Abstract
Searching for biomarkers has been a chief pursuit of the field of psychiatry. Toward this end, studies have catalogued candidate resting-state biomarkers in nearly all forms of mental disorder. However, it is becoming increasingly clear that these biomarkers lack specificity, limiting their capacity to yield clinical impact. We discuss three avenues of research that are overcoming this limitation: (i) the adoption of transdiagnostic research designs, which involve studying and explicitly comparing multiple disorders from distinct diagnostic axes of psychiatry; (ii) dimensional models of psychopathology that map the full spectrum of symptomatology and that cut across traditional disorder boundaries; and (iii) modeling individuals’ unique functional connectomes throughout development. We provide a framework for tying these subfields together that draws on tools from machine learning and network science.
Introduction
Precision medicine refers to the idea that diagnosis and treatment strategies for disease processes are optimized when an individual’s unique biology is taken into account [1,2]. Critical to this paradigm is the biomarker, which is broadly defined as any biological signature that provides an objective indication of an individual’s disease status and that ideally predicts clinical outcomes [3]. In recent decades, detecting biomarkers of psychiatric disorders has become a central goal for neuroimaging research as a means to drive psychiatry towards precision medicine [1]. To this end, a large body of literature has emerged using resting-state functional magnetic resonance imaging (rs-fMRI) to characterize the brain dysfunction accompanying psychiatric conditions [4]. However, a combination of high symptomatic and biological heterogeneity within disorders as well as comorbidity amongst disorders [5,6] has resulted in a lack of disorder specificity in candidate biomarkers. This lack of specificity has rendered the field unable to translate knowledge reliably into clinical practice [7]. Here, we discuss three recent developments in rs-fMRI research that hold promise for improving our capacity to generate biomarkers for mental health. First, we highlight a shift in focus from studying single disorders in isolation to transdiagnostic paradigms that examine multiple disorders in a single study. Second, we discuss insights from studies that take a dimensional, as opposed to categorical, approach to measuring psychopathology. Third, we briefly review how understanding the developing functional connectome is providing new directions into studying brain dysfunction. Finally, we provide a framework for tying these three areas of research together drawing on machine learning models of normative neurodevelopment [8,9] and tools from network science [10,11].
1. A shift toward transdiagnostic research
The standard paradigm for biomarker detection in psychiatry is the case-control design, a categorical approach wherein case refers to a group of individuals experiencing a given mental disorder, and control refers to a group of individuals not experiencing the disorder [12]. Typically, individuals in the control group are selected because they are not experiencing any mental disorder (i.e., healthy control), hence any observed group differences are attributed to the disease process. Since the inception of the rs-fMRI paradigm [13], case-control rs-fMRI studies have been conducted in almost every major disorder described by psychiatric nosologies (e.g., DSM-5, ICD-11). However, within a single study, the case-control paradigm is typically applied to a single disorder; rarely are multiple disorders studied concurrently. Consequently, recent meta-analytic work has revealed that many of the field’s candidate biomarkers lack disorder specificity. Sha et al. [14] examined 182 rs-fMRI studies spanning attention deficit hyperactivity disorder (ADHD), autism spectrum disorder, bipolar disorder, depression, obsessive-compulsive disorder (OCD), post-traumatic stress disorder, and schizophrenia (as well as several neurological disorders) and found that dysfunction in the default-mode, sensorimotor, fronto-parietal, and subcortical systems was common to all disorders. This result challenges the idea that large-scale dysconnectivity between these systems represents a disorder-specific biomarker (see Dong et al. [15] for a recent example in schizophrenia). Instead, this result demonstrates that in order to attain specificity, the field may need to transition to transdiagnostic designs, wherein multiple disorders are examined concurrently, and differential diagnostic sensitivity can be directly assessed.
One candidate biomarker where progress toward disorder specificity has shown recent promise is dysfunction in the frontostriatal circuits [16]. Frontostriatal circuits are a set of parallel circuits that topographically connect different regions of the frontal cortex to different subregions of the striatum [17,18]. Using a combination of resting-state activity within the striatum, as well as intra- and extra- striatal connectivity, Li et al. [16] trained a support vector machine (SVM) to separate schizophrenia patients from healthy controls. Then, the authors took the distance between each individual and the separating SVM hyperplane as an index for frontostriatal abnormality (FSA). Next, they compared FSA scores from individuals with schizophrenia, bipolar disorder, depression, OCD, and ADHD against healthy controls and found that only the schizophrenia and bipolar groups showed significant differences in FSA scores, with differences being greater for schizophrenia relative to bipolar. This pattern of results suggests that the author’s SVM learned multivariate patterns of frontostriatal dysfunction that selectively separated schizophrenia and bipolar from healthy controls. However, such selectivity does not imply that disorder-specific frontostriatal dysfunction is absent in other disorders (e.g., OCD [19,20]). Li et al. trained their SVM to separate schizophrenia and healthy controls, and then used that model to generate FSA scores for other disorder groups. Thus, a different SVM could be trained to separate OCD from healthy controls and to test for selective frontostriatal dysfunction in OCD. Further, this OCD-trained SVM could be compared to the schizophrenia-trained SVM to characterize the potentially qualitatively distinct patterns of frontostriatal dysfunction that support differential diagnosis. Note, such an approach is not limited to the study of frontostriatal circuits. Equally, such transdiagnostic designs could also be applied to test for whole-brain connectome differences using tools from network science (see van den Heuvel & Sporns [21] for a review).
