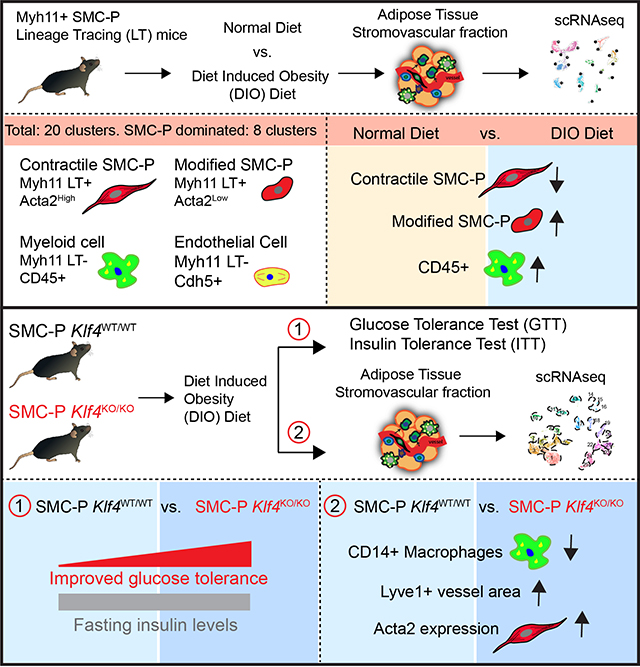
Abstract
Objective:
Smooth muscle cells and pericytes (SMC-P) display remarkable plasticity during injury and disease progression. Here, we tested the hypothesis that perivascular cells give rise to Klf4-dependent macrophage (MФ)-like cells that augment adipose tissue inflammation and metabolic dysfunction associated with diet-induced obesity (DIO).
Approach and Results:
Using Myh11-CreERT2eYFP mice and flow cytometry of the stromovascular fraction (SVF) of epididymal adipose tissue we observed a large fraction of SMC-P lineage traced eYFP+ cells expressing MФ markers. Subsequent single cell RNA sequencing, however, showed that the majority of these cells had no detectable eYFP transcript. Further exploration revealed that intraperitoneal injection of tamoxifen in peanut oil, used for generating conditional knockout or reporter mice in thousands of previous studies, resulted in large increase in the autofluorescence and false identification of MФs within epididymal adipose tissue as being eYFP+; and unintended pro-inflammatory consequences. Using newly generated Myh11-DreERT2tdTomato mice given oral tamoxifen we virtually eliminated the problem with autofluorescence and identified eight perivascular cell dominated clusters, half of which were altered upon DIO. Given that perivascular cell KLF4 can have beneficial or detrimental effects, we tested its role in obesity associated adipose tissue inflammation. While SMC-P-specific Klf4 knockout (SMC-P Klf4Δ/Δ) mice were not protected from DIO, they displayed improved glucose tolerance upon DIO, and showed marked decreases in pro-inflammatory MФs and increases in LYVE1+ lymphatic endothelial cells in the epididymal adipose tissue.
Conclusions:
Perivascular cells within the adipose tissue microvasculature dynamically respond to DIO and modulate tissue inflammation and metabolism in a KLF4-dependent manner.
Graphical Abstract
INTRODUCTION
Vascular smooth muscle cells and pericytes (SMC-P or perivascular cells) are not terminally differentiated but instead display remarkable plasticity in response to injury, atherosclerosis, or angiogenic stimuli1–3. Using an SMC-P specific lineage tracing mouse model, Myh11-CreERT2eYFP4,5, in which administration of tamoxifen leads to expression of eYFP exclusively in cells that express smooth muscle myosin heavy chain 11 (Myh11), our group and others demonstrated that the majority of SMC-P derived cells within advanced atherosclerotic lesions lack expression of typical SMC markers such as ACTA25, and activate markers of mesenchymal stem cells, myofibroblasts, presumptive macrophages (MФs)5,6, and osteogenic phenotypes5–9. Furthermore, recent studies by our laboratory, using combined SMC-P lineage tracing and knockout (KO) mouse models, showed that some of these transitions were regulated by the stem cell pluripotency genes Klf45 and Oct47. SMC-P-specific loss of Klf4 resulted in multiple beneficial effects in atherosclerotic lesions including formation of smaller lesions with fewer SMC-P-derived LGALS3+ cells and a thicker fibrous cap5. Since the clinical consequences of atherosclerosis, including myocardial infarction and stroke, typically occur well after our reproductive years, we hypothesized that KLF4 and OCT4 likely have beneficial roles in perivascular cells that are critical for survival and reproduction and thus are evolutionarily conserved. Consistent with this, we demonstrated that KLF4 plays a key role in maintenance of perivascular coverage and appropriate permeability properties of terminal arterioles10. Moreover, our laboratory showed that SMC-P-specific expression of Oct4 was required for perivascular cell investment of newly formed endothelial tubes and formation of functional vascular networks following injury- or ischemia-induced angiogenesis11. Taken together, the preceding studies provide compelling evidence that perivascular cells retain extensive plasticity that is normally beneficial7,10,11 but can become maladaptive in the context of disease2.
Obesity is associated with chronic adipose tissue inflammation, insulin resistance, type 2 diabetes, as well as development and exacerbation of cardiovascular disease12–14. There is evidence that perivascular cells play a key role in regulating adipose tissue inflammation in response to DIO15,16. For example, smooth muscle selective KO of IKKβ using a SM22-Cre IKKβ Flox mouse model prevents DIO and associated metabolic complications at least in part by preventing adipocyte precursor cell differentiation15. Unfortunately, the SM22-Cre mouse model system would have also resulted in KO of IKKβ in cardiomyocytes, adipocytes, and fibroblasts17 thus making it unclear if the striking metabolic improvements observed were due to the loss of pro-inflammatory signaling exclusively in perivascular cells. Another group profiled PDGFRβ lineage traced cells using scRNAseq of adipose tissue SVF cells of young lean mice18. They found that, PDGFRβ+ cells gave rise to fibro-inflammatory and adipogenic phenotypes18. However, PDGFRβ is not only expressed in perivascular cells, but also many other cells including fibroblasts19, myofibroblasts, MФs20, and endothelial cells21 that have undergone endothelial to mesenchymal transition (EndoMT)22. Moreover, using a Myh11-CreERT2 mouse model, Long et al. demonstrated that SMC-P derived cells give rise to beige adipocytes as a beneficial adaptive response to cold stress23. Taken together, these data indicate that perivascular cells may exhibit diverse phenotypes or cell transitions within adipose tissue. However, the specific nature of changes in perivascular phenotypes and how these modify obesity-induced adipose tissue inflammation and metabolic dysfunction remain poorly understood.
Activation of Galectin-3 (LGALS3 or MAC2) in perivascular cells was originally thought to reflect transition of these cells to a MФ-like state5,6 However, recent scRNAseq studies in combination with SMC-lineage tracing from our group24 and Thomas Quertermous’ group25 showed that LGALS3 activation did not mark a terminal differentiation pathway towards a MФ phenotype, but rather a transition state from which cells can undergo multiple phenotypic changes that can be either beneficial or detrimental. The Quertermous group identified a single atheroprotective fibroblast-like SMC phenotype, “fibromyocytes”. However, these scRNAseq analyses were performed on whole aortic root segments, including medial and adventitial layers, which likely resulted in limited sensitivity in detecting SMC-P-derived MФ-like cells within lesions. Moreover, results claiming that SMC give rise to a single beneficial lesion phenotype are incompatible with numerous studies from our5 and many other laboratories6,9,26–28. Our recent scRNAseq studies using micro-dissected atherosclerotic lesion samples, identified 4 clusters of Myh11 negative SMC-derived lesion cells, including inflammatory/immune-regulatory, extracellular matrix-rich similar to “fibromyocytes”, and osteogenic clusters, demonstrating considerable phenotypic plasticity of SMC within lesions. In addition, we also observed some SMC-derived cells expressing classical МФ markers, although their number was lower than we expected based on our previous flow cytometry and microscopy studies with a very limited number of presumptive marker genes. Moreover, there is compelling evidence from iterative comparisons of bulk RNAseq and scRNAseq data that typical methods used to isolate single cell suspensions from aortas of atherosclerotic mice are associated with a >70% reduction in the frequency of MФs29. Indeed, using specialized methods for preserving sensitive foam cells by pre-fixation, Gordon Francis’ laboratory28 reported that 60–70% of MФ-like foam cells in advanced lesions are of SMC origin based on flow cytometric analysis of aortic lesion samples from Myh11CreERT2tdTomApoE−/− mice. Unfortunately, fixation is incompatible with scRNAseq analysis. Thus, there is extensive controversy in the literature as to the abundance and functions of SMC-P derived MФ-like foam cells and whether these cells exhibit unique transcriptomes as compared to myeloid- or tissue resident yolk sac-derived MФs.
Herein, we tested the hypothesis that adipose tissue perivascular cells exhibit extensive phenotypic plasticity with subsets of KLF4-dependent MФ-like cells playing a key role in adipose tissue inflammation and the metabolic response to DIO. Flow cytometry and scRNAseq analyses of the SVF of epididymal adipose tissue from Myh11-CreERT2eYFP mice given intraperitoneal (IP) tamoxifen in peanut oil showed that between 15–25% of eYFP+ cells expressed multiple MФ markers (CD45+CD11b+F4/80+) and showed a transcriptome indistinguishable from adipose tissue MФs. However, the majority of these presumptive SMC-P-derived MФs had no detectable eYFP transcript expression and were revealed to be Myh11−eYFP− autofluorescent epididymal adipose tissue MФs engorged with peanut oil. To eliminate the problem with MФ autofluorescence within the eYFP channel, we generated a new SMC-P lineage tracing mouse model wherein Myh11-DreERT2tdTomato mice were given tamoxifen in a normal diet. We then repeated our scRNAseq analysis and identified eight cell clusters dominated by Myh11-lineage tagged perivascular phenotypes including four that were significantly altered upon DIO and found that only about 2% of perivascular cells within adipose tissue exhibit a MФ transcriptome. Finally, we showed that SMC-P-specific Klf4 KO mice were not resistant to DIO, but exhibited improved glucose tolerance possibly due to marked reductions in the frequency of pro-inflammatory MФ subtypes and increases in LYVE1+ lymphatic endothelial cells.
METHODS
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Animals
All animal experiments were approved by the University of Virginia Animal Care and Use Committee (Protocol #2400 and #4195). Experiments were conducted according to the guidelines about reporting sex in preclinical studies30 formulated by the American Heart Association. Myh11-CreERT2eYFP, Myh11-CreERT2eYFPKlf4WT/WT and Myh11-CreERT2eYFPKlf4Δ/Δ mice were described previously4,5. Although previously thought to target exclusively SMC (>95% efficiency in healthy animals)4,5, we subsequently showed Myh11-Cre targets both SMC and >80% of pericytes11. Cre recombinase was activated in male mice with a series of 10 daily IP injections of 1 mg tamoxifen (Sigma, cat. no. T-5648) dissolved in peanut oil or by feeding mice Tamoxifen Diet (250 mg/kg TD.130856 Envigo) from 6 to 8 weeks of age. Myh11-DreERT2 mice were made by Cyagen Laboratory using traditional transgenic mice generation methodologies as previously described24. In brief, the myosin heavy chain 11 (Myh11) promoter was placed upstream of a Dre recombinase fused to an estrogen receptor 2(ERT2) domain. Mouse embryos were injected with the transgene that randomly inserted into the genome. The founder line was crossed with ROSA-roxed tdTomato-floxed eGFP mice from The Jackson Laboratory (Stock # 026931). Tamoxifen induced >90% labeling of SMC-P in Myh11-DreERT2tdTomato mice (Supplemental. Fig. III). Starting at 10 weeks of age (2 weeks after tamoxifen treatment) mice were either fed a normal diet (Envigo, Teklad #7912) or a high fat diet with 60 kcal% fat (Diet Induced Obesity (DIO) diet from Research Diets, D12492) for indicated times. For studies involving Myh11-CreERT2, only male mice were analyzed since the transgene is located on the Y chromosome. For Myh11-DreERT2tdTomato, both male and female mice were analyzed. Myh11-CreERT2eYFPKlf4WT/FL breeders were mated to produce Myh11-CreERT2eYFPKlf4FL/FL and Myh11-CreERT2eYFPKlf4WT/WT littermates for experiments.
