Abstract
The ongoing pandemic of coronavirus disease 2019 threatens the whole world, which catalyzes a variety of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nucleic acid test (NAT) kits. To monitor test quality and evaluate NAT kits, quality control materials that best simulate real clinical samples are needed. In this study, the performance of SARS-CoV-2 cell culture supernatant, PCDH-based pseudovirus, and MS2-based pseudovirus as quality control materials was compared. PCDH-based pseudovirus was found to be more similar in characteristics to SARS-CoV-2 particle, and more suitable for evaluating SARS-CoV-2 NAT kits than MS2-based pseudovirus. Proper detection using sensitive and precise NAT kits is essential to guarantee diagnosis. Thus, limit of detection, precision, anti-inference ability, and cross-reactivity of NAT kits from PerkinElmer, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd, Shanghai Kehua Bio-Engineering Co, Ltd, Sansure Biotech Inc., Da An Gene Co, Ltd, Shanghai BioGerm Medical Biotechnology Co, Ltd, and Applied Biological Technologies Co, Ltd, were compared using PCDH-based pseudovirus. For the seven kits evaluated, N gene was more sensitive than ORF1ab gene in most kits, whereas E gene was most sensitive among the three genes in Shanghai Kehua Bio-Engineering Co, Ltd, and Applied Biological Technologies Co, Ltd. PerkinElmer got the lowest limit of detection for N gene at 11.61 copies/mL, and the value was 34.66 copies/mL for ORF1ab gene. All of the kits showed good precision, with CV values less than 5%, as well as acceptable anti-interference ability of 2 mg/L human genomic DNA. No cross-reactivity was observed with other respiratory viruses.
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative virus of coronavirus disease 2019 (COVID-19), emerged. On March 11, 2020, the World Health Organization declared COVID-19 a pandemic. As of December 14, 2020, there have been 71,051,805 confirmed cases of COVID-19 and 1,608,648 deaths [World Health Organization Coronavirus Disease (COVID-19) Dashboard, https://covid19.who.int]. At present, SARS-CoV-2 is still spreading and posing a threat to global public health.
Rapid and accurate diagnosis of SARS-CoV-2 is essential for early discovery and early treatment. Because of its easy methods and high sensitivity and specificity, real-time RT-PCR is becoming the optimal and most widely used method for detecting nucleic acid, and even the gold standard for viral detection.1 Multiple commercial real-time RT-PCR assays have been developed to meet diagnostic needs for SARS-CoV-2. However, clinically confirmed patients with negative nucleic acid test (NAT) results have been reported.2 , 3 Comparing the performance of commonly used NAT kits and providing evidence for choosing sensitive, specific, and precise NAT kits are the key to obtaining reliable test results.
Quality control materials are needed for evaluating the performance of SARS-CoV-2 NAT kits. Although patient samples are most in line with the actual situation, difficulty in obtaining samples and the risk of transmission limit their application as quality control materials. Therefore, construction of pseudoviruses similar to SARS-CoV-2 virus is necessary to evaluate the performance of NAT kits, as well as to control the quality of experimental procedure. Vectors of lentivirus and bacteriophage have been widely used to produce stable quality control materials.4, 5, 6, 7 These vectors vary in their characteristics, including size, packaging capacity, and stability. To best simulate real patient samples, the performance of different quality control materials needs to be compared.
In this study, the applicability of MS2- and PCDH-based pseudovirus, containing the amplification regions of SARS-CoV-2 NAT kits, was compared with that of SARS-CoV-2 cell culture supernatant. Furthermore, limit of detection (LoD), precision, anti-inference ability, and cross-reactivity of seven Conformité Européene marked and widely used SARS-CoV-2 NAT kits were evaluated using PCDH-based pseudovirus.
Materials and Methods
Quality Control Materials
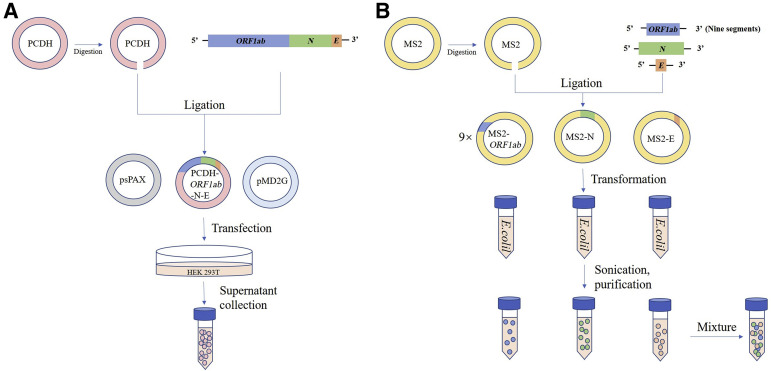
Three kinds of quality control materials were constructed. SARS-CoV-2 cell culture supernatant was kindly gifted by Chinese Academy of Military Medical Sciences (GenBank, https://www.ncbi.nlm.nih.gov/nuccore; accession number MT135042.1). Briefly, virus was cultured in Vero-E6 cells for 96 hours, and then the supernatant was collected and disinfected by heating at 65°C for 30 minutes.8 For constructing PCDH-based pseudovirus (Figure 1 A), PCDH-CMV-MCS-EF1-copGFP vector was digested with enzymes NotI and SalI, and the product was gel purified. N gene, E gene, and segments of ORF1ab gene were synthesized with overlapping ends by PCR, and further cloned into digested PCDH vector using one step of Gibson assembly (New England Biolabs, Ipswich, MA) at 50°C for 1 hour. The resultant plasmid was electroporated into ER2738 electrocompetent cells (Lucigen, Middleton, WI). After recovery in SOC medium at 37°C for 1 hour, the ER2738 cells were spread onto LB solid medium with 8 mg/L glucose and 50 mg/L ampicillin and incubated at 37°C overnight. Positive clones were sequenced, and successfully constructed plasmid was extracted. Then, HEK293T cells were seeded in 10-cm dish. After 24 hours, PCDH plasmid and two packaging plasmids (psPAX and pMD2G) were transfected at the ratio of 4:3:1 using polyethylenimine (Polysciences, Warrington, PA). Supernatant was gathered 48 hours after transfection, and centrifugated at 800 g for 5 minutes to remove cell debris. For constructing MS2-based pseudovirus (Figure 1B), pGSI-MS2 vector was digested with NotI and BamHI, and gel purified. N gene, E gene, and nine segments of ORF1ab gene were synthesized, ligated into MS2 vector, and transformed into competent Escherichia coli cells. Products were harvested, sonicated, and purified, as before.9 Finally, MS2-based pseudovirus, containing different regions of SARS-CoV-2 genome, was mixed together with the same volume ratio so that it can contain all of the target sequences of all reagents evaluated. After production, materials were stored at −40°C.
