Abstract
Dozens of genes contribute to the vast variation in human pigmentation. Many of these encode proteins that localize to the melanosome, the lysosome-related organelle that synthesizes pigment, but have unclear functions 1,2. Here, we describe the MelanoIP method for rapidly isolating melanosomes and profiling their labile metabolite contents. We use it to study MFSD12, a transmembrane protein of unknown molecular function that when suppressed causes darker pigmentation in mice and humans 3,4. We find that MFSD12 is required to maintain normal levels of cystine, the oxidized dimer of cysteine, in melanosomes, and to produce cysteinyldopas, the precursors of pheomelanin synthesis made in melanosomes via cysteine oxidation 5,6. Tracing and biochemical analyses show that MFSD12 is necessary for the import of cysteine into melanosomes, and, in non-pigmented cells, lysosomes. Indeed, loss of MFSD12 reduced the accumulation of cystine in lysosomes of fibroblasts from patients with cystinosis, a lysosomal storage disease caused by inactivation of the lysosomal cystine exporter CTNS (Cystinosin)7–9. Thus, MFSD12 is an essential component of the long-sought cysteine importer for melanosomes and lysosomes.
Pigment producing cells like melanocytes synthesize melanin in membrane-bound, lysosome-related organelles (LRO) called melanosomes. Within melanosomes, tyrosinase catalyzes the oxidation of tyrosine to dopaquinone, which is then further oxidized and polymerized into a melanin pigment 5,6. While in cell-free systems tyrosinase, tyrosine, and oxygen are sufficient to produce simple melanin polymers, in vivo additional melanosomal proteins influence the chemical composition and quantity of the melanin synthesized. Many of these proteins have been implicated in human pigment variation, but in many cases their biochemical functions remain unknown or unclear 1,2. Our current understanding of melanin synthesis has been in large part driven by the study of cell-free melanin synthesis reactions and the chemical degradation of isolated melanins 5,6. We reasoned that the direct measurement of melanosomal metabolites would enable a better understanding of the function of melanosomal proteins in cells. However, established methods for isolating pure melanosomes are lengthy and would allow for metabolites of interest to oxidize or diffuse out of the organelle 10.
Method for rapid melanosome purification
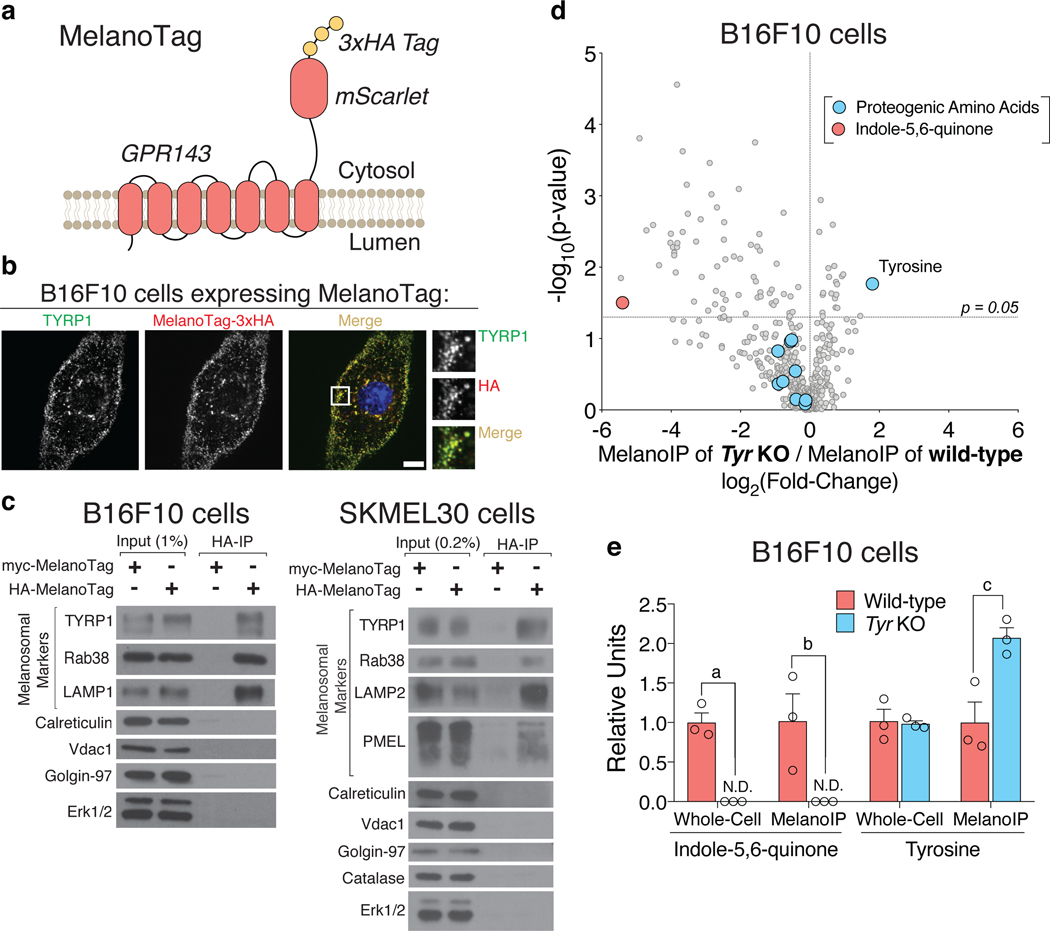
To overcome this challenge, we developed a method for the rapid isolation of melanosomes that is compatible with the metabolite profiling of their contents. This method, termed ‘MelanoIP,’ adapts previously developed techniques for the isolation of mitochondria, lysosomes, and peroxisomes in which recombinant proteins that serve as specific tags for organelles enable their rapid immunopurification (Fig. 1a) 11–13. We engineered a melanosome-specific tag (MelanoTag) based on the melanosomal marker GPR143 14 (Fig. 1a) and validated it co-localizes with the melanosomal protein TYRP1, when expressed in cultured melanoma cells (Fig. 1b). The MelanoIP method successfully purified melanosomes from murine B16F10 and human SKMEL30 melanoma cell lines as judged by the enrichment of melanosomal markers TYRP1, PMEL, and Rab38 over of those for the Golgi, endoplasmic reticulum, peroxisomes, and mitochondria (Fig. 1c, Supplementary Fig. 1). As many lysosomal proteins also localize to melanosomes 1,15, it was not surprising that the purified melanosomes also contained the lysosomal marker proteins LAMP1 and LAMP2 (Fig. 1c, Supplementary Fig. 1). In SKMEL30 cells, we could also see the enrichment of processed PMEL over its full length form, consistent with the method capturing the luminal contents of the organelles 16.
Fig. 1. MelanoIP enables the rapid isolation of pure melanosomes.
a, Schematic diagrams of the MelanoTag (GPR143-mScarlet-3xHA) and the MelanoIP workflow. b, Immunofluorescence analyses of B16F10 cells expressing the MelanoTag using antibodies to TYRP1 and the HA epitope tag. Cells were treated for two days with 10 μM forskolin before immunofluorescent labeling. TYRP1 and MelanoTag positive puncta are concentrated on the cell periphery, consistent with staining patterns observed previously for both markers 14. Micrographs are representative of the majority staining pattern and experiments were replicated in more than three independent experiments. Scale bar, 10 μm. c, Immunoblot analyses of whole-cell lysates, control immunoprecipitates (from cells expressing a myc-MelanoTag), and purified melanosomes (from cells expressing the HA-MelanoTag) prepared from murine B16F10 and human SKMEL30 cells. Melanosomal marker selection was guided by the ability of selected antibodies to react with either murine or human target proteins. Immunoblots are representative of more than three independent replicates. d, Untargeted polar metabolite profiling of melanosomes isolated from wild-type and Tyr knock-out cells. The red datapoint highlights a eumelanin synthesis intermediate with m/z and in silico MS/MS data consistent with indole-5,6-quinone 34,35, and the blue datapoints highlight all the detected proteogenic amino acids, including tyrosine, which is labelled (n = 3 independently prepared metabolite extracts). e, Metabolite profiling analysis of an annotated indole-5,6-quinone species and tyrosine in whole-cell lysates and purified melanosomes from wild-type and Tyr knock-out B16F10 cells. (n = 3 independently prepared metabolite extracts, aP = 1.2×10−3, bP = 4.2×10−2, cP = 2.1×10−2, N.D. = not detected). Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
To validate the MelanoIP approach, we used liquid chromatography/mass spectrometry (LC/MS) to profile metabolites in melanosomes from wild-type and Tyr-deficient B16F10 cells. Tyr encodes the aforementioned tyrosinase, an essential and rate-limiting enzyme in melanin synthesis (Extended Data Fig. 1a). Unlike wild-type samples, Tyr knock-out cells and melanosomes lacked detectable melanin synthesis intermediates (Fig. 1d and e). Loss of tyrosinase did not impact whole-cell tyrosine levels, but did cause a 2-fold accumulation of tyrosine in melanosomes (Fig. 1d and e), consistent with tyrosine being the tyrosinase substrate. No other proteogenic amino acids were affected to the same degree (Fig. 1d and Extended Data Fig. 1b). These results highlight the value of the MelanoIP for detecting compartmentalized changes in metabolites that have only a small fraction of their whole-cell pool represented in the melanosome.
