Abstract
Affinity maturation of U33, a recombinant Fab inhibitor of uPA, was used to improve the affinity and the inhibitory effect compared to the parental Fab. Arginine scanning of the six CDR loops of U33 was done to identify initial binding determinants since uPA prefers arginine in its primary substrate binding pocket. Two CDR loops were selected to create an engineered affinity maturation library of U33 that was diversified around ArgL91 (CDR L3) and ArgH52 (CDR H2). Biopanning of the randomized U33 library under stringent conditions resulted in eight Fabs with improved binding properties. One of the most potent inhibitors, AB2, exhibited a 13-fold decrease in IC50 when compared to U33 largely due to a decrease in its off rate. To identify contributions of interfacial residues that might undergo structural rearrangement upon interface formation we used X-ray footprinting and mass spectrometry (XFMS). Four residues showed a pronounced decrease in solvent accessibility, and their clustering suggests that AB2 targets the active site and also engages residues in an adjacent pocket unique to human uPA. The 2.9 Å resolution crystal structure of AB2-bound to uPA shows a binding mode in which the CDR L1 loop inserts into the active site cleft and acts as a determinant of inhibition. The selectivity determinant of this binding mode is unlike previously identified inhibitory Fabs against uPA related serine proteases, MTSP-1, HGFA and FXIa. CDRs H2 and L3 loops aid in interface formation and provide critical salt-bridges to remodel loops surrounding the active site of uPA providing specificity and further evidence that antibodies can be potent and selective inhibitors of proteolytic enzymes.
Introduction
Proteases make up 2% of the human proteome [1], and their functional activity is a unique, irreversible post-translational modification of their target substrates [2]. Proteases are involved in a number of important physiological functions from blood coagulation to digestion and pathophysiological activities necessary for activation and propagation of cancer cascades and Alzheimer’s disease [3]. More information regarding the biology and pathophysiology of proteolysis in addition to new selective therapeutics are important for understanding and treating human disease.
The omnipresence of proteases in cancer make them a promising class of enzymes for cancer detection [4] and also as activators of prodrugs [5]. Proteolytic processing is necessary in nearly every stage of cancer growth and progression including angiogenesis, extracellular matrix remodeling, cellular signaling, apoptosis and metastasis [6]. Changes in relative levels of proteases or their cognate inhibitors are often associated with cancer, suggesting that dysregulation of proteolysis can contribute to malignant growth. Although localization of proteases can be studied using various experimental approaches, understanding and determining protease activity in particular pathways cannot be achieved with simple labels of protease expression. Since proteolytic activity is finely regulated through activation, compartmentalization, changes in pH and also interactions with endogenous inhibitors, methods to selectively monitor their activity in vivo are needed.
Studying individual proteases and the particular role that each play in different physiological and disease pathways has been a challenging endeavor without specific probes. Attempts to create selective small molecule tools for proteases have been difficult and are oftentimes without additional non-specific inhibition of other protease targets due to the general conserved structural features of the protease domain and active site. Antibodies (Abs) are ideal tools for selectively targeting and studying individual proteases even within the same family since Abs can generate greater binding footprints against targets than small molecules. In addition, conformationally selective antibodies that recognize the active form of the enzyme over its inactive state provide a powerful approach to monitor enzyme activity in vivo [7,8]. The development of specific and inhibitory Abs allows pinpointing of the protease of interest to determine what effect protease activity has in the protease signaling pathway. Additionally, these inhibitory Abs provide much-needed preclinical tools for validating proteases as potential targets for pharmaceuticals in human diseases.
Well-validated and specific Abs are scarce since many commercially available antibodies suffer from lot-to-lot variation and provide varying results [9]. While hybridoma technology that involves animal immunization is often productive, many antigen targets do not yield useful Abs, particularly if unique states or critical non-immunogenic sequences need to be recognized by the Abs. Renewable Abs from a recombinant source that are highly reproducible have the potential to increase the access to high quality, reproducible Ab reagents that target specific states. We provided initial proof of principle for identifying and characterizing recombinant Abs (rAbs) that specifically bind and inhibit particular protease targets. Our results for protease production and identification of rAbs to MT-SP1 (aka, matriptase), and urokinase plasminogen activator (uPA) showed that conformationally selective inhibitory antibodies can be identified from a naïve human Fab library displayed on bacteriophage. Several inhibitory antibodies have been developed to other proteases including HGFA [10], Factor XIa [11], MMP9 [12] and β-tryptase [13]. Another advantage of rAbs is that the antibody sequence of the binder allows for in vitro affinity maturation, mimicking an immune response, to select Abs with higher affinity and lower off-rate [14].
Accurately imaging metastatic prostate cancer in soft tissue remains an unmet clinical need. The plasminogen activation system (PAS) consists of the serine protease urokinase plasminogen activator (uPA), the membrane-localized receptor uPAR and the SERPIN-type inhibitor PAI-1. All three are overexpressed in primary and metastatic prostate cancer. Plasmin-mediated activation of the secreted uPA precursor pro-uPA cascades a series of events resulting in dissolution of the cellular membrane. Cleavage of PAI-1 inactivates uPA and leads to internalization of the entire uPA-uPAR-PAI-1 complex. Previously an active-site binding recombinant antibody termed U33 (Supplemental Figure 1) was discovered by phage-display panning against the uPA protease domain using a fully human naïve Fab library [15]. U33 mimics PAI-1 because it both inhibits uPA and is internalized with the uPA-uPAR complex. However, the fast off-rate of U33 (Supplemental Figure 1B) made structural characterization of the U33-uPA complex difficult. Various rAbs we previously isolated against MTSP-1 were shown to have differing molecular mechanisms of inhibition, which motivated us to uncover how an antibody could inhibit uPA. Therefore, we initiated an affinity maturation campaign to isolate antibodies with better binding kinetics to enable structural studies.
Here we describe the discovery and the mechanistic properties of the inhibitory Fab, AB2. A series of affinity matured Fabs were generated, derived from the parental U33 Fab. After characterization of binding affinity and steady-state inhibition kinetics towards uPA, AB2 was chosen from the affinity matured pool for further structural characterization using XFMS and X-ray crystallography.
Results
Affinity maturation
An anti-uPA Fab, U33, was previously identified from a fully human naïve Fab phage display library. U33 Fab is a competitive inhibitor of uPA with a KD value of 300 nM [15].