Flow Cytometry Analysis
Mice were euthanized by CO2 asphyxiation and then perfused manually with 10 ml PBS via the left heart ventricle. Epididymal and subcutaneous adipose tissue depots, aorta, heart, and lung were excised and placed into FACS buffer (1% BSA in PBS). Tissues were chopped with scissors and digested in a cocktail of 0.75 mg/ml Liberase TM (Roche, #355374) and 57 IU per 10 ml Elastase (Worthington Bio. Corp. LS002279) in RPMI for 60 min at 37°C, passed through 19 gauge needle multiple times to allow for homogenization, and incubated for an additional 30 minutes at 37°C. Digested samples were collected in 15 ml conical tubes and FACS Buffer was added making a total volume of 13 ml and tubes centrifuged at 1000g for 10 min. The supernatant with the layer of floating adipocytes was discarded and red blood cells in the pellet were lysed using 1–3 ml 1X BD Pharm Lyse Buffer (#555899) for a maximum of 5 minutes. Pellets were washed with excess FACS buffer and subsequently filtered through 70 μm filters. Remaining cells were stained with antibodies against CD45-BV650, CD11b-PerCP Cy5.5, F4/80-PE Cy7, Lgals3-AF647, CD86-PerCP, Viability dye (Viability Far Red or Live/Dead Red or Live/Dead Violet) and either fixed with 4% paraformaldehyde or fixed and permeabilized using Fix & Perm kit (Thermo Fisher Scientific #GAS003) for intracellular CD206 and GFP staining. Samples were acquired on BD LSR Fortessa using BD FACS Diva 6.0 software and analyzed by Flow Jo V10.6.1.
Flow Sorting for scRNA Sequencing Library Preparation
Samples were prepared similarly to above protocol except that mice were perfused with 1X PBS with 1 μg/ml actinomycin D (Gibco #11805017) and excised tissues were placed into FACS buffer with 1 μg/ml actinomycin D to stop further transcription including artifactual activation of stress response genes during the cell isolation process. Samples were then digested in the Liberase TM cocktail with actinomycin D. Digested samples were collected in 15 ml conical tubes and FACS buffer (without actinomycin D henceforth) was added to yield a total volume of 13 ml. Tubes were then centrifuged twice: once at 400g for 10 min saving the pellet and transferring the supernatant to a new tube which was centrifuged again at 1000g for 10 min. Both pellets were then combined. LoBind pipet tips (Xtip Biotix #R-1000–9FC) were used for transferring samples whereas regular tips were used for resuspending pellets. RBC Lysis and staining was performed as described above. After staining, samples were washed and resuspended in 1X PBS plus 0.04% Ultra-Pure non-acetylated BSA (Thermo Fisher #AM2616). Samples were filtered through 35 μm filters (Falcon #352235) on ice and sorted on a BD Influx sorter with the 100 μm nozzle into LoBind tubes (Eppendorf #022431048) containing 100 μl PBS with non-acetylated BSA. In addition to obtaining cell numbers from the cell sorter, the cells were counted again using a BioRad TC20. To generate Chromium 10X genomic libraries, up to 7000 cells/sample were submitted to target 2000 cells/sample to the Genome Analysis Technology Core at University of Virginia. After barcoding, cells were pooled and sequenced on the Illumina NextSeq system 150 cycle high output. Quality control was conducted by Qubit and Agilent DNA high sensitivity tape stations after 10X library and Next Generation Sequencing (NGS) library preparation.
Single Cell RNA Sequencing
FastQC (Andrew S, 2010. FastQC: a quality control tool for high throughput sequence data, available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc), was used to perform quality control of Illumina NextSeq reads. After QC check, alignment of reads was performed using the cell ranger software (10x Genomics) against a modified mouse mm10 genome that included a custom eYFP chromosome. Libraries were then integrated and normalized using Seurat v3.1.031 in R (https://www.R-project.org/). Cells were excluded from analysis if they presented less than 200 genes (indicating low cell viability-RNA degradation), more than 5000 genes (indicating possible doublets), more than 10% mitochondrial gene content, or more than 5% hemoglobin gene content. Significant principal components (PCs) were calculated using JackStraw with 10000 repetitions. The number of dimensions used for cluster definition was 30 in the Myh11-CreERT2eYFP mouse model and libraries, and 20 in the Myh11-DreERT2tdTomato mouse model and libraries, with a 0.6 resolution for both analyses. FindAllMarkers function in Seurat was used to identify marker genes associated with each cluster. Raw data is available to download at GEO (submission pending) and the code is available upon request.
Bulk RNA Sequencing
Total RNA was isolated using Trizol (Invitrogen) from the mesenteric arcade of Myh11-CreERT2eYFPKlf4WT/WT and Myh11-CreERT2eYFPKlf4Δ/Δ mice (n=3 for each strain, each sample representing a pool of 3 mice) 2 weeks after the last tamoxifen injection. An RNA library, with ribosomal RNA reduction, was prepared according to Illumina RNA Seq library kit instructions. Agilent 2100 Bioanalyzer and Kapa Library Quantification Kit (Kapa Biosystems) were used to perform quality control and quantification of RNA and library according to manufacturer’s protocol. Sequencing of the libraries was performed by HudsonAlpha Institute for Biotechnology with the Illumina HiSeq2000 (2×100bp). STAR software version 2.4332 was used to align reads (one hundred-nucleotide paired-end reads) to the mouse genome M21 (GRCm38.p6) from Gencode (https://www.gencodegenes.org/). Following alignment, FeatureCounts33 was used to generate a table of gene counts and differential gene expression analysis was performed using the DESeq234 R package. Gene set enrichment analysis was performed using Ingenuity Pathway Analysis35 or MSigDB GSEA36. Significantly enriched pathways were identified using a 5% false discovery rate cutoff, and enrichment significance was presented using −log10 of padj. Data are presented as pathways downregulated or upregulated in knockout mice as compared to wild type mice.
RT-PCR
Epidydimal adipose tissue samples were harvested from Myh11-CreERT2eYFPKlf4WT/WT and Myh11-CreERT2eYFPKlf4Δ/Δ mice and snap frozen in liquid nitrogen. Total RNA was isolated using Trizol (Invitrogen) or Qiagen RNeasy kit. cDNA was prepared using iScript cDNA synthesis kit. RT-PCR reactions were prepared using Biorad IQ-Sybr Green supermix. The list of primers used are provided in Supplemental Table II. The expression of genes were normalized to GAPDH as housekeeping gene, and subsequently SMC-P Klf4Δ/Δ data was normalized to expression in SMC-P Klf4WT/WT mice.
Glucose and Insulin Tolerance Tests
Myh11-CreERT2eYFPKlf4WT/WT and Myh11-CreERT2eYFPKlf4Δ/Δ mice were subjected to intraperitoneal glucose and insulin tolerance tests (GTT and ITT) as described37,38. In brief, mice were fasted overnight in cages with sanichip bedding, and provided free access to drinking water. In the morning, baseline blood glucose values were measured in tail vein blood using a hand-held glucometer (AimStrip Plus #37350). The mice were injected with glucose at 2 mg/g body weight (Sigma, G8270) or at 1 mg/g body weight for 16 W DIO mice. Blood glucose was measured at indicated times. For insulin tolerance tests (ITT), random fed mice were injected with 0.75 U/kg Humulin-R from Eli Lilly (# NDC 0002-8215-17) in saline at ~2 pm (time 0) and blood glucose was measured at indicated times. STELLUX® Chemi Rodent Insulin ELISA (Alpco, # 80-INSMR-CH01) was used to measure insulin levels.
Immunofluorescence and immunohistochemistry
Epidydimal adipose tissue samples were harvested from Myh11-CreERT2eYFPKlf4WT/WT and Myh11-CreERT2eYFPKlf4Δ/Δ mice and fixed in 4% freshly diluted PFA overnight. The next day samples were switched to 70% ethanol and processed for paraffin embedding. 10 um thick sections were either subjected to Hematoxylin and Eosin (H&E) staining or utilized in immunofluorescence experiments. In brief, the sections were deparaffinized, rehydrated and subjected to heat-mediated antigen retrieval steps as described24. Sectioned tissues were blocked in horse serum containing 1X PBS and fish skin gelatin. The slides were stained using primary antibodies for LYVE1 (Abcam ab33682, 1:50) or CD31 (Abcam ab124432, 1:500) and costained for ACTA2-FITC (Sigma F3777, 1:500) and DAPI. Following mounting, the slides were imaged on a Zeiss 880 confocal microscope. Z-stacks were acquired using 20x objective and maximum intensity projections using same numbers of stacks were generated. H&E stained sections were imaged using Axio Vision light microscope and analyzed using Image J and Adipocount software as described39.
Statistical Analysis
Quantitative data were plotted and statistical analysis performed using GraphPad Prism (version 8.3). Data is presented as mean ± standard error of the mean (SEM), as indicated in figure legends. Assuming Gaussian distribution, unpaired two-tailed t-test with Welch’s correction was used for the comparison of two bars, and an ordinary two-way ANOVA with alpha=0.05 followed by Sidak’s multiple post-test was used for plots with multiple bars and repeated measures two-way ANOVA was used for comparisons of the two genotypes over time. P values smaller than 0.05 were considered significant. Experiments were not randomized and investigators were not blinded to animal distribution, but were blinded to outcome analysis.
RESULTS
Intraperitoneal injections of tamoxifen in peanut oil vehicle resulted in marked increases in autofluorescence of adipose tissue macrophages and their false identification as Myh11-eYFP+.
We hypothesized that adipose tissue perivascular cells exhibit extensive phenotypic plasticity and can, in a KLF4-dependent manner, differentiate into MФ-like cells that play key roles in adipose tissue inflammation and impaired whole body metabolism in response to DIO. To identify perivascular-derived MФ-like cells at baseline and in DIO, we performed flow cytometry of SVF cells isolated from epididymal and subcutaneous adipose tissues and heart of normal diet-fed Myh11-CreERT2eYFP mice (Fig. 1A, B) at 12 weeks of age. Surprisingly, within epidydimal adipose tissue, we found that a high percentage (15–25%) of perivascular-derived eYFP+ cells were CD45+, and ~60% of these were also positive for the MФ markers CD11b and F4/80 (Fig. 1C, D), accounting for approximately 5% of all MФs (Supp. Fig. I A). In contrast, the numbers of eYFP+ MФ marker+ cells in subcutaneous adipose tissue and heart were much lower (< 5 %). Interestingly, the abundance of SMC-P-derived MФ marker positive cells in adipose tissue depots increased roughly 2-fold after 6 weeks on a high fat diet (60% fat in diet, labeled diet induced obesity [DIO]) (Fig. 1E – G) concomitant with increased body and adipose tissue weights (Fig. 1F–G and Supp. Fig I B, C). The eYFP+ MФ-like cells appeared to exhibit an anti-inflammatory M2-like polarization in that they were CD206+ but CD86− by flow cytometry (Supp. Fig. I D). These observations suggested that a surprisingly large fraction of perivascular cells transdifferentiated into MФ-like cells within the microvasculature upon DIO.
