Abstract
With the increasing prevalence of antibiotic resistance, the need to develop antimicrobial susceptibility testing (AST) technologies is urgent. The current challenge has been to perform the antibiotic susceptibility testing in short time, directly with clinical samples, and with antibiotics over a broad dynamic range of clinically relevant concentrations. Here we develop a technology for point-of-care diagnosis of antimicrobial-resistant bacteria in urinary tract infections (UTI), by imaging the clinical urine samples directly with an innovative large-image-volume solution scattering imaging (LVSi) system and analyzing the image sequences with a single-cell division tracking method. The high sensitivity of single-cell division tracking associated with large volume imaging enabled rapid antibiotic susceptibility testing directly on the clinical urine samples. The results demonstrated direct detection of bacterial infections in 60 clinical urine samples with a 60 min LVSi video, and digital AST of 30 positive clinical samples with 100% categorical agreement with both the clinical culture results and the on-site agar plating validation results. This technology provides opportunities for precise antibiotic prescription and prompt proper treatment of the patient within a single clinic visit.
Keywords: antimicrobial susceptibility testing, AST, optical imaging, intrinsic features, single-cell division tracking, antibiotic resistance, E. coli, urinary tract infections, UTI
Antimicrobial resistance is a rapidly growing threat to global public health, affecting millions of people annually.1 An important cause for this global concern is the misuse and overuse of antimicrobials.2, 3 To combat this threat, a technology that can quickly identify pathogen infection and perform antimicrobial susceptibility testing (AST) is needed. The gold standard methods used in clinical labs rely on overnight cell culture for microorganism growth and isolation and additional sub-culture steps for AST.4, 5 Various emerging AST technologies generally fit into two categories: genotypic and phenotypic approaches.6, 7 The former detects antibiotic resistance genes.7–11 While sensitive,12 it requires prior knowledge of the bacteria, which can lead to a false negative result when a new resistant mechanism emerges, and false positive results when resistance genes are present but are not expressed or are not contributing to resistant phenotypes13, 14. The latter measures a phenotypic feature more directly, such as size or number of bacterial cells, an approach used in today’s gold standard AST methods. Despite recent advances,15–22 most of the AST technologies still require culture, isolation, and enrichment of the bacteria. A fast AST must remove these time-consuming steps and must be functional directly with clinical samples with minimal pre-processing.
One example of a direct AST method is nucleic acid amplification testing (NAT), such as loop-mediated isothermal amplification (LAMP).23 This technique has been used to detect single bacterial cells at different times after antibiotic exposure yielding a quantification of the target sequence. However, since LAMP is a NAT-based detection method, it requires a series of sample preparation steps and use of primers and enzymes. Single bacterial cells can be also measured by trapping them in microfluidic channels, but it typically requires culture or isolation of the bacterial cells.10, 15, 17, 24–27 A recent advance in microfluidics is the introduction of an adaptable mechanism to enable fast AST with clinical urine samples.25 This method flows a clinical sample along microfluidic channels. Once a bacterial cell is detected with a high-resolution optical microscope, pressure within the channel traps the cell, which allows cell length measurements upon antibiotic exposure over time. Trapping of bacterial cells requires prior knowledge of bacteria size and need longer loading time for low bacteria concentration samples. This mothed also cannot differentiate normal bacteria growth from those antibiotics that promotes cell elongation.
Here, we introduce a fast AST with single cell tracking capability that works directly on urine samples in a cuvette. The method images, tracks, and counts individual division events of single bacterial cells. Division is the most universal phenotypic feature for AST to determine if a cell is viable. Compared to the LAMP-based AST, the present method provides real time tracking of individual cell growth and division without the need for DNA primers, reagents, or long incubation periods. Direct image in a cuvette also provide several advantages over microfluidic-based approach: simpler setup by eliminating microfluidics and associated pumps and valves, more reliable measurement by avoiding clogging of microfluidic channels by bubbles or impurity particles in urine samples. and improved throughput enabled by simultaneous tracking of many cells.
Traditional optical microscopy can image bacterial cells but requires immobilization of the cells on a surface. These limitations, combined with the small field of view of high-resolution optical microscopy, necessitate bacterial enrichment in low concentration samples. Our AST method features digitally counting single cell division events recorded by large volume scattering imaging (LVSi). LVSi illuminates and images a large sample volume with low optical zoom, which eliminates the need of sample enrichment, and enables tracking bacterial cells in free solution. We have shown that this strategy can direct image and count urine samples with clinically relevant bacterial concentration, e.g., 104 cells/mL (equivalent to 104 CFU in agar plating method) for fast AST.28 To precisely track and count single division events of bacterial cells in the presence of various particles (e.g., crystals and cellular debris) in patient urine samples, we further introduced a forward scattering optical imaging configuration and an imaging processing algorithm. We describe the optical setup and principle of the digital LVSi-AST, validate it with cultured stationary phase Escherichia coli, and apply it to 60 clinical urine samples from patients with suspected urinary tract infections (UTIs). We focus on UTI because it affects millions of people annually, the pathogens causing UTIs pose the highest threat of antimicrobial resistance, and the sample type is already a liquid.29, 30
PRINCIPLE
Detection Principle
Today’s gold standard AST technologies measure changes in optical density associated with bacterial growth in a cultured isolate, which typically requires overnight incubation to reliably detect growth. Optical density is an average feature that depends on the number and size of the cells, as well as impurity particles in the sample. Intrinsic phenotypic features of single cells, including the cellular size, motion, and morphology have been used for AST, but these features are not always reliable in rapid detection technologies. For example, ampicillin promotes cell elongation prior to cell death, so bacterial cell length measurements are not reliable in early stages of antibiotic exposure. The most reliable and universal feature is the division of bacterial cells, which is a primarily intrinsic feature used in the present work.
Tracking the division of single bacterial cells in a patient urine samples is challenging with traditional optical microscopy. Clinically relevant bacterial concentrations that signify a UTI are ~105 cells/mL. However, for a typical 20x optical microscope with an image volume of 2.5 nL, less than one bacterial cell will be present in such a small volume. We developed LVSi to image bacterial cells directly from urine samples with an image volume of 5 μL, allowing imaging of sufficient numbers of bacterial cells simultaneously for reliable AST (Fig. 1A and Supporting Information Fig. S1). LVSi uses a forward scattering geometry to minimize image intensity blinking associated with the rotation of cells in the solution. Furthermore, a beam block prevents the incident light from directly entering the camera, enabling high contrast and low background images of the bacterial cells.
Figure 1. Principle and setup of digital LVSi-AST.
