Abstract
Arabinogalactan-proteins (AGPs) are a large, complex, and highly diverse class of heavily glycosylated proteins that belong to the family of cell wall hydroxyproline-rich glycoproteins. Approximately 90% of the molecules consist of arabinogalactan polysaccharides, which are composed of arabinose and galactose as major sugars and minor sugars such as glucuronic acid, fucose, and rhamnose. About half of the AGP family members contain a glycosylphosphatidylinositol (GPI) lipid anchor, which allows for an association with the outer leaflet of the plasma membrane. The mysterious AGP family has captivated the attention of plant biologists for several decades. This diverse family of glycoproteins is widely distributed in the plant kingdom, including many algae, where they play fundamental roles in growth and development processes. The journey of AGP biosynthesis begins with the assembly of amino acids into peptide chains of proteins. An N-terminal signal peptide directs AGPs toward the endoplasmic reticulum, where proline hydroxylation occurs and a GPI anchor may be added. GPI-anchored AGPs, as well as unanchored AGPs, are then transferred to the Golgi apparatus, where extensive glycosylation occurs by the action of a variety glycosyltransferase enzymes. Following glycosylation, AGPs are transported by secretory vesicles to the cell wall or to the extracellular face of the plasma membrane (in the case of GPI-anchored AGPs). GPI-anchored proteins can be released from the plasma membrane into the cell wall by phospholipases. In this review, we present an overview of the accumulated knowledge on AGP biosynthesis over the past three decades. Particular emphasis is placed on the glycosylation of AGPs as the sugar moiety is essential to their function. Recent genetics and genomics approaches have significantly contributed to a broader knowledge of AGP biosynthesis. However, many questions remain to be elucidated in the decades ahead.
Keywords: arabinogalactan-proteins, arabinogalactan-proteinbiosynthesis; glypiation; proline hydroxylation; glycosylation; glycosyltransferases; hydroxyproline; cell wall
The Plant Cell Wall: the Protector of the Realm
Plant growth and development are essential processes that are regulated and supported by the cell wall (Serpe and Nothnagel, 1999). The plant cell wall is a dynamic and complex structure composed of different components that act together and contribute to cell wall architecture and function (Somerville et al., 2004). The cell wall plays different key roles in plant growth and development, cell differentiation, environmental sensing and signaling, intercellular communication, water movement, and defense against invading pathogens (Cosgrove, 2005; Jamet et al., 2008; Malinovsky et al., 2014; Bacete et al., 2018). Typically, the primary cell wall is a thin (0.1–1 μm) and flexible layer composed of three major classes of polysaccharides, namely cellulose, hemicelluloses, and pectins, along with cell wall proteins (CWPs; Somerville et al., 2004; Cosgrove, 2005; Atmodjo et al., 2013; Anderson and Kieber, 2020). Some specific cell types also include lignins (Jamet et al., 2006). Approximately 15% of the 27,000 Arabidopsis thaliana genes are involved in cell wall synthesis, modification, and turnover (Carpita et al., 2001; Wang et al., 2012).
Polysaccharides constitute up to 90–95% of the cell wall mass, whereas CWPs account for 5–10% (Cassab and Varner, 1988; Jamet et al., 2008). CWPs are dedicated to cell wall support, structure, signaling, and interactions with other proteins (Jamet et al., 2006). CWPs are classified into nine functional categories in Arabidopsis: (1) proteins acting on carbohydrates, which include glycoside hydrolases (GHs), glycosyltransferases (GTs), carbohydrate esterases, polysaccharide lyases, and expansins; (2) oxido-reductases such as peroxidases, multicopper oxidases, berberine bridge enzyme, and blue copper binding proteins; (3) proteases like serine carboxypeptidases and aspartic, cysteine, and serine proteases; (4) proteins with interaction domains, for example, lectins, leucine-rich repeat domains, and enzyme inhibitors; (5) proteins possibly involved in signaling, which include arabinogalactan-proteins (AGPs) and receptors; (6) structural proteins like glycine-rich proteins, extensins (EXTs), leucine-rich-repeat extensins, and proline-rich proteins (PRPs); (7) proteins related to lipid metabolism, for instance, glycine, aspartic acid, serine, leucine (GDSL) lipases and lipid transfer proteins; (8) miscellaneous proteins such as purple acid phosphatases, phosphate-induced proteins, and germin; and (9) unknown function proteins with domains of unknown function (DUFs; Showalter, 1993; Jamet et al., 2008; Albenne et al., 2013).
The cell wall hydroxyproline-rich glycoproteins (HRGPs) superfamily is subdivided into three families: lightly glycosylated PRPs, moderately glycosylated EXTs, and highly glycosylated AGPs (Showalter, 1993; Nothnagel, 1997; Showalter et al., 2010; Hijazi et al., 2014; Johnson et al., 2017b). HRGPs are extracellular intrinsically disordered proteins (IDPs) as they are rich in Pro, the most disorder-promoting residue due to its rigid conformation and have motifs that direct posttranslational modifications (Johnson et al., 2017a).
Arabinogalactan-Proteins: All You Need is Sugar
AGPs are one of the most complex and diverse families of glycoproteins found in plants. AGPs may have a core-protein backbone rich in Pro/Hyp, Ala, Ser, and Thr (PAST) decorated by a diversity of carbohydrates (Showalter, 1993; Chasan, 1994; Schultz et al., 2002; Showalter et al., 2010). The amino acids of AGPs are often arranged in characteristic dipeptide repeats: Ala-Hyp, Ser-Hyp, Thr-Hyp, introduced as AG glycomodules (Tan et al., 2003; Ellis et al., 2010). The carbohydrate moiety of AGPs represents more than 90% of their total molecular mass, and it is composed mainly of arabinose (Ara) and galactose (Gal), as well as minor sugars such as glucuronic acid (GlcA), fucose (Fuc), rhamnose (Rha), and xylose (Xyl; Clarke et al., 1979; Fincher et al., 1983; Showalter, 1993; Chasan, 1994). AGPs have different molecular weights that reflect different extents of glycosylation of their specific protein cores. The extensive glycosylation of the protein backbone confers resistance to proteolysis (Showalter, 1993). Approximately half of the AGP family members are predicted to be tethered to the plasma membrane by a glycosylphosphatidylinositol (GPI) lipid anchor, being perfect candidates for signal perception and transduction (Youl et al., 1998; Oxley and Bacic, 1999; Sherrier et al., 1999; Svetek et al., 1999; Borner et al., 2002, 2003; Seifert and Roberts, 2007).
Positive reactions with β-Yariv reagent, a chemical reagent that specifically binds to the β-(1→3)-linked D-Galp backbone of AGPs (Yariv et al., 1967; Kitazawa et al., 2013), and immunolocalization studies with monoclonal antibodies that recognize AGP epitopes have shown that AGPs are ubiquitous in the plant kingdom, from bryophytes to angiosperms, and are also present in many algae (Clarke et al., 1979; Fincher et al., 1983; Showalter, 1993; Serpe and Nothnagel, 1999; Lee et al., 2005; Hervé et al., 2016). These glycoproteins were conserved during evolution, presumably because of their vital roles in plants (Serpe and Nothnagel, 1999). AGPs are found in plant cell walls, plasma membranes, apoplastic spaces, secretions, and intracellular multivesicular bodies (Herman and Lamb, 1992; Serpe and Nothnagel, 1999; Majewska-Sawka and Nothnagel, 2000; Ellis et al., 2010; Nguema-Ona et al., 2012).
AGPs are expressed in distinct cells and tissues and at particular stages of development. AGPs are a heterogenous family with their members, individually or collectively, implicated to function in many plant growth and development processes, such as cell proliferation and programed cell death, cell-cell signaling, embryo and postembryonic pattern formation, somatic embryogenesis, female and male gametophyte development, cell wall plasticizers, pollen tube growth and guidance, pollen incompatibility, root growth, xylem differentiation, secondary wall deposition, hormone signaling pathways, plant microbe interactions, and abiotic stress responses (reviewed in Showalter, 2001; Gaspar et al., 2004; Seifert and Roberts, 2007; Ellis et al., 2010; Nguema-Ona et al., 2013; Pereira et al., 2016). Recently, classical AGPs have been proposed to make a three-fold contribution: as a primary source of cytosolic Ca2+, as a pectic plasticizer and as Ca2+ signposts to the ovule (Lamport et al., 2018).
Finding and Classifying AGPs: A History Overview
AGPs were initially discovered as polysaccharides isolated from suspension-cultured sycamore (Acer pseudoplatanus L.) cells (Aspinall et al., 1969). Since then, AGPs have fascinated and challenged researchers with their huge diversity of protein backbones discovered by genome sequencing. Two decades ago, the term AGP was defined as a group of molecules that presents three criteria (Clarke et al., 1979; Du et al., 1996; Knox, 1999): a core-protein backbone rich in Hyp; type II arabino-3,6-galactan polysaccharides (AGs); and the ability to bind to Yariv reagents (Yariv et al., 1962, 1967). However, since then, investigators have discovered that several AGPs do not fit these criteria (Du et al., 1996), as it is the case of AG-peptides from wheat (Fincher et al., 1974) and two glycoproteins from Nicotiana alata style that do not bind the Yariv reagent (Lind et al., 1994; Sommer-Knudsen et al., 1996). Moreover, other AGPs are histidine-rich (Kieliszewski et al., 1992) or have short oligoarabinosides (Qi et al., 1991).
AGPs cDNA were first isolated, cloned, and sequenced in 1994 from cultured pear (Pyrus communis) cells (Chen et al., 1994) and from N. alata styles (Du et al., 1994). AGPs were first classified based on the amino acid sequence composition as classical AGPs and non-classical AGPs (Mau et al., 1995; Du et al., 1996). Classical AGPs are characterized by the presence of an N-terminal hydrophobic secretion signal sequence, a central domain rich in PAST residues (including sites for Hyp-O-glycosylation) and potentially a hydrophobic C-terminal anchor addition sequence that directs the attachment of a GPI anchor (Du et al., 1996). Non-classical AGPs contain an N-terminal hydrophobic secretion signal sequence followed by one or more PAST-rich regions, which can be Hyp-O-glycosylated, along with other non-PAST rich regions, such as hydrophilic C-terminal Asn-rich domains (Mau et al., 1995; Du et al., 1996).
After the sequencing of the Arabidopsis genome in 2000 by the Arabidopsis Genome Initiative (AGI; Kaul et al., 2000), Borner et al. (2002) identified on the basis of sequence analysis 210 predicted GPI-anchored proteins and over 40% of these proteins had putative AG glycomodules, including 13 classical AGPs, 9 AG peptides, 18 fasciclin-like proteins, 8 phytocyanin-like proteins, 8 early nodulin-like proteins, and 9 lipid transfer protein-like proteins. In the same year, Schultz et al. (2002) conducted the first bioinformatics identification of AGPs based on sequence analysis, the amino acid bias method identified and classified 47 candidate AGP genes in four different classes: classical AGPs (13), Lys-rich AGPs (3), AG peptides (10), and fasciclin-like AGPs (FLAs; 21). The other classes are structurally similar to the classical AGPs but have different lengths and domains in their polypeptide core. AG peptides are short classical AGPs with only 10–13 amino acids (Schultz et al., 2000). Lys-rich AGPs contain a Lys-rich domain of approximately 16 amino acid residues between a Pro-rich domain and the C-terminus (Schultz et al., 2002; Sun et al., 2005). FLAs can have one or two fasciclin-like (FAS1) cell adhesion domains with 110–150 amino acids and one or two AGP domains (Gaspar et al., 2001; Schultz et al., 2002; Johnson et al., 2003). Then, Showalter et al. (2010) conducted a bioinformatics approach using the BIO OHIO program to identify HRGPs in the Arabidopsis genome based on the amino acid composition and specific motifs. This analysis allowed the identification of 166 HRGPs: 85 AGPs, 59 EXTs, 18 PRPs, and four hybrid AGP/EXTs (HAEs), which contain modules characteristic of AGPs and EXTs.
Nowadays, with the improvement of technologies, whole genome sequences of several plant species have been annotated, which allowed bioinformatic identification of AGPs and consequently, an update on AGPs classification. A bioinformatics approach named as finding-AGP based on AG glycosylation has been performed in 47 plant species from Chlorophyta to Eudicot, enabling the identification of thousands of putative AGPs. The number of AGPs varied between 48 in Amborella trichopoda and 313 in Glycine max, with 151 AGPs being identified in Arabidopsis (Ma et al., 2017). According to their polypeptide core and based on the presence/absence of particular motif/domains, AGPs are now classified into classical AGPs, AG peptides, Lys-rich AGPs, FLAs, early nodulin-like AGPs (ENODLs) with plastocyanin-like domains, xylogen-like AGPs (XYLPs) with non-specific lipid transfer protein (nsLTP) domains, other chimeric AGPs that do not belong in any of the mentioned groups and HAEs (Borner et al., 2002, 2003; Schultz et al., 2002; Johnson et al., 2003; Mashiguchi et al., 2009; Ma and Zhao, 2010; Showalter et al., 2010; Kobayashi et al., 2011; Ma et al., 2017). Therefore, we propose that AGPs should be defined as a large family of glycoproteins, which can share common features, such as the typical protein domain rich in PAST, the occurrence of Ala-Hyp, Ser-Hyp, and/or Thr-Hyp dipeptide repeats, the occurrence of an N-terminal hydrophobic secretion signal sequence, the presence of type II AGs attached to Hyp residues, the ability to interact with the synthetic chemical dye β-Yariv reagent (Yariv et al., 1962, 1967), which recognizes the β-1,3-Gal main chains of type II AGs (Kitazawa et al., 2013), the possibility to be attached to the plasma membrane by a C-terminal GPI anchor and the potential presence of other functional domains.