Figure 1.
Schematic diagram of producing PCDH-based pseudovirus (A) and MS2-based pseudovirus (B). Nine regions of ORF1ab were inserted into MS2 vector, and one of them was drawn as representative.
Digital PCR
Quality control materials were diluted to about 105 copies/mL, and sent to reagent manufacturers. Digital PCR was conducted using the same primers and probes as in their NAT kits, and the median of all results was taken as the exact concentration of quality control materials. The detailed information of digital PCR conducted by each manufacturer was summarized in Supplemental Table S1. The process of digital PCR was indicated before.10
Real-Time RT-PCR
Seven SARS-CoV-2 NAT kits were included. They were from PerkinElmer, Inc. (Waltham, MA), Beijing Wantai Biological Pharmacy Enterprise Co, Ltd (Wantai; Beijing, China), Shanghai Kehua Bio-Engineering Co, Ltd (KHB; Beijing, China), Sansure Biotech Inc. (Sansure; Changsha, China), Da An Gene Co, Ltd, of Sun Yat-sen University (Da An; Guangzhou, China), Shanghai BioGerm Medical Biotechnology Co, Ltd (BioGerm; Shanghai, China), and Beijing Applied Biological Technologies Co, Ltd (ABT; Beijing, China). The kits were all Conformité Européene marked. Among them, the kits of Sansure, Da An, BioGerm, and ABT have been approved by the China National Medical Products Administration. The kits of PerkinElmer, Sansure, and Wantai have been authorized for emergency use by the US Food and Drug Administration. The kits of PerkinElmer, KHB, Wantai, Da An, and ABT are on the list of World Health Organization emergency use listing. PCDH-based pseudovirus was sent to each manufacturer, and nucleic acids were extracted and real-time RT-PCR was conducted following the protocol. KHB and ABT tested N, ORF1ab, and E genes, whereas other kits only tested N and ORF1ab genes. The information of extraction and amplification is summarized in Supplemental Table S2.
Stability Test
For stability test, three quality control materials were diluted to about 1000 copies/mL using Dulbecco's modified Eagle’s medium (Corning, NY) containing 0.1% ProClin 300. Each material was thoroughly mixed and divided into four groups. Control group was stored at −40°C for 2 weeks. To assess freeze-thaw stability, materials were stored at −40°C for 2 weeks, during which they were frozen and thawed three times repeatedly. To evaluate whether the materials are stable after opening the tubes, materials were stored at −40°C for a week and tubes were open for a while, and then they were stored at 2°C to 8°C for a week. For fundamental stability, materials were stored at room temperature for a week and then put at 2°C to 8°C for another week. After processing, the nucleic acids were extracted using PerkinElmer PreNAT II, and tested using PerkinElmer's SARS-CoV-2 NAT kit on ABI7500 in sextuplicate. Results were analyzed following criteria from China National Accreditation Service for Conformity Assessment GL003: Guidance on Evaluating the Homogeneity and Stability of Samples Used for Proficiency Testing. Considering the bias of real-time RT-PCR assay, σ was identified as 0.15 in this study.
Amplification Efficiency
SARS-CoV-2 cell culture supernatant, MS2-based pseudovirus, and PCDH-based pseudovirus were diluted to 100,000, 20,000, 4000, and 800 copies/mL. Each concentration of each material was extracted and tested three times in a single run. Standard curve was performed with log of concentration and mean cycle threshold (Ct) value of three repeats. Amplification efficiency of each kit on each material was calculated as follow: E% = [10(−1/slope) − 1] × 100%.
Limit of Detection
LoD was evaluated in accordance with EP17-A2 guideline (Evaluation of Detection Capability for Clinical Laboratory Measurement Procedures; Approved Guideline—Second Edition) of Clinical Laboratory Standards Institute. Briefly, PCDH-based pseudovirus was gradiently diluted with Dulbecco's modified Eagle’s medium containing 0.1% ProClin 300. Each sample was extracted and tested eight times per day in a single run for 3 consecutive days. Positive rate of each dilution was counted, and LoD was calculated from a probit regression model as the measurand concentration at which measurement results yielded a positive classification with a 95% probability. Result determination was following manufacturer’s instructions. For BioGerm and ABT, Ct value ≤38 was considered positive. For PerkinElmer, Ct value ≤42 was considered positive. For other kits, Ct value ≤40 was considered positive.
Precision
Precision was evaluated in accordance with EP15-A3 guideline (User Verification of Precision and Estimation of Bias; Approved Guideline) of Clinical Laboratory Standards Institute. Briefly, two concentrations of PCDH-based pseudovirus were extracted and tested five times per day in a single run for 5 consecutive days, and the within-run precision (repeatability) and within-laboratory precision were calculated following the guideline.
Interference Testing
Interference testing was evaluated in accordance with EP07-A2 guideline (Interference Testing in Clinical Chemistry; Approved Guideline) of Clinical Laboratory Standards Institute. Briefly, two concentrations of PCDH-based pseudovirus were prepared, and a final concentration of 2 mg/L human genomic DNA or double-distilled water was added into pseudovirus and thoroughly mixed. Nucleic acids were extracted and amplificated 20 times in a single run.
