Abstract
Nowadays there is an ongoing acute respiratory outbreak caused by the novel highly contagious coronavirus (COVID-19). The diagnostic protocol is based on quantitative reverse-transcription polymerase chain reaction (RT-PCR) and chests CT scan, with uncertain accuracy. This meta-analysis study determines the diagnostic value of an initial chest CT scan in patients with COVID-19 infection in comparison with RT-PCR. Three main databases; PubMed (MEDLINE), Scopus, and EMBASE were systematically searched for all published literature from January 1st, 2019, to the 21st May 2020 with the keywords "COVID19 virus", "2019 novel coronavirus", "Wuhan coronavirus", "2019-nCoV", "X-Ray Computed Tomography", "Polymerase Chain Reaction", "Reverse Transcriptase PCR", and "PCR Reverse Transcriptase". All relevant case-series, cross-sectional, and cohort studies were selected. Data extraction and analysis were performed using STATA v.14.0SE (College Station, TX, USA) and RevMan 5. Among 1022 articles, 60 studies were eligible for totalizing 5744 patients. The overall sensitivity, specificity, positive predictive value, and negative predictive value of chest CT scan compared to RT-PCR were 87% (95% CI 85–90%), 46% (95% CI 29–63%), 69% (95% CI 56–72%), and 89% (95% CI 82–96%), respectively. It is important to rely on the repeated RT-PCR three times to give 99% accuracy, especially in negative samples. Regarding the overall diagnostic sensitivity of 87% for chest CT, the RT-PCR testing is essential and should be repeated to escape misdiagnosis.
Subject terms: Cell biology, Genetics, Health care
Introduction
In late December of 2019, a cluster of patients was diagnosed with a strange viral pneumonia in Wuhan City, Hubei Province, China, which later was confirmed to be caused by the novel coronavirus (the disease named COVID-19)1. Up to now, millions of cases have been identified, causing thousands of deaths at an alarming pace worldwide. Officially, the World Health Organization has declared the pandemic of COVID-192 and due to the non-existence of effective antiviral drug or vaccine, both detecting patient at an early stage and immediate patient isolation play a mandatory role in the fighting against COVID-193.
The chest computed tomography (CT) scan plays a central role on the disease staging and checking the treatment efficacy, while the reverse transcription-polymerase chain reaction (RT-PCR) remains the mainstay of COVID-19 diagnosis4,5, though limited to identify the virus, which poses important restrictions6.
Recent studies claim that initial chest CT may enable the detection of the disease with higher sensitivity in comparison to RT-PCR7. This systematic review and meta-analysis were performed to determine the diagnostic accuracy of the initial chest CT scan compared to RT-PCR in COVID-19 patients.
Materials and methods
All stages of this study followed the PRISMA guidelines and all relevant English, Chinese, and other language case-series, cross-sectional, and cohort studies were selected and checked for scientific validity.
Inclusion criteria: observational epidemiological study design, clear report of the number of positive cases by PCR and chest CT, and the ability to calculate accuracy indicators.
Exclusion criteria: case reports or not meeting one or more inclusion criteria.
Search strategy
All relevant literature from three main databases: MEDLINE (PubMed), Scopus, and EMBASE were explored from January 1st, 2019, to the 21th May 2020, using the keywords “COVID19 virus”, “2019 novel coronavirus”, “Wuhan coronavirus”, “2019-nCoV”, “X-Ray Computed Tomography”, “Polymerase Chain Reaction”, “Reverse Transcriptase PCR”, and “PCR Reverse Transcriptase” (Supplementary file 1). The references of the selected articles were also reviewed.
The variables extracted included the first author name, publication year, country and city of the study, subjects average age, gender, study design, total sample size, true positives, true negatives, false positives, and false negatives.
Data extraction and statistical analysis
Two researchers (SZA and SSTZ) screened articles separately by checking titles and abstracts. Disagreements were solved by a third one (FKH). Included articles had data of confirmed COVID-19 patients by chest CT scan and quantitative real-time polymerase chain reaction (RT-PCR) and were accessed in full text. The quality assessment was performed by the Newcastle–Ottawa Scale (NOS) assessment tool. The papers that receive scores more than 6 were reflected as the “high quality” and underwent additional meta-analysis steps.
The outcomes of interest, including the CT-scan to identify COVID-19 were submitted to summary receiver operating characteristic (SROC) curve by the random effect model for sensitivity and specificity.
Sensitivity, indicating the capacity of index test to identify patients, considered by “Sensitivity = TP/(TP + FN)”. Specificity as the examination to remove disease-free, calculated by “Specificity = TN/(FP + TN)”. The Metaprop command to calculate sensitivity and specificity excluded studies that have reported 100% sensitivity or specificity.
Positive Predictive Value (PPV) is the probability of disease if the test is positive calculated by "Positive predictive Value = TP/(FP + TP)". Negative Predictive Value (NPV) is the probability of disease-free if the test is negative calculated by "negative predictive value = TN/(FN + TN)".
The Cochran's Q-test of heterogeneity at 5% was used to evaluate statistical heterogeneity based on the Higgins classification in which an I2 > 75% means significant heterogeneity.
Deeks’ funnel plot was used to evaluate publication bias by the “Metafunnel”. Briefly, to create the funnel plot, the odds ratio were first calculated using the equations of (TP/FN)/(FP/TN) and after estimating the odds ratio logarithm, the standard error of odds ratio was calculated. Extracted data were collected in Excel 2007 (Microsoft Corporation, Redmond, CA) and analysis was done by using STATA v.14.0SE (College Station, TX, USA) and RevMan 5.
Results
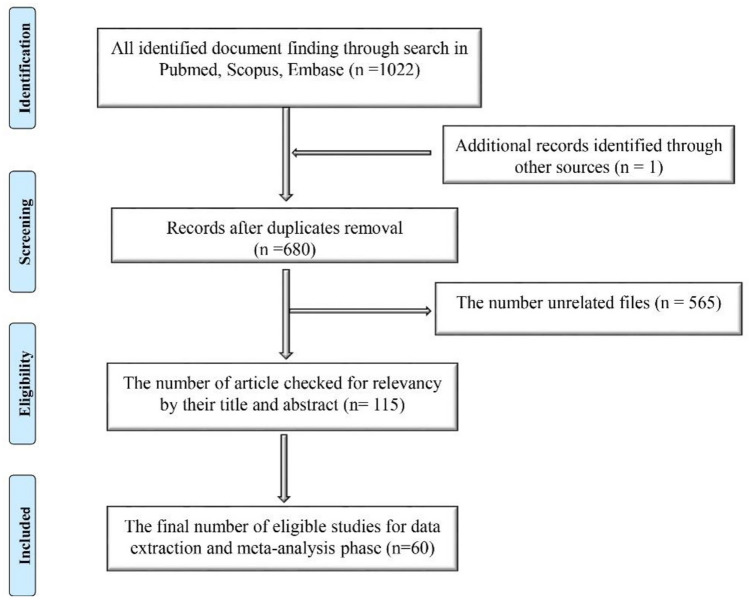
Among 1022 identified articles, 115 were considered eligible and after the NOS screening, 60 articles, including 5744 subjects were included, all published in the first quarter of 2020 (Fig. 1).
