Abstract
Objective
Atherosclerosis is the main cause of the cardiovascular disease (CVD). Elevated blood cholesterol and inflammation of the endothelium are two major mechanisms contributing to the establishment of atherosclerotic plaques. Statins, such as pravastatin, are blood‐cholesterol lowering drugs commonly prescribed for patients with or at risk for CVDs. In addition to lowering blood cholesterols, statins have recently been shown to improve endothelial function in both hyper‐ and normocholesterolemic patients with atherosclerosis. To understand the molecular mechanisms underlying the endothelial function improvement by statins, we assessed the RNA profile of pravastatin‐treated endothelial cells, particularly their mRNAs and long non‐coding RNAs (lncRNAs).
Methods
Human umbilical vein endothelial cells (HUVECs) treated with pravastatin (10 µM) for 24 hr were profiled for lncRNAs and mRNAs using the Arraystar Human lncRNA Expression Microarray V3.0.
Results
Of the 30,584 different lncRNAs screened, 95 were significantly upregulated, while 86 were downregulated in HUVECs responding to pravastatin. LINC00281 and BC045663 were the most upregulated (~8‐fold) and downregulated (~3.5‐fold) lncRNAs, respectively. Of the 26,106 different mRNAs screened in the pravastatin‐treated HUVEC samples, 190 were significantly upregulated, while 90 were downregulated. Assigning the differentially expressed genes by bioinformatics into functional groups revealed their molecular signaling involvement in the following physiological processes: osteoclast differentiation, Rap1 signaling pathway, hematopoiesis, immunity, and neurotrophin signaling pathway.
Conclusions
This is the first lncRNA and mRNA expression profiling of pravastatin‐mediated changes in human endothelial cells. Our results reveal potential novel targets and mechanisms for pravastatin‐mediated vascular protection in atherosclerosis.
Keywords: coronary artery disease, endothelial dysfunction, lncRNA, pravastatin
First lncRNA and mRNA expression profiling of pravastatin‐mediated changes in human endothelial cells. We provide the basis for mechanisms associated with pravastatin to improve endothelial function.
1. INTRODUCTION
Evidence from epidemiological, genetics and basic science studies associate elevated levels of plasma cholesterol with increased risk of atherosclerosis (Robinson et al., (2018); Kannel et al., 1964). Elevated cholesterol levels, also known as hypercholesterolemia, lead to endothelial dysfunction, which is now known to play a critical role in the majority of cardiovascular diseases including coronary artery disease (CAD) (Libby, 2001). The anatomical location of the endothelium enables it to sense hemodynamic and chemical changes within the blood to maintain vascular homeostasis by regulating the balance between vasoconstriction and vasodilatation. LDL (low‐density lipoprotein) cholesterol disturbs this homeostasis via several means that include the impairing production of the vasodilator nitric oxide (NO) by inactivating endothelial nitric oxide synthase (eNOS) in endothelial cells (ECs), disrupting the vasomotor tone (Hermida & Balligand, 2014; Noor et al., 2007; Steinberg et al., 1997). Accordingly, cholesterol‐lowering drugs have been shown to reduce the burden of CAD, in part due to improved endothelial vasomotor tone and reduced coronary artery dilatation (Anderson et al., 1995; Ward et al., 2019).
Statins are a class of drugs designed to lower blood cholesterol levels. While different statins vary in bioavailability and half‐life, they are widely prescribed for hypercholesterolemic patients who are at risk for cardiovascular diseases (CVDs) (Meor Anuar Shuhaili et al., 2017). The statin agents antagonize and inhibit the enzyme hepatic 3‐Hydroxy‐3‐Methylglutaryl‐CoA Reductase (HMG‐CoA reductase), which is normally responsible for an early rate‐limiting step in the biosynthesis of cholesterol and LDL. There are six statin drugs in use, including pitavastatin, atorvastatin, rosuvastatin, simvastatin, fluvastatin, and pravastatin. Except pravastatin, all other statins are metabolized by the cytochrome p450 group of enzymes and any drug interference with these enzymes can lead to an enhanced level of statin, thereby increasing the risk for toxicity (Ramkumar et al., 2016). Pravastatin has the least drug interaction among other statins and is the preferred statin for patients on immunosuppressants or protease inhibitors (Ramkumar et al., 2016). By blocking cholesterol synthesis in the liver, statins, such as pravastatin, not only reduce plasma LDL levels but also upregulate the hepatic LDL receptors, encouraging the liver to uptake and clear blood cholesterols (Pinal‐Fernandez et al., 2018). A lower level of plasma LDL is associated with a reduced risk for CVDs. Recently, a new perspective has emerged suggesting that statins‐mediated CVD risk reduction is not solely due to the lowering of blood cholesterols. Statins have been shown to restore or improve many aspects of endothelial function, which includes statins upregulating eNOS expression and activity through preventing the post‐translational modification of Rho and activating protein kinase Akt (Kureishi et al., 2000; Rikitake & Liao, 2005). In addition, potent vasoconstrictors dysregulated in atherosclerosis, such as endothelin‐1 and angiotensin‐II, were shown to be attenuated by statin treatments (Hernandez‐Perera et al., 1998; Ichiki et al., 2001). Statins can also control the vascular oxidative level and mitigate the induction of inflammatory mechanisms by downregulating endothelial inflammatory markers (Niwa et al., 1996; Rezaie‐Majd et al., 2002; Wassmann et al., 2001), thereby decreasing leukocyte recruitment.
Another new prospect of regulating endothelial function involves long‐non‐coding RNAs (lncRNAs). These transcripts are >200 nucleotides long and, although not as conserved, are transcribed like protein‐coding transcripts, similarly polyadenylated and capped, but are devoid of open reading frames (ORFs) (Djebali et al., 2012; Ulitsky & Bartel, 2013). Functionally, most lncRNAs interact with nuclear chromatin‐modifying ribonucleoprotein complexes as ligands and guide them through complementary base‐pairing to the targeted genomic sequences for epigenetic regulation. LncRNA’s mode of action is also characterized by whether the target sequence is within the vicinity of the lncRNA’s gene (cis‐acting) or not (trans‐acting) (Rinn et al., 2007). Previously regarded as transcriptional noise due to their low expression and poor evolutionary conservation (Hangauer et al., 2013). LncRNAs have recently emerged as potential molecular determinants of a wide range of diseases (Batista & Chang, 2013; Wapinski & Chang, 2011). Increasing evidence suggests the involvement of lncRNAs in a variety of cellular functions, including survival, proliferation, migration, invasion, angiogenesis and differentiation (Beltran et al., 2008; Fiedler et al., 2015; Guenther et al., 2007; Rinn et al., 2007; Wahlestedt, 2013; Zhao et al., 2008). Indeed, several lncRNAs have been notably characterized in cardiovascular pathophysiology: metastasis‐associated lung adenocarcinoma transcript 1 (MALAT1), one of the more conserved and abundant lncRNAs (Zhang et al., 2012) and Tie‐1‐AS were shown to regulate endothelial function (Wapinski & Chang, 2011; Zhao et al., 2008). Antisense noncoding RNA gene at the INK4 locus (ANRIL) and Apolipoprotein A1 antisense transcript (APOA1‐AS) were shown to have crucial roles in atherosclerosis (Holdt et al., 2013; Lund‐Katz & Phillips, 2010).
Given that statins and lncRNAs have been shown to impact endothelial function and cardiovascular status, it remains to be understood whether the endothelial processes regulated by lncRNAs are connected to statin's actions on ECs. Therefore, we aim to profile the variations in lncRNAs and mRNAs expression upon pravastatin treatment in ECs. Our approach would identify novel lncRNA and mRNA targets and associated pathways affected by pravastatin in ECs, thereby providing insights into the mechanisms behind pravastatin‐associated cardiovascular protection. This is the first transcriptome profiling of pravastatin‐mediated changes in human ECs.
2. MATERIALS AND METHODS
2.1. Cell culture and treatment
Human umbilical vein ECs (Pooled HUVECs, Lonza) were grown in the EC growth medium supplemented with growth factors, 5% fetal bovine serum (FBS), and antibiotics (EGMTM‐2 BulletkitTM; Lonza) at 37oC and 5% CO2. Passage 4–6 HUVECs were used for the experiments. Cells were (60%–70%) confluent and were starved over‐night in MCDB‐131 basal medium with 1% FBS, followed by treatment with pravastatin sodium salt hydrate (10 µM; Sigma) (Abe et al., 2006; Panczel et al., 2019) in the same MCDB‐131 basal medium with 1% FBS for 24 hr. Controls were treated with vehicle phosphate‐buffered saline. To validate the expression level of top up‐ or downregulated lncRNAs and mRNAs, HUVECs were cultured and treated as previously described.
