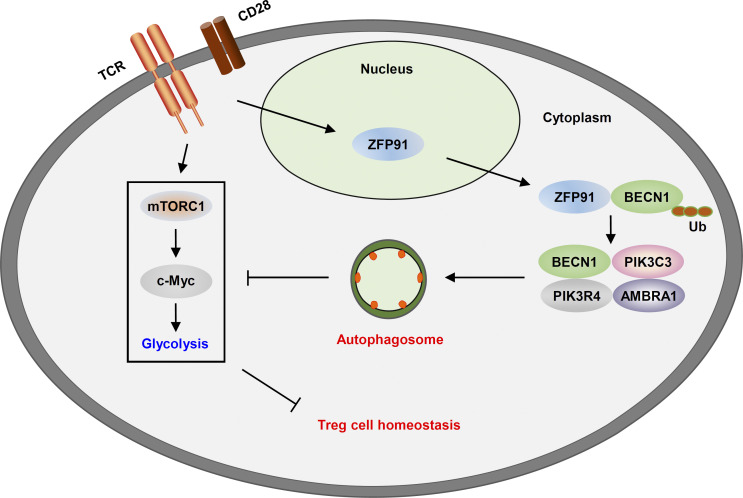
The mechanisms governing autophagy initiation in T reg cells remain unclear. Wang et al. identify a ZFP91-dependent mechanism promoting TCR-initiated autophagosome maturation to restrict hyperglycolysis and maintain T reg cell homeostasis and function.
Abstract
Autophagy programs the metabolic and functional fitness of regulatory T (T reg) cells to establish immune tolerance, yet the mechanisms governing autophagy initiation in T reg cells remain unclear. Here, we show that the E3 ubiquitin ligase ZFP91 facilitates autophagy activation to sustain T reg cell metabolic programming and functional integrity. T reg cell–specific deletion of Zfp91 caused T reg cell dysfunction and exacerbated colonic inflammation and inflammation-driven colon carcinogenesis. TCR-triggered autophagy induction largely relied on T reg cell–derived ZFP91 to restrict hyperglycolysis, which is required for the maintenance of T reg cell homeostasis. Mechanistically, ZFP91 rapidly translocated from the nucleus to the cytoplasm in response to TCR stimulation and then mediated BECN1 ubiquitination to promote BECN1–PIK3C3 complex formation. Therefore, our results highlight a ZFP91-dependent mechanism promoting TCR-initiated autophagosome maturation to maintain T reg cell homeostasis and function.
Graphical Abstract
Introduction
Regulatory T (T reg) cells play central roles in establishing immune tolerance and preventing immunological disorders (Campbell and Rudensky, 2020; Wing et al., 2019). Cellular metabolism is indispensable for T reg cell identity maintenance, and metabolic influences on the lineage stability, function, and migration of T reg cells have been established (Kishore et al., 2017; Newton et al., 2016; Yu et al., 2018; Zeng and Chi, 2017). Unlike CD4+ effector T cells, T reg cells mainly employ oxidative phosphorylation (OXPHOS) to support their bioenergetic needs, whereas elevated glycolytic activity leads to T reg cell instability and dysfunction (Newton et al., 2016; Yu et al., 2018; Zeng and Chi, 2017). However, T reg cells lacking the glycolytic pathway are functionally suppressive but fail to migrate to inflamed tissue (Alon, 2017; Kishore et al., 2017). Thus, the molecular mechanisms balancing OXPHOS and glycolysis are important in maintaining T reg cell functional features.
Macroautophagy (herein referred to as “autophagy”), which involves the degradation and recycling of intracellular substrates in the lysosome, has emerged as a critical process in T cell metabolism and immune responses (Galluzzi et al., 2014; Saravia et al., 2020; Xu et al., 2014). Autophagy is induced after TCR and cytokine stimulation to promote conventional T cell proliferation and survival (Hubbard et al., 2010; Li et al., 2006; Pua et al., 2007). Actively regulated autophagy in T reg cells restrains excessive apoptotic and metabolic activities by inhibiting mechanistic target of rapamycin complex 1 (mTORC1) activity and c-Myc expression (Wei et al., 2016). Consequently, T reg cell–specific deletion of Atg5 and Atg7, two crucial genes in autophagy, causes disturbed immune homeostasis, inflammatory disorder development, and increased tumor resistance in mice (Wei et al., 2016). Consistently, T cell–specific ablation of Pik3c3 or Atg16l1, other essential genes in autophagy, leads to the defective homeostasis and functions in T reg cells (Kabat et al., 2016; Parekh et al., 2013). However, how autophagy is activated in T reg cells remains unclear.
Zinc finger protein 91 (ZFP91) has been identified as an E3 ubiquitin ligase that ubiquitinates NF-κB–inducing kinase (NIK), FOXA1, heterogeneous nuclear ribonucleoprotein (hnRNP) A1, and hnRNP F (Chen et al., 2020; Jin et al., 2010; Tang et al., 2020; Xu et al., 2018). ZFP91 contains five zinc finger domains, one coiled-coil structure, one leucine zipper pattern, and several nuclear localization signals and participates in tumor growth and metastasis (Jin et al., 2010; Unoki et al., 2003; Xu et al., 2018). Blocking ZFP91-mediated hnRNP F degradation accelerates cancer metastasis (Xu et al., 2018). ZFP91 suppresses tumor cell apoptosis and promotes tumor growth in acute myelogenous leukemia (Unoki et al., 2003). A recent study shows that ZFP91 positively regulates the activation of MAPK signaling and the noncanonical caspase-8 inflammasome to control active IL-1β production in macrophages (Mi et al., 2018). Of note, the physiological function of ZFP91 in T cell immunity, especially in T reg cell–mediated immune tolerance, has not been clarified.
In this study, we demonstrated that ZFP91 promoted the TCR-initiated autophagosome maturation, allowing autophagy to restrict mTORC1-mediated hyperglycolysis and maintain T reg cell homeostasis and function. T reg cell–specific deletion of Zfp91 resulted in T reg cell dysfunction and aggravated disease severity in chemically induced colitis and colitis-associated cancer (CAC) models. Mechanistically, ZFP91 rapidly translocated from the nucleus to the cytoplasm in response to TCR stimulation and then mediated BECN1 ubiquitination and BECN1–PIK3C3 complex formation, thereby facilitating TCR-induced autophagy and associated downstream signaling. These findings identify ZFP91 as a pivotal regulator of autophagy-mediated T reg cell metabolic and functional fitness and suggest that enhancing ZFP91 activity in T reg cells may be a valuable strategy for the prevention and treatment of colonic inflammation and inflammation-driven colorectal cancer.
Results
ZFP91 is indispensable for T reg cell function in vivo
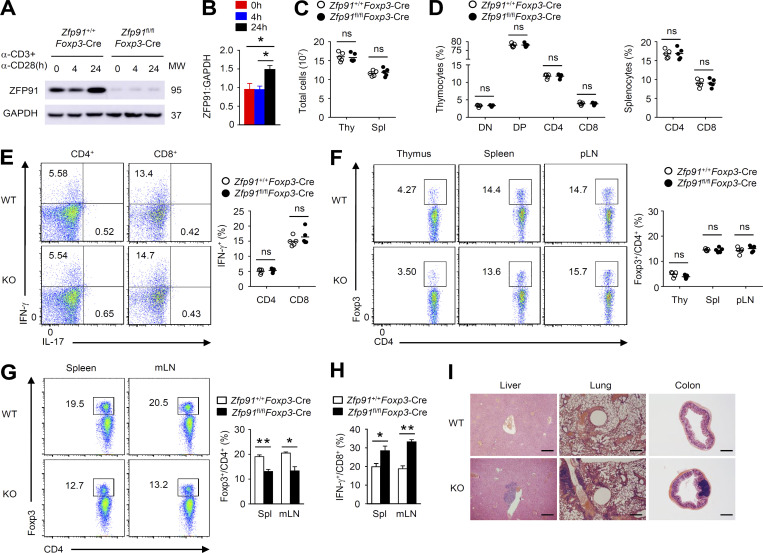
To investigate the physiological function of ZFP91 in T reg cells, we crossed Zfp91-flox mice with Foxp3-YFP-Cre mice to generate Zfp91 T reg cell–conditional KO mice (Zfp91fl/flFoxp3-Cre). Immunoblot (IB) analysis revealed that ZFP91 was greatly accumulated in WT T reg cells stimulated with anti-CD3 and anti-CD28 for 24 h (Fig. 1, A and B). Efficient deletion of ZFP91 in resting or activated Zfp91fl/flFoxp3-Cre T reg cells was confirmed (Fig. 1 A). T reg cell–specific deletion of Zfp91 did not affect T cell development or peripheral T cell homeostasis in 6-wk-old mice (Fig. 1, C–E). In addition, the T reg cell percentages in the thymus, spleen (Spl), and peripheral LNs of 6-wk-old Zfp91+/+Foxp3-Cre mice and Zfp91fl/flFoxp3-Cre mice were comparable (Fig. 1 F). However, low T reg cell percentages and elevated IFN-γ–producing CD8+ effector T cell proportions in the Spl and mesenteric LNs (mLNs) were observed in 10-mo-old Zfp91fl/flFoxp3-Cre mice (Fig. 1, G and H). Compared with Zfp91+/+Foxp3-Cre mice, Zfp91fl/flFoxp3-Cre mice at 10 mo of age displayed severe autoimmune symptoms with lymphocytic infiltration in many nonlymphoid organs (Fig. 1 I). These results suggest that ZFP91 is required for T reg cell function in vivo.