Figure 1.
The number of articles during several steps based on the PRISMA flow diagram (2009). (http://www.prisma-statement.org/PRISMAStatement/CitingAndUsingPRISMA.aspx).
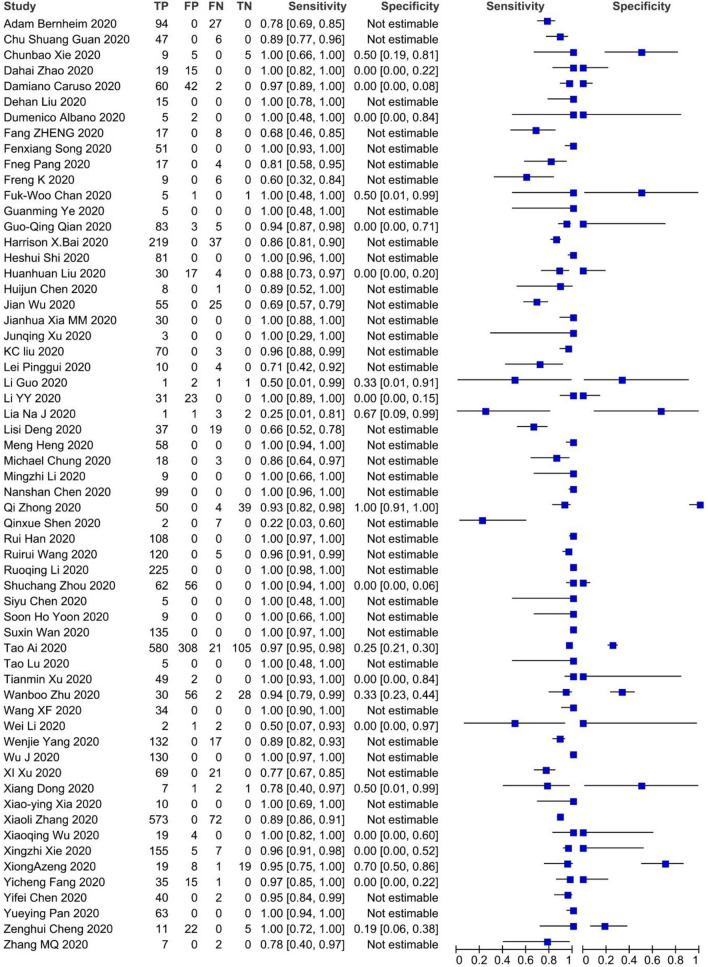
A summary of the information of included articles is shown in Table 1. The forest plot, False Positives (FP), False Negatives (FN), True Positives (TP), True Negatives (TN), Sensitivity, Specificity, and 95% Confidence Intervals (CI) of each study are shown in Fig. 2 .
Table 1.
Data of 60 included studies in the data extraction step.
| N. | 1st author | Country (city) | Gender% male | Age (mean, range, year) | Type of study | Sample size |
|---|---|---|---|---|---|---|
| 1 | Adam Bernheim1 | United States (New York) | 50% | 45, 18–80 | Case-series | 121 |
| 2 | Chun Shuang Guan8 | China (Beijing) | 47% | 42, 1–86 | Case-series | 53 |
| 3 | Chunbao Xie8 | China (Chengdu) | 58% | 33, 8–62 | Cross-sectional | 19 |
| 4 | Dahai Zhao9 | China (Anhui) | 50% | 42, 27–56 | Cohort | 34 |
| 5 | Damiano Caruso10 | Italy (Rome) | 53% | 57, 18–89 | Case-series | 158 |
| 6 | Dehan Liu11 | China (Wuhan) | 0% | 32, 23–40 | Case-series | 15 |
| 7 | Domenico Albano12 | Italy (Brescia) | 29% | 65, 55–79 | Case-series | 7 |
| 8 | Fang Zheng11 | China (Wuhan) | 56% | 3, 2–9 | Case-series | 25 |
| 9 | Fengxiang Song13 | China (Shanghai) | 49% | 49, 33–65 | Case-series | 51 |
| 10 | Fneg Pang14 | China (Wuhan) | 29% | 40, 25–63 | Case-series | 21 |
| 11 | Feng Kai15 | China (Shenzhen) | 33% | 8, 4–14 | Case-series | 15 |
| 12 | Jasper Fuk-Woo Chan16 | China (Hong Kong) | 50% | 46, 33–66 | Case-series | 6 |
| 13 | Guangming Ye17 | China (Wuhan) | 40% | 32, 27–42 | Case-series | 5 |
| 14 | Guo-Qing Qian18 | China (Ningbo) | 41% | 50, 5–96 | Case-series | 91 |
| 15 | Harrison X. Bai19 | China (Changsha) | NS | NS | Cohort | 256 |
| 16 | Heshui Shi18 | China (Wuhan) | 52% | 49.5, 39–61 | Case-series | 81 |
| 17 | Huanhuan Liu20 | China (Shanghai) | 14% | 20 (2 month–58 years) | Case-series | 51 |
| 18 | Huijun Chen21 | China (Wuhan) | 0% | 30, 26–40 | Case-series | 9 |
| 19 | Jian Wu22 | China (Yuncheng) | 49% | 46, 4–> 65 | Case-series | 80 |
| 20 | Jianhua Xia MM23 | China (Zhejiang) | 70% | 54.5, 13–74 | Cross-sectional | 30 |
| 21 | Junqing Xu24 | China (Shenzhen) | 0% | 52, 45–65 | Case-series | 3 |
| 22 | KC Liu22 | China (Hefei) | 51% | 42, 5–86 | Cohort | 73 |
| 23 | Pinggui Lei25 | China (Guiyang) | 57% | 47 (12–83) | Case-series | 14 |
| 24 | Li Guo23 | China (Beijing) | 50% | 35, 2–64 | Cross-sectional | 6 |
| 25 | Li Yuanyuan26 | China (Wuhan) | 46% | 52, 25–82 | Cross-sectional | 54 |
| 26 | Lia Na Ji27 | China (Beijing) | NS | NS | Case-series | 7 |
| 27 | Lisi Deng28 | china (Zhuha) | NS | ≥ 18 year | Cohort | 56 |
| 28 | Heng Meng26 | China (Wuhan) | 45% | 43 | Case-series | 58 |
| 29 | Michael Chung29 | United States (New York) | 61% | 51, 29–77 | Case-series | 21 |
