Abstract
Capicua (CIC) is a highly conserved transcriptional repressor that is differentially regulated through MAPK signaling or genetic alteration across human cancer. CIC contributes to tumor progression and metastasis through direct transcriptional control of effector target genes. Recent findings indicate that CIC dysregulation is mechanistically linked and restricted to specific cancer subtypes, yet convergence on key downstream transcriptional nodes are critical for CIC-regulated oncogenesis across these cancers. In this review, we focus on how differential regulation of CIC through functional and genetic mechanisms contributes to subtype-specific cancer phenotypes, and we propose new therapeutic strategies to effectively target CIC-altered cancers.
The Evolving Role of Capicua in Cancer.
Developmentally regulated transcription factors (TFs) are often dysregulated in cancer and result in aberrant expression of key target genes. One key developmental regulator is Capicua (CIC), an evolutionarily conserved high-mobility group (HMG)-box transcriptional repressor initially discovered in Drosophila, where it controls terminal (head and tail) specification. Thus, CIC derives its name from the Catalan term “head-and-tail” [1, 2].
Structurally, CIC exists as CIC-S (short) and CIC-L (long) with CIC-S being the more well-characterized isoform in mammals [3-5]. Functionally, CIC-mediated transcriptional repression is regulated by receptor tyrosine kinases (RTK) through direct interactions with downstream RTK effectors, ERK and c-SRC [6-8]. Specifically, ERK and c-SRC physically bind and phosphorylate key residues in the C-terminal region of CIC, promoting its nuclear export and relieving target gene expression [8-11]. Mechanistically, CIC leverages both its HMG-box and C1 domains to bind DNA, recognizing an evolutionary conserved sequence T(G/C)AATG(G/A)A [12]. This bipartite mode of DNA-binding ensures target gene specificity and partially explains why somatic events that disrupt or rearrange the HMG-box or the C1 domains are prevalent in cancer [1, 12, 13].
The initial role of CIC in human cancer was described in 2006 with the discovery of the CIC-DUX4 fusion in a subset of sarcomas [14]. The CIC-DUX4 chimera retained wild-type (WT) CIC DNA-binding specificity while replacing its C-terminal domain with the transactivating domain of DUX4, a double homeodomain gene [14]. Consequently, CIC-DUX4 functions as a transcriptional activator instead of a repressor, driving sarcomagenesis through increased expression of key CIC target genes.
Over the past five years, there has been a rapid expansion of the role of CIC in the context of cancer. The discovery of genetic and functional alterations that lead to CIC dysregulation across diverse histological subsets of human cancer have increased our understanding of how CIC contributes to tumor progression and metastasis [4]. In this review, we highlight 1) how CIC is differentially dysregulated across human cancer subtypes; 2) how CIC mechanistically functions to promote cancer-relevant phenotypes; and 3) therapeutic strategies to overcome CIC alterations in human cancer.
CIC Dysregulation Across Human Cancer Subtypes.
Aberrant TF control arises through diverse mechanisms, including altered gene expression, genetic alterations, and post-translational modification [15]. A key unanswered question remains whether the mode of TF dysregulation uniquely contributes to defined phenotypes across human cancer subtypes. Using CIC as a model system, we provide a mechanistic framework to understand how diverse genetic and functional mechanisms converge on specific transcriptional programs to control tumor progression across distinct histological cancer subsets (Figure 1).
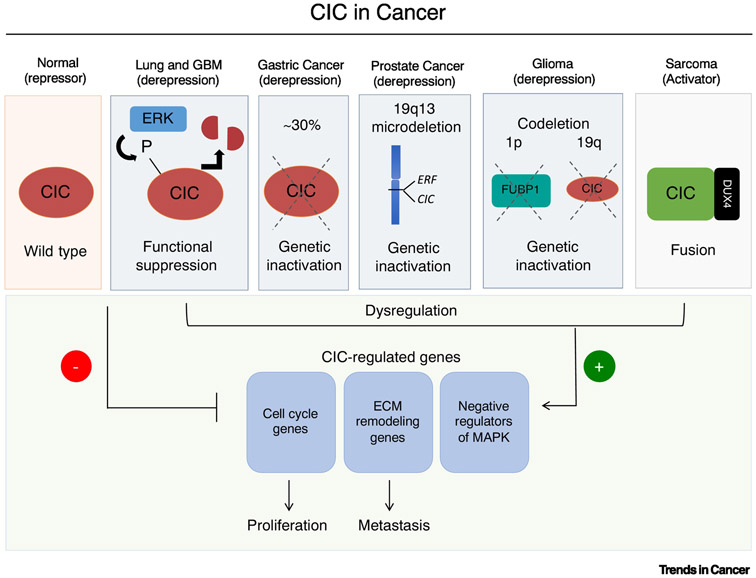
Figure 1. Histological subtype-specific CIC alterations in human cancer.
Wild-type CIC represses target gene transcription that suppresses tumor progression and metastasis. CIC is de-repressed through 1) MAPK-ERK activation in lung adenocarcinoma and glioblastoma multiforme (GBM); 2) genetic inactivation in gastric adenocarcinoma; 3) 19q13 microdeletion in a subset of prostate cancer; and 4) 1p19q co-deletion in oligodendroglioma. CIC is transformed into a transcriptional activator when fused with the DUX4 transactivating domain. The CIC-DUX4 fusion oncoprotein retains CIC DNA-binding specificity but gains activating capacity to increase transcriptional output.
Gliomas.