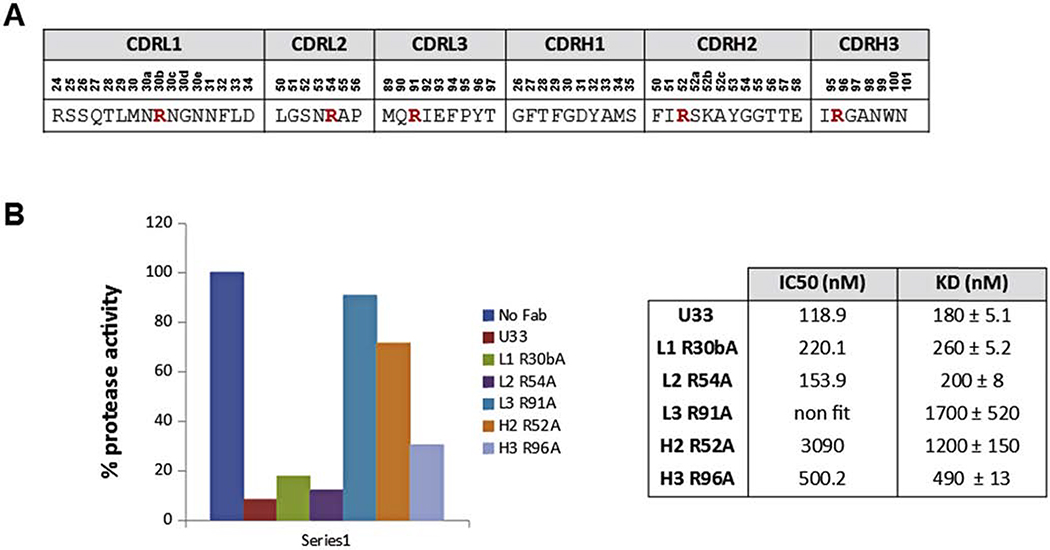
The identification of the U33 Fab loops involved in binding and/or inhibition of uPA is crucial for successful affinity maturation; the arginine residues in the CDRs loops of Fab U33 were substituted individually by alanine and a panel of U33 alanine mutants were generated. Arg residues were selected because uPA is a trypsin-like serine protease with a preference for basic residues in the P1 position of the substrate. Furthermore, in the previously described anti-MT-SP1 rAbs, Arg residues in the CDRH3 loop are essential for the inhibition of the protease. Based on the KD and IC50 values of the generated U33 mutants, the CDRL3 and CDRH2 loops were chosen as targets for affinity improvement of Fab U33 (Figure 1).
Figure 1: Identification of U33 Fab loops involved in binding and/or inhibition of uPA.
A. Sequence of the CDR loops of Fab U33, the arginine residues mutated to alanine are shown in red. B. Effect of U33 point-mutations on the inhibition of uPA protease activity. The proteolytic activity of human uPA was read in absence and presence of 1 μM of each U33 alanine mutants. The enzyme activity is expressed as percentage of the uPA activity in absence of Fab (100 %). Enzymatic assays were done in triplicate with all standard deviations being 10% or less of the reported values. IC50 values were calculated on the GraphPad Software 5 using a non linear log(inhibitor) vs. response fitting.
A phage displayed, affinity maturation library was constructed by randomizing the CDRH2 and CDRL3 loops of the U33 parental Fab. The theoretical diversity of the library was 1.45 × 1015, and the constructed library had a diversity of ~ 9 × 109. Semi-random degenerate codons were introduced that encode the original amino acid plus a small set of residues in each of the positions mutated [16,17]. The arginine residue in the two loops was not mutated as the mutation of these residues to alanine reduces the inhibitory effect of the parental antibody U33.
After three rounds of off-rate selections [18] with the generated affinity maturation library using the catalytic domain of uPA as the antigen, 188 individual clones were screened by ELISA. Eleven clones showed higher ELISA signal than the parental U33 Fab. A preliminary off-rate analysis prior to antibody purification, using Escherichia coli supernatants, was performed by biolayer interferometry (BLI) using an Octet RED384 instrument [19]. All clones showed slower dissociation rates than Fab U33 (data not shown).
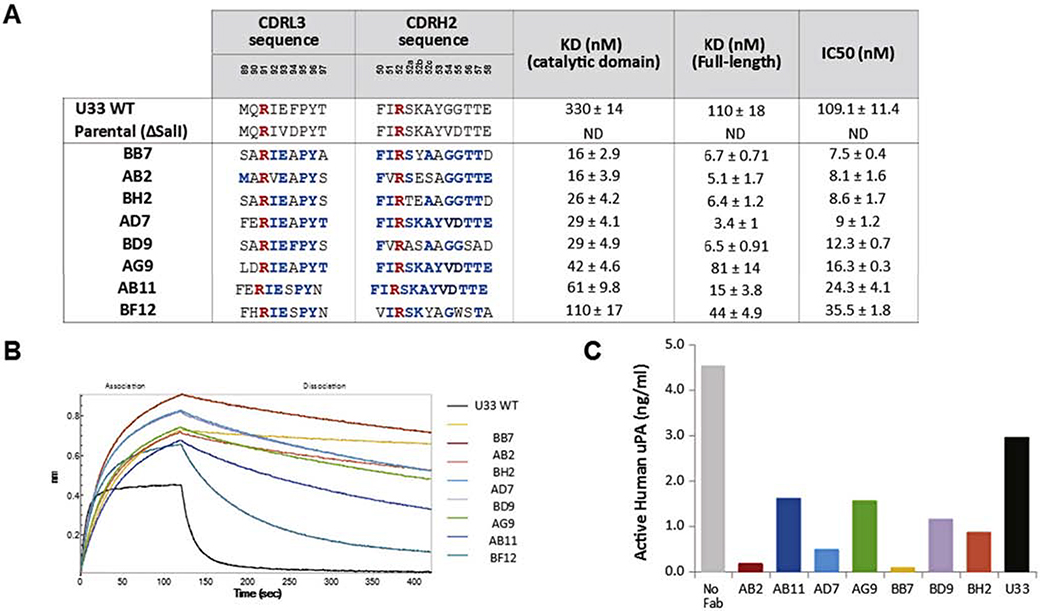
Sequencing of the positive clones identified only eight unique Fab sequences (Figure 2A). Sequence analysis showed some positions highly conserved in all the identified clones: the isoleucine in position L91, the isoleucine in position H51, the glycines in positions H54 and H55 and the threonines in positions H56 and H57. Some amino acids were totally conserved in all the clones: the glutamic acid in position L93, the proline in position L95 and the tyrosine in position L96 indicating an essential role of these residues in the binding to uPA.
Figure 2. Characterization of U33 affinity matured Fabs.
A. Sequences of CDRL3 and CDRH2 loops of affinity maturation Fabs: residues not mutated are shown in red and residues identical to the parental U33 Fab are shown in blue. The KD values of affinity matured Fabs values for Fabs were determined by bioluminescence interferometry using an Octet RED384 instrument. Samples were run in triplicate and IC50 values were calculated on the GraphPad Software 5 using a non linear log (inhibitor) vs. response fitting. B. BLI traces of U33 affinity matured Fabs compared with U33 parental Fab. C. U33 affinity matured Fabs prevent uPA binding to PAI-1 coated plates: 1μM of each Fab was pre-incubated overnight with active human uPA (5 ng/mL) and added to PAI-1 coated plates. All samples were run in triplicate. The amount of uPA bound to PAI was determined by ELISA and standard deviations were 10% or less of the reported values.
The eight unique clones were expressed, purified, and tested for binding to uPA (full-length and catalytic domain) and also tested for inhibition of uPA. All the identified clones showed off-rate improvements over the parental U33 Fab, therefore all of them have improved KD values. All the clones also showed more potent inhibition of uPA compared with parental U33 Fab (Figure 2A). All the affinity matured Fabs block binding of uPA to its endogenous inhibitor PAI-1 more potently than parental U33 Fab (Figure 2C). Clones BB7 and AB2 had the lowest KD to the catalytic domain of uPA, and because AB2 had a lower KD to full-length uPA between the two, it was selected for crystallization in complex with the catalytic domain of uPA.