Despite progress, there are still challenges that potentially limit the insights derived from case-control designs, irrespective of whether they are deployed transdiagnostically or to study single disorders. First, it remains common practice in the case-control literature to recruit patients that are without comorbidities for multiple disorders. However, it is well established that psychiatric comorbidities are pervasive in the general population [5,22]. This ubiquity suggests that even if researchers could find disorder-specific biomarkers using transdiagnostic designs, their utility may be limited to patients who only appear infrequently in a clinical setting. Second, there is mounting evidence suggesting that, when it comes to the underlying neurobiology, clean separation between patient groups or between patients and healthy controls may not exist [23–25]. This indistinctness is particularly evident in the biotyping literature, which involves applying unsupervised clustering techniques to brain data with the goal of finding neurobiologically-informed subgroups of patients [26]. A prominent example in the rs-fMRI literature is that of Drysdale et al. [27], who reported four biotypes of depression based on functional connectivity that showed differential responsiveness to repetitive transcranial magnetic stimulation. However, these biotypes did not represent unique, cleanly separable, subgroups of patients, but rather a partitioning of a single group of patients that included many edge cases. This issue of a lack of separability is not specific to the analysis of functional connectivity data, but is rather also present in the structural neuroimaging literature [28–30]. The central problem is that clustering algorithms will always produce clusters, even if such a partition is not supported by the data [31]. Furthermore, work by Dinga et al. [24] demonstrated that the methods used by Drysdale et al. have the potential to yield unstable patient clusters. Thus, while clustering approaches may be a step in the right direction — insofar as they move away from a system of classification based solely on symptoms —, the lack of clean separability in the biotyping literature suggests that the pathways linking the brain to psychiatric symptoms may be better elucidated through the study of dimensional variance in both brain and symptom data.
2. Dimensions of psychopathology
While transdiagnostic case-control designs may assist in identifying which biomarkers are disorder-specific or disorder-general, these study designs remain focused on the group-level signatures revealed by comparing healthy individuals with those that meet criteria for a mental disorder. Consequently, they fail to capture subclinical variation in symptomatology that is important for understanding the link between brain dysfunction and psychiatry [32,33]. In response to this limitation, rs-fMRI studies have begun taking an explicitly dimensional approach. A dimensional approach suggests that mental disorders lay at the end of continuous spectrums of symptom severity, ranging from the absence of symptoms at one end to frank disorder at the other. Thus, in contrast to case-control designs, dimensional approaches characterize the full range of variation -- normal, subclinical, and abnormal -- in symptomatology and brain function.
Recent rs-fMRI studies deploying dimensional models have varied in terms of the number and scope of the estimated dimensions of psychopathology. On the one hand, some studies have aimed to quantify dimensions of psychopathology that cut across a broad range of diagnostic entities, with a view to teasing apart which biomarkers are unique to, or common across, most diagnostic axes of psychiatry. These studies have often drawn on the idea of the p-factor [34,35], a statistical concept that posits that the endorsement of any psychiatric symptom increases the probability of endorsing any other symptom (so-called overall psychopathology). Studies typically estimate the p-factor alongside multiple specific dimensions of psychopathology (e.g., depression, psychosis) in a hierarchical fashion, rendering the specific dimensions orthogonal to each other and the p-factor. This model structure allows researchers to uncover both disorder-general and disorder-specific biomarkers. On the other hand, some studies have focused on decomposing a single diagnostic axis of psychiatry (e.g., psychosis) into multiple subcomponents (e.g., positive and negative psychosis dimensions), aiming instead to discover multiple distinct biomarkers within select disorders.
At the multi-diagnosis level, recent studies by Elliot et al. [36] and Kebets et al. [37] found that greater scores on the p-factor were associated with hypo-connectivity within sensory and somatomotor systems, as well as with hyper-connectivity between these systems and the executive systems. These results point toward a disorder-general biomarker of dysconnectivity between lower- and higher-order systems in the cortical hierarchy. Notably, convergence between these studies was present despite the use of distinct statistical methods (for a review of the methods see Kaczkurkin et al. [26]). Briefly, Elliot et al. modeled latent psychopathology dimensions independently of the rs-fMRI data (i.e., a so-called single-view approach), and then related the p-factor to functional connectivity. By contrast, Kebets et al. deployed a multi-view approach that jointly estimated latent dimensions of psychopathology and functional connectivity in a single model and labeled one of their dimensions as a p-factor post hoc. However, another multi-view study by Xia et al. [38] found instead that dysconnectivity between the higher-order default mode and executive systems was common across dimensions of mood, psychosis, fear, and externalizing behavior. That Xia et al.’s results implicated the higher-order cortical systems while Kebets et al. and Elliot et al.’s results implicated the lower-order cortical systems may be due to the fact that Xia et al. did not find a p-factor in their data. Instead, Xia et al. derived disorder-general biomarkers by examining the spatial correspondence between patterns of dysconnectivity observed for their specific dimensions. This pattern of results suggests that the presence or absence of a p-factor impacts the ensuing biomarkers. Indeed, how to best model broad dimensions of psychopathology that span multiple diagnostic categories remains open to debate in the field [39].
At the single-diagnosis level, dimensional models have been used to uncover differences in functional connectivity across subdimensions of specific forms of psychopathology. Sabaroedin et al. [40] examined the extent to which normal and subclinical variation in negative and positive psychosis-like experiences (PLE) tracked differences in resting-state frontostriatal connectivity in a large population cohort. Convergent with previous literature in schizophrenia, greater positive PLEs were associated with reduced connectivity between the dorsal striatum and prefrontal cortex, demonstrating that frontostriatal dysfunction in schizophrenia may generalize to subclinical levels of psychotic phenomenology. Taking a dimensional and transdiagnostic approach, Parkes et al. [19] examined frontostriatal dysfunction in individuals with OCD and individuals with gambling disorder (GD). The authors examined whether dimensional measures of impulsivity and compulsivity, two constructs with joint relevance to OCD and GD, tracked variation in resting-state effective connectivity in frontostriatal circuits. Compared to pairwise case-control and case-case comparisons, Parkes et al. found that inter-individual differences in compulsivity better explained frontostriatal dysconnectivity when patients and controls were pooled into a single group. These studies show that splitting specific dimensions of psychopathology into subdimensions reveal addition insights in brain dysfunction and, together with the broad multi-diagnosis studies, highlight the importance of considering dimensional models of psychopathology at multiple scales of specificity.
