Abstract
Stress granules (SGs) are membrane-less ribonucleoprotein-based cellular compartments that form in the cytoplasm of a cell upon exposure to various environmental stressors. SGs contain a large set of proteins, as well as mRNAs that have been stalled in translation as a result of stress-induced polysome disassembly. Despite the fact that SGs have been extensively studied for many years, their function is still not clear. They presumably help the cell to cope with the encountered stress, and facilitate the recovery process after stress removal, upon which SGs disassemble. Aberrant formation of SGs and impaired SG disassembly majorly contributes to various pathological phenomena in cancer, viral infections, and neurodegeneration.
The assembly of SGs is largely driven by liquid-liquid phase separation (LLPS), however the molecular mechanisms behind that are not fully understood. Recent studies have proposed a novel mechanism for SG formation that involves the interplay of a large interaction network of mRNAs and proteins. Here we review this novel concept for the assembly of SGs, and discuss the current insights into SG disassembly.
I. Introduction: General introduction to RNA granules and SGs
Throughout their life, cellular mRNAs are coated with proteins and exist as messenger ribonucleoprotein (mRNP) complexes (Erickson and Lykke-Andersen, 2011; Ivanov et al., 2019). The mRNP composition determines the fate of the mRNA and controls different processes such as transport, degradation and translational regulation. Remodeling of mRNPs via changes in the protein composition dictates whether the mRNA can actively engage in translation, or if it is translationally silenced and consequently prone to storage or degradation (Anderson and Kedersha, 2002a, 2008; Erickson and Lykke-Andersen, 2011; Kedersha and Anderson, 2002; Zhang et al., 2011). Within cells, mRNP complexes can organize to form microscopically visible cytoplasmic RNA granules (Anderson and Kedersha, 2009a). Specific types of RNA granules occur in different cell types and have varying functions, e.g. neuronal transport RNP granules, U bodies, or germline granules (Anderson and Kedersha, 2006; Banani et al., 2017; Moser and Fritzler, 2010). To date, the best characterized RNA granules are processing bodies (PBs) and stress granules (SGs) (Anderson and Kedersha, 2002a, 2009b; Buchan and Parker, 2009; Ivanov et al., 2019; Kedersha and Anderson, 2009; Markmiller et al., 2018; Parker and Sheth, 2007; Youn et al., 2018).
SGs are non-membraneous cytoplasmic foci ranging in size from 100 – 2000 nm (Moser and Fritzler, 2010). They form in cells exposed to environmental stress conditions such as heat shock, oxidative stress, viral infection, osmotic stress, UV irradiation, or cold shock (Anderson and Kedersha, 2009b; Hofmann et al., 2012; Moser and Fritzler, 2010; Zeng et al., 2020). Components of SGs include stalled 48S preinitiation complexes (PICs) consisting of translationally arrested mRNAs, small ribosomal subunits, translation initiation factors, and the RNA-binding proteins PolyA-binding protein (PABP), RasGAPSH3-binding protein (G3BP) and TIA-1 (Kedersha et al., 2002, 1999; Moser and Fritzler, 2010; Tourrière et al., 2003). Many additional SG-related proteins have been identified over the last 20 years (Anderson and Kedersha, 2008; Buchan and Parker, 2009; Kedersha and Anderson, 2009), and the invention of proximity-labeling techniques has revealed an even bigger SG interactome (Cao et al., 2020; Markmiller et al., 2018; Youn et al., 2019, 2018).
Interestingly, not only the composition of SGs, but also the exchange dynamics of individual SG components vary greatly (Advani and Ivanov, 2019; Kedersha et al., 2005). Fluorescent recovery after photobleaching (FRAP) analysis, which measures the recovery time of any given protein at distinct locations in the cell, shows that some proteins such as TIA1, TTP, PABP, and G3BP rapidly shuttle between SGs and the cytoplasm, while others (e.g. FAST, IGF2BP1, YB1 or HUR) exchange very slowly (Bley et al., 2015; Kedersha et al., 2005). In many cases, the distribution of different proteins within the granule was shown to be heterogeneous (Guillén-Boixet et al., 2020; Jain et al., 2016; Niewidok et al., 2018; Wheeler et al., 2016). Furthermore, it has been reported that SGs are comprised of a stable core substructure that is surrounded by a more dynamic shell, wherein components are in a dynamic equilibrium with polysomes and can rapidly shuttle between these structures (Banani et al., 2017; Jain et al., 2016; Zhang et al., 2011). In a recent study by Niewidok et al. using single-molecule fluorescence microscopy, individual G3BP molecules have even been shown to exhibit two different populations; one that resembles the free, diffusing state (presumably in the SG shell), and another immobile (presumably in the SG core (Niewidok et al., 2018). SG assembly is reported to be nucleated by the stable core, followed by the assembly of the shell (Jain et al., 2016; Niewidok et al., 2018).
Besides the growing list of SG-associated proteins, SGs also contain translationally stalled mRNAs, which mostly encode housekeeping proteins, while a distinct subset of excluded transcripts (e.g. HSP70) is selectively translated under stress (Kedersha and Anderson, 2002; Stöhr et al., 2006). Thus, the assembly of SGs may be part of an adaptation process in response to unfavorable environmental conditions. However, the function of SGs still remains unclear (Erickson and Lykke-Andersen, 2011; Guzikowski et al., 2019; Riggs et al., 2020). Many studies have reported a connection between the ability of cells to assemble SGs and their survival during stress (Arimoto et al., 2008; Eisinger-Mathason et al., 2008; Kim et al., 2012; Lavut and Raveh, 2012; Zou et al., 2012, 2011). Although numerous translational repressors are SG components, SG formation per se is not required for translation inhibition during stress (Anderson and Kedersha, 2009a; Buchan et al., 2008; Fujimura et al., 2009; Kwon et al., 2007; Loschi et al., 2009; Mokas et al., 2009; Ohn et al., 2008). Global stabilization of mRNAs, which occurs under stress conditions, also does also not require assembly of SGs (Buchan et al., 2008). Yet the selective exclusion of stress-induced transcripts from SGs and their preferential translation under stress conditions suggest that SGs enable translation of mRNAs involved in the adaptation response (Anderson and Kedersha, 2009b; Buchan and Parker, 2009).
The sequestration of specific proteins into SGs upon exposure to stress leads to an altered concentration and composition of cytosolic proteins, and thereby, the course of biochemical reactions in the cytosol is changed (Buchan and Parker, 2009; Riggs et al., 2020). For example, under mild stress conditions, the pro-apoptotic protein Receptor for activated C kinase 1 (RACK1) localizes to SGs resulting in the suppression of the MTK1-SAPK signaling pathway, which in turn prevents apoptosis (Arimoto et al., 2008). Similarly, TNFα receptor associated factor 2 (TRAF2) is trapped in heat shock-induced SGs, whereby the NF-κB-dependent, pro-inflammatory, and pro-apoptotic response is impaired (Buchan and Parker, 2009; Kim et al., 2005). Along the same lines, the steroid receptor coactivator-3 (SRC-3) is a nuclear coactivator of several transcription factors involved in the inflammatory response, and promotes the expression of inflammatory cytokines upon LPS stimulation (Anderson and Kedersha, 2007; Yu et al., 2007). Under stress conditions, SRC-3 is exported to the cytoplasm where it binds to TIA-1 and localizes to SGs (Anderson and Kedersha, 2007; Kedersha and Anderson, 2009), coincident with the silencing of pro-inflammatory genes. Many other SG-associated proteins, e.g. TIA-1/TIAR, G3BP, HuR, and FMRP/FXR, are also nuclear-cytoplasmic shuttling proteins. Their stress-induced cytoplasmic localization and sequestration into SGs precludes their nuclear functions such as alternative splicing, transcriptional regulation or mRNA processing (Kedersha and Anderson, 2009). Thus, the formation of SGs under various stress conditions alters specific nuclear events and provides a link between nuclear and cytoplasmic processes. For a more comprehensive review about the role of SGs as signaling hubs, see (Kedersha et al., 2013).
SGs are proposed to be sites of mRNA triage (Kedersha and Anderson, 2002), wherein individual transcripts are sorted and routed to either reinitiation, degradation or packaging into stable, non-polysomal mRNPs (Anderson and Kedersha, 2009a, 2008; Kedersha and Anderson, 2009, 2002; Somasekharan et al., 2020). The spatial and functional link between SGs and PBs supports this notion (Kedersha et al., 2005; Mollet et al., 2008; Riggs et al., 2020). SGs and PBs are distinct and independent cytoplasmic structures, but share several proteins and RNA components (Anderson and Kedersha, 2008, 2006). Both SGs and PBs are dynamic structures that constantly exchange mRNAs with the cytosol, polysomes and other RNA granules (Kedersha and Anderson, 2009; Parker and Sheth, 2007). Drugs that stabilize polysomes (e.g. cycloheximide (CHX), emetine) and thus disrupt this equilibrium cause the disassembly of SGs and PBs (Anderson and Kedersha, 2006; Kedersha et al., 2000). In contrast, drugs that enhance polysome disassembly (e.g. puromycin) promote the formation of SGs and PBs (Anderson and Kedersha, 2006; Kedersha et al., 2000).
The formation of SGs is (at least) biphasic. First, translation initiation is inhibited, and stalled mRNAs accumulate (Kedersha et al., 2002). Secondly, these mRNAs and their associated proteins condense into distinct cytoplasmic foci. In most physiological cases, translation inhibition is initiated through phosphorylation of eIF2α by one or more of the four eIF2α kinases: PKR, PERK/PEK, HRI and GCN2 (Anderson and Kedersha, 2009b, 2006; Kedersha et al., 2002, 1999). Consequently, the depletion of eIF2-GTP-tRNAiMet ternary complexes disrupts translation initiation, which leads to polysome disassembly and concomitant recruitment of stalled 48S PICs into SGs (Anderson and Kedersha, 2002b; Kedersha et al., 2002, 1999). However, pharmacologically-induced SG formation occurs independently of phospho-eIF2α. For example, the eIF4A inhibitors Pateamine A and hippuristanol bind to eIF4A, and thereby disrupt the association of eIF4A with eIF4G, which prevents formation of the eIF4F complex and causes inhibition of translation initiation downstream of joining of the ternary complex (Bordeleau et al., 2006, 2005; Low et al., 2007b, 2007a, 2005).
Treatment of mammalian cells with Pateamine A or hippuristanol causes SG formation without inducing or requiring eIF2α phosphorylation (Dang et al., 2006; Mazroui et al., 2006). In addition, Farny et al. showed that in Drosophila S2R+ cells, SG assembly in response to heat shock does not require the activity of eIF2α kinases (Farny et al., 2009). In fission yeast, the formation of SGs upon glucose starvation requires protein kinase A (PKA) and is also independent of eIF2α phosphorylation (Nilsson and Sunnerhagen, 2011). Furthermore, cold shock causes phosphorylation of eIF2α in both mammalian cells and in yeast cells, but the formation of cold shock SGs is independent of eIF2α phosphorylation (Hofmann et al., 2012). Therefore, the universal trigger of SG formation appears to be the disassembly of polysomes, which can be mediated by either inhibition of mTOR, phosphorylation of eIF2 α or interference with the eIF4F complex (Panas et al., 2016).