2.2. RNA extraction
HUVEC cultures were washed with ice‐cold PBS and then TRIzolTM (Invitrogen) was directly added and incubated for 10 min at room temperature to extract total RNAs from HUVECs as instructed by the manufacturer. RNA was quantified using the NanoDrop ND‐1000 spectrophotometer and confirmed for integrity by standard denaturing agarose gel electrophoresis. Validation was performed for selected top up‐ or downregulated lncRNAs and mRNAs by extracting total RNA as previously described (Singh, Nguyen, et al., 2020). Later, cDNA was synthesized using 1μg of total RNA with the QuantiTect® Reverse Transcription kit (Qiagen) according to the manufacturer's instructions. Quantitative PCR (qPCR) was performed using primers for lncRNAs RP11‐469H8.6, LINC00281, BC045663, RP11‐791G16.2 and AC009948.5, and mRNAs DMKN, PIEGO2, APOLD1, ABI1, SND1, TMED2, SCAF8, SNX10, and GAPDH (Murugavel et al., 2018) (Table 1) using SYBR Select Master Mix (Applied Biosystems) and QuantStudio 3 Real‐Time PCR System (Applied Biosystems) (N = 3 in triplicates for both conditions). Fold‐change gene expression was calculated by the 2‐ΔΔCt method, and the expression differences between the two groups were calculated using Student's t test. Validation data are presented as mean ± SD. A p‐value of less than 0.05 was considered significant.
Table 1.
Primers used to perform qPCR
| Nr | Gene symbol | Forward Primer ( 5′ to 3′) | Reverse Primer ( 5′ to 3′) |
|---|---|---|---|
| 1 | RP11‐469H8.6 | tgaggtgatgggagaggtg | acctccttggggtaggtcat |
| 2 | LINC00281 | aaagaagatgctgccaatgag | ggaagtgagttatttcagggtacg |
| 3 | BC045663 | acaagccatggacaacagc | gcacatgttggagattgcac |
| 4 | RP11‐791G16.2 | gcctgcctgttacattcctg | tgcaaccacgaactaattttctc |
| 5 | AC009948.5 | gagtagcggtggctgaaga | ggtgccatcaagttccaaaa |
| 6 | DMKN | cagagcggagaggaaagcac | gcctcactgactttagagccag |
| 7 | PIEGO2 | atggcctcagaagtggtgtg | atgtccttgcatcgtcgtttt |
| 8 | APOLD1 | agagatgtaacccaactcgttca | caggggaaggtgcatcctc |
| 9 | ABI1 | accagtcctgctaggcttg | actgttttctcgacttccacttc |
| 10 | SND1 | cctgagcggcagatcaacc | aggtagatcatgccatactctcg |
| 11 | TMED2 | catcgacgtggagattacagg | ggtggacatccggttactaaaac |
| 12 | SCAF8 | gtgcgacaatcccgacatca | tccccagggcaacgatataaa |
| 13 | SNX10 | cacttttgctttcagatagcagc | acacacgcctcaatgtcttct |
2.3. Microarray profiling
We established an expression profile of 30,584 human lncRNAs and 26,106 protein‐coding transcripts using the Arraystar Human LncRNA Microarray V3.0. Three replicates were used for pravastatin‐treated and vehicle‐treated groups. Total RNAs were amplified and transcribed into fluorescent complimentary RNA (cRNAs) by the Arraystar Flash RNA Labeling Kit (Arraystar). One microgram of each cRNA was labeled and hybridized onto the microarray slide using the Agilent Array platform. Hybridized arrays were washed and fixed before scanning with the Agilent DNA Microarray Scanner (Product#G2505C).
2.4. Array analysis
The Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the images acquired from scanning the hybridized array. Quantile normalization was performed using the GeneSpring GX v11.5.1 software package (Agilent Technologies) and p‐values were calculated for the differentially expressed genes by student's t test. p‐values were then subjected to multiple testing by the Benjamini Hochberg method for minimizing false discovery rate. Volcano‐plot filtering set at a threshold of ≥2.0‐folds was used to screen the lncRNAs and mRNAs exhibiting significantly different expression levels between the two experimental groups (adjusted p < .05; unpaired t test). Pathway analyses were conducted using the current Kyoto Encyclopedia of Genes & Genomes (KEGG) database.
3. RESULTS
3.1. Quality and expression assessment of lncRNAs and mRNAs
We first evaluated the integrity and purity of the total RNAs extracted from HUVECs. Employing the standard denaturing agarose gel electrophoresis, RNAs from all samples showed an upper 28S ribosomal RNA band that was twice as intense compared to the lower 18S band, validating the integrity of our RNA extracts (Supp. Figure S1). RNA purity was also verified also by these upper 28S ribosomal RNA bands presenting without any surrounding smears, as well as by the optical density (OD) A260/A280 and A260/A230 ratios close to 2.0 and exceeding 1.8, respectively, obtained from the NanoDrop ND‐1000 (Singh, Adam, et al., 2020). Cluster analysis of the distribution of fluorescent intensities for lncRNAs and mRNAs demonstrated similar patterns in global gene expression between samples and groups across the board that is illustrated by box and whisker plot (10th, 90th percentile) (Supp. Figure S2).
Our analysis presented a profile of significantly upregulated and downregulated lncRNAs (Figure 1a) and mRNAs (Figure 1b) in response to pravastatin in HUVECs. We also examined the differential expression degree and observed similar average fold‐changes between lncRNAs and mRNAs (Figure 1c). From this profile, we uncovered 95 significantly upregulated and 86 significantly downregulated lncRNAs in pravastatin‐treated HUVECs compared to controls (Figure 1d), of which the lncRNAs ranged from 10,350 to 156 bp in size. Particularly, LINC00281 (RNA length: 3,215 bp, chromosome 22) and BC045663 (RNA length: 1823 bp, chromosome 20) in pravastatin‐treated HUVECs were the most upregulated (~8‐fold) and downregulated (~3‐fold) lncRNAs, respectively (Tables 2 and 3). We validated our findings by performing qPCR for selected upregulated lncRNAs LINC00281 and RP11‐469H8.6, and downregulated lncRNAs BC045663, RP11‐791G16.2, and AC009948.5. Among upregulated lncRNAs, RP11‐469H8.6 was 4.42 ± 1.48 (p = .0021) fold upregulated in the pravastatin‐treated group; however, qPCR for LINC00281 did not provide quantifiable data due to extremely low expression in the control group. Validation for all selected downregulated lncRNAs [BC045663 (0.288 ± 0.177‐fold, p = .0018), RP11‐791G16.2 (0.399 ± 0.008‐fold, p = .0008) and AC009948.5 (0.669 ± 0.050‐fold, p = .01850)] showed similar patterns as observed in the lncRNA array (Table 4). At the mRNA level, 190 differentially expressed mRNAs were upregulated, while 90 differentially expressed mRNAs were downregulated (Figure 1e) in HUVECs in response to pravastatin treatment, with the endocytosis‐associated protein DMKN (Dermokine) and cell growth‐associated ABI1 (ABL Interactor 1) (Biesova et al., 1997) being the most upregulated (~11‐fold) and downregulated (~ 5‐fold) gene, respectively (Tables 5 and 6). Validation qPCR performed for selected upregulated (DMKN, PIEGO2, and APOLD1) or downregulated (AB1I, SND1, TMED2, SCAF8, and SNX10) genes demonstrated a similar expression profile as observed in the array. Among upregulated genes, DMKN (5.49 ± 1.19‐fold, p = .001), PIEGO2 (1.755 ± 0.428‐fold, p = .033) and APOLD1 (1.296 ± 0.179‐fold, p = .020) were significantly upregulated genes in the pravastatin‐treated group in comparison to the control group. Similarly, among downregulated genes, AB1I (0.643 ± 0.179, p = .044), SCAF8 (0.728 ± 0.114, p = .028) and SNX10 (0.536 ± 0.313, p = .022) were significantly downregulated in the pravastatin‐treated group, but SND1 (0.813 ± 0.431, p = .436) and TMED2 (1.174 ± 0.367, p = .344) appeared to be unaffected by pravastatin in our validation qPCR (Table 4). As a supplement, we also provide the complete array data on our differentially expressed mRNAs and lncRNAs, their respective sequences, exact chromosomal locations, fold changes, and known target genes in Supp. Table S1 (https://figshare.com/s/82bb5cdf645b549ae416).
Figure 1.