Oligodendroglioma (OD). The 1p/19q co-deletion represents a diagnostic hallmark of OD which was identified in 1994 [16]. Despite its prognostic and predictive significance, the corresponding genes that comprise this molecular event was not revealed until 2011 when recurrent somatic alterations in FUBP1 (1p) and CIC (19q) were identified on the residual alleles of 1p19q co-deleted OD tumors [17]. Subsequent studies demonstrated that somatic alterations in CIC occur in approximately 70% of OD and are nearly exclusively found in the context of the 1p19q co-deletion [18-20]. These findings suggested that CIC could potentially suppress gliomagenesis. To-date, multiple neural progenitor-specific CIC knockout (KO) mice have been established [21-23]. In these in vivo systems, CIC loss enhances premalignant phenotypes (e.g. proliferation, self-renewal), but overt glioma formation has not been observed [21-23]. These findings, coupled with phylogenetic reconstruction of primary OD tumors, indicate that co-occurring events are necessary for full transformation in CIC-deficient neural precursor cells [24].
Glioblastoma Multiforme (GBM). In contrast to ODs, genetic alterations in CIC have not been detected in GBM [25]. Interestingly, CIC protein expression was recently found to be constitutively suppressed due to tonic EGFR activation through amplification or variant III mutations (EGFRvIII) in GBM tumors [6, 13]. Specifically, ERK-mediated phosphorylation of CIC enabled ubiquitin-mediated degradation through E3 ubiquitin ligase, PJA1. Genetic inhibition of PJA1 in GBM cells did not only restore CIC protein expression but also decreased CIC target gene (ETV1/4/5) expression and cellular proliferation in vitro and in vivo. To mechanistically link MAPK-ERK activity to CIC degradation, the ERK-binding interface on CIC was deleted, which stabilized CIC protein expression and reduced the proliferative and transformation capacity of GBM cells [6]. Collectively, these findings highlight a context-specific mechanism to post-translationally regulate CIC expression.
Sarcoma.
In addition to genetic and functional inactivation, chromosomal rearrangements that produce oncogenic CIC-fusions have also been characterized in a subset of sarcomas [14]. The prototypical CIC-DUX4 (t(4;19) (q35;q13)) oncoprotein fuses over 90% of native CIC to the C-terminal end of the double homeodomain protein, DUX4 [14]. In this context, the CIC-DUX4 oncoprotein retains WT CIC DNA-binding specificity, its ERK-binding domain, and nuclear localization, but the addition of the DUX4 transactivating domain transforms its repressor function into a transcriptional activator [14, 26, 27]. The mechanistic underpinnings of how the addition of DUX4 confers activating capacity to the CIC-DUX4 fusion remains unclear. One potential mechanism is through DUX4-mediated recruitment of histone acetyltransferases (P300 and CBP) to the CIC-DUX4 transcriptional complex, enabling transcriptional activation [28].
Clinically, CIC-DUX4 sarcomas are chemo-insensitive, have high metastatic rates, and poor survival outcomes [29]. In vitro and in vivo mouse models have recapitulated these CIC-DUX4-associated clinicopathologic traits [26, 27]. In the first study, embryonic mesenchymal cells expressing human CIC-DUX4 cDNA were transplanted into immunodeficient mice, which generated highly aggressive undifferentiated sarcomas that upregulated known CIC target (ETV1/4/5) genes that regulate extracellular matrix (ECM) remodeling and cell-cycle progression [26]. Using conventional subcutaneous and orthotopic mouse xenografts, the second study molecularly dissected the functional role of specific CIC-DUX4 target genes, including ETV4 and the cell-cycle regulator CCNE1 [27]. Through these studies, distinct functional roles for ETV4 and CCNE1 in regulating CIC-DUX4-mediated metastasis and tumor growth were defined. Collectively, these studies indicate that CIC-DUX4 drives sarcomagenesis through distinct downstream transcriptional repertoires.
Importantly, novel chimeras including CIC-FOXO4, CIC-NUTM1, and CIC-LEUTX have recently been identified in sarcomas [30-33]. These non-CIC-DUX4 fusions all retain the WT CIC DNA-binding domain while replacing the C-terminal region with a known transactivating domain from another TF. Thus, similar to CIC-DUX4, it is likely that these CIC-fusions work as CIC-specific transcriptional activators.
Prostate Cancer.
In 2015, an anticorrelation between CIC protein expression and prostate cancer progression was observed [34]. Specifically, abundant nuclear CIC expression was observed in normal human prostate tissue while reduced in primary tumors and ablated in advanced prostatic adenocarcinoma [35]. These findings suggested that CIC expression could potentially suppress prostate cancer progression. Consistent with this, genetic alterations, including CIC copy number loss, were found at increased frequency in metastatic prostate cancer (23%) compared to primary prostate tumors (8%) [35]. Recent molecular profiling studies in prostate cancers revealed recurrent focal deletions at chromosome 19q13.2, a region that encompasses CIC and ERF, an ETS transcriptional repressor that can suppress prostate cancer progression [36]. Interestingly, CIC and ERF are physically adjacent to one another on chromosome 19q13, and in a prostate cancer-specific manner, focal genomic loss creates a CIC-ERF co-deletion. The functional and genetic interplay between these two TFs in prostate cancer is currently under investigation.
Lung (LA) and Gastric Adenocarcinoma (GA).