MelanoIP uncovers MFSD12 function
We next tested the value of the MelanoIP method for deorphaning a melanosomal protein of unknown function. GWAS analysis recently identified MFSD12 as a gene associated with pigment variation in African and Venezuelan populations 3,4. MFSD12 loss of function mutations result in darker pigmentation because MFSD12 is required for the synthesis of red pheomelanin pigment, but not brown-black eumelanin pigment 4. However, no direct biochemical function has been suggested for MFSD12, and it has been reported to localize mostly to lysosomes in cultured pigmented cells 4. Given that lysosomal proteins are often also melanosomal, we hypothesized that a population of MFSD12 could exist at the melanosome and directly affect melanosomal metabolism.
As MFSD12 is part of the large MFS superfamily of 12-transmembrane domain proteins, many of which are small molecule transporters 17, we thought it likely that MFSD12 has an unknown transport activity. Untargeted metabolite profiling of melanosomes from wild-type and Mfsd12 knock-out B16F10 melanoma cells revealed that loss of MFSD12 greatly reduced melanosomal cystine levels (Fig. 2a). In follow-up studies, cystine dropped 11-fold in melanosomes from B16F10 cells upon loss of MFSD12, a phenotype fully reversed by its re-expression (Fig. 2b). We reasoned that the decrease in cystine, the oxidized dimeric form of cysteine, might result from a decrease in cysteine in melanosomes. Consistent with this possibility, loss of MFSD12 also greatly reduced the levels of cysteinyldopas (Fig 2c, Extended Data Fig. 2a-d), which are precursors for pheomelanin synthesis and made in melanosomes by the co-oxidation of cysteine and tyrosine 5,6. Even with chemical derivatization we were unable to detect cysteine in melanosomes, likely because of its rapid conversion to cystine and the cysteinyldopas. Consistently, like in the murine cells, in human SKMEL30 cells loss of MFSD12 also reduced melanosomal cystine and cysteinyldopas (Fig. 2d and e). Interestingly, the MFSD12-null SKMEL30 cells are visibly darker than their wild-type counterparts, as are the Mfsd12 mutant mice (Fig. 2f) 4,18. It has been known for decades that isolated melanosomes can influx cysteine, but the responsible transporter has not yet been identified 19. Our results suggested that MFSD12 is a component of this melanosomal cysteine import system (Fig. 2g).
Fig. 2. MFSD12 is necessary to maintain melanosomal cystine levels and produce cysteinyldopas.
a, Untargeted polar metabolite profiling of melanosomes isolated from Mfsd12 knock-out and wild-type B16F10 cells. The red datapoint highlights cystine (n = 3 independently prepared extracts, P values by two-sided Student’s t-test). b, Follow-up metabolite analysis of cystine in B16F10 whole-cell lysates and purified melanosomes (n = 3 independently prepared metabolite extracts, aP = 4.8×10−5; bP = 1.0× 10−5). c, Metabolite profiling of cysteinyldopas in B16F10 whole-cell lysates. The ‘Minor Isomer’ and ‘Major Isomer’ are distinguished by retention time, and we annotate them to represent 2’-cysteinyldopa and 5’-cysteinyldopa, respectively (n = 3 independently prepared metabolite extracts, aP = 2.2×10−4, bP = 5.4×10−4, cP = 4.3×10−4, dP = 7.7×10−4, N.D. = not detected). d, Cystine levels in SKMEL30 whole-cell lysates and purified melanosomes (n = 3 independently prepared metabolite extracts, aP = 4.4×10−4). e, Levels of cysteinyldopas in SKMEL30 cells (n = 4 independently prepared metabolite extracts, aP =3.4×10−8, bP = 4.3×10−7). f, SKMEL30 cells lacking MFSD12 are darker than their wild-type counterparts. Photographs are of pellets containing 3 million SKMEL30 cells at the bottom of 1.5 mL centrifuge tubes. g, Schematic of proposed function for MFSD12 in melanosomes, whereby it controls the influx of cysteine into melanosomes, enabling the production of cystine, cysteinyldopas, and pheomelanin. Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
MFSD12 also functions in lysosomes
Many genes involved in pigmentation, such as TYR and SLC45A2, are only expressed in pigmented tissues and cells, but MFSD12 is expressed body-wide (Fig. 3a, Extended Data Fig. 3a)20,21. Consistent with MFSD12 also functioning in non-pigmented cells, lysosomes from HEK-293T cells lacking MFSD12 had strongly reduced levels of cystine as detected by untargeted metabolite profiling (Fig. 3b). Follow-up work showed that MFSD12 loss decreased lysosomal cystine levels over 20-fold, which was rescued by MFSD12 re-expression (Fig. 3c). Importantly, MFSD12 loss did not cause any increases in alanine, proline, or other amino acids that tend to accumulate in lysosomes upon inhibition of the V-ATPase, which is required to acidify the lysosomal lumen (Extended Data Fig. 3b) 11, suggesting that MFSD12 impacts cystine levels independently of an effect on the lysosomal pH. In whole-cell samples from HEK-293T cells, we detected a 2.4-fold decrease in cystine upon MFSD12 deletion, consistent with previous data suggesting that a large fraction of total cellular cystine is in lysosomes 22. In contrast to melanosomes in which cysteine was undetectable, we were able to readily measure cysteine in lysosomes from HEK-293T cells and found that MFSD12 loss reduced it by 41-fold (Fig. 3c). As with melanosomes, cysteine transport into isolated lysosomes was first reported decades ago 22, and our data pointed to MFSD12 as a long-sought component of such an import system.
Fig. 3. MFSD12 is necessary to maintain lysosomal cystine and cysteine levels.
