How cells delay completion of cytokinesis (abscission) in the presence of chromatin bridges is unclear. Petsalaki and Zachos describe an ATM–Chk2–INCENP signaling pathway that activates the abscission checkpoint to prevent chromatin breakage in cytokinesis.
Abstract
During cell division, in response to chromatin bridges, the chromosomal passenger complex (CPC) delays abscission to prevent chromosome breakage or tetraploidization. Here, we show that inhibition of ATM or Chk2 kinases impairs CPC localization to the midbody center, accelerates midbody resolution in normally segregating cells, and correlates with premature abscission and chromatin breakage in cytokinesis with trapped chromatin. In cultured human cells, ATM activates Chk2 at late midbodies. In turn, Chk2 phosphorylates human INCENP-Ser91 to promote INCENP binding to Mklp2 kinesin and CPC localization to the midbody center through Mklp2 association with Cep55. Expression of truncated Mklp2 that does not bind to Cep55 or nonphosphorylatable INCENP-Ser91A impairs CPC midbody localization and accelerates abscission. In contrast, expression of phosphomimetic INCENP-Ser91D or a chimeric INCENP protein that is targeted to the midbody center rescues the abscission delay in Chk2-deficient or ATM-deficient cells. Furthermore, the Mre11–Rad50–Nbs1 complex is required for ATM activation at the midbody in cytokinesis with chromatin bridges. These results identify an ATM–Chk2–INCENP pathway that imposes the abscission checkpoint by regulating CPC midbody localization.
Introduction
To ensure faithful inheritance of the genetic material during cell division, completion of cytokinesis (abscission) is tightly coordinated with chromosome segregation (Mierzwa and Gerlich, 2014). In response to chromosome segregation defects giving rise to chromatin bridges or lagging chromosomes trapped inside the intercellular canal (Gisselsson, 2008), eukaryotic cells delay abscission to prevent chromatin breakage and tetraploidization by regression of the cleavage furrow (Bai et al., 2020; Carlton et al., 2012; Gisselsson, 2008; Norden et al., 2006; Steigemann et al., 2009; Thoresen et al., 2014), which are associated with genomic instability and cancer predisposition (Ganem and Pellman, 2012; Lens and Medema, 2019; Sadler et al., 2018). In mammalian cells, this abscission delay is called the “abscission checkpoint” and is dependent on Aurora B kinase activity at the midbody (Petsalaki and Zachos, 2016, 2019; Steigemann et al., 2009).
The activity and targeting of Aurora B depend on its partners in the chromosomal passenger complex (CPC) that also includes the scaffolding protein INCENP and the nonenzymatic subunits Survivin and Borealin (Carmena et al., 2012; Honda et al., 2003). In human cells, CPC localization to central spindle microtubules requires INCENP binding to Mklp2 kinesin (Adriaans et al., 2020; Gruneberg et al., 2004; Hümmer and Mayer, 2009; Kitagawa et al., 2014; van der Horst et al., 2015). At the midbody, the CPC localizes to the midbody arms (Cooke et al., 1987; Hu et al., 2012; Vagnarelli and Earnshaw, 2004). More recently, a relatively small population of Aurora B was detected inside the Flemming body (FB), the narrow region at the midbody center where tubulin staining by immunofluorescence is blocked (Hu et al., 2012), in late cytokinesis (Petsalaki and Zachos, 2016); however, the significance of this localization for the abscission delay is incompletely understood. Cdc-like kinases (Clk’s) 1, 2, and 4 phosphorylate Aurora B serine 331 (Ser331) to fully induce Aurora B catalytic activity at the midbody (Petsalaki and Zachos, 2016). Active Aurora B imposes the abscission checkpoint by phosphorylating the endosomal sorting complex required for transport III (ESCRT-III) subunit charged multivesicular body protein 4C (Chmp4c) to target Chmp4c to the midbody center (Capalbo et al., 2012; Carlton et al., 2012; Petsalaki and Zachos, 2016) to inhibit the ATPase Vps4 and prevent its activity on ESCRT-III filaments thereby delaying abscission (Caballe et al., 2015; Mierzwa et al., 2017; Thoresen et al., 2014). Inhibition of Aurora B also accelerates abscission in normally segregating cells, i.e., in the absence of trapped chromatin, indicating the abscission checkpoint functions more generally as an abscission timer (Carlton et al., 2012; Petsalaki and Zachos, 2016; Steigemann et al., 2009). However, the molecular mechanisms that relay cytoplasmic stresses, such as chromatin bridges, to the CPC have not been previously identified.
The ataxia-telangiectasia mutated (ATM) and Chk2 kinases are master regulators of the DNA damage response (Smith et al., 2010). ATM activation requires the MRN (Mre11–Rad50–Nbs1) double-strand break sensor complex that processes DNA ends and recruits ATM to broken DNA molecules (Bakkenist and Kastan, 2003; Falck et al., 2005; Lee and Paull, 2005; Uziel et al., 2003). In turn, active ATM phosphorylates Chk2 threonine 68 (Thr68) to promote Chk2 activation and stimulate downstream responses (Ahn et al., 2000, 2002). In the absence of DNA damage, ATM and Chk2 are required for spindle checkpoint signaling (Petsalaki and Zachos, 2014; Yang et al., 2011). ATM and Chk2 also localize to the midbody in cytokinesis (Tsvetkov et al., 2003; Yang et al., 2011); however, a role for ATM or Chk2 in the abscission checkpoint has not been previously reported. In this study, we show that ATM phosphorylates and activates Chk2 at the midbody center in late cytokinesis in normally segregating cells. In turn, active Chk2 phosphorylates human INCENP at the newly identified site Ser91 to promote INCENP binding to Mklp2, resulting in recruitment of the INCENP–Mklp2 complex to the midbody center through Mklp2’s interaction with the midbody protein Cep55 to delay abscission. We also show that in the presence of a chromatin bridge, the MRN complex activates ATM and the downstream Chk2–INCENP signaling pathway at the midbody to delay abscission and prevent chromatin bridge breakage in cytokinesis.
Results
Chk2 inhibition accelerates midbody disassembly
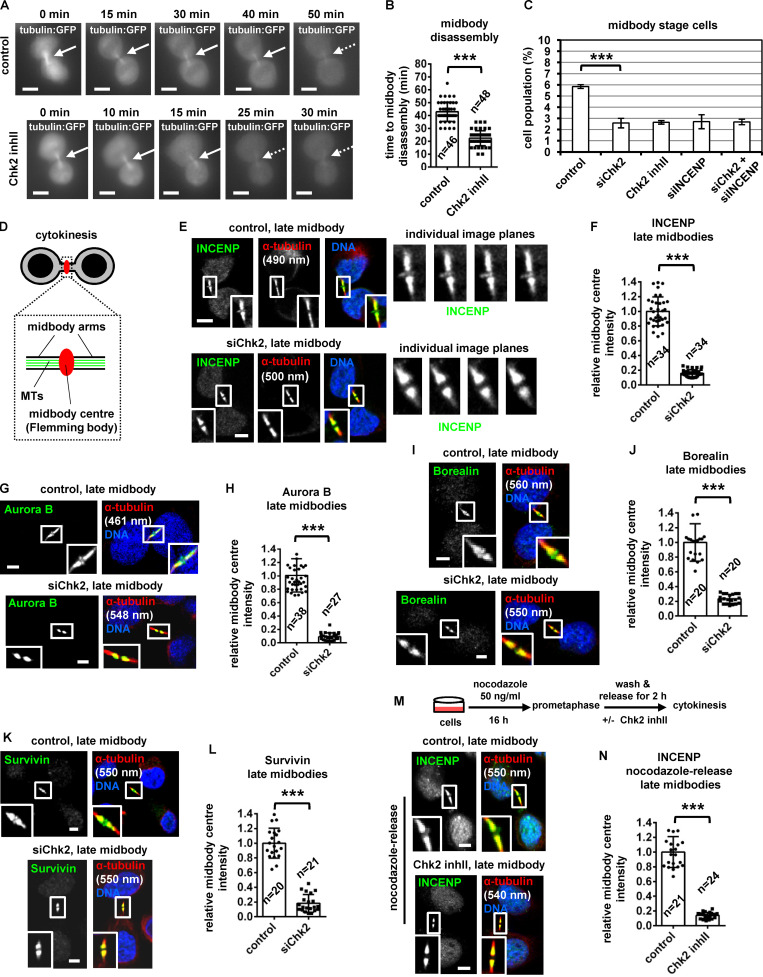
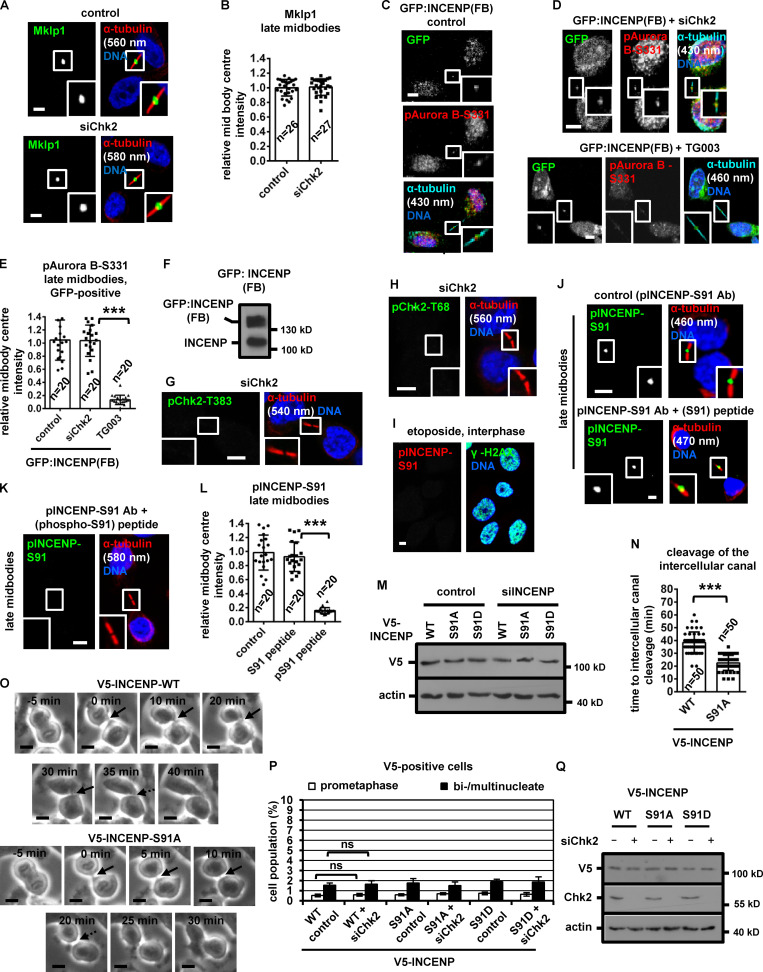
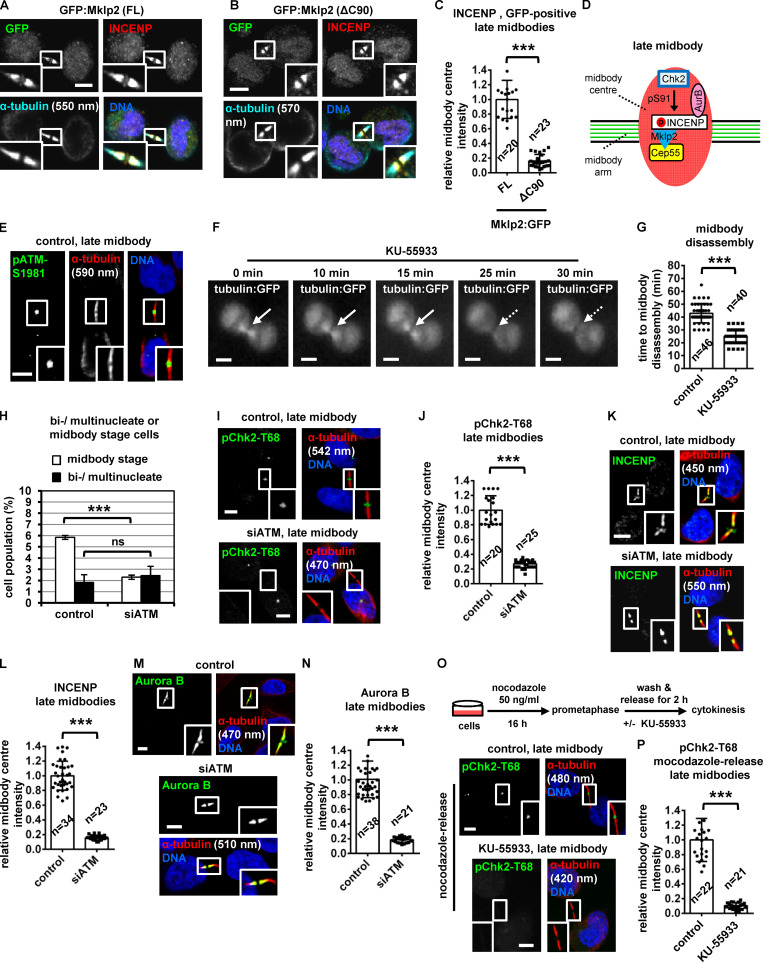
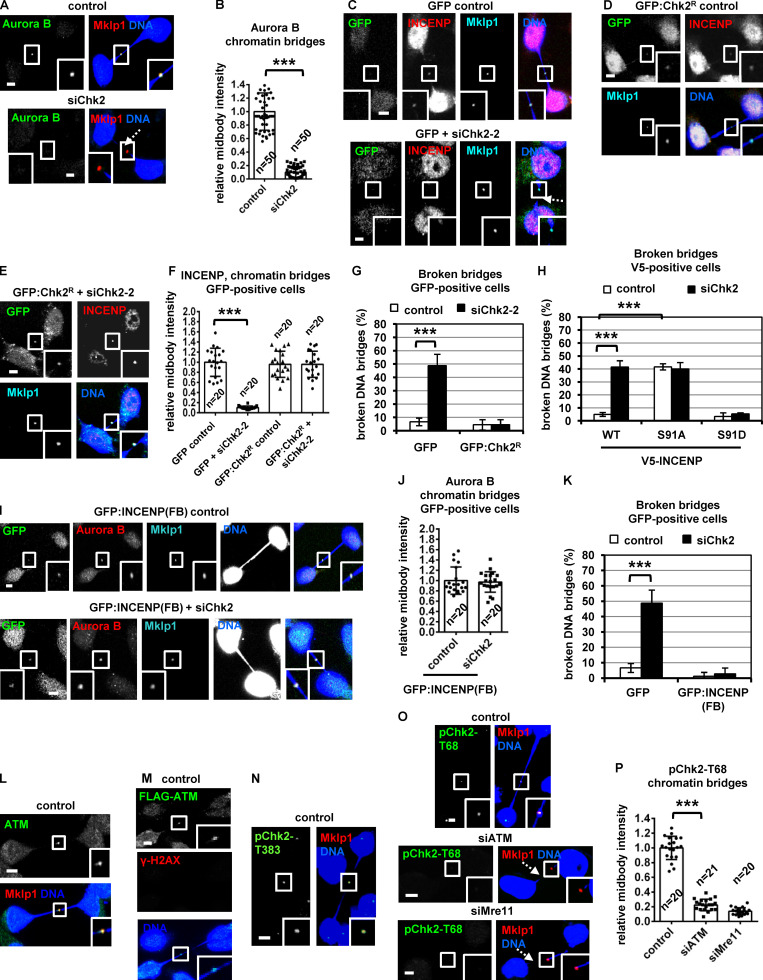
To investigate a role for Chk2 in midbody resolution, HeLa cells stably expressing α-tubulin fused to GFP (tubulin:GFP) were monitored by time-lapse microscopy and the kinetics of tubulin disassembly at the midbody determined. In control cells, the midbody microtubules remained visible for a median time of 43 ± 7 min after formation. In contrast, treatment of cells in cytokinesis with 10 µM Chk2 inhibitor II accelerated midbody disassembly (t = 22 ± 6 min) compared with controls (P < 0.001; Fig. 1, A and B; Video 1; and Video 2). Furthermore, Chk2-deficient cells exhibited faster cleavage of the intercellular canal compared with controls by phase-contrast live-cell imaging, indicating premature abscission (P < 0.001; Fig. S1, A and B; Video 3; and Video 4). These findings correlated with reduced frequency of human colon carcinoma BE cells at midbody stage (midbody index) after Chk2 inhibition or Chk2 depletion by siRNA (siChk2), but not with an increase in binucleate/multinucleate or prometaphase cells, suggesting Chk2 inhibition does not prevent mitotic entry and that Chk2-deficient cells can progress through abscission and disassemble their midbodies more rapidly than controls (Fig. 1 C; and Fig. S1, C–E). Depletion of INCENP by an siRNA located in the 3′ untranslated region of INCENP (siINCENP) also reduced the frequency of cells at midbody stage compared with controls transfected with negative siRNA; furthermore, simultaneous depletion of Chk2 and INCENP did not further diminish the midbody index, suggesting Chk2 and INCENP regulate abscission timing by acting through the same pathway (Fig. 1 C; and Fig. S1, C and E). We propose that Chk2 regulates proper abscission timing in normally segregating cells.
Figure 1.