Figure 1.
ZFP91 is indispensable for T reg cell function in vivo. (A) IB analysis of ZFP91 in whole-cell lysates of Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre CD4+CD25+YFP+ T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 24 h. MW, molecular weight. (B) Quantifications of ZFP91:GAPDH levels in WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 24 h are shown as the mean ± SEM of three experiments. (C) Total cell numbers in the thymus (Thy) and Spl from 6-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 5 mice for each group). (D) Flow cytometric analysis of the percentage of CD4–CD8– (DN), CD4+CD8+ (DP), CD4+CD8– (CD4), and CD4–CD8+ (CD8) in the thymus and CD4+ and CD8+ T cells in the Spl of 6-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 5 mice for each group). (E) Flow cytometric analysis of the percentage of IFN-γ–producing CD4+ and CD8+ T cells in the Spl from 6-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 5 mice for each group). (F) Flow cytometric analysis of the percentage of Foxp3+ T reg cells in the thymus, Spl, and peripheral LNs (pLN) from 6-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 5 mice for each group). (G) Flow cytometric analysis of the percentage of Foxp3+ T reg cells in the Spl and mLNs from 10-mo-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 3 mice for each group). (H) Flow cytometric analysis of the percentage of IFN-γ–producing CD8+ T cells in the Spl and mLNs from 10-mo-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (n = 3 mice for each group). (I) Hematoxylin and eosin staining of the indicated tissue sections of 10-mo-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice. Scale bars, 500 µm. Data are representative of three independent experiments. Data are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test.
ZFP91 sustains T reg cell transcriptional programs
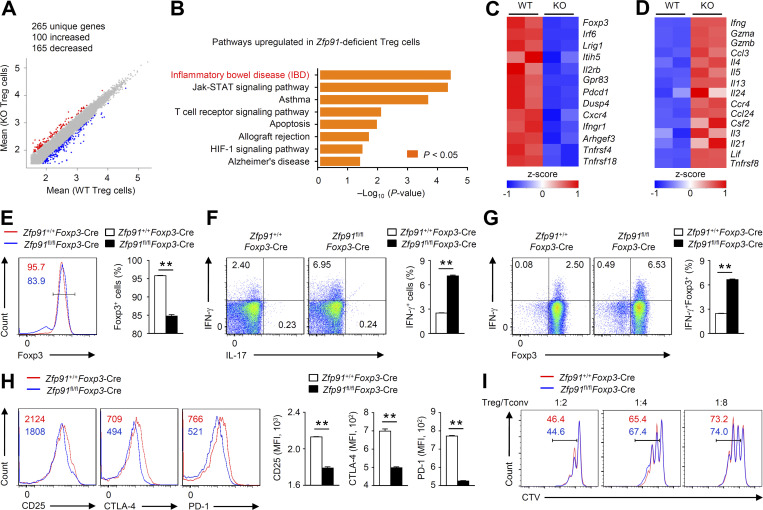
To determine whether ZFP91 contributes to T reg cell transcriptional programs, we performed transcriptomic analysis using Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h in the presence of IL-2. Compared with Zfp91+/+Foxp3-Cre T reg cells, Zfp91fl/flFoxp3-Cre T reg cells showed up-regulation of 100 genes and down-regulation of 165 genes (fold change >2 and adjusted P value <0.01; Fig. 2 A). Functional pathway enrichment analysis revealed a significant up-regulation of autoimmune disease–associated gene expression in ZFP91-deficient T reg cells (Fig. 2 B). Moreover, ZFP91-deficient T reg cells failed to express T reg cell signature genes, such as Foxp3, Pdcd1, and Tnfrsf18 (Fig. 2 C). In contrast, Zfp91fl/flFoxp3-Cre T reg cells exhibited the ability to express genes associated with effector T cell differentiation, including Ifng, Il4, Il13, and Il21 (Fig. 2 D). Consistently, Zfp91fl/flFoxp3-Cre T reg cells activated in vitro showed much lower Foxp3 expression (Fig. 2 E) and higher IFN-γ effector cytokine expression (Fig. 2, F and G). In addition, Zfp91fl/flFoxp3-Cre Foxp3+ T reg cells had lower expression of several T reg cell markers, including CD25, CTLA-4, and PD-1 (Fig. 2 H). In vitro assays of T reg cell function showed that WT and ZFP91-deficient T reg cells had a similar ability to suppress effector T cell proliferation (Fig. 2 I). These results indicate a crucial role for ZFP91 in sustaining T reg cell transcriptional programs.
Figure 2.
ZFP91 sustains T reg cell transcriptional programs. (A) Scatter plot comparing global gene expression profiles of Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of 200 U/ml IL-2 for 24 h. Transcripts with a log2 (fold change) >1 and adjusted P value <0.01 in Zfp91fl/flFoxp3-Cre (KO) T reg cells are shown in red (increased expression) or blue (decreased expression). (B) Functional enrichment analysis of Kyoto Encyclopedia of Genes and Genomes pathways up-regulated in ZFP91-deficient T reg cells compared with WT T reg cells. (C and D) Clustering of down-regulated (C) or up-regulated (D) genes identified in ZFP91-deficient T reg cells relative to that in WT T reg cells. (E–G) Flow cytometric analysis of Foxp3 expression (E) or cytokine expression (F and G) in Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of 200 U/ml IL-2 for 24 h (n = 4 mice for each group). (H) Flow cytometric analysis of CD25, CTLA-4, and PD-1 expression levels in Foxp3+ T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of 200 U/ml IL-2 for 24 h (n = 3 mice for each group). (I) Zfp91+/+Foxp3-Cre or Zfp91fl/flFoxp3-Cre T reg cells were cocultured with naive CD4+ T (Tconv) cells labeled with CTV at the indicated ratios and were activated with Dynabeads Mouse T-activator CD3/CD28. CTV signal was assessed after 3 d. Representative data are shown from two (H) and three (E–G and I) independent experiments. Data are presented as mean ± SEM. **, P < 0.01. Two-tailed Student’s t test. MFI, mean fluorescence intensity.
Zfp91fl/flFoxp3-Cre mice are susceptible to experimental colitis and CAC
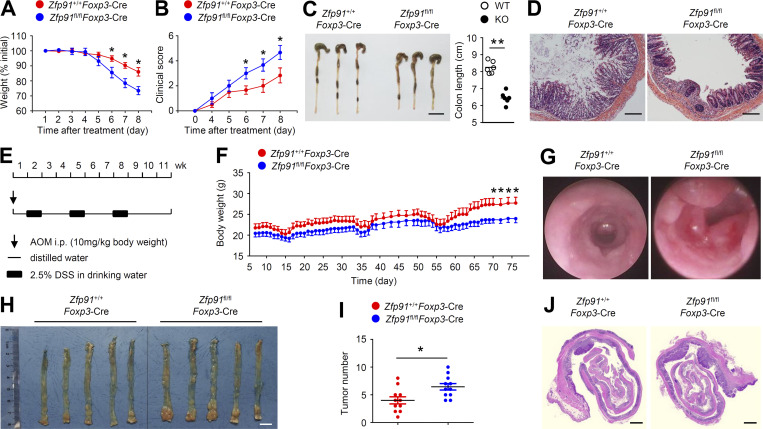
Transcriptomic analysis revealed that the inflammatory bowel disease (IBD) pathway was the most significantly up-regulated pathway in ZFP91-deficient T reg cells (Fig. 2 B). To verify these results, we evaluated the colonic inflammatory response using a dextran sodium sulfate (DSS)–induced acute colitis model. Zfp91fl/flFoxp3-Cre mice showed rapid body weight loss and increased clinical scores after DSS treatment (Fig. 3, A and B). Moreover, DSS-treated Zfp91fl/flFoxp3-Cre mice displayed exacerbated colon length shortening (Fig. 3 C). Histological analysis revealed increased infiltration of inflammatory cells into the colon of DSS-treated Zfp91fl/flFoxp3-Cre mice (Fig. 3 D). These data suggest that the loss of ZFP91 in T reg cells aggravates DSS-induced colitis.
Figure 3.
Loss of ZFP91 in T reg cells aggravates experimental colitis and CAC. (A–D) 6–8-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice were provided with drinking water containing 3% DSS for 7 d, followed by regular water. Body weight loss (A; n = 5 mice for each group) and clinical score (B; n = 6 mice for each group) of Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice after induction of colitis using DSS. Mice were killed at day 9. They were analyzed for colon length (C) and hematoxylin and eosin staining of colon section (D). Scale bars, 2 cm (C) and 100 µm (D). (E–J) 6–8-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice received a single intraperitoneal injection of AOM followed by three cycles of 2.5% DSS exposure. Schematic of AOM/DSS-induced CAC model (E). Body weight of Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice after induction of CAC using AOM/DSS (F; n = 8 mice for each group). Statistical analysis was performed using the data of body weight from Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice at day 70, 72, 74, or 76. Mice were killed at day 80. They were analyzed for images of miniendoscopic colon (G), macroscopic polyps in the colon (H), tumor numbers in the whole colon (I), and hematoxylin and eosin staining of colon section (J). Scale bars, 1 cm (H) and 1,000 µm (J). Data are representative of three independent experiments and are presented as mean ± SEM. *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test.