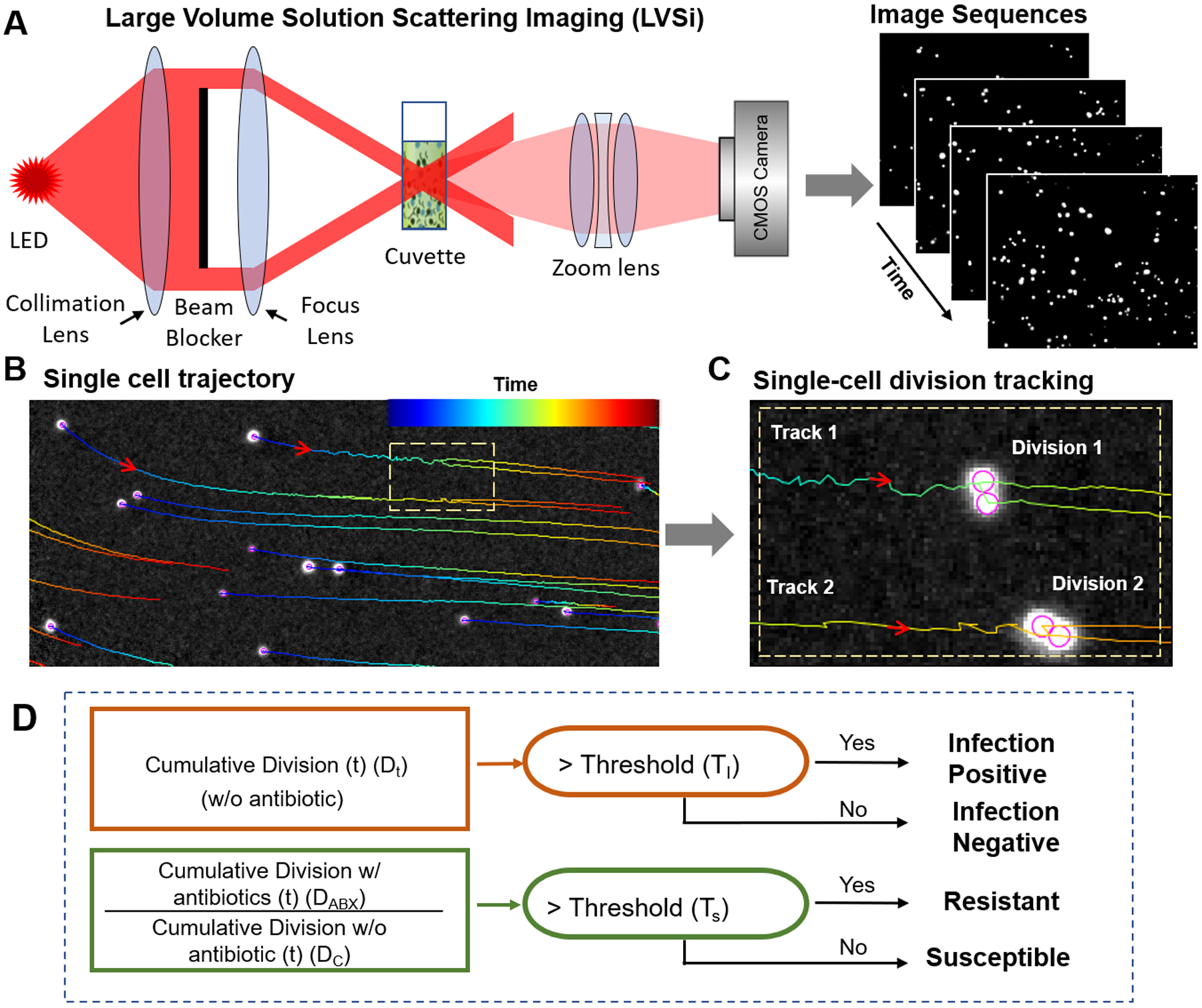
(A) Schematic illustration of one channel of the dual channel LVSi setup that allows imaging single bacterial cells in urine, supplemented with bacterial culture media, loaded in a cuvette. (B) From the video captured with LVSi, the trajectory of each bacterial cell or impurity particle is determined to represent the time course of the cell or particle in the sample, where the color of the lines are the trajectories and colors, from blue to red, represent the time sequence (minimally 15 sec). (C) Representative example of two division events for two cells tracked with the imaging processing algorithm described in the text. (D) Algorithm for infection detection and AST based on the division counts in samples with and without antibiotics, where Dc and DABX are the cumulative division counts at time t for antibiotic-untreated (control) and -treated samples.
LVSi images bacterial cells, as well as impurities, in a urine sample as individual bright spots that move in and out of the view dynamically due to Brownian motion and thermal reflux. The thermal reflex (due to heating of the sample from bottom to keep it at 37°C) also helps circulating the cells and prevents sedimentation. We developed an automated imaging processing algorithm that identifies each particle (bacterial cell or impurity), determines the trajectory of the particle (Fig. 1B), and then detects and counts each division event from all trajectories (Fig. 1C). First, common background noise and image drift are corrected with temporal local minimal subtraction, which improves the image contrast. Second, a Laplace of Gaussian (LOG) filter is used to detect individual cell particles (bright spots in the image). Third, directional linking of spots in adjacent frames is performed using the Kalman filter to obtain single cell tracking trajectories (Fig. 1B). Finally, a division detection filter is applied to detect the division events of bacterial cells. The division detection filter looks for both splitting of a trajectory and intensity decreases in the two resulting trajectories, as expected for bacterial cell division (Fig. 1C).
The division counting allows for both bacterial detection and for AST (if bacteria are detected above the threshold) with the following algorithm (Fig. 1D). A patient’s urine sample is measured in two conditions simultaneously: a control sample without antibiotics and an AST sample with an antibiotic added. To detect infection, bacterial division in the control sample is counted over time (e.g., accumulation every 5 min for a total of 60 min). If the cumulative division count (DC) is above an infection threshold (TI) (see Materials and Methods), or DC > TI, the sample is identified as infection positive (Fig. 1D). Otherwise, the sample is determined as infection negative (Fig. 1D). To test antibiotic susceptibility, the ratio of the division count (DC) in the raw urine sample to that of an antibiotic treated sample (DABX) is determined. If the ratio is above a susceptible threshold (TS), which indicates no bacterial inhibition by the antibiotic, the sample is identified as resistant (Fig. 1D). If the ratio is at or below TS, bacteria are inhibited by the antibiotic, and the sample is susceptible to that antibiotic.
RESULTS
Testing of digital LVSi-AST with pure E. coli cultures
To establish the method, E. coli cultures (see Materials and Methods) with and without antibiotics were imaged via LVSi. To minimize additional sub-culture steps and speed up AST, we diluted E. coli stationary phase cultures in fresh culture medium to a concentration of ~105 cells/mL before imaging. The individual bacterial cells were imaged as bright spots moving dynamically in the video and tracked continuously for 60 min (Fig. 2). The time sequence images were then processed with the method described above to detect and count the division events.
Figure 2. Validation of digital LVSi-AST with pure E. coli cultures.