Mapping the binding epitope of AB2 Using XFMS
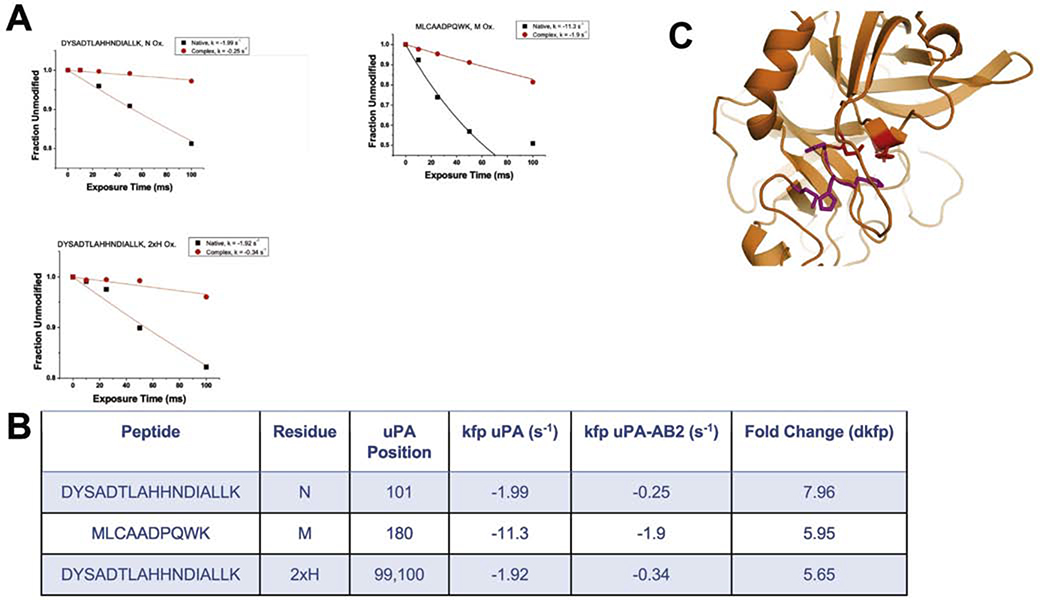
Comparative analysis of XFMS data from the uPA-AB2 complex and uPA alone suggests that AB2 interacts with the uPA active site and engages other residues that are not conserved in similar proteases. The rate constant of footprinting (kfp) was identified for 11 residues distributed on the uPA surface (Supplementary Figure 4). From a pattern of protection from radical attack, four residues, His99, His100, Asn101, and Met180 were selected as possible participants in the uPA-AB2 binding interface because they had the highest change in kfp (dkfp) and clustered near the uPA active site (Figure 3B). The dose-response plots for these residues (Figure 3A) demonstrate that they are dramatically protected from modification when AB2 is bound to uPA. A crystal structure of the AB2-uPA complex was solved to elucidate a potentially novel mode of inhibition.
Figure 3: Mapping of the binding epitope of AB2 using XFMS.
A. Dose-response graphs of the four residues with highest dkfp for AB2-uPA complex (red) and native uPA (black). Oxidation of His99 and His100 was detected on the same peptide. B. Residues from 3A (pink) mapped to structure of uPA and residues forming the catalytic triad (red).
Mode of inhibition - Structural Determinants
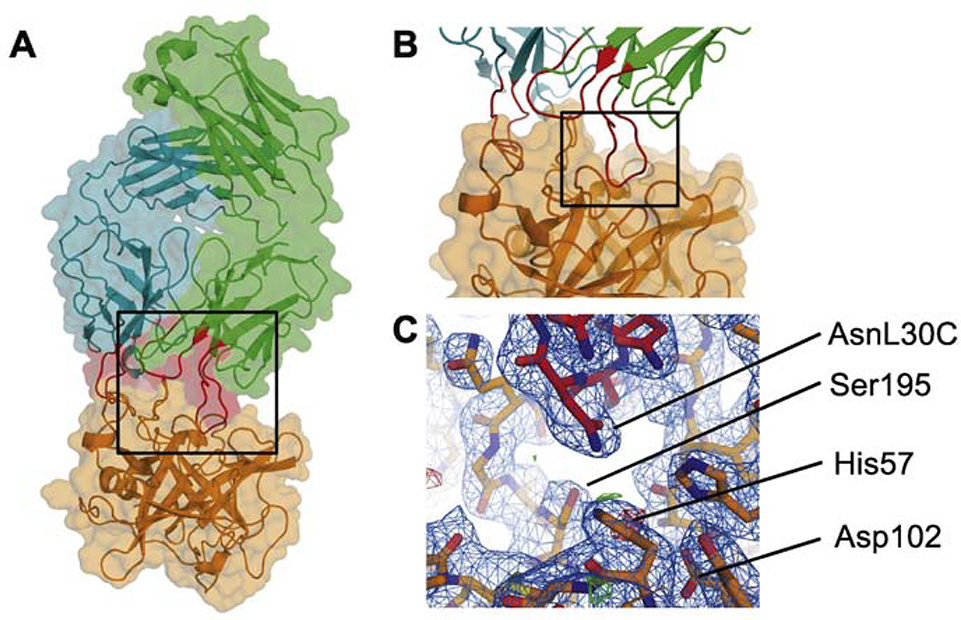
Modes of inhibition exhibited by antibodies against serine proteases fall into two broad subcategories: allosteric or active-site directed. The uPA-AB2 co-crystal structure revealed that AB2 recognizes a surface-contiguous, three-dimensional epitope encompassing the active site of uPA (Figure 4A). Critical contacts are made by light-chain variable loops CDR L1 and CDR L3 as well as heavy-chain loop CDR H2 (Figure 4B). The resulting interface covers a surface area of 564 Å2 on uPA. The interface involves five distinct elements of secondary structure on the uPA surface. Residues from loop 90–104, loop 168–179, loop 185–197, loop 215–225 and His57 are directly involved in forming an intermolecular interface.
Figure 4: Co-crystal structure of AB2-uPA complex.
A. Cartoon and surface representation of the AB2-uPA complex. Region emphasized by black square shown in B. B. Close-up of the AB2-uPA interface with critical loops CDRL1, CDRL2 and CDRH3 highlighted in red. Region emphasized by black square shown in C. C. Electron density and stick representation of the interface between the inhibitory loop CDRL1 (specifically residue Asn30C) and the catalytic site of uPA.
In the substrate binding pocket, the L1 loop inserts in a substrate-like manner and occupies the S1 subsite using ArgL30B (Figure 5A, lower panel). The active-site carboxyl oxygen donated by Ser195 is 5.7Å removed from the carbonyl carbon of what might serve as the scissile bond in a bound substrate, rendering the enzyme unable to catalyze proteolysis. A system consisting of adjacent asparagine residues in CDR L1 positions amine side chains as a main chain surrogate, enabling donation of a hydrogen bond by AsnL30C to His57 and Ser195 (Figure 5A). In this unique mode of inhibition, the gamma-carbon of AsnL30C, being only 4.7 Å removed from the carboxyl oxygen of Ser195, appears to serve as a surrogate for the carbonyl carbon. uPA residue Asp102, which is part of the catalytic triad and normally serves as a proton acceptor during substrate cleavage, is not directly involved in interface formation.