Chk2 inhibition diminishes localization of CPC proteins to the midbody center in late midbodies. (A and B) Time-lapse microscopy analysis of HeLa cells expressing tubulin:GFP. Cells were untreated (control) or treated with 10 µM Chk2 inhibitor II immediately before filming. Midbodies are shown by solid arrows. Time is from midbody formation to midbody disassembly (dotted arrows). Related to Video 1 and Video 2. (C) Frequency of midbody stage cells. BE cells were transfected as indicated, or treated with 10 µM Chk2 inhibitor II (inhII) for 4 h. Values represent mean ± SD from three independent experiments (n > 900). ***, P < 0.001 (ANOVA and Student’s t test). (D) Midbody cartoon. MTs, microtubules. (E–L) Localization and mean intensity of CPC proteins at the midbody center in BE cells. Individual image planes show 3.5× magnification of the midbodies. (M and N) INCENP localization and mean intensity after treatment of BE cells with 10 µM Chk2 inhII in cytokinesis. Values represent mean ± SD from n cells. Values in control were set to 1. ***, P < 0.001 (Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. Scale bars, 5 µm.
Video 1.
Midbody disassembly in control cells. HeLa cells stably expressing tubulin:GFP (white) were analyzed by time-lapse fluorescence microscopy in cytokinesis. Frames were taken every 5 min for 50 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. 1 A.
Video 2.
Midbody disassembly in Chk2-deficient cells. HeLa cells stably expressing tubulin:GFP (white) were treated with 10 µM Chk2 inhibitor II and analyzed by time-lapse fluorescence microscopy in cytokinesis. Frames were taken every 5 min for 30 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. 1 A.
Figure S1.
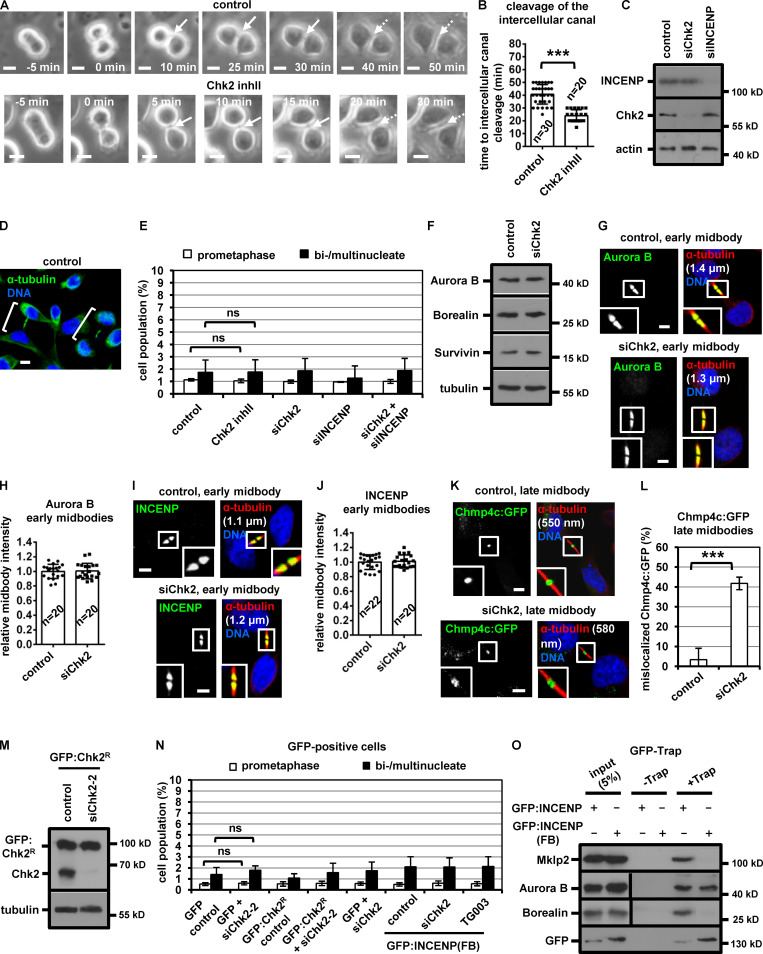
Chk2-depletion correlates with mislocalization of Chmp4c:GFP in late midbodies. (A and B) Phase-contrast live-cell microscopy analysis of HeLa tubulin:GFP cells. Cells were untreated (control) or treated with 10 µM Chk2 inhibitor II (inhII) immediately before filming. Intercellular canals are shown by solid arrows. Time is from formation of an intercellular canal to canal cleavage (dotted arrows). Related to Video 3 and Video 4. (C and F) Western blot analysis of total CPC proteins, Chk2, actin and tubulin from BE cell extracts. (D) Examples of BE cells at midbody stage (shown in brackets). (E and N) Frequency of bi/multinucleate or prometaphase BE cells. Cells were transfected as indicated or treated with 10 µM Chk2 inhibitor II or TG003 for 4 h. Values represent mean ± SD from three independent experiments (n > 90). (G–J) Localization of Aurora B or INCENP and mean intensity at the midbody in BE cells in early midbodies. Values represent mean ± SD from n cells. Values in control were set to 1. (K and L) Chmp4c:GFP localization and frequency of BE cells exhibiting mislocalized Chmp4c:GFP (two dots) at late midbodies. Values represent mean ± SD from three independent experiments (n > 90). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. (M) Western blot analysis of total Chk2 and tubulin in cells transfected with siRNA-resistant GFP:Chk2R. (O) GFP-Trap assay. BE cell lysates were incubated with GFP-Trap (+Trap) or agarose beads only (−Trap). Precipitated proteins were analyzed by Western blotting. ns, not statistically significant; ***, P < 0.001 (Student’s t test). Scale bars, 5 µm.
Video 3.
Cleavage of the intercellular canal in control cells. HeLa cells stably expressing tubulin:GFP were analyzed by phase-contrast time-lapse microscopy in cytokinesis. Frames were taken every 5 min for 60 min. The intercellular canal is shown by an arrow. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. S1 A.
Video 4.
Cleavage of the intercellular canal in Chk2-deficient cells. HeLa cells stably expressing tubulin:GFP were treated with 10 µM Chk2 inhibitor II and analyzed by phase-contrast time-lapse microscopy in cytokinesis. Frames were taken every 5 min for 60 min. The intercellular canal is shown by an arrow. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. S1 A.
Chk2 is required for CPC localization to the midbody center
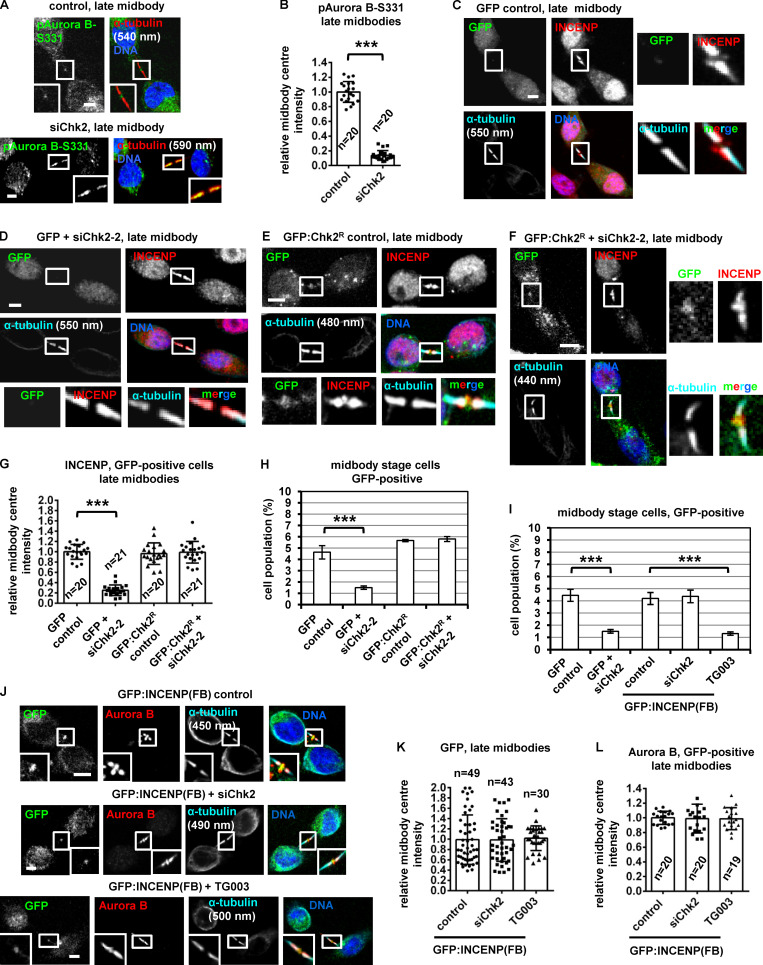
During cytokinesis, the microtubule bundles at the midbody progressively get narrower (Carlton et al., 2012; Elia et al., 2011), and midbody thickness ranges from ∼400 to 1,400 nm in BE cells (Petsalaki and Zachos, 2016). In relative “late” midbodies exhibiting midbody thickness of 400–700 nm, the CPC proteins INCENP, Aurora B, Borealin, and Survivin mainly localize to the midbody arms; however, a relatively small population of CPC proteins was also detectable at the midbody center (FB) in control cells (Fig. 1, D, E, G, I, and K). Depletion of Chk2 diminished localization of CPC proteins to the midbody center in late midbodies (Fig. 1, E–L); however, Chk2 depletion did not reduce the total levels of CPC proteins and did not impair localization of the CPC proteins on the midbody arms in relatively early midbodies with midbody thickness of 800–1,400 nm (Fig. S1, F–J). Also, inhibition of Chk2 by Chk2 inhibitor II in cytokinesis after release of cells from a nocodazole-induced prometaphase block diminished INCENP localization to the midbody center in late midbodies (Fig. 1, M and N). Localization of phosphorylated Aurora B–Ser331 (active Aurora B; Petsalaki et al., 2011) or Chmp4c fused to GFP (Chmp4c:GFP) to the midbody center (which correlates with Aurora B catalytic activity at the midbody; Petsalaki and Zachos, 2016) was impaired in Chk2-deficient cells compared with controls; furthermore, phosphorylated Aurora B–Ser331 or Chmp4c:GFP mislocalized on the midbody arms in late midbodies (Fig. 2, A and B; and Fig. S1, K and L). Expression of GFP:Chk2R resistant to degradation by a second Chk2 siRNA (siChk2-2), but not GFP only, rescued INCENP localization to the midbody center and restored the frequency of cells at midbody stage after Chk2 depletion compared with controls (Fig. 2, C–H; and Fig. S1, M and N). We propose that Chk2 is required for CPC localization to the midbody center and optimal localization of Chmp4c to late midbodies.
Figure 2.
Expression of GFP:INCENP(FB) rescues the frequency of cells at midbody stage after Chk2 inhibition. (A and B) Localization and mean intensity of phosphorylated Aurora B-Ser331 (pAurora B-Ser331) at the midbody center in BE cells. ***, P < 0.001 (Student’s t test). (C–G and J–L) Localization and mean intensity of GFP, INCENP, and Aurora B proteins at the midbody center in BE cells transfected with siRNA or treated with 10 µM TG003 for 4 h. Values represent mean ± SD from n cells. Values in control were set to 1. (H and I) Frequency of midbody stage BE cells. Values represent mean ± SD from three independent experiments (n > 900). ***, P < 0.001 (ANOVA and Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× (A and J) or 3.5× (C–F) magnification of the midbodies. Scale bars, 5 µm.
INCENP targeting to the midbody center delays midbody resolution in Chk2-deficient cells
To investigate the significance of CPC localization inside the FB for abscission timing, a truncated human INCENP protein (amino acids 49–918) fused to the C-terminal region (amino acids 456–858) of Mklp1 and GFP, GFP:INCENP(FB), was constructed. GFP:INCENP(FB) localizes to the midbody center through its Mklp1 sequence (Hu et al., 2012) but exhibits relatively diminished localization to the midbody arms, because it lacks the centromere-targeting domain of INCENP (amino acids 1–48) that binds to Mklp2 (Fig. 2 J; also compare with the endogenous INCENP in Fig. 1 E [control] and transfected INCENP in Fig. 4 A [WT control]; Kitagawa et al., 2014). GFP:INCENP(FB) binds to Aurora B in cell extracts, but does not bind to Mklp2 or Borealin compared with GFP:INCENP (Fig. S1 O). Furthermore, both the endogenous Mklp1 and GFP:INCENP(FB) localize to the midbody center in Chk2-deficient cells similar to controls (Fig. 2, J and K; and Fig. S2, A and B). Expression of GFP:INCENP(FB), but not GFP only, rescued the frequency of cells at midbody stage after Chk2 depletion (Fig. 2 I and Fig. S1 N). Furthermore, Chk2-deficient cells expressing GFP:INCENP(FB) exhibited total and phosphorylated Ser331 (active) Aurora B at the midbody center similar to controls (Fig. 2, J and L; and Fig. S2, C–F). In contrast, treatment of GFP:INCENP(FB) cells with 1 µM TG003, an inhibitor of Clk catalytic activity, reduced the frequency of cells at midbody stage and diminished localization of phosphorylated Ser331, but not total, Aurora B at the midbody center compared with controls, indicating Chk2 regulates midbody disassembly independently of Aurora B activation by Clk (Fig. 2, I–L; Fig. S1 N; and Fig. S2, C–E). These results suggest that INCENP targeting to the midbody center by Chk2 is required for proper abscission timing in normally segregating cells.
Figure 4.
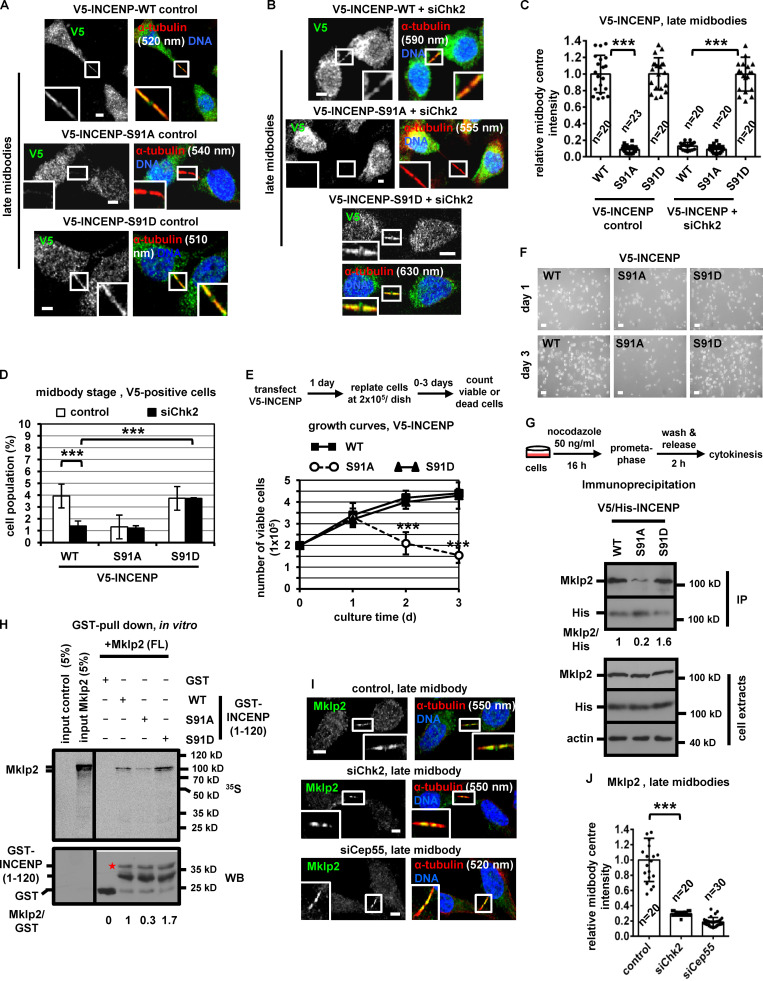
Expression of INCENP-Ser91A reduces the frequency of midbody stage cells and diminishes cell proliferation. (A–C) Localization and mean intensity of V5-INCENP at the midbody center in BE cells. Scale bars, 5 µm. (D) Frequency of midbody stage BE cells. Values represent mean ± SD from three independent experiments (n > 900). (E and F) Growth curves and phase-contrast images in BE cells expressing V5-INCENP plasmids. Values represent mean ± SD from three independent experiments (n = 6). Scale bars, 50 µm. (G) Immunoprecipitation (IP) of V5/His-INCENP and Mklp2 from cytokinesis-enriched BE cell extracts. (H) GST pull-down. Radiolabeled FL Mklp2 generated by in vitro transcription-translation was incubated with GST-INCENP or GST. Phosphorimager (35S, top) and Western blot analysis (WB; bottom) of the same gel. Asterisk indicates predicted molecular weight. (I and J) Localization and mean intensity of Mklp2 at the midbody center in BE cells. Values represent mean ± SD from n cells. Values in control were set to 1. ***, P < 0.001 (ANOVA and Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. S, serine. Scale bars, 5 µm.
Figure S2.
Expression of Ser91A V5-INCENP accelerates cleavage of the intercellular canal in cytokinesis. (A and B) Localization of Mklp1 and mean intensity at the midbody center in BE cells in late midbodies. (C–E) Localization of phosphorylated Aurora B-Ser331 (pAurora B-Ser331) and mean intensity at the midbody center in BE cells. (F, M, and Q) Western blot analysis of total INCENP, V5, Chk2 and actin in BE cells expressing GFP:INCENP(FB) or V5-INCENP. (G and H) Specificity of the anti-phospho-Chk2-Thr383 (pChk2-Thr383) and phospho-Chk2-Thr68 (pChk2-Thr68) antibodies. (I) Localization of phosphorylated INCENP-Ser91 (pINCENP-Ser91). BE cells were treated with 10 µM etoposide for 4 h. Damaged DNA is evidenced by γ-H2AX-staining. (J and K) Specificity of the anti-phospho-Ser91 antiserum. Where indicated, the anti-pINCENP-Ser91 antiserum (Ab) was incubated with the phosphorylated Ser91 peptide (phospho-Ser91, pSer91) or with the unphosphorylated (Ser91) synthetic peptide. Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. (L) Mean pINCENP-Ser91 intensity at the midbody center in BE cells. Values in control were set to 1. (N and O) Phase-contrast live-cell microscopy analysis of BE cells expressing WT or Ser91A V5-INCENP. Intercellular canals are shown by solid arrows. Time is from formation of an intercellular canal to canal cleavage (dotted arrows). Values represent mean ± SD from n cells. (P) Frequency of bi/multinucleate or prometaphase BE cells. Values represent mean ± SD from three independent experiments (n > 90). ***, P < 0.001 (ANOVA and Student’s t test). S, serine. Scale bars, 5 µm.