3. Modeling the connectome throughout development
In addition to transdiagnostic and dimensional designs, linking mental disorder to rs-fMRI has also been facilitated by progress in modeling the whole-brain functional connectome throughout development. Here, we define the functional connectome as a network of macroscopic brain regions whose functional connections are instantiated as the correlations between regional timeseries measured with fMRI. In a connectome identifiability [41] study (Box 1), Kaufmann et al. [42] showed that individuals’ connectomes became increasingly distinct from one another throughout childhood and adolescence, demonstrating that the whole-brain functional connectome encodes individualized fingerprints that emerge with age. Critically, individuals with greater scores on the p-factor showed reduced connectome distinctiveness compared to healthy individuals, indicating that disruptions to the formation of these fingerprints is relevant to mental health. Subsequent longitudinal work showed that whole-brain connectomes could be used to identify participants across time intervals of several months and up to 2 years [43,44], demonstrating that connectome fingerprints are stable over long periods.
Box 1. Individuals have unique connectome fingerprints.
Studying individual differences in resting-state connectivity has been aided by a shift away from the mass-univariate study of edge-level functional connectivity towards studying the whole-brain connectome as a single multivariate object. In this context, the brain is represented as a symmetric N×N adjacency matrix, A, where N represents a number of discrete brain regions referred to as nodes. Within this matrix, elements Aij take on a weighted value corresponding to the functional connectivity between nodes i and j. This weight can be estimated by a simple Pearson correlation coefficient between rs-fMRI timeseries, or by other more sophisticated measures [64]. One promising approach for analyzing multivariate inter-individual differences in the connectome is known as identifiability. Popularized by Finn et al. [41], identifiability involves correlating connectivity estimates within pairs of whole-brain connectomes. The simplest setup involves generating at least two whole-brain connectomes for each individual in a sample (e.g., across multiple scans/sessions) and then calculating the correlation between all possible connectome pairs in a sample. Multiple studies have found that correlation coefficients are higher for connectome pairs taken from the same individual than for connectome pairs taken across different individuals, allowing for the identification of individuals over repeated scans with near perfect accuracy [41,65,66]. This phenomenon illustrates that the multivariate pattern of whole brain functional connectivity is highly unique to each individual.
The above studies have also illustrated that different brain systems have a differential impact on identifiability. Horien et al. [43] and Miranda-Dominguez et al. [44] each found, across multiple datasets, that higher-order executive systems offered higher identifiability than lower-order sensory and motor systems; the latter only outperformed subcortical systems [43]. This differential system effect remained throughout childhood and adolescence, and into adulthood [42,45]. Thus, whereas higher-order systems develop into increasingly individualized functional fingerprints, lower-order systems remain stable and more homogenous across individuals. Critically, Kaufmann et al. [42] found that the reduced connectome distinctiveness associated with greater p-factor scores was more pronounced in the executive systems, suggesting that disruptions to an individual’s higher-order functional fingerprint is indicative of mental disorder.
The above neurodevelopmental work stands in contrast to the studies demonstrating that greater p-factor scores were predominantly associated with dysconnectivity centered on the lower-order sensory and somatomotor systems [36,37]. This discrepancy may be explained by differences between edge-level modeling and distance-based modeling of inter-individual differences in the connectome. For example, Kebets et al. [37] deployed an edge-level approach testing whether dimensions of psychopathology covaried directly with edge strength and found that the strongest signals originated from lower-order cortical systems. In contrast, identifiability studies effectively score individuals based on the distance between their whole-brain connectomes across scans in an N-dimensional space (where N denotes the number of edges). Subsequently, covariation with psychopathology indexes inter-individual differences in the extent to which connectomes are individualized, signals for which appear concentrated in higher-order systems [42,43]. This finding suggests that mental disorders affect different parts of the cortical hierarchy in qualitatively distinct ways, with edge-level modeling being sensitive to lower-order systems that are relatively stable and homogenous across people, and the distance-based modeling being sensitive to higher-order systems that are highly and increasingly individualized throughout development.
Future directions
The intersection between transdiagnostic research, dimensional models of psychopathology, and connectome development has the potential to uncover novel biomarkers in psychiatry. Synthesizing these approaches will require the joint application of tools from machine learning and network neuroscience [46]. In particular, recent developments in machine learning have provided new tools for modeling, and detecting abnormalities in, neurodevelopment [8]. This approach, known as normative modeling, involves estimating growth charts of brain development that can track the extent to which an individual’s biology deviates from what is expected given their age. This estimation is achieved through Bayesian regression techniques that provide estimates of percentiles of variation within a healthy cohort [9]. In contrast to case-control designs, which are restricted to estimating brain abnormalities at the group level, these percentiles of variation allow researchers to quantify the extent to which patients deviate from the normative age trajectory on an individual basis. In psychiatry research, normative models have already been used to illustrate the large amount of heterogeneity in brain structure present within disorders such as schizophrenia [47], bipolar [47], ADHD [48], and autism [49,50]. These studies have shown that few patients, if any, have spatially consistent patterns of age-related brain abnormalities, supporting the notion that case-control paradigms provide little utility in biomarker detection.