Through analysis of an orthotopic lung metastasis model, CIC was found to suppress spontaneous metastasis [37]. Specifically, genetic inactivation of CIC drives LA metastasis by derepressing its downstream effectors, ETV4 and MMP24. Since genetic inactivation of CIC is an infrequent event in LA and MEK-ERK activation leads to rapid CIC protein degradation [6, 37, 38], these models suggested that the predominant mode of CIC suppression in LA is through hyperactivation of MAPK-ERK signaling, which is present in ~60% of LA cases [39].
CIC is genetically altered through copy number loss or mutation in 26% of GA [40]. The frequency of deleterious CIC alterations were increased in advanced stage GA, and decreased nuclear CIC protein expression correlated with GA progression in clinical samples [37]. Unlike LA, genetic alterations in CIC did not co-occur with MAPK-activating mutations in the TCGA GA dataset. Interestingly, whether CIC inactivation occurred through genetic loss (GA) or functional suppression (LA), consequent activation of conserved target genes, including ETV4, was observed across these distinct histological subsets [37].
Lymphoid Malignancies.
Despite the low incidence of CIC alterations in human T-cell acute lymphoblastic leukemia/lymphoma (T-ALL), two independent studies revealed that CIC ablation in mice is sufficient to induce T-ALL [41, 42]. The first study engineered a conditional loss-of-function CIC allele through targeted deletion of the HMG-box domain. Systemic expression of this DNA-binding deficient CIC allele caused T-ALL in adult mice [41]. Using a tamoxifen inducible cre-driven system (UBC-cre/ERT2; CICflox/flox) to KO CIC in adult mice, a second study observed disruption of early T-cell maturation and T-ALL development [42]. Notably, these studies are the first to validate CIC as a tumor suppressor in murine models of cancer.
Hepatocellular (HCC), Colorectal (CRC), and Breast Cancer (BC).
There is emerging data that CIC also contributes to the progression of other solid tumor malignancies including, HCC, CRC, and BC [43-45]. In HCC and CRC, genetic inhibition of CIC increased proliferation, migration, and invasion [43, 44]. Moreover, decreased CIC expression was observed in patient-derived HCC and CRC tumors relative to normal tissue [43, 44]. In BC, CIC deficiency enhances cancer cell self-renewal without impacting tumor growth or invasion [45]. While early, these studies continue to highlight a critical role for CIC-mediated repression of a core subset of effectors that drive tumor progression and metastasis in histologically distinct cancer subtypes.
CIC Regulates Tumor Progression and Metastasis through Target Gene Expression.
Invasion and Metastasis.
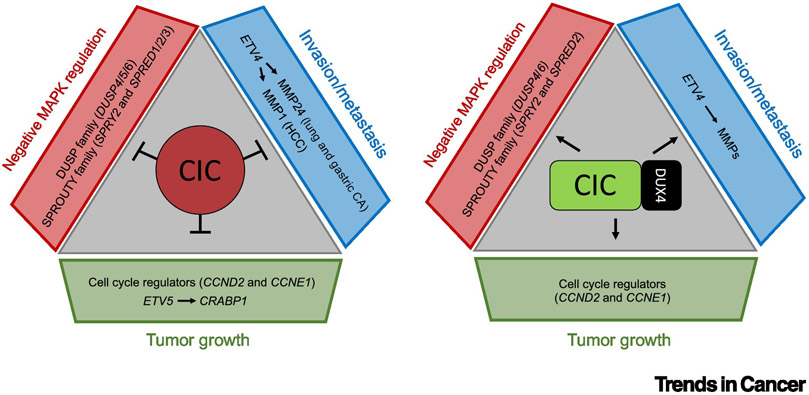
TFs orchestrate complex physiological phenotypes through target gene expression [15]. The most well-characterized CIC-target genes include PEA3 (ETV1/4/5) family members of ETS TFs [46]. CIC binds to PEA3 genes at T(G/C)AATG(G/A)A motifs to modulate metastatic phenotypes in multiple histological subtypes of human cancer. Specifically, inactivation of WT CIC leads to ETV4-mediated upregulation of multiple matrix metalloproteases (MMP) genes and consequent ECM remodeling, driving metastasis in lung, gastric, liver, and breast cancers [9, 37, 43, 45, 47]. This highly conserved mechanism to enhance invasion and promote metastasis has been recapitulated using multiple independent model systems. In LA and GA for example, CIC loss derepresses ETV4 to enhance MMP24 expression, which is necessary and sufficient to promote metastasis in vivo [37]. The pro-metastatic effects of CIC suppression through an ETV4-MMP1 axis has also been reported in CIC-deficient HCC [43]. Through a similar mechanism, CIC deficiency increased ETV1/4/5 expression which consequently enhanced invasion in CRC and melanoma [9, 44]. Thus, while dependence on effector MMPs that act downstream of the CIC-ETV4 can be subtype specific, the role of CIC-mediated ETV4 repression to suppress metastasis is firmly established across histologically distinct cancers. To further support the pro-metastatic function of the ETV4-MMP axis, it was recently shown through in vivo orthotopic models that the CIC-DUX4 oncoprotein transcriptionally activates ETV4 to drive metastasis [27]. Interestingly, while CIC-DUX4-mediated upregulation of the ETV4-MMP24 axis accelerated pulmonary metastases, it did not significantly impact tumor growth [27].
CIC has also been associated with epithelial-to-mesenchymal transition (EMT), which has been linked to enhanced migratory and invasive properties in development and cancer [48]. The precise CIC-regulated target genes that drive this process are not well-defined [48]. However, there is emerging data that the PEA3 TFs can upregulate EMT-promoting genes, including SPARC, Has2, and Twist1 [49, 50]. Efforts to establish a more direct mechanistic link between CIC and EMT-inducing genes remains an area of active investigation.
