Abstract
Background:
A critical challenge in genomic medicine is identifying the genetic and environmental risk factors for disease. Currently, the available data links a majority of known coding human genes to phenotypes, but the environmental component of human disease is extremely underrepresented in these linked data sets. Without environmental exposure information, our ability to realize precision health is limited, even with the promise of modern genomics. Achieving integration of gene, phenotype, and environment will require extensive translation of data into a standard, computable form and the extension of the existing gene/phenotype data model. The data standards and models needed to achieve this integration do not currently exist.
Objectives:
Our objective is to foster development of community-driven data-reporting standards and a computational model that will facilitate the inclusion of exposure data in computational analysis of human disease. To this end, we present a preliminary semantic data model and use cases and competency questions for further community-driven model development and refinement.
Discussion:
There is a real desire by the exposure science, epidemiology, and toxicology communities to use informatics approaches to improve their research workflow, gain new insights, and increase data reuse. Critical to success is the development of a community-driven data model for describing environmental exposures and linking them to existing models of human disease. https://doi.org/10.1289/EHP7215
Introduction
Background
In the rapidly advancing era of genomic medicine, a critical challenge is to identify the genetic etiologies and environmental factors in disease in order to translate basic science to inform interventions and advance health care (Vineis and Russo 2018). Currently, available human data associates of known human coding genes with phenotype data (Amberger et al. 2015; Davis et al. 2019; Landrum et al. 2016; MacArthur et al. 2017; Rath et al. 2012). By including gene and phenotype relationships derived using model organisms, coverage is closer to 83% (Mungall et al. 2017), but the environmental component of these relationships is extremely underrepresented in these and other public databases (Vineis 2004). Without considering the effect of the environment, our ability to understand human disease and realize precision health is limited, even with the promise of modern genomics (Wesseling et al. 2020). Achieving integration of phenotype, genotype, and environment information will require an extensive translation of data into a computable form and the extension of the gene/phenotype data model, both to ensure discovery of extant data and to provide structure that encourages new data and analyses (NRC 2011). The informatics approaches needed to address these challenges have historically been focused on genomics, with less attention paid to additional types of data streams.
Recent efforts to integrate biomedical databases have resulted in the creation of several related, but often unconnected, biomedical knowledge graphs (Nicholson and Greene 2020), a type of data structure that prioritizes relationships between data points. Because of their structure, knowledge graphs have great potential to find patterns in data that are hidden due to the volume, heterogeneity, and complexity of that data (Bakal et al. 2018). One such biological knowledge graph has been developed by the Monarch Initiative (Mungall et al. 2017; Shefchek et al. 2020) and contains information about genes, variants, phenotypes, biological processes, proteins, and diseases; however, information about the effect of environmental exposures on these entities is still very much underrepresented. This is for two reasons: a) there are few curated data sources that associate environmental exposures to phenotypic outcomes in a structured manner (Davis et al. 2019; Richarz 2019); and b) the complexity of exposure science has not yet been modeled sufficiently using modern semantic structures so as to allow large-scale data integration (Richarz 2019). In this context, interactions between specific genotypes and environmental factors cannot be determined. Consequently, the types of algorithms used for genetic diagnostics that rely on the Monarch knowledge graph are not accessible for diseases that have critical environmental components (Smedley et al. 2015), such as a spectrum of environmental causes, exacerbations, compensatory mechanisms, repair, and potential therapeutic interventions. Thus, the authors propose that it is essential that environmental exposure data be included in translational science modeling efforts in order to make laboratory insights available for mechanistic and etiologic discovery tools.
Including exposure data in disease models is nontrivial because of the metadata needed to characterize key environmental factors and their interactions with mechanistic biological events that likely contribute to the pathogenesis of specific diseases (Richarz 2019). To use this data for risk assessment, issues surrounding the temporality of exposure, such as timing relative to a life cycle (Aaseth et al. 2020), duration of exposure (Lee and Steemers 2017), frequency of exposure (Spear 2020), and latency (Gwinn et al. 2011), as well as the route of exposure (Custer et al. 2016) and point of contact are critical components of the metadata (U.S. EPA 2019). Furthermore, research on environmental exposures and human health often does not include a binary (yes or no) health measure but, instead, relies on continuous measures of function such as blood pressure or intelligence quotient, or it may be based on surrogate panels to represent specific disease indicators (Manisalidis et al. 2020; Patel et al. 2017; Schwarzenbach et al. 2010; Fritschi 2011). Evidence codes (Rogers and Ben-Hur 2009) and probability measures (Kluxen 2020) are needed to correctly weigh a single piece of evidence in an integrated data set. All of these metadata increase the size and complexity of the model but are essential for correct interpretation of the data.