Chk2 phosphorylates human INCENP-Ser91 in vitro
Phosphorylated (active) Chk2-Thr68 and Chk2-Thr383 (Ahn et al., 2000, 2002) localized to the midbody center in late cytokinesis (Fig. 3 A). Depletion of Chk2 by siRNA abolished phospho-Chk2-Thr68 and phospho-Chk2-Thr383 staining, indicating the antibodies were specific (Fig. S2, G and H). Chk2:GFP colocalized with INCENP at the midbody center in late midbodies (Fig. 2, E and F); furthermore, endogenous Chk2 associated with INCENP by coimmunoprecipitation experiments in cytokinesis-enriched cell extracts (Fig. 3 B). Recombinant Chk2 phosphorylated human GST–INCENP (1–120 amino acids) compared with other GST-INCENP fragments or GST alone in vitro and this phosphorylation was diminished when Chk2 inhibitor II was included in the kinase reaction (Fig. 3, C and D). By mutating serine/threonine residues conforming to the minimum Chk1/Chk2 phosphorylation motif to alanine (Hutchins et al., 2000), we found that Ser91A GST-INCENP (1–120) exhibited reduced phosphorylation by Chk2 compared with the WT fragment, suggesting Chk2 phosphorylates INCENP–Ser91 in vitro (Fig. 3 E). This site is conserved as serine or threonine in mammals (Fig. 3 F).
Figure 3.
Chk2 phosphorylates INCENP-Ser91 in late midbodies. (A) Localization of phospho-Chk2-Thr383 (pChk2-Thr383) or phospho-Chk2-Thr68 (pChk2-Thr68) in BE cells. (B) Immunoprecipitation (IP) of Chk2 and INCENP in cytokinesis-enriched BE cell extracts. (C–E) Chk2 in vitro kinase assays. Autoradiography (32P, top) and Western blot analysis (WB) or Ponceau staining (bottom) of the same gel. Asterisks, predicted molecular weights. (F) Alignment of INCENP protein sequences. Human Ser91 is boxed and marked by asterisk. (G–O) Localization and mean intensity of phosphorylated INCENP-Ser91 (pINCENP-Ser91) or V5-INCENP in asynchronous BE cells, or after treatment with 10 µM Chk2 inhibitor II (inhII) in cytokinesis (L and M). Values represent mean ± SD from n cells. Values in control were set to 1. ***, P < 0.001 (Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. S, serine. Scale bars, 5 µm.
Chk2 is required for INCENP-Ser91 phosphorylation inside the FB
To investigate INCENP-Ser91 phosphorylation in cultured cells, an anti-phospho-INCENP-Ser91 antiserum was raised against the human protein sequence. Phosphorylated INCENP-Ser91 (pINCENP-Ser91) localized to the midzone in anaphase, on the midbody arms in early midbodies, and to the midbody center in late midbodies in control cells but was undetectable in interphase, prometaphase, or metaphase or after treatment of cells with the DNA damage agent etoposide (Fig. 3, G, H, and J; and Fig. S2 I). Phosphorylated Ser91 staining was impaired after incubation of the anti-pINCENP-Ser91 antiserum with the phosphorylated (phospho-Ser91) peptide compared with the unphosphorylated (Ser91) synthetic peptide by immunofluorescence, showing this reagent is specific for the phosphorylation (Fig. S2, J–L). Depletion of INCENP diminished phospho-Ser91 staining compared with control cells (Fig. 3, J and K). Also, depletion of Chk2 by siRNA or inhibition of Chk2 by Chk2 inhibitor II in cytokinesis after release of cells from a nocodazole block reduced pINCENP-Ser91 staining inside the FB and associated with mislocalized pINCENP-Ser91 on the midbody arms in late midbodies (Fig. 3, J–M). However, Chk2 was dispensable for Ser91 phosphorylation on the midbody arms in early midbodies, suggesting a different kinase phosphorylates INCENP-Ser91 in early cytokinesis (Fig. 3, H and I). We propose that Chk2 phosphorylates INCENP-Ser91 at the midbody center in late cytokinesis.
Ser91 phosphorylation is required for INCENP localization to the midbody, proper midbody resolution timing, and cell proliferation
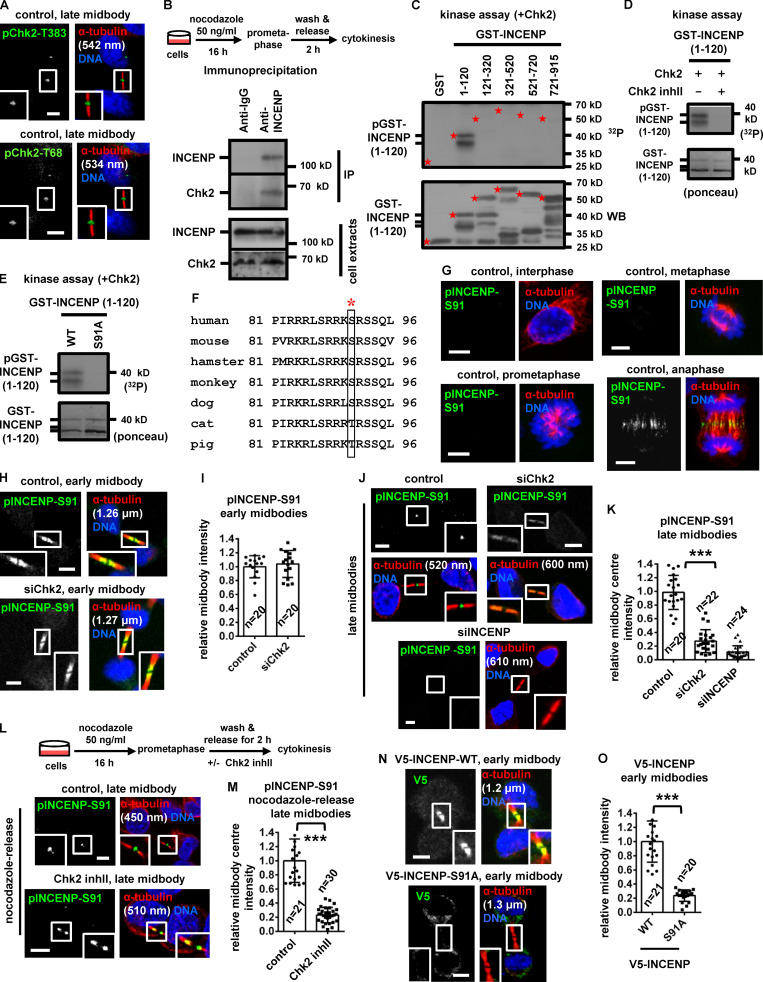
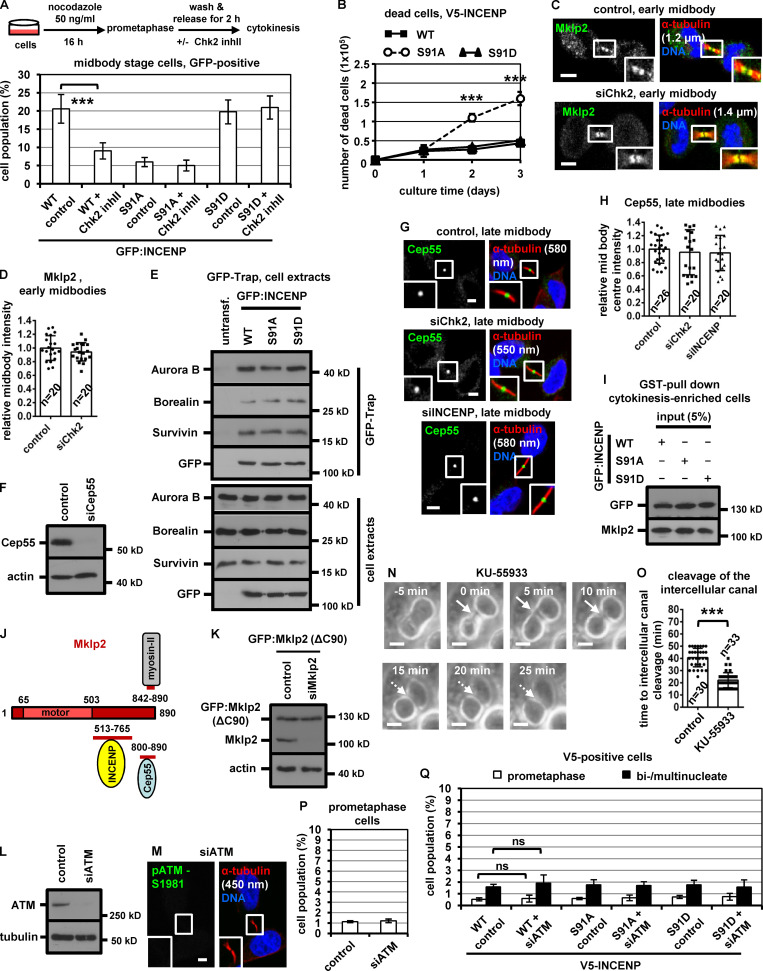
To investigate the significance of INCENP-Ser91 phosphorylation, BE cells transiently expressing V5/His-tagged, siRNA-resistant versions of WT, nonphosphorylatable Ser91 to alanine (Ser91A) or a phosphomimetic Ser91 to aspartic acid (Ser91D) mutant INCENP were analyzed after depletion of the endogenous protein by siRNA (Fig. S2 M). The nonphosphorylatable Ser91A V5/His-INCENP exhibited impaired localization to early or late midbodies compared with the WT protein (Fig. 3, N and O; and Fig. 4, A–C). Expression of Ser91A V5/His-INCENP accelerated cleavage of the intercellular canal compared with the WT V5/His-INCENP in cytokinesis by time-lapse microscopy, indicating premature abscission (Fig. S2, N and O). Furthermore, expression of Ser91A V5/His-INCENP correlated with reduced frequency of cells at midbody stage compared with WT controls (Fig. 4 D; and Fig. S2, P and Q). In contrast, the phosphomimetic Ser91D, but not WT, V5/His-INCENP, restored localization of V5/His-INCENP to the midbody center and rescued the midbody index after Chk2 depletion by siRNA (Fig. 4, A–D; and Fig. S2, P and Q), or after Chk2-inhibition by Chk2 inhibitor II in cytokinesis after release from a nocodazole block (Fig. S3 A). Also, expression of Ser91A V5/His-INCENP reduced cell proliferation and increased cell death compared with WT or Ser91D V5/His-INCENP (Fig. 4, E and F; and Fig. S3 B). We propose that INCENP-Ser91 phosphorylation by Chk2 is required for INCENP localization to the midbody center and proper abscission timing in normally segregating cells. We also propose that Ser91 phosphorylation promotes cell proliferation.
Figure S3.
Mutation of Ser91 to alanine does not impair INCENP-association with CPC proteins. (A) Frequency of midbody stage cells. BE cells were released from a prometaphase block and treated with 10 µM Chk2 inhibitor II (inhII) in cytokinesis, or left untreated. Values represent mean ± SD from three independent experiments (n > 900). (B) Accumulation of dead BE cells with time as determined by trypan blue staining. Mean ± SD from three independent experiments (n = 6). (A and B) ***, P < 0.001 (ANOVA and Student’s t test). (C, D, G, and H) Localization of Mklp2 or Cep55 and mean intensity at the midbody or midbody center in BE cells. (E) GFP-Trap assay. BE cells were untransfected (untransf.) or transfected with GFP:INCENP. Precipitated (top) or total proteins (bottom) were analyzed by Western blotting. (F and L) Western blot analysis of total Cep55, ATM, actin and tubulin. (I) Total GFP and Mklp2 from Fig. 5 I. (J) Cartoon indicating the regions of human Mklp2 protein interacting with INCENP, Cep55 and myosin-II. Numbers show amino-acid residues. motor, kinesin motor domain. (K) Western blot analysis of total Mklp2 and actin in BE cells expressing GFP:Mklp2(ΔC90). (M) Specificity of the anti-phospho-ATM-Ser1981 (pATM-Ser1981) antibody. Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. (N and O) Phase-contrast live-cell microscopy analysis of HeLa tubulin:GFP cells treated with 10 µM KU-55933 in cytokinesis. Intercellular canals are shown by solid arrows. Time is from formation of an intercellular canal to canal cleavage (dotted arrows). Related to Video 5. Values represent mean ± SD from n cells. ***, P < 0.001 (Student’s t test). Scale bars, 5 µm. (P and Q) Frequency of prometaphase or bi/multinucleate BE cells. Values represent mean ± SD from three independent experiments (n > 900). ns, not statistically significant. S, serine.
Ser91 phosphorylation promotes INCENP binding to Mklp2
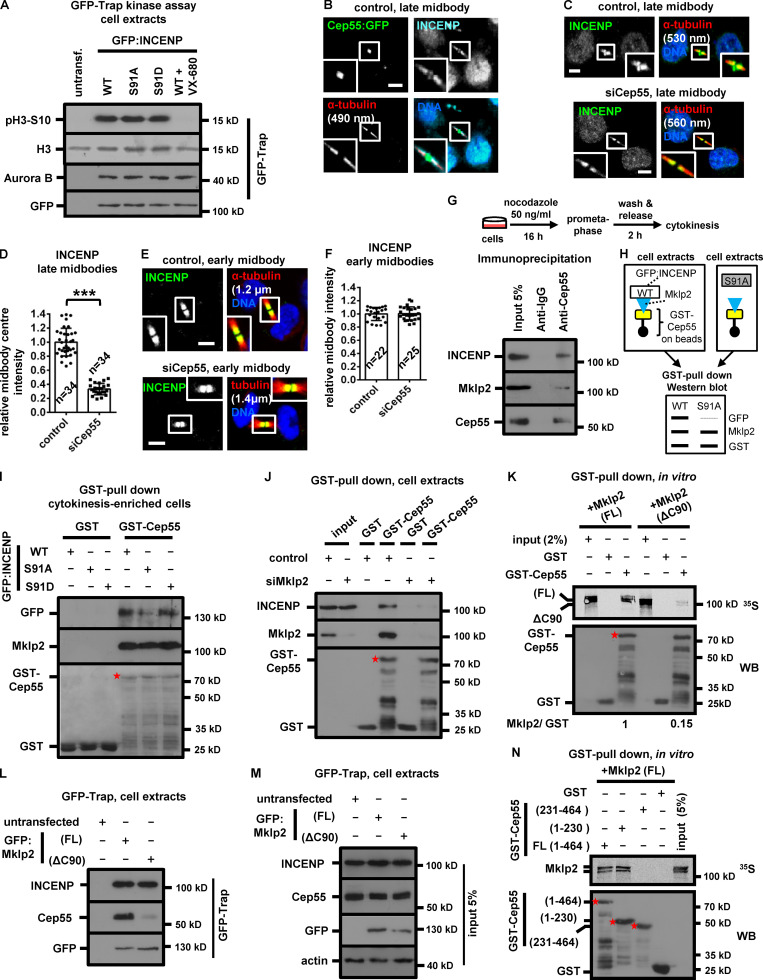
INCENP binding to Mklp2 is required for CPC and Mklp2 localization to the midbody (Hümmer and Mayer, 2009). The nonphosphorylatable V5/His-INCENP-Ser91A exhibited reduced association with the endogenous Mklp2 compared with WT or phosphomimetic Ser91D V5/His-INCENP by coimmunoprecipitation experiments in cytokinesis-enriched extracts (Fig. 4 G). Also, recombinant Mklp2 generated by in vitro transcription-translation exhibited reduced binding to purified Ser91A compared with WT or Ser91D GST-INCENP (1–120) by GST pull-down (Fig. 4 H). Chk2-depleted cells exhibited mislocalized Mklp2 on the midbody arms and reduced Mklp2-staining at the midbody center in late midbodies compared with controls (Fig. 4, I and J), suggesting Chk2 promotes the INCENP–Mklp2 interaction inside the FB. However, Chk2-depleted or control cells exhibited similar staining of Mklp2 on the midbody arms in early midbodies (Fig. S3, C and D). Cells expressing WT, Ser91A, or Ser91D GFP:INCENP exhibited similar levels of GFP-associated Aurora B, Survivin, and Borealin and similar histone H3-serine 10 phosphorylation (pH3-Ser10; a marker of Aurora B catalytic activity) by GFP-Trap-kinase assay using recombinant histone H3 as substrate (Fig. 5 A and Fig. S3 E). These results show that Ser91 phosphorylation is required for optimal INCENP binding to Mklp2, but not for CPC formation or Aurora B catalytic activity.
Figure 5.