Tumor Proliferation.
It is well-established that CIC is post-translationally regulated by RTK/Ras signaling, a major proliferative pathway in human cancer [1, 51, 52]. Until recently, identification of CIC-regulated genes contributing to RAS-mediated proliferation has been based largely on non-cancerous Drosophila models [51, 53]. These studies have nominated cell-cycle regulators, including CCNE1, as a potential CIC-target gene in cancer. Consistent with this, recent studies have credentialed CCNE1 and CCND2 as direct downstream targets of CIC-DUX4 [26, 27]. Specifically, in vitro functional studies revealed that genetic silencing of CCNE1 or CCND2 in CIC-DUX4 expressing cells reduced tumor growth [26, 27]. Moreover, inhibition of CCNE1 in CIC-DUX4 transformed mouse fibroblasts reduced tumor growth in vivo. Additional studies targeting the CCNE1 binding partner, CDK2, decreased growth of CIC-DUX4 patient-derived tumor xenografts. These studies reveal components of the cell-cycle machinery as key regulators of CIC-DUX4-mediated proliferation. In the context of WT CIC inactivation, others have observed increased expression of cell-cycle genes in GA, prostate adenocarcinoma, and astrocytoma patients [54, 55], Transcriptomic analyses of these histological subtypes demonstrated that CIC loss increased mitotic cell-cycle genes, CCND1/2, and CCNE1/2 [54-56]. Whether these genes functionally influence tumor growth in WT CIC-deficient tumors is relatively unknown and should be explored.
CIC-ETS factors also play a suppressive role against cancer proliferation. In prostate cancer for example, CIC regulates cell proliferation through the repression of ETV5 in a CRABP1-dependent manner [34]. Regulatory functions of CIC and ETV4 in cell growth have also been demonstrated in CRC with the use of CIC-deficient CRC cell lines and mouse xenografts [44]. In addition to direct CIC-mediated repression of PEA3 family members, other cell proliferation-related genes have been reported to be regulated by ETV1/4/5, including CCND1 and genes involved in the Wnt/β-catenin pathway in gastrointestinal stromal tumor [57, 58]. Despite these findings, the mechanistic relevance of these putative targets in CIC-deficient tumors is still poorly defined. In summary, CIC either directly (CIC binds target genes) or indirectly (through PEA3 family members) regulates key cell-cycle genes to control tumor proliferation (Figure 2).
Figure 2. CIC controls tumor progression and metastasis through effector target genes.
A) Wild-type CIC represses ETV1 and ETV4 to suppresses invasion and metastasis in multiple human cancer subsets. CIC regulates tumor growth, in part, through direct or indirect control of the cell cycle. CIC modulates MAPK-flux by suppressing negative regulators (DUSP and SPROUTY family members) of MAPK activity. B) The CIC-DUX4 fusion oncoprotein activates highly conserved CIC-specific target genes to drive sarcomagenesis.
Drug resistance.
Two recent independent genetic screens identified CIC as a major regulator of MAPK inhibitor resistance across several subsets of human cancer [59, 60]. The first study revealed that CIC loss could suppress the effect of EGFR inhibition in EGFR-mutant lung cancer through partial restoration of an EGFR-associated gene expression program [59]. The second study employed CRIPSR-based screening to identify genetic mechanisms of resistance to MEK inhibitors in multiple cell line-based models and demonstrated that CIC-loss conferred resistance to MEK inhibitors, in KRASG12-mutated pancreatic and colorectal adenocarcinoma [60]. At the clinical level, a recent study identified a BRAFV600E-mutant multiple myeloma (MM) patient who acquired a CICA984P mutation on combined BRAF-MEK inhibitor therapy. Functional studies revealed that CIC silencing decreased sensitivity of MM cells to BRAF-MEK inhibition in vitro [60, 61]. In HCC, the multi-kinase inhibitor, sorafenib, has been effectively used as first-line treatment [62]. A recent study revealed that CIC expression in sorafenib-resistant cells was decreased and genetic inhibition of CIC led to sorafenib resistance [63]. These results suggest that CIC expression could be utilized as a predictive marker for sorafenib response in HCC patients. Additional studies that correlate CIC mutational status to sorafenib response are needed. Beyond targeted therapies, emerging data suggests that CIC loss decreases ovarian cancer response to conventional chemotherapy, including paclitaxel, which suggests that CIC deficiency may confer broad therapeutic resistance across cancer subsets [64]. Interestingly, genetic reconstitution of CIC into drug-resistant, CIC-deficient cancer cells does not re-sensitize these cells to therapy [37, 59, 60]. These findings indicate that inactivation of CIC may irreversibly influence therapeutic response through presently unknown mechanisms. Thus, it will be important to identify the CIC targets that mediate this broad drug resistance mechanism in human cancer.
One interesting area of investigation leverages the highly conserved role of CIC in negative feedback of MAPK signaling. Specifically, multiple negative regulators of MAPK pathway activation, including DUSPs and SPROUTY family members have recently been shown to be directly regulated by CIC [54, 65]. Since DUSP family members, including DUSP6, have direct negative effects on ERK phosphorylation, it is plausible that CIC loss can constitutively suppress ERK-mediated signaling. Consequently, dampened ERK activity may potentially decrease MAPK-ERK mediated dependence in some cancers. Future studies are needed to elucidate the functional role of these negative regulators of MAPK activity in CIC-deficient cancers.