Figure 1: A large subset of Myh11-CreERT2eYFP+ cells from the stromovascular fraction of epididymal adipose tissue appear to express multiple macrophage markers:
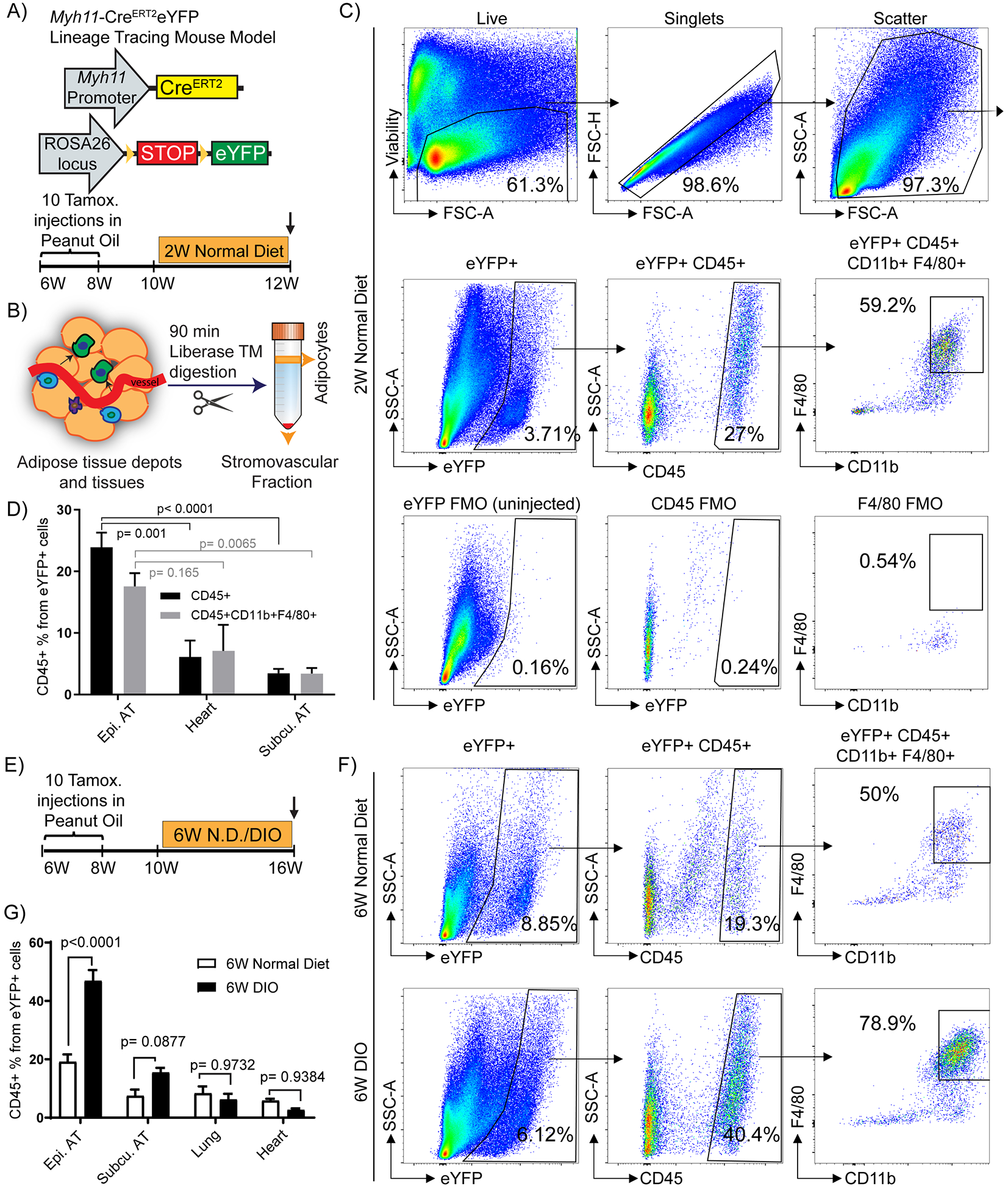
(A and B) Schematic of the mouse model and experimental design. (A) Myh11-CreERT2eYFP mice received ten intraperitoneal (IP) tamoxifen injections in peanut oil between 6 – 8 weeks of age to label SMC-P with eYFP followed by a 2-week washout period. Mice were fed a normal diet and euthanized at 12 weeks of age and tissues harvested (W – weeks). (B) Adipose tissues from Myh11-CreERT2eYFP SMC-P lineage-tracing mice were digested and processed for flow cytometric analyses. (C) Representative flow cytometry plots showing the gating strategy used to identify and quantity eYFP+ MФ marker positive cells within the adipose tissue SVF. Viability-dye negative cells were gated to exclude dead cells. Subsequently, forward scatter-height (FSC-H) versus forward scatter-area (FSC-A) gating was applied to exclude doublets. FSC-A versus side scatter-area (SSC-A) gating was applied to exclude debris. The example shown is an SVF sample from epididymal adipose tissue of a 12 week old normal diet-fed Myh11-CreERT2eYFP SMC-P lineage tracing mouse. (D) Quantification of the percentage of eYFP+CD45+ and eYFP+CD45+CD11b+F4/80+ cells in different tissues based on flow cytometry. Values represent mean ± SEM. P values were determined using an ordinary Two-way ANOVA with alpha = 0.05 followed by Sidak’s multiple comparisons post-test. Epididymal adipose tissue (n = 12), subcutaneous adipose tissue (n = 5), heart (n = 4). (E) Schematic for six weeks of DIO diet experiments. Myh11-CreERT2eYFP mice received ten tamoxifen injections in peanut oil between 6 – 8 weeks of age, followed by a 2-week washout period. The mice were then fed a normal diet for the course of the experiment (6W N.D. group) or switched to a DIO diet for 6 weeks at 10 weeks of age (6W DIO group). (F) Representative flow plots showing that SMC-P lineage-tracing mice fed a DIO diet for 6-weeks to induce obesity, have an increased fraction of eYFP+ SVF cells in the epididymal adipose tissue positive for the MФ markers, CD45, CD11b, and F4/80. (C and F) The eYFP FMO gate was set using epididymal adipose tissue SVF cells from a Myh11-CreERT2eYFP control mouse not given IP tamoxifen in peanut oil. (G) Quantification of the percentage of eYFP+CD45+ cells in different tissues from Myh11-CreERT2eYFP SMC-P lineage-tracing mice fed either a normal diet or DIO diet for 6-weeks. Values represent mean ± SEM. P values were determined using an ordinary Two-way ANOVA with alpha=0.05 followed by Sidak’s multiple comparisons post-test. Epididymal adipose tissue (n = 10 6W N.D., n = 16 6W DIO), subcutaneous adipose tissue (n = 10 6W N.D., n = 15 6W DIO), lung (n = 9), heart (n = 13 6W N.D., n = 4 6W DIO). P values are given in plots. AT: adipose tissue.
To test whether perivascular origin MФ marker positive cells exhibit a similar transcriptome to non-perivascular origin MФ in adipose tissue we performed scRNAseq using 10X Genomics technology on flow-sorted populations (Fig. 2A, B and Supp. Fig. II) of epididymal adipose tissue SVF. We also aimed to identify SMC-P phenotypic transitions within adipose tissue and changes upon DIO +/− perivascular cell KO of KLF4 (Fig. 2A). Our initial Uniform Manifold Approximation and Projection (UMAP) analysis of integrated Seurat libraries consisting of 25,356 cells identified 23 transcriptomic clusters of adipose tissue SVF cells (Fig. 2C –E). Based on dot plots and feature plots for the top differentially expressed genes and traditional marker genes in Figs. 2F. clusters 1–7 appeared to represent MФ clusters, whereas five clusters (15, 16, 21, 22, and 23) exhibited SMC-P transcriptomic signatures (Fig. 2G), and two clusters 12 and 14 exhibited endothelial cell phenotypes. Strikingly, a large fraction of SMC-P-derived (eYFP+) cells appeared within the MФ island (Fig. 2D), suggesting that perivascular origin MФ marker positive cells and non-perivascular origin MФ were transcriptionally indistinguishable. We were intrigued but also suspicious of this observation and further scrutinized our data by including a step of eYFP transcript detection, via inserting an artificial chromosome containing the ROSA eYFP sequence, in our sequence alignment procedure. We found that the numbers of SMC-P derived cells in the MФ island based on eYFP transcript detection was markedly reduced, suggesting that the majority of eYFP fluorescence+ MФ-like cells were eYFP transcript negative (Fig. 2E) indicating that a high rate of false positive cells were detected in the eYFP channel.
Figure 2: A significant fraction of the flow-sorted eYFP+ cells from Myh11-CreERT2eYFP mice treated with tamoxifen in peanut oil exhibit a MФ transcriptomic signature but have no detectable eYFP transcript expression.
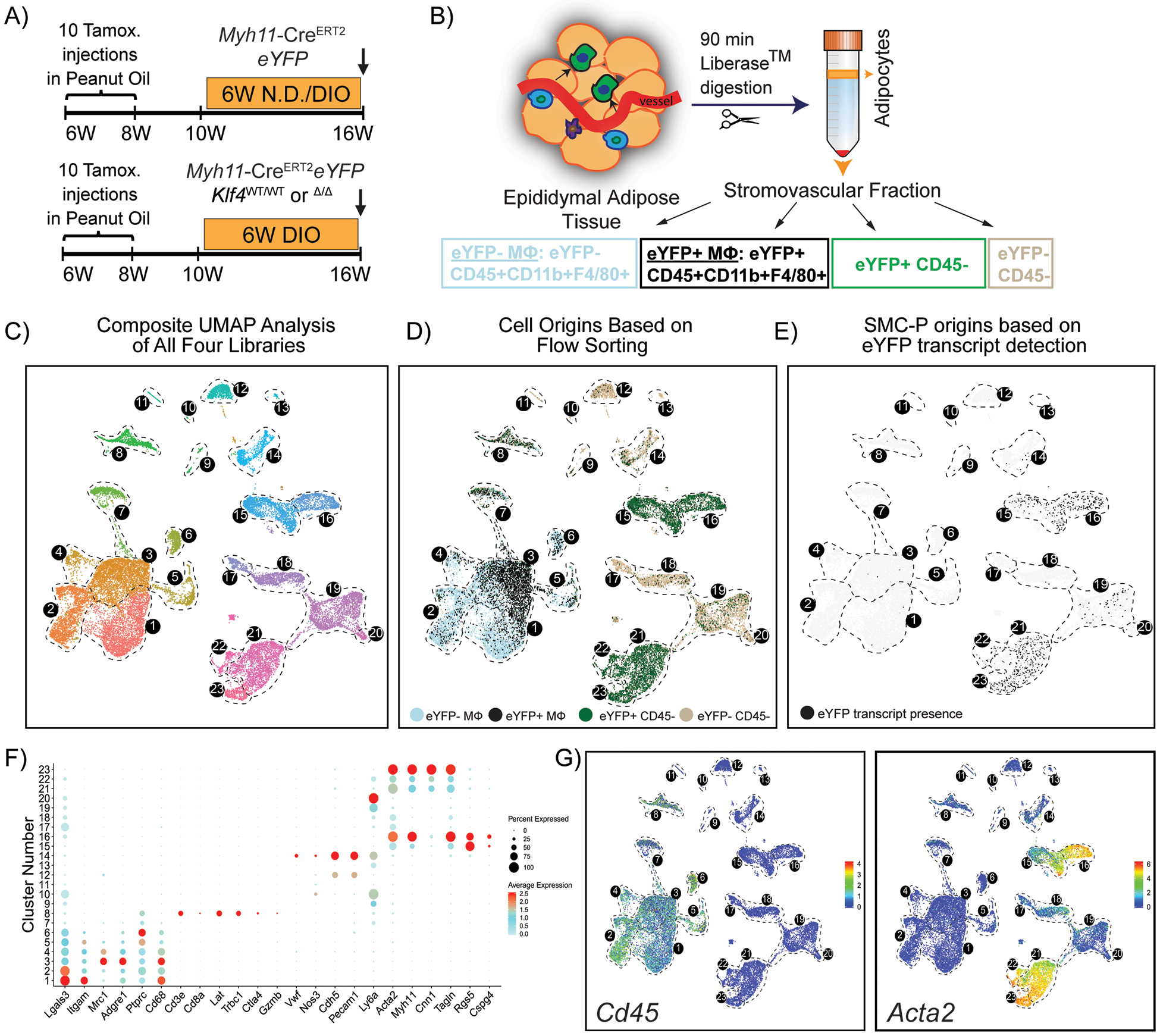
(A) Schematic of the mouse model and experimental design. Four experimental animal groups were analyzed: Myh11-CreERT2eYFP SMC-P lineage-tracing mice on normal diet (Group 1, N.D.), or DIO diet (Group 2, DIO) for 6 weeks, as well as SMC-P Klf4WT/WT mice on DIO diet (SMC-P Klf4WT/WT DIO, Group 3), and SMC-P Klf4Δ/Δ mice on DIO diet (SMC-P Klf4Δ/Δ DIO, Group 4) for 6 weeks. (B) For each experimental group, SVF cells of epididymal adipose tissue were sorted for four cell-type groups, including eYFP+CD45−, eYFP+CD45+CD11b+F4/80+ (eYFP+MФ), eYFP‒CD45+CD11b+F4/80+ (eYFP−MФ), and eYFP−CD45− cells followed by scRNAseq library preparation resulting in a total of 16 libraries. The eYFP FMO gate was set using epididymal SVF cells from a Myh11-CreERT2eYFP control mouse not given IP tamoxifen in peanut oil. (C – E) Integrated UMAP plots of 25,356 cells from four experimental groups: N.D. (4,516 cells), DIO (3,995 cells), SMC-P Klf4WT/WT DIO (8,506 cells), SMC-P Klf4Δ/Δ DIO (8,339 cells). (C) Color-coded UMAP plot of integrated libraries shows 23 clusters. (D) UMAP plot of integrated libraries showing cell distribution based on the sorted cell-types. (E) UMAP feature plot of the eYFP transcript distribution. (F) Dot plot depicting the expression levels and percentages of cells expressing a pre-determined list of traditional marker genes in each cluster. (G) Feature plots showing expression levels of the traditional myeloid cell marker Cd45, and SMC marker Acta2.
To explore the underlying mechanism for false positive eYFP+ cells, we evaluated the set of fluorescence minus one controls (FMO), used to draw gates, in our flow cytometry experiments. We postulated that peanut oil, the vehicle for IP delivery of tamoxifen, could be the confounding factor for flow cytometry analyses particularly in tissue samples that are in contact with the peritoneal cavity including epididymal adipose tissue. We hypothesized that the IP delivered peanut oil is taken up by phagocytic cells thereby increasing their auto-fluorescence and false identification as Myh11-eYFP+ cells. Consistent with this possibility, we found that peanut oil injection alone induced appearance of auto fluorescent MФ relative to control mice (Fig. 3A). In addition, feeding tamoxifen in the diet, instead of injecting tamoxifen IP in peanut oil, dramatically reduced the abundance of eYFP+CD45+ cells from 33.6% to approximately 2% (Fig. 3A, B). Furthermore, Imagestream analysis revealed that autofluorescent single nuclei MФ with distinct compartmentalized fluorescence in the eYFP channel were anti-GFP/eYFP antibody negative (Fig. 3C). Taken together, these results indicate that IP peanut oil injections result in marked increases in the auto-fluorescence of peritoneal MФ and their detection in the eYFP channel.
