Abstract
Extracellular vesicles (EVs) are heterogeneous membranous particles released from the cells through different biogenetic and secretory mechanisms. We now conceive EVs as shuttles mediating cellular communication, carrying a variety of molecules resulting from intracellular homeostatic mechanisms. The RNA is a widely detected cargo and, impressively, a recognized functional intermediate that elects EVs as modulators of cancer cell phenotypes, determinants of disease spreading, cell surrogates in regenerative medicine, and a source for non‐invasive molecular diagnostics. The mechanistic elucidation of the intracellular events responsible for the engagement of RNA into EVs will significantly improve the comprehension and possibly the prediction of EV “quality” in association with cell physiology. Interestingly, the application of multidisciplinary approaches, including biochemical as well as cell‐based and computational strategies, is increasingly revealing an active RNA‐packaging process implicating RNA‐binding proteins (RBPs) in the sorting of coding and non‐coding RNAs. In this review, we provide a comprehensive view of RBPs recently emerging as part of the EV biology, considering the scenarios where: (i) individual RBPs were detected in EVs along with their RNA substrates, (ii) RBPs were detected in EVs with inferred RNA targets, and (iii) EV‐transcripts were found to harbour sequence motifs mirroring the activity of RBPs. Proteins so far identified are members of the hnRNP family (hnRNPA2B1, hnRNPC1, hnRNPG, hnRNPH1, hnRNPK, and hnRNPQ), as well as YBX1, HuR, AGO2, IGF2BP1, MEX3C, ANXA2, ALIX, NCL, FUS, TDP‐43, MVP, LIN28, SRP9/14, QKI, and TERT. We describe the RBPs based on protein domain features, current knowledge on the association with human diseases, recognition of RNA consensus motifs, and the need to clarify the functional significance in different cellular contexts. We also summarize data on previously identified RBP inhibitor small molecules that could also be introduced in EV research as potential modulators of vesicular RNA sorting.
Keywords: extracellular vesicles, EVs, inhibitors, protein domains, RBPs, RNA, RNA‐binding proteins, RNA consensus
1. INTRODUCTION
Extracellular vesicles are cell‐secreted membranous particles showing heterogeneous size and molecular composition. EVs can originate from multi‐vesicular bodies (MVBs), giving rise to vesicles referred to as exosomes, or from the plasma membrane budding, in this case referred to as microvesicles (MVs) or ectosomes (Yáñez‐Mó et al., 2015). The mean diameter of EVs ranges from nanometres to few micrometres, and the variety of their biological content includes lipids, proteins, and nucleic acids. It is now widely recognized that cells secrete EVs as vehicles to communicate, exchanging factors that can modulate cell signalling pathways as well as transcriptional and post‐transcriptional processes (Sork et al., 2018). Indeed, seminal studies indicated that alterations in EV abundance and quality might influence the phenotype of tumour cells, the cross‐talk with immune cells, and the spreading of the tumour in different organs (Becker et al., 2016; Choi et al., 2017; Cianciaruso et al., 2019). Moreover, EVs are also taking place in the field of regenerative medicine, acting as cell surrogates in a‐cellular biocompatible scaffolds (Cabral et al., 2018), or in targeted therapy, acting as modifiable cell‐derived vectors (Armstrong et al., 2017). Nevertheless, the role of EV‐RNA is also emerging in liquid biopsy, promising as an alternative non‐invasive source for molecular testing, possibly complementing the cell‐free DNA and circulating tumour cells (CTCs) (Heitzer et al., 2019).
The RNA is part of the EV cargo and the focus of many functions ascribed to EVs. Currently, the Vesiclepedia database includes more than 27,000 entries for mRNAs and more than 10,000 entries for non‐coding RNAs (http://microvesicles.org). The expansion of our knowledge on the different classes of RNAs populating the EV cargo, including coding and non‐coding species (spliceosomal RNAs, lncRNAs, Y RNAs, tRNA‐derived small RNAs, miRNAs, tiny transcription‐initiation RNAs, promoter‐associated short RNAs, termini‐associated short RNAs, TASRs, antisense TASRs, 3ʹUTR‐derived RNAs, and tRNA‐derived short RNAs) results from the recent broad applicability of sensitive RNA detection technologies and computational tools. However, we are still far from predicting a specific RNA sorting into bulk or sub‐populations of EVs according to the cell physiology.
Very recently, the activity of competitive RNA‐binding proteins is emerging as a crucial determinant in diversifying the enrichment of selected transcripts into EVs. Interestingly, the application of biochemical, cell‐based, and computational approaches revealed in several models the enrichment of transcript sequences, defining ‘RNA motifs’, recurrently found in EVs and validated as consensus substrate for RNA‐binding proteins (RBPs). In this review, we provide a comprehensive view of the current knowledge about RBPs described as part of the EV biology. We included the scenarios where (i) individual RBPs were detected in EVs along with their substrate RNAs, (ii) RBPs were detected in EVs but the substrate RNA targets are inferred only, and (iii) transcripts found in EVs which harboured sequence motifs mirroring the activity of RBPs.
Irrespective of the methodologies used to isolate EVs, we highlight the protein domain features of RBPs detected into EVs (Figure 1), the previous knowledge on their association with human diseases, the EV‐RNA consensus sequences responsible for the interaction between the RBPs, and known small molecules inhibitors of RBP activity and interactions.
FIGURE 1.
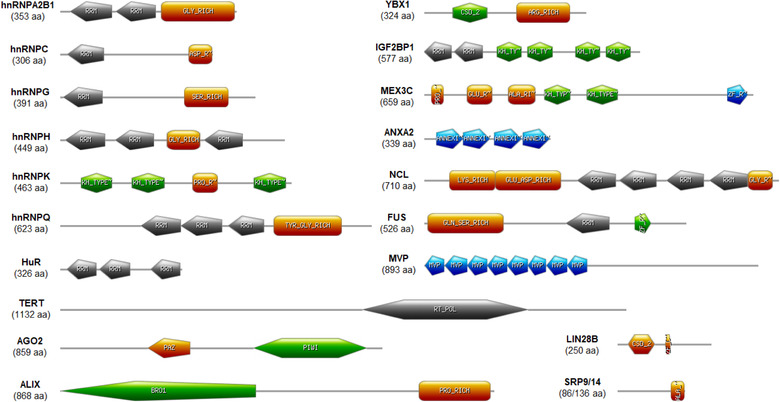
RBPs identified in association with EVs and principal protein domain features. Secondary structures were recognized using primary sequence analysis by ProScan. RRM (RNA recognition motif), GLY_RICH (Glycine‐rich region profile) KH_TY (hnTNP K Homology domain type 1), ZF_RANBP2_2 (Zinc finger RanBP2 type profile), CSD_2 (Cold‐shock domain profile), ZF_CCHC (Zinc finger CCHC‐type profile), ANNEXIN_2 (Annexin repeat profile), BRO1 (BRO1 domain profile), PAZ (PAZ domain profile), PIWI (Piwi domain profile), RT_POL (Reverse transcriptase catalytic domain profile). QKI protein domains were not detected in ProScan
Here, we describe members of the hnRNP family (hnRNPA2B1, hnRNPC1, hnRNPG, hnRNPH1, hnRNPK, and hnRNPQ), as well as YBX1, HuR, AGO2, IGF2BP1, MEX3C, ANXA2, ALIX, NCL, FUS, MVP, LIN28, SRP9/14, QKI, and TERT RNA‐binding proteins.
2. THE hnRNP FAMILY
Heterogeneous nuclear ribonucleoproteins (hnRNPs) represent a large family of 20 major and minor proteins in humans with differential RNA‐binding capacities (Dreyfuss, 1993). hnRNPs are characterized by a typical structure of four unique RNA‐binding domains (RBDs): the RNA recognition motif (RRM), the quasi‐RRM (qRRM), a glycine‐rich domain constituting an RGG box, and a KH domain. The RRM is the most common, while the glycine‐, proline‐, or acid‐rich domains seem auxiliary structures responsible for homologous or heterologous interactions with other hnRNPs (Dreyfuss et al., 2002). As also highlighted in Figure 1, all RBDs are not always present, and the RNA‐recognition specificity is mediated by flexible protein conformations and post‐translational modifications that finely tune the RNA‐protein interactions (Geuens et al., 2016).
Six members of this RBP family emerged in association with EVs: hnRNPA2B1, hnRNPC1, hnRNPG, hnRNPH1, hnRNPK, and hnRNPQ. These RBPs are ubiquitously expressed factors involved to a different extent in the maturation, transport, stability, and translational efficiency of mRNAs and the metabolism of miRNAs. Altered expression of these proteins has been reported in several cancer types and in associations with clinical outcomes. Table 1 summarizes the relevant intracellular functions and the specific involvement in disease described so far for these proteins. In the text, we highlight the recent studies implicating individual hnRNP members' activity in association with extracellular vesicles.
| Member | Functions | Involvement in disease |
|---|---|---|
| hnRNPA2B1 |
‐Alternative splicing (Martinez‐Contreras et al., 2007) ‐mRNA transport (Han et al., 2010) ‐Pri‐miRNA maturation (Alarcón et al., 2015) ‐Telomere maintenance (Moran‐Jones et al., 2019) ‐Innate immunity against DNA viruses (Wang et al., 2019) |
‐ Recognized as autoantigen in RA and SLE (Wu et al., 2018) ‐ Prion‐like properties in ALS, FTD, AD (Harrison & Shorter, 2017) ‐ Overexpressed in lung cancer, pancreatic adenocarcinoma, breast cancer (Barceló et al., 2014; Hung et al., 2017; Klinge et al., 2019) |
| hnRNPC1 |
‐ Formation of RNP complex with pre‐mRNA (Rech et al., 1995) ‐ Splicing regulation (König et al., 2010; Venables et al., 2008) ‐ Transport of mRNA and snRNAs (Mccloskey et al., 2012) ‐ Translation enhancer (Cieniková et al., 2015) |
‐ hnRNPC1 targets pri‐miR‐21 in glioblastoma multiforme, controlling its metastatic potential (Park et al., 2012) ‐ Negative prognostic factor in advanced gastric cancer (Huang et al., 2016) |
| hnRNPG (RBMX) |
‐ Alternative splicing (Heinrich et al., 2009) ‐ Chromosome integrity and repair (Matsunaga et al., 2012) ‐ Chromosome segregation (Cho et al., 2018) ‐ Genome stability (Munschauer et al., 2018) |
‐ RBMX somatic mutations and altered expression levels associate with lung, endometrial and other solid tumours (Elliott et al., 2019) ‐ Overexpression is associated to poor prognosis in oral squamous cell carcinoma (Guo et al., 2020) |
| hnRNPH1 |
‐ Alternative splicing (Alkan et al., 2006) ‐ DNA replication (Barrack, 1987) ‐ RNA transcription (Barrack, 1987) ‐ Inhibition of cell differentiation (Liu et al., 2001) |
‐ Overexpression associates with poorly differentiated solid tumours (head, neck, hepatocellular, pancreatic, colon, laryngeal, oesophageal, and prostate) (Sun et al., 2016) ‐ hnRNPH1 mRNA in exosomes was suggested as a biomarker for HCC (Xu et al., 2018) and advanced prostate cancer (Yang et al., 2016) |
| hnRNPK | ‐ Regulation of mRNA stability and translation (Xu et al., 2019) |
‐ hnRNPK targets lncRNAs involved in cancer progression: SCAT7, pancEts (Ali et al., 2018) and SLINKY (Gong et al., 2017) ‐ Frequently overexpressed and associated with poor prognosis in various malignancies (Chen et al., 2008; Hong et al., 2014) ‐ Downregulation of hnRNPK limits tumour growth (Tang et al., 2014) |
| hnRNPQ (SYNCRIP) |
‐ Alternative splicing (Chen et al., 2008) ‐ mRNA stability (Williams et al., 2016) ‐ RNA transport (Rossoll et al., 2002) |
‐ SYNCRIP, overexpression correlates with late‐onset SMA III, as it interacts with SMA‐related mutant SMN1 (Helmken et al., 2003; Yoo et al., 2009) ‐ Downregulation associated with decreased proliferation and chemoresistance in colon cancer (Lai et al., 2017) |
TABLE 1.
Technical and biological indications about RBPs identified in EVs
| Protein | EV isolation method | PTMs in EVs | Main interactors | RNA motifs | Biological indication in EVs | Inhibitors |
|---|---|---|---|---|---|---|
| hnRNPA2B1 |
Differential ultracentrifugation Polymer precipitation |
Sumoylation O‐GlcNacylation |
PTBP1, HNRNPL, HNRNPH1, SRSF1, TRA2B, HNRNPC, HNRNPA1, HNRNPF, HNRNPK, SRSF3 |
A/G‐rich motifs 5′‐AGG 5′‐UAG 5′‐GGAG |
RBP and RNA substrates |
Epirubicin MO‐460 Compound 2155‐14 Several planar aromatic compounds |
| hnRNPC1 | Differential ultracentrifugation | ND | HNRNPA1, HNRNPA2B1, ALYREF, HNRNPL, HNRNPM, CDC5L, HNRNPK, ELAVL1, SRSF1, HNRNPH1 | AU‐rich elements (AREs) | RBP and RNA substrates | ND |
| hnRNPG | Differential ultracentrifugation | ND | HNRNPK, CDC5L, HNRNPH1, TR2B, HNRNPA1, HNRNPL, PTBP1, HNRNPC, SRSF3, HNRNPR | 5’‐CC[A/C]‐rich | RBP and RNA substrates | ND |
| hnRNPH1 | Differential ultracentrifugation | ND | HNRNPA1, HNRNPK, HNRNPA2B1, HNRNPM, HNRNPA0, SRSF1, HNRNPF, HNRNPC, TRA2B | 5′‐GGGA | RBP and RNA substrates | Compound 2155‐14 |
| hnRNPK |
Differential ultracentrifugation Polymer precipitation |
ND | HNRNPM, HNRNPA1, HNRNPH1, PTBP1, RBMX, HNRNPL, HNRNPF, ELAVL1, HNRNPA2B1, HNRNPC | 5′‐UC3–4(U/A)2 | RBPs and RNA substrates |
Nujiangexathone Quinoline derivatives Psammaplysene A |
| hnRNPQ (SYNCRIP) |
Differential ultracentrifugation Polymer precipitation Ultrafiltration |
ND | APOBEC1, A1CF, PAPC1, PAIP1, CSDE1, HNRNPD, HNRNPR, SMN, HABP4, DHX9, HNRNPU, IGF2BP1, YBX1, ELAVL1 |
5′‐AYAAYY 5′‐UAUYRR 5′‐GGCU |
RBP and RNA substrates | ND |
| YBX1 |
Differential ultracentrifugation Polymer precipitation Ultrafiltration Density gradient ultracentrifugation |
Dephosphorylation (T271) | SRNPD1,HNRNP, SF3A2, HNRNPA2B1, HSPA8, SYNCRIP, LSM3, HNRNPK, ZCRB1, HNRNPA1 |
5′‐ACCAGCCU 5′‐CAGUGAGC 5′‐UAAUCCCA |
RBP and RNA substrates |
RUS0207‐A006, RUS0202‐G005 K0395‐B007 Fisetin HSc025 |
| HuR | Differential ultracentrifugation | Ubiquitination | PTBP1, hnRNPK, hnRNPC, CDC5L, hnRNPL, hnRNPA2B1, SRSF3, TRA2B, hnRNPA1, SRSF1 | AU‐rich elements (AREs) | RBP and RNA substrates |
Among others, DHTS MS‐444 |
| AGO2 |
Differential ultracentrifugation Density gradient ultracentrifugation |
Phosphorylation (S387) | DICER1, TNRC6A, TARBP2,TNRC6C, TNRC6B, XPO5, DROSHA, MOV10, DHX9, PRKRA | G‐rich sequences 5′‐GCACUU | RBP and RNA substrates |
4c and 4e BCI‐137 acriflavine aurintricarboxylic acid, suramin oxidopamin |
| IGF2BP1 |
Differential ultracentrifugation Density gradient ultracentrifugation |
ND | IGF2BP3, SYNCRIP, YBX1, HNRNPU, DHX9, MAPK4, CSDE1, ELAVL4, IGF2, LIN28B |
5′‐GGACU 5′‐ACACCC 5′‐AAGCACCCGUU 5′‐GG(m6A)C |
RBP and RNA substrates | BTNYB |
| MEX3C |
Differential ultracentrifugation Density gradient ultracentrifugation |
ND | RBBP6, FBXW, 2DZIP, 3SPSB, 2LRRC41, KLHL3, MGRN1, SPSB1, TRIM4, ARIH2 | 5′‐CAGAGUUUAG | RBP and RNA substrates | Triptolide |
| ANXA2 | Differential ultracentrifugation | Phosphorylation (T23) | S100A10, S100A4, PLG, DYSF, PLAT, RPS6KA1, TRIM72, ANXA1, SDCBP, MAPK1 | 5’‐AA(C/G)(A/U)G | RBP and RNA substrates |
1,2,4‐triazole analogues G‐Rg5 G‐Rk1 |
| ALIX | Differential ultracentrifugation | Palmitoylation | CHMP4B, TSG101, CHMP4A, CHMP4C, VPS4B, VPS4A, VPS28, CHMP6, CHMP2A, CHMP5 | ND | RBP and RNA substrates | ND |
| NCL | Polymer precipitation | ND | NPM1, FBL, WDR43, NSUS2, NOP56, NOP58, WDR46, RPS6, HSP90AA1, WDR75 |
5'‐UUAGGG 5'‐UCCCGA |
RBP and RNA substrates |
Oridonin Curcumol |
| FUS | Differential ultracentrifugation | ND | CDC5L, hnRNPD, TNP01, SRSF10, hnRNPA3, SF3A2, SRF2, hnRNPA1, hnRNPC, PTBP1 | ND | RBP | ND |
| MVP |
Immunobeads Differential ultracentrifugation Density gradient ultracentrifugation Polymer precipitation Ultrafiltration |
ND |
TEP1, PTEN, MAPK1, PTPN11, SRC, APEX1 CDC5L, PARP4, TEP1, PARP9, C19ORF48, VWF, PRPF19, PARP10, VIT, ABCC1 |
Aspecific, RNA‐mediated | Mostly RBP | Silvestrol |
| LIN28 |
Differential ultracentrifugation Ultrafiltration |
ND |
EIF3S2, NCL, DHX9, TUT4, ZCCHC6, TRIM71, DROSHA, DICER1, MYCN, HMGA2, SOX2, IGF2BP1, ZCCHC11, DIS3L2 POU5F1, SOX2, NANOG, SMAD2, SMAD4, KLF4, SALL4, STAT3, PRDM14, ZSCAN10 |
5’‐GGAGG/GGAGA | RNA substrates |
Among others, N‐methyl‐N‐[3‐(3‐methyl[1,2,4]triazolo[4,3‐b]pyridazin‐6‐yl) phenyl]acetamide 6‐hydroxy‐dl‐DOPA SB/ZW/0065 TPEN (LI38) |
| SRP9/14 |
Differential ultracentrifugation Polymer precipitation |
ND |
SRP19, SRP54, SRP68, SRP72, RPL30 with SRP14: RPS17, RPS25, RPS27, RPL27 with SRP9: RPL23A, RPL21, RPL15, RPL23 |
ND | RBP and RNA substrates | Scutellaria barbata polysaccharides (SBPS) |
| QKI | Differential ultracentrifugation | ND | CARM1, FUBP3, HNRNPK, HNRNPLL, NABP1, PCBP1, PRKAB2, PTBP1, RBFOX1, RBFOX2, RBM11, RBPMS, SNRPA, TIAL1, MED25, MED15, MED14, MED4, MED24, MED12, MED17, MED27, MED1, MED16 | 5’‐NACUAAY‐N(1‐20)‐UAAY | RNA substrates | ND |
| TERT |
Differential ultracentrifugation Polymer precipitation |
ND | DKC1, SMARCA4, PINX1, MYC, XRCC5, XRCC6, PIF1, NHP2, AKT1, SMG6 | ND | RBP‐encoding mRNA | GRN163L |
EV isolation methods, post‐translational modifications (PTMs), main interactors (by STRING Network Database; https://string-db.org), RNA motifs, biological indication in EVs, and targeting inhibitors are reported. ND: Not yet detected.
Recently, hnRNPA2B1 emerged as a key player in the sorting of specific miRNAs into exosomes and microvesicles (MVs). The protein was found responsible for direct interaction with a GGAG motif present in the 3′‐end of miRNAs (i.e., miR‐198) enriched in vesicles. Notably, the relevant activity of hnRNPA2B1 was associated with its sumoylation status (Villarroya‐Beltri et al., 2013). Another study showed that the entrapment of hnRNPA2B1‐bound miRNAs into epithelial cell‐derived MVs is mediated by the membranous protein caveolin‐1, which interacted with hnRNPA2B1 upon phosphorylation (Lee et al., 2019). The complex formation induced an O‐GlcNacylation in hnRNPA2B1, essential for the sorting of AGG and UAG motifs‐containing miR‐17 and ‐93 into MVs, guiding hnRNPA2B1 towards MVs rather than exosomes.
On the other hand, hnRNPA2B1 was recently identified as an inhibitor of the exosomal sorting of miR‐503 in endothelial cells (Pérez‐Boza et al., 2020). This group determined that epirubicin increases the exosomal export of miR‐503 in endothelial cells by disrupting the interaction between hnRNPA2B1 and the miRNA, probably mediated by ANXA2 (below described).
Other studies instead highlighted the hnRNPA2B1‐dependent accumulation of long non‐coding RNAs (lncRNAs) within exosomes. Lei et al., (2018) demonstrated that lncRNA H19 is secreted through hnRNPA2B1‐mediated packaging into exosomes, promoting resistance to gefitinib (an EGFR‐tyrosine kinase inhibitor) in NSCLC cells. H19 was significantly reduced in the presence of RNase and Triton X‐100, demonstrating its encapsulation in intact vesicles. The authors found a GGAG motif at the 5’‐end region of H19, which, when mutated, impaired the hnRNPA2B1‐binding ability. hnRNPA2B1 overexpression induced an increase of exosomal H19, while its knockdown decreased H19 expression in NSCLC cells. Han et al., (2020) showed a role for the exosomal AFAP1‐AS1 lncRNA in acquired trastuzumab resistance. hnRNPA2B1 was found crucial for the AFAP1‐AS1 loading into exosomes. This study corroborated the work of Zheng et al., (2019), who demonstrated an increased encapsulation of AGAP2 antisense RNA 1 mediated by hnRNPA2B1 in SKBR‐3R and BT474R cells in association with trastuzumab resistance. Chen et al., (2020) described a direct interaction between hnRNPA2B1 and lncRNA LNMAT2 in bladder cancer (BCa) cells, promoting lymphatic metastasis in a VEGFC–independent manner. They identified the specific exo‐motif GGAG as a determinant for direct interaction with hnRNPA2B1, further confirmed by site‐directed mutagenesis experiments. These data suggest a pivotal role of hnRNPA2B1 in the selection of cancer‐associated miRNAs and lncRNAs, emphasizing the function of this RBP in connection with the vesicular trafficking.
Given the multiple roles of hnRNPA2B1 in disease, some screening campaigns identified small molecules proposed to interact with the RNA‐binding activity of the protein. MO‐460, an analogue of the natural product (R)‐(‐)moracin‐O, suppresses the accumulation in Hep3B cells of HIF‐1α (Hypoxia‐inducible factor‐1α), involved in tumour cell adaptation to hypoxic conditions. MO‐460 binds to the C‐terminal glycine‐rich domain of hnRNPA2B1, inhibiting its subsequent binding to the 3'‐untranslated region of HIF‐1α mRNA (Soung et al., 2019). Fang et al. identified daunorubicin, pyrvinium, and pararosaniline as active planar aromatic moieties able to disrupt the RNA‐dependent recruitment of the ALS‐associated TDP‐34, FUS, and hnRNPA2B1 into stress granules, reducing progression to pathological protein aggregates (Fang et al., 2019).
Balaguer et al. identified hnRNPC1 as a protein influencing the exosome loading of miR‐30d, a transcript involved in embryonic adhesion processes (Vilella et al., 2015). Co‐localization studies of hnRNPC1 and CD63 indicated exosomes carrying hnRNPC1, whose level associated with miR‐30d biogenesis in epithelial‐like cells. Interestingly, co‐culture assays of miR‐30d KO and silenced hnRNPC1 revealed a decrease in embryo‐miR‐30d acquisition during the adhesion and invasion stages. Mechanistically, a putative direct binding of hnRNPC1 to miR‐30d was proposed at the intracellular level before the loading into the inner cavity of exosomes (Balaguer et al., 2018).
The chromosome‐linked RNA‐Binding Motif X (RBMX), also known as hnRNPG, as an ARTS‐1 (aminopeptidase regulator of TNFR1 shedding)‐associated protein, is involved in two mechanisms of TNFR1 secretion: the constitutive release through exosome‐like vesicles and the IL‐1β‐mediated inducible proteolytic cleavage of TNFR1 ectodomains. The RBMX RNAi reduces, in fact, TNFR1 release through both mechanisms (Adamik et al., 2008). An altered abundance of RBMX was shown in endothelial cell‐derived exosomes upon cellular stress, and the presence of the protein was also observed by proteomic analysis in exosomes isolated from ovarian cancer cells (de Jong et al., 2012; Liang et al., 2013).
More recently, Ahadi et al. investigated the presence of miRNA seed regions and specific RBP binding sites on lncRNAs in exosomes derived from four prostate cancer cell lines (VCaP, LNCaP, DU145, PC3) and healthy prostatic cells (Ahadi et al., 2016). They found hundreds of lncRNAs enriched in cancer exosomes and 38 predicted RBPs with putative binding sites. Among them, RBMX, SFRS1, and ELAVL1 resulted in having the highest number of predicted binding sites. Through their algorithm, the group identified 126 distinct six‐base motifs exclusively enriched in the four prostate cancer cell lines: RBMX gave positive matching in VCaP, PC3, and LNCaP cell lines.