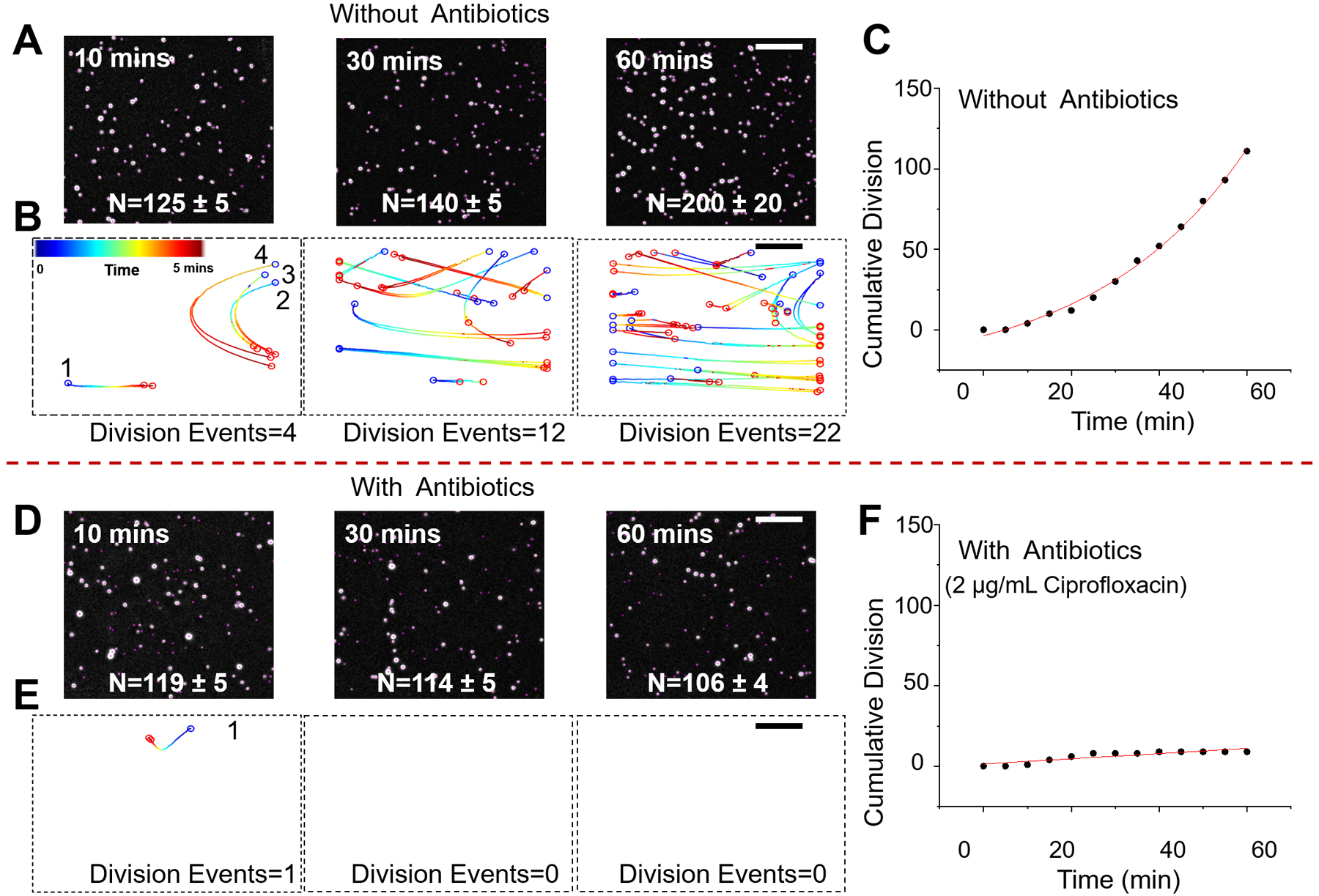
Snapshots of E. coli cells after 10, 30, and 60 min in growth medium without antibiotics (A) and with antibiotics (2 μg/mL ciprofloxacin) (D). Division events over a 5-min time interval at the different time points with (B) and without ciprofloxacin (E). Cumulative division events over 60 min with (C) and without ciprofloxacin (F). Scale bar, 400 μm.
In the absence of antibiotics, E. coli multiplies over 60 min as indicated by the increase in the cell count (Fig. 2A). E. coli growth is tracked precisely by counting the number of division events of single bacterial cells every 5 min (Fig. 2B). Each division event starts with a parent cell, marked as a blue circle at the starting position, that moves along the trajectory and splits into two daughter cells (marked as red circles in the end positions) (Fig. 2B). The cumulative division count shows a rapid increase over time (Fig. 2C). In contrast, for the antibiotic-treated sample (2 μg/mL ciprofloxacin), very few division events were detected over 60 min (Figs. 2D, 2E and F).
To validate the robustness of the method, we performed five biological replicates with at least two technical replicates. To compare the results from different experiments, the cumulative division events are normalized to the initial spots number (N0) to generate the fold increase of cell growth. Five representative cumulative division tracking results of E. coli samples with and without antibiotics are plotted in Fig. 3A (ciprofloxacin) and Fig. 3C (ampicillin). Without antibiotics, all the samples show significant increase in the divisions detected, indicating the existence of viable bacterial cells in each of the sample. Although the fold increase in the division events varies from sample to sample, in the presence of either ciprofloxacin or ampicillin, the fold increase over 60 min is significantly lower than all control samples (Fig. 3). Statistical analysis of all the samples (n = 11) at different time points (0, 30, 60 min) indicates one can reliably differentiate the inhibition of the bacterial cells as fast as 30 min, and the statistical significance increases with time (Figs. 3B and 3D). Traditional plating and colony forming unit (CFU) quantitation were performed simultaneously on the same samples to verify the bacterial growth at each time point, which validated the present digital LVSi-AST (Supporting information S2).
Figure 3. E. coli division tracking for digital AST with different antibiotics.
