas flagellar genes are repressed in planta, and flagellin expression can be detected only in the vector (Andrade et al., 2020; Yan et al., 2013). On one hand, motility mutants of S. citri that display only twitching movement are not impaired in pathogenicity. On the other hand, loss of helicity and motility seems to occur naturally at a very low rate in this pathogen (Duret et al., 2003; Fletcher et al., 2006). In spiroplasmas and other mollicutes, specialized tip structures consisting of exposed surface proteins are important pathogenicity determinants required for host membrane attachment and pathogen movement (Ammar el et al., 2004). Similar surface proteins were also found in phytoplasmas and may play an analogous role (Kakizawa et al., 2006a, 2006b). Liberibacters and mollicutes use their pleomorphic shape to cross the tight space between sieve plates, even when they are fortified by callose (Figure 1) (Achor et al., 2020) (Waters and Hunt, 1980). Phytoplasmas have been shown to adhere to the inner surface of the sieve tube membranes (Marcone, 2009), and liberibacters often aggregate near sieve pores (Achor et al., 2020), a feature that may facilitate movement (Figure 1).
The ability of the Xanthomonadaceae family to colonize the xylem has been linked to the presence of CbsA, a cell-wall-degrading cellobiohydrolase (Gluck-Thaler et al., 2020). Deletion of CbsA in non-vector-borne vascular Xanthomonas oryzae pv. oryzae increased apoplastic colonization and symptom development. By contrast, X. fastidiosa cbsA mutants were severely reduced in vascular colonization and symptom development (Gluck-Thaler et al., 2020). CbsA expression correlates with the planktonic lifestyle and cell proliferation of X. fastidiosa, and it is also highly expressed in biofilm-deficient mutants (Gouran et al., 2016). Tracking studies of fluorescent-tagged cell-degrading enzymes, including CbsA, permit the study of movement in degradative enzyme mutants and will be required to understand the role of these enzymes in bacterial movement from primary to secondary xylem and between xylem vessels.
Our inability to culture Ca. Liberibacters and phytoplasmas present a significant challenge to the identification of important loci required for host or vector tropism. Other than Liberibacter crescens BT-1, no liberibacters have been obtained in pure culture despite extensive efforts (Ha et al., 2019; Naranjo et al., 2019). Spiroplasma represents an excellent model for the investigation of phloem-limited bacteria because of its culturability, broad host range, and potential for genetic manipulation (Renaudin et al., 2014). The development of high throughput genetic screens such as those that use TNseq will quickly advance our understanding of general mechanisms of host colonization in phloem-limited bacteria. The structure of the plant phloem may play a role in CLas acquisition and transmission by the psyllid vector by acting as a determinant for the establishment of plant feeding sites (George et al., 2017). Major unanswered questions also include the influence of genetic variation on pathogen tropism. Do tropism patterns observed under laboratory conditions with defined bacterial isolates reflect field conditions? What are the roles of pathogen and host genetic diversity in the colonization of particular vascular niches?
PLANT DEFENSE RESPONSE TO VECTOR-BORNE DISEASES
Plants possess sophisticated immune systems capable of perceiving conserved pathogen, damage, and insect features using surface-localized receptors and intracellular nucleotide-binding leucine-rich repeat (NLR) immune receptors that recognize secreted pathogen and insect proteins (Boissot et al., 2016; Boutrot and Zipfel, 2017; Lolle et al., 2020). Immediate plant defenses are triggered by vector feeding. Hemipteran insects secrete watery saliva as they probe into the apoplast and different cell types to acquire cell contents until the stylet reaches the phloem (Jaouannet et al., 2014). Insect effectors secreted during cell penetration can be perceived by NLR immune receptors (Boissot et al., 2016). Watery saliva contains insect features (herbivore-associated molecular patterns [HAMPs]) as well as microbial features from bacterial pathogens or symbionts (microbial-associated molecular patterns [MAMPs]) (Chaudhary et al., 2014; Prince et al., 2014). Honeydew is also known to contain HAMPs and MAMPs and to activate immunity (Schwartzberg and Tumlinson, 2014; Wari et al., 2019). These features are perceived by surface-localized pattern recognition receptors, resulting in the induction of plant immune responses (Chaudhary et al., 2014; Jaouannet et al., 2014; Steinbrenner et al., 2019).
The stylets of hemipteran insects induce defense responses and Ca2+ bursts during probing (Vincent et al., 2017), and watery saliva also contains hemipteran effectors that can suppress immune responses, illustrating the importance of plant immunity in vector-borne disease (Mugford et al., 2016). HAMP and MAMP release during initial vector probing can activate immune responses that affect the phloem environment. The phloem itself is an important highway for systemic signaling in response to wounding and immune perception, which result in rapid waves of Ca2+ (Toyota et al., 2018). Many questions remain concerning innate immunity in the phloem. Do canonical plant immune receptors function in phloem tissues? Can bacterial effector proteins that move cell-to-cell via plasmodesmata be recognized by intracellular plant immune receptors?
Common phloem defense responses include the production of Ca2+, callose deposition, and activation of phloem-specific forisomes and P proteins (Figure 1) (Jiang et al., 2019). Callose deposition in sieve pores is a common response to infection by phloem-limited pathogens. CLas infection induces the expression of multiple citrus callose synthases, including CalS3, CalS7, CalS8, CalS9, CalS11, and CalS12 (Granato et al., 2019). Callose hydrogel complexes have distinct biophysical properties that contribute to elasticity, which may account for the inability of callose to impair mass flow (Hunter et al., 2019; Abou-Saleh et al., 2018). Activation of Ca2+-dependent sieve proteins, including Sieve Element Occluding Relatives (SEOR) P proteins and forisomes, occurs during vector feeding (Figure 1) (Knoblauch et al., 2014). SEOR proteins form highly organized aggregates that disperse upon Ca2+ activation, surround the site of vector feeding, and block phloem acquisition to play an important role in resistance against phloem-feeding insects (Garzo et al., 2018; Peng and Walker, 2020). Surprisingly, Ca. Phytoplasma asteris exhibits reduced growth in Arabidopsis seor1 mutants, indicating that AtSEOR1 may also be a susceptibility gene (Pagliari et al., 2017). Development of genetic tools for investigating phloem responses will be necessary to provide clear genetic evidence linking phloem-specific defenses with disease resistance.
Plant genome sequencing projects have enabled quantitative gene and protein expression studies of host–pathogen interactions in vector-borne disease. For CLas, these studies have identified citrus genotype-specific and, importantly, common responses to the bacterium during disease progression. Both Washington Navel orange and Lisbon lemon showed perturbations in carbohydrate metabolism and differential expression of proteins involved in plant defense (Chin et al., 2019; Ramsey et al., 2020). The two aforementioned studies captured the plant response in the entire leaf tissue, possibly masking vascular tissue-specific responses. Advances in single-cell RNA sequencing and proteomics, coupled with RNA aptamers capable of capturing specific vascular cell types, can facilitate an understanding of host responses in cells that are directly in contact with and adjacent to pathogen cells (Denyer et al., 2019; Solanki et al., 2020).
A common xylem defense response is the formation of tyloses, which are extensions of living parenchyma cells. Despite being a common and visible defense response, tylose formation occurs late in infection and fails to prevent bacterial movement in the X. fastidiosa–grape interaction; instead, it exacerbates disease symptoms (Stevenson et al., 2004). Both intracellular and surface-localized immune receptors that recognize bacterial and fungal xylem-colonizing pathogens such as Fusarium oxysporum and Ralstonia solanacearum have been cloned (Narusaka et al., 2009; Takken and Rep, 2010). Both Fusarium and Ralstonia must invade the root before reaching the xylem. There is evidence that immune perception is important in the activation of plant responses to X. fastidiosa infection, but perception is delayed by the presence of cell surface lipopolysaccharides (Rapicavoli et al., 2018).
CRITICAL PATHOGENICITY DETERMINANTS
Bacterial Vector-Borne Pathogens Have Different Retained and Acquired Virulence Capacities
Bacterial vector-borne pathogens appear to have differentially retained or acquired virulence genes. X. fastidiosa colonizes the plant xylem and the foregut of its insect vectors, which are xylem sap-feeding leafhoppers (sharpshooters) and spittlebugs, through biofilm formation on the cuticular surface of the foregut lumen (Figure 2) (Killiny et al., 2013; Ionescu et al., 2016). Virulence factors involved in X. fastidiosa migration, rather than in biofilm formation in the xylem and the insect, are homologs of proteins from related free-living and plant pathogenic Xanthomonads (Almeida et al., 2012; Caserta et al., 2017). The latter possess type III secretion systems (TTSSs) for the penetration of plant cell membranes and the deposition in cells of an arsenal of protein effectors that modulate plant processes, but such systems are lacking in the X. fastidiosa genome (Lu et al., 2008; Toruño et al., 2016). Presumably, being xylem limited, X. fastidiosa does not require TTSSs and associated effectors, although it possesses other systems for the secretion of virulence factors that may unload from the xylem and may differ from those of plant pathogenic Xanthomonads (Lu et al., 2008).