Because IBD is an important factor in the induction of CAC (Lasry et al., 2016; West et al., 2015), we next explored the possible roles of T reg cell–derived ZFP91 in the initiation and progression of CAC. An inflammation-driven CAC model was induced with azoxymethane (AOM) plus three cycles of 2.5% DSS (Fig. 3 E). Over the entire course of the CAC model, Zfp91fl/flFoxp3-Cre mice showed significantly exacerbated weight loss (Fig. 3 F). In addition, Zfp91fl/flFoxp3-Cre mice had significantly higher tumor numbers in the colon than WT mice (Fig. 3, G–J). Collectively, these results indicate critical roles for T reg cell–derived ZFP91 in inhibiting experimental colitis and colitis-associated tumorigenesis.
ZFP91 maintains T reg cell homeostasis and function in vivo
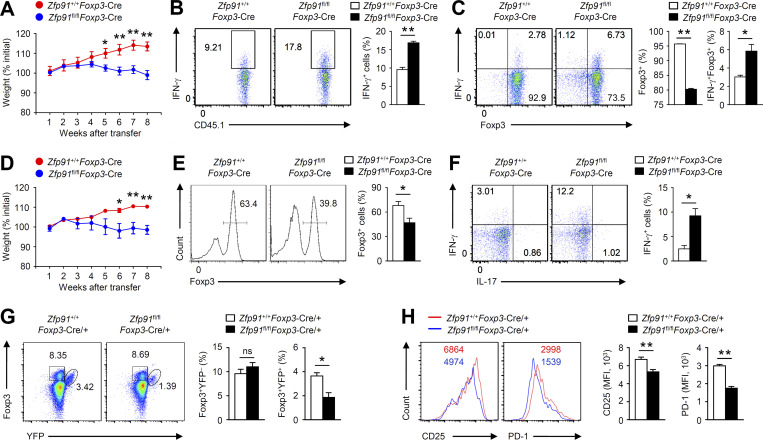
To examine the in vivo function of ZFP91-deficient T reg cells, we transferred WT or ZFP91-deficient T reg cells together with naive CD45RBhiCD4+ T cells into RAG-1–deficient mice. The transfer of ZFP91-deficient T reg cells along with naive CD45RBhiCD4+ T cells resulted in gradual weight loss and a greater frequency of IFN-γ–producing effector CD4+ T cells (Fig. 4, A and B), but the transfer of WT T reg cells along with naive CD45RBhiCD4+ T cells did not (Fig. 4, A and B), suggesting considerable attenuation of the suppressive activity of ZFP91-deficient T reg cells in vivo. Importantly, the fraction of Foxp3-positive cells from mice transferred with ZFP91-deficient T reg cells and naive CD45RBhiCD4+ T cells was significantly decreased compared with those from mice transferred with WT T reg cells and naive CD45RBhiCD4+ T cells (Fig. 4 C). In addition, transferred ZFP91-deficient T reg cells produced higher amounts of IFN-γ (Fig. 4 C). We further investigated the fate of T reg cells in lymphopenic conditions (Takahashi et al., 2011). WT or ZFP91-deficient T reg cells were transferred into RAG-1–deficient mice. Unlike WT T reg cells, ZFP91-deficient T reg cells caused gradual weight loss (Fig. 4 D). Notably, ZFP91-deficient T reg cells exhibited a more profound loss of Foxp3 expression than did WT T reg cells (Fig. 4 E). Moreover, the Foxp3-maintaining fraction of ZFP91-deficient T reg cells produced higher amounts of IFN-γ (Fig. 4 F). These results indicate that ZFP91-deficient T reg cells tend to lose Foxp3 expression and produce effector cytokine IFN-γ, thus suggesting that ZFP91 is necessary for T reg cell homeostasis in vivo.
Figure 4.
ZFP91 maintains T reg cell homeostasis and function in vivo. (A–C) 6-wk-old RAG-1–deficient mice were given adoptive transfer of WT (CD45.1+) naive CD45RBhi CD4+ T cells together with T reg cells isolated from 6-wk-old Zfp91+/+Foxp3-Cre (WT) or Zfp91fl/flFoxp3-Cre (KO) mice. Body weight is presented relative to initial weight (A; Zfp91+/+Foxp3-Cre mice: n = 5; Zfp91fl/flFoxp3-Cre mice: n = 6). Frequency of IFN-γ–producing effector CD4+ T cells in the mLNs by gating on CD45.1+ cells (B; n = 4 mice for each group). Flow cytometric analysis of Foxp3 expression or IFN-γ expression in CD45.2+ T reg cells from the mLNs (C; n = 4 mice for each group). (D–F) 6-wk-old RAG-1–deficient mice were given adoptive transfer of T reg cells isolated from 6-wk-old Zfp91+/+Foxp3-Cre (WT) or Zfp91fl/flFoxp3-Cre (KO) mice. Body weight is presented relative to initial weight (D; n = 6 mice for each group). Flow cytometric analysis of Foxp3 expression in CD45.2+ T reg cells from the mLNs (E; Zfp91+/+Foxp3-Cre mice: n = 3; Zfp91fl/flFoxp3-Cre mice: n = 4). Flow cytometric analysis of effector cytokine expression in CD45.2+Foxp3+ T reg cells from the mLNs (F; Zfp91+/+Foxp3-Cre mice: n = 3; Zfp91fl/flFoxp3-Cre mice: n = 4). (G) Flow cytometric analysis of the percentages of Foxp3+YFP+ and Foxp3+YFP– T reg cells in the Spl from 4-mo-old Zfp91+/+Foxp3-Cre/+ female mice and Zfp91fl/flFoxp3-Cre/+ female mice (n = 4 mice for each group). (H) Flow cytometric analysis of CD25 and PD-1 expressions in splenic Foxp3+YFP+ T reg cells from 4-mo-old Zfp91+/+Foxp3-Cre/+ female mice and Zfp91fl/flFoxp3-Cre/+ female mice (n = 4 mice). Representative data are shown from two (G and H) and three (A–F) independent experiments. Data are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test. MFI, mean fluorescence intensity.
To determine whether ZFP91 regulates T reg cell homeostasis in an intrinsic manner, we took advantage of Zfp91fl/flFoxp3-Cre/+ female mice, in which around half of the T reg cells are ZFP91 sufficient, while the other half of the T reg cells are ZFP91 deficient due to the random X chromosome inactivation (Ren et al., 2019). The Zfp91+/+Foxp3-Cre/+ female mice and Zfp91fl/flFoxp3-Cre/+ female mice had similar proportions of Foxp3+YFP– T reg cells (Fig. 4 G). In contrast, the percentage of Foxp3+YFP+ T reg cells in Zfp91fl/flFoxp3-Cre/+ female mice was lower than that in Zfp91+/+Foxp3-Cre/+ female mice (Fig. 4 G). In addition, Foxp3+YFP+ T reg cells from Zfp91fl/flFoxp3-Cre/+ female mice had lower expressions of CD25 and PD-1 than did Foxp3+YFP+ T reg cells from Zfp91+/+Foxp3-Cre/+ female mice (Fig. 4 H). Therefore, these data suggest that ZFP91 is required for T reg cell homeostasis, thereby maintaining T reg cell suppressive function in vivo.
ZFP91 restricts excessive c-Myc activity and hyperglycolysis in T reg cells
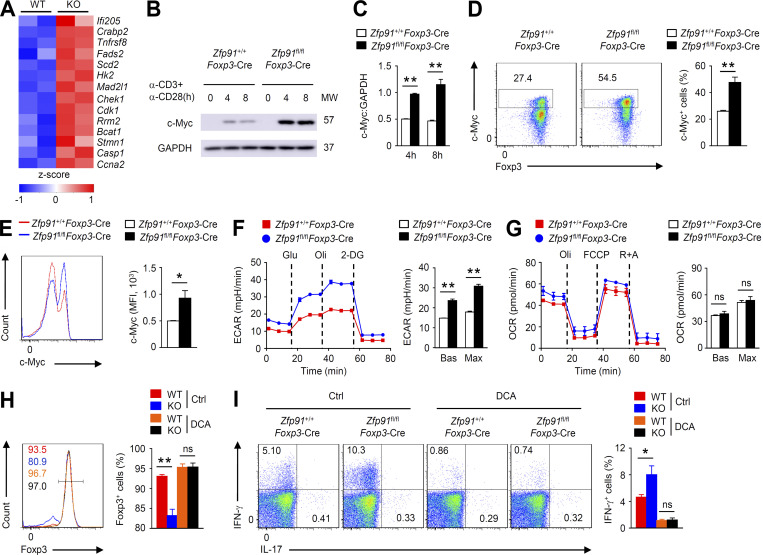
The maintenance of T reg cell homeostasis and function is tightly controlled by cellular metabolism (Wei et al., 2016; Yu et al., 2018). Interestingly, the expression of c-Myc–associated genes, which is associated with T reg cell hyperglycolytic metabolism (Wei et al., 2016), was up-regulated in ZFP91-deficient T reg cells (Fig. 5 A). In addition, TCR- and CD28-induced c-Myc expression in Zfp91fl/flFoxp3-Cre T reg cells was remarkably augmented (Fig. 5, B–E). The aberrant c-Myc activity in ZFP91-deficient T reg cells prompted us to measure extracellular acidification rate (ECAR) and mitochondrial oxygen consumption rate (OCR), which reflect glycolysis and OXPHOS, respectively. Interestingly, ZFP91-deficient T reg cells had significantly higher baseline and maximum glycolytic rates than WT T reg cells (Fig. 5 F), although the Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells had similar baseline and maximum OXPHOS rates (Fig. 5 G). To confirm the functional importance of hyperglycolysis in ZFP91-deficient T reg cell homeostasis, we treated T reg cells with dichloroacetic acid (DCA), which shifts glycolysis toward OXPHOS (Gerriets et al., 2015). As expected, DCA treatment efficiently restored Foxp3 expression but diminished IFN-γ production in Zfp91fl/flFoxp3-Cre T reg cells (Fig. 5, H and I). Therefore, ZFP91 restricts excessive c-Myc activity and hyperglycolysis in T reg cells.
