Abstract
Healthy pancreatic β-cells well adapt to systemic insulin resistance to maintain normal blood glucose levels, and failure to this process develops type 2 diabetes in humans. Genome-wide association studies uncovered genetic variants that are associated with type 2 diabetes. However, it is still insufficient to explain the high prevalence of this disease. Epigenetics is the study of gene expression changes that do not involve DNA sequence alterations such as DNA methylation, histone modification, and non-coding RNAs. In recent decades, a large number of studies demonstrated the roles of epigenetics in β-cell biology. In this review, we summarize the epigenetic mechanisms in β-cell adaptation and type 2 diabetes, which include three-dimensional chromatin structure and RNA modification.
Keywords: Pancreatic β-cell, Compensation, Adaptation, Type 2 diabetes, Epigenetics
Introduction
Type 2 diabetes (T2D) is a chronic disease that is characterized by insulin resistance and hyperglycemia [1]. Pancreatic β-cell is the unique cell-type that secretes physiological amounts of insulin to regulate blood glucose levels [2]. In response to systemic insulin resistance, β-cells adapt by expanding its mass, promoting its survival, increasing its function and maintaining its mature identity [3-5]. Collectively, this process so-called β-cell compensation is determined by a total amount of functional β-cell mass. Failure of β-cell compensation leads to the development of T2D in humans [6].
Recent improvements in next-generation sequencing (NGS), cell sorting, and singlecell technologies motivated researchers to identify target genes for β-cell dysfunction in T2D patients [7-9]. However, due to the limitation of human samples and complex nature of T2D, there is yet no unique or definite transcriptional signature that represents dysfunctional β-cells in T2D [10].
Nevertheless, it is evident that proper regulation of multiple genes is essential for successful β-cell compensation [11]. Indeed, multiple β-cell transcriptional factors (e.g. PDX1, NKX6.1 and MAFA) are inactivated or reduced in the β-cells of T2D patients [12]. These genes cover not only essential β-cell genes that should be expressed during β-cell adaptation, but also β-cell disallowed genes that should be repressed in functional β-cells [13].
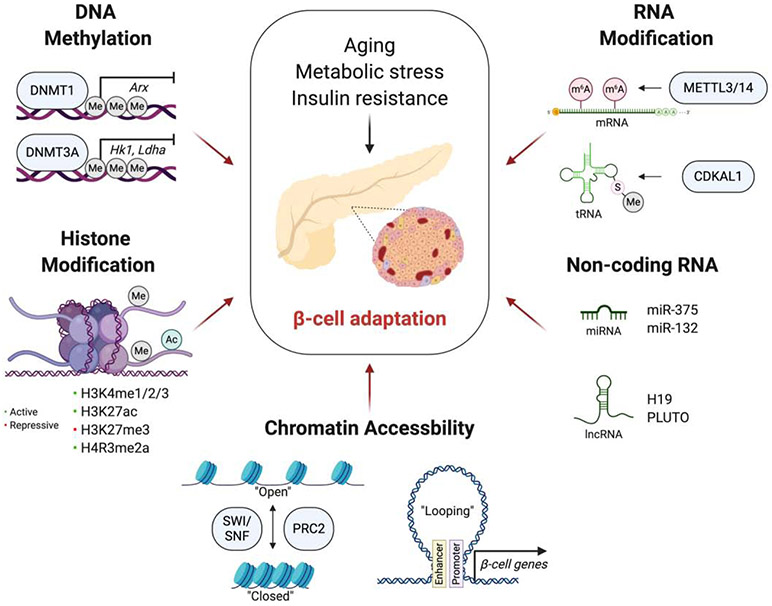
Genome-wide association studies (GWAS) have revealed genetic predisposition in T2D population [14]. However, genetic variant is still insufficient to explain the high prevalence and heritability of T2D [15,16]. Epigenetics is a field that studies heritable traits of cells, especially gene expression, without the changes in a DNA sequence. In this context, understanding epigenetics in metabolic tissues can be a promising way to find the environmental factors and a missing heritability in T2D population. Here, we highlight the current studies on the epigenetic regulation of β-cell adaptation (Figure 1) and epigenetic features of β-cells linked with T2D.
Figure 1.