Recently, the mRNA of hnRNPH1 was detected in exosomes isolated from the serum of hepatocellular carcinoma (HCC), chronic hepatitis B (CHB), and liver cirrhosis (LC) patients, resulting to significantly higher levels in patients compared to control groups. Also, high exosomal hnRNPH1 mRNA levels were associated with worse overall survival (OS) in HCC patients (Xu et al., 2018). Together with other proteins, Statello et al. identified hnRNPH1 in their exosome extracts capable of interacting with exo‐RNAs and cell mRNAs. Preliminary experiments revealed an unbalanced trend in the relative abundance of total RNA at the intracellular level in human epithelial cells and in derivative exosomes. For this inverse correlation with hnRNPH1 protein levels, the authors hypothesized a possible inhibitory pathway sustained by hnRNPH1 to retain cytoplasmic RNAs at the intracellular level.
These observations exist in parallel with data reported by Datta et al., who identified hnRNPH1 as a partial mediator of the RAS‐dependent Manumycin‐A (MA) suppression of exosome biogenesis and release (Datta et al., 2017). In this study, the natural microbial metabolite MA inhibited exosome release in castration‐resistant prostate cancer (CRPC) cells through the reduction of RAF and ERK and also the attenuation of ESCRT‐0, ALIX, and Rab27a proteins. The authors found hnRNPH1 as an intermediate factor inhibited by MA and responsible for the decreased levels of Alix and Rab27a transcripts. The silencing of hnRNPH1 through shRNA confirmed the decreased release of EVs along with a reduced expression of cellular Ras, pERK, Alix, Rab27a, and CD9 proteins. Of note, the hnRNPH1 mRNA in blood exosomes was suggested as a biomarker for HCC (Xu et al., 2018) or advanced prostate cancer (Yang et al., 2016).
The compound 2155‐14, recently found as an inducer of autophagy in melanoma cells, was identified as a direct interactor of the RNA helicase DDX1, as well as of hnRNPH1, H2, and hnRNPA2/B1 (Palrasu et al., 2019). Given the demonstrated effect of this drug on ER‐stress marker RNAs regulated by these RBPs, it could be plausible a potential investigation of this compound as a function of hnRNP modulation and RNA sorting into EVs.
Gao et al. indicated a role of hnRNPK in exosomes mediated transfer of lncRNA 91H in the context of colorectal cancer (CRC) progression. The overexpression of lncRNA 91H was also observed in exosomes from HCT‐8, HCT‐116 cell lines, and in patients’ serum. Mass spectrometry analysis revealed a physical interaction between hnRNPK and lncRNA 91, further confirmed by RNA interference (Gao et al., 2018). Statello et al. also found this protein in their biochemical assays using RNA extracts from cells and exosomes (Statello et al., 2018). Recently, Leidal et al. identified the LC3‐dependent EV loading and secretion (LDELS) of RBPs, such as hnRNPK and SAFB. This process employed a pool of LC3‐II (endogenous LC3) located at the MVB limiting membrane to directly capture RBPs and package them into ILV for the subsequent release as EVs via MVB fusion with the plasma membrane (Leidal et al., 2020).
In the context of small molecules, the natural compound nujiangexathone was found to repress tumour growth by targeting hnRNPK (Zhang et al., 2016). Moreover, Shu et al. identified the quinoline derivatives as able to disrupt hnRNPK binding to c‐MYC gene promoter and a promising approach for cancer therapy (Shu et al., 2019). Psammaplysene A (PA), a neuroprotective drug, was found to interact with hnRNPK, suggesting a possible role for HNRNPK in neuroprotection (Boccitto et al., 2017). A biochemical understanding of the interaction between hnRNPK and its protein partners has been proposed as necessary to establish hnRNPK as a potential therapeutic target in different cellular contexts (Wang et al., 2020).
Synaptotagmin‐binding Cytoplasmic RNA Interacting Protein (SYNCRIP, also known as hnRNPQ or NSAP1) has been described as a secreted factor in extracellular vesicles. The presence of SYNCRIP in hepatocellular exosomes was linked to the control of specific miRNAs (Santangelo et al., 2016), such as miR‐3470a and miR‐194‐2‐3p, previously associated with liver regeneration and hepatocellular carcinoma, respectively (Schug et al., 2013; Zhao et al., 2015). Additionally, Santangelo et al. identified an exosome‐enriched GGCU consensus motif responsible for the packaging of miRNAs into the vesicles. Remarkably, the SYNCRIP‐dependent RNA cargo did not overlap with the hnRNPA2B1‐dependent one, as demonstrated from the analysis of miRnome upon RBP silencing (Santangelo et al., 2016).
Summarizing, these data indicate that the intracellular balance of RBP isoforms can be responsible for a selective entrapment of RNA species into EVs, with a potential impact of post‐transcriptional control in the physiological tissue homeostasis.
3. YBX1
The Y‐box binding protein 1 (YBX1) is a RNA‐binding protein composed of an A/P rich domain (alanine/proline‐rich domain), a cold shock domain (CSD), and a long C‐terminal domain with alternating positively and negatively charged amino acids (Eliseeva et al., 2011). The human protein is composed of 324 residues (Figure 1) that can recognize the Y box motif (5'‐CTGATTGGCCAA) in the promoter of the major histocompatibility complex II (HLA‐DRαgene), the single‐stranded motif GGGG, as well as either single‐ or double‐stranded forms of the motifs CACC and CATC. The YBX1 A/P domain interacts with SRp30c, p53, and cyclin D1, whereas it's CSD interacts with AKT kinase and E3 ubiquitin ligase. The CTD is involved in protein homodimerization. YBX1 directly interacts with proliferating cell nuclear antigen (PCNA), mutS homolog 2 (MSH2), X‐ray repair cross complementing 5 (XRCC5), and DNA ligase III, clearly indicating its involvement in DNA repair (Eliseeva et al., 2011; Gaudreault, 2004). YBX1 is subjected to post‐translational modifications, such as phosphorylation, ubiquitination, and acetylation, that tune the presence of the YBX1 in mRNPs and polysomes to regulate the translation of many genes involved in cancer (Suresh et al., 2018).
The YBX1‐regulated non‐coding transcriptome includes a large number of RNA classes, such as spliceosomal RNAs, lncRNAs, Y RNAs, tRNA‐derived small RNAs, miRNAs, tiny transcription‐initiation RNAs, promoter‐associated short RNAs, termini‐associated short RNAs (TASRs), antisense TASRs, 3ʹUTR‐derived RNAs, and tRNA‐derived short RNAs (tsRNAs and tRFs) (Lee et al., 2009; Prensner & Chinnaiyan, 2011).
YBX1 can co‐localize with cytoplasmic P‐bodies containing members of the RISC complex, including GW182, which can be found in exosomes (Gallois‐Montbrun et al., 2007; Goodier et al., 2007). Interestingly, YBX1 can be found in exosomes secreted by normal and malignant cells as well, like B cells, hepatocytes, prostate, colorectal, and ovarian cancer cells (Suresh et al., 2018). There is biochemical evidence that YBX1 is secreted in a form that resists trypsin in the absence, but not in the presence, of non‐ionic detergent (Triton X‐100), consistently with a location in vesicles, probably exosomes (Frye et al., 2009; Rauen et al., 2009). There is evidence that YBX1 may undergo dephosphorylation at T271 to reach exosomes as an RNP form (Kossinova et al., 2017). A very recent study employed ssODN‐library to target exosomes secreted by VCaP and LNCaP prostate cancer cell lines. Using the adaptive dynamic artificial polyligand targeting (ADAPT), the authors were able to differentiate exosomes from cancer cell subtypes. Specifically, using an easily detectable sequence as a probe for target capturing, the authors identified proteins known to be involved in the formation of the endosomal sorting complexes required for transport (ESCRT) machinery. In these settings, YBX1 was identified as part of this interactome, specifically in complex with ubiquitinylated TSG101 (Hornung et al., 2020).
Shurtleff et al. demonstrated that YBX1 is required for miR‐223 packaging into exosomes released from HEK293T cells. In the absence of statistically significant primary sequence motifs for miRNAs in those vesicles, the authors suggested that sorting mechanisms could be based on secondary RNA structures that YBX1 could escort into exosomes (Shurtleff et al., 2016). In this line, Yanshina et al. evidenced recurrent hairpins in the 3’‐UTR of exosomal transcripts recognized by YBX1 (Yanshina et al., 2018). Subsequent analyses, aimed at defining the YBX1‐dependent exosomal RNA composition, revealed that YBX1 is required to sort other small ncRNAs such as tRNAs, YRNAs, and vault RNAs. Indeed, through TGIRT‐seq on YBX1‐null exosomes from HEK293T cells, Shurtleff et al. observed a proportional reduction of these RNAs in YBX‐1 null exosomes, while the matching YBX1‐null cells were displaying an increase in the corresponding proportion of reads. These data suggested that a YBX1‐mediated blocking of cellular export via the exosome pathway resulted in the accumulation of these transcripts at the intracellular level (Shurtleff et al., 2017).
A recent study by Lin et al. shows that miR‐133 is sorted into hypoxia/reoxygenation (H/R)‐induced human endothelial progenitor cell (EPC)‐derived exosomes via YBX1, increasing fibroblast angiogenesis and mesenchymal‐endothelial transition (MEndoT) (Lin et al., 2019). Another work highlights the interplay of YBX1 and NSUN2 proteins in recognizing exosomal RNA through the binding to ACCAGCCU, CAGUGAGC, and UAAUCCCA motifs in the HEK293 S100 extract. Interestingly, these proteins appeared in exosomes together with mRNAs containing these motifs and the degree of enrichment in exosomes correlated with the relative position of the motifs in the mRNA primary structure (Kossinova et al., 2017).
The high expression level of YBX1 is a poor prognostic marker in patients with breast cancer (Habibi et al., 2008), ovarian cancer (Kamura et al., 1999), prostate cancer (Giménez‐Bonafé et al., 2004), and other human malignancies (Kosnopfel et al., 2014). Therefore, YBX1 is an attractive target to develop novel therapies (Kosnopfel et al., 2014). A drug screening campaign carried by Trevarton et al. identified RUS0207‐A006, RUS0202‐G005, and JK0395‐B007 as small molecules interfering with the transcriptional functions of YBX1 (Trevarton et al., 2019). By using the phenotypic Epithelial‐to‐Mesenchymal Transition (EMT) readout in prostate cancer (PCa) models, Khan et al. identified the fisetin compound as a drug to inhibit YBX1 activity (Khan et al., 2014). In another disease context, Higashi et al. identified the small molecule HSc025 as a direct ligand of YBX1 able to disrupt the interaction with PABP, resulting in accelerated nuclear translocation of YBX1 with an improvement from liver injury and hepatic fibrosis in mice (Higashi et al., 2011).
4. HuR (ELAVL1)
Human antigen R (HuR or ELAVL1) is a member of the embryonic lethal abnormal vision (ELAV) family of RNA‐binding proteins (Simone & Keene, 2013). HuR is one of the most widely studied RBPs involved in the post‐transcriptional control of a plethora of RNA targets in different cellular contexts. From the nucleus to the cytoplasm, it regulates the splicing, transport, stability, and translation of thousands of coding and noncoding RNAs bearing AU‐rich elements (AREs) (Zucal et al., 2015). These sequences, mainly found in the 3’‐untranslated region, mediate the affinity of the protein and are present in genes involved in inflammation, cell division, angiogenesis, senescence, apoptosis, immune, and hypoxia response. Interestingly, the interplay among different RNA species highlighted the involvement of HuR in regulating critical miRNAs involved in development (miR‐126), cell homeostasis (miR‐126, miR‐92a), and pathological angiogenesis (miR‐200b, miR‐132), as reviewed by Chang and Hla (Chang & Hla, 2014).
Structurally, HuR contains two RRMs at the N‐terminal region, determining the recognition of the RNA, while a third RRM (RRM3) in the C‐terminal region seems implicated to poly‐A recognition and protein oligomerization (Wang et al., 2013). In cancer, the overexpression of HuR was directly involved in the balance of transcripts that increase cell proliferation and survival. Consistently, the upregulation of HuR, along with its cytoplasmic re‐localization, is associated with disease progression and poor prognosis in patients diagnosed with several types of solid tumours, including breast, colon, ovarian, prostate, pancreatic, and oral cancer (Schultz et al., 2020).
A recent study describing the tumour microenvironment of carcinoma‐associated fibroblasts (CAFs) demonstrated an association between CAFs oxaliplatin resistance and the horizontal transfer of CCAL (colorectal cancer‐associated lncRNA) lncRNA from neighbouring cancer cells (Deng et al., 2020). In this study, Deng et al. showed that this lncRNA, shuttled by colon cancer‐derived extracellular vesicles, directly interacts with HuR in recipient fibroblasts leading to an increase in β‐catenin mRNA and protein levels, triggering survival and a chemoresistant phenotype. A variety of lncRNAs, in turn substrate for several miRNAs, was also identified in prostate cancer exosomes. These lncRNAs displayed an over‐representation of cis‐elements inferring the activity of HuR and RBMX RBPs (Ahadi et al., 2016).
Recently, Mukherjee et al. demonstrated that HuR accelerates an export of miRNAs inside EVs (Mukherjee et al., 2016). In this work, the authors show a counter‐interplay of HuR and AGO2 for the binding to miRNAs, balanced by a ubiquitination step of HuR triggered by cellular stress. This PTM was associated with an HuR‐unloaded RNA and a consequent association to MVBs in human hepatocytes. Under conditions of cellular stress, such as starvation, the HuR‐dependent extracellular export of miR‐122 was observed along with CD63‐positive EVs. Interestingly, the loss of blood‐circulating miR‐122 was reported in mice starved for 12 h.
The exploratory approach conducted by Shi et al., who exploited the exosomal RNA in pull‐down assays, revealed that miR‐1246, previously found significantly upregulated in the serum of gastric cancer patients, is packaged into exosomes with HuR involvement, suggesting a rationale to detect miR‐1246 as a potential biomarker for the disease (Shi et al., 2020).
As recently reviewed by Schultz et al., various strategies have been proposed to target HuR, including the inhibition of the shuttling from the nucleus to the cytoplasm, the interference during the RNA recognition step, and the reduction of protein expression levels (Schultz et al., 2020). In the context of small molecules, besides the small molecule inhibitors recently reviewed (D'agostino et al., 2019), we highlight novel indole derivatives tested in models of retinal endothelial cells (Platania et al., 2020), the suramin tested in oral cancer cells (Kakuguchi et al., 2018), the azaphilone‐9 tested in biochemical settings (Kaur et al., 2017), and tanshinone mimics tested in different cancer cells (Manzoni et al., 2018) using specific RNA targets as a readout of HuR activity. Regarding the small molecules with a reported follow‐up testing, we highlight the MS‐444 compound in adenomatous polyposis (Lang et al., 2017) and the DHTS, previously identified to inhibit HuR:RNA interaction (D'agostino et al., 2015), very recently found as able to reduce autoimmune neuroinflammation in mouse models (Chen et al., 2020).
5. AGO2
Argonaute proteins constitute an evolutionarily conserved protein family, including two subclasses: AGO proteins and PIWI proteins. These proteins can bind to small non‐coding RNAs, such as shRNAs, miRNAs, and Piwi‐interacting RNAs (piRNAs), mediating gene‐silencing effects on their RNA targets (Elkayam et al., 2012; Höck & Meister, 2008; Müller et al., 2020). In humans, four AGO proteins (hAGO1 ‐ 4) are highly expressed and share ∼85% of sequence identity with four conserved structural domains: The N‐terminal domain (N), the PIWI/Argonaute/Zwille (PAZ) domain, the MID domain, and the P‐element‐induced wimpy testis (PIWI) domain (Höck & Meister, 2008; Meister, 2013).
In somatic cells, AGO proteins are localized in the cytoplasm and take part in processing bodies (P‐bodies) and stress granules (Höck & Meister, 2008; Meister, 2013). It has been extensively reported that eukaryotic AGOs play a crucial role in transcriptional and post‐transcriptional silencing processes, as components of the RNA‐induced silencing complex (RISC). Small RNAs, such as miRNAs, serve as guide to AGO proteins to recognize partially complementary sequences on mRNA targets, facilitating the downstream silencing effects (Höck & Meister, 2008; Meister, 2013). Moreover, AGO proteins can interact with GW182 protein family (also known as TNRC6 proteins), which recruits AGOs to target RNAs (Meister, 2013). At this point, AGO proteins can coordinate the RNA inducing silencing processes by repressing the target mRNA through RNA degradation or translation inhibition (Höck & Meister, 2008; Müller et al., 2020). AGO proteins have also been found involved in alternative splicing and DNA double‐strand break repair mechanisms (Meister, 2013; Müller et al., 2020) in cancer (Ye et al., 2015) or other diseases like diabetes (Florijn et al., 2019).
AGO2 is the most characterized AGO protein. Through the interaction with the endoribonuclease Dicer1 and TRBP (transactivation response element RNA‐binding) proteins to form the RISC loading complex, it processes precursor miRNAs into mature miRNAs (Macrae et al., 2008).
Given the remarkable enrichment of small‐sized RNA in a typical profile of extracellular vesicle lysates, multiple studies focused on the association between AGO2 activity and the potential sorting of miRNAs. Interestingly, AGO2 was found in complex with miR‐223 in microvesicles released by platelets, further proved to be functional in regulating miR‐223 in recipient endothelial cells (Ye et al., 2015). Gibbins et al. showed that AGO2, together with GW182 protein, localizes to endosomes and MVBs, suggesting that the sorting of miRNAs inside EVs might be mediated by the RISC complex (Gibbings et al., 2009). Later, a study from McKenzie et al. confirmed the association of AGO2 with MVBs in colon cancer cells and demonstrated that the activation of KRAS‐dependent MEK‐ERK signalling was able to hamper the sorting of AGO2 and AGO2‐regulated miRNAs inside extracellular vesicles (Gibbings et al., 2009). AGO2 protein is highly subjected to post‐translational modifications, such as phosphorylation, which influences its cellular localization (Müller et al., 2020). McKenzie et al. observed that the phosphorylation of AGO2 at Serine 387 prevented the secretion of AGO2 and the sorting of specific miRNAs inside vesicles (Mckenzie et al., 2016; Müller et al., 2020). However, other studies pointed out that a form of AGO2 is also highly present in the extracellular space in a vesicle‐free form, bound to circulating miRNAs, even more represented than EVs (Arroyo et al., 2011; Turchinovich et al., 2011; Weaver & Patton, 2020). Indeed, the detection of AGO2 inside vesicles could be dependent on the EV isolation procedure and/or better revealed in certain cell types (Weaver & Patton, 2020). An interesting study by Arroyo et al in 2011 also highlighted how, among a subset of circulating miRNA associated with AGO in human plasma and serum, only a minority was found in vesicles (Arroyo et al., 2011).
Recently, compounds able to bind AGO2 were described. In the hypothesis that an interference with microRNA's seed region could inhibit the recognition of a target mRNA and the cargo mediated by AGO2, Schmidt et al. combined a short nucleotide sequence with a small molecule moiety targeting the AGO2 active site (Schmidt et al., 2013). This strategy evolved a PNA‐based approach to target microRNA‐122 (Schmidt et al., 2013). In another study, Masciarelli et al. employed a structure‐based molecular design approach and identified a cell‐permeable small molecules (BCI‐137) able to compete with AGO2:miRNAs interaction (Masciarelli et al., 2014). This compound mimicked the silencing of AGO2 by lentiviral vector shRNAs in acute promyelocytic leukaemia models favouring granulocytic differentiation of that cells in response to retinoic acid.
6. IGF2BP1
The insulin‐like growth factor 2 mRNA‐binding protein 1 (IGF2BP1, alias IMP1, ZBP1, CRD‐BP, VICKZ1) is a member of the insulin‐like growth 2 proteins, a family of RBPs conserved from insects to mammals (Nielsen et al., 1999). IGF2BP1 consists of two RRM domains at the N‐terminus and four hnRNPK homology (KH) domains (Nielsen et al., 2002) (Figure 1). IGF2BP1 mainly localizes in the perinuclear region, with dynamic distribution towards lamellipodia, RNP granules, and microtubular network dependent on KH domains (Farina et al., 2003; Nielsen et al., 2002). The RNA is recognized upon dimerization also in heterodimers with IMP2 and IMP3 members of the IMP family (Chao et al., 2010; Nielsen, 2004). IGF2BP1 is involved in the maintenance of cell polarity as it binds to the zipcode sequence in the 3’UTR of β‐actin mRNA, resulting in transcript translocation to lamellipodia (Farina et al., 2003; Vikesaa et al., 2006). A role of IGF2BP1 was also reported in mRNA stability, as the knockdown of this protein prevents mRNA decay in stress granules during integrated stress response (StöHr et al., 2006). Jønson et al. showed that IGF2BP1 forms specific mRNPs granules unrelated with stress conditions and exploits microtubules to translocate transcripts outside the perinuclear regions (Jønson et al., 2007). IGF2BP1 participates in the autoregulatory translational control of poly‐A‐binding protein as it forms ribonucleic complexes with containing 5’UTRs (Patel, 2005). Interestingly, IGF2BP1 also binds to non‐coding RNAs, such as oncofetal H19, which contains four IGF2BP1‐high affinity recognition sites at the 3’ end (Runge et al., 2000). Electrophoretic mobility shift assays revealed the importance of GGACU and ACACCC motifs in the 54‐nucleotides element zipcode at the 3’UTR of ACTB mRNA (Chao et al., 2010; Ross et al., 1997). Farina et al. used RNA selection amplification (SELEX) to identify in ACTB 3’UTR an 11‐mer consensus AAGCACCCGUU after eight rounds of selection, recently demonstrated to be IGF2BP1 target‐specific due to the recognition of few amino acids in the variable loops (Biswas et al., 2019; Farina et al., 2003). Enhanced CLIP (eCLIP) experiments allowed the detection of thousands of IGF2BP1 binding sites within RNA targets in human pluripotent stem cells, mainly occurring in 3’UTR of genes related to cell adhesion and extracellular matrix reorganization (Conway et al., 2016).
7. I
IGF2BP1 is overexpressed in non‐small cell lung cancer (NSCLC) and associated with poor prognosis (in lung adenocarcinoma) at earlier onset: more than 3‐fold increased IGF2BP1 expression can be detected in 70% of NSCLCs where it positively correlates with a 30% reduction in tumour‐specific 5‐years survival after surgery (Kato et al., 2007; Shi et al., 2017). Rosenfeld et al. reported a synergistic phenomenon between oncogenic KRAS mutation and IGF2BP1 overexpression in murine and human lung carcinoma models: KRASG12V mutant/IGF2BP1 transgenic mice presented an accelerated tumour progression related with KRAS mRNA association to IGF2BP1 (Rosenfeld et al., 2019). Additionally, IGF2BP1 overexpression is associated with poor prognosis also in ovarian carcinoma (Köbel et al., 2007) in colon cancer (Dimitriadis et al., 2007), and hepatocellular carcinoma (Gutschner et al., 2014). Unexpectedly, IGF2BP1 has also been correlated with a tumour‐suppressive role in breast cancer (Lapidus et al., 2007) and gallbladder carcinoma (Kessler et al., 2017), suggesting the existence of a cell‐specific post‐transcriptional regulation.
IGF2BP1 was reported as top enriched protein in exosomes derived from murine metastatic pancreatic cell line (Panc02‐H7) compared to exosomes derived from less aggressive cells (Panc02) (Yu et al., 2017). In this study, Panc02‐H7 exosomes enhanced the formation of pre‐metastatic niches in the liver of C57BL/6 mice (Yu et al., 2017).
A biological effect in recipient cells was reported by Nguyen et al., who used EVs derived from mice and human atherogenic macrophages enriched in miR‐146a. The exposure of miR‐146a‐carrying exosomes to naïve macrophages resulted in the inhibition of the migratory pathways through the repression of IGF2BP1 expression (Nguyen et al., 2018). This observation is consistent with a recent finding on mesenchymal stem cells (MSC)‐derived exosomes and the formation of atherosclerotic plaque. MSC exosome‐associated miR‐let7 inhibited M2 macrophages infiltration and polarization in apoE‐/‐ mice aortic root as a consequence of HMGA2 and IGF2BP1‐mediated mRNA post‐transcriptional repression (Li et al., 2019).
A recent article reported the role of IGF2BP1 in the selection of RNA cargo in metastatic melanoma‐derived EVs. Ghoshal et al. showed, in two different mouse models, that the shRNA‐mediated knockdown of IGF2BP1 altered the total gene expression profile of pro‐metastatic genes in EVs. The authors provided a list of differently enriched RNA motifs between the upregulated (top enriched motif: (A/C)AGGGG) and the downregulated (top enriched motif: AUGACGUA) genes in the transcriptomes which could implicate a direct or indirect sorting mechanism in EVs upon IGF2BP1 depletion (Ghoshal et al., 2019).
An interference with IGF2BP1 cellular function was in the meantime obtained using BTYNB, a recently identified small molecule inhibitor which impaired the binding of the protein to c‐MYC mRNA (Mahapatra et al., 2017).