The protein composition of mammalian SGs can slightly vary depending on which step of the mRNA translational cycle the translation stalling point happens. At least three different subtypes of SGs have been identified - for details see previously published reviews (Advani and Ivanov, 2019; Ivanov et al., 2019; Riggs et al., 2020). Type I canonical SGs form e.g. upon exposure to oxidative stress, ER stress or viral infection as a result of stress-related phosphorylation of eIF2α. Type I SGs require G3BP and contain 48S PICs, but lack eIF2 and eIF5 (Kedersha et al., 2002). In contrast, Type II SGs form upon phospho-eIF2α-independent inhibition of translation, mainly through inactivation of the eIF4A helicase by eIF4A inhibitors, but still require G3BP. In this case, the translation initiation stalling point is downstream of joining of the eIF2-loaded ternary complex to the 48S PIC, and thus, Type II SGs contain eIF2 and eIF5. While both Type I and Type II SGs contain eIF3, Type III SGs are characterized by their lack of eIF3 (Anderson et al., 2015; Aulas et al., 2018; Fujimura et al., 2012). Their formation is triggered by sodium selenite, UV, nitric oxide, glucose starvation or other xenobiotic agents through an unknown mechanism (Advani and Ivanov, 2019).
Finally, osmotic stress causes the assembly of another type of non-canonical SG that forms independently of both phospho-eIF2α and G3BP. It has been reported that molecular crowding plays a role in their formation, but the exact mechanism is unclear (Bevilacqua et al., 2010; Bounedjah et al., 2012; Ivanov et al., 2019; Zeng et al., 2020). The distinct functions and properties of the different SG subtypes are not known yet, but the differences in composition and dynamics of the various subtypes might give insights into their pathological nature (Advani and Ivanov, 2019).
II. LLPS
Molecular crowding and multivalency-driven LLPS
The cytoplasm of a living cell is very complex, and harbors many biochemical reactions and critical functions (e.g. mRNA translation, etc.). It is remarkable that around 20–30% of the volume in the cytoplasm is, in fact, occupied by macromolecules such as RNA, proteins, or polysaccharides, and therefore a large fraction of the cytoplasm is physically unavailable for all the other hundreds of cytosolic molecules and constituents, including metabolites and small ions (Ellis, 2001). This excluded volume effect, also referred to as macromolecular crowding, may affect various enzymatic activities within the cytoplasm, as well as the binding affinities of different molecular interactors (Banani et al., 2017; Kuznetsova et al., 2015). The total concentration of macromolecules inside a cell is extremely high (e.g. the concentration of total protein is in the range of 200–300 g/l, and the total concentration of RNA in the range of 75–150 g/l (Ellis, 2001).
Besides macromolecules, ribosomes are by far one of the most dominant constituents in a cell, and ribosomal RNAs make up about 85% of the cellular RNA pool at any given time (Scott et al., 2010). There are about 70,000 ribosomes in a single bacterial cell (Bremer and Dennis, 2008), which accounts for one third of the total dry mass of the cell (Bremer and Dennis, 2008; Brunschede et al., 1977). Thus, ribosomes (and the translation machinery in general) take up a significant fraction of the cell interior (Milo and Philips, 2016). In 1999 Johansson et al. predicted that the high intracellular concentration of any two incompatible macromolecules should drive demixing in the cytoplasm (Johansson et al., 2000). The Flory-Huggins model, that was first postulated in the 1940ś, predicts that molecular crowding promotes liquid-liquid phase separation (LLPS) in the cellular cytosol (Johansson et al., 2000). Twenty years later we now know that LLPS does indeed occur within the crowded cytoplasm, and several membraneless compartments, also called biomolecular condensates, of various different functions have been identified both in the cytoplasm and in the nucleus (Alberti and Dormann, 2019; Banani et al., 2017; Boeynaems et al., 2018; Shin and Brangwynne, 2017; Yoshizawa et al., 2020).
There is evidence that spontaneous, passive, simple phase separation mediated through strong, non-specific interactions of macromolecules accounts for the formation of condensates, e.g. the formation of SGs upon exposure to osmotic stress (Bounedjah et al., 2012; Ivanov et al., 2019). However, most condensates are assembled from weak, specific interactions between certain macromolecules, and phase separation is an active process driven by multivalency of particular central interactors (Alberti and Dormann, 2019; Banani et al., 2017; Sanders et al., 2020).
Protein interaction domains and molecular interactions that contribute to multivalency-driven LLPS in SG dynamics
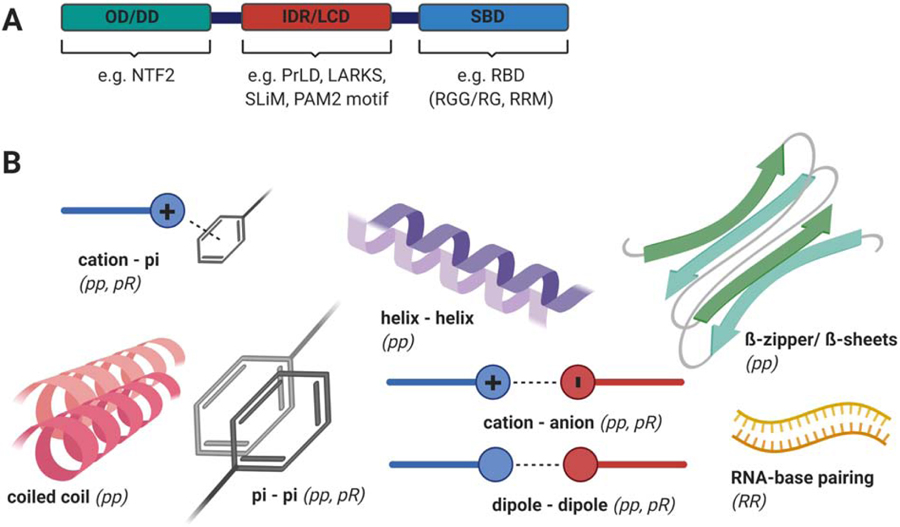
Proteins essential to multivalency-driven LLPS are multi-domain proteins that carry oligomerization domains, intrinsically-disordered regions (IDRs), and/or substrate-binding domains (SBDs, e.g. RNA-binding domain (RBD)) (see Fig. 1A). The more interaction domains that a protein contains, the higher its valence (v). Examples for protein-interaction domains are Prion-like domains (PrLDs), low complexity domains (LCDs), short linear motifs (SLiMs), low complexity aromatic-rich kinked segments (LARKS), or the PABP-interacting motif, protospacer adjacent motif 2 (PAM2) (see Fig. 1A, glossary and previously published reviews (Boeynaems et al., 2018; Cao et al., 2020)). Interactions between different macromolecules can occur at the level of protein-protein, protein-RNA, or RNA-RNA interactions (Cao et al., 2020; Protter et al., 2018; Van Treeck and Parker, 2018). The different types of molecular interactions are summarized in Fig. 1B, and are reviewed in detail elsewhere (Boeynaems et al., 2018; Cao et al., 2020).
Figure 1:
(A) generic model of a central SG protein, depicting an oligomerization domain (OD) or dimerization domain (DD), intrinsically disordered regions (IDRs) or low-complexity domains (LCDs), and a substrate binding domain (SBD), such as RNA-recognition motif (RRM) or RGG/RG domain. (B) Different types of protein-protein (pp), protein-RNA (pR), and RNA-RNA (RR) interaction that are implicated in SG assembly. The schematic was created in ©BioRender (biorender.com) and does not resemble actual sizes and ratios.
As mentioned above, SG assembly requires two steps, the first being the inhibition of translation initiation coupled with the subsequent disassembly of polysomes. The second step, which is downstream of translational inhibition and polysome disassembly, leads to the visible appearance of distinct cytoplasmic foci and is regulated by a variety of different proteins (Anderson and Kedersha, 2006) called SG nucleators, since they induce the formation of SGs upon overexpression even in the absence of stress (Anderson and Kedersha, 2009a; Buchan and Parker, 2009; Erickson and Lykke-Andersen, 2011; Gilks et al., 2004; Kedersha et al., 1999; Ohn et al., 2008; Tourrière et al., 2003). SG nucleators typically contain IDRs or PrLDs in conjunction with RNA-binding domains (RBDs), and are prone to self-aggregation (Anderson and Kedersha, 2006; Erickson and Lykke-Andersen, 2011) (see Fig. 2). See Kedersha et al. (Kedersha et al., 2013) for extensive list of SG nucleators and SG-associated proteins that contain IDRs and LCDs.
Figure 2:
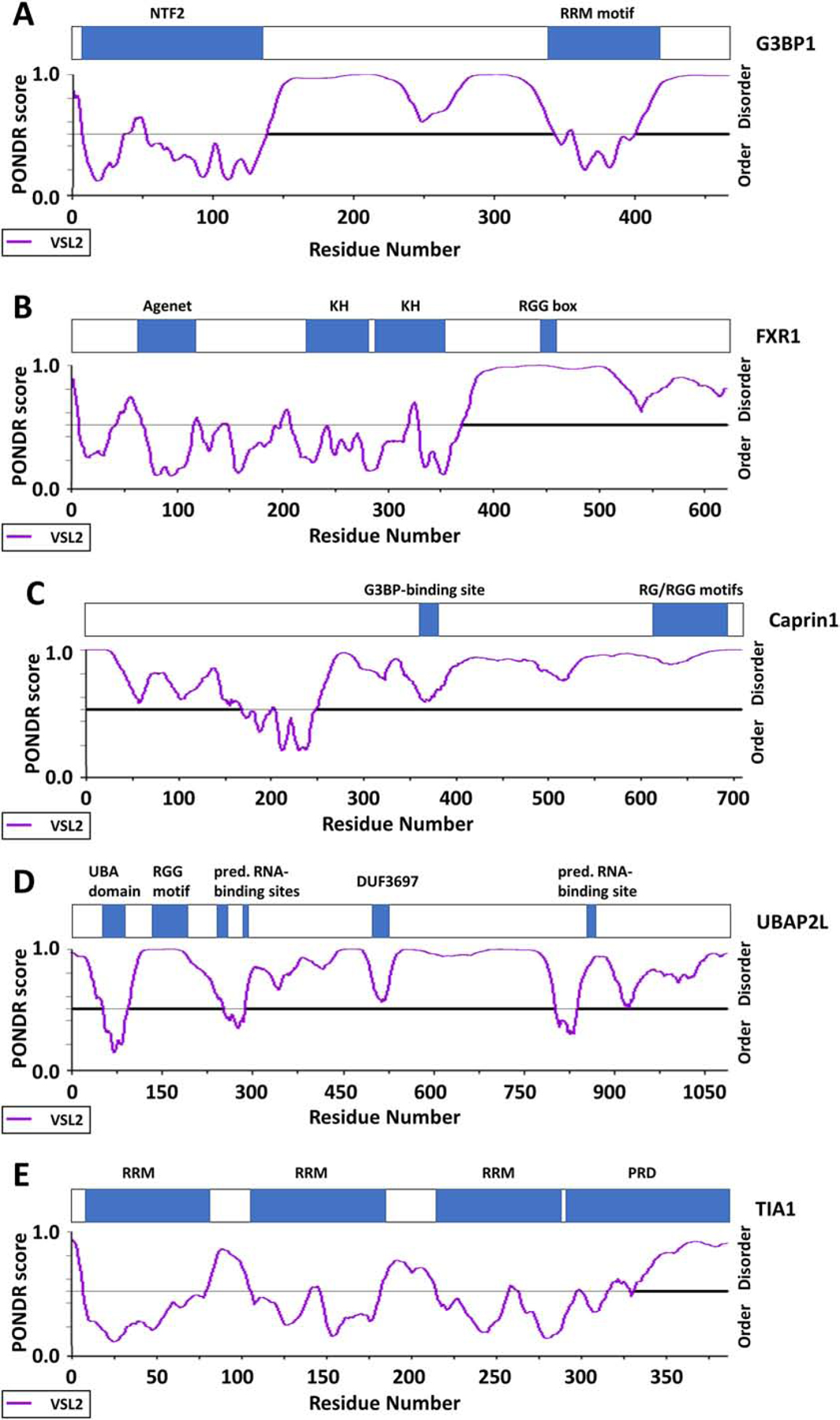
Localization of key motifs and domains, as well as predicted disordered regions calculated using the PONDR-VSL2 algorithm of (A) G3BP1, (B) FXR1, (C) Caprin1, (D) UBAP2L, and (E) TIA1.