Epigenetic regulation of β-cell adaptation
Schematic representation summarizing the epigenetic mechanisms in β-cell adaptation. In response to systemic insulin resistance such as aging and obesity, β-cells adapt to external conditions by self-duplication, enhancing insulin secretion, and maintaining its mature identity. The figure created with BioRender.com.
DNA methylation
DNA methylation at the C5 position of cytosines induces heterochromatin structure, thus decreases chromatin accessibility and gene expression.
Whole-genome bisulfite sequencing (WGBS) in the islets of control and T2D donors revealed that genes involved in β-cell function and development, such as PDX1 and TCF7L2 are heavily methylated in the islets of T2D patients [17].
The Bhushan group reported the different roles of DNA methyltransferases (DNMTs) in β-cell biology [18,19]. DNMT1 represses α-cell gene Arx during β-cell replication [18]. This provides an important explanation on how β-cell maintains its identity while it enters a cell-cycle for an expansion. Somewhat different from DNMT1, DNMT3a is required for functional maturation of β-cells [19]. Genetic deletion of Dnmt3a in β-cells resulted in demethylation and upregulation of Hk1 and Ldha genes, which lead to impaired glucose-stimulated insulin secretion (GSIS). Taken together, DNMTs play important role in expansion of functional β-cell mass. Aging is one of the significant drivers of insulin resistance. Genome-wide DNA methylome analysis from purified β-cells of young (5-week-old) and old (18-month-old) mice indicated that the functional adaptation of β-cells in the aging process might be associated with DNA methylation [20]. In this study, authors showed that promoters of proliferation genes (e.g. Mki67, Plk1 and Ccnd3) become hypermethylated with aging, while enhancers of genes that are involved in β-cell function (e.g. Foxa2, Nkx6.1 and Neurod1) become demethylated. Interestingly, these methylation changes in β-cell function genes were correlated with actual insulin secretory function, as islets of old mice showed increased GSIS compared to those of young mice.
Recently, Hall et al. reported that long term exposure to high glucose and palmitate levels significantly altered DNA methylation and correlative gene expression in human islets [21]. Among genes that were differentially methylated and expressed, FICD and TPX2 were shown to repress GSIS function in the human β-cell line, EndoC-βH1 cells. These results indicate that glucolipotoxicity-induced DNA methylation may be linked to impaired β-cell function in T2D patients.
Histone modification
Histones are highly basic proteins that package DNA into the nucleosome. N-terminal tails of histones are subject to various post-translational modifications including acetylation, citrullination and methylation that affect chromatin accessibility and gene expression.
Genome-wide profiling of histone modifications using chromatin immunoprecipitation with sequencing (ChIP-seq) in human islets revealed the islet-specific enrichment of active (H3K4me1, H3K4me2, H3K4me3 and H3K27ac) and repressive (H3K27me3) histone marks at different genomic regions [22,23]. Importantly, islet enhancers occupied by these active histone marks were associated with T2D susceptibility variants in humans [23].
Histone acetylation is one of the most studied histone modifications in β-cell adaptation. Active enhancer mark, histone H3 lysine 27 acetylation (H3K27ac) is increased in the promoter regions of fatty acid β-oxidation genes in the islets of diet-induced obesity (DIO) mice [24]. Meanwhile, both pharmacological and genetic inhibition of histone deacetylases (HDACs) increased the insulin secretory function in β-cells [25-27]. Several studies indicated that histone acetylation could be regulated by PDX1, a critical TF for β-cell compensation [28]. Interestingly, enzymes that act conversely toward histone acetylation are recruited to PDX1 in a glucose-dependent manner [29,30]. In low glucose concentration, PDX1 interacts with HDAC1 and HDAC2 to repress insulin gene expression [29]. On the other hand, PDX1 form complex with histone acetyltransferase p300 in high glucose condition and induces insulin gene expression [30].
Histone methylation also regulates β-cell function. Set7/9 is a histone lysine methyltransferase that is responsible for generating active histone mark, histone H3 lysine 4 methylation (H3K4me). Deering et al. reported that Set7/9 is required for H3K4me formation within the promoters of important β-cell function genes, such as Pdx1 and Slc2a2 [31]. Indeed, knocking out Set7/9 in mouse β-cells resulted in downregulation of these genes followed by impaired GSIS [32]. Protein arginine methyltransferase 1 (PRMT1) generates an active histone mark, histone H4 arginine 3 asymmetric dimethylation (H4R3me2a) in β-cell. Knocking out PRMT1 in mature β-cells induced loss of mature identity and exacerbated glucose intolerance in DIO mice, suggesting that its activity is required for β-cell adaptation in insulin resistance [33].