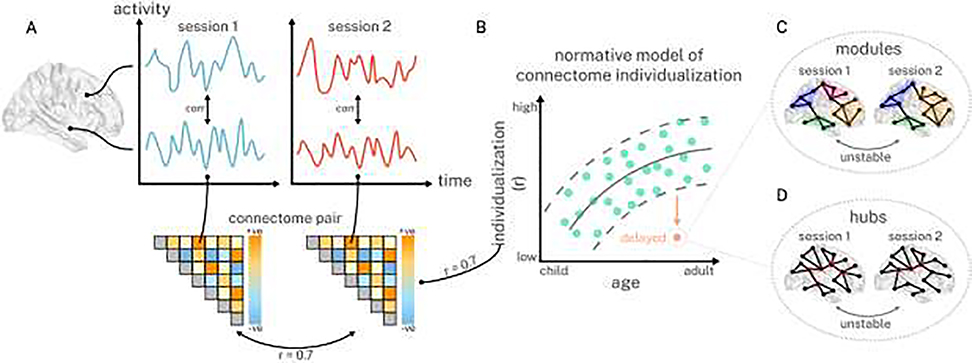
The extant normative modeling literature has so far focused on indices of brain structure, in part because normative models depend upon a robust underlying age effect in order to yield interpretable abnormalities. Robust regional age effects are trivially obtained using structural indices [51,52] (e.g., gray matter volume), but this is not true of edge-level estimates of resting-state functional connectivity. Given the age effects reported in the identifiability literature [42,53], normative models may be well suited to identifying individual patients with abnormally reduced connectome individualization for their age (Figure 1). Using pairs of rs-fMRI (or task-based fMRI) scans from longitudinal developmental cohorts (Figure 1A), normative models of connectome individualization can be estimated in typically developing individuals (Figure 1B,  ). Next, deviations can be estimated in a separate cohort (Figure 1B,
). Next, deviations can be estimated in a separate cohort (Figure 1B,  ) and correlated with dimensional psychopathology models. Such models may assist in identifying which axes of psychiatry are associated with the abnormal formation of individual connectomes, which may be underpinned by disorganization in higher-order brain systems.
) and correlated with dimensional psychopathology models. Such models may assist in identifying which axes of psychiatry are associated with the abnormal formation of individual connectomes, which may be underpinned by disorganization in higher-order brain systems.
Figure 1. Abnormally delayed connectome individualization may be characterized using a combination of normative modeling and tools from network science.
A, For each individual, pairs of whole-brain resting-state connectomes may be obtained from different scan sessions (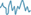 ,
, 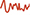 ). Correlations between connectome-pairs may be used to index individualization (e.g., r = 0.7). B, Normative models of age-related increases in individualization (
). Correlations between connectome-pairs may be used to index individualization (e.g., r = 0.7). B, Normative models of age-related increases in individualization ( ) can then be specified, and the individual’s deviating connectomes (
) can then be specified, and the individual’s deviating connectomes ( ) can be further characterized using tools from network science (C, D).
) can be further characterized using tools from network science (C, D).
The issue of biological heterogeneity in psychiatry reveals a clear ‘many to one’ problem, where multiple pathophysiological pathways converge on similar forms of psychopathology. Thus, normative models of connectome individualization may give rise to individuals with quantitatively similar deviations in individualization, and levels of psychopathology, that correspond to qualitatively distinct underlying disruptions in their connectome architecture. We anticipate that tools from network science will be critical to uncovering these unique disruptions [54,55]. For example, brain regions in the connectome are organized into functional modules [21] — groups of brain regions that are more densely connected amongst themselves than they are to the rest of the brain — and abnormal individualization may be reflected in the instability of an individual’s modular structure across scans (Figure 1C). That is, the regions that constitute specific functional modules in an individual’s brain may be reassigned across timepoints. Similarly, hub nodes [56,57] — brain regions whose densely connected nature are thought to support the integration of information across functional modules — may be less stable across an individual’s scans (Figure 1D). These examples are not exhaustive and we note that many other avenues may also be of interest, including developments in the areas of time-varying functional connectivity [58,59] and individual-specific parcellations [60–63]. Critically, the nature of an individual’s connectome instability (e.g., modular and hub instability), as indexed by network science, need not be the same across individuals with similarly high scores on dimensions of psychopathology, accommodating the notion that unique patterns of pathophysiology give rise to similar symptom profiles.
Conclusions
While initially fruitful, single disorder case-control studies in psychiatry are unlikely to continue to yield advances in biomarker research in the field of psychiatry. Here, we briefly discussed recent rs-fMRI work in transdiagnostic psychiatry, dimensional psychopathology, and neurodevelopment that we believe are driving the field towards more precise biomarkers in mental health. While some differences in methodologies remain to be thoroughly examined, results suggest that edge-level and distance-based models (i.e., identifiability) yield complementary information relevant to mental health. We suggest that combining normative models of connectome individualization with transdiagnostic dimensional models of psychopathology and tools from network science hold great promise for understanding the link between brain dysfunction and mental disorder.
Citation diversity statement
Recent work in several fields of science has identified a bias in citation practices such that papers from women and other minority scholars are under-cited relative to the number of such papers in the field [67–71]. Here we sought to proactively consider choosing references that reflect the diversity of the field in thought, form of contribution, gender, race, ethnicity, and other factors. First, we obtained the predicted gender of the first and last author of each reference by using databases that store the probability of a first name being carried by a woman [71,72]. By this measure (and excluding self-citations to the first and last authors of our current paper), our references contain 8.7% woman(first)/woman(last), 16.13% man/woman, 18.72% woman/man, and 56.45% man/man. This method is limited in that a) names, pronouns, and social media profiles used to construct the databases may not, in every case, be indicative of gender identity and b) it cannot account for intersex, non-binary, or transgender people. Second, we obtained predicted racial/ethnic category of the first and last author of each reference by databases that store the probability of a first and last name being carried by an author of color [73,74]. By this measure (and excluding self-citations), our references contain 14.03% author of color (first)/author of color(last), 19.37% white author/author of color, 16.58% author of color/white author, and 50.02% white author/white author. This method is limited in that a) names and Wikipedia profiles used to make the predictions may not be indicative of racial/ethnic identity, and b) it cannot account for Indigenous and mixed-race authors, or those who may face differential biases due to the ambiguous racialization or ethnicization of their names. We look forward to future work that could help us to better understand how to support equitable practices in science.
Highlights.