8. MEX3C
MEX‐3 RNA‐binding family member C (MEX3C), also known as RKHD2, BM‐013or RNF194, is a member of an evolutionarily conserved family of four RNA‐binding E3 ubiquitin ligase, known as MEX3 proteins, initially identified in Caenorhabditis elegans (Buchet‐Poyau et al., 2007). A study of Li et al. identified isoforms of human MEX3C proteins resulting from alternative splicing or alternative transcription initiation (Li et al., 2016). These proteins are characterized by two heterogeneous K homology (KH) RNA‐binding domains and one carboxy‐terminal RING‐finger domain (Li et al., 2016). Yang et al. identified the motif 5'‐CAGAGUUUAG‐3' as MEX3C‐recognition element in target RNAs (Yang et al., 2017). MEX3C is mostly known to regulate MHC‐I through a post‐transcriptional mechanism (Cano et al., 2012). In particular, a study from Cano et al. in 2012 identified the interaction of MEX3C‐KH domains with the 3'UTR of HLA‐A2 mRNA as responsible for its degradation (Cano et al., 2012). The predominant localization of MEX3C is cytoplasmic, organized in dot‐like structures, when the export receptor CRM1 (also known as exportin1 or Xpo1) does not bind to a leucine‐rich nuclear export signal (NES) causing the shuttling to the cytoplasm (Buchet‐Poyau et al., 2007; Li et al., 2016).
MEX3C1, the longest isoform with 659 amino acid residues, was found overexpressed in cells of the innate immune system and in different types of tumours, such as breast, liver, and bladder cancer, where it displayed oncogenic properties by regulating lipid metabolism (Chao et al., 2019). Other evidence suggested it may play a role in regulating energy balance (Han et al., 2014), stem cell self‐renewal and differentiation, and it contributed to hypertension susceptibility (Guzmán et al., 2006).
A recent study indicated the presence of MEX3C within exosomes isolated by hepatocarcinoma cells and normal hepatocytes (He et al., 2015). Later, the role of MEX3C in the sorting of microRNA inside EVs was reinforced by its colocalization with the endolysosomal adaptor‐related protein complex 2 (AP‐2), a cargo adaptor in clathrin‐mediated endocytosis, and AGO2. Analyzing the exosomes derived by silenced MEX3C or AP‐2 cells, they detected a lower expression of mir‐451, whose downregulation was not ultimately ascribed to the activity of MEX3C alone (Lu et al., 2017). These data suggest the existence of a synergy among RBPs for the selection of miRNAs to be packaged in vesicles.
Li et al. identified the small molecule triptolide as an inhibitor of the enzymatic activity of MEX3C, out of 2027 bioactive compounds screened. This natural compound targeted the UBE2S‐MEX3C interaction leading to inhibition of epithelial‐mesenchymal transition in renal epithelial cells (Li et al., 2019).
9. ANNEXIN A2 (ANXA2)
The annexins constitute a family of widely distributed, phospholipid‐binding peripheral membrane proteins characterized by the annexin ‘fold’. This motif, which appears either four or eight times in all annexins, has the form G‐X‐G‐T‐(38)‐(D/E), and allows the annexins to shuttle between water‐soluble and membrane compartments in response to fluctuations in calcium concentration. This property suggests an ability to influence membrane‐related events in response to the cell's homeostasis (Hajjar, 1999). Structurally, the annexins have a variable N‐terminal tail and a highly conserved C‐terminal core structure. The core contains four homologous domains (repeats) (I–IV) (except for AnxA6, which has eight repeats), each consisting of five α‐helices (helices A through E) generating a right‐handed super‐helix (Aukrust et al., 2007; Gerke & Moss, 2002). Generally, the AB‐loop and the DE‐loop form the Ca (2+−) binding sites in the annexins (type II and type III), which differ from the well‐characterized EF‐hand motif (Aukrust et al., 2007; Rosengarth & Luecke, 2004; Rosengarth et al., 2001; Thiel et al., 1991). The human annexin II gene spans approximately 40 kb on the long arm of chromosome 15 (15q21) (Hajjar, 1999; Huebner et al., 1988) and codifies for ANXA2 (also known as p36 or annexin II), which is a 36 kDa calcium‐dependent phospholipid‐binding protein (Hajjar, 1999; Xiu et al., 2016). In addition to Ca2+, the core structure of ANXA2 binds different extracellular and intracellular ligands, including heparin (Shao et al., 2006) and F‐actin (Filipenko & Waisman, 2001) and both ligands interact with domain IV (Aukrust et al., 2007; Filipenko & Waisman, 2001; Shao et al., 2006). ANXA2 also binds to specific mRNAs translated on cytoskeleton‐attached polysomes (Vedeler & Hollås, 2000) and therefore it is identified as an RNA‐binding protein (Aukrust et al., 2007). ANXA2 binds to a ∼100 nucleotides region in the 3′‐untranslated regions of its cognate (Hollås et al., 2006). Among transcripts, the c‐MYC mRNA (Mickleburgh et al., 2005) contains the consensus sequence 5’‐AA(C/G)(A/U)G, although this stretch alone is not sufficient to ensure binding to ANXA2, which requires an ‘ordered’ RNA structure (Hollås et al., 2006; Vedeler et al., 2012).
ANXA2 is found in the cytoplasm as a monomer or in the cell surface as a heterotetramer consisting of two ANXA2 monomers bridged non‐covalently to an S100A10 dimer (Gerke & Moss, 2002; Woodham et al., 2014). ANXA2 is involved in endocytosis (Emans et al., 1993; Morel & Gruenberg, 2007) and membrane trafficking (Babiychuk & Draeger, 2000). Its role has been also highlighted in cancer, enhancing proliferation, adhesion, migration, invasion, and angiogenesis of different tumour cells (Lokman et al., 2011; Shiozawa et al., 2008; Wang et al., 2012; Xiu et al., 2016). ANXA2 knockdown upregulated p53 and caused cell cycle arrest of non‐small‐cell lung carcinoma cells (Wang et al., 2012). Interestingly, ANXA2 acted as a downstream effector of EGFR signalling in breast cancer cells (Ackland et al., 2003; Grewal & Enrich, 2009), inhibiting the pathway leading to the activation of survival proteins (pAKT, pERK, pSTAT3) (Shetty et al., 2012).
Recently, there is an increasingly recognized role of annexins in influencing the transport and metabolism of coding and non‐coding RNAs (Monastyrskaya, 2018). They can directly or indirectly associate with RNAs and are horizontally transferred between cells through exosomes and microvesicles (Monastyrskaya, 2018). The post‐translational modifications in ANXA2, such as phosphorylation of Tyr23 (Glenney & Tack, 1985), phosphorylation of Ser11 (Jost & Gerke, 1996), and Ser25 (Gould et al., 1986; Johnsson et al., 1986) in the N‐terminal domain, as well as Ser1 acetylation (Johnsson et al., 1986), S‐Glutathionylation of Cys8 (Caplan et al., 2004; Sullivan et al., 2000) and Cys132 (Caplan et al., 2004), determine the interaction with proteins and RNAs. Tyr23 phosphorylation of ANXA2 is required for its Ca2+‐dependent association with endosomes and lipid rafts before entering exosomes (Morel & Gruenberg, 2009; Valapala & Vishwanatha, 2011; Vedeler et al., 2012). On the other hand, Tyr phosphorylation could be involved in the regulation of mRNA transport and translation by acting as a negative modulator of ANXA2‐mRNA interaction (Filipenko & Waisman, 2001; Vedeler et al., 2012), although the extent of these interactions is not fully elucidated (Monastyrskaya, 2018).
Hagiwara et al., in 2015, demonstrated that ANXA2 plays a significant role in the loading of miRNAs into EVs (Hagiwara et al., 2015). In this work, they examined a set of miRNAs (miR‐16, miR‐21, miR‐24, miR‐29a, miR‐100, miR‐125, let‐7a, and let‐7b) previously identified in PC3‐derived EVs (Kosaka et al., 2012), and found that their presence in vesicles associated with the levels of intracellular ANXA2 protein. In control experiments overexpressing ANXA2, the protein was found increased in both cells and EVs, with enrichment of miR‐16 in EVs and no significant effects of this transcript in cells, corroborating the evidence that ANXA2 regulates the packaging process of miRNAs into secreted EVs. Interestingly, Ca2+ modulated the binding efficiency of ANXA2 to miRNAs and highlighted the relevance of this factor in influencing the loading process of miRNAs in EVs (Hagiwara et al., 2015).
Another study in 2020 reported the interplay between ANXA2 and hnRNPA2B1 as key mediators of the exosomal export of miR‐503 (Pérez‐Boza et al., 2020). The authors demonstrated that, in endothelial cells, the chemotherapeutic drug epirubicin induced the increased exosomal export of miR‐503 by disrupting the interaction between hnRNPA2B1 and miR‐503. They also observed that, upon treatment, hnRNPA2B1 relocates in the nucleus while a fraction of the initial MicroRNA exporting complex (MEC), composed by ANXA2 and miR‐503, is remarkably sorted into exosomes despite the modest variation at intracellular level (Pérez‐Boza et al., 2020).
Other examples underlined of small molecules in blocking the interaction of ANXA2 with other proteins and the cell secretory mechanism. Reddy et al., in 2012, identified substituted 1,2,4‐triazole analogues as inhibitors of the ANXA2–S100A10 protein interaction (Reddy et al., 2012). These molecules resulted able to compete with the binding of the ANXA2 N terminus to S100A10, disrupting protein‐protein interaction (PPI) (Reddy et al., 2012; Woodham et al., 2014). Moreover, it has been shown by Jung et al. that ANXA2 promotes NF‐κB activation by binding to NF‐κB p50 subunit with its N terminal sequences, increasing its transcriptional activity and upregulating the transcription of several target genes downstream of NF‐κB, including interleukin (IL)‐6, which contributes to anti‐apoptotic signalling (Jung et al., 2015). Wang et al., in 2018, demonstrated that two ginsenosides Rg5 (G‐Rg5) and Rk1 (G‐Rk1), with similar structure, directly bound to ANXA2 (Wang et al., 2018), inhibiting its interaction with the NF‐κB p50 subunit and leading to caspase activation and apoptosis. These studies demonstrated that inhibition of ANXA2 provided a new regulatory tool on NF‐κB activity and supported the notion that the inhibition of ANXA2 may be a suitable anti‐cancer therapy (Wang et al., 2018).
10. ALIX
The apoptosis‐linked gene 2 (ALG‐2)‐Interacting protein X (ALIX) is one of the established protein markers enriched in exosomes (Vanessa et al., 2019). It is a cytosolic adaptor protein in mammalian cells, ubiquitously expressed, and concentrated in phagosomes and exosomes (Chatellard‐Causse et al., 2002; Odorizzi, 2006; Vanessa et al., 2019). ALIX was initially identified based on its association with the pro‐apoptotic signalling component ALG‐2 (Mahul‐Mellier et al., 2008; Missotten et al., 1999; Trioulier et al., 2004). Currently, we know that this protein has a pivotal role in regulating the endocytic membrane trafficking (Matsuo, 2004; Odorizzi, 2006), the virus entry process (Martin‐Serrano et al., 2003; Morita & Sundquist, 2004), the integrin‐mediated cell adhesions, the extracellular matrix assembly (Pan et al., 2008), and repair of the plasma membrane (Jimenez et al., 2014; Scheffer et al., 2014). This protein presents a tripartite domain organization consisting of an N‐terminal Bro1 domain, which is followed by coiled‐coils (CC) that build the V‐domain, and by a proline‐rich C‐terminal domain (Odorizzi, 2006). Bro1 mediates the localization to endosomes, while the proline‐rich region determines interaction promiscuity with proteins (Odorizzi, 2006). For instance, ALIX interacts with several ESCRT proteins in the formation of exosomes (Fujii et al., 2007; Odorizzi, 2006; Vanessa et al., 2019).
Iavello et al. evaluated the contribution of ALIX in the packaging of miRNAs in EVs released by human liver stem‐like cells (HLSCs) (Iavello et al., 2016). They found those EVs enriched in miRNAs eliciting antitumor as well as regenerative activity (miR ‐24, miR‐31, miR‐125b, miR‐99b, miR‐221, miR‐16, and miR‐451). Of relevance, authors highlighted the interaction between ALIX and AGO2 and the influence of relative miRNA abundance without affecting the number of released EVs (Iavello et al., 2016).
Overall, these observations indicated a synergistic activity of ALIX in determining the miRNA cargo into EVs. A dynamic analysis of its RNA‐dependent interactome might contribute to our understanding of the selective enrichment of EV‐miRNAs.
11. NUCLEOLIN (NCL)
Nucleolin (NCL), discovered in 1973, is a highly conserved RBP and one of the major nucleolar phosphoproteins in eukaryotic cells (Orrick et al., 1973). Three main structural domains form it: N‐terminal domain, enriched in acid glutamate/aspartate repeats, the central domain, containing four RRMs that mediate the interactions with nucleic acids, and the C‐terminal RGG region, that allows protein‐protein interaction (Figure 1). Acetylation and phosphorylation at N‐term domain contribute to its localization and RNA‐binding (Belenguer et al., 1990; Das et al., 2013; Ghisolfi‐Nieto et al., 1996; Salvetti et al., 2016; Ugrinova et al., 2018). NCL binds to RNA stem loop structures containing a UCCCGA consensus sequence (Bouvet et al., 2001) and associates with RNA oligonucleotides containing UUAGGG repeats more efficiently than the corresponding telomeric single‐stranded DNA (Ishikawa et al., 1993). NCL mostly localizes in nucleoli, and to some extent in nucleoplasm, cytoplasm, and cell membrane. NCL is involved in ribosome biogenesis, DNA repair and genome stability, cell division and survival, angiogenesis, and EMT. Its ability to form complexes with several different proteins could explain the pleiotropic function of NCL, still not completely understood (Birmpas et al., 2012; Jia et al., 2017; Ugrinova et al., 2018). It has been recently described as an antigen of cancer cells and cancer‐associated endothelial cells, mediating the biogenesis of miRNAs involved in tumour development, aggressiveness, and drug‐resistance. Of note, NCL post‐transcriptionally regulated miR‐21, ‐221, and ‐222, enhancing their maturation from primary transcript state (Pichiorri et al., 2013).
D'Avino et al. tested a novel NCL‐targeting immunoconjugate (4LB5‐HP‐RNase), engineered by fusion of the novel human anti‐NCL scFv 4LB5 and a human pancreatic RNase, on exosomes from derivatives of colorectal cancer cells (D'avino et al., 2016). Using miR‐21 as readout, their results suggested that, possibly, the binding disruption between NCL and miR‐21, mediated by 4LB5 scFv moiety, allows miR‐21 free‐form to be degraded by the RNase. More recently, Wang et al. designed a nanoplatform able to target NCL‐positive BC cells, exploiting the DNA aptamer AS1411 affinity to NCL. Reduced NCL expression levels and change of aptamer sequence impaired the binding efficiency (Wang et al., 2017).
NCL was found as the most abundant host cell natural protein within influenza virus‐like particles, VLPs, produced in suspension and serum‐free media by transient transfection of an inducible clone of HEK‐293SF cells (Venereo‐Sánchez et al., 2019). In the same work, NCL was not detected in EVs produced independently from non‐transformed HEK‐293SF cell line. The group, through this comparative study, identified NCL as a specific VLP marker to potentially guide VLP purification.
Since NCL functions are implicated in cancer, inflammation, and viral infection, the protein has been recently proposed as a druggable target. The first evidence of a small NCL inhibitor was identified in the anti‐tumour diterpene oridonin in Jurkat cells (Vasaturo et al., 2018). Moreover, NCL was validated as a target of curcumol in nasopharyngeal carcinoma (NPC) cells, through cellular thermal shift assay (CETSA), molecular docking (7.8 kcal/mol binding free energy) and cell‐based assay. The results revealed that the anti‐tumour effects of curcumol in NPC cells are mediated, at least partially, by NCL inhibition (Wang et al., 2018). A novel hybrid chalcone (3',4',5'‐trimethoxy‐5‐chloro‐isatinylchalcone, 3MCIC) interacts with different protein targets, confirmed by LAC (ligand‐affinity chromatography)‐MS, including NCL (Cao et al., 2016).
12. FUS/TDP‐43
Fused in Sarcoma/Translocated in Liposarcoma (FUS/TLS) is a predominantly nuclear RNA‐binding protein that belongs to the FET (FUS, EWSR1, TAF15) protein family together with EWS and TAF15RBPs. FUS contains an N‐terminal domain with a glutamine‐glycine‐serine‐tyrosine‐rich (QGSY‐rich) region, a highly conserved RRM domain, a zinc finger motif, and multiple RGG amino acid repeats at the C‐terminal (Figure 1). Analogously to TDP‐43, another ALS‐associated protein, FUS is a nucleo‐cytoplasmic shuttling factor working at transcriptional and post‐transcriptional levels. It has a role in DNA repair, transcription, RNA splicing, dendritic RNA transport, and miRNA biogenesis (Loughlin & Wilce, 2019). The binding of FUS to nascent pri‐miRNAs can recruit Drosha at chromatin sites of active transcription, promoting pri‐miRNA processing. In fact, in human neuroblastoma cells, the depletion of FUS reduced the level of several miRNAs, including miR‐9, miR‐125b, and miR‐132, which have important roles in neuronal metabolism and differentiation (Morlando et al., 2012). On the other hand, the over‐expression of FUS was directly associated with increased biogenesis of miR141 and miR200a (Dini Modigliani et al., 2014). Kapeli et al. demonstrated a stabilization effect of up to 330 mRNAs or degradation of 44 transcripts in human neural progenitor cells (Kapeli et al., 2016). Besides links with liposarcoma and myeloid leukaemia, this protein is involved in neurodegenerative disorders, such as ALS and frontotemporal lobar degeneration (FTLD) (Hortobágyi & Cairns, 2018).
Recent reports indicated an association between an imbalance in the nucleo‐cytoplasmic shuttling process and the presence of ribonucleoparticle aggregates in exosomes. Kamelgarn et al. detected FUS along with the interaction partners DDX3X and hnRNPA1 in the exosome fraction, and reported that the ALS‐associated FUS mutant R495X was present in a significantly higher level (Kamelgarn et al., 2016). Sproviero et al. showed that MVs of ALS patients were enriched with potentially aggregation‐prone FUS compared to control subjects. These observations lead to the hypothesis that EV‐carrying mutant FUS could promote the spreading of FUS‐mediated toxicity by a prion‐like mechanism (Kamelgarn et al., 2016; Sproviero et al., 2018).
Consistent observations in the same line of FUS, also the ALS‐associated TDP‐43 protein was reported to be present in exosomes from primary neurons of human amyotrophic lateral sclerosis brains (Iguchi et al., 2016) and plasma of ALS patients (Sproviero et al., 2018). Interestingly, the exposure of Neuro2a cells to those exosomes caused a cytoplasmic redistribution of TDP‐43, indicating a possible TDP‐43‐dependent propagation of proteinopathy through exosomes (Iguchi et al., 2016).
13. MVP (MAJOR VAULT PROTEIN)
Major Vault protein, also known as lung resistance‐related protein, is a ribonucleoprotein of 100 kDa constituting the main fraction (70%) of the multimeric vault ribonucleoproteins (Scheffer et al., 2000). The vault complex is composed of three proteins: MVP, vault poly (ADP‐ribose) polymerase (vPARP), and the telomerase‐associated protein 1 (TEP1), which binds to short non‐coding RNAs named vault RNAs (vRNA) (Berger et al., 2009). These large RNA–protein structures serve as intracellular shuttles for proteins and other small molecules between the nucleus and cytoplasm (Scheffer et al., 2000). MVP appears equally distributed in both nucleus and cytoplasm (Ryu et al., 2008). The MVP core of the vault complex is a polypeptide composed of proteins organized in a barrel‐like shape, with an invaginated waist and two protruding caps that can open in a flower‐like shape (Kedersha et al., 1991). MVP sequence consists of a ∼55 amino acid residue domain at the N‐terminal, which is the main repeat element in human MVP that folds in three‐stranded antiparallel β‐sheets, a central region and a coiled‐coil domain at the C‐terminal, which is fundamental for maintaining the structure of the Vault complex and the interaction with the other vault proteins (Kedersha et al., 1991; Kozlov et al., 2006; Van Zon et al., 2002). It is still unclear the mechanism underlying MVP ability to bind calcium ions, previously explained by the presence of three EF‐hand motifs in MVP sequence (Van Zon et al., 2002), later excluded following the most recent studies (Kozlov et al., 2006).
MVP is subjected to post‐translational modifications, being phosphorylated on Tyr residues after EGF stimulation, and is dephosphorylated by SHP‐2 (Kolli et al., 2004). Other studies also reported its interaction with the phosphatase PTEN, suggesting a role for MVP as a scaffold in signal transduction pathways (Kolli et al., 2004; Minaguchi et al., 2006). Importantly, MVP expression correlates with an increased multi‐drug resistance in cell lines, possibly mediated by its role in the intracellular transport of molecules. Notably, it is considered as a negative predictive marker of chemotherapy (Kickhoefer et al., 1998; Kong et al., 2000; Van Zon et al., 2001).
MVP was identified in exosomes derived from multiple colorectal cancer cell lines of (Xu et al., 2015), hepatocellular carcinoma and hepatocytes (He et al., 2015), ovarian cancer (Liang et al., 2013), neuroblastoma (Keerthikumar et al., 2015), melanoma (Peinado et al., 2012), bladder cancer (Welton et al., 2010), and nasopharyngeal carcinoma cells (Chan et al., 2015). Moreover, it has been found in exosomes isolated from human breast milk (Admyre et al., 2007), B cells (Buschow et al., 2010), thymus (Skogberg et al., 2013), platelets (Pienimaeki‐Roemer et al., 2017), saliva (Gonzalez‐Begne et al., 2009), and urine (Gonzales et al., 2009).
A sort of ‘a‐specific’ RNA‐binding activity of MVP has been observed in the study carried out by Statello et al. They reported a lower amount of total exosome‐derived RNA (of about 50%) upon MVP silencing. An RNA‐pull‐down assay validated the role of MVP on exosomes derived from HEK293 cells transfected with biotinylated full‐length MVP, whose RNA was later extracted and quantified (Statello et al., 2018). The relatively high abundance of RNA in exosomes as a function of MVP modulation indicated a potential crucial involvement of MVP in the sorting of miRNA and mRNA species into vesicles (Statello et al., 2018). The potential RNA‐binding selectivity of MVP has not been exhaustively investigated yet. An exception is a recent study which highlighted that MVP can sort miR‐193a inside exosomes (Teng et al., 2017). In detail, Teng et al. analyzed a distribution pattern of miRNA expressed by three types of exosomes isolated from primary mouse colon cancer, liver metastasis of colon cancer, and naive colon tissues. The metastasis‐derived exosomes carried the highest level of tumour‐suppressive miRNAs (miR‐193A), while the donor cells mostly expressed tumorigenic miRNAs. As further validation, MVP knockout caused miR‐193a accumulation in cells instead of exosomes, which lead to inhibition of tumour growth. These results suggested that the sorting of miRNA inside exosomes was not a passive phenomenon, but possibly mediated by RBPs, and that circulating exosomes could exert a sort of tumour suppressive effect on surrounding tissues (Teng et al., 2017).
Interestingly, a recent study on Zika virus showed that silvestrol, a natural compound of therocaglate family known to inhibit eIF4A (DEAD‐box RNA helicase eukaryotic initiation factor‐4A) was able to downregulate the expression of MVP in A549 cells (Elgner et al., 2018).
14. LIN28
LIN28, discovered in the nematode Caenorhabditis elegans, is an evolutionary conserved RNA‐binding protein of approximately 22 kDa, which in mammals has 2 paralogs: LIN28 (or LIN28A) and LIN28B. In vertebrates, these ribonucleoproteins share a high sequence identity (73%) and three conserved structural regions composed of an N‐terminal cold shock domain (CSD) and two knuckle‐type cysteine‐cysteine‐histidine‐cysteine (CCHC) zinc finger (ZnF) domains (Loughlin et al., 2012; Wang et al., 2016). LIN28A is mainly cytoplasmatic, while LIN28B is almost exclusively present in the nucleus and nucleoli (Balzeau et al., 2017; Piskounova et al., 2011). The family of Lethal‐7 (let‐7) microRNAs, involved in the regulation of differentiation pathways, is the main target of LIN28, which post‐transcriptionally suppresses the biogenesis of let‐7 precursors (pri and pre‐let 7) (Wang et al., 2012). LIN28 is a recognized stemness marker, essential to promote pluripotency with OCT4, SOX2, NANOG, and play a role in the regulation of de‐differentiation in both let‐7 dependent and independent manners (Tsialikas & Romer‐Seibert, 2015; Yu et al., 2007).
A single‐nucleotide‐resolution mapping of LIN28 binding sites, made by Ustianenko et al., showed that the presence of a (U)GAU sequence on RNA targets is necessary for their recognition by the CSD domain (Ustianenko et al., 2018), while ZnF domains bind to GGAG‐like motifs on the terminal loop of precursors of let‐7 miRNAs. Upon binding, it has been reported that the two LIN28 forms repress the maturation of let‐7 miRNAs with distinct mechanisms. Conserved GGAG/GGUG motifs located within exons and the 3′‐UTR of mRNAs were defined as binding sites with LIN28A/B, and multiple studies suggested that LIN28A/B might play a role in modulating the translation efficiency and splicing of target transcripts, although these mechanisms have not been fully characterized yet (Enriquez et al., 2015; Huang et al., 2012; Peters et al., 2016; Wilbert et al., 2012).
Several studies have shown that LIN28 proteins are overexpressed in different types of cancers (colorectal, breast, and ovarian), where the function of let‐7 as tumour suppressor against RAS or MYC is lost (Balzeau et al., 2017; Enriquez et al., 2015; Guo et al., 2006; Viswanathan et al., 2009; Wang et al., 2016). LIN28 proteins promoted tumour progression, the formation of metastasis and their overexpression correlated with a poor prognosis (Hamano et al., 2012; Viswanathan et al., 2009).