Figure 5.
ZFP91 restricts excessive c-Myc activity and hyperglycolysis. (A) A heatmap of up-regulated genes associated with c-Myc in ZFP91-deficient (KO) T reg cells relative to that in WT T reg cells. (B) IB analysis of c-Myc in Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 8 h. (C) Quantifications of c-Myc:GAPDH levels in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 or 8 h are shown as the mean ± SEM of three experiments. (D and E) Flow cytometric analysis of c-Myc expression and Foxp3 expression in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 h (n = 4 mice for each group). (F) ECAR of Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 3 h under basal conditions (Bas) or at maximum (Max) with the addition of glucose (Glu), oligomycin (Oli), and 2-deoxy-D-glucose (2-DG). ECAR is reported in milli-pH units (mpH) per minute. (G) Extracellular flux analysis of the OCRs of Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 3 h with the addition of oligomycin (Oli), the mitochondrial uncoupler FCCP, and rotenone plus antimycin A (R+A). (H and I) Flow cytometric analysis of Foxp3 expression (H) or cytokine expression (I) in Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells activated in vitro with anti-CD3, anti-CD28, and IL-2 for 24 h in the presence of DMSO (Ctrl) or DCA (n = 4 mice for each group). The data shown are representative of three independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test. MFI, mean fluorescence intensity; MW, molecular weight.
ZFP91 restrains mTORC1 signaling in T reg cells
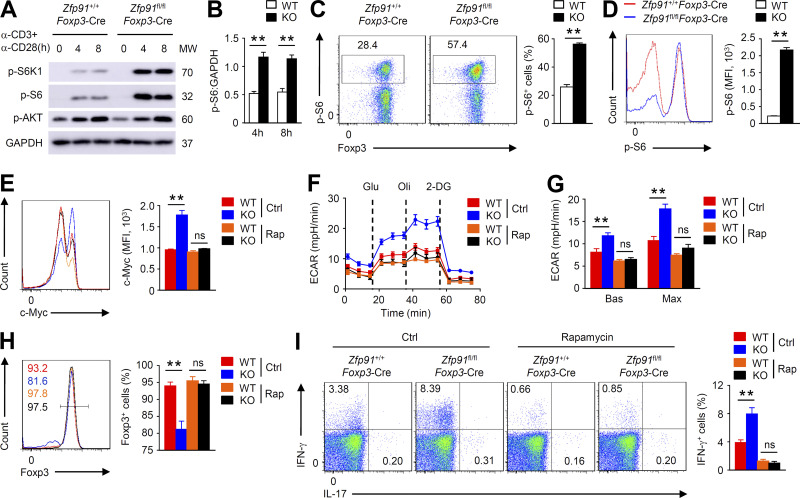
Since mTOR signaling mediates glycolytic metabolism by regulating the cellular level of c-Myc (Masui et al., 2013; Wei et al., 2016), we sought to determine whether ZFP91 regulates mTOR activity in T reg cells. IB and flow cytometric analyses revealed increased phosphorylation of the ribosomal protein S6 kinase β-1 and ribosomal protein S6, which is indicative of mTORC1 activation, in ZFP91-deficient T reg cells stimulated with anti-CD3 and anti-CD28 antibodies (Fig. 6, A–D). In contrast, the mTORC2 activity (monitored by measuring the phosphorylation of serine/threonine protein kinase AKT at Ser473) in ZFP91-deficient T reg cells was unaltered (Fig. 6 A). To validate the contribution of this elevated mTORC1 activity to the ZFP91-deficient T reg cell phenotypes observed, we stimulated T reg cells with anti-CD3 and anti-CD28 antibodies in the presence or absence of the mTORC1 inhibitor rapamycin. Rapamycin-treated ZFP91-deficient T reg cells showed a substantial decrease in c-Myc expression (Fig. 6 E). Importantly, rapamycin treatment considerably lowered the ECAR and erased the differences between ZFP91-deficient and ZFP91-sufficient T reg cells (Fig. 6, F and G), indicating that aberrant mTORC1 activity leads to ZFP91-deficient T reg cell hyperglycolysis. Moreover, rapamycin greatly restored Foxp3 expression (Fig. 6 H) and diminished IFN-γ production (Fig. 6 I) in ZFP91-deficient T reg cells. Thus, ZFP91 maintains T reg cell metabolic programs by restraining mTORC1 signaling.
Figure 6.
ZFP91 restrains mTORC1 signaling in T reg cells. (A) IB analysis of the indicated proteins in Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with antibodies against CD3 and CD28. (B) Quantifications of p-S6:GAPDH levels in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 or 8 h are shown as the mean ± SEM of three experiments. (C and D) Flow cytometric analysis of p-S6 expression and Foxp3 expression in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 h (n = 4 mice for each group). (E) Flow cytometric analysis of c-Myc expression in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of DMSO (Ctrl) or rapamycin (Rap) for 4 h (n = 4 mice for each group). (F and G) ECAR of Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of DMSO (Ctrl) or rapamycin (Rap) for 3 h. (H and I) Flow cytometry analyzing Foxp3 expression (H) or cytokine expression (I) in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells activated in vitro with anti-CD3, anti-CD28, and IL-2 for 24 h in the presence of DMSO (Ctrl) or rapamycin (Rap; n = 4 mice for each group). The data shown are representative of three independent experiments and are presented as mean ± SEM. ns, not statistically significant; **, P < 0.01. Two-tailed Student’s t test. 2-DG, 2-deoxy-D-glucose; MFI, mean fluorescence intensity; MW, molecular weight. mph, milli-pH units.
ZFP91 mediates BECN1 ubiquitination and autophagy induction in T reg cells
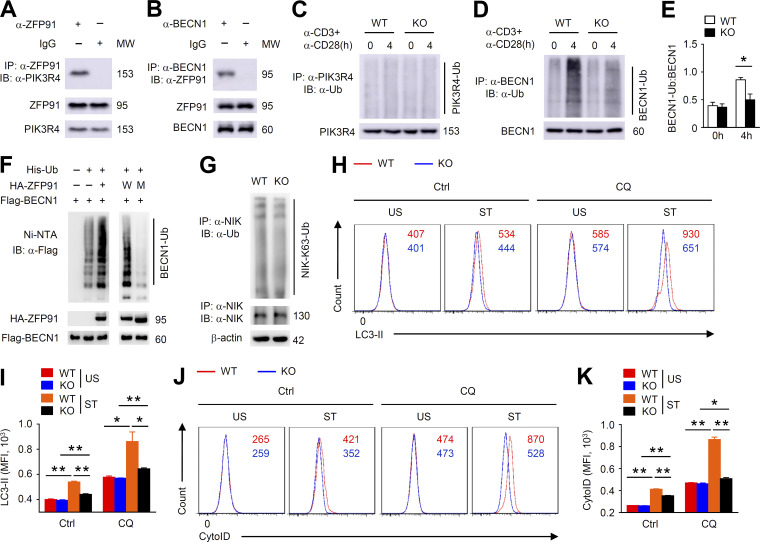
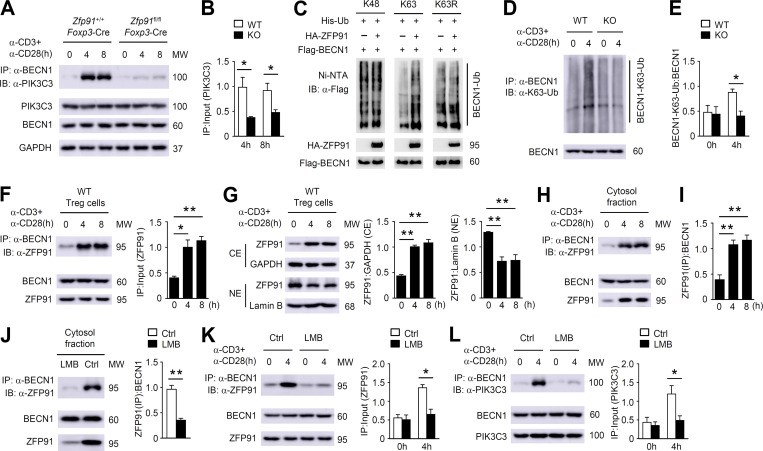
Autophagy can protect the maintenance of T reg cell homeostasis and function by restricting TCR-dependent mTORC1 signaling (Wei et al., 2016). Mass spectrometry analysis has indicated that one of the ZFP91-binding proteins is PIK3R4, an essential component of the autophagosome initiation complex including BECN1, PIK3C3, PIK3R4, and AMBRA1 (Hurley and Young, 2017). We confirmed the endogenous ZFP91–PIK3R4 interaction with coimmunoprecipitation (co-IP) assays (Fig. 7 A). Consistent with the ZFP91–PIK3R4 association, ZFP91 also interacted with BECN1 in T reg cells (Fig. 7 B). A ubiquitination assay revealed that loss of ZFP91 did not alter the ubiquitination status of PIK3R4 (Fig. 7 C). Interestingly, we observed abundant ubiquitination of BECN1 in activated WT T reg cells but not in activated ZFP91-deficient T reg cells (Fig. 7, D and E). Indeed, overexpression of ZFP91 induced strong ubiquitination of BECN1 in HEK293T cells (Fig. 7 F). ZFP91 has five consecutive zinc finger (ZnF) domains, and ZnF domain 2 of ZFP91 is critical for its E3 ligase activity (Jin et al., 2010). To examine whether ZnF domain 2 is required for ZFP91-mediated BECN1 ubiquitination, we constructed the vector encoding WT ZFP91 and mutant ZFP91 lacking ZnF domain 2. When the ZnF domain 2 deletion of ZFP91 was expressed with BECN1, BECN1 ubiquitination was almost lost (Fig. 7 F), indicating that ZFP91-mediated BECN1 ubiquitination relies on its ZnF domain 2. Although ZFP91 mediates K63-linked ubiquitination of NIK in HEK293 cells (Jin et al., 2010), loss of ZFP91 did not alter the K63-linked ubiquitination status of NIK in the activated T reg cells (Fig. 7 G). We next monitored autophagic flux in T reg cells using flow cytometry by quantifying LC3-II mean fluorescence intensity (Puleston et al., 2014). WT T reg cells exhibited increased autophagosome formation after anti-CD3 and anti-CD28 stimulation (Fig. 7, H and I), as measured by intracellular membrane-bound LC3-II staining in the presence or absence of a lysosomal inhibitor chloroquine. However, autophagy induction in stimulated ZFP91-deficient T reg cells was greatly inhibited (Fig. 7, H and I). Furthermore, autophagy levels (CytoID) were significantly decreased in ZFP91-deficient T reg cells compared with WT T reg cells stimulated with anti-CD3 and anti-CD28 in the presence or absence of chloroquine (Fig. 7, J and K), suggesting a crucial role for ZFP91 in regulating TCR-induced autophagy. Thus, ZFP91 mediates BECN1 ubiquitination and autophagy induction in the activated T reg cells.