Resting-state fMRI biomarkers in psychiatry often lack disorder specificity
Transdiagnostic study designs allow direct assessment of biomarker specificity
p-factor models reveal biomarkers common across most diagnostic axes of psychiatry
The functional connectome develops into an individualized fingerprint
Delayed individualization is a biomarker that may be characterized using networks
Acknowledgements
LP, TDS, and DSB primarily acknowledge financial support from the National Institute of Mental Health through R01 award MH113550 to TDS and DSB. Additional support was provided by the National Institutes of Mental Health through awards 1R01MH119219 (PI: Gur) and 1R01MH120482-01 (PI: TDS), and the Army Research Laboratory through CPNF Subcontract PO #182200 (PI: DSB). The authors acknowledge Dr. Eli J. Cornblath and Dr. Arun Mahadevan for valuable discussions during the writing of the manuscript.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Collins FS, Varmus H, A New Initiative on Precision Medicine, N Engl J Med. 372 (2015) 793–795. 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Insel TR, Cuthbert BN, Brain disorders? Precisely, Science. 348 (2015) 499–500. 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- [3].Strimbu K, Tavel JA, What are biomarkers?, Current Opinion in HIV and AIDS. 5 (2010) 463–466. 10.1097/COH.0b013e32833ed177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Woodward ND, Cascio CJ, Resting-State Functional Connectivity in Psychiatric Disorders, JAMA Psychiatry. 72 (2015) 743 10.1001/jamapsychiatry.2015.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Plana-Ripoll O, Pedersen CB, Holtz Y, Benros ME, Dalsgaard S, de Jonge P, Fan CC, Degenhardt L, Ganna A, Greve AN, Gunn J, Iburg KM, Kessing LV, Lee BK, Lim CCW, Mors O, Nordentoft M, Prior A, Roest AM, Saha S, Schork A, Scott JG, Scott KM, Stedman T, Sørensen HJ, Werge T, Whiteford HA, Laursen TM, Agerbo E, Kessler RC, Mortensen PB, McGrath JJ, Exploring Comorbidity Within Mental Disorders Among a Danish National Population, JAMA Psychiatry. 76 (2019) 259 10.1001/jamapsychiatry.2018.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Merikangas KR, Calkins ME, Burstein M, He J-P, Chiavacci R, Lateef T, Ruparel K, Gur RC, Lehner T, Hakonarson H, Gur RE, Comorbidity of Physical and Mental Disorders in the Neurodevelopmental Genomics Cohort Study, 135 (2015) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Casey BJ, Craddock N, Cuthbert BN, Hyman SE, Lee FS, Ressler KJ, DSM-5 and RDoC: progress in psychiatry research?, Nat Rev Neurosci. 14 (2013) 810–814. 10.1038/nrn3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Marquand AF, Rezek I, Buitelaar J, Beckmann CF, Understanding Heterogeneity in Clinical Cohorts Using Normative Models: Beyond Case-Control Studies, Biological Psychiatry. 80 (2016) 552–561. 10.1016/j.biopsych.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marquand AF, Kia SM, Zabihi M, Wolfers T, Buitelaar JK, Beckmann CF, Conceptualizing mental disorders as deviations from normative functioning, Mol Psychiatry. (2019). 10.1038/s41380-019-0441-1.* This article discusses the concept of identifying abnormal brain signatures on an individual rather than a group basis. It provides an overview of the normative modeling methodology and how this approach will help to disentangle biological heterogeneity in psychiatry.
- [10].Bassett DS, Sporns O, Network neuroscience, Nat Neurosci. 20 (2017) 353–364. 10.1038/nn.4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Bassett DS, Zurn P, Gold JI, On the nature and use of models in network neuroscience, Nat Rev Neurosci. 19 (2018) 566–578. 10.1038/s41583-018-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lewis G, Pelosi AJ, The Case-Control Study in Psychiatry, Br J Psychiatry. 157 (1990) 197–207. 10.1192/bjp.157.2.197. [DOI] [PubMed] [Google Scholar]
- [13].Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS, Functional connectivity in the motor cortex of resting human brain using echo-planar mri, Magn. Reson. Med. 34 (1995) 537–541. 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- [14].Sha Z, Xia M, Lin Q, Cao M, Tang Y, Xu K, Song H, Wang Z, Wang F, Fox PT, Evans AC, He Y, Meta-Connectomic Analysis Reveals Commonly Disrupted Functional Architectures in Network Modules and Connectors across Brain Disorders, Cerebral Cortex. 28 (2018) 4179–4194. 10.1093/cercor/bhx273.** This study performed a comprehensive meta-analysis of rs-fMRI connectome data and showed that patterns of dysfunction were common across multiple psychiatric populations.
- [15].Dong D, Wang Y, Chang X, Luo C, Yao D, Dysfunction of Large-Scale Brain Networks in Schizophrenia: A Meta-analysis of Resting-State Functional Connectivity, Schizophrenia Bulletin. 44 (2018) 168–181. 10.1093/schbul/sbx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li A, Zalesky A, Yue W, Howes O, Yan H, Liu Y, Fan L, Whitaker KJ, Xu K, Rao G, Li J, Liu S, Wang M, Sun Y, Song M, Li P, Chen J, Chen Y, Wang H, Liu W, Li Z, Yang Y, Guo H, Wan P, Lv L, Lu L, Yan J, Song Y, Wang H, Zhang H, Wu H, Ning Y, Du Y, Cheng Y, Xu J, Xu X, Zhang D, Wang X, Jiang T, Liu B, A neuroimaging biomarker for striatal dysfunction in schizophrenia, Nat Med. 26 (2020) 558–565. 10.1038/s41591-020-0793-8.** This study trained a support vector machine to separate patients with schizophrenia from healthy controls and showed that their model did not generalize to other psychiatric groups, demonstrating disorder-specificity.