Cross-Reactivity
The potential cross-reactivity of organisms was tested in both the presence and absence of PCDH-based SARS-CoV-2 pseudovirus by seven kits. SARS-CoV-2–positive samples were prepared by mixing each of the viruses with 900 copies/mL PCDH-based SARS-CoV-2 pseudovirus. For reagents of PerkinElmer, Wantai, and KHB, cross-reactivity was also determined at the level of 180 copies/mL. Human coronavirus 229E [ATCC (Manassas, VA) VR-740], OC43 (ATCC VR-1558), influenza A virus seasonal H1N1 (ATCC VR-1520), influenza A virus H3N2 (ATCC VR-1679), and influenza B virus (ATCC VR-1807) were kindly gifted by PerkinElmer, and were tested at 1 × 105 copies/mL. PUC57 plasmid vector, inserting 28,267 to 29,535 bp of SARS virus Urbani isolate icSARS-C7-MA (GenBank, https://www.ncbi.nlm.nih.gov/nuccore; accession number MK062184.1), and MS2-based pseudovirus, containing 28,564 to 29,807 bp of Middle East respiratory syndrome virus isolate HCoV-EMC/2012 (GenBank, https://www.ncbi.nlm.nih.gov/nuccore; accession number NC_019843.3), were kindly gifted by Da An, which were tested at 1 × 105 median tissue culture infective dose (TCID50)/mL. All of the tests were performed in triplicate in a single run.
Statistical Analysis
Statistical analysis was performed on IBM (Armonk, NY) SPSS version 21.0. Wilcoxon matched-pairs signed rank test was performed to compare the differences between two groups.
Results
Quantification of Three Quality Control Materials
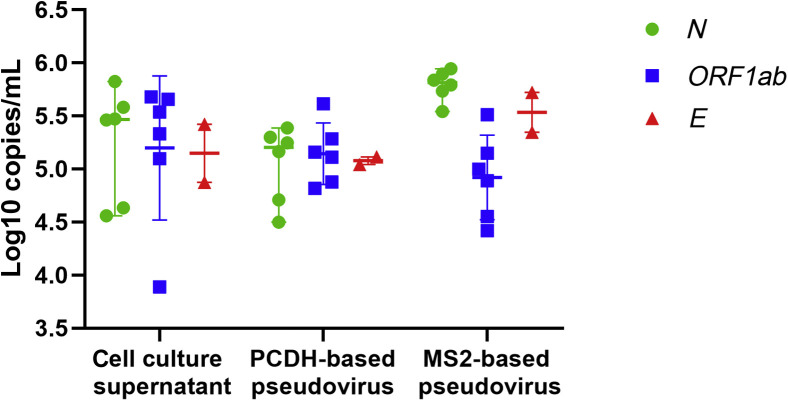
SARS-CoV-2 cell culture supernatant, MS2-based pseudovirus, and PCDH-based pseudovirus were diluted 10-fold, 1000-fold, and 10,000-fold, respectively. Digital PCR was performed to quantify N gene, E gene, and ORF1ab gene by six reagent manufacturers, except BioGerm. Concentration of quality control materials varied from 4 to 6 log10 copies/mL, so that median was taken as the final concentration (Figure 2 ). Among the three quality control materials, PCDH-based pseudovirus showed the smallest variation among both reagents and genes.
Figure 2.
Concentration of three quality control materials. Digital PCR was performed to quantify the concentration of N gene, E gene, and ORF1ab gene by six reagent manufacturers. Data are presented as median with range.
Stability of Quality Control Materials
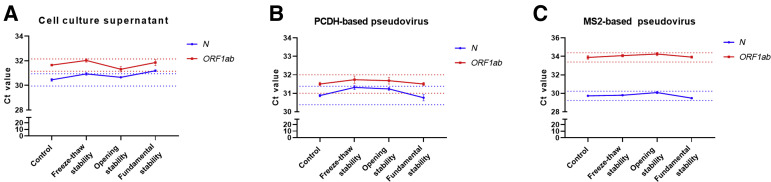
Three kinds of quality control materials were processed at four conditions indicated before. Real-time RT-PCR using the PerkinElmer kit was performed thereafter. The difference in mean Ct values of ORF1ab gene between each experimental group and control group was all <0.5 (Figure 3 and Supplemental Table S3). However, when evaluating fundamental stability of cell culture supernatant, the Ct value of N gene was 0.76 higher than control group, indicating that it might be unstable at this condition.
Figure 3.
Stability of severe acute respiratory syndrome coronavirus 2 cell culture supernatant (A), PCDH-based pseudovirus (B), and MS2-based pseudovirus (C). Materials were thoroughly mixed, divided, and processed with four different conditions. The Ct values of N and ORF1ab genes were tested. Red dotted line represents the level of mean Ct value of ORF1ab gene in control group ±0.5. Blue dotted line represents the level of mean Ct value of N gene in control group ±0.5. Data are presented as means ± SEM (A–C).
Amplification Efficiency among Quality Control Materials
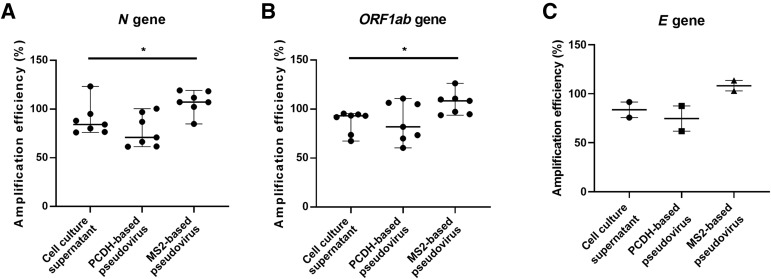
Three kinds of quality control materials were diluted in a fivefold gradient between 100,000 and 800 copies/mL, and real-time RT-PCR was performed using seven kits evaluated. The amplification efficiencies of N gene and ORF1ab gene in PCDH-based pseudovirus had no difference with those in cell culture supernatant, whereas the amplification efficiencies of MS2-based pseudovirus were significantly higher than those of cell culture supernatant (Figure 4 ). Although only KHB and ABT detected E gene, it could also be seen that the E gene amplification efficiency of PCDH-based pseudovirus was closer to cell culture supernatant than MS2-based pseudovirus. These results suggest that PCDH-based pseudovirus is more similar to SARS-CoV-2 particle, and is more suitable for evaluating SARS-CoV-2 NAT kits than MS2-based pseudovirus.
Figure 4.
Amplification efficiency of seven severe acute respiratory syndrome coronavirus 2 nucleic acid test kits when amplifying N gene (A), ORF1ab gene (B), and E gene (C) in cell culture supernatant, PCDH-based pseudovirus, and MS2-based pseudovirus. Wilcoxon matched-pair signed rank test was performed to compare the differences between two groups. ∗P < 0.05.