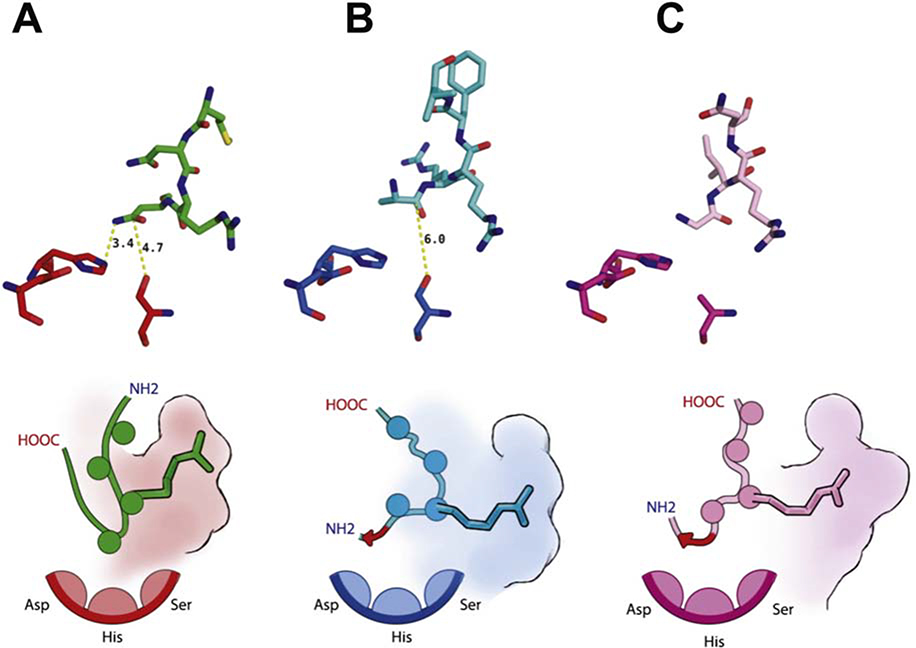
Figure 5: Modes of serine protease inhibition exhibited by active-site targeting antibody fragments.
A. Inhibitory loop L1 of the Fab AB2 shown in relation to the catalytic triad of uPA. B. Inhibitory loop H3 of the Fab A11 shown in relation to the catalytic triad of MT-SP1. Illustration highlighting the reverse binding motif. C. Inhibitory loop L1 of the Fab DEF shown in relation to the catalytic triad of FXIa. Schematic illustration highlighting conserved placement of Arginine residues in S1 subsite and the reverse binding motifs (red arrows). In all panels the catalytic triad comprises residues His57, Asp102 and Ser195.
Effect of Affinity Maturation – Determinants of Specificity
Select residues in CDR L3 and CDR H2 aid in extending the binding surface beyond the substrate binding pocket. The crystal structure of uPA-AB2 provides additional insights into the effects of affinity maturation. Affinity maturation did not introduce new residues making interactions and expanding the binding surface but instead aids in optimization of contacts made by CDRs L3 and H2 and truncation of residues with bulky side-chains relative to the ancestral Fab U33. Residues GluL93 in CDR L3 and ArgH52 in CDR H2 form critical contacts with uPA residues Arg217 and Glu175, respectively. Those interactions remained conserved through the process of affinity maturation. In U33 CDR L3, GluL93 is flanked by PheL94, a rather bulky side chain that needs to be accommodated within the interface. During affinity maturation AlaL94 was selected to replace PheL94, increasing flexibility in positioning of GluL93. A similar observation can be made in CDR H2, where the loop containing ArgH52 is centered around TyrH53 which was selected against in favor of AlaH53.
Discussion
There is a significant interest in specific antibodies to proteases based on the success of anti-proteolytic agents in the clinic and the lack of specific small molecule drugs and reliable western blot and antibody reagents. Identifying inhibitory rAbs is often a bottleneck and novel selection methodology is aiming to specifically identify functionally relevant candidates. Using one of our recombinant antibody phage-displayed libraries, we have identified and characterized a number of antibody binders and inhibitors of protease targets and protease receptors. The development of specific and inhibitory rAbs allows pinpointing of the protease of interest to determine what effect protease activity has in the protease signaling pathway. Additionally, these inhibitory rAbs have potential to be translated into much-needed clinical therapeutics in human diseases.
The urokinase plasminogen activator (uPA) system is functionally involved in many steps in cancer progression. The over-expression of uPA and uPAR has been documented in numerous cancer types and is highly associated with poor disease prognosis [20,21]. uPA is a diagnostic marker for breast and prostate cancers. Inhibition of uPA was proposed as an efficient strategy for cancer treatments [22].
The development of potent and selective inhibitors of the serine protease domain of urokinase has been a difficult process [23]. Several uPA inhibitors have been identified as potential treatment of cancer and other pathologies, mainly small molecule uPA inhibitors (amino acid derivatives and small peptides) [22–25]. We have used several strategies to identify specific inhibitory antibodies of uPA. Based on previously identified potent antibody inhibitors against serine proteases [26,27], and matrix metallopeptidases (MMPs) [12], where the main driver of inhibition is the long CDRH3 loop inserted into the protease active site [27,28], we designed a set of synthetic antibody libraries based on the long CDRH3 loops of two potent MTSP-1 inhibitory Fabs (Unpublished results). Several uPA antibodies were identified using these biased antibody libraries but none showed an inhibitory effect, and some behaved as substrates of urokinase (Supplemental Figure 2).
We also used a fully human naïve Fab phage display library as a source of antibodies and several Fabs were identified with only one being an inhibitor of uPA namely, Fab U33. U33 selectively binds the active form of human uPA and is a potent inhibitor of soluble uPA and uPA bound to uPAR. This anti-uPA antibody prevents uPA binding to PAI-1 and after binding to uPA the uPA-U33 complex internalizes. The IgG form of U33 was shown to be useful tool for detecting active uPA in vivo [15]. The analysis of the Fab U33 sequence showed that the CDRH3 loop is shorter than the CDRH3 loop of previously identified serine protease inhibitors (Supplemental Figure 1A) suggesting that the mechanism of inhibition could be different.
Since structural efforts with U33 and uPA were thwarted by non-diffracting crystals, we generated an affinity matured U33 antibody to improve its off-rate with the hopes of stabilizing the complex. The H2 and L3 CDRs loops were shown to be critical to the binding and inhibition of uPA in single point mutation experiments. Since uPA prefers arginine in the S1 site we focused our mutational analysis on these residues. Mutation of the arginine residue in these loops showed increasing KD values and a concomitant decrease of the inhibitory effect in the variants generated (Figure 1). One affinity matured library with randomized H2 and L3 CDR loops was constructed. After three rounds of affinity-driven selections using the biotinylated catalytic domain of uPA and with off-rate washes of 1–5 hours with the non- biotinylated catalytic domain of uPA, eight unique Fabs with slower dissociation rates and lower KD values than the parental Fab U33 were identified. All of them are more potent inhibitors of uPA (Figure 2). All the affinity matured Fabs prevent uPA binding to PAI-1 coated plates as the parental U33 Fab, indicating that the mechanism of action would be the same (Figure 2C).