Kuechler et al. identified a comprehensive set of molecular features in SG proteins (Kuechler et al., 2020). The authors analyzed recently published proteomics data from yeast and mammalian SGs, and assign a mammalian granule z-score (MaGS) that predicts the likelihood that a certain protein will partition into SGs (Kuechler et al., 2020). The features that contribute to the MaGS include total amount of protein disorder/IDRs within a protein, as well as the PScore which determines the enrichment of π-π interactions for potential protein-protein or RNA-protein interactions. Moreover, the CamSol score indicates the predicted solubility of a protein. Kuechler et al. found that SG-associated proteins have a higher CamSol score and thus, have a higher propensity to remain in solution than other proteins within the cytosol (Kuechler et al., 2020). This is particularly interesting since another feature of SG proteins is their high abundance. Despite their supersaturation, they are actively being kept in solution through still unknown mechanisms. Furthermore, increased potential for post-translational modifications (PTMs), as well as RNA-binding capacity also contributes to a higher MaGS and presents another interesting feature of proteins associated with SGs.
Role of RNA-RNA interactions in SG formation and LLPS
Besides the importance of molecular interactions of multivalent proteins in the formation of macromolecular condensates through LLPS, intermolecular interactions between RNA molecules have lately also gained attention (Fay and Anderson, 2018; Van Treeck and Parker, 2018). RNA itself can self-separate (Jain and Vale, 2017), but its sequence-specific and structural features specifically contribute to it (Fay and Anderson, 2018). Specifically, RNAs containing GGGGCC (rG4C2), CAG, and CUG repeats can phase separate in vitro and in cells (Fay et al., 2017; Jain and Vale, 2017). This phase separation is dependent on the RNA length (Fay et al., 2017; Jain and Vale, 2017), but also on the secondary structure of the RNA molecules (Langdon et al., 2018). For example, rG4C2 repeats in the ALS/FTD-associated C9ORF72 RNA form a G-quadruplex (G4) structure, and manipulating the G4 structure leads to impaired LLPS and RNA granule formation in vitro (Fay et al., 2017). G4-containing RNAs act as molecular scaffolds to sequester specific RBPs (such as G3BP1) thus increasing their local concentrations and promoting SG nucleation (Fay et al., 2017). Likewise, the intermolecular G4 structure formed from the 5′ fragments of cleaved tRNAs Ala/Cys have also play an essential role in LLPS, and specifically in SG formation, in a structure-dependent manner (Fay et al., 2017; Fay and Anderson, 2018; Ivanov et al., 2011; Lyons et al., 2017).
RNA molecules have a tendency to self-assemble in in vitro systems (Bounedjah et al., 2012; Langdon et al., 2018; Van Treeck et al., 2018). Direct RNA-RNA interactions such as Watson-Crick base-pairing, non-canonical base-pairing, or helical stacking, therefore facilitate the formation of macromolecular condensates by LLPS - see previous review (Van Treeck and Parker, 2018).
Methodology to study LLPS and SGs
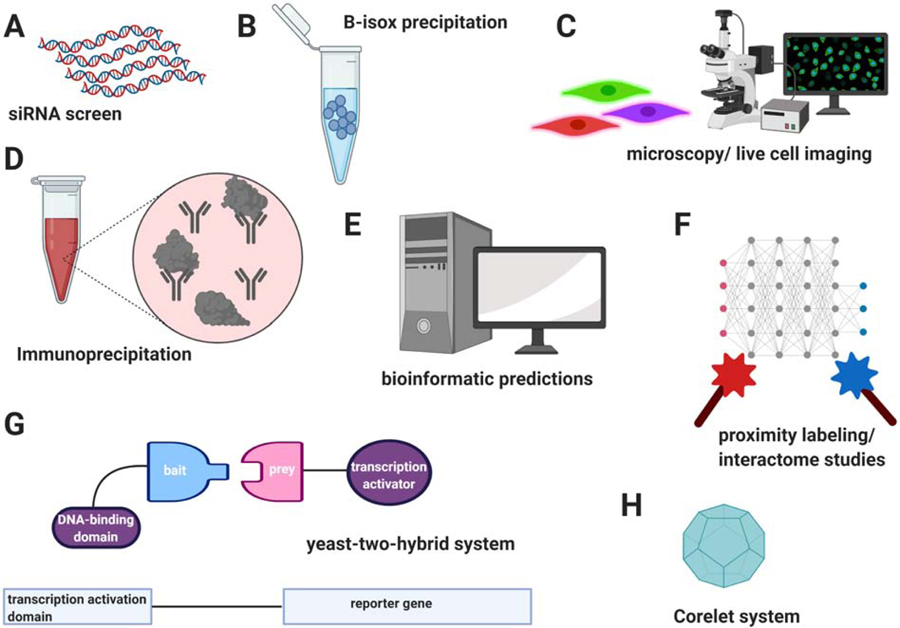
There are a wide variety of methods that are used to study LLPS and/or SG formation (see Fig. 3). A classical way to investigate the role of a certain gene in a specific phenomenon is to perform knockdown or knockout assays and to monitor the outcome. The first such study by Ohn et al. (Ohn et al., 2008) used an siRNA screen against around 7000 druggable targets to identify a wide variety of genes regulating Type I SG assembly triggered by arsenite (Ohn et al., 2008). Subsequently several other studies have been published and further broadened the list of SG regulators (Kim et al., 2012; Mokas et al., 2009; Wheeler et al., 2020; Youn et al., 2018). Most recently Yang et al. conducted another screen using siRNAs and reported a set of at least 274 genes whose knockdown impaired the formation of SGs (Yang et al., 2020), and which significantly overlapped with the previous study by Ohn et al (Ohn et al., 2008).
Figure 3:
Methodology to study SGs. (A) siRNAs screen to identify genes whose knockdown impairs assembly (or disassembly) of SGs. (B) Use of molecular crowding agents (e.g. B-isox, PEG) causes LLPS within a cell lysate, and allows separation of soluble and non-soluble fractions. (C) Both live imaging and fixed cell imaging present a microscopy-based approach to study LLPS and SGs. (D) An affinity purification approach (e.g. via GFP-tagged G3BP) can be utilized in combination with mass spectrometry analysis to identify proteins that are enriched in SGs. (E) Bioinformatic studies that can predict whether a particular protein is localized to SGs. (F) Interactome analyses using proximity labeling studies reveals a wide network of SG-associated proteins. (G) Yeast-two hybrid screen can be used to identify protein-protein interactions with SG-associated proteins as bait. (H) The Corelet system visualizes condensate formation in live cells using imaging as a readout. The schematic was created in ©BioRender (biorender.com) and does not resemble actual sizes and ratios.
A biochemical approach to study LLPS or SG formation employs molecular crowding agents (e.g. B-isox, PEG), addition of which cause LLPS within cell lysates, allowing the physical separation of pellet and supernatant from which the RNA or protein composition can be analyzed (Han et al., 2012; Kato et al., 2012). Kedersha et al. utilized the application of B-isox precipitation for the study of SG formation (Kedersha et al., 2016). An affinity purification approach (e.g. via GFP-tagged G3BP) can be utilized in combination with mass spectrometry analysis, in order to identify proteins that are enriched in SGs (Jain et al., 2016). Similarly, Goodier et al. used a yeast-two hybrid screen to identify several RNA-binding proteins and SG-associated proteins that coimmunoprecipitate with the SG protein LINE-1 ORF1p (Goodier et al., 2007). A more recent methodology for interactome analyses is provided by proximity labeling, which detects an even wider network of SG-associated proteins (Markmiller et al., 2018; Marmor-Kollet et al., 2020; Youn et al., 2018).
In addition, bioinformatic methods predicting whether a particular protein is localized to SGs have become more recently available. While previous predictions mainly focused on whether a particular protein exhibits low complexity domains or intrinsically disordered regions (Kedersha et al., 2013), a newer study by Kuechler et al. describes several distinct, characteristic features that predict the localization to SGs (Kuechler et al., 2020) (see above for more details).
The most classically used method, however, is a microscopy-based imaging approach. Both live and fixed cell imaging provide the most direct experimental data on LLPS and SG formation. Moreover, the Corelet system, an optogenetic protein system that forms an oligomer structure within cells upon induction, and which was invented by Bracha et al. provides a powerful novel tool (Bracha et al., 2018). Its latest adaption allowing its use for condensate formation under live imaging conditions (Sanders et al., 2020).
III. Regulation of SG assembly
Novel model of protein interaction networks that control multivalency-driven LLPS in SG assembly
Several critical SG regulator proteins have been identified. For example, the TIA proteins TIA-1 and TIAR exhibit Q/N-rich domains that are required for the stress-induced aggregation of TIA proteins and thereby promote the formation of SGs (Gilks et al., 2004). In addition, TIA-1 contains two RNA-recognition motifs (RRM), which are needed for the recruitment of TIA-1 to mRNPs and for the subsequent assembly of SGs (Erickson and Lykke-Andersen, 2011; Kedersha et al., 1999). Other examples are TTP and BRF1 (Stoecklin et al., 2004), Ago2 (Leung et al., 2006), fragile X mental retardation protein (FMRP), fragile X mental retardation-related protein 1 (FXR1) (Antar et al., 2005) and Fas-activated Ser/Thr phosphoprotein (FAST) (Anderson and Kedersha, 2008; Kedersha et al., 2005).
More recently, G3BP has become recognized as the central regulator of SG assembly (Gal et al., 2016; Guillén-Boixet et al., 2020; Kedersha et al., 2016; Sanders et al., 2020; Tourrière et al., 2003; Tsai et al., 2016; Yang et al., 2020). Double knockout cells lacking both G3BP1 and G3BP2 (ΔΔG1/G2 cells) grow normally, but cannot form Type I (phospho-eIF2α-dependent) or Type II (eIF4A dependent) SGs (Kedersha et al., 2016). G3BP-mediated SG assembly is regulated by G3BP’s two major binding partners: the SG nucleator Caprin1 and Ubiquitin carboxyl-terminal hydrolase 10 (USP10) (Kedersha et al., 2016). The binding sites of Caprin1 and USP10 on G3BP are adjacent or overlapping, and the two proteins compete for mutually exclusive binding to G3BP with opposite outcomes (Kedersha et al., 2016; Panas et al., 2015; Vognsen et al., 2013). Caprin1 binding to G3BP promotes the formation of SGs, whereas the binding of USP10 to G3BP abolishes SG assembly. A mutant of G3BP incapable of binding to either Caprin1 or USP10 is fully SG competent, whereas overexpression of Caprin1 in the ΔΔG1/G2 cells does not restore SGs (Kedersha et al., 2016; Sanders et al., 2020). One explanation for the central role of the G3BP-Caprin1-USP10 axis is that binding of Caprin1 to G3BP causes a conformational shift within the G3BP-Caprin1 complex that facilitates LLPS mediated through the IDRs of G3BP. In contrast, binding of USP10 to G3BP promotes the soluble confirmation of G3BP, and thereby induces SG disassembly (Kedersha et al., 2016).
Sanders et al., Yang et al., and Guillen-Boixet et al. recently proposed a novel network model to describe the mechanisms underlying G3BP’s role in SG assembly (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). Protein-interaction networks are viewed as interconnected complexes (= nodes) of modular proteins, and the valence of each node is determined by the number of available interaction domains. Stoichiometry-dependent competition, either within one protein network or between different networks, critically regulates the assembly process. G3BP is depicted as the central node, as it exhibits a modular domain architecture which includes a dimerization domain and an RNA-binding domain, connected by a disordered region comprised of two compositionally distinct IDRs (IDR1, IDR2). Yang et al. claim that G3BP has the highest centrality within the core SG network (Yang et al., 2020). Despite the common assertion that IDRs universally drive phase separation, all three papers showed that both IDRs are dispensable for the role of G3BP in SG assembly (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). These show that intrinsic properties of G3BP, specifically its ability to undergo dimerization coupled with its the high binding affinities towards RNA, drive LLPS upon polysome disassembly.