Figure 3: Intraperitoneal delivery of tamoxifen in peanut oil results in marked increases in macrophage autofluorescence.
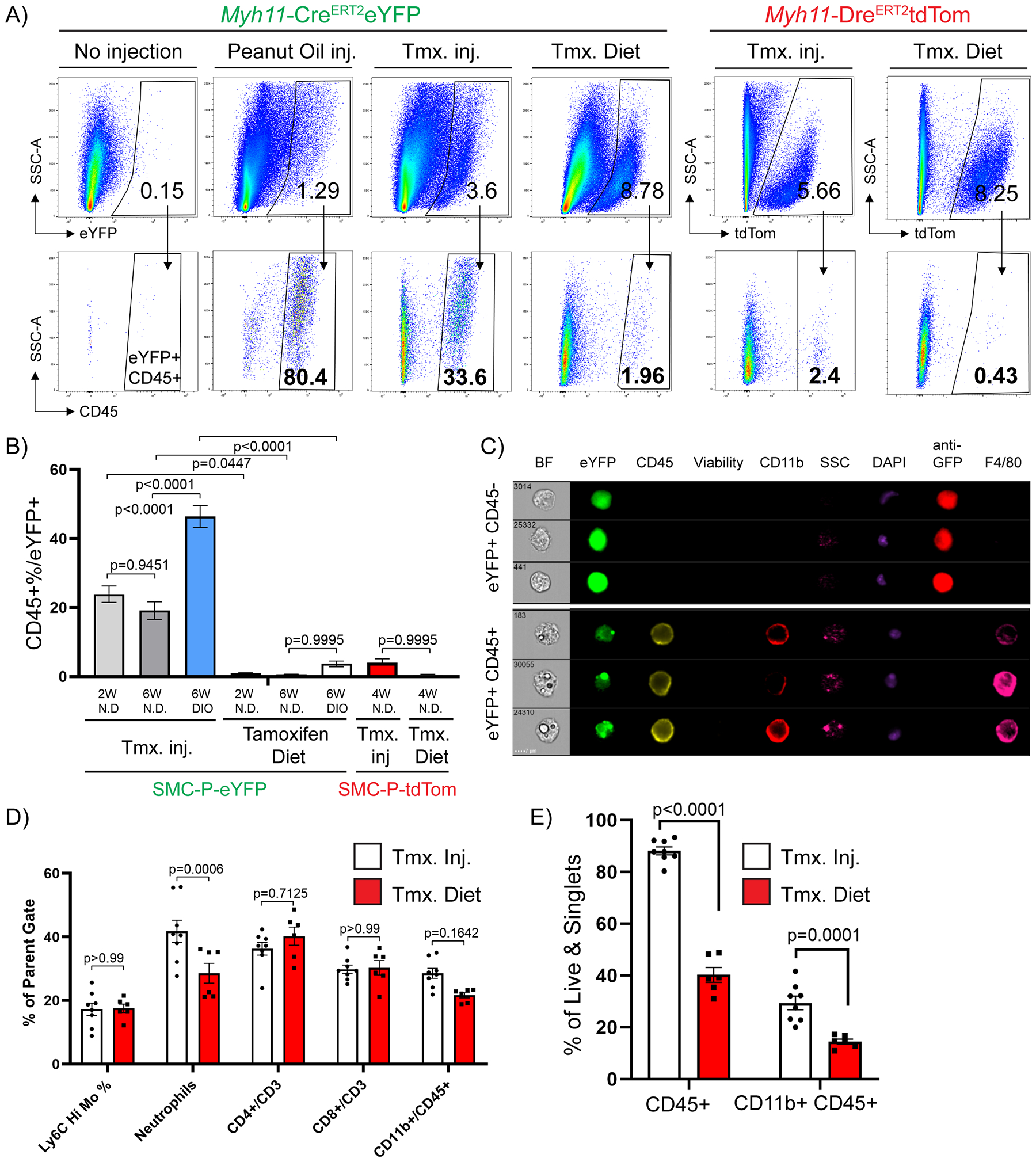
Between 6–8 weeks of age, Myh11-CreERT2eYFP or Myh11-DreERT2tdTom mice received either 10 tamoxifen injections in peanut oil (Tmx inj), or pure peanut oil injections (Peanut Oil inj), or tamoxifen-containing diet (Tmx. Diet), or nothing (No injection, negative control) followed by a 2-week washout period. The mice were fed a normal diet or DIO diet for indicated times (W – weeks), tissues were harvested and epididymal adipose tissue SVF cells were subjected to flow cytometry. (A) Representative plots showing the frequency of CD45+ within eYFP (Myh11-CreERT2eYFP panels) or tdTomato (Myh11-DreERT2tdTom panels) gates based on eYFP or tdTomato FMOs. The eYFP and tdTomato FMO gates were set using SVF cells of epididymal adipose tissue from Myh11-CreERT2eYFP or Myh11-DreERT2tdTom control mice that received neither injections nor tamoxifen in the diet. (B) Quantification of flow cytometric analyses (Mean ± SEM) using different tamoxifen delivery methods and reporter lines. Myh11-CreERT2eYFP (SMC-P-eYFP) mouse line – Tamoxifen injection in peanut oil (2W normal diet n = 12, 6W normal diet n = 10, 6W DIO n = 19), Tamoxifen Diet (normal diet (2W) n = 7, normal diet (6W) n = 3, DIO (6W) n = 4); Myh11-DreERT2tdTom (SMC-P tdTom) mouse line – Tamoxifen injections in peanut oil (normal diet (4W) n = 6), Tamoxifen Diet (normal diet (4W) n = 5). (C) Epididymal adipose tissue SVF cells were stained with antibodies and subjected to ImageStream analysis. Live single cells excluding debris were gated for analysis. Representative cells were picked to illustrate differences in eYFP signals among single nuclei cells. Columns show endogenous eYFP signal (green), staining for GFP (anti-GFP), CD45, Viability dye, CD11b, DAPI, and F4/80. Side scatter (SSC) indicates granularity of the cells. (D) Myh11-CreERT2eYFP mice received 10 tamoxifen injections in peanut oil or were fed tamoxifen in normal diet for 2 weeks followed by a 2-week washout period. Mice were fed a normal diet and euthanized at 12 weeks of age and tissues were harvested. Blood samples were stained with antibodies and subjected to flow cytometry. Circulating blood cell types are gated as follows: live/singlets/scatter gates were initially applied to remove dead cells, doublets and debris. CD45+ cells were gated for CD3+, then frequency of CD4 or CD8 single positive cells plotted. From the same CD45+ gate, CD11b+ cells are gated and plotted. From the CD11b+ gate the frequency of Ly6chi monocytes are plotted as well as Ly6G+ neutrophils (mean ± SEM). (E) Similar to (D), Epididymal adipose tissue SVF cell suspensions from the same mice were harvested and stained for flow cytometry. The percentage of CD45+ cells (mean ± SEM) among live and single cells are indicated, depicting the increased abundance of immune cells following peanut oil injection as compared to tamoxifen diet fed mice. (B, D, and E) P values were determined using an ordinary Two-way ANOVA with alpha=0.05 followed by Sidak’s multiple comparisons post-test.
To test if this phenomenon was specific to eYFP, we generated a new SMC-P lineage tracing Myh11-DreERT2 mouse line and crossed it to a ROSA-roxed-tdTom-floxed eGFP mouse. Myh11-DreERT2tdTom mice have >92% labeling efficiency of medial SMC in brachiocephalic artery (BCA) sections (Supp. Fig III A, B) as well as high efficiency labeling of SMC and pericytes in the microvasculature upon tamoxifen treatment similar to the Myh11-CreERT2eYFP mice (Supp. Fig. III C–F). Of major significance, the number of Myh11 derived tdTomato+ perivascular cells that were also CD45+ was dramatically reduced when compared to our previous analysis using the Myh11-CreERT2eYFP reporter mouse (2.4% with tamoxifen IP injections in peanut oil, and 0.43% using a tamoxifen diet, Fig. 3A–B), suggesting that similarities in eYFP and lipid auto-fluorescence spectra contributed to detection of false eYFP+ MФ. To test the hypothesis that peanut oil injections induce systemic inflammation we assessed changes in circulating cytokine levels or blood cell types. Surprisingly, IP peanut oil injections caused little systemic inflammation with no changes in circulating cytokines and no changes in circulating immune cells except for neutrophils (Fig. 3D and Supp. Fig. IV). However, the effect on epididymal adipose tissue inflammation was pronounced; including a two-fold increase in the abundance of CD45+ cells after peanut oil injections (Fig. 3E). Thus, these results indicate that the effects of IP peanut oil injections were likely primarily on epididymal adipose tissue within peritoneal cavity, and not on circulating factors.
Perivascular cells demonstrated remarkable plasticity in adipose tissue and dynamically responded to DIO diet feeding.
With this knowledge, to rigorously identify perivascular phenotypes within the adipose tissue microvasculature and to avoid peanut oil effects, we repeated scRNAseq using Myh11-DreERT2tdTom mice labeled with tamoxifen in the diet. We fed mice either 6 weeks of normal diet or DIO and enriched for tdTom+ cells versus unsorted SVF cell populations (Fig. 4A and Supp. Fig. V). Remarkably, we identified 20 clusters within integrated populations, of which 8 clusters (13–20) were dominated by perivascular derived cells (Fig. 4B–C). Using traditional markers, we observed eight clusters (13–20) with high expression of Acta2, five clusters (1–5) with Cd45 expression, and two clusters (6 and 8) with Cdh5 expression (Fig. 4D–E). Importantly, DIO caused major shifts in perivascular cell dominated clusters. Specifically, the frequency of tdTomato+ sorted cells in cluster 13 was increased by >2-fold upon DIO, whereas the frequency of these cells in clusters 14, 15 and 16 were decreased by 60–100% (Fig. 4F, left panel). Pathway analysis of cluster 13 revealed enrichment of not only classical SMC contractile genes, but also cardiac and skeletal muscle contractile genes relevant to lymphatic muscle cells40. For instance, top differentially expressed genes include cardiac troponin (Tnnt2) and skeletal muscle troponin (Tnnt1), as well as other cardiac and skeletal muscle genes, including Acta1, Etp1a2, Casq2, Kcnd3. In addition, collagen biosynthesis, cartilage development and several embryonic/developmental cell differentiation and morphogenesis pathways, and negative regulation of contraction were associated with the top 100 significantly differentially expressed genes (including Tnnt1, Col4a4, Wnt6, Tcf21, Pdgfrα) in Cluster 13. It is intriguing to speculate that this perivascular cell type might be involved in not only contraction of lymphatic vessels, but maybe also extracellular matrix remodeling, regulation of insulin-like growth factor (IGF) transport and uptake based on pathway analysis. Considering that not only insulin resistance, but also IGF resistance and alterations in the growth hormone/IGF-1 axis lead to metabolic dysfunction41–44, cluster 13 cells might play a key role in the harmful response to obesity. Meanwhile, pathway analysis of cluster 14 identified SMC migration and contraction, regulation of actin filament assembly, regulation of arterial blood pressure and angiogenesis associated with the top 100 genes (including Cnn1, Tagln, Myh11, Acta2). Collectively, these results suggest that a significant portion of perivascular cells lose (or significantly decrease) their contractile properties in response to DIO and this might be one of the possible mechanisms related to vascular dysfunction associated with obesity45. In addition, DIO resulted in marked changes in a number of other clusters (Fig. 4F, right panel) including increased abundance of MФs expressing Cd14+Emr1+ (Cluster 3), Cd204+Il1b+ (Cluster 4), Lyz2+Itgam+ (Cluster 5) and decreases in Dcn+ and Lum+ cell types (clusters 9 and 11). Taken together, we present the first rigorous assessment of perivascular cell plasticity within adipose tissue and identify several clusters of predominantly perivascular origin that dynamically respond to obesity.
Figure 4: Single-cell RNA sequencing using Myh11-DreERT2tdTom lineage-tracing mice treated with tamoxifen in the diet reveals novel perivascular cell phenotypes and obesity-induced changes in epididymal adipose SVF cells.