Chromatin remodeling
Transcriptional regulation is a dynamic process which is mediated by multi-subunit chromatin remodeling complexes. Several studies reported the potential roles of these complexes in β-cell adaptation.
Switch/sucrose non-fermentable (SWI/SNF) complex is an ATP-dependent chromatin remodeler that has been extensively studied in the epigenetics research field. Similar to the action mechanism with histone acetylases (HATs) and HDACs, the SWI/SNF complex associates with PDX1 in a glucose-dependent manner [34]. The mammalian SWI/SNF complex contain either one of these two mutually exclusive ATPase subunits: BRG1 and BRM. In mouse β-cell, PDX1 interacts with BRG1 in a high glucose concentration and enhances its target genes (e.g. Ins1, Slc2a2 and Ucn3), while it associates with BRM in a low glucose concentration and represses these genes [34]. Indeed, knocking out BRG1 and BRM in β-cells induced glucose intolerance and impaired GSIS in mice [35]. Interestingly, a physical interaction between BRG1 and PDX1 was decreased in the islets of T2D patients [34]. Meanwhile, Wei et al. demonstrated that vitamin D switches SWI/SNF complex (BRD9-SWI/SNF to BRD7-SWI/SNF) and changes chromatin accessibility to induce pro-survival gene expression and protect β-cells from metabolic stress condition [36]. Collectively, these data indicate that SWI/SNF complex play an important role in β-cell adaptation.
Polycomb repressive complex 2 (PRC2) generates histone H3 lysine 27 trimethylation (H3K27me3) mark, mediating the formation of bivalent chromatin regions. Lu et al. demonstrated that the loss of PRC2-dependent gene silencing may be a potential mechanism for β-cell dedifferentiation in T2D patients [37]. In this study, they performed a chromatin-state segmentation analysis in mouse islets using ChIP-seq data for various histone marks, including H3K27ac, H3K4me1, and H2K27me3. Single-cell RNA-seq in the islets of normal chow- and high-fat diet (HFD)-fed mice revealed that HFD triggers β-cell population into the poorly differentiated states. Knocking out EED, a core subunit of PRC2, in β-cells induced upregulation of bivalent genes (e.g. Barx1, Hoxb7 and Pitx1) and progressive diabetes in mice due to β-cell dedifferentiation. Importantly, these bivalent genes were upregulated, and H3K27me3 signal was decreased in the β-cells of T2D patients.
Chromatin accessibility
Euchromatin, often called accessible chromatin regions, harbor binding sites for TFs, chromatin modifiers and RNA polymerase. Chromatin accessibility is largely dependent on DNA methylation and histone modifications, hence useful information for studying epigenetic mechanism in gene expression. With rapid advances in epigenetic technologies, including assay for transposase-accessible chromatin with sequencing (ATAC-seq) and chromatin conformation capture techniques, it became possible to reveal genome-wide chromatin accessibility and structure in a high-resolution.
Khetan et al. demonstrated that T2D-associated single-nucleotide polymorphisms (SNPs) are associated with chromatin accessibility in human β-cells [38]. Using chromatin accessibility quantitative trait loci (caQTL) analysis, they identified 2,949 SNPs that are associated with cis-regulatory elements in human islets (n=19). Among the variants tested for enhancer activity using luciferase reporter assays in MIN6 β-cells, more than half exhibited consistent effects with in vivo chromatin accessibility changes in human islets. Importantly, this caQTL analysis proposed 13 T2D-associated GWAS SNPs that are predicted to affect chromatin accessibility in human islets.