Figure 7.
ZFP91 mediates BECN1 ubiquitination and autophagy induction in T reg cells. (A and B) Lysates from WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 h were subjected to IP using an antibody against ZFP91 (A) or BECN1 (B). (C) IB analysis of PIK3R4 ubiquitination in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with antibodies against CD3 and CD28. (D) IB analysis of BECN1 ubiquitination in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with antibodies against CD3 and CD28. (E) Quantification of BECN1-Ub:BECN1 levels in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0 or 4 h are shown as the mean ± SEM of three experiments. (F) IB analysis of BECN1 ubiquitination using HEK293T cells transfected with the indicated expression vectors. W, WT ZFP91. M, mutant ZFP91 lacking the ZnF domain 2. (G) IB analysis of NIK K63 ubiquitination in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with antibodies against CD3 and CD28 for 4 h. (H and I) Flow cytometry analyzing LC3-II level in CD4+CD25+ T reg cells from Zfp91+/+Cd4-Cre (WT) and Zfp91fl/flCd4-Cre (KO) mice after overnight stimulation with anti-CD3 and anti-CD28 stimulation. Where indicated, a lysosomal inhibitor chloroquine was added (CQ) or not added (Ctrl) during the last 4 h. US, unstimulated with anti-CD3 and anti-CD28; ST, stimulated with anti-CD3 and anti-CD28 (n = 3 mice for each group). (J and K) Flow cytometry analyzing CytoID level in CD4+CD25+ T reg cells from Zfp91+/+Cd4-Cre (WT) and Zfp91fl/flCd4-Cre (KO) mice after overnight stimulation with anti-CD3 and anti-CD28 stimulation. Chloroquine was added (CQ) or not added (Ctrl) during the last 4 h. US, unstimulated; ST, stimulated (n = 3 mice for each group). Representative data are shown from two (G–K) and three (A–F) independent experiments. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test. HA, hemagglutinin; MFI, mean fluorescence intensity; MW, molecular weight; Ni-NTA, Ni–nitrilotriacetic acid.
ZFP91 promotes autophagy-mediated T reg cell metabolism and homeostasis
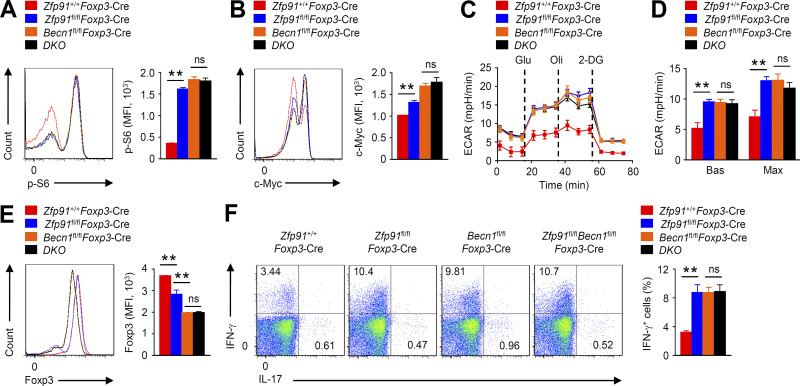
To confirm the functional significance of BECN1 activity in the ZFP91-mediated T reg cell homeostasis, we crossed Zfp91fl/flFoxp3-Cre mice with Becn1fl/fl mice to generate Zfp91fl/flBecn1fl/flFoxp3-Cre mice. Compared with WT T reg cells, BECN1-deficient T reg cells showed markedly enhanced mTORC1 activity and c-Myc expression (Fig. 8, A and B). Moreover, BECN1-deficient T reg cells had significantly higher baseline and maximum glycolytic rates than WT T reg cells (Fig. 8, C and D). In addition, BECN1-deficient T reg cells showed much lower Foxp3 expression but higher IFN-γ expression (Fig. 8, E and F). Importantly, BECN1 deficiency distinctly erased the differences of mTORC1 activity, c-Myc expression, glycolytic rates, Foxp3 expression, and IFN-γ production between ZFP91-deficient and ZFP91-sufficient T reg cells (Fig. 8, A–F). Therefore, ZFP91 acts in a BECN1-dependent manner to promote autophagy-mediated T reg cell metabolism and homeostasis.
Figure 8.
ZFP91 promotes autophagy-mediated T reg cell metabolism and homeostasis. (A and B) Flow cytometric analysis of p-S6 (A; n = 3 mice for each group) and c-Myc (B; n = 4 mice for each group) expression in Zfp91+/+Foxp3-Cre, Becn1fl/flFoxp3-Cre, Zfp91fl/flFoxp3-Cre, and Zfp91fl/flBecn1fl/flFoxp3-Cre (DKO) T reg cells stimulated for 4 h with antibodies against CD3 and CD28. (C and D) ECAR of Zfp91+/+Foxp3-Cre, Becn1fl/flFoxp3-Cre, Zfp91fl/flFoxp3-Cre, and Zfp91fl/flBecn1fl/flFoxp3-Cre (DKO) T reg cells stimulated for 3 h with antibodies against CD3 and CD28. (E and F) Flow cytometry analyzing Foxp3 expression (E) or cytokine expression (F) in Zfp91+/+Foxp3-Cre, Becn1fl/flFoxp3-Cre, Zfp91fl/flFoxp3-Cre, and Zfp91fl/flBecn1fl/flFoxp3-Cre (DKO) T reg cells activated in vitro with anti-CD3, anti-CD28, and IL-2 for 24 h (n = 4 mice for each group). Data are representative of three independent experiments and are presented as mean ± SEM. ns, not statistically significant; **, P < 0.01. Two-tailed Student’s t test. 2-DG, 2-deoxy-D-glucose; MFI, mean fluorescence intensity. mph, milli-pH units.
T reg cell–derived ZFP91 facilitates TCR-induced BECN1–PIK3C3 complex assembly
Ubiquitin chains are essential for mediating protein degradation and protein–protein interactions (Popovic et al., 2014; Swatek and Komander, 2016). Our IB analyses revealed that WT and ZFP91-deficient T reg cells exhibited similar levels of the BECN1 and PIK3C3 proteins (Fig. 9 A), but the amount of BECN1-associated PIK3C3 protein was dramatically decreased in ZFP91-deficient T reg cells (Fig. 9, A and B), suggesting that ZFP91-mediated BECN1 ubiquitination is involved in the association of BECN1 with PIK3C3. In addition, overexpression of ZFP91 with K63-ubiquitin (K63), but not with K48-ubiquitin (K48) or K63R-ubiquitin (K63R), induced strong ubiquitination of BECN1 in HEK293T cells (Fig. 9 C). Consistently, abundant K63-linked ubiquitination (K63-Ub) of BECN1 was observed in activated WT T reg cells but not in activated ZFP91-deficient T reg cells (Fig. 9, D and E). These data indicate that T reg cell–derived ZFP91 mediates BECN1 K63–linked ubiquitination to facilitate the assembly of the BECN1–PIK3C3 complex.
Figure 9.