Comparison with other inhibitory antibodies
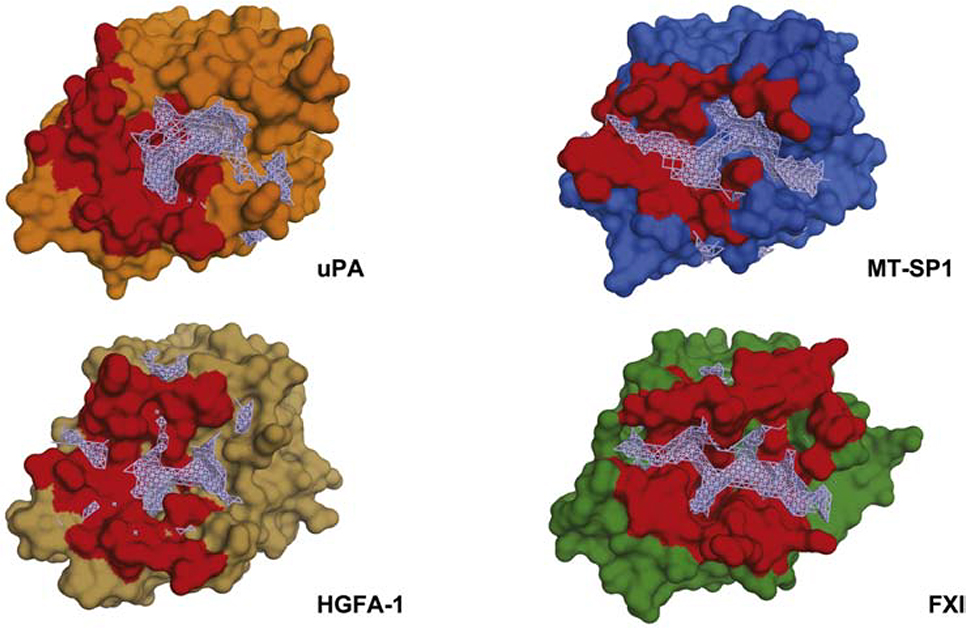
In recent years several crystal structures of antibody fragments inhibiting serine proteases in an active-site directed manner have been determined. Comparing buried surface areas among different complexes of serine proteases and active-site directed inhibitory antibody fragments reveals that contributions made by heavy and light chains form a surface-contiguous interface (Figure 6). The largest interface by buried surface area is made between FXIa and the inhibitory Fab called DEF (976.4 Å2 and 276.6 Å2 made by the light and heavy chains, respectively). The comparatively small surface area covered by AB2 correlates with the weaker binding affinity exhibited in biophysical assays when compared with other inhibitory antibody fragments. A comparative analysis of the druggable pocket shape between uPA, MT-SP1, HGFA1 and FXIa substrate binding sites allows assessment of variations in modes of inhibiting different serine proteases (Figure 6). While the overall fold between the proteases is conserved, the pockets surrounding the active sites differ in shape, markedly. For all proteases except uPA, the pockets are almost fully enclosed within the binding epitope of the respective inhibitory antibody fragment. The pocket analysis shows all other proteases besides uPA feature a contiguous groove spanning the active site, yet in the case of uPa is interrupted by a ridge. XFMS revealed four residues in distinct pockets separated by this ridge. This suggested a binding mode bridging critical determinants across both pockets. A notable exception to this active-site directed paradigm is the anti-uPA Fab-112, which was shown by Jiang et al. [29] to “rezymogenyze” uPA by closing its active site via an allosteric rearrangement. Shared between binding modes of the different complexes is occupation of the S1 subsite with an arginine in a substrate-like manner. Both, the anti-MT-SP1 fragment A11 as well as the anti-FXIa fragment DEF use a reverse binding motif to present the putative scissile bond in an inverted configuration (see Figure 5). The main difference in modes of inhibition between A11 and DEF is in the use of loop H3 and L1 to present the reverse binding motif, respectively. Similar to DEF, AB2 uses loop L1, but presents it in a substrate-like but non-cleavable configuration. Instead of a scissile bond, an asparagine head group serves as a surrogate peptide backbone to engage the catalytic triad non-productively.
Figure 6: Epitopes targeted by active-site directed inhibitory antibody fragments and their relation to druggable pockets.
Binding epitopes shown in red (defined as protease residues within 4 Å of the antibody fragment), druggable pockets centered on S1 substrate pocket identified using sitemap shown in wireframe (grey). PDB codes are 3SO3, 2R0K and 6AOD.
The structure of AB2 bound to uPA provides an understanding at the atomic level of how antibodies can use different mechanisms to inhibit proteolytic enzymes. Recombinant antibodies are versatile and specific binders and by showcasing another example of an anti-proteolytic antibody, it is clear that a variety of CDR loops can be employed to inactivate proteases. Whether it is uPA or other members of this class, these principles can be used to design highly selective inhibitors to the numerous proteases involved in different biological functions to better understand them.
Materials and Methods
uPA expression and purification
We produced a LMWuPA (low molecular weight uPA) mutant C122A that lose the A-chain after activation [30]. LMWuPA-C122A was expressed in Shuffle cells (C3026H, New England Biolabs). Briefly, transformed cells were grown in 1 L of LB containing 100 μg/mL ampicillin at 37 °C and 250 rpm to an OD600 of 0.8. The enzyme expression was induced with 1 mM IPTG and the induced cultures were grown 16h at 20°C. The bacteria were harvested by centrifugation and resuspended in lysis buffer (50 mM Tris base, 300 mM NaCl, pH 7.8) containing protease inhibitor cocktail (complete EDTA-free, Roche). The cells were lysed by sonication and the cell debris was removed by centrifugation at 4°C for 20 min at 18,000 g. The LMWuPA-C122A was purified by affinity chromatography using Ni2+-NTA agarose resin (QIAGEN) and a standard protocol recommended by the manufacturer. Pure LMWuPA-C122A was dialyzed overnight at 4 °C against activation buffer (50 mM NaCl, 15 mM Tris-HCl pH 8.0, 1mM BME).
LMWuPA-C122A was activated with Plasmin (Roche) during 4 h at 37°C. Active LMWuPA-C122A was purified by affinity chromatography with benzamidine agarose beads (Sigma) using 0.1 M Tris-HCl, 0.5M NaCl pH 8.0 as binding and washing buffer: 0.1 M Tris-HCl, 0.5M NaCl pH 8.0. and eluting with 0.1 M Glycine-HCl, 0.05M NaCl pH 2.0–2.5. LMWuPA-C122A was then further purified by size exclusion chromatography on a Superdex 75 column in PBS. The yield was around 0.1mg for 1L of cells.
The purified uPA was analyzed by SDS-PAGE and the enzymatic activity was tested with Spectrozyme®uPA substrate, the activity values were similar to the commercial uPA (Supplemental Figure 2).
uPA biotinylation
Pure uPA was biotinylated using EZ-Link NHS-Chromogenic-Biotin (Pierce) as described previously [31].
Affinity maturation
The CDRL3 and CDRH2 loops were chosen as targets for affinity improvement of Fab U33. One U33 affinity maturation library with randomized CDRH2 and CDRL3 loops was constructed by Kunkel mutagenesis as described previously [32].
Two primers containing degenerate codons were designed in order to introduce mutations in selected positions on the CDRs following the four-amino-acid code [16,17], so each selected amino acid can be replaced by Ala, Ser, Tyr, Asp or the parental amino acid. While other amino acids may also be introduced due to degenerate codon usage, these ones were chosen because they represent a broad range of chemical diversity and introducing only a handful of amino acids keeps the library size smaller. The arginine residues on the CDRs were not mutated (Table 1). To avoid the parental U33 Fab in the final library, unique SalI restriction sites were introduced to each CDR-loop in the template gene, so parental clone was eliminated by restriction enzyme digestion of the plasmid before the transformation of the library in TG1 cells.
Table 1:
Primers used for generation of affinity maturation library.
| Primer name | Sequence (IUB code) |
|---|---|
| CDRL3 Degenerate Codon | DHK BMK CGT DHT KMK KHT BMT KMT DMT |
| CDRH2 Degenerate Codon | KHT DHT AGA DMT DMK KMT KMT KVT KVK DMT DMT KMK |
Off-rate selections [18] were performed in three rounds using streptavidin magnetic beads (Invitrogen) coated with biotinylated-uPA and off-rate washes with non-biotinylated-uPA. The stringency of panning was increased after each round by decreasing the concentration of biotinylated-uPA (200 nM in first round, 100 nM in second round and 10 nM in third round) and by increasing the concentration excess of non-biotinylated-uPA and duration of the off-rate washes (10 μM in first and second rounds and 5 μM in the third round, with incubation times of 1, 3, and 5 h respectively).