GFP:INCENP binds to GST-Cep55 through Mklp2. (A) GFP-Trap kinase assay from BE cell extracts using histone H3 as substrate. GFP-associated histone H3-Ser10 phosphorylation (pH3-Ser10), immunoprecipitated Aurora B, and GFP were analyzed by Western blotting. Histone H3 was analyzed by Ponceau staining. 300 nM VX-680 was added to the kinase reaction to inhibit Aurora B catalytic activity. (B) Cep55:GFP colocalizes with INCENP at the midbody center in BE cells. (C–F) Localization and mean intensity of INCENP at the midbody or midbody center in BE cells. Mean ± SD from n cells. Values in control were set to 1. ***, P < 0.001 (Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. Scale bars, 5 µm. (G) Immunoprecipitation from cytokinesis-enriched BE cell extracts. (H) Predicted experimental outcome. In the Western blot, the width of the lines indicates band signals intensity. (I and J) Cell lysates from BE cells were incubated with GST-Cep55 or GST. Associated proteins were detected by Western blotting. (K and N) Radiolabeled FL or truncated Mklp2 (ΔC90) was incubated with GST-Cep55 or GST. Phosphorimager (35S, top) and Western blot analysis (WB; bottom) of the same gel. Asterisks, predicted molecular weights. (L and M) GFP-Trap assay. Precipitated (L) or input proteins (M) were analyzed by Western blotting. S, serine.
INCENP–Mklp2 localizes to the midbody center through Mklp2 binding to Cep55
Because CPC localization inside the FB is required for proper abscission timing, we investigated the mechanism by which the INCENP–Mklp2 complex localizes to the midbody center. The midbody adaptor protein Cep55 also localizes inside the FB (Zhao et al., 2006); furthermore, Cep55 fused to GFP (Cep55:GFP) colocalizes with INCENP at the midbody center in late midbodies (Fig. 5 B). Depletion of Cep55 impaired INCENP or Mklp2 localization to the midbody center in late midbodies (Fig. 5, C and D; Fig. 4, I and J; and Fig. S3 F), but did not interfere with INCENP localization on the midbody arms in early midbodies (Fig. 5, E and F). Cep55 localized inside the FB with similar intensity in INCENP-deficient, Chk2-deficient, or control cells (Fig. S3, G and H). These results suggest that Cep55 is required for INCENP–Mklp2 localization to the midbody center.
Cep55 coprecipitated with Mklp2 and INCENP in cytokinesis-enriched cell extracts (Fig. 5 G). Importantly, GFP:INCENP-Ser91A exhibited reduced association with GST-Cep55 compared with the WT or Ser91D GFP:INCENP by GST pull-down, whereas Mklp2 binding to GST-Cep55 was similar in all treatments, suggesting INCENP binding to Cep55 is mediated by Mklp2 (Fig. 5, H and I; and Fig. S3 I). Consistently, Mklp2-depleted cells exhibited diminished INCENP binding to GST-Cep55 compared with controls (Fig. 5 J). Recombinant full-length (FL; 1–890 amino acids) Mklp2 generated by in vitro transcription-translation associated with purified GST-Cep55; furthermore, deletion of the C-terminal 90 amino acids of Mklp2 (ΔC90) diminished the Cep55–Mklp2 interaction in vitro and in cell extracts (Fig. 5, K–M). Also, the N-terminal (amino acids 1–230) half of Cep55 was required for binding to Mklp2 in vitro (Fig. 5 N). These results suggest that Cep55 directly binds to Mklp2 at the C-terminus (Fig. S3 J). Importantly, after depletion of the endogenous Mklp2, cells expressing siRNA-resistant GFP:Mklp2 (ΔC90) that does not efficiently bind to Cep55 exhibited reduced localization of INCENP to the midbody center compared with cells expressing the FL GFP:Mklp2 (Fig. 6, A–C; and Fig. S3 K), and this was not caused by impaired INCENP binding to GFP:Mklp2 (ΔC90; Fig. 5, L and M). We propose that the INCENP–Mklp2 complex localizes to the midbody center through Mklp2 binding to Cep55 and that Ser91 phosphorylation by Chk2 promotes INCENP binding to Mklp2 at the midbody center in late midbodies (Fig. 6 D).
Figure 6.
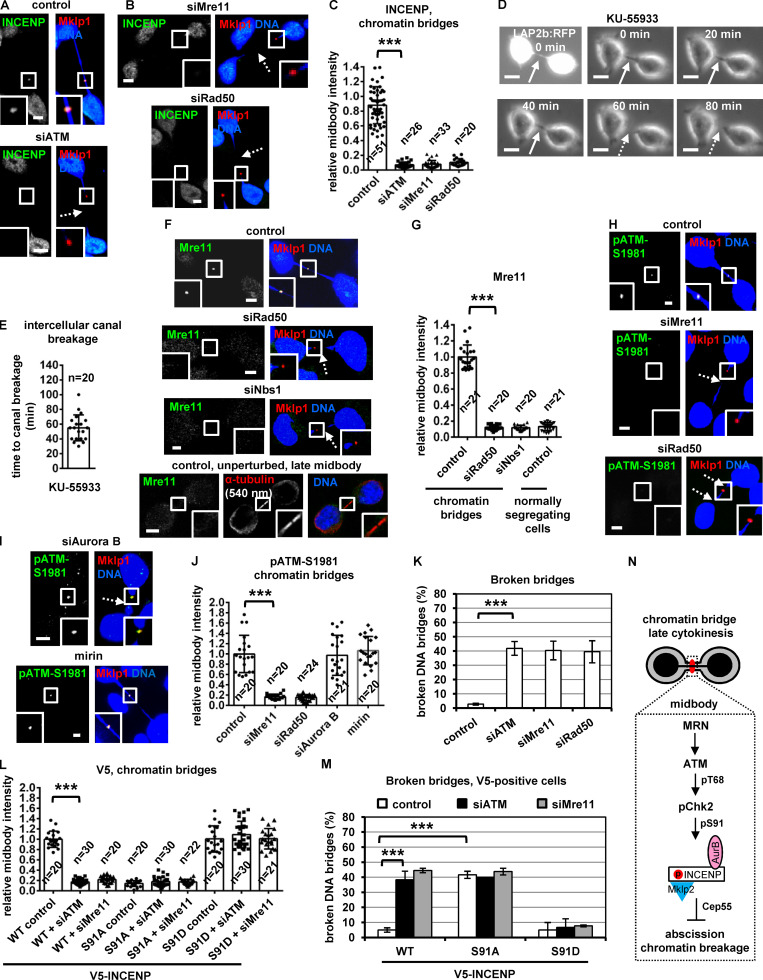
ATM inhibition accelerates midbody disassembly. (A–C) Localization and mean intensity of INCENP at the midbody center in BE cells expressing FL or truncated (ΔC90) GFP:Mkp2. (D) Mechanism of INCENP-Aurora B localization to the midbody center. p, phosphorylation. (E) Phosphorylated ATM-Ser1981 (pATM-Ser1981) localizes to the midbody center in BE cells. (F and G) Time-lapse microscopy analysis of HeLa cells expressing tubulin:GFP. Cells were untreated (control) or treated with 10 µM KU-55933 immediately before filming. Midbodies are shown by solid arrows. Time from midbody formation to midbody disassembly (dotted arrows) is indicated. (H) Frequency of bi/multinucleate or midbody stage BE cells. Values represent mean ± SD from three independent experiments (n > 900). (I–N) Localization and mean intensity of phospho-Chk2-Thr68 (pChk2-Thr68), INCENP or Aurora B at the midbody center in BE cells. (O and P) Localization and mean intensity of pChk2-Thr68 after treatment of BE cells with 10 µM KU-55933 in cytokinesis. Values represent mean ± SD from n cells. Values in control were set to 1. Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. S, serine. ns, not statistically significant; ***, P < 0.001 (Student’s t test). Scale bars, 5 µm.
ATM inhibition accelerates midbody resolution
Total ATM and phosphorylated ATM-Ser1981 (active ATM; Bakkenist and Kastan, 2003) localized to the midbody center in late midbodies in control cells (Fig. 6 E and Fig. 7 K). Depletion of ATM abolished phospho-ATM-Ser1981 staining by immunofluorescence, confirming the antibody is specific (Fig. S3, L and M). Treatment of HeLa tubulin:GFP cells in cytokinesis with 10 µM KU-55933, an ATM kinase inhibitor, accelerated midbody disassembly compared with controls (Fig. 6, F and G). Furthermore, HeLa tubulin:GFP cells treated with KU-55933 in cytokinesis exhibited faster cleavage of the intercellular canal compared with controls by phase-contrast live-cell imaging, indicating premature abscission (P < 0.001; Fig. S3, N and O; and Video 5). Also, BE cells exhibited reduced frequency of cells at midbody stage after ATM depletion compared with controls, and the frequency of bi/multinucleate or prometaphase cells was similar in both treatments (Fig. 6 H and Fig. S3 P). These results suggest that ATM catalytic activity delays abscission in normally segregating cells.
Figure 7.
Aurora B depletion impairs ATM activation at late midbodies. (A and B) Localization and mean intensity of phosphorylated INCENP-Ser91 (pINCENP-Ser91) at the midbody center after treatment of BE cells with 10 µM KU-55933 in cytokinesis. (C) Frequency of midbody stage BE cells. Values represent mean ± SD from three independent experiments (n > 900). (D) Model for the role of ATM and Chk2 in abscission delay in normally segregating cells. Dashed arrow indicates feedback loop. p, phosphorylation. (E) Mre11 does not localize to late midbodies. As a positive control, Mre11 forms DNA foci after treatment of BE cells with 1 mM hydroxyurea (HU) for 4 h. (F, G, and J–M) Localization and mean intensity of phospho-ATM-Ser1981 (pATM-Ser1981), phospho-Chk2-Thr68 (pChk2-Thr68), or ATM at the midbody center in BE cells. Values represent mean ± SD, from n cells. Values in control were set to 1. ***, P < 0.001 (ANOVA and Student’s t test). Tubulin values indicate midbody thickness. Insets show 1.6× magnification of the midbodies. Scale bars, 5 µm. (H and I) Western blot analysis of total Mre11, actin, Aurora B (AurB), and tubulin. T, threonine.
Video 5.
Cleavage of the intercellular canal in ATM-deficient cells. HeLa cells stably expressing tubulin:GFP were treated with 10 µM KU-55933 and analyzed by phase-contrast time-lapse microscopy in cytokinesis. Frames were taken every 5 min for 60 min. The intercellular canal is shown by an arrow. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. S3 N.
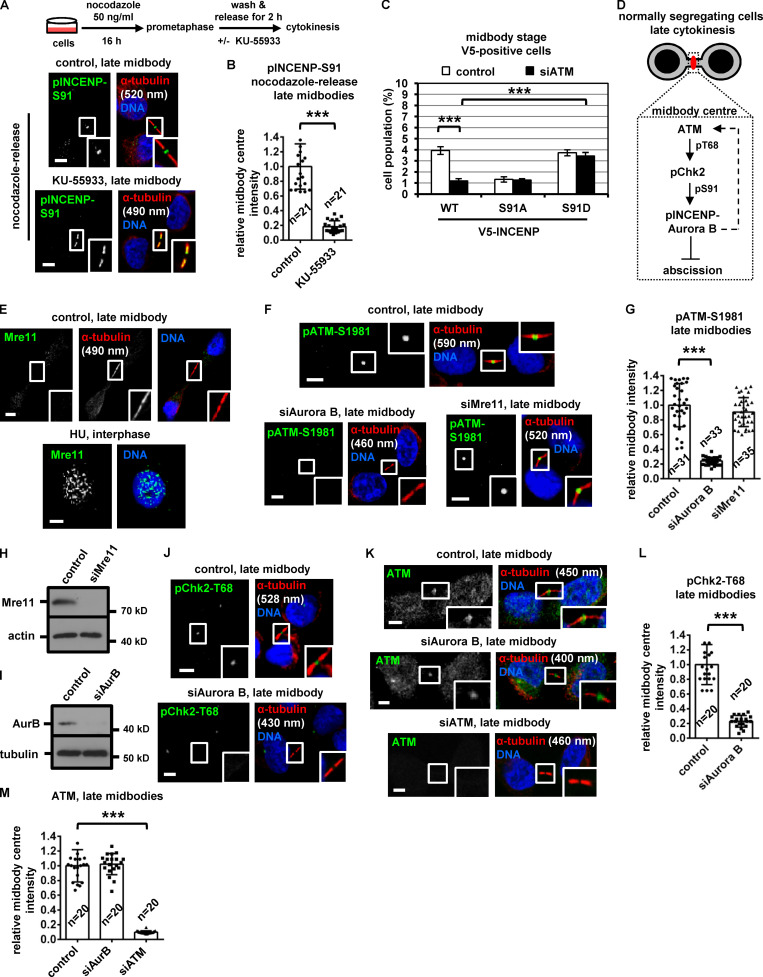
ATM is required for Chk2 activation at late midbodies
Phosphorylation of Thr68 is required for Chk2 activation (Ahn et al., 2000, 2002). Depletion of ATM by siRNA or inhibition of ATM by KU-55933 in cytokinesis after release of cells from a nocodazole block, diminished localization of phosphorylated Chk2-Thr68, total INCENP, total Aurora B, or phosphorylated INCENP-Ser91 to the midbody center compared with controls in late midbodies (Fig. 6, I–P; and Fig. 7, A and B). Also, after depletion of the endogenous INCENP, expression of Ser91D, but not WT or Ser91A, V5/His-INCENP rescued the frequency of cells at midbody stage in ATM-deficient cells compared with WT controls (Fig. 7 C and Fig. S3 Q). We propose that ATM phosphorylates Chk2-Thr68 in late cytokinesis; in turn, phosphorylated (active) Chk2 phosphorylates INCENP-Ser91 to promote INCENP–Aurora B localization to the midbody center to delay midbody resolution in normally segregating cells (Fig. 7 D).
Aurora B, but not Mre11, is required for ATM activation in normally segregating cells
Mre11 was not detected at late midbodies; furthermore, Mre11 depletion did not reduce ATM-Ser1981 phosphorylation, suggesting the MRN complex is dispensable for ATM activation (Fig. 7, E–H). In contrast, Aurora B depletion diminished localization of phosphorylated ATM-Ser1981 and Chk2-Thr68, but not total ATM, to late midbodies (Fig. 7, I–M), suggesting Aurora B is required for ATM activation at the midbody in normally segregating cells (Fig. 7 D, dashed arrow).
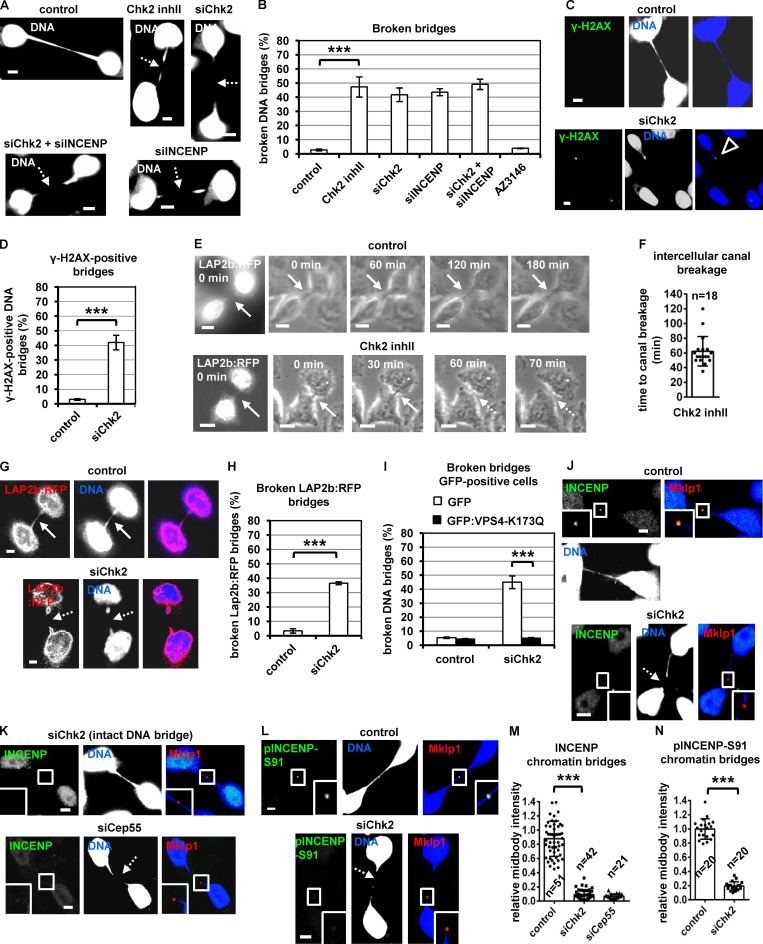
Chk2 prevents chromatin breakage in cytokinesis
We next investigated a potential role of Chk2 in cytokinesis with chromatin bridges. Chk2-deficient cells exhibited increased frequency of broken chromatin bridges compared with controls as judged by DNA or γ-H2AX staining; furthermore, simultaneous depletion of Chk2 and INCENP did not exacerbate broken chromatin bridges in Chk2-deficient cells (Fig. 8, A–D). Inhibition of the spindle checkpoint kinase Mps1 by 10 µM AZ3146 that fully inhibits Mps1 catalytic activity did not induce broken chromatin bridges (Fig. 8 B), suggesting inhibition of the mitotic spindle checkpoint does not lead to defective activation of the abscission checkpoint (Abrieu et al., 2001; Petsalaki and Zachos, 2014).
Figure 8.