ZFP91 facilitates TCR-induced BECN1–PIK3C3 complex assembly. (A) Lysates from Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 8 h were subjected to IP using an antibody against BECN1. (B) Quantifications of immunoprecipitated PIK3C3:PIK3C3 levels in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 or 8 h are shown as the mean ± SEM of three experiments. (C) IB analysis of BECN1 ubiquitination using HEK293T cells transfected with the indicated expression vectors. (D) IB analysis of BECN1 K63–linked ubiquitination in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with antibodies against CD3 and CD28. (E) Quantifications of BECN1-K63-Ub:BECN1 levels in Zfp91+/+Foxp3-Cre (WT) and Zfp91fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0 or 4 h are shown as the mean ± SEM of three experiments. (F) Lysates from WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies were subjected to IP using an antibody against BECN1. Quantifications of immunoprecipitated ZFP91:ZFP91 levels are shown as the mean ± SEM of three experiments. (G) IB analysis of the indicated proteins in nuclear (NE) and cytoplasmic (CE) extracts of WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 8 h. Quantifications of ZFP91:GAPDH levels and ZFP91:lamin B levels are shown as the mean ± SEM of three experiments. (H) IB and co-IP assays using the cytosol fractions of WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 8 h. (I) Quantifications of immunoprecipitated ZFP91:BECN1 levels in the cytosol fractions of WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 0, 4, or 8 h are shown as the mean ± SEM of three experiments. (J) IB and co-IP assays using the cytosol fractions of WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of LMB for 4 h. Ctrl, control. Quantifications of immunoprecipitated ZFP91:BECN1 levels are shown as the mean ± SEM of three experiments. (K and L) Lysates from WT T reg cells stimulated with anti-CD3 and anti-CD28 antibodies in the presence of LMB for 4 h were subjected to IP using an antibody against BECN1. Quantifications of immunoprecipitated ZFP91:ZFP91 levels (K) and immunoprecipitated PIK3C3:PIK3C3 levels (L) are shown as the mean ± SEM of three experiments. Data are representative of three independent experiments. Data are presented as mean ± SEM. *, P < 0.05; **, P < 0.01. Two-tailed Student’s t test. MW, molecular weight.
To clarify the mechanism underlying the ZFP91-dependent, TCR-induced BECN1–PIK3C3 interaction, we detected the expression and subcellular localization of ZFP91 in WT T reg cells upon TCR stimulation. Although the protein level of ZFP91 was not appreciably affected, ZFP91 rapidly bound BECN1 in response to TCR stimulation (Fig. 9 F), suggesting an inducible interaction between ZFP91 and BECN1. Notably, subcellular fractionation assays revealed that TCR stimulation induced cytosolic localization of ZFP91 (Fig. 9 G). Importantly, the ZFP91–BECN1 interaction was associated with the TCR-induced translocation of ZFP91 to the cytoplasm (Fig. 9, H and I). Consistent with this, in response to treatment with leptomycin B (LMB), an inhibitor of nuclear export, ZFP91 failed to localize to the cytoplasm (Fig. 9 J), resulting in the impaired interactions between BECN1 and ZFP91 or PIK3C3 (Fig. 9, J–L). Collectively, these data suggest that the TCR-induced cytosolic translocation of ZFP91 facilitates BECN1 ubiquitination and BECN1–PIK3C3 complex formation, thereby promoting autophagy activation and controlling T reg cell homeostasis and function.
Discussion
Here, we show that ZFP91 facilitates TCR-dependent autophagosome formation to sustain T reg cell metabolic programming and functional integrity. ZFP91 deficiency attenuated the activation of autophagy and associated downstream pathways and impaired T reg cell homeostasis and function, thereby rendering mice sensitive to colonic inflammation and inflammation-driven colon carcinogenesis. TCR-triggered autophagosome maturation in T reg cells greatly relied on ZFP91-mediated BECN1 K63–linked ubiquitination and BECN1–PIK3C3 complex assembly. Our studies therefore establish ZFP91 as an important regulator of autophagy initiation in the maintenance of T reg cell homeostasis and function.
Autophagy has emerged as a key process that regulates T reg cell identity, and deletion of Atg16l1, Pik3c3, Atg5, or Atg7 specifically in T reg cells results in T reg cell instability and dysfunction (Kabat et al., 2016; Parekh et al., 2013; Wei et al., 2016). In line with these findings, we found that T reg cells from Becn1fl/flFoxp3-Cre mice had excessive mTORC1, c-Myc, and glycolytic activities; increased loss of Foxp3 expression; and acquisition of IFN-γ expression. Of note, the mechanisms governing autophagy activation in T reg cells remain unclear. Our results show that TCR stimulation triggers BECN1–PIK3C3 complex assembly to initiate autophagosome maturation and associated downstream signaling induction in T reg cells. Importantly, T reg cell–derived ZFP91 rapidly translocated from the nucleus to the cytoplasm in response to TCR stimulation, which promoted BECN1 K63–linked ubiquitination to strengthen the association between BECN1 and PIK3C3. On the basis of these data, we propose a model of TCR-dependent autophagy activation in T reg cells, in which TCR-induced ZFP91 cytosolic translocation facilitates autophagosome formation by accelerating BECN1 ubiquitination and the BECN1–PIK3C3 interaction.
Autophagy is completely abrogated in T reg cells lacking Atg7, resulting in a gradual loss of Foxp3 expression (Wei et al., 2016). In agreement, our current data show that T reg cells lacking Becn1 also showed a gradual loss of Foxp3 expression. Although autophagy levels were significantly decreased in ZFP91-deficient T reg cells, autophagy induction after TCR stimulation was severely but not completely blocked in ZFP91-deficient T reg cells, which is likely responsible for the complete loss of Foxp3 expression in a small fraction of T reg cells. However, ZFP91-deficient T reg cells lacking Becn1 further displayed a gradual loss of Foxp3 expression. These different phenomena are probably due to the different intensity of autophagy in the activated T reg cells. Therefore, the functional importance of different intensity of autophagy in mTORC1 activation and T reg cell homeostasis remains to be further studied.
The impact of autophagy on T reg cells in the setting of autoimmune diseases has been characterized (Kabat et al., 2016; Parekh et al., 2013; Wei et al., 2016). Selective deletion of Atg16l1 or Pik3c3 in T cells in mice causes chronic intestinal inflammation along with decreased T reg cell numbers (Kabat et al., 2016; Parekh et al., 2013). Atg16l1 or Atg7 ablation in T reg cells in mice leads to loss of T reg cells and development of autoimmune symptoms (Kabat et al., 2016; Wei et al., 2016). In our study, ZFP91 deficiency in T reg cells caused profound defects in autophagic flux, resulting in severe systemic inflammatory disorders. Consistently, our transcriptomic analysis revealed a significant up-regulation of IBD and asthma-associated gene expression in ZFP91-deficient T reg cells. Indeed, loss of ZFP91 in T reg cells aggravated DSS-induced colitis. Together, these findings suggest that activation of autophagy in T reg cells may be beneficial to autoimmune diseases, including colonic inflammation and allergic asthma. Thus, modulating ZFP91 activity in T reg cells may be a valuable strategy for the prevention and treatment of autoimmune inflammation.
Chronic colonic disorders promote the development of inflammation-driven colon carcinogenesis (Li et al., 2017; Zheng et al., 2018). In our study, we showed that T reg cell–derived ZFP91 strengthened autophagy initiation to enforce T reg cell functional integrity and maintain colonic immune homeostasis, thereby inhibiting colitis-associated tumorigenesis. These results indicate that actively regulated autophagy in T reg cells is crucial for the maintenance of immune tolerance and prevention of CAC. However, tumor growth has been shown to be severely suppressed in Atg7fl/flFoxp3-Cre mice inoculated with MC38 colon adenocarcinoma cells (Wei et al., 2016), suggesting an important role for autophagy in T reg cells in suppressing antitumor immune responses in a transplantable tumor model. These seemingly opposing results are likely due to the major differences in the formation of spontaneous and transplantable tumors. In particular, the formation of transplantable tumors bypasses the early steps of tumorigenesis, in which chronic colonic inflammation initiates colon carcinogenesis. Differences between spontaneous and transplantable tumor models have also been reported in previous studies (Swann et al., 2008; Zou et al., 2014, 2015).
Prior studies suggest that ZFP91 contains several nuclear localization signals and localizes in the nucleus (Unoki et al., 2003). Indeed, our data showed the predominant nuclear localization of ZFP91 in resting T reg cells. In activated T reg cells, TCR stimulation triggered the translocation of ZFP91 from the nucleus to the cytoplasm, which promoted ZFP91-mediated autophagy activation. Nevertheless, how the TCR signal induces the cytosolic translocation of ZFP91 in T reg cells remains to be determined. It is still unclear whether ZFP91 regulates T helper 1 (Th1), Th2, Th17, or CD8+ T cell metabolic and functional activities by manipulating autophagy initiation. Therefore, future studies are needed to examine the potential mechanism underlying TCR-induced ZFP91 cytosolic translocation in T reg cells and the functional importance of ZFP91 in different types of T cells and disease models.
In summary, our results highlight a ZFP91-dependent mechanism promoting TCR-initiated autophagosome maturation to restrict mTORC1-dependent hyperglycolysis and maintain T reg cell homeostasis and function. Our study indicates that T reg cell–derived ZFP91 mediates BECN1 K63–linked ubiquitination and the BECN1–PIK3C3 association to boost the TCR-induced activation of autophagy and associated downstream pathways. Our findings also suggest that enhancing ZFP91 activity in T reg cells represents a promising strategy for the treatment of colonic inflammation and inflammation-driven colorectal cancer.
Materials and methods
Mice
Zfp91-floxed mice (in C57BL/6 background) were generated at the Shanghai Model Organisms Center, Inc., using a LoxP targeting system. The Zfp91-floxed mice were crossed with Foxp3-YFP-Cre transgenic mice (The Jackson Laboratory) in B6 background to produce age-matched Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice for experiments. The Zfp91-floxed mice were also crossed with Cd4-Cre transgenic mice to obtain age-matched Zfp91+/+Cd4-Cre and Zfp91fl/flCd4-Cre mice for LC3-II and CytoID detection experiments. The Becn1-floxed mice (in a C57BL/6 background) were generated at the Shanghai Model Organisms Center, Inc., and crossed with Zfp91fl/flFoxp3-Cre mice to produce Zfp91+/+Becn1fl/flFoxp3-Cre and Zfp91fl/flBecn1fl/flFoxp3-Cre mice. B6.SJL mice and RAG-1–deficient mice in C57BL/6 background were purchased from The Jackson Laboratory. Mice were maintained in a specific pathogen–free facility, and all animal experiments were in accordance with protocols approved by the Institutional Animal care and Use Committee of Shanghai Jiao Tong University School of Medicine.