ELISA
Binding ELISA screening was performed with phage free Fabs that leaked into the cell culture media as described previously [31].
Fab expression and purification
Fabs were expressed in Escherichia coli BL21 (DE3) Gold (Stratagene) and purified from the periplasmic fraction as described previously [31]. Briefly, cultures were grown in 1 L of 2x YT containing 100 μg/mL ampicillin and 0.1% glucose at 37 °C and 200 rpm to an OD600 of 0.6. The protein expression was induced with the addition of 1mM IPTG and grow overnight at 20°C. The cells were harvested by centrifugation and the cell pellet was resuspended in ice-cold 1X TES (0.2 M Tris pH 8, 0.5 mM EDTA, 0.5 M sucrose). The cell suspension was mixed with the same volume of ice cold ddH2O and incubated on ice for 30 min. The solution was then pelleted and the supernatant (periplasmic fraction) was used for the purification.
The Fabs were purified from the periplasmic fraction by affinity chromatography using Ni2+-NTA agarose resin (QIAGEN) and a standard protocol recommended by the manufacturer. Pure Fabs were dialyzed overnight at 4 °C against PBS buffer pH 7.4 and analyzed by SDS-PAGE.
KD calculation
Kinetic constants for uPA-Fabs were determined using an Octet RED384 biolayer interferometer (BLI) (ForteBio). Five concentrations of each Fab (500 nM, 200 nM, 100 nM, 50 nM and 10 nM) was tested for binding to the biotinylated antigen (human uPA) immobilized on ForteBio streptavidin SA biosensors. All measurements were performed at room temperature in 384-well microplates and the running buffer was PBS with 0.1% (w/v) bovine serum albumin (BSA) and 0.02% (v/v) Tween 20. Biotinylated human uPA (full-length or catalytic domain) was loaded for 180 s from a solution of 150 nM, baseline was equilibrated for 60 s, and then the Fabs were associated for 120 s followed by 300 s disassociation. Between each Fab sample, the biosensor surfaces were regenerated three times by exposing them to 10 mM glycine, pH 1.5 for 5 s followed by PBS for 5 s. Data were analyzed using a 1:1 interaction model on the ForteBio data analysis software 8.2.
IC50 calculation
For IC50 calculations, 6.2 nM of human uPA were incubated with serial dilutions of Fab (from 1μM to 0.12 nM) in assay buffer (50 mM Tris pH 8.8, 0.01% Tween 20), after 1 hour of incubation at room temperature, the enzyme activity was initiated by adding the chromogenic substrate Spectrozyme®uPA (American Diagnostica Inc) at a final concentration of 50 μM. The reaction velocity was monitored by reading the absorbance at 405 nm during 30 min. The Vmax values were then plotted against log of Fab concentration to obtain the IC50. Enzymatic assays were done in triplicate with all standard deviations being 10% or less of the reported values. The data were analyzed using Graphpad Prism version 5.0 for Windows, GraphPad Software, San Diego, CA, USA.
PAI-1 competitive ELISA
Active human uPA (5 ng/mL) was pre-incubated overnight with 1 μM of each Fab, HUPAKT kit (Molecular Innovations) was used for measure active uPA in the samples following the manufacturer’s instructions. Only free active uPA reacts with the biotinylated PAI-1 immobilized on plates, inactive or complexed uPA not binds to the plate and is not detected. The uPA bound to the plate is detected with an anti-uPA antibody and a secondary antibody conjugated to peroxidase, TMB is used for color development at 450nm. The absorbance at 450 nm is directly proportional to the concentration of active uPA in the sample.
Crystallography
For crystallization purposes, uPA was co-incubated with AB2 in a 1:1 stoichiometric ratio for 1h and co-purified using size-exclusion chromatography. The complex co-eluted was concentrated to 15 mg/mL. Crystallization drops were produced by mixing 0.1 μL of uPA-AB2 solution with 0.1 μL of the respective crystallization solution. A single crystal was produced using a solution containing 0.2 M (NH4)2HCit (Salt) and 20% PEG 3350 and incubating the experiment for 14 days at room temperature. The crystal was flash frozen in liquid nitrogen and diffracted at beamline 8.3.1 at the Advanced Light Source. Reflections were processed using xia2 in space group P43 and a 2.9 Å structure could be solved using the anti-MT-SP1 Fab (PDB code: 3SO3 [27]) and a structure of uPA (PDB code: 4DW2 [29]) as a search model for molecular replacement. Iterative rounds of model building and refinement with phenix.refine [33] were necessary to unambiguously place interfacing loops and residue sidechains. Model statistics are shown in table 2.
Table 2.
Crystallographic statistics for uPa-AB2 co-crystal structure
| uPa_AB2 | |
|---|---|
| Resolution range | 86.7 – 2.9 (3.0 – 2.9) |
| Space group | P 43 |
| Unit cell | 86.74 86.74 172.42 90 90 90 |
| Total reflections | 2,062,816 (96556) |
| Unique reflections | 28,202 (2814) |
| Multiplicity | 73.1 (34.3) |
| Completeness (%) | 93.57 (94.31) |
| Mean I/sigma(I) | 10.81 (1.47) |
| Wilson B-factor | 69.5 |
| R-merge | 0.36 (2.36) |
| R-meas | 0.36 (2.39) |
| R-pim | 0.04 (0.40) |
| CC1/2 | 0.998 (0.833) |
| CC* | 0.999 (0.953) |
| Reflections used in refinement | 26,388 (2654) |
| Reflections used for R-free | 1270 (140) |
| R-work | 0.23 (0.40) |
| R-free | 0.30 (0.46) |
| CC(work) | 0.91 (0.75) |
| CC(free) | 0.92 (0.70) |
| Number of non-hydrogen atoms | 7644 |
| macromolecules | 7600 |
| solvent | 44 |
| Protein residues | 1105 |
| RMS(bonds) | 0.011 |
| RMS(angles) | 1.41 |
| Ramachandran favored (%) | 80.26 |
| Ramachandran allowed (%) | 13.04 |
| Ramachandran outliers (%) | 6.7 |
| Rotamer outliers (%) | 21.43 |
| Clashscore | 22.38 |
| Average B-factor | 93.55 |
| macromolecules | 93.79 |
| solvent | 51.7 |
| Number of TLS groups | 31 |
Data Analysis
Interfaces were analyzed using the pisa web server [34]. Coordinates of other serine protease models in complex with inhibitory antibody fragments were found under PDB codes 3SO3, 2R0K and 6AOD. Binding pockets were analyzed using Schrodinger SiteMap (Schrodinger, Inc.) within a 10 Å radius of the catalytic residue [35]. Cartoons were generated using PyMOL (Schrodinger, Inc.).
XFMS
All samples were buffer exchanged into Dulbecco’s phosphate buffered saline (Gibco) using a 10k MWCO concentrator (Millipore). Protein concentrations and beam parameters were optimized using an Alexa-488 fluorophore assay [36]. Purified uPA and uPA-AB2 complex at concentrations of 0.1–1 μM were exposed to a focused synchrotron X-ray white beam for 0–150 ms at beamline 3.2.1 at the Advanced Light Source in Berkeley, CA. Samples were immediately quenched post exposure and overnight digestion with trypsin or glu-c was initiated on the same day. Oxidation sites were detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS) carried out at Joint BioEnergy Institute, Berkeley, CA and analyzed as previously described [37]. The amount of unmodified peptide is calculated by dividing the unmodified peak area by the sum of unmodified and modified peak areas. As the dose of X-ray radiation is increased, the fraction of unmodified peptide decreases, and fitting this dose-response relationship to single exponential decay reveals the rate of footprinting (dkfp). This rate is dependent on both the intrinsic reactivity of the specific side chain and its accessibility to solvent. The ratio R of the rate constants at specific amino acid sites between samples reveals changes in relative solvent accessibility. The R values determined in this study were mapped onto the presented crystal structure for comparative structural analysis.