Chk2 inhibition correlates with chromatin breakage in cytokinesis. (A) Telophase BE cells with chromatin bridges. (B) Frequency of cells with broken DNA bridges. BE cells were transfected and treated with 10 µM Chk2 inhibitor II (inhII), or 10 µM AZ3146 for 4 h. (C and D) Example of a chromatin bridge that is positive for γ-H2AX staining (open arrowhead) and frequency of γ-H2AX–positive chromatin bridges. Values represent mean ± SD from three independent experiments (n > 150). (E and F) Fluorescence (t = 0 min) and phase-contrast live-cell imaging of HeLa cells expressing LAP2b:RFP. Cells were untreated (control) or treated with 10 µM Chk2 inhibitor II immediately before filming. Time is from the detection of the intercellular canals containing LAP2b:RFP bridges. Intact intercellular canals are indicated by solid arrows, and broken canals are indicated by dotted arrows. Related to Video 6 and Video 7. (G) HeLa LAP2b:RFP cells exhibiting LAP2b:RFP bridges were analyzed by fluorescence microscopy of fixed samples. (H) Frequency of broken LAP2b:RFP bridges from G. (I) Percentage of broken DNA bridges in BE cells transfected with GFP or GFP:Vps4-K173Q. Values represent mean ± SD from three independent experiments (n > 150). (J–N) Localization and mean intensity of phosphorylated INCENP-Ser91 (pINCENP-Ser91) or total INCENP at the midbody in BE cells with chromatin bridges. Values represent mean ± SD from n cells. Values in control were set to 1. ***, P < 0.001 (ANOVA and Student’s t test). Broken chromatin or LAP2b:RFP bridges are indicated by dotted arrows. Insets show 1.6× magnification of the midbodies. S, serine. Scale bars, 5 µm.
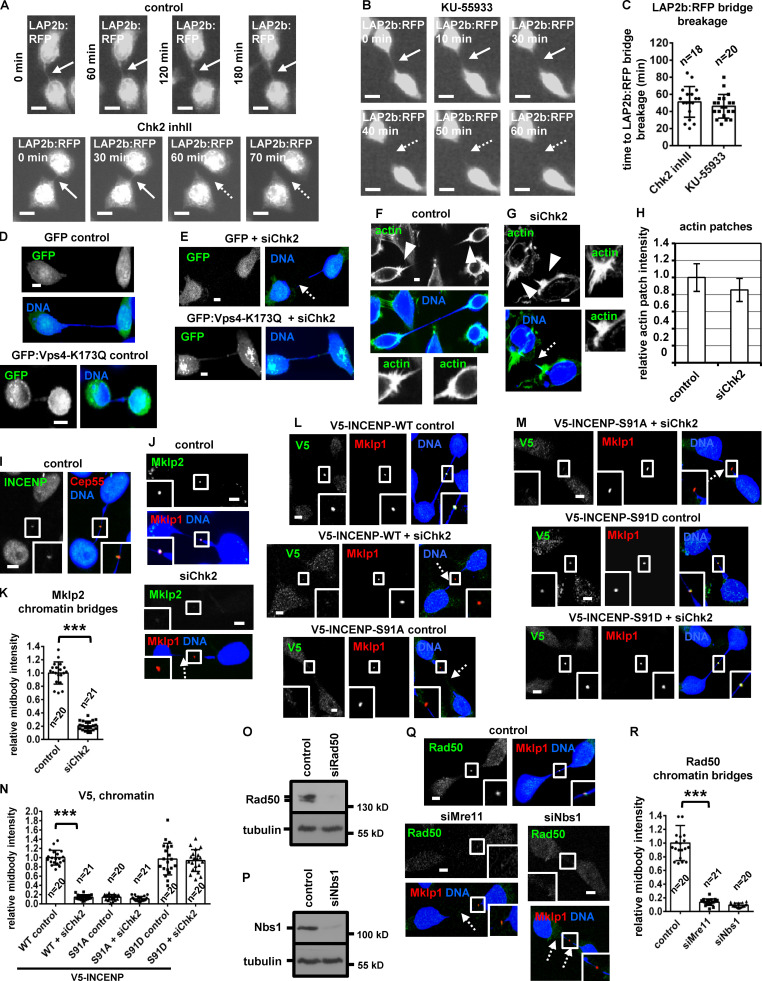
To further investigate the role of Chk2 in chromatin breakage, HeLa cells expressing the inner nuclear envelope marker LAP2b fused to RFP (LAP2b:RFP) that correlates with chromatin bridges (Steigemann et al., 2009) and displaying LAP2b:RFP bridges in cytokinesis were monitored for up to 180 min by time-lapse microscopy (Fig. 8 E). We found that 20 of 20 control cells exhibiting intercellular canals with LAP2b:RFP bridges sustained those canals and the LAP2b:RFP bridges for the duration of the experiment. In contrast, 18 of 28 cells (64%) treated with Chk2 inhibitor II in cytokinesis exhibited breakage of LAP2b:RFP-positive intercellular canals and LAP2b:RFP bridges after ∼52–62 min (n = 18; Fig. 8, E and F; Fig. S4, A and C; and Video 6, Video 7, and Video 8). Consistently, Chk2-deficient HeLa LAP2b:RFP cells exhibited increased frequency of broken LAP2b:RFP bridges compared with controls by confocal microscopy analysis of fixed samples (Fig. 8, G and H). Expression of dominant-negative mutant GFP:Vps4-K173Q that inhibits abscission (Morita et al., 2007) diminished broken chromatin bridges after Chk2 depletion compared with GFP-only controls in BE cells by confocal microscopy analysis of fixed samples (Fig. 8 I; and Fig. S4, D and E). Also, Chk2 was not required for formation of actin patches at the bases of the intercellular canal that stabilize chromatin bridges (Fig. S4, F–H; Dandoulaki et al., 2018). We propose that Chk2 is required for the abscission checkpoint in cytokinesis with chromatin bridges.
Figure S4.
Chk2 depletion correlates with breakage of LAP2b:RFP bridges in cytokinesis. (A–C) Fluorescence live-cell imaging of HeLa cells expressing LAP2b:RFP. Cells were untreated (control) or treated with 10 µM Chk2 inhibitor II (inhII) or 10 µM KU-55933 immediately before filming. Time is from the detection of the LAP2b:RFP bridges. Intact LAP2b:RFP bridges are indicated by solid arrows, and broken LAP2b:RFP bridges are indicated by dotted arrows. Related to Video 8 and Video 10. Mean ± SD from n cells. (D and E) BE cells transfected with GFP or GFP:Vps4-K173Q. (F and G) Actin patches (arrowheads) in BE cells in cytokinesis with chromatin bridges. Insets show magnification of the canals bases. (H) Relative actin-patch intensity values. Values represent mean ± SD from three independent experiments (n > 90). (I) Localization of INCENP. (J–N, Q, and R) Localization of Mklp2, V5-INCENP, or Rad50 and mean intensity at the midbody in BE cells, in cytokinesis with chromatin bridges. Insets show 1.6× magnification of the midbodies. Broken chromatin bridges are indicated by dotted arrows. Values represent mean ± SD from n cells. ***, P < 0.001 (ANOVA and Student’s t test). Scale bars, 5 µm. (O and P) Western blot analysis of total Rad50, Nbs1, and tubulin. S, serine.
Video 6.
Control cells exhibit stable intercellular canals in cytokinesis. HeLa cells stably expressing LAP2b:RFP were analyzed by phase-contrast time-lapse microscopy. Frames were taken every 10 min for 180 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. 8 E.
Video 7.
Breakage of intercellular canals in Chk2-deficient cells. HeLa cells stably expressing LAP2b:RFP were treated with 10 µM Chk2 inhibitor II and analyzed by phase-contrast time-lapse microscopy. Frames were taken every 5 min for 80 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. 8 E.
Video 8.
Breakage of LAP2b:RFP bridges in Chk2-deficient cells. HeLa cells stably expressing LAP2b:RFP (white) were treated with 10 µM Chk2 inhibitor II and analyzed by time-lapse fluorescence microscopy. Frames were taken every 5 min for 65 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. S4 A.
INCENP-Ser91 phosphorylation by Chk2 promotes INCENP localization to the midbody and prevents chromatin breakage
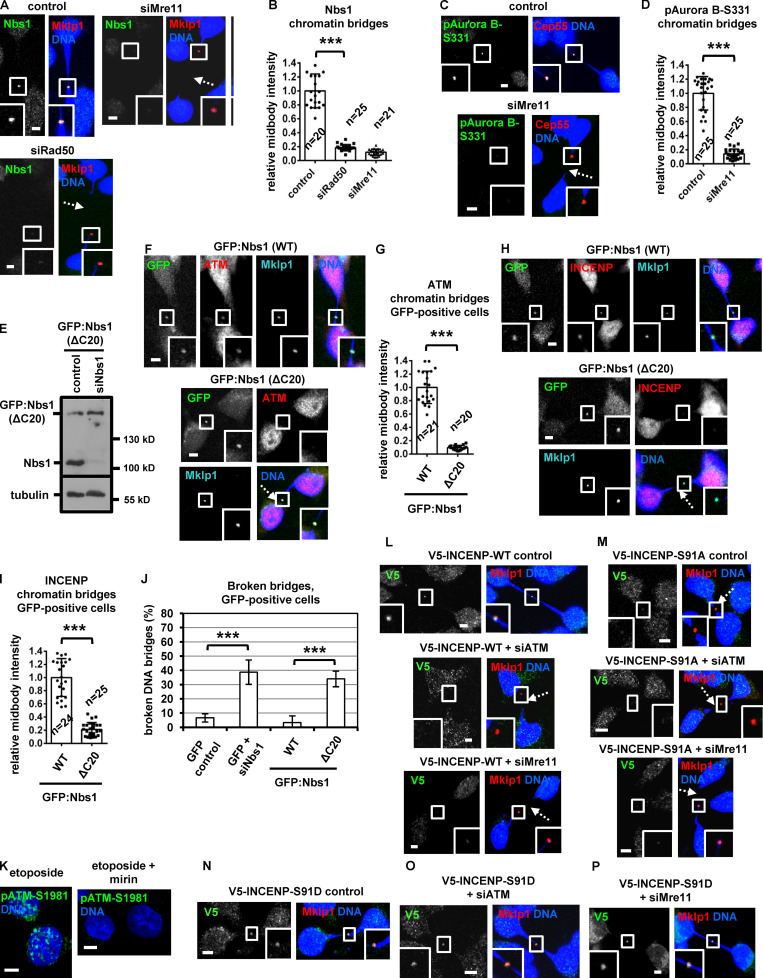
Chk2 depletion diminished localization of total INCENP, phosphorylated INCENP-Ser91, total Aurora B, or Mklp2 to the midbody remnant, marked by Mklp1 or Cep55 staining, compared with control cells in cytokinesis with chromatin bridges (Fig. 8, J–N; Fig. 9, A and B; and Fig. S4, I–K). Expression of siRNA-resistant GFP:Chk2R, phosphomimetic Ser91D V5/His-INCENP, or FB-targeted GFP:INCENP(FB), but not GFP-only, WT or Ser91A V5/His-INCENP, restored localization of INCENP or Aurora B to the midbody and reduced broken chromatin bridges in Chk2-deficient cells compared with controls (Fig. 9, C–K; and Fig. S4, L–N). Also, depletion of Cep55 diminished INCENP staining at the midbody (Fig. 8, K and M), in agreement with our findings in normally segregating cells. We propose that INCENP-Ser91 phosphorylation by Chk2 promotes INCENP and Aurora B localization to the midbody to prevent chromatin breakage in cytokinesis.
Figure 9.
Expression of INCENP-Ser91A correlates with chromatin bridge breakage in cytokinesis. (A, B, O, and P) Localization and mean intensity of Aurora B or phospho-Chk2-Thr68 (pChk2-Thr68) at the midbody in telophase BE cells with chromatin bridges. (C–F, I, and J) Localization and mean intensity of INCENP and Aurora B at the midbody in cells expressing GFP, siRNA-resistant GFP:Chk2R, or GFP:INCENP(FB). Values represent mean ± SD from n cells. Values in control were set to 1. Broken chromatin bridges are indicated by dotted arrows. (G, H, and K) Percentage of broken DNA bridges in BE cells. Values represent mean ± SD from three independent experiments (n > 150). ***, P < 0.001 (ANOVA and Student’s t test). (L–N) Localization of ATM, FLAG-ATM, or phosphorylated Chk2-Thr383 (pChk2-Thr383) in BE cells. Insets show 1.6× magnification of the midbodies. S, serine; T, threonine. Scale bars, 5 µm.
ATM activates Chk2 at the midbody in cytokinesis with chromatin bridges
Phosphorylated (active) Chk2-Thr68 or Chk2-Thr383, total ATM, or phosphorylated (active) ATM-Ser1981 localized to the midbody in control cells with chromatin bridges in the absence of double-strand DNA breaks as evidenced by lack of γ-H2AX staining (Fig. 9, L–O). Depletion of ATM diminished localization of phosphorylated Chk2-Thr68 or total INCENP to the midbody and increased broken chromatin bridges compared with control cells (Fig. 9, O and P; and Fig. 10, A, C, and K). Also, 20 of 30 HeLa LAP2b:RFP cells (67%) exhibited breakage of LAP2b:RFP-positive intercellular canals and LAP2b:RFP bridges after treatment with the ATM inhibitor KU-55933 in cytokinesis by live-cell imaging (Fig. 10, D and E; Fig. S4, A–C; and Video 9 and Video 10). These results suggest that ATM is required for Chk2 activation and stable chromatin bridges in cytokinesis.
Figure 10.
Depletion of MRN proteins correlates with chromatin breakage. (A–C, F–J, and L) Localization and mean intensity of INCENP, Mre11, phospho-ATM-Ser1981 (pATM-Ser1981) or V5-INCENP at the midbody in telophase BE cells with chromatin bridges. Cells were transfected or treated with 25 µM mirin for 4 h. Values represent mean ± SD from n cells. Values in control or WT control were set to 1. Broken chromatin bridges are indicated by dotted arrows. Insets show 1.6× magnification of the midbodies. Scale bars, 5 µm. (D and E) Fluorescence (t = 0 min) and phase-contrast live-cell imaging of HeLa cells expressing LAP2b:RFP. Cells were treated with 10 µM KU-55933 immediately before filming. Time is from the detection of the intercellular canals containing LAP2b:RFP bridges. Intact intercellular canals are indicated by solid arrows and broken canals are indicated by dotted arrows. Related to Video 9. (K and M) Frequency of broken chromatin bridges in BE cells in cytokinesis. Values represent mean ± SD from three independent experiments (n > 150). ***, P < 0.001 (ANOVA and Student’s t test). (N) Proposed mechanism by which the MRN–ATM–Chk2–INCENP pathway delays abscission in cytokinesis with chromatin bridges. AurB, Aurora B; p, phosphorylation; S, serine.
Video 9.
Breakage of intercellular canals in ATM-deficient cells. HeLa cells stably expressing LAP2b:RFP were treated with 10 µM KU-55933 and analyzed by phase-contrast time-lapse microscopy. Frames were taken every 10 min for 100 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. 10 D.
Video 10.
Breakage of LAP2b:RFP bridges in ATM-deficient cells. HeLa cells stably expressing LAP2b:RFP (white) were treated with 10 µM KU-55933 and analyzed by time-lapse fluorescence microscopy. Frames were taken every 10 min for 90 min. Time counters show minutes:seconds. Display rate is one frame per second. Related image stills are shown in Fig. S4 B.
The MRN complex is required for ATM activation in cytokinesis with chromatin bridges
The MRN complex proteins Mre11, Rad50, and Nbs1 localized to the midbody in BE control cells in cytokinesis with chromatin bridges; furthermore, depletion of any single Mre11, Rad50, or Nbs1 protein diminished localization of the remaining two MRN proteins at the midbody, suggesting MRN proteins localize to the midbody in an interdependent manner (Fig. 10, F and G; Fig. S4, O–R; and Fig. S5, A and B; also compare with the Mre11 intensity at the midbody in normally segregating control cells). Depletion of Mre11 or Rad50 reduced localization of phosphorylated Chk2-Thr68, phosphorylated ATM-Ser1981, phospho–Aurora B–Ser331, or total INCENP at the midbody and increased the frequency of broken chromatin bridges compared with control cells (Fig. 9, O and P; Fig. 10, B, C, and H–K; and Fig. S5, C and D). Furthermore, after depletion of the endogenous Nbs1, expression of siRNA-resistant GFP:Nbs1 (ΔC20) lacking the C-terminal 20 amino acids of human Nbs1 that are required for ATM binding (Falck et al., 2005) diminished localization of total ATM or INCENP to the midbody and increased chromatin breakage compared with cells expressing WT GFP:Nbs1 (Fig. S5, E–J). Depletion of Aurora B by siRNA or treatment of cells with 25 µM mirin, an inhibitor of Mre11 nuclease activity that is required for DNA break processing, did not reduce ATM-Ser1981 phosphorylation at the midbody compared with control cells (Fig. 10, H–J; and Fig. S5 K). Also, after depletion of the endogenous INCENP, expression of Ser91D, but not WT or Ser91A, V5/His-INCENP rescued localization of V5/His-INCENP to the midbody and prevented chromatin breakage in ATM or Mre11-deficient cells compared with WT controls (Fig. 10, L and M; and Fig. S5, L–P). On the basis of these findings, we propose the following model (Fig. 10 N): in cytokinesis with chromatin bridges, the MRN complex activates ATM at the midbody. Active ATM phosphorylates Chk2-Thr68; in turn, active Chk2 phosphorylates INCENP-Ser91 to promote INCENP binding to Mklp2 and localization of the Mklp2–INCENP–Aurora B complex to the midbody through Mklp2’s association with Cep55 to delay abscission and prevent chromatin breakage.
Figure S5.