| 30 | Mingzhi Li30 | China (Nanchang) | 55.5% | 43, 31–68 | Case-series | 9 |
| 31 | Nanshan Chen31 | China (Wuhan) | 68% | 55.5, 21–82 | Case-series | 99 |
| 32 | Qi Zhong32 | China (Wuhan) | 23% | 32, 28–35 | Cohort | 93 |
| 33 | Qinxue Shen33 | China (Hunan) | 33% | 8, 1–12 | Case-series | 9 |
| 34 | Rui Han34 | China (Wuhan) | 35% | 45, 21–95 | Case-series | 108 |
| 35 | Ruirui Wang33 | China (Anhui) | 57% | 39, 1–80 | Case-series | 125 |
| 36 | Ruoqing Li35 | China (Chongqing) | 53% | 50 | Case-series | 225 |
| 37 | Shuchang Zhou34 | China (Wuhan) | 63% | 53, 30–77 | Case-series | 118 |
| 38 | Siyu Chen36 | China (Chongqing) | 0% | 29, 25–31 | Case-series | 5 |
| 39 | Soon Ho Yoon37 | Korea (Seoul) | 44% | 54 | Case-series | 9 |
| 40 | Suxin Wan38 | China (Chongqing) | 53% | 47, 36–55 | Cross-sectional | 135 |
| 41 | Tao Ai36 | China (Wuhan) | 46% | 51, 2–95 | Cross-sectional | 1014 |
| 42 | Tao Lu32 | China (Sichuan) | 20% | 52, 41–62 | Case-series | 5 |
| 43 | Tianmin Xu39 | China (Changzhou) | 49% | 42, 24–65 | Cohort | 51 |
| 44 | Wanbo Zhu34 | China (Hefei) | 48% | 40, 27–53 | Case-series | 116 |
| 45 | Wang XF33 | China (Shenzhen) | 41% | 9 | Case-series | 34 |
| 46 | Wei Li18 | China (Zhuhai) | 80% | 3 (10 month–6 years) | Case-series | 5 |
| 47 | Wenjie Yang38 | China (Shanghai) | 54% | 45 | Case-series | 149 |
| 48 | Wu Jing40 | China (Nanjing) | 40% | 52, 25–80 | Case-series | 130 |
| 49 | Xi Xu41 | China (Guangzhou) | 43% | 50, 18–86 | Case-series | 90 |
| 50 | Xiang Dong31 | China (Wuhan) | 45% | 37(2–69) | Case-series | 11 |
| 51 | Xiao-ying Xia42 | China (Chongqing) | 60% | 56.5, 43–71 | Case-series | 10 |
| 52 | Xiaoli Zhang43 | China (Zhejiang) | 51% | 46 | Cross-sectional | 645 |
| 53 | Xiaoqing Wu43 | China (Wuhan) | 0% | 29, 21–36 | Case-series | 23 |
| 54 | Xingzhi Xie44 | China (Changsha) | NS | NS | Cross-sectional | 167 |
| 55 | Xiong Zeng40 | China (Changsha) | NS | NS | Cross-sectional | 47 |
| 56 | Yicheng Fang44 | China (Shanghai) | 57% | 45, 39–55 | Case-series | 51 |
| 57 | Yifei Chen45 | China (Wuhan) | 36% | 51 (42–62) | Case-series | 42 |
| 58 | Yueying Pan46 | China (Wuhan) | 52% | 45 | Case-series | 63 |
| 59 | Zenghui Cheng46 | China (Shanghai) | NS | NS | Cross-sectional | 38 |
| 60 | Zhang MQ47 | China (Beijing) | 56% | 36, 15–49 | Case-series | 9 |
NS not stated.
Figure 2.
Sensitivity and specificity of 60 included studies.
Sensitivity ranged from 25 to 100% and the specificity was estimated to vary from 19 to 70%.
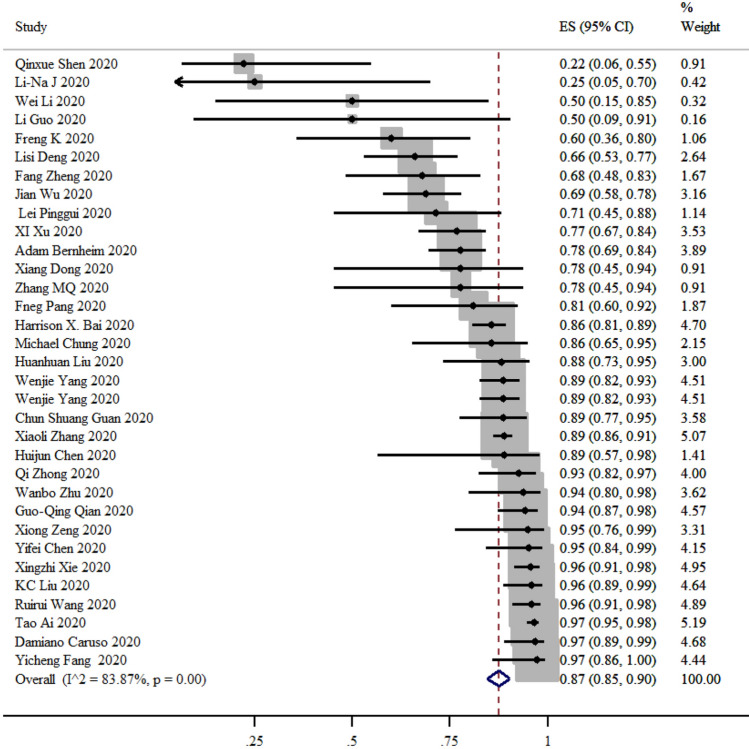
After excluding studies reporting 100% sensitivity or specificity, the sensitivity was ranging from 25 to 97% and specificity from 25 to 70% (Figs. 3, 4). Based on 37 studies, the sensitivity of CT compared to PCR was 87% (95% CI 85–90%) and based on seven studies the specificity of CT was 46% (95% CI 29–63%).
Figure 3.
Summary of sensitivity and 95% CI, generated by the STATA.
Figure 4.
Summary of specificity and 95% CI, generated by the STATA.