Plasmids, antibodies, and reagents
Flag-tagged mouse WT BECN1 and hemagglutinin-tagged mouse ZFP91 were cloned into the pcDNA3 vector. His-tagged K48, K63, and K63R mutant ubiquitin were cloned into the pcDNA3 vector. Antibodies for GAPDH, lamin B, p-S6K, p-S6, p-AKT (S473), c-Myc, BECN1, PIK3R4, PIK3C3, and K63 ubiquitin were purchased from Cell Signaling Technology, Inc. HRP-conjugated anti-hemagglutinin antibody (3F10) was from Roche, Inc. Anti-Flag (M2) antibody and anti–β-actin antibody were from Sigma-Aldrich, Inc. Ni–nitrilotriacetic acid agarose was from QIAGEN, Inc. Anti-ZFP91 antibody was from Bethyl Laboratories, Inc. Anti-LC3B antibody was purchased from Abcam, Inc. Anti-p62 antibody was from Proteintech Group, Inc. Antibodies for ubiquitin (P4D1) and NIK (A-12) were from Santa Cruz Biotechnology, Inc. The fluorochrome-conjugated antibodies for CD4 (GK1.5), CD8 (53-6.7), CD44 (IM7), CD62L (MEL-14), Foxp3 (FJK-16s), IFN-γ (XMG1.2), IL-17 (eBio17B7), CD25 (PC61.5), CTLA-4 (UC10-4B9), and PD-1 (J43) were purchased from Thermo Fisher Scientific, Inc. c-Myc (D84C12) rabbit mAb (PE conjugate) and phospho-S6 ribosomal protein (Ser235/236; D57.2.2E) XP Rabbit mAb (allophycocyanin conjugate) were purchased from Cell Signaling Technology, Inc. Rapamycin (R0395), chloroquine diphosphate salt (C6628), leptomycin B solution (L2913), and DCA (D54702) were purchased from Sigma-Aldrich, Inc.
Flow cytometry
To determine surface marker expression, cells were stained in PBS containing 2% FBS with indicated fluorochrome-conjugated antibodies for flow cytometric analysis. To determine intracellular IFN-γ and IL-17 expressions, cells were stimulated with PMA and ionomycin in the presence of monensin for 5 h. After in vitro stimulation, cells were stained with the indicated fluorochrome-conjugated antibodies according to the manufacturer’s instructions (Thermo Fisher Scientific, Inc.). Foxp3 or c-Myc staining was performed according to the manufacturer’s instructions (Thermo Fisher Scientific, Inc.). For intracellular staining of p-S6, stimulated cells were fixed in Fix Buffer I (BD Biosciences) for 10 min, followed by incubation in cold Perm Buffer III (BD Biosciences) for 20 min, and then subjected to the indicated fluorochrome-conjugated antibody staining and flow cytometric analysis. For Foxp3 staining in the presence of YFP, splenic cells were fixed with 3.7% formaldehyde following cell surface staining and then were permeabilized with 0.2% Triton X-100, followed by staining with Pacific Blue–anti-Foxp3 at 4°C for 2 h. Autophagic flux detection by flow cytometry was measured using the CytoID Autophagy Detection Kit (Enzo Life Sciences) and the Guava Autophagy LC3 Antibody-based Assay Kit (FCCH100171; Luminex) according to the manufacturer's instructions. The Guava Autophagy LC3 Antibody-based Assay Kit involves a permeabilization step to wash out cytosolic LC3-I, leaving only membrane-bound LC3-II before staining.
T reg cell isolation and stimulation
T reg cells (CD4+CD25+YFP+) were isolated from the Spl of age- and sex-matched Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (6–8 wk old). The purified T reg cells were stimulated with plate-bound anti-CD3 (1 µg/ml) and anti-CD28 (1 µg/ml) in replicate wells for flow cytometric, IB, or RNA-sequencing analysis.
Histology
Organs were removed from age- and sex-matched 10-mo-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice or age- and sex-matched DSS-treated or AOM/DSS-treated Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice, fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned for staining with hematoxylin and eosin.
Colitis and CAC animal model
For the DSS-induced colitis model, age- and sex-matched Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (6–8 wk old) were subjected to one cycle of 3% DSS in sterile drinking water for 7 d followed with normal drinking water until the end of the experiment. For induction of CAC, age- and sex-matched Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice (6–8 wk old) received a single intraperitoneal injection of AOM (10 mg/kg body weight) followed by three cycles of 2.5% DSS exposure. Colon lengths and histopathology were measured for each mouse at the completion of each study. The clinical score was calculated according to the characteristics of stool consistency and rectal bleeding as previously described (Zheng et al., 2018). In brief, stool score was determined as: 0 = normal stool; 1 = soft stool; 2 = soft stool that adheres to the anus; 3 = diarrhea. Bleeding score was determined as: 0 = no bleed; 1 = positive hemoccult; 2 = visible blood in stool; 3 = gross rectal bleeding. Stool score and bleeding score were added as the clinical score. Analysis of clinical score was done in a blinded fashion.
RNA-sequencing analysis
Fresh splenic T reg cells (CD4+CD25+YFP+) were isolated from 6-wk-old Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre mice and stimulated with anti-CD3 and anti-CD28 antibodies in the presence of 200 U/ml IL-2 for 24 h. Stimulated T reg cells were used for total RNA isolation with TRIzol (Invitrogen) and subjected to RNA sequencing using Illumina NextSeq 500 system (75-bp paired-end reads). Using HISAT2 RNA-sequencing alignment software, the raw reads were aligned to the mouse reference genome (version mm10). The mapping rate was 96%, on average, across all the samples in the dataset. The alignment files (bam files) were processed to generate read counts for every gene using HISAT and HTSeq. The read counts were normalized using the R package DESeq2 and then were centered and scaled for each gene, generating z-scores. P values obtained from multiple tests were adjusted using the Benjamini-Hochberg correction. Kyoto Encyclopedia of Genes and Genomes pathway analysis was performed using OmicsBean software. The RNA-sequencing data have been deposited in the Sequence Read Archive database with BioProject identifier no. PRJNA627357.
In vitro suppression assay
Naive CD4+ T cells were isolated from WT mice and were labeled with 2.5 µM CellTrace Violet (CTV; Invitrogen) for 20 min at 37°C. T reg cells isolated from Zfp91+/+Foxp3-Cre or Zfp91fl/flFoxp3-Cre mice were cocultured with CTV-labeled CD4+ T cells and were activated by Dynabeads Mouse T-activator CD3/CD28 (Invitrogen). Three days later, CTV-labeled CD4+ T cell proliferation was detected by flow cytometry.
Adoptive transfer of T cells
Naive CD4+CD45RBhi cells from B6.SJL (CD45.1+) mice and CD4+CD25+YFP+ T reg cells from Zfp91+/+Foxp3-Cre or Zfp91fl/flFoxp3-Cre (CD45.2+) mice were isolated by flow cytometry. RAG-1–deficient mice were given an intraperitoneal injection of Zfp91+/+Foxp3-Cre or Zfp91fl/flFoxp3-Cre CD4+CD25+YFP+ T reg cells (2 × 105) either alone or together with naive CD4+CD45RBhi cells (4 × 105).
Glycolytic and mitochondrial respiration rate measurement
The Seahorse XFe96 Extracellular Flux Analyzer (Agilent Technologies) was used for the detection of glycolytic and OXPHOS rates. Isolated Zfp91+/+Foxp3-Cre and Zfp91fl/flFoxp3-Cre T reg cells were stimulated for 3 h with anti-CD3 and anti-CD28. In vitro stimulated T reg cells were seeded at a density of 4 × 105 per well. The ECAR and the OCR for each well were calculated, while the cells were subjected to the XF Cell Mito or the XF Glycolytic stress test using the following concentrations of injected compounds: 10 mM glucose, 2 µM oligomycin, 50 mM 2-deoxy-D-glucose, 1 µM FCCP, and 0.5 µM rotenone/antimycin A. The XF Cell Mito and the XF Glycolytic stress test kits were purchased from Agilent Technologies, Inc.
IB and ubiquitination assays
T reg cells were washed with ice-cold PBS and lysed on ice for 30 min in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 135 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 1 mM EDTA, and 10% glycerol) containing protease inhibitor (1:100, P8340; Sigma-Aldrich, Inc.), 1 mM NaF, and 1 mM PMSF. Cell lysates were cleared by centrifugation, and supernatants were immunoprecipitated with the appropriate antibodies using protein A/G–agarose beads. Samples were then used for IB analysis with indicated antibodies. For ubiquitination assays, BECN1 was isolated from T reg cells by IP under denaturing conditions, and the ubiquitinated BECN1 was detected by IB using ubiquitin antibodies. For transfection models, the expression vector encoding BECN1 was transfected into HEK293T cells along with the His-ubiquitin expression vector; the His-ubiquitin was isolated by Ni–nitrilotriacetic acid agarose; and the ubiquitinated BECN1 was detected by anti-Flag IB.
Statistical analysis
Statistical analysis was performed using Prism software (version 6.01; GraphPad Software). Two-tailed unpaired Student’s t tests were performed, and data are presented as mean ± SEM. P < 0.05 is considered statistically significant, and the level of significance was indicated as ns, not statistically significant; *, P < 0.05; and **, P < 0.01.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81930040, 31922025, 31870884, 31900631), the Shanghai Municipal Health and Family Planning Commission (2018YQ08), and the Natural Science Foundation of Shanghai (19ZR1445700).