In contrast to X. fastidiosa, Ca. Liberibacter species, phytoplasmas, and spiroplasma plant pathogens all colonize the phloem and, moreover, are invasive colonizers of their insect vectors. Ca. Liberibacters are transmitted by phloem-feeding psyllids, spiroplasmas by phloem-feeding leafhoppers, and phytoplasmas by phloem-feeding leafhoppers, planthoppers, and psyllids (Figure 2). All three bacteria can be pathogenic to their insect vectors as well as their plant hosts (Bové, 1997; Marina Mann et al., 2018; Koinuma et al., 2020). S. citri takes advantage of uncommon post-translational mechanisms to increase the antigenic diversity at its surface, which may play a role in insect colonization by this mollicute (Dubrana et al., 2017). Moreover, the virulence genes of phytoplasmas lie within large pathogenicity islands of ± 20 kb that are comprised of composite transposon-like elements and named potential mobile units (PMUs) (Bai et al., 2006). These PMU-encoded virulence genes are differentially expressed between phytoplasmas in the plant and vector (Toruño et al., 2010). The pathogenicity islands are prone to frequent mutations and deletions, recombine with each other, and can form extrachromosomal units, making phytoplasma genomes highly unstable but probably also giving rise to more opportunities for adaptation (Bai et al., 2006; Arashida et al., 2008; Music et al., 2019). PMU genes include effectors that modulate plant development (discussed below). There is evidence of horizontal DNA transfer, including that of PMU-like elements, among distantly related phytoplasmas and between spiroplasmas and phytoplasmas (Bai et al., 2004; Arashida et al., 2008; Music et al., 2019). Spiroplasma genomes are also highly repeat rich; the repeats include phage-like elements and multiple plasmids with repeat-rich adhesin-like genes that are involved in virulence (Beven et al., 2015). Taken together, these findings suggest that there are ongoing acquisition and recombination of genomic virulence clusters that have integrated into the minimal genomes of vector-borne plant pathogens.
However, most interesting for pathologists are the minimal requirements for pathogenesis. The minimal requirement for pathogenesis appears to be defined by genomic areas different from those involved in metabolism, although mechanisms that detect the processing of specific metabolites may be part of a larger machinery that controls the regulation of virulence gene expression. A genomic area that encodes virulence factors, such as a phytoplasma PMU, may be isolated and integrated into another culturable bacterium to assess mechanisms that enable the colonization of plants versus insect vectors.
Effector Biology
The ability to secrete proteins, called effectors, is required for diverse organisms to colonize a host and/or cause disease (Toruño et al., 2016). Among bacterial vector-borne pathogens, phytoplasma effectors are well characterized. Although phytoplasmas are restricted to the phloem, effectors secreted into the cytoplasm of sieve cells can be unloaded from the phloem and move systemically, possibly via plasmodesmata (Bai et al., 2009; Hoshi et al., 2009; MacLean et al., 2011a). Phytoplasma effectors have dramatic impacts on plant development and induce several distinct symptoms characteristic of diseases. For example, SAP11 effectors from diverse phytoplasmas induce a range of phenotypes when stably expressed in plants, including crinkled leaves and siliques, increased stem proliferation, altered root architecture, and abnormal glandular trichome development (Sugio et al., 2011b; Lu et al., 2014; Tan et al., 2016; Chang et al., 2018; Wang et al., 2018; Pecher et al., 2019). The role of these diverse plant phenotypes induced by phytoplasma effectors remains an important subject of future research.
Effector targeting of transcription factors is a strategy for targeting multiple plant processes simultaneously. SAP11 proteins differentially interact with and destabilize members of the plant TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1 (TCP) transcription factor family, which regulates diverse plant developmental processes. Another phytoplasma effector, SAP54/phyllogen, is responsible for the conversion of plant flowers into leaf-like tissues, a phenomenon known as phyllody that is commonly seen in infected plants (MacLean et al., 2011a, 2014; Maejima et al., 2014). SAP54/phyllogen targets plant homeotic MADS-box transcription factors that control floral development for destabilization through the host 26S proteasome (MacLean et al., 2014). A third phytoplasma effector, TENGU, induces dwarfism, stem proliferation, and sterility in plants by unknown mechanisms that suppress auxin and jasmonic acid signaling (Hoshi et al., 2009; Minato et al., 2014). Phytoplasma effectors appear to manipulate the entire host to their advantage. Not only do they maintain newly infected tissues in a juvenile or vegetative state that facilitates nutrient acquisition, but they also render plant tissues more attractive or less hostile to vectors that feed on infected plants (Sugio et al., 2011c; Orlovskis and Hogenhout, 2016; Tomkins et al., 2018).
In contrast to phytoplasmas, Ca. Liberibacter effectors are under-explored, and Spiroplasma effector functions remain completely elusive. Effectors from Ca. Liberibacter can inhibit immune-related protease activity (SDE1), suppress host cell death (HPE1), and suppress ROS production through peroxidase activity (Jain et al., 2015; Clark et al., 2018; Levy et al., 2020). It is likely that the immune-suppressing activity of liberibacter effectors is just the tip of the iceberg. For example, the Ca. Liberibacter effector SDE1 is multifunctional, localizes to diverse subcellular compartments, induces cell death in the model plant Nicotiana, and targets diverse host genes (e.g., DEAD-box RNA helicase DDX3 and immune proteases) to manipulate different plant responses (Pitino et al., 2016; Clark et al., 2018; Zhou et al., 2020). The moonlighting properties of SDE1 are reminiscent of viral proteins, which are capable of manipulating multiple cellular processes simultaneously through interactions with diverse host targets (DeBlasio et al., 2016).
Effectors from vector-borne pathogens exhibit variation in expression across host and vector and are influenced by plant genotype. Liberibacters and phytoplasmas deploy specific suites of effectors to differentially manipulate each organism and enable the switch between vector and host (Thapa et al., 2020). For example, 63% of CLas and 60% of phytoplasma effectors were primarily expressed in citrus and Arabidopsis, and 7.4% of CLas and 39% of phytoplasma effectors were primarily expressed in the insect vector (Hogenhout et al., 2009; MacLean et al., 2011b). In addition, effector expression profiles differ among host genotypes with varying levels of tolerance and resistance for both CLas and phytoplasma (Bertazzon et al., 2019; Shi et al., 2019). Filamentous pathogens spatially and temporally regulate suites of effectors during different infection stages (Toruño et al., 2016). Liberibacters, phytoplasmas, and spiroplasmas can infect perennial plants, including trees, residing in their hosts for years. Changes in effector expression over the course of long-term infection is an unexplored area. Is effector expression influenced by host, pathogen, and vector genotype? To what extent do bacterial effector proteins regulate tissue tropism during infection?
Functional characterization of effector candidates is key to understanding the tripartite interaction between plants, bacteria, and insects (Figure 3). Robust effector prediction pipelines will enable more accurate identification of effector candidates. Effectors from different vector-borne bacteria probably have divergent roles compared with those from apoplastic colonizing pathogens. The first challenge in characterizing effector function is to determine where these proteins function inside the host. For instance, both SAP11 and TENGU were demonstrated to accumulate in different cell types beyond the phloem using immunohistochemical analysis (Bai et al., 2009; Hoshi et al., 2009). The CLas SDE1 effector can also be detected in plant tissues by immunoblot when bacteria are undetectable by PCR (Pagliaccia et al., 2017). Depending on their localization, effectors can directly or indirectly affect bacterial growth and colonization. Because most bacterial effectors are small, it remains difficult to visualize effector localization and movement in vivo. Large tags, such as fluorescent proteins, may interfere with effector localization, movement, or function. In this regard, the development of new fluorescent-labeling methods with smaller tags for visualizing effector proteins in the hosts is highly desirable (Figure 3).
Figure 3. Identification and Functional Analysis of Vector-Borne Bacterial Effectors.
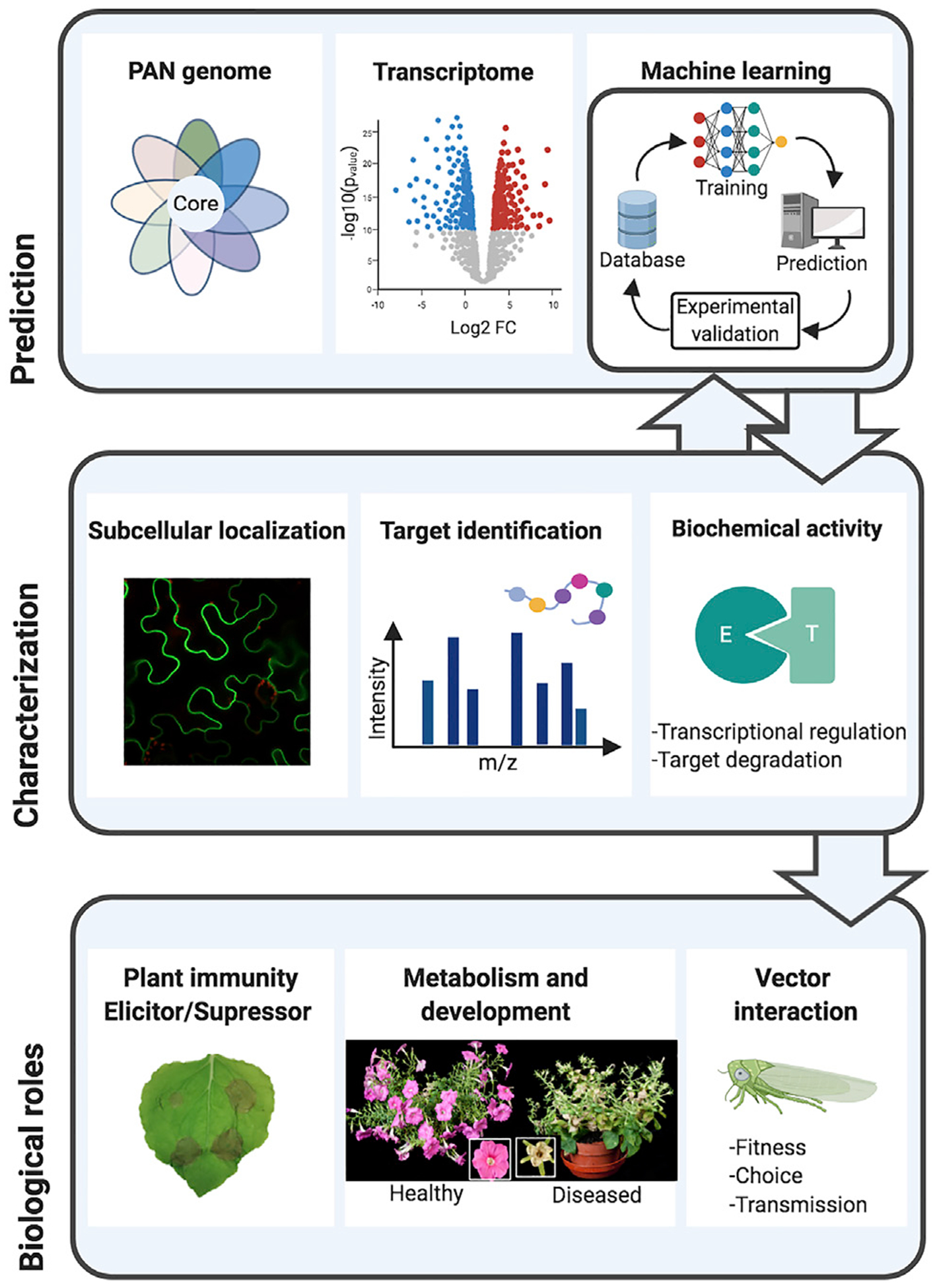
(A) Prediction of effector candidates by machine learning, PAN genome analysis, and host-specific gene expression analysis.