New Approaches to Therapeutically Target CIC Deficiency in Human Cancer.
The therapeutic approaches outlined below (Figure 3) leverage our mechanistic understanding of CIC biology and aim to exploit direct convergence on CIC-regulated transcriptional programs across human cancer.
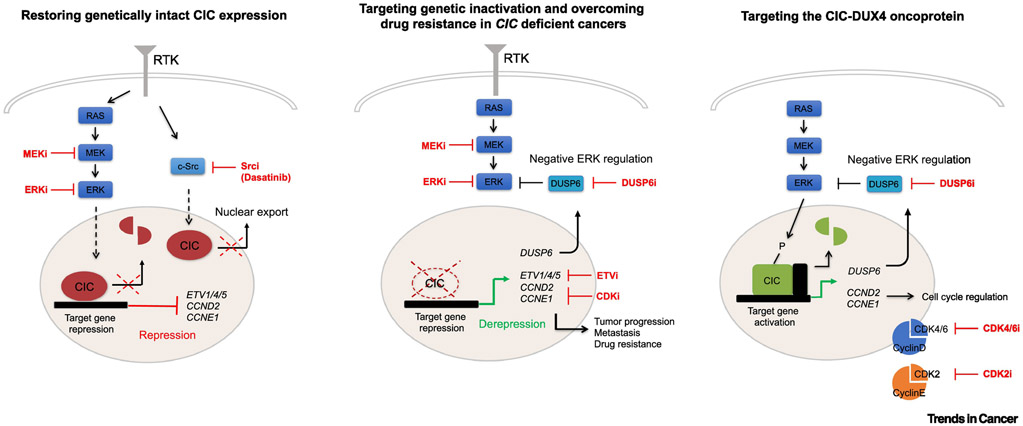
Figure 3. Targeting CIC deficiencies in human cancer.
A) In human cancers with hyperactive RTK-MAPK-ERK signaling and genetically intact CIC, pharmacologic MEK and/or ERK inhibition can potentially restore CIC protein expression to limit tumor progression and metastasis. Using a parallel approach to inhibit c-Src can also increase nuclear CIC expression. B) Genetic inactivation of CIC uncouples ERK from CIC target gene regulation, leading to tumor progression and drug resistance in RAS-MEK-ERK driven cancers. Repurposing or developing new drugs to target CIC downstream effectors, including ETV1/4/5, CCND2 and CCNE1 are potential therapeutic approaches to target CIC-deficient cancers. DUSP6 inhibition in CIC-deficient cancers may increase the dependence on ERK activity and enhance sensitivity to MEK-ERK inhibitors in some drug resistant cancers. C) Cell-cycle regulators, including CCND2 and CCNE1, are direct transcriptional targets of the CIC-DUX4 fusion oncoprotein. Targeting these cyclin-CDK complexes can overcome CIC-DUX4 dependence in undifferentiated sarcomas. Pharmacologic activation of MAPK-ERK signaling through DUSP6 inhibition results in direct degradation of the CIC-DUX4 fusion and apoptosis.
Genetically Intact CIC.
Hyperactive MAPK-ERK signaling leads to CIC protein degradation, contributing to tumor progression and metastasis [6, 37]. Decreased CIC protein expression in the context of hyperactive ERK signaling can potentially identify a subset of patients who may benefit from anti-MEK or anti-ERK therapies. Thus, using clinically approved MEK inhibitors to block ERK activity and functionally restore CIC expression may limit tumor and metastatic progression in cancers with genetically intact CIC. This therapeutic approach can potentially benefit a significant fraction of cancers that harbor hyperactive MAPK signaling. Using GBM models, it has been proposed that chronic, long-term MEK-ERK blockade can paradoxically lead to decreased CIC mRNA expression through transcriptional downregulation [6]. These data are highly informative and should be integrated into future CIC-directed MEK-ERK inhibitor studies to further understand the cell context-specific effects of long-term MEK-ERK inhibition. Utilizing an intermittent MEK-ERK inhibitor dosing schema could act as an alternative approach to therapeutically rescue CIC protein expression while avoiding transcriptional downregulation of CIC mRNA.
Genetic Alterations of CIC.
In cancers with genetic CIC alterations where WT CIC protein expression cannot be restored, we propose mechanism-informed therapeutic strategies. For example, intercepting downstream CIC targets that control the cell-cycle (CCND2 and CCNE1) with clinically relevant CDK inhibitors is one attractive approach. As proof-of-principle, blocking the CCNE1-CDK2 complex with CDK2 inhibitors in patient-derived CIC-DUX4 expressing cells induces apoptosis [27]. Additional studies have demonstrated the therapeutic efficacy of targeting CCND2 through inhibition of its binding partner, CDK4, in CIC-DUX4 sarcomas [26]. Despite these encouraging findings in CIC-DUX4 sarcomas, it remains unclear if cancers with WT CIC deficiency that upregulate cell-cycle genes are also sensitive to CDK blockade. Future studies should explore the therapeutic impact of cell-cycle inhibition in cancers with genetic loss of CIC, such as GA where CIC alterations are observed in 26% of patients. An alternative strategy is to pharmacologically target CIC downstream effectors ETV1/4/5, which drive key malignant hallmarks in CIC-deficient tumors. Unfortunately, to our knowledge, BRD32048 is the only available chemical inhibitor that effectively targets the PEA3 family member, ETV1 [66]. Further development of pan-PEA3 TF (ETV1/4/5) inhibitors may provide an alternative therapeutic approach to overcome CIC loss.