- [17].Parkes L, Fulcher BD, Yücel M, Fornito A, Transcriptional signatures of connectomic subregions of the human striatum, Genes, Brain and Behavior. 16 (2017) 647–663. 10.1111/gbb.12386. [DOI] [PubMed] [Google Scholar]
- [18].Marquand AF, Haak KV, Beckmann CF, Functional corticostriatal connection topographies predict goal-directed behaviour in humans, Nature Human Behaviour. 1 (2017) 0146 10.1038/s41562-017-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Parkes L, Tiego J, Aquino K, Braganza L, Chamberlain SR, Fontenelle LF, Harrison BJ, Lorenzetti V, Paton B, Razi A, Fornito A, Yücel M, Transdiagnostic variations in impulsivity and compulsivity in obsessive-compulsive disorder and gambling disorder correlate with effective connectivity in cortical-striatal-thalamic-cortical circuits, NeuroImage. 202 (2019) 116070 10.1016/j.neuroimage.2019.116070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jung WH, Yücel M, Yun J, Yoon YB, Cho KIK, Parkes L, Kim SN, Kwon JS, Altered functional network architecture in orbitofronto-striato-thalamic circuit of unmedicated patients with obsessive-compulsive disorder, Hum. Brain Mapp. 38 (2017) 109–119. 10.1002/hbm.23347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].van den Heuvel MP, Sporns O, A cross-disorder connectome landscape of brain dysconnectivity, Nat Rev Neurosci. (2019). 10.1038/s41583-019-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kessler RC, Chiu WT, Demler O, Walters EE, Prevalence, Severity, and Comorbidity of 12-Month DSM-IV Disorders in the National Comorbidity Survey Replication, ARCH GEN PSYCHIATRY. 62 (2005) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Marquand AF, Wolfers T, Dinga R, Phenomapping: Methods and Measures for Deconstructing Diagnosis in Psychiatry, in: Passos IC, Mwangi B, Kapczinski F (Eds.), Personalized Psychiatry, Springer International Publishing, Cham, 2019: pp. 119–134. 10.1007/978-3-030-03553-2_7. [DOI] [Google Scholar]
- [24].Dinga R, Schmaal L, Penninx BWJH, van Tol MJ, Veltman DJ, van Velzen L, Mennes M, van der Wee NJA, Marquand AF, Evaluating the evidence for biotypes of depression: Methodological replication and extension of, NeuroImage: Clinical. (2019) 101796 10.1016/j.nicl.2019.101796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Marquand AF, Wolfers T, Mennes M, Buitelaar J, Beckmann CF, Beyond Lumping and Splitting: A Review of Computational Approaches for Stratifying Psychiatric Disorders, Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. 1 (2016) 433–447. 10.1016/j.bpsc.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kaczkurkin AN, Moore TM, Sotiras A, Xia CH, Shinohara RT, Satterthwaite TD, Approaches to Defining Common and Dissociable Neurobiological Deficits Associated With Psychopathology in Youth, Biological Psychiatry. (2019) S0006322319319560 10.1016/j.biopsych.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, Fetcho RN, Zebley B, Oathes DJ, Etkin A, Schatzberg AF, Sudheimer K, Keller J, Mayberg HS, Gunning FM, Alexopoulos GS, Fox MD, Pascual-Leone A, Voss HU, Casey B, Dubin MJ, Liston C, Resting-state connectivity biomarkers define neurophysiological subtypes of depression, Nat Med. 23 (2017) 28–38. 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ivleva EI, Clementz BA, Dutcher AM, Arnold SJM, Jeon-Slaughter H, Aslan S, Witte B, Poudyal G, Lu H, Meda SA, Pearlson GD, Sweeney JA, Keshavan MS, Tamminga CA, Brain Structure Biomarkers in the Psychosis Biotypes: Findings From the Bipolar-Schizophrenia Network for Intermediate Phenotypes, Biological Psychiatry. 82 (2017) 26–39. 10.1016/j.biopsych.2016.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, Keshavan MS, Tamminga CA, Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers, AJP. 173 (2016) 373–384. 10.1176/appi.ajp.2015.14091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Barch DM, Biotypes: Promise and Pitfalls, Biological Psychiatry. 82 (2017) 2–3. 10.1016/j.biopsych.2017.04.012. [DOI] [PubMed] [Google Scholar]
- [31].Liu Y, Hayes DN, Nobel A, Marron JS, Statistical Significance of Clustering for High-Dimension, Low–Sample Size Data, Journal of the American Statistical Association. 103 (2008) 1281–1293. 10.1198/016214508000000454. [DOI] [Google Scholar]
- [32].Cuthbert BN, Insel TR, Toward the future of psychiatric diagnosis: the seven pillars of RDoC, BMC Med. 11 (2013) 126 10.1186/1741-7015-11-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, Sanislow C, Wang P, Research Domain Criteria (RDoC): Toward a New Classification Framework for Research on Mental Disorders, AJP. 167 (2010) 748–751. 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- [34].Caspi A, Houts RM, Belsky DW, Goldman-Mellor SJ, Harrington H, Israel S, Meier MH, Ramrakha S, Shalev I, Poulton R, Moffitt TE, The p Factor: One General Psychopathology Factor in the Structure of Psychiatric Disorders?, Clinical Psychological Science. 2 (2014) 119–137. 10.1177/2167702613497473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Moore TM, Calkins ME, Satterthwaite TD, Roalf DR, Rosen AFG, Gur RC, Gur RE, Development of a computerized adaptive screening tool for overall psychopathology (“p”), Journal of Psychiatric Research. 116 (2019) 26–33. 10.1016/j.jpsychires.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Elliott ML, Romer A, Knodt AR, Hariri AR, A Connectome-wide Functional Signature of Transdiagnostic Risk for Mental Illness, Biological Psychiatry. 84 (2018) 452–459. 10.1016/j.biopsych.2018.03.012.* This study combined a dimensional model of psychopathology with connectome-wide association analysis to demonstrate that dysconnectivity between the visual cortex and higher-order executive systems was associated with greater scores on the p-factor.
- [37].Kebets V, Holmes AJ, Orban C, Tang S, Li J, Sun N, Kong R, Poldrack RA, Yeo BTT, Somatosensory-Motor Dysconnectivity Spans Multiple Transdiagnostic Dimensions of Psychopathology, Biological Psychiatry. 86 (2019) 779–791. 10.1016/j.biopsych.2019.06.013.** This study used partial least squares to demonstrate that dysconnectivity within lower-order cortical systems and between lower- and higher-order systems may be a common biomarker across many mental disorders.