T2D-associated variants are enriched in enhancer regions, but their actual target genes were not well known. Recently, researchers identified global enhancer looping and enhancer-promoter interactions in human islets using chromatin conformation capture techniques [39,40]. Greenwald et al. performed genome-wide Hi-C assay in human islets to generate high-resolution map of three-dimensional (3D) chromatin architecture [39]. Using islet Hi-C, RNA-seq, ATAC-seq and enhancer mark ChIP-seq data, they identified potential target genes for thousands of islet enhancers that were correlated with islet-specific gene expression. Furthermore, they fine-mapped T2D variant loci that affect chromatin looping and target gene expression, revealing that these regions are enriched for protein transport and secretion. Meanwhile, Miguel-Escalada et al. used a promoter capture Hi-C (pcHi-C) to make a genome-scale map of promoter-enhancer interactions in human islets [40]. The pcHi-C analysis uncovered more than 1,300 groups of islet enhancers that form clusters in 3D space, in which they call islet hub. Interestingly, the islet hubs were enriched for T2D-associated loci, and perturbation of these chromatin regions affected target gene expression in EndoC-βH3 cells. Importantly, they demonstrated that genetic variation in islet hubs impact insulin secretion. Thus, it can be used for polygenic risk scores to predict T2D susceptibility in humans.
Non-coding RNAs
Non-coding RNAs (ncRNAs) cover various nonprotein-coding RNAs including microRNAs (miRNAs), ribosomal RNAs (rRNAs), transfer RNA (tRNA), and long noncoding RNAs (lncRNAs). Mounting evidence demonstrate that ncRNAs play essential roles in diverse cellular processes.
Once cleaved by endonucleases Drosha and Dicer, miRNAs exert their function by blocking their target mRNA expression. Numerous cell line and mouse studies indicated that miRNAs regulate stress responses in β-cells [41]. More than 30 different types of miRNAs have been reported to mediate β-cell adaptation or be linked with T2D development [41,42].
As one of the most highly expressed miRNAs in mouse and human β-cells, miR-375 is required for β-cell mass expansion in vivo [43]. Knocking out miR-375 in mouse induced overt diabetes with reduced β-cell mass which was exacerbated in the background of ob/ob mouse [43]. Several targets including Pdpk1, Cav1, Cadmn1 and Id3 have been proposed to mediate the function of miR-375 in β-cell growth and proliferation [43,44]. Recently, Mziaut et al. reported that miR-132 promotes β-cell survival in vitro and in vivo [45]. Mechanistically, miR-132 downregulates Pten in β-cells, activating AKT (pro-proliferative) and inactivating Foxo3a (pro-apoptotic) signaling pathways.
Meanwhile, Belgardt et al. reported that miRNA-200 is linked to β-cell death in vivo [46]. Overexpression of miR-200 in β-cells induced β-cell apoptosis and overt diabetes in wild-type mice, while genetic ablation of miR-200 ameliorated β-cell death and diabetic phenotype in Akita mice. Quantitative PCR analysis in islets of these mice and luciferase activity assays in MIN6 β-cells identified multiple anti-apoptotic genes (e.g. Jazf1, Dnajc3 and Xiap) as direct target of miR-200. Independent mouse studies demonstrated that miR-7 is associated with decreased β-cell function and immature β-cell identity [47,48]. Overexpressing miR-7 in β-cells resulted in β-cell dedifferentiation, while miR-7 KO in β-cells improved insulin secretion by enhancing insulin granule exocytosis [47,48]. Interestingly, plasma miR-7 levels were reduced in mouse models of successful β-cell compensation (ob/ob, C57BL/6; DIO mice) and were elevated in β-cell decompensation rodent models (db/db, BLKS; GK diabetic rats), suggesting that its plasma level is linked to the β-cell failure in T2D [47,49]. Other miRNAs, including miR-184 [50] and miR-802 [51], have also been reported to suppress insulin secretion in insulin-resistant mouse models.
LncRNA is broadly defined as a ncRNA that is larger than 200 nucleotides and takes most abundant portion among all ncRNAs [52]. Motterle et al. identified β-cell-enriched lncRNAs in mice: βlinc2 and βlinc3, which levels significantly alter in the islets of insulin-resistant conditions [53]. The expression level of βlinc2 increased while the βlinc3 level decreased in the islets of DIO and db/db mice. Although the action mechanisms remain elusive, these findings suggest that there may be specific functions of individual lncRNAs in β-cell response to insulin resistance.