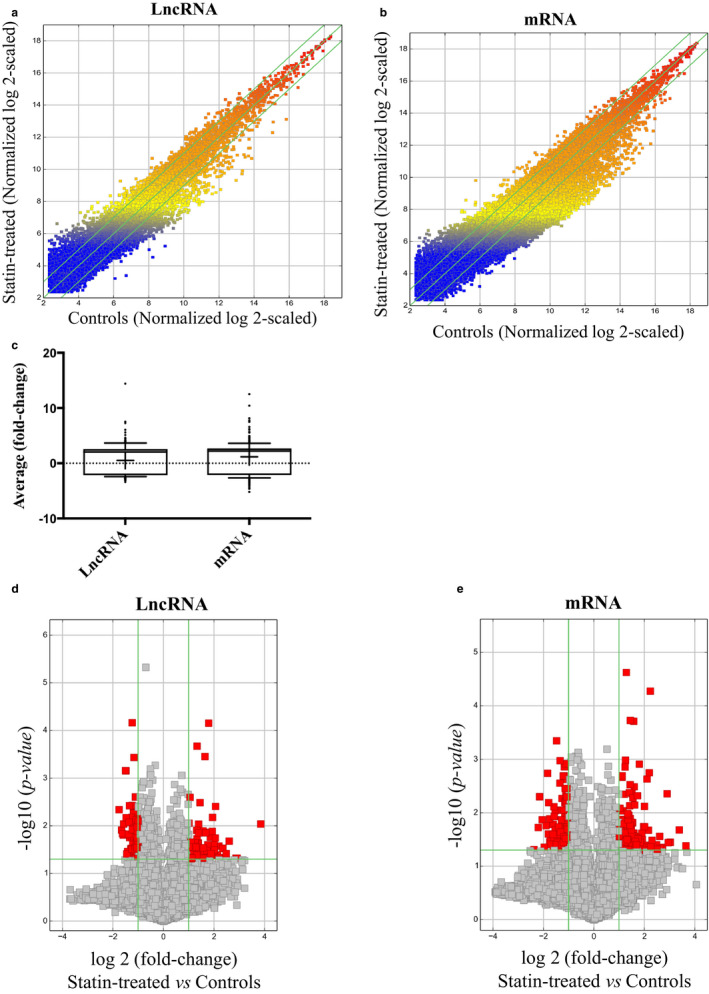
LncRNA and mRNA expression profiles in HUVECs treated with pravastatin (10 µM) versus control. (a and b) Scatter plots comparing the variation in lncRNA and mRNA expression. The values plotted are the averaged normalized signal values (log2‐scaled) for the control (x‐axis) and the pravastatin‐treatment (y‐axis) groups. The green lines indicate fold‐change. LncRNAs and mRNAs above the top green line and below the bottom green line exhibit at least a 2.0‐fold difference between the two study groups. (c) Box‐and‐Whisker plots (10th, 90th percentile) showing average fold‐change of lncRNAs and mRNAs. Median intensity is denoted with a “‐” and mean intensity denoted with a “+” sign. (d & e) Volcano plots detailing the magnitude of expression difference. The vertical green lines correspond to 2.0‐fold upregulation and 2.0‐fold downregulation of expression. The horizontal green line indicates an adjusted p‐value of ≤ 0.05. Red points represent lncRNAs and mRNAs with statistically significant differential expression (fold‐change ≥ 2.0, adjusted p ≤ .05)
Table 2.
Ten most upregulated lncRNAs in HUVECs upon pravastatin (10 µM) stimulation in comparison to controls
| Top 10 upregulated differentially expressed lncRNAs in Statin‐treated versus Control endothelial cells | ||||||
|---|---|---|---|---|---|---|
| Nr | Gene symbol | Fold change | Adj. p‐value | Associated gene | RNA length | Chr/ Strand |
| 1 | LINC00281 | 7.5390834 | .0476663 | 3,215 | 22/− | |
| 2 | AK125078 | 7.2998619 | .0477598 | 847 | 1/‐ | |
| 3 | XLOC_007368 | 6.0619879 | .0209871 | 218 | 9/+ | |
| 4 | XLOC_004244 | 5.5920067 | .0383514 | 690 | 5/+ | |
| 5 | RP11‐469H8.6 | 5.1033278 | .0296308 | AQP2 | 592 | 12/‐ |
| 6 | RP11‐493L12.2 | 4.5832587 | .046438631 | PCED1B | 359 | 12/‐ |
| 7 | RP11‐983C2.2 | 4.5736328 | .026541316 | 482 | 16/‐ | |
| 8 | RP11‐473M20.9 | 4.3979907 | .04118165 | IL32 | 1,207 | 2/‐ |
| 9 | XLOC_001747 | 4.1675991 | .003911454 | 383 | 5/+ | |
| 10 | XLOC_005737 | 4.0376803 | .032008961 | RUNX2 | 2,243 | 6/‐ |
Abbreviations: Adj, Adjusted; AQP2, Aquaporin 2; IL32, Interleukin 32; PCED1B, PC‐Esterase Domain Containing 1B; RUNX2, Runt‐related Transcription Factor 2.
Table 3.
Ten most downregulated lncRNAs in HUVECs upon pravastatin (10 µM) stimulation in comparison to controls
| Top 10 downregulated differentially expressed lncRNAs in Statin‐treated versus Control endothelial cells | ||||||
|---|---|---|---|---|---|---|
| Nr | Gene symbol | Fold change | Adj. p‐value | Associated gene | RNA length | Chr/ Strand |
| 1 | BC045663 | 3.3799237 | .0045743 | 1823 | 20/‐ | |
| 2 | RP11‐876N24.2 | 3.1491515 | .0122195 | CIITA | 575 | 16/‐ |
| 3 | XLOC_001373 | 3.0753019 | .0146288 | 387 | 2/+ | |
| 4 | RP11‐791G16.2 | 3.0002025 | .0156631 | TMEM150C | 1,493 | 4/‐ |
| 5 | RPS11P6 | 2.9800209 | .0091166 | C12orf56 | 1,352 | 12/+ |
| 6 | XLOC_008645 | 2.8372396 | .000696542 | 310 | 10/+ | |
| 7 | UTY | 2.8021482 | .00069716 | UTY | 6,761 | Y/‐ |
| 8 | AC009948.5 | 2.7780046 | .00671064 | PRKRA | 563 | 2/+ |
| 9 | REV3L‐IT1 | 2.7320222 | .038811723 | REV3L | 382 | 6/‐ |
| 10 | RP11‐542B15.1 | 2.6786082 | .034661639 | 559 | 12/+ | |
Abbreviations: Adj, Adjusted; C12orf56, Chromosome 12 Open Reading Frame 56; CIITA, Major Histocompatibility Complex Class II, Transactivator; PRKRA, Protein Kinase, Interferon‐Inducible Double‐Stranded RNA‐Dependent Activator; REV3L, Rev3, S. Cerevisiae, Homolog Of; TMEM150C, Transmembrane Protein 150C; UTY, Ubiquitously Transcribed Tetratricopeptide Repeat Gene on Y Chromosome.
Table 4.
Validation was performed for selected top up or downregulated lncRNAs and mRNAs isolated from HUVECs upon pravastatin (10 µM) stimulation and controls
| Nr | Gene symbol |
Array Data (Fold change) |
p‐value (Adj) |
qPCR Data (Fold change) |
p‐value |
|---|---|---|---|---|---|
| 1 | RP11‐469H8.6 | 5.1033278 | .0296308 | 4.42 ± 1.48 | .002 |
| 2 | BC045663 | −3.3799237 | .0045743 | 0.288 ± 0.177 | .001 |
| 3 | RP11‐791G16.2 | −3.0002025 | .0156631 | 0.399 ± 0.008 | .000 |
| 4 | AC009948.5 | −2.7780046 | .00671064 | 0.669 ± 0.050 | .018 |
| 5 | DMKN | 10.4331054 | .020871174 | 5.49 ± 1.19 | .001 |
| 6 | PIEGO2 | 7.7281947 | .036009804 | 1.755 ± 0.428 | .033 |
| 7 | APOLD1 | 7.4822837 | .00441241 | 1.296 ± 0.179 | .020 |
| 8 | ABI1 | −4.6038017 | .018848386 | 0.643 ± 0.179 | .044 |
| 9 | SND1 | −4.4388801 | .004988181 | 0.813 ± 0.431 | .436 |
| 10 | TMED2 | −4.135675 | .013520699 | 1.174 ± 0.367 | .344 |
| 11 | SCAF8 | −3.7269421 | .017383289 | 0.728 ± 0.114 | .028 |
| 12 | SNX10 | −3.566869 | .001821224 | 0.536 ± 0.313 | .022 |
The “‐” sign indicates downregulation and qPCR data are presented as mean ± SD.
Table 5.
Ten most upregulated mRNAs in HUVECs upon pravastatin (10 µM) stimulation in comparison to controls
| Top 10 upregulated differentially expressed mRNAs in Statin‐treated versus Control endothelial cells | |||
|---|---|---|---|
| Nr | Gene symbol | Fold change | Adj p‐value |
| 1 | DMKN | 10.4331054 | .020871174 |
| 2 | TNNT2 | 8.1375169 | .035583326 |
| 3 | PIEZO2 | 7.7281947 | .036009804 |
| 4 | APOLD1 | 7.4822837 | .00441241 |
| 5 | C7orf34 | 6.5808857 | .044510939 |
| 6 | EIF2B4 | 6.1793913 | .045905061 |
| 7 | TMEM191B | 6.0818645 | .027576263 |
| 8 | ENTHD2 | 5.685957 | .047950255 |
| 9 | A1CF | 5.5672248 | .03958813 |
| 10 | LOC100287177 | 4.9306594 | .032827533 |
Abbreviations: A1CF, Apobec1 Complementation Factor; Adj, Adjusted; APOLD1, Apolipoprotein L Domain‐Containing 1; C7orf34, Chromosome 7 Open Reading Frame 34; DMKN, Dermokine; EIF2B4, Eukaryotic Translation Initiation Factor 2b, Subunit 4; PIEZO2, Piezo‐Type Mechanosensitive Ion Channel Component 2; TMEM191B, Transmembrane Protein 191B; TNNT2, Troponin T2, Cardiac.
Table 6.