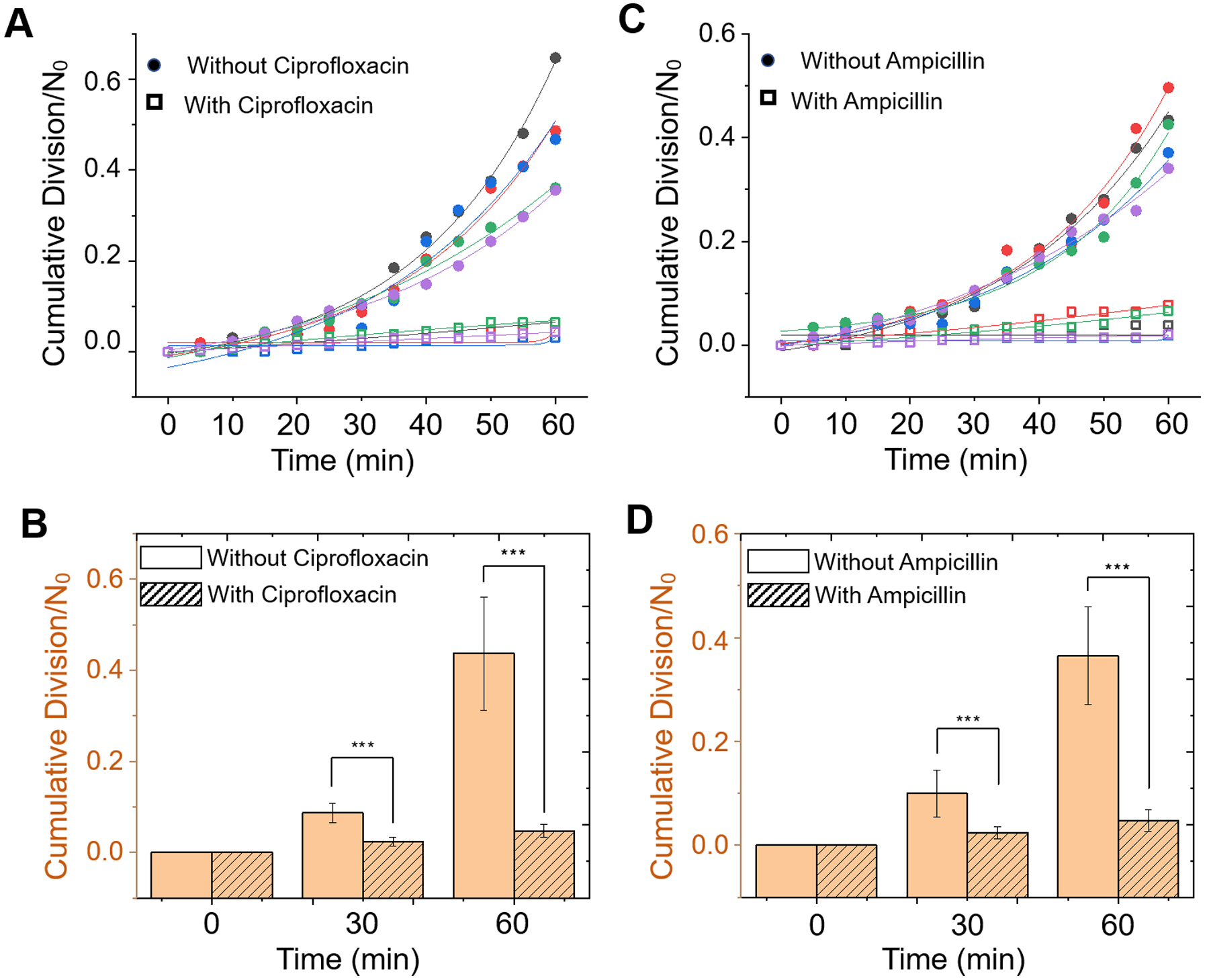
Five representative cumulative division-over-time plots for E. coli with and without antibiotics 2 μg/ml ciprofloxacin (A) and 16 μg/ml ampicillin (B). Each color represents an individual test. Dots are measured data, and the solid lines are the exponential fitting to the data. Analysis of bacterial growth in the absence of antibiotics or presence of 2 μg/ml ciprofloxacin (C) and 16 μg/ml ampicillin (D) over 60 min. *** p<0.001.
Digital LVSi-AST with clinical urine samples
After validation of digital LVSi-AST with pure E. coli cultures, we applied it to clinical urine samples for both detection of UTI and AST of the UTI-causing bacteria. Infection detection tracks cellular division events in the control sample, signaling the existence of viable bacterial cells, while AST tracks and compares division events in samples incubated with and without antibiotics. Sixty de-identified clinical urine samples were collected from patients and subjected to LVSi-AST. In parallel, the urine samples were subjected to agar plating and CFU quantitation.
UTI detection:
We tested 60 clinical urine samples and identified samples with UTI (Fig. 4). Samples showing substantial numbers of division events are UTI positive, while samples showing few division events are UTI negative (Fig. 4A). Half of the 60 clinical samples show minimal division activity (Fig. 4B), and the other 30 samples exhibit significant division activity despite sample-to-sample variability (Fig. 4C, Supporting Information S3). Due to the high density of particles in clinical samples, the final division results are calibrated to remove the over-counting error due to cell density (Materials and Methods). Statistical analyses were performed by determining and plotting the cumulative division counts of each sample at different time points (Figs. 4D–F). LVSi-AST results were compared with the BD Phoenix gold standard method results obtained in the Microbiology Lab at Mayo Clinic, where the samples were collected. At 30 min, the cumulative division counts fall into two separated clusters: one cluster of samples with less than 1 division event and one cluster of samples with 2 – 200 division events (Fig. 4D). The two clusters were further separated with increased incubation times, indicating increasing UTI detection accuracy with time (Figs. 4E and 4F). By setting the infection threshold at 5 cumulative division events (Materials and Methods), 30 samples were determined infection negative and 30 samples were infection positive with agreement accuracy (consistent samples/total samples, compared with the clinical results) that increased from ~90% at 30 min to ~93% at 45 min to ~97% at 60 min (Fig. 4G). Two false negative samples were determined from the 60 samples tested, revealing 97% accuracy at 60 min (Fig. 4G). To assess these false negative sample, we subjected the urine sample to a parallel microbiological agar plating and failed to detect viable cells, indicating that the false negative samples may be due to sample handling and/or sample processing. Furthermore, the bacterial density of the samples can be calculated by the parallel plating validation. For one sample with bacterial density at 2.0×104 (~100 cells in view), the cumulative division events within 60 min is ~90, while for another sample with 1.12×105 (~500 cells in view) bacterial density, the cumulative division events in 60 min is tracked to be ~270.