(B) Functional characterization of effector candidates using localization, target identification, and mode-of-action studies.
(C) Potential roles of effectors in modulating plant–bacteria–insect interactions. Images of healthy and AY-WB phytoplasma-infected petunia plants. Images are close-ups of a healthy flower and an infected flower exhibiting virescence (floral greening).
A second challenge lies in the identification of effector targets. Because the host range of some pathogens can be quite specific, the tools and resources for identifying effector targets may not be as readily available as in model organisms such as Arabidopsis. One potential solution is the establishment of more pathosystems or surrogate systems for studying vector-borne bacterial diseases. For example, Nicotiana has been used to screen for CLas effectors that induce cell death and could therefore be useful in the identification of plant receptors for engineering resistance in citrus (Pitino et al., 2016) (Figure 3). The next challenge is to understand how effectors interfere with host processes and facilitate pathogen spread. As mentioned above, the reprograming of plant development by phytoplasma effectors SAP11 and SAP54 also modulates plant–insect interactions and promotes insect vector attraction and fecundity (Sugio et al., 2011a; MacLean et al., 2014; Orlovskis and Hogenhout, 2016). However, whether bacterial effectors modulate plant–insect interaction more generally requires further exploration. Moreover, current studies on vector-borne bacteria largely focus on the function of an effector within one organism, either a plant or insect. It remains to be seen how effector-triggered changes in one organism may interfere with processes in another, resulting in effective disease spread.
VECTOR COMPATIBILITY AND VECTOR IMMUNITY
Does the Vector Actively Limit Pathogen Colonization and Tropism?
The most important hemipteran vectors that transmit bacterial pathogens are members of Sternorrhyncha, including psyllids (Psylloidea) and Auchenorrhyncha, including leafhoppers (Membracoidea), froghoppers/spittlebugs (Cercopoidea), and planthoppers (Fulgoroidea) (Figure 2) (Bové and Garnier, 2003; Perilla-Henao and Casteel, 2016; Sicard et al., 2018). Vector-borne bacterial pathogens depend on their insect vectors for spread and transmission to plants, and insect–bacteria interactions are as intricate as those between plant and bacteria. Some pathogens provide fitness benefits to their vectors; however, neutral or positive effects are not always observed. Similar to the plant host, the tissue tropism of the pathogen within the vector defines the mode of insect transmission.
Transmission occurs in distinct phases: acquisition of the bacteria from infected plants, translocation through the body tissues (which includes bacterial propagation; X. fastidiosa has a different mode of insect colonization), and inoculation of a new recipient host plant (Ammar et al., 2018; Heck and Brault, 2018; Koinuma et al., 2020). The anterior midgut of the alimentary canal and type III cells of salivary glands were identified as the major sites of onion yellows phytoplasma infection in a leafhopper vector (Koinuma et al., 2020). For CLas acquisition, the insect developmental stage is critical, but how this relates to tissue tropism is not well understood. CLas must be acquired during the nymphal stage for adults to transmit efficiently (Inoue et al., 2009; Ammar et al., 2016; George et al., 2018). Moreover, young citrus tissue (called flush) where the nymphs hatch and develop becomes infectious just 15 days after inoculation with CLas (Lee et al., 2015). Thus, asymptomatic spread coincides with the vector’s reproductive cycle. This pattern of spread underscores the need for research on early detection and removal of infected plant material to prevent disease outbreaks.
Molecular methods, such as in situ hybridization, confocal microscopy, electron microscopy, and qPCR, have been used to localize bacteria to specific insect tissues (Ammar et al., 2011; Ghanim et al., 2016, 2017; Kruse et al., 2017; Mann et al., 2018) and have identified apoptosis as involved in the developmental regulation of CLas acquisition. The gut is a physical barrier to acquisition and the first site of proliferation for propagative pathogens (Figure 2). In adult D. citri, CLas induces extreme apoptosis and nuclear disruption in the midgut and malpighian tubules, whereas nuclei in foreguts and hindguts appear normal (Ghanim et al., 2016; Mann et al., 2018). Comparative transcriptome analyses of CLas infected and uninfected D. citri midguts revealed extensive transcriptional changes, including multiple differentially expressed genes involved in the ubiquitin-proteasome and immune system (Yu et al., 2020). Collectively, these data highlight extensive cellular and transcriptional changes in adult D. citri midguts in response to CLas acquisition.
Whereas adult D. citri midguts are severely affected by CLas, nymphal midgut nuclei are not affected (Mann et al., 2018). Evidence for midgut cell apoptosis as a barrier to CLas acquisition by adult D. citri comes from comparative studies on Bactericera cockerelli, the potato psyllid. In this psyllid, Ca. Liberibacter solanacearum (CLso) does not cause apoptotic nuclear disruption in any cells of the gut of adult psyllids, and adult psyllids are able to acquire and transmit CLso (Tang and Tamborindeguy, 2019). Development of strategies that induce apoptosis in the psyllid midgut, by manipulation of caspase genes, for example, may be a novel means to control acquisition and transmission of liberibacters by psyllids. For example, the legume lectin concanavalin A has been reported to induce apoptosis in the potato psyllid midgut when ingested from an artificial diet (Tang et al., 2020).
The Relationship between Vector-Borne Pathogens and Their Vectors
Studies on the coevolution of two stunting mollicutes, the Dalbulus leafhopper spp., and maize-related grasses eloquently highlight how host associations in nature have favored positive relationships between plant pathogenic bacteria and their insect vectors. Maize bushy stunt phytoplasma (MBSP) and corn stunt spiroplasma (S. kunkelli) are two major maize stunting pathogens primarily transmitted by two maize specialists, Dalbulus maidis and D. elimatus. These two species belong to a larger genus that also includes D. gelbus and D. quinquenotatus, which feed on teosinte and gamagrass. Although these insects can be force-fed on maize, in nature they are rarely found in maize. When D. gelbus and D. quinquenotatus are infected by MBSP or S. kunkelli, the insects often die before or at the time of pathogen transmission to maize. By contrast, D. maidis and D. elimatus seem to be less affected by bacterial colonization and in some cases even benefit from the infection (Madden et al., 1984; Nault et al., 1984; Moya-Raygoza and Nault, 1998). Therefore, Dalbulus spp. that have been exposed to MBSP and S. kunkelli for a longer time are likely to develop better tolerance than those that have had little exposure to these bacteria.
A bacterium’s association with specific vector(s) probably determines not only the tissue where it can become established in vector and host, but also its host range. Liberibacter species associate exclusively with psyllid vectors, and both vector and bacteria associate with a limited number of hosts (Figure 2). By contrast, phytoplasmas infect nearly 200 plant species and can be transmitted by at least 30 polyphagous species of phloem-feeding insects, including psyllids, leafhoppers, and planthoppers (Weintraub and Beanland, 2006; Ku et al., 2013) (Figures 1 and 2). The host range of spiroplasmas is more restricted than that of phytoplasmas, and spiroplasmas are transmitted by several species of leafhoppers (Figures 1 and 2). The case of X. fastidiosa is substantially different. There is no evidence of X. fastidiosa–vector specificity, meaning that insects in the abovementioned groups should be considered vectors until proven otherwise (Sicard et al., 2018). Most xylem sap-sucking insects are generalists, and this may be linked to the ability of X. fastidiosa to colonize hundreds of host plant species, usually without causing disease (Sicard et al., 2018).
GOING BEYOND MINIMAL SYSTEMS: HOW DOES THE GENETIC DIVERSITY OF THE PATHOGEN, VECTOR, AND HOST INFLUENCE DISEASE?
Technical limitations, including culturability, insectary infrastructure, and a lack of studies on biology under field settings, have led to important gaps in our understanding of bacterial vector-borne disease. How diverse are bacterial and vector populations in the field and natural ecosystems? How does this diversity affect epidemiology and capacity to cause plant disease? Can laboratory findings be translated to the field?
Vector Diversity
Natural variation in the ability of insect vectors to transmit plant and animal pathogens has been demonstrated extensively in many species, including the psyllid D. citri, which transmits CLas. This natural variation has been used as a powerful tool to dissect the genetic and molecular mechanisms of vector competence and vector–pathogen interactions (Guo et al., 1996; Habekuss et al., 1999; Terradot et al., 1999; Bencharki et al., 2000; Lucio-Zavaleta et al., 2001; Burrows et al., 2006, 2007; Gray et al., 2007; Yang et al., 2008; Cilia et al., 2011; Ogada et al., 2016; Ammar et al., 2018). Variability in transmission efficiency has also been found for phytoplasmas and spiroplasmas (Gonella et al., 2019). For phytoplasmas, the interaction between phytoplasma membrane proteins and insect factors determines vector specificity (Suzuki et al., 2006). It is well known that pathogens encounter barriers to transmission within the insect vector, and movement of pathogens across the gut and salivary tissues is also variable across populations and is genetically regulated (Ammar et al., 2018). Future research should leverage the natural variation in D. citri populations for CLas acquisition and transmission (Coy and Stelinski, 2015; Ammar et al., 2018; Hall, 2018), genomic resources for D. citri (Hosmani et al., 2019), and genome-wide association studies to identify psyllid genes that regulate CLas acquisition and transmission. By understanding individual-level diversity instead of generalizing across populations, we can also begin to understand why lab-generated management strategies may not always work in the field.