Ten most downregulated mRNAs in HUVECs upon pravastatin (10 µM) stimulation in comparison to controls
| Top 10 downregulated differentially expressed mRNAs in Statin‐treated versus Control endothelial cells | |||
|---|---|---|---|
| Nr | Gene symbol | Fold change | Adj. p‐value |
| 1 | ABI1 | 4.6038017 | .018848386 |
| 2 | SND1 | 4.4388801 | .004988181 |
| 3 | TMED2 | 4.135675 | .013520699 |
| 4 | RAB23 | 3.9035519 | .044063826 |
| 5 | SCAF8 | 3.7269421 | .017383289 |
| 6 | SNX10 | 3.566869 | .001821224 |
| 7 | EMD | 3.4986437 | .024932582 |
| 8 | SGK1 | 3.4659766 | .006466557 |
| 9 | B4GALT6 | 3.3686982 | .013007055 |
| 10 | PRIM2 | 3.3550803 | .035131682 |
Abbreviations: ABI1, Abl Interactor 1; Adj, Adjusted; B4GALT6, Udp‐Gal:Beta‐Glcnac Beta‐1,4‐Galactosyltransferase, Polypeptide 6; EMD, Emerin; PRIM2, Primase Polypeptide 2a; RAB23, Ras‐Associated Protein Rab23; SCAF8, Sr‐Related C‐Terminal Domain‐Associated Factor 8; SGK1, Serum/Glucocorticoid‐Regulated Kinase 1; SND1, Staphylococcal Nuclease Domain‐ And Tudor Domain‐Containing Protein 1; SNX10, Sorting Nexin 10; TMED2, Transmembrane emp24 Domain‐containing Protein 2
3.2. LncRNA chromosomal distribution and subtype analysis
We proceeded to further examine the lncRNAs profile of pravastatin‐treated HUVECs with respect to the vehicle. After evaluating every chromosome, pravastatin‐induced differentially expressed lncRNAs were most abundant on chromosomes 2, 12, and 17 (Figure 2a). These lncRNAs were expressed along the entire length of the chromosomes with a notable clustering pattern (Figure 2b). LncRNAs are generally classified into six different categories based on the origin of their transcription and the arrangement of their neighboring genes. These categories are sense, natural antisense, intronic, intergenic, bidirectional promoter, and enhancer lncRNA. Subgroup analysis revealed that pravastatin‐induced differentially expressed lncRNAs were intergenic, bidirectional, natural and intronic antisense, exon‐ and intron sense‐overlapping. The majority were functionally intergenic in origin, followed by natural and intronic antisense (Figure 2c).
Figure 2.
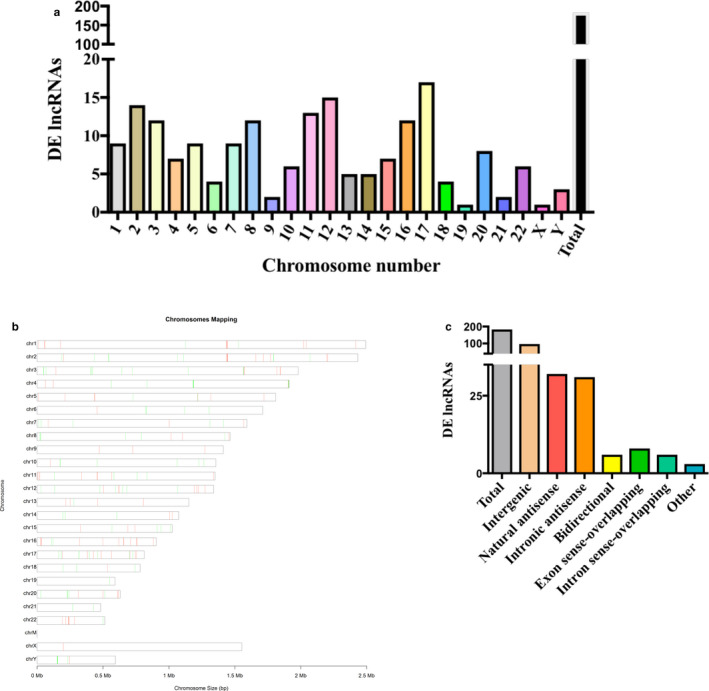
Distribution, location, and classification of differentially expressed lncRNAs in HUVECs treated with pravastatin (10 µM) versus control. Demonstration of (a) numbers and (b) chromosomal location of differentially expressed (DE) lncRNAs on different chromosomes. (c) Bar‐graph representing types of differently expressed lncRNAs, depending upon their genomic location
3.3. Bioinformatics analyses
To functionally associate the pravastatin‐induced differentially expressed lncRNAs, pathway analysis using the current KEGG database was conducted on the concomitant pravastatin‐induced differentially expressed mRNAs. We observed that in response to pravastatin, the genes upregulated in HUVECs were associated with the Rap1‐signaling pathway and osteoclast differentiation (Table 7), while those downregulated are involved in pathways noted in cancer and infection (Table 8).
Table 7.
Results of bioinformatics analyses on upregulated pathways in HUVECs after pravastatin (10 µM) stimulation in comparison to controls
| Upregulated pathways in Statin‐treated versus Control endothelial cells | ||||
|---|---|---|---|---|
| Nr | Pathways | Count | Adj. p‐value | Genes |
| 1 | Rap1 signaling pathway | 5 | .03996061 | FLT4//LCP2//PRKCI//RAP1GAP//VEGFB |
| 2 | Osteoclast differentiation | 4 | .02855664 | FOSB//LCP2//LILRB1//OSCAR |
| 3 | Maturity onset diabetes of the young | 2 | .02041033 | FOXA3//MNX1 |
Abbreviation: Adj, Adjusted.
Table 8.
Results of bioinformatics analyses on downregulated pathways in HUVECs after pravastatin (10 µM) stimulation in comparison to controls
| Downregulated pathways in Statin‐treated versus Control endothelial cells | ||||
|---|---|---|---|---|
| Nr | Pathways | Count | Adj. p‐value | Genes |
| 1 | Pathways in cancer | 5 | .04629817 | CCDC6//JAK1//KIT//TRAF6//XIAP |
| 2 | Epstein‐Barr virus infection | 4 | .03368377 | EIF2AK4//JAK1//SND1//TRAF6 |
| 3 | Hematopoietic cell‐lineage | 3 | .01593718 | IL5//ITGA1//KIT |
| 4 | Neurotrophin signaling pathway | 3 | .03571419 | IRAK3//RPS6KA2//TRAF6 |
| 5 | Toxoplasmosis | 3 | .03571419 | JAK1//TRAF6//XIAP |
| 6 | Hepatitis C | 3 | .0461964 | EIF2AK4//JAK1//TRAF6 |
| 7 | Measles | 3 | .04705986 | EIF2AK4//JAK1//TRAF6 |
| 8 | Ubiquitin mediated proteolysis | 3 | .04969846 | TRAF6//UBE3A//XIAP |
| 9 | Autoimmune thyroid disease | 2 | .04216142 | IL5//TSHR |
| 10 | NOD‐like receptor signaling pathway | 2 | .04649291 | NOD2//TRAF6 |
Abbreviations Adj, Adjusted.
4. DISCUSSION
LncRNAs have been shown to exhibit multiple functions within the cell by modulating gene expression through their interactions with a diverse array of RNA and proteins. A number of lncRNAs have been implicated in the regulation of endothelial function such as endothelial‐to‐mesenchymal transition (Neumann et al., 2018), migration, proliferation, apoptosis (Man et al., 2018), hypoxia‐induced angiogenesis (Voellenkle et al., 2016), and regulation of eNOS expression (Cheng et al., 2019). Despite the growing body of work on lncRNAs in cardiovascular disease, their role in regulating endothelial function in response to lipid‐lowering therapy by statins is unknown. Previous studies have evaluated the effect of statins on individual lncRNAs such as LASER in hepatocytes and RP1‐13D10.2 in lymphoblastoid cells (Li et al., 2019; Mitchel et al., 2016). However, to our knowledge, this is the first study highlighting the differential expression of endothelial lncRNAs and mRNAs following statin treatment.
Using a dose of 10 μM pravastatin (Abe et al., 2006; Panczel et al., 2019), we were able to profile all differentially expressed mRNAs and lncRNAs in human ECs in vitro. A total of 181 lncRNAs and 280 mRNAs were significantly differentially expressed in pravastatin‐treated versus control ECs (log 2‐fold > 2, adjusted p < .05). Of the 30,854 lncRNAs, 95 were upregulated, and 86 were downregulated following pravastatin treatment. Validation qPCR performed for the selected four out of five most up‐ or downregulated lncRNAs demonstrated similar expression patterns in pravastatin‐treated in comparison to control ECs. In the same samples, 190 out of 280 differentially expressed mRNAs were upregulated and 90 mRNAs were downregulated. Validation qPCR performed for six out of eight selected most up‐ or downregulated genes confirmed a similar expression profile in the pravastatin‐treated ECs. The overall expression of lncRNAs was lower than mRNAs, which is consistent with other studies (Chen et al., 2018). Differentially expressed lncRNA and their class distribution were in order with other previously published reports on endothelial lncRNAs, with the majority being intergenic in nature (Singh, Mantella, et al., 2016; Singh, Matkar, Muhammad, et al., 2016; Singh et al., 2017; Singh, Matkar, Quan, et al., 2016). All the differentially regulated lncRNAs were evenly distributed on all chromosomes; however, it was interesting to note that five differentially expressed lncRNAs among the top 10 differentially upregulated and downregulated lncRNAs were located on chromosome 12, which has been previously linked by the whole‐genome scan with prevention and treatment of CVDs (Ganesh et al., 2013; Gong et al., 2003). Our data on chromosomal distribution and location will be of immense interest to geneticists as it provides a base to identify associated lncRNAs if present in the linked locus of the genome‐wide association study.