a, Expression of pigmentation genes across tissues. FANTOM5 expression profiling data from Human Protein Atlas displayed as scaled tags per million 20,21. Full labels of tissues and additional pigmentation genes are displayed in Extended Fig. 3a. b, Untargeted polar metabolite profiling of lysosomes isolated from MFSD12 knock-out and wild-type HEK-293T cells (n = 6 independently prepared metabolite extracts per condition) c, Follow-up metabolite analysis of cystine and cysteine in whole-cell extracts and purified lysosomes. Samples were split and run via standard methods for cystine and derivatized with Ellman’s reagent to enable cysteine detection (n = 3 independently prepared metabolite extracts per condition, aP = 4.5×10−2, bP = 3.6×10−2, cP = 2.5×10−6, dP = 1.3×10−2, eP = 1.3×10−3, fP = 2.9×10−4). d, Cystine measurements in whole-cell extracts and purified lysosomes. To generate MFSD12&CTNS combined knock-out cells, MFSD12 was deleted from the CTNS knock-out cells (n = 3 independently prepared metabolite extracts per condition, aP = 1.4×10−2, bP = 3.0×10−2, cP = 1.9×10−3, dP = 1.9×10−3, eP = 2.1×10−3). e, Cystine measurements in whole-cell extracts and purified lysosomes from patient-derived fibroblasts. Matched unaffected parental (GM00906) and cystinotic patient (GM00379) fibroblast cell lines were obtained from the Coriell institute. Cells were grown to confluence to arrest division before cystine was measured (n= 4 – 6 independently prepared metabolite extracts per condition, aP = 1.5×10−7, bP = 5.0×10−6, cP = 1.1×10−3, dP = 1.5×10−7, eP = 1.7×10−4, fP = 4.2×10−5). f, Schematic of proposed role of MFSD12 in lysosomes in which it is upstream of CTNS in the lysosomal cystine/cysteine cycle. Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
While how cysteine enters melanosomes and lysosomes is unknown, it is well appreciated that cystine effluxes out of these organelles through the transporter CTNS (cystinosin)7–9. Humans with biallelic loss-of-function mutations in CTNS manifest cystinosis, a debilitating lysosomal storage disorder driven by cystine accumulation in tissues across the body, particularly the kidney 9,23. We reasoned that a major source of lysosomal cystine is the oxidation of cysteine brought into lysosomes in an MFSD12-dependent fashion. To test this possibility, we measured cystine in whole-cell and lysosomal samples from HEK-293T cells lacking MFSD12, CTNS, or both. Loss of CTNS increased cystine levels 15- and 8-fold in whole-cell and lysosomal samples, respectively (Fig. 3d). The concomitant loss of MFSD12 reversed these increases, normalizing them to approximately the levels seen in wild-type cells (Fig. 3d). The higher amount of cystine in lysosomes from the double knock-out cells compared to those just lacking MFSD12 suggests that other processes, such as lysosomal proteolysis or vesicular traffic, also contribute to maintaining lysosomal cystine levels, albeit to much smaller extents than MFSD12. We obtained consistent results using RNAi to reduce MFSD12 levels in fibroblasts derived from patients with cystinosis (Fig. 3e and Extended Data Fig. 3c). The reduction in lysosomal cystine was more modest than what we observed in the knock-out HEK-293T cells, likely because the RNAi reduced the MFSD12 mRNA by only 75–85% so that residual MFSD12 protein remained. These data are consistent with MFSD12 functioning upstream of CTNS (Figure 3f).
MFSD12 mediates cysteine import
Mammalian cells acquire most of their cysteine by importing extracellular cystine via the xCT plasma membrane transporter and rapidly reducing it to cysteine in the cytosol 24,25. Loss of MFSD12 had no impact on the plasma membrane transport of [14C]-cystine in B16F10 or HEK-293T cells (Fig. 4a and b). In contrast, in cells lacking MFSD12 and given [14C]-cystine in an extracellular buffer, the amount of [14C] recovered in melanosomes and lysosomes captured via the rapid immunoprecipitation methods was severely blunted. It was undetectable over background in the case of melanosomes and more than 7.5-fold lower in lysosomes (Fig 4. a and b). While these results were consistent with MFSD12 being necessary for the uptake of cysteine into melanosomes and lysosomes, we could not rule out that the cysteine derived from the labeled cystine was converted into something else before its MFSD12-dependent import into these organelles.
Fig. 4. MFSD12 is necessary and likely sufficient for the import of cysteine into melanosomes and lysosomes.
a and b, Measurements of [14C] in melanosomes and lysosomes from cells exposed to [14C]-cystine. MelanoTag-expressing B16F10 cells or LysoTag-expressing HEK-293T cells were incubated with 1.2μM [14C]-cystine for 1 hour, upon which organelles were isolated via the MelanoIP or LysoIP methods. For the lysosomal assays (b), whole-cell and immunoprecipitated samples were normalized using hexosaminidase assay (n = 3 independently prepared purifications per condition, aP =1.7×10−2, bP = 1.2×10−3). c, Cysteine uptake assay into isolated lysosomes prepared by differential centrifugation. To track import, 20μM unlabeled cysteine was added with trace amounts of [35S]-cysteine (n = 3 independently performed assays per condition, aP = 3.5×10−4). d, Cysteine and tyrosine uptake assays into isolated lysosomes. Lysosome preparation and cysteine import was performed as in c. [3H]-tyrosine was added at a concentration of 500nM. ‘Competitor’ refers to an unlabeled analog of the radiolabeled compound used for each assay (n = 3 independently performed assays per condition, aP = 1.7×10−3, bP = 2.4×10−3, cP = 2.7×10−3, dP =1.9×10−3, NS = not significant). e, Immunofluorescence of HEK-293T cells expressing wild-type MFSD12 or MFSD12LL253−254AA mutant (MFSD12PM). MFSD12–3xHA was visualized with an anti-HA epitope tag antibody. Wheat-germ agglutinin (WGA) was used to mark the plasma membrane. Micrographs shown are representative of the majority staining pattern from three independent experiments. Scale bar, 10μm. f, Cysteine uptake in whole-cells expressing MFSD12PM. Cells expressing the indicated proteins were incubated in a buffer containing inhibitors of native cysteine transport before the addition of cysteine. Under the non-competed conditions, 10μM unlabeled cysteine was added with a trace amount of [35S]-cysteine (n = 3 independently performed assays per condition, aP = 2.0×10−2, NS = not significant). Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
Thus, to directly test the necessity of MFSD12 in lysosomal cysteine import we used differential centrifugation to isolate large quantities of lysosomes from wild-type and MFSD12 knock-out HEK-293T cells (Fig. 4c). Lysosomes containing MFSD12 sequestered 11-fold more [35S]-cysteine than those without it during a 30-minute transport assay (Fig. 4c and d). Lysosomes with and without MFSD12 sequestered [3H]-tyrosine to similar extents, emphasizing the specificity of the cysteine transport deficit (Fig. 4d). Consistent with carrier-mediated transport of [35S]-cysteine, preloading of lysosomes with cysteine methyl ester stimulated the influx of [35S]-cysteine into lysosomes, but only when they contained MFSD12 (Extended Data Fig 4a) 19,26. In cells, cystine and glutathione are other candidate sources of lysosomal cysteine. However, lysosomes sequestered the same amount of [14C]-cystine regardless of the genotype of the cells they were isolated from (Extended Data Fig 4b), and reduced glutathione did not compete for [35S]-cysteine transport in our isolated lysosomes (Extended Data Fig. 4c). These results are consistent with MFSD12 impacting melanosomal and lysosomal cysteine and cystine solely through the import of cysteine.
As a test of sufficiency, we asked if MFSD12 could promote the transport of cysteine between a different pair of compartments, namely from the extracellular space into the cytosol. We identified a single dileucine motif (LL253–254) in MFSD12 that when mutated reduced its localization to lysosomes and re-directed it to the plasma membrane (Fig. 4e). In a buffer containing inhibitors of other systems with the potential to transport cysteine and cystine, HEK-293T cells expressing plasma membrane-localized MFSD12 had an enhanced rate of [35S]-cysteine uptake compared to cells expressing a plasma membrane-localized mutant of the lysosomal transmembrane protein TMEM192 (Fig. 4f) 27. Critically, unlabeled cysteine completely competed MFSD12-induced cysteine transport (Fig. 4f). These results suggest that MFSD12 is sufficient to transport cysteine, but a definitive test of this will require future work to directly measure the transport activity of purified MFSD12 reconstituted into artificial liposomes.
Our work indicates that the pigmentation protein MFSD12 is necessary and likely sufficient for the transport of cysteine into melanosomes and lysosomes, processes that were described decades ago 19,22, but for which no protein component of the import system had been convincingly identified. In melanosomes, MFSD12 provides cysteine for the production of the cysteinyldopas used in pheomelanin synthesis, giving a molecular explanation for how genetic variation in MFSD12 influences human pigmentation 3,4. In lysosomes, cysteine is thought to promote the activity of lysosomal proteases 28,29, and our findings provide a potential explanation for why MFSD12 has scored in a genetic screen that indirectly assays lysosomal function 30. Cysteine might represent a primary reducing ‘currency’ in the lysosome 28, and the identification of MFSD12 provides a handle to study the cysteine to cystine cycle in lysosomes in vivo. MFSD12 inhibitors may represent a new therapeutic class for the treatment of cystinosis, a disease for which cysteamine has remained the standard of care since it was developed in 1976 31,32. Given the function of MFSD12 in melanosomes, such inhibitors may also darken skin pigmentation and thus function as sunless tanning agents 33. It is striking that while the core functions of melanosomes and lysosomes are so different, MFSD12 plays important roles in both. Our work provides a roadmap for how the MelanoIP method along with untargeted metabolite profiling can be used to deorphan the function of other melanosomal proteins with roles in human pigmentation.