Limit of Detection
PCDH-based pseudovirus was gradiently diluted. Each sample was extracted and tested eight times per day in a single run for 3 consecutive days. A total of 24 tests were performed in each concentration, and LoDs were calculated by probit analysis. The N gene was more sensitive than ORF1ab gene in the kits from PerkinElmer, Wantai, KHB, Sansure, and Da An (Supplemental Tables S4 through S6). The LoDs of ORF1ab were slightly lower than N gene in the kits of BioGerm and ABT. Among all of the kits, only KHB and ABT detected E gene, which held better sensitivity than N gene and ORF1ab gene. The PerkinElmer kit had the lowest LoD for N gene at 11.61 copies/mL, and the value was 34.66 copies/mL for ORF1ab gene.
Specificity
Twelve negative oropharyngeal swabs mixed with 900 or 180 copies/mL PCDH-based SARS-CoV-2 pseudovirus and 24 negative oropharyngeal swabs were tested for specificity (Supplemental Table S7). For these seven kits, all of the positive samples yielded positive results, and all of the negative swabs were tested negative, showing a specificity of 100%.
Precision
Two concentrations of PCDH-based pseudovirus were prepared for each reagent to evaluate precision. Concentrations of three regions were calculated by droplet digital PCR results and dilution multiples. High concentration was the same to all reagents (ie, 900 copies/mL of N gene, 760 copies/mL of ORF1ab gene, and 670 copies/mL of E gene). Low concentration varied according to the LoD of each kit. PerkinElmer holds the lowest LoD, so the low level to evaluate precision of PerkinElmer was 90 copies/mL for N gene and 76 copies/mL for ORF1ab gene. For Wantai and KHB, the low level was twice that for PerkinElmer. For other reagents, low concentration was three times that for PerkinElmer. Within-run CV% and within-laboratory CV% of all reagents were <5%, indicating that all these reagents had good precision (Table 1 ).
Table 1.
Precision of SARS-CoV-2 NAT Kits
| Region | Reagent manufacturer | Low concentration |
High concentration |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean Ct | Within-run SD | Within-run CV, % | Within-laboratory SD | Within-laboratory CV, % | Mean Ct | Within-run SD | Within-run CV, % | Within-laboratory SD | Within-laboratory CV, % | ||
| N | PerkinElmer | 35.32 | 0.76 | 2.15 | 1.05 | 2.97 | 31.88 | 0.27 | 0.85 | 0.73 | 2.30 |
| Wantai | 34.80 | 0.69 | 1.99 | 0.71 | 2.04 | 32.07 | 0.33 | 1.03 | 0.50 | 1.57 | |
| KHB | 31.26 | 0.58 | 1.84 | 0.83 | 2.66 | 28.49 | 0.31 | 1.10 | 0.53 | 1.86 | |
| Sansure | 35.75 | 0.47 | 1.31 | 0.62 | 1.73 | 34.26 | 0.37 | 1.08 | 0.60 | 1.76 | |
| Da An | 36.37 | 0.57 | 1.58 | 0.71 | 1.96 | 34.56 | 0.36 | 1.05 | 0.34 | 0.98 | |
| BioGerm | 35.99 | 0.63 | 1.75 | 0.84 | 2.33 | 34.31 | 0.49 | 1.42 | 0.83 | 2.42 | |
| ABT | 36.70 | 0.65 | 1.77 | 1.03 | 2.80 | 35.46 | 0.47 | 1.33 | 0.65 | 1.84 | |
| ORF1ab | PerkinElmer | 35.05 | 0.63 | 1.79 | 0.84 | 2.41 | 31.42 | 0.22 | 0.69 | 0.49 | 1.55 |
| Wantai | 35.62 | 1.20 | 3.37 | 1.16 | 3.25 | 32.54 | 0.40 | 1.23 | 0.53 | 1.62 | |
| KHB | 32.76 | 1.19 | 3.65 | 1.31 | 4.00 | 30.08 | 0.53 | 1.76 | 0.85 | 2.82 | |
| Sansure | 38.01 | 0.70 | 1.85 | 0.84 | 2.21 | 36.89 | 0.94 | 2.55 | 1.20 | 3.26 | |
| Da An | 38.68 | 0.57 | 1.47 | 0.62 | 1.60 | 36.82 | 0.39 | 1.05 | 0.48 | 1.29 | |
| BioGerm | 35.54 | 0.74 | 2.09 | 0.75 | 2.11 | 33.80 | 0.49 | 1.45 | 0.75 | 2.23 | |
| ABT | 37.26 | 1.05 | 2.82 | 1.38 | 3.70 | 35.66 | 0.63 | 1.76 | 0.80 | 2.25 | |
| E | KHB | 31.04 | 0.55 | 1.78 | 0.71 | 2.29 | 28.47 | 0.27 | 0.96 | 0.53 | 1.88 |
| ABT | 36.08 | 0.77 | 2.12 | 0.87 | 2.40 | 34.41 | 0.46 | 1.32 | 0.65 | 1.90 | |
High concentration was 900 copies/mL of N gene, 760 copies/mL of ORF1ab gene, and 670 copies/mL of E gene. For N gene, low concentration was 90 copies/mL for PerkinElmer, 180 copies/mL for Wantai and KHB, and 360 copies/mL for other reagents. For ORF1ab gene, low concentration was 76 copies/mL for PerkinElmer, 152 copies/mL for Wantai and KHB, and 304 copies/mL for other reagents. For E gene, low concentration was 134 copies/mL for KHB and 268 copies/mL for ABT.
ABT, Beijing Applied Biological Technologies Co, Ltd; BioGerm, Shanghai BioGerm Medical Biotechnology Co, Ltd; Da An, Da An Gene Co, Ltd; KHB, Shanghai Kehua Bio-Engineering Co, Ltd; NAT, nucleic acid test; Sansure, Sansure Biotech Inc.; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; Wantai, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd.