Expression of GFP:Nbs1 (ΔC20) reduces ATM localization to the midbody and induces chromatin bridge breakage in cytokinesis. (A–D and F–I) Localization of Nbs1, phospho-Aurora B-Ser331 (pAurora B-Ser331), ATM, or INCENP and mean intensity at the midbody in BE cells in cytokinesis with chromatin bridges. Values represent mean ± SD from n cells. (E) Western blot analysis of total Nbs1 and tubulin in BE cells expressing GFP:Nbs1 (ΔC20). (J) Frequency of BE cells with broken chromatin bridges. Values represent mean ± SD from three independent experiments (n > 150). ***, P < 0.001 (ANOVA and Student’s t test). (K) Treatment with mirin impairs ATM-Ser1981 phosphorylation (pATM-Ser1981) by etoposide. BE cells were treated with 10 µM etoposide in the absence or presence of 25 µM mirin for 4 h. (L–P) Localization of WT, Ser91A or Ser91D V5-INCENP in BE cells. Insets show 1.6× magnification of the midbodies. Broken chromatin bridges are indicated by dotted arrows. Scale bars, 5 µm.
Discussion
In response to chromatin bridges in cytokinesis, the CPC localizes to the midbody to delay abscission to prevent chromatin breakage or tetraploidization (Carlton et al., 2012; Petsalaki and Zachos, 2016; Steigemann et al., 2009; Thoresen et al., 2014). However, the molecular mechanisms that relay chromatin bridges to the CPC have not been previously described. In this study, we show that the DNA damage kinases ATM and Chk2 delay abscission by promoting CPC midbody localization in late cytokinesis. More specifically, in the absence of missegregated chromatin, ATM phosphorylates and activates Chk2 at the midbody center. In turn, active Chk2 phosphorylates INCENP-Ser91 to promote INCENP binding to Mklp2 kinesin and localization of the CPC–Mklp2 complex to the midbody center to delay abscission.
Inhibition of Chk2 impairs CPC or Mklp2 localization inside the FB in late midbodies; furthermore, expression of nonphosphorylatable INCENP-Ser91A that does not efficiently bind to Mklp2 impairs INCENP or Mklp2 localization to the midbody arms and the midbody center in early or late midbodies. These results are consistent with previous reports that CPC and Mklp2 mutually depend on each other for midzone/midbody localization by Mklp2 transporting CPC along central spindle microtubules and CPC binding promoting the microtubule processivity of Mklp2 in cytokinesis (Adriaans et al., 2020; Hümmer and Mayer, 2009; Kitagawa et al., 2013; Serena et al., 2020). Of note, however, INCENP binding to Mklp2 is not required for Mklp2’s interaction with the midbody protein Cep55 by GST pull-downs when proteins can freely interact in cell extracts, indicating the CPC–Mklp2 complex localizes to the midbody center through Mklp2 binding to Cep55. Deletion of human Mklp2 (amino acids 800–890) C-terminal region diminishes the Mklp2–Cep55 interaction in cell extracts and in vitro and impairs INCENP localization to the midbody center. Because Mklp2 (amino acids 800–890) overlaps with a myosin-II binding site (Fig. S3 J; Kitagawa et al., 2013), a potential role for myosin-II in stabilizing the Mklp2–Cep55 interaction in cell extracts cannot be formally excluded and requires further investigation. Also, because the CPC and Mklp2 are interdependent for their localization to the midbody, a direct role for Mklp2 in the abscission delay, beyond targeting of the CPC to the midbody, cannot be excluded (Fung et al., 2017). However, because expression of midbody-targeted GFP:INCENP(FB) rescues the frequency of cells at midbody stage after Chk2 depletion and GFP:INCENP(FB) exhibits impaired binding to Mklp2, we believe accelerated abscission in Chk2-deficient cells can be attributed to impaired CPC localization to the midbody center.
Phosphorylated INCENP-Ser91 localizes to the midzone in anaphase, on the midbody arms in early midbodies, and inside the FB in late midbodies. Chk2-deficient cells exhibit impaired Ser91 phosphorylation inside the FB; however, Chk2 is dispensable for Ser91 phosphorylation on the midbody arms, suggesting a different kinase phosphorylates INCENP-Ser91 in early midbodies. Because Chk2 localizes to the midbody center in both early and late midbodies, this division of labor likely reflects different intracellular compartments in which the Ser91-targeting kinases localize and/or are activated. This kinase promiscuity is perhaps reminiscent of the Aurora B–Ser331 phosphorylation at kinetochores by two different kinases (Chk2 and Chk1) in, respectively, early or late prometaphase (Petsalaki et al., 2011; Petsalaki and Zachos, 2014). It also implies that Ser91 undergoes a rapid phospho-cycling, perhaps with the involvement of a phosphatase. Recruitment of phosphatases to the midbody during abscission has been previously reported (Bhowmick et al., 2019; Fung et al., 2017), and this notion is consistent with our using of the phosphatase inhibitor microcystin during cell fixation to visualize Ser91 phosphorylation by immunofluorescence (please also see Materials and methods).
We also show that Aurora B is required for ATM and Chk2 activation at the midbody in normally segregating cells. Our results suggest that Aurora B catalytic activity is required for the interdependent Mklp2 and CPC recruitment to the midbody (Kitagawa et al., 2013) by governing Chk2 activation, thus imposing INCENP-Ser91 phosphorylation at the FB. Also, these findings are in agreement with previous data showing that active ATM phosphorylated by Aurora B localizes to the midbody where it colocalizes with Aurora B (Yang et al., 2011).
In contrast, in cytokinesis with chromatin bridges, the MRN complex is required for activation of the ATM–Chk2–INCENP pathway at the midbody to delay abscission and prevent chromatin breakage. It should perhaps be noted that Steigemann et al. (2009) had observed binucleation instead of chromatin breakage after Aurora B inhibition by chemical inhibitors using the same HeLa LAP2b:RFP cell line. Because Chk2 regulates CPC localization to the midbody center in late cytokinesis, but not in early midbodies, one possibility is that potent Aurora B inhibition at relatively early midbodies with chromatin bridges in the study by Steigemann et al. (2009) destabilizes the anchoring of the plasma membrane to the midbody, leading to binucleation, whereas impaired CPC localization to the midbody remnant in late midbodies after Chk2 inhibition in our study leads to premature abscission and chromatin breakage. Better understanding of the midbody morphology in cytokinesis with chromatin bridges is required to address these possibilities.
Inhibition of ATM or Chk2 did not accelerate midbody resolution in normally segregating cells in a previous study based on live-cell imaging of a HeLa cell line (Mackay and Ullman, 2015). This appears to be at odds with findings from our study showing that ATM or Chk2 inhibition, or expression of a nonphosphorylatable Ser91A INCENP, accelerates abscission compared with control cells, as judged by timing of midbody disassembly and cleavage of the intercellular canal in HeLa and BE cells by time-lapse microscopy and also by midbody index analysis of fixed cells. Although the reason for the difference between the results of Mackay and Ullman (2015) and our study is unknown, it could be due to genomic differences in the cell lines used resulting in varying levels of ATM or Chk2 inhibition (Liu et al., 2019b).
Our findings are the first to describe an MRN–ATM–Chk2–INCENP pathway that activates the abscission checkpoint by signaling chromatin bridges to the CPC and identify the molecular mechanism by which the CPC localizes to the midbody in cytokinesis with DNA bridges through Mklp2-mediated association with Cep55. Our findings also highlight a previously undescribed mechanism by which ATM and Chk2 could protect against genomic instability and tumorigenesis (Antoni et al., 2007; Smith et al., 2010) through their role in abscission checkpoint signaling, and we propose that the abscission checkpoint machinery and DNA damage pathways crosstalk to maintain genome integrity (Petsalaki and Zachos, 2020). Although unchecked ATM activity on DNA after DNA repair may lead to cytokinesis defects (Liu et al., 2019a), our results show that localized ATM activity on the midbody is required for proper abscission timing in the absence of DNA damage. Also, the INCENP-Ser91 consensus site is conserved only in mammals, suggesting that this mechanism might have emerged late in evolution and indicating potential differences between the mechanisms for chromosome segregation sensing in higher eukaryotes and the budding yeast, where the CPC can activate the abscission delay (“NoCut”) by physically binding to acetylated chromatin at the spindle midzone (Mendoza et al., 2009).
How Aurora B (which mainly localizes to the midbody arms) sustains formation of an abscission-control protein complex inside the midbody center in normally segregating cells has not been fully understood (Carlton et al., 2012; Petsalaki and Zachos, 2019; Thoresen et al., 2014). Our observations that a relatively small population of CPC molecules localizes to the midbody center in late cytokinesis and that this localization is required for the abscission delay provide an apparent explanation and are in agreement with previous findings that Aurora B phosphorylation by Clk inside the FB is required for the abscission delay (Petsalaki and Zachos, 2016).
The morphology of the midbody appears different in the presence of a chromatin bridge compared with “unperturbed” midbodies in normally segregating cells, based for example on CPC proteins or Mklp2 colocalizing with Mklp1 as a single dot on the midbody remnant and not exhibiting “midbody arms” localization in control cells with chromatin bridges. One possibility is that in cytokinesis with chromatin bridges exhibiting a relatively long and narrow intercellular canal, midbody arms/microtubules are quickly disassembled, giving rise to midbody remnants that correspond to the midbody center/FB (Connell et al., 2009). Although a more systematic analysis of the midbody structure in the presence of chromatin bridges is required, our results are consistent with the midbody pool of ATM, Chk2, Mklp2, or CPC proteins in the presence of a chromatin bridge corresponding to the “midbody center/FB” identified in unperturbed late midbodies. Such differences in midbody structure may also explain why the mislocalization of the FB pool of phosphorylated Aurora B Ser331 or phospho-INCENP-Ser91 on the midbody arms after Chk2 inhibition that is observed in unperturbed late midbodies is not apparent in the presence of a chromatin bridge.
It is perhaps striking that ATM can be activated in two different ways at the midbody, by the MRN complex or Aurora B kinase, depending on whether there is a chromatin bridge inside the intercellular canal or not. Because there is no pool of midbody arms/CPC adjacent to the midbody remnant in the presence of a chromatin bridge as discussed in the previous paragraph, a different signal (generated by the MRN complex) may be required to control recruitment of the checkpoint machinery to the FB compared with the unperturbed midbody. Our results argue that the Aurora B–ATM–Chk2–INCENP–Chmp4c pathway functions as an abscission timer (as it does not function in the presence of a chromatin bridge), whereas the MRN–ATM–Chk2–INCENP–Chmp4c pathway, activated in the presence of a bridge, generates a robust abscission delay signal and constitutes the actual checkpoint.
A more detailed mechanism by which ATM is activated at the midbody requires further investigation. Aurora B activates ATM in mitosis through ATM-Ser1403 phosphorylation (Yang et al., 2011). Therefore, one possibility is that midbody proteins function as scaffold to promote or regulate the Aurora B–ATM interaction in cytokinesis in normally segregating cells. How does the MRN activate ATM in cytokinesis with chromatin bridges? It was previously reported that continuous immobilization of MRN components to chromatin can activate the ATM–Chk2 signaling pathway in the absence of damaged DNA (Soutoglou and Misteli, 2008). One possibility is that chromatin remodeling enzymes acting to resolve the intertwined DNA strands can promote stable binding of the MRN complex to the chromatin bridge and subsequent ATM activation at the midbody (Chan et al., 2007).
Whether or how the abscission checkpoint signaling can be exploited to improve cancer therapy has remained an open question, at least in part because inhibition of abscission-control proteins such as Aurora B or Chmp4c also impairs important cell functions in early mitosis (Petsalaki et al., 2018a, 2018b; Sadler et al., 2018). Because expression of nonphosphorylatable INCENP-Ser91A increases cell death and diminishes cell proliferation and Ser91 is only phosphorylated in cytokinesis, our results support targeting abscission-control proteins as a potential therapeutic strategy to kill proliferating tumor cells (Petsalaki and Zachos, 2019). Chk2-deficient cells exhibit relatively mild proliferation defects compared with cells expressing INCENP-Ser91A (Rainey et al., 2008); however, this could be due to Chk2 regulating Ser91 phosphorylation only in the late stages of cytokinesis. Also, because Chk2 is not an essential gene and Chk2-knockout mice develop normally (Takai et al., 2002), it would be interesting to examine whether Chk2 is required for the abscission checkpoint only in somatic cells and whether the abscission checkpoint is wired differently in embryonic cells. In conclusion, our study describes a molecular mechanism that prevents chromatin breakage in cytokinesis by imposing the abscission checkpoint in human cells by regulating CPC midbody localization.
Materials and methods
Antibodies
Anti-pSer91 polyclonal antiserum was generated in rabbits by immunization against the phosphorylated peptide phospho-Ser91 (LSRRK{pS}RSSQLSSR) of human INCENP (Proteogenix). The unphosphorylated peptide Ser91 (LSRRKSRSSQLSSR) was used in competition experiments as appropriate.
Mouse monoclonal antibodies against GST (B-14; sc-138), Cep55 (B-8; sc-374051; used in Western blotting), Mre11 (18; sc-135992), Rad50 (G-2; sc-74460), Survivin (D-8; sc-17779; used in Western blotting), Borealin (A-5; sc-376635), ATM (1B10; sc-135663), phospho-ATM-Ser1981 (10H11.E12; sc-47739), and Mklp2 (KIF20A, D-3; sc-374508) were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against GFP (FL; sc-8334), phospho-histone H3-Ser10 (pH3; sc-8656-R), Chk2 (H-300; sc-9064), Nbs1 (Nibrin; H-300; sc-11431), and Mklp1 (N-19; sc-867) were also from Santa Cruz Biotechnology. Rabbit polyclonal anti-Survivin (ab469; used in immunofluorescence), anti–α-tubulin (ab18251; used in immunofluorescence), anti-INCENP (ab12183), anti-Chk2-pThr383 (ab59408), and anti–Aurora B (ab2254; used in immunofluorescence) antibodies were from Abcam. Mouse monoclonal anti-phospho-Histone H2A.X (S139; clone JBW301; γ-H2AX) and rabbit polyclonal anti-V5 epitope (AB3792) antibodies were from Millipore. Mouse monoclonal antibodies against α-tubulin (DM1A; used in Western blotting and immunofluorescence) and actin (AC-40) were from Sigma. Rabbit polyclonal anti-phospho-Chk2-Thr68 (2661) antibody was obtained from Cell Signaling Technology, rabbit polyclonal anti-Cep55 (N2C3; GTX112190; used in immunoprecipitations and immunofluorescence) antibody was from Genetex, mouse monoclonal penta-His (34660) antibody was from Qiagen, and mouse monoclonal anti-AIM1 (Aurora B; 611082; used in Western blotting) antibody was from BD Biosciences. Anti-pSer331 rabbit polyclonal antiserum against phosphorylated Ser331 of human Aurora B was previously described (Petsalaki et al., 2011).
Secondary antibodies
For immunofluorescence, goat anti-rabbit IgG Alexa Fluor 633–conjugated (A21070) and goat anti-mouse IgG Alexa Fluor 633–conjugated (A21050) antibodies were from Thermo Fisher Scientific. Goat anti-rabbit IgG FITC (fluorescein)-conjugated (111–096-047), goat anti-mouse IgG FITC (fluorescein)-conjugated (115–096-072), goat anti-rabbit IgG Rhodamine (TRITC)–conjugated (111–025-046) and sheep anti-mouse IgG Rhodamine (TRITC)–conjugated (515–025-072) antibodies were from Jackson ImmunoResearch. For Western blotting, horse anti-mouse IgG HRP-linked (7076) and goat anti-rabbit IgG HRP-linked (7074) antibodies were from Cell Signaling.
Plasmids and cloning
Plasmid pcDNA3.1/V5-INCENP-WT encoding WT human INCENP into the pcDNA3.1/V5-His vector (Invitrogen) was previously described (Petsalaki et al., 2011). For plasmids pcDNA3.1/V5-INCENP-Ser91A or pcDNA3.1/V5-INCENP-Ser91D encoding, respectively, Ser91A or Ser91D V5/His-INCENP, please see Mutagenesis section below. To generate the pEGFPN1/INCENP-WT or pEGFPN1/INCENP-Ser91A vectors encoding WT or Ser91A GFP:INCENP, the FL human INCENP cDNA sequence was amplified by PCR by using, respectively, the pcDNA3.1/V5-INCENP-WT or pcDNA3.1/V5-INCENP-Ser91A plasmid as template and cloned into the pEGFPN1 (Clontech) as a XhoI–BamHI fragment. To generate vector pEGFPN1/INCENP-Ser91D encoding Ser91D GFP:INCENP, the FL human INCENP cDNA sequence was amplified by PCR from the pcDNA3.1/V5-INCENP-Ser91D template and cloned into the pEGFPN1 (Clontech) as a XhoI–KpnI fragment. To generate pGEX4T1/INCENP vectors encoding GST-tagged WT or Ser91 mutant fragments of human INCENP, INCENP cDNA sequences were amplified by PCR by using the appropriate pcDNA3.1/V5-INCENP plasmid as template and cloned into the pGEX4T1 vector (GE Healthcare) as BamHI–XhoI fragments.
To generate pEGFPN1/INCENP(FB) encoding GFP-tagged human INCENP (amino acids 49–919) that is targeted to the FB, the following two-step strategy was used. First, the 145-nt to 2,757-nt sequence of human INCENP was amplified by PCR by using pcDNA3.1/V5-INCENP-WT plasmid as template and cloned into the pEFGP-N1 vector (Clontech) as an EcoRI–BamHI fragment (plasmid pEGFPN1/INCENP [49–919]). Second, the 1,368–2,574-nt sequence of human Mklp1 was amplified by PCR by using pmCherry-C1-Mklp1 plasmid as a template (plasmid #70154, coding Mklp1 isoform 2 that is 858 amino acids long; Addgene; Joseph et al., 2012) and inserted into the pEGFPN1/INCENP (49–919) plasmid as a XhoI–EcoRI fragment to generate pEGFPN1/INCENP(FB).