Supplementary Material
Figure 4. Table of kfp values for each residue detected by XFMS on uPA
Figure 3. Characterization of uPA Fabs from biased libraries. A. Mechanism of inhibition of E2 and A11 MTSP-1 (matriptase) inhibitory Fabs, the sequence of long CDRH3 loops used as templates for biased libraries construction is shown. B. Sequence of CDRs loops of uPA Fabs identified from biased libraries. The KD values were determined by bioluminescence interferometry using an Octet RED384 instrument. C. The proteolytic activity of human uPA was read in absence and presence of 1 μM of each Fab, the enzyme activity is expressed as percentage of the uPA activity in absence of Fab (100 %). D. uPA-Fabs from biased libraries are cleaved by uPA.
Figure 2. Recombinant expression of the catalytic domain of uPA. The enzyme activity of 6.2 nM of human uPA (from R&D Systems or own produced) was initiated by adding the chromogenic substrate Spectrozyme®uPA at 50 μM. The reaction velocity was monitored by reading the absorbance at 405 nm during 30 min.
Figure 1. Parental anti-uPA U33Fab. A. Sequence of parental U33 Fab, in red are shown the sequences of the CDRs loops. B. Off-rate profile of U33 parental Fab measured by BLI.
Acknowledgements
The XFMS is supported in part by NIH 1R01GM126218. Advanced Light Source and Joint BioEnergy Institute were supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. DOE under contract DE-AC02-05CH11231. Beamline 8.3.1 at the Advanced Light Source is operated by the University of California Office of the President, Multicampus Research Programs and Initiatives grant MR-15-328599 the National Institutes of Health (R01 GM124149 and P30 GM124169), Plexxikon Inc. and the Integrated Diffraction Analysis Technologies program of the US Department of Energy Office of Biological and Environmental Research. The Advanced Light Source (Berkeley, CA) is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy under contract number DE-AC02-05CH11231, Office of Basic Energy Sciences. This research was supported by NIH/NIGMS funding for the HIV Accessory & Regulatory Complexes (HARC) Center (P50 GM082250) and NIH/NCI funding for the Antibody Reseach Technology Center (P41 CA196276).
Footnotes
Declaration of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Puente XS, Sánchez LM, Gutiérrez-Fernández A, Velasco G, López-Otín C, A genomic view of the complexity of mammalian proteolytic systems, Biochem Soc Trans. 33 (2005) 331–334. 10.1042/BST0330331. [DOI] [PubMed] [Google Scholar]
- [2].Turk B, Targeting proteases: successes, failures and future prospects, Nat Rev Drug Discov. 5 (2006) 785–799. 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- [3].De Strooper B, Proteases and Proteolysis in Alzheimer Disease: A Multifactorial View on the Disease Process, Physiological Reviews. 90 (2010) 465–494. 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- [4].Funovics M, Weissleder R, Tung C-H, Protease sensors for bioimaging, Anal Bioanal Chem. 377 (2003) 956–963. 10.1007/s00216-003-2199-0. [DOI] [PubMed] [Google Scholar]
- [5].Weidle UH, Tiefenthaler G, Georges G, Proteases as Activators for Cytotoxic Prodrugs in Antitumor Therapy, Cancer Genomics Proteomics. 11 (2014) 67–79. [PubMed] [Google Scholar]
- [6].Duffy MJ, McGowan PM, Gallagher WM, Cancer invasion and metastasis: changing views, The Journal of Pathology. 214 (2008) 283–293. 10.1002/path.2282. [DOI] [PubMed] [Google Scholar]
- [7].Paulick MG, Bogyo M, Application of activity-based probes to the study of enzymes involved in cancer progression, Current Opinion in Genetics & Development. 18 (2008) 97–106. 10.1016/j.gde.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Darragh MR, Schneider EL, Lou J, Phojanakong PJ, Farady CJ, Marks JD, Hann BC, Craik CS, Tumor Detection by Imaging Proteolytic Activity, Cancer Res. 70 (2010) 1505–1512. 10.1158/0008-5472.CAN-09-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoffman R, Chauret C, Standridge J, Peterson L, Evaluation of four commercial antibodies, Journal - AWWA. 91 (1999) 69–78. 10.1002/j.1551-8833.1999.tb08698.x. [DOI] [Google Scholar]
- [10].Wu Y, Eigenbrot C, Liang W-C, Stawicki S, Shia S, Fan B, Ganesan R, Lipari MT, Kirchhofer D, Structural insight into distinct mechanisms of protease inhibition by antibodies, PNAS. 104 (2007) 19784–19789. 10.1073/pnas.0708251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ely LK, Lolicato M, David T, Lowe K, Kim YC, Samuel D, Bessette P, Garcia JL, Mikita T, Minor DL, Coughlin SR, Structural Basis for Activity and Specificity of an Anticoagulant Anti-FXIa Monoclonal Antibody and a Reversal Agent, Structure. 26 (2018) 187–198.e4. 10.1016/j.str.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nam DH, Ge X, Generation of Highly Selective MMP Antibody Inhibitors, in: Cal S, Obaya AJ (Eds.), Proteases and Cancer: Methods and Protocols, Springer, New York, NY, 2018: pp. 307–324. 10.1007/978-1-4939-7595-2_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Maun HR, Jackman JK, Choy DF, Loyet KM, Staton TL, Jia G, Dressen A, Hackney JA, Bremer M, Walters BT, Vij R, Chen X, Trivedi NN, Morando A, Lipari MT, Franke Y, Wu X, Zhang J, Liu J, Wu P, Chang D, Orozco LD, Christensen E, Wong M, Corpuz R, Hang JQ, Lutman J, Sukumaran S, Wu Y, Ubhayakar S, Liang X, Schwartz LB, Babina M, Woodruff PG, Fahy JV, Ahuja R, Caughey GH, Kusi A, Dennis MS, Eigenbrot C, Kirchhofer D, Austin CD, Wu LC, Koerber JT, Lee WP, Yaspan BL, Alatsis KR, Arron JR, Lazarus RA, Yi T, An Allosteric Anti-tryptase Antibody for the Treatment of Mast Cell-Mediated Severe Asthma, Cell. 179 (2019) 417–431.e19. 10.1016/j.cell.2019.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hawkins RE, Russell SJ, Winter G, Selection of phage antibodies by binding affinity: Mimicking affinity maturation, Journal of Molecular Biology. 226 (1992) 889–896. 10.1016/0022-2836(92)90639-2. [DOI] [PubMed] [Google Scholar]
- [15].LeBeau AM, Sevillano N, Markham K, Winter MB, Murphy ST, Hostetter DR, West J, Lowman H, Craik CS, VanBrocklin HF, Imaging Active Urokinase Plasminogen Activator in Prostate Cancer, Cancer Res. 75 (2015) 1225–1235. 10.1158/0008-5472.CAN-14-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fellouse FA, Wiesmann C, Sidhu SS, Synthetic antibodies from a four-amino-acid code: A dominant role for tyrosine in antigen recognition, PNAS. 101 (2004) 12467–12472. 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goh AX, Bertin-Maghit S, Yeo SP, Ho A, Derks H, Mortellaro A, Wang C-I, A novel human anti-interleukin-1β neutralizing monoclonal antibody showing in vivo efficacy, MAbs. 6 (2014) 764–772. 10.4161/mabs.28614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Altshuler EP, Serebryanaya DV, Katrukha AG, Generation of recombinant antibodies and means for increasing their affinity, Biochemistry Moscow. 75 (2010) 1584–1605. 10.1134/S0006297910130067. [DOI] [PubMed] [Google Scholar]