Author contributions: A. Wang designed and performed the experiments, interpreted the results, and prepared the manuscript. L. Ding helped with experimental design and analyzed RNA-sequencing data. Z. Wu, R. Ding, X.-L. Teng, F. Wang, and Z. Hu helped perform several experiments. L. Chen and X. Yu helped with experimental design and research supervision. Q. Zou designed experiments, supervised the research, and prepared the manuscript.
References
- Alon, R. 2017. A sweet solution: glycolysis-dependent Treg cell migration. Immunity. 47:805–807. 10.1016/j.immuni.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Campbell, C., and Rudensky A.. 2020. Roles of regulatory T cells in tissue pathophysiology and metabolism. Cell Metab. 31:18–25. 10.1016/j.cmet.2019.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, D., Wang Y., Lu R., Jiang X., Chen X., Meng N., Chen M., Xie S., and Yan G.R.. 2020. E3 ligase ZFP91 inhibits Hepatocellular Carcinoma Metabolism Reprogramming by regulating PKM splicing. Theranostics. 10:8558–8572. 10.7150/thno.44873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi, L., Pietrocola F., Levine B., and Kroemer G.. 2014. Metabolic control of autophagy. Cell. 159:1263–1276. 10.1016/j.cell.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets, V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., et al. 2015. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125:194–207. 10.1172/JCI76012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, V.M., Valdor R., Patel B., Singh R., Cuervo A.M., and Macian F.. 2010. Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 185:7349–7357. 10.4049/jimmunol.1000576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley, J.H., and Young L.N.. 2017. Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86:225–244. 10.1146/annurev-biochem-061516-044820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, X., Jin H.R., Jung H.S., Lee S.J., Lee J.H., and Lee J.J.. 2010. An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-kappaB-inducing kinase via Lys63-linked ubiquitination. J. Biol. Chem. 285:30539–30547. 10.1074/jbc.M110.129551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat, A.M., Harrison O.J., Riffelmacher T., Moghaddam A.E., Pearson C.F., Laing A., Abeler-Dörner L., Forman S.P., Grencis R.K., Sattentau Q., et al. 2016. The autophagy gene Atg16l1 differentially regulates Treg and TH2 cells to control intestinal inflammation. eLife. 5:e12444 10.7554/eLife.12444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore, M., Cheung K.C.P., Fu H., Bonacina F., Wang G., Coe D., Ward E.J., Colamatteo A., Jangani M., Baragetti A., et al. 2017. Regulatory T cell migration is dependent on glucokinase-mediated glycolysis. Immunity. 47:875–889.e10. 10.1016/j.immuni.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasry, A., Zinger A., and Ben-Neriah Y.. 2016. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 17:230–240. 10.1038/ni.3384 [DOI] [PubMed] [Google Scholar]
- Li, C., Capan E., Zhao Y., Zhao J., Stolz D., Watkins S.C., Jin S., and Lu B.. 2006. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J. Immunol. 177:5163–5168. 10.4049/jimmunol.177.8.5163 [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang Z., Li L., Gong W., Lazenby A.J., Swanson B.J., Herring L.E., Asara J.M., Singer J.D., and Wen H.. 2017. Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. J. Exp. Med. 214:1093–1109. 10.1084/jem.20161105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui, K., Tanaka K., Akhavan D., Babic I., Gini B., Matsutani T., Iwanami A., Liu F., Villa G.R., Gu Y., et al. 2013. mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 18:726–739. 10.1016/j.cmet.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, C., Wang Z., Li M.Y., Zhang Z.H., Ma J., and Jin X.. 2018. Zinc finger protein 91 positively regulates the production of IL-1β in macrophages by activation of MAPKs and non-canonical caspase-8 inflammasome. Br. J. Pharmacol. 175:4338–4352. 10.1111/bph.14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, R., Priyadharshini B., and Turka L.A.. 2016. Immunometabolism of regulatory T cells. Nat. Immunol. 17:618–625. 10.1038/ni.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh, V.V., Wu L., Boyd K.L., Williams J.A., Gaddy J.A., Olivares-Villagómez D., Cover T.L., Zong W.X., Zhang J., and Van Kaer L.. 2013. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J. Immunol. 190:5086–5101. 10.4049/jimmunol.1202071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic, D., Vucic D., and Dikic I.. 2014. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20:1242–1253. 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- Pua, H.H., Dzhagalov I., Chuck M., Mizushima N., and He Y.W.. 2007. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J. Exp. Med. 204:25–31. 10.1084/jem.20061303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puleston, D.J., Zhang H., Powell T.J., Lipina E., Sims S., Panse I., Watson A.S., Cerundolo V., Townsend A.R., Klenerman P., and Simon A.K.. 2014. Autophagy is a critical regulator of memory CD8(+) T cell formation. eLife. 3:e03706 10.7554/eLife.03706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, J., Han L., Tang J., Liu Y., Deng X., Liu Q., Hao P., Feng X., Li B., Hu H., and Wang H.. 2019. Foxp1 is critical for the maintenance of regulatory T-cell homeostasis and suppressive function. PLoS Biol. 17:e3000270 10.1371/journal.pbio.3000270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravia, J., Raynor J.L., Chapman N.M., Lim S.A., and Chi H.. 2020. Signaling networks in immunometabolism. Cell Res. 30:328–342. 10.1038/s41422-020-0301-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann, J.B., Vesely M.D., Silva A., Sharkey J., Akira S., Schreiber R.D., and Smyth M.J.. 2008. Demonstration of inflammation-induced cancer and cancer immunoediting during primary tumorigenesis. Proc. Natl. Acad. Sci. USA. 105:652–656. 10.1073/pnas.0708594105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek, K.N., and Komander D.. 2016. Ubiquitin modifications. Cell Res. 26:399–422. 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, R., Nishimoto S., Muto G., Sekiya T., Tamiya T., Kimura A., Morita R., Asakawa M., Chinen T., and Yoshimura A.. 2011. SOCS1 is essential for regulatory T cell functions by preventing loss of Foxp3 expression as well as IFN-gamma and IL-17A production. J. Exp. Med. 208:2055–2067. 10.1084/jem.20110428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, D.E., Dai Y., Xu Y., Lin L.W., Liu D.Z., Hong X.P., Ou M.L., Jiang H.W., and Xu S.H.. 2020. The ubiquitinase ZFP91 promotes tumor cell survival and confers chemoresistance through FOXA1 destabilization. Carcinogenesis. 41:56–66. [DOI] [PubMed] [Google Scholar]
- Unoki, M., Okutsu J., and Nakamura Y.. 2003. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. Int. J. Oncol. 22:1217–1223. 10.3892/ijo.22.6.1217 [DOI] [PubMed] [Google Scholar]
- Wei, J., Long L., Yang K., Guy C., Shrestha S., Chen Z., Wu C., Vogel P., Neale G., Green D.R., and Chi H.. 2016. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17:277–285. 10.1038/ni.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, N.R., McCuaig S., Franchini F., and Powrie F.. 2015. Emerging cytokine networks in colorectal cancer. Nat. Rev. Immunol. 15:615–629. 10.1038/nri3896 [DOI] [PubMed] [Google Scholar]
- Wing, J.B., Tanaka A., and Sakaguchi S.. 2019. Human FOXP3+ regulatory T cell heterogeneity and function in autoimmunity and cancer. Immunity. 50:302–316. 10.1016/j.immuni.2019.01.020 [DOI] [PubMed] [Google Scholar]
- Xu, X., Araki K., Li S., Han J.H., Ye L., Tan W.G., Konieczny B.T., Bruinsma M.W., Martinez J., Pearce E.L., et al. 2014. Autophagy is essential for effector CD8+ T cell survival and memory formation. Nat. Immunol. 15:1152–1161. 10.1038/ni.3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S.H., Zhu S., Wang Y., Huang J.Z., Chen M., Wu Q.X., He Y.T., Chen D., and Yan G.R.. 2018. ECD promotes gastric cancer metastasis by blocking E3 ligase ZFP91-mediated hnRNP F ubiquitination and degradation. Cell Death Dis. 9:479 10.1038/s41419-018-0525-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Teng X.L., Wang F., Zheng Y., Qu G., Zhou Y., Hu Z., Wu Z., Chang Y., Chen L., et al. 2018. Metabolic control of regulatory T cell stability and function by TRAF3IP3 at the lysosome. J. Exp. Med. 215:2463–2476. 10.1084/jem.20180397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, H., and Chi H.. 2017. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr. Opin. Immunol. 46:103–111. 10.1016/j.coi.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, T., Zhang B., Chen C., Ma J., Meng D., Huang J., Hu R., Liu X., Otsu K., Liu A.C., et al. 2018. Protein kinase p38α signaling in dendritic cells regulates colon inflammation and tumorigenesis. Proc. Natl. Acad. Sci. USA. 115:E12313–E12322. 10.1073/pnas.1814705115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Q., Jin J., Hu H., Li H.S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., et al. 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 15:562–570. 10.1038/ni.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Q., Jin J., Xiao Y., Zhou X., Hu H., Cheng X., Kazimi N., Ullrich S.E., and Sun S.C.. 2015. T cell intrinsic USP15 deficiency promotes excessive IFN-γ production and an immunosuppressive tumor microenvironment in MCA-induced fibrosarcoma. Cell Rep. 13:2470–2479. 10.1016/j.celrep.2015.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]