METHODS
Cell culture
HEK-293T,B16F10, and SKMEL30 cells were from ATCC. The unaffected parental (GM00906) and cystinotic patient (GM00379) fibroblasts were from the Coriell institute and were immortalized with retrovirus generated with pBABE-hTERT (Addgene: #1773). HEK-293T cells, B16F10 cells, and patient-derived fibroblasts were propagated in IMDM (Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS, Gibco). SKMEL30 cells were propagated in RPMI (Gibco) supplemented with 10% FCS (Gibco). Suspension cultures of HEK-293T and B16F10 cells were generated by transferring adherent cells to FreeStyle media (Gibco) supplemented with 1% FCS in a rotating shaker. All medias were supplemented with penicillin/streptomycin (50 U/mL, Gibco). Cells were tested for mycoplasma contamination. Unique biological materials in the form of cell lines are available from the authors by request.
Virus production and generation of cells stably-expressing cDNAs
Lentiviruses were produced by co-transfection of HEK-293T cells with a target lentiviral transfer vector (1 μg), pCMV-VSV-G (100 ng, Addgene #8454), and pCMV-dR8.2 (900 ng, Addgene #8455) plasmids. Retrovirus was generated by substituting pCMV-dR8.2 with pCL-Eco (Addgene #12371). Virus containing supernatnts were harvested forty-eight hours post-transfection and stored at −80°C. For infection, ~1 million cells were incubated with 50–1000 μL viral supernatant, 10 μg/ml polybrene, and 2 mL of culture media in a 6-well plate before spinning at 1,200 x g for 45 minutes at 37°C. Infected cells were passaged into media containing the selection agent 36–48 hours post-transduction. Selection was maintained for 3 to 5 days before use of the cells.
Knock-out cell line generation
Knock-out cell lines were generated by infection with lentiCRISPRv2-Opti (Addgene # Pending) vectors encoding single guide RNAs (sgRNAs) directed against early exonic regions of genes of interest. Clonal knock-out cell lines were isolated through fluorescence-activated cell sorting and frameshifted alleles were confirmed by deep-sequencing of each locus. Double-knockouts were isolated by the same process, starting with clonal single knock-out cells. The following oligonucleotides were used for sgRNA cloning and include cloning overhangs for ligation after BsmBI digest of lentiCRISPRv2-Opti vector:
Tyr (Mouse):
Sense: caccgATGGGTGATGGGAGTCCCTG
Anti-sense: aaacCAGGGACTCCCATCACCCATc
Mfsd12 (Mouse):
Sense: caccGAAGCTCAGCCGCGCGGCGA
Anti-sense: aaacTCGCCGCGCGGCTGAGCTTC
MFSD12 (Human):
Sense: caccgCAAAGGCCAGGATCACCAGG
Anti-sense: aaacCCTGGTGATCCTGGCCTTTGc
CTNS (Human):
Sense: caccGATTTCAAAAGTGATCACCA
Anti-sense: aaacTGGTGATCACTTTTGAAATC
Generation of cDNA constructs
HA-MelanoTag (Addgene # Pending) and myc-MelanoTag (Addgene # Pending) lentiviral constructs: the GPR143 cDNA was isolated via PCR from a cDNA library generated from SKMEL30 cells and cloned directly into pLJC6 (Addgene #104435) containing a contiguous reading frame with c-terminal mScarlet-3xHA or mScarlet-myc. pRK5-HA-MFSD12 (Addgene # Pending): A codon-optimized MFSD12 nucleotide sequence was synthesized as a gBlock from IDT and cloned into pRK5 (Addgene #46326) with a c-terminal TEV-3xFLAG-3xHA tag. MFSD12 mutants (LL253-254AA) were generated with this construct using an adapted QuikChange protocol. pRK5-HA-TMEMPM: TMEM192 from a LysoTag lentiviral vector (Addgene # 102930) was transferred to pRK5 (Addgene #46326) and mutagenized with an adapted QuikChange protocol. pCHA1.1-MFSD12-V5 (Addgene # Pending): the MFSD12 coding sequence was transferred from pRK5 to the pLJC6 (Addgene #104435) variant pCHA1 with a c-terminal V5-tag encoded on its 3’ primer.
The recoded MFSD12 nucleotide sequence is:
ATGGGTCCTGGTCCACCTGCCGCTGGCGCGGCTCCATCTCCACGGCCCCTGAGTCTCGTAGCCCGGTTGTCCTATGCCGTGGGCCATTTCCTTAATGACCTTTGTGCCAGTATGTGGTTTACCTACTTGTTGCTTTATCTCCATAGCGTGAGAGCTTATTCTTCTCGAGGAGCGGGTTTGCTCCTGTTGCTGGGCCAAGTTGCAGATGGCCTGTGCACCCCTCTTGTGGGTTATGAGGCCGATAGAGCGGCAAGTTGCTGTGCAAGGTATGGTCCGCGAAAGGCATGGCATTTGGTTGGCACCGTATGCGTACTTCTTTCTTTCCCATTTATCTTCTCTCCATGTCTCGGCTGCGGCGCAGCCACCCCCGAATGGGCTGCTTTGTTGTATTACGGACCTTTCATCGTAATATTCCAATTCGGCTGGGCGTCAACGCAAATTAGCCATCTCAGTCTCATTCCGGAACTTGTTACTAATGATCACGAAAAAGTAGAGCTTCGCTATGCCTTCACCGTTGTAGCCAACATAACGGTAcACGGCGCTGCCTGGCTTCTCTTGCACCTCCAGGGGAGCTCAAGGGTCGAGCCAACGCAGGATATTTCAATATCAGACCAGCTCGGTGGTCAGGATGTTCCGGTGTTCCGGAATCTGAGTTTGCTCGTCGTCGGAGTTGGAGCTGTCTTCAGCCTTCTTTTCCACCTTGGAACAAGGGAACGCAGACGGCCTCACGCTGAAGAGCCAGGTGAGCACACTCCGCTTCTGGCCCCTGCTACAGCGCAACCCTTGCTTCTCTGGAAGCATTGGCTGAGAGAACCCGCTTTCTATCAAGTGGGCATACTGTATATGACAACGAGGCTTATTGTGAATCTGAGTCAAACCTATATGGCCATGTATTTGACATATTCTCTTCACTTGCCTAAAAAGTTCATCGCCAcAATTCCGCTTGTTATGTATCTGAGTGGTTTCTTGAGTAGCTTTCTGATGAAGCCGATCAACAAGTGCATTGGACGGAACATGACGTACTTCAGTGGCCTTTTGGTCATCCTGGCTTTTGCAGCATGGGTTGCTCTTGCGGAGGGCCTGGGAGTAGCAGTGTATGCGGCTGCTGTTCTGTTGGGGGCCGGGTGTGCAACAATCCTCGTGACGTCCCTTGCGATGACGGCAGATCTGATTGGGCCTCACACGAACTCCGGAGCCTTCGTTTACGGTTCTATGTCCTTCTTGGACAAGGTTGCTAATGGGCTTGCCGTGATGGCAATTCAATCCCTTCACCCGTGCCCTTCTGAGTTGTGCTGCAGAGCGTGTGTGTCCTTTTATCATTGGGCTATGGTCGCTGTGACGGGTGGAGTAGGGGTGGCAGCAGCCCTCTGCCTCTGTAGTCTTTTGCTGTGGCCGACTAGGCTTCGCCGCTGGGACCGGGACGCCCGCCCG
RNAi-mediated knock-down cell lines
Constructs for shRNA-mediated knock-down were accessed via the Broad Institute RNAi Consortium and were in pLKO.1 lentiviral backbones with the following targeting sequences:
shMFSD12_1: CCCATTCTCAACTCTAATCCA
shMFSD12_8: CTTCTTGTCCTCCTTCCTCAT
Knock-down was quantified by qPCR. RNA was isolated via RNAeasy Plus Micro Kit (Qiagen) and cDNA was generated with qScript cDNA Supermix (Quantabio). 100 ng RNA equivalents of cDNA were assayed for MFSD12 and ATCB transcript levels with SYBR green qPCR mix (Roche), utilizing the following primer sets:
qPCR_MFSD12_F: TCCACACAGATCTCCCACCT
qPCR_MFSD12_R: GCCGTAGACGGTGATGTTG
qPCR_ACTB_F: GGTTCCGCTGCCCTGAGG
qPCR_ACTB_R: GAAGGTAGTTTCGTGGATGCC
Immunofluorescence