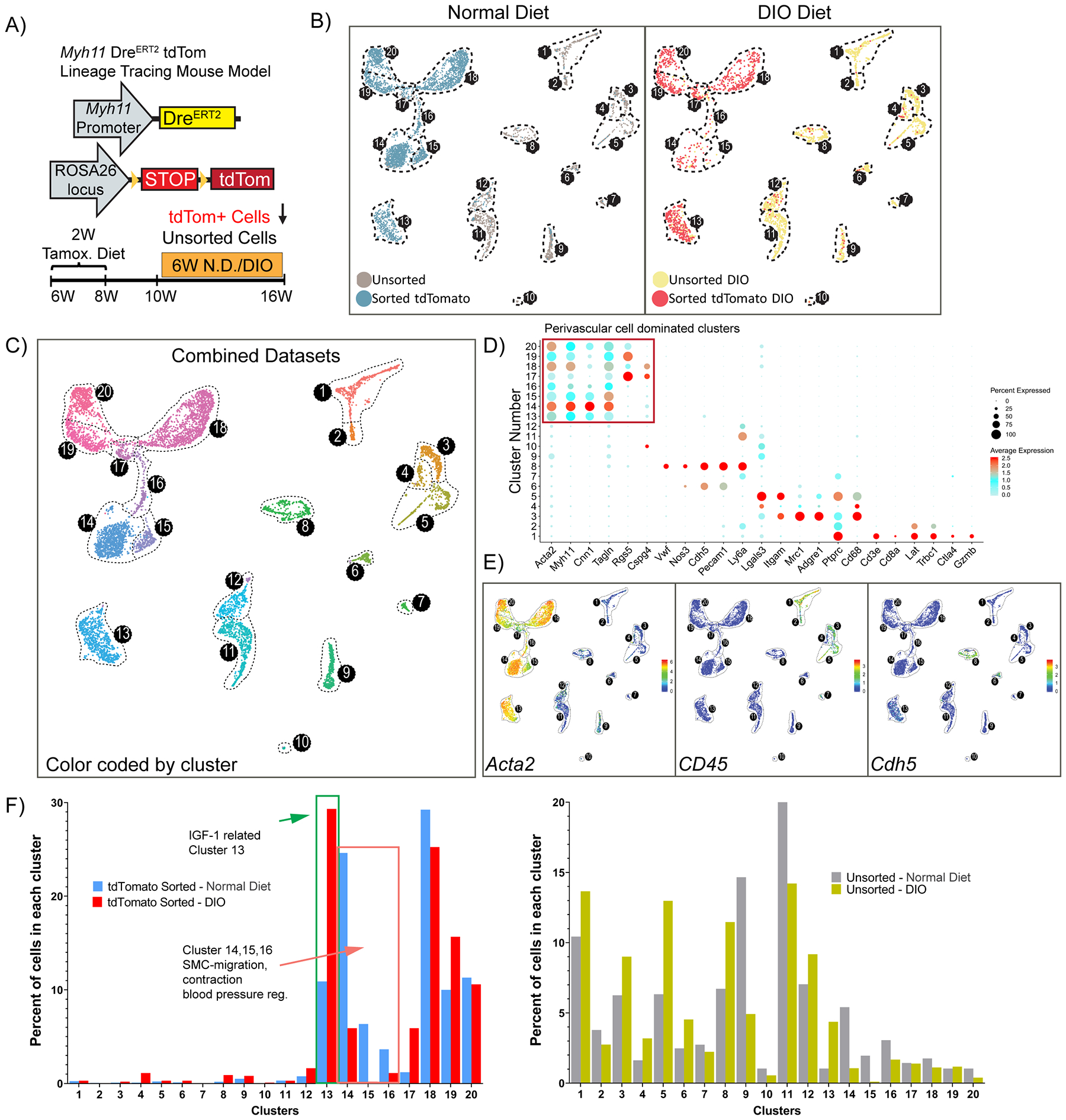
(A) Schematic of a mouse model and experimental design. Myh11-DreERT2tdTom lineage-tracing mice were fed a tamoxifen diet for 2 weeks, followed by a 2-week washout period. Subsequently, mice were fed either a normal diet or DIO diet for 6 weeks and then tissues were harvested. Two libraries were prepared for each condition: flow-sorted tdTomato positive cells or unsorted SVF cells. (B) Color-coded UMAP analysis of integrated libraries shows 20 unique clusters. A total of 7,449 cells were analyzed including N.D. (4,680 cells, left panel), DIO (2,769 cells, right panel). (C) UMAP plot depicting the integrated dataset color-coded by cluster. (D) Dot plot analysis depicting the expression level and percentage of cells expressing a pre-determined list of traditional marker genes in each cluster. (E) Feature plots showing the expression level of the traditional MФ marker Cd45, SMC marker Acta2, and endothelial marker Cdh5. (F) Quantitative analysis for the frequency distribution of cells within each library among different clusters. (Total number of cells in each library are: 3,146 tdTom+, 1,534 unsorted N.D.; 982 tdTom+, 1,787 unsorted DIO).
Genetic inactivation of Klf4 in SMC-P results in improved glucose tolerance upon DIO diet.
We previously showed that the pluripotency gene Klf4 promotes atherosclerosis within advanced BCA lesions in SMC-P Apoe−/− mice that were fed a Western diet (WD) for 18 weeks5. Chromatin immunoprecipitation-sequencing (ChIP-Seq) analysis identified enrichment of >800 putative SMC-P Klf4 target genes in aortic arch/BCA region samples of SMC-P Klf4WT/WT Apoe−/− mice versus SMC-P Klf4Δ/Δ Apoe−/− mice5. Among these putative SMC-P-specific KLF4 target genes were >80 genes associated with inflammation, antigen presentation, and the immune response, and a number of pro-inflammatory cytokines5. In contrast, our laboratory recently showed that KLF4 plays a critical beneficial role in the maintenance of perivascular cell coverage and the permeability properties of resistance vessels10. We therefore tested if Klf4-dependent plasticity of perivascular cells is beneficial or detrimental to adipose tissue inflammation and metabolic dysfunction in the context of DIO. To understand the role of perivascular Klf4 in DIO, we fed Myh11-CreERT2 eYFPKlf4Δ/Δ (SMC-P Klf4Δ/Δ) and control Myh11-CreERT2eYFPKlf4WT/WT (SMC-P Klf4WT/WT) mice either a normal diet or DIO diet. SMC-P Klf4Δ/Δ mice were not resistant to DIO, and had similar body and individual adipose tissue weights (Fig. 5A, B and Supp. Fig. VI A). SMC-P Klf4Δ/Δ mice also exhibited no changes in fasting blood glucose (Fig. 5C), insulin levels and HOMA-IR (Fig. 5D), insulin tolerance (Fig. 5E), or the number of circulating blood cells and immune cell types within epididymal adipose tissue (Supp. Fig. VI B–C). KLF4 has been shown to transcriptionally regulate adipogenic differentiation of 3T3-L1 cells in culture46. Therefore, we tested the adipocyte size and the abundance of crown like structures. We did not find significant changes (Supp. Fig. VI D). In addition, circulating cytokines in the blood or expression analysis of select genes in epidydimal adipose tissue did not reveal significant changes (Supp. Fig. VI E). However, of major interest, SMC-P Klf4Δ/Δ mice exhibited improved glucose tolerance as compared to SMC-P Klf4WT/WT controls upon DIO diet (Fig. 5F). This was not the case when mice were fed a normal diet (Fig. 5G).
Figure 5: SMC-P Klf4Δ/Δ mice are not resistant to DIO, but display improved glucose tolerance.
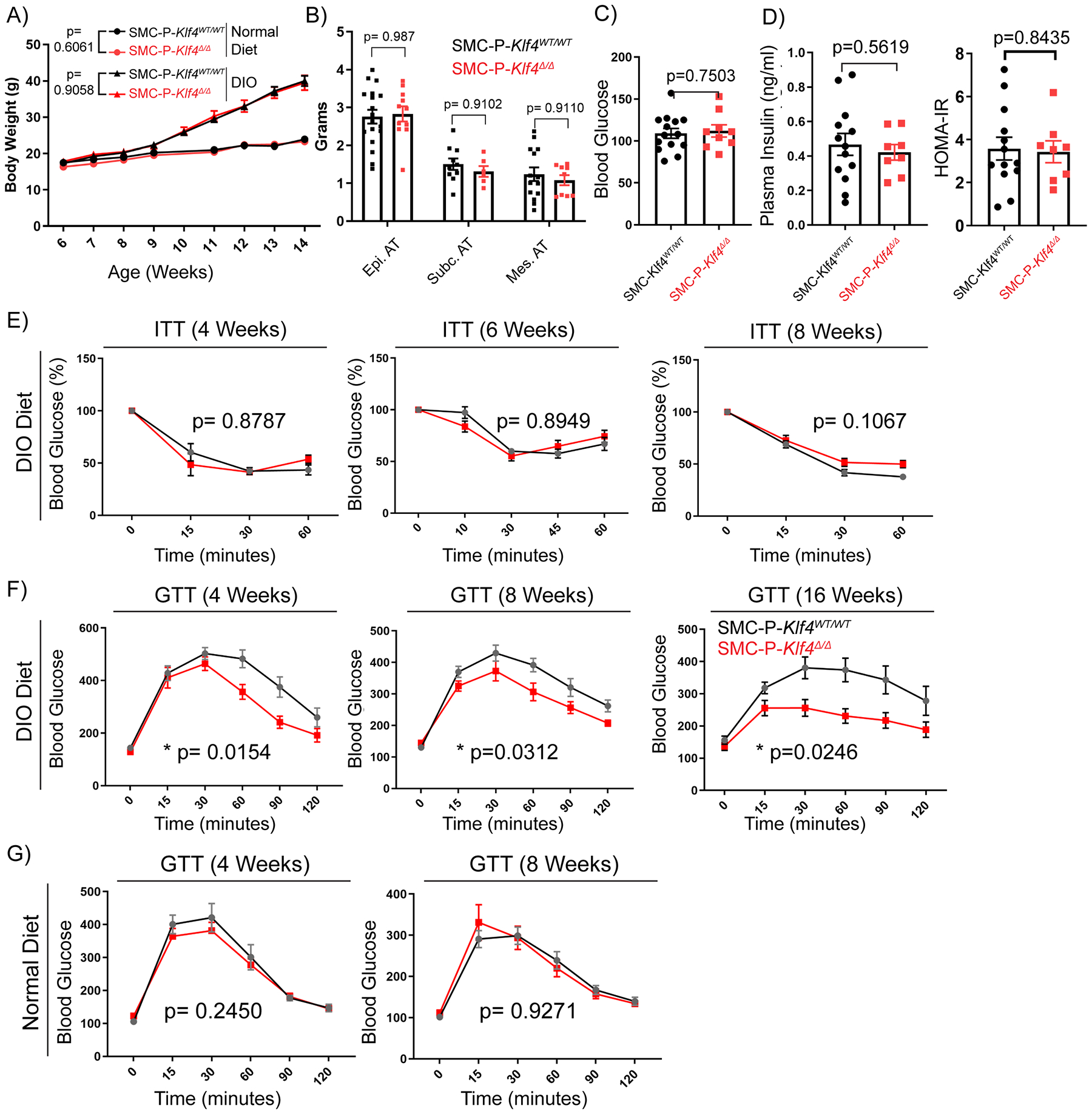
SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice were injected with tamoxifen in peanut oil between 6 – 8 weeks of age, followed by a 2-week washout period, and then fed either a normal diet or DIO diet for six weeks. (A) Graph of body weight changes over time (weeks of age) for SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice showing mean ± SEM N.D.: SMC-P Klf4WT/WT n = 4, SMC-P Klf4Δ/Δ n = 5; DIO: SMC-P Klf4WT/WT n = 11, SMC-P Klf4Δ/Δ n = 11. No significant differences were observed between genotypes using two-way repeated measures ANOVA, P values as indicated. (B) Adipose tissue weights of mice receiving 6–7 weeks of high fat diet were plotted (mean ± SEM). P values were determined using an ordinary Two-way ANOVA with alpha=0.05 followed by Sidak’s multiple comparisons post-test. No significant differences were observed between genotypes (P values given above bar graphs). (C) SMC-P Klf4WT/WT (n = 14) or SMC-P Klf4Δ/Δ (n = 9) mice were injected with tamoxifen in peanut oil and fed high fat diet for six weeks. After overnight fasting, plasma glucose levels were measured (mean ± SEM). An unpaired two-tailed Welch’s t-test, with alpha = 0.05 without assuming a consistent standard deviation yielded P=0.7503 (D) Plasma insulin levels were measured using the STELLUX® Chemi Rodent Insulin ELISA after six weeks of DIO feeding followed by overnight fasting (SMC-P Klf4WT/WT n = 13; SMC-P Klf4Δ/Δ n = 8). HOMA-IR is calculated by multiplying fasting plasma insulin (μU/ml) by fasting blood glucose (mM) and dividing by 22.5. Values represent mean ± SEM. An unpaired two-tailed Welch’s t-test, with alpha = 0.05 without assuming a consistent standard deviation yielded P=0.5619 for plasma insulin, P=0.8435 for HOMA-IR. (E) SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice were injected with tamoxifen in peanut oil followed by a 2-week washout period, and then fed DIO diet for indicated times. Then mice were subjected to insulin tolerance tests (ITT) under random-fed conditions at 2 pm. Blood glucose values were plotted as percent of values at time 0. Values represent means ± SEM. (ITT 4-weeks DIO: SMC-P Klf4WT/WT n = 5, SMC-P Klf4Δ/Δ n = 2; ITT 6-weeks DIO: SMC-P Klf4WT/WT n = 4, SMC-P Klf4Δ/Δ n = 10; ITT 8-weeks DIO: SMC-P Klf4WT/WT n = 4, SMC-P Klf4Δ/Δ n = 7). (F) SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice were injected with tamoxifen in peanut oil and fed DIO diet for indicated times. They were then subjected to glucose tolerance tests (GTT) following overnight fasting. Values represent means ± SEM. (GTT 4-weeks DIO: SMC-P Klf4WT/WT n = 10, SMC-P Klf4Δ/Δ n = 10; GTT 8-weeks DIO: SMC-P Klf4WT/WT n = 10, SMC-P Klf4Δ/Δ n = 11; GTT 16-weeks DIO: SMC-P Klf4WT/WT n = 8, SMC-P Klf4Δ/Δ n = 5.) (G) Similar to D, GTT were performed with mice fed a normal diet for indicated times. GTT 4-weeks Normal Diet: SMC-P Klf4WT/WT n = 9, SMC-P Klf4Δ/Δ n = 8; GTT 8-weeks Normal Diet: SMC-P Klf4WT/WT n = 11, SMC-P Klf4Δ/Δ n = 9. Statistical analysis for (E, F and G): For GTT and ITT, repeated measures two-way ANOVA with the Geisser-Greenhouse correction was used for comparing data between genotypes, P values are indicated in plots.