LncRNA H19 has been reported to contribute to the β-cell mass expansion in rodents [54]. Overexpression of H19 in INS1 β-cells led to increased proliferation and reduced cytokine-induced apoptosis. In parallel, the silencing of H19 in primary rat β-cells resulted in a robust decrease in proliferation. Moreover, the expression levels of H19 were increased in the islets of DIO and ob/ob mice, indicating that it may play an essential role in β-cell adaptation. Meanwhile, Akerman et al. demonstrated the function of lncRNA PLUTO in human β-cells [55]. PLUTO is enriched in human β-cells and its level is downregulated in the islets of T2D patients. Interestingly, it regulates β-cell-specific transcriptional networks by modifying 3D structure of β-cell chromatin. In particular, PLUTO facilitates promoter-enhancer interaction, thus enhancing the gene expression of PDX1, a critical TF for β-cell compensation.
RNA modification
Modification on tRNAs and mRNAs affects stability and translational efficiency of the transcript. CDK5 regulatory subunit associated protein 1-like 1 (CDKAL1), one of the important risk genes for T2D in humans, encodes methylthiotransferase (MTTase) that mediates methylthiolation of N6-threonylcarbmoyl adenosine (t6A) to produce 2-methylthio-N6-threonylcarbmoyl adenosine (ms2t6A) at position 37 adenosine of lysine tRNA (anticodon: UUU) [56]. β-cell-specific Cdkal1 KO mice exhibited significantly reduced insulin biosynthesis and secretion due to the decreased lysine incorporation within the cleavage site of proinsulin [57].
Recent studies reported the importance of mRNA methylation in β-cell adaptation [58,59]. De Jesus et al. performed N6-methyladenosine (m6A) sequencing in the islets from controls and T2D donors, demonstrating that gene transcripts that are essential for β-cell compensation (e.g. IGF1R, IRS2 and PDX1) are hypomethylated in T2D patients [58]. Knocking down m6A writers methyltransferase like 3 (METTL3) and/or methyltransferase like 14 (METTL14) in EndoC-βH1 cells resulted in decreased AKT phosphorylation and PDX1 level along with cell cycle arrest [58]. Moreover, Mettl14 KO in both fetal and adult β-cells resulted in β-cell dysfunction and overt glucose intolerance in mice, indicating that m6A modification is essential for β-cell adaptation in vivo [58,59].
Conclusions
Rapid advances in research technologies made epigenetics an exciting field to study the pathophysiology of T2D. However, several things may be considered when carrying out studies.
First, overinterpretation of the results should be avoided in the studies. As well known, the characteristics between cell-lines and primary cells, the condition between in vitro and in vivo, the meaning between association and causality are entirely different. Given that, a conclusion should be carefully addressed in each study, not to mislead the direction of the research field.
Another consideration may be a practical translation of epigenetic mechanisms into the diagnostics and therapeutics. Such examples can be 1) using a circulating methylated-DNAs as biomarkers for T2D diagnosis [60]; 2) developing an epigenetic manipulation technique for cell-based therapy [61]. Considering these points will advance epigenetics to develop novel approaches to prevent and cure T2D.
Acknowledgements
The authors acknowledge funding support from NIH Grants R01 DK067536 and DK103215.
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Halban PA, Polonsky KS, Bowden DW, Hawkins MA, Ling C, Mather KJ, Powers AC, Rhodes CJ, Sussel L, Weir GC: β-Cell Failure in Type 2 Diabetes: Postulated Mechanisms and Prospects for Prevention and Treatment. Diabetes Care 2014, 37:1751 LP–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prentki M, Nolan CJ: Islet β-cell failure in type 2 diabetes. J Clin Invest 2006, 116:1802–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.*.Weir GC, Gaglia J, Bonner-Weir S: Inadequate β-cell mass is essential for the pathogenesis of type 2 diabetes. Lancet Diabetes Endocrinol 2020, 8:249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donath MY, Ehses JA, Maedler K, Schumann DM, Ellingsgaard H, Eppler E, Reinecke M: Mechanisms of β-Cell Death in Type 2 Diabetes. Diabetes 2005, 54:S108 LP–S113. [DOI] [PubMed] [Google Scholar]