Inhibiting Negative MAPK-ERK Regulators to Degrade the CIC-DUX4 Fusion Oncoprotein and Overcome Therapeutic Resistance
Since MAPK regulates native CIC expression through direct ERK-mediated phosphorylation, a recent study developed a mechanism-based therapeutic approach to directly degrade the CIC-DUX4 oncoprotein [67]. Specifically, direct ERK activation or inhibition of negative regulators of MAPK-ERK signaling, including DUSP6, leads to sustained CIC-DUX4 degradation and apoptosis of CIC-DUX4 cells. Mechanistically, pharmacological inhibition of DUSP6 increases ERK activity, which in turn, leads to direct CIC-DUX4 degradation in an ERK-dependent manner [67-69]. One inadvertent outcome of ERK activation is increased WT CIC degradation. Importantly, one human-derived CIC-DUX4 model did not express WT CIC RNA transcript, suggesting that other CIC-DUX4 cancers may also silence the WT CIC copy [27, 67]. Furthermore, with the exception of rare lymphomas in mice, tumor initiation through targeted CIC suppression in multiple genetic models has not yet been shown to induce solid tumor malignancies. Thus, genetic or functional suppression of CIC is not a potent tumor initiating event and likely requires additional co-occurring genetic or non-genetic changes. Thus, these preclinical results provide rationale to further develop clinical-grade DUSP6 inhibitors in CIC-DUX4 patients.
A recent study provided an intriguing approach to potentially overcome CIC-mediated drug resistance. Specifically, expression of an ERK-unresponsive mutant CIC isoform sensitized GBM cells to MEK inhibition [6]. Mechanistically, this enhanced sensitivity may in part be dependent on DUSP6 repression which, through feedback mechanisms, would enhance ERK activity, increasing dependence on the MAPK-ERK pathway. One potential combinatorial strategy would be to use a DUSP6 inhibitor with MEK-ERK blockade to enhance therapeutic responses.
Mechanism-based Combination Therapies.
Recent findings demonstrate that c-Src cooperates with ERK in a parallel and complementary fashion to modulate CIC expression in GBM [7]. Thus, using the Src family kinase inhibitor, dasatinib, in combination with MEK-ERK blockade can attenuate EGF-mediated CIC phosphorylation, decreasing ETV1/5 expression and inhibiting GBM growth [7]. Further investigations are required to fully elucidate the efficacy of combined MEK-ERK and c-Src inhibition in other CIC-deficient cancers.
Concluding Remarks
The first direct evidence that CIC contributed to cancer was in 2006 when the CIC-DUX4 oncoprotein was identified in a rare subset of sarcoma [14]. Since this time, the presence of cancer-associated CIC alterations has greatly expanded and, in some cases, now represent subtype-specific genetic events. Importantly, the functional impact of CIC alterations is being explored through multiple cell-based and animal model systems, leading to new provocative questions (see Outstanding Questions) and the potential development of novel therapies to target CIC-altered cancers. One critical discovery that may lead to more rapid therapies to overcome CIC deficiency is the highly conserved regulatory pathway that links MAPK signaling to CIC expression. Clinically approved inhibitors that block MAPK-ERK flux can be employed to restore WT CIC expression. Finally, modeling CIC dynamics in response to MAPK-ERK signaling can provide new insight into how other ERK-responsive transcriptional repressors, including ERF [70] and TLE-1 [71], independently or collectively contribute to malignant progression across human cancers.
Outstanding Questions.
In solid tumors, what are the co-occurring genetic and epigenetic events required for full cellular transformation in CIC-deficient cells?
What are the functional and genetic interactions between CIC and known co-occurring events, including FUBP1 in OD, MAPK signaling in GBM and LA, and ERF in prostate cancer?
Can wild-type CIC expression be efficiently restored in genetically intact CIC cells through MAPK-signaling blockade to limit cancer progression?
What are the underlying mechanisms of drug resistance in CIC-deficient cancers?
Box 1.
Capicua operates as a nuclear sensor of RTK-MAPK-ERK signaling in Drosophila and mammals. Activation of MAPK-ERK signaling leads to direct ERK-mediated CIC phosphorylation and nuclear export, leading to de-repression of CIC target genes. Through target gene expression, CIC regulates extracellular matrix remodeling, cell-cycle machinery, and MAPK signaling flux.
CIC binds TG/CAATGA/GA DNA motifs through a mechanism involving its HMG-box (N-terminus) and C1 (C-terminus) domains. This bipartite mechanism ensures sequence-specific recognition of CIC targets. Thus, dysregulation of CIC through post-translational modification or through genetic mechanisms can derepress a highly conserved CIC-regulated transcriptional network in development and cancer.
Highlights.
The mode of CIC dysregulation is restricted to specific cancer subsets and in select histology’s represent subtype-specific events.
New cell-based and animal models demonstrate convergence on key CIC target genes that directly contribute to malignant progression.
Therapeutic interception of key CIC-regulated transcriptional targets in CIC-altered cancers can block tumor progression and metastasis.
Mechanism-informed therapeutic strategies to either rescue wild-type CIC protein expression or degrade CIC-fusion oncoproteins can potentially limit CIC-mediated cancer progression.