Loss of Klf4 in perivascular cells leads to decreases in pro-inflammatory MФs and increases in lymphatic endothelial cells in adipose tissue.
To elucidate potential mechanisms by which loss of Klf4 in perivascular cells resulted in improved glucose tolerance, we performed bulk RNAseq on the SVF of the mesenteric arcade from SMC-P Klf4Δ/Δ and SMC-P Klf4WT/WT mice (Fig. 6A, B). Importantly, we chose to do these RNAseq analyses on normal diet not DIO-fed mice two weeks following completion of IP tamoxifen injections (Fig. 6A) in order to detect baseline phenotypic changes resulting from perivascular cell-specific Klf4 KO independent of complexities associated with DIO. Ingenuity Pathway Analysis showed that loss of Klf4 in perivascular cells resulted in increased expression of genes involved in oxidative phosphorylation and mitochondrial dysfunction but marked decreases of genes in pathways associated with inflammation and MФ activation (Fig. 6B) as compared to SMC-P Klf4WT/WT control. Notably, these pathway alterations represent the summation of direct and indirect changes resulting from initial loss of KLF4 exclusively in perivascular cells. We then hypothesized that KLF4 expression by perivascular cells augments adipose tissue inflammation in response to DIO leading to increasing numbers of MФ and other CD45+ immune cells. To test this hypothesis, we turned back to the scRNAseq analysis on epididymal adipose SVF from SMC-P Klf4Δ/Δ and SMC-P Klf4WT/WT mice following 6-weeks of DIO (Fig. 2A, Fig. 6C, D, Supp. Fig. VII). These experiments were performed using tamoxifen injections in peanut oil as these SMC-P Klf4 KO studies were completed prior to discovering the peanut oil effects. We considered repeating the SMC-P Klf4 KO studies using tamoxifen in the diet but did not for the following reasons. First, as has been the case for all SMC lineage tracing and conditional gene KO experiments in our laboratory, we employ a highly rigorous single variable experimental design wherein SMC-P Klf4Δ/Δ and SMC-P Klf4WT/WT mice are C57BL6 congenic littermates that differ only by whether they have wild type or floxed Klf4 alleles. All mice were treated identically with both control and experimental mice receiving IP tamoxifen injections. As such, the IP tamoxifen is an inherent part of the model system not an undefined experimental variable between the experimental and littermate control mice. Second, in spite of the profound effects of the IP peanut oil on autofluorescence of peritoneal macrophages (Fig.3), we saw no changes in circulating cytokines (Supp. Fig. IV) suggesting that there were not sustained systemic effects of IP peanut oil. However, we cannot rule out that IP peanut oil injections might have influenced the consequences of genetic loss of Klf4 in perivascular cells. Third, although oral delivery of tamoxifen minimized the problem with macrophage autofluorescence, it can theoretically introduce its own experimental complications including unknown effects on gastro-epithelial cells, gastrointestinal immune cells and/or the microbiome. We conclude that the differences between SMC-P Klf4Δ/Δ and SMC-P Klf4WT/WT mice would represent the sum of all KLF4-dependent events following the loss of Klf4 only in perivascular cells, including a potential difference in responding to peanut oil treatment.
Figure 6: Genetic inactivation of Klf4 specifically in SMC-P does not result in marked changes in the distribution of cells within perivascular clusters but impacted transcriptomic clusters of MФs and endothelial cells.
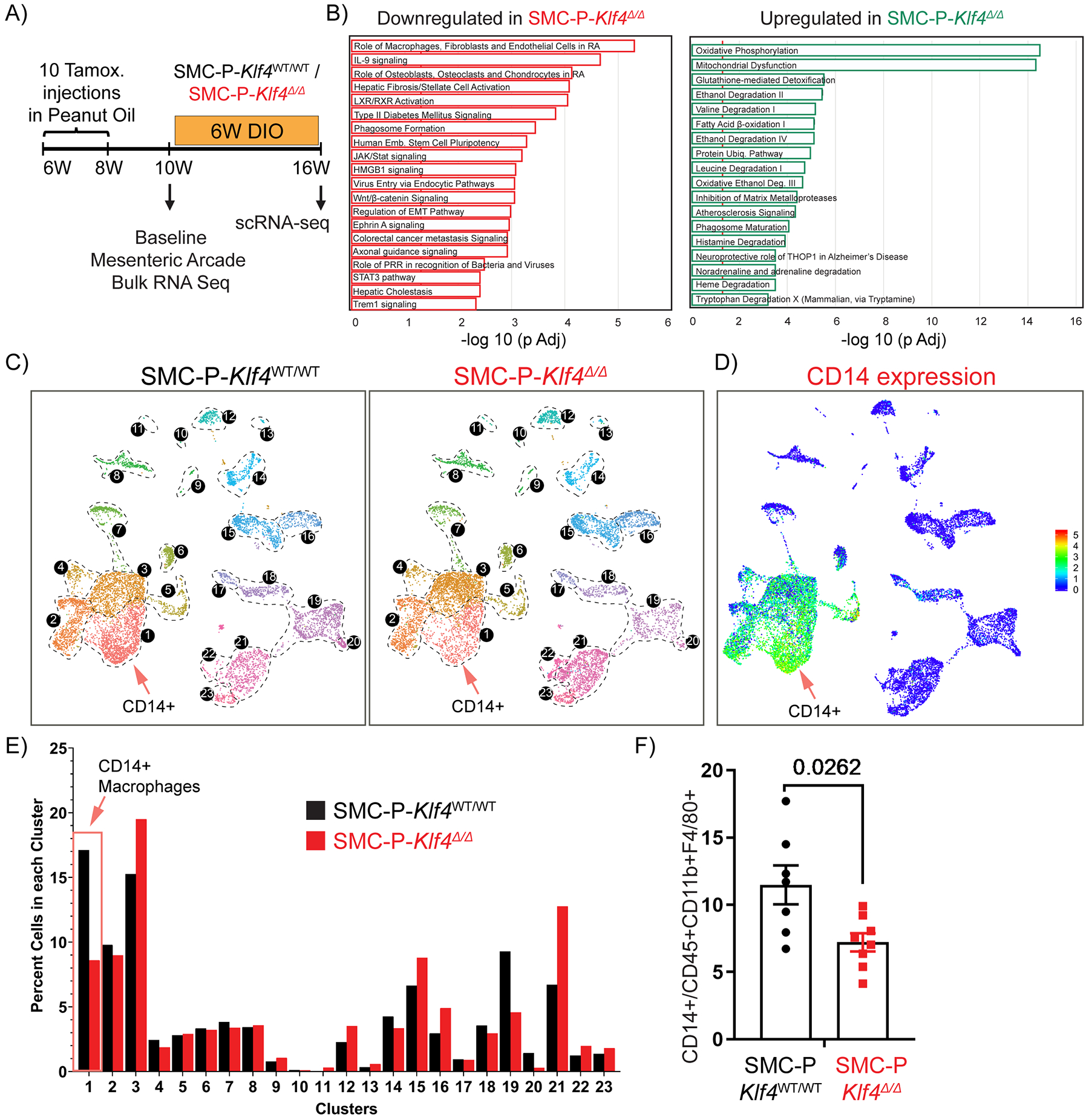
(A) Schematic of experimental design. SMC-P Klf4WT/WT or SMC-P Klf4Δ/Δ mice were injected with tamoxifen in peanut oil and following a 2-week washout period either tissues were harvested for baseline bulk RNA-sequencing experiment or mice fed DIO diet for 6 weeks before subsequent scRNAseq. (B) Mesenteric arcades, including arteries of variable sizes and surrounding adipose tissue (excluding intestines), were harvested, and SVF cells were prepared for total RNA isolation. Bulk RNA-seq and Ingenuity Pathway Analysis were performed. Significantly enriched pathways were identified using a 5% false discovery rate cutoff. Data are presented as pathways downregulated or upregulated in SMC-P Klf4Δ/Δ mice when compared to SMC-P Klf4WT/WT mice. Enrichment is shown as −log10 of Padj values. (C) UMAP plots showing integrated Seurat libraries depicting 23 clusters. Four libraries were included in the analysis: eYFP+CD45‒, eYFP+CD45+CD11b+F4/80+ (eYFP+ MФ), eYFP‒CD45+CD11b+F4/80+ (eYFP‒ MФ), and eYFP‒CD45‒ from 4 different conditions as in Figure 2. Cells from the libraries from 6-weeks DIO diet-fed SMC-P Klf4WT/WT mice (left panel) and SMC-P Klf4Δ/Δ mice (right panel) are color-coded by clusters as described in Figure 2. (D) Feature plot for Cd14, one of the top significantly differentially expressed genes in cluster 1. (E) Quantitative analysis for frequency distribution of cells from SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice among different clusters not-segregated based on the library of flow sorting origin. (F) SMC-P Klf4WT/WT (n = 7) and SMC-P Klf4Δ/Δ mice (n = 8) were fed tamoxifen in diet and following a 2-week washout were fed a high fat diet for six weeks. Epididymal adipose tissue SVF cell suspensions were stained with antibodies for CD14. The frequency of CD14+ MФs among CD45+CD11b+F4/80+ MФs was plotted as mean ± SEM. An unpaired t-test with Welch’s correction yielded P = 0.0262.
To avoid potential artifacts associated with peanut oil and autofluorescent MФs appearing as false eYFP+ cells, we present results for the entire dataset (Fig. 6C) without segregating libraries based on cell lineage information. Interestingly, perivascular Klf4 deletion altered multiple non-SMC-P clusters including MФ and endothelial cell marker positive clusters (Fig. 6C – E). Most notably, loss of Klf4 in perivascular cells resulted in a marked decrease in the frequency of cells within cluster 1 that exhibits a pro-inflammatory MФ subtype. One of the top differentially expressed genes for cluster 1 was the monocyte differentiation antigen CD14. Importantly, flow cytometric evaluation of SVF from SMC-P Klf4Δ/Δ vs SMC-P Klf4WT/WT mice showed a decrease in CD14+ CD45+ CD11b+ F4/80+ MФ (Fig. 6F). Interestingly, multifunctional receptor CD14 has been shown to modulate insulin resistance upon high fat diet feeding, specifically CD14 macrophages originating from the bone marrow47. Our results might suggest a crosstalk between perivascular KLF4-mediated signaling events and retention or amplification of CD14+ macrophages in adipose tissue.
We also reevaluated our dataset (from Fig. 2 and Fig. 6) based on eYFP transcript detection to identify cells of perivascular origin (Supp. Fig. VII A). We eliminated cells from the eYFP+ library if eYFP mRNA transcript was absent or eliminated cells from the eYFP− library if eYFP mRNA transcript was present. A dot plot is presented describing overall clusters with traditional marker genes (Supp. Fig. VII B). UMAP analysis of these integrated libraries yielded 25 clusters, two of which were dominated by perivascular phenotypes (Cluster 18 and 19). Replotting UMAPs comparing cell origins based on library preparation and eYFP transcript presence on a feature plot resulted in overlapping profiles (Supp. Fig. VII C). Next we compared SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice SVF profiles (Supp. Fig. VII D). We found that in DIO, perivascular loss of Klf4 did not result in changes in the distribution of perivascular dominated clusters but rather impacted the frequency of pro-inflammatory MФs and increased the abundance of Lyve1+ lymphatic endothelial cells in adipose tissue (Supp. Fig. VII E). Immunohistochemical evaluation of epididymal adipose tissue sections from SMC-P Klf4Δ/Δ and SMC-P Klf4WT/WT mice, also revealed significant increases in LYVE1+ lymphatic vessel area and ACTA2+ vessel area, despite no changes in CD31+ endothelial cell coverage (Supp. Fig. VII and VIII). Based on these results, it is intriguing to speculate that overall increased lymphatic vessel area and ACTA2+ vessel area might contribute to improved glucose tolerance in SMC-P Klf4Δ/Δ mice upon DIO. In addition, we compared differentially expressed genes among all cells or cluster-by-cluster in our scRNAseq datasets of SMC-P Klf4WT/WT and SMC-P Klf4Δ/Δ mice (Supp. Table I). This table shows increased expression of genes such as Acta2 and Cldn5 in SMC-P Klf4Δ/Δ cells, whereas expression of several inflammation and MФ-related genes such as Cd14 were decreased in all cells. Taken together, SMC-P specific deletion of KLF4 resulted in decreased adipose tissue inflammation possibly via modulation of macrophage populations and lymphatic vessel density.