AGP Biosynthesis Pathway: The Sugar Factory
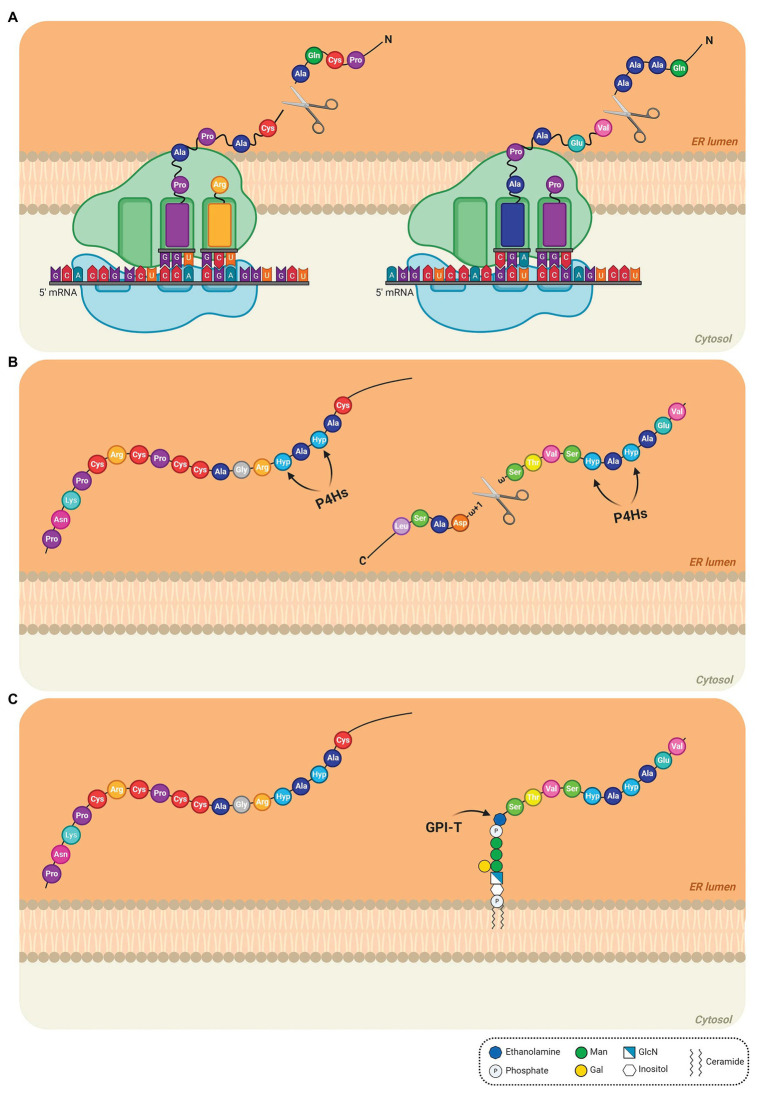
The biosynthesis of AGPs comprises the addition and removal of amino acids, lipids, and carbohydrates. As some AGPs are composed of approximately 90% sugar, the AGP biosynthetic pathway resembles a candy factory occurring inside the cells. The production line involves sequential operations and begins with the translation of the N-terminal signal sequence of AGPs on ribosomes, which allows entry into the endoplasmic reticulum (ER) and endomembrane system (Figure 1A). Translation continues of the AGPs with concomitant removal of the signal peptide (Figure 1A) and hydroxylation of Pro residues by prolyl hydroxylase enzymes located in the ER (Walter and Johnson, 1994; Schatz and Dobberstein, 1996; Faye et al., 2005; Figure 1B). The resulting AGP backbone can be further modified in the ER if it contains a GPI anchor addition sequence (Schultz et al., 1998; Yeats et al., 2018; Figure 1C). Subsequently, AGPs, either free in the ER lumen or tethered to the ER membrane by a GPI anchor (Figure 1C), are transported to the Golgi apparatus to allow for the successive addition of various monosaccharide units to the protein backbone (Figure 2A). Several enzymes constitute the required machinery for these consecutive sugar additions, and once the process is finished, the final products are transported to the plasma membrane for direct release to the cell wall (if no GPI anchor is present) or immobilized to the outer leaflet of the plasma membrane via a GPI anchor, with the bulk of these AGPs occurring in the periplasmic space (Figure 2B). Subsequent proteolytic processing can cleave the GPI anchor and release the AGP to the cell wall (Schultz et al., 1998).
Figure 1.
Detailed steps of the biosynthesis of AGPs in the endoplasmic reticulum (ER). (A) The N-terminal sequence is translated on the ribosomes, allowing the entry of the AGP into the ER. The N-terminal signal is removed and the AGP mRNA continues to be translated to produce the AGP protein backbone. (B) In the ER lumen, proline (Pro) residues are converted to hydroxyproline (Hyp) residues by prolyl-4-hydroxylases (P4Hs; Pro hydroxylation) and the C-terminal GPI anchor signal sequence is removed. The arrows indicate the site of action of P4Hs. (C) The preassembled GPI anchor is attached to the ω site of the mature protein via a transamidation reaction catalyzed by the transamidase complex (GPI-T). AtAGP13 (At4g26320; NM_118765) and AtAGP42 (At1g51915; NM_104072), two AG peptides were used as models in the schematic (left and right AGP, respectively). AtAGP13 (Q9STQ3-1) and AtAGP42 (Q8L9S8-1) have the smallest amino-acid sequence predicted to have or not a GPI anchor, respectively (Showalter et al., 2010). The extended Pro hydroxylation code was applied to determine, which Pro residues are hydroxylated (Canut et al., 2016; Duruflé et al., 2017). In these cases, only Pro residues after alanine (Ala) residues were converted to Hyp. Signal peptides and C-terminal anchor addition sequence positions were determined using UniProt (The UniProt Consortium, 2019). The GPI model structure was based on the GPI of PcAGP1 (Oxley and Bacic, 1999), which consists of phosphoethanolamine attached to the protein, three mannoses, one galactose, and glucosamine-inositol linked to phosphoceramide. Created by BioRender.com.
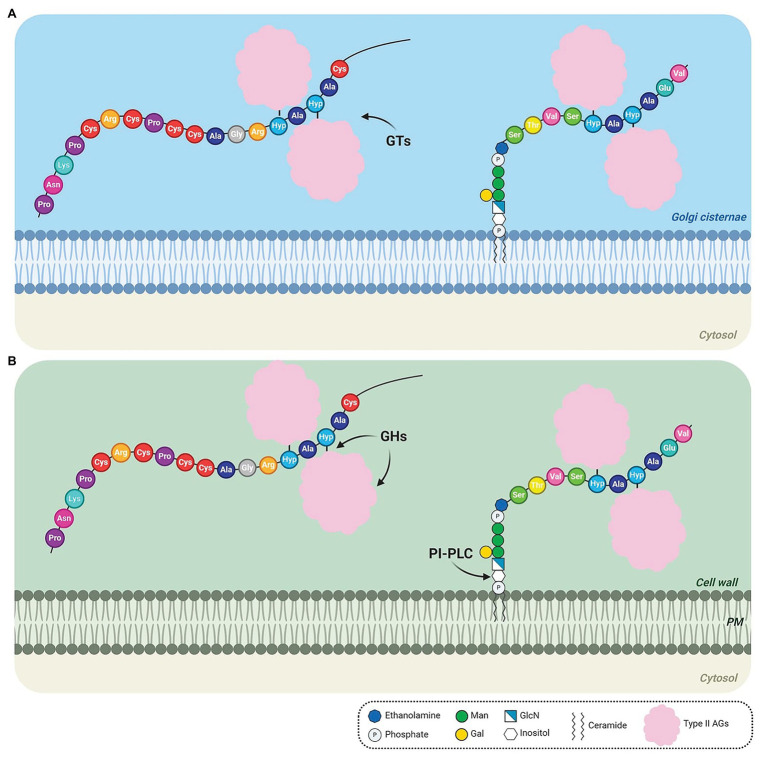
Figure 2.
Detailed steps of the biosynthesis of AGPs after transport from the endoplasmic reticulum (ER). (A) AGPs are transported to the Golgi, where type II arabinogalactan polysaccharides (AGs) are O-glycosidically linked to hydroxyproline (Hyp) residues by glycosyltransferases (GTs). (B) O-glycosylated AGPs are transported via Golgi vesicles to the cell wall, where they remain temporarily attached to the outer leaflet of the plasma membrane (PM) in the case of GPI-anchored AGPs. GPI-anchored AGPs may be released from the PM by PI-PLC phospholipase. The AGs may be cleaved by glycoside hydrolases (GHs). The arrows indicate the site of action of GTs (A), GHs and PI-PLC (B). AtAGP13 (At4g26320; NM_118765) and AtAGP42 (At1g51915; NM_104072), two AG peptides were used as models in the schematic (left and right AGP, respectively). AtAGP13 (Q9STQ3-1) and AtAGP42 (Q8L9S8-1) have the smallest amino-acid sequence predicted to have or not a GPI anchor, respectively (Showalter et al., 2010). Based on the Hyp contiguity hypothesis, non-contiguous Hyp residues are arabinogalactosylated. The GPI model structure was based on the GPI of PcAGP1 (Oxley and Bacic, 1999), which consists of phosphoethanolamine attached to the protein, three mannoses, one galactose, and glucosamine-inositol linked to phosphoceramide. Created by BioRender.com.
Pro Hydroxylation: Getting Ready
The N-terminal signal sequence, recognized by the signal recognition particle, is proteolytically removed, directing AGPs into the ER, where extensive post-translational modifications begin (Walter and Johnson, 1994; Schatz and Dobberstein, 1996; Figure 1A). Pro hydroxylation is a prerequisite for O-glycosylation of CWPs on Hyp residues (Faye et al., 2005). Pro hydroxylation begins in the ER and selected Pro residues are converted to Hyp residues by a multigene family of enzymes, prolyl-4-hydroxylases (P4Hs) providing reactive hydroxyl groups for O-glycosylation (Faye et al., 2005; Yuasa et al., 2005; Hijazi et al., 2014; Nguema-Ona et al., 2014; Figure 1B). P4Hs (EC 1.14.11.2) belong to the family of 2-oxoglutarate-dependent dioxygenases and catalyze the formation of 4-Hyp requiring 2-oxoglutarate and O2 as co-substrates, Fe2+ as cofactor and ascorbate (Kivirikko and Myllyharju, 1998; Vlad et al., 2007). Several plant P4Hs have been partially characterized in vitro and in vivo: AtP4H1 (Hieta and Myllyharju, 2002; Asif et al., 2009), AtP4H2 (Tiainen et al., 2005; Velasquez et al., 2011b), AtP4H5 and AtP4H13 (Velasquez et al., 2011a,b 2015), NtP4H (Yuasa et al., 2005), DcP4H1 and DcP4H2 (Vlad et al., 2010), and CrP4H1 (Keskiaho et al., 2007). Only the activity of AtP4H1 and AtP4H2 from the 13 member P4H family in Arabidopsis has been fully characterized (Velasquez et al., 2015). Recent evidence indicates that P4Hs may form protein complexes required for Pro hydroxylation as AtP4H5 forms homodimers/heterodimers with AtP4H2 and AtP4H13 in the Golgi (Velasquez et al., 2015).
To date it remains difficult to predict with confidence, which Pro residues will be hydroxylated (Duruflé et al., 2017). The extended Pro hydroxylation code, based on the initial Pro hydroxylation rules (Kieliszewski and Lamport, 1994) and additional experimental LC-MS/MS and Edman sequencing data, indicates that Pro residues are typically hydroxylated when they occur after Ala, Gln, Hyp, Pro, Ser, Thr, and Val residues (Canut et al., 2016; Duruflé et al., 2017).
Glypiation: GPI Anchoring
The addition of GPI-anchors, also referred to as glypiation, is a post-translational modification allowing AGPs to be attached to the plasma membrane (Duruflé et al., 2017). In Arabidopsis, 55 of the 85 identified AGPs are predicted to contain a GPI-anchor addition motif (Showalter et al., 2010). GPI-anchors allow attachment of proteins to the cell surface and may increase their lateral mobility in the membrane, exclusion from clathrin-coated pits, targeting to membrane microdomains/lipid rafts, and function in signal transduction pathways (Schultz et al., 1998; Ellis et al., 2010; Desnoyer and Palanivelu, 2020). The GPI-attachment signal (GAS) in the C-terminal region of the protein consists of ~11 polar residues, followed by the ω region of ~4 small residues containing the ω site, a spacer region of ~6 moderately polar residues and a C-terminal hydrophobic region of variable length between 9 and 24 residues (Schultz et al., 1998; Eisenhaber et al., 2003; Ellis et al., 2010; Desnoyer and Palanivelu, 2020). The highly conserved GPI moiety is initially synthesized on the cytosolic surface of the ER via the sequential addition of glucosamine, three α-linked mannosyl (Man) residues, and phosphoethanolamine to phosphatidylinositol (Schultz et al., 1998; Yeats et al., 2018; Beihammer et al., 2020; Desnoyer and Palanivelu, 2020). Although GPI anchor biosynthesis has not been biochemically studied in plants, the proteins that catalyze this pathway have been well studied in mammalian cells, yeast, and protozoa (Schultz et al., 1998; Pittet and Conzelmann, 2007; Morotti et al., 2017; Desnoyer and Palanivelu, 2020; Kinoshita, 2020). Nevertheless, this process may be conserved as orthologs of GPI biosynthetic genes are found in plant genomes (Schultz et al., 1998; Ellis et al., 2010; Desnoyer and Palanivelu, 2020).