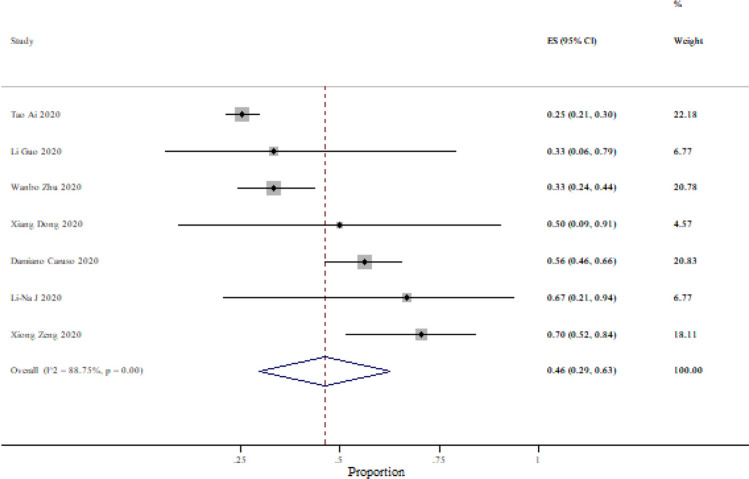
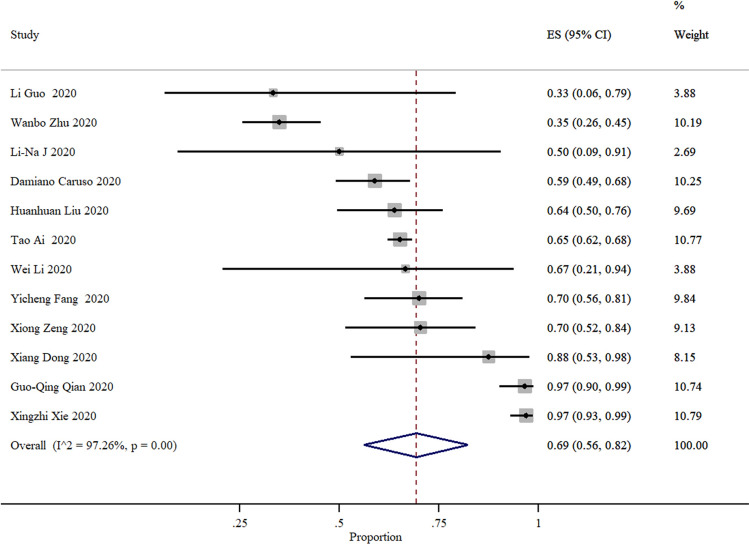
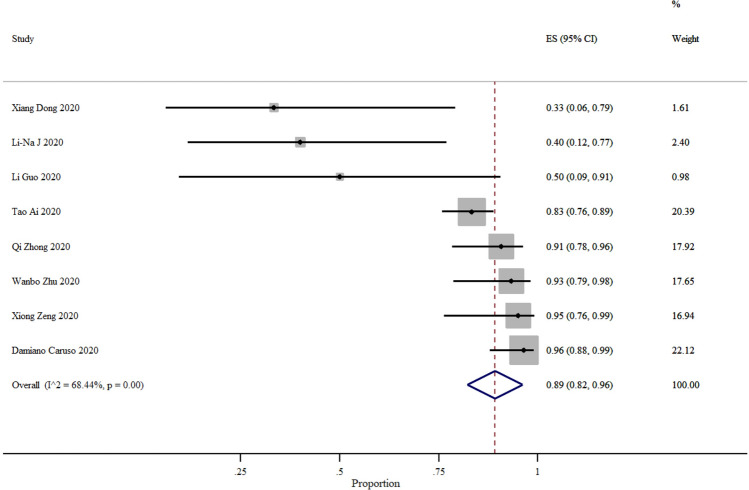
The positive predictive value of CT was 69% (95% CI 56–72%) and the negative predictive value was 89% (95% CI 82–96%) and the variation of the estimated numbers were 33% to 97% and 33% to 96% for PPV and NPV, respectively (Figs. 5, 6).
Figure 5.
Summary of positive predictive value and 95% CI of 18 studies, generated by the STATA.
Figure 6.
Summary of negative predictive value and 95% CI of six studies, generated by the STATA.
The symmetry between the two sides of the funnel plot regression line indicates that the included publications are not biased. However, due to a large number of zeros in the FP and TN cells, it was possible to calculate the odds ratio for six studies only, and the interpretation of this plot in our study should be done with caution (Fig. 7).
Figure 7.
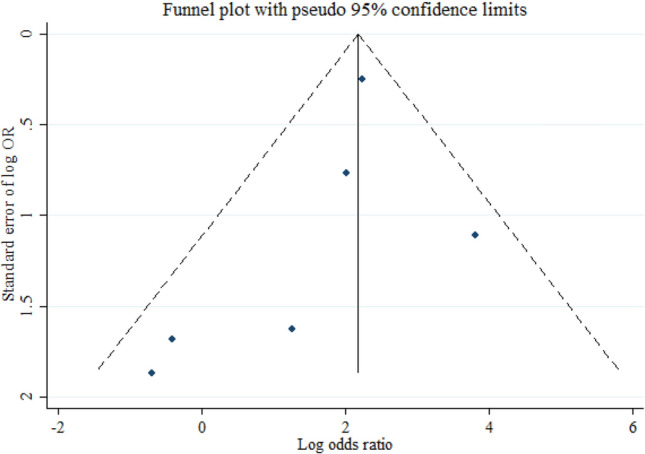
The Deeks’ funnel plot curve for assessment of publication bias.
Table 2 includes 35 studies with the first RT-PCR test of the suspected patients available (subsequent RT-PCR data were included if available). Moreover, the swabs should have been taken from sputum, nasopharyngeal, oropharyngeal, nose, or throat and if a combination was used, nasopharyngeal or throat swab was considered as the primary.
Table 2.
The number of positive test results in RT-PCR testing and the number of confirmed ones.