In our array data, most of the pravastatin‐induced significantly differentially expressed lncRNAs are novel and have not been characterized. However, based on prediction analyses and other relevant studies, we can hypothesize their potential roles in statin‐mediated changes and EC biology. The most significantly upregulated lncRNAs include XLOC_007368, XLOC_004244, RP11‐469H8.6, and RP11‐493L12.2 among others. LncRNA XLOC_007368 is predicted to have a binding site for microRNA 22‐3p, (Paraskevopoulou et al., 2016) which inhibits endothelial inflammation in vitro by targeting ICAM‐1, indicating a role for lncRNA XLOC_007368 in inflammation (Gidlof et al., 2015). LncRNA XLOC_004244 acts as a competitively endogenous RNA for the protein‐coding gene Acyl‐CoA Synthetase Long‐Chain Family Member 3 (ACSL3). ACSL3 belongs to a family of proteins that serve a key role in lipid biosynthesis and fatty acid degradation by converting free long‐chain fatty acids into Fatty Acyl‐CoA Esters (Li et al., 2010). Knockdown of ACSL3 inhibits fatty acid synthesis in primary hepatocytes (Bu et al., 2009) and its suppression perturbs fatty acid oxidation in cancer cells (Padanad et al., 2016). Although statins are known for the regulation of fatty acid metabolism, there are no studies linking statin use to direct changes in the ACSL3 expression. These findings highlight that the upregulation of XLOC_004244 would result in reduced ACSL3 protein expression leading to reduced fatty acid synthesis. LncRNA XLOC_004244 is also associated with the hsa‐miR‐485‐3p (Su et al., 2019). MiR‐485‐3p directly inhibits the expression of PGC‐1α to regulate mitochondrial respiration and cell migration (Lou et al., 2016). LncRNA RP11‐469H8.6 is predicted to target the expression of the aquaporin (AQP2) (Perron et al., 2017), which is known primarily for the regulation of water absorption via collecting duct principal cells, but little is known about AQP2 role in ECs (Kwon et al., 2013). This implies that statins may lead to a decrease in AQP2 expression in endothelial cells. LncRNA RP11‐493L12.2 expression is significantly downregulated in macrophages following mycobacterium tuberculosis and is thought to have a role in the regulation of macrophage apoptosis (Yang et al., 2016). However, its role in endothelial cells is unknown.
The most significantly downregulated lncRNAs include RP11‐876N24.2, XLOC_001373, and XLOC_008645, among others. RP11‐876N24.2 is located near the protein‐coding gene of class II MHC trans‐activator (CIITA). CIITA plays a key role in T cell‐mediated acute rejection following transplant as highlighted by the suppression of CD4 alloresponse following EC‐specific knockdown of CIITA (Abrahimi et al., 2016). The downregulation of RP11‐876N24.2 would, therefore, suggest increased CD4 alloresponse following statin use. LncRNA XLOC_001373, another downregulated lncRNA, has not been characterized but the role of its predicted interactor hsa‐miR‐345‐5p has been well studied in the cardiovascular system (Paraskevopoulou et al., 2016). MiR‐345‐3p directly targets TRAF6, thereby inhibiting oxLDL‐mediated apoptosis and inflammation in ECs by suppression of the TAK1/p38/NF‐kB signaling pathway (Wei et al., 2020). MiR‐345‐3p expression is also decreased in the apolipoprotein‐E deficient mouse model of atherosclerosis, further supporting this hypothesis (Chen et al., 2019). This is in line with statin‐mediated treatment as the downregulation of XLOC_001373 would lead to the upregulation of miR‐345‐3p leading to reduced inflammation in ECs. In addition, circulating miR‐345‐3p levels are elevated in early heart failure ( Wang et al., 2017), highlighting the potential role of XLOC_001373 in the cardiovascular system. LncRNA XLOC_008645 is predicted to interact with hsa‐miR‐127‐3p (Paraskevopoulou et al., 2016), which directly targets α6 Integrin (ITGA6). ITGA6 plays a key role in vascular endothelial growth factor‐A and fibroblast growth factor‐2‐driven angiogenesis (Primo et al., 2010). Therefore, this downregulation of XLOC_008645 would lead to a reduction in angiogenesis, which is relevant as statins, depending on their dose, have also been shown to inhibit and promote angiogenesis (Weis et al., 2002).
Transcriptome analyses following treatment with pravastatin revealed differential expressions of genes associated with endothelial function. The most upregulated genes included Dermokine (DMKN), Apolipoprotein L domain containing 1 (APOLD1), and Piezo‐Type Mechanosensitive Ion Channel Component 2 (PIEZO2), among others. DMKN, the most upregulated gene, promotes vascular endothelial growth factor (VEGF) production and is also associated with high‐density lipid (HDL) proteome within liver cells (Pamir et al., 2019; Pentecost & Yudkin, 1996). APOLD1 is highly expressed in ECs of developing tissues (Regard et al., 2004). Endothelial PIEZO2 plays a role in mechanosensing, which is required for the maintenance of vascular homeostasis (Zhong et al., 2018).
The most downregulated genes included ABL‐Interactor 1 (ABI1), Transmembrane p24 Trafficking Protein 2 (TMED2), and RAS‐Associated Protein 23 (RAB23), among others. ABI1 is critical for coordinating cytoskeletal organization and actin polymerization, and plays a role in cardiovascular development (Kotula, 2012; Ring et al., 2011). Reduced expression of TMED2, a protein involved in vesicular trafficking (Jerome‐Majewska et al., 2010), is in line with other findings where statins are shown to inhibit vesicular trafficking in other cell types (Sakamoto et al., 2011; Wade, 2011). RAB23 belongs to the superfamily of small GTPase, is a negative regulator of Sonic hedgehog signaling (Shh), and plays a critical role in the development of multiple organs (Eggenschwiler et al., 2001, 2006). The role of RAB23 in ECs is unknown; however, statins have been shown to inhibit Shh signaling in hepatic stellate cells. This is due to the cholesterol‐lowering ability of statins as Shh signaling is required for cholesterol biosynthesis (Gordon et al., 2018; Uschner et al., 2015). These findings suggest that Rab23 might represent an important signaling protein in regulating Shh signaling within ECs. Statin‐induced differentially expressed genes that are not characterized in ECs and have also not been studied in relation to statin, warrant further investigation. However, most of the observed changes do support the mechanisms by which statins exert their effect in ECs and other cell types.
Pathway analysis revealed upregulated Rap1 (Ras‐associated protein‐1) signaling pathway, osteoclast differentiation, and maturity‐onset diabetes of the young. Rap1 is a small GTPase that has diverse functionality regulating basic cellular functions such as cell adhesions and junctions, cell migration, and polarization (Zhang et al., 2017). In ECs, Rap1 plays a role in stabilizing newly formed vessels, promotes angiogenesis and endothelial barrier function, and is critical for NO production, therefore, controls the vascular tone and stabilizes blood pressure (Chrzanowska‐Wodnicka et al., 2008; Lakshmikanthan et al., 2015; Yan et al., 2008). The impaired NO production in Rap1 EC‐KO mice is due to the inability of ECs to sense shear stress in the absence of Rap1. Interestingly, statins inhibit EC Rap1 activity in a dose‐dependent manner (Kou et al., 2012). While the actions of statins on the Rap1 pathway have been highlighted before, no functional studies to date have linked lncRNAs and the regulation of the Rap1 signaling pathway. The downregulated pathways after statin‐treatment in EC include pathways in cancer, Epstein‐Barr virus infection, hematopoietic cell‐lineage, and neurotrophin signaling. The cancer‐associated pathway can be attributed to the overlap of cell regulatory genes and lncRNAs that are common to both cardiovascular diseases and cancer. The linkage of the downregulated genes to infection‐related pathways is in line with statin treatment. Statins have been shown to have an anti‐inflammatory effect on ECs as they inhibit the expression of cell adhesion molecules intercellular adhesion molecule‐1 (ICAM‐1) (Chung et al., 2002), vascular cell adhesion molecule (VCAM‐1), and E‐selectin (Rasmussen et al., 2001). While there is not much available information on other significant up‐ or downregulated pathways in the endothelium, our data provide the first hint toward the significant roles of these pathways in the endothelium, particularly after statin‐treatment, and warrant future investigation.
There are some limitations to the present work. Although HUVECs are an established representative cell type for endothelial research in vitro, it is important to validate our findings in other EC types, such as in human coronary artery ECs and human microvascular ECs to confirm the non‐specificity of our findings. Our conclusions cannot be generalized for all statins because we only used pravastatin and different statins can have varying therapeutic and toxic effects both in vitro and in vivo (Ward et al., 2019). One of the strengths, which is also a weakness in our report, is the novelty since most of the identified differentially expressed lncRNA are not characterized; therefore, it is difficult to associate the relevance of these changes with pravastatin and the endothelium. Thus, we strongly recommend further characterization of these novel lncRNAs.