Figure 4. Detection of UTI in 60 clinical urine samples.
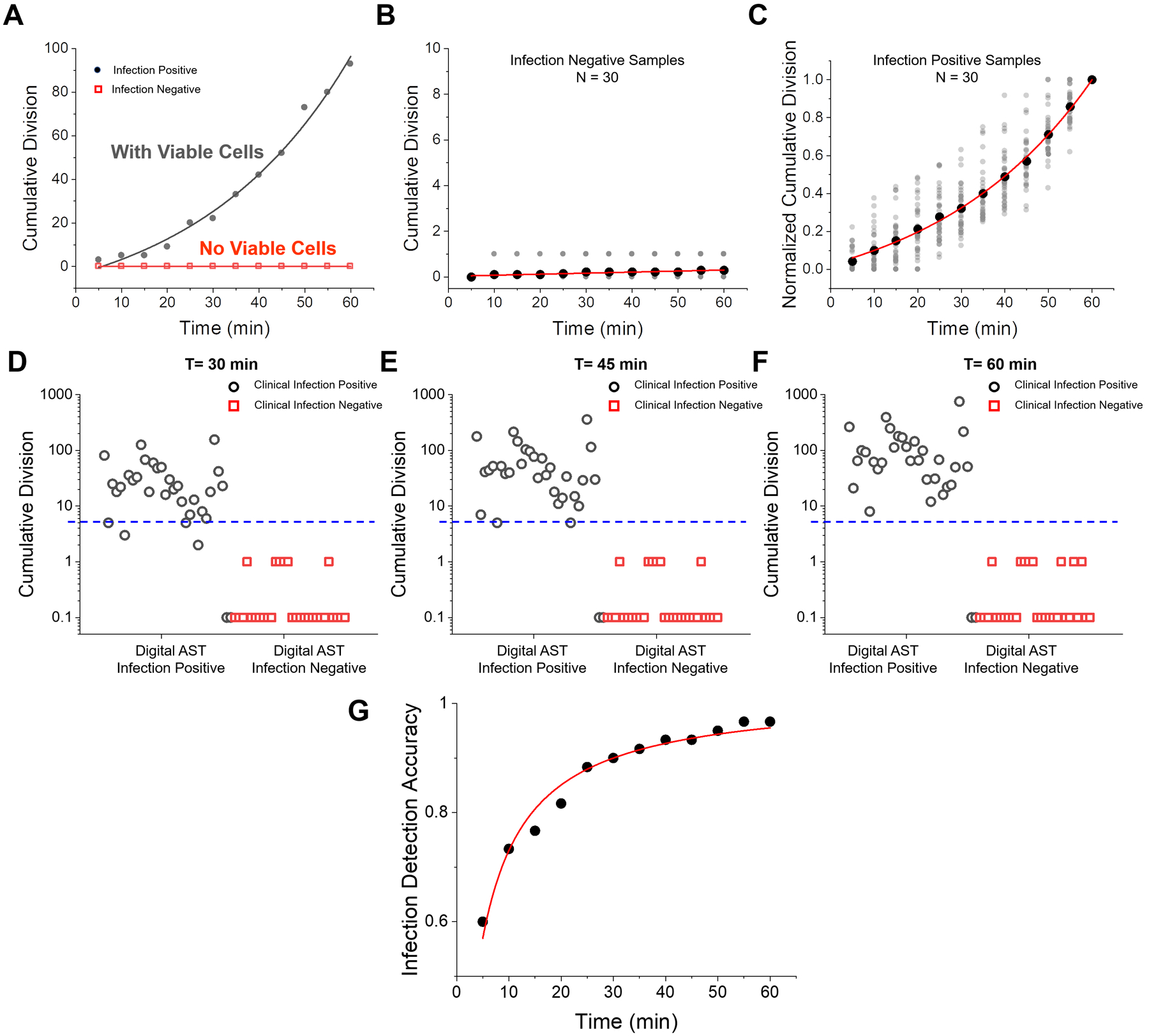
(A) Representative division vs. time plots for an infection negative (red, sample ATU090319_9) and an infection positive (black, sample ATU090319_10) samples. (B) Division counts over 60 min for 30 infection-negative clinical samples. (C) Division counts (normalized by the division events at 60 min) over 60 min of 30 infection-positive clinical samples. (D-F) Cumulative divisions for all the 60 clinical samples at 30, 45 and 60 min, respectively, where open circles correspond to infection positive and open squares correspond to infection negative samples measured by BD Phoenix, and 0 division counts are marked at 0.1 on the logarithmic scale y axis. (G) Infection detection accuracy vs. detection time.
Rapid AST:
We performed AST on the 30 UTI positive clinical samples by comparing cumulative division in antibiotic-treated samples (DABX) with the control samples (DC) following the algorithm defined in Fig. 1D. When DABX is consistently lower than DC, the sample is identified as an antibiotic susceptible sample (Fig. 5A, Supporting Information Fig. S4A). In contrast, when DABX and DC values are similar, the sample is identified as a resistant sample (Fig. 5B, Supporting Information Fig. S4B). To explore LVSi-AST accuracy over time, we set the susceptibility threshold (TS) at 0.5, corresponding to inhibition of 50% of the bacterial cells, and plotted comparisons of the reference method (BD Phoenix) and digital AST at the time points of 30, 45 and 60 min (Fig. 5C–E). With the 30-min detection, four susceptible samples localized within the resistance zone or at the TS, demonstrating a category accuracy of ~87% (Fig. 5C), while the 45 min detection increased the category accuracy to 94% (Fig. 5D). At 60 min, all susceptibility profiles were correctly determined with 8 samples identified as resistant to ciprofloxacin and the remaining 22 were susceptible (Fig. 5E). These results were in 100% agreement with BD Phoenix results from both clinical microbiology testing and the parallel agar plating validation experiments (Supporting Information S5). Fig. 5F plots the AST accuracy over the 60 min, with a hyperbolic increase observed.
Figure 5. Digital AST with infection positive clinical samples.
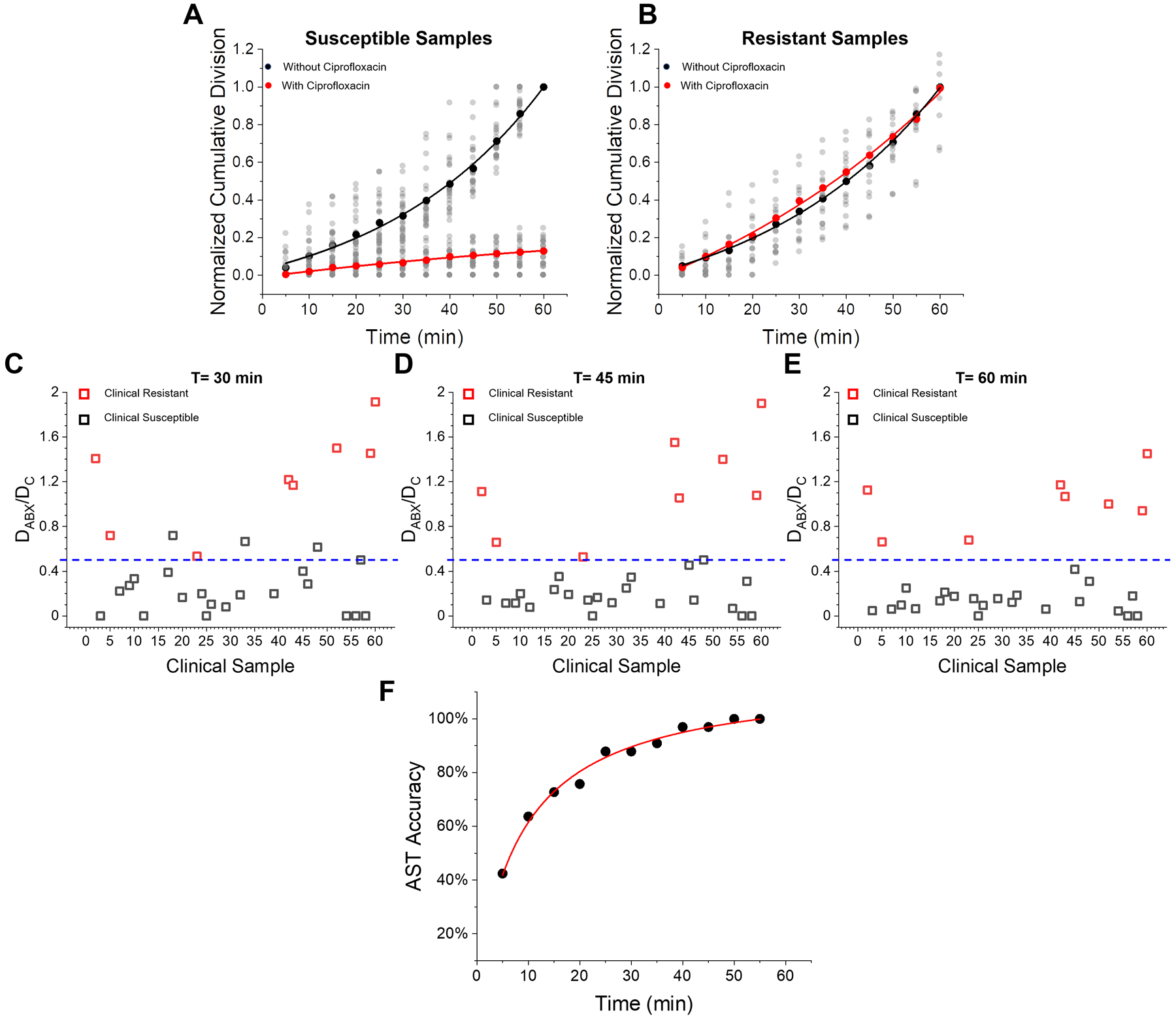
(A) Normalized cumulative division counting results for all 22 susceptible samples determined by LVSi_AST. (B) Normalized cumulative division counting results for all 8 ciprofloxacin-resistant samples determined by LVSi_AST. Comparison of reference method (clinical results measured by BD Phoenix) and digital AST for susceptibility determination with (C) 30 min detection, (D) 45 min detection, and (E) 60 min detection. (F) Digital AST accuracy over detection time.