Interference
To simulate real patient samples and evaluate whether human genomic DNA in patient samples can interfere with detection result, 2 mg/L human genomic DNA or double-distilled water was added into PCDH-based pseudovirus, and samples were tested 20 times in a single PCR run. The concentrations of pseudovirus were the same with the levels to evaluate precision. When conducting interference testing at high concentration, all of the 20 tests of each reagent were positive in the existence of 2 mg/L human genomic DNA, indicating this interfering substance did not interfere with the determination of positive results (Table 2 ). When comparing the Ct values of interfering group and control group by Wilcoxon matched-pairs signed rank test, N gene and E gene of KHB, as well as ORF1ab gene of ABT, showed significant differences between two groups. This result suggests that 2 mg/L human genomic DNA might still interfere with the PCR process, but the interference is acceptable for qualitative reagents because it does not change the determination of results.
Table 2.
Interference Testing at High Concentration of PCDH-Based Pseudovirus
| Reagent manufacturer |
N gene |
ORF1ab gene |
E gene |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control |
2 mg/L DNA |
P value | Control |
2 mg/L DNA |
P value | Control |
2 mg/L DNA |
P value | |||||||
| Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | ||||
| PerkinElmer | 32.07 | 20/20 | 31.79 | 20/20 | 0.26 | 31.62 | 20/20 | 31.70 | 20/20 | 0.39 | |||||
| Wantai | 33.33 | 20/20 | 33.16 | 20/20 | 0.21 | 33.46 | 20/20 | 33.13 | 20/20 | 0.06 | |||||
| KHB | 28.53 | 20/20 | 28.41 | 20/20 | 0.03 | 29.92 | 20/20 | 30.02 | 20/20 | 0.38 | 28.52 | 20/20 | 28.34 | 20/20 | 0.02 |
| Sansure | 36.85 | 20/20 | 36.65 | 20/20 | 0.44 | 37.45 | 20/20 | 37.87 | 20/20 | 0.14 | |||||
| Da An | 35.58 | 20/20 | 35.78 | 20/20 | 0.20 | 37.26 | 20/20 | 37.37 | 20/20 | 0.46 | |||||
| BioGerm | 34.56 | 20/20 | 34.46 | 20/20 | 0.70 | 34.89 | 20/20 | 34.32 | 20/20 | 0.11 | |||||
| ABT | 36.02 | 20/20 | 36.15 | 20/20 | 0.23 | 36.99 | 20/20 | 36.16 | 20/20 | 0.01 | 35.55 | 20/20 | 35.55 | 20/20 | 0.89 |
The concentration was 900 copies/mL of N gene, 760 copies/mL of ORF1ab gene, and 670 copies/mL of E gene.
ABT, Beijing Applied Biological Technologies Co, Ltd; BioGerm, Shanghai BioGerm Medical Biotechnology Co, Ltd; Da An, Da An Gene Co, Ltd; KHB, Shanghai Kehua Bio-Engineering Co, Ltd; Sansure, Sansure Biotech Inc.; Wantai, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd.
Because low concentration was near the LoD of several reagents, the positive rate was not 100% for all reagents. 2 mg/L human genomic DNA did not interfere with the determination of positive results of the Wantai kit (Table 3 ). As for PerkinElmer, the positive rate of ORF1ab gene slightly reduced from 100% to 90%. For the kits with less sensitivity, positive rates were compared. For ORF1ab gene of KHB, and both N and ORF1ab gene of ABT, the positive rate of interfering group is a bit lower than that of control group. For ORF1ab gene of Sansure and BioGerm, the positive rate was higher in interfering group than in control group.
Table 3.
Interference Testing at Low Concentration of PCDH-Based Pseudovirus
| Reagent manufacturer |
N gene |
ORF1ab gene |
E gene |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control |
2 mg/L DNA |
P value | Control |
2 mg/L DNA |
P value | Control |
2 mg/L DNA |
P value | |||||||
| Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | Mean Ct | Positive rate | ||||
| PerkinElmer | 34.44 | 20/20 | 35.40 | 20/20 | 0.004 | 35.24 | 20/20 | 34.48 | 18/20 | 0.006 | |||||
| Wantai | 36.07 | 20/20 | 35.75 | 20/20 | 0.26 | 36.21 | 19/20 | 35.73 | 20/20 | 0.45 | |||||
| KHB | 31.14 | 20/20 | 30.78 | 20/20 | 0.02 | 32.55 | 20/20 | 33.42 | 18/20 | 0.14 | 31.3 | 20/20 | 30.77 | 20/20 | 0.01 |
| Sansure | 38.66 | 19/20 | 37.56 | 20/20 | 0.001 | 38.96 | 12/20 | 38.51 | 16/20 | 0.17 | |||||
| Da An | 37.19 | 18/20 | 37.83 | 19/20 | 0.17 | 38.92 | 16/20 | 39.04 | 17/20 | 0.61 | |||||
| BioGerm | 36.62 | 16/20 | 36.17 | 18/20 | 0.25 | 35.68 | 13/20 | 35.85 | 17/20 | 0.31 | |||||
| ABT | 37.56 | 12/20 | 38.42 | 9/20 | 0.02 | 38.71 | 12/20 | 38.00 | 10/20 | 0.28 | 37.11 | 17/20 | 37.17 | 19/20 | 0.95 |
For N gene, the concentration was 90 copies/mL for PerkinElmer, 180 copies/mL for Wantai and KHB, and 360 copies/mL for other reagents. For ORF1ab gene, the concentration was 76 copies/mL for PerkinElmer, 152 copies/mL for Wantai and KHB, and 304 copies/mL for other reagents. For E gene, the concentration was 134 copies/mL for KHB and 268 copies/mL for ABT. Undetermined results were excluded.
ABT, Beijing Applied Biological Technologies Co, Ltd; BioGerm, Shanghai BioGerm Medical Biotechnology Co, Ltd; Da An, Da An Gene Co, Ltd; KHB, Shanghai Kehua Bio-Engineering Co, Ltd; Sansure, Sansure Biotech Inc.; Wantai, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd.
Cross-Reactivity
When mixing 229E, OC43, seasonal H1N1, H3N2, influenza B, SARS, and Middle East respiratory syndrome with 900 or 180 copies/mL SARS-CoV-2 pseudovirus, all seven kits yielded expected results, indicating that these respiratory viruses do not cause false-negative results of detecting SARS-CoV-2 (Table 4 ). Organisms were also tested in the absence of SARS-CoV-2, and no false-positive test results were obtained. These results suggest all of the kits are designed for the specific detection of SARS-CoV-2, with no expected cross-reactivity to other coronaviruses or human influenza viruses mentioned.