In mammals, the first step involves the transfer of β-N-acetylglucosamine (GlcNAc), from the nucleotide sugar UDP-GlcNAc, to phosphatidylinositol (PI) to generate GlcNAc-PI by the GPI-N acetylglucosaminyltransferase (GPI-GnT) complex, which contains seven subunits (PIG-A, PIG-C, PIG-H, PIG-Q, PIG-P, PIG-Y, and DPM2). The product formed is de-N-acetylated subsequently by PIG-L, a GPI deacetylase, to yield GlcN-PI. At some point in the process, the synthesis switches from the cytoplasmic to the luminal face of the ER by an unknown flippase enzyme. In the ER lumen, GlcN-PI is acylated by the acyltransferase PIG-W to generate GlcN-(acyl)PI. Then, three Man residues, donated by dolichol-phospho-mannose (Dol-P-Man), are sequentially added to GlcN-(acyl)PI by the GPI-mannosyltransferases PIG-M, PIG-V, and PIG-B. The addition of phosphoethanolamine (PEtN) on the third Man residue is catalyzed by PIG-O and PIG-F (Schultz et al., 1998; Ellis et al., 2010; Beihammer et al., 2020; Desnoyer and Palanivelu, 2020; Kinoshita, 2020). The resulting GPI structure is ready to be transferred to proteins but other side chains may be added, such as phosphoethanolamines or sugars may be linked to Man and acyl groups linked to inositol (Luschnig and Seifert, 2010; Beihammer et al., 2020).
The GPI biosynthetic process converges with the cotranslational insertion of the protein backbone into the ER. The GAS is recognized and proteolytically cleaved between the ω and ω + 1 sites by the GPI transamidase (GPI-T) complex and the GPI anchor is attached to the ω site of the mature protein by a transamidation reaction (Schultz et al., 1998; Ellis et al., 2010; Yeats et al., 2018; Beihammer et al., 2020; Desnoyer and Palanivelu, 2020; Figure 1C). The mammal GPI-T consists of five subunits, PIG-K, GPAA1, PIG-S, PIG-T, and PIG-U (Kinoshita, 2020).
Some GPI biosynthesis enzymes in mammals have homologs already characterized in the Arabidopsis genome: SETH1 for PIG-C, SETH2 for PIG-A, PEANUT1 (PNT1) for PIG-M, PEANUT 5 (PNT5) for PIG-W, ABNORMAL POLLEN TUBE GUIDANCE1 (APTG1) for PIG-B, AtGPI8, and AtPIG-S (Lalanne et al., 2004; Gillmor et al., 2005; Dai et al., 2014; Bundy et al., 2016; Liu et al., 2016; Beihammer et al., 2020; Desnoyer et al., 2020; Desnoyer and Palanivelu, 2020). Although several different structures of GPI anchors have been observed in several kingdoms, to date, in plants, the only known structure was determined on PcAGP1 isolated from pear (Oxley and Bacic, 1999). This structure contains a glycan moiety conserved in all eukaryotic GPI anchors, D-Manα(1–2)-D-Manα(1–6)-D-Manα(1–4)-D-GlcN-inositol with a specific β(1–4)-galactosyl substitution of the 6-linked Man residue, it is devoid of PEtN side chains and contains a phosphoceramide, composed of phytosphingosine and tetracosanoic acid, instead of the common glycerolipid (Oxley and Bacic, 1999; Ellis et al., 2010; Yeats et al., 2018; Desnoyer and Palanivelu, 2020; Figure 1C). Moreover, an AGP isolated from Rosa sp. cell suspension cultures also included a ceramide, composed of tetracosanoic acid and 4-hydroxysphinganine, as the GPI lipid component (Svetek et al., 1999). These results suggest the PI glycerolipid moiety of some GPI-anchored proteins is remodeled to contain ceramide, as described in yeast (Bosson and Conzelmann, 2007). Discovering other plant GPI structures will identify possible forms of GPI and determine the conservation of these structures in plants (Desnoyer and Palanivelu, 2020).
Glycosylation: Becoming Sweeter
AGPs free in the ER and AGPs anchored to the ER membrane are then transferred to the Golgi apparatus, where they undergo glycosylation (Figure 2A). Glycosylation is one of the major post-translational modifications found in almost every living organism. It is performed by GTs and includes N-glycosylation, O-glycosylation, and glypiation (Hurtado-Guerrero and Davies, 2012; Duruflé et al., 2017). Each of these post-translational modifications occurs on specific amino acid sequences (Duruflé et al., 2017). The addition of carbohydrates on a polypeptide backbone may affect the physico-chemical properties of a protein, including resistance to thermal denaturation, protection from proteolytic degradation, solubility, and it can alter essential biological functions (Faye et al., 2005).
N-glycosylation occurs on Asn residues in Asn-X-Ser/Thr specific sequences, where X can be any amino acid except Pro (Faye et al., 2005; Duruflé et al., 2017). N-glycosylation starts in the ER by the co-translational transfer of Glc3Man9GlcNAc2, an oligosaccharide precursor, onto the amide nitrogen of Asn residues. During the transportation of the glycoprotein along the secretory pathway, the N-glycan undergoes a maturation process that involves the removal and the addition of sugar residues in the ER and the Golgi (Faye et al., 2005; Nguema-Ona et al., 2014). Classical AGPs do not contain this conserved sequence but many chimeric AGPs, including FLAs, contain the consensus sequence for N-glycosylation (Du et al., 1996; Johnson et al., 2003; Ellis et al., 2010).
O-glycosylation is the most complex type of glycosylation and, in plants, it occurs predominantly on Hyp residues and less often on Ser and Thr residues in the Golgi apparatus (Nothnagel, 1997; Faye et al., 2005; Duruflé et al., 2017). The Golgi apparatus, the central organelle in the secretory pathway, is responsible for glycosylation, protein sorting, and secretion; it contains a diverse group of membrane-bound GTs required for synthesis of a variety of linkage types (Nikolovski et al., 2012). O-glycosylation of Hyp is a complex mechanism unique to plants that involves the transfer of a glycan from the donor substrate to the acceptor hydroxyl group of Hyp residues (Faye et al., 2005). Gal can be linked to Ser and Hyp residues, whereas Ara can only be linked to Hyp residues (Duruflé et al., 2017). HRGPs are glycosylated by two types of O-glycosylation on their Hyp residues in the Golgi apparatus: Hyp arabinosylation and Hyp arabinogalactosylation (Kieliszewski, 2001; Nguema-Ona et al., 2014). The Hyp contiguity hypothesis (Kieliszewski and Lamport, 1994) predicts that contiguous Hyp residues are arabinosylated, adding short 4-6 residue long oligoarabinoside chains, as occurs in EXTs, whereas non-contiguous Hyp residues are arabinogalactosylated, adding larger 30–150 residue acidic or neutral AGs polysaccharides, as occurs in AGPs (Shpak et al., 1999, 2001; Kieliszewski, 2001; Faye et al., 2005).
AG glycans have a complex and heterogenous structure that changes throughout development (Tryfona et al., 2014). AG polysaccharides may affect cell surface protein trafficking and stability, act as chaperoning polysaccharides or Ca2+ chelators and generate signaling molecules (Borner et al., 2003; Lamport and Várnai, 2013; Tryfona et al., 2014). In AGPs, type II AGs are O-glycosidically linked to Hyp residues (Ellis et al., 2010; Figure 2A). The structure of type II AGs is not fully resolved but it seems to consist of a β-(1→3)-linked D-galactopyranosyl (Galp) backbone substituted at O6 positions by side-chains of β-(1→6)-linked D-Galp, which are further decorated with α-(1→3)-l-arabinofuranosyl residues (Araf) and less frequently with other sugars, such as GlcA, Rha, Fuc, and Xyl (Clarke et al., 1979; Gaspar et al., 2001; Showalter, 2001; Tan et al., 2004, 2010; Tryfona et al., 2010, 2012; Figure 3). The β-1,6-galactan side chains often terminate with β-1,6-GlcA or 4-O-Me-GlcA, giving the molecules an overall negative charge (Ellis et al., 2010; Tryfona et al., 2012; Temple et al., 2019). Two other types of AG polysaccharides have been described in plants (Clarke et al., 1979; Hijazi et al., 2014). Type I AGs are formed by a linear chain of β-(1→4)-linked D-Galp with lateral chains of α-Araf and β-(1→4)-linked D-Galp (Clarke et al., 1979; Hijazi et al., 2014). Leonard et al. (2005) defined the Artemisia vulgaris Art v 1 polysaccharide, formed by a short linear β-(1→6)-linked D-Galp chain, which contains Gal and Ara residues and large branched Ara chains, as a new AG type, the type III AG. However, this structure is inconsistent with the finding that it binds to β-Yariv, as chains of β-(1→3)-linked D-Galp7 are required for β-Yariv binding (Kitazawa et al., 2013). Therefore, this structure may need to be revised.
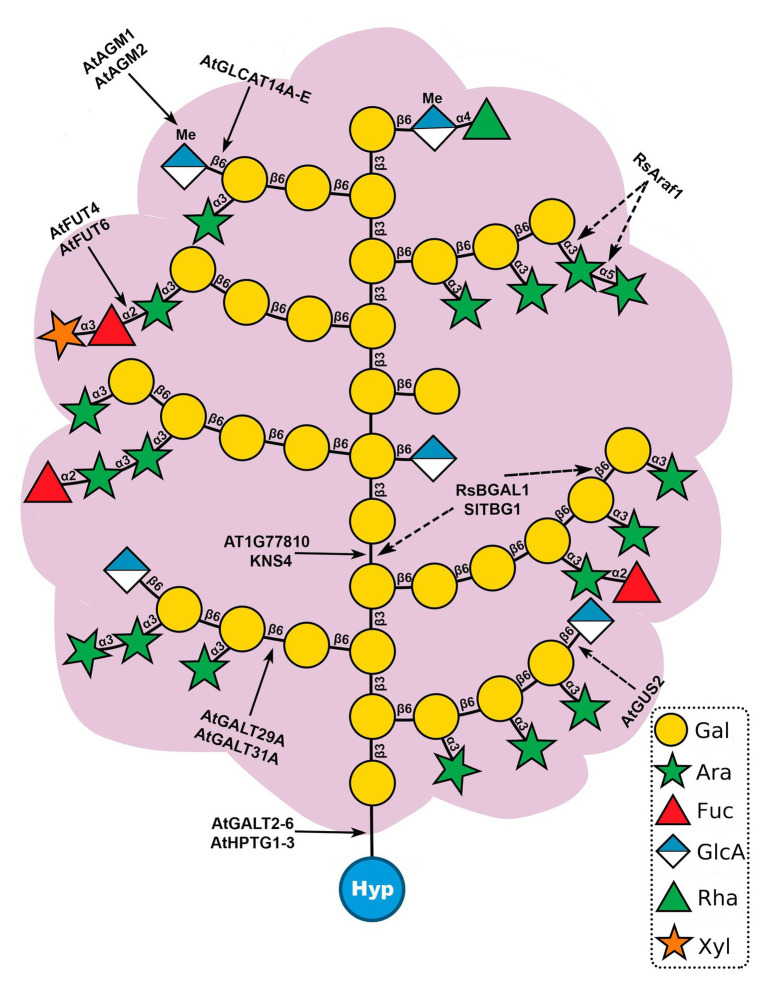
Figure 3.
Model structure of type II arabinogalactan polysaccharides (AGs) and sites of action of known glycosyltransferases (GTs) and glycoside hydrolases (GHs) acting on AGPs. Type II AGs are O-glycosidically linked to hydroxyproline (Hyp) and consist of a β-(1→3)-linked backbone of galactose (Gal) with β-(1→6)-galactan side chains. Further modifications involve the addition of arabinose (Ara), fucose (Fuc), rhamnose (Rha), glucuronic acid (GlcA), 4-O-methylglucuronosyl (4-O-MeGlcA), and xylose (Xyl). Solid line arrows (left) represent sites of action of GTs and dotted line arrows (right) indicate sites of action of GHs. This structure is based on AGs analyzed from Arabidopsis leaves (Tryfona et al., 2012).
Three models have been proposed for the molecular structure of AGPs: the wattle-blossom, the twisted hairy rope, and the necklace (Fincher et al., 1983; Qi et al., 1991; Du et al., 1996; Lamport et al., 2014). The wattle blossom model predicts globular units of polysaccharide chains anchored to a protein core of a spheroidal molecule, and the twisted hairy rope model foresees an alignment of AG chains along the protein backbone, whereas the recent necklace model compares AGP structure to an ancient gold necklace from Afghanistan with pendant glycomodules. Detailed analysis of different AGPs will determine the number, length, and sequence of polysaccharide chains, allowing the improvement of the existing models.
AGPs may be connected to other cell wall components such as pectins and hemicelluloses (Tan et al., 2013). Keegstra et al. (1973) hypothesized that type II AGs could be linked to rhamnogalacturonan (RGI) by the Rha residues. This linkage has been demonstrated in the complex arabinoxylan pectin arabinogalactan protein 1 (APAP1). This complex also has a link between arabinoxylan and an arabinose residue in the type II AGs (Tan et al., 2013). A FLA, SALT-OVERLY SENSITIVE 5 (SOS5) was proposed to mediate seed coat mucilage adherence by interacting with pectins (Griffiths et al., 2014). These results support the hypothesis that AGPs may serve as cross-linkers in the cell wall and act as polysaccharide plasticizers (Lamport, 2001; Lamport et al., 2006). The binding of the Yariv reagent specifically to AGs supports the view that AGs bind specific glycans (Kitazawa et al., 2013).