In conclusion, the present study is the first, to our knowledge, to demonstrate the expression profile of lncRNAs and mRNAs in ECs following treatment with pravastatin. We identified several lncRNAs that have previously not been associated with signaling pathways in the endothelium and therefore present as novel targets. Our bioinformatic analyses shed light on some of the pathways that might govern endothelial function following pravastatin treatment. Our findings identify novel biomarkers and potential therapeutic targets in addition to providing insights into the mechanisms underlying the effects of pravastatin in ECs. Future investigations in these directions are warranted.
CONFLICTS OF INTEREST
None.
AUTHOR CONTRIBUTIONS
KS conceived and designed the study. KS and HN carried out the experiments and analyzed the data. SS, HN, ME, DM, and MQ helped improve the discussion. SS, HN, PS, ME and KS wrote, assembled, and revised the manuscript with final figures.
Supporting information
Fig S1‐S2
ACKNOWLEDGMENTS
The funding for this project was provided by the Project Grant (FRN # 153216), Canadian Institutes of Health Research, Canada to KS. KS is also the recipient of the 2018/19 National New Investigator Award‐ Salary Support from the Heart and Stroke Foundation of Canada, Canada.
Singh S, Nguyen HC, Ehsan M, et al. Pravastatin‐induced changes in expression of long non‐coding and coding RNAs in endothelial cells. Physiological Reports. 2020;9:e14661 10.14814/phy2.14661
REFERENCES
- Abe, Y. , Izumi, T. , Urabe, A. , Nagai, M. , Taniguchi, I. , Ikewaki, K. , & Mochizuki, S. (2006). Pravastatin prevents myocardium from ischemia‐induced fibrosis by protecting vascular endothelial cells exposed to oxidative stress. Cardiovascular Drugs and Therapy, 20, 273–280. 10.1007/s10557-006-9525-7 [DOI] [PubMed] [Google Scholar]
- Abrahimi, P. , Qin, L. , Chang, W. G. , Bothwell, A. L. , Tellides, G. , Saltzman, W. M. , & Pober, J. S. (2016). Blocking MHC class II on human endothelium mitigates acute rejection. JCI Insight, 1(1), 10.1172/jci.insight.85293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, T. J. , Meredith, I. T. , Yeung, A. C. , Frei, B. , Selwyn, A. P. , & Ganz, P. (1995). The effect of cholesterol‐lowering and antioxidant therapy on endothelium‐dependent coronary vasomotion. New England Journal of Medicine, 332, 488–493. 10.1056/NEJM199502233320802 [DOI] [PubMed] [Google Scholar]
- Batista, P. J. , & Chang, H. Y. (2013). Long noncoding RNAs: Cellular address codes in development and disease. Cell, 152, 1298–1307. 10.1016/j.cell.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran, M. , Puig, I. , Pena, C. , Garcia, J. M. , Alvarez, A. B. , Pena, R. , …, de Herreros, A. G. (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1‐induced epithelial‐mesenchymal transition. Genes & Development, 22, 756–769. 10.1101/gad.455708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesova, Z. , Piccoli, C. , & Wong, W. T. (1997). Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene, 14, 233–241. 10.1038/sj.onc.1200822 [DOI] [PubMed] [Google Scholar]
- Bu, S. Y. , Mashek, M. T. , & Mashek, D. G. (2009). Suppression of long chain acyl‐CoA synthetase 3 decreases hepatic de novo fatty acid synthesis through decreased transcriptional activity. Journal of Biological Chemistry, 284, 30474–30483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zhang, Y. H. , Pan, X. , Liu, M. , Wang, S. , Huang, T. , & Cai, Y. D. (2018). Tissue Expression Difference between mRNAs and lncRNAs. International Journal of Molecular Sciences, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Li, X. , Wang, J. , Song, N. , Zhu, A. , & Jia, L. (2019). miR‐378a Modulates Macrophage Phagocytosis and Differentiation through Targeting CD47‐SIRPalpha Axis in Atherosclerosis. Scandinavian Journal of Immunology, 90, e12766. [DOI] [PubMed] [Google Scholar]
- Cheng, X. W. , Chen, Z. F. , Wan, Y. F. , Zhou, Q. , Wang, H. , & Zhu, H. Q. (2019). Long non‐coding RNA H19 suppression protects the endothelium against hyperglycemic‐induced inflammation via inhibiting expression of miR‐29b target gene vascular endothelial growth factor a through activation of the protein kinase B/Endothelial nitric oxide synthase pathway. Frontiers in Cell and Developmental Biology, 7, 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska‐Wodnicka, M. , Kraus, A. E. , Gale, D. , White, G. C. 2nd , & Vansluys, J. (2008). Defective angiogenesis, endothelial migration, proliferation, and MAPK signaling in Rap1b‐deficient mice. Blood, 111, 2647–2656. 10.1182/blood-2007-08-109710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, H. K. , Lee, I. K. , Kang, H. , Suh, J. M. , Kim, H. , Park, K. C. , …, Shong, M. (2002). Statin inhibits interferon‐gamma‐induced expression of intercellular adhesion molecule‐1 (ICAM‐1) in vascular endothelial and smooth muscle cells. Experimental & Molecular Medicine, 34, 451–461. [DOI] [PubMed] [Google Scholar]
- Djebali, S. , Davis, C. A. , Merkel, A. , Dobin, A. , Lassmann, T. , Mortazavi, A. , … Gingeras, T. R. (2012). Landscape of transcription in human cells. Nature, 489, 101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler, J. T. , Bulgakov, O. V. , Qin, J. , Li, T. , & Anderson, K. V. (2006). Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Developmental Biology, 290, 1–12. 10.1016/j.ydbio.2005.09.022 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler, J. T. , Espinoza, E. , & Anderson, K. V. (2001). Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature, 412, 194–198. 10.1038/35084089 [DOI] [PubMed] [Google Scholar]
- Fiedler, J. , Breckwoldt, K. , Remmele, C. W. , Hartmann, D. , Dittrich, M. , Pfanne, A. , …, Thum, T. (2015). Development of Long Noncoding RNA‐Based Strategies to Modulate Tissue Vascularization. Journal of the American College of Cardiology, 66, 2005–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh, S. K. , Arnett, D. K. , Assimes, T. L. , Basson, C. T. , Chakravarti, A. , Ellinor, P. T. , … Waldman, S. A. ; American Heart Association Council on Functional G, Translational B, American Heart Association Council on E, Prevention, American Heart Association Council on Basic Cardiovascular S, American Heart Association Council on Cardiovascular Disease in the Y, American Heart Association Council on C, Stroke N and American Heart Association Stroke C . (2013). Genetics and genomics for the prevention and treatment of cardiovascular disease: update: a scientific statement from the American Heart Association. Circulation. 128:2813–2851. [DOI] [PubMed] [Google Scholar]
- Gidlof, O. , Sathanoori, R. , Magistri, M. , Faghihi, M. A. , Wahlestedt, C. , Olde, B. , & Erlinge, D. (2015). Extracellular uridine triphosphate and adenosine triphosphate attenuate endothelial inflammation through miR‐22‐mediated ICAM‐1 inhibition. Journal of Vascular Research, 52, 71–80. 10.1159/000431367 [DOI] [PubMed] [Google Scholar]
- Gong, M. , Zhang, H. , Schulz, H. , Lee, Y. A. , Sun, K. , Bahring, S. , …, Hubner, N. (2003). Genome‐wide linkage reveals a locus for human essential (primary) hypertension on chromosome 12p. Human Molecular Genetics, 12, 1273–1277. 10.1093/hmg/ddg135 [DOI] [PubMed] [Google Scholar]
- Gordon, R. E. , Zhang, L. , Peri, S. , Kuo, Y. M. , Du, F. , Egleston, B. L. , …, Yang, Z. J. (2018). Statins synergize with hedgehog pathway inhibitors for treatment of medulloblastoma. Clinical Cancer Research, 24, 1375–1388. 10.1158/1078-0432.CCR-17-2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther, M. G. , Levine, S. S. , Boyer, L. A. , Jaenisch, R. , & Young, R. A. (2007). A chromatin landmark and transcription initiation at most promoters in human cells. Cell, 130, 77–88. 10.1016/j.cell.2007.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangauer, M. J. , Vaughn, I. W. , & McManus, M. T. (2013). Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genetics., 9, e1003569 10.1371/journal.pgen.1003569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida, N. , & Balligand, J. L. (2014). Low‐density lipoprotein‐cholesterol‐induced endothelial dysfunction and oxidative stress: The role of statins. Antioxidants & Redox Signaling, 20, 1216–1237. 10.1089/ars.2013.5537 [DOI] [PubMed] [Google Scholar]