Table 4.
Cross-Reactivity of SARS-CoV-2 NAT Kits
| Region | Reagent manufacturer | High concentration |
Low concentration |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 229E | OC43 | Seasonal H1N1 | H3N2 | Influenza B | SARS | MERS | Control | 229E | OC43 | Seasonal H1N1 | H3N2 | Influenza B | SARS | MERS | ||
| N | PerkinElmer | 32.54 | 32.36 | 32.60 | 32.59 | 32.54 | 33.01 | 32.75 | 32.37 | 35.50 | 35.60 | 35.46 | 35.65 | 35.72 | 35.12 | 35.60 | 36.18 |
| Wantai | 32.06 | 32.19 | 32.07 | 32.03 | 31.92 | 32.41 | 32.57 | 32.18 | 36.17 | 36.68 | 36.06 | 35.70 | 34.80 | 35.57 | 36.30 | 35.61 | |
| KHB | 26.31 | 26.80 | 26.39 | 26.48 | 26.37 | 26.78 | 26.19 | 26.89 | 29.30 | 30.02 | 29.73 | 29.48 | 29.44 | 29.17 | 28.95 | 29.25 | |
| Sansure | 33.01 | 32.12 | 33.15 | 32.88 | 32.76 | 33.03 | 33.08 | 32.97 | |||||||||
| Da An | 34.81 | 35.23 | 34.79 | 35.05 | 34.74 | 35.70 | 34.86 | 35.34 | |||||||||
| BioGerm | 33.45 | 33.27 | 33.13 | 33.11 | 33.24 | 33.33 | 33.73 | 34.11 | |||||||||
| ABT | 33.17 | 33.30 | 33.42 | 33.37 | 33.14 | 33.37 | 32.56 | 34.34 | |||||||||
| ORF1ab | PerkinElmer | 32.40 | 32.39 | 32.43 | 32.76 | 32.53 | 32.60 | 32.61 | 32.38 | 35.47 | 35.37 | 35.16 | 35.07 | 35.49 | 35.49 | 35.33 | 35.99 |
| Wantai | 31.82 | 31.55 | 31.94 | 31.84 | 32.10 | 32.13 | 31.90 | 32.03 | 35.04 | 34.68 | 34.62 | 35.56 | 35.31 | 34.39 | 35.17 | 34.76 | |
| KHB | 29.10 | 27.69 | 28.44 | 28.31 | 28.33 | 28.54 | 28.37 | 28.85 | 31.71 | 33.76 | 32.52 | 30.88 | 32.31 | 31.76 | 30.54 | 32.85 | |
| Sansure | 33.95 | 33.64 | 34.18 | 34.37 | 34.06 | 33.70 | 34.40 | 34.10 | |||||||||
| Da An | 36.05 | 36.22 | 35.60 | 35.95 | 35.63 | 36.53 | 35.72 | 35.74 | |||||||||
| BioGerm | 33.03 | 32.72 | 33.46 | 32.71 | 33.32 | 33.36 | 32.47 | 33.80 | |||||||||
| ABT | 34.08 | 34.35 | 32.85 | 34.88 | 33.37 | 34.09 | 32.89 | 35.50 | |||||||||
| E | KHB | 26.56 | 26.70 | 26.26 | 26.39 | 26.47 | 26.92 | 26.33 | 26.86 | 29.42 | 30.37 | 30.12 | 29.53 | 29.48 | 29.24 | 28.74 | 29.20 |
| ABT | 33.14 | 34.08 | 33.22 | 34.20 | 32.51 | 34.12 | 32.97 | 34.66 | |||||||||
A total of 1 × 105 copies/mL of human coronavirus 229E (ATCC VR-740), OC43 (ATCC VR-1558), influenza A virus seasonal H1N1 (ATCC VR-1520), influenza A virus H3N2 (ATCC VR-1679), and influenza B virus (ATCC VR-1807), as well as 1 × 105 median tissue culture infective dose (TCID50)/mL of pUC57 plasmid containing SARS coronavirus fragments and MS2-based MERS coronavirus pseudovirus, were mixed with PCDH-based SARS-CoV-2 pseudovirus. High concentration was 900 copies/mL of N gene, 760 copies/mL of ORF1ab gene, and 670 copies/mL of E gene. Low concentration was 180 copies/mL of N gene, 152 copies/mL of ORF1ab gene, and 134 copies/mL of E gene. Each concentration was extracted and tested three times in a single run, and mean Ct value was presented.
ABT, Beijing Applied Biological Technologies Co, Ltd; BioGerm, Shanghai BioGerm Medical Biotechnology Co, Ltd; Da An, Da An Gene Co, Ltd; KHB, Shanghai Kehua Bio-Engineering Co, Ltd; MERS, Middle East respiratory syndrome; NAT, nucleic acid test; Sansure, Sansure Biotech Inc.; SARS, severe acute respiratory syndrome; SARS-CoV-2, SARS coronavirus 2; Wantai, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd.
Discussion
SARS-CoV-2 is an enveloped single-stranded positive-sense RNA virus with a genome length of about 29.9 kb.4 To construct quality control materials that best simulate SARS-CoV-2 particles, vectors of lentivirus and phage are used in this study. Recombinant lentivirus vector is developed on the genome of HIV-1, in which the parental HIV-1 envelope is substituted with vesicular stomatitis virus G glycoprotein.11 According to the instruction, a PCDH lentivector can hold 3 to 5 kb of insert, and high-titer (about 109 infectious units/mL) concentrated viral particles can be generated. In this study, N gene, E gene, and parts of ORF1ab gene of SARS-CoV-2, with a total length of 4276 bp, were inserted into the PCDH vector and packaged into 293T cells. The titer of pseudovirus produced was about 1 × 109 copies/mL. Because the three regions were inserted together, it is assumed that the concentrations of three regions are the same. However, although droplet digital PCR can precisely quantify nucleic acids, the process is subject to bias and variance.12 Therefore, the concentrations of three regions are not identical.