DISCUSSION
Nearly all previous efforts to characterize SMC-P phenotypic switching have been conducted in the context of atherosclerosis or vascular injury48. In this study we sought to define phenotypic transitions of SMC-P within the microvasculature of adipose tissue and how they change during DIO with and without SMC-P KO of KLF4. Using scRNAseq, several studies9,25,49 have taken SMC-P-rich tissues and defined unique populations of phenotypically modulated SMC-P based on their transcriptional profile. The number of distinct populations of SMC-P identified was greatly dependent on the location from which they were isolated, and whether or not the mice were fed a WD. For instance, in a recent study Dobnikar et al9 isolated advanced atherosclerotic lesions from aortas of SMC-lineage tracing mice and found nine unique SMC-P clusters. In contrast, Wirka et al25 reported only one unique “atherosclerosis-associated” SMC-P transcriptomic cluster within aortic root segments from SMC-lineage tracing WD diet fed ApoE−/− mice. This latter claim that perivascular cells give rise to a single “fibromyocyte” population during plaque pathogenesis, however, is most likely due to the fact that samples were dominated by medial not lesion SMC. The observation is clearly at odds with numerous previous studies5,6,9,26–28 that identified several classes of SMCs within atherosclerotic lesions, including our recent scRNAseq study on the lesion SMC24. Herein, we also identified eight transcriptionally unique SMC-P clusters in adipose tissue SVF demonstrating the incredible diversity of perivascular cells (Fig. 4D) and adding to a growing body of evidence that these cells retain extensive plasticity even in adult animals. Of interest, we observed that clusters 20–25 to express some of the known adipocyte progenitor markers in our scRNAseq datasets (Supp. Fig. IX) where clusters 18 and 19 contained the majority of eYFP transcript+ perivascular origin cells. Our findings further support the critical role of perivascular cells in processes essential for survival and reproduction including angiogenesis and blood vessel repair and remodeling11. However, this plasticity also is likely to make them susceptible to undergoing maladaptive changes in the context of disease.
Obesity and related metabolic and cardiovascular complications are a growing worldwide problem, incurring over 340 billion dollars in direct health care costs in the United States alone. Within adipose tissue, obesity progression results in hypertrophy and functional abnormalities of adipocytes leading to recruitment of pro-inflammatory MФs, chronic low-grade inflammation, and insulin resistance45. We observed that DIO was associated with an increase in a cluster dominated by perivascular derived cells (cluster 13) whose distinguishing marker genes include Inhba, Tnnt1, Col4a4, Tcf21, Pdgfra and Wnt6. Pathway and literature analysis using the top 100 differentially expressed genes revealed these cells could be relevant to lymphatic muscle cells, IGF-1 transport and uptake, as well as modification of extracellular matrix components such as collagen. Results suggest that the cells in cluster 13 may play an important role in extracellular matrix remodeling upon DIO. We also observed decreased cell numbers in three other clusters (14, 15, and 16). According to pathway analysis, cells within these clusters are likely involved in regulation of SMC migration, contraction, actin filament bundle assembly, and regulation of arterial blood pressure. In addition, we observed that the frequency of SMC-P lineage traced tdTomato+ cells within the epididymal adipose tissue SVF were decreased with DIO (7.82% vs 4.33% of live singlets, Supp. Fig. V). This observation is in line with previous studies showing decreased microvessel density in adipose tissue with obesity50, which was shown to result in disproportional angiogenesis and hypoxia, as well as obesity induced blood pressure dysregulation51. In line with published literature, we also find perivascular origin cells expressing Rgs552, Notch353, and Kcnj8 (clusters 17–20), Lum, Dcn, and extracellular matrix components in clusters 11 and 12, reported to be perivascular fibroblast like cells43 or fibromyocytes19. Taken together our results show that SMC-P in adipose tissue markedly alter their transcriptome in response to a high fat diet, suggesting that changes in the balance of contractile SMC/pericyte and de-differentiated SMC phenotypes might contribute to obesity associated vascular dysfunction.
Our lab and others have previously shown that KLF4 in SMC-P can have either detrimental effects during atherosclerosis progression5 or beneficial effects for maintenance of perivascular coverage and permeability of terminal arterioles10,55. Therefore, we decided to analyze the effect of loss of KLF4 in SMC-P within adipose tissue. Results of our scRNAseq analyses provide evidence that SMC-P Klf4 KO resulted in reduced adipose tissue inflammation as indicated by a reduction in MФs exhibiting a proinflammatory phenotype. Consistent with this, we previously showed that MCP-1 secretion by cultured SMC in response to cholesterol loading was KLF4-dependent5. Furthermore, KLF4 ChIP-seq analysis identified many other putative KLF4 target genes linked to inflammation, antigen presentation, and immune responses5. Unexpectedly, SMC-P Klf4Δ/Δ mice, despite decreased adipose tissue inflammation, did not show improved whole body insulin sensitivity as indicated by insulin tolerance, fasting insulin levels, and HOMA-IR similar to SMC-P Klf4WT/WT mice. However, SMC-P Klf4 KO mice exhibited improved glucose tolerance. This result is unlikely due to the loss of a single KLF4-regulated gene but rather is the summation of many complex changes, both direct and indirect, resulting from initial loss of KLF4 exclusively in perivascular cells. Possible mechanisms could include increased glucose leakage into the interstitium, enhanced uptake of glucose in different tissue beds, or mitochondrial homeostasis defects56 leading to increased glucose demand that is exacerbated upon DIO. In addition, although we found no changes in overall insulin tolerance, we cannot rule out the possibility of increased insulin sensitivity with improved insulin signaling within specific tissues57.
Single cell RNAseq analysis revealed an increased frequency of Lyve1+ lymphatic endothelial cells in SMC-P Klf4 knockout mice. LYVE1 is the lymphatic vessel endothelial hyaluronan receptor 158 and a marker for lymphatic endothelial cells59. Immunohistochemical analysis indeed revealed an enlarged lymphatic vessel area in adipose tissue of SMC-P Klf4 Δ/Δ mice. Interestingly, obesity is associated with impaired lymphatic function and decreased lymphatic vessel density60,61. Increased lymphatic density with improved microcirculation in SMC-P Klf4Δ/Δ mice might thus contribute to improved glucose tolerance upon DIO. Interestingly, our laboratory previously showed increased leakage of corneal limbal arterioles in SMC-P Klf4Δ/Δ mice10. Thus, SMC-P Klf4Δ/Δ mice may have an increase in lymphatic vessel density in response to increased baseline glucose leakage from arterioles. Taken together, our results suggest that perivascular cells within the adipose tissue microcirculation possess remarkable plasticity and dynamically respond to obesity-induced changes. Klf4-regulated transitions thereby may impact cellular SVF composition and glucose tolerance. Results indicate that Klf4 dependent changes in SMC-P plasticity, that as we previously showed are detrimental in the pathogenesis of atherosclerosis, may also be harmful to whole body metabolism.
Our studies further showcase a problem with using IP injections of tamoxifen reconstituted in peanut oil to excise floxed alleles. This method has been used in thousands of high impact studies over the last decade to induce conditional gene knockouts and/or in lineage tracing. Herein, we found that it is associated with a marked increase in the autofluorescence of peritoneal adipose tissue MФs thereby leading to their false identification as eYFP+ cells using flow cytometry. It also led to the activation of proinflammatory responses in peritoneal MФs that may have confounded results believed to be exclusively a function of the conditional knockout of a candidate gene of interest. We believe that the increase in autofluorescence is due to phagocytosis and clearance of the peanut oil by MФs. Further studies will be needed to show if this phenomenon is unique to peanut oil or includes other lipids. By providing tamoxifen in normal diet and switching to a tdTomato reporter with greater spectral separation, we were able to virtually eliminate this confounding variable. Our results are consistent with previous studies showing that sunflower, corn or peanut oil injections elicit angiogenic and lymphangiogenic responses in the avascular mouse mesentery62. In addition, peanut allergens induce production of specific IgE anitbodies63. Although we reduced the autofluorescence problem by using Myh11-DreERT2tdTom mice fed a tamoxifen-containing diet, we cannot rule out the possibility that oral delivery of tamoxifen may result in inflammatory responses along the gastrointestinal tract. Taken together, our study highlights that IP tamoxifen injections in peanut oil have unintended consequences that may confound interpretation of results from studies that did not use an appropriate genetic control mouse (a congenic wild type littermate that only differes by not having floxed gene KO alleles) that is treated identically experimentally including being injected IP with tamoxifen in peanut oil. If non-tamoxifen treated mice are used as controls this MФ response becomes a huge undefined and unintended experimental variable. Importantly, we did not detect significant differences in the number of eYFP+ or TdTomato+ cells in aortas either after IP injections in peanut oil or tamoxifen diet (Supp. Fig. III F) indicating that the eYFP/oil macrophage autofluorescence effect is not present in aortas. Nevertheless, our results with peritoneal MФ call for increased attention to the use of correct and potentially multiple controls in all animal experiments that involve inducible Cre/Dre recombinases.
Finally, our results add to a growing body of evidence that pro-inflammatory signaling in SMC-P cells plays a key role in obesity-associated complications and atherosclerosis. For example, we recently showed that SMC-P specific deletion of the Interleukin-1 receptor 1 (Il1r1) resulted in dramatic reductions in atherosclerotic lesion size and impaired investment of SMC into the protective fibrous cap64. In contrast, deletion of the Il1r1 in MФs and granulocytes using a LysM Cre system had no effect on lesion size or composition. Moreover, of major relevance to the present studies, Sui et al. showed that Tagln-Cre-IKKβΔ/Δ mice were resistant to developing diet-induced obesity and insulin resistance, as well as, atherosclerosis15. These latter results are highly intriguing and unexpected since the general dogma is that pro-inflammatory responses of immune cells, not perivascular cells, play a dominant role in initiating and sustaining chronic inflammation associated with DIO. Unfortunately, the constitutive Tagln(SM22α)-Cre mouse line used by Sui et al. not only targets SMC-P but also a number of other cell types including cardiomyocytes, adipocytes, myofibroblasts, and platelets that express SM22α during development, wound healing, or disease states17. Interestingly, our dataset provides a valuable resource for generating novel hypothesis on molecular mechanisms that may induce perivascular pro-inflammatory phenotypes including those associated with increased expression of Il6, Il1r1, or the senescence marker Cdkn1a. Thus going forward, it will be of interest to study loss of function models to define the contribution of SMC-P pro-inflammatory responses to adipose tissue inflammation. This research may also help to identify novel potential therapeutic targets for treating or preventing microvascular disease and metabolic dysfunction.
Supplementary Material
Highlights.
Intraperitoneal injection of tamoxifen in peanut oil increases autofluorescence of adipose tissue macrophages causing false identification as eYFP+ cells.
Single cell RNAseq using Myh11-DreERT2tdTomato mice given oral tamoxifen identifies eight perivascular cell dominated clusters, half of which are altered upon diet induced obesity.
Smooth muscle cell/pericyte-specific Klf4 knockout mice display marked decreases in pro-inflammatory macrophages and increases in LYVE1+ lymphatic endothelial cells in epididymal adipose tissue and exhibit improved glucose tolerance.
ACKNOWLEDGMENTS
We are thankful for helpful discussions with Vlad Serbulea and Hema Kothari and technical assistance of Angie Washington, Amber Saenz, Rupa Tripathi, Ana Tsiskarishvili and Sophia Kirmani. We are also very thankful to The University of Virginia Flow Cytometry Core facility, Genome Technologies Core facility and Advanced Microscopy Facility.
SOURCES OF FUNDING
This work was supported by funding from NIH Funding T32 HL007284, R01 HL132904, R01 HL135018, R01 HL141425, and R01 HL136314 to G.K.O; as well as AHA Postdoctoral Fellowship 19POST34380085 to G.B.