- Hernandez‐Perera, O. , Perez‐Sala, D. , Navarro‐Antolin, J. , Sanchez‐Pascuala, R. , Hernandez, G. , Diaz, C. , & Lamas, S. (1998). Effects of the 3‐hydroxy‐3‐methylglutaryl‐CoA reductase inhibitors, atorvastatin and simvastatin, on the expression of endothelin‐1 and endothelial nitric oxide synthase in vascular endothelial cells. Journal of Clinical Investigation, 101, 2711–2719. 10.1172/JCI1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdt, L. M. , Hoffmann, S. , Sass, K. , Langenberger, D. , Scholz, M. , Krohn, K. , …, Teupser, D. (2013). Alu elements in ANRIL non‐coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans‐regulation of gene networks. PLoS Genetics., 9, e1003588 10.1371/journal.pgen.1003588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiki, T. , Takeda, K. , Tokunou, T. , Iino, N. , Egashira, K. , Shimokawa, H. , …, Takeshita, A. (2001). Downregulation of angiotensin II type 1 receptor by hydrophobic 3‐hydroxy‐3‐methylglutaryl coenzyme A reductase inhibitors in vascular smooth muscle cells. Arteriosclerosis, Thrombosis, and Vascular Biology, 21, 1896–1901. 10.1161/hq1201.099430 [DOI] [PubMed] [Google Scholar]
- Jerome‐Majewska, L. A. , Achkar, T. , Luo, L. , Lupu, F. , & Lacy, E. (2010). The trafficking protein Tmed2/p24beta(1) is required for morphogenesis of the mouse embryo and placenta. Developmental Biology, 341, 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannel, W. B. , Dawber, T. R. , Friedman, G. D. , Glennon, W. E. , & McNamara, P. M. (1964). Risk factors in coronary heart disease. An evaluation of several serum lipids as predictors of coronary heart disease; the framingham study. Annals of Internal Medicine, 61, 888–899. 10.7326/0003-4819-61-5-888 [DOI] [PubMed] [Google Scholar]
- Kotula, L. (2012). Abi1, a critical molecule coordinating actin cytoskeleton reorganization with PI‐3 kinase and growth signaling. FEBS Letters, 586, 2790–2794. 10.1016/j.febslet.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou, R. , Shiroto, T. , Sartoretto, J. L. , & Michel, T. (2012). Suppression of Galphas synthesis by simvastatin treatment of vascular endothelial cells. Journal of Biological Chemistry, 287, 2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kureishi, Y. , Luo, Z. , Shiojima, I. , Bialik, A. , Fulton, D. , Lefer, D. J. , …, Walsh, K. (2000). The HMG‐CoA reductase inhibitor simvastatin activates the protein kinase Akt and promotes angiogenesis in normocholesterolemic animals. Nature Medicine, 6, 1004–1010. 10.1038/79510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, T. H. , Frokiaer, J. , & Nielsen, S. (2013). Regulation of aquaporin‐2 in the kidney: A molecular mechanism of body‐water homeostasis. Kidney Research and Clinical Practice, 32, 96–102. 10.1016/j.krcp.2013.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmikanthan, S. , Zheng, X. , Nishijima, Y. , Sobczak, M. , Szabo, A. , Vasquez‐Vivar, J. , …, Chrzanowska‐Wodnicka, M. (2015). Rap1 promotes endothelial mechanosensing complex formation, NO release and normal endothelial function. EMBO Reports, 16, 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Hu, Z. , Zhang, W. , Yu, J. , Yang, Y. , Xu, Z. , …, Zeng, C. (2019). Regulation of cholesterol homeostasis by a novel long non‐coding RNA LASER. Scientific Reports, 9, 7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L. O. , Klett, E. L. , & Coleman, R. A. (2010). Acyl‐CoA synthesis, lipid metabolism and lipotoxicity. Biochimica et Biophysica Acta, 1801, 246–251. 10.1016/j.bbalip.2009.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby, P. (2001). Current concepts of the pathogenesis of the acute coronary syndromes. Circulation, 104, 365–372. 10.1161/01.CIR.104.3.365 [DOI] [PubMed] [Google Scholar]
- Lou, C. , Xiao, M. , Cheng, S. , Lu, X. , Jia, S. , Ren, Y. , & Li, Z. (2016). MiR‐485‐3p and miR‐485‐5p suppress breast cancer cell metastasis by inhibiting PGC‐1alpha expression. Cell Death & Disease, 7, e2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund‐Katz, S. , & Phillips, M. C. (2010). High density lipoprotein structure‐function and role in reverse cholesterol transport. Sub‐cellular Biochemistry, 51, 183–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man, H. S. J. , Sukumar, A. N. , Lam, G. C. , Turgeon, P. J. , Yan, M. S. , Ku, K. H. , …, Marsden, P. A. (2018). Angiogenic patterning by STEEL, an endothelial‐enriched long noncoding RNA. Proceedings of the National Academy of Sciences of the United States of America, 115, 2401–2406. 10.1073/pnas.1715182115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meor Anuar Shuhaili, M. F. R. , Samsudin, I. N. , Stanslas, J. , Hasan, S. , & Thambiah, S. C. (2017). Effects of different types of statins on lipid profile: A perspective on Asians. International Journal of Endocrinology and Metabolism, 15, e43319 10.5812/ijem.43319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel, K. , Theusch, E. , Cubitt, C. , Dose, A. C. , Stevens, K. , Naidoo, D. , & Medina, M. W. (2016). RP1‐13D10.2 Is a Novel Modulator of Statin‐Induced Changes in Cholesterol. Circulation: Cardiovascular Genetics, 9, 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugavel, S. , Bugyei‐Twum, A. , Matkar, P. N. , Al‐Mubarak, H. , Chen, H. H. , Adam, M. , …, Singh, K. K. (2018). Valproic acid induces endothelial‐to‐mesenchymal transition‐like phenotypic switching. Frontiers in Pharmacology, 9, 737 10.3389/fphar.2018.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, P. , Jae, N. , Knau, A. , Glaser, S. F. , Fouani, Y. , Rossbach, O. , …, Dimmeler, S. (2018). The lncRNA GATA6‐AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nature Communications, 9, 237 10.1038/s41467-017-02431-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, S. , Totsuka, T. , & Hayashi, S. (1996). Inhibitory effect of fluvastatin, an HMG‐CoA reductase inhibitor, on the expression of adhesion molecules on human monocyte cell line. International Journal of Immunopharmacology, 18, 669–675. 10.1016/S0192-0561(96)00068-9 [DOI] [PubMed] [Google Scholar]
- Noor, R. , Shuaib, U. , Wang, C. X. , Todd, K. , Ghani, U. , Schwindt, B. , & Shuaib, A. (2007). High‐density lipoprotein cholesterol regulates endothelial progenitor cells by increasing eNOS and preventing apoptosis. Atherosclerosis, 192, 92–99. 10.1016/j.atherosclerosis.2006.06.023 [DOI] [PubMed] [Google Scholar]
- Padanad, M. S. , Konstantinidou, G. , Venkateswaran, N. , Melegari, M. , Rindhe, S. , Mitsche, M. , …, Scaglioni, P. P. (2016). Fatty acid oxidation mediated by Acyl‐CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Reports, 16(6), 1614–1628. 10.1016/j.celrep.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamir, N. , Pan, C. , Plubell, D. L. , Hutchins, P. M. , Tang, C. , Wimberger, J. , …, Lusis, A. J. (2019). Genetic control of the mouse HDL proteome defines HDL traits, function, and heterogeneity. Journal of Lipid Research, 60, 594–608. 10.1194/jlr.M090555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panczel, Z. , Kukor, Z. , Supak, D. , Kovacs, B. , Kecskemeti, A. , Czizel, R. , …, Valent, S. (2019). Pravastatin induces NO synthesis by enhancing microsomal arginine uptake in healthy and preeclamptic placentas. BMC Pregnancy Childbirth, 19, 426 10.1186/s12884-019-2507-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou, M. D. , Vlachos, I. S. , Karagkouni, D. , Georgakilas, G. , Kanellos, I. , Vergoulis, T. , …, Hatzigeorgiou, A. G. (2016). DIANA‐LncBase v2: Indexing microRNA targets on non‐coding transcripts. Nucleic Acids Research, 44, D231–D238. 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentecost, B. , & Yudkin, J. S. (1996). The St Vincent Task Force for diabetes: Report of the cardiovascular disease subgroup. Heart, 76, 107–108. 10.1136/hrt.76.2.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, U. , Provero, P. , & Molineris, I. (2017). In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinformatics, 18, 144 10.1186/s12859-017-1535-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal‐Fernandez, I. , Casal‐Dominguez, M. , & Mammen, A. L. (2018). Statins: Pros and cons. Medicina Clínica, 150, 398–402. 10.1016/j.medcli.2017.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primo, L. , Seano, G. , Roca, C. , Maione, F. , Gagliardi, P. A. , Sessa, R. , …, Bussolino, F. (2010). Increased expression of alpha6 integrin in endothelial cells unveils a proangiogenic role for basement membrane. Cancer Research, 70, 5759–5769. [DOI] [PubMed] [Google Scholar]