Immunofluorescence-compatible antibodies to LAMP2 (sc-18822) and TYRP1 (sc-166857) were from Santa Cruz Biotechnology; HA (CST-3724) was from Cell Signaling Technology. Fluorophore-labeled secondary antibodies (goat anti-mouse AlexaFluor488 and goat anti-rabbit AlexaFluor594) were from Thermo Fisher Scientific. For B16F10 cells, 10,000 cells were plated in a 24-well plate onto glass coverings and treated with 10 μM forskolin (LC Labs) the next day. The Immunofluorescence assay was performed 48 hours after plating. For HEK-293T cells, 10,000 cells were plated onto fibronectin-coated glass coverings in a 24-well plate and transfected with 50 ng plasmid and 1 μg salmon sperm carrier DNA with polyethylenimine (PEI, 3 μg per 1 μg plasmid). The immunofluorescence assay was performed 48 hours after transfection. Cells were washed with ice cold PBS and fixed in 4% paraformaldehyde in Phosphate Buffered Saline (PBS, Gibco) for 15 minutes at room temperature before rinsing three times with PBS. Fixed cells were then incubated in Blocking Buffer (5% BSA with 0.1% Tween-20 in PBS) for 1 hour at room temperature. Primary antibodies were diluted 1:400 in Blocking Buffer and applied to cells for 1 hour at room temperature. Cells were then washed three times with PBS before incubation with secondary antibodies, diluted 1:400 in Blocking Buffer, for 1 hour at room temperature. Cells were washed three times with PBS and mounted in Vectashield mounting media. For wheat germ agglutinin (WGA) labeling, 5 μg/mL Alex Fluor 594 WGA conjugate (Thermo Fisher) in PBS was applied directly after fixation and before permeabilization and blocking. Cell images were captured with a 63x objective lens mounted on a spinning-disk confocal microscope (Perkin Elmer) using MetaMorph 7 software for acquisition. Images were processed and prepared in Fiji.
Rapid organellar immunoprecipitation
For B16F10 cells, 3 million MelanoTag-expressing cells were plated per 15 cm plate and treated with 10 μM forskolin (LC Labs) the next day. Two days later, cells were harvested for MelanoIP. For SKMEL30 cells, 10 million cells were plated per 15 cm plate and harvested two days after plating. The cell culture media was exchanged two hours before cell harvest. Cells were washed and harvested by scraping in ice cold KPBS (136 mM KCl, 10 mM KH2PO4, pH 7.25 adjusted with KOH)12 before pelleting by centrifugation at 2,000 x g for 1 minutes at 4°C. Pellets were resuspended in 200 μL ice cold KPBS and 10 μL of the cell suspension was taken as a whole-cell control. Cells were homogenized with a hand-held, rotary pestle (Kimble-Chase) for 45 seconds before the further addition of 800 μL of KPBS and centrifugation at 3,000 x g for 2 minutes at 4°C. Supernatants were transferred into new 1.5 mL tubes containing 100 μL of KBPS-washed anti-HA magnetic beads (Thermo Fisher) and incubated rocking for 4 minutes at 4°C. Beads were washed 3 times with 1000 μL ice cold KPBS, transferring to a new Eppendorf tube each time. Whole-cell and immunoprecipitated samples were then lysed in 50 μL Lysis Buffer (40 mM HEPES, pH 7.4; 1% Triton-X100; 2.5 mM MgCl2; 10 mM β-glycerol phosphate, 10 mM pyrophosphate, and Complete EDTA-free Protease Inhibitor Cocktail (Roche)) for protein analysis, or 50 μL Extraction Buffer (80% methanol v/v supplemented with 500 nM internal extraction standards (Cambridge Isotope Laboratories, MSK-A2-1.2)) for metabolite analyses. For LysoIP experiments, the same procedure was followed as above, but with 10 million LysoTag-expressing (Addgene # 102930 and # 104435) HEK-293T cells plated into a 15 cm dish one day before immunoprecipitation and without forskolin treatment.
Immunoblotting
Immunoblotting antibodies to LAMP2 (sc-18822), TYRP1 (sc-166857), Rab38 (sc-390176), and PMEL (sc-377325) were from Santa Cruz Biotechnology; Vdac1 (CST-4661), Calreticulin (CST-12238), Golgin-97 (CST-13192), Erk1/2 (CST-137F5), and Catalase (CST-12980) were from Cell Signaling Technologies; LAMP1 (1D4B) was from the Developmental Studies Hybridoma Bank at the University of Iowa. Species-specific, HRP-conjugated secondary antibodies were from Cell Signaling Technology. Cells and organelles, immobilized on beads, were lysed on ice in Lysis Buffer (40 mM HEPES pH 7.4, 1% Triton X-100, 2.5 mM MgCl2 and Complete EDTA-free Protease Inhibitor Cocktail (Roche). Cell lysates were clarified by spinning 17,000 x g for 8 minutes at 4°C while organellar lysates were cleared of magnetic beads before adding Laemelli buffer. Lysates were resolved by SDS-PAGE at 120 V. Proteins were transferred for 2 hours at 40 V to PVDF membranes. Membranes were blocked with 5% nonfat dry milk in TBST (Tris-buffered saline with 0.1% Tween-20) before incubation with primary antibodies, diluted 1:1000 in 5% BSA in TBST. Membranes were washed three times in TBST before incubation in 1:3000 species specific HRP-conjugated antibody (Cell Signaling Technologies). Membranes were washed again three times in TBST before visualization with ECL (Thermo Fisher) substrate.
Tissue expression
FANTOM5 expression data (Fig. 3a and Extended Data Fig. 3a) were accessed via the Human Protein Atlas at the following address: https://www.proteinatlas.org/about/assays+annotation#fantom 20,21.
Polar metabolite profiling by LC/MS
LC/MS analysis of polar metabolite content of whole-cell and organellar isolations has been described before 12. LC/MS-based analyses were conducted on a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and HESI II probe, which was coupled to a Dionex UltiMate 3000 ultra-high performance liquid chromatography system (Thermo). External mass calibration was performed using a standard calibration mixture every 7 days.
Microliter volumes (generally 2.5 μL) of sample were injected into a SeQuant ZIC-pHILIC Polymeric column (2.1 × 150 mm; MilliporeSigma) connected with a guard column (2.1 × 20 mm; MilliporeSigma). Both analytical and guard columns were of 5 μm particle size. Column oven and autosampler tray were held at 25°C and 4°C, respectively. Mobile Phase A consisted of 20 mM ammonium carbonate, 0.1% ammonium hydroxide. Mobile Phase B was acetonitrile. The flow rate was 0.150 ml minutes−1 with the following gradient programming: (1) 0–20 minutes: linear gradient from 80% to 20% B; (2) 20–20.5 minutes: linear gradient from 20% to 80% B; (3) 20.5–28 minutes: hold at 80% B.
The mass spectrometer was operated in full scan, polarity switching mode with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HESI probe was held at 350°C. Sheath gas flow was set to 40 units, and the auxiliary gas flow was set to 15 units. The MS data acquisition was performed in a range of 70–1000 m/z, with the resolution set to 70,000, the AGC target at 106, and the maximum injection time at 20 msec.
For untargeted metabolomics experiments, data were acquired as described above, with additional data-dependent (dd) MS/MS collected on pooled samples to aid with unknown metabolite identification. For ddMS/MS, the top 10 ions in each full scan were isolated with a 1.0-Da window, fragmented with a step-wise collision energy of 15, 30, and 45 units, and analyzed at a resolution of 17,500 with an AGC target of 2×105 and a maximum injection time of 100 msec. The underfill ratio was set to 0. The selection of the top 10 ions was set to isotopic exclusion, a dynamic exclusion window of 5.0 sec, and an exclusion list of background ions based on a solvent bank.