All three publications highlight the importance of the NTF2 dimerization domain in SG assembly, as cells that express ΔNTF2 mutants of G3BP (in ΔΔG1/G2 cells) fail to form SGs upon arsenite stress (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). Likewise, G3BP mutants with impaired RNA-binding capacity show reduced (Yang et al., 2020) or even complete loss (Sanders et al., 2020) of SGs. The authors conclude that multivalency for RNA binding is a critical factor in SG assembly (Yang et al., 2020), and the G3BP-RNA condensates act as recruiting platforms for various other RNA-binding proteins (Guillén-Boixet et al., 2020).
Another novel high-affinity binding partner of G3BP, UBAP2L, was recently found through proteomics studies to be required for SG formation (Markmiller et al., 2018; Youn et al., 2018), and might act upstream of G3BP in nucleating SG assembly (Cirillo et al., 2020). Overexpression of UBAP2L in HeLa cells caused the formation of SGs in the absence of stress (Huang et al., 2020). As in the case of G3BP, deletion of the (RNA-binding) RGG motif of UBAP2L abolishes SG formation upon arsenite stress (Huang et al., 2020). In addition, UBAP2L contains a self-associative IDR that is likely critical for SG formation (Sanders et al., 2020). UBAP2L has been found to interact with G3BP via its DUF domain, and DUF deletion disrupted the interaction between G3BP and UBAP2L, and compromised SG assembly (Huang et al., 2020). Thus, it is fair to assume that UBAP2L serves as an interaction node to collectively confer the high RNA-binding capacity (“RBD valence”) of G3BP that is needed to form a condensed RNP network following stress-dependent mRNA influx. In the context of this networking model, G3BP represents a central node in the SG network, and the G3BP dimerization domain (NTF2) serves as the major interaction site for RBD-containing bridges (e.g. Caprin1) and secondary nodes (e.g. UBAP2L) (Sanders et al., 2020; Yang et al., 2020). Upon stress-dependent influx of RNA, the increase in valence of the G3BP node, mediates the multivalency-driven LLPS assembly of SGs.
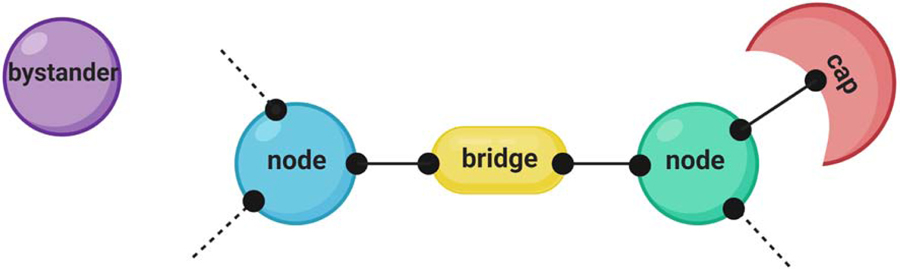
Furthermore, the networking model also describes a so called “valence capping” mechanism for regulating phase separation, which explains USP10-mediated SG inhibition. Hereby, USP10 acts as a v=1 interactor (= cap), and upon binding to G3BP, USP10 decreases the overall valence of the G3BP complex, turning it from a v>3 node into a v=2 bridge, which thus inhibits the formation of SGs (Fig. 4). Valence capping could provide a powerful mechanism for temporal regulation of multiphase structuring and substrate processing, particularly given that effective valence can depend on sensitively or stoichiometry.
Figure 4:
Novel model depicting a SG protein and RNA interaction network: Macromolecules with more than three interaction domains (v ≥ 3) are defined as nodes, and constitute the essential drivers for LLPS. Proteins that lack interactions with other particles are bystanders (v = 0), whereas v = 1 proteins are called caps (modified from Sanders et al., 2020). This model depicts a mechanistic framework for network-based cellular condensation. The schematic was created in ©BioRender (biorender.com) and does not resemble actual sizes and ratios.
Thus, the combined studies by Sanders et al., Yang et al. and Guillen-Boixet et al. show that protein network-based interactions directly control multiphase condensation through finely tuned interaction strengths of interconnected caps, bridges, and nodes to allow substrate-dependent “phase switches” (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). In G3BP this “switch” arises from competition between intra- and inter-molecular interactions (Guillén-Boixet et al., 2020; Yang et al., 2020). Under physiological conditions, when the concentration of “free”, unfolded RNA in the cytoplasm is low, intramolecular interactions between the acidic IDR1 and the basic IDR3/RG rich region in G3BP are favoured, which creates a compact, “closed” conformation. Upon stress-induced translational arrest and polysome disassembly, once a threshold concentration of protein-free, unfolded mRNA sequences is reached, RNA binds to G3BP and outcompetes the intramolecular interactions with G3BP. This induces a conformational shift in G3BP, and exposes an expanded, “open” confirmation, which allows the engagement in other protein-RNA or protein-protein interactions. As a result, the valence of the G3BP-RNA complex increases, and thereby initiates LLPS and subsequent SG assembly.
Speculative: Interaction of SG proteins (e.g. G3BP) with ribosomes
While considerable evidence shows that translational arrest and SG formation are two distinct, uncoupled processes, it still remains unclear how the disassembled polysomes are being recognized, how exactly they are recruited to the SG condensates, and what role certain SG proteins play. It has been shown that G3BP1/2 can bind to dissociated 40S subunits via its RGG motif, and that both Caprin1 and USP10 can be recruited to the 40S-associated G3BP1 (Kedersha et al., 2016). Meyer et al recently confirmed these earlier findings, and showed that G3BP1/2 directly crosslinks with the 18S rRNA of the 40S subunit, (but not with the 60S subunit) (Meyer et al., 2020). The authors claim that the G3BP1/2-USP10 complex is specifically recruited to dissociated 40S subunits, due to the fact that the G3BP binding site on the 40S subunit is near or at the interaction site between the 40S and 60S subunits. Upon binding, USP10 causes site-specific deubiquitination of rpS2, rpS3, and rpS10, and thus prevents lysosomal degradation of the 40S subunit (Meyer et al., 2020).
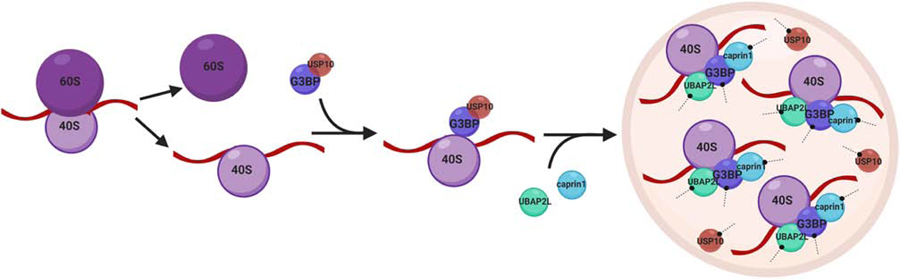
These findings invite speculation that G3BP-USP10 complexes are the cellular sensors that detect disassembled polysomes (Fig. 5). Once G3BP has been recruited to the stalled 40S subunits, USP10 dissociates from the complex, and thus allows UBAP2L, Caprin1, and other node proteins to bind to G3BP. Interestingly, neither Caprin1 nor USP10 binding to G3BP is needed for its association with the 40S subunit (Kedersha et al., 2016). However, it was recently shown that knockdown of UBAP2L abolished the association of G3BP with PABPC1 and rpS6 (Huang et al., 2020). It is not known whether the interaction between G3BP and ribosomal proteins happens directly via G3BP, or whether it might occur though UBAP2L binding. Therefore, one can imagine a model where the G3BP-USP10 complex is first recruited to dissociated 40S subunits upon stress, then USP10 (which acts as a cap) is replaced by Caprin1 and UBAP2L, where the latter serves as a bridge for additional binding to ribosomal proteins. Subsequently, the increase in multivalency causes LLPS and the formation of visible SGs.
Figure 5:
Speculative model
Upon polysome disassembly, G3BP-USP10 complexes bind to dissociated 40S subunits. Prior to its dissociation from G3BP, USP10 mediates deubiquitination of specific ribosomal proteins to prevent their degradation through the lysosome. Binding of UBAP2L and caprin1 to G3BP leads to increased valency, which promotes LLPS and subsequent SG formation. The schematic was created in ©BioRender (biorender.com); it is not time-resolved and does not resemble actual sizes and ratios.
Posttranslational modifications (PTMs) of node proteins/ SG regulator proteins in the regulation of SG assembly.
The network model proposed by Sanders et al., Yang et al. and Guillen-Boixet et al. convincingly describes how multivalency is the main driver for LLPS. It also provides an explanation of how the assembly of stimulus-responsive condensates can be regulated: minor changes in the network stoichiometry can have drastic effects in influencing the valence of a central node. Given that many key components of SGs have been identified as substrates for posttranslational modifications (PTMs) (Cao et al., 2020; Kuechler et al., 2020), PTMs likely also regulate SG formation (Hofweber and Dormann, 2019; Kim et al., 2019; Ohn and Anderson, 2010). Of the various PTMs, phosphorylation events are known to be key in SG assembly and disassembly: phosphorylation of eIF2α at Ser51 is critical for translational arrest and subsequent SG formation in response to many types of stress (Anderson and Kedersha, 2008; Kedersha et al., 1999). Secondly, phosphorylation of TTP regulates the interaction between SGs and PBs by preventing the recruitment of TTP to SGs (Kedersha et al., 2005). Finally, phosphorylation of growth factor receptor-bound protein 7 (Grb7) promotes the disassembly of heat shock-induced SGs (Tsai et al., 2008).
Another PTM influencing SG formation is the reversible addition of O-linked N-acetylglucosamine (O-GlcNAc) to serine and threonine residues of target proteins, which is crucial for sensing environmental conditions and regulating signaling cascades (Hart et al., 2007; Slawson et al., 2006). Many SG proteins are O-GlcNAc-modified, and glycosylation by O-GlcNAc is generally increased upon arsenite-induced oxidative stress (Ohn and Anderson, 2010). Knockdown of the O-GlcNAc transferase (OGT) did not affect translational arrest upon arsenite treatment, but inhibited the formation of SGs (Ohn et al., 2008). Thus, O-GlcNAc-modification seems to be required for modulating the aggregation process during SG formation (Ohn et al., 2008; Ohn and Anderson, 2010).
The translation factor eIF5A is post-translationally modified by covalent conjugation of hypusine, a derivative of spermidine, to Lys50 via the polyamine biosynthetic pathway (Dever et al., 2014; Murphey and Gerner, 1987; Ohn and Anderson, 2010). Knockdown of eIF5A or factors involved in the polyamine synthesis pathway leads to inhibition of arsenite-induced polysome disassembly and SG formation (Li et al., 2010; Ohn et al., 2008). Mechanistically, cells lacking hypusine-modified eIF5A show decrease in the rate of stress-induced ribosome run-off and translation elongation. Mechanistically, eIF5A increases the rate of peptidyl-tRNA hydrolysis during translation termination and specifically enhances translation of ribosomes stalled at some sequences, e.g. proline stretches, during translation elongation (Schuller and Green, 2017).