A recent study by Enriquez et al. investigated the oncogenic properties of exosomes secreted by ovarian cells overexpressing LIN28 (Alicka et al., 2019). An increased invasive phenotype was observed in HEK293 cells upon uptake of exosomes with high LIN28 transcript levels (Alicka et al., 2019). In another study focused on genes involved in the control of the glucose–lipid metabolism, the expression of LIN28 transcripts was investigated in tissue‐derived mesenchymal stem cells (ASCs) obtained from patients with Type‐2 Diabetes (T2D) and compared with the content in deriving EVs. Also in this model, no evidence of LIN28 protein in EVs was reported, in contrast to its mRNA (Alicka et al., 2019). A recent work from McDaniel et al. investigated the potential therapeutic properties of liver stem cell‐derived EVs (LSCEVs) to treat cholangiopathies. They found a high expression of let‐7 miRNAs in LSCEVs (let‐7a and let‐7b), which suppressed the activity of their preferred downstream targets, as LIN28A/B expression decreased upon treatment of total liver isolates from multidrug resistance protein 2 (MDR2‐/‐) mice, with LSCEVs. In turn, LIN28 proteins reduced overall liver damage, acting on NF‐κB and IL‐13 signalling pathways, suggesting a therapeutic effect of LSCEVs in liver diseases mediated by the let‐7/LIN28 axis (Mcdaniel et al., 2019).
Among the inhibitors of LIN28 recently reviewed (D'agostino et al., 2019), a recent study investigated more on C1632 (N‐Methyl‐N‐[3‐(3‐methyl[1,2,4]triazolo[4,3‐b]pyridazin‐6‐yl) phenyl]acetamide), a compound already known to block the interaction between LIN28 and let‐7 (Chen et al., 2019; Roos et al., 2016). In particular, the authors found that C1632 was able to downregulate the programmed death ligand‐1 (PD‐L1) and inhibit the tumour growth in vitro and in vivo, suggesting that C1632 and its derivatives could be employed as multifunctional antitumor drugs (Chen et al., 2019).
15. SRP9/14
The signal recognition particle (SRP) is a cytoplasmic ribonucleoprotein complex, which in eukaryotes consists of six polypeptides (SRPs 72, 68, 54, 19, 14, and 9) and a 300‐nucleotide 7S RNA (RN7SL1) (Walter & Blobel, 1983). In Archaea, SRP19 and SRP54 represent the only two SRP‐constituting protein subunits (Bhuiyan, 2000). In eukaryotes, SRP9 and SRP14 bind to the 5’ and 3' terminal sequences of 7S RNA (47 and 39 nucleotides, respectively), forming the so‐called Alu domain (Weichenrieder et al., 2000). SRP RNA initially interacts with SRP9/14, SRP19, and SRP68/72 in the nucleolus, forming a pre‐SRP that is subsequently transported to the cytosol where it associates with protein SRP54 (Politz et al., 2000). At cytoplasmic level, SRP, via its conjugate receptor, is involved in targeting secretory proteins to the RER membrane in eukaryotes, or the plasma membrane in prokaryotes (Reyes et al., 2007; Römisch et al., 2006). SRP S domain recognizes the signal sequence of the ribosome‐associated nascent polypeptide, and the Alu domain promotes SRP docking of the ribosome‐polypeptide complex to the ER membrane (Hsu et al., 1995; Koch et al., 2003). RN7SL1 RNA catalyzes the SRP‐receptor interaction, directly mediated by conserved active site residues in the GTPase domains of both SRP and its receptor (Bradshaw & Walter, 2007; Keenan et al., 2001).
A few indications about SRP9/14 association in EVs were so far reported. Nabet et al. showed that stromal fibroblasts, triggered by breast cancer cells upon abnormal juxtacrine signalling, secrete exosomes with increased RN7SL1 RNA (Nabet et al., 2017). The group reported that exosomes mediate the RN7SL1 transfer from stromal cells to breast cancer cells: 5‐ethynyl uridine‐modification by azide‐linked fluorescein allowed to see the horizontal acquisition of stromal cell RNA, represented by transcripts regulated by POL3 besides RN7SL1 (White, 2011). The authors also showed that the SRP9/14:RN7SL1 RNA stoichiometry controls the accumulation of unshielded RN7SL1 in the cytoplasm and secreted exosomes.
A study published in 2019 introduced the Scutellariabarbata polysaccharides (SBPS) as potential SRP9/14 inhibitors in a hepatocellular carcinoma mouse model. After SBPS treatment at different doses, the examination of the serum proteins revealed reduced SRP9/14 as compared to its levels in tumour‐bearing mice (Li et al., 2019).
16. QKI
Quaking proteins (QKI) are spliced‐proteins encoded by the quaking gene (QKI) and part of an evolutionarily conserved class of RNA‐binding proteins called STAR (signal transduction and activation of RNA) proteins (Vernet & Artzt, 1997). The spliced isoforms encoded by QKI (QKI‐5, QKI‐6, QKI‐7, and QKI‐7b) are mainly expressed in the central nervous system (CNS), glial cells, oligodendrocytes, and neuronal progenitors (Larocque et al., 2009), where they act as regulators of brain development (Ebersole et al., 1996). In particular, QKI‐5 is mostly expressed during embryogenesis, while the other isoforms are found involved in the formation and maintenance of myelination (Zhao et al., 2010). These isoforms also differ in their subcellular localization and, while QKI‐5 is mainly located in the nucleus, QKI‐6, and QKI‐7 shuttle between cytoplasm and nucleus (Chénard & Richard, 2008).
Structurally, all STAR proteins share a tripartite domain of 200 amino acids composed by a single, large KH domain (maxi‐KH), surrounded by QUA1 and QUA2 domains (Vernet & Artzt, 1997) and Tyrosine residues in the C‐terminal region. The maxi‐KH and QUA2 domains are the regions involved in RNA binding, to RNA targets enclosing a 5'‐NACUAAY‐N (Barceló et al., 2014; Yáñez‐Mó et al., 2015)‐UAAY‐3' RNA recognition motif, while the QUA1 region facilitates the recognition of target RNAs by KH‐QUA1 domains (Galarneau & Richard, 2009; Ryder, 2004; Teplova et al., 2013). Multiple studies reported that QKI plays a central role as post‐transcriptional regulators of mRNA stability and export (Teplova et al., 2013) and alternative splicing of target messenger RNAs (Wu et al., 2002). QKI is involved in the regulation of the mRNA of major myelin structural genes, in particular, QKI affects the alternative splicing of MAG (myelin‐associated glycoprotein), inducing a demyelinating phenotype (Wu et al., 2002). It drives the nuclear export of MBP (myelin basic protein) mRNA in the myelin compartment preserving myelinogenesis (Ryder, 2004). Importantly, quaking mutant mice displayed a phenotypic pattern of neuronal dysfunctions resembling schizophrenia, explained by the lacking of expression QKI‐6 and QKI‐7 isoforms in oligodendrocytes of mutant mice (Galarneau & Richard, 2009; Hardy, 1998). Zhang et al. showed that the phosphorylation of Tyr‐rich ends of QKI hampers the MBP‐RNA binding activity of QKI (Zhang, 2003). In support of this study, a lower expression of QKI was found in the brain of 55 schizophrenia patients (Aberg et al., 2006). Other studies showed that QKI‐6 and QKI‐7 isoforms were promoters of oligodendrocyte differentiation and regulators of neural progenitors fate (Larocque et al., 2009). Alterations of the quaking gene in gliomas detected during a mutational screening on primary brain tumours also exist (Li et al., 2002) high expression of QKI in tissues of breast cancer patients correlated with shorter overall survival time, proposing QKI as a prognostic marker in synergy with SLUG, a pivotal player in the EMT process (Gu et al., 2019). Interestingly, QKI was identified as the principal regulator of biogenesis of circular RNAs during epithelial to mesenchymal transition (EMT) (Conn et al., 2015).
Studying EVs, the group of Wang et al. investigated the expression of miR‐208a and miR‐208b in vesicles released by hypoxic cardiomyocytes (CMs), mimicking the environment of hypoxia/reoxygenation (H/R) injury. They found an enrichment of miR‐208a and miR‐208b miRNA. Since QKI is known to inhibit the apoptosis of cardiomyocytes (CMs) and an in silico analysis predicted the binding site of miR‐208a and miR‐208b to 3’UTR of QKI, the authors tested the effect of this RNA‐RBP interaction by exposing hypoxic cardiomyocyte to EVs. Notably, the EV‐mediated transfer of miR‐208a and miR‐208b suppressed the protective influence of QKI on apoptosis, suggesting QKI as a target to regulate CM apoptosis (Wang et al., 2017).
17. TERT
Human telomerase reverse transcriptase catalytic subunit, TERT, is a ribonucleoprotein enzyme that is part of the telomerase complex, mainly involved in elongation of telomeres. Albeit ascribed as a ‘non‐canonical RBP’, the role of TERT is increasingly recognized in the binding of different RNA molecules (Bryan & Cech, 1999; Lingner, 1997). TERT interacts with the non‐coding RNA hTR that provides the template for the reverse transcription of TTAGGG repeats at the ends of chromosomes. The TERT subunit consists of a 127 kDa protein with an N‐terminal RNA‐binding domain and a C‐terminal RT‐like domain (Bryan & Cech, 1999; Lingner, 1997). Studies focused on Tetrahymena thermophila homolog evaluated TERT‐specific motifs, T1 and T2, as essential for the binding with two sequences of RNA component, hTR (nucleotides 1–209) and the Inserted Hairpin Element 1 (IH1), independently from the C‐terminal active site (Lai et al., 2001; Rouda & Skordalakes, 2007). Of relevance, the RBPs hnRNPF/H have been reported to regulate the activity of TERT upon binding to both enzyme and RNA components, contributing to cancer and human mesenchymal stem cell proliferation and senescence (Xu et al., 2020). In the nucleus, TERT can also act as a transcriptional co‐factor, as demonstrated for the Wnt/β‐catenin pathway member BRG1 (Yang et al., 2017) or the IL6, IL8, and TNFα cytokines (Ghosh et al., 2012).
As a consequence of cell stress, TERT also shuttles to the cytoplasm and mitochondria where it exerts non‐canonical extra‐telomeric functions (Chiodi & Mondello, 2012; Sharma et al., 2012). In particular, the overexpression of TERT coupled with the induction of reactive oxygen species (ROS) might trigger a translocation in the mitochondrial matrix where it could protect from oxidative stress, repairing stress derived mtDNA damage (Haendeler et al., 2009; Santos et al., 2006). Post‐translational modifications heavily influence the sub‐cellular localization of TERT (Seimiya, 2000).
The telomerase upregulation during cancer progression is one of the hallmarks of neoplastic transformation, as it sustains the increased rate of genome replication (Hanahan & Weinberg, 2000). Several studies have therefore investigated and proposed a role of TERT transcript as a serum biomarker in breast (Qi Chen et al., 2000), prostate (March‐Villalba et al., 2012), lung (Miura et al., 2006), gastric (Kang et al., 2013), colorectal (Terrin et al., 2008), and hepatocellular carcinomas (Miura et al., 2003). Especially in gastric and prostate cancer, the high level of TERT mRNA correlated with advanced stages of disease (Kang et al., 2013; March‐Villalba et al., 2012).
Skog et al. detected TERT mRNA in microvesicles derived from glioblastoma patients (Skog et al., 2008). More recently, Gutkin et al. detected this transcript in exosomes isolated from Jurkat (T cell leukaemia), MCF‐7 (breast carcinoma), K562 (chronic myeloid leukaemia), and HCT116 (colon carcinoma) cell lines. Remarkably, these vesicles were reported as able to induce the expression of TERT in telomerase‐negative fibroblasts and induce malignant phenotype (Gutkin et al., 2016). An exploratory study involving heterogeneous cohorts of cancer patients indicated that ∼60% of cases expressed detectable levels of TERT (Goldvaser et al., 2017).
The role of EVs as vehicle of TERT protein was addressed by Radeghieri et al., who tested primary amniotic fluid cells isolated from 25 subjects and found TERT in ∼30% of exosome samples (8 out of 25) (Radeghieri et al., 2017). More recently, the group detected TERT protein in rat bone marrow‐MSC‐derived EVs (Gissi et al., 2020). An interesting finding from Wang et al. associated telomeric repeat‐containing RNA (TERRA) fragments to EVs. TERRA results from the stress‐induced transcription of telomeric DNA repeats and is implicated in structured nucleic acid hybrids and ribonucleoprotein particles (Arora et al., 2014; Azzalin & Lingner, 2015). Cell‐free TERRA was co‐fractionated with human lymphoblastoid cell lines‐derived exosomes. Exosomal TERRA was able to induce the transcription of inflammatory cytokines in recipient peripheral blood mononuclear cells (PBMCs). A similar response was inducible with HCT116 exosomes engineered to express TERRA (Wang et al., 2015).
A broad panel of TERT inhibitors is currently available for telomerase targeting, ranging from small molecules, antisense oligonucleotides, immunotherapeutic peptides, or trans‐acting repressors (Ruden & Puri, 2013). GRN163L/Imetelstat is an established telomerase inhibitor consisting of a thio‐phosphoramidate TAGGGTTAGACAA oligonucleotide complementary to the template region of hTR element. It was reported as an efficient tool to interfere with the RNA‐binding activity of TERT (Gryaznov et al., 2007).
18. CONCLUSIONS
The data so far described indicate an important coverage of qualitative aspects, ranging from the mere detection of the protein to the inference of bound RNAs, linking RNA‐binding proteins to EV biology. As shown in Figure 1, a qualitative comparison of shared domain features among EV‐associated RBPs does not reveal enrichment of molecular clues preferentially involved in vesicular packaging. Nevertheless, a substantial proportion of hnRNP family members emerges as a well‐represented hub between the RNA post‐transcriptional control and vesicular trafficking. hnRNPs are well‐known factors involved in RNA metabolism, from maturation to translation, and the elucidation of their networks of protein:protein/RNA interactions could represent an interesting level of analysis in relation to the endosomal compartment. In different cellular contexts, caveolin‐1 was shown as a direct interactor of hnRNPA2B1 (Lee et al., 2019), while Rab27a was found modulated by hnRNPH1 (Datta et al., 2017), and hnRNPK directly involved in LC3‐depended EV loading and secretion (Leidal et al., 2020). Relevantly, other RBPs, such as YBX1 and MEX3C, were found in complex with central nodes of the ESCRT machinery (Hornung et al., 2020) or clathrin‐mediated endocytosis (Lu et al., 2017), respectively, underlying the existence of a significant role in the transport of, at least sub‐populations, of vesicles. These relationships gain mechanistic values when specific PTMs are associated with a cytoplasmic activity of the RBP and the EV‐RNA results consequently modulated. The sumoylation status of hnRNPA2B1 was found associated with enrichment of the protein in exosomes along with a plethora of miRNAs defined by a detectable motif (Villarroya‐Beltri et al., 2013). At the same time, its phosphorylation/O‐GlcNAcylation associated with a localization of the protein into distinct EV sub‐populations (Lee et al., 2019). Analogously, the accumulation of cytoplasmic YBX1 upon its phosphorylation induced secretion of proangiogenic factors in vivo (Gopal et al., 2015), in a context where the protein was revealed important in the vesicular packaging of abundant small ncRNAs (Shurtleff et al., 2016).
Several studies performed in different cellular contexts with case‐by‐case biochemical and statistical approaches, identified some vesicle‐enriched RNA motifs variable in lengths and complexity. The EV‐RNA consensus at a glance could propose sequence subsets recognized by different RBPs, such as the A/G‐rich stretches by hnRNPA2B1, hnRNPH1, hnRNPQ, AGO2, and ANXA2, AU‐rich elements by HuR and hnRNPC1; C‐rich stretches by hnRNPG, hnRNPK, YBX1, and NCL. These subsets can find consistency from the reported, experimentally validated protein:protein interactions, where the RNA mediates these networks. These observations indicate that RNA packaging into EVs might significantly be impacted by a competitive interplay of RBPs. Sparse data on AGO2 protein could represent just an example of a dynamics where the cooperation with other RBPs and the endosomal compartment appeared in some works crucial to convey different miRNAs inside EVs (Gibbings et al., 2009; Mckenzie et al., 2016). Transcribed precursor miRNAs are processed by Drosha complex, then exported into the cytoplasm and digested by Dicer complex to reach maturation. As recently reviewed (Zhang et al., 2015), four potential mechanisms of miRNA sorting into EVs have been described: (i) The neural sphingomyelinase 2 (nSMase2)‐dependent pathway; (ii) The sumoylated hnRNPs‐dependent pathway; (iii) The 3′ miRNA sequence‐dependent pathway; iv) The AGO2/miRISC‐related pathway. Besides the relative dosage/equilibrium of single miRNAs that could be indirectly co‐regulated by messenger RNAs and potentially described by the competing endogenous RNA (ceRNA) model (Salmena et al., 2011), the contribution of the phospholipid bilayer of forming vesicles has been recently described as part of the RNA selection mechanism, where the fluid membrane environment can electrostatically favour specific RNA sequences, as also demonstrated in terms of the affinity of different RNA EXOmotif sequences (Janas, 2006). In this frame, the interactions of RNAs with raft‐like regions of the lipid bilayer, for instance influenced by sphingomyelin, cholesterol, and ceramide composition, could be part of a mechanism contributing the RNA sorting into vesicles. Intriguingly, the connection of RBPs with the phospholipid bilayer, as already demonstrated for hnRNP or AGO2 proteins (Villarroya‐Beltri et al., 2013), could significantly elucidate the mechanism of trafficking and functional delivery of small non‐coding RNAs from the nucleus to secreted vesicles.
In cancer, the observed up‐regulation of most RBPs here described was found to induce an altered post‐transcriptional control, which could, in turn, generate transcript unbalance in the secreted vesicles. When mechanistically validated, this paradigm provides the rationale to search for specific RNA sequences in biological fluids and EVs to investigate them as potential biomarkers. Interestingly, Xu et al. detected higher levels of hnRNPH1 mRNA in vesicles from the serum of hepatocellular carcinoma patients (Xu et al., 2018), in a context where an aberrant expression of hnRNPH1 was reported in different solid tumours, including hepatocellular carcinomas. Therefore, EVs could offer at least two distinct layers of investigation by including the detection of RBP mRNAs and RBP‐bound transcripts in a liquid biopsy test.
However, the connection of RBPs’ activity with EV biology requires comparative studies based on quantitative characterizations of protein localization, dosage, and RNA‐binding efficiency both at the intracellular and vesicular levels. Nevertheless, systematic EV isolation, characterization, and RBP/RNA detection methods could be successfully integrated with a frame of normalization‐based approaches to describe the biological matching between cells and secreted EVs, and among EVs recovered from different sources. These approaches should also include strategies to get insights on secondary RNA structures that could lead to discovering non‐obvious, key trans‐acting factors. In this view, we could elucidate a potential ‘asymmetric relevance’ of RBPs and identify new mechanisms to explain and predict the outcomes of EV‐RNA content from the vesicular transport.
Table 2 summarizes the EV isolation methods and biological indication in EVs, the post‐translational modifications (PTMs), main RBP interactors, RNA motifs, and includes identified RBP small molecule inhibitors. Some of these inhibitors could represent potential tools that could be successfully used as complementary approaches to study an RNA‐packaging effect into EVs.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
We thank the Italian Ministry of Health (GR‐2016‐02361552), the Italian MIUR (PRIN 2017F2A2C5), and Fondazione Caritro to VGD for supporting this work.
Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA packaging into extracellular vesicles: An orchestra of RNA‐binding proteins?. J Extracell Vesicles. 2020;10:e12043 10.1002/jev2.12043
Fabrizio Fabbiano and Jessica Corsi equally contributed to this work.