Generally, packaging 500 bp of RNA is efficient using MS2, whereas it does not work well when packaging 1.5 kb of RNA.13 Although MS2-based armored L-RNA technology could package >2 kb RNA sequence,14 the packaging capacity of MS2 is still not as good as lentivirus. Because of the limitation, N gene, E gene, and nine parts of ORF1ab gene were inserted into the MS2 vector separately, and different MS2-based pseudoviruses were mixed together to form quality control materials. This raised the problem that the packaging efficiencies of different gene fragments were not exactly the same, so that the concentrations of pseudovirus containing different regions varied widely when mixing in the same volume ratio. In this study, digital PCR showed that the concentration of ORF1ab gene was about sixfold lower than N gene, and fourfold lower than E gene. The big difference between the levels of different genes made it not suitable to evaluate NAT kits using mixed MS2-based pseudoviruses.
SARS-CoV-2 particles are generally spherical, with some pleomorphism, and diameter varies from about 60 to 140 nm.15 Lentivector is a spherical virus between 80 and 120 nm in diameter, encapsulating a single-stranded positive-sense RNA of 7 to 12 kb in length.16 , 17 MS2 is icosahedral capsid self-assembled from 180 copies of a single coat protein and measures 22 to 29 nm in diameter.18 Lentivirus-based pseudovirus is closer in size to SARS-CoV-2 particle, which makes it better to mimic SARS-CoV-2 virus during extraction and amplification. Consistent with this, the amplification efficiencies of PCDH-based pseudovirus in the seven kits evaluated are close to that of SARS-CoV-2 cell culture supernatant, suggesting that PCDH-based pseudoviruses are more similar to SARS-CoV-2 virus presented in real patient samples. For laboratories having the need to evaluate SARS-CoV-2 NAT kits, the characteristics among different pseudoviruses should be considered. Different evaluation materials may get different results.
SARS-CoV-2 is highly pathogenic, and must be isolated and cultured in Biosafety level-3 laboratory. This means that most laboratories cannot produce cell culture supernatant as quality control materials. Moreover, virus needs to be inactivated sufficiently before using in BSL-2. Potential biological hazard of cell culture supernatant makes pseudovirus safer as quality control material. To use as quality control material, supernatant was heated at 65°C for 30 minutes. However, 1000 copies/mL of heated SARS-CoV-2 cell culture supernatant was unstable when staying at room temperature for a week and then putting at 2°C to 8°C for another week. It has been proven that the Ct values in specimens from diagnosed COVID-19 patients increase after thermal incubation. Furthermore, about half of weak-positive samples containing about 700 copies/mL SARS-CoV-2 RNA tested negative after heating at 56°C for 30 minutes.19 Combining these lines of evidence, heat inactivation might not be suitable for producing quality control materials with SARS-CoV-2 cell culture supernatant.
With the outbreak of COVID-19, commercial kits detecting SARS-CoV-2 RNA also burst. Sensitivity is one of the most concerning questions about NAT kits. The median viral load in posterior oropharyngeal saliva or endotracheal aspirate was 5.2 log10 copies/mL.20 Salivary viral load was highest during the first week after symptom onset and subsequently declined, with a slope of –0.15 log10 copies/mL per day.20 For detecting viral RNA at the late time of disease, kits with adequate sensitivities are needed. Of the seven kits in this study, Da An and BioGerm reached the analytical sensitivity claimed in their instructions, which is 500 and 1000 copies/mL, respectively. LoD of Sansure, KHB, and ABT was higher than LoD in their instructions, which is claimed to be 200, 20, and 200 copies/mL, respectively. The analytical sensitivity declared of PerkinElmer and Wantai is 30 and 50 copies/mL, respectively, which is between the LoD of N and ORF1ab genes performed in our study. The sensitivities of ORF1ab gene were generally less than N and E genes. In addition, SARS-CoV-2 expresses nine subgenomic mRNAs containing N and E genes, whereas ORF1ab can only transcript from the whole genome.21 The subgenome results in more N and E gene copies than the less sensitive ORF1ab gene. Therefore, when detecting patient samples with low viral titers, it is normal to detect positive N or E gene with a negative result of ORF1ab gene. Samples should be retested in this situation following the instructions. The extraction systems were different in this study, so the LoD reflected the combined performance of both the extraction process and PCR process. Most kits do not restrict the matching extraction system. Therefore, extraction systems should be taken into consideration for sensitivity in laboratories detecting SARS-CoV-2 RNA. To increase analytical sensitivity, extraction systems can be optimized by increasing the volume ratio of elution to extraction or by increasing the extraction efficiency.
All of the seven kits showed high within-run precision and within-laboratory precision with the CV value of <5%, indicating a good repeatability of results. This result prompts that an indeterminate result should be confirmed with another more sensitive reagent, rather than multiple tests with one reagent. Specificity of each reagent was 100%, demonstrating that false-positive results are unlikely to appear. However, real patient samples are more complicated, and interference of various substances may result in false-positive results.
Sample type is diverse in COVID-19 patients. Virus RNA can be detected in nasopharyngeal and oropharyngeal swabs, sputum, bronchoalveolar lavage, endotracheal aspirate, blood, stool, and urine.4 In all these samples, swabs contain more cells that might be more susceptible by human genomic DNA interference. A total of 2 mg/L human genomic DNA showed no interference with the determination of positive results at the level of 900 copies/mL. For ORF1ab gene of KHB, and N gene of ABT, interfering group got a lower positive rate and a higher mean Ct value of detectable repeats than that of control group, suggesting that 2 mg/L human genomic DNA might interfere with the determination of result at levels near LoD in these two reagents. For ORF1ab gene of Sansure and BioGerm, the positive rate was higher in interfering group. However, interference substances are complicated in real patient samples; it is hard to say whether they would be better in detecting patient specimens with SARS-CoV-2 RNA at concentrations near LoD.
In summary, PCDH-based pseudovirus was characterized more closely to SARS-CoV-2, and was more suitable to evaluate SARS-CoV-2 NAT kits than mixed MS2-based pseudovirus. To produce quality control materials using SARS-CoV-2 cell culture supernatant, it should be considered that fundamental stability might be affected after heating inactivation. As for the seven kits evaluated, N gene was more sensitive than ORF1ab gene in most kits, whereas E gene was most sensitive among the three genes in KHB and ABT. PerkinElmer got the lowest LoD for N gene at 11.61 copies/mL, and the value was 34.66 copies/mL for ORF1ab gene. All of the kits showed good precision, with CV values of <5%, as well as acceptable anti-interference ability of 2 mg/L human genomic DNA. No cross-reactivity was observed with other respiratory viruses.