Several SG proteins, such as FMRP, G3BP, UBAP2L, and PABP contain an RGG/RRM domain where arginine methylation can occur (Huang et al., 2020; Tsai et al., 2016; Xie and Denman, 2011). Methylation of the arginine residue in FMRP is required for the assembly of SGs (De Leeuw et al., 2007; Dolzhanskaya et al., 2006; Goulet et al., 2008), whereas for G3BP and UBAP2L arginine demethylation was reported to promote SG formation (Huang et al., 2020; Tsai et al., 2016). During recovery from stress, UBAP2L was found to be methylated again. Interestingly, overexpression of the arginine methyltransferase PRMT1, which prevents demethylation of UBAP2L upon arsenite stress, inhibited the formation of SGs (Huang et al., 2020). In addition, acetylation/deacetylation (Jedrusik-Bode et al., 2013; Kwon et al., 2007), ubiquitination (Kwon et al., 2007; Mazroui et al., 2007) and neddylation (Jayabalan et al., 2016; Xirodimas, 2008) of specific target proteins might also play a role in SG formation (Markmiller et al., 2019; Ohn and Anderson, 2010; Xie and Denman, 2011).
Another type of PTM that has been shown to play a role in SG formation is Poly(ADP)-ribosylation (Duan et al., 2019; Leung, 2020). Thereby, Poly(ADP)-ribose (PAR), a polymer of two or more ADP-ribose units, is added onto target proteins by one or several PAR polymerases (PARPs), and the removal of PAR mainly involves PAR glycohydrolase (PARG) (Leung, 2020, 2014). Several PARPs and PARG isoforms are localized into SGs, and the overexpression of some PARPs has been shown to induce SG formation (Leung et al., 2012, 2011; Leung, 2020). Duan et al. recently characterized the PARylation site of the SG-associated protein hnRNP A1, as well as a PAR-binding motif (PBM) of hnRNP A1 which can bind PAR and PARylated proteins (Duan et al., 2019). The authors further showed that a decrease in the overall cellular PARylation levels prevents the localization of hnRNP A1 to SGs, that PARylation at K298 regulates the nucleocytoplasmic transport of hnRNP A1, and that the PBM is indispensable for LLPS of hnRNP A1 in vitro and thus for its recruitment to SGs in vivo (Duan et al., 2019).
Some recent studies point to the involvement of SUMOylation in the regulation of SG assembly (Jongjitwimol et al., 2016; Keiten-Schmitz et al., 2020; Sanders et al., 2020). Most recently, proximity labeling studies demonstrated that several SUMO ligases (RANBP2, UBE2I) are recruited to SGs (Marmor-Kollet et al., 2020). Furthermore, knockdown of several factors of the SUMO-targeted ubiquitin ligase (StUbL) pathway caused impairment of SG disassembly (Keiten-Schmitz et al., 2020). The underlying mechanism is not clear yet, but the authors claim a significant role for SUMOylation in SG dynamics (see section on SG disassembly below).
Interestingly, PTMs are reported to preferentially affect IDRs (Bah and Forman-Kay, 2016). This could complicate the hypothesis that PTMs fine tune the LLPS properties of node and bridge proteins, and are thus provide a switch in the regulation of SG assembly (Guillén-Boixet et al., 2020; Sanders et al., 2020; Yang et al., 2020). Yang et al., and Guillen-Boixet et al. have investigated the role of G3BP phosphorylation, and found that site-specific phosphorylation of G3BP at Ser149 and Ser232 provides a potential switch for regulating phase separation, presumably by affecting intra- and intermolecular interactions between acidic regions, charged regions or IDRs (Guillén-Boixet et al., 2020; Yang et al., 2020). This contradicts an earlier report by Panas et al. which showed that phosphorylation of G3BP at Ser149 does not influence SG formation (Panas et al., 2019).
Finally, an emerging theme in SG research is the role of RNA modifications and their effects on SG assembly. SG-associated mRNAs contain multiple N6-methyladenosine (m6A) modifications, which specifically distinguishes them from mRNAs that are not localized to SGs (Anders et al., 2018). The m6A “reader” proteins, the YTH domain family (YTHDF) proteins, undergo LLPS upon binding to the mRNAs, and are thereby promoting the formation of SGs (Fu and Zhuang, 2020; Ries et al., 2019).
In conclusion, while it is now widely accepted and proved by many studies that the physics of LLPS underlies the formation of SGs, the exact molecular mechanisms about how the assembly process is regulated are not fully understood. However, it appears likely that there is a whole network of interconnected SG regulator proteins, with G3BP as the central node.
Finally, it needs to be pointed out that viral infection also causes the formation of SGs, and that viruses have developed distinct strategies to suppress SG assembly. While it is beyond the scope of this review, it is worth mentioning that viruses have various different mechanisms in play to modulate SG regulation and SG dynamics - for detailed reviews see (Eiermann et al., 2020; Lloyd, 2012; Reineke and Lloyd, 2013; White and Lloyd, 2012).
IV. Regulation of SG disassembly
While the mechanisms of SG assembly in response to various stressors and in different model systems have been extensively studied, less attention has been paid to the SG disassembly process. Apart from cold shock-induced SGs, which disassemble within minutes after return to normal temperature (Hofmann et al., 2012), SG recovery after arsenite stress, H2O2 treatment, sorbitol exposure, or heat shock occurs between 60–120 minutes (Anderson and Kedersha, 2002a; Cherkasov et al., 2013; Huang et al., 2020; Kedersha et al., 2002; Marmor-Kollet et al., 2020). The reversibility of SGs after stress removal or upon co-treatment with translation elongation inhibitors cycloheximide or emetine, as a critical characteristic of bona fide SGs, hints on the important roles of polysome dynamics in this process, although how polysome formation affects phase separation is not known.
It was recently shown by Huang et al. that UBAP2L could be a critical regulator in SG dynamics. Both knockdown and overexpression of UBAP2L impair SG disassembly (Huang et al., 2020). The underlying mechanism for that is not clear yet. However, as UBAP2L is regulated by arginine methylation, this data may point to an important role of PTMs in the disassembly process, consistent with reports that phosphorylation of growth factor receptor-bound protein 7 (Grb7) promotes the disassembly of heat shock-induced SGs (Tsai et al., 2008), and chemical inhibition of PARylation or knockdown of the cytoplasmic PAR glycohydrolase (PARG), which impairs the removal of PAR from its target proteins, cause a delay in SG disassembly upon stress relief (Duan et al., 2019; Leung et al., 2012, 2011).
Chaperones/disaggregases such as VCP (Buchan et al., 2013; Seguin et al., 2014; Wang et al., 2019) and HSP70 (Cherkasov et al., 2013; Kroschwald et al., 2015; Mazroui et al., 2007) may be important for SG disassembly. Ganassi et al. proposed a SG surveillance system, wherein HSPB8 is recruited to SGs and prevents the irreversible aggregation of aberrant proteins within SGs (Ganassi et al., 2016). Under normal conditions, the incidence of having misfolded proteins within a SGs is very low, thus the HSPB8-mediated neutralization of misfolded proteins, as well as the recruitment of the BAG3-HSP70 subcomplex, and the subsequent degradation of aberrant SG components ensure SG fluidity and proper SG disassembly (Ganassi et al., 2016).
In 2016, Wheeler et al. postulated that SG disassembly occurs through multiple steps, wherein the stalled mRNAs are titrated out of SGs, thereby causing structural instability of the protein complexes, and subsequently stepwise disassembly of the visible SGs (Wheeler et al., 2016). This hypothesis is in line with more recent experimental data. In fact, given the network multivalency model of SG formation, the easiest explanation would be that SG disassembly happens as a consequence of a decrease in multivalency. The release from stress, allows replenishment and restoration of functional eIFs and/or ternary complexes, e.g. upon dephosphorylation of eIF2α. This most likely involves other signaling cascades, which might alter various PTMs on essential SG node proteins. Indeed, G3BP1 is reported to be acetylated by CBP/p300 upon release from stress (Gal et al., 2019) decreasing its binding affinity for RNA (Gal et al., 2019). Other PTMs such as arginine methylation could similarly cause a decrease in RNA binding and/or conformational changes within the core SG proteins, which would then allow to “re-mixing” of the SG components with the cytoplasm, and would reverse the LLPS. As a result, the sequestered mRNAs would be released to allow the resumption of translation.
A stepwise model for the disassembly process of SGs, and the coordinated interplay between SG disassembly and restoration/recovery of the translational machinery is supported by an often-overlooked study on cold shock-induced SG formation (Hofmann et al., 2012). Upon return to ambient temperatures, SGs disassemble within a few minutes, proceeding polysome reassembly, which supports reversed LLPS rather than complex and slower degradation processes.
In addition, the speed with which cold-shock SGs resolve may reflect the fact that enzymatic activities can instantly be resumed coincident with optimal temperature, and that protein conformational changes occur very quickly (potentially mediated by PTMs). In the case of cold shock-induced SG disassembly, it is clear that the disassembly process precedes the recovery of mRNA translation.
Another interesting insight into the mechanisms of SG disassembly come from two new studies by Marmor-Kollet et al. and Keiten-Schmitz et al. (Marmor-Kollet et al., 2020). Marmor-Kollet et al conducted proximity labeling of several core SG proteins, and identified distinct subsets of proteins that associate with these core proteins during the recovery phase (Marmor-Kollet et al., 2020). The appearance of these unique subsets of proteins, termed Disassembly Engaged Proteins (DEPs), indicates that the disassembly of SGs is indeed a controlled process occurring in a stepwise manner and not merely passive dissolution of SG components into the cytoplasm. Furthermore, Marmor et al. identified several SUMO ligases as DEPs that are specifically recruited to SGs during recovery from stress, and they showed that there is broad SUMOylation of SG proteins (Marmor-Kollet et al., 2020). SUMOylation could thus be another important PTM regulating the valence of SG proteins and thus SG disassembly. Likewise, Keiten-Schmitz et al. show that the nuclear SUMO-targeted ubiquitin ligase (StUbL) pathway is involved in SG disassembly (Keiten-Schmitz et al., 2020). Knockdown of key components of the StUbL pathway (e.g. SUMO2, RNF4) significantly impair SG disassembly after stress. This provides a first notion that nuclear and cytosolic stress response pathways are connected.
Finally, we must say that even though the mechanisms for SG disassembly are not yet widely studied, the resolution of SGs is important for maintaining viability and protein homeostasis. Impaired clearance of SGs results in persistent protein aggregates, which are implicated in several neurodegenerative diseases, e.g. ALS (Aulas et al., 2012; Baron et al., 2013; Bosco et al., 2010; Dammer et al., 2012; Gal et al., 2016, 2011; Li et al., 2013; Liu-Yesucevitz et al., 2010), Parkinsońs disease (Repici et al., 2019), and Alzheimeŕs (Vanderweyde et al., 2012). Some studies report that aberrant SGs are degraded by autophagy, Cdc48/VCP function, and the HSPB8-BAG3-HSP70 chaperone complex (Buchan et al., 2013; Ganassi et al., 2016; Seguin et al., 2014). Whether or not it is indeed an autophagy-dependent process, is still controversial. However, it is clear that resolution of any type of protein aggregate is required for maintaining cellular proteostasis, and the interplay between restoration of polysomes, rerouting of mRNAs into active translation, and disassembly of the remaining protein-protein and protein-RNA complexes is critical for enduring viability.
Conclusions
Understanding the molecular mechanisms of SG assembly and disassembly is an ongoing challenge. Remarkable genetic evidence connects neurological diseases with mutations in specific proteins related to RNA metabolism, proteostasis, and cytoskeleton maintenance, and link these to the regulation of SG dynamics and clearance.
Recent advances in proteomics have identified factors that contribute to SG assembly, primarily via interaction with the hallmark node protein G3BP1 (Fig. 6). Whether other proteins constitute the central node in all cell types remains unknown, and will be especially interesting to ascertain in cells such as neurons and stem cells. Lagging behind is our understanding of the molecular mechanisms of SG disassembly. In theory, upon stress removal, altering PTMs at key sites on critical node/bridge or cap proteins may decrease overall multivalency, and lead to stepwise disaggregation of larger microscopically visible SGs into smaller SGs. Subsequently, the smaller SGs disperse into RNP complexes (Fig. 6). This hypothesis demands further testing.