REFERENCES
- Aberg, K. , Saetre, P. , Jareborg, N. , & Jazin, E. (2006). Human QKI, a potential regulator of mRNA expression of human oligodendrocyte‐related genes involved in schizophrenia. Proceedings of the National Academy of Sciences of the United States of America, 103(19), 7482–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackland, M. L. , Newgreen, D. F. , Fridman, M. , Waltham, M. C. , Arvanitis, A. , Minichiello, J. , Price, J. T. , & Thompson, E. W. (2003). Epidermal growth factor‐induced epithelio‐mesenchymal transition in human breast carcinoma cells. Laboratory Investigation; a Journal of Technical Methods and Pathology, 83(3), 435–448. [DOI] [PubMed] [Google Scholar]
- Adamik, B. , Islam, A. , Rouhani, F. N. , Hawari, F. I. , Zhang, J. , & Levine, S. J. (2008). An association between RBMX, a heterogeneous nuclear ribonucleoprotein, and ARTS‐1 regulates extracellular TNFR1 release. Biochemical and Biophysical Research Communications, 371(3), 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Admyre, C. , Johansson, S. M. , Qazi, K. R. , Filén, J.‐J. , Lahesmaa, R. , Norman, M. , Neve, E. P. A. , Scheynius, A. , & Gabrielsson, S. (2007). Exosomes with immune modulatory features are present in human breast milk. Journal of Immunology, 179(3), 1969–1978. [DOI] [PubMed] [Google Scholar]
- Ahadi, A. , Brennan, S. , Kennedy, P. J. , Hutvagner, G. , & Tran, N. (2016). Long non‐coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Scientific Reports, 6, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahadi, A. , Brennan, S. , Kennedy, P. J. , Hutvagner, G. , & Tran, N. (2016). Long non‐coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Scientific Reports, 6:24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón, C. R. , Goodarzi, H. , Lee, H. , Liu, X. , Tavazoie, S. , & Tavazoie, S. F. (2015). HNRNPA2B1 is a mediator of m6A‐dependent nuclear RNA processing events. Cell, 162(6), 1299–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, M. M. , Akhade, V. S. , Kosalai, S. T. , Subhash, S. , Statello, L. , Meryet‐Figuiere, M. , Abrahamsson, J. , Mondal, T. , & Kanduri, C. (2018). PAN‐cancer analysis of S‐phase enriched lncRNAs identifies oncogenic drivers and biomarkers. Nature Communications, 9(1), 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alicka, M. , Major, P. , Wysocki, M. , & Marycz, K. (2019). Adipose‐derived mesenchymal stem cells isolated from patients with type 2 diabetes show reduced “Stemness” through an altered secretome profile, impaired anti‐oxidative protection, and mitochondrial dynamics deterioration. Journal of Clinical Medicine, 8(6), 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan, S. A. , Martincic, K. , & Milcarek, C. (2006). The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochemical Journal, 393(1), 361–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, J. P. K. , Holme, M. N. , & Stevens, M. M. (2017). Re‐engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano, 11(1), 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, R. , Lee, Y. , Wischnewski, H. , Brun, C. M. , Schwarz, T. , & Azzalin, C. M. (2014). RNaseH1 regulates TERRA‐telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nature Communications, 5, 5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo, J. D. , Chevillet, J. R. , Kroh, E. M. , Ruf, I. K. , Pritchard, C. C. , Gibson, D. F. , Mitchell, P. S. , Bennett, C. F. , Pogosova‐Agadjanyan, E. L. , Stirewalt, D. L. , Tait, J. F. , & Tewari, M. (2011). Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proceedings of the National Academy of Sciences of the United States of America, 108(12), 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukrust, I. , Hollås, H. , Strand, E. , Evensen, L. , Travé, G. , Flatmark, T. , & Vedeler, A. (2007). The mRNA‐binding site of Annexin A2 resides in helices C‐D of its domain IV. Journal of Molecular Biology, 368(5), 1367–1378. [DOI] [PubMed] [Google Scholar]
- Azzalin, C. M. , & Lingner, J. (2015). Telomere functions grounding on TERRA firma. Trends in Cell Biology, 25(1), 29–36. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E. B. , & Draeger, A. (2000). Annexins in cell membrane dynamics: Ca2+‐regulated association of lipid microdomains. Journal of Cell Biology, 150(5), 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaguer, N. , Moreno, I. , Herrero, M. , González, M. , Simón, C. , & Vilella, F. (2018). Heterogeneous nuclear ribonucleoprotein C1may control miR‐30d levels in endometrial exosomes affecting early embryo implantation. Molecular Human Reproduction, 24(8), 411–425. [DOI] [PubMed] [Google Scholar]
- Balzeau, J. , Menezes, M. R. , Cao, S. , & Hagan, J. P. (2017). The LIN28/let‐7 pathway in cancer. Frontiers in Genetics, 8:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barceló, C. , Etchin, J. , Mansour, M. R. , Sanda, T. , Ginesta, M. M. , Sanchez‐Arévalo Lobo, V. J. , Real, F. X. , Capellà, G. , Estanyol, J. M. , Jaumot, M. , Look, A. T. , & Agell, N. (2014). Ribonucleoprotein HNRNPA2B1 interacts with and regulates oncogenic KRAS in pancreatic ductal adenocarcinoma cells. Gastroenterology, 147(4), 882–892.e8. [DOI] [PubMed] [Google Scholar]
- Barrack, E. R. (1987). Steroid hormone receptor localization in the nuclear matrix: Interaction with acceptor sites. Journal of Steroid Biochemistry, 27(1‐3), 115–121. [DOI] [PubMed] [Google Scholar]
- Becker, A. , Thakur, B. K. , Weiss, J. M. , Kim, H. S. , Peinado, H. , & Lyden, D. (2016). Extracellular vesicles in cancer: cell‐to‐cell mediators of metastasis. Cancer Cell, 30(6), 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer, P. , Caizergues‐Ferrer, M. , Labbé, J. C. , Dorée, M. , & Amalric, F. (1990). Mitosis‐specific phosphorylation of nucleolin by p34cdc2 protein kinase. Molecular and Cellular Biology, 10(7), 3607–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, W. , Steiner, E. , Grusch, M. , Elbling, L. , & Micksche, M. (2009). Vaults and the major vault protein: novel roles in signal pathway regulation and immunity. Cellular and Molecular Life Sciences, 66(1), 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan, S. H. (2000). Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Research, 28(6), 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmpas, C. , Briand, J. P. , Courty, J. , & Katsoris, P. (2012). Nucleolin mediates the antiangiogenesis effect of the pseudopeptide N6L. Bmc Cell Biology [Electronic Resource], 13:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, J. , Patel, V. L. , Bhaskar, V. , Chao, J. A. , Singer, R. H. , & Eliscovich, C. (2019). The structural basis for RNA selectivity by the IMP family of RNA‐binding proteins. Nature Communications, 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccitto, M. , Lee, N. , Sakamoto, S. , Spruce, L. , Handa, H. , Clardy, J. , Seeholzer, S. , & Kalb, R. (2017). The neuroprotective marine compound psammaplysene A binds the RNA‐binding protein HNRNPK. Marine Drugs, 15(8), 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet, P. , Allain, F. H.‐T. , Finger, L. D. , Dieckmann, T. , & Feigon, J. (2001). Recognition of pre‐formed and flexible elements of an RNA stem‐loop by nucleolin. Journal of Molecular Biology, 309(3), 763–775. [DOI] [PubMed] [Google Scholar]
- Bradshaw, N. , & Walter, P. (2007). The signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Molecular Biology of the Cell, 18(7), 2728–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan, T. M. , & Cech, T. R. (1999). Telomerase and the maintenance of chromosome ends. Current Opinion in Cell Biology, 11(3), 318–324. [DOI] [PubMed] [Google Scholar]
- Buchet‐Poyau, K. , Courchet, J. , Hir, H. L. , Seraphin, B. , Scoazec, J.‐Y. , Duret, L. , Domon‐Dell, C. , Freund, J.‐N. , & Billaud, M. (2007). Identification and characterization of human Mex‐3 proteins, a novel family of evolutionarily conserved RNA‐binding proteins differentially localized to processing bodies. Nucleic acids research, 35(4), 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschow, S. I. , Balkom, B. W. M. , Aalberts, M. , Heck, A. J. R. , Wauben, M. , & Stoorvogel, W. (2010). MHC class II‐associated proteins in B‐cell exosomes and potential functional implications for exosome biogenesis. Immunology and Cell Biology, 88(8), 851–856. [DOI] [PubMed] [Google Scholar]
- Cabral, J. , Ryan, A. E. , Griffin, M. D. , & Ritter, T. (2018). Extracellular vesicles as modulators of wound healing. Advanced Drug Delivery Reviews, 129:394–406. [DOI] [PubMed] [Google Scholar]
- Cano, F. , Bye, H. , Duncan, L. M. , Buchet‐Poyau, K. , Billaud, M. , Wills, M. R. , & Lehner, P. J. (2012). The RNA‐binding E3 ubiquitin ligase MEX‐3C links ubiquitination with MHC‐I mRNA degradation. Embo Journal, 31(17), 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, L. , Zhang, L. , Zhao, X. , & Zhang, Y. (2016). A hybrid chalcone combining the trimethoxyphenyl and isatinyl groups targets multiple oncogenic proteins and pathways in hepatocellular carcinoma cells. Plos One, 11(8), e0161025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J. F. , Filipenko, N. R. , Fitzpatrick, S. L. , & Waisman, D. M. (2004). Regulation of Annexin A2 by reversible glutathionylation. Journal of Biological Chemistry, 279(9), 7740–7750. [DOI] [PubMed] [Google Scholar]
- Chan, Y.‐K. , Zhang, H. , Liu, P. , Tsao, S.‐W. , Lung, M. L.i , Mak, N.‐K. , Ngok‐Shun Wong, R. , & Ying‐Kit Yue, P. (2015). Proteomic analysis of exosomes from nasopharyngeal carcinoma cell identifies intercellular transfer of angiogenic proteins. International Journal of Cancer, 137(8), 1830–1841. [DOI] [PubMed] [Google Scholar]
- Chang, S.‐H. , & Hla, T. (2014). Post‐transcriptional gene regulation by HuR and microRNAs in angiogenesis. Current Opinion in Hematology, 21(3), 235–240. https://journals.lww.com/co-hematology/Fulltext/2014/05000/Post_transcriptional_gene_regulation_by_HuR_and.13.aspx [DOI] [PubMed] [Google Scholar]
- Chao, H. , Deng, L. , Xu, F. , Yu, Z. , Xu, X. , Huang, J. , & Zeng, T. (2019). MEX3C regulates lipid metabolism to promote bladder tumorigenesis through JNK pathway. Onco Targets Therapy, 12, 3285–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, J. A. , Patskovsky, Y. , Patel, V. , Levy, M. , Almo, S. C. , & Singer, R. H. (2010). ZBP1 recognition of β‐actin zipcode induces RNA looping. Genes & Development, 24(2), 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatellard‐Causse, C. , Blot, B. , Cristina, N. , Torch, S. , Missotten, M. , & Sadoul, R. (2002). Alix (ALG‐2‐interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. Journal of Biological Chemistry, 277(32), 29108–29115. [DOI] [PubMed] [Google Scholar]
- Chen, C. , Luo, Y. , He, W. , Zhao, Y. , Kong, Y. , Liu, H. , Zhong, G. , Li, Y. , Li, J. , Huang, J. , Chen, R. , & Lin, T. (2020). Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. Journal of Clinical Investigation, 130(1), 404–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.‐H. , Chang, J.‐G. , Lu, R.‐M. , Peng, T.‐Y. , & Tarn, W.‐Y. (2008). The RNA Binding Protein hnRNP Q Modulates the Utilization of Exon 7 in the Survival Motor Neuron 2 (SMN2) Gene. Molecular and Cellular Biology, 28(22), 6929–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Martindale, J. L. , Abdelmohsen, K. , Kumar, G. , Fortina, P. M. , Gorospe, M. , Rostami, A. , & Yu, S. (2020). RNA‐Binding Protein HuR Promotes Th17 Cell Differentiation and Can Be Targeted to Reduce Autoimmune Neuroinflammation. Journal of Immunology, 204(8), 2076–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.‐C. , Hsueh, C. , Tsang, N.‐M. , Liang, Y. , Chang, K.‐P. , Hao, S.‐P. , Yu, J.‐S. , & Chang, Y.‐S. (2008). Heterogeneous ribonucleoprotein K and thymidine phosphorylase are independent prognostic and therapeutic markers for nasopharyngeal carcinoma. Clinical Cancer Research, 14(12), 3807–3813. [DOI] [PubMed] [Google Scholar]
- Chen, Y. , Xie, C. , Zheng, X. , Nie, X. , Wang, Z. , Liu, H. , & Zhao, Y. (2019). LIN28/let‐7/PD‐L1 pathway as a target for cancer immunotherapy. Cancer immunology research, 7(3), 487–497. [DOI] [PubMed] [Google Scholar]
- Chénard, C. A. , & Richard, S. (2008). New implications for the QUAKING RNA binding protein in human disease. Journal of Neuroscience Research, 86(2), 233–242. [DOI] [PubMed] [Google Scholar]
- Chiodi, I. , & Mondello, C. (2012). Telomere‐independent functions of telomerase in nuclei, cytoplasm, and mitochondria. Frontiers in Oncology, 2:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Ideue, T. , Nagayama, M. , Araki, N. , & Tani, T. (2018). RBMX is a component of the centromere noncoding RNP complex involved in cohesion regulation. Genes to Cells, 23(3), 172–184. [DOI] [PubMed] [Google Scholar]
- Choi, D. , Lee, T. H. , Spinelli, C. , Chennakrishnaiah, S. , D'asti, E. , & Rak, J. (2017). Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation. Seminars in Cell & Developmental Biology, 67, 11–22. [DOI] [PubMed] [Google Scholar]
- Cianciaruso, C. , Beltraminelli, T. , Duval, F. , Nassiri, S. , Hamelin, R. , Mozes, A. , Gallart‐Ayala, H. , Ceada Torres, G. , Torchia, B. , Ries, C. H. , Ivanisevic, J. , & De Palma, M. (2019). Molecular profiling and functional analysis of macrophage‐derived tumor extracellular vesicles. Cell reports, 27(10), 3062–3080.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieniková, Z. , Jayne, S. , Damberger, F. F. , Allain, F. H.‐.T. , & Maris, C. (2015). Evidence for cooperative tandem binding of hnRNP C RRMs in mRNA processing. RNA, 21(11), 1931–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn, S. J. , Pillman, K. A. , Toubia, J. , Conn, V. M. , Salmanidis, M. , Phillips, C. A. , Roslan, S. , Schreiber, A. W. , Gregory, P. A. , & Goodall, G. J. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell, 160(6), 1125–1134. [DOI] [PubMed] [Google Scholar]
- Conway, A. E. , van Nostrand, E. L. , Pratt, G. A. , Aigner, S. , Wilbert, M. L. , Sundararaman, B. , Freese, P. , Lambert, N. J. , Sathe, S. , Liang, T. Y. , Essex, A. , Landais, S. , Burge, C. B. , Jones, D. L , Yeo, G. W. (2016). Enhanced CLIP uncovers IMP Protein‐RNA targets in human pluripotent stem cells important for cell adhesion and survival. Cell Reports, 15(3), 666–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'agostino, V. G. , Lal, P. , Mantelli, B. , Tiedje, C. , Zucal, C. , Thongon, N. , Gaestel, M. , Latorre, E. , Marinelli, L. , Seneci, P. , Amadio, M. , & Provenzani, A. (2015). Dihydrotanshinone‐I interferes with the RNA‐binding activity of HuR affecting its post‐transcriptional function. Scientific Reports, 5, 16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'agostino, V. G. , Sighel, D. , Zucal, C. , Bonomo, I. , Micaelli, M. , Lolli, G. , Provenzani, A. , Quattrone, A. , & Adami, V. (2019). Screening approaches for targeting ribonucleoprotein complexes: a new dimension for drug discovery. SLAS Discovery: Advancing Life Sciences R & D, 24(3), 314–331. [DOI] [PubMed] [Google Scholar]
- Das, S. , Cong, R. , Shandilya, J. , Senapati, P. , Moindrot, B. , Monier, K. , Delage, H. , Mongelard, F. , Kumar, S. , Kundu, T. K. , & Bouvet, P. (2013). Characterization of nucleolin K88 acetylation defines a new pool of nucleolin colocalizing with pre‐mRNA splicing factors. Febs Letters, 587(5), 417–424. [DOI] [PubMed] [Google Scholar]
- Datta, A. , Kim, H. , Lal, M. , McGee, L. , Johnson, A. , Moustafa, A. A. , Jones, J. C. , Mondal, D. , Ferrer, M. , & Abdel‐Mageed, A. B. (2017). Manumycin A suppresses exosome biogenesis and secretion via targeted inhibition of Ras/Raf/ERK1/2 signaling and hnRNP H1 in castration‐resistant prostate cancer cells. Cancer Letters, 408, 73‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'avino, C. , Palmieri, D. , Braddom, A. , Zanesi, N. , James, C. , Cole, S. , Salvatore, F. , Croce, C. M. , & De Lorenzo, C. (2016). A novel fully human anti‐NCL immunoRNase for triple‐negative breast cancer therapy. Oncotarget, 7(52), 87016–87030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, O. G. , Verhaar, M. C. , Chen, Y. , Vader, P. , Gremmels, H. , Posthuma, G. , Schiffelers, R. M. , Gucek, M. , & van Balkom, B. W. (2012). Cellular stress conditions are reflected in the protein and RNA content of endothelial cell‐derived exosomes. Journal of Extracellular Vesicles, 1(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X. , Ruan, H. , Zhang, X. , Xu, X. , Zhu, Y. , Peng, H. , Zhang, X. , Kong, F. , & Guan, M. (2020). Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. International Journal of Cancer, 146(6), 1700–1716. [DOI] [PubMed] [Google Scholar]
- Dimitriadis, E. , Trangas, T. , Milatos, S. , Foukas, P. G. , Gioulbasanis, I. , Courtis, N. , Nielsen, F. C. , Pandis, N. , Dafni, U. , Bardi, G. , & Ioannidis, P. (2007). Expression of oncofetal RNA‐binding protein CRD‐BP/IMP1 predicts clinical outcome in colon cancer. International Journal of Cancer, 121(3), 486–494. [DOI] [PubMed] [Google Scholar]
- Dini Modigliani, S. , Morlando, M. , Errichelli, L. , Sabatelli, M. , & Bozzoni, I. (2014). An ALS‐associated mutation in the FUS 3′‐UTR disrupts a microRNA–FUS regulatory circuitry. Nature Communications, 5(1), 4335. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G. (1993). hnRNP Proteins and the Biogenesis of mRNA. Annual Review of Biochemistry, 62(1), 289–321. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G. , Kim, V. N. , & Kataoka, N. (2002). Messenger‐RNA‐binding proteins and the messages they carry. Nature Reviews Molecular Cell Biology, 3(3), 195–205. [DOI] [PubMed] [Google Scholar]
- Ebersole, T. A. , Chen, Q. , Justice, M. J. , & Artzt, K. (1996). The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nature Genetics, 12(3), 260–265. [DOI] [PubMed] [Google Scholar]
- Elgner, F. , Sabino, C. , Basic, M. , Ploen, D. , Grünweller, A. , & Hildt, E. (2018). Inhibition of Zika virus replication by silvestrol. Viruses, 10(4), 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliseeva, I. A. , Kim, E. R. , Guryanov, S. G. , Ovchinnikov, L. P. , & Lyabin, D. N. (2011). REVIEW Y Box Binding Protein 1 (YB 1) and Its Functions, 76(13), 1402–1433. [DOI] [PubMed] [Google Scholar]
- Elkayam, E. , Kuhn, C.‐D. , Tocilj, A. , Haase, A. D. , Greene, E. M. , Hannon, G. J. , & Joshua‐Tor, L. (2012). The structure of human argonaute‐2 in complex with miR‐20a. Cell, 150(1), 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, D. J. , Dalgliesh, C. , Hysenaj, G. , & Ehrmann, I. (2019). RBMX family proteins connect the fields of nuclear RNA processing, disease and sex chromosome biology. International Journal of Biochemistry & Cell Biology, 108, 1–6. [DOI] [PubMed] [Google Scholar]
- Emans, N. , Gorvel, J. P. , Walter, C. , Gerke, V. , Kellner, R. , Griffiths, G. , & Gruenberg, J. (1993). Annexin II is a major component of fusogenic endosomal vesicles. Journal of Cell Biology, 120(6), 1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enriquez, V. A. , Cleys, E. R. , Da Silveira, J. C. , Spillman, M. A. , Winger, Q. A. , & Bouma, G. J. (2015). High LIN28A expressing ovarian cancer cells secrete exosomes that induce invasion and migration in HEK293 cells. BioMed Research International, 2015, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, M. Y. , Markmiller, S. , Vu, A. Q. , Javaherian, A. , Dowdle, W. E. , Jolivet, P. , Bushway, P. J. , Castello, N. A. , Baral, A. , Chan, M. Y. , Linsley, J. W. , Linsley, D. , Mercola, M. , Finkbeiner, S. , Lecuyer, E. , Lewcock, J. W. , Yeo, G. W. (2019). Small‐Molecule Modulation of TDP‐43 Recruitment to Stress Granules Prevents Persistent TDP‐43 Accumulation in ALS/FTD. Neuron, 103(5), 802–819.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina, K. L. , HüTtelmaier, S. , Musunuru, K. , Darnell, R. , & Singer, R. H. (2003). Two ZBP1 KH domains facilitate beta‐actin mRNA localization, granule formation, and cytoskeletal attachment. Journal of Cell Biology, 160(1), 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipenko, N. R. , & Waisman, D. M. (2001). The C terminus of Annexin II mediates binding to F‐actin. Journal of Biological Chemistry, 276(7), 5310–5315. [DOI] [PubMed] [Google Scholar]
- Florijn, B. W. , Duijs, J. M.G.J. , Levels, J. H. , Dallinga‐Thie, G. M. , Wang, Y. , Boing, A. N. , Yuana, Y. , Stam, W. , Limpens, R. W.A.L. , Au, Y. W. , Nieuwland, R. , Rabelink, T. J. , Reinders, M. E.J. , Van Zonneveld, A. J. , & Bijkerk, R. (2019). Diabetic nephropathy alters the distribution of circulating angiogenic MicroRNAs among extracellular vesicles, HDL, and Ago‐2. Diabetes, 68(12), 2287–2300. [DOI] [PubMed] [Google Scholar]
- Frye, B. C. , Halfter, S. , Djudjaj, S. , Muehlenberg, P. , Weber, S. , Raffetseder, U. , En‐Nia, A. , Knott, H. , Baron, J. M. , Dooley, S. , Bernhagen, J. , Mertens, P. R. (2009). Y‐box protein‐1 is actively secreted through a non‐classical pathway and acts as an extracellular mitogen. Embo Reports, 10(7), 783–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, K. , Hurley, J. H. , & Freed, E. O. (2007). Beyond Tsg101: the role of Alix in “ESCRTing” HIV‐1. Nature Reviews Microbiology, 5(12), 912–916. [DOI] [PubMed] [Google Scholar]
- Galarneau, A. , & Richard, S. (2009). The STAR RNA binding proteins GLD‐1, QKI, SAM68 and SLM‐2 bind bipartite RNA motifs. Bmc Molecular Biology [Electronic Resource], 10:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois‐Montbrun, S. , Kramer, B. , Swanson, C. M. , Byers, H. , Lynham, S. , Ward, M. , & Malim, M. H. (2007). Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. Journal of Virology, 81(5), 2165–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, T. , Liu, X. , He, B. , Nie, Z. , Zhu, C. , Zhang, P. , & Wang, S. (2018). Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer cell international, 18(1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault, I. (2004). YB‐1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Research, 32(1), 316–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, V. , & Moss, S. E. (2002). Annexins: from structure to function. Physiological Reviews, 82(2), 331–371. [DOI] [PubMed] [Google Scholar]
- Geuens, T. , Bouhy, D. , & Timmerman, V. (2016). The hnRNP family: insights into their role in health and disease. Human Genetics, 135(8), 851–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi‐Nieto, L. , Joseph, G. , Puvion‐Dutilleul, F. , Amalric, F. , & Bouvet, P. (1996). Nucleolin is a sequence‐specific RNA‐binding protein: Characterization of targets on pre‐ribosomal RNA. Journal of Molecular Biology, 260(1), 34–53. [DOI] [PubMed] [Google Scholar]
- Ghosh, A. , Saginc, G. , Leow, S. C. , Khattar, E. , Shin, E. M. , Yan, T. D. , Wong, M. , Zhang, Z. , Li, G. , Sung, W.‐K. , Zhou, J. , Chng, W. J. , Li, S. , Liu, E. , & Tergaonkar, V. (2012). Telomerase directly regulates NF‐B‐dependent transcription. Nature Cell Biology, 14(12), 1270–1281. [DOI] [PubMed] [Google Scholar]
- Ghoshal, A. , Rodrigues, L. C. , Gowda, C. P. , Elcheva, I. A. , Liu, Z. , Abraham, T. , & Spiegelman, V. S. (2019). Extracellular vesicle‐dependent effect of RNA‐binding protein IGF2BP1 on melanoma metastasis. Oncogene, 38(21), 4182–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings, D. J. , Ciaudo, C. , Erhardt, M. , & Voinnet, O. (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nature Cell Biology, 11(9), 1143–1149. [DOI] [PubMed] [Google Scholar]
- Giménez‐Bonafé, P. , Fedoruk, M. N. , Whitmore, T. G. , Akbari, M. , Ralph, J. L. , Ettinger, S. , Gleave, M. E. , & Nelson, C. C. (2004). YB‐1 is upregulated during prostate cancer tumor progression and increases P‐glycoprotein activity. The Prostate, 59(3), 337–349. [DOI] [PubMed] [Google Scholar]