Another modeling concern is that environmental exposures do not occur in isolation—interactions among the multiple exposures that characterize the real world can be critical to exposure health impacts, but there are limited human (or experimental) data upon which to comprehensively model exposure mixtures (Vineis and Russo 2018). In reality, multiple heterogeneous stressors, as well as potential mitigating factors, combine to determine the actual vulnerability to disease (U.S. EPA 2003). For example, an observable effect of a single toxic exposure also includes the effect of an organism’s intrinsic traits, living conditions, and the accumulation of any previous exposures [described by the exposome (Wild 2005)]. Conceptual site models used in cumulative risk assessment attempts to capture those complexities by mapping stressors and the propagation of their impacts with some success (Abt et al. 2010; Menzie et al. 2007; U.S. EPA 2008); however, most of these approaches identify qualitative associations and patterns rather than quantitative, causal interactions. A workflow that simultaneously integrates data from source to outcome has recently been proposed to address this need and may serve to guide the integration of ontologies for exposure and health outcomes (Jarabek and Hines 2019).
Challenges
Here the authors propose the concept of a computable exposure to refer to a representation of exposure data and metadata that can be used by an algorithm or a piece of software to answer a question. Translating exposure data from a human-readable to a computable format is nontrivial primarily because the cause-and-effect relationships being studied can be complex (as discussed above) and laboratories producing these data are not using a community-wide standard (Boyles et al. 2019). Several toxicology-focused databases and data repositories exist [such as the Chemical Effects in Biological Systems (CEBS) (Lea et al. 2017), Toxicology Data Network (TOXNET) (Fowler and Schnall 2014), Comparative Toxicogenomics Database (CTD) (Davis et al. 2017), National Health and Nutrition Examination Survey (NHANES), ExpoCast, and Aggregated Computational Toxicology Resource (ACToR) databases (Judson et al. 2012)] that can provide semi-structured information about the effects of environmental exposures on several species (Davis et al. 2020). Although providing quality data, no resource can present the full picture of exposure effects on human health; thus, integration across databases is needed. Without a common standard, there is no way to quickly and easily link toxicology-focused databases with each other or the body of related biological data (Boyles et al. 2019). Environmental exposure data remains extremely difficult to search, integrate, and compute upon—not only because of the complexity but also because of the lack of convergence on exposure standards and terminologies (Watford et al. 2019). Semantic approaches can provide an automated solution to connecting databases if a minimal set of standards are applied.
The specific aim of this commentary is to put forward a proposed semantic model for exposures. This model was developed collaboratively during the “Computable Exposures Workshop” convened for that purpose and attended by exposure scientists, ontologists, toxicologists, computer scientists, epidemiologists, librarians, ecotoxicologists, and data scientists. During the workshop, participants developed use cases and competency questions to inform the model (see the Supplemental Material). Use cases are narratives written from the perspective of a hypothetical user trying to complete a specific task in the proposed infrastructure. They are used to help constrain the features of the system so that development effort is focused on a user need. Competency questions are specific queries with known answers that function as a test of the infrastructure. They could be used to identify missing functionality and data. This effort expands on previous community-building efforts (Mattingly et al. 2016) and advances the field of translational public health.
Those previous efforts failed to gain momentum for multiple reasons. First, individual researchers are often not convinced of the use of standards or semantics in their own research. It is only across research groups, data sets, and perspectives that the specific utility can be realized and recognized. Second, technology and data infrastructure have advanced enough that projects such as the Monarch Initiative (Shefchek et al. 2020) and Exomiser (Smedley et al. 2015) have been able to demonstrate significant benefits of semantic technology in medicine (e.g., diagnosing disease); thus demonstrating the return on investing in standards and semantics development. Third, with the advent of the Findability, Accessibility, Interoperability, and Reusability (FAIR) principles (Wilkinson et al. 2016), funding agencies are now investing more heavily in standards and semantic technologies, thereby providing necessary resources that were not available in the past. We anticipate a higher chance of success now because of a convergence of demonstrated utility, maturation of the technology, funding agency expectations for FAIR data, and financial investments that have been able to catch the interest of researchers who want to take their work to the next level using semantic technology. Whereas before, semantic technology was perceived as a solution looking for a problem, now that enough infrastructure is in place to have a visible impact, users are interested. There is a critical mass of users who are already using this technology in their research and in the clinic (Jovanović and Bagheri 2017; Kamdar et al. 2019; Roos et al. 2017; Zhang et al. 2018; Losko and Heumann 2017; Iyappan et al. 2016; Rashid et al. 2020).
Discussion
Researchers are beginning to explore the role of the entire genome in human disease, and in doing so, numerous additional opportunities arise, for example, defining the role of environmental exposures in the phenotypic spectrum of common disease (Vineis and Russo 2018). Recent developments in semantic data structures for geospatial information have added new integration possibilities for heterogeneously labeled data that may impact health outcomes (e.g., ZIP code, city, latitude, and longitude) (Hu 2018). The biomedical informatics community has been interested in incorporating environmental exposures into existing data models for several years (Martin Sanchez et al. 2014). The concepts of computational exposure science (Egeghy et al. 2016) and exposome informatics (Martin Sanchez et al. 2014; Sarigiannis et al. 2018) have been introduced to a wider community. A basic semantic model for an exposure event [Exposure Ontology (ExO)] has been proposed and incorporated into the CTD and the Environmental Conditions, Treatments, and Exposures Ontology (ECTO) (Davis et al. 2017; Grondin et al. 2016; Mattingly et al. 2012). The exposure science and toxicology communities see promise in data science methods and are ready to explore them to further their research (Mattingly et al. 2016). These multiple, isolated community efforts are ready for combined collaboration to advance the field. The assertions and semantic data model for exposure events presented below reflect the combined work of this interdisciplinary community.