- [38].Xia CH, Ma Z, Ciric R, Gu S, Betzel RF, Kaczkurkin AN, Calkins ME, Cook PA, García de la Garza A, Vandekar SN, Cui Z, Moore TM, Roalf DR, Ruparel K, Wolf DH, Davatzikos C, Gur RC, Gur RE, Shinohara RT, Bassett DS, Satterthwaite TD, Linked dimensions of psychopathology and connectivity in functional brain networks, Nature Communications. 9 (2018). 10.1038/s41467-018-05317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Feczko E, Fair DA, Methods and Challenges for Assessing Heterogeneity, Biological Psychiatry. (2020) S0006322320301104 10.1016/j.biopsych.2020.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sabaroedin K, Tiego J, Parkes L, Sforazzini F, Finlay A, Johnson B, Pinar A, Cropley V, Harrison BJ, Zalesky A, Pantelis C, Bellgrove M, Fornito A, Functional Connectivity of Corticostriatal Circuitry and Psychosis-like Experiences in the General Community, Biological Psychiatry. 86 (2019) 16–24. 10.1016/j.biopsych.2019.02.013. [DOI] [PubMed] [Google Scholar]
- [41].Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity, Nat Neurosci. 18 (2015) 1664–1671. 10.1038/nn.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kaufmann T, Alnæs D, Doan NT, Brandt CL, Andreassen OA, Westlye LT, Delayed stabilization and individualization in connectome development are related to psychiatric disorders, Nature Neuroscience. 20 (2017) 513–515. 10.1038/nn.4511.* This study showed that abnormal patterns of connectome distinctiveness throughout development was common to multiple mental disorders, and that this effect was concentrated in higher-order systems.
- [43].Horien C, Shen X, Scheinost D, Constable RT, The individual functional connectome is unique and stable over months to years, NeuroImage. 189 (2019) 676–687. 10.1016/j.neuroimage.2019.02.002.** This study demonstrated that individual’s unique functional fingerprints were stable over months to years, and that higher-order systems drove this uniqueness to a greater extent than lower-order systems.
- [44].Miranda-Dominguez O, Feczko E, Grayson DS, Walum H, Nigg JT, Fair DA, Heritability of the human connectome: A connectotyping study, Network Neuroscience. 2 (2018) 175– 199. 10.1162/netn_a_00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Demeter DV, Engelhardt LE, Mallett R, Gordon EM, Nugiel T, Harden KP, Tucker-Drob EM, Lewis-Peacock JA, Church JA, Functional Connectivity Fingerprints at Rest Are Similar across Youths and Adults and Vary with Genetic Similarity, IScience. 23 (2020) 100801 10.1016/j.isci.2019.100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang X, Braun U, Tost H, Bassett DS, Data-Driven Approaches to Neuroimaging Analysis to Enhance Psychiatric Diagnosis and Therapy, Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. (2020) S2451902219303556 10.1016/j.bpsc.2019.12.015. [DOI] [PubMed] [Google Scholar]
- [47].Wolfers T, Doan NT, Kaufmann T, Alnæs D, Moberget T, Agartz I, Buitelaar JK, Ueland T, Melle I, Franke B, Andreassen OA, Beckmann CF, Westlye LT, Marquand AF, Mapping the Heterogeneous Phenotype of Schizophrenia and Bipolar Disorder Using Normative Models, JAMA Psychiatry. 75 (2018) 1146 10.1001/jamapsychiatry.2018.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wolfers T, Beckmann CF, Hoogman M, Buitelaar JK, Franke B, Marquand AF, Individual differences v. the average patient: mapping the heterogeneity in ADHD using normative models, Psychological Medicine. (2019) 1–10. 10.1017/S0033291719000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zabihi M, Oldehinkel M, Wolfers T, Frouin V, Goyard D, Loth E, Charman T, Tillmann J, Banaschewski T, Dumas G, Holt R, Baron-Cohen S, Durston S, Bölte S, Murphy D, Ecker C, Buitelaar JK, Beckmann CF, Marquand AF, Dissecting the Heterogeneous Cortical Anatomy of Autism Spectrum Disorder Using Normative Models, Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. (2018). 10.1016/j.bpsc.2018.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Floris DL, Wolfer T, Zabihi M, Holz NE, Zwiers MP, Charman T, Tillmann J, Ecker C, Dell’Acqua F, Banaschewski T, Moessnang C, Baron-Cohen S, Holt R, Durston S, Loth E, Murphy DGM, Marquand A, Buitelaar JK, Beckmann CF, Atypical brain asymmetry in autism - a candidate for clinically meaningful stratification, Neuroscience, 2020. 10.1101/2020.03.24.000349. [DOI] [PubMed] [Google Scholar]
- [51].Mills KL, Goddings A-L, Herting MM, Meuwese R, Blakemore S-J, Crone EA, Dahl RE, Güroğlu B, Raznahan A, Sowell ER, Tamnes CK, Structural brain development between childhood and adulthood: Convergence across four longitudinal samples, NeuroImage. 141 (2016) 273–281. 10.1016/j.neuroimage.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tamnes CK, Herting MM, Goddings A-L, Meuwese R, Blakemore S-J, Dahl RE, Güroğlu B, Raznahan A, Sowell ER, Crone EA, Mills KL, Development of the Cerebral Cortex across Adolescence: A Multisample Study of Inter-Related Longitudinal Changes in Cortical Volume, Surface Area, and Thickness, J. Neurosci. 37 (2017) 3402–3412. 10.1523/JNEUROSCI.3302-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kaufmann T, Alnæs D, Brandt CL, Bettella F, Djurovic S, Andreassen OA, Westlye LT, Stability of the Brain Functional Connectome Fingerprint in Individuals With Schizophrenia, JAMA Psychiatry. 75 (2018) 749 10.1001/jamapsychiatry.2018.0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bassett DS, Xia CH, Satterthwaite TD, Understanding the Emergence of Neuropsychiatric Disorders With Network Neuroscience, Biological Psychiatry: Cognitive Neuroscience and Neuroimaging. 