- Gissi, C. , Radeghieri, A. , Antonetti Lamorgese Passeri, C. , Gallorini, M. , Calciano, L. , Oliva, F. , Veronesi, F. , Zendrini, A. , Cataldi, A. , Bergese, P. , Maffulli, N. , & Berardi, A. C. (2020). Extracellular vesicles from rat‐bone‐marrow mesenchymal stromal/stem cells improve tendon repair in rat Achilles tendon injury model in dose‐dependent manner: a pilot study. Plos One, 15(3), e0229914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenney, J. R. , & Tack, B. F. (1985). Amino‐terminal sequence of p36 and associated p10: Identification of the site of tyrosine phosphorylation and homology with S‐100. Proceedings of the National Academy of Sciences of the United States of America, 82(23), 7884–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldvaser, H. , Gutkin, A. , Beery, E. , Edel, Y. , Nordenberg, J. , Wolach, O. , Rabizadeh, E. , Uziel, O. , & Lahav, M. (2017). Characterisation of blood‐derived exosomal hTERT mRNA secretion in cancer patients: A potential pan‐cancer marker. British Journal of Cancer, 117(3), 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, X. , Siprashvili, Z. , Eminaga, O. , Shen, Z. , Sato, Y. , Kume, H. , Homma, Y. , Ogawa, S. , Khavari, P. A. , Pollack, J. R. , & Brooks, J. D. (2017). Novel lincRNA SLINKY is a prognostic biomarker in kidney cancer. Oncotarget, 8(12), 18657–18669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales, P. A. , Pisitkun, T. , Hoffert, J. D. , Tchapyjnikov, D. , Star, R. A. , Kleta, R. , Wang, N. S. , & Knepper, M. A. (2009). Large‐scale proteomics and phosphoproteomics of urinary exosomes. Journal of the American Society of Nephrology, 20(2), 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez‐Begne, M. , Lu, B. , Han, X. , Hagen, F. K. , Hand, A. R. , Melvin, J. E. , & Yates,, J. R. (2009). Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). Journal of Proteome Research, 8(3), 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier, J. L. , Zhang, L. , Vetter, M. R. , & Kazazian, H. H. (2007). LINE‐1 ORF1 protein localizes in stress granules with other RNA‐binding proteins, including components of RNA interference RNA‐induced silencing complex. Molecular and Cellular Biology, 27(18), 6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, S. K. , Greening, D. W. , Mathias, R. A. , Ji, H. , Rai, A. , Chen, M. , Zhu, H.‐J. , & Simpson, R. J. (2015). YBX1/YB‐1 induces partial EMT and tumourigenicity through secretion of angiogenic factors into the extracellular microenvironment. Oncotarget, 6(15), 13718–13730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, K. L. , Woodgett, J. R. , Isacke, C. M. , & Hunter, T. (1986). The protein‐tyrosine kinase substrate p36 is also a substrate for protein kinase C in vitro and in vivo. Molecular and Cellular Biology, 6(7), 2738–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, T. , & Enrich, C. (2009). Annexins ‐ Modulators of EGF receptor signalling and trafficking. Cellular Signalling, 21(6), 847–858. [DOI] [PubMed] [Google Scholar]
- Gryaznov, S. M. , Jackson, S. , Dikmen, G. , Harley, C. , Herbert, B.‐S. , Wright, W. E. , & Shay, J. W. (2007). Oligonucleotide conjugate GRN163L targeting human telomerase as potential anticancer and antimetastatic agent. Nucleosides, Nucleotides & Nucleic Acids, 26(10‐12), 1577–1579. [DOI] [PubMed] [Google Scholar]
- Gu, S. , Chu, C. , Chen, W. , Ren, H. , Cao, Y. , Li, X. , He, J. , Wang, Y. , Jin, Y. , Liu, X. , & Zou, Q. (2019). Prognostic value of epithelial‐mesenchymal transition related genes: SLUG and QKI in breast cancer patients. International Journal of Clinical and Experimental Pathology, 12(6), 2009–2021. [PMC free article] [PubMed] [Google Scholar]
- Guo, J. , Wang, X. , Jia, J. , & Jia, R. (2020). Underexpression of SRSF3 and its target gene RBMX predicts good prognosis in patients with head and neck cancer. Journal of Oral Science, 62(2), 175–179. [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Chen, Y. , Ito, H. , Watanabe, A. , Ge, X. , Kodama, T. , & Aburatani, H. (2006). Identification and characterization of lin‐28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene, 384(1‐2), 51–61. [DOI] [PubMed] [Google Scholar]
- Gutkin, A. , Uziel, O. , Beery, E. , Nordenberg, J. , Pinchasi, M. , Goldvaser, H. , Henick, S. , Goldberg, M. , & Lahav, M. (2016). Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget, 7(37), 59173–59188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner, T. , Hämmerle, M. , Pazaitis, N. , Bley, N. , Fiskin, E. , Uckelmann, H. , Heim, A. , Groβ, M. , Hofmann, N. , Geffers, R. , Skawran, B. , Longerich, T. , Breuhahn, K. , Schirmacher, P. , Mühleck, B. , Hüttelmaier, S. , & Diederichs, S. (2014). Insulin‐like growth factor 2 mRNA‐binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology, 59(5), 1900–1911. [DOI] [PubMed] [Google Scholar]
- Guzmán, B. , Cormand, B. , Ribasés, M. , González‐Núñez, D. , Botey, A. , & Poch, E. (2006). Implication of chromosome 18 in hypertension by sibling pair and association analyses: Putative involvement of the RKHD2 gene. Hypertension, 48(5), 883–891. [DOI] [PubMed] [Google Scholar]
- Habibi, G. , Leung, S. , Law, J. H. , Gelmon, K. , Masoudi, H. , Turbin, D. , Pollak, M. , Nielsen, T. O. , Huntsman, D. , & Dunn, S. E. (2008). Redefining prognostic factors for breast cancer: YB‐1 is a stronger predictor of relapse and disease‐specific survival than estrogen receptor or HER‐2 across all tumor subtypes. Breast Cancer Research, 10(5), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haendeler, J. , DröSe, S. , BüChner, N. , Jakob, S. , Altschmied, J. , Goy, C. , Spyridopoulos, I. , Zeiher, A. M. , Brandt, U. , & Dimmeler, S. (2009). Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arteriosclerosis, Thrombosis, and Vascular Biology, 29(6), 929–935. [DOI] [PubMed] [Google Scholar]
- Hagiwara, K. , Katsuda, T. , Gailhouste, L. , Kosaka, N. , & Ochiya, T. (2015). Commitment of Annexin A2 in recruitment of microRNAs into extracellular vesicles. Febs Letters, 589(24), 4071–4078. [DOI] [PubMed] [Google Scholar]
- Hajjar, K. (1999). Annexin II: a mediator of the plasmin/plasminogen activator system. Trends in Cardiovascular Medicine, 9(5), 128–138. [DOI] [PubMed] [Google Scholar]
- Hamano, R. , Miyata, H. , Yamasaki, M. , Sugimura, K. , Tanaka, K. , Kurokawa, Y. , Nakajima, K. , Takiguchi, S. , Fujiwara, Y. , Mori, M. , & Doki, Y. (2012). High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. British Journal of Cancer, 106(8), 1415–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, C. , Jiao, Y. , Zhao, Q. , & Lu, B. (2014). Mex3c mutation reduces adiposity partially through increasing physical activity. Journal of Endocrinology, 221(3), 457–468. [DOI] [PubMed] [Google Scholar]
- Han, M. , Gu, Y. , Lu, P. , Li, J. , Cao, H. , Li, X. , Qian, X. , Yu, C. , Yang, Y. , Yang, X. , Han, N. , Dou, D. , Hu, J. , & Dong, H. (2020). Exosome‐mediated lncRNA AFAP1‐AS1 promotes trastuzumab resistance through binding with AUF1 and activating ERBB2 translation. Molecular Cancer [Electronic Resource], 19(1), 26. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Han, S. P. , Tang, Y. H. , & Smith, R. (2010). Functional diversity of the hnRNPs: Past, present and perspectives. Biochemical Journal, 430(3), 379–392. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. , & Weinberg, R. A. (2000). The hallmarks of cancer. Cell, 100(1), 57–70. [DOI] [PubMed] [Google Scholar]
- Hardy, R. J. (1998). Molecular defects in the dysmyelinating mutant quaking. Journal of Neuroscience Research, 51(4), 417–422. [DOI] [PubMed] [Google Scholar]
- Harrison, A. F. , & Shorter, J. (2017). RNA‐binding proteins with prion‐like domains in health and disease Protein misfolding unites diverse neurodegenerative diseases. Biochemical Journal, 474(8), 1417–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, M. , Qin, H. , Poon, T. C.W. , Sze, S.‐C. , Ding, X. , Co, N. N.a , Ngai, S.‐M. , Chan, T.‐F. , & Wong, N. (2015). Hepatocellular carcinoma‐derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis, 36(9), 1008–1018. [DOI] [PubMed] [Google Scholar]
- Heinrich, B. , Zhang, Z. , Raitskin, O. , Hiller, M. , Benderska, N. , Hartmann, A. M. , Bracco, L. , Elliott, D. , Ben‐Ari, S. , Soreq, H. , Sperling, J. , Sperling, R. , & Stamm, S. (2009). Heterogeneous nuclear ribonucleoprotein G regulates splice site selection by binding to CC(A/C)‐rich regionsin pre‐mRNA. Journal of Biological Chemistry, 284(21), 14303–14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer, E. , Haque, I. S. , Roberts, C. E. S. , & Speicher, M. R. (2019). Current and future perspectives of liquid biopsies in genomics‐driven oncology. Nature Reviews Genetics, 20(2), 71–88. [DOI] [PubMed] [Google Scholar]
- Helmken, C. , Hofmann, Y. , Schoenen, F. , Oprea, G. , Raschke, H. , Rudnik‐Schöneborn, S. , Zerres, K. , & Wirth, B. (2003). Evidence for a modifying pathway in SMA discordant families: Reduced SMN level decreases the amount of its interacting partners and Htra2‐beta1. Human Genetics, 114(1), 11–21. [DOI] [PubMed] [Google Scholar]
- Higashi, K. , Tomigahara, Y. , Shiraki, H. , Miyata, K. , Mikami, T. , Kimura, T. , Moro, T. , Inagaki, Y. , & Kaneko, H. (2011). A novel small compound that promotes nuclear translocation of YB‐1 ameliorates experimental hepatic fibrosis in mice. Journal of Biological Chemistry, 286(6), 4485–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck, J. , & Meister, G. (2008). The Argonaute protein family. Genome Biology, 9(2), 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollås, H. , Aukrust, I. , Grimmer, S. , Strand, E. , Flatmark, T. , & Vedeler, A. (2006). Annexin A2 recognises a specific region in the 3′‐UTR of its cognate messenger RNA. Biochimica et biophysica acta, 1763(11), 1325–1334. [DOI] [PubMed] [Google Scholar]
- Hong, X. , Song, R. , Song, H. , Zheng, T. , Wang, J. , Liang, Y. , Qi, S. , Lu, Z. , Song, X. , Jiang, H. , Liu, L. , & Zhang, Z. (2014). PTEN antagonises Tcl1/hnRNPK‐mediated G6PD pre‐mRNA splicing which contributes to hepatocarcinogenesis. Gut, 63(10), 1635–1647. [DOI] [PubMed] [Google Scholar]
- Hornung, T. , O'neill, H. A. , Logie, S. C. , Fowler, K. M. , Duncan, J. E. , Rosenow, M. , Bondre, A. S. , Tinder, T. , Maher, V. , Zarkovic, J. , Zhong, Z. , Richards, M. N. , Wei, X. , Miglarese, M. R. , Mayer, G. , Famulok, M. , & Spetzler, D. (2020). ADAPT identifies an ESCRT complex composition that discriminates VCaP from LNCaP prostate cancer cell exosomes. Nucleic Acids Research, 48(8), 4013–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortobágyi, T. , & Cairns, N. J. (2018). Amyotrophic lateral sclerosis and non‐tau frontotemporal lobar degeneration. Handbook of Clinical Neurology, 145:369–381. [DOI] [PubMed] [Google Scholar]
- Hsu, K. , Chang, D.‐Y. , & Maraia, R. J. (1995). Human signal recognition particle (SRP) Alu‐associated protein also binds Alu interspersed repeat sequence RNAs. Characterization of human SRP9. Journal of Biological Chemistry, 270(17), 10179–10186. [DOI] [PubMed] [Google Scholar]
- Huang, H. , Han, Y. , Zhang, C. , Wu, J. , Feng, J. , Qu, L. , & Shou, C. (2016). HNRNPC as a candidate biomarker for chemoresistance in gastric cancer. Tumour Biology : The Journal of the International Society for Oncodevelopmental Biology and Medicine, 37(3), 3527–3534. [DOI] [PubMed] [Google Scholar]
- Huang, Y.‐W. A. , Ruiz, C. R. , Eyler, E. C. H. , Lin, K. , & Meffert, M. K. (2012). Dual regulation of miRNA biogenesis generates target specificity in neurotrophin‐induced protein synthesis. Cell, 148(5), 933–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner, K. , Cannizzaro, L. A. , Frey, A. L. , Hecht, B. K. , Hecht, F. , Croce, C. M. , & Wallner, B. P. (1988). Chromosomal localization of the human genes for lipocortin I and lipocortin II. Oncogene Research, 2(4), 299–310. https://www.ncbi.nlm.nih.gov/pubmed/?term=Huebner++K%2C++Cannizzaro++LA%2C++Frey++AZ%2C++et++al.%3A+1988.++Chromosomal++localization++of++the+genes++for++lipocortin++I++and++lipocortin++II.+Oncogene+Res+2%3A299-310 Accessed 30 April 2020. [PubMed] [Google Scholar]
- Hung, C.‐Y. , Wang, Y.‐C. , Chuang, J.‐Y. , Young, M.‐J. , Liaw, H. , Chang, W.‐C. , & Hung, J.‐J. (2017). Nm23‐H1‐stabilized hnRNPA2/B1 promotes internal ribosomal entry site (IRES)‐mediated translation of Sp1 in the lung cancer progression. Scientific Reports, 7(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavello, A. , Frech, V. S.L. , Gai, C. , Deregibus, M. C. , Quesenberry, P. J. , & Camussi, G. (2016). Role of Alix in miRNA packaging during extracellular vesicle biogenesis. International Journal of Molecular Medicine, 37(4), 958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi, Y. , Eid, L. , Parent, M. , Soucy, G. , Bareil, C. , Riku, Y. , Kawai, K. , Takagi, S. , Yoshida, M. , Katsuno, M. , Sobue, G. , & Julien, J.‐P. (2016). Exosome secretion is a key pathway for clearance of pathological TDP‐43. Brain, 139(12), 3187–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, F. , Matunis, M. J. , Dreyfuss, G. , & Cech, T. R. (1993). Nuclear proteins that bind the pre‐mRNA 3’ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Molecular and Cellular Biology, 13(7), 4301–4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas, T. (2006). Specific RNA binding to ordered phospholipid bilayers. Nucleic acids research, 34(7), 2128–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, W. , Yao, Z. , Zhao, J. , Guan, Q. , & Gao, L. (2017). New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sciences, 186, 1–10. [DOI] [PubMed] [Google Scholar]
- Jimenez, A. J. , Maiuri, P. , Lafaurie‐Janvore, J. , Divoux, S. , Piel, M. , & Perez, F. (2014). ESCRT machinery is required for plasma membrane repair. Science, 343(6174), 1247136. [DOI] [PubMed] [Google Scholar]
- Johnsson, N. , Nguyen Van, P. , Söling, H. D. , & Weber, K. (1986). Functionally distinct serine phosphorylation sites of p36, the cellular substrate of retroviral protein kinase; differential inhibition of reassociation with p11. Embo Journal, 5(13), 3455–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønson, L. , Vikesaa, J. , Krogh, A. , Nielsen, L. K. , Hansen, T. vO , Borup, R. , Johnsen, A. H. , Christiansen, J. , & Nielsen, F. C. (2007). Molecular composition of IMP1 ribonucleoprotein granules. Molecular and Cellular Proteomics, 6(5), 798–811. [DOI] [PubMed] [Google Scholar]
- Jost, M. , & Gerke, V. (1996). Mapping of a regulatory important site for protein kinase C phosphorylation in the N‐terminal domain of annexin II In: Biochimica et Biophysica Acta ‐ Molecular Cell Research. Vol 1313 Elsevier B.V., 283–289. [DOI] [PubMed] [Google Scholar]
- Jung, H. , Kim, J. S. , Kim, W. K. , Oh, K.‐J. , Kim, J.‐M. , Lee, H. J. , Han, B. S. , Kim, D. S. , Seo, Y. S. , Lee, S. C. , Park, S. G. , & Bae, K.‐H. (2015). Intracellular annexin A2 regulates Nf‐kB signaling by binding to the p50 subunit: implications for gemcitabine resistance in pancreatic cancer. Cell Death & Disease, 6(1), e1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuguchi, W. , Nomura, T. , Kitamura, T. , Otsuguro, S. , Matsushita, K. , Sakaitani, M. , Maenaka, K. , & Tei, K. (2018). Suramin, screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells. Cancer Medicine, 7(12), 6269–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamelgarn, M. , Chen, J. , Kuang, L. , Arenas, A. , Zhai, J. , Zhu, H. , & Gal, J. (2016). Proteomic analysis of FUS interacting proteins provides insights into FUS function and its role in ALS. Biochimica et Biophysica Acta, 1862(10), 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T. , Yahata, H. , Amada, S. , Ogawa, S. , Sonoda, T. , Kobayashi, H. , Mitsumoto, M. , Kohno, K. , Kuwano, M. , & Nakano, H. (1999). Is nuclear expression of Y box‐binding protein‐1 a new prognostic factor in ovarian serous adenocarcinoma? Cancer, 85(11), 2450–2454. [DOI] [PubMed] [Google Scholar]
- Kang, Y. , Zhang, J. , Sun, P. , & Shang, J. (2013). Circulating cell‐free human telomerase reverse transcriptase mRNA in plasma and its potential diagnostic and prognostic value for gastric cancer. International Journal of Clinical Oncology, 18(3), 478–486. [DOI] [PubMed] [Google Scholar]
- Kapeli, K. , Pratt, G. A. , Vu, A. Q. , Hutt, K. R. , Martinez, F. J. , Sundararaman, B. , Batra, R. , Freese, P. , Lambert, N. J. , Huelga, S. C. , Chun, S. J. , Liang, T. Y. , Chang, J. , Donohue, J. P. , Shiue, L. , Zhang, J. , Zhu, H. , Cambi, F. , Kasarskis, E. … Yeo, G. W. (2016). Distinct and shared functions of ALS‐associated proteins TDP‐43, FUS and TAF15 revealed by multisystem analyses. Nature Communications, 7(1), 12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, T. , Hayama, S. , Yamabuki, T. , Ishikawa, N. , Miyamoto, M. , Ito, T. , Tsuchiya, E. , Kondo, S. , Nakamura, Y. , Daigo, Y. (2007). Increased expression of insulin‐like growth factor‐II messenger RNA‐binding protein 1 is associated with tumor progression in patients with lung cancer. Clinical Cancer Research, 13(2 I), 434–442. [DOI] [PubMed] [Google Scholar]
- Kaur, K. , Wu, X. , Fields, J. K. , Johnson, D. K. , Lan, L. , Pratt, M. , Somoza, A. D. , Wang, C. C. C. , Karanicolas, J. , Oakley, B. R. , Xu, L. , & De Guzman, R. N. (2017). The fungal natural product azaphilone‐9 binds to HuR and inhibits HuR‐RNA interaction in vitro. Kontoyiannis D. L., ed. Plos One, 12(4), e0175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha, N. L. , Heuser, J. E. , Chugani, D. C. , Rome, L. H. (1991). Vaults. III. Vault ribonucleoprotein particles open into flower‐like structures with octagonal symmetry. Journal of Cell Biology, 112(2):225–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, R. J. , Freymann, D. M. , Stroud, R. M. , & Walter, P. (2001). The Signal Recognition Particle. Annual Review of Biochemistry, 70(1), 755–775. [DOI] [PubMed] [Google Scholar]
- Keerthikumar, S. , Gangoda, L. , Liem, M. , Fonseka, P. , Atukorala, I. , Ozcitti, C. , Mechler, A. , Adda, C. G. , Ang, C.‐S. , & Mathivanan, S. (2015). Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget, 6(17), 15375–15396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, S. M. , Lederer, E. , Laggai, S. , Golob‐Schwarzl, N. , Hosseini, K. , Petzold, J. , Schweiger, C. , Reihs, R. , Keil, M. , Hoffmann, J. , Mayr, C. , Kiesslich, T. , Pichler, M. , Kim, K. S. , Rhee, H. , Park, Y. N. , Lax, S. , Obrist, P. , Kiemer, A. K. , & Haybaeck, J. (2017). IMP2/IGF2BP2 expression, but not IMP1 and IMP3, predicts poor outcome in patients and high tumor growth rate in xenograft models of gallbladder cancer. Oncotarget, 8(52), 89736–89745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, M. I. , Adhami, V. M. , Lall, R. K. , Sechi, M. , Joshi, D. C. , Haidar, O. M. , Syed, D. N. , Siddiqui, I. A. , Chiu, S.‐Y. , & Mukhtar, H. (2014). YB‐1 expression promotes epithelial‐to‐mesenchymal transition in prostate cancer that is inhibited by a small molecule fisetin. Oncotarget, 5(9), 2462–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kickhoefer, V. A. , Rajavel, K. S. , Scheffer, G. L. , Dalton, W. S. , Scheper, R. J. , & Rome, L. H. (1998). Vaults are up‐regulated in multidrug‐resistant cancer cell lines. Journal of Biological Chemistry, 273(15), 8971–8974. [DOI] [PubMed] [Google Scholar]
- Klinge, C M. , Piell, K M. , Tooley, C. S. , Rouchka, E C. (2019). HNRNPA2/B1 is upregulated in endocrine‐resistant LCC9 breast cancer cells and alters the miRNA transcriptome when overexpressed in MCF‐7 cells. Scientific Reports, 9(1), 9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köbel, M. , Weidensdorfer, D. , Reinke, C. , Lederer, M. , Schmitt, W. D. , Zeng, K. , Thomssen, C. , Hauptmann, S. , & Hüttelmaier, S. (2007). Expression of the RNA‐binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene, 26(54), 7584–7589. [DOI] [PubMed] [Google Scholar]
- Koch, H. G. , Moser, M. , & Müller, M. (2003). Signal recognition particle‐dependent protein targeting, universal to all kingdoms of life. Reviews of Physiology Biochemistry and Pharmacology, 146, 55–94. [DOI] [PubMed] [Google Scholar]
- Kolli, S. , Zito, C. I. , Mossink, M. H. , Wiemer, E. A. C. , & Bennett, A. M. (2004). The major vault protein is a novel substrate for the tyrosine phosphatase SHP‐2 and scaffold protein in epidermal growth factor signaling. Journal of Biological Chemistry, 279(28), 29374–29385. [DOI] [PubMed] [Google Scholar]
- Kong, L. B. , Siva, A. C. , Kickhoefer, V. A. , Rome, L. H. , & Stewart, P. L. (2000). RNA location and modeling of a WD40 repeat domain within the vault. RNA, 6(6), 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König, J. , Zarnack, K. , Rot, G. , Curk, T. , Kayikci, M. , Zupan, B., Turner, D. J., Luscombe, N. M., & Ule, J. (2010). iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature Structural & Molecular Biology, 17(7), 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka, N. , Iguchi, H. , Yoshioka, Y. , Hagiwara, K. , Takeshita, F. , & Ochiya, T. (2012). Competitive interactions of cancer cells and normal cells via secretory microRNAs. Journal of Biological Chemistry, 287(2), 1397–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosnopfel, C. , Sinnberg, T. , & Schittek, B. (2014). Y‐box binding protein 1 ‐ A prognostic marker and target in tumour therapy. European Journal of Cell Biology, 93(1‐2), 61–70. [DOI] [PubMed] [Google Scholar]
- Kossinova, O. A. , Gopanenko, A. V. , Tamkovich, S. N. , Krasheninina, O. A. , Tupikin, A. E. , Kiseleva, E. , Yanshina, D. D. , Malygin, A. A. , Ven'yaminova, A. G. , Kabilov, M. R. , & Karpova, G. G. (2017). Cytosolic YB‐1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochimica et Biophysica Acta. Proteins and Proteomics, 1865(6), 664–673. [DOI] [PubMed] [Google Scholar]
- Kozlov, G. , Vavelyuk, O. , Minailiuc, O. , Banville, D. , Gehring, K. , Ekiel, I. (2006). Solution structure of a two‐repeat fragment of major vault protein. Journal of Molecular Biology, 356(2), 444–452. [DOI] [PubMed] [Google Scholar]
- Lai, C. K. , Mitchell, J. R. , & Collins, K. (2001). RNA binding domain of telomerase reverse transcriptase. Molecular and Cellular Biology, 21(4), 990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.‐H. , Huang, Y.‐C. , Lee, J.‐C. , Tseng, J. T.a.‐C. , Chang, K.‐C. , Chen, Y.‐J. , Ding, N.‐J. , Huang, P.‐H. , Chang, W.‐C. , Lin, B.‐W. , Chen, R.‐Y. , Wang, Y.‐C. , Lai, Y.‐C. , & Hung, L.‐Y. (2017). Translational upregulation of Aurora‐A by hnRNP Q1 contributes to cell proliferation and tumorigenesis in colorectal cancer. Cell Death & Disease, 8(1), e2555–e2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, M. , Berry, D. , Passecker, K. , Mesteri, I. , Bhuju, S. , Ebner, F. , Sedlyarov, V. , Evstatiev, R. , Dammann, K. , Loy, A. , Kuzyk, O. , Kovarik, P. , Khare, V. , Beibel, M. , Roma, G. , Meisner‐Kober, N. , & Gasche, C. (2017). HuR Small‐Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer Research, 77(9), 2424 LP – 2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus, K. , Wyckoff, J. , Mouneimne, G. , Lorenz, M. , Soon, L. , Condeelis, J. S. , & Singer, R. H. (2007). ZBP1 enhances cell polarity and reduces chemotaxis. Journal of Cell Science, 120(18), 3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larocque, D. , Fragoso, G. , Huang, J. , Mushynski, W. E. , Loignon, M. , Richard, S. , & Almazan, G. (2009). The QKI‐6 and QKI‐7 RNA binding proteins block proliferation and promote Schwann cell myelination. Plos One, 4(6), e5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Li, C. , Zhang, Y. , Zhang, D. , Otterbein, L. E. , & Jin, Y. (2019). Caveolin‐1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. Journal of Experimental Medicine, 216(9), 2202–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S. , Shibata, Y. , Malhotra, A. , & Dutta, A. (2009). A novel class of small RNAs: TRNA‐derived RNA fragments (tRFs). Genes & Development, 23(22), 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, Y. , Guo, W. , Chen, B. , Chen, L. , Gong, J. , & Li, W. (2018). Tumor‐released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non‐small cell lung cancer. Oncology Reports, 40(6), 3438–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal, A. M. , Huang, H. H. , Marsh, T. , Solvik, T. , Zhang, D. , Ye, J. , Kai, F. , Goldsmith, J. , Liu, J. Y. , Huang, Y.‐H. , Monkkonen, T. , Vlahakis, A. , Huang, E. J. , Goodarzi, H. , Yu, L. , Wiita, A. P. , & Debnath, J. (2020). The LC3‐conjugation machinery specifies the loading of RNA‐binding proteins into extracellular vesicles. Nature Cell Biology, 22(2), 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Xue, H. , Li, T. , Chu, X. , Xin, D. , Xiong, Y. , Qiu, W. , Gao, X. , Qian, M. , Xu, J. , Wang, Z. , & Li, G. (2019). Exosomes derived from mesenchymal stem cells attenuate the progression of atherosclerosis in ApoE −/‐ mice via miR‐let7 mediated infiltration and polarization of M2 macrophage. Biochemical and Biophysical Research Communications, 510(4), 565–572. [DOI] [PubMed] [Google Scholar]
- Li, L. , Xu, X. , Wu, L. , Zhu, H. , He, Z. , Zhang, B. , Chi, Y. , & Song, G. (2019). Scutellaria barbata polysaccharides inhibit tumor growth and affect the serum proteomic profiling of hepatoma H22‐bearing mice. Molecular Medicine Reports, 19(3), 2254–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Li, Y. , Liu, C. , Jin, M. , & Lu, B. (2016). Oocyte‐specific expression of mouse MEX3C652AA in the ovary and its potential role in regulating maternal Fos mRNA1. Biology of Reproduction, 94(5). [DOI] [PubMed] [Google Scholar]