Although vector diversity is likely to significantly impact the vector transmission efficiency of phloem-limited bacterial pathogens, the role of within-species vector diversity in X. fastidiosa transmission is unknown and, if existent, likely to be linked to vector behavior and ecology. This may be explained by X. fastidiosa colonization of the cuticular surface of the foregut, which is a conserved substrate among these insect groups. X. fastidiosa does not interact with live insect host cells, such as the phloem-limited bacteria discussed here (Sicard et al., 2018). Evidence has shown that an array of factors—including host tissue preferences in relation to pathogen distribution within plants (Daugherty et al., 2011)—influence transmission rates and among-species differences in vector transmission of X. fastidiosa (Sicard et al., 2018), although evidence of within-species differences is so far absent or limited (Almeida and Purcell, 2003; Lopes et al., 2009; Krugner et al., 2012).
Pathogen and Host Diversity
Molecular tools facilitate the investigation of bacterial diversity. However, much of this work has been limited in scope, biased toward agricultural crops, and reliant on a limited number of genetic markers. The widespread advent of next-generation sequencing tools has enabled whole-genome sequences to be generated. Because there are more sequences available for X. fastidiosa (~350 isolates at the time of writing), we focus on this pathogen to illustrate aspects of pathogen diversity that require investigation.
There are currently two species of Xylella, the widely studied fastidiosa and the less-known taiwanensis. Because X. fastidiosa studies have focused on diseased crops, we do not understand what diversity may exist within X. taiwanensis or X. fastidiosa in natural ecosystems or whether there are other species of Xylella (Almeida and Nunney, 2015). It is unreasonable to assume that the only Xylella not considered to be endemic to the Americas is found in an island in Asia; even within the Americas, there is evidence of unsampled clades of X. fastidiosa (Castillo et al., 2020). Similarly, in the Americas, and more recently in Europe, the vast majority of X. fastidiosa for which there are genetic data are linked to crops of economic importance. X. fastidiosa genetic diversity can be found within regions, orchards, trees, and even branches (Coletta-Filho et al., 2014). There is little knowledge of what this variation, particularly at the within-crop and local spatial levels, may mean biologically. Finally, when extrapolated to other vector-borne plant pathogenic bacteria, these knowledge gaps only become more extreme.
Available genomic data are now beginning to be used for an array of evolutionary and ecological questions that were previously impossible to address. Genome sequences provide sufficient genetic resolution to study recent pathogen dispersal pathways, with implications for a greater understanding of pathogen biology and clues for how to avoid future introductions (Landa et al., 2019). Similarly, genomic data have elucidated how endemic pathogens have adapted to an introduced insect vector, leading to a novel phytoplasma disease (Malembic-Maher et al., 2020). Plant pathogen population genomic studies, including one focused on X. fastidiosa that colonizes grapevines in California, are resulting in novel interpretations of the geographical structure of pathogens in the landscape, as well as possible adaptive changes associated with temperature gradients (Vanhove et al., 2020). We expect this emerging field to continue yielding novel and relevant insights into the biology of vector-borne plant pathogens, but the difficulty of obtaining large populations of clonal isolates for genome sequencing remains.
Variations in host responses are also an important component of vector-borne disease. For example, CLas can proliferate on multiple plant hosts within the subfamily Aurantioideae to which citrus belongs. While all cultivated citrus is susceptible to HLB, other members of the Aurantioideae subfamily exhibit a gradient of responses (Ramadugu et al., 2016). Some genotypes are not suitable hosts for D. citri (white sapote, Casimiroa edulis), others lack high CLas populations despite being preferred hosts for the psyllid (curry leaf, Bergera koeniggi; pink wampee, Clausena excavata), and some exhibit tolerance to HLB despite higher CLas populations (orange jasmine, Murraya paniculata) (Westbrook et al., 2011; Ramadugu et al., 2016). Similar variability in plant responses to X. fastidiosa infection exists. For example, although all sweet orange varieties are susceptible to X. fastidiosa infection, leading to citrus variegated chlorosis, varieties of lemon, mandarin, grapefruit, and other citrus fruits are tolerant or resistant (Coletta-Filho et al., 2020). European grapevine varieties (Vitis vinifera) express a range of susceptibility to X. fastidiosa (Rashed et al., 2013), but many American hybrids and species of Vitis are tolerant to this pathogen (Krivanek et al., 2005).
Insect–plant–pathogen interactions at the ecological level and their consequences for disease incidence and pathogen spread are poorly understood. The main challenges are associated with the need to extrapolate controlled experimental manipulations to the field or landscape scales. This is usually performed with mathematical modeling, yet experimental work on these interactions is rarely designed with models in mind. Thus, captured data are often inadequate for downstream inferences. Similarly, manipulations are usually performed at single time points and may not measure the role of vector choice on pathogen transmission. The impact of disease symptoms on vector behavior, for example, is a function of time after infection (Daugherty et al., 2017), and the incorporation of time into experiments is therefore essential for understanding disease incidence. Although there are data supporting the hypothesis that insect vectors of phloem-limited bacterial pathogens are attracted to infected plants, the opposite has been demonstrated for X. fastidiosa (Daugherty et al., 2011). In the case of X. fastidiosa, it is clear that time after infection is key to determining vector behavior, ultimately driving a decrease in pathogen transmission: plants become better sources of inoculum, but that is offset by vector discrimination against infected and symptomatic plants (Daugherty et al., 2017). In addition, experimental and modeling work has revealed distinct transmission dynamics in tolerant and susceptible plants (Zeilinger et al., 2018). Studying how time after plant infection with a pathogen affects vector behavior and the design of experiments with mathematical models in mind should provide a more holistic understanding of disease dynamics and translate results more effectively to field situations.
CONCLUDING REMARKS
Bacterial vector-borne diseases represent some of the most economically important and invasive agricultural diseases. Because of their unique biological attributes, including the colonization of both host vascular tissue and their insect vector, the study of these diseases is poised to uncover novel biological mechanisms. We hope the ideas presented in this perspective serve as an invitation for new generations of scientists to investigate many unanswered questions about vector-borne diseases and push the field forward.
ACKNOWLEDGMENTS
No conflict of interest declared.
FUNDING
P.R.-C., S.S., and G.C. are supported by grants from the USDA CDRE (2019-70016-29796, 2016-70016-24833) awarded to G.C. L.B. is supported by an INRAE Department of Plant Health and Environment (SPE) grant (VMP-ADAPT), grants from the University of Bordeaux, and SFR Integrative Biology and Ecology. M.H. and M.M. are supported by a grant from the USDA ARS CRIS (8062-22410-006-00-D) awarded to M.H. W.H. and S.A.H. were funded from the BBSRC Institute Strategy Program (BB/P012574/1) and the John Innes Foundation. M.H. and M.M. are funded by USDA ARS project 8062-22410-006-00-D. R.P.P.A. was funded by the Pierce’s Disease Research Program, California Department of Food and Agriculture.
REFERENCES
- Abou-Saleh RH, Hernandez-Gomez MC, Amsbury S, Paniagua C, Bourdon M, Miyashima S, Helariutta Y, Fuller M, Budtova T, Connell SD, and Ries ME (2018). Interactions between callose and cellulose revealed through the analysis of biopolymer mixtures. Nat. Commun 9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achor D, Welker S, Ben-Mahmoud S, Wang C, Folimonova SY, Dutt M, Gowda S, and Levy A (2020). Dynamics of Candidatus Liberibacter asiaticus movement and sieve-pore plugging in citrus sink cells. Plant Physiol. 182:882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida RP, Killiny N, Newman KL, Chatterjee S, Ionescu M, and Lindow SE (2012). Contribution of rpfB to cell-to-cell signal synthesis, virulence, and vector transmission of Xylella fastidiosa. Mol. Plant Microbe Interact 25:453–462. [DOI] [PubMed] [Google Scholar]
- Almeida RP, and Purcell AH (2003). Transmission of Xylella fastidiosa to grapevines by Homalodisca coagulata (Hemiptera: Cicadellidae). J. Econ. Entomol 96:264–271. [DOI] [PubMed] [Google Scholar]
- Almeida RPP, and Nunney L (2015). How do plant diseases caused by Xylella fastidiosa emerge? Plant Dis. 99:1457–1467. [DOI] [PubMed] [Google Scholar]
- Ammar E-D, Ramos JE, Hall DG, Dawson WO, and Shatters RG Jr. (2016). Acquisition, replication and inoculation of Candidatus Liberibacter asiaticus following various acquisition periods on huanglongbing-infected citrus by nymphs and adults of the Asian citrus psyllid. PLoS One 11:e0159594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar ED, Hall DG, Hosseinzadeh S, and Heck M (2018). The quest for a non-vector psyllid: natural variation in acquisition and transmission of the huanglongbing pathogen ‘Candidatus Liberibacter asiaticus’ by Asian citrus psyllid isofemale lines. PLoS One 13:e0195804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar ED, Shatters R, Lynch C, and Hall D (2011). Detection and relative titer of Candidatus Liberibacter asiaticus in the salivary glands and alimentary canal of Diaphorina citri (Hemiptera: Psyllidae) vector of citrus Huanglongbing disease. Ann. Entomol. Soc. Am 104:526–533. [Google Scholar]