3 (2018) 742–753. 10.1016/j.bpsc.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Braun U, Schaefer A, Betzel RF, Tost H, Meyer-Lindenberg A, Bassett DS, From Maps to Multi-dimensional Network Mechanisms of Mental Disorders, Neuron. 97 (2018) 14–31. 10.1016/j.neuron.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].van den Heuvel MP, Sporns O, Network hubs in the human brain, Trends in Cognitive Sciences. 17 (2013) 683–696. 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- [57].Sporns O, Honey CJ, Kötter R, Identification and Classification of Hubs in Brain Networks, PLoS ONE. 2 (2007) e1049 10.1371/journal.pone.0001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lurie DJ, Kessler D, Bassett DS, Betzel RF, Breakspear M, Kheilholz S, Kucyi A, Liégeois R, Lindquist MA, McIntosh AR, Poldrack RA, Shine JM, Thompson WH, Bielczyk NZ, Douw L, Kraft D, Miller RL, Muthuraman M, Pasquini L, Razi A, Vidaurre D, Xie H, Calhoun VD, Questions and controversies in the study of time-varying functional connectivity in resting fMRI, Network Neuroscience. 4 (2020) 30–69. 10.1162/netn_a_00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Braun U, Schäfer A, Bassett DS, Rausch F, Schweiger JI, Bilek E, Erk S, Romanczuk-Seiferth N, Grimm O, Geiger LS, Haddad L, Otto K, Mohnke S, Heinz A, Zink M, Walter H, Schwarz E, Meyer-Lindenberg A, Tost H, Dynamic brain network reconfiguration as a potential schizophrenia genetic risk mechanism modulated by NMDA receptor function, Proc Natl Acad Sci USA. 113 (2016) 12568–12573. 10.1073/pnas.1608819113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Cui Z, Li H, Xia CH, Larsen B, Adebimpe A, Baum GL, Cieslak M, Gur RE, Gur RC, Moore TM, Oathes DJ, Alexander-Bloch AF, Raznahan A, Roalf DR, Shinohara RT, Wolf DH, Davatzikos C, Bassett DS, Fair DA, Fan Y, Satterthwaite TD, Individual Variation in Functional Topography of Association Networks in Youth, Neuron. 106 (2020) 340–353.e8. 10.1016/j.neuron.2020.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gratton C, Kraus BT, Greene DJ, Gordon EM, Laumann TO, Nelson SM, Dosenbach NUF, Petersen SE, Defining Individual-Specific Functional Neuroanatomy for Precision Psychiatry, Biological Psychiatry. (2019) S0006322319318293 10.1016/j.biopsych.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Gratton C, Smith DM, Dorn M, Digging Deeper to Chart the Landscape of Human Brain Development, Neuron. 106 (2020) 209–211. 10.1016/j.neuron.2020.03.030. [DOI] [PubMed] [Google Scholar]
- [63].Salehi M, Greene AS, Karbasi A, Shen X, Scheinost D, Constable RT, There is no single functional atlas even for a single individual: Functional parcel definitions change with task, NeuroImage. 208 (2020) 116366 10.1016/j.neuroimage.2019.116366. [DOI] [PubMed] [Google Scholar]
- [64].Mahadevan AS, Tooley UA, Bertolero MA, Mackey AP, Bassett DS, Evaluating the sensitivity of functional connectivity measures to motion artifact in resting-state fMRI data, Neuroscience, 2020. 10.1101/2020.05.04.072868. [DOI] [PubMed] [Google Scholar]
- [65].Horien C, Noble S, Finn ES, Shen X, Scheinost D, Constable RT, Considering factors affecting the connectome-based identification process: Comment on Waller et al., NeuroImage. 169 (2018) 172–175. 10.1016/j.neuroimage.2017.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Waller L, Walter H, Kruschwitz JD, Reuter L, Müller S, Erk S, Veer IM, Evaluating the replicability, specificity, and generalizability of connectome fingerprints, NeuroImage. 158 (2017) 371–377. 10.1016/j.neuroimage.2017.07.016. [DOI] [PubMed] [Google Scholar]
- [67].Mitchell SM, Lange S, Brus H, Gendered Citation Patterns in International Relations Journals, Int Stud Perspect. 14 (2013) 485–492. 10.1111/insp.12026. [DOI] [Google Scholar]
- [68].Maliniak D, Powers R, Walter BF, The Gender Citation Gap in International Relations, Int Org. 67 (2013) 889–922. 10.1017/S0020818313000209. [DOI] [Google Scholar]
- [69].Caplar N, Tacchella S, Birrer S, Quantitative evaluation of gender bias in astronomical publications from citation counts, Nat Astron. 1 (2017) 0141 10.1038/s41550-017-0141. [DOI] [Google Scholar]
- [70].Dion ML, Sumner JL, Mitchell SM, Gendered Citation Patterns across Political Science and Social Science Methodology Fields, Polit. Anal. 26 (2018) 312–327. 10.1017/pan.2018.12. [DOI] [Google Scholar]
- [71].Dworkin JD, Linn KA, Teich EG, Zurn P, Shinohara RT, Bassett DS, The extent and drivers of gender imbalance in neuroscience reference lists, Nat Neurosci. 23 (2020) 918–926. 10.1038/s41593-020-0658-y. [DOI] [PubMed] [Google Scholar]
- [72].Zhou D, Cornblath EJ, Stiso J, Teich EG, Dworkin JD, Blevins AS, Bassett DS, Gender Diversity Statement and Code Notebook v1.0, (2020). 10.5281/zenodo.3672110. [DOI] [Google Scholar]
- [73].Ambekar A, Ward C, Mohammed J, Male S, Skiena S, Name-ethnicity classification from open sources, in: Proceedings of the 15th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining - KDD ‘09, ACM Press, Paris, France, 2009: p. 49 10.1145/1557019.1557032. [DOI] [Google Scholar]
- [74].Sood G, Laohaprapanon S, Predicting Race and Ethnicity From the Sequence of Characters in a Name, ArXiv:1805.02109 [Stat]. (2018). http://arxiv.org/abs/1805.02109 (accessed September 25, 2020). [Google Scholar]