Glycosyltransferases: Meet the Sugar Workers
Glycoproteins have a complex structure and, therefore, plants require the action of numerous GTs families to assist in their biosynthesis. AGP glycosylation is catalyzed by a large number of GTs (EC 2.4.x.y) in the secretory pathway. GTs are enzymes that catalyze glycosidic bond formation between a sugar moiety and a specific acceptor molecule (sugars, proteins, lipids, or small molecules), creating a diverse collection of oligosaccharides and glycoconjugates in nature (Hansen et al., 2012; Gloster, 2014). GTs have been classified into 111 families in the carbohydrate active enzymes (CAZy) database1 (Coutinho et al., 2003; Lombard et al., 2014). GTs are very specific and, thus, each different linkage may require a distinct GT. With AGP glycosylation, several GTs work together to regulate the density, length, and sequences of AG chains (Qu et al., 2008). Almost 500 putative GTs sequences have been identified in the Arabidopsis genome and classified into 42 different CAZy families (Coutinho et al., 2003). In order to understand the structure of AGPs, the discovery of GTs involved in the initiation and elongation of AG chains is a priority. GTs can be type I membrane proteins located in the ER, type II membrane proteins located in the Golgi or integral membrane proteins such as cellulose synthases (Hansen et al., 2009). GTs generally are localized in the Golgi and have a type II membrane protein topology with a short N-terminal fragment facing the cytosol, one helical transmembrane domain, and a hydrophilic C-terminal catalytic domain containing the active site attached to a flexible stem region facing the luminal side (Perrin et al., 2001; Hansen et al., 2009; Chou et al., 2012).
The biosynthesis of AGP glycans requires at least 10 functionally distinct enzymes, e.g., galactosyltransferases (GALTs), arabinosyltransferases, fucosyltransferases (FUTs), rhamnosyltransferases, xylosyltransferases, glucuronosyltransferases (GLCATs), and glucuronic acid methyltransferases. To date, 22 transferases responsible for AGP glycosylation have been identified in Arabidopsis (Tables 1 and 2): eight hydroxyproline-O-β-GALTs (Hyp-O-GALTs; AtGALT2, AtGALT3, AtGALT4, AtGALT5, and AtGALT6, Basu et al., 2013, 2015a,b; Showalter and Basu, 2016; AtHPTG1, AtHPTG2, and AtHPTG3, Ogawa-Ohnishi and Matsubayashi, 2015), two β-1,3-GALTs (At1g77810, Qu et al., 2008 and AtKSN4, Suzuki et al., 2017), two β-1,6-GALTs (AtGALT31A, Geshi et al., 2013 and AtGALT29A; Dilokpimol et al., 2014), five β-1,6-GLCATs (AtGLCAT14A, AtGLCAT14B, AtGLCAT14C, AtGLCAT14D, and AtGLCAT14E, Knoch et al., 2013; Dilokpimol et al., 2014; Lopez-Hernandez et al., 2020; Zhang et al., 2020), two α-1,2-FUTs (AtFUT4 and AtFUT6, Wu et al., 2010; Liang et al., 2013; Tryfona et al., 2014), a putative β-arabinosyltransferase (AtRAY1, Gille et al., 2013), and two glucuronic acid methyltransferases (AtAGM1 and AtAGM2, Temple et al., 2019; Figure 3). However, several enzymes remain to be identified, including α-arabinofuranosyltransferases, β-arabinopyranosyltransferases, α-rhamnosyltransferases, α-xylosyltransferases, an α-GALTs, and other β-GALTs, β-GLCATs, α-FUTs, and glucuronic acid methyltransferases.
Table 1.
Information about the characterized transferases involved in the biosynthesis of AGP glycans in Arabidopsis thaliana.
| CAZy GT family | Enzyme | Locus | Enzyme Activity | Subcellular localization | References |
|---|---|---|---|---|---|
| GT14 | GLCAT14A | At5g39990 | β-1,6-GLCAT | Golgi Unique subcellular compartments |
Knoch et al., 2013; Dilokpimol and Geshi, 2014; Lopez-Hernandez et al., 2020; Zhang et al., 2020 |
| GLCAT14B | At5g15050 | - | |||
| GLCAT14C | At2g37585 | - | |||
| GLCAT14D | At3g24040 | - | |||
| GLCAT14E | At3g15350 | - | |||
| GT29 | GALT29A | At1g08280 | β-1,6-GALT | Golgi Unique subcellular compartments |
Dilokpimol et al., 2014 |
| GT31 | GALT2 | At4g21060 | Hyp-O-β-GALT | ER and Golgi | Basu et al., 2013, 2015a,b |
| GALT3 | At3g06440 | Golgi | |||
| GALT4 | At1g27120 | Golgi | |||
| GALT5 | At1g74800 | Golgi | |||
| GALT6 | At5g62620 | Golgi | |||
| HPGT1 | At5g53340 | Golgi | Ogawa-Ohnishi and Matsubayashi, 2015 | ||
| HPGT2 | At4g32120 | Golgi | |||
| HPGT3 | At2g25300 | Golgi | |||
| At1g77810 | At1g77810 | β-1,3-GALT | Golgi | Qu et al., 2008 | |
| KNS4 | At1g33430 | - | Suzuki et al., 2017 | ||
| GALT31A | At1g32930 | β-1,6-GALT | Golgi Unique subcellular compartments |
Geshi et al., 2013 | |
| GT37 | FUT4 | At2g15390 | α-1,2-FUT | - | Wu et al., 2010; Liang et al., 2013; Tryfona et al., 2014 |
| FUT6 | At1g14080 | Golgi | |||
| GT77 | RAY1 | At1g70630 | β-arabinofuranosyltransferase | Golgi | Gille et al., 2013 |
| - | AGM1 | At1g27930 | GlcA O-methyltransferase | Golgi | Temple et al., 2019 |
| AGM2 | At1g67330 | - |
Table 2.
Information about the mutants of the characterized transferases involved in the biosynthesis of AGP glycans in Arabidopsis thaliana.
| Genes | Mutants | Mutation Localization | Mutant Phenotypes | References |
|---|---|---|---|---|
| GLCAT14A | glcat14a-1 (SALK_064313) | Exon | Reduced of GlcA substitution on AGPs (glcat14a, glcat14a-2, glcat14b-1, glcat14d-1, glcat14e-1, glcat14aglcat14b, glcat14a-2/b-1, glcat14bglcat14c, glcat14aglcat14bglcat14c, glcat14a-2/b-1/d-1, glcat14a-2/b-1/d-2, and glcat14a-2/b-1/e-1); increase of Gal and Ara in AGPs (glcat14c, glcat14aglcat14b, glcat14bglcat14c, and glcat14aglcat14bglcat14c); reduction of Ara in AGPs (glcat14a-1 and glcat14a-2); reduced amount of glycosylated AGPs (glcat14c, glcat14aglcat14b, and glcat14aglcat14bglcat14c); reduction in Ca2+ binding capacity of AGPs (glcat14b, glcat14c, glcat14aglcat14b, glcat14bglcat14c, glcat14aglcat14bglcat14c, and glcat14a-2/b-1/d-1); enhanced cell elongation in seedlings (glcat14a-1 and glcat14a-2); reduced seed coat mucilage (glcat14b, glcat14c, glcat14aglcat14b, glcat14bglcat14c, and glcat14aglcat14bglcat14c); delayed seed germination and reduced root hair length (glcat14aglcat14b and glcat14aglcat14bglcat14c); reduced trichome branching (glcat14aglcat14b, glcat14a-2/b-1, glcat14aglcat14bglcat14c, glcat14a-2/b-1/d-1, and glcat14a-2/b-1/d-2); shorter siliques and reduced seed production (glcat14bglcat14c and glcat14aglcat14bglcat14c); small and defective pollen that failed to germinate (glcat14aglcat14b and glcat14aglcat14bglcat14c); shorter inflorescence stems (glcat14a-2/b-1 and glcat14a-2/b-1/d-1); smaller etiolated hypocotyls and altered [Ca2+]cyt signature on roots (glcat14a-2/b-1/e-1) | Knoch et al., 2013; Dilokpimol and Geshi, 2014; Lopez-Hernandez et al., 2020; Zhang et al., 2020 |
| glcat14a-2 (SALK_043905) | Exon | |||
| glcat14a (CRISPR-Cas9) | Exon | |||
| GLCAT14B | glcat14b (CRISPR-Cas9) | Exon | ||
| glcat14b-1 (SALK_080923) | Exon | |||
| GLCAT14C | glcat14c (CRISPR-Cas9) | Exon | ||
| GLCAT14D | glcat14d-1 (GK363F05.01) | Exon | ||
| glcat14d-2 (GK508D01) | Intron | |||
| GLCAT14E | glcat14e-1 (SALK_022820) | Intron | ||
| GALT29A | - | - | - | Dilokpimol et al., 2014 |
| GALT2 | galt2-1 (SALK_117233) | Exon | Lower GALT activities and reduced β-Yariv-precipitable AGPs; root and pollen tubes sensitivity reduction to β-Yariv reagent; reduced growth and root tip swelling in response to salt and sucrose; reduced root hair length and density (galt2, galt3, galt5, and galt2galt5), seed production (galt4 and galt6), and seed coat mucilage (galt3, galt6, and galt2galt5); premature senescence (galt6); and large number of rosette leaves, delayed flowering, and shorter siliques (galt2galt5) | Basu et al., 2013, 2015a,b |
| galt2-2 (SALK_141126) | Exon | |||
| GALT3 | galt3-1 (SALK_085633) | Promoter | ||
| galt3-2 (SALK_005178) | Promoter | |||
| GALT4 | galt4-1 (SALK_136251) | Exon | ||
| galt4-2 (SALK_131723) | Exon | |||
| GALT5 | galt5-1 (SALK_105404) | 5' UTR | ||
| galt5-2 (SALK_115741) | Exon | |||
| GALT6 | galt6-1 (SAIL_59_D08) | Exon | ||
| galt6-2 (SAIL_70_B02) | Exon | |||
| HPGT1 | hpgt1-1 (SALK_007547) | Intron | Longer lateral roots, longer and denser root hairs, shorter inflorescence stems, and shorter siliques (hpgt2-1, hpgt3-1, and hpgt1hpgt2hpgt3) and thicker roots, smaller rosette leaves, shorter petioles, and reduced fertility (hpgt1hpgt2hpgt3) | Ogawa-Ohnishi and Matsubayashi, 2015 |
| HPGT2 | hpgt2-1 (SALK_070368) | Exon | ||
| HPGT3 | hpgt3-1 (SALK_009405) | Exon | ||
| At1g77810 | - | - | - | Qu et al., 2008 |
| KNS4 | - | - | - | Suzuki et al., 2017 |
| GALT31A | galt31a (FLAG_379B06) | Exon | Embryo development arrested at the globular stage | Geshi et al., 2013 |
| FUT4 | fut4 (SAIL_284_B05) | Exon | Reduced root growth under salt stress (fut4, fut6, and fut4fut6) and lack of fucose in leaf and root AGPs (fut4fut6) | Wu et al., 2010; Liang et al., 2013; Tryfona et al., 2014 |
| fut4-2 (SALK_125310) | Exon | |||
| FUT6 | fut6 (SALK_099500) | Exon | ||
| fut6-2 (SALK_078257) | Exon | |||
| RAY1 | ray1-1 (SALK_053158) | Exon | Reduced level of arabinose in their AGPs in etiolated seedlings, roots and rosette leaves; reduced root growth; and reduced rosette and inflorescence size | Gille et al., 2013 |
| ray1-2 (GK_001C09) | Exon | |||
| AGM1 | agm1-1 (SALK_000253) | Exon | Reduction on GlcA methylation (agm1); absence of GlcA methylation (agm1agm2) | Temple et al., 2019 |
| AGM2 | agm2-1 (GABI_054A04) | Exon | ||
| agm2-2 (SALK_057182) | 5' UTR |
Galactosyltransferases: The First Ones to Arrive
AGP glycosylation is initiated by the action of Hyp-O-GALTs that add the first Gal onto the hydroxyl group of Hyp residues in the protein backbone (Ogawa-Ohnishi and Matsubayashi, 2015; Showalter and Basu, 2016). This process allows the subsequent addition of different sugars by other GTs. The eight GALTs specific for Hyp identified in Arabidopsis belong to the CAZy GT31 family. AtGALT2, AtGALT3, AtGALT4, AtGALT5, and AtGALT6 encode a GALT domain as well as a GALECTIN domain, whereas AtHPGT1, AtHPGT2, and AtHPGT3 lack a GALECTIN domain (Showalter and Basu, 2016). Their activity was demonstrated by heterologous expression in Pichia pastoris (AtGALT2, AtGALT5; Basu et al., 2013, 2015b), in Nicotiana tabacum leaf epidermal cells (AtGALT2, AtGALT3, AtGALT4, AtGALT5, and AtGALT6; Basu et al., 2015a) and BY-2 cells (AtHPGT1, AtHPGT2, and AtHPGT3; Ogawa-Ohnishi and Matsubayashi, 2015). galt2, galt3, galt4, galt5, galt6, galt2galt5, and hpgt1hpgt2hpgt3 mutants demonstrated lower GALT activities and reduced β-Yariv-precipitable AGPs compared to wild-type plants. AtGALT3-6 and AtHPGT1 transiently expressed in N. tabacum and Arabidopsis T87 protoplasts, respectively, localized to the Golgi, whereas AtGALT2 was found in both ER and Golgi when expressed in tobacco (Basu et al., 2013, 2015a,b; Ogawa-Ohnishi and Matsubayashi, 2015). These results may indicate that AGP glycosylation may start in the ER, but predominantly occurs in the Golgi (Basu et al., 2013, 2016). Nevertheless, further studies are required to explore this hypothesis.