Gaps in Exposure Science Data Services
Exposure science and toxicology research now requires the integration of multiple data types and studies (Canzler et al. 2020). The use cases developed during the workshop (see the section “Use Cases” in the Supplemental Material) represent the types of translational questions researchers would like to answer but currently cannot. For example, “What genetic variants are found in leukemias associated with exposure to benzene in the air?” requires disease and genetic data from patients and measurements of benzene from their environment. These data, if they exist, likely require significant normalization in order to be analyzed together. In the experience of the authors, the time needed to find, access, and integrate all the data makes answering a question like this intractable. The functionality needed to normalize and transform at scale does not currently exist. Another important point to make regarding this use case is that there likely will not be direct measurements addressing the specific case in question. When needed, data gaps can be filled using formal-logic reasoning over a knowledge graph. In this way, information about benzene in one data set and information about model organisms in another data set can be reasoned over to form a hypothesis about genetic variants and organisms of interest.
This degree of data integration requires shared standards, a good persistent identifier system, quality control, good data practices, and open data (Stathias et al. 2018). None of these things are uniformly practiced across the exposure science or toxicology communities. This lack of shared standards creates an inefficient research practice of having to continuously recreate models and workflows, spend huge amounts of time looking for data, and recollect data. Computable exposures can fill these gaps through their requirements for standards and their integration with other data types via the knowledge graph. Community-identified requirements for exposure science data services are enabled by computable exposures, but computable exposures require a foundation of good data practices.
Four important gaps were identified by workshop participants that prevent the adoption and use of computable exposures.
A minimum reporting standard for exposure science and toxicology. A major barrier to the reuse of exposure data is sparse metadata. A reporting standard would ensure that at least a minimum set of metadata are reported. This would help with reuse and integration.
Curated mappings across chemical authorities. Chemical-naming authorities do not all agree on how to name chemicals or chemical mixtures. It is very difficult to judge which authority to use and the quality of mappings across authorities. Curated mappings across chemical authorities would lead to less ambiguity and make it easier for researchers to integrate data via automation.
A semantic model for exposure data. Inferencing requires a semantic data model that can capture the necessary metadata and link computable exposures to the rest of the knowledge graph. Having the semantics in place solves terminology and granularity problems, in addition to the benefits of using logical inference for hypothesis generation.
Ontology terms. Existing ontologies do not contain all needed classes for every type of common exposure. Some development effort will be needed to add terms as the community needs them.
Once these gaps are filled, we can apply modern data science to discover, access, and integrate exposure data at scale.
Community Development
The objective of the “Computable Exposures Workshop” was to foster the development of data reporting standards and a computational model that will facilitate the inclusion of exposure data in computational analysis of human disease. To this end, the workshop brought together a diverse community to discuss the nature of exposure data and how it might best be integrated with genomic and phenomic data. The technologists were asked to consider the science that motivated the data and questions. The toxicologists were asked to think about their workflows, data management practices, and pain points from an external perspective. The use cases represented the hopes for functions that would be enabled by computable exposures. These included large-scale integration of multiple data types and discovery of new insights using semantic models and inferencing. The competency questions represented basic functionality. The answers to these questions are known and test the breadth and depth of the data and semantic model. According to the vast model development experience of the authors, understanding the aspirations and expectations of the community is the first step in creating a semantic model to serve their research needs, which is an essential requirement for adoption of the resulting model. The use cases and competency questions will help ensure that the infrastructure is fit for purpose to meet real-world user needs (Copeland et al. 2012). The outcomes of this workshop were presented virtually at the Society of Toxicology meeting and at the U.S. Semantic Technologies Symposium (US2TS) semantic technology meeting in 2020. Buy-in from the community of stakeholders is necessary for the lasting change in data culture needed to make computable exposures a reality (Copeland et al. 2012). Closing the gaps must be done by the community, facilitated by data science (Copeland et al. 2012).