The pCS2GFP/Mklp2 plasmid encoding GFP-tagged human Mklp2 into the pCS2-GFP vector (Amersham) was from Thomas Mayer (University of Kostanz, Kostanz, Germany; Hümmer and Mayer, 2009). To generate the pcDNA3.1/Mklp2 and pcDNA3.1/Mklp2(ΔC90) plasmids used for in vitro transcription-translation, the FL human Mklp2 cDNA or the 1–2,400-nt sequence of human Mklp2, respectively, was amplified by PCR by using pCS2GFP/Mklp2 plasmid as a template and cloned into the pcDNA3.1 zeo vector (Invitrogen) as HindIII–XbaI fragments. To generate pEGFPN1/Mklp2(ΔC90) encoding GFP-tagged Mklp2 (amino acids 1–800), nucleotides 1–2,400 of human Mklp2 were amplified by PCR by using pCS2GFP/Mklp2 plasmid as template and cloned into the pEGFPN1 vector (Clontech) as a HindIII–ApaI fragment.
Plasmid pEGFPC1/Cep55 encoding human Cep55 into pEGFP-C1 (Clontech) was from Chi-Kuo Hu (Stanford University, Stanford, CA). To generate pGEX4T1/Cep55 plasmids encoding GST-tagged fragments of human Cep55, Cep55 cDNA sequences were amplified by PCR by using pEGFPC1/Cep55 plasmid as template and cloned into the pGEX4T1 vector (GE Healthcare) as EcoRI–XhoI fragments. Plasmid EGFPx2-N1-Nbs1 encoding GFP-tagged human Nbs1 was a gift from Jiri Lukas (University of Copenhagen, Copenhagen, Denmark). Briefly, FL Nbs1 was cloned into the XhoI site of EGFP-N1 (Clontech), and an additional GFP was amplified by PCR and cloned into the BamHI site.
Plasmid pEGFP-N1 coding for GFP under cytomegalovirus promoter was obtained from Takara Bio, and plasmid pcDNA3.1/Flag-His-ATM WT encoding FL FLAG-tagged human ATM was from Addgene (plasmid 31985; Canman et al., 1998). Plasmid pEGFP-vps4-K173Q encoding human Vps4 harboring the K173Q point mutation fused to EGFP into pEGFP-C1 vector (Clontech) was a gift from Wesley Sundquist (University of Utah, Salt Lake City, UT; Morita et al., 2007). Plasmid pCR3.1 GFP-EXN/chmp4c encoding human Chmp4c fused to GFP was from Paul Beniasz (The Aaron Diamond AIDS Research Center, New York, NY; Jouvenet et al., 2011). All plasmids were completely sequenced.
Mutagenesis
Point mutations were generated by using the Q5 site-directed mutagenesis kit (New England Biolabs). To generate an siRNA-resistant form of Chk2:GFP, the pEGFPN1/Chk2 plasmid coding for human Chk2:GFP was used to introduce C483T, A486G and C489T point mutations giving resistance to the Chk2-2 siRNA. To generate pcDNA3.1/V5-INCENP-Ser91A, the pcDNA3.1/V5-INCENP-WT plasmid was used to introduce a T217G point mutation changing Ser91 to alanine. To generate pcDNA3.1/V5-INCENP-Ser91D, the pcDNA3.1/V5-INCENP-WT plasmid was used to introduce T217G and C272A point mutations changing Ser91 to aspartic acid. To generate EGFPx2-N1-Nbs1(ΔC20) encoding GFP:Nbs1 (ΔC20), the EGFPx2-N1-Nbs1 plasmid was used as template to delete nucleotides 2,206–2,265 of human Nbs1 by using the Q5 site-directed mutagenesis kit.
siRNA sequences
Negative (control) siRNA (a pool of four different siRNAs: 5′-UAAGGCUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-AUGAACGUGAAUUGCUCAA-3′, and 5′-UGGUUUACAUGUCGACUAA-3′) was from Thermo Fisher Scientific–GE Healthcare. Human Mre11 (a pool of three different siRNAs: 5′-CAGAACAGAUGGCUAAUGA-3′, 5′-GUGAGGGAAUGGUCACUAA-3′, and 5′-GUAGGGAAUUCUUCUGUUA-3′), ATM (a pool of three different siRNAs: 5′-GUAGCAACAUACUACUCAA-3′, 5′-GCAACAUACUACUCAAAGA-3′, and 5′-GCAACCCAAUUAAUAUCAA-3′), Rad50 (a pool of three different siRNAs: 5′-CAAGGUUGCUCAAGAAACA-3′, 5′-CCAACCAACUGAUGAAUGA-3′, and 5′-CAACAGAACUUGUGAACAA-3′), and Cep55 (a pool of three different siRNAs: 5′-CUACCGCAUUGCUUGAACA-3′, 5′-CAGGUCCAGUUUCUUUACA-3′, and 5′-GCAUGCUAGUGAAUCAUGU-3′) siRNAs were from Santa Cruz Biotechnology. Human Nbs1 located in the 3′ untranslated region (5′-CCAGAACCAAAUUAACUUA-3′), INCENP located in the 3′ untranslated region (5′-GCAUGAGGUCUGAGUAGAA-3′), and Chk2 (5′-GAACCUGAGGACCAAGAACTT-3′) siRNAs were also from Santa Cruz Biotechnology. Human Chk2-2 (5′-GGGACAAAAGCUGUGAAUA-3′) and Mklp2 (5′-CCACCAGGGAAGAAACCAU-3′) siRNAs were from Ambion (Thermo Fischer Scientific). Only the sense sequences of the siRNA duplexes are shown.
Bacteria culture and GST protein purification
BL21 (DE3) cells (Agilent Technologies) were grown in 100–200 ml LB Broth (1% wt/vol tryptone, 0.5% wt/vol yeast extract, 171 mM NaCl) supplemented with the appropriate selection antibiotic. At OD600 = 0.5, cells were induced with 0.1 mM IPTG at 16°C for 16–18 h. Afterward, cells were spun down at 1,000 g for 10 min, and the pellet was resuspended in 3 ml NETN (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA, and 0.5% NP-40) for every 100 ml of culture. The suspension was sonicated for 3 × 10 s and cleared by centrifugation at 12,000 g for 10 min to produce the supernatant (crude extract) containing the GST fusion protein. To prepare GST fusion protein bound to glutathione agarose beads, 150 µl glutathione agarose slurry (sc-2009; Santa Cruz Biotechnology) was added for every 3 ml crude extract and mixed for 1 h at 4°C using a spiramix; beads were spun down at 1,000 g for 5 min and washed three times with 500 µl NETN, and an equal volume of NETN (150 µl) was added. GST proteins on glutathione agarose beads were stored at 4°C for up to 1 wk.
Cell culture and treatments
Human colon carcinoma BE cells (diploid cells that contain an oncogenic Kras-G13D mutation as well as the BRAF-G463V oncogenic mutation; a gift from Simon Wilkinson and Christopher Marshall, Institute of Cancer Research, London, UK; Petsalaki et al., 2018b), cervical carcinoma HeLa cells stably expressing LAP2b fused to RFP (a gift from Daniel Gerlich, Institute of Molecular Biotechnology, Vienna, Austria; Steigemann et al., 2009), or HeLa cells stably expressing α-tubulin fused to GFP (a gift from Jan-Michael Peters, Institute of Molecular Pathology, Vienna, Austria) were grown in DMEM (Gibco) containing 10% FBS at 37°C in 5% CO2. Cells were treated with 10 µM Chk2 inhibitor II (C3742; Millipore Sigma), 10 µM KU-55933 (sc-202963; Santa Cruz Biotechnology), 1 mM hydroxyurea (Sigma), 10 µM etoposide (Sigma), or 25 µM mirin (ab141182; Abcam) as appropriate. siRNA duplexes were transfected into BE cells 24 h before analysis using Lipofectamine 2000 (Invitrogen). For expression of GFP proteins, plasmids were transfected into cells in the absence or presence of appropriate siRNA duplexes 24 h before analysis or further drug treatment using Turbofect (Thermo Fisher Scientific). All cell lines used exhibited consistent morphology and growth properties and were negative for mycoplasma contamination.
Enrichment of cells in cytokinesis
BE cells were treated with 50 ng/ml nocodazole (Sigma) for 16 h, washed twice with prewarmed (37°C) PBS, and released in prewarmed fresh medium for 2 h before cell lysis or fixation. Microscopic examination had shown ∼30% of untransfected control cells were at midbody stage after this treatment.
Time-lapse imaging
HeLa LAP2b:RFP or tubulin:GFP cells were seeded onto Petri dishes with a 30-mm glass base (Greiner), and an inverted fluorescence microscope (Observer D1; Zeiss) was used. Fluorescence or phase-contrast images were taken by using a 63× Plan Neofluor 0.75 NA Ph2 dry objective (Zeiss). Imaging was performed at 37°C in 5% CO2 by using a Zeiss AxioCam MRm camera and Zeiss ZEN 2 acquisition software. Drugs were added to the medium immediately before filming as appropriate.
Indirect immunofluorescence microscopy and microscope image acquisition
For phospho-Aurora B-Ser331 or phospho-INCENP-Ser91 staining, cells were extracted in prewarmed (37°C) Phem buffer (60 mM Pipes, 25 mM Hepes, pH 7.0, 10 mM EGTA, and 4 mM MgSO4) supplemented with 0.5% CHAPS and 100 nM microcystin (Sigma-Aldrich) for 5 min at room temperature, fixed with prewarmed (37°C) 4% paraformaldehyde in Phem buffer for 10 min at room temperature, permeabilized in 0.5% Triton X-100 in Phem buffer for 2 min at room temperature, washed twice with PBS, and immunostained. For phospho-ATM-1981 or total Chk2 staining, cells were fixed in ice-cold methanol for 5 min at −20°C, washed twice with PBS at room temperature, and immunostained. For all other fluorescence microscopy applications, cells were fixed in 4% paraformaldehyde in cytoskeleton buffer (1.1 M Na2HPO4, 0.4 M KH2PO4, 137 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM EGTA, 5 mM Pipes, and 5 mM glucose, pH 6.1) for 5 min at 37°C, permeabilized in 0.5% Triton X-100 in cytoskeleton buffer, washed twice with PBS at room temperature, and immunostained. For peptide competitions, 2 µg of the anti-pSer91 polyclonal antiserum was incubated with 100 µg peptide at 37°C for 1 h, and then used for immunostaining. FITC- or rhodamine-TRITC-conjugated (Jackson ImmunoResearch Laboratories) or Alexa Fluor 633-conjugated (Thermo Fisher Scientific) secondary antibodies were used as appropriate. DNA was stained with 10 µg/ml DAPI (Biotium), and cells were mounted in Vectashield medium (Vector Laboratories). Images were collected by using a laser-scanning spectral confocal microscope (TCS SP8; Leica Microsystems), LASX software (Leica Microsystems), and a 63× Apochromat 1.40 NA oil objective. The low-fluorescence immersion oil (11513859; Leica Microsystems) was used, and imaging was performed at room temperature. Mean projections of image stacks were obtained by using the LASX software.
Multinucleate or midbody stage cells
BE cells were fixed and stained with α-tubulin. Multinucleate cells or cells connected by midbodies were scored visually by immunofluorescence microscopy as described in the previous paragraph.
Broken bridges
Cells were fixed and stained for immunofluorescence microscopy as appropriate. The percentage of broken chromatin (DAPI) or LAP2b:RFP bridges was calculated as follows: (number of bridges that appear broken / total number of bridges) × 100%.
Quantification of fluorescence signals
Fluorescence intensity signals at midbodies were quantified using the LASX polygon tool by analyzing an image area of 2 µm2 around each FB, and intensity values were normalized versus background values obtained by analyzing an identical area within the cell immediately adjacent on the midbody by subtracting the background-signal value from the midbody value (Petsalaki and Zachos, 2016; Waters, 2009). After subtraction of the background, the average values from control or mutant midbodies were calculated and were then all divided with the control midbodies-average value to obtain the relative midbody intensity values plotted (i.e., relative to control = 1), for comparison reasons.
To analyze midbody thickness, the diameter of each microtubule bundle at the midbody was measured using the LASX line tool and the average value calculated.
Actin-patch fluorescence intensity signals were quantified using the LCS Lite polygon tool by analyzing an image area of 40 µm2 around the base of the DNA bridge, and intensity values were normalized versus background values obtained by analyzing an identical area within the cell near the base of the DNA bridge, by subtracting the background-signal value from the actin-patch value (Dandoulaki et al., 2018). After background subtraction, the average values from control or mutant actin patches were calculated and then all divided with the control actin patch average value to obtain the relative intensity values plotted (i.e., relative to control = 1).
Chk2 in vitro kinase assay
For Chk2 in vitro kinase assays, 0.5 µg recombinant human Chk2 (14–347; EMD Millipore) was incubated with ∼1 µg purified GST protein substrate in 20 µl kinase buffer (50 mM Hepes, pH 8.0, 2.5 mM EDTA, 10 mM MgCl2, 1 mM DTT, 10 mM sodium β-glycerophosphate, 0.1 mM sodium vanadate, 0.1 mM PMSF, 1 mM sodium fluoride, 100 µM ATP, and 2 µCi [γ-32P]-ATP) for 30 min at 30°C before analysis by SDS-PAGE and autoradiography. Where indicated, 10 µM Chk2 inhibitor II was included as a negative control in the kinase reaction.
GFP-Trap, coimmunoprecipitation, and GST pull-down
To prepare the cell lysates, cells were sonicated three times for 10 s in ice-cold immunoprecipitation-kinase buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 10% glycerol, 0.1% Tween 20, 0.1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin, 1 mM sodium fluoride, 10 mM sodium β-glycerophosphate, and 0.1 mM sodium vanadate), incubated for 30 min on ice, and lysates were cleared by centrifugation at 15,000 g for 10 min
For GFP-Trap immunoprecipitation, 1 mg cell lysate was incubated with 10 µl GFP-Trap (Chromotek) for 16 h at 4°C.
For coimmunoprecipitation, 1 mg cell lysate was incubated with 1 µg antibody for 16 h followed by the addition of 10 µl protein A/G PLUS–agarose beads (Santa Cruz Biotechnology) for 1 h at 4°C.
For GST pull-down, 1 mg cell lysate or 50 µl in vitro transcription-translation reactions (Fig. 4 H; and Fig. 5, K and N) was incubated with 1 µg GST protein on glutathione-agarose beads (sc-2009; Santa Cruz Biotechnology) for 4 h at 4°C.
After incubation with the beads, samples were spun down and washed three times with immunoprecipitation kinase buffer, and precipitated proteins on beads were analyzed by SDS-PAGE, followed by Western blotting, autoradiography (32P), or phosphorimaging (35S).
Aurora B kinase assay
GFP-Trap-precipitated proteins on beads (Fig. 5 A) were included in a 20 µl kinase reaction containing 1 µg histone H3, 50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 1 mM EGTA, 1 mM DTT, 5 mM NaF, 5 mM sodium β-glycerophosphate, 50 µM sodium vanadate, and 0.1 mM ATP. Where indicated, 300 nM VX-680 (Selleckchem), an Aurora B inhibitor, was included in the kinase reaction. Reactions were incubated for 30 min at 30°C, stopped by addition of 2 µl 10× gel sample buffer, and analyzed by SDS-PAGE and Western blotting using a polyclonal antibody against phosphorylated histone H3-Ser10.
In vitro transcription-translation and phosphorimager analysis
For in vitro transcription-translation, the TnT Quick Coupled Transcription/Translation System (Promega) was used. Briefly, 0.5 µg plasmid DNA template was incubated with 40 µl TnT T7 Quick Master Mix and 10 µCi 35S-methionine per 50 µl reaction for 60 min at 30°C. Radiolabeled (35S) samples transferred onto nitrocellulose membrane were analyzed by phosphorimaging using the Sapphire Biomolecular Imager (Azure Biosystems).
Growth curves
BE cells were transfected with WT, Ser91A, or Ser91D pcDNA3.1/V5-INCENP plasmids for 24 h. Cells were then replated in 30-mm dishes at 2 × 105 cells per dish for another 0–3 d and counted with a hemocytometer. Viable or dead cells were distinguished by trypan blue exclusion and optical microscopy.
Western blotting
Cells were lysed in ice-cold, whole-cell extract buffer (20 mM Hepes, 5 mM EDTA, 10 mM EGTA, 0.4 M KCl, 0.4% Triton X-100, 10% glycerol, 5 mM NaF, 1 mM DTT, 5 µg/ml leupeptin, 50 µg/ml PMSF, 1 mM benzamidine, 5 µg/ml aprotinin, and 1 mM Na3VO4) for 30 min on ice. Lysates were cleared by centrifugation at 15,000 g for 10 min, analyzed by SDS-PAGE, and transferred onto a nitrocellulose membrane (Santa Cruz Biotechnology).
Densitometry
Densitometric analysis of bands was performed using ImageJ (National Institutes of Health).