- Ammar el D, Fulton D, Bai X, Meulia T, and Hogenhout SA (2004). An attachment tip and pili-like structures in insect- and plant-pathogenic spiroplasmas of the class Mollicutes. Arch. Microbiol 181:97–105. [DOI] [PubMed] [Google Scholar]
- Andrade MO, Pang Z, Achor DS, Wang H, Yao T, Singer BH, and Wang N (2020). The flagella of ‘Candidatus Liberibacter asiaticus’ and its movement in planta. Mol. Plant Pathol 21:109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arashida R, Kakizawa S, Hoshi A, Ishii Y, Jung HY, Kagiwada S, Yamaji Y, Oshima K, and Namba S (2008). Heterogeneic dynamics of the structures of multiple gene clusters in two pathogenetically different lines originating from the same phytoplasma. DNA Cel. Biol 27:209–217. [DOI] [PubMed] [Google Scholar]
- Bai X, Correa VR, Toruno TY, Ammar el D, Kamoun S, and Hogenhout SA (2009). AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol. Plant Microbe Interact 22:18–30. [DOI] [PubMed] [Google Scholar]
- Bai X, Zhang J, Ewing A, Miller SA, Jancso Radek A, Shevchenko DV, Tsukerman K, Walunas T, Lapidus A, Campbell JW, et al. (2006). Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol 188:3682–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Zhang J, Holford IR, and Hogenhout SA (2004). Comparative genomics identifies genes shared by distantly related insect-transmitted plant pathogenic mollicutes. FEMS Microbiol. Lett 235:249–258. [DOI] [PubMed] [Google Scholar]
- Bencharki B, El Yamani M, and Zaoui D (2000). Assessment of transmission ability of barley yellow dwarf virus-PAV isolates by different populations of Rhopalosiphum padi and Sitobion avenae. Eur. J. Plant Pathol 106:455–464. [Google Scholar]
- Bertazzon N, Bagnaresi P, Forte V, Mazzucotelli E, Filippin L, Guerra D, Zechini A, Cattivelli L, and Angelini E.J.B.g. (2019). Grapevine comparative early transcriptomic profiling suggests that Flavescence dorée phytoplasma represses plant responses induced by vector feeding in susceptible varieties. BMC Genom. 20:526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beven L, Arricau-Bouvery N, Renaudin J, and Saillard C (2015). Pathogenicity, Virulence, and Transmission of Plant Spiroplasmas (APS press; ). [Google Scholar]
- Blaustein RA, Lorca GL, Meyer JL, Gonzalez CF, and Teplitski M (2017). Defining the core citrus leaf- and root-associated microbiota: factors associated with community structure and implications for managing huanglongbing (citrus greening) disease. Appl. Environ. Microbiol 83, e00210–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissot N, Schoeny A, and Vanlerberghe-Masutti F (2016). Vat, an amazing gene conferring resistance to aphids and viruses they carry: from molecular structure to field effects. Front. Plant Sci 7:1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, and Zipfel C (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol 55:257–286. [DOI] [PubMed] [Google Scholar]
- Bové J, and Garnier M (2003). Phloem-and xylem-restricted plant pathogenic bacteria. Plant Sci. 164:423–438. [Google Scholar]
- Bové JM (1997). Spiroplasmas: infectious agents of plants, arthropods and vertebrates. Wiener Klinische Wochenschrift 109:604–612. [PubMed] [Google Scholar]
- Bové JM, Renaudin J, Saillard C, Foissac X, and Garnier M (2003). Spiroplasma citri, a plant pathogenic mollecute: relationships with its two hosts, the plant and the leafhopper vector. Annu. Rev. Phytopathol 41:483–500. [DOI] [PubMed] [Google Scholar]
- Burrows ME, Caillaud MC, Smith DM, Benson EC, Gildow FE, and Gray SM (2006). Genetic regulation of polerovirus and luteovirus transmission in the aphid Schizaphis graminum. Phytopathology 96:828–837. [DOI] [PubMed] [Google Scholar]
- Burrows ME, Caillaud MC, Smith DM, and Gray SM (2007). Biometrical genetic analysis of luteovirus transmission in the aphid Schizaphis graminum. Heredity 98:106–113. [DOI] [PubMed] [Google Scholar]
- Cao Y, Trivellone V, and Dietrich CH (2020). A timetree for phytoplasmas (Mollicutes) with new insights on patterns of evolution and diversification. Mol. Phylogenet. Evol 149:106826. [DOI] [PubMed] [Google Scholar]
- Caserta R, Souza-Neto RR, Takita MA, Lindow SE, and De Souza AA (2017). Ectopic expression of Xylella fastidiosa rpfF conferring production of diffusible signal factor in transgenic tobacco and citrus alters pathogen behavior and reduces disease severity. Mol. Plant Microbe Interact 30:866–875. [DOI] [PubMed] [Google Scholar]
- Castillo AI, Chacón-díaz C, Rodríguez-murillo N, Coletta HD, Almeida RPP, and Rica C (2020). Impacts of local population history and ecology on the evolution of a globally dispersed pathogen. BMC Genom. 21:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Tan CM, Wu CT, Lin TH, Jiang SY, Liu RC, Tsai MC, Su LW, and Yang JY (2018). Alterations of plant architecture and phase transition by the phytoplasma virulence factor SAP11. J. Exp. Bot 69:5389–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary R, Atamian HS, Shen Z, Briggs SP, and Kaloshian I (2014). GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc. Natl. Acad. Sci. U S A 111:8919–8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Chung WC, Lin CP, and Kuo CH (2012). Comparative analysis of gene content evolution in phytoplasmas and mycoplasmas. PLoS One 7:e34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin EL, Ramsey JS, Mishchuk DO, Saha S, Foster E, Chavez JD, Howe K, Zhong X, Polek M, Godfrey KE, et al. (2019). Longitudinal transcriptomic, proteomic, and metabolomic analyses of Citrus sinensis (L.) Osbeck graft-inoculated with “Candidatus Liberibacter asiaticus”. J. Proteome Res 19:719–732. [DOI] [PubMed] [Google Scholar]
- Cilia M, Tamborindeguy C, Fish T, Howe K, Thannhauser TW, and Gray S (2011). Genetics coupled to quantitative intact proteomics links heritable aphid and endosymbiont protein expression to circulative polerovirus transmission. J. Virol 85:2148–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Franco JY, Schwizer S, Pang Z, Hawara E, Liebrand TWH, Pagliaccia D, Zeng L, Gurung FB, Wang P, et al. (2018). An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat. Commun 9:1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coletta-Filho HD, Castillo AI, Laranjeira FF, de Andrade EC, Silva NT, de Souza AA, Bossi ME, Almeida RPP, and Lopes JRS (2020). Citrus variegated chlorosis: an overview of 30 years of research and disease management. Trop. Plant Pathol 10.1007/s40858-020-00358-5. [DOI] [Google Scholar]
- Coletta-Filho HD, Francisco CS, and Almeida RP (2014). Temporal and spatial scaling of the genetic structure of a vector-borne plant pathogen. Phytopathology 104:120–125. [DOI] [PubMed] [Google Scholar]
- Court C, Hodges A, Rahmani M, and Spreen T (2018). Economic Contributions of the Florida Citrus Industry in 2015–16: FE1021, 7/2017 (UF/IFAS Extension; ). [Google Scholar]
- Coy M, and Stelinski LL (2015). Great variability in the infection rate of ‘Candidatus Liberibacter asiaticus’ in field populations of Diaphorina citri (Hemiptera: Liviidae) in Florida. Fla. Entomol 98:356–357. [Google Scholar]
- Daugherty MP, Rashed A, Almeida RP, and Perring TM (2011). Vector preference for hosts differing in infection status: sharpshooter movement and Xylella fastidiosa transmission. Ecol. Entomol 36:654–662. [Google Scholar]
- Daugherty MP, Zeilinger AR, and Almeida RP (2017). Conflicting effects of climate and vector behavior on the spread of a plant pathogen. Phytobiomes 1:46–53. [Google Scholar]
- Dawson WO, Bar-Joseph M, Garnsey SM, and Moreno P (2015). Citrus tristeza virus: making an ally from an enemy. Annu. Rev. Phytopathol 53:137–155. [DOI] [PubMed] [Google Scholar]
- DeBlasio SL, Chavez JD, Alexander MM, Ramsey J, Eng JK, Mahoney J, Gray SM, Bruce JE, and Cilia M (2016). Visualization of host-polerovirus interaction topologies using protein interaction reporter technology. J. Virol 90:1973–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, and Timmermans MC (2019). Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA sequencing. Dev. Cell 48:840–852.e5. [DOI] [PubMed] [Google Scholar]
- Donkersley P, Blanford JM, Queiroz RB, Silva FWS, Carvalho CM, Al-Sadi AM, and Elliot SL (2018). Witch’s broom disease of lime (Candidatus Phytoplasma aurantifolia): identifying high-risk areas by climatic mapping. J. Econ. Entomol 111:2553–2561. [DOI] [PubMed] [Google Scholar]
- Dubrana MP, Guegueniat J, Bertin C, Duret S, Arricau-Bouvery N, Claverol S, Lartigue C, Blanchard A, Renaudin J, and Beven L (2017). Proteolytic post-translational processing of adhesins in a pathogenic bacterium. J. Mol. Biol 429:1889–1902. [DOI] [PubMed] [Google Scholar]
- Duret S, Berho N, Danet JL, Garnier M, and Renaudin J (2003). Spiralin is not essential for helicity, motility, or pathogenicity but is required for efficient transmission of spiroplasma citri by its leafhopper vector Circulifer haematoceps. Appl. Environ. Microbiol 69:6225–6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher J, Melcher U, and Wayadande A (2006). The Phytopathogenic Spiroplasmas, Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, and Stackebrandt E, eds. (New York, NY: Springer US; ), pp. 905–947. [Google Scholar]
- Garzo E, Fernández-Pascual M, Morcillo C, Fereres A, Gómez-Guillamón ML, and Tjallingii WF (2018). Ultrastructure of compatible and incompatible interactions in phloem sieve elements during the stylet penetration by cotton aphids in melon. Insect Sci. 25:631–642. [DOI] [PubMed] [Google Scholar]