Some physiological phenotypes were revealed under normal growth conditions in the GALT mutants: galt2, galt3, galt4, galt5, and galt2galt5 displayed reduced root hair length and density, galt4 and galt6 showed reduced seed production, galt3, galt6, and galt2galt5 presented reduced seed coat mucilage and galt6 revealed accelerated leaf senescence. Additionally, all GALT mutants (galt2, galt3, galt4, galt5, galt6, and galt2galt5) roots and pollen tubes exhibited less sensitivity to β-Yariv reagent, and root growth and root tip swelling were impaired under salt stress and in elevated levels of sucrose. galt2galt5 displayed a large number of rosette leaves, delayed flowering time, reduced silique length, and plant height (Basu et al., 2015a,b). The galt2galt5 double mutant phenocopies the root swelling, the reduced seed coat mucilage, and the reduced cellulose phenotypes of sos5 and fei1fei2, a double mutant of two cell wall-associated leucine-rich repeat receptor-like kinases, and sos5fei1fei2, indicating that the carbohydrate moiety of SOS5 is important for signaling in the cell (Basu et al., 2015b). The analysis of the quintuple mutant galt2galt5sos5fei1fei2 showed that these five genes act in a single and linear genetic pathway, and it was hypothesized that SOS5 glycosylation by GALT2 and GALT5 was required for its function in the SOS5/FEI1-FEI2 signaling pathway (Basu et al., 2016). In addition, HPGT mutants exhibited several pleiotropic phenotypes such as longer lateral roots, increased root hair length, and density, shorter inflorescence stems, shorter siliques (hpgt2hpgt3 and hpgt1hpgt2hpgt3), thicker primary roots, smaller rosette leaves, shorter petioles, and reduced fertility in the lower portion of the inflorescence (hpgt1hpgt2hpgt3; Ogawa-Ohnishi and Matsubayashi, 2015).
Four additional GALTs were identified. At1g77810 encodes a β-1,3-GALT that belongs to the GT31 family and likely functions in β-1,3-galactan backbone synthesis. This Golgi membrane-located enzyme was demonstrated to add Gal to a synthetic β-1,3-Gal disaccharide using heterologous expression in COS cells (Qu et al., 2008). AtKSN4 (KAONASHI4) is also a member of GT31 family. Heterologous expression of AtKSN4 in Nicotiana benthamiana showed β-1,3-GALT activity on AG glycans from AGPs and pectins. ksn4 mutants present an abnormality in the exine layer of developing microspores. Immunolabeling showed that ksn4 mutants have reduced AGP content in the primexine of developing microspores. Furthermore, ksn4 mutants exhibit pollen aggregation and reduced fertility (shorter fruit lengths and lower seed set compared to wild type; Suzuki et al., 2017). Another GT31 member, AtGALT31A, is involved in elongation of β-1,6-galactan side chains. Its activity was demonstrated by heterologous expression in Escherichia coli and N. benthamiana. AtGALT31A accumulates in the Golgi apparatus and unidentified organelles in N. benthamiana. A mutation in this gene showed an abnormal asymmetric cell division in the hypophysis causing the arrest of embryo development at the globular stage. This phenotype reveals the importance of AG glycans in embryo development (Geshi et al., 2013). AtGALT29A co-expresses with AtGALT31A and AtGLCAT14A. AtGALT29A resides in the GT29 family and this enzyme recombinantly expressed in N. benthamiana possesses β-1,6-GALT elongation and branch initiation activities. Dilokpimol et al. (2014) showed that AtGALT29A was localized in the Golgi in N. benthamiana, where it interacted with AtGALT31A, as indicated by Förster resonance energy transfer. AtGALT31A was also targeted in tobacco to uncharacterized small compartments, which are not part of the trans-Golgi network, cis-Golgi network, or endosomes and that colocalized with EXO70E2, a marker for exocyst-positive organelles that mediate an unconventional protein secretory pathway in plants (Poulsen et al., 2014). Moreover, the enzyme complex containing AtGALT31A and AtGALT29A exhibited enhanced β-1,6-GALT activity when compared to AtGALT29 alone.
Glucuronosyltransferases: Adding the Special Sugar
AtGLCAT14A, AtGLCAT14B, AtGLCAT14C AtGLCAT14D, and AtGLCAT14E are glucuronosyltransferases that belong to the GT14 family and add glucuronic acid to β-1,6‐ and β-1,3-galactose chains of AGPs. The activity of AtGLCAT14A, AtGLCAT14B, and AtGLCAT14C enzymes was confirmed by recombinant expression in P. pastoris and in vitro enzyme assays (Knoch et al., 2013; Dilokpimol and Geshi, 2014). AtGLCAT14A was transiently expressed in N. benthamiana and localized in the Golgi apparatus. AtGLCAT14A is co-localized with AtGALT31A in the Golgi and in uncharacterized small compartments, but the two enzymes do not interact (Knoch et al., 2013).
AtGLCAT14 mutants have defective synthesis of AGs. glcat14a, glcat14b, glcat14d, glcat14e, glcat14aglcat14b, glcat14bglcat14c, glcat14aglcat14bglcat14c, glcat14aglcat14bglcat14d, and glcat14aglcat14bglcat14e mutants have reduced the content of GlcA on AGPs when compared to wild type (Knoch et al., 2013; Lopez-Hernandez et al., 2020; Zhang et al., 2020). Although this suggests some redundancy between these enzymes, the mutants show some preferential changes in GlcA on specific branch lengths of AG and, therefore, may have roles glucuronidating different parts of the AG or different AGPs (Lopez-Hernandez et al., 2020). An increase of Gal and Ara was detected in AG extracts of glcat14c, glcat14aglcat14b, glcat14bglcat14c, and glcat14aglcat14bglcat14c (Zhang et al., 2020). However, in glcat14a, an increase of Gal and a reduction of Ara were detected. The increase of galactosylation may result from the increase of O6 acceptor sites, which are shared by AtGLCAT14A and GALTs and, thus, the addition of GlcA may terminate Gal chain extension (Knoch et al., 2013). Indeed, the amount of glycosylated AGPs was increased in the mutants glcat14c, glcat14aglcat14b, and glcat14aglcat14bglcat14c, which is a result of the loss of GlcA residues, allowing for the elongation of the branched β-(1,3)‐ and β-(1,6)-galactans. On the other hand, Lopez-Hernandez et al. (2020) did not find evidence for an increase in galactan side chain length in the glcat14 mutants.
AtGLCAT14 mutants present several phenotypes. glcat14a knockout mutants showed enhanced cell elongation during seedling growth (Knoch et al., 2013), glcat14b, glcat14c, glcat14aglcat14b, glcat14bglcat14c, and glcat14aglcat14bglcat14c presented reduced seed coat mucilage, glcat14aglcat14b and glcat14aglcat14bglcat14c exhibited delayed seed germination, reduced root hair length, glcat14aglcat14b, glcat14aglcat14bglcat14c, and glcat14aglcat14bglcat14d reduced trichome branching, glcat14aglcat14b and glcat14aglcat14bglcat14d presented shorter inflorescences, glcat14bglcat14c and glcat14aglcat14bglcat14c presented reduced silique length and seed set, and glcat14aglcat14b and glcat14aglcat14bglcat14c displayed a significant percentage of small and defective pollen that failed to germinate (Lopez-Hernandez et al., 2020; Zhang et al., 2020). glcat14aglcat14bglcat14e triple mutant plants had severely limited seedling growth and were sterile (Lopez-Hernandez et al., 2020).
In addition, glcat14b, glcat14c, glcat14aglcat14b, glcat14bglcat14c, glcat14aglcat14bglcat14c, and glcat14aglcat14bglcat14d showed a reduction in Ca2+ binding in AGP extracts compared to wild type (Lopez-Hernandez et al., 2020; Zhang et al., 2020), consistent with the model proposed by Lamport and Várnai (2013) in which AGPs can bind and store Ca2+ through GlcA in a reversible and pH-dependent way at the plasma membrane. It was demonstrated that in vitro AGPs could hold Ca2+ in a pH range of 4–5 and as the pH was lowered Ca2+ was released. AGPs fully released Ca2+ at pH 2.5 (Lamport and Várnai, 2013). This bound Ca2+ may be important for intracellular signaling. Indeed, many of the plant developmental phenotypes in glcat14 mutants can be suppressed by raising the Ca2+ concentration in the growth medium (Lopez-Hernandez et al., 2020). Intracellular Ca2+ signals were disrupted in the glcat14aglcat14bglcat14e mutant plants, which showed altered movement of the Ca2+ signal through the roots. Thus, GLCATs may play an important role in Ca2+ signaling as they determine the presence of GlcA on AGPs (Dilokpimol and Geshi, 2014).
Glucuronic Acid Methyltransferases: Changing GlcA
In type II AGs from Arabidopsis, most of the GlcA substituted is 4-O-methylglucuronosyl (4-O-Me-GlcA; Tryfona et al., 2012). Recently, two GlcA-specific methyltransferases have been identified in Arabidopsis (Temple et al., 2019). Arabinogalactan methyltransferases 1 (AGM1) and 2 (AGM2), two family members of DUF579 family, have GlcA-O-methylation activity on AGPs. AGM1 fused to GFP localized in the Golgi apparatus when transiently expressed in tobacco leaves. An agm1 mutant showed reduced methylation of GlcA on root AGPs and in the double mutant agm1agm2 there was no AG GlcA methylation of root AGPs. AGM1 and AGM2 are involved in methylation of GlcA of AG in root AGPs. An agm1agm2 double mutant did not exhibit a growth or fertility phenotype, showing that GlcA methylation is not essential for viability. The biological role of GlcA methylation modification on AGPs is still unclear (Temple et al., 2019). However, it has been shown that the methyl group on GlcA is essential for the effectiveness of a signaling molecule in pollen tube guidance (Mizukami et al., 2016). In addition, 4-O-methylation of GlcA may change the Ca2+ binding affinity to GlcA thus modulating the calcium release response to pH (Lamport and Várnai, 2013). Moreover, the addition of the methyl group to GlcA prevents the addition of 4-linked sugars, such as Rha, and the extension of 4-linked side chains to the GlcA of AG as seen in APAP1 (Tan et al., 2013; Temple et al., 2019).
Fucosyltransferases: Continuing the Hard Work
AtFUT4 and AtFUT6, two AGP-α-(1,2)-FUT of AGPs, belong to the plant GT37 CAZy family. Their enzymatic activities were demonstrated by heterologous expression in N. tabacum BY2 cells (Wu et al., 2010). Tobacco BY2 cells contain nonfucosylated AGPs but heterologous expression of these genes resulted in fucosylated AGPs (Wu et al., 2010). Knock-out mutants (fut4, fut6, and fut4fut6) grown under salt stress showed reduced root growth (Liang et al., 2013; Tryfona et al., 2014). This conditional phenotype reveals the importance of this sugar in root growth and salt sensitivity. Fuc was absent in fut4 leaf AGPs, whereas the fut4fut6 double mutant lacks fucose in both leaf and root AGPs (Liang et al., 2013; Tryfona et al., 2014). As AtFUT4 is expressed in roots and leaves while AtFUT6 is expressed mostly in roots, it is likely that AtFUT4 and AtFUT6 are both responsible for AGP fucosylation in roots while AtFUT4 is also responsible for fucosylation in leaves (Wu et al., 2010; Liang et al., 2013; Tryfona et al., 2014). The fut4 and fut6 single mutants had reduced Fuc content in root AGPs. The fut6 mutant was not stained by eel lectin that binds specifically to terminal α-L-Fuc, indicating that AtFUT6 likely adds terminal Fuc residues to AG polysaccharides (Liang et al., 2013). AtFUT6-GFP was transiently expressed in tobacco leaves and localized to the Golgi apparatus (Wu et al., 2010).
Arabinosyltransferases: The Ghost Workers
Finally, ray1 mutants showed a reduced level of Ara in their AGPs in etiolated seedlings, roots, and rosette leaves of Arabidopsis, leading to its name REDUCED ARABINOSE YARIV 1. Knockout mutants also exhibit reduced root growth, reduced rosette diameter, and delayed development of the inflorescence. AtRAY1 belongs to the GT77 family, and its heterologous expression in N. benthamiana demonstrated β-arabinofuranosyltransferase activity (Gille et al., 2013). However, only α-1,3-linked Ara, and not β-1,3-linked Ara, has been reported in AGPs so it remains unclear whether and how RAY1 functions in the biosynthesis of AGPs glycans (Showalter and Basu, 2016). There are no published candidates for the α-arabinofuranosyltransferases that transfer the main arabinose decoration on AGs.
Glycoside Hydrolases: Cutting All the Sugar
GHs (EC 3.2.1.x) hydrolyze the glycosidic bond between two sugars and are likely important for the metabolism of AGPs (Henrissat and Davies, 1997; Knoch et al., 2014). GHs are classified into 167 families in the CAZy database (Coutinho et al., 2003; Lombard et al., 2014). Hydrolysis of AGP glycans requires several GHs, such as β-galactosidases, β-galactanases, α-arabinofuranosidases, β-arabinopyranosidases, β-glucuronidases, α-fucosidases, α-rhamnosidases, and β-xylosidases. These enzymes can be a useful tool to analyze AG sugars decorating AGPs (Knoch et al., 2014).