Figure 6:
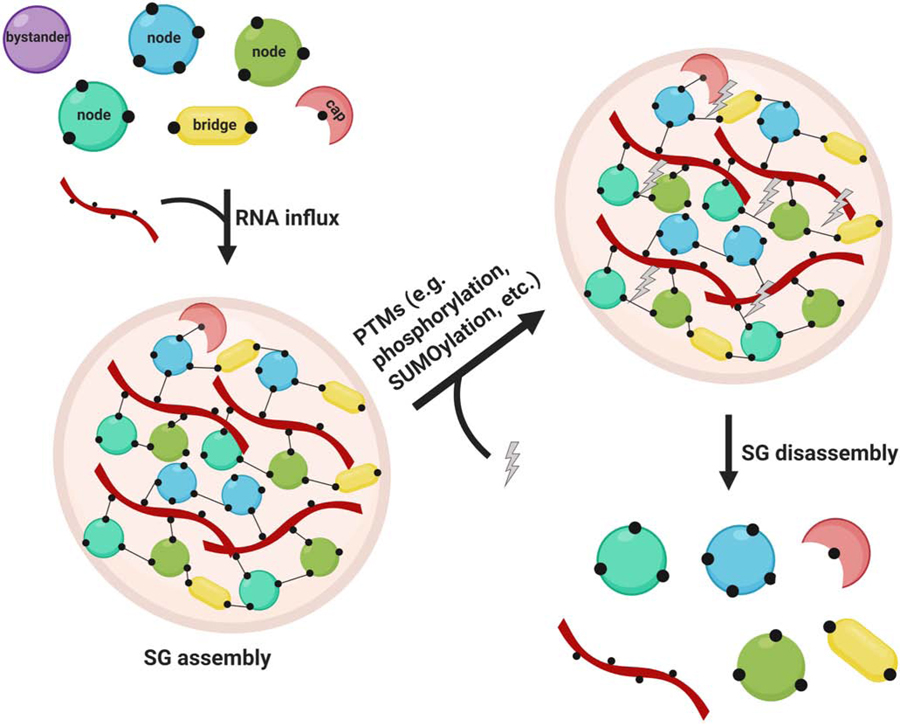
Model of SG assembly and disassembly. Upon RNA influx, key SG node and bridge proteins form a multi-interaction network, which causes LLPS and the assembly of SGs. Following stress removal, PTMs potentially lead to the loss of protein-protein, protein-RNA, and RNA-RNA interactions, leading to a decrease in valence and allowing subsequent SG disassembly. The schematic was created in ©BioRender (biorender.com); it is not time-resolved and does not resemble actual sizes and ratios.
The network multivalency model may also explain how mutations in specific SG-associated proteins can cause abnormalities in SG dynamics, which, at least in neurodegenerative diseases, culminate in the pathological aggregation or coalescing mutated RBPs into SGs. We speculate that by introduction of mutations, protein factors can change their biophysical properties or interaction pattern thereby changing their natural valence (e.g., from bystander into bridge, or from bridge to cap). Mutations in some proteins (e.g., FUS/TLS or TDP43) can also influence their subcellular localization that, in turn, change their nuclear-cytoplasmic distribution leading to changes in relative abundance of SG-associated factors in the cytosol. Again, this awaits further examination in the in vivo context.
In conclusion, and possibly most importantly, we expect significant advances in the development of new therapies towards neurodegenerative diseases based on the investigation of basic mechanisms of LLPS and SG dynamics. Current data reflecting the complex nature of RNA granule dynamics also provide us with a roadmap to define critical targets for therapy. Most critically, we are in the urgent need in the development of in vivo models that ultimately will examine the relevance of these findings in the physiological context.
Highlights.
Multivalency is the main driver for LLPS leading to SG assembly
Proteins essential for multivalency-driven LLPS are multi-domain proteins
PTMs play a role in regulating SG formation and disassembly.
Acknowledgments:
We thank members of Ivanov and Anderson labs for discussion and helpful critique.
Funding: This work was supported by the National Institutes of Health [R35 GM126901 to P.A., RO1 GM126150 to P.I.]
Glossary:
- Liquid-liquid phase separation (LLPS)
Physical process by which a homogeneous liquid solution (phase) of mixed molecules separates into two distinct phases, one phase that is enriched for a certain subset of molecules and another phase that is depleted of the same molecules
- Liquid phase transition
Describes the switch from one phase to another (e.g. liquid to solid) of one particular molecule
- Molecular crowding
Excluded volume effect
- RNA condensate/Biomolecule condensate
Cellular structures that contain RNA, and/or other biomolecules (such as proteins) and that were formed by LLPS
- RNP granule/RNA granule
Biomolecule condensate consisting of RNA and proteins
- Intrinsically disordered region (IDR)/IDR protein (IDRP)
IDRs contain amino acid sequences that promote structural flexibility and instability. IDRPs lack a stable structure and rather thus exist as conformational ensembles
- Low complexity domain (LCD)
Consists of amino acid sequences that contain repeats of a particular amino acid, or amino acid motifs, and lacks diversity in its amino acid composition
- Prion-like domains (PrLDs)
Low complexity domain in RNA-binding proteins
- Short linear motifs (SLiMs)
Regulate low-affinity interactions, modulated by PTM
- low complexity aromatic-rich kinked segments (LARKS)
Weak interactions through polar atoms and aromatic side chains
- PABP-interacting motif, protospacer adjacent motif 2 (PAM2)
A 12 amino acids peptide motif that specifically interacts with MLLE motif (Mademoiselle motif) found in PABP
- Valence (v)
Number of potential interaction domains within a protein
- Node protein
Protein with v >3
- Bridge protein
Protein with v = 2
- Cap protein
Protein with v = 1
- SG nucleator
Proteins that causes the formation of SGs when overexpressed in a cell, even in the absence of stress
- SG regulators
Proteins whose knockout/ knockdown prevents the assembly of SGs upon stress
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest: The authors declare no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Advani VM, Ivanov P, 2019. Translational Control under Stress: Reshaping the Translatome. BioEssays News Rev. Mol. Cell. Dev. Biol 41, e1900009 10.1002/bies.201900009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S, Dormann D, 2019. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet 53, 171–194. 10.1146/annurev-genet-112618-043527 [DOI] [PubMed] [Google Scholar]
- Anders M, Chelysheva I, Goebel I, Trenkner T, Zhou J, Mao Y, Verzini S, Qian S-B, Ignatova Z, 2018. Dynamic m6A methylation facilitates mRNA triaging to stress granules. Life Sci. Alliance 1, e201800113 10.26508/lsa.201800113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2009a. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol 10, 430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2009b. Stress granules. Curr. Biol. CB 19, R397–398. 10.1016/j.cub.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci 33, 141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2007. On again, off again: the SRC-3 transcriptional coactivator moonlights as a translational corepressor. Mol. Cell 25, 796–797. 10.1016/j.molcel.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2006. RNA granules. J. Cell Biol 172, 803–808. 10.1083/jcb.200512082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2002a. Stressful initiations. J. Cell Sci 115, 3227–3234. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, 2002b. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N, Ivanov P, 2015. Stress granules, P-bodies and cancer. Biochim. Biophys. Acta 1849, 861–870. 10.1016/j.bbagrm.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ, 2005. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav 4, 350–359. 10.1111/j.1601-183X.2005.00128.x [DOI] [PubMed] [Google Scholar]
- Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M, 2008. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol 10, 1324–1332. 10.1038/ncb1791 [DOI] [PubMed] [Google Scholar]
- Aulas A, Lyons SM, Fay MM, Anderson P, Ivanov P, 2018. Nitric oxide triggers the assembly of “type II” stress granules linked to decreased cell viability. Cell Death Dis 9, 1129 10.1038/s41419-018-1173-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas A, Stabile S, Vande Velde C, 2012. Endogenous TDP-43, but not FUS, contributes to stress granule assembly via G3BP. Mol. Neurodegener 7, 54 10.1186/1750-1326-7-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bah A, Forman-Kay JD, 2016. Modulation of Intrinsically Disordered Protein Function by Post-translational Modifications. J. Biol. Chem 291, 6696–6705. 10.1074/jbc.R115.695056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK, 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron DM, Kaushansky LJ, Ward CL, Sama RRK, Chian R-J, Boggio KJ, Quaresma AJC, Nickerson JA, Bosco DA, 2013. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol. Neurodegener 8, 30 10.1186/1750-1326-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua E, Wang X, Majumder M, Gaccioli F, Yuan CL, Wang C, Zhu X, Jordan LE, Scheuner D, Kaufman RJ, Koromilas AE, Snider MD, Holcik M, Hatzoglou M, 2010. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem 285, 17098–17111. 10.1074/jbc.M110.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N, Lederer M, Pfalz B, Reinke C, Fuchs T, Glaß M, Möller B, Hüttelmaier S, 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res 43, e26 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M, 2018. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol 28, 420–435. 10.1016/j.tcb.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M-E, Cencic R, Lindqvist L, Oberer M, Northcote P, Wagner G, Pelletier J, 2006. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol 13, 1287–1295. 10.1016/j.chembiol.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Bordeleau M-E, Matthews J, Wojnar JM, Lindqvist L, Novac O, Jankowsky E, Sonenberg N, Northcote P, Teesdale-Spittle P, Pelletier J, 2005. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. U. S. A 102, 10460–10465. 10.1073/pnas.0504249102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Sapp P, McKenna-Yasek D, Brown RH, Hayward LJ, 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum. Mol. Genet 19, 4160–4175. 10.1093/hmg/ddq335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O, Hamon L, Savarin P, Desforges B, Curmi PA, Pastré D, 2012. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J. Biol. Chem 287, 2446–2458. 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha D, Walls MT, Wei M-T, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP, 2018. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 175, 1467–1480.e13. 10.1016/j.cell.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP, 2008. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus 3 10.1128/ecosal.5.2.3 [DOI] [PubMed] [Google Scholar]
- Brunschede H, Dove TL, Bremer H, 1977. Establishment of exponential growth after a nutritional shift-up in Escherichia coli B/r: accumulation of deoxyribonucleic acid, ribonucleic acid, and protein. J. Bacteriol 129, 1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis R-M, Taylor JP, Parker R, 2013. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474. 10.1016/j.cell.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R, 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol 183, 441–455. 10.1083/jcb.200807043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R, 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Jin X, Liu B, 2020. The involvement of stress granules in aging and aging-associated diseases. Aging Cell 19, e13136 10.1111/acel.13136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G, Bukau B, 2013. Coordination of translational control and protein homeostasis during severe heat stress. Curr. Biol. CB 23, 2452–2462. 10.1016/j.cub.2013.09.058 [DOI] [PubMed] [Google Scholar]
- Cirillo L, Cieren A, Barbieri S, Khong A, Schwager F, Parker R, Gotta M, 2020. UBAP2L Forms Distinct Cores that Act in Nucleating Stress Granules Upstream of G3BP1. Curr. Biol. CB 30, 698–707.e6. 10.1016/j.cub.2019.12.020 [DOI] [PubMed] [Google Scholar]
- Dammer EB, Fallini C, Gozal YM, Duong DM, Rossoll W, Xu P, Lah JJ, Levey AI, Peng J, Bassell GJ, Seyfried NT, 2012. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PloS One 7, e38658 10.1371/journal.pone.0038658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Kedersha N, Low W-K, Romo D, Gorospe M, Kaufman R, Anderson P, Liu JO, 2006. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem 281, 32870–32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C, 2007. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res 313, 4130–4144. 10.1016/j.yexcr.2007.09.017 [DOI] [PubMed] [Google Scholar]
- Dever TE, Gutierrez E, Shin B-S, 2014. The hypusine-containing translation factor eIF5A. Crit. Rev. Biochem. Mol. Biol 49, 413–425. 10.3109/10409238.2014.939608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolzhanskaya N, Merz G, Aletta JM, Denman RB, 2006. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci 119, 1933–1946. 10.1242/jcs.02882 [DOI] [PubMed] [Google Scholar]
- Duan Y, Du A, Gu J, Duan G, Wang C, Gui X, Ma Z, Qian B, Deng X, Zhang K, Sun L, Tian K, Zhang Y, Jiang H, Liu C, Fang Y, 2019. PARylation regulates stress granule dynamics, phase separation, and neurotoxicity of disease-related RNA-binding proteins. Cell Res 29, 233–247. 10.1038/s41422-019-0141-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A, 2020. Dance with the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses 12 10.3390/v12090984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TSK, Andrade J, Groehler AL, Clark DE, Muratore-Schroeder TL, Pasic L, Smith JA, Shabanowitz J, Hunt DF, Macara IG, Lannigan DA, 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31, 722–736. 10.1016/j.molcel.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RJ, 2001. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol 11, 114–119. 10.1016/s0959-440x(00)00172-x [DOI] [PubMed] [Google Scholar]
- Erickson SL, Lykke-Andersen J, 2011. Cytoplasmic mRNP granules at a glance. J. Cell Sci 124, 293–297. 10.1242/jcs.072140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farny NG, Kedersha NL, Silver PA, 2009. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA N. Y. N 15, 1814–1821. 10.1261/rna.1684009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, 2018. The Role of RNA in Biological Phase Separations. J. Mol. Biol 430, 4685–4701. 10.1016/j.jmb.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay MM, Anderson PJ, Ivanov P, 2017. ALS/FTD-Associated C9ORF72 Repeat RNA Promotes Phase Transitions In Vitro and in Cells. Cell Rep 21, 3573–3584. 10.1016/j.celrep.2017.11.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhuang X, 2020. m6A-binding YTHDF proteins promote stress granule formation. Nat. Chem. Biol 10.1038/s41589-020-0524-y [DOI] [PMC free article] [PubMed]
- Fujimura K, Katahira J, Kano F, Yoneda Y, Murata M, 2009. Microscopic dissection of the process of stress granule assembly. Biochim. Biophys. Acta 1793, 1728–1737. 10.1016/j.bbamcr.2009.08.010 [DOI] [PubMed] [Google Scholar]