- George J, Ammar E-D, Hall DG, and Lapointe SL (2017). Sclerenchymatous ring as a barrier to phloem feeding by Asian citrus psyllid: evidence from electrical penetration graph and visualization of stylet pathways. PLoS One 12:e0173520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Ammar E-D, Hall DG, Shatters RG, and Lapointe SL (2018). Prolonged phloem ingestion by Diaphorina citri nymphs compared to adults is correlated with increased acquisition of the citrus greening pathogen. Sci. Rep 8 (1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim M, Achor D, Ghosh S, Kontsedalov S, Lebedev G, and Levy A (2017). Candidatus Liberibacter asiaticus’ accumulates inside endoplasmic reticulum associated vacuoles in the gut cells of Diaphorina citri. Sci. Rep 7:16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim M, Fattah-Hosseini S, Levy A, and Cilia M (2016). Morphological abnormalities and cell death in the Asian citrus psyllid (Diaphorina citri) midgut associated with Candidatus Liberibacter asiaticus. Sci. Rep 6:33418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck-Thaler E, Cerutti A, Perez-Quintero A, Butchacas J, Roman-Reyna V, Madhaven VN, Shantharaj D, Merfa MV, Pesce C, Jauneau A, et al. (2020). Repeated gain and loss of a single gene modulates the evolution of vascular pathogen lifestyles. bioRxiv 10.1101/2020.04.24.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonella E, Tedeschi R, Crotti E, and Alma A (2019). Multiple guests in a single host: interactions across symbiotic and phytopathogenic bacteria in phloem-feeding vectors—a review. Entomol. Exp. Appl 167:171–185. [Google Scholar]
- Gouran H, Gillespie H, Nascimento R, Chakraborty S, Zaini PA, Jacobson A, Phinney BS, Dolan D, Durbin-Johnson BP, Antonova ES, et al. (2016). The secreted protease PrtA controls cell growth, biofilm formation and pathogenicity in Xylella fastidiosa. Sci. Rep 6:31098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato LM, Galdeano DM, Nathália Da Roz DA, Breton MC, and Machado MA (2019). Callose synthase family genes plays an important role in the Citrus defense response to Candidatus Liberibacter asiaticus. Eur. J. Plant Pathol 155:25–38. [Google Scholar]
- Gray SM, Caillaud MC, Burrows M, and Smith DM (2007). Transmission of two viruses that cause Barley Yellow Dwarf is controlled by different loci in the aphid, Schizaphis graminum. J. Insect Sci 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen DE, Lee IM, Rehner SA, Davis RE, and Kingsbury DT (1994). Phylogeny of mycoplasmalike organisms (phytoplasmas): a basis for their classification. J. Bacteriol 176:5244–5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JQ, Moreau JP, and Lapierre H (1996). Variability among aphid clones of Rhopalosiphum padi L and Sitobion avenae Fabr (Homoptera: Aphididae) in transmission of three PAV isolates of barley yellow dwarf viruses. Can. Entomol 128:209–217. [Google Scholar]
- Gupta RS, Son J, and Oren A (2019). A phylogenomic and molecular markers based taxonomic framework for members of the order Entomoplasmatales: proposal for an emended order Mycoplasmatales containing the family Spiroplasmataceae and emended family Mycoplasmataceae comprised of six genera. Antonie van Leeuwenhoek 112:561–588. [DOI] [PubMed] [Google Scholar]
- Gurr GM, Johnson AC, Ash GJ, Wilson BA, Ero MM, Pilotti CA, Dewhurst CF, and You MS (2016). Coconut lethal yellowing diseases: a phytoplasma threat to palms of global economic and social significance. Front. Plant Sci 7:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha PT, He R, Killiny N, Brown JK, Omsland A, Gang DR, and Beyenal H (2019). Host-free biofilm culture of “Candidatus Liberibacter asiaticus,” the bacterium associated with Huanglongbing. J. Biofilm 1:100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habekuss A, Leistner HU, and Schliephake C (1999). Characterization of Rhopalosiphum padi genotypes differing in the geographical origin by transmission efficiency of Barley yellow dwarf viruses and molecular markers. Z. Pflanzenkrankheiten Pflanzenschutz-Journal Plant Dis. Prot 106:437–443. [Google Scholar]
- Hall D (2018). Incidence of “Candidatus Liberibacter asiaticus” in a Florida population of Asian citrus psyllid. J. Appl. Entomol 142:97–103. [Google Scholar]
- Hartung JS, Paul C, Achor D, and Brlansky RH (2010). Colonization of dodder, Cuscuta indecora, by ‘Candidatus Liberibacter asiaticus’ and ‘Ca. L. americanus. Phytopathology 100:756–762. [DOI] [PubMed] [Google Scholar]
- Heck M, and Brault V (2018). Targeted disruption of aphid transmission: a vision for the management of crop diseases caused by Luteoviridae members. Curr. Opin. Virol 33:24–32. [DOI] [PubMed] [Google Scholar]
- Hilf ME, and Luo W (2020). Inoculation period and citrus host effect establishment of new infections of ‘Candidatus Liberibacter asiaticus’ transmitted via vegetative grafting. Plant Dis. 10.1094/PDIS-1009-1019-2022-RE. [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Van der Hoorn RA, Terauchi R, and Kamoun S (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact 22:115–122. [DOI] [PubMed] [Google Scholar]
- Hoshi A, Oshima K, Kakizawa S, Ishii Y, Ozeki J, Hashimoto M, Komatsu K, Kagiwada S, Yamaji Y, and Namba S (2009). A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. U S A 106:6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani PS, Flores-Gonzalez M, Shippy T, Vosburg C, Massimino C, Tank W, Reynolds M, Tamayo B, Miller S, Norus J, et al. (2019). Chromosomal length reference assembly for Diaphorina citri using single-molecule sequencing and Hi-C proximity ligation with manually curated genes in developmental, structural and immune pathways. bioRxiv 10.1101/869685. [DOI] [Google Scholar]
- Hunter K, Kimura S, Rokka A, Tran HC, Toyota M, Kukkonen JP, and Wrzaczek M (2019). CRK2 enhances salt tolerance by regulating callose deposition in connection with PLDα1. Plant Physiol. 180:2004–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingel B, Jeske DR, Sun Q, Grosskopf J, and Roper MC (2019). Xylella fastidiosa endoglucanases mediate the rate of Pierce’s disease development in Vitis vinifera in a cultivar-dependent manner. Mol. Plant Microbe Interact 32:1402–1414. [DOI] [PubMed] [Google Scholar]
- Inoue H, Ohnishi J, Ito T, Tomimura K, Miyata S, Iwanami T, and Ashihara W (2009). Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol 155:29–36. [Google Scholar]
- Ionescu M, Yokota K, Antonova E, Garcia A, Beaulieu E, Hayes T, Iavarone AT, and Lindow SE (2016). Promiscuous diffusible signal factor production and responsiveness of the Xylella fastidiosa Rpf system. mBio 7 10.1128/mBio.01054-01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Fleites LA, and Gabriel DW (2015). Prophage-encoded peroxidase in ‘Candidatus Liberibacter asiaticus’ is a secreted effector that suppresses plant defenses. Mol. Plant Microbe Interact 28:1330–1337. [DOI] [PubMed] [Google Scholar]
- Jaouannet M, Rodriguez PA, Thorpe P, Lenoir CJ, MacLeod R, Escudero-Martinez C, and Bos JI (2014). Plant immunity in plant–aphid interactions. Front. Plant Sci 5:663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Zhang C-X, Chen R, and He SY (2019). Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U S A 116:23390–23397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, Jung H-Y, Suzuki S, Nishigawa H, Arashida R, Miyata S. i., Ugaki M, Kishino H, and Namba S (2006a). Positive selection acting on a surface membrane protein of the plant-pathogenic phytoplasmas. J. Bacteriol 188:3424–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa S, Oshima K, and Namba S (2006b). Diversity and functional importance of phytoplasma membrane proteins. Trends Microbiol. 14:254–256. [DOI] [PubMed] [Google Scholar]
- Killiny N, Martinez RH, Dumenyo CK, Cooksey DA, and Almeida RP (2013). The exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Mol. Plant Microbe Interact 26:1044–1053. [DOI] [PubMed] [Google Scholar]
- Knoblauch M, Froelich DR, Pickard WF, and Peters WS (2014). SEORious business: structural proteins in sieve tubes and their involvement in sieve element occlusion. J. Exp. Bot 65 (7):1879–1893. [DOI] [PubMed] [Google Scholar]
- Koinuma H, Maejima K, Tokuda R, Kitazawa Y, Nijo T, Wei W, Kumita K, Miyazaki A, Namba S, and Yamaji Y (2020). Spatiotemporal dynamics and quantitative analysis of phytoplasmas in insect vectors. Sci. Rep 10:4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivanek A, Stevenson J, and Walker M (2005). Development and comparison of symptom indices for quantifying grapevine resistance to Pierce’s disease. Phytopathology 95:36–43. [DOI] [PubMed] [Google Scholar]
- Krugner R, Sisterson MS, and Lin H (2012). Effects of gender, origin, and age on transmission of Xylella fastidiosa to grapevines by Homalodisca vitripennis (Hemiptera: Cicadellidae). Ann. Entomol. Soc. Am 105:280–286. [Google Scholar]
- Kruse A, Fattah-Hosseini S, Saha S, Johnson R, Warwick E, Sturgeon K, Mueller L, MacCoss MJ, Shatters RG Jr., and Cilia Heck M (2017). Combining ‘omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS One 12:e0179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse A, Fleites LA, and Heck M (2019). Lessons from one fastidious bacterium to another: what can we learn about liberibacter species from Xylella fastidiosa. Insects 10:300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku C, Lo WS, and Kuo CH (2013). Horizontal transfer of potential mobile units in phytoplasmas. Mob Genet Elements 3:e26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube M, Mitrovic J, Duduk B, Rabus R, and Seemuller E (2012). Current view on phytoplasma genomes and encoded metabolism. Sci. World J 2012:185942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kube M, Siewert C, Migdoll AM, Duduk B, Holz S, Rabus R, Seemüller E, Mitrovic J, Müller I, Büttner C, et al. (2014). Analysis of the complete genomes of Acholeplasma brassicae, A. palmae and A. laidlawii and their comparison to the obligate parasites from ‘Candidatus Phytoplasma’. J. Mol. Microbiol. Biotechnol 24:19–36. [DOI] [PubMed] [Google Scholar]