- 5.*.Bensellam M, Jonas JC, Laybutt DR: Mechanisms of β;-cell dedifferentiation in diabetes: Recent findings and future research directions. J Endocrinol 2018, 236: R109–R143. [DOI] [PubMed] [Google Scholar]
- 6.*.Mezza T, Cinti F, Cefalo CMA, Pontecorvi A, Kulkarni RN, Giaccari A: β-cell fate in human insulin resistance and type 2 diabetes: A perspective on islet plasticity. Diabetes 2019, 68:1121–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, Gromada J: RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes. Cell Metab 2016, 24:608–615. [DOI] [PubMed] [Google Scholar]
- 8.Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, et al. : Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes. Cell Metab 2016, 24:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawlor N, George J, Bolisetty M, Kursawe R, Sun L, Sivakamasundari V, Kycia I, Robson P, Stitzel ML: Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res 2017, 27:208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.*.Wang YJ, Kaestner KH: Single-Cell RNA-Seq of the Pancreatic Islets—a Promise Not yet Fulfilled? Cell Metab 2019, 29:539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weir GC, Laybutt DR, Kaneto H, Bonner-Weir S, Sharma A: Beta-cell adaptation and decompensation during the progression of diabetes. Diabetes 2001, 50: S154–S159. [DOI] [PubMed] [Google Scholar]
- 12.Guo S, Powers AC, Stein R, Guo S, Dai C, Guo M, Taylor B, Harmon JS, Sander M, Robertson RP, et al. : Inactivation of specific β cell transcription factors in type 2 diabetes. J Clin Invest 2013, 123:3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pullen TJ, Rutter GA: When less is more: the forbidden fruits of gene repression in the adult β-cell. Diabetes, Obes Metab 2013, 15:503–512. [DOI] [PubMed] [Google Scholar]
- 14.Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, et al. : The genetic architecture of type 2 diabetes. Nature 2016, 536:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.*.Ling C, Rönn T: Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab 2019, 29:1028–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.*.De Jesus DF, Kulkarni RN: “Omics” and “epi-omics” underlying the β-cell adaptation to insulin resistance. Mol Metab 2019, 27:S42–S48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkov P, Bacos K, Ofori JK, Esguerra JLS, Eliasson L, Rönn T, Ling C: Whole-Genome bisulfite sequencing of human pancreatic islets reveals novel differentially methylated regions in type 2 diabetes pathogenesis. Diabetes 2017, 66:1074–1085. [DOI] [PubMed] [Google Scholar]
- 18.Dhawan S, Georgia S, Tschen Sing, Fan G, Bhushan A: Pancreatic β Cell Identity Is Maintained by DNA Methylation-Mediated Repression of Arx. Dev Cell 2011, 20:419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhawan S, Tschen S-I, Zeng C, Guo T, Hebrok M, Matveyenko A, Bhushan A: DNA methylation directs functional maturation of pancreatic β cells. J Clin Invest 2015, 125:2851–2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avrahami D, Li C, Zhang J, Schug J, Avrahami R, Rao S, Stadler MB, Burger L, Schübeler D, Glaser B, et al. : Aging-dependent demethylation of regulatory elements correlates with chromatin state and improved β cell function. Cell Metab 2015, 22:619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.*.Hall E, Jönsson J, Ofori JK, Volkov P, Perfilyev A, Nitert MD, Eliasson L, Ling C, Bacos K: Glucolipotoxicity alters insulin secretion via epigenetic changes in human islets. Diabetes 2019, 68:1965–1974. [DOI] [PubMed] [Google Scholar]
- 22.Bhandare R, Schug J, Le Lay J, Fox A, Smirnova O, Liu C, Naji A, Kaestner KH: Genome-wide analysis of histone modifications in human pancreatic islets. Genome Res 2010, 20:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morán I, Gómez-Marín C, van de Bunt M, et al. : Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 2014, 46:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.*.Nammo T, Udagawa H, Funahashi N, Kawaguchi M, Uebanso T, Hiramoto M, Nishimura W, Yasuda K: Genome-wide profiling of histone H3K27 acetylation featured fatty acid signalling in pancreatic beta cells in diet-induced obesity in mice. Diabetologia 2018, 61:2608–2620. [DOI] [PubMed] [Google Scholar]