So far, five GHs of AGPs have been reported in plants (Figure 3). RsBGAL1, a β-galactosidase from Raphanus sativus, resides in the GH35 family and the protein when heterologously expressed in P. pastoris hydrolyzed β-(1,3)‐ and β-(1,6)-galactosyl residues (Kotake et al., 2005). RsAraf1 from R. sativus immature seeds encodes an α-arabinofuranosidase in the GH3 family. When expressed in Arabidopsis, RsAraf1 hydrolyzed α-arabinofuranosyl residues of AGPs (Kotake et al., 2006). AtGUS2 is a β-glucuronidase that belongs to the GH79 family. Knockout mutants showed increased levels of GlcA while plants over-expressing AtGUS2 showed the opposite phenotype, no GlcA in seedlings AGP fractions and increased cell elongation in seedlings, a similar phenotype of atglcat14a (Eudes et al., 2008). SlTBG1, a tomato (Solanum lycopersicum) β-galactosidase belonging to the GH35 family, has activity against β-(1,3)-linkages and β-(1,6)-linkages in a galactooligosaccharide, which may be involved in the hydrolysis of AGs from AGPs (Eda et al., 2014). AtAPSE (β-l-ARAPASE) is a β-l-arabinopyranosidase in the GH27 family. In an apse mutant, the amount of β-l-Arap residues of AGPs was higher compared to wild type, confirming its activity (Imaizumi et al., 2017).
AGPs in the Extracellular Space: Facing the Challenge
O-glycosylated GPI-anchored AGPs are transferred to the extracellular space, or more precisely to the cell surface apoplast, through the endomembrane system via post-Golgi vesicles, where they remain temporarily attached to the outer face of the plasma membrane (Schultz et al., 1998; Showalter, 2001). In response to intercellular or intracellular signals, GPI-anchored AGPs can be released from the plasma membrane by phosphatidylinositol-specific phospholipases that removes inositol from the diacylglycerol moiety (Schultz et al., 1998; Oxley and Bacic, 1999; Gillmor et al., 2005; Desnoyer and Palanivelu, 2020; Figure 2B). Up to the present, only one inositol phospholipid-specific phospholipase C (PI-PLC) has been identified, partially purified and characterized in plants (Butikofer and Brodbeck, 1993). Several GPI-anchored proteins such as FLAs, COBRA-like, and receptor-like proteins are released by phospholipases in Arabidopsis (Borner et al., 2003). This process may serve to regulate the amount of these glycoproteins in the plasma membrane and control the release of soluble AGPs into the extracellular matrix, where they may act as soluble signals for neighboring cells (Schultz et al., 1998). Nevertheless, some AGP members are not GPI-anchored. The sugars may be cleaved by GHs and released into the extracellular medium, where they may function as signaling molecules binding to specific receptors and triggering signaling pathways (Showalter, 2001; Figure 2B). Furthermore, AGPs may also be processed by proteolytic activities (Fincher et al., 1983; Faye et al., 2005) or transported by endocytotic multivesicular bodies to the vacuole, where they are degraded (Herman and Lamb, 1992; Šamaj et al., 2000).
AGP Research Overview and Future Directions
In this review, information on AGP biosynthesis accumulated in the last three decades was presented. Much has been accomplished, but there is even more to be done. With the sequencing of several plant genomes, bioinformatics opened doors to identify several candidate genes involved in AGP biosynthesis. However, a huge amount of data is generated and much of it is waiting for further analysis. A major challenge will be to conduct the more laborious biochemical analyses to test and support the findings from bioinformatic and genomic analyses, which moves at such a rapid pace.
This review has largely focused on AGP glycosylation given the cumulative supporting information of the importance of the 90% sugar moiety of AGPs. There is no doubt that AGPs play a range of important functions in plants. The new challenge is to define how AGPs, after all the processes involved with their biosynthesis, act in the cells. Specifically, it will be important to elucidate the mechanism of action by which AGPs, and specifically their sugar epitopes, function. This will mean discovering the molecules, which interact with these sugar residues.
Several other important and stimulating questions remain to be answered: do the enzymes involved in AGP synthesis act on a specific single AGP, a subset of AGPs or all AGPs? How many GTs are involved in AGP glycosylation and what is the substrate specificity for each GT? Are the GTs involved in AGP biosynthesis associated with one another in enzyme complexes? The structure of AG is very variable, but is the pattern of glycosylation of AGPs cell/tissue-specific, stage-specific, or AGP-specific? What is the precise glycan structure of each AGP? Are the different patterns of glycosylation determined by the diverse amino acid sequences of AGPs? What implications do the different sugar compositions have in AGP function? Do AGPs follow a conventional or unconventional protein secretory pathway? Which AGPs localize in the plasma membrane? How does AGP turnover occur? These major challenges will be a stimulus for new future research developments on AGPs through the next 30 years.
Author Contributions
JS organized and wrote the manuscript. RF assisted in writing the manuscript. SC proposed and corrected the manuscript. PD and AMS assisted in writing and revising the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The work described in this review was supported by the EU project 690946 – SexSeed – Sexual Plant Reproduction – Seed Formation, funded by H2020-MSCA-RISE-2015 and SeedWheels FCT Project – POCI-01-0145-FEDER-027839.
References
- Albenne C., Canut H., Jamet E. (2013). Plant cell wall proteomics: the leadership of Arabidopsis thaliana. Front. Plant Sci. 4:111. 10.3389/fpls.2013.00111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. T., Kieber J. J. (2020). Dynamic construction, perception, and remodeling of plant cell walls. Annu. Rev. Plant Biol. 71, 39–69. 10.1146/annurev-arplant-081519-035846, PMID: [DOI] [PubMed] [Google Scholar]
- Asif M. H., Trivedi P. K., Misra P., Nath P. (2009). Prolyl-4-hydroxylase (AtP4H1) mediates and mimics low oxygen response in Arabidopsis thaliana. Funct. Integr. Genomics 9, 525–535. 10.1007/s10142-009-0118-y, PMID: [DOI] [PubMed] [Google Scholar]
- Aspinall G. O., Molloy J. A., Craig J. W. T. (1969). Extracellular polysaccharides from suspension-cultured sycamore cells. Can. J. Biochem. 47, 1063–1070. 10.1139/o69-170, PMID: [DOI] [PubMed] [Google Scholar]
- Atmodjo M. A., Hao Z., Mohnen D. (2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64, 747–779. 10.1146/annurev-arplant-042811-105534, PMID: [DOI] [PubMed] [Google Scholar]
- Bacete L., Mélida H., Miedes E., Molina A. (2018). Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636. 10.1111/tpj.13807, PMID: [DOI] [PubMed] [Google Scholar]
- Basu D., Liang Y., Liu X., Himmeldirk K., Faik A., Kieliszewski M., et al. (2013). Functional identification of a hydroxyproline-O-galactosyltransferase specific for arabinogalactan protein biosynthesis in Arabidopsis. J. Biol. Chem. 288, 10132–10143. 10.1074/jbc.M112.432609, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., Tian L., DeBrosse T., Poirier E., Emch K., Herock H., et al. (2016). Glycosylation of a fasciclin-like arabinogalactan-protein (SOS5) mediates root growth and seed mucilage adherence via a cell wall receptor-like kinase (FEI1/FEI2) pathway in Arabidopsis. PLoS One 11:e0145092. 10.1371/journal.pone.0145092, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., Tian L., Wang W., Bobbs S., Herock H., Travers A., et al. (2015a). A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol. 15:295. 10.1186/s12870-015-0670-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu D., Wang W., Ma S., DeBrosse T., Poirier E., Emch K., et al. (2015b). Two hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS One 10:e0125624. 10.1371/journal.pone.0125624, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beihammer G., Maresch D., Altmann F., Strasser R. (2020). Glycosylphosphatidylinositol-anchor synthesis in plants: a glycobiology perspective. Front. Plant Sci. 11:611188. 10.3389/fpls.2020.611188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G. H. H., Lilley K. S., Stevens T. J., Dupree P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. 10.1104/pp.103.021170, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G. H., Sherrier D. J., Stevens T. J., Arkin I. T., Dupree P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499. 10.1104/pp.010884, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson R., Conzelmann A. (2007). Multiple functions of inositolphosphorylceramides in the formation and intracellular transport of glycosylphosphatidylinositol-anchored proteins in yeast. Biochem. Soc. Symp. 74, 199–209. 10.1042/BSS0740199, PMID: [DOI] [PubMed] [Google Scholar]
- Bundy M. G. R., Kosentka P. Z., Willet A. H., Zhang L., Miller E., Shpak E. D. (2016). A mutation in the catalytic subunit of the glycosylphosphatidylinositol transamidase disrupts growth, fertility, and stomata formation. Plant Physiol. 171, 974–985. 10.1104/pp.16.00339, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butikofer P., Brodbeck U. (1993). Partial purification and characterization of a (glycosyl) inositol phospholipid-specific phospholipase C from peanut. J. Biol. Chem. 268, 17794–177802. PMID: [PubMed] [Google Scholar]
- Canut H., Albenne C., Jamet E. (2016). Post-translational modifications of plant cell wall proteins and peptides: a survey from a proteomics point of view. Biochim. Biophys. Acta 1864, 983–990. 10.1016/j.bbapap.2016.02.022, PMID: [DOI] [PubMed] [Google Scholar]
- Carpita N., Tierney M., Campbell M. (2001). Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol. Biol. 47, 1–5. 10.1023/A:1010603527077, PMID: [DOI] [PubMed] [Google Scholar]
- Cassab G. I., Varner J. E. (1988). Cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 321–353. 10.1146/annurev.pp.39.060188.001541 [DOI] [PubMed] [Google Scholar]
- Chasan R. (1994). Arabinogalactan-proteins: getting to the core. Plant Cell 6, 1519–1521. 10.1105/tpc.6.11.1519 [DOI] [Google Scholar]
- Chen C. G., Pu Z. Y., Moritz R. L., Simpson R. J., Bacic A., Clarke A. E., et al. (1994). Molecular cloning of a gene encoding an arabinogalactan-protein from pear (Pyrus communis) cell suspension culture. Proc. Natl. Acad. Sci. U. S. A. 91, 10305–10309. 10.1073/pnas.91.22.10305, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y. H., Pogorelko G., Zabotina O. A. (2012). Xyloglucan xylosyltransferases XXT1, XXT2, and XXT5 and the glucan synthase CSLC4 form Golgi-localized multiprotein complexes. Plant Physiol. 159, 1355–1366. 10.1104/pp.112.199356, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. E., Anderson R. L., Stone B. A. (1979). Form and function of arabinogalactans and arabinogalactan-proteins. Phytochemistry 18, 521–540. 10.1016/S0031-9422(00)84255-7 [DOI] [Google Scholar]
- Cosgrove D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. 10.1038/nrm1746, PMID: [DOI] [PubMed] [Google Scholar]
- Coutinho P. M., Deleury E., Davies G. J., Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328, 307–317. 10.1016/S0022-2836(03)00307-3, PMID: [DOI] [PubMed] [Google Scholar]
- Dai X. R., Gao X. Q., Chen G. H., Tang L. L., Wang H., Zhang X. S. (2014). ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyltransferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR biosynthesis b in human, is required for arabidopsis pollen tube micropylar guidance and embryo development. Plant Physiol. 165, 1544–1556. 10.1104/pp.114.236133, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyer N., Howard G., Jong E., Palanivelu R. (2020). AtPIG-S, a predicted glycosylphosphatidylinositol transamidase subunit, is critical for pollen tube growth in Arabidopsis. BMC Plant Biol. 20:380. 10.1186/s12870-020-02587-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyer N., Palanivelu R. (2020). Bridging the GAPs in plant reproduction: a comparison of plant and animal GPI-anchored proteins. Plant Reprod. 33, 129–142. 10.1007/s00497-020-00395-9, PMID: [DOI] [PubMed] [Google Scholar]
- Dilokpimol A., Geshi N. (2014). Arabidopsis thaliana glucuronosyltransferase in family GT14. Plant Signal. Behav. 9:e28891. 10.4161/psb.28891, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilokpimol A., Poulsen C. P., Vereb G., Kaneko S., Schulz A., Geshi N. (2014). Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol. 14:90. 10.1186/1471-2229-14-90, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Clarke A. E., Bacic A. (1996). Arabinogalactan proteins: a class of extracellular matrix proteoglycans involved in plant growth and development. Trends Cell Biol. 6, 411–414. 10.1016/S0962-8924(96)20036-4, PMID: [DOI] [PubMed] [Google Scholar]
- Du H., Simpson R. J., Moritz R. L., Clarke A. E., Bacic A. (1994). Isolation of the protein backbone of an arabinogalactan-pttein from the styles of Nicotiana alata and characterization of a corresponding cDNA. Plant Cell 6, 1643–1653. 10.1105/tpc.6.11.1643, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duruflé H., Hervé V., Balliau T., Zivy M., Dunand C., Jamet E. (2017). Proline hydroxylation in cell wall proteins: is it yet possible to define rules? Front. Plant Sci. 8:1802. 10.3389/fpls.2017.01802, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda M., Ishimaru M., Tada T., Sakamoto T., Kotake T., Tsumuraya Y., et al. (2014). Enzymatic activity and substrate specificity of the recombinant tomato β-galactosidase 1. J. Plant Physiol. 171, 1454–1460. 10.1016/j.jplph.2014.06.010, PMID: [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Wildpaner M., Schultz C. J., Borner G. H. H., Dupree P., Eisenhaber F. (2003). Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence‐ and genome-wide studies for Arabidopsis and rice. Plant Physiol. 133, 1691–1701. 10.1104/pp.103.023580, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M., Egelund J., Schultz C. J., Bacic A. (2010). Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419. 10.1104/pp.110.156000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A., Mouille G., Thévenin J., Goyallon A., Minic Z., Jouanin L. (2008). Purification, cloning and functional characterization of an endogenous beta-glucuronidase in Arabidopsis thaliana. Plant Cell Physiol. 49, 1331–1341. 10.1093/pcp/pcn108, PMID: [DOI] [PubMed] [Google Scholar]