Statistical analysis and repetitions
For midbody fluorescence intensity, a minimum of 10 cells per experiment from two independent experiments were analyzed per treatment (n ≥ 20), and SD was calculated. For the frequency of cells at midbody stage or in prometaphase, SD from the mean from three independent experiments was calculated, and at least 300 cells per experiment and per treatment (n > 900) were analyzed. For growth curves, two replicates from three independent experiments (n = 6) were analyzed and the SD from the mean was calculated. For the frequency of broken chromatin bridges or γ-H2AX–positive or LAP2b:RFP bridges, at least 50 cells with bridges per experiment from three independent experiments were scored blindly, and the SD was calculated (n > 150). For the frequency of cells exhibiting Chmp4c:GFP-mislocalization (Fig. S1 L), at least 30 cells per experiment from three independent experiments (n > 90) were analyzed and the SD was calculated. For actin-patches fluorescence (Fig. S4 H), at least 30 cells per experiment from three independent experiments were examined (n > 90), and the SD was calculated. Statistically significant differences among three or more groups were determined by one-way ANOVA followed by the Student’s t test between two groups. No statistical method was used to predetermine sample size.
Immunoprecipitations (Figs. 3 B, 4 G, and 5 G) and GST pull-downs (Fig. 4 H; and Fig. 5, I–K and N) were repeated three times. GFP-Trap, kinase assays, and all other gels were done twice, and representative gels are shown.
Online supplemental material
Fig. S1 shows faster cleavage of the intercellular canal and mislocalized Chmp4c:GFP at the midbody in Chk2-deficient cells compared with controls, and localization of Aurora B or INCENP to early midbodies in normally segregating cells. Fig. S2 shows faster cleavage of the intercellular canal in cells expressing Ser91A-INCENP compared with controls, localization of phosphorylated Aurora B-Ser331 at the midbody in cells expressing GFP:INCENP(FB), and specificity of the anti-pChk2-T68, anti-pChk2-T68, or anti-pINCENP-Ser91 phospho-specific antibodies. Fig. S3 shows midbody indices in cells expressing Ser91D GFP:INCENP, localization of Mklp2 or Cep55 to early or late midbodies, binding of Aurora B, Survivin or Borealin with WT or mutant GFP:INCENP, and accelerated cleavage of the intercellular canal after ATM-inhibition in cytokinesis compared with control cells. Fig. S4 shows breakage of LAP2b:RFP bridges in cytokinesis, formation of actin patches, localization of WT, Ser91A, or Ser91D V5-INCENP, and localization of Rad50 to the midbody in cytokinesis with chromatin bridges. Fig. S5 shows ATM or INCENP localization in cells expressing ΔC20 GFP:Nbs1, and localization of Nbs1, phospho-Aurora B-Ser331, or V5-INCENP to the midbody in ATM-deficient or MRN-deficient cells in cytokinesis with chromatin bridges. Video 1 shows a control HeLa tubulin:GFP cell exhibiting midbody disassembly in cytokinesis. Video 2 shows a HeLa tubulin:GFP cell exhibiting midbody disassembly in the presence of Chk2 inhibitor II. Video 3 shows a control HeLa tubulin:GFP cell exhibiting cleavage of the intercellular canal in cytokinesis. Video 3 and Video 4 show HeLa tubulin:GFP cells exhibiting cleavage of the intercellular canal in the presence of, respectively, Chk2 inhibitor II or the ATM inhibitor KU-55933. Video 5 shows cleavage of the intercellular canal in ATM-deficient cells. Video 6 shows a control HeLa LAP2b:RFP cell exhibiting a stable LAP2b-positive intercellular canal in cytokinesis. Video 7 shows a HeLa LAP2b:RFP cell exhibiting breakage of the LAP2b-positive intercellular canal in the presence of Chk2 inhibitor II. Video 8 shows a HeLa LAP2b:RFP cell exhibiting breakage of the LAP2b:RFP bridge in the presence of Chk2 inhibitor II. Video 9 shows a HeLa LAP2b:RFP cell exhibiting breakage of the LAP2b-positive intercellular canal in the presence of KU-55933. Video 10 shows a HeLa LAP2b:RFP cell exhibiting breakage of the LAP2b:RFP bridge in the presence of KU-55933.
Supplementary Material
Acknowledgments
We thank P. Beniasz, D. Gerlich, Chi-Kuo Hu, J. Lukas, T. Mayer, J.-M. Peters, and W. Sundquist for generously sharing reagents. We also thank M. Dandoulaki, P. Fragkiadaki, and E. Kouvidi for assisting with cloning. We also thank the Special Account for Research Funds of the University of Crete for paying the publication costs.
Work in our laboratory was supported by Fondation Santé. E. Petsalaki was supported by the State Scholarships Foundation (project “Reinforcement of Postdoctoral Researchers - 2nd Cycle” grant MIS-5033021), cofinanced by Greece and the European Union (European Social Fund) through the Operational Program Human Resources Development, Education and Lifelong Learning.
The authors declare no competing financial interests.
Author contributions: E. Petsalaki performed the experiments and analyzed the results. G. Zachos designed the study and wrote the paper.
References
- Abrieu, A., Magnaghi-Jaulin L., Kahana J.A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D.W., and Labbé J.C.. 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 106:83–93. 10.1016/S0092-8674(01)00410-X [DOI] [PubMed] [Google Scholar]
- Adriaans, I.E., Hooikaas P.J., Aher A., Vromans M.J.M., van Es R.M., Grigoriev I., Akhmanova A., and Lens S.M.A.. 2020. MKLP2 Is a Motile Kinesin that Transports the Chromosomal Passenger Complex during Anaphase. Curr. Biol. 30:2628–2637.e9. 10.1016/j.cub.2020.04.081 [DOI] [PubMed] [Google Scholar]
- Ahn, J.Y., Schwarz J.K., Piwnica-Worms H., and Canman C.E.. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60:5934–5936. [PubMed] [Google Scholar]
- Ahn, J.Y., Li X., Davis H.L., and Canman C.E.. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 277:19389–19395. 10.1074/jbc.M200822200 [DOI] [PubMed] [Google Scholar]
- Antoni, L., Sodha N., Collins I., and Garrett M.D.. 2007. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat. Rev. Cancer. 7:925–936. 10.1038/nrc2251 [DOI] [PubMed] [Google Scholar]
- Bai, J., Wioland H., Advedissian T., Cuvelier F., Romet-Lemonne G., and Echard A.. 2020. Actin reduction by MsrB2 is a key component of the cytokinetic abscission checkpoint and prevents tetraploidy. Proc. Natl. Acad. Sci. USA. 117:4169–4179. 10.1073/pnas.1911629117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist, C.J., and Kastan M.B.. 2003. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 421:499–506. 10.1038/nature01368 [DOI] [PubMed] [Google Scholar]
- Bhowmick, R., Thakur R.S., Venegas A.B., Liu Y., Nilsson J., Barisic M., and Hickson I.D.. 2019. The RIF1-PP1 axis controls abscission timing in human cells. Curr. Biol. 29:1232–1242.e5. 10.1016/j.cub.2019.02.037 [DOI] [PubMed] [Google Scholar]
- Caballe, A., Wenzel D.M., Agromayor M., Alam S.L., Skalicky J.J., Kloc M., Carlton J.G., Labrador L., Sundquist W.I., and Martin-Serrano J.. 2015. ULK3 regulates cytokinetic abscission by phosphorylating ESCRT-III proteins. eLife. 4:e06547 10.7554/eLife.06547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman, C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., and Siliciano J.D.. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 281:1677–1679. 10.1126/science.281.5383.1677 [DOI] [PubMed] [Google Scholar]
- Capalbo, L., Montembault E., Takeda T., Bassi Z.I., Glover D.M., and D’Avino P.P.. 2012. The chromosomal passenger complex controls the function of endosomal sorting complex required for transport-III Snf7 proteins during cytokinesis. Open Biol. 2:120070 10.1098/rsob.120070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J.G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J.. 2012. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 336:220–225. 10.1126/science.1217180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena, M., Wheelock M., Funabiki H., and Earnshaw W.C.. 2012. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 13:789–803. 10.1038/nrm3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K.L., North P.S., and Hickson I.D.. 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26:3397–3409. 10.1038/sj.emboj.7601777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell, J.W., Lindon C., Luzio J.P., and Reid E.. 2009. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 10:42–56. 10.1111/j.1600-0854.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke, C.A., Heck M.M.S., and Earnshaw W.C.. 1987. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105:2053–2067. 10.1083/jcb.105.5.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandoulaki, M., Petsalaki E., Sumpton D., Zanivan S., and Zachos G.. 2018. Src activation by Chk1 promotes actin patch formation and prevents chromatin bridge breakage in cytokinesis. J. Cell Biol. 217:3071–3089. 10.1083/jcb.201802102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, N., Sougrat R., Spurlin T.A., Hurley J.H., and Lippincott-Schwartz J.. 2011. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. USA. 108:4846–4851. 10.1073/pnas.1102714108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck, J., Coates J., and Jackson S.P.. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 434:605–611. 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- Fung, S.Y.S., Kitagawa M., Liao P.J., Wong J., and Lee S.H.. 2017. Opposing Activities of Aurora B Kinase and B56-PP2A Phosphatase on MKlp2 Determine Abscission Timing. Curr. Biol. 27:78–86. 10.1016/j.cub.2016.10.042 [DOI] [PubMed] [Google Scholar]
- Ganem, N.J., and Pellman D.. 2012. Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 199:871–881. 10.1083/jcb.201210040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson, D. 2008. Classification of chromosome segregation errors in cancer. Chromosoma. 117:511–519. 10.1007/s00412-008-0169-1 [DOI] [PubMed] [Google Scholar]
- Gruneberg, U., Neef R., Honda R., Nigg E.A., and Barr F.A.. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172. 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, R., Körner R., and Nigg E.A.. 2003. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 14:3325–3341. 10.1091/mbc.e02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, C.K., Coughlin M., and Mitchison T.J.. 2012. Midbody assembly and its regulation during cytokinesis. Mol. Biol. Cell. 23:1024–1034. 10.1091/mbc.e11-08-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümmer, S., and Mayer T.U.. 2009. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19:607–612. 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- Hutchins, J.R., Hughes M., and Clarke P.R.. 2000. Substrate specificity determinants of the checkpoint protein kinase Chk1. FEBS Lett. 466:91–95. 10.1016/S0014-5793(99)01763-9 [DOI] [PubMed] [Google Scholar]
- Joseph, N., Hutterer A., Poser I., and Mishima M.. 2012. ARF6 GTPase protects the post-mitotic midbody from 14-3-3-mediated disintegration. EMBO J. 31:2604–2614. 10.1038/emboj.2012.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet, N., Zhadina M., Bieniasz P.D., and Simon S.M.. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401. 10.1038/ncb2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, M., Fung S.Y.S., Onishi N., Saya H., and Lee S.H.. 2013. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One. 8:e64826 10.1371/journal.pone.0064826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa, M., Fung S.Y.S., Hameed U.F.S., Goto H., Inagaki M., and Lee S.H.. 2014. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Rep. 7:166–179. 10.1016/j.celrep.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Lee, J.H., and Paull T.T.. 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 308:551–554. 10.1126/science.1108297 [DOI] [PubMed] [Google Scholar]
- Lens, S.M.A., and Medema R.H.. 2019. Cytokinesis defects and cancer. Nat. Rev. Cancer. 19:32–45. 10.1038/s41568-018-0084-6 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Efimova E.V., Ramamurthy A., and Kron S.J.. 2019a Repair-independent functions of DNA-PKcs protect irradiated cells from mitotic slippage and accelerated senescence. J. Cell Sci. 132:jcs229385 10.1242/jcs.229385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Mi Y., Mueller T., Kreibich S., Williams E.G., Van Drogen A., Borel C., Frank M., Germain P.L., Bludau I., et al. 2019b Multi-omic measurements of heterogeneity in HeLa cells across laboratories. Nat. Biotechnol. 37:314–322. 10.1038/s41587-019-0037-y [DOI] [PubMed] [Google Scholar]
- Mackay, D.R., and Ullman K.S.. 2015. ATR and a Chk1-Aurora B pathway coordinate postmitotic genome surveillance with cytokinetic abscission. Mol. Biol. Cell. 26:2217–2226. 10.1091/mbc.E14-11-1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza, M., Norden C., Durrer K., Rauter H., Uhlmann F., and Barral Y.. 2009. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 11:477–483. 10.1038/ncb1855 [DOI] [PubMed] [Google Scholar]
- Mierzwa, B., and Gerlich D.W.. 2014. Cytokinetic abscission: molecular mechanisms and temporal control. Dev. Cell. 31:525–538. 10.1016/j.devcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Mierzwa, B.E., Chiaruttini N., Redondo-Morata L., von Filseck J.M., König J., Larios J., Poser I., Müller-Reichert T., Scheuring S., Roux A., and Gerlich D.W.. 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19:787–798. 10.1038/ncb3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, E., Sandrin V., Chung H.Y., Morham S.G., Gygi S.P., Rodesch C.K., and Sundquist W.I.. 2007. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 26:4215–4227. 10.1038/sj.emboj.7601850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden, C., Mendoza M., Dobbelaere J., Kotwaliwale C.V., Biggins S., and Barral Y.. 2006. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 125:85–98. 10.1016/j.cell.2006.01.045 [DOI] [PubMed] [Google Scholar]
- Petsalaki, E., and Zachos G.. 2014. Chk2 prevents mitotic exit when the majority of kinetochores are unattached. J. Cell Biol. 205:339–356. 10.1083/jcb.201310071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki, E., and Zachos G.. 2016. Clks 1, 2 and 4 prevent chromatin breakage by regulating the Aurora B-dependent abscission checkpoint. Nat. Commun. 7:11451 10.1038/ncomms11451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki, E., and Zachos G.. 2019. Building bridges between chromosomes: novel insights into the abscission checkpoint. Cell. Mol. Life Sci. 76:4291–4307. 10.1007/s00018-019-03224-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki, E., and Zachos G.. 2020. DNA damage response proteins regulating mitotic cell division: double agents preserving genome stability. FEBS J. 287:1700–1721. 10.1111/febs.15240 [DOI] [PubMed] [Google Scholar]
- Petsalaki, E., Akoumianaki T., Black E.J., Gillespie D.A., and Zachos G.. 2011. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 195:449–466. 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki, E., Dandoulaki M., and Zachos G.. 2018a Chmp4c is required for stable kinetochore-microtubule attachments. Chromosoma. 127:461–473. 10.1007/s00412-018-0675-8 [DOI] [PubMed] [Google Scholar]
- Petsalaki, E., Dandoulaki M., and Zachos G.. 2018b The ESCRT protein Chmp4c regulates mitotic spindle checkpoint signaling. J. Cell Biol. 217:861–876. 10.1083/jcb.201709005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, M.D., Black E.J., Zachos G., and Gillespie D.A.. 2008. Chk2 is required for optimal mitotic delay in response to irradiation-induced DNA damage incurred in G2 phase. Oncogene. 27:896–906. 10.1038/sj.onc.1210702 [DOI] [PubMed] [Google Scholar]
- Sadler, J.B.A., Wenzel D.M., Williams L.K., Guindo-Martínez M., Alam S.L., Mercader J.M., Torrents D., Ullman K.S., Sundquist W.I., and Martin-Serrano J.. 2018. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc. Natl. Acad. Sci. USA. 115:E8900–E8908. 10.1073/pnas.1805504115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serena, M., Bastos R.N., Elliott P.R., and Barr F.A.. 2020. Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle. J. Cell Biol. 219:e201910059 10.1083/jcb.201910059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J., Tho L.M., Xu N., and Gillespie D.A.. 2010. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108:73–112. 10.1016/B978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- Soutoglou, E., and Misteli T.. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510. 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steigemann, P., Wurzenberger C., Schmitz M.H., Held M., Guizetti J., Maar S., and Gerlich D.W.. 2009. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 136:473–484. 10.1016/j.cell.2008.12.020 [DOI] [PubMed] [Google Scholar]
- Takai, H., Naka K., Okada Y., Watanabe M., Harada N., Saito S., Anderson C.W., Appella E., Nakanishi M., Suzuki H., et al. 2002. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21:5195–5205. 10.1093/emboj/cdf506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen, S.B., Campsteijn C., Vietri M., Schink K.O., Liestøl K., Andersen J.S., Raiborg C., and Stenmark H.. 2014. ANCHR mediates Aurora-B-dependent abscission checkpoint control through retention of VPS4. Nat. Cell Biol. 16:550–560. 10.1038/ncb2959 [DOI] [PubMed] [Google Scholar]
- Tsvetkov, L., Xu X., Li J., and Stern D.F.. 2003. Polo-like kinase 1 and Chk2 interact and co-localize to centrosomes and the midbody. J. Biol. Chem. 278:8468–8475. 10.1074/jbc.M211202200 [DOI] [PubMed] [Google Scholar]
- Uziel, T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., and Shiloh Y.. 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22:5612–5621. 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli, P., and Earnshaw W.C.. 2004. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 113:211–222. 10.1007/s00412-004-0307-3 [DOI] [PubMed] [Google Scholar]
- van der Horst, A., Vromans M.J.M., Bouwman K., van der Waal M.S., Hadders M.A., and Lens S.M.A.. 2015. Inter-domain Cooperation in INCENP Promotes Aurora B Relocation from Centromeres to Microtubules. Cell Rep. 12:380–387. 10.1016/j.celrep.2015.06.038 [DOI] [PubMed] [Google Scholar]
- Waters, J.C. 2009. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185:1135–1148. 10.1083/jcb.200903097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C., Tang X., Guo X., Niikura Y., Kitagawa K., Cui K., Wong S.T., Fu L., and Xu B.. 2011. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol. Cell. 44:597–608. 10.1016/j.molcel.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, W.M., Seki A., and Fang G.. 2006. Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol. Biol. Cell. 17:3881–3896. 10.1091/mbc.e06-01-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.