Acknowledgements
This work is supported through an NIH-NCI K08 grant CA222625 to RAO. The authors deeply apologize to our colleagues whose work was not cited in this manuscript due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Jiménez G et al. (2000) Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev 14 (2), 224–31. [PMC free article] [PubMed] [Google Scholar]
- 2.Chittaranjan S et al. (2014) Mutations in CIC and IDH1 cooperatively regulate 2-hydroxyglutarate levels and cell clonogenicity. Oncotarget 5 (17), 7960–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y et al. (2020) Regulation and function of capicua in mammals. Exp Mol Med 52 (4), 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong D and Yip S (2020) Making heads or tails - the emergence of capicua (CIC) as an important multifunctional tumour suppressor. J Pathol 250 (5), 532–540. [DOI] [PubMed] [Google Scholar]
- 5.Simón-Carrasco L et al. (2018) The Capicua tumor suppressor: a gatekeeper of Ras signaling in development and cancer. Cell Cycle 17 (6), 702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunda S et al. (2019) CIC protein instability contributes to tumorigenesis in glioblastoma. Nat Commun 10 (1), 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunda S et al. (2020) c-Src Phosphorylates and Inhibits the Function of the CIC Tumor Suppressor Protein. Mol Cancer Res 18 (5), 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Futran AS et al. (2015) Mapping the binding interface of ERK and transcriptional repressor Capicua using photocrosslinking. Proc Natl Acad Sci U S A 112 (28), 8590–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dissanayake K et al. (2011) ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capúa. Biochem J 433 (3), 515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul S et al. (2020) Activation-induced substrate engagement in ERK signaling. Mol Biol Cell 31 (4), 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keenan SE et al. (2020) Rapid Dynamics of Signal-Dependent Transcriptional Repression by Capicua. Dev Cell 52 (6), 794–801.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forés M et al. (2017) A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet 13 (3), e1006622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Padul V et al. (2015) ETV/Pea3 family transcription factor-encoding genes are overexpressed in CIC-mutant oligodendrogliomas. Genes Chromosomes Cancer 54 (12), 725–33. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura-Saito M et al. (2006) Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet 15 (13), 2125–37. [DOI] [PubMed] [Google Scholar]
- 15.Bushweller JH (2019) Targeting transcription factors in cancer - from undruggable to reality. Nat Rev Cancer 19 (11), 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reifenberger J et al. (1994) Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol 145 (5), 1175–90. [PMC free article] [PubMed] [Google Scholar]
- 17.Bettegowda C et al. (2011) Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333 (6048), 1453–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffin CA et al. (2006) Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 65 (10), 988–94. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins RB et al. (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66 (20), 9852–61. [DOI] [PubMed] [Google Scholar]
- 20.Bromberg JE and van den Bent MJ (2009) Oligodendrogliomas: molecular biology and treatment. Oncologist 14 (2), 155–63. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad ST et al. (2019) Capicua regulates neural stem cell proliferation and lineage specification through control of Ets factors. Nat Commun 10 (1), 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang R et al. (2017) Cic Loss Promotes Gliomagenesis via Aberrant Neural Stem Cell Proliferation and Differentiation. Cancer Res 77 (22), 6097–6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang I et al. (2020) CIC is a critical regulator of neuronal differentiation. JCI Insight 5 (9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tirosh I et al. (2016) Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539 (7628), 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Network CGAR (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455 (7216), 1061–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimoto T et al. (2017) CIC-DUX4 Induces Small Round Cell Sarcomas Distinct from Ewing Sarcoma. Cancer Res 77 (11), 2927–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okimoto RA et al. (2019) CIC-DUX4 oncoprotein drives sarcoma metastasis and tumorigenesis via distinct regulatory programs. J Clin Invest 129 (8), 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi SH et al. (2016) DUX4 recruits p300/CBP through its C-terminus and induces global H3K27 acetylation changes. Nucleic Acids Res 44 (11), 5161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Antonescu CR et al. (2017) Sarcomas With CIC-rearrangements Are a Distinct Pathologic Entity With Aggressive Outcome: A Clinicopathologic and Molecular Study of 115 Cases. Am J Surg Pathol 41 (7), 941–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lake JA et al. (2020) Targeted fusion analysis can aid in the classification and treatment of pediatric glioma, ependymoma, and glioneuronal tumors. Pediatr Blood Cancer 67 (1), e28028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugita S et al. (2014) A novel CIC-FOXO4 gene fusion in undifferentiated small round cell sarcoma: a genetically distinct variant of Ewing-like sarcoma. Am J Surg Pathol 38 (11), 1571–6. [DOI] [PubMed] [Google Scholar]
- 32.Le Loarer F et al. (2019) Clinicopathologic Features of CIC-NUTM1 Sarcomas, a New Molecular Variant of the Family of CIC-Fused Sarcomas. Am J Surg Pathol 43 (2), 268–276. [DOI] [PubMed] [Google Scholar]