- Faye L., Boulaflous A., Benchabane M., Gomord V., Michaud D. (2005). Protein modifications in the plant secretory pathway: current status and practical implications in molecular pharming. Vaccine 23, 1770–1778. 10.1016/j.vaccine.2004.11.003, PMID: [DOI] [PubMed] [Google Scholar]
- Fincher G. B., Sawyer W. H., Stone B. A. (1974). Chemical and physical properties of an arabinogalactan peptide from wheat endosperm. Biochem. J. 139, 535–545. 10.1042/bj1390535, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincher G. B., Stone B. A., Clarke A. E. (1983). Arabinogalactan-proteins: structure, biosynthesis, and function. Annu. Rev. Plant Physiol. 34, 47–70. 10.1146/annurev.pp.34.060183.000403 [DOI] [Google Scholar]
- Gaspar Y., Johnson K. L., Mckenna J. A., Bacic A., Schultz C. J. (2001). The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol. Biol. 47, 161–176. 10.1023/A:1010683432529, PMID: [DOI] [PubMed] [Google Scholar]
- Gaspar Y. M., Nam J., Schultz C. J., Lee L. Y., Gilson P. R., Gelvin S. B., et al. (2004). Characterization of the arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiol. 135, 2162–2171. 10.1104/pp.104.045542, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geshi N., Johansen J. N., Dilokpimol A., Rolland A., Belcram K., Verger S., et al. (2013). A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 76, 128–137. 10.1111/tpj.12281, PMID: [DOI] [PubMed] [Google Scholar]
- Gille S., Sharma V., Baidoo E. E. K., Keasling J. D., Scheller H. V., Pauly M. (2013). Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the arabidopsis glycosyltransferase mutant ray1. Mol. Plant 6, 1369–1372. 10.1093/mp/sst029, PMID: [DOI] [PubMed] [Google Scholar]
- Gillmor C. S., Lukowitz W., Brininstool G., Sedbrook J. C., Hamann T., Poindexter P., et al. (2005). Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17, 1128–1140. 10.1105/tpc.105.031815, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloster T. M. (2014). Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 28, 131–141. 10.1016/j.sbi.2014.08.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J. S., Tsai A. Y. -L., Xue H., Voiniciuc C., Sola K., Seifert G. J., et al. (2014). SALT-OVERLY SENSITIVE5 mediates Arabidopsis seed coat mucilage adherence and organization through pectins. Plant Physiol. 165, 991–1004. 10.1104/pp.114.239400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S. F., Bettler E., Wimmerová M., Imberty A., Lerouxel O., Breton C. (2009). Combination of several bioinformatics approaches for the identification of new putative glycosyltransferases in Arabidopsis. J. Proteome Res. 8, 743–753. 10.1021/pr800808m, PMID: [DOI] [PubMed] [Google Scholar]
- Hansen S. F., Harholt J., Oikawa A., Scheller H. V. (2012). Plant glycosyltransferases beyond CAZy: a perspective on DUF families. Front. Plant Sci. 3:59. 10.3389/fpls.2012.00059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Davies G. (1997). Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644. 10.1016/S0959-440X(97)80072-3, PMID: [DOI] [PubMed] [Google Scholar]
- Herman E. M., Lamb C. J. (1992). Arabinogalactan-rich glycoproteins are localized on the cell surface and in intravacuolar multivesicular bodies. Plant Physiol. 98, 264–272. 10.1104/pp.98.1.264, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervé C., Siméon A., Jam M., Cassin A., Johnson K. L., Salmeán A. A., et al. (2016). Arabinogalactan proteins have deep roots in eukaryotes: identification of genes and epitopes in brown algae and their role in Fucus serratus embryo development. New Phytol. 209, 1428–1441. 10.1111/nph.13786, PMID: [DOI] [PubMed] [Google Scholar]
- Hieta R., Myllyharju J. (2002). Cloning and characterization of a low molecular weight prolyl 4-hydroxylase from Arabidopsis thaliana: effective hydroxylation of proline-rich, collagen-like, and hypoxia-inducible transcription factor α-like peptides. J. Biol. Chem. 277, 23965–23971. 10.1074/jbc.M201865200, PMID: [DOI] [PubMed] [Google Scholar]
- Hijazi M., Velasquez S. M., Jamet E., Estevez J. M., Albenne C. (2014). An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front. Plant Sci. 5:395. 10.3389/fpls.2014.00395, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Guerrero R., Davies G. J. (2012). Recent structural and mechanistic insights into post-translational enzymatic glycosylation. Curr. Opin. Chem. Biol. 16, 479–487. 10.1016/j.cbpa.2012.10.013, PMID: [DOI] [PubMed] [Google Scholar]
- Imaizumi C., Tomatsu H., Kitazawa K., Yoshimi Y., Shibano S., Kikuchi K., et al. (2017). Heterologous expression and characterization of an Arabidopsis β-l-arabinopyranosidase and α-d-galactosidases acting on β-l-arabinopyranosyl residues. J. Exp. Bot. 68, 4651–4661. 10.1093/jxb/erx279, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E., Albenne C., Boudart G., Irshad M., Canut H., Pont-Lezica R. (2008). Recent advances in plant cell wall proteomics. Proteomics 8, 893–908. 10.1002/pmic.200700938, PMID: [DOI] [PubMed] [Google Scholar]
- Jamet E., Canut H., Boudart G., Pont-Lezica R. F. (2006). Cell wall proteins: a new insight through proteomics. Trends Plant Sci. 11, 33–39. 10.1016/j.tplants.2005.11.006, PMID: [DOI] [PubMed] [Google Scholar]
- Johnson K. L., Cassin A. M., Lonsdale A., Bacic A., Doblin M. S., Schultz C. J. (2017a). A motif and amino acid bias bioinformatics pipeline to identify hydroxyproline-rich glycoproteins. Plant Physiol. 174, 886–903. 10.1104/pp.17.00294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. L., Cassin A. M., Lonsdale A., Wong G. K. S., Soltis D. E., Miles N. W., et al. (2017b). Insights into the evolution of hydroxyproline-rich glycoproteins from 1000 plant transcriptomes. Plant Physiol. 174, 904–921. 10.1104/pp.17.00295, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. L., Jones B. J., Bacic A., Schultz C. J. (2003). The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiol. 133, 1911–1925. 10.1104/pp.103.031237, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul S., Koo H. L., Jenkins J., Rizzo M., Rooney T., Tallon L. J., et al. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. 10.1038/35048692, PMID: [DOI] [PubMed] [Google Scholar]
- Keegstra K., Talmadge K. W., Bauer W. D., Albersheim P. (1973). The structure of plant cell walls: III. A model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51, 188–197. 10.1104/pp.51.1.188, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskiaho K., Hieta R., Sormunen R., Myllyharju J. (2007). Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell 19, 256–269. 10.1105/tpc.106.042739, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M. J. (2001). The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57, 319–323. 10.1016/S0031-9422(01)00029-2, PMID: [DOI] [PubMed] [Google Scholar]
- Kieliszewski M. J., Kamyab A., Leykam J. F., Lamport D. T. A. (1992). A histidine-rich extensin from Zea mays is an arabinogalactan protein. Plant Physiol. 99, 538–547. 10.1104/pp.99.2.538, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski M. J., Lamport D. T. A. (1994). Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 5, 157–172. 10.1046/j.1365-313X.1994.05020157.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kinoshita T. (2020). Biosynthesis and biology of mammalian GPI-anchored proteins. Open Biol. 10:190290. 10.1098/rsob.190290, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K., Tryfona T., Yoshimi Y., Hayashi Y., Kawauchi S., Antonov L., et al. (2013). β-Galactosyl yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol. 161, 1117–1126. 10.1104/pp.112.211722, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivirikko K. I., Myllyharju J. (1998). Prolyl 4-hydroxylases and their protein disulfide isomerase subunit. Matrix Biol. 16, 357–368. 10.1016/S0945-053X(98)90009-9, PMID: [DOI] [PubMed] [Google Scholar]
- Knoch E., Dilokpimol A., Geshi N. (2014). Arabinogalactan proteins: focus on carbohydrate active enzymes. Front. Plant Sci. 5:198. 10.3389/fpls.2014.00198, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch E., Dilokpimol A., Tryfona T., Poulsen C. P., Xiong G., Harholt J., et al. (2013). A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 76, 1016–1029. 10.1111/tpj.12353, PMID: [DOI] [PubMed] [Google Scholar]
- Knox P. (1999). Intriguing, complex and everywhere: getting to grips with arabinogalactan-proteins. Trends Plant Sci. 4, 123–125. 10.1016/S1360-1385(99)01392-8 [DOI] [Google Scholar]
- Kobayashi Y., Motose H., Iwamoto K., Fukuda H. (2011). Expression and genome-wide analysis of the xylogen-type gene family. Plant Cell Physiol. 52, 1095–1106. 10.1093/pcp/pcr060, PMID: [DOI] [PubMed] [Google Scholar]
- Kotake T., Dina S., Konishi T., Kaneko S., Igarashi K., Samejima M., et al. (2005). Molecular cloning of a β-galactosidase from radish that specifically hydrolyzes β-(1→3)‐ and →-(1→6)-galactosyl residues of arabinogalactan protein. Plant Physiol. 138, 1563–1576. 10.1104/pp.105.062562, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake T., Tsuchiya K., Aohara T., Konishi T., Kaneko S., Igarashi K., et al. (2006). An α-L-arabinofuranosidase/β-D-xylosidase from immature seeds of radish (Raphanus sativus L.). J. Exp. Bot. 57, 2353–2362. 10.1093/jxb/erj206, PMID: [DOI] [PubMed] [Google Scholar]
- Lalanne E., Honys D., Johnson A., Borner G. H. H., Lilley K. S., Dupree P., et al. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16, 229–240. 10.1105/tpc.014407, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T. A. (2001). Life behind cell walls: paradigm lost, paradigm regained. Cell. Mol. Life Sci. 58, 1363–1385. 10.1007/PL00000782, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamport D. T. A., Kieliszewski M. J., Showalter A. M. (2006). Salt stress upregulates periplasmic arabinogalactan proteins: using salt stress to analyse AGP function. New Phytol. 169, 479–492. 10.1111/j.1469-8137.2005.01591.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lamport D. T. A., Tan L., Held M. A., Kieliszewski M. J. (2018). Pollen tube growth and guidance: Occam’s razor sharpened on a molecular arabinogalactan glycoprotein Rosetta Stone. New Phytol. 217, 491–500. 10.1111/nph.14845, PMID: [DOI] [PubMed] [Google Scholar]
- Lamport D. T. A., Várnai P. (2013). Periplasmic arabinogalactan glycoproteins act as a calcium capacitor that regulates plant growth and development. New Phytol. 197, 58–64. 10.1111/nph.12005, PMID: [DOI] [PubMed] [Google Scholar]
- Lamport D. T. A., Varnai P., Seal C. E. (2014). Back to the future with the AGP-Ca2+ flux capacitor. Ann. Bot. 114, 1069–1085. 10.1093/aob/mcu161, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. J. D., Sakata Y., Mau S. L., Pettolino F., Bacic A., Quatrano R. S., et al. (2005). Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17, 3051–3065. 10.1105/tpc.105.034413, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard R., Petersen B. O., Himly M., Kaar W., Wopfner N., Kolarich D., et al. (2005). Two novel types of O-glycans on the mugwort pollen allergen Art v 1 and their role in antibody binding. J. Biol. Chem. 280, 7932–7940. 10.1074/jbc.M410407200, PMID: [DOI] [PubMed] [Google Scholar]
- Liang Y., Basu D., Pattathil S., Xu W. L., Venetos A., Martin S. L., et al. (2013). Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. J. Exp. Bot. 64, 5537–5551. 10.1093/jxb/ert321, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind J. L., Bacic A., Clarke A. E., Anderson M. A. (1994). A style-specific hydroxyproline-rich glycoprotein with properties of both extensins and arabinogalactan proteins. Plant J. 6, 491–502. 10.1046/j.1365-313x.1994.6040491.x, PMID: [DOI] [PubMed] [Google Scholar]
- Liu X., Castro C., Wang Y., Noble J., Ponvert N., Bundy M., et al. (2016). The role of LORELEI in pollen tube reception at the interface of the synergid cell and pollen tube requires the modified eight-cysteine motif and the receptor-like kinase FERONIA. Plant Cell 28, 1035–1052. 10.1105/tpc.15.00703, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495. 10.1093/nar/gkt1178, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Hernandez F., Tryfona T., Rizza A., Yu X., Harris M. O., Webb A. A., et al. (2020). Calcium binding by arabinogalactan polysaccharides is important for normal plant development. Plant Cell 32, 3346–3369. 10.1105/tpc.20.00027, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Seifert G. J. (2010). Posttranslational modifications of plasma membrane proteins and their implications for plant growth and development. Plant Cell Monogr. 19, 109–128. 10.1007/978-3-642-13431-9_5 [DOI] [Google Scholar]