Proposed Data Model
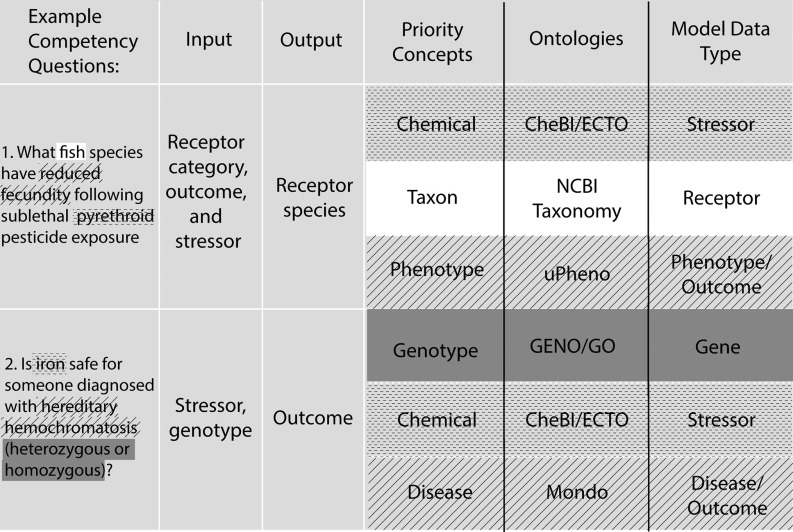
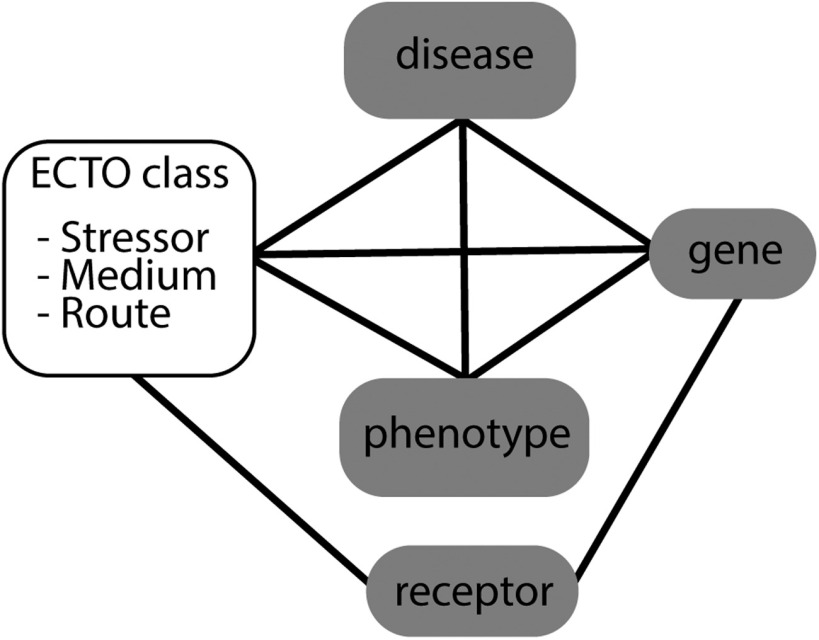
The competency questions (see the section “Competency Questions” in the Supplemental Material) developed during the workshop were analyzed by the authors to inform development of the proposed data model. The authors identified priority concepts in each question, and mapped them to an ontology and a model data type (Figure 1). Each question was categorized according to inputs and outputs given and requested (respectively) by a hypothetical user as implied by the competency question. This exercise revealed that priority concepts could be described by existing ontologies even if specific classes were not already available in those ontologies. The structure of these questions formed the basis of a data model that incorporated all priority concepts (Figure 2). Some outstanding modeling issues remain:
The outcome can be a change in risk, susceptibility, or severity of a preexisting condition (See Figure 1, question 2).
The outcome can be another exposure.
Exposures and outcomes can occur across generations.
Outcomes can be linked to deficiencies.
Exposures rarely have single stressors.
Priority concepts can be implied rather than explicitly stated.
Figure 1.
Example competency questions. Competency questions were developed by workshop participants (see the section “Competency Questions” in the Supplemental Material) to help guide the data model and expose deficiencies in ontological coverage. The input indicates the type of information provided by the hypothetical user in the example question. The output indicates the type of information the hypothetical user is requesting. The semantic types of the inputs and outputs are listed as priority concepts that can be represented by the listed ontologies, such as chemical (dashed line), taxon (white), phenotype or disease (hashed lines), and genotype (gray). These semantic types and are represented in the semantic model (Figure 2) as the model data type. Note: CheBI, Chemicals of Biological Interest; ECTO, Environmental Conditions, Treatments, and Exposures Ontology; GENO, Genotype Ontology; GO, Gene Ontology; Mondo, Mondo Disease Ontology; NCBI, National Center for Biotechnology Information; uPheno, Unified Phenotype Ontology.
Figure 2.
Semantic model for representing environmental exposures. Exposure events (open box, represented by terms in the ECTO) can be incorporated into the Monarch knowledge graph via direct links to the diseases and phenotypes resulting from the exposure, the genes being affected, and the organism(s) or organism parts that are being exposed. Note: ECTO, Environmental Conditions, Treatments, and Exposures Ontology.