- Li, Y. , Hu, Q. , Li, C. , Liang, K. , Xiang, Y. , Hsiao, H. , Nguyen, T. K. , Park, P. K. , Egranov, S. D. , Ambati, C. R. , Putluri, N. , Hawke, D. H. , Han, L. , Hung, M.‐C. , Danesh, F. R. , Yang, L. , & Lin, C. (2019). PTEN‐induced partial epithelial‐mesenchymal transition drives diabetic kidney disease. Journal of Clinical Investigation, 129(3), 1129–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z. Z. , Kondo, T. , Murata, T. , Ebersole, T A. , Nishi, T. , Tada, K. , Ushio, Y. , Yamamura, K.‐I. , Abe, K. (2002). Expression of Hqk Encoding a KH RNA Binding Protein Is Altered in Human Glioma. Japanese Journal of Cancer Research: Gann, 93(2), 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, B. , Peng, P. , Chen, S. , Li, L. , Zhang, M. , Cao, D. , Yang, J. , Li, H. , Gui, T. , Li, X. , & Shen, K. (2013). Characterization and proteomic analysis of ovarian cancer‐derived exosomes. Journal of Proteomics, 80, 171–182. [DOI] [PubMed] [Google Scholar]
- Lin, F. , Zeng, Z. , Song, Y. , Li, L. , Wu, Z. , Zhang, X. , Li, Z. , Ke, X. , & Hu, X. (2019). YBX‐1 mediated sorting of miR‐133 into hypoxia/reoxygenation‐induced EPC‐derived exosomes to increase fibroblast angiogenesis and MEndoT. Current Stem cell Research & Therapy, 10(1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J. (1997). Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276(5312), 561–567. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Beqaj, S. , Yang, Y. , Honoré, B. , & Schuger, L. (2001). Heterogeneous nuclear ribonucleoprotein‐H plays a suppressive role in visceral myogenesis. Mechanisms of Development, 104(1‐2), 79–87. [DOI] [PubMed] [Google Scholar]
- Lokman, N. A. , Ween, M. P. , Oehler, M. K. , & Ricciardelli, C. (2011). The role of annexin A2 in tumorigenesis and cancer progression. Cancer Microenvironment: Official journal of the International Cancer Microenvironment Society, 4(2), 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin, F. E. , & Wilce, J. A. (2019). TDP‐43 and FUS–structural insights into RNA recognition and self‐association. Current Opinion in Structural Biology, 59, 134–142. [DOI] [PubMed] [Google Scholar]
- Loughlin, F. , Brunschweiger, A. , Allain, E. T. H. , & Zurich, F. H. (2012). Structural basis of pre‐let‐7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28 Screening of DNA‐encoded libraries View project Automated microfluidic reaction screening platform View project. Nature Structural & Molecular Biology, 19, 84–89 [DOI] [PubMed]
- Lu, P. , Li, H. , Li, N. , Singh, R. N. , Bishop, C. E. , Chen, X. , & Lu, B. (2017). MEX3C interacts with adaptor‐related protein complex 2 and involves in miR‐451a exosomal sorting. Plos One, 12(10), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P. , Li, H. , Li, N. , Singh, R. N. , Bishop, C. E. , Chen, X. , & Lu, B. (2017). MEX3C interacts with adaptor‐related protein complex 2 and involves in miR‐451a exosomal sorting. Hakami R. M., ed. Plos One, 12(10), e0185992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae, I. J. , Ma, E. , Zhou, M. , Robinson, C. V. , & Doudna, J. A. (2008). In vitro reconstitution of the human RISC‐loading complex. Proceedings of the National Academy of Sciences of the United States of America, 105(2), 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra, L. , Andruska, N. , Mao, C. , Le, J. , & Shapiro, D. J. (2017). A novel IMP1 inhibitor, BTYNB, targets c‐Myc and inhibits melanoma and ovarian cancer cell proliferation. Translational Oncology, 10(5), 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul‐Mellier, A.‐L. , Strappazzon, F. , Petiot, A. , Chatellard‐Causse, C. , Torch, S. , Blot, B. , Freeman, K. , Kuhn, L. , Garin, J. , Verna, J.‐M. , Fraboulet, S. , & Sadoul, R. (2008). Alix and ALG‐2 are involved in tumor necrosis factor receptor 1‐induced cell death. Journal of Biological Chemistry, 283(50), 34954–34965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoni, L. , Zucal, C. , Maio, D. D.i , D'agostino, V. G. , Thongon, N. , Bonomo, I. , Lal, P. , Miceli, M. , Baj, V. , Brambilla, M. , Cerofolini, L. , Elezgarai, S. , Biasini, E. , Luchinat, C. , Novellino, E. , Fragai, M. , Marinelli, L. , Provenzani, A. , & Seneci, P. (2018). Interfering with HuR‐RNA interaction: design, synthesis and biological characterization of tanshinone mimics as novel, effective HuR inhibitors. Journal of Medicinal Chemistry, 61(4), 1483–1498. [DOI] [PubMed] [Google Scholar]
- March‐Villalba, J. A. , Martínez‐Jabaloyas, J. M. , Herrero, M. J. , Santamaría, J. , Aliño, S. F. , & Dasí, F. (2012). Plasma hTERT mRNA discriminates between clinically localized and locally advanced disease and is a predictor of recurrence in prostate cancer patients. Expert Opinion on Biological Therapy, 12(Suppl 1), S69–S77. [DOI] [PubMed] [Google Scholar]
- Martinez‐Contreras, R. , Cloutier, P. , Shkreta, L. , Fisette, J. F. , Revil, T. , & Chabot, B. (2007). hnRNP proteins and splicing control. Advances in Experimental Medicine and Biology, 623:123–147. [DOI] [PubMed] [Google Scholar]
- Martin‐Serrano, J. , Yaravoy, A. , Perez‐Caballero, D. , & Bieniasz, P. D. (2003). Divergent retroviral late‐budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proceedings of the National Academy of Sciences of the United States of America, 100(21), 12414–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciarelli, S. , Quaranta, R. , Iosue, I. , Colotti, G. , Padula, F. , Varchi, G. , Fazi, F. , & Del Rio, A. (2014). A small‐molecule targeting the microRNA binding domain of argonaute 2 improves the retinoic acid differentiation response of the acute promyelocytic leukemia cell line NB4. Acs Chemical Biology, 9(8), 1674–1679. [DOI] [PubMed] [Google Scholar]
- Matsunaga, S. , Takata, H. , Morimoto, A. , Hayashihara, K. , Higashi, T. , Akatsuchi, K. , Mizusawa, E. , Yamakawa, M. , Ashida, M. , Matsunaga, T. M. , Azuma, T. , Uchiyama, S. , & Fukui, K. (2012). RBMX: A Regulator for Maintenance and Centromeric Protection of Sister Chromatid Cohesion. Cell Reports, 1(4), 299–308. [DOI] [PubMed] [Google Scholar]
- Matsuo, H. (2004). Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science, 303(5657), 531–534. [DOI] [PubMed] [Google Scholar]
- Mccloskey, A. , Taniguchi, I. , Shinmyozu, K. , & Ohno, M. (2012). Length to classify RNA polymerase II transcripts for export. Science, 335, 1643–1646. [DOI] [PubMed] [Google Scholar]
- Mcdaniel, K. , Wu, N. , Zhou, T. , Huang, L. , Sato, K. , Venter, J. , Ceci, L. , Chen, D. , Ramos‐Lorenzo, S. , Invernizzi, P. , Bernuzzi, F. , Wu, C. , Francis, H. , Glaser, S. , Alpini, G. , & Meng, F. (2019). Amelioration of ductular reaction by stem cell derived extracellular vesicles in MDR2 knockout mice via lethal‐7 microRNA. Hepatology, 69(6), 2562–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mckenzie, A. J. , Hoshino, D. , Hong, N. H. , Cha, D. J. , Franklin, J. L. , Coffey, R. J. , Patton, J. G. , & Weaver, A. M. (2016). KRAS‐MEK signaling controls Ago2 sorting into exosomes. Cell reports, 15(5), 978–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G. (2013). Argonaute proteins: functional insights and emerging roles. Nature Reviews Genetics, 14(7), 447–459. [DOI] [PubMed] [Google Scholar]
- Mickleburgh, I. , Burtle, B. , Hollås, H. , Campbell, G. , Chrzanowska‐Lightowlers, Z. , Vedeler, A. , & Hesketh, J. (2005). Annexin A2 binds to the localization signal in the 3′ untranslated region of c‐myc mRNA. The Febs Journal, 272(2), 413–421. [DOI] [PubMed] [Google Scholar]
- Minaguchi, T. , Waite, K. A. , & Eng, C. (2006). Nuclear localization of PTEN is regulated by Ca2+ through a tyrosil phosphorylation‐independent conformational modification in major vault protein. Cancer Research, 66(24), 11677–11682. [DOI] [PubMed] [Google Scholar]
- Missotten, M. , Nichols, A. , Rieger, K. , & Sadoul, R. (1999). Alix, a novel mouse protein undergoing calcium‐dependent interaction with the apoptosis‐linked‐gene 2 (ALG‐2) protein. Cell Death and Differentiation, 6(2), 124–129. [DOI] [PubMed] [Google Scholar]
- Miura, N. , Nakamura, H. , Sato, R. , Tsukamoto, T. , Harada, T. , Takahashi, S. , Adachi, Y. , Shomori, K. , Sano, A. , Kishimoto, Y. , Ito, H. , Hasegawa, J. , & Shiota, G. (2006). Clinical usefulness of serum telomerase reverse transcriptase (hTERT) mRNA and epidermal growth factor receptor (EGFR) mRNA as a novel tumor marker for lung cancer. Cancer Science, 97(12), 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, N. , Shiota, G. , Nakagawa, T. , Maeda, Y. , Sano, A. , Marumoto, A. , Kishimoto, Y. , Murawaki, Y. , & Hasegawa, J. (2003). Sensitive detection of human telomerase reverse transcriptase mRNA in the serum of patients with hepatocellular carcinoma. Oncology, 64(4), 430–434. [DOI] [PubMed] [Google Scholar]
- Monastyrskaya, K. (2018). Functional association between regulatory RNAs and the annexins. International Journal of Molecular Sciences, 19(2), 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran‐Jones, K. , Wayman, L. , Kennedy, D. D. , Reddel, R. R. , Sara, S. , Snee, M. J. , & Smith, R. (2005). hnRNP A2, a potential ssDNA/RNA molecular adapter at the telomere. Nucleic Acids Research, 33(2), 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, E. , & Gruenberg, J. (2007). The p11/S100A10 light chain of annexin A2 is dispensable for annexin A2 association to endosomes and functions in endosomal transport. Plos One, 2(10), e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel, E. , & Gruenberg, J. (2009). Annexin A2 Binding to endosomes and functions in endosomal transport are regulated by tyrosine 23 phosphorylation. Journal of Biological Chemistry, 284(3), 1604–1611. [DOI] [PubMed] [Google Scholar]
- Morita, E. , & Sundquist, W. I. (2004). Retrovirus budding. Annual Review of Cell and Developmental Biology, 20(1), 395–425. [DOI] [PubMed] [Google Scholar]
- Morlando, M. , Dini Modigliani, S. , Torrelli, G. , Rosa, A. , Di Carlo, V. , Caffarelli, E. , & Bozzoni, I. (2012). FUS stimulates microRNA biogenesis by facilitating co‐transcriptional Drosha recruitment. Embo Journal, 31(24), 4502–4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, K. , Ghoshal, B. , Ghosh, S. , Chakrabarty, Y. , Shwetha, S. , Das, S. , & Bhattacharyya, S. N. (2016). Reversible HuR‐micro RNA binding controls extracellular export of miR‐122 and augments stress response . Embo Reports, 17(8), 1184–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, M. , Fazi, F. , & Ciaudo, C. (2020). Argonaute proteins: from structure to function in development and pathological cell fate determination. Frontiers in Cell and Developmental Biology, 7:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munschauer, M. , Nguyen, C. T. , Sirokman, K. , Hartigan, C. R. , Hogstrom, L. , Engreitz, J. M. , Ulirsch, J. C. , Fulco, C. P. , Subramanian, V. , Chen, J. , Schenone, M. , Guttman, M. , Carr, S. A. , & Lander, E. S. (2018). The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature, 561(7721), 132–136. [DOI] [PubMed] [Google Scholar]
- Nabet, B. Y. , Qiu, Y. , Shabason, J. E. , Wu, T. J. , Yoon, T. , Kim, B. C. , Benci, J. L. , Demichele, A. M. , Tchou, J. , Marcotrigiano, J. , & Minn, A. J. (2017). Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell, 170(2), 352–366.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, M.‐A. , Karunakaran, D. , Geoffrion, M. , Cheng, H S. , Tandoc, K. , Perisic Matic, L. , Hedin, U. , Maegdefessel, L. , Fish, J E. , Rayner, K J. Extracellular vesicles secreted by atherogenic macrophages transfer microRNA to inhibit cell migration. Arteriosclerosis, Thrombosis, and Vascular Biology. 2018;38(1):49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, F. C. , Nielsen, J. , Kristensen, M. A. , Koch, G. , & Christiansen, J. (2002). Cytoplasmic trafficking of IGF‐II mRNA‐binding protein by conserved KH domains. Journal of Cell Science, 115(10), 2087–2097. [DOI] [PubMed] [Google Scholar]
- Nielsen, J. (2004). Sequential dimerization of human zipcode‐binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Research, 32(14), 4368–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, J. , Christiansen, J. , Lykke‐Andersen, J. , Johnsen, A. H. , Wewer, U. M. , & Nielsen, F. C. (1999). A family of insulin‐like growth factor II mRNA‐binding proteins represses translation in late development. Molecular and Cellular Biology, 19(2), 1262–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G. (2006). The multiple personalities of Alix. Journal of Cell Science, 119(15), 3025–3032. [DOI] [PubMed] [Google Scholar]
- Orrick, L. R. , Olson, M. O. J. , & Busch, H. (1973). Comparison of nucleolar proteins of normal rat liver and Novikoff hepatoma ascites cells by two dimensional polyacrylamide gel electrophoresis. Proceedings of the National Academy of Sciences of the United States of America, 70(5), 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palrasu, M. , Knapinska, A. M. , Diez, J. , Smith, L. , LaVoi, T. , Giulianotti, M. , Houghten, R. A. , Fields, G. B. , & Minond, D. (2019). A novel probe for spliceosomal proteins that induces autophagy and death of melanoma cells reveals new targets for melanoma drug discovery. Cellular Physiology and Biochemistry, 53(4), 656–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, S. , Wang, R. , Zhou, X. , Corvera, J. , Kloc, M. , Sifers, R. , Gallick, G. E. , Lin, S.‐H. , & Kuang, J. (2008). Extracellular Alix regulates integrin‐mediated cell adhesions and extracellular matrix assembly. Embo Journal, 27(15), 2077–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, Y. M. , Hwang, S. J. , Masuda, K. , Choi, K.‐M. , Jeong, M.‐R. , Nam, D.‐H. , Gorospe, M. , & Kim, H. H. (2012). Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Molecular and Cellular Biology, 32(20), 4237–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, G. P. (2005). The autoregulatory translational control element of poly(A)‐binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Research, 33(22), 7074–7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado, H. , Alečković, M. , Lavotshkin, S. , Matei, I. , Costa‐Silva, B. , Moreno‐Bueno, G. , Hergueta‐Redondo, M. , Williams, C. , García‐Santos, G. , Ghajar, C. M. , Nitadori‐Hoshino, A. , Hoffman, C. , Badal, K. , Garcia, B. A. , Callahan, M. K. , Yuan, J. , Martins, V. R. , Skog, J. , Kaplan, R. N. … Lyden, D. (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nature Medicine, 18(6), 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez‐Boza, J. , Boeckx, A. , Lion, M. , Dequiedt, F. , & Struman, I. (2020). hnRNPA2B1 inhibits the exosomal export of miR‐503 in endothelial cells. Cellular and Molecular Life Sciences. 77(21), 4413–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, D. T. , Fung, H. K. H. , Levdikov, V. M. , Irmscher, T. , Warrander, F. C. , Greive, S. J. , Kovalevskiy, O. , Isaacs, H. V. , Coles, M. , & Antson, A. A. (2016). Human Lin28 forms a high‐affinity 1:1 complex with the 106∼363 cluster miRNA miR‐363. Biochemistry, 55(36), 5021–5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichiorri, F. , Palmieri, D. , De Luca, L. , Consiglio, J. , You, J. , Rocci, A. , Talabere, T. , Piovan, C. , Lagana, A. , Cascione, L. , Guan, J. , Gasparini, P. , Balatti, V. , Nuovo, G. , Coppola, V. , Hofmeister, C. C. , Marcucci, G. , Byrd, J C. , Volinia, S. … Croce, C M. (2013). In vivo NCL targeting affects breast cancer aggressiveness through miRNA regulation. Journal of Experimental Medicine, 210(5), 951–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pienimaeki‐Roemer, A. , Konovalova, T. , Musri, M. M. , Sigruener, A. , Boettcher, A. , Meister, G. , & Schmitz, G. (2017). Transcriptomic profiling of platelet senescence and platelet extracellular vesicles. Transfusion, 57(1), 144–156. [DOI] [PubMed] [Google Scholar]
- Piskounova, E. , Polytarchou, C. , Thornton, J. E. , Lapierre, R. J. , Pothoulakis, C. , Hagan, J. P. , Iliopoulos, D. , & Gregory, R. I. (2011). Lin28A and Lin28B inhibit let‐7 MicroRNA biogenesis by distinct mechanisms. Cell, 147(5), 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platania, C. B. M. , Pittalà, V. , Pascale, A. , Marchesi, N. , Anfuso, C. D. , Lupo, G. , Cristaldi, M. , Olivieri, M. , Lazzara, F. , Di Paola, L. , Drago, F. , & Bucolo, C. (2020). Novel indole derivatives targeting HuR‐mRNA complex to counteract high glucose damage in retinal endothelial cells. Biochemical Pharmacology, 175, 113908. [DOI] [PubMed] [Google Scholar]
- Politz, J. C. , Yarovoi, S. , Kilroy, S. M. , Gowda, K. , Zwieb, C. , & Pederson, T. (2000). Signal recognition particle components in the nucleolus. Proceedings of the National Academy of Sciences of the United States of America, 97(1), 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner, J. R. , & Chinnaiyan, A. M. (2011). The emergence of lncRNAs in cancer biology. Cancer discovery, 1(5), 391–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Chen, X. , Bonnefoi, H. , Pelte, M. F. , Lyautey, J. , Lederrey, C. , Movarekhi, S. , Schaeffer, P. , Mulcahy, H. E. , Meyer, P. , Stroun, M. , & Anker, P. (2000). Telomerase RNA as a detection marker in the serum of breast cancer patients. Clinical Cancer Research, 6(10), 3823–3826. [PubMed] [Google Scholar]
- Radeghieri, A. , Savio, G. , Zendrini, A. , Di Noto, G. , Salvi, A. , Bergese, P. , & Piovani, G. (2017). Cultured human amniocytes express hTERT, which is distributed between nucleus and cytoplasm and is secreted in extracellular vesicles. Biochemical and Biophysical Research Communications, 483(1), 706–711. [DOI] [PubMed] [Google Scholar]
- Rauen, T. , Raffetseder, U. , Frye, B. C. , Djudjaj, S. , Mühlenberg, P. J. T. , Eitner, F. , Lendahl, U. , Bernhagen, J. , Dooley, S. , & Mertens, P. R. (2009). YB‐1 acts as a ligand for notch‐3 receptors and modulates receptor activation. Journal of Biological Chemistry, 284(39), 26928–26940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech, J. E. , Lestourgeon, W. M. , & Flicker, P. F. (1995). Ultrastructural morphology of the hnRNP C protein tetramer. Journal of Structural Biology, 114(2), 77–83. [DOI] [PubMed] [Google Scholar]
- Reddy, T. R. K. , Li, C. , Fischer, P. M. , & Dekker, L. V. (2012). Three‐dimensional pharmacophore design and biochemical screening identifies substituted 1,2,4‐Triazoles as inhibitors of the AnnexinA2‐S100A10 protein interaction. Chemmedchem, 7(8), 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes, C. L. , Rutenber, E. , Walter, P. , & Stroud, R. M. (2007). X‐ray structures of the signal recognition particle receptor reveal targeting cycle intermediates. Plos One, 2(7), e607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römisch, K. , Miller, F. , Dobberstein, B. , & High, S. (2006). Human autoantibodies against the 54 kDa protein of the signal recognition particle block function at multiple stages. Arthritis Research & Therapy, 8(2), R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos, M. , Pradère, U. , Ngondo, R. P. , Behera, A. , Allegrini, S. , Civenni, G. , Zagalak, J. A. , Marchand, J.‐R. , Menzi, M. , Towbin, H. , Scheuermann, J. , Neri, D. , Caflisch, A. , Catapano, C. V. , Ciaudo, C. , & Hall, J. (2016). A Small‐Molecule Inhibitor of Lin28. Acs Chemical Biology, 11(10), 2773–2781. [DOI] [PubMed] [Google Scholar]
- Rosenfeld, Y. B.‐.Z. , Krumbein, M. , Yeffet, A. , Schiffmann, N. , Mishalian, I. , Pikarsky, E. , Oberman, F. , Fridlender, Z. , & Yisraeli, J. K. (2019). VICKZ1 enhances tumor progression and metastasis in lung adenocarcinomas in mice. Oncogene, 38(21), 4169–4181. [DOI] [PubMed] [Google Scholar]
- Rosengarth, A. , & Luecke, H. (2004). Annexin A2: Does it induce membrane aggregation by a new multimeric state of the protein? Annexins, 1, 129–136, https://www.rcsb.org/structure/1XJL Published Accessed 1 May 2020. [Google Scholar]
- Rosengarth, A. , Rösgen, J. , Hinz, H.‐J. , & Gerke, V. (2001). Folding energetics of ligand binding proteins II. Cooperative binding of Ca2+ to Annexin I. Journal of Molecular Biology, 306(4), 825–835. [DOI] [PubMed] [Google Scholar]
- Ross, A. F. , Oleynikov, Y. , Kislauskis, E. H. , Taneja, K. L. , & Singer, R. H. (1997). Characterization of a beta‐actin mRNA zipcode‐binding protein. Molecular and Cellular Biology, 17(4), 2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossoll, W. , Kröning, A.‐K. , Ohndorf, U.‐M. , Steegborn, C. , Jablonka, S. , & Sendtner, M. (2002). Specific Interaction of Smn, the Spinal Muscular Atrophy Determining Gene Product, with HnRNP‐R and Gry‐Rbp/HnRNP‐Q: A Role for Smn in RNA Processing in Motor Axons? Human Molecular Genetics, 11(1), 93–105. [DOI] [PubMed] [Google Scholar]
- Rouda, S. , & Skordalakes, E. (2007). Structure of the RNA‐binding domain of telomerase: implications for RNA recognition and binding. Structure (London, England), 15(11), 1403–1412. [DOI] [PubMed] [Google Scholar]
- Ruden, M. , & Puri, N. (2013). Novel anticancer therapeutics targeting telomerase. Cancer Treatment Reviews, 39(5), 444–456. [DOI] [PubMed] [Google Scholar]
- Runge, S. , Nielsen, F. C. , Nielsen, J. , Lykke‐Andersen, J. , Wewer, U. M. , & Christiansen, J. (2000). H19 RNA binds four molecules of insulin‐like growth factor II mRNA‐binding protein. Journal of Biological Chemistry, 275(38), 29562–29569. [DOI] [PubMed] [Google Scholar]
- Ryder, S. P. (2004). Specificity of the STAR/GSG domain protein Qk1: Implications for the regulation of myelination. Rna, 10(9), 1449–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, S. J. , An, H. J. , Oh, Y. S. , Choi, H. R. , Ha, M. K. , & Park, S. C. (2008). On the role of major vault protein in the resistance of senescent human diploid fibroblasts to apoptosis. Cell Death and Differentiation, 15, 1673–1680. [DOI] [PubMed] [Google Scholar]
- Salmena, L. , Poliseno, L. , Tay, Y. , Kats, L. , & Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3), 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti, A. , Couté, Y. , Epstein, A. , Arata, L. , Kraut, A. , Navratil, V. , Bouvet, P. , & Greco, A. (2016). Nuclear functions of nucleolin through global proteomics and interactomic approaches. Journal of Proteome Research, 15(5), 1659–1669. [DOI] [PubMed] [Google Scholar]
- Santangelo, L. , Giurato, G. , Cicchini, C. , Montaldo, C. , Mancone, C. , Tarallo, R. , Battistelli, C. , Alonzi, T. , Weisz, A. , & Tripodi, M. (2016). The RNA‐binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell reports, 17(3), 799–808. [DOI] [PubMed] [Google Scholar]
- Santos, J. H. , Meyer, J. N. , & Van Houten, B. (2006). Mitochondrial localization of telomerase as a determinant for hydrogen peroxide‐induced mitochondrial DNA damage and apoptosis. Human Molecular Genetics, 15(11), 1757–1768. [DOI] [PubMed] [Google Scholar]
- Scheffer, G. L. , Schroeijers, A. B. , Izquierdo, M. A. , Wiemer, E. A.C. , & Scheper, R. J. (2000). Lung resistance‐related protein/major vault protein and vaults in multidrug‐resistant cancer. Current Opinion in Oncology, 12(6), 550–556. [DOI] [PubMed] [Google Scholar]
- Scheffer, L. L. , Sreetama, S. C. , Sharma, N. , Medikayala, S. , Brown, K. J. , Defour, A. , & Jaiswal, J. K. (2014). Mechanism of Ca2+‐triggered ESCRT assembly and regulation of cell membrane repair. Nature Communications, 5, 5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, M. F. , Korb, O. , & Abell, C. (2013). MicroRNA‐specific argonaute 2 protein inhibitors. Acs Chemical Biology, 8(10), 2122–2126. [DOI] [PubMed] [Google Scholar]
- Schug, J. , Mckenna, L. B. , Walton, G. , Hand, N. , Mukherjee, S. , Essuman, K. , Shi, Z. , Gao, Y. , Markley, K. , Nakagawa, M. , Kameswaran, V. , Vourekas, A. , Friedman, J. R. , Kaestner, K. H. , & Greenbaum, L. E. (2013). Dynamic recruitment of microRNAs to their mRNA targets in the regenerating liver. Bmc Genomics [Electronic Resource], 14(1), 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C. W. , Preet, R. , Dhir, T. , Dixon, D. A. , & Brody, J. R. (2020). Understanding and targeting the disease‐related RNA binding protein human antigen R (HuR). WIREs RNA, 11(3), e1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seimiya, H. (2000). Involvement of 14‐3‐3 proteins in nuclear localization of telomerase. Embo Journal, 19(11), 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, C. , Zhang, F. , Kemp, M. M. , Linhardt, R. J. , Waisman, D. M. , Head, J. F. , & Seaton, B. A. (2006). Crystallographic analysis of calcium‐dependent heparin binding to annexin A2. Journal of Biological Chemistry, 281(42), 31689–31695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, N. K. , Reyes, A. , Green, P. , Caron, M. J. , Bonini, M. G. , Gordon, D. M. , Holt, I. J. , & Santos, J. H. (2012). Human telomerase acts as a hTR‐independent reverse transcriptase in mitochondria. Nucleic Acids Research, 40(2), 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty, P. K. , Thamake, S. I. , Biswas, S. , Johansson, S. L. , & Vishwanatha, J. K. (2012). Reciprocal Regulation of Annexin A2 and EGFR with Her‐2 in Her‐2 Negative and Herceptin‐Resistant Breast Cancer. Plos One, 7(9), e44299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, R. , Yu, X. , Wang, Y. , Sun, J. , Sun, Q. , Xia, W. , Dong, G. , Wang, A. , Gao, Z. , Jiang, F. , & Xu, L. (2017). Expression profile, clinical significance, and biological function of insulin‐like growth factor 2 messenger RNA‐binding proteins in non–small cell lung cancer. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 39(4), 101042831769592. [DOI] [PubMed] [Google Scholar]