- Ma Y., Yan C., Li H., Wu W., Liu Y., Wang Y., et al. (2017). Bioinformatics prediction and evolution analysis of arabinogalactan proteins in the plant kingdom. Front. Plant Sci. 8:66. 10.3389/fpls.2017.00066, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Zhao J. (2010). Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 61, 2647–2668. 10.1093/jxb/erq104, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewska-Sawka A., Nothnagel E. A. (2000). The multiple roles of arabinogalactan proteins in plant development. Plant Physiol. 122, 3–10. 10.1104/pp.122.1.3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky F. G., Fangel J. U., Willats W. G. T. (2014). The role of the cell wall in plant immunity. Front. Plant Sci. 5:178. 10.3389/fpls.2014.00178, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K., Asami T., Suzuki Y. (2009). Genome-wide identification, structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Biosci. Biotechnol. Biochem. 73, 2452–2459. 10.1271/bbb.90407, PMID: [DOI] [PubMed] [Google Scholar]
- Mau S. L., Chen C. G., Pu Z. Y., Moritz R. L., Simpson R. J., Bacic A., et al. (1995). Molecular cloning of cDNAs encoding the protein backbones of arabinogalactan-proteins from the filtrate of suspension-cultured cells of Pyrus communis and Nicotiana alata. Plant J. 8, 269–281. 10.1046/j.1365-313x.1995.08020269.x, PMID: [DOI] [PubMed] [Google Scholar]
- Mizukami A. G., Inatsugi R., Jiao J., Kotake T., Kuwata K., Ootani K., et al. (2016). The AMOR arabinogalactan sugar chain induces pollen-tube competency to respond to ovular guidance. Curr. Biol. 26, 1091–1097. 10.1016/j.cub.2016.02.040, PMID: [DOI] [PubMed] [Google Scholar]
- Morotti A. L. M., Martins-Teixeira M. B., Carvalho I. (2017). Protozoan parasites glycosylphosphatidylinositol anchors: structures, functions and trends for drug discovery. Curr. Med. Chem. 26, 4301–4322. 10.2174/0929867324666170727110801, PMID: [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Coimbra S., Vicré-Gibouin M., Mollet J. C., Driouich A. (2012). Arabinogalactan proteins in root and pollen-tube cells: distribution and functional aspects. Ann. Bot. 110, 383–404. 10.1093/aob/mcs143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Cannesan M. A., Driouich A. (2013). Arabinogalactan proteins in root-microbe interactions. Trends Plant Sci. 18, 440–449. 10.1016/j.tplants.2013.03.006, PMID: [DOI] [PubMed] [Google Scholar]
- Nguema-Ona E., Vicré-Gibouin M., Gotté M., Plancot B., Lerouge P., Bardor M., et al. (2014). Cell wall O-glycoproteins and N-glycoproteins: aspects of biosynthesis and function. Front. Plant Sci. 5:499. 10.3389/fpls.2014.00499, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolovski N., Rubtsov D., Segura M. P., Miles G. P., Stevens T. J., Dunkley T. P. J., et al. (2012). Putative glycosyltransferases and other plant Golgi apparatus proteins are revealed by LOPIT proteomics. Plant Physiol. 60, 1037–1051. 10.1104/pp.112.204263, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel E. A. (1997). Proteoglycans and related components in plant cells. Int. Rev. Cytol. 174, 195–291. 10.1016/s0074-7696(08)62118-x, PMID: [DOI] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M., Matsubayashi Y. (2015). Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J. 81, 735–746. 10.1111/tpj.12764, PMID: [DOI] [PubMed] [Google Scholar]
- Oxley D., Bacic A. (1999). Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc. Natl. Acad. Sci. U. S. A. 96, 14246–14251. 10.1073/pnas.96.25.14246, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. M., Lopes A. L., Coimbra S. (2016). Arabinogalactan proteins as interactors along the crosstalk between the pollen tube and the female tissues. Front. Plant Sci. 7:1895. 10.3389/fpls.2016.01895, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin R., Wilkerson C., Keegstra K. (2001). Golgi enzymes that synthesize plant cell wall polysaccharides: finding and evaluating candidates in the genomic era. Plant Mol. Biol. 47, 115–130. 10.1023/A:1010675213873, PMID: [DOI] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A. (2007). Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1771, 405–420. 10.1016/j.bbalip.2006.05.015, PMID: [DOI] [PubMed] [Google Scholar]
- Poulsen C. P., Dilokpimol A., Mouille G., Burow M., Geshi N. (2014). Arabinogalactan glycosyltransferases target to a unique subcellular compartment that may function in unconventional secretion in plants. Traffic 15, 1219–1234. 10.1111/tra.12203, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Fong C., Lamport D. T. A. (1991). Gum arabic glycoprotein is a twisted hairy rope: a new model based on o-galactosylhydroxyproline as the polysaccharide attachment site. Plant Physiol. 96, 848–855. 10.1104/pp.96.3.848, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Egelund J., Gilson P. R., Houghton F., Gleeson P. A., Schultz C. J., et al. (2008). Identification of a novel group of putative Arabidopsis thaliana β-(1,3)-galactosyltransferases. Plant Mol. Biol. 68, 43–59. 10.1007/s11103-008-9351-3, PMID: [DOI] [PubMed] [Google Scholar]
- Šamaj J., Šamajová O., Peters M., Baluška F., Lichtscheidl I., Knox J. P., et al. (2000). Immunolocalization of LM2 arabinogalactan protein epitope associated with endomembranes of plant cells. Protoplasma 212, 186–196. 10.1007/bf01282919 [DOI] [Google Scholar]
- Schatz G., Dobberstein B. (1996). Common principles of protein translocation across membranes. Science 271, 1519–1526. 10.1126/science.271.5255.1519, PMID: [DOI] [PubMed] [Google Scholar]
- Schultz C., Gilson P., Oxley D., Youl J., Bacic A. (1998). GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci. 3, 426–431. 10.1016/S1360-1385(98)01328-4 [DOI] [Google Scholar]
- Schultz C. J., Johnson K. L., Currie G., Bacic A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1768. 10.1105/tpc.12.9.1751, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C. J., Rumsewicz M. P., Johnson K. L., Jones B. J., Gaspar Y. M., Bacic A. (2002). Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiol. 129, 1448–1463. 10.1104/pp.003459, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G. J., Roberts K. (2007). The biology of arabinogalactan proteins. Annu. Rev. Plant Biol. 58, 137–161. 10.1146/annurev.arplant.58.032806.103801, PMID: [DOI] [PubMed] [Google Scholar]
- Serpe M. D., Nothnagel E. A. (1999). Arabinogalactan-proteins in the multiple domains of the plant cell surface. Adv. Bot. Res. 30, 207–289. 10.1016/S0065-2296(08)60229-3, PMID: 29857505 [DOI] [Google Scholar]
- Sherrier D. J., Prime T. A., Dupree P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. , PMID: [DOI] [PubMed] [Google Scholar]
- Showalter A. M. (1993). Structure and function of plant cell wall proteins. Plant Cell 5, 9–23. 10.2307/3869424, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M. (2001). Arabinogalactan-proteins: structure, expression and function. Cell. Mol. Life Sci. 58, 1399–1417. 10.1007/PL00000784, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M., Basu D. (2016). Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Front. Plant Sci. 7:814. 10.3389/fpls.2016.00814, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter A. M., Keppler B., Lichtenberg J., Gu D., Welch L. R. (2010). A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513. 10.1104/pp.110.156554, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E., Barbar E., Leykam J. F., Kieliszewski M. J. (2001). Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. J. Biol. Chem. 276, 11272–11278. 10.1074/jbc.M011323200, PMID: [DOI] [PubMed] [Google Scholar]
- Shpak E., Leykam J. F., Kieliszewski M. J. (1999). Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proc. Natl. Acad. Sci. U. S. A. 96, 14736–14741. 10.1073/pnas.96.26.14736, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., et al. (2004). Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211. 10.1126/science.1102765, PMID: [DOI] [PubMed] [Google Scholar]
- Sommer-Knudsen J., Clarke A. E., Bacic A. (1996). A galactose-rich, cell-wall glycoprotein from styles of Nicotiana alata. Plant J. 9, 71–83. 10.1046/j.1365-313x.1996.09010071.x, PMID: [DOI] [PubMed] [Google Scholar]
- Sun W., Xu J., Yang J., Kieliszewski M. J., Showalter A. M. (2005). The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant Cell Physiol. 46, 975–984. 10.1093/pcp/pci106, PMID: [DOI] [PubMed] [Google Scholar]
- Suzuki T., Narciso J. O., Zeng W., van de Meene A., Yasutomi M., Takemura S., et al. (2017). KNS4/UPEX1: a type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173, 183–205. 10.1104/pp.16.01385, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetek J., Yadav M. P., Nothnagel E. A. (1999). Presence of a glycosylphosphatidylinositol lipid anchor on rose arabinogalactan proteins. J. Biol. Chem. 274, 14724–14733. 10.1074/jbc.274.21.14724, PMID: [DOI] [PubMed] [Google Scholar]
- Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287. 10.1105/tpc.112.107334, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Leykam J. F., Kieliszewski M. J. (2003). Glycosylation motifs that direct arabinogalactan addition to arabinogalactan-proteins. Plant Physiol. 132, 1362–1369. 10.1104/pp.103.021766, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Qiu F., Lamport D. T. A., Kieliszewski M. J. (2004). Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165. 10.1074/jbc.M311864200, PMID: [DOI] [PubMed] [Google Scholar]
- Tan L., Varnai P., Lamport D. T. A., Yuan C., Xu J., Qiu F., et al. (2010). Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583. 10.1074/jbc.M109.100149, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple H., Mortimer J. C., Tryfona T., Yu X., Lopez-Hernandez F., Sorieul M., et al. (2019). Two members of the DUF579 family are responsible for arabinogalactan methylation in Arabidopsis. Plant Direct 3, 1–4. 10.1002/pld3.117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515. 10.1093/nar/gky1049, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiainen P., Myllyharju J., Koivunen P. (2005). Characterization of a second Arabidopsis thaliana prolyl 4-hydroxylase with distinct substrate specificity. J. Biol. Chem. 280, 1142–1148. 10.1074/jbc.M411109200, PMID: [DOI] [PubMed] [Google Scholar]
- Tryfona T., Liang H. C., Kotake T., Kaneko S., Marsh J., Ichinose H., et al. (2010). Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr. Res. 345, 2648–2656. 10.1016/j.carres.2010.09.018, PMID: [DOI] [PubMed] [Google Scholar]
- Tryfona T., Liang H. C., Kotake T., Tsumuraya Y., Stephens E., Dupree P. (2012). Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 160, 653–666. 10.1104/pp.112.202309, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryfona T., Theys T. E., Wagner T., Stott K., Keegstra K., Dupree P. (2014). Characterisation of FUT4 and FUT6 α-(1→2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases arabidopsis root growth salt sensitivity. PLoS One 9:e93291. 10.1371/journal.pone.0093291, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S. M., Iusem N. D., Estevez J. M. (2011a). Root hair sweet growth. Plant Signal. Behav. 6, 1600–1602. 10.4161/psb.6.10.17135, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez S. M., Ricardi M. M., Dorosz J. G., Fernandez P. V., Nadra A. D., Pol-Fachin L., et al. (2011b). O-glycosylated cell wall proteins are essential in root hair growth. Science 332, 1401–1403. 10.1126/science.1206657, PMID: [DOI] [PubMed] [Google Scholar]
- Velasquez S. M., Ricardi M. M., Poulsen C. P., Oikawa A., Dilokpimol A., Halim A., et al. (2015). Complex regulation of prolyl-4-hydroxylases impacts root hair expansion. Mol. Plant 8, 734–746. 10.1016/j.molp.2014.11.017, PMID: [DOI] [PubMed] [Google Scholar]
- Vlad F., Spano T., Vlad D., Daher F. B., Ouelhadj A., Fragkostefanakis S., et al. (2007). Involvement of arabidopsis prolyl 4 hydroxylases in hypoxia, anoxia and mechanical wounding. Plant Signal. Behav. 2, 368–369. 10.4161/psb.2.5.4462, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F., Tiainen P., Owen C., Spano T., Daher F. B., Oualid F., et al. (2010). Characterization of two carnation petal prolyl 4 hydroxylases. Physiol. Plant. 140, 199–207. 10.1111/j.1399-3054.2010.01390.x, PMID: [DOI] [PubMed] [Google Scholar]
- Walter P., Johnson A. E. (1994). Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10, 87–119. 10.1146/annurev.cb.10.110194.000511, PMID: [DOI] [PubMed] [Google Scholar]
- Wang S., Yin Y., Ma Q., Tang X., Hao D., Xu Y. (2012). Genome-scale identification of cell-wall related genes in Arabidopsis based on co-expression network analysis. BMC Plant Biol. 12:138. 10.1186/1471-2229-12-138, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Williams M., Bernard S., Driouich A., Showalter A. M., Faik A. (2010). Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285, 13638–13645. 10.1074/jbc.M110.102715, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J., Lis H., Katchalski E. (1967). Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem. J. 105, 1C–2C. 10.1042/bj1050001C, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yariv J., Rapport M. M., Graf L. (1962). The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochem. J. 85, 383–388. 10.1042/bj0850383, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats T. H., Bacic A., Johnson K. L. (2018). Plant glycosylphosphatidylinositol anchored proteins at the plasma membrane-cell wall nexus. J. Integr. Plant Biol. 60, 649–669. 10.1111/jipb.12659, PMID: [DOI] [PubMed] [Google Scholar]
- Youl J. J., Bacic A., Oxley D. (1998). Arabinogalactan-proteins from Nicotiana alata and Pyrus communis contain glycosylphosphatidylinositol membrane anchors. Proc. Natl. Acad. Sci. U. S. A. 95, 7921–7926. 10.1073/pnas.95.14.7921, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa K., Toyooka K., Fukuda H., Matsuoka K. (2005). Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 41, 81–94. 10.1111/j.1365-313X.2004.02279.x, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang Y., Held M. A., Showalter A. M. (2020). Elucidating the roles of three β-glucuronosyltransferases (GLCATs) acting on arabinogalactan-proteins using a CRISPR-Cas9 multiplexing approach in Arabidopsis. BMC Plant Biol. 20:221. 10.1186/s12870-020-02420-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]