| N. | First author | N. of total cases | Number of total confirmed patients | N. of patients confirmed with the first RT-PCR test (perc.a) | N. of patients confirmed with the second RT-PCR test (perc.b) | N. of patients confirmed with the third RT-PCR test (perc.c) | N. of patients confirmed with the fourth RT-PCR test (perc.d) | N. of Patients confirmed later (perc.e) |
|---|---|---|---|---|---|---|---|---|
| 1 | Tao Ai36 | 1014 | 909 | 343 (37.7%) | 205 (22.6%) | 45 (5%) | 8 (0.9%) | 308 (33.9%) |
| 2 | Xingzhi Xie44 | 167 | 167 | 162 (97%) | 2 (1.2%) | 2 (1.2%) | 0 (0%) | 1 (0.6%) |
| 3 | Jian Wu22 | 80 | 80 | 41 (51.2%) | 30 (37.5%) | 9 (11.3%) | 0 (0%) | 0 (0%) |
| 4 | Anne Kimball48 | 82 | 76 | 23 (30%) | _f | _ | _ | 53 (70%) |
| 5 | Li Yuanyuan49 | 54 | 54 | 31 (57%) | _ | _ | _ | 23 (42.6%) |
| 6 | Chenyao Lin50 | 52 | 52 | 23 (44%) | _ | _ | _ | 29 (56%) |
| 7 | Yicheng Fang44 | 51 | 51 | 36 (70.6%) | 12 (23.6%) | 2 (7.8%) | 1 (2%) | 0 (0%) |
| 8 | Qi Zhong32 | 49 | 49 | 13 (26.5%) | _ | _ | _ | 36 (73.5%) |
| 9 | Lorenzo Azzi51 | 25 | 25 | 23 (92%) | 2 (8%) | - | - | 0 (0%) |
| 10 | Xiaoqing Wu43 | 23 | 23 | 19 (83%) | _ | _ | _ | 4 (17%) |
| 11 | Qing Chen52 | 9 | 9 | 9 (100%) | _ | _ | _ | 0 (0%) |
| 12 | Li-Na Ji27 | 7 | 5 | 4 (80%) | _ | _ | _ | 1 (20%) |
| 13 | Jasper Fuk-Woo Chan16 | 6 | 5 | 4 (80%) | _ | _ | _ | 1 (20%) |
| 14 | YajunYuan53 | 6 | 6 | 6 (100%) | _ | _ | _ | 0 (0%) |
| 15 | Wei Li18 | 5 | 5 | 4 (80%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (20%) |
| 16 | Zohre Khodamoradi54 | 4 | 4 | 4 (100%) | _ | _ | _ | 0 (0%) |
| 17 | Li Ni55 | 3 | 3 | 1 (33.3%) | 0 (0%) | 0 (0%) | _ | 2 (66.7%) |
| 18 | Junqing Xu 24 | 3 | 3 | 0 (0%) | 2 (67%) | 1 (33%) | _ | 0 (0%) |
| 19 | Yuanzhe Li41 | 2 | 2 | 1 (50%) | 1 (50%) | _ | _ | 0 (0%) |
| 20 | Michal Paret56 | 2 | 2 | 2 (100%) | _ | _ | _ | 0 (0%) |
| 21 | Zhi-Qun Mao57 | 2 | 2 | 2 (100%) | _ | _ | _ | 0 (0%) |
| 22 | Wendong Hao44 | 1 | 1 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (100%) | 0 (0%) |
| 23 | Peikai Huang40 | 1 | 1 | 0 (0%) | 0 (0%) | 1 (100%) | _ | 0 (0%) |
| 24 | Jinrong Qu58 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 25 | Xavier Marchand-Senécal59 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 26 | Takeshi Arashiro55 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 27 | Hao Feng30 | 1 | 1 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (100%) |
| 28 | Ryota Hase60 | 1 | 1 | 0 (0%) | 1 (100%) | _ | _ | 0 (0%) |
| 29 | Yosuke Hirotsu61 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 30 | E. Kalafat62 | 1 | 1 | 0 (0%) | 1 (100%) | _ | _ | 0 (0%) |
| 31 | Mojtaba Kamali Aghdam63 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 32 | Parisa Karami64 | 1 | 1 | 1 (100%) | _ | _ | _ | 0 (0%) |
| 33 | Dasheng Li7 | 1 | 1 | 0 (0%) | 0 (0%) | 1 (100%) | _ | 0 (0%) |
| 34 | Ding-feng Lv65 | 1 | 1 | 0 (0%) | 0 (0%) | 1 (100%) | 1 (100%) | 1 (100%) |
| 35 | Chaisith Sivakorn66 | 1 | 1 | 0 (0%) | 1 (100%) | _ | _ | 0 (0%) |
aNumber of primarily confirmed patients divided total confirmed patients.
bNumber of secondary confirmed patients divided total confirmed patients.
cNumber of thirdly confirmed patients divided total confirmed patients.
dNumber of fourthly confirmed patients divided total confirmed patients.
eNumber of patients who were confirmed later divided by total confirmed patients.
fThis means the test has not been conducted or reported.
The COVID-19 diagnosis was confirmed by positive result of the first, second, third, and fourth RT-PCR tests and also information of patients who had negative results until the fourth test or no more than one test conducted, but considered as confirmed or most likely ill later according to more RT-PCR tests or examining other swabs, clinical manifestations, typical chest CT scan's features or developmental changes in the series of CT scans or a mixture of prior methods.
In the articles with more than 10 total confirmed patients (first 10 articles included), the RT-PCR test could diagnose 58.9% of the COVID-19 infected patients in the first test, and about 41.1% of infected patients could not be recognized in the first place by RT-PCR test. Among these 10 articles, 5 included the information of second tests (number 1, 2, 3, 7 and 9). In these five articles, the mean percentage of secondary diagnosed patients divided by total confirmed patients is 18.6%. Out of 4 articles (number 1, 2, 3 and 7) with the exact data of thirdly and fourthly conducted tests, the mean percentage of positivity are 6.3% and 0.7%, respectively. Moreover, the percentage of patients who were not diagnosed after 4 times of repeating the test is 8.6% (in the previous 4 articles). The numbers and sequences of primers and probes could be influential on PCR sensitivity and specificity which were surveyed in Table 3.
Table 3.
From the information illustrated in Table 2, those which their primer and probe's data were available.
| N. | First author | Total N. of cases | N. of patients confirmed with the first RT-PCR test | N. patients confirmed later | Primarily confirmed patients divided total confirmed patients (%) | Number of sets (primer and probe) | Type of genes |
|---|---|---|---|---|---|---|---|
| 1 | Jian Wu22 | 80 | 41 | 39 | 51.2 | 2 | N and ORF1ab |
| 2 | Anne Kimball48 | 82 | 23 | 53 | 30 | 2 | N |
| 3 | Chenyao Lin50 | 52 | 23 | 29 | 44 | 2 | N and ORF1ab |
| 4 | Qing Chen52 | 9 | 9 | 0 | 100 | 3 | RdRP, E and N |
| 5 | Jasper Fuk-Woo Chan16 | 6 | 4 | 1 | 80 | 2 | RdRP and S |
| 6 | Wendong Hao44 | 1 | 0 | 1 | 0 | 1 | ORF1ab |
| 7 | Xavier Marchand-Senécal59 | 1 | 1 | 0 | 100 | 1 | RdRP |
| 8 | Yosuke Hirotsu61 | 1 | 1 | 0 | 100 | 7 | N gene |
| 9 | Ding-feng Lv65 | 1 | 0 | 1 | 0 | 1 | ORF1ab or N gene |
As we can see in Table 3, in the case report by Wendong Hao44, using one pair of primer and probe did not indicate a positive result at first but in the fourth repeated test. In another case report by Feng Ly65, the oropharyngeal swab by detection of N gene showed a positive result in the third, fourth, and fifth time, whereas ORF1ab detection showed a positive result in the fifth examination. On the other hand, Xavier Marchand-Senécal et al.67 and Yosuke Hirotsu et al.61 reported 2 cases that were diagnosed initially with one pair of primer and probe of PCR test.