- [19].Ylera F, Harth S, Waldherr D, Frisch C, Knappik A, Off-rate screening for selection of high-affinity anti-drug antibodies, Analytical Biochemistry. 441 (2013) 208–213. 10.1016/j.ab.2013.07.025. [DOI] [PubMed] [Google Scholar]
- [20].Su S-C, Lin C-W, Yang W-E, Fan W-L, Yang S-F, The urokinase-type plasminogen activator (uPA) system as a biomarker and therapeutic target in human malignancies, Expert Opinion on Therapeutic Targets. 20 (2016) 551–566. 10.1517/14728222.2016.1113260. [DOI] [PubMed] [Google Scholar]
- [21].Mahmood N, Mihalcioiu C, Rabbani SA, Multifaceted Role of the Urokinase-Type Plasminogen Activator (uPA) and Its Receptor (uPAR): Diagnostic, Prognostic, and Therapeutic Applications, Front. Oncol. 8 (2018). 10.3389/fonc.2018.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang D, Yang Y, Jiang L, Wang Y, Li J, Andreasen PA, Chen Z, Huang M, Xu P, Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides, J. Med. Chem. 62 (2019) 2172–2183. 10.1021/acs.jmedchem.8b01908. [DOI] [PubMed] [Google Scholar]
- [23].Rockway TW, Nienaber V, Giranda VL, Inhibitors of the protease domain of urokinase-type plasminogen activator, Curr. Pharm. Des. 8 (2002) 2541–2558. 10.2174/1381612023392676. [DOI] [PubMed] [Google Scholar]
- [24].Sulimov VB, Katkova EV, Oferkin IV, Sulimov AV, Romanov AN, Roschin AI, Beloglazova IB, Plekhanova OS, Tkachuk VA, Sadovnichiy VA, Application of molecular modeling to urokinase inhibitors development, Biomed Res Int. 2014 (2014) 625176 10.1155/2014/625176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Islam I, Yuan S, West CW, Adler M, Bothe U, Bryant J, Chang Z, Chu K, Emayan K, Gualtieri G, Ho E, Light D, Mallari C, Morser J, Phillips G, Schaefer C, Sukovich D, Whitlow M, Chen D, Buckman BO, Discovery of selective urokinase plasminogen activator (uPA) inhibitors as a potential treatment for multiple sclerosis, Bioorganic & Medicinal Chemistry Letters. 28 (2018) 3372–3375. 10.1016/j.bmcl.2018.09.001. [DOI] [PubMed] [Google Scholar]
- [26].Sun J, Pons J, Craik CS, Potent and Selective Inhibition of Membrane-Type Serine Protease 1 by Human Single-Chain Antibodies, Biochemistry. 42 (2003) 892–900. 10.1021/bi026878f. [DOI] [PubMed] [Google Scholar]
- [27].Schneider EL, Lee MS, Baharuddin A, Goetz DH, Farady CJ, Ward M, Wang C-I, Craik CS, A Reverse Binding Motif That Contributes to Specific Protease Inhibition by Antibodies, Journal of Molecular Biology. 415 (2012) 699–715. 10.1016/j.jmb.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Farady CJ, Egea PF, Schneider EL, Darragh MR, Craik CS, Structure of an Fab–Protease Complex Reveals a Highly Specific Non-canonical Mechanism of Inhibition, Journal of Molecular Biology. 380 (2008) 351–360. 10.1016/j.jmb.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jiang L, Botkjaer KA, Andersen LM, Yuan C, Andreasen PA, Huang M, Rezymogenation of active urokinase induced by an inhibitory antibody, Biochem J. 449 (2013) 161–166. 10.1042/BJ20121132. [DOI] [PubMed] [Google Scholar]
- [30].Zeslawska E, Schweinitz A, Karcher A, Sondermann P, Sperl S, Stürzebecher J, Jacob U, Crystals of the urokinase type plasminogen activator variant βc-uPA in complex with small molecule inhibitors open the way towards structure-based drug design11Edited by A. Fersht, Journal of Molecular Biology. 301 (2000) 465–475. 10.1006/jmbi.2000.3966. [DOI] [PubMed] [Google Scholar]
- [31].Kim J, Stroud RM, Craik CS, Rapid identification of recombinant Fabs that bind to membrane proteins, Methods. 55 (2011) 303–309. 10.1016/j.ymeth.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chen G, Sidhu SS, Design and Generation of Synthetic Antibody Libraries for Phage Display, in: Ossipow V, Fischer N (Eds.), Monoclonal Antibodies: Methods and Protocols, Humana Press, Totowa, NJ, 2014: pp. 113–131. 10.1007/978-1-62703-992-5_8. [DOI] [PubMed] [Google Scholar]
- [33].Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung L-W, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD, Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix, Acta Cryst D. 75 (2019) 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state, J. Mol. Biol. 372 (2007) 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- [35].Halgren TA, Identifying and Characterizing Binding Sites and Assessing Druggability, J. Chem. Inf. Model. 49 (2009) 377–389. 10.1021/ci800324m. [DOI] [PubMed] [Google Scholar]
- [36].Gupta S, Sullivan M, Toomey J, Kiselar J, Chance MR, The Beamline X28C of the Center for Synchrotron Biosciences: a National Resource for Biomolecular Structure and Dynamics Experiments Using Synchrotron Footprinting, J Synchrotron Rad. 14 (2007) 233–243. 10.1107/S0909049507013118. [DOI] [PubMed] [Google Scholar]
- [37].Gupta S, Guttman M, Leverenz RL, Zhumadilova K, Pawlowski EG, Petzold CJ, Lee KK, Ralston CY, Kerfeld CA, Local and global structural drivers for the photoactivation of the orange carotenoid protein, Proc. Natl. Acad. Sci. U.S.A. 112 (2015) E5567–5574. 10.1073/pnas.1512240112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 4. Table of kfp values for each residue detected by XFMS on uPA
Figure 3. Characterization of uPA Fabs from biased libraries. A. Mechanism of inhibition of E2 and A11 MTSP-1 (matriptase) inhibitory Fabs, the sequence of long CDRH3 loops used as templates for biased libraries construction is shown. B. Sequence of CDRs loops of uPA Fabs identified from biased libraries. The KD values were determined by bioluminescence interferometry using an Octet RED384 instrument. C. The proteolytic activity of human uPA was read in absence and presence of 1 μM of each Fab, the enzyme activity is expressed as percentage of the uPA activity in absence of Fab (100 %). D. uPA-Fabs from biased libraries are cleaved by uPA.
Figure 2. Recombinant expression of the catalytic domain of uPA. The enzyme activity of 6.2 nM of human uPA (from R&D Systems or own produced) was initiated by adding the chromogenic substrate Spectrozyme®uPA at 50 μM. The reaction velocity was monitored by reading the absorbance at 405 nm during 30 min.
Figure 1. Parental anti-uPA U33Fab. A. Sequence of parental U33 Fab, in red are shown the sequences of the CDRs loops. B. Off-rate profile of U33 parental Fab measured by BLI.