Discussion
Ciprofloxacin kills bacteria by inhibiting DNA replication, while ampicillin disrupts cell wall synthesis.31, 32 In contrasts to tracking other phenotypic features, such as cellular morphology, length and motion that depends on antibiotic working mechanisms, division tracking is more general and provides reliable AST detection regardless of the antibiotic mechanism of action. Furthermore, with the single-cell division tracking capability, we observed some cells still dividing in the presence of antibiotics, showing the cell to cell heterogeneity in resistance within a sample. Thus, our division counting method provides a mechanism for early detection of drug resistance or tracking the progress of resistance development.
We present tracking division events of single cells as a rapid and accurate method to determine the presence of bacteria in urine samples at clinically relevant concentrations. While simple cell counting work well with pure or culture isolated bacterial samples with LSVi, they are problematic for clinical urine samples where a large number of nonbacterial and/or non-viable particles are present. These particles are hard to distinguish from bacterial cells at low zoom and overwhelm the small number of bacterial cells and mask out increases in bacterial numbers. In fact, we found that many urine samples contain so many particles that some precipitate and fall out of the imaging volume during the test, leading to false detection of bacterial cell growth by counting bacterial numbers (Supporting Information S6, S7). Tracking of the division events of single bacterial cells is more sensitive and more robust in the presence of a large number of impurity particles, thereby improving accuracy and shortening detection time.
The present LVSi and imaging processing algorithm has an image volume of 5 μL. We estimate a minimum of 50 bacterial cells are needed for reliable tracking results, which corresponds to a minimum bacterial cell concentration of 1×104 cells/mL, which is 10-fold less than the threshold for most clinically relevant samples. However, if the particle concentration (impurities + bacterial cells) is too high, overlapping particles in the LVSi compromises the tracking accuracy. The current setup can track up to 1,000 particles per image frame, corresponding to ~2×105 particles/mL. If patient urine samples have greater than ~2×105 particles or cells/mL, additional sample dilutions will be necessary. A quick count of the particles in image view is needed, which takes about 2–5 minutes for a quick imaging, counting and dilution. This limitation may be improved with better optical imaging resolution and more robust division tracking algorithm. Additionally, because we currently can only obtain refrigerated urine samples, a 30 min prewarm was performed to mimic the condition of fresh and warm urine sample, as the technology is targeting for rapid detection in point of care settings. Therefore, the current total assay time for direct AST in clinical sample includes 30 min sample pre-warming, 2 to 5 min sample pre-treatment (filtration/dilution), 60 min LVSi imaging, and 10 min division tracking processing. The total assay time can be shorten to ~ 60 min with improved sample collection and software automation for measuring fresh urine sample in point-of-care settings. Despite the limitation, digital LVSi-AST successfully tracked real clinical urine samples.
Due to the throughput limit of current prototype setup, we focused on E. coli infection detection (the most common cause of UTI) and AST with ciprofloxacin antibiotic for proof of concept demonstration of cell division counting based rapid bacteria ID and AST. Our future development plan includes improvement of detection throughput with multiplexed sample detections, and study of additional strains of bacteria and different types of antibiotics for complete coverage of UTI/AST diagnosis. As an intrinsic feature for cell growth estimation, single cell division tracking is a universal method for all categories of antibiotics. However, when work with bacterial strains that does not split, such as Staphylococcal bacteria in UTI, the division tracking will not work. This limitation can be solved by analysis additional phenotypic features in LSVi images, such as single particle intensity. The coccus cell growth induces cell aggregation and increases the spots intensity in LSVi image and can be easily tracked. Furthermore, the present work focus on direct AST without specific bacterial strains identification, which is different from the current practice. To be consistent with the clinical protocol, we are also working on the bacteria identification with LVSi, and have successfully identified the cells with different shapes (rod vs. cocci), which is out of the scope of this paper. Therefore, by improving the system throughput and integrating multiple phenotypic features (e.g. counts, intensity and division), we anticipate this technique can be used for bacterial cell identification and multiplex AST directly on the raw urine samples.
Conclusions
We described a large volume solution scattering imaging (LVSi) technique to detect the presence of bacteria and determine antimicrobial susceptibility directly in clinical urine samples. By tracking single-cell division events, we quantified the growth of the bacterial cells with high sensitivity in a 60 min LVSi video. For pure E. coli cultures, we have achieved direct AST with stationary phase bacteria in 60 min. Our results revealed the variability in the growth rate of cells from different populations, indicating the existence of persistent cells in the presence of antibiotics. The method offers single cell detection capability, which enable studies of cell heterogeneity response to antibiotics and the antibiotic resistance evolution. We then applied the technique to 60 clinical urine samples and accurately predicted 94% of the infection-positive samples. We also performed AST on 30 infection positive patient samples with ciprofloxacin and achieved 100% categorical agreements with culture-based commercial reference method. The technique can test raw clinical samples without overnight culturing and can track the division events of individual viable bacterial cells in real time. Collectively, LVSi-AST simplifies testing procedures, improves precision, and shortens the turnaround time from sample receipt to result determination. As the division tracking quantifies the bacterial cell growth, a universal phenotypic feature for AST, we anticipate that the technique can be expanded to applications beyond UTIs.
Materials and Methods
Materials.
E. coli ATCC 25922 was purchased from American Type Culture Collection (ATCC) and stored at −80°C in 5% glycerol. Ciprofloxacin and ampicillin were purchased from Sigma-Aldrich. The antibiotic powders were stored in the dark at 2 to 8°C.
Bacterial preparation.
E. coli was grown overnight (~15 h) in Luria−Bertani (LB) broth (per liter: 10 g peptone 140, 5 g yeast extract, and 5 g sodium chloride) at 37°C and 150 rpm. E. coli cultures were diluted in fresh LB broth to ~105 cells/mL. With LVSi system, cells/mL or particles/mL is used for cell density quantification, which is equivalent to CFU unit for pure cultured samples. An antibiotic at the standard breakpoint concentration was added to one of two preparations. Each bacterial suspension (70 μL), one with and one without antibiotic, was transferred into a cuvette at 37 °C for imaging.
Clinical urine samples.