The preliminary semantic data model uses existing ontologies to represent exposure events and links environmental exposures to other data types (Figure 2). Exposure events are represented using classes from ECTO (Figure 3). ECTO can be used for both humans and research organisms, and integrates a number of resources such as the CTD’s ExO (Grondin et al. 2016; Mattingly et al. 2012) and the Zebrafish Information Network’s Zebrafish Experimental Conditions Ontology (ZECO) into a logically consistent and integrated ontology. The workshop participants made the importance of contextual data very clear, providing the application domain and explicit limitation on reuse for other applications. ECTO classes are precomposed and model the stressor (the exposure agent), the route (e.g., oral), and medium (e.g., air, water, food). Other types of contextual data, such as information about the receptor [the organism(s) being exposed] and the frequency and duration of the exposure event, can be added as annotations on the instance of the exposure event. By using ECTO classes to represent exposure events, we can use the Monarch knowledge graph to integrate exposures, phenotypes, genes, and diseases in a queryable resource. The workshop participants identified three high-priority modeling needs for designing computable exposure data: a) highly granular metadata annotations, b) modeling multi-stressor exposures; and c) linking combinations of exposures and genotypes to a particular phenotype. Including exposures in the knowledge graph can better reveal how they influence genetic diseases, provide causal evidence for nongenetic diseases and phenotypes, and place into context exposures that have buffering properties.
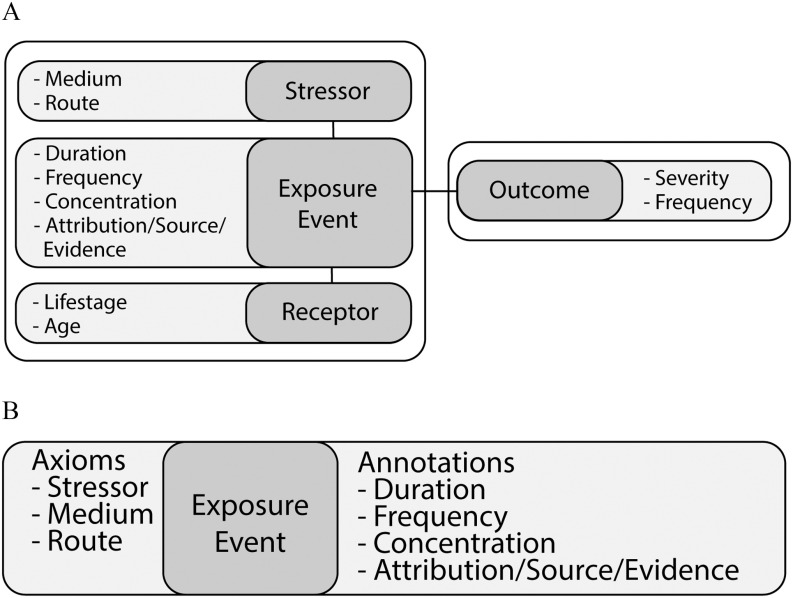
Figure 3.
ExO and ECTO exposure data model. (A) The ExO (Mattingly et al. 2012) model combines stressors and receptors in the representation of the exposure event, which is linked to an outcome. Each element (dark gray) has associated metadata (light gray). (B) Exposure events in ECTO are precomposed with the stressor, medium, and route (when known) contained in the axiomatic definition: “exposure event” and (“has exposure stimulus” some “stressor”) and (“has exposure medium” some “medium”) and (“has exposure route” some “route”). Additional metadata about the exposure event (dark gray) are added as annotations on the event (light gray). Information about the receptor and the outcome are linked to the exposure event in the larger knowledge graph as shown in Figure 2. Note: ECTO, Environmental Conditions, Treatments, and Exposures Ontology; ExO, Exposure Ontology.
ECTO builds on the previous work of Mattingly et al. (2012) on ExO to create the specific exposure classes that are needed for inferencing (Figure 3) but are considered out of scope for ExO (Davis et al. 2017; Mattingly et al. 2012). Further, ExO was designed prior to the modern logical interoperability capabilities that ECTO is well positioned to exploit. Toward these ends, ECTO, with its modern semantic engineering, acts as a unifying ontology by incorporating classes from other ontologies—such as the Environment Ontology (EnvO) (Buttigieg et al. 2016), Medical Action Ontology (MAxO), and Chemical Entities of Biological Interest (ChEBI) (Hastings et al. 2016)—to precompose exposure classes. This reuse of ontology terms that is out of scope for ExO helps to ensure data integration to support applications that adhere to Open Biomedical Ontologies (OBO) Foundry principles (Smith et al. 2007), now and into the future.
In summary, informatics approaches can help the toxicology and exposure science community to standardize their research workflow, gain new insights, and increase data reuse and to increase their contribution to translational science. Existing data repositories contain a wealth of information, but these data are difficult to aggregate across databases or integrate with other data types because there are no community-endorsed best practices or data standards. In the “Computable Exposures Workshop,” our community-development efforts brought together an interdisciplinary group of researchers to identify gaps in exposure data services and developed use cases and competency questions to inform a proposed semantic model for integrating exposure data into a larger biomedical knowledge graph.
Supplementary Material
Acknowledgments
The authors and participants of the “Computable Exposures Workshop” thank the National Institutes of Health for funding provided through their grant U13# 5U13CA221044-03.