By using 2 pairs of primer and probe, the mean of initially detecting patients divided by total confirmed patients is 51.3% in the 4 studies above. Also, Qing Chen52 findings with the utilization of 3 pairs of primer and probe caused 100% of initially discovering COVID-19 patients52. Moreover, using 7 sets of primers and probes also resulted positively for the first test61. Based on limited data available, it seems that the greater the number of primer and probe, more likely to initially detect patients, although more specific information is needed from future studies.
Discussion
Considering the outcomes of RT-PCR as a reference, in our meta-analysis, the sensitivity and specificity of initial chest CT scan for detecting patients, who were highly suspicious for COVID-19, were 87% and 43% respectively. The PPV and NPV of CT scans were 67% and 84% respectively.
It means that 67% of individuals with positive chest CT scans had positive RT-PCR and 84% of individuals with negative chest CT scans had negative RT-PCR. So, a chest CT scan may have beneficial diagnostic features as adjuvant diagnostic tool compared to RT-PCR36,68.
Tao Ai and colleagues studying 1014 patients, 888 (88%) with a positive chest CT scan and 601 with a positive RT-PCR for COVID-19, described 97%, 25%, 65%, and 83% of sensitivity, specificity, PPV, and NPV for the CT scan, respectively. The relatively high sensitivity and low specificity in this study might be related to the low odds ratio of positive RT-PCR, considered as the reference test69, as suggested by the World Health Organization (WHO)70.
Some patients have typical chest CT scan findings and symptoms for COVID-19 but their initial RT-PCR results were negative agreeing with previous research reports19,36,44. Fang et al. described 15 out of 51 patients who have an initial negative RT-PCR while their chest CT scan was positive36,44, so it is very important to pay attention to chest CT scan, epidemiologic features, and clinical symptoms. Furthermore, a combination of humoral (IgG-IgM antibody) and cellular immunity, in addition to RT-PCR could refine the detection of COVID-1923,49.
The results of Chan and colleagues indicated that among 273 specimens (15 COVID-19 positives), the RdRp-P2 test showed 77 positive specimens and the RdRp/Hel test showed 42 positives. Moreover, RdRp/Hel analysis did not cross-reacted with any human coronaviruses or other respiratory pathogens while RdRp-P2 analysis reacted to SARS-CoV either71. Another study expressed that the sensitivity of N gene assay in finding the positive samples is 10 times higher than the ORF-1b gene assay72.
In February, 280 suspected patients with clinical manifestations of COVID-19 were tested in the Marseille hospital. None of the patients were positive for SARS-COV-273. Guo-Qing Qian et al. reported that all the patients, except three of them, were confirmed with the second RT-PCR test18. In the study by Tao Ai et al., it is highlighted that out of 1014 patients, 308 patients with negative PCR results, were strongly perceived as infected by clinical manifestations and CT scans. The percentage of a positive test for the first, second, third, and fourth tests were 37.7%, 22.6%, 5%, and 0.9%, respectively. Unfortunately, about 33.9% of the patients could not be diagnosed even with the fourth test. CT scans were positive in 580 of 601 (97%) COVID-19 confirmed patients by the RT-PCR test and in 308 of 413(74.6%) patients with negative RT-PCR assay74. In a retrospective analysis, among 51 patients, 98% (50/51) had abnormal CT, besides 70.6% (36/51) had positive PCR assay initially and about 30% of the tests became positive after second, third, and fourth scanning44.
A study of five children suggested that four children had positive PCR outcomes within the first assay, but one with COVID-19 suggestive CT findings turned positive after six times of examining35. It is also possible that negative RT-PCR with three times of repeating, turns positive on the fourth test, while CT demonstrated typical features such as GGO44. Chest CT presented rapidly developing multiple patchy consolidations and GGOs in both lungs of a case reported by J Wei, while in the later stage, there was the development of fibrosis. So with high-resolution CT, it will be easier to find GGOs in the early stage75.
What stands out from Table 2 is that 58.9% of the infected COVID-19 patients could be recognized in the first test and about 18.6%, 6.3% and 0.7% could be diagnosed in the further second, third and fourth tests, respectively. Besides, about 9% of the infected patients have not been detected, even after the fourth test. According to the results and due to the PCR's cost and time consumption, it seems that repeating the test up to 3 times is reasonable in patients with initially negative results (with 24 h to 3 days' time interval based on literature). Also, CT scan findings and clinical manifestation should be encountered in all patients, especially in suspected ones with multiple negative PCRs.
About 80% of COVID-19 patients have mild disease and just about 15% of them will reach severe stages. Positive RT-PCR results usually have a high positive predictive value, but negative RT-PCR should be repeated three times to increase the negative predictive value up to 98% (57% at first test, 34% at second one, and 7% at third time).
If the patient's death is due to COVID-19, but their PCR is negative, even if their chest CT is positive, their cause of death would not report COVID-19. Some patients with negative PCR result die, but based on our results, 87% of them are Covid-19 positive and their disease should be confirmed by repeating PCR for up to three times.
On the other hand, the exact place of chest CT is for staging the COVID-19 disease as mild, moderate, and severe, instead of being a screening tool. Some antibody and serology testing can support the RT-PCR test. A study in two patients with COVID-19 pneumonia by Lin and colleagues indicated the presence of ground-glass lesions and patchy consolidations in repeated chest CT76. Also, the lesions were classically accompanied by bronchial bundles or subpleural lesions. In patients who have a fever but not having the previous contact with the epidemic area, the appropriate finding of the COVID-19 RNA is compulsory to guarantee the high efficacy of treatment76.
We acknowledge that our study had some limitations: (1) the specificity of CT scan was not as reliable as the sensitivity, due to the majority of studies' nature, which were case-series and the number of true negative patients in those studies were zero. (2) It has been postulated in Bernheim et al.'s study that the chance of detecting lung involvement in chest CT scan will be increased if the duration between symptom onset and initial chest CT scan rises and this duration was different among 60 studies.