- Labroussaa F, Lebaudy A, Baby V, Gourgues G, Matteau D, Vashee S, Sirand-Pugnet P, Rodrigue S, and Lartigue C (2016). Impact of donor-recipient phylogenetic distance on bacterial genome transplantation. Nucleic Acids Res. 44:8501–8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa BB, Castillo AI, Giampetruzzi A, Kahn A, Román-Écija M, Velasco-Amo MP, Navas-Cortés JA, Marco-Noales E, Barbé S, Moralejo E, et al. (2019). Multiple intercontinental introductions associated with the emergence of a plant pathogen in Europe. Appl. Environ. Microbiol 86:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA 3rd, Smith HO, and Venter JC (2007). Genome transplantation in bacteria: changing one species to another. Science 317:632–638. [DOI] [PubMed] [Google Scholar]
- Lee JA, Halbert SE, Dawson WO, Robertson CJ, Keesling JE, and Singer BH (2015). Asymptomatic spread of huanglongbing and implications for disease control. Proc. Natl. Acad. Sci. U S A 112:7605–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy JG, Gross R, Mendoza-Herrera A, Tang X, Babilonia K, Shan L, Kuhl JC, Dibble MS, Xiao F, and Tamborindeguy C (2020). Lso-HPE1, an effector of ‘Candidatus Liberibacter solanacearum’, can repress plant immune response. Phytopathology 110:648–655. [DOI] [PubMed] [Google Scholar]
- Lolle S, Stevens D, and Coaker G (2020). Plant NLR-triggered immunity: from receptor activation to downstream signaling. Curr. Opin. Immunol 62:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes JR, Daugherty MP, and Almeida RP (2009). Context-dependent transmission of a generalist plant pathogen: host species and pathogen strain mediate insect vector competence. Entomol. Exp. Appl 131:216–224. [Google Scholar]
- Lu H, Patil P, Van Sluys MA, White FF, Ryan RP, Dow JM, Rabinowicz P, Salzberg SL, Leach JE, Sonti R, et al. (2008). Acquisition and evolution of plant pathogenesis-associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas. PLoS One 3:e3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YT, Li MY, Cheng KT, Tan CM, Su LW, Lin WY, Shih HT, Chiou TJ, and Yang JY (2014). Transgenic plants that express the phytoplasma effector SAP11 show altered phosphate starvation and defense responses. Plant Physiol. 164:1456–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucio-Zavaleta E, Smith DM, and Gray SM (2001). Variation in transmission efficiency among barley yellow dwarf virus-RMV isolates and clones of the normally inefficient aphid vector, Rhopalosiphum padi. Phytopatholog 91:792–796. [DOI] [PubMed] [Google Scholar]
- MacLean AM, Orlovskis Z, Kowitwanich K, Zdziarska AM, Angenent GC, Immink RG, and Hogenhout SA (2014). Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 12:e1001835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Toth R, Nicolaisen M, and Hogenhout SA (2011a). Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 157:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean AM, Sugio A, Makarova OV, Findlay KC, Grieve VM, Tóth R, Nicolaisen M, and Hogenhout SA (2011b). Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 157:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden LV, Nault LR, Heady SE, and Styler WE (1984). Effect of maize stunting mollicutes on survival and fecundity of Dalbulus leafhopper vectors. Ann. Appl. Biol 105:431–441. [Google Scholar]
- Maejima K, Iwai R, Himeno M, Komatsu K, Kitazawa Y, Fujita N, Ishikawa K, Fukuoka M, Minato N, Yamaji Y, et al. (2014). Recognition of floral homeotic MADS domain transcription factors by a phytoplasmal effector, phyllogen, induces phyllody. Plant J. 78:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malembic-Maher S, Desqué D, Khalil D, Salar P, Bergey B, Danet JL, Duret S, Dubrana-Ourabah MP, Beven L, Ember I, et al. (2020). When a Palearctic bacterium meets a Nearctic insect vector: genetic and ecological insights into the emergence of the grapevine Flavescence dorée epidemics in Europe. PLoS Pathog. 16:1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Fattah-Hosseini S, Ammar ED, Stange R, Warrick E, Sturgeon K, Shatters R, and Heck M (2018). Diaphorina citri nymphs are resistant to morphological changes induced by “Candidatus Liberibacter asiaticus” in midgut epithelial cells. Infect. Immun 86, e00889–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcone C (2009). Phytoplasmas: genomes, plant hosts and vectors In Movement of Phytoplasmas and the Development of Disease in the Plant, Weintraub PG and Jones P, eds. (Wallingford: CABI; ), pp. 114–131. [Google Scholar]
- Marina Mann SF-H, Ammar El-Desouky, Stange Richard, Warrick EricaRose, Sturgeon Kasie, Shatters Robert, and Heck Michelle (2018). Diaphorina citri nymphs are resistant to morphological changes induced by “Candidatus Liberibacter asiaticus” in midgut epithelial cells. Infect. Immun 86, e00889–00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minato N, Himeno M, Hoshi A, Maejima K, Komatsu K, Takebayashi Y, Kasahara H, Yusa A, Yamaji Y, Oshima K, et al. (2014). The phytoplasmal virulence factor TENGU causes plant sterility by downregulating of the jasmonic acid and auxin pathways. Sci. Rep 4:7399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya-Raygoza G, and Nault LR (1998). Transmission biology of maize bushy stunt phytoplasma by the corn leafhopper (Homoptera: Cicadellidae). Ann. Entomol. Soc. Am 91:668–676. [Google Scholar]
- Mugford ST, Barclay E, Drurey C, Findlay KC, and Hogenhout SA (2016). An immuno-suppressive aphid saliva protein is delivered into the cytosol of plant mesophyll cells during feeding. Mol. Plant Microbe Interact 29:854–861. [DOI] [PubMed] [Google Scholar]
- Musetti R, Paolacci A, Ciaffi M, Tanzarella OA, Polizzotto R, Tubaro F, Mizzau M, Ermacora P, Badiani M, and Osler R (2010). Phloem cytochemical modification and gene expression following the recovery of apple plants from apple proliferation disease. Phytopathology 100:390–399. [DOI] [PubMed] [Google Scholar]
- Music MS, Samarzija I, Hogenhout SA, Haryono M, Cho ST, and Kuo CH (2019). The genome of ‘Candidatus Phytoplasma solani’ strain SA-1 is highly dynamic and prone to adopting foreign sequences. Syst. Appl. Microbiol 42:117–127. [DOI] [PubMed] [Google Scholar]
- Naranjo E, Merfa MV, Ferreira V, Jain M, Davis MJ, Bahar O, Gabriel DW, and De La Fuente L (2019). Liberibacter crescens biofilm formation in vitro: establishment of a model system for pathogenic ‘Candidatus Liberibacter spp.’. Sci. Rep 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, and Narusaka Y (2009). RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J. 60:218–226. [DOI] [PubMed] [Google Scholar]
- Nault LR, Madden LV, Styer WE, Triplehorn BW, Shambaugh GF, and Heady SE (1984). Pathogenicity of corn stunt spiroplasma and maize bushy stunt mycoplasma to their vector, Dalbubus longulus. Phytopathology 74:977–979. [Google Scholar]
- Newman KL, Almeida RP, Purcell AH, and Lindow SE (2003). Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol 69:7319–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niza B, Coletta-Filho HD, Merfa MV, Takita MA, and de Souza AA (2015). Differential colonization patterns of Xylella fastidiosa infecting citrus genotypes. Plant Pathol. 64:1259–1269. [Google Scholar]
- Ogada PA, Debener T, and Poehling H-M (2016). Inheritance genetics of the trait vector competence in Frankiniella occidentalis (Western flower thrips) in the transmission of Tomato spotted wilt virus. Ecol. Evol 6:7911–7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlovskis Z, and Hogenhout SA (2016). A bacterial parasite effector mediates insect vector attraction in host plants independently of developmental changes. Front. Plant Sci 7:885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Kakizawa S, Nishigawa H, Jung H-Y, Wei W, Suzuki S, Arashida R, Nakata D, Miyata S. i., Ugaki M, et al. (2004). Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet 36:27–29. [DOI] [PubMed] [Google Scholar]
- Pagliaccia D, Shi J, Pang Z, Hawara E, Clark K, Thapa SP, De Francesco AD, Liu J, Tran T-T, Bodaghi S, et al. (2017). A pathogen secreted protein as a detection marker for citrus huanglongbing. Front. Microbiol 8:2041. 10.3389/fmicb.2017.02041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliari L, Buoso S, Santi S, Furch ACU, Martini M, Degola F, Loschi A, Van Bel AJE, and Musetti R (2017). Filamentous sieve element proteins are able to limit phloem mass flow, but not phytoplasma spread. J. Exp. Bot 68:3673–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecher P, Moro G, Canale MC, Capdevielle S, Singh A, MacLean A, Sugio A, Kuo CH, Lopes JRS, and Hogenhout SA (2019). Phytoplasma SAP11 effector destabilization of TCP transcription factors differentially impact development and defence of Arabidopsis versus maize. PLoS Pathog. 15:e1008035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng HC, and Walker GP (2020). Sieve element occlusion provides resistance against Aphis gossypii in TGR-1551 melons. Insect Sci. 27:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilla-Henao LM, and Casteel CL (2016). Vector-borne bacterial plant pathogens: interactions with hemipteran insects and plants. Front. Plant Sci 7:1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter KA, Gildow F, Palukaitis P, and Gray SM (2009). The C terminus of the polerovirus p5 readthrough domain limits virus infection to the phloem. J. Virol 83:5419–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Armstrong CM, Cano LM, and Duan Y (2016). Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci 7:982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince DC, Drurey C, Zipfel C, and Hogenhout SA (2014). The leucine-rich repeat receptor-like kinase BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiol. 164:2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadugu C, Keremane ML, Halbert SE, Duan YP, Roose ML, Stover E, and Lee RF (2016). Long-term field evaluation reveals huanglongbing resistance in Citrus relatives. Plant Dis. 100:1858–1869. [DOI] [PubMed] [Google Scholar]