Metabolomics data processing and analyses
Untargeted metabolic analysis was performed with CompoundDiscoverer v3.1. MS1 data were collected on individual samples, while representative samples, pooled according to genotype, were used to collect ddMS2 data to aid metabolite identification. We have included these analyses to illustrate how we initially made observations. Data were analyzed using Compound Discoverer 3.1 (Thermo Fisher Scientific) and by including an in-house mass-list with retention time library. Normalization was performed using the default total ion count (TIC) parameter. Identification or annotation of metabolite species was confirmed by matching retention times and MS/MS data to authentic standards run on the same instrument. For specifics regarding identification and annotation of melanin intermediate and the cysteinyldopas, see sections below. In many cases, we were unable to identify the metabolites detected. Therefore, p-values are not multiple hypothesis corrected. Instead, every metabolite highlighted in an untargeted analysis has an accompanying panel from a targeted analysis performed by extracting ion chromatograms using XCalibur 4.0 with a mass tolerance of 5 p.p.m. For relative quantification, the raw peak area for each metabolite was divided by the raw peak area of the relevant isotope-labeled internal standard to calculate the relative abundance.
Follow-up metabolite identification and quantification were performed by extracting ion chromatograms using XCalibur 4.0 (Thermo Fisher) with a mass tolerance of 5 p.p.m and referencing an in-house library of authentic standards. For relative quantification, the raw peak area for each metabolite was divided by the raw peak area of the relevant isotope-labeled internal standard to calculate the relative abundance.
Melanin intermediate validation and detection
Indole-5,6-quinone in untargeted analysis (Fig. 1d) was annotated by matching to in silico MS/MS 34,35. A dihydroxyindolequinone (DHI) chemical standard (Cayman) was used for follow-up compound validation. When dissolved in water, this standard contained two species with m/z values consistent with DHI and indole-5,6-quinone 6. Consistent with this, indole-5,6-quinone is known to be generated from the spontaneous oxidation of DHI. As this annotated indole-5,6-quinone species was more readily detectable in biological samples, we have quantified here and included it in figures (Fig. 1d and e).
Cysteine measurements with Ellman labeling
Ellman labeling for cysteine quantification was adapted from previously described methods for measuring thiol containing compounds in biological extracts36. Whole-cell and immunopurified samples were extracted in 25 μL 80% v/v methanol without internal standards. An internal control of 100 nM [U-13C, 15N]-cysteine (Cambridge Isotopes) was added to each sample before the addition of a 1:1 volume of 10 mM solution of Ellman’s reagent (Thermo Fisher) prepared in 80% v/v methanol. Samples were incubated on ice for 1 hour before storage at −80°C. Samples were run as above with a targeted selected ion monitoring scan (tSIM) in positive mode centered on m/z 319.00530 and m/z 323.01240 to increase signal for cysteine-TNB and [U-13C, 15N]-cysteine-TNB, respectively.
Cysteinyldopas measurements with solid phase extraction
The cysteinyldopa extraction protocol was adapted from previously described methods for measuring cysteinyldopas in serum37. Cells were plated under the same conditions used for the MelanoIP protocols. After scraping and washing, cells were lysed via rotary homogenizer (Kimble-Chase) in 0.1 N HCl in water before spinning 18,000 x g for 5 minutes at 4°C. 100 nM [15N]-phenylalanine (Cambridge Isotopes) was added to lysate supernatant as an internal control. MCX cartridges (1 mL, Oasis) were conditioned with 1.0 mL of 100% methanol then 1.0 mL of 0.1 N HCl before applying lysate supernatant and washing with 1.0 mL of 0.1 N HCl then 1.0 mL of 100% methanol. Elution was performed with an 80:10:10 v/v/v mixture of methanol/water/~30% ammonia with 0.1% ascorbic acid (MilliporeSigma) added as an antioxidant. Samples were acidified after elution with 100 μL formic acid and dried under nitrogen stream before storage at −80°C. Before analysis, extracts were resuspended in 250 μL 0.1% formic acid with 0.1% ascorbic acid and debris were cleared by centrifugation with 18,000 x g for 5 minutes at 4°C before LC/MS analysis.
Detection of cysteinyldopas by LC/MS was performed on the same instrumentation as above, with the following modifications: A 10 μL sample volume was injected onto an Ascentis Express dC18 HPLC column (2.7 μM x 150 mm x 2.1 mm; MilliporeSigma). Column oven and autosampler tray were held at 20°C and 4°C, respectively. Mobile Phase A consisted of 0.1% formic acid in water. Mobile Phase B was 0.1% formic acid in 100% methanol. The flow rate was 0.250 ml minutes−1 with the following gradient programming: 0–12 minutes: linear gradient from 0% to 10% B. The mass spectrometer was operated in positive mode an MS data acquisition range from 100 – 350 m/z. ddMS/MS data were collected in positive mode as described above.
Cysteinyldopa detection was validated with standards synthesized from an adapted protocol for cysteinyldopa synthesis 38. L-dopa (8.5 mM) and cysteine (17.0 mM) were prepared fresh in 60 mL of 50 mM sodium phosphate buffer, pH 6.8. Reactions were run with the addition of ~ 250 U of mushroom tyrosinase (MilliporeSigma), stirring in well aerated conditions at room temperature. Aliquots were quenched by dilution 1:10 in 1 N HCl. After quenching, standards were extracted and analyzed like biological samples. Mirror plots comparing MS/MS spectra between the standards and representative biological samples were generated using an in-house R script using a threshold of 4%.
Hexosaminidase assay
Samples were diluted multiple times to ensure signal was quantified within a linear range. Each analyte was then diluted 1:10 in Substrate Solution (90 mM potassium acetate, pH 5.0; 2 mM p-nitrophenyl hexosaminidase substrate; 0.5% Triton X-100) and briefly vortexed to induce lysis before incubation for 30 minutes at 37°C. Samples were quenched with 1 part 0.25 M NaOH to 2 parts of the substrate solution. Absorbance was quantified at 405 nm with a SpectraMax iD5 plate reader (Molecular Devices) and SoftMax Pro 7 acquisition software (Molecular Devices).
Cellular cystine uptake assays
50 million suspension cells were washed twice with HBSS, and resuspended in 200 μL HBSS. Upon addition of 1.2 μM [14C]-cystine (Perkin-Elmer), cell suspensions were mixed and incubated at 37°C with periodic mixing for 1 hour. Cells were then washed twice in ice cold KPBS and organelles were isolated via organellar immunoprecipitation protocols as detailed above. For background quantification samples, 1% Triton X-100 was added to KPBS during the immunoprecipitation and washing steps. Beads were resuspended in 1 % Triton X-100 and water and pipetted directly into scintillation fluid before reading scintillation signal (TriCarb T9000TR, Perkin-Elmer). For lysosomal cellular cystine uptake assays, aliquots of whole-cell and immunoprecipitated samples were taken and measured by hexosaminidase assay. Hexosaminidase assay values were used to normalize scintillation counts of cellular input and lysosomal capture.