References
- Aaseth J, Wallace DR, Vejrup K, Alexander J. 2020. Methylmercury and developmental neurotoxicity: a global concern. Curr Opin Toxicol 19:80–87, 10.1016/j.cotox.2020.01.005. [DOI] [Google Scholar]
- Abt E, Rodricks JV, Levy JI, Zeise L, Burke TA. 2010. Science and decisions: advancing risk assessment. Risk Anal 30(7):1028–1036, PMID: 20497395, 10.1111/j.1539-6924.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. 2015. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 43(database issue):D789–D798, PMID: 25428349, 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal G, Talari P, Kakani EV, Kavuluru R. 2018. Exploiting semantic patterns over biomedical knowledge graphs for predicting treatment and causative relations. J Biomed Inform 82:189–199, PMID: 29763706, 10.1016/j.jbi.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles RR, Thessen AE, Waldrop A, Haendel MA. 2019. Ontology-based data integration for advancing toxicological knowledge. Curr Opin Toxicol 16:67–74, 10.1016/j.cotox.2019.05.005. [DOI] [Google Scholar]
- Buttigieg PL, Pafilis E, Lewis SE, Schildhauer MP, Walls RL, Mungall CJ. 2016. The environment ontology in 2016: bridging domains with increased scope, semantic density, and interoperation. J Biomed Semantics 7(1):57, PMID: 27664130, 10.1186/s13326-016-0097-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canzler S, Schor J, Busch W, Schubert K, Rolle-Kampczyk UE, Seitz H, et al. 2020. Prospects and challenges of multi-omics data integration in toxicology. Arch Toxicol 94(2):371–388, PMID: 32034435, 10.1007/s00204-020-02656-y. [DOI] [PubMed] [Google Scholar]
- Copeland M, Brown A, Parkinson H, Stevens R, Malone J. 2012. The SWO project: a case study of applying agile ontology engineering methods in community driven ontologies. In: Proceedings of the Third International Conference on Biomedical Ontology. Cornet R, Stevens R, editors. 21–25 July 2012. Aachen, Germany: Sun SITE Central Europe, CEUR-Workshop Proceedings; http://staff.cs.manchester.ac.uk/∼browna/publications/icbo2012-swo.pdf [accessed 13 December 2020]. [Google Scholar]
- Custer KW, Hammerschmidt CR, Burton GA Jr.. 2016. Nickel toxicity to benthic organisms: the role of dissolved organic carbon, suspended solids, and route of exposure. Environ Pollut 208(pt B):309–317, PMID: 26552544, 10.1016/j.envpol.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, et al. 2017. The comparative toxicogenomics database: update 2017. Nucleic Acids Res 45(D1):D972–D978, PMID: 27651457, 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers TC, Grondin CJ, Johnson RJ, Sciaky D, Wiegers J, et al. 2020. Leveraging the comparative toxicogenomics database to fill in knowledge gaps for environmental health: a test case for air pollution-induced cardiovascular disease. Toxicol Sci 177(2):392–404, PMID: 32663284, 10.1093/toxsci/kfaa113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Wiegers J, Wiegers TC, Mattingly CJ. 2019. Public data sources to support systems toxicology applications. Curr Opin Toxicol 16:17–24, 10.1016/j.cotox.2019.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeghy PP, Sheldon LS, Isaacs KK, Özkaynak H, Goldsmith MR, Wambaugh JF, et al. 2016. Computational exposure science: an emerging discipline to support 21st-century risk assessment. Environ Health Perspect 124(6):697–702, PMID: 26545029, 10.1289/ehp.1509748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschi L. 2011. Burden of Disease from Environmental Noise: Quantification of Healthy Life Years Lost in Europe. Copenhagen, Denmark: World Health Organization. Regional Office for Europe, https://apps.who.int/iris/handle/10665/326424. [Google Scholar]
- Fowler S, Schnall JG. 2014. TOXNET: information on toxicology and environmental health. Am J Nurs 114(2):61–63, PMID: 24481372, 10.1097/01.NAJ.0000443783.75162.79. [DOI] [PubMed] [Google Scholar]
- Grondin CJ, Davis AP, Wiegers TC, King BL, Wiegers JA, Reif DM, et al. 2016. Advancing exposure science through chemical data curation and integration in the comparative toxicogenomics database. Environ Health Perspect 124(10):1592–1599, PMID: 27170236, 10.1289/EHP174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn MR, DeVoney D, Jarabek AM, Sonawane B, Wheeler J, Weissman DN, et al. 2011. Meeting report: mode(s) of action of asbestos and related mineral fibers. Environ Health Perspect 119(12):1806–1810, PMID: 21807578, 10.1289/ehp.1003240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, et al. 2016. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res 44(D1):D1214–D1219, PMID: 26467479, 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y. 2018. Geospatial Semantics. In: Comprehensive Geographic Information Systems. Huang B, ed. Amsterdam, Netherlands: Elsevier, 80–94. [Google Scholar]