- 25.Lundh M, Galbo T, Poulsen SS, Mandrup-Poulsen T: Histone deacetylase 3 inhibition improves glycaemia and insulin. Diabetes, Obes Metab 2015, 17:703–707. [DOI] [PubMed] [Google Scholar]
- 26.Remsberg JR, Ediger BN, Ho WY, Damle M, Li Z, Teng C, Lanzillotta C, Stoffers DA, Lazar MA: Deletion of histone deacetylase 3 in adult beta cells improves glucose tolerance via increased insulin secretion. Mol Metab 2017, 6:30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.*.Daneshpajooh M, Eliasson L, Bacos K, Ling C: MC1568 improves insulin secretion in islets from type 2 diabetes patients and rescues β-cell dysfunction caused by Hdac7 upregulation. Acta Diabetol 2018, 55:1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spaeth JM, Walker EM, Stein R: Impact of Pdx1-associated chromatin modifiers on islet β-cells. Diabetes, Obes Metab 2016, 18:123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosley AL, Özcan S: The pancreatic duodenal homeobox-1 protein (Pdx-1) interacts with histone deacetylases Hdac-1 and Hdac-2 on low levels of glucose. J Biol Chem 2004, 279:54241–54247. [DOI] [PubMed] [Google Scholar]
- 30.Qiu Y, Guo M, Huang S, Stein R: Insulin Gene Transcription Is Mediated by Interactions between the p300 Coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002, 22:412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deering TG, Ogihara T, Trace AP, Maier B, Mirmira RG: Methyltransferase set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 2009, 58:185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maganti A V, Maier B, Tersey SA, Sampley ML, Mosley AL, Özcan S, Pachaiyappan B, Woster PM, Hunter CS, Stein R, et al. : Transcriptional activity of the islet β cell factor Pdx1 Is augmented by lysine methylation catalyzed by the methyltransferase Set7/9. J Biol Chem 2015, 290:9812–9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.*.Kim H, Yoon BH, Oh CM, Lee J, Lee K, Song H, Kim E, Yi K, Kim MY, Kim H, et al. : PRMT1 Is Required for the Maintenance of Mature β-Cell Identity. Diabetes 2020, 69:355–368. [DOI] [PubMed] [Google Scholar]
- 34.McKenna B, Guo M, Reynolds A, Hara M, Stein R: Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in Islet β cells. Cell Rep 2015, 10:2032–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.*.Spaeth JM, Liu JH, Peters D, Guo M, Osipovich AB, Mohammadi F, Roy N, Bhushan A, Magnuson MA, Hebrok M, et al. : The Pdx1-bound Swi/Snf chromatin remodeling complex regulates pancreatic progenitor cell proliferation and mature islet β-cell function. Diabetes 2019, 68:1806–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.*.Wei Z, Yoshihara E, He N, Hah N, Fan W, Pinto AFM, Huddy T, Wang Y, Ross B, Estepa G, et al. : Vitamin D Switches BAF Complexes to Protect β Cells. Cell 2018, 173:1135–1149.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.*.Lu TTH, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang CH, Sagar, Arrigoni L, Dalgaard K, et al. : The Polycomb-Dependent Epigenome Controls β Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab 2018, 27:1294–1308.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.*.Khetan S, Kursawe R, Youn A, Lawlor N, Jillette A, Marquez EJ, Ucar D, Stitzel ML: Type 2 diabetes-associated genetic variants regulate chromatin accessibility in human islets. Diabetes 2018, 67:2466–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.*.Greenwald WW, Chiou J, Yan J, Qiu Y, Dai N, Wang A, Nariai N, Aylward A, Han JY, Kadakia N, et al. : Pancreatic islet chromatin accessibility and conformation reveals distal enhancer networks of type 2 diabetes risk. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.*.Miguel-Escalada I, Bonàs-Guarch S, Cebola I, Ponsa-Cobas J, Mendieta-Esteban J, Atla G, Javierre BM, Rolando DMY, Farabella I, Morgan CC, et al. : Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genet 2019, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaPierre MP, Stoffel M: MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol Metab 2017, 6:1010–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filios SR, Shalev A: β-cell microRNAs: Small but powerful. Diabetes 2015, 64:3631–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P, Zavolan M, Stoffel M: miR-375 maintains normal pancreatic α- and β-cell mass. Proc Natl Acad Sci U S A 2009, 106:5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouaamari A El, Baroukh N, Martens GA, Lebrun P, Pipeleers D, Van Obberghen E: MiR-375 targets 3′l-Phosphoinositide-Dependent protein Kinase-1 and regulates Glucose-Induced biological responses in pancreatic β-Cells. Diabetes 2008, 57:2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.