- Fujimura K, Sasaki AT, Anderson P, 2012. Selenite targets eIF4E-binding protein-1 to inhibit translation initiation and induce the assembly of non-canonical stress granules. Nucleic Acids Res 40, 8099–8110. 10.1093/nar/gks566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Chen J, Na D-Y, Tichacek L, Barnett KR, Zhu H, 2019. The Acetylation of Lysine-376 of G3BP1 Regulates RNA Binding and Stress Granule Dynamics. Mol. Cell. Biol 39 10.1128/MCB.00052-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Kuang L, Barnett KR, Zhu BZ, Shissler SC, Korotkov KV, Hayward LJ, Kasarskis EJ, Zhu H, 2016. ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol. (Berl.) 132, 563–576. 10.1007/s00401-016-1601-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal J, Zhang J, Kwinter DM, Zhai J, Jia H, Jia J, Zhu H, 2011. Nuclear localization sequence of FUS and induction of stress granules by ALS mutants. Neurobiol. Aging 32, 2323.e27–40. 10.1016/j.neurobiolaging.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganassi M, Mateju D, Bigi I, Mediani L, Poser I, Lee HO, Seguin SJ, Morelli FF, Vinet J, Leo G, Pansarasa O, Cereda C, Poletti A, Alberti S, Carra S, 2016. A Surveillance Function of the HSPB8-BAG3-HSP70 Chaperone Complex Ensures Stress Granule Integrity and Dynamism. Mol. Cell 63, 796–810. 10.1016/j.molcel.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P, 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15, 5383–5398. 10.1091/mbc.e04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Zhang L, Vetter MR, Kazazian HH, 2007. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol 27, 6469–6483. 10.1128/MCB.00332-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I, Boisvenue S, Mokas S, Mazroui R, Côté J, 2008. TDRD3, a novel Tudor domain-containing protein, localizes to cytoplasmic stress granules. Hum. Mol. Genet 17, 3055–3074. 10.1093/hmg/ddn203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillén-Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlüßler R, Kim K, Trussina IREA, Wang J, Mateju D, Poser I, Maharana S, Ruer-Gruß M, Richter D, Zhang X, Chang Y-T, Guck J, Honigmann A, Mahamid J, Hyman AA, Pappu RV, Alberti S, Franzmann TM, 2020. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 181, 346–361.e17. 10.1016/j.cell.2020.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzikowski AR, Chen YS, Zid BM, 2019. Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA 10, e1524 10.1002/wrna.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL, 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C, 2007. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022. 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]
- Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G, 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell 23, 3786–3800. 10.1091/mbc.E12-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofweber M, Dormann D, 2019. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem 294, 7137–7150. 10.1074/jbc.TM118.001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Chen Y, Dai H, Zhang, Huan, Xie M, Zhang, Hanbin, Chen F, Kang X, Bai X, Chen Z, 2020. UBAP2L arginine methylation by PRMT1 modulates stress granule assembly. Cell Death Differ 27, 227–241. 10.1038/s41418-019-0350-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P, 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov P, Kedersha N, Anderson P, 2019. Stress Granules and Processing Bodies in Translational Control. Cold Spring Harb. Perspect. Biol 11 10.1101/cshperspect.a032813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vale RD, 2017. RNA phase transitions in repeat expansion disorders. Nature 546, 243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R, 2016. ATPase-Modulated Stress Granules Contain a Diverse Proteome and Substructure. Cell 164, 487–498. 10.1016/j.cell.2015.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayabalan AK, Sanchez A, Park RY, Yoon SP, Kang G-Y, Baek J-H, Anderson P, Kee Y, Ohn T, 2016. NEDDylation promotes stress granule assembly. Nat. Commun 7, 12125 10.1038/ncomms12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedrusik-Bode M, Studencka M, Smolka C, Baumann T, Schmidt H, Kampf J, Paap F, Martin S, Tazi J, Müller KM, Krüger M, Braun T, Bober E, 2013. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J. Cell Sci 126, 5166–5177. 10.1242/jcs.130708 [DOI] [PubMed] [Google Scholar]
- Johansson HO, Brooks DE, Haynes CA, 2000. Macromolecular crowding and its consequences. Int. Rev. Cytol 192, 155–170. 10.1016/s0074-7696(08)60525-2 [DOI] [PubMed] [Google Scholar]
- Jongjitwimol J, Baldock RA, Morley SJ, Watts FZ, 2016. Sumoylation of eIF4A2 affects stress granule formation. J. Cell Sci 129, 2407–2415. 10.1242/jcs.184614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL, 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P, 2009. Regulation of translation by stress granules and processing bodies. Prog. Mol. Biol. Transl. Sci 90, 155–185. 10.1016/S1877-1173(09)90004-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P, 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans 30, 963–969. 10.1042/bst0300963 [DOI] [PubMed] [Google Scholar]
- Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P, 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell 13, 195–210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P, 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol 151, 1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Ivanov P, Anderson P, 2013. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci 38, 494–506. 10.1016/j.tibs.2013.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T, Thomas M, Lieberman J, McInerney GM, Ivanov P, Anderson P, 2016. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J. Cell Biol 212, 845–860. 10.1083/jcb.201508028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P, 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol 169, 871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P, 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol 147, 1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiten-Schmitz J, Wagner K, Piller T, Kaulich M, Alberti S, Müller S, 2020. The Nuclear SUMO-Targeted Ubiquitin Quality Control Network Regulates the Dynamics of Cytoplasmic Stress Granules. Mol. Cell 79, 54–67.e7. 10.1016/j.molcel.2020.05.017 [DOI] [PubMed] [Google Scholar]
- Kim B, Cooke HJ, Rhee K, 2012. DAZL is essential for stress granule formation implicated in germ cell survival upon heat stress. Dev. Camb. Engl 139, 568–578. 10.1242/dev.075846 [DOI] [PubMed] [Google Scholar]
- Kim TH, Tsang B, Vernon RM, Sonenberg N, Kay LE, Forman-Kay JD, 2019. Phospho-dependent phase separation of FMRP and CAPRIN1 recapitulates regulation of translation and deadenylation. Science 365, 825–829. 10.1126/science.aax4240 [DOI] [PubMed] [Google Scholar]
- Kim WJ, Back SH, Kim V, Ryu I, Jang SK, 2005. Sequestration of TRAF2 into stress granules interrupts tumor necrosis factor signaling under stress conditions. Mol. Cell. Biol 25, 2450–2462. 10.1128/MCB.25.6.2450-2462.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, Alberti S, 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife 4, e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuechler ER, Budzyńska PM, Bernardini JP, Gsponer J, Mayor T, 2020. Distinct Features of Stress Granule Proteins Predict Localization in Membraneless Organelles. J. Mol. Biol 432, 2349–2368. 10.1016/j.jmb.2020.02.020 [DOI] [PubMed] [Google Scholar]
- Kuznetsova IM, Zaslavsky BY, Breydo L, Turoverov KK, Uversky VN, 2015. Beyond the excluded volume effects: mechanistic complexity of the crowded milieu. Mol. Basel Switz 20, 1377–1409. 10.3390/molecules20011377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S, Zhang Y, Matthias P, 2007. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev 21, 3381–3394. 10.1101/gad.461107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon EM, Qiu Y, Ghanbari Niaki A, McLaughlin GA, Weidmann CA, Gerbich TM, Smith JA, Crutchley JM, Termini CM, Weeks KM, Myong S, Gladfelter AS, 2018. mRNA structure determines specificity of a polyQ-driven phase separation. Science 360, 922–927. 10.1126/science.aar7432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavut A, Raveh D, 2012. Sequestration of highly expressed mRNAs in cytoplasmic granules, P-bodies, and stress granules enhances cell viability. PLoS Genet 8, e1002527 10.1371/journal.pgen.1002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A, Todorova T, Ando Y, Chang P, 2012. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol 9, 542–548. 10.4161/rna.19899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, 2020. Poly(ADP-ribose): A Dynamic Trigger for Biomolecular Condensate Formation. Trends Cell Biol 30, 370–383. 10.1016/j.tcb.2020.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, 2014. Poly(ADP-ribose): an organizer of cellular architecture. J. Cell Biol 205, 613–619. 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Calabrese JM, Sharp PA, 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. U. S. A 103, 18125–18130. 10.1073/pnas.0608845103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P, 2011. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol. Cell 42, 489–499. 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CH, Ohn T, Ivanov P, Tisdale S, Anderson P, 2010. eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PloS One 5, e9942 10.1371/journal.pone.0009942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD, 2013. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol 201, 361–372. 10.1083/jcb.201302044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang Y-J, Vanderweyde T, Vanderwyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, Petrucelli L, Wolozin B, 2010. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PloS One 5, e13250 10.1371/journal.pone.0013250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd RE, 2012. How do viruses interact with stress-associated RNA granules? PLoS Pathog 8, e1002741 10.1371/journal.ppat.1002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschi M, Leishman CC, Berardone N, Boccaccio GL, 2009. Dynein and kinesin regulate stress-granule and P-body dynamics. J. Cell Sci 122, 3973–3982. 10.1242/jcs.051383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low W-K, Dang Y, Bhat S, Romo D, Liu JO, 2007a. Substrate-dependent targeting of eukaryotic translation initiation factor 4A by pateamine A: negation of domain-linker regulation of activity. Chem. Biol 14, 715–727. 10.1016/j.chembiol.2007.05.012 [DOI] [PubMed] [Google Scholar]
- Low W-K, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Merrick WC, Romo D, Liu JO, 2005. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell 20, 709–722. 10.1016/j.molcel.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Low W-K, Dang Y, Schneider-Poetsch T, Shi Z, Choi NS, Rzasa RM, Shea HA, Li S, Park K, Ma G, Romo D, Liu JO, 2007b. Isolation and identification of eukaryotic initiation factor 4A as a molecular target for the marine natural product Pateamine A. Methods Enzymol 431, 303–324. 10.1016/S0076-6879(07)31014-8 [DOI] [PubMed] [Google Scholar]
- Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P, 2017. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun 8, 1127 10.1038/s41467-017-01278-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Fulzele A, Higgins R, Leonard M, Yeo GW, Bennett EJ, 2019. Active Protein Neddylation or Ubiquitylation Is Dispensable for Stress Granule Dynamics. Cell Rep 27, 1356–1363.e3. 10.1016/j.celrep.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo E-C, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao F-B, Bennett EJ, Lécuyer E, Yeo GW, 2018. Context-Dependent and Disease-Specific Diversity in Protein Interactions within Stress Granules. Cell 172, 590–604.e13. 10.1016/j.cell.2017.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor-Kollet H, Siany A, Kedersha N, Knafo N, Rivkin N, Danino YM, Olender T, Cohen N, Moens T, Higginbottom A, Knock JC-, Eitan C, Cohen BT, Bosch LVD, Anderson P, Ivanov P, Geiger T, Hornstein E, 2020. Spatio-temporal Proteomic Analysis of Stress Granule Disassembly Using APEX Reveals Regulation by SUMOylation and Links to ALS Pathogenesis. bioRxiv 2020.01.29.830133 10.1101/2020.01.29.830133 [DOI] [PMC free article] [PubMed]
- Mazroui R, Di Marco S, Kaufman RJ, Gallouzi I-E, 2007. Inhibition of the ubiquitin-proteasome system induces stress granule formation. Mol. Biol. Cell 18, 2603–2618. 10.1091/mbc.e06-12-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R, Sukarieh R, Bordeleau M-E, Kaufman RJ, Northcote P, Tanaka J, Gallouzi I, Pelletier J, 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell 17, 4212–4219. 10.1091/mbc.e06-04-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Garzia A, Morozov P, Molina H, Tuschl T, 2020. The G3BP1-Family-USP10 Deubiquitinase Complex Rescues Ubiquitinated 40S Subunits of Ribosomes Stalled in Translation from Lysosomal Degradation. Mol. Cell 77, 1193–1205.e5. 10.1016/j.molcel.2019.12.024 [DOI] [PubMed] [Google Scholar]
- Milo R, Philips R, 2016. Cell Biology by the Numbers URL http://book.bionumbers.org/how-many-ribosomes-are-in-a-cell/ (accessed 7.7.20).