- Iyappan A, Kawalia SB, Raschka T, Hofmann-Apitius M, Senger P. 2016. NeuroRDF: semantic integration of highly curated data to prioritize biomarker candidates in Alzheimer’s disease. J Biomed Semantics 7(July):45, PMID: 27392431, 10.1186/s13326-016-0079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarabek AM, Hines DE. 2019. Mechanistic integration of exposure and effects: advances to apply systems toxicology in support of regulatory decision-making. Curr Opin Toxicol 16:83–92, 10.1016/j.cotox.2019.09.001. [DOI] [Google Scholar]
- Jovanović J, Bagheri E. 2017. Semantic annotation in biomedicine: the current landscape. J Biomed Semantics 8(1):44, PMID: 28938912, 10.1186/s13326-017-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Martin MT, Egeghy P, Gangwal S, Reif DM, Kothiya P, et al. 2012. Aggregating data for computational toxicology applications: the U.S. Environmental Protection Agency (EPA) Aggregated Computational Toxicology Resource (ACToR) system. Int J Mol Sci 13(2):1805–1831, PMID: 22408426, 10.3390/ijms13021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar MR, Fernández JD, Polleres A, Tudorache T, Musen MA. 2019. Enabling Web-scale data integration in biomedicine through Linked Open Data. NPJ Digit Med 2(September):90, PMID: 31531395, 10.1038/s41746-019-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluxen FM. 2020. “New statistics” in regulatory toxicology? Regul Toxicol Pharmacol 117: 104763, PMID: 32781239, 10.1016/j.yrtph.2020.104763. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, et al. 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(D1):D862–D868, PMID: 26582918, 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea IA, Gong H, Paleja A, Rashid A, Fostel J. 2017. CEBS: a comprehensive annotated database of toxicological data. Nucleic Acids Res 45(D1):D964–D971, PMID: 27899660, 10.1093/nar/gkw1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WV, Steemers K. 2017. Exposure duration in overheating assessments: a retrofit modelling study. Build Res Inf 45(1–2):60–82, 10.1080/09613218.2017.1252614. [DOI] [Google Scholar]
- Losko S, Heumann K. 2017. Semantic data integration and knowledge management to represent biological network associations. Methods Mol Biol 1613:403–423, PMID: 28849570, 10.1007/978-1-4939-7027-8_16. [DOI] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. 2017. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45(D1):D896–D901, PMID: 27899670, 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manisalidis I, Stavropoulou E, Stavropoulos A, Bezirtzoglou E. 2020. Environmental and health impacts of air pollution: a review. Front Public Health 8:14, PMID: 32154200, 10.3389/fpubh.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Sanchez F, Gray K, Bellazzi R, Lopez-Campos G. 2014. Exposome informatics: considerations for the design of future biomedical research information systems. J Am Med Inform Assoc 21(3):386–390, PMID: 24186958, 10.1136/amiajnl-2013-001772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, Boyles R, Lawler CP, Haugen AC, Dearry A, Haendel M. 2016. Laying a community-based foundation for data-driven semantic standards in environmental health sciences. Environ Health Perspect 124(8):1136–1140, PMID: 26871594, 10.1289/ehp.1510438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattingly CJ, McKone TE, Callahan MA, Blake JA, Hubal EAC. 2012. Providing the missing link: the exposure science ontology ExO. Environ Sci Technol 46(6):3046–3053, PMID: 22324457, 10.1021/es2033857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzie CA, MacDonell MM, Mumtaz M. 2007. A phased approach for assessing combined effects from multiple stressors. Environ Health Perspect 115(5):807–816, PMID: 17520072, 10.1289/ehp.9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, McMurry JA, Köhler S, Balhoff JP, Borromeo C, Brush M, et al. 2017. The Monarch Initiative: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 45(D1):D712–D722, PMID: 27899636, 10.1093/nar/gkw1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NRC (National Research Council). 2011. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. Washington, DC: National Academies Press. [PubMed] [Google Scholar]
- Nicholson DN, Greene CS. 2020. Constructing knowledge graphs and their biomedical applications. Comput Struct Biotechnol J 18:1414–1428, PMID: 32637040, 10.1016/j.csbj.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel CJ, Kerr J, Thomas DC, Mukherjee B, Ritz B, Chatterjee N, et al. 2017. Opportunities and challenges for environmental exposure assessment in population-based studies. Cancer Epidemiol Biomarkers Prev 26(9):1370–1380, PMID: 28710076, 10.1158/1055-9965.EPI-17-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid SM, McCusker JP, Pinheiro P, Bax MP, Santos H, Stingone JA, et al. 2020. The Semantic Data Dictionary—an approach for describing and annotating data. Data Intell 2(4):443–486, PMID: 33103120, 10.1162/dint_a_00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, Ayme S. 2012. Representation of rare diseases in health information systems: the ORPHANET approach to serve a wide range of end users. Hum Mutat 33(5):803–808, PMID: 22422702, 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- Richarz AN. 2019. Big data in predictive toxicology: challenges, opportunities and perspectives. In: Big Data in Predictive Toxicology. Neagu D, Richarz, AN, eds. Cambridge, UK: Cambridge Royal Society of Chemistry, RSC Publishing, 1–37. [Google Scholar]