De-identified clinical urine samples were obtained from the clinical microbiology laboratory at Mayo Clinic Hospital, Phoenix, Arizona. Clinical urine samples were stored at 4°C and transported in an insulated box with ice packs. Prior to processing, urine samples were pre-warmed for 30 min at 37°C and passed through a 5 μm filter to remove large substances. To provide nutritional supplementation, each urine sample was then diluted 1:10 with LB broth. Additional 10-fold dilutions with LB broth were performed as needed to make the final concentration of detectable particles less than 2 × 105 particles/mL. A quick count of the particles in image view is needed, which takes about 2–5 minutes for imaging, counting and dilution. For AST test, another identical sample was prepared with the addition of ciprofloxacin (2 μg/mL, final concentrations). After mixing, diluted samples (70 μL) were transferred to cuvettes (Uvette, Eppendorf, Germany), and measured by LVSi. A total of 60 urine samples were tested using both optical division tracking and on-site validating agar plating (Supporting Information S8). Urine samples were prepared and transferred to researchers in a blinded fashion without providing any clinical testing results. The LVSi-AST, and microbiological plating results were compared with clinical microbiology culture results (Supporting Information S9) from the Mayo Clinic Hospital lab after the completion of the experiments.
LVSi.
The dual channel LVSi imaging system (Fig. 1A and Supporting Information Fig. S1) consists of two 800 mW, 780 nm infrared (IR) LEDs (M780LP1, Thorlabs, Inc., USA), each with collimating and focusing lens and a central blocking aperture to focus a ring-shaped illumination through the sample or the reference cuvettes. Wide-view and deep field depth scattering images were recorded by two CMOS camera (BFS-U3–16S2M-CS, Point Grey Research Inc., Canada) at 10 fps through two variable zoom lenses (NAVITAR 12X, Navitar, USA) with zoom factors set at 2.0X for the sample and reference cuvettes. The image volume was determined by the viewing size and focal depth of the optics. For the experiments described in this study, the viewing volume of 2.5 mm × 1.9 mm × 1.0 mm was equivalent to 4.8 μL at 2.0X magnifying power. The imaging system was enclosed in a thermally isolated housing unit with a controlled temperature (37°C).
Biosafety.
All sample preparations and measurements were performed in biosafety level 2 (BSL2) laboratories following an IBC-approved BSL2 protocol.
Single-cell division tracking.
Individual cells recorded by LVSi are resolved as a bright spot and the image sequences were taken as stacks and analyzed by ImageJ plug-in TrackMate.33 Before division tracking, each spot was detected with a Laplacian of Gaussian (LoG) filter with a defined radius and threshold. Then, the spots from adjacent time frames were connected with a Karman filter for directional linking, so that each bright spot became a single-cell trajectory. After filtering out the short tracks, the average length of the valid cell trajectory is about 2 min. For a sample with 5.0 × 104 cell density, there are several hundreds of valid tracks for division detection. These steps take ~ 5 min processing time for each sample. Then, to track the division events, the single-cell trajectories were imported into Matlab and subjected to division filtering. First, the newly appeared cell trajectories from the image edges were excluded, and the nearby original trajectories that appear before the division event were checked. If the daughter cell trajectory candidate is traced back to an original single trajectory and a splitting event is identified, then a potential division event was tracked. As cells float past each other, there is potential to mistake these passing/crossing cells as divisions thus, these ‘spot merging’ events were also checked and filtered out from the potential division events. Finally, an intensity filter was used to evaluate the remaining division events, ensuring the parent cell intensity is near the summation of the two resulting daughter cells, and the two daughter cells are similar in size (Supporting Information Fig. S10). The bacterial morphological changes induced splitting, such as breaking, lysis, or rupture, are not equal division in most cases, and can be easily filter out. The division filter process takes another 5 min for each sample. Fig. 1C shows two representative examples of single-cell division tracking. Each parent cell travels with time and increases in size, before dividing into two smaller daughter cells.
To validate that the tracked division events are due to bacterial cell multiplication and not an artifact of the particle merging or crossing, a division over-counting calibration test was designed. To rule out the valid division events, the E. coli cells were heat-inactivated at ~ 65°C for 15 min. Then, 5-min videos of the dead E. coli cells at different concentrations (2.0 × 104 – ~2.0 × 105 cells/mL, corresponding to 100–1000 spots in image view) were analyzed for single-cell division tracking. Each concentration test was repeated three times. Then, the division over-counting calibration curve was extracted from the tracking results (Supporting information Fig. S11). When the cell number is below 500, corresponding to the bacterial concentration of <1.0 × 105 particles or cells/mL, the risk of division over-counting is low. When the cell/particle number is > 500 per view, overcrowding causes some division events to be miscounted. Final division events in all clinical samples are calibrated by subtracting the density-associated division over-counting events.
Setting thresholds for data interpretation.
The statistical error of the division tracking was estimated by the division over-counting calibration discussed above. The final division events were calibrated by subtracting the fitted mean value of the division over-counting, in which the averaged standard error of the mean is calculated to be ~1 in every 5-min video. To establish a 95% confidence interval, we multiplied the error by 2. For a 60-min detection, the cumulative standard error of the mean is ~ 24. Since standard error of the mean in each 5-min is random, the final statistical error of the tracking was estimated by , where N is the cumulative standard error of the mean. Therefore, the final threshold for infection identification (TI) was set as 5 with a 95% confidence for the calibration. The susceptible threshold (TS) was set as 0.5, corresponding to 50% growth inhibition in the antibiotic-treated samples.
Statistical analysis
An unpaired two-sided student t-test was used to compare the group differences. A p value of <0.05 was considered as statistically significant.
Supplementary Material
ACKNOWLEDGMENT
Financial support from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01AI138993) is acknowledged. We thank Noel De Lucia and Kathrine McAulay at Mayo Clinic Arizona for clinical sample coordination.
Footnotes
Supporting Information
Supporting Information Available: dual-optical system; comparison of division tracking and traditional agar plating with pure E. coli samples; clinical ID results; normalized cumulative division curves of all susceptible and resistant samples; clinical AST results; comparison of division tracking and cell counting with pure E. coli samples; comparison of division tracking and cell counting with clinical samples; on-site validating method; clinical reference method; division tracking flowchart; and calibration of the division over-counting.
The authors declare no competing financial interest.