In conclusion, the results of the present systematic review and meta analysis shed new light on the comparison between chest-CT scan and rRT-PCR validity in terms of diagnosis in patients with COVID-19. Due to lower diagnostic sensitivity of chest-CT scan in comparison to rRT-PCR, performing rRT-PCR is mandatory for any individuals with suspicious symptoms. Nevertheless, the initial negative rRT-PCR result is not fully able to roll out COVID-19 in all cases and because of that, repeating the test for three times is vital to roll out COVID-19.
Supplementary Information
Acknowledgements
Special thanks to Urology Research Center, Tehran University of Medical Sciences, Tehran, Iran.
Author contributions
S.M.K.A. was the principal investigator and supervisor of the project. M.S. was the epidemiologist who runs the statistical analysis of data and provides figures. S.S.T.Z. and S.Z.A. individually screened the data and make the data extraction sheet and tables of the article. F.K.H. had design the search strategy and wrote the manuscript. A.N.S. co-supervised the project and data curation. L.O.R. edited the manuscript.
Data availability
Information, data, and photos will be provided if they are requested.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-020-80061-2.
References
- 1.Bernheim A, et al. Chest CT findings in coronavirus disease-19 (COVID-19): Relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Organization, W. H. Coronavirus disease 2019 ( COVID-19): situation report, 92 (2020).
- 3.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 4.Corman VM, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rubin EJ, Baden LR, Morrissey S, Campion EW. Medical journals and the 2019-nCoV outbreak. N. Engl. J. Med. 2020;382:10–15. doi: 10.1056/NEJMe2024117. [DOI] [PubMed] [Google Scholar]
- 6.Loeffelholz MJ, Tang YW. Laboratory diagnosis of emerging human coronavirus infections–the state of the art. Emerg. Microbes. Infect. 2020;9(1):747–756. doi: 10.1080/22221751.2020.1745095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young BE, et al. Epidemiologic features and clinical course of patients infected with SARS-CoV-2 in Singapore. JAMA. 2020 doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan CS, et al. Imaging features of coronavirus disease 2019 (COVID-19): Evaluation on thin-section CT. Acad. Radiol. 2020 doi: 10.1016/j.acra.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao D, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caruso D, et al. Chest CT features of COVID-19 in Rome, Italy. Radiology. 2020 doi: 10.1148/radiol.2020201237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu D, et al. Pregnancy and perinatal outcomes of women with coronavirus disease (COVID-19) pneumonia: A preliminary analysis. Am. J. Roentgenol. 2020 doi: 10.2214/ajr.20.23072. [DOI] [PubMed] [Google Scholar]
- 12.Albano D, et al. Incidental findings suggestive of COVID-19 in asymptomatic patients undergoing nuclear medicine procedures in a high prevalence region. J. Nucl. Med. 2020 doi: 10.2967/jnumed.120.246256. [DOI] [PubMed] [Google Scholar]
- 13.Song F, et al. Emerging coronavirus 2019-nCoV pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan F, et al. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020 doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng K, et al. Analysis of CT features of 15 children with 2019 novel coronavirus infection. Zhonghua er ke za zhi = Chin. J. Pediatr. 2020;58:E007. doi: 10.3760/cma.j.issn.0578-1310.2020.0007. [DOI] [PubMed] [Google Scholar]
- 16.Chan JF, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye G, et al. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian GQ, et al. Epidemiologic and clinical characteristics of 91 hospitalized patients with COVID-19 in Zhejiang, China: A retrospective, multi-centre case series. QJM Monthly J. Assoc. Physicians. 2020 doi: 10.1093/qjmed/hcaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai HX, et al. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H, et al. Clinical and CT imaging features of the COVID-19 pneumonia: Focus on pregnant women and children. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet. 2020 doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, et al. Clinical characteristics of imported cases of COVID-19 in Jiangsu province: A multicenter descriptive study. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia J, Tong J, Liu M, Shen Y, Guo D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020 doi: 10.1002/jmv.25725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, et al. Computed tomographic imaging of 3 patients with coronavirus disease 2019 pneumonia with negative virus real-time reverse-transcription polymerase chain reaction test. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei P, et al. Clinical and computed tomographic (CT) images characteristics in the patients with COVID-19 infection: What should radiologists need to know? J. X-ray Sci. Technol. 2020 doi: 10.3233/xst-200670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng H, et al. CT imaging and clinical course of asymptomatic cases with COVID-19 pneumonia at admission in Wuhan, China. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji LN, et al. Clinical features of pediatric patients with COVID-19: A report of two family cluster cases. World J. Pediatr. 2020 doi: 10.1007/s12519-020-00356-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng L, et al. Arbidol combined with LPV/r versus LPV/r alone against Corona Virus Disease 2019: A retrospective cohort study. J. Infect. 2020 doi: 10.1016/j.jinf.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung M, et al. CT imaging features of 2019 novel coronavirus (2019-nCoV) Radiology. 2020 doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng H, Liu Y, Lv M, Zhong J. A case report of COVID-19 with false negative RT-PCR test: Necessity of chest CT. Jpn. J. Radiol. 2020 doi: 10.1007/s11604-020-00967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, et al. Clinical characteristics of non-critically ill patients with novel coronavirus infection (COVID-19) in a Fangcang Hospital. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong Q, et al. Spinal anaesthesia for patients with coronavirus disease 2019 and possible transmission rates in anaesthetists: Retrospective, single-centre, observational cohort study. Br. J. Anaesth. 2020 doi: 10.1016/j.bja.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, et al. Epidemiological and clinical features of 125 hospitalized patients with COVID-19 in Fuyang, Anhui, China. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, Wang Y, Zhu T, Xia L. CT features of coronavirus disease 2019 (COVID-19) pneumonia in 62 patients in Wuhan, China. Am. J. Roentgenol. 2020 doi: 10.2214/ajr.20.22975. [DOI] [PubMed] [Google Scholar]
- 35.Li R, et al. Clinical characteristics of 225 patients with COVID-19 in a tertiary Hospital near Wuhan, China. J. Clin. Virol. 2020;127:104363. doi: 10.1016/j.jcv.2020.104363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ai T, et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon SH, et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19): Analysis of nine patients treated in Korea. Korean J. Radiol. 2020 doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang W, et al. Clinical characteristics and imaging manifestations of the 2019 novel coronavirus disease (COVID-19): A multi-center study in Wenzhou city, Zhejiang, China. J. Infect. 2020 doi: 10.1016/j.jinf.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu T, et al. Clinical features and dynamics of viral load in imported and non-imported patients with COVID-19. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Z, et al. Construction and evaluation of a novel diagnosis process for 2019-Corona Virus Disease. Zhonghua yi xue za zhi. 2020;100:E019. doi: 10.3760/cma.j.cn112137-20200228-00499. [DOI] [PubMed] [Google Scholar]