Acknowledgments
We thank Chinese Academy of Military Medical Sciences for providing severe acute respiratory syndrome coronavirus 2 cell culture supernatant; PerkinElmer, Beijing Wantai Biological Pharmacy Enterprise Co, Ltd, Shanghai Kehua Bio-Engineering Co., Ltd., Sansure Biotech Inc., Da An Gene Co., Ltd., Shanghai BioGerm Medical Biotechnology Co., Ltd., and Beijing Applied Biological Technologies Co., Ltd. for supplying nucleic acid test kits and performing these tests; and PerkinElmer and Da An Gene Co., Ltd. for providing other respiratory viruses.
Footnotes
Supported by the Ministry of Science and Technology of the People's Republic of China (National Key Research and Development Program of China number 2020YFC0848200).
Disclosures: PerkinElmer, Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., Shanghai Kehua Bio-Engineering Co., Ltd., Sansure Biotech Inc., Da An Gene Co., Ltd., Shanghai BioGerm Medical Biotechnology Co., Ltd., and Beijing Applied Biological Technologies Co., Ltd. supplied nucleic acid test kits and performed testing; PerkinElmer and Da An Gene Co., Ltd. also provided other respiratory viruses.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.12.002.
Supplemental Data
References
- 1.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 2.Winichakoon P., Chaiwarith R., Liwsrisakun C., Salee P., Goonna A., Limsukon A., Kaewpoowat Q. Negative nasopharyngeal and oropharyngeal swabs do not rule out COVID-19. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00297-20. e00297-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahamtan A., Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y., Chang L., Wang L. Laboratory testing of SARS-CoV, MERS-CoV, and SARS-CoV-2 (2019-nCoV): current status, challenges, and countermeasures. Rev Med Virol. 2020;30:e2106. doi: 10.1002/rmv.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D., Li Y., Li J., Yu J., Yang H., Wei H. Construction of lentivirus-based reference material for RT-PCR detection of Middle East respiratory syndrome coronavirus and its application in external quality assessment. J Nanosci Nanotechnol. 2019;19:5510–5516. doi: 10.1166/jnn.2019.16591. [DOI] [PubMed] [Google Scholar]
- 6.Fu Y., Wang G., Wu Q., Yang X., Zhang R., Zhang K., Lin G., Han Y., Bao L., Li Z., Li J. Preparation of MS2-based nanoparticles as control and standard materials for the molecular detection of dengue virus serotypes. Virus Res. 2017;233:42–50. doi: 10.1016/j.virusres.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Hao M., Zhang K., Zhang R., Lin G., Jia T., Zhang D., Chang L., Xie J., Li J. External quality assessment for the molecular detection of MERS-CoV in China. J Clin Virol. 2016;75:5–9. doi: 10.1016/j.jcv.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y.I., Casel M.A.B., Kim S.M., Kim S.G., Park S.J., Kim E.H., Jeong H.W., Poo H., Choi Y.K. Development of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) thermal inactivation method with preservation of diagnostic sensitivity. J Microbiol. 2020;58:886–891. doi: 10.1007/s12275-020-0335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang L., Wang G., Jia T., Zhang L., Li Y., Han Y., Zhang K., Lin G., Zhang R., Li J., Wang L. Armored long non-coding RNA MEG3 targeting EGFR based on recombinant MS2 bacteriophage virus-like particles against hepatocellular carcinoma. Oncotarget. 2016;7:23988–24004. doi: 10.18632/oncotarget.8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang L.Y.Y., Zhao L., Hu G., Deng L., Su D., Peng D., Nie X., Wang S., Li Y., Wang J., Ruan Z., Gao S., Yang H., Guo F., Wang L. No evidence of SARS-CoV-2 RNA among blood donors: a multicenter study in Hubei, China. Transfusion. 2020;60:2038–2046. doi: 10.1111/trf.15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockrell A.S., Kafri T. Gene delivery by lentivirus vectors. Mol Biotechnol. 2007;36:184–204. doi: 10.1007/s12033-007-0010-8. [DOI] [PubMed] [Google Scholar]
- 12.Huggett J.F., Cowen S., Foy C.A. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61:79–88. doi: 10.1373/clinchem.2014.221366. [DOI] [PubMed] [Google Scholar]
- 13.Pasloske B.L., Walkerpeach C.R., Obermoeller R.D., Winkler M., DuBois D.B. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J Clin Microbiol. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Y., Yang C., Wei B., Huang J., Wang L., Meng S., Zhang R., Li J. RNase-resistant virus-like particles containing long chimeric RNA sequences produced by two-plasmid coexpression system. J Clin Microbiol. 2008;46:1734–1740. doi: 10.1128/JCM.02248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogt V.M., Simon M.N. Mass determination of rous sarcoma virus virions by scanning transmission electron microscopy. J Virol. 1999;73:7050–7055. doi: 10.1128/jvi.73.8.7050-7055.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segura M.M., Mangion M., Gaillet B., Garnier A. New developments in lentiviral vector design, production and purification. Expert Opin Biol Ther. 2013;13:987–1011. doi: 10.1517/14712598.2013.779249. [DOI] [PubMed] [Google Scholar]
- 18.Fu Y., Li J. A novel delivery platform based on bacteriophage MS2 virus-like particles. Virus Res. 2016;211:9–16. doi: 10.1016/j.virusres.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan Y., Long L., Zhang D., Yan T., Cui S., Yang P., Wang Q., Ren S. Potential false-negative nucleic acid testing results for severe acute respiratory syndrome coronavirus 2 from thermal inactivation of samples with low viral loads. Clin Chem. 2020;66:794–801. doi: 10.1093/clinchem/hvaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., Yip C.C., Cai J.P., Chan J.M., Chik T.S., Lau D.P., Choi C.Y., Chen L.L., Chan W.M., Chan K.H., Ip J.D., Ng A.C., Poon R.W., Luo C.T., Cheng V.C., Chan J.F., Hung I.F., Chen Z., Chen H., Yuen K.Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e910. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.