- Mokas S, Mills JR, Garreau C, Fournier M-J, Robert F, Arya P, Kaufman RJ, Pelletier J, Mazroui R, 2009. Uncoupling stress granule assembly and translation initiation inhibition. Mol. Biol. Cell 20, 2673–2683. 10.1091/mbc.e08-10-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S, Cougot N, Wilczynska A, Dautry F, Kress M, Bertrand E, Weil D, 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell 19, 4469–4479. 10.1091/mbc.e08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser JJ, Fritzler MJ, 2010. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int. J. Biochem. Cell Biol. 42, 828–843. 10.1016/j.biocel.2009.11.018 [DOI] [PubMed] [Google Scholar]
- Murphey RJ, Gerner EW, 1987. Hypusine formation in protein by a two-step process in cell lysates. J. Biol. Chem 262, 15033–15036. [PubMed] [Google Scholar]
- Niewidok B, Igaev M, Pereira da Graca A, Strassner A, Lenzen C, Richter CP, Piehler J, Kurre R, Brandt R, 2018. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol 217, 1303–1318. 10.1083/jcb.201709007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson D, Sunnerhagen P, 2011. Cellular stress induces cytoplasmic RNA granules in fission yeast. RNA N. Y. N 17, 120–133. 10.1261/rna.2268111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Anderson P, 2010. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip. Rev. RNA 1, 486–493. 10.1002/wrna.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P, 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol 10, 1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Ivanov P, Anderson P, 2016. Mechanistic insights into mammalian stress granule dynamics. J. Cell Biol 215, 313–323. 10.1083/jcb.201609081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Kedersha N, Schulte T, Branca RM, Ivanov P, Anderson P, 2019. Phosphorylation of G3BP1-S149 does not influence stress granule assembly. J. Cell Biol 218, 2425–2432. 10.1083/jcb.201801214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas MD, Schulte T, Thaa B, Sandalova T, Kedersha N, Achour A, McInerney GM, 2015. Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog 11, e1004659 10.1371/journal.ppat.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Sheth U, 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646. 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Protter DSW, Rao BS, Van Treeck B, Lin Y, Mizoue L, Rosen MK, Parker R, 2018. Intrinsically Disordered Regions Can Contribute Promiscuous Interactions to RNP Granule Assembly. Cell Rep 22, 1401–1412. 10.1016/j.celrep.2018.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE, 2013. Diversion of stress granules and P-bodies during viral infection. Virology 436, 255–267. 10.1016/j.virol.2012.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repici M, Hassanjani M, Maddison DC, Garção P, Cimini S, Patel B, Szegö ÉM, Straatman KR, Lilley KS, Borsello T, Outeiro TF, Panman L, Giorgini F, 2019. The Parkinson’s Disease-Linked Protein DJ-1 Associates with Cytoplasmic mRNP Granules During Stress and Neurodegeneration. Mol. Neurobiol 56, 61–77. 10.1007/s12035-018-1084-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR, 2019. m6A enhances the phase separation potential of mRNA. Nature 571, 424–428. 10.1038/s41586-019-1374-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs CL, Kedersha N, Ivanov P, Anderson P, 2020. Mammalian stress granules and P bodies at a glance. J. Cell Sci 133 10.1242/jcs.242487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A, Wei M-T, Whitney G, Lyons SM, Anderson P, Jacobs WM, Ivanov P, Brangwynne CP, 2020. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 181, 306–324.e28. 10.1016/j.cell.2020.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Green R, 2017. The ABC(E1)s of Ribosome Recycling and Reinitiation. Mol. Cell 66, 578–580. 10.1016/j.molcel.2017.05.017 [DOI] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T, 2010. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102. 10.1126/science.1192588 [DOI] [PubMed] [Google Scholar]
- Seguin SJ, Morelli FF, Vinet J, Amore D, De Biasi S, Poletti A, Rubinsztein DC, Carra S, 2014. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ 21, 1838–1851. 10.1038/cdd.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP, 2017. Liquid phase condensation in cell physiology and disease. Science 357 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Slawson C, Housley MP, Hart GW, 2006. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem 97, 71–83. 10.1002/jcb.20676 [DOI] [PubMed] [Google Scholar]
- Somasekharan SP, Zhang F, Saxena N, Huang JN, Kuo I-C, Low C, Bell R, Adomat H, Stoynov N, Foster L, Gleave M, Sorensen PH, 2020. G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress. Nucleic Acids Res 48, 6855–6873. 10.1093/nar/gkaa376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WFC, Blackwell TK, Anderson P, 2004. MK2-induced tristetraprolin:14–3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23, 1313–1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöhr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Hüttelmaier S, 2006. ZBP1 regulates mRNA stability during cellular stress. J. Cell Biol 175, 527–534. 10.1083/jcb.200608071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J, 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol 160, 823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Tsai N-P, Ho P-C, Wei L-N, 2008. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J 27, 715–726. 10.1038/emboj.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai W-C, Gayatri S, Reineke LC, Sbardella G, Bedford MT, Lloyd RE, 2016. Arginine Demethylation of G3BP1 Promotes Stress Granule Assembly. J. Biol. Chem 291, 22671–22685. 10.1074/jbc.M116.739573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Parker R, 2018. Emerging Roles for Intermolecular RNA-RNA Interactions in RNP Assemblies. Cell 174, 791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R, 2018. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc. Natl. Acad. Sci. U. S. A 115, 2734–2739. 10.1073/pnas.1800038115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderweyde T, Yu H, Varnum M, Liu-Yesucevitz L, Citro A, Ikezu T, Duff K, Wolozin B, 2012. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J. Neurosci. Off. J. Soc. Neurosci 32, 8270–8283. 10.1523/JNEUROSCI.1592-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vognsen T, Møller IR, Kristensen O, 2013. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PloS One 8, e80947 10.1371/journal.pone.0080947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Maxwell BA, Joo JH, Gwon Y, Messing J, Mishra A, Shaw TI, Ward AL, Quan H, Sakurada SM, Pruett-Miller SM, Bertorini T, Vogel P, Kim HJ, Peng J, Taylor JP, Kundu M, 2019. ULK1 and ULK2 Regulate Stress Granule Disassembly Through Phosphorylation and Activation of VCP/p97. Mol. Cell 74, 742–757.e8. 10.1016/j.molcel.2019.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Vu AQ, Einstein JM, DiSalvo M, Ahmed N, Van Nostrand EL, Shishkin AA, Jin W, Allbritton NL, Yeo GW, 2020. Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors. Nat. Methods 17, 636–642. 10.1038/s41592-020-0826-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R, 2016. Distinct stages in stress granule assembly and disassembly. eLife 5 10.7554/eLife.18413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Lloyd RE, 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20, 175–183. 10.1016/j.tim.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Denman RB, 2011. Protein methylation and stress granules: posttranslational remodeler or innocent bystander? Mol. Biol. Int 2011, 137459 10.4061/2011/137459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP, 2008. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans 36, 802–806. 10.1042/BST0360802 [DOI] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis R-M, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q, Yu J, Martin EW, Mittag T, Kim HJ, Taylor JP, 2020. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 181, 325–345.e28. 10.1016/j.cell.2020.03.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Nozawa R-S, Jia TZ, Saio T, Mori E, 2020. Biological phase separation: cell biology meets biophysics. Biophys. Rev 12, 519–539. 10.1007/s12551-020-00680-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn J-Y, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, Angers S, Morris Q, Fabian M, Côté J-F, Gingras A-C, 2018. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 69, 517–532.e11. 10.1016/j.molcel.2017.12.020 [DOI] [PubMed] [Google Scholar]
- Youn J-Y, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD, Gingras A-C, 2019. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 76, 286–294. 10.1016/j.molcel.2019.09.014 [DOI] [PubMed] [Google Scholar]
- Yu C, York B, Wang S, Feng Q, Xu J, O’Malley BW, 2007. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol. Cell 25, 765–778. 10.1016/j.molcel.2007.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Lu C, Shi Y, Wu C, Chen X, Li C, Yao J, 2020. Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. Biochim. Biophys. Acta BBA - Mol. Cell Res 1867, 118795 10.1016/j.bbamcr.2020.118795 [DOI] [PubMed] [Google Scholar]
- Zhang J, Okabe K, Tani T, Funatsu T, 2011. Dynamic association-dissociation and harboring of endogenous mRNAs in stress granules. J. Cell Sci 124, 4087–4095. 10.1242/jcs.090951 [DOI] [PubMed] [Google Scholar]
- Zou T, Rao JN, Liu L, Xiao L, Cui Y-H, Jiang Z, Ouyang M, Donahue JM, Wang J-Y, 2012. Polyamines inhibit the assembly of stress granules in normal intestinal epithelial cells regulating apoptosis. Am. J. Physiol. Cell Physiol 303, C102–111. 10.1152/ajpcell.00009.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Yang X, Pan D, Huang J, Sahin M, Zhou J, 2011. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell. Mol. Neurobiol 31, 541–550. 10.1007/s10571-011-9647-8 [DOI] [PMC free article] [PubMed] [Google Scholar]