- Shi, Y. , Wang, Z. , Zhu, X. , Chen, L. , Ma, Y. , Wang, J. , Yang, X. , & Liu, Z. (2020). Exosomal miR‐1246 in serum as a potential biomarker for early diagnosis of gastric cancer. International Journal of Clinical Oncology, 25(1), 89–99. [DOI] [PubMed] [Google Scholar]
- Shiozawa, Y. , Havens, A. M. , Jung, Y. , Ziegler, A. M. , Pedersen, E. A. , Wang, J. , Wang, J. , Lu, G. , Roodman, G. D. , Loberg, R. D. , Pienta, K. J. , & Taichman, R. S. (2008). Annexin II/annexin II receptor axis regulates adhesion, migration, homing, and growth of prostate cancer. Journal of Cellular Biochemistry, 105(2), 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, B. , Zeng, P. , Kang, S. , Li, P.‐H. , Hu, D. , Kuang, G. , Cao, J. , Li, X. , Zhang, M. , An, L.‐K. , Huang, Z.‐S. , Li, D. (2019). Syntheses and evaluation of new Quinoline derivatives for inhibition of hnRNP K in regulating oncogene c‐myc transcription. Bioorganic Chemistry, 85, 1–17. [DOI] [PubMed] [Google Scholar]
- Shurtleff, M. J. , Yao, J. , Qin, Y. , Nottingham, R. M. , Temoche‐Diaz, M. M. , Schekman, R. , & Lambowitz, A. M. (2017). Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proceedings of the National Academy of Sciences of the United States of America, 114(43), E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff, M. J. , Temoche‐Diaz, M. M. , Karfilis, K. V. , Ri, S. , & Schekman, R. (2016). Y‐box protein 1 is required to sort microRNAs into exosomes in cells and in a cell‐free reaction. Elife, 5:e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone, L. E. , & Keene, J. D. (2013). Mechanisms coordinating ELAV/Hu mRNA regulons. Current Opinion in Genetics & Development, 23(1), 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog, J. , Würdinger, T. , Van Rijn, S. , Meijer, D. H. , Gainche, L. , Curry, W. T. , Carter, B. S. , Krichevsky, A. M. , & Breakefield, X. O. (2008). Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology, 10(12), 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogberg, G. , Gudmundsdottir, J. , Van Der Post, S. , Sandström, K. , Bruhn, S. , Benson, M. , Mincheva‐Nilsson, L. , Baranov, V. , Telemo, E. , & Ekwall, O. (2013). Characterization of human thymic exosomes. Stoddart CA, ed. Plos One, 8(7), e67554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sork, H. , Corso, G. , Krjutskov, K. , Johansson, H. J. , Nordin, J. Z. , Wiklander, O. P. B. , Lee, Y. X. F. , Westholm, J. O. , Lehtiö, J. , Wood, M. J. A. , Mäger, I. , & El Andaloussi, S. (2018). Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Scientific Reports, 8(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung, N.‐K. , Kim, H.‐M. , Asami, Y. , Kim, D. H. , Cho, Y. , Naik, R. , Jang, Y. , Jang, K. , Han, H. J. , Ganipisetti, S. R. , Cha‐Molstad, H. , Hwang, J. , Lee, K. H.o , Ko, S.‐K. , Jang, J.‐H. , Ryoo, I.‐J. , Kwon, Y. T. , Lee, K. S. , Osada, H. … Ahn, J. S. (2019). Mechanism of the natural product moracin‐O derived MO‐460 and its targeting protein hnRNPA2B1 on HIF‐1α inhibition. Experimental & Molecular Medicine, 51(2), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproviero, D. , La Salvia, S. , Giannini, M. , Crippa, V. , Gagliardi, S. , Bernuzzi, S. , Diamanti, L. , Ceroni, M. , Pansarasa, O. , Poletti, A. , & Cereda, C. (2018). Pathological proteins are transported by extracellular vesicles of sporadic amyotrophic lateral sclerosis patients. Front Neurosci, 12, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello, L. , Maugeri, M. , Garre, E. , Nawaz, M. , Wahlgren, J. , Papadimitriou, A. , Lundqvist, C. , Lindfors, L. , Collén, A. , Sunnerhagen, P. , Ragusa, M. , Purrello, M. , Di Pietro, C. , Tigue, N. , & Valadi, H. (2018). Identification of RNA‐binding proteins in exosomes capable of interacting with different types of RNA: RBP‐facilitated transport of RNAs into exosomes. Plos One, 13(4), e0195969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StöHr, N. , Lederer, M. , Reinke, C. , Meyer, S. , Hatzfeld, M. , Singer, R. H. , & HüTtelmaier, S. (2006). ZBP1 regulates mRNA stability during cellular stress. Journal of Cell Biology, 175(4), 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, D. M. , Wehr, N. B. , Fergusson, M. M. , Levine, R. L. , & Finkel, T. (2000). Identification of oxidant‐sensitive proteins: TNF‐α induces protein glutathiolation. Biochemistry, 39(36), 11121–11128. [DOI] [PubMed] [Google Scholar]
- Sun, Y. L. , Liu, F. , Liu, F. , & Zhao, X. H. (2016). Protein and gene expression characteristics of heterogeneous nuclear ribonucleoprotein H1 in esophageal squamous cell carcinoma. World Journal of Gastroenterology: Wjg, 22(32), 7322–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh, P. S. , Tsutsumi, R. , & Venkatesh, T. (2018). YBX1 at the crossroads of non‐coding transcriptome, exosomal, and cytoplasmic granular signaling. European Journal of Cell Biology, 97(3), 163–167. [DOI] [PubMed] [Google Scholar]
- Tang, F. , Li, W. , Chen, Y. , Wang, D. , Han, J. , & Liu, D. (2014). Downregulation of hnRNP K by RNAi inhibits growth of human lung carcinoma cells. Oncology Letters, 7(4), 1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, Y. , Ren, Y. , Hu, X. , Mu, J. , Samykutty, A. , Zhuang, X. , Deng, Z. , Kumar, A. , Zhang, L. , Merchant, M. L. , Yan, J. , Miller, D. M. , & Zhang, H.‐G. (2017). MVP‐mediated exosomal sorting of miR‐193a promotes colon cancer progression. Nature Communications, 8, 14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova, M. , Hafner, M. , Teplov, D. , Essig, K. , Tuschl, T. , & Patel, D. J. (2013). Structure‐function studies of STAR family quaking proteins bound to their in vivo RNA target sites. Genes & Development, 27(8), 928–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrin, L. , Rampazzo, E. , Pucciarelli, S. , Agostini, M. , Bertorelle, R. , Esposito, G. , Delbianco, P. , Nitti, D. , & De Rossi, A. (2008). Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: Implications for monitoring of neoplastic disease. Clinical Cancer Research, 14(22), 7444–7451. [DOI] [PubMed] [Google Scholar]
- Thiel, C. , Weber, K. , & Gerke, V. (1991). Characterization of a Ca2+‐binding site in human annexin II by site‐directed mutagenesis. Journal of Biological Chemistry, 266(22), 14732–14739. https://www.ncbi.nlm.nih.gov/pubmed/1830590 Accessed 30 April 2020. [PubMed] [Google Scholar]
- Trevarton, A. , Zhou, Y. , Yang, D. , Rewcastle, G. W. , Flanagan, J. U. , Braithwaite, A. , Shepherd, P. R. , Print, C. G. , Wang, M.‐W. , & Lasham, A. (2019). Orthogonal assays for the identification of inhibitors of the single‐stranded nucleic acid binding protein YB‐1. Acta Pharmaceutica Sinica B, 9(5), 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trioulier, Y. , Torch, S. , Blot, B. , Cristina, N. , Chatellard‐Causse, C. , Verna, J.‐M. , & Sadoul, R. (2004). Alix, a protein regulating endosomal trafficking, is involved in neuronal death. Journal of Biological Chemistry, 279(3), 2046–2052. [DOI] [PubMed] [Google Scholar]
- Tsialikas, J. , & Romer‐Seibert, J. (2015). LIN28: roles and regulation in development and beyond. Development, 142(14), 2397–2404. [DOI] [PubMed] [Google Scholar]
- Turchinovich, A. , Weiz, L. , Langheinz, A. , & Burwinkel, B. (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Research, 39(16), 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugrinova, I. , Petrova, M. , Chalabi‐Dchar, M. , & Bouvet, P. (2018). Multifaceted Nucleolin Protein and Its Molecular Partners in Oncogenesis In: Advances in Protein Chemistry and Structural Biology. Vol 111 Academic Press Inc, 133–164. [DOI] [PubMed] [Google Scholar]
- Ustianenko, D. , Chiu, H.‐S. , Treiber, T. , Weyn‐Vanhentenryck, S. M. , Treiber, N. , Meister, G. , Sumazin, P. , & Zhang, C. (2018). LIN28 selectively modulates a subclass of Let‐7 microRNAs. Molecular Cell, 71(2), 271–283.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valapala, M. , & Vishwanatha, J. K. (2011). Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2. Journal of Biological Chemistry, 286(35), 30911–30925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zon, A. , Mossink, M. H. , Schoester, M. , Scheffer, G. L. , Scheper, R. J. , Sonneveld, P. , & Wiemer, E. A. C. (2001). Multiple human vault RNAs: expression and association with the vault complex. Journal of Biological Chemistry, 276(40), 37715–37721. [DOI] [PubMed] [Google Scholar]
- Van Zon, A. , Mossink, M. H. , Schoester, M. , Scheffer, G. L. , Scheper, R. J. , Sonneveld, P. , & Wiemer, E. A.C. (2002). Structural domains of vault proteins: a role for the coiled coil domain in vault assembly. Biochemical and Biophysical Research Communications, 291(3), 535–541. [DOI] [PubMed] [Google Scholar]
- Vanessa, L.‐R. , Cristina, P. R. X. , Diana, S. , Hugo, O. , Yehuda, G. A. , Raquel, T. L. , & M., H. V. (2019). ALIX protein analysis: Storage temperature may impair results. Journal of Molecular and Clinical Medicine, 2(2), 29. [Google Scholar]
- Vasaturo, M. , Cotugno, R. , Fiengo, L. , Vinegoni, C. , Dal Piaz, F. , & De Tommasi, N. (2018). The anti‐tumor diterpene oridonin is a direct inhibitor of Nucleolin in cancer cells. Scientific Reports, 8(1), 16735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedeler, A. , & Hollås, H. (2000). Annexin II is associated with mRNAs which may constitute a distinct subpopulation. Biochemical Journal, 348(3), 565–572. [PMC free article] [PubMed] [Google Scholar]
- Vedeler, A. , Hollas, H. , Kari Grindheim, A. , & Raddum, A. M. (2012). Multiple roles of Annexin A2 in post‐transcriptional regulation of gene expression. Current Protein & Peptide Science, 13(4), 401–412. [DOI] [PubMed] [Google Scholar]
- Venables, J. P. , Koh, C.‐S. , Froehlich, U. , Lapointe, E. , Couture, S. , Inkel, L. , Bramard, A. , Paquet, É R. , Watier, V. , Durand, M. , Lucier, J.‐F. , Gervais‐Bird, J. , Tremblay, K. , Prinos, P. , Klinck, R. , Elela, S. A. , & Chabot, B. (2008). Multiple and specific mRNA processing targets for the major human hnRNP proteins. Molecular and Cellular Biology, 28(19), 6033–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venereo‐Sánchez, A. , Fulton, K. , Koczka, K. , Twine, S. , Chahal, P. , Ansorge, S. , Gilbert, R. , Henry, O. , & Kamen, A. (2019). Characterization of influenza H1N1 Gag virus‐like particles and extracellular vesicles co‐produced in HEK‐293SF. Vaccine, 37(47), 7100–7107. [DOI] [PubMed] [Google Scholar]
- Vernet, C. , & Artzt, K. (1997). STAR, a gene family involved in signal transduction and activation of RNA. Trends in Genetics, 13(12), 479–484. [DOI] [PubMed] [Google Scholar]
- Vikesaa, J. , Hansen, T. V. O. , Jønson, L. , Borup, R. , Wewer, U. M. , Christiansen, J. , & Nielsen, F. C. (2006). RNA‐binding IMPs promote cell adhesion and invadopodia formation. Embo Journal, 25(7), 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella, F. , Moreno‐Moya, J. M. , Balaguer, N. , Grasso, A. , Herrero, M. , Martinez, S. , Marcilla, A. , & Simon, C. (2015). Hsa‐miR‐30d, secreted by the human endometrium, is taken up by the pre‐implantation embryo and might modify its transcriptome. Development, 142(18), 3210–3221. [DOI] [PubMed] [Google Scholar]
- Villarroya‐Beltri, C. , Gutiérrez‐Vázquez, C. , Sánchez‐Cabo, F. , Pérez‐Hernández, D. , Vázquez, J. , Martin‐Cofreces, N. , Martinez‐Herrera, D. J. , Pascual‐Montano, A. , Mittelbrunn, M. , & Sánchez‐Madrid, F. (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications, 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan, S. R. , Powers, J. T. , Einhorn, W. , Hoshida, Y. , Ng, T. L. , Toffanin, S. , O'sullivan, M. , Lu, J. , Phillips, L. A. , Lockhart, V. L. , Shah, S. P. , Tanwar, P. S. , Mermel, C. H. , Beroukhim, R. , Azam, M. , Teixeira, J. , Meyerson, M. , Hughes, T. P. , Llovet, J. M. … Daley, G. Q. (2009). Lin28 promotes transformation and is associated with advanced human malignancies. Nature Genetics, 41(7), 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P. , & Blobel, G. (1983). Disassembly and reconstitution of signal recognition particle. Cell, 34(2), 525–533. [DOI] [PubMed] [Google Scholar]
- Wang, F. , Yuan, Y. , Yang, P. , & Li, X. (2017). Extracellular vesicles‐mediated transfer of miR‐208a/b exaggerate hypoxia/reoxygenation injury in cardiomyocytes by reducing QKI expression. Molecular and Cellular Biochemistry, 431(1‐2), 187–195. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Zeng, F. , Liu, Q. , Liu, H. , Liu, Z. , Niu, L. , Teng, M. , & Li, X. (2013). The structure of the ARE‐binding domains of Hu antigen R (HuR) undergoes conformational changes during RNA binding. Acta Crystallographica Section D: Biological Crystallography, 69(3), 373–380. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Wu, J. , Li, X. , Liu, H. , Qin, J. , Bai, Z. , Chi, B. , & Chen, X. (2018). Identification and validation nucleolin as a target of curcumol in nasopharyngeal carcinoma cells. Journal of Proteomics, 182, 1–11. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Wen, M. , & Cao, X. (2019). Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science, 365(6454), eaav0758. [DOI] [PubMed] [Google Scholar]
- Wang, T. , Han, P. , He, Y. , Zhao, C. , Wang, G. , Yang, W. , Shan, M. , Zhu, Y. , Yang, C. , Weng, M. , Wu, D. , Gao, L. , Jin, X. , Wei, Y. , Cui, B. , Shen, G. , & Li, X. (2016). Lin28A enhances chemosensitivity of colon cancer cells to 5‐FU by promoting apoptosis in a let‐7 independent manner. Tumour Biology: The Journal of the International Society for Oncodevelopmental Biology and Medicine, 37(6), 7657–7665. [DOI] [PubMed] [Google Scholar]
- Wang, T. , He, Y. , Zhu, Y. , Chen, M. , Weng, M. , Yang, C. , Zhang, Y. , Ning, N. , Zhao, R. , Yang, W. , Jin, Y. , Li, J. , Redpath, R. J. R. E. , Zhang, L. , Jin, X. , Zhong, Z. , Zhang, F. , Wei, Y. , Shen, G. … Li, X. (2016). Comparison of the expression and function of Lin28A and Lin28B in colon cancer. Oncotarget, 7(48), 79605–79616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Cao, L. , Wang, Y. , Wang, X. , Liu, N. , & You, Y. (2012). Regulation of let‐7 and its target oncogenes (Review). Oncology letters, 3(5), 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. Q. , Zhang, F. , Tian, R. , Ji, W. , Zhou, Y. , Sun, X. M. , Liu, Y. , Wang, Z. Y. , & Niu, R. F. (2012). Tyrosine 23 phosphorylation of Annexin A2 promotes proliferation, invasion, and Stat3 phosphorylation in the nucleus of human breast cancer SK‐BR‐3 cells. Cancer Biology & Medicine, 9(4), 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Chen, X. , Tian, B. , Liu, J. , Yang, L. , Zeng, L. , Chen, T. , Hong, A. , & Wang, X. (2017). Nucleolin‐targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics, 7(5), 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y.‐S. , Li, H. , Li, Y. , Zhu, H. , & Jin, Y.‐H. (2018). Identification of natural compounds targeting Annexin A2 with an anti‐cancer effect. Protein Cell, 9(6), 568–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Deng, Z. , Dahmane, N. , Tsai, K. , Wang, P. , Williams, D. R. , Kossenkov, A. V. , Showe, L. C. , Zhang, R. , Huang, Q. , Conejo‐Garcia, J. R. , & Lieberman, P. M. (2015). Telomeric repeat‐containing RNA (TERRA) constitutes a nucleoprotein component of extracellular inflammatory exosomes. Proceedings of the National Academy of Sciences of the United States of America, 112(46), E6293–E6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Qiu, H. , He, J. , Liu, L. , Xue, W. , Fox, A. , Tickner, J. , & Xu, J. (2020). The emerging roles of hnRNPK. Journal of Cellular Physiology, 235(3), 1995–2008. [DOI] [PubMed] [Google Scholar]
- Weaver, A. M. , & Patton, J. G. (2020). Argonautes in extracellular vesicles: Artifact or selected cargo?. Cancer Research, 80(3), 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder, O. , Wild, K. , Strub, K. , & Cusack, S. (2000). Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature, 408(6809), 167–173. [DOI] [PubMed] [Google Scholar]
- Welton, J. L. , Khanna, S. , Giles, P. J. , Brennan, P. , Brewis, I. A. , Staffurth, J. , Mason, M. D. , & Clayton, A. (2010). Proteomics analysis of bladder cancer exosomes. Molecular and Cellular Proteomics, 9(6), 1324–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, R. J. (2011). Transcription by RNA polymerase III: More complex than we thought. Nature Reviews Genetics, 12(7), 459–463. [DOI] [PubMed] [Google Scholar]
- Wilbert, M. L. , Huelga, S. C. , Kapeli, K. , Stark, T. J. , Liang, T. Y. , Chen, S. X. , Yan, B. Y. , Nathanson, J. L. , Hutt, K. R. , Lovci, M. T. , Kazan, H. , Vu, A. Q. , Massirer, K. B. , Morris, Q. , Hoon, S. , & Yeo, G. W. (2012). LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Molecular Cell, 48(2), 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, K. R. , Mcaninch, D. S. , Stefanovic, S. , Xing, L. , Allen, M. , Li, W. , Feng, Y. , Mihailescu, M. R. , Bassell, G. J. (2016). HnRNP‐Q1 represses nascent axon growth in cortical neurons by inhibiting Gap‐43 mRNA translation. Molecular Biology of the Cell, 27(3), 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodham, A. W. , Taylor, J. R. , Jimenez, A. I. , Skeate, J. G. , Schmidt, T. , Brand, H. E. , Da Silva, D. M. , & Kast, W. M. (2014). Small molecule inhibitors of the annexin A2 heterotetramer prevent human papillomavirus type 16 infection. Journal of Antimicrobial Chemotherapy, 70(6), 1686–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, B. , Su, S. , Patil, D. P. , Liu, H. , Gan, J. , Jaffrey, S. R. , & Ma, J. (2018). Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nature communications, 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. I. , Reed, R. B. , Grabowski, P. J. , & Artzt, K. (2002). Function of quaking in myelination: Regulation of alternative splicing. Proceedings of the National Academy of Sciences of the United States of America, 99(7), 4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu, D. , Liu, L. , Qiao, F. , Yang, H. , Cui, L. , & Liu, G. (2016). Annexin A2 coordinates STAT3 to regulate the invasion and migration of colorectal cancer cells in vitro. Gastroenterology Research and Practice, 2016, 1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. , Xie, N. , Su, Y. , Sun, Z. , Liang, Y. , Zhang, N. , Liu, D. , Jia, S. , Xing, X. , Han, L. , Li, G. , Tong, T. , & Chen, J. (2020). HnRNP F/H associate with hTERC and telomerase holoenzyme to modulate telomerase function and promote cell proliferation. Cell Death and Differentiation, 27(6), 1998–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Dong, X. , Chen, Y. , & Wang, X. (2018). Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clinical Chemistry and Laboratory Medicine, 56(3), 479–484. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Greening, D. W. , Rai, A. , Ji, H. , & Simpson, R. J. (2015). Highly‐purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods (San Diego, Calif.), 87:11‐25. [DOI] [PubMed] [Google Scholar]
- Xu, Y. , Wu, W. , Han, Q. , Wang, Y. , Li, C. , Zhang, P. , & Xu, H. (2019). Post‐translational modification control of RNA‐binding protein hnRNPK function. Open Biol, 9(3), 180239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles, 4(2015), 1–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Wang, C. , Li, F. , Zhang, J. , Nayab, A. , Wu, J. , Shi, Y. , & Gong, Q. (2017). The human RNA‐binding protein and E3 ligase MEX‐3C binds the MEX‐3–recognition element (MRE) motif with high affinity. Journal of Biological Chemistry, 292(39), 16221–16234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, T.‐L. B. , Chen, Q. , Deng, J. T. , Jagannathan, G. , Tobias, J. W. , Schultz, D. C. , Wang, S. , Lengner, C. J. , Rustgi, A. K. , Lynch, J. P. , & Johnson, F. B. (2017). Mutual reinforcement between telomere capping and canonical Wnt signalling in the intestinal stem cell niche. Nature Communications, 8, 14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. , Jia, D. , Kim, H. , Abd Elmageed, Z. Y. , Datta, A. , Davis, R. , Srivastav, S. , Moroz, K. , Crawford, B. E. , Moparty, K. , Thomas, R. , Hudson, R. S. , Ambs, S. , Abdel‐Mageed, A. B. (2016). Dysregulation of miR‐212 promotes castration resistance through hnRNPH1‐mediated regulation of AR and AR‐V7: Implications for racial disparity of prostate cancer. Clinical Cancer Research, 22(7), 1744–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanshina, D. D. , Kossinova, O. A. , Gopanenko, A. V. , Krasheninina, O. A. , Malygin, A. A. , Venyaminova, A. G. , & Karpova, G. G. (2018). Structural features of the interaction of the 3′‐untranslated region of mRNA containing exosomal RNA‐specific motifs with YB‐1, a potential mediator of mRNA sorting. Biochimie, 144, 134–143. [DOI] [PubMed] [Google Scholar]
- Ye, Z. , Jin, H. , Qian, Q. (2015). Argonaute 2: A novel rising star in cancer research. Journal of Cancer, 6(9), 877–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, B. C. , Hong, S.‐H. , Ku, J.‐L. , Kim, Y.‐H. , Shin, Y.‐K. , Jang, S.‐G. , Kim, I.‐J. , Jeong, S.‐Y. , & Park, J.‐G. (2009). Galectin‐3 stabilizes heterogeneous nuclear ribonucleoprotein Q to maintain proliferation of human colon cancer cells. Cellular and Molecular Life Sciences, 66(2), 350–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. , Vodyanik, M. A. , Smuga‐Otto, K. , Antosiewicz‐Bourget, J. , Frane, J. L. , Tian, S. , Nie, J. , Jonsdottir, G. A. , Ruotti, V. , Stewart, R. , Slukvin, I. I. , & Thomson, J. A. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858), 1917–1920. [DOI] [PubMed] [Google Scholar]
- Yu, Z. , Zhao, S. , Ren, L. , Wang, L. , Chen, Z. , Hoffman, R. M. , & Zhou, J. (2017). Pancreatic cancer‐derived exosomes promote tumor metastasis and liver pre‐metastatic niche formation. Oncotarget, 8(38), 63461–63483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Li, S. , Li, L. , Li, M. , Guo, C. , Yao, J. , & Mi, S. (2015). Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genomics, Proteomics & Bioinformatics, 13(1), 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Feng, J. , Kong, S. , Wu, M. , Xi, Z. , Zhang, B. , Fu, W. , Lao, Y. , Tan, H. , & Xu, H. (2016). Nujiangexathone A, a novel compound from Garcinia nujiangensis, suppresses cervical cancer growth by targeting hnRNPK. Cancer Letters, 380(2), 447–456. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. (2003). Tyrosine phosphorylation of QKI mediates developmental signals to regulate mRNA metabolism. Embo Journal, 22(8), 1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L. , Mandler, M. D. , Yi, H. , & Feng, Y. (2010). Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin‐associated glycoprotein. Proceedings of the National Academy of Sciences of the United States of America, 107(44), 19061–19066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Li, F. , Zhang, X. , Liu, A. , Qi, J. , Cui, H. , & Zhao, P. (2015). MicroRNA‐194 acts as a prognostic marker and inhibits proliferation in hepatocellular carcinoma by targeting MAP4K4. International Journal of Clinical and Experimental Pathology, 8(10), 12446. [PMC free article] [PubMed] [Google Scholar]
- Zheng, Z. , Chen, M. , Xing, P. , Yan, X. , & Xie, B. (2019). Increased expression of exosomal AGAP2‐AS1 (AGAP2 antisense RNA 1) in breast cancer cells inhibits trastuzumab‐induced cell cytotoxicity. Medical Science Monitor, 25:2211–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucal, C. , D'agostino, V. , Loffredo, R. , Mantelli, B. , Natthakanthongon, Lal, P. , Latorre, E. , & Provenzani, A. (2015). Targeting the Multifaceted HuR Protein, Benefits and Caveats. Current Drug Targets, 16(5), 499–515. [DOI] [PubMed] [Google Scholar]