Nonstandard Abbreviations and Acronyms:
- ACTA2
actin alpha 2, smooth muscle
- APOE
apolipoprotein E
- AT
adipose tissue
- BCA
brachiocephalic artery
- DIO
diet induced obesity
- EndoMT
endothelial to mesenchymal transition
- eYFP
enhanced yellow fluorescent protein
- FMO
fluorescence minus one
- G-CSF
granulocyte-colony stimulating factor
- GTT
glucose tolerance test
- IGF
insulin-like growth factor
- IKKβ
IκB kinase β
- IL-1R1
interleukin-1 receptor 1
- IP
intraperitoneal
- ITT
insulin tolerance test
- KLF4
kruppel like factor 4
- LGALS3
Galectin-3
- LYVE1
lymphatic vessel endothelial hyaluronan receptor 1
- MCP-1
monocyte chemotactic protein
- MYH11
myosin heavy chain 11
- MФ
macrophages
- OCT4
octamer-binding transcription factor 4
- PDGFRβ
platelet derived growth factor receptor β
- scRNAseq
single cell RNA sequencing
- SD
standard deviation
- SEM
standard error of the mean
- SM22
smooth muscle protein 22-alpha
- SMC-P
smooth muscle cells and pericytes
- SVF
stromovascular fraction
- UMAP
uniform manifold approximation and projection
- WD
western diet
Footnotes
DISCLOSURES
None
REFERENCES
- 1.Liu M, Gomez D. Smooth Muscle Cell Phenotypic Diversity. Arterioscler Thromb Vasc Biol. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basatemur GL, Jørgensen HF, Clarke MCH, Bennett MR, Mallat Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol 2019; [DOI] [PubMed] [Google Scholar]
- 4.Gomez D, Shankman LS, Nguyen AT, Owens GK. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat Methods. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014; [DOI] [PubMed] [Google Scholar]
- 7.Cherepanova OA, Gomez D, Shankman LS, Swiatlowska P, Williams J, Sarmento OF, Alencar GF, Hess DL, Bevard MH, Greene ES, Murgai M, Turner SD, Geng YJ, Bekiranov S, Connelly JJ, Tomilin A, Owens GK. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat Med. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speer MY, Yang HY, Brabb T, Leaf E, Look A, Lin WL, Frutkin A, Dichek D, Giachelli CM. Smooth muscle cells give rise to osteochondrogenic precursors and chondrocytes in calcifying arteries. Circ Res. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haskins RM, Nguyen AT, Alencar GF, Billaud M, Kelly-Goss MR, Good ME, Bottermann K, Klibanov AL, French BA, Harris TE, Peirce SM, Isakson BE, Owens GK. Klf4 has an unexpected protective role in perivascular cells within the microvasculature. Am J Physiol Circ Physiol. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess DL, Kelly-Goss MR, Cherepanova OA, Nguyen AT, Baylis RA, Tkachenko S, Annex BH, Peirce SM, Owens GK. Perivascular cell-specific knockout of the stem cell pluripotency gene Oct4 inhibits angiogenesis. Nat Commun [Internet]. 2019;1–15. Available from: 10.1038/s41467-019-08811-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006; [DOI] [PubMed] [Google Scholar]
- 13.Van Gaal LF, Mertens IL, De Block CE. Mechanisms linking obesity with cardiovascular disease. Nature. 2006; [DOI] [PubMed] [Google Scholar]
- 14.Czech MP. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sui Y, Park SH, Xu J, Monette S, Helsley RN, Han SS, Zhou C. IKKβ links vascular inflammation to obesity and atherosclerosis. J Exp Med. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson LE, Soriano P. PDGFRβ signaling regulates mural cell plasticity and inhibits fat development. Dev Cell. 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty R, Saddouk FZ, Carrao AC, Krause DS, Greif DM, Martin KA. Promoters to Study Vascular Smooth Muscle. Arterioscler Thromb Vasc Biol. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hepler C, Shan B, Zhang Q, Henry GH, Shao M, Vishvanath L, Ghaben AL, Mobley AB, Strand D, Hon GC, Gupta RK. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattaneo P, Mukherjee D, Spinozzi S, Zhang L, Larcher V, Stallcup WB, Kataoka H, Chen J, Dimmeler S, Evans SM, Guimarães-Camboa N. Parallel Lineage-Tracing Studies Establish Fibroblasts as the Prevailing In Vivo Adipocyte Progenitor. Cell Rep. 2020; [DOI] [PubMed] [Google Scholar]
- 20.Inaba T, Shimano H, Gotoda T, Harada K, Shimada M, Ohsuga JI, Watanabe Y, Kawamura M, Yazaki Y, Yamada N. Expression of platelet-derived growth factor β receptor on human monocyte-derived macrophages and effects of platelet-derived growth factor BB dimer on the cellular function. J Biol Chem. 1993; [PubMed] [Google Scholar]
- 21.Beitz JG, Kim IS, Calabresi P, Frackelton AR. Human microvascular endothelial cells express receptors for platelet-derived growth factor. Proc Natl Acad Sci U S A. 1991; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiong J, Kawagishi H, Yan Y, Liu J, Wells QS, Edmunds LR, Fergusson MM, Yu ZX, Rovira II, Brittain EL, Wolfgang MJ, Jurczak MJ, Fessel JP, Finkel T. A Metabolic Basis for Endothelial-to-Mesenchymal Transition. Mol Cell. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, Castellot JJ, Rosen ED, Spiegelman BM. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alencar GF, Owsiany KM, K S, Sukhavasi K, Mocci G, Nguyen A, Williams CM, Shamsuzzaman S, Mokry M, Henderson CA, Haskins R, Baylis RA, Finn AV, McNamara CA, Zunder ER, Venkata V, Pasterkamp G, Björkegren J, Bekiranov S, Owens GK. The Stem Cell Pluripotency Genes Klf4 and Oct4 Regulate Complex SMC Phenotypic Changes Critical in Late-Stage Atherosclerotic Lesion Pathogenesis. Circulation. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, Quertermous T. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation. 2014; [DOI] [PubMed] [Google Scholar]
- 27.Misra A, Feng Z, Chandran RR, Kabir I, Rotllan N, Aryal B, Sheikh AQ, Ding L, Qin L, Fernández-Hernando C, Tellides G, Greif DM. Integrin beta3 regulates clonality and fate of smooth muscle-derived atherosclerotic plaque cells. Nat Commun. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Dubland JA, Allahverdian S, Asonye E, Sahin B, Jaw JE, Sin DD, Seidman MA, Leeper NJ, Francis GA. Smooth Muscle Cells Contribute the Majority of Foam Cells in ApoE (Apolipoprotein E)-Deficient Mouse Atherosclerosis. Arterioscler Thromb Vasc Biol. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, Hamers AAJ, Cochain C, Vafadarnejad E, Saliba AE, Zernecke A, Pramod AB, Ghosh AK, Michel NA, Hoppe N, Hilgendorf I, Zirlik A, Hedrick CC, Ley K, Wolf D. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinet P, Milewicz DM, Cassis LA, Leeper NJ, Lu HS, Smith JD. Consideration of Sex Differences in Design and Reporting of Experimental Arterial Pathology Studies-Statement from ATVB Council. Arterioscler Thromb Vasc Biol. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; [DOI] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu SCH, Wang Q, Lienhard GE, Keller SR. Insulin receptor substrate 3 is not essential for growth or glucose homeostasis. J Biol Chem. 1999; [DOI] [PubMed] [Google Scholar]
- 38.Fantin VR, Keller SR, Lienhard GE, Wang LM. Insulin receptor substrate 4 supports insulin- and interleukin 4-stimulated proliferation of hematopoietic cells. Biochem Biophys Res Commun. 1999; [DOI] [PubMed] [Google Scholar]
- 39.Zhi X, Wang J, Lu P, Jia J, Shen H Bin, Ning G. AdipoCount: A new software for automatic adipocyte counting. Front Physiol. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver G, Kipnis J, Randolph GJ, Harvey NL. The Lymphatic Vasculature in the 21st Century: Novel Functional Roles in Homeostasis and Disease. Cell. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imrie H, Abbas A, Viswambharan H, Rajwani A, Cubbon RM, Gage M, Kahn M, Ezzat VA, Duncan ER, Grant PJ, Ajjan R, Wheatcroft SB, Kearney MT. Vascular insulin-like growth factor-I resistance and diet-induced obesity. Endocrinology. 2009; [DOI] [PubMed] [Google Scholar]
- 42.Mughal RS, Bridge K, Buza I, Slaaby R, Worm J, Klitgaard-Povlsen G, Hvid H, Schiødt M, Cubbon R, Yuldasheva N, Skromna A, Makava N, Skytte-Olsen G, Kearney MT. Effects of obesity on insulin: insulin-like growth factor 1 hybrid receptor expression and Akt phosphorylation in conduit and resistance arteries. Diabetes Vasc Dis Res. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berryman DE, Glad CAM, List EO, Johannsson G. The GH/IGF-1 axis in obesity: Pathophysiology and therapeutic considerations. Nat. Rev. Endocrinol 2013; [DOI] [PubMed] [Google Scholar]
- 44.AsghariHanjani N, Vafa M. The role of IGF-1 in obesity, cardiovascular disease, and cancer. Med J Islam Repub Iran. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat. Rev. Endocrinol 2017; [DOI] [PubMed] [Google Scholar]
- 46.Birsoy K, Chen Z, Friedman J. Transcriptional Regulation of Adipogenesis by KLF4. Cell Metab. 2008;7:339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernández-Real JM, Del Pulgar SP, Luche E, Moreno-Navarrete JM, Waget A, Serino M, Sorianello E, Sánchez-Pla A, Pontaque FC, Vendrell J, Chacón MR, Ricart W, Burcelin R, Zorzano A. CD14 modulates inflammation-driven insulin resistance. Diabetes. 2011;60:2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Chen W, Li W, Li Y, Priest JR, Zhou B, Wang J, Zhou Z. Single-Cell RNA-Seq of the Developing Cardiac Outflow Tract Reveals Convergent Development of the Vascular Smooth Muscle Cells. Cell Rep. 2019; [DOI] [PubMed] [Google Scholar]
- 50.Frisbee JC. Obesity, insulin resistance, and microvessel density. Microcirculation. 2007; [DOI] [PubMed] [Google Scholar]
- 51.Ottolini M, Hong K, Cope EL, Daneva Z, DeLalio LJ, Sokolowski JD, Marziano C, Nguyen NY, Altschmied J, Haendeler J, Johnstone SR, Kalani MY, Park MS, Patel RP, Liedtke W, Isakson BE, Sonkusare SK. Local Peroxynitrite Impairs Endothelial TRPV4 Channels and Elevates Blood Pressure in Obesity. Circulation. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bondjers C, Kalén M, Hellström M, Scheidl SJ, Abramsson A, Renner O, Lindahl P, Cho H, Kehrl J, Betsholtz C. Transcription profiling of platelet-derived growth factor-B-deficient mouse embryos identifies RGS5 as a novel marker for pericytes and vascular smooth muscle cells. Am J Pathol. 2003; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu H, Kennard S, Lilly B. NOTCH3 expression is induced in mural cells through an autoregulatory loop that requires Endothelial-expressed JAGGED1. Circ Res. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018; [DOI] [PubMed] [Google Scholar]
- 55.Murgai M, Ju W, Eason M, Kline J, Beury DW, Kaczanowska S, Miettinen MM, Kruhlak M, Lei H, Shern JF, Cherepanova OA, Owens GK, Kaplan RN. KLF4-dependent perivascular cell plasticity mediates pre-metastatic niche formation and metastasis. Nat Med. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao X, Zhang R, Lu Y, Prosdocimo DA, Sangwung P, Zhang L, Zhou G, Anand P, Lai L, Leone TC, Fujioka H, Ye F, Rosca MG, Hoppel CL, Christian Schulze P, Dale Abel E, Stamler JS, Kelly DP, Jain MK. Kruppel-like factor 4 is critical for transcriptional control of cardiac mitochondrial homeostasis. J Clin Invest. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Luca C, Olefsky JM. Stressed out about obesity and insulin resistance. Nat. Med 2006; [DOI] [PubMed] [Google Scholar]
- 58.Jackson DG. Immunological functions of hyaluronan and its receptors in the lymphatics. Immunol. Rev 2009; [DOI] [PubMed] [Google Scholar]
- 59.Lokmic Z Utilizing lymphatic cell markers to visualize human lymphatic abnormalities. J. Biophotonics 2018; [DOI] [PubMed] [Google Scholar]
- 60.Savetsky IL, Torrisi JS, Cuzzone DA, Ghanta S, Albano NJ, Gardenier JC, Joseph WJ, Mehrara BJ. Obesity increases inflammation and impairs lymphatic function in a mouse model of lymphedema. Am J Physiol - Hear Circ Physiol. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Escobedo N, Oliver G. The Lymphatic Vasculature: Its Role in Adipose Metabolism and Obesity. Cell Metab. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suarez-Martinez AD, Peirce SM, Isakson BE, Nice M, Wang J, Lounsbury KM, Scallan JP, Murfee WL. Induction of microvascular network growth in the mouse mesentery. Microcirculation. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palladino C, Breiteneder H. Peanut allergens. Mol Immunol. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gomez D, Baylis RA, Durgin BG, Newman AAC, Alencar GF, Mahan S, St. Hilaire C, Müller W, Waisman A, Francis SE, Pinteaux E, Randolph GJ, Gram H, Owens GK. Interleukin-1β has atheroprotective effects in advanced atherosclerotic lesions of mice. Nat Med. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