- 33.Huang SC et al. (2016) Recurrent CIC Gene Abnormalities in Angiosarcomas: A Molecular Study of 120 Cases With Concurrent Investigation of PLCG1, KDR, MYC, and FLT4 Gene Alterations. Am J Surg Pathol 40 (5), 645–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi N et al. (2015) miR-93/miR-106b/miR-375-CIC-CRABP1: a novel regulatory axis in prostate cancer progression. Oncotarget 6 (27), 23533–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seim I et al. (2017) Whole-Genome Sequence of the Metastatic PC3 and LNCaP Human Prostate Cancer Cell Lines. G3 (Bethesda) 7 (6), 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang FW et al. (2017) Exome Sequencing of African-American Prostate Cancer Reveals Loss-of-Function. Cancer Discov 7 (9), 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okimoto RA et al. (2017) Inactivation of Capicua drives cancer metastasis. Nat Genet 49 (1), 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimm O et al. (2012) Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139 (21), 3962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Network CGAR (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511 (7511), 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Network CGAR (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513 (7517), 202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simón-Carrasco L et al. (2017) Inactivation of Capicua in adult mice causes T-cell lymphoblastic lymphoma. Genes Dev 31 (14), 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan Q et al. (2018) Loss of Capicua alters early T cell development and predisposes mice to T cell lymphoblastic leukemia/lymphoma. Proc Natl Acad Sci U S A 115 (7), E1511–E1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E et al. (2018) Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology 67 (6), 2287–2301. [DOI] [PubMed] [Google Scholar]
- 44.Lee JS et al. (2020) Capicua suppresses colorectal cancer progression via repression of ETV4 expression. Cancer Cell Int 20, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoe J et al. (2020) Capicua restricts cancer stem cell-like properties in breast cancer cells. Oncogene 39 (17), 3489–3506. [DOI] [PubMed] [Google Scholar]
- 46.Jiménez G et al. (2012) The Capicua repressor--a general sensor of RTK signaling in development and disease. J Cell Sci 125 (Pt 6), 1383–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee Y et al. (2011) ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell 21 (4), 746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao LJ et al. (2019) miR-106b promotes proliferation and invasion by targeting Capicua through MAPK signaling in renal carcinoma cancer. Onco Targets Ther 12, 3595–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heeg S et al. (2016) ETS-Transcription Factor ETV1 Regulates Stromal Expansion and Metastasis in Pancreatic Cancer. Gastroenterology 151 (3), 540–553.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khatiwada P et al. (2020) Androgen up-regulation of Twist1 gene expression is mediated by ETV1. PeerJ 8, e8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin Y et al. (2015) EGFR/Ras Signaling Controls Drosophila Intestinal Stem Cell Proliferation via Capicua-Regulated Genes. PLoS Genet 11 (12), e1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santarpia L et al. (2012) Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets 16 (1), 103–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krivy K et al. (2013) Capicua regulates proliferation and survival of RB-deficient cells in Drosophila. Biol Open 2 (2), 183–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong D et al. (2019) Transcriptomic analysis of CIC and ATXN1L reveal a functional relationship exploited by cancer. Oncogene 38 (2), 273–290. [DOI] [PubMed] [Google Scholar]
- 55.LeBlanc VG et al. (2017) Comparative transcriptome analysis of isogenic cell line models and primary cancers links capicua (CIC) loss to activation of the MAPK signalling cascade. J Pathol 242 (2), 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gleize V et al. (2015) CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol 78 (3), 355–74. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y et al. (2020) ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol Carcinog 59 (1), 73–86. [DOI] [PubMed] [Google Scholar]
- 58.Zeng S et al. (2017) ETV4 collaborates with Wnt/β-catenin signaling to alter cell cycle activity and promote tumor aggressiveness in gastrointestinal stromal tumor. Oncotarget 8 (69), 114195–114209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liao S et al. (2017) A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev 31 (2), 184–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang B et al. (2017) ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Rep 18 (6), 1543–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Da Vià MC et al. (2020) CIC Mutation as a Molecular Mechanism of Acquired Resistance to Combined BRAF-MEK Inhibition in Extramedullary Multiple Myeloma with Central Nervous System Involvement. Oncologist 25 (2), 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Llovet JM et al. (2008) Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 359 (4), 378–90. [DOI] [PubMed] [Google Scholar]
- 63.Hashiba T et al. (2020) Inactivation of Transcriptional Repressor Capicua Confers Sorafenib Resistance in Human Hepatocellular Carcinoma. Cell Mol Gastroenterol Hepatol. [DOI] [PMC free article] [PubMed]
- 64.Zhou Y et al. (2019) MiR-1307 influences the chemotherapeutic sensitivity in ovarian cancer cells through the regulation of the CIC transcriptional repressor. Pathol Res Pract 215 (10), 152606. [DOI] [PubMed] [Google Scholar]
- 65.Weissmann S et al. (2018) The Tumor Suppressor CIC Directly Regulates MAPK Pathway Genes via Histone Deacetylation. Cancer Res 78 (15), 4114–4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pop MS et al. (2014) A small molecule that binds and inhibits the ETV1 transcription factor oncoprotein. Mol Cancer Ther 13 (6), 1492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin YK et al. (2020) Negative MAPK-ERK regulation sustains CIC-DUX4 oncoprotein expression in undifferentiated sarcoma. Proc Natl Acad Sci U S A 117 (34), 20776–20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu QN et al. (2018) Pharmacological inhibition of DUSP6 suppresses gastric cancer growth and metastasis and overcomes cisplatin resistance. Cancer Lett 412, 243–255. [DOI] [PubMed] [Google Scholar]
- 69.James NE et al. (2019) Inhibition of DUSP6 sensitizes ovarian cancer cells to chemotherapeutic agents via regulation of ERK signaling response genes. Oncotarget 10 (36), 3315–3327. [PMC free article] [PubMed] [Google Scholar]
- 70.Le Gallic L et al. (1999) Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol Cell Biol 19 (6), 4121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zahavi T et al. (2017) Ras-Erk signaling induces phosphorylation of human TLE1 and downregulates its repressor function. Oncogene 36 (26), 3729–3739. [DOI] [PubMed] [Google Scholar]