- Rogers MF, Ben-Hur A. 2009. The use of gene ontology evidence codes in preventing classifier assessment bias. Bioinformatics 25(9):1173–1177, PMID: 19254922, 10.1093/bioinformatics/btp122. [DOI] [PubMed] [Google Scholar]
- Roos M, López Martin E, Wilkinson MD. 2017. Preparing data at the source to foster interoperability across rare disease resources. Adv Exp Med Biol 1031:165–179, PMID: 29214571, 10.1007/978-3-319-67144-4_9. [DOI] [PubMed] [Google Scholar]
- Sarigiannis DA, Karakitsios SP, Handakas E, Papadaki K, Chapizanis D, Gotti A. 2018. Informatics and data analytics to support exposome-based discovery: part 1—assessment of external and internal exposure. In: Applying Big Data Analytics in Bioinformatics and Medicine. Lytras MD, Papadopoulou P, eds. Hershey, PA: IGI Global, 115–144. [Google Scholar]
- Schwarzenbach RP, Egli T, Hofstetter TB, von Gunten U, Wehrli B. 2010. Global water pollution and human health. Annu Rev Environ Resour 35:109–136, 10.1146/annurev-environ-100809-125342. [DOI] [Google Scholar]
- Shefchek KA, Harris NL, Gargano M, Matentzoglu N, Unni D, Brush M, et al. 2020. The Monarch Initiative in 2019: an integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res 48(D1):D704–D715, PMID: 31701156, 10.1093/nar/gkz997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Jacobsen JOB, Jäger M, Köhler S, Holtgrewe M, Schubach M, et al. 2015. Next-generation diagnostics and disease-gene discovery with the exomiser. Nat Protoc 10(12):2004–2015, PMID: 26562621, 10.1038/nprot.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. 2007. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol 25(11):1251–1255, PMID: 17989687, 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear LP. 2020. Timing eclipses amount: the critical importance of intermittency in alcohol exposure teffects. Alcohol Clin Exp Res 44(4):806–813, PMID: 32056231, 10.1111/acer.14307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathias V, Koleti A, Vidović D, Cooper DJ, Jagodnik KM, Terryn R, et al. 2018. Sustainable data and metadata management at the BD2K-LINCS data coordination and integration center. Sci Data 5:180117, PMID: 29917015, 10.1038/sdata.2018.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. EPA (U.S. Environmental Protection Agency). 2003. Framework for Cumulative Risk Assessment. EPA/600/P-02/001F. Washington, DC: U.S. EPA; https://www.epa.gov/sites/production/files/2014-11/documents/frmwrk_cum_risk_assmnt.pdf [13 December 2020]. [Google Scholar]
- U.S. EPA. 2008. Concepts, Methods, and Data Sources for Cumulative Health Risk Assessment of Multiple Chemicals, Exposures and Effects: A Resource Document (Final Report, 2008). Washington, DC: U.S. EPA; https://cfpub.epa.gov/si/si_public_record_report.cfm?Lab=NCEA&dirEntryId=190187 [13 December 2020]. [Google Scholar]
- U.S. EPA. 2019. Guidelines for Human Health Exposure Assessment. EPA/100/B-19/001. Washington, DC: U.S. EPA; https://www.epa.gov/sites/production/files/2020-01/documents/guidelines_for_human_exposure_assessment_final2019.pdf [13 December 2020]. [Google Scholar]
- Vineis P. 2004. A self-fulfilling prophecy: are we underestimating the role of the environment in gene–environment interaction research? Int J Epidemiol 33(5):945–946, PMID: 15319401, 10.1093/ije/dyh277. [DOI] [PubMed] [Google Scholar]
- Vineis P, Russo F, eds. 2018. Epigenetics and the exposome: environmental exposure in disease etiology. In: Oxford Research Encyclopedia of Environmental Science. New York, NY: Oxford University Press, 10.1093/acrefore/9780199389414.013.325. [DOI] [Google Scholar]
- Watford S, Edwards S, Angrish M, Judson RS, Paul Friedman K. 2019. Progress in data interoperability to support computational toxicology and chemical safety evaluation. Toxicol Appl Pharmacol 380:114707, PMID: 31404555, 10.1016/j.taap.2019.114707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling C, Glaser J, Rodríguez-Guzmán J, Weiss I, Lucas R, Peraza S, et al. 2020. Chronic kidney disease of non-traditional origin in Mesoamerica: a disease primarily driven by occupational heat stress. Rev Panam Salud Publica 44:e15, PMID: 31998376, 10.26633/RPSP.2020.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild CP. 2005. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 14(8):1847–1850, PMID: 16103423, 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. 2016. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3:160018, PMID: 26978244, 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Guo Y, Li Q, George TJ, Shenkman E, Modave F, et al. 2018. An ontology-guided semantic data integration framework to support integrative data analysis of cancer survival. BMC Med Inform Decis Mak 18(suppl 2):41, PMID: 30066664, 10.1186/s12911-018-0636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.