- Ramkumar, S. , Raghunath, A. , & Raghunath, S. (2016). Statin Therapy: Review of safety and potential side effects. Acta Cardiol Sin., 32, 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, L. M. , Hansen, P. R. , Nabipour, M. T. , Olesen, P. , Kristiansen, M. T. , & Ledet, T. (2001). Diverse effects of inhibition of 3‐hydroxy‐3‐methylglutaryl‐CoA reductase on the expression of VCAM‐1 and E‐selectin in endothelial cells. The Biochemical Journal, 360, 363–370. 10.1042/bj3600363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard, J. B. , Scheek, S. , Borbiev, T. , Lanahan, A. A. , Schneider, A. , Demetriades, A. M. , …, Worley, P. F. (2004). Verge: A novel vascular early response gene. Journal of Neuroscience, 24, 4092–4103. 10.1523/JNEUROSCI.4252-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaie‐Majd, A. , Maca, T. , Bucek, R. A. , Valent, P. , Muller, M. R. , Husslein, P. , …, Baghestanian, M. (2002). Simvastatin reduces expression of cytokines interleukin‐6, interleukin‐8, and monocyte chemoattractant protein‐1 in circulating monocytes from hypercholesterolemic patients. Arteriosclerosis, Thrombosis, and Vascular Biology, 22, 1194–1199. 10.1161/01.ATV.0000022694.16328.CC [DOI] [PubMed] [Google Scholar]
- Rikitake, Y. , & Liao, J. K. (2005). Rho GTPases, statins, and nitric oxide. Circulation Research, 97, 1232–1235. 10.1161/01.RES.0000196564.18314.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring, C. , Ginsberg, M. H. , Haling, J. , & Pendergast, A. M. (2011). Abl‐interactor‐1 (Abi1) has a role in cardiovascular and placental development and is a binding partner of the alpha4 integrin. Proceedings of the National Academy of Sciences of the United States of America, 108, 149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn, J. L. , Kertesz, M. , Wang, J. K. , Squazzo, S. L. , Xu, X. , Brugmann, S. A. , …, Chang, H. Y. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell, 129, 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. G. , Williams, K. J. , Gidding, S. , Boren, J. , Tabas, I. , Fisher, E. A. , …, Sniderman, A. (2018). Eradicating the burden of atherosclerotic cardiovascular disease by lowering apolipoprotein B lipoproteins earlier in life. Journal of the American Heart Association, 7, e009778 10.1161/JAHA.118.009778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, K. , Wada, I. , & Kimura, J. (2011). Inhibition of Rab1 GTPase and endoplasmic reticulum‐to‐Golgi trafficking underlies statin's toxicity in rat skeletal myofibers. Journal of Pharmacology and Experimental Therapeutics, 338, 62–69. 10.1124/jpet.111.179762 [DOI] [PubMed] [Google Scholar]
- Singh, K. K. , Mantella, L. E. , Pan, Y. , Quan, A. , Sabongui, S. , Sandhu, P. , …, Verma, S. (2016). A global profile of glucose‐sensitive endothelial‐expressed long non‐coding RNAs. Canadian Journal of Physiology and Pharmacology, 94, 1007–1014. [DOI] [PubMed] [Google Scholar]
- Singh, K. K. , Matkar, P. N. , Muhammad, S. , Quan, A. , Gupta, V. , Teoh, H. , …, Verma, S. (2016). Investigation of novel LPS‐induced differentially expressed long non‐coding RNAs in endothelial cells. Molecular and Cellular Biochemistry, 421, 157–168. [DOI] [PubMed] [Google Scholar]
- Singh, K. K. , Matkar, P. N. , Pan, Y. , Quan, A. , Gupta, V. , Teoh, H. , …, Verma, S. (2017). Endothelial long non‐coding RNAs regulated by oxidized LDL. Molecular and Cellular Biochemistry, 431, 139–149. [DOI] [PubMed] [Google Scholar]
- Singh, K. K. , Matkar, P. N. , Quan, A. , Mantella, L. E. , Teoh, H. , Al‐Omran, M. , & Verma, S. (2016). Investigation of TGFbeta1‐Induced Long Noncoding RNAs in Endothelial Cells. International Journal of Vascular Medicin, 2016, 2459687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Adam, M. , Matkar, P. N. , Bugyei‐Twum, A. , Desjardins, J. F. , Chen, H. H. , …, Singh, K. K. (2020). Endothelial‐specific Loss of IFT88 promotes endothelial‐to‐mesenchymal transition and exacerbates bleomycin‐induced pulmonary fibrosis. Scientific Reports, 10, 4466 10.1038/s41598-020-61292-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Nguyen, H. , Michels, D. , Bazinet, H. , Matkar, P. N. , Liu, Z. , …, Singh, K. K. (2020). BReast CAncer susceptibility gene 2 deficiency exacerbates oxidized LDL‐induced DNA damage and endothelial apoptosis. Physiol Rep., 8, e14481 10.14814/phy2.14481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, H. O. , Bayazeed, B. , Hook, G. , Johnson, A. , Cronin, J. , & Baron, A. D. (1997). Endothelial dysfunction is associated with cholesterol levels in the high normal range in humans. Circulation, 96, 3287–3293. 10.1161/01.CIR.96.10.3287 [DOI] [PubMed] [Google Scholar]
- Su, L. , Chen, S. , Zheng, C. , Wei, H. , & Song, X. (2019). Meta‐analysis of gene expression and identification of biological regulatory mechanisms in Alzheimer's disease. Frontiers in Neuroscience, 13, 633 10.3389/fnins.2019.00633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky, I. , & Bartel, D. P. (2013). lincRNAs: Genomics, evolution, and mechanisms. Cell, 154, 26–46. 10.1016/j.cell.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uschner, F. E. , Ranabhat, G. , Choi, S. S. , Granzow, M. , Klein, S. , Schierwagen, R. , …, Trebicka, J. (2015). Statins activate the canonical hedgehog‐signaling and aggravate non‐cirrhotic portal hypertension, but inhibit the non‐canonical hedgehog signaling and cirrhotic portal hypertension. Scientific Reports, 5, 14573 10.1038/srep14573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellenkle, C. , Garcia‐Manteiga, J. M. , Pedrotti, S. , Perfetti, A. , De Toma, I. , Da Silva, D. , …, Martelli, F. (2016). Implication of Long noncoding RNAs in the endothelial cell response to hypoxia revealed by RNA‐sequencing. Scientific Reports, 6, 24141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade, J. B. (2011). Statins affect AQP2 traffic. American Journal of Physiology. Renal Physiology, 301, F308 10.1152/ajprenal.00248.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlestedt, C. (2013). Targeting long non‐coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov., 12, 433–446. 10.1038/nrd4018 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Song, C. , Zhou, X. , Han, X. , Li, J. , Wang, Z. , …, Cao, H. (2017). Mitochondria Associated MicroRNA Expression Profiling of Heart Failure. BioMed Research International, 2017, 4042509 10.1155/2017/4042509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski, O. , & Chang, H. Y. (2011). Long noncoding RNAs and human disease. Trends in Cell Biology, 21, 354–361. 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Ward, N. C. , Watts, G. F. , & Eckel, R. H. (2019). Statin Toxicity. Circulation Research, 124, 328–350. 10.1161/CIRCRESAHA.118.312782 [DOI] [PubMed] [Google Scholar]
- Wassmann, S. , Laufs, U. , Baumer, A. T. , Muller, K. , Konkol, C. , Sauer, H. , …, Nickenig, G. (2001). Inhibition of geranylgeranylation reduces angiotensin II‐mediated free radical production in vascular smooth muscle cells: Involvement of angiotensin AT1 receptor expression and Rac1 GTPase. Molecular Pharmacology, 59, 646–654. 10.1124/mol.59.3.646 [DOI] [PubMed] [Google Scholar]
- Wei, Q. , Tu, Y. , Zuo, L. , Zhao, J. , Chang, Z. , Zou, Y. , & Qiu, J. (2020). MiR‐345‐3p attenuates apoptosis and inflammation caused by oxidized low‐density lipoprotein by targeting TRAF6 via TAK1/p38/NF‐kB signaling in endothelial cells. Life Sciences, 241, 117142 10.1016/j.lfs.2019.117142 [DOI] [PubMed] [Google Scholar]
- Weis, M. , Heeschen, C. , Glassford, A. J. , & Cooke, J. P. (2002). Statins have biphasic effects on angiogenesis. Circulation, 105, 739–745. 10.1161/hc0602.103393 [DOI] [PubMed] [Google Scholar]
- Yan, J. , Li, F. , Ingram, D. A. , & Quilliam, L. A. (2008). Rap1a is a key regulator of fibroblast growth factor 2‐induced angiogenesis and together with Rap1b controls human endothelial cell functions. Molecular and Cellular Biology, 28, 5803–5810. 10.1128/MCB.00393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X. , Yang, J. , Wang, J. , Wen, Q. , Wang, H. , He, J. , …, Ma, L. (2016). Microarray analysis of long noncoding RNA and mRNA expression profiles in human macrophages infected with Mycobacterium tuberculosis. Scientific Reports, 6, 38963 10.1038/srep38963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B. , Arun, G. , Mao, Y. S. , Lazar, Z. , Hung, G. , Bhattacharjee, G. , …, Spector, D. L. (2012). The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis‐regulatory role in the adult. Cell Reports, 2, 111–123. 10.1016/j.celrep.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. L. , Wang, R. C. , Cheng, K. , Ring, B. Z. , & Su, L. (2017). Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol Med., 14, 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Sun, B. K. , Erwin, J. A. , Song, J. J. , & Lee, J. T. (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science, 322, 750–756. 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, M. , Komarova, Y. , Rehman, J. , & Malik, A. B. (2018). Mechanosensing Piezo channels in tissue homeostasis including their role in lungs. Pulmonary Circulation, 8, 2045894018767393 10.1177/2045894018767393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2