- 41.Xu X, et al. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging. 2020 doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia XY, et al. Epidemiological and initial clinical characteristics of patients with family aggregation of COVID-19. J. Clin. Virol. 2020;127:104360. doi: 10.1016/j.jcv.2020.104360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu X, et al. Radiological findings and clinical characteristics of pregnant women with COVID-19 pneumonia. Int. J. Gynaecol. Obstet. 2020 doi: 10.1002/ijgo.13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang Y, et al. Sensitivity of chest CT for COVID-19: Comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y, et al. The presence of SARS-CoV-2 RNA in feces of COVID-19 patients. J. Med. Virol. 2020 doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 46.Cheng Z, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a Single-Center Study in Shanghai, China. Am. J. Roentgenol. 2020 doi: 10.2214/ajr.20.22959. [DOI] [PubMed] [Google Scholar]
- 47.Zhang MQ, et al. Clinical features of 2019 novel coronavirus pneumonia in the early stage from a fever clinic in Beijing. Zhonghua jie he he hu xi za zhi = Zhonghua jiehe he huxi zazhi = Chin. J. Tubercul. Respir. Dis. 2020;43:E013. doi: 10.3760/cma.j.issn.1001-0939.2020.0013. [DOI] [PubMed] [Google Scholar]
- 48.Kimball A, et al. Asymptomatic and presymptomatic SARS-CoV-2 infections in residents of a long-term care skilled nursing facility—King County, Washington, March 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:377–381. doi: 10.15585/mmwr.mm6913e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Filiberto DM, et al. Radiographic predictors of therapeutic operative intervention after blunt abdominal trauma: The RAPTOR score. Eur. J. Trauma Emerg. Surg. 2020 doi: 10.1007/s00068-020-01371-8. [DOI] [PubMed] [Google Scholar]
- 50.Lin C, et al. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19) Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- 51.Azzi L, et al. Saliva is a reliable tool to detect SARS-CoV-2. J. Infect. 2020 doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang P, et al. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020 doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan Y, Wang N, Ou X. Caution should be exercised for the detection of SARS-CoV-2, especially in the elderly. J. Med. Virol. 2020 doi: 10.1002/jmv.25796. [DOI] [PubMed] [Google Scholar]
- 54.Khodamoradi Z, Moghadami M, Lotfi M. Co-infection of coronavirus disease 2019 and influenza A: A report from Iran. Arch. Iran. Med. 2020;23:239–243. doi: 10.34172/aim.2020.04. [DOI] [PubMed] [Google Scholar]
- 55.Arashiro T, et al. SARS-CoV-2 and Legionella co-infection in a person returning from a Nile Cruise. J. Travel Med. 2020 doi: 10.1093/jtm/taaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paret M, et al. SARS-CoV-2 infection (COVID-19) in febrile infants without respiratory distress. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao ZQ, Wan R, He LY, Hu YC, Chen W. The enlightenment from two cases of asymptomatic infection with SARS-CoV-2: Is it safe after 14 days of isolation? Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qu J, Yang R, Song L, Kamel IR. Atypical lung feature on chest CT in a lung adenocarcinoma cancer patient infected with COVID-19. Ann. Oncol. 2020 doi: 10.1016/j.annonc.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marchand-Senecal X, et al. Diagnosis and management of first case of COVID-19 in Canada: Lessons applied from SARS. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hase R, et al. A case of imported COVID-19 diagnosed by PCR-positive lower respiratory specimen but with PCR-negative throat swabs. Infect. Dis. 2020 doi: 10.1080/23744235.2020.1744711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirotsu Y, Maejima M, Nakajima M, Mochizuki H, Omata M. Environmental cleaning is effective for the eradication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in contaminated hospital rooms: A patient from the Diamond Princess cruise ship. Infect. Control Hosp. Epidemiol. 2020 doi: 10.1017/ice.2020.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalafat E, et al. Lung ultrasound and computed tomographic findings in pregnant woman with COVID-19. Ultrasound Obstet. Gynecol. 2020 doi: 10.1002/uog.22034. [DOI] [PubMed] [Google Scholar]
- 63.Kamali Aghdam M, Jafari N, Eftekhari K. Novel coronavirus in a 15-day-old neonate with clinical signs of sepsis, a case report. Infect. Dis. 2020 doi: 10.1080/23744235.2020.1747634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karami P, et al. Mortality of a pregnant patient diagnosed with COVID-19: A case report with clinical, radiological, and histopathological findings. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lv DF, et al. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease 2019 patient. Clinica chimica acta Int. J. Clin. Chem. 2020;506:172–175. doi: 10.1016/j.cca.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sivakorn C, et al. Case report: Walking pneumonia in novel coronavirus disease (COVID-19): Mild symptoms with marked abnormalities on chest imaging. Am. J. Trop. Med. Hyg. 2020 doi: 10.4269/ajtmh.20-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchand-Senécal X, et al. Diagnosis and management of first case of COVID-19 in Canada: Lessons applied from SARS. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiri I, et al. Next-generation radiogenomics sequencing for prediction of EGFR and KRAS mutation status in NSCLC patients using multimodal imaging and machine learning algorithms. Mol. Imaging. Biol. 2020 doi: 10.1007/s11307-020-01487-8. [DOI] [PubMed] [Google Scholar]
- 69.Zheng F, et al. Clinical characteristics of children with coronavirus disease 2019 in Hubei, China. Curr. Med. Sci. 2020 doi: 10.1007/s11596-020-2172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mehdi A, Riazalhosseini Y. Epigenome aberrations: Emerging driving factors of the clear cell renal cell carcinoma. Int. J. Mol. Sci. 2017 doi: 10.3390/ijms18081774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan JF, et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J. Clin. Microbiol. 2020 doi: 10.1128/jcm.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chu DKW, et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Amrane S, et al. Rapid viral diagnosis and ambulatory management of suspected COVID-19 cases presenting at the infectious diseases referral hospital in Marseille, France,—January 31st to March 1st, 2020: A respiratory virus snapshot. Travel Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stranieri A, et al. Preliminary investigation on feline coronavirus presence in the reproductive tract of the tom cat as a potential route of viral transmission. J. Feline Med. Surg. 2020;22:178–185. doi: 10.1177/1098612x19837114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wei J, et al. 2019 novel coronavirus (COVID-19) pneumonia: Serial computed tomography findings. Korean J. Radiol. 2020;21:501–504. doi: 10.3348/kjr.2020.0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin X, et al. Novel coronavirus pneumonia outbreak in 2019: Computed tomographic findings in two cases. Korean J. Radiol. 2020;21:365–368. doi: 10.3348/kjr.2020.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Information, data, and photos will be provided if they are requested.