*.Mziaut H, Henniger G, Ganss K, Hempel S, Wolk S, McChord J, Chowdhury K, Ravassard P, Knoch KP, Krautz C, et al. : MiR-132 controls pancreatic beta cell proliferation and survival through Pten/Akt/Foxo3 signaling. Mol Metab 2020, 31:150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belgardt BF, Ahmed K, Spranger M, Latreille M, Denzler R, Kondratiuk N, Von Meyenn F, Villena FN, Herrmanns K, Bosco D, et al. : The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med 2015, 21:619–627. [DOI] [PubMed] [Google Scholar]
- 47.Latreille M, Hausser J, Stützer I, Zhang Q, Hastoy B, Gargani S, Kerr-Conte J, Pattou F, Zavolan M, Esguerra JLS, et al. : MicroRNA-7a regulates pancreatic β cell function. J Clin Invest 2014, 124:2722–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kredo-Russo S, Mandelbaum AD, Ness A, Alon I, Lennox KA, Behlke MA, Hornstein E: Pancreas-enriched miRNA refines endocrine cell differentiation. Dev 2012, 139:3021–3031. [DOI] [PubMed] [Google Scholar]
- 49.Esguerra JLS, Bolmeson C, Cilio CM, Eliasson L: Differential glucose-regulation of microRNAs in pancreatic islets of non-obese type 2 diabetes model Goto-Kakizaki rat. PLoS One 2011, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tattikota SG, Rathjen T, Hausser J, Khedkar A, Kabra UD, Pandey V, Sury M, Wessels HH, Mollet IG, Eliasson L, et al. : miR-184 regulates pancreatic β-cell function according to glucose metabolism. J Biol Chem 2015, 290:20284–20294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.*.Zhang F, Ma D, Zhao W, Wang D, Liu T, Liu Y, Yang Y, Liu Y, Mu J, Li B, et al. : Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat Commun 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.*.Singer RA, Sussel L: Islet long noncoding RNAs: A playbook for discovery and characterization. Diabetes 2018, 67:1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Motterle A, Gattesco S, Peyot ML, Esguerra JLS, Gomez-Ruiz A, Laybutt DR, Gilon P, Burdet F, Ibberson M, Eliasson L, et al. : Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol Metab 2017, 6:1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.*.Sanchez-Parra C, Jacovetti C, Dumortier O, Lee K, Peyot ML, Guay C, Prentki M, Laybutt DR, Van Obberghen E, Regazzi R: Contribution of the long noncoding RNA H19 to β-cell mass expansion in neonatal and adult rodents. Diabetes 2018, 67:2254–2267. [DOI] [PubMed] [Google Scholar]
- 55.Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L, et al. : Human Pancreatic β Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab 2017, 25:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.*.Wei FY, Tomizawa K: tRNA modifications and islet function. Diabetes, Obes Metab 2018, 20:20–27. [DOI] [PubMed] [Google Scholar]
- 57.Wei FY, Suzuki T, Watanabe S, Kimura S, Kaitsuka T, Fujimura A, Matsui H, Atta M, Michiue H, Fontecave M, et al. : Deficit of tRNALys modification by Cdkal1 causes the development of type 2 diabetes in mice. J Clin Invest 2011, 121:3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.*.De Jesus DF, Zhang Z, Kahraman S, Brown NK, Chen M, Hu J, Gupta MK, He C, Kulkarni RN: m6A mRNA methylation regulates human β-cell biology in physiological states and in type 2 diabetes. Nat Metab 2019, 1:765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.*.Men L, Sun J, Luo G, Ren D: Acute Deletion of METTL14 in β-Cells of Adult Mice Results in Glucose Intolerance. Endocrinology 2019, 160:2388–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.*.Willmer T, Johnson R, Louw J, Pheiffer C: Blood-Based DNA Methylation Biomarkers for Type 2 Diabetes: Potential for Clinical Applications. Front Endocrinol (Lausanne) 2018, 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.*.Ou K, Yu M, Moss NG, Wang YJ, Wang AW, Nguyen SC, Jiang C, Feleke E, Kameswaran V, Joyce EF, et al. : Targeted demethylation at the CDKN1C/p57 locus induces human β cell replication. J Clin Invest 2019, 129:209–214. [DOI] [PMC free article] [PubMed] [Google Scholar]