REFERENCES
- 1.CDC. Antibiotic Resistance Threats in the United States; Atlanta, GA: U.S., 2019. [Google Scholar]
- 2.Laxminarayan R; Duse A; Wattal C; Zaidi AK; Wertheim HF; Sumpradit N; Vlieghe E; Hara GL; Gould IM; Goossens H; Greko C; So AD; Bigdeli M; Tomson G; Woodhouse W; Ombaka E; Peralta AQ; Qamar FN; Mir F; Kariuki S; Bhutta ZA; Coates A; Bergstrom R; Wright GD; Brown ED; Cars O, Antibiotic resistance-the need for global solutions. Lancet Infect Dis 2013, 13 (12), 1057–98. [DOI] [PubMed] [Google Scholar]
- 3.Tacconelli E; Carrara E; Savoldi A; Harbarth S; Mendelson M; Monnet DL; Pulcini C; Kahlmeter G; Kluytmans J; Carmeli Y; Ouellette M; Outterson K; Patel J; Cavaleri M; Cox EM; Houchens CR; Grayson ML; Hansen P; Singh N; Theuretzbacher U; Magrini N; Group WHOPPLW, Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 2018, 18 (3), 318–327. [DOI] [PubMed] [Google Scholar]
- 4.Jorgensen JH; Ferraro MJ, Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 2009, 49 (11), 1749–55. [DOI] [PubMed] [Google Scholar]
- 5.Bauer AW; Kirby WM; Sherris JC; Turck M, Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966, 45 (4), 493–6. [PubMed] [Google Scholar]
- 6.Bauer KA; Perez KK; Forrest GN; Goff DA, Review of rapid diagnostic tests used by antimicrobial stewardship programs. Clin Infect Dis 2014, 59 Suppl 3, S134–45. [DOI] [PubMed] [Google Scholar]
- 7.Khan ZA; Siddiqui MF; Park S, Current and Emerging Methods of Antibiotic Susceptibility Testing. Diagnostics 2019, 9 (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fluit AC; Visser MR; Schmitz FJ, Molecular detection of antimicrobial resistance. Clin Microbiol Rev 2001, 14 (4), 836–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frye JG; Lindsey RL; Rondeau G; Porwollik S; Long F; McClelland M; Jackson CR; Englen MD; Meinersmann RJ; Berrang ME; Davis JA; Barrett JB; Turpin JB; Thitaram SN; Fedorka-Cray PJ, Development of a DNA microarray to detect antimicrobial resistance genes identified in the National Center for Biotechnology Information database. Microb Drug Resist 2010, 16 (1), 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park S; Zhang Y; Lin S; Wang TH; Yang S, Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol Adv 2011, 29 (6), 830–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Athamanolap P; Hsieh K; Chen LB; Yang S; Wang TH, Integrated Bacterial Identification and Antimicrobial Susceptibility Testing Using PCR and High-Resolution Melt. Anal Chem 2017, 89 (21), 11529–11536. [DOI] [PubMed] [Google Scholar]
- 12.Machowski EE; Kana BD, Genetic Mimetics of Mycobacterium tuberculosis and Methicillin-Resistant Staphylococcus aureus as Verification Standards for Molecular Diagnostics. Journal of Clinical Microbiology 2017, 55 (12), 3384–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicasio AM; Kuti JL; Nicolau DP, The current state of multidrug-resistant gram-negative bacilli in North America. Pharmacotherapy 2008, 28 (2), 235–49. [DOI] [PubMed] [Google Scholar]
- 14.Bard JD; Lee F, Why Can’t We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing. Clin Microbiol Newsl 2018, 40 (11), 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi J; Jung YG; Kim J; Kim S; Jung Y; Na H; Kwon S, Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13 (2), 280–7. [DOI] [PubMed] [Google Scholar]
- 16.Douglas IS; Price CS; Overdier KH; Wolken RF; Metzger SW; Hance KR; Howson DC, Rapid automated microscopy for microbiological surveillance of ventilator-associated pneumonia. Am J Respir Crit Care Med 2015, 191 (5), 566–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y; Gao J; Zhang DD; Gau V; Liao JC; Wong PK, Single cell antimicrobial susceptibility testing by confined microchannels and electrokinetic loading. Anal Chem 2013, 85 (8), 3971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ertl P; Robello E; Battaglini F; Mikkelsen SR, Rapid antibiotic susceptibility testing via electrochemical measurement of ferricyanide reduction by Escherichia coli and Clostridium sporogenes. Anal Chem 2000, 72 (20), 4957–64. [DOI] [PubMed] [Google Scholar]
- 19.Longo G; Alonso-Sarduy L; Rio LM; Bizzini A; Trampuz A; Notz J; Dietler G; Kasas S, Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors. Nat Nanotechnol 2013, 8 (7), 522–526. [DOI] [PubMed] [Google Scholar]
- 20.Lissandrello C; Inci F; Francom M; Paul MR; Demirci U; Ekinci KL, Nanomechanical motion of Escherichia coli adhered to a surface. Appl Phys Lett 2014, 105 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syal K; Iriya R; Yang Y; Yu H; Wang S; Haydel SE; Chen HY; Tao N, Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano 2016, 10 (1), 845–52. [DOI] [PubMed] [Google Scholar]
- 22.Shifman O; Steinberger-Levy I; Aloni-Grinstein R; Gur D; Aftalion M; Ron I; Mamroud E; Ber R; Rotem S, A Rapid Antimicrobial Susceptibility Test for Determining Yersinia pestis Susceptibility to Doxycycline by RT-PCR Quantification of RNA Markers. Front Microbiol 2019, 10, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoepp NG; Schlappi TS; Curtis MS; Butkovich SS; Miller S; Humphries RM; Ismagilov RF, Rapid pathogen-specific phenotypic antibiotic susceptibility testing using digital LAMP quantification in clinical samples. Science Translational Medicine 2017, 9 (410). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baltekin O; Boucharin A; Tano E; Andersson DI; Elf J, Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. P Natl Acad Sci USA 2017, 114 (34), 9170–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H; Torab P; Mach KE; Surrette C; England MR; Craft DW; Thomas NJ; Liao JC; Puleo C; Wong PK, Adaptable microfluidic system for single-cell pathogen classification and antimicrobial susceptibility testing. Proc Natl Acad Sci U S A 2019, 116 (21), 10270–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baltekin Ö; Boucharin A; Tano E; Andersson DI; Elf J, Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proceedings of the National Academy of Sciences 2017, 114 (34), 9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cermak N; Olcum S; Delgado FF; Wasserman SC; Payer KR; A Murakami M; Knudsen SM; Kimmerling RJ; Stevens MM; Kikuchi Y; Sandikci A; Ogawa M; Agache V; Baléras F; Weinstock DM; Manalis SR, High-throughput measurement of single-cell growth rates using serial microfluidic mass sensor arrays. Nat Biotechnol 2016, 34 (10), 1052–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mo MN; Yang YZ; Zhang FN; Jing WW; Iriya R; Popovich J; Wang SP; Grys T; Haydel SE; Tao NJ, Rapid Antimicrobial Susceptibility Testing of Patient Urine Samples Using Large Volume Free-Solution Light Scattering Microscopy. Anal Chem 2019, 91 (15), 10164–10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foxman B, Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014, 28 (1), 1–13. [DOI] [PubMed] [Google Scholar]
- 30.Flores-Mireles AL; Walker JN; Caparon M; Hultgren SJ, Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 2015, 13 (5), 269–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campoli-Richards DM; Monk JP; Price A; Benfield P; Todd PA; Ward A, Ciprofloxacin. A review of its antibacterial activity, pharmacokinetic properties and therapeutic use. Drugs 1988, 35 (4), 373–447. [DOI] [PubMed] [Google Scholar]
- 32.Acred P; Brown DM; Turner DH; Wilson MJ, Pharmacology and chemotherapy of ampicillin--a new broad-spectrum penicillin. Br J Pharmacol Chemother 1962, 18, 356–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tinevez JY; Perry N; Schindelin J; Hoopes GM; Reynolds GD; Laplantine E; Bednarek SY; Shorte SL; Eliceiri KW, TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