Transport assays with isolated lysosomes
Lysosomal purification protocols were adapted from previous protocols 39. 500 million suspension-adapted HEK-293T cells were spun down, washed in ice-cold PBS, and resuspended in 15 mL Homogenization Buffer (250 mM sucrose; 20 mM HEPES, pH 7.4; 1 mM EDTA) supplemented with protease inhibitor tablets (Roche). Cells were dounced 5 times with a loose-fitting glass dounce and the resulting lysate was centrifuged at 1,500 x g for 10 minutes at 4°C. After transferring the supernatant, 15 mL Homogenization Buffer was used to resuspend the pellet, and douncing and centrifugation were repeated. Supernatants from three successive rounds of homogenization and centrifugation were pooled and centrifuged a final time before transferring the ‘post-nuclear supernatant’ to the subsequent step. ‘Post-nuclear supernatant’ was centrifuged at 20,000 x g for 10 minutes at 4°C to pellet organelles. A working stock of 30% OptiPrep (Millipore Sigma) was prepared by mixing equal volumes of 60% OptiPrep and Dilution Buffer (250 mM sucrose; 40 mM HEPES, pH 7.4; 2 mM EDTA). All subsequent dilutions were prepared by mixing this stock and Homogenization Buffer. The resulting pellet or ‘light mitochondrial fraction’ was resuspended in 9% OptiPrep and overlaid on top of a discontinuous gradient of 18, 16, 14, 12, and 10% OptiPrep. After centrifugation at 145,000 x g for 2 hours at 4°C, 1 mL fractions were taken and assayed for lysosomal activity with a hexosaminidase assay. High activity fractions were diluted 1:10 in homogenization buffer and spun 8,000 x g for 10 min at 4°C to pellet lysosomes before resuspending in Uptake Buffer (KPBS, 250 mM sucrose, 2 mM ATP, 2 mM MgCl2, and 3 mM DTT). The lysosomal content of each sample was normalized with a hexosaminidase assay and samples were equilibrated for 30 minutes on ice. Assays were started with the addition of 20 μM cold cysteine and 50 μCi/mL (< 100 pM) [35S]-cysteine (Perkin-Elmer). Samples were incubated at 30°C with periodic mixing. Aliquots were taken at indicated time points and passed over glass fiber (GFA) filters (MilliporeSigma) that had been blocked overnight with 1% BSA in PBS. Filters were washed two times in ice-cold PBS, dried, and submerged in scintillation fluid before measuring scintillation counts (TriCarb T9000TR, Perkin-Elmer).
Lysosomal uptake assays with other radioactive compounds ([3H]-tyrosine and [14C]-cystine) were performed under identical conditions as the [35S]-cysteine uptake experiments, except where otherwise specified. [3H]-tyrosine (American Radiolabeled Chemicals) was incubated at a concentration of 500 nM for 10 minutes. Lysosomal uptake assays in Fig. 4d compare cysteine and tyrosine uptake into identical lysosome preparations.
For [14C]-cystine, in ‘Unreduced’ conditions,’ 1 μM [14C]-cystine was incubated for 10 minutes in Uptake Buffer without DTT. In ‘Reduced’ conditions, the same amount of [14C]-cystine was added, but it was pre-treated with 10 mM DTT for 1 hour at 50°C before addition to the assay and Uptake Buffer had DTT included.
Cysteine methyl ester loading was performed by incubating lysosomes with 1 mM cysteine methyl ester (MilliporeSigma) for 20 minutes at 37°C in Uptake Buffer. Lysosomes were washed three times in Uptake Buffer before performing [35S]-cysteine uptake assays as described above. Samples were harvested after 5 minutes, a pre-steady state time point.
Background scintillation counts for each condition were determined by performing an identical assay but using lysosomes pretreated with 20 mM glycine methyl ester (Sigma) for 10 min at 37°C. This treatment has been previously shown to selectively permeabilize lysosomes40,41. To obtain the assay signal, scintillation counts from these background assays were subtracted from counts from untreated preparations.
Plasma membrane cellular uptake assays
3 million HEK-293T cells were plated in 10 cm dishes. Cells were transfected with 3 μg of plasmid with PEI (3 μg per 1 μg plasmid) and scraped 48 hours post-transfection into ice cold KPBS. After washing twice, cells were resuspended in KPBS supplemented with 125 mM sucrose, 2.5 μM Erastin (MilliporeSigma), 4 mM leucine, and 3 mM serine. Assays were started with the addition of cold cysteine (10 μM for transport conditions and 500 μM for competition) and 50 μCi/mL (< 100 pM) [35S]-cysteine. After samples were incubated at 30°C for 10 minutes, cells were harvested by centrifugation at 1,000 x g for 30 seconds at 4°C and washed twice in ice-cold PBS. Cell pellets were resuspended in water before mixing into scintillation fluid and measuring scintillation counts (TriCarb T9000TR, Perkin-Elmer).
Data preparation and statistics
Displays of quantitative data were prepared in Microsoft Excel and GraphPad Prism 8. Statistical comparisons were via two-tailed unpaired t-tests, performed in Prism. All measurements displayed represent samples generated independently, or biological replicates. Immunoblot and immunofluorescence data are representative of experiments repeated at least three times.
Extended Data
Extended Data Fig. 1. MelanoIP analysis detects changes in Tyr dependent melanosomal metabolites.
a, Schematic of melanin synthesis. The common pathway elements for eumelanin and pheomelanin synthesis have a gray backdrop. The brown and red backdrops highlight unique portions of eumelanin and pheomelanin synthesis, respectively. Enzymes proposed to catalyze each step are shown in green. Synthetic intermediates annotated and validated in biological samples in this study are in blue. b, Follow-up analysis with standard validated m/z and internal normalization of ‘proteogenic amino acids’ highlighted in untargeted metabolite profiling of wild-type and Tyr knock-out melanosomes (Fig. 1d). Amino acids are presented in order of increasing retention time (n = 3 independently prepared extracts, aP = 3.9×10−2, bP = 2.0×10−3, cP = 6.5×10−3, dP = 3.8×10−2, eP = 1.9×10−2). Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
Extended Data Fig. 2. In vitro synthesis and biological detection of cysteinyldopas.
a, Cysteinyldopas were synthesized according to an adapted protocol from Ito and Prota, 197738. Two species, distinguished by retention time, were generated at the expected m/z for cysteinyldopas. It has been shown that 5’-cysteinyldopa is produced in greater abundance than 2’-cysteinyldopa in this reaction. Taking MS1 peak intensity to approximate abundance, we putatively annotate the ‘Minor Isomer’ as 2’ substituted, and the ‘Major Isomer’ as 5’ substituted. b, Mirror plot of ddMS2 data comparing 2’- and 5’-cysteinyldopa in synthetic cysteinyldopas. c and d, Mirror plots of ddMS2 peaks displaying similarities in ddMS2 spectra of 2’- and 5’-cysteinyldopa species in biological samples (B16F10 extracts) and synthetic standards.
Extended Data Fig. 3. MFSD12 maintains lysosomal cystine in non-pigmented cells.
a, FANTOM5 CAGE profiling data accessed via Human Protein Atlas 20,21. Six representative pigmentation genes, including MFSD12, are shown. b, Metabolite profiling of LysoIP samples from HEK-293T cells comparing lysosomes from wild-type and MFSD12 knock-out cells. ‘Accumulates upon inhibition of:’ has been previously reported 11 (n = 4 independently prepared metabolite extracts, aP = 7.0×10−4, bP = 3.0×10−3, cP = 4.1×10−2, dP = 4.2×10−2, eP = 1.7×10−4). c, Lentiviral shRNA knock-down of MFSD12 quantified via qPCR and normalized to ACTB levels (n = 3 assays on independently prepared cDNA libraries, aP = 1.97×10−3, bP = 3.0×10−3). Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
Extended Data Fig. 4. MFSD12 mediated cysteine transport is cysteine specific.
a, Test of lysosomal counter-transport. Lysosomes were purified by differential centrifugation and incubated with water or 1 mM cysteine methyl ester before washing, resuspension, and incubated for 5 minutes with 20 μM cysteine and trace amounts of [35S]-cysteine (n = 3 independently performed assays per condition, aP =2.5×10−3, bP = 3.3×10−2, NS = not significant). b, Lysosomal import of [14C]-cystine. Lysosomes were purified by differential centrifugation and incubated for 10 min with 1 μM [14C]-cystine, either untreated (Unreduced) or pre-treated with 10 mM DTT (Reduced, n = 6 independently performed assays per condition, aP = 2.1×10−7, bP = 1.3×10−6, cP = 3.8×10−8, NS = not significant). c, Competition for [35S]-cysteine transport. Lysosomes were purified by differential centrifugation and incubated for 10 minutes with 20 μM cysteine and trace amounts of [35S]-cysteine with 500 μM competitor where indicated (n = 3 independently performed assays per condition, P values compare competition condition versus water control condition (red), aP = 2.7×10−4, bP =2.5×10−4, cP = 3.2×10−4, NS = not significant). Error bars are mean ± s.e.m, P-values by two-sided Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank P. Budde, M. Abu-Remaileh, W.W. Chen, L. Bar-Peled, and J.G. Bryan, as well as all current members of the Sabatini laboratory for helpful insights. This work was supported by grants from the Leo Foundation (LF18057) and NIH (R01 CA103866, R01 CA129105, and R01 AI047389), as well as fellowship support from the NIH (NRSA F31 CA228241-01) to C.H.A., Marshall Plan Foundation to A.K.T., HHMI XROP to B.C., and NSF (2016197106) to K.J.C. D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society Research Professor.