- Ramsey JS, Chin EL, Chavez JD, Saha S, Mischuk D, Mahoney J, Mohr J, Robison FM, Mitrovic E, Xu Y, et al. (2020). Longitudinal transcriptomic, proteomic, and metabolomic analysis of citrus limon response to graft inoculation by Candidatus Liberibacter asiaticus. J. Proteome Res 19:2247–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapicavoli JN, Blanco-Ulate B, Muszyński A, Figueroa-Balderas R, Morales-Cruz A, Azadi P, Dobruchowska JM, Castro C, Cantu D, and Roper MC (2018). Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun 9:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed A, Kwan J, Baraff B, Ling D, Daugherty MP, Killiny N, and Almeida RPP (2013). Relative susceptibility of vitis vinifera cultivars to vector-borne Xylella fastidiosa through time. PLoS One 8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J, Beven L, Batailler B, Duret S, Desque D, Arricau-Bouvery N, Malembic-Maher S, and Foissac X (2015). Heterologous expression and processing of the flavescence doree phytoplasma variable membrane protein VmpA in Spiroplasma citri. BMC Microbiol. 15:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin J, Breton M, and Citti C (2014). Molecular Genetic Tools for Mollicutes Mollicutes: Molecular Biology and Pathogenesis (Caister Academic Press; ), pp. 55–76. [Google Scholar]
- Roberts MAJ, Cranenburgh RM, Stevens MP, and Oyston PCF (2013). Synthetic biology: biology by design. Microbiology 159:1219–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper U, and Seemüller E (1984). Recolonization of the stem of apple proliferation and pear decline-diseased trees by the causal organisms in spring. J. Plant Dis. Prot 91:608–613. [Google Scholar]
- Schneider K, van der Werf W, Cendoya M, Mourits M, Navas-Cortés JA, Vicent A, and Oude Lansink A (2020). Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. U S A 117:9250–9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg EG, and Tumlinson JH (2014). Aphid honeydew alters plant defence responses. Funct. Ecol 28:386–394. [Google Scholar]
- Shi Q, Pitino M, Zhang S, Krystel J, Cano LM, Shatters RG, Hall DG, and Stover E (2019). Temporal and spatial detection of Candidatus Liberibacter asiaticus putative effector transcripts during interaction with Huanglongbing-susceptible, tolerant, and-resistant citrus hosts. BMC Plant Biol. 19:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Zeilinger AR, Vanhove M, Schartel TE, Beal DJ, Daugherty MP, and Almeida RP (2018). Xylella fastidiosa: insights into an emerging plant pathogen. Annu. Rev. Phytopathol 56:181–202. [DOI] [PubMed] [Google Scholar]
- Solanki S, Ameen G, Zhao J, Flaten J, Borowicz P, and Brueggeman RS (2020). Visualization of spatial gene expression in plants by modified RNAscope fluorescent in situ hybridization. Plant Methods 16:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner AD, Muñoz-Amatriaín M, Venegas JMA, Lo S, Shi D, Holton N, Zipfel C, Abagyan R, Huffaker A, and Close TJ (2019). A receptor for herbivore-associated molecular patterns mediates plant immunity. Biorxiv 10.1101/679803. [DOI] [Google Scholar]
- Stevenson JF, Matthews MA, and Rost TL (2004). Grapevine susceptibility to Pierce’s disease I: relevance of hydraulic architecture. Am. J. Enol. Viticult 55:228–237. [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, and Hogenhout SA (2011a). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U S A 108:E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, and Hogenhout SA (2011b). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U S A 108:E1254–E1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, MacLean AM, Kingdom HN, Grieve VM, Manimekalai R, and Hogenhout SA (2011c). Diverse targets of phytoplasma effectors: from plant development to defense against insects. Annu. Rev. Phytopathol 49:175–195. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Oshima K, Kakizawa S, Arashida R, Jung HY, Yamaji Y, Nishigawa H, Ugaki M, and Namba S (2006). Interaction between the membrane protein of a pathogen and insect microfilament complex determines insect-vector specificity. Proc. Natl. Acad. Sci. U S A 103:4252–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F, and Rep M (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol 11:309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CM, Li CH, Tsao NW, Su LW, Lu YT, Chang SH, Lin YY, Liou JC, Hsieh LC, Yu JZ, et al. (2016). Phytoplasma SAP11 alters 3-isobutyl-2-methoxypyrazine biosynthesis in Nicotiana benthamiana by suppressing NbOMT1. J. Exp. Bot 67:4415–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X-T, Ibanezy F, and Tamborindeguy C (2020). Concanavalin A toxicity towards potato psyllid and apoptosis induction in midgut cells. Insects 11 10.3390/insects11040243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X-T, and Tamborindeguy C (2019). No evidence of apoptotic response of the potato psyllid Bactericera cockerelli to “Candidatus Liberibacter solanacearum” at the gut interface. Infect. Immun 88, e00242–00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Simon JC, Leterme N, Bourdin D, Wilson ACC, Gauthier JP, and Robert Y (1999). Molecular characterization of clones of the Myzus persicae complex (Hemiptera: Aphididae) differing in their ability to transmit the potato leafroll luteovirus (PLRV). Entomol. Res 89:355–363. [Google Scholar]
- Thapa SP, De Francesco A, Trinh J, Gurung FB, Pang Z, Vidalakis G, Wang N, Ancona V, Ma W, and Coaker G (2020). Genome-wide analyses of Liberibacter species provides insights into evolution, phylogenetic relationships, and virulence factors. Mol. Plant Pathol 21:716–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins M, Kliot A, Maree AF, and Hogenhout SA (2018). A multi-layered mechanistic modelling approach to understand how effector genes extend beyond phytoplasma to modulate plant hosts, insect vectors and the environment. Curr. Opin. Plant Biol 44:39–48. [DOI] [PubMed] [Google Scholar]
- Toruño TY, Musić MS, Simi S, Nicolaisen M, and Hogenhout SA (2010). Phytoplasma PMU1 exists as linear chromosomal and circular extrachromosomal elements and has enhanced expression in insect vectors compared with plant hosts. Mol. Microbiol 77:1406–1415. [DOI] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, and Coaker G (2016). Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol 54:419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Spencer D, Sawai-Toyota S, Jiaqi W, Zhang T, Koo AJ, Howe GA, and Gilroy S (2018). Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361:1112–1115. [DOI] [PubMed] [Google Scholar]
- Tully JG, Whitcomb RF, Clark HF, and Williamson DL (1977). Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892–894. [DOI] [PubMed] [Google Scholar]
- Tumber KP, Alston JM, and Fuller KB (2014). Pierce’s disease costs California $104 million per year. Calif. Agric 68:20–29. [Google Scholar]
- Vanhove M, Sicard A, Ezennia J, Leviten N, and Almeida RPP (2020). Population structure and adaptation of a bacterial pathogen in California grapevines. Environ. Microbiol 10.1111/1462-2920.14965. [DOI] [PubMed] [Google Scholar]
- Vincent TR, Avramova M, Canham J, Higgins P, Bilkey N, Mugford ST, Pitino M, Toyota M, Gilroy S, Miller AJ, et al. (2017). Interplay of plasma membrane and vacuolar ion channels, together with BAK1, elicits rapid cytosolic calcium elevations in Arabidopsis during aphid feeding. Plant Cell 29:1460–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang H, Yin Z, Liu W, Sun L, and Wu Y (2018). Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol 19:2623–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wari D, Kabir MA, Mujiono K, Hojo Y, Shinya T, Tani A, Nakatani H, and Galis I (2019). Honeydew-associated microbes elicit defense responses against brown planthopper in rice. J. Exp. Bot 70:1683–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters H, and Hunt P (1980). The in vivo three-dimensional form of a plant mycoplasma-like organism by the analysis of serial ultrathin sections. J. Gen. Microbiol 116:111–131. [Google Scholar]
- Wei W, Kakizawa S, Suzuki S, Jung HY, Nishigawa H, Miyata S, Oshima K, Ugaki M, Hibi T, and Namba S (2004). In planta dynamic analysis of onion yellows phytoplasma using localized inoculation by insect transmission. Phytopathology 94:244–250. [DOI] [PubMed] [Google Scholar]
- Weintraub PG, and Beanland L (2006). Insect vectors of phytoplasmas. Annu. Rev. Entomol 51:91–111. [DOI] [PubMed] [Google Scholar]
- Westbrook CJ, Hall DG, Stover E, Duan YP, and Lee RFJH (2011). Colonization of citrus and citrus-related germplasm by Diaphorina citri (Hemiptera: Psyllidae). Am. Soc. Hortic. Sci 46:997–1005. [Google Scholar]
- Yan Q, Sreedharan A, Wei S, Wang J, Pelz-Stelinski K, Folimonova S, and Wang N (2013). Global gene expression changes in Candidatus Liberibacter asiaticus during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol 14:391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Thannhauser TW, Burrows M, Cox-Foster D, Gildow FE, and Gray SM (2008). Coupling genetics and proteomics to identify aphid proteins associated with vector-specific transmission of polerovirus (luteoviridae). J. Virol 82:291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H-Z, Li N-Y, Zeng X-D, Song J-C, Yu X-D, Su H-N, Chen C-X, Yi L, and Lu Z-JJI (2020). Transcriptome analyses of Diaphorina citri midgut responses to Candidatus Liberibacter asiaticus infection. Insects 11:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilinger AR, Turek D, Cornara D, Sicard A, Lindow SE, and Almeida RP (2018). Bayesian vector transmission model detects conflicting interactions from transgenic disease-resistant grapevines. Ecosphere 9:e02494. [Google Scholar]
- Zhou Y, Li Y, Liu Z, Zhao X, Xu X, Duan Y, and Zou H (2020). Transient expression of ‘Candidatus Liberibacter asiaticus’ effector SDE1 induces chlorosis by suppressing NbDDX3 gene in Nicotiana benthamiana: research Square. 10.21203/rs.21203.rs-19409/v21201. [DOI]