Footnotes
DATA AVAILABILITY
Source data for figures are provided with the paper.
Fig. 3a and Extended Data Fig. 3a were generated from FANTOM5 expression data, accessed via the Human Protein Atlas at the following address: https://www.proteinatlas.org/about/assays+annotation#fantom 20,21
Raw values from this accession are included in the source data for Fig. 3a and Extended Data Figure 3a.
Unique biological materials in the form of plasmids are be available from AddGene. Unique biological materials in the form of cell lines are available from the authors by request.
COMPETING INTEREST
The authors declare the following competing interests: C.H.A, A.K.T., and D.M.S are listed on a patent application based on the work described here.
REFERENCES
- 1.Basrur V. et al. Proteomic Analysis of Early Melanosomes: Identification of Novel Melanosomal Proteins. J. Proteome Res. 2, 69–79 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Sturm RA Molecular genetics of human pigmentation diversity. Human Molecular Genetics 18, R9–R17 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Adhikari K. et al. A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford NG et al. Loci associated with skin pigmentation identified in African populations. Science 358, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Alba L. & Shawkey MD Melanosomes: Biogenesis, Properties, and Evolution of an Ancient Organelle. Physiological Reviews 99, 1–19 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Prota G. Melanins and Melanogenesis. (Academic, 1992). [Google Scholar]
- 7.Gahl WA, Bashan N, Tietze F, Bernardini I. & Schulman JD Cystine transport is defective in isolated leukocyte lysosomes from patients with cystinosis. Science 217, 1263–1265 (1982). [DOI] [PubMed] [Google Scholar]
- 8.Jonas AJ, Smith ML & Schneider JA ATP-dependent lysosomal cystine efflux is defective in cystinosis. J. Biol. Chem. 257, 13185–13188 (1982). [PubMed] [Google Scholar]
- 9.Town M. et al. A novel gene encoding an integral membrane protein is mutated in nephropathic cystinosis. Nature Genetics 18, 319–324 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Watabe H, Kushimoto T, Valencia JC & Hearing VJ Isolation of Melanosomes in Current Protocols in Cell Biology (eds. Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J. & Yamada KM) (John Wiley & Sons, Inc., 2005). doi: 10.1002/0471143030.cb0314s26. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Remaileh M. et al. Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen WW, Freinkman E, Wang T, Birsoy K. & Sabatini DM Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ray GJ et al. A PEROXO-Tag Enables Rapid Isolation of Peroxisomes from Human Cells. iScience 23, 101109 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruder JM et al. Melanosomal Dynamics Assessed with a Live-Cell Fluorescent Melanosomal Marker. PLOS ONE 7, e43465 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diment S, Eidelman M, Rodriguez GM & Orlow SJ Lysosomal Hydrolases Are Present in Melanosomes and Are Elevated in Melanizing Cells. J. Biol. Chem. 270, 4213–4215 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Bissig C, Rochin L. & van Niel G. PMEL Amyloid Fibril Formation: The Bright Steps of Pigmentation. Int J Mol Sci 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reddy VS, Shlykov MA, Castillo R, Sun EI & Saier MH The major facilitator superfamily (MFS) revisited. FEBS J. 279, 2022–2035 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloom JL & Falconer DS ‘Grizzled’, a mutant in linkage group X of the mouse. Genetics Research 7, 159–167 (1966). [Google Scholar]
- 19.Potterf SB et al. Cysteine Transport in Melanosomes from Murine Melanocytes. Pigment Cell Research 12, 4–12 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Kawaji H, Kasukawa T, Forrest A, Carninci P. & Hayashizaki Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Scientific Data 4, 1–3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlén M. et al. Tissue-based map of the human proteome. Science 347, (2015). [DOI] [PubMed] [Google Scholar]
- 22.Pisoni RL, Acker TL, Lisowski KM, Lemons RM & Thoene JG A cysteine-specific lysosomal transport system provides a major route for the delivery of thiol to human fibroblast lysosomes: possible role in supporting lysosomal proteolysis. J. Cell Biol. 110, 327–335 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gahl WA, Thoene JG & Schneider JA Cystinosis. N. Engl. J. Med. 347, 111–121 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Oshima RG, Rhead WJ, Thoene JG & Schneider JA Cystine metabolism in human fibroblasts. Comparison of normal, cystinotic, and gamma-glutamylcysteine synethetase-deficient cells. J. Biol. Chem. 251, 4287–4293 (1976). [PubMed] [Google Scholar]
- 25.Sato H, Tamba M, Ishii T. & Bannai S. Cloning and expression of a plasma membrane cystine/glutamate exchange transporter composed of two distinct proteins. J. Biol. Chem. 274, 11455–11458 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Wilbrandt W. & Rosenberg T. The Concept of Carrier Transport and Its Corollaries in Pharmacology. Pharmacol Rev 13, 109–183 (1961). [PubMed] [Google Scholar]
- 27.Behnke J, Eskelinen E-L, Saftig P. & Schröder B. Two dileucine motifs mediate late endosomal/lysosomal targeting of transmembrane protein 192 (TMEM192) and a C-terminal cysteine residue is responsible for disulfide bond formation in TMEM192 homodimers. Biochemical Journal 434, 219–231 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Lloyd JB Disulphide reduction in lysosomes. The role of cysteine. Biochem J 237, 271–272 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mego JL Role of thiols, pH and cathepsin D in the lysosomal catabolism of serum albumin. Biochem J 218, 775–783 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsui CK et al. CRISPR-Cas9 screens identify regulators of antibody–drug conjugate toxicity. Nat Chem Biol 15, 949–958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gahl WA et al. Cysteamine Therapy for Children with Nephropathic Cystinosis. New England Journal of Medicine 316, 971–977 (1987). [DOI] [PubMed] [Google Scholar]
- 32.Thoene JG, Oshima RG, Crawhall JC, Olson DL & Schneider JA Cystinosis. Intracellular cystine depletion by aminothiols in vitro and in vivo. J Clin Invest 58, 180–189 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mujahid N. et al. A UV-Independent Topical Small-Molecule Approach for Melanin Production in Human Skin. Cell Rep 19, 2177–2184 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 34.Djoumbou-Feunang Y. et al. CFM-ID 3.0: Significantly Improved ESI-MS/MS Prediction and Compound Identification. Metabolites 9, 72 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wishart DS et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35, D521–526 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan X, Hoffman B, Dwivedi C. & Matthees DP A simultaneous liquid chromatography/mass spectrometric assay of glutathione, cysteine, homocysteine and their disulfides in biological samples. Journal of Pharmaceutical and Biomedical Analysis 31, 251–261 (2003). [DOI] [PubMed] [Google Scholar]
- 37.Martin GB et al. Development of a mass spectrometry method for the determination of a melanoma biomarker, 5-S-cysteinyldopa, in human plasma using solid phase extraction for sample clean-up. Journal of Chromatography A 1156, 141–148 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Ito S. & Prota GMA A facile one-step synthesis of cysteinyldopas using mushroom tyrosinase. Experientia 33, 1118–1119 (1977). [DOI] [PubMed] [Google Scholar]
- 39.Graham JM Isolation of lysosomes from tissues and cells by differential and density gradient centrifugation. Curr Protoc Cell Biol Chapter 3, Unit 3.6 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Goldman R. & Kaplan A. Rupture of rat liver lysosomes mediated by l-amino acid esters. Biochimica et Biophysica Acta (BBA) - Biomembranes 318, 205–216 (1973). [DOI] [PubMed] [Google Scholar]
- 41.Verdon Q. et al. SNAT7 is the primary lysosomal glutamine exporter required for extracellular protein-dependent growth of cancer cells. Proceedings of the National Academy of Sciences 114, E3602–E3611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.