Abstract
Background
The process of wound healing is complex. Increasing evidences have shown that lncRNA MALAT1 is abundant in fibroblasts and may be engaged in wound healing process. Therefore, we explored the mechanism of MALAT1 affecting wound healing.
Methods
The expression levels of MALAT1, miR-141-3p as well as ZNF217 in human fibroblast cells (HFF-1) were quantified by qRT-PCR. HFF-1 proliferation was measured by MTT, while migration was detected by wound healing assay. SMAD2 activation and matrix proteins expression were detected by western blotting. The interaction between miR-141-3p and MALAT1 or ZNF217 was further confirmed using the luciferase reporter gene assay. In vivo wound healing was assessed by full-thickness wound healing model on C57BL/6 mice.
Result
Knockdown of MALAT1 as well as overexpression miR-141-3p remarkably inhibited the proliferation, migration and matrix protein expression in HFF-1 cells. MALAT1 directly targeted and inhibited the expression of miR-141-3p. MiR-141-3p suppressed the activation of TGF-β2/SMAD2 signaling pathway by targeting ZNF217. Knockdown of MALAT1 inhibited wound healing process in mice.
Conclusions
MALAT1 up-regulates ZNF217 expression by targeting miR-141-3p, thus enhances the activity of TGF-β2/SMAD2 signaling pathway and promotes wound healing process. This investigation shed new light on the understanding of the role of MALAT1 in wound healing, and may provide potential target for the diagnosis or therapy of chronic wounds.
Keywords: MALAT1, miR-141-3p, ZNF217, Wound healing
Abbreviations: lncRNA, long non-coding RNA; HFF-1, human fibroblast cells; TGF-β2, Transforming Growth Factor-β2; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide; ELISA, enzyme linked immunosorbent assay; ECM, extra cellular matrix; MALAT1, metastasis-associated lung adenocarcinoma transcript 1; ZEB1, E-box binding homeobox 1; EMT, epithelial mesenchymal transition; ZNF217, zinc-finger protein 217; qRT-PCR, quantitative real-time PCR; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; PVDF, polyvinylidene fluoride
1. Introduction
Wound healing involves the collaboration of many different cells and cytokines, and is considered to be one of the most complex physiological process [1]. Abnormal wound healing may affect millions of people around the world and lead to serious complications such as chronic wounds and fibrosis [2]. Many steps are involved in wound healing process, including inflammation, tissue proliferation and remodeling, which are spatially and temporally intertwined during tissue repair [3]. Fibroblasts play critical roles in wound healing including fibrin clot degradation, extra cellular matrix (ECM) generation, myofibroblasts formation and wound contraction [4]. Thus, further investigation of the factors that affect the function of fibroblast may provide important clues for the diagnosis/treatment of abnormal wound healing.
Long non-coding RNAs (lncRNAs) are gene transcripts that contain more than 200 nucleotides and not translated into any proteins. Many studies highlighted the role of lncRNAs in wound healing process, such as H19 and Gas5 could accelerate the process of chronic wound healing caused by diabetes [[5], [6], [7]]. The metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA, which was proved to be important in cancer metastasis [8,9]. MALAT1 facilitates the proliferation and metastasis of pancreatic cancer cell via activating autophagy [10]. However, MALAT1 suppresses breast cancer metastasis via inactivates the prometastatic transcription factor TEAD [11]. The opposite results may be caused by different tumor types. Increasing evidences have shown that MALAT1 is abundant in fibroblasts [12], and promotes human dermal fibroblast migration and wound healing [13,14]. In addition, knockdown MALAT1 prevented angiotensin II-induced fibroblast proliferation, collagen production, and α-SMA expression in cardiac fibroblasts [15]. However, it is not fully understood how MALAT1 affect wound healing.
MicroRNAs (miRNAs) are short non-coding RNAs (20–24 nt) that are involved in regulating of gene expression by affecting the stability or translation of mRNAs [16]. MicroRNA-141 (miR-141), which is widely expressed in many cells/tissues, has been reported to be related to various human malignancies [17]. Recent studies have demonstrated that miR-141-3p inhibits cancer cell [18,19] and epithelial cell migration [20]. Moreover, miR-141-3p was also detected in many fibroblasts [21,22] and was found to induce apoptosis and inhibit proliferation and migration of fibroblasts in keloids via negatively regulating GAB1 expression [23]. Since fibroblasts are the key cells in wound healing [4], the working mechanism of miR-141-3p on wound healing needs further investigation.
The zinc-finger protein 217 (ZNF217) is an oncogenic protein which directly binds to the promoter of transforming growth factor-β (TGF-β) and promotes TGF-β expression and epithelial–mesenchymal transition (EMT) [24]. TGF-β family is considered to be one of the key regulator of cell proliferation, differentiation as well as metabolism in wound healing and tissue repair process [25,26]. Transduction of TGF-β signal after TGF-β receptor activation triggers the phosphorylation of SMAD family proteins, thus subsequently accelerates wound healing [27,28]. Previous study demonstrates that ZNF217 positively regulates wound healing process by regulating TGF-β/SMAD signaling and enhances collagen (collagen I, collagen III), Fibronectin and other matrix proteins expression, thus promoting the reconstruction of extracellular matrix and accelerating the healing of skin wounds [25,29]. However, the regulator of ZNF217 in fibroblasts needs further elucidation.
This study investigated the mechanism by which MALAT1 promotes wound healing process, and provides novel insights into the understanding of MALAT1 in wound healing process.
2. Material and methods
2.1. Cell culture
Human foreskin fibroblast cell line (HFF-1) was got from American Type Culture Collection (ATCC, VA, USA) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, California, USA) supplemented with 15% fetal calf serum (FCS, Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified atmosphere at 37 °C with 5% CO2.
2.2. Cell transfection
ZNF217 cDNA was synthesized and cloned into pcDNA3.1 plasmid for overexpression and pcDNA3.1 plasmid contains a scrambled sequence was used as negative control (NC). Sh-MALAT1, sh-NC, pcDNA3.1, pcDNA-ZNF217, miR-141-3p mimics (sense 5′-UAACACUGUCUGGUAAAGAUGG-3′ and antisense 5′-AUCUUUACCAGACAGUGUUAUU-3′), mimics NC (5′- UUCUCCGAACGUGUCACGUTT-3′), miR-141-3p inhibitor (5′-CCAUCUUUACCAGACAGUGUUA-3′) and inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′) were all constructed or synthesized by GenePharma (Shanghai, China). The pGMLV-SC5 RNAi lentivirus plasmid (GeneChem) was used to knock down MALAT1 expression in mice model. In brief, HFF1 cells were plated overnight to reach 50% confluence before transfection. Sh-MALAT1, miR-141-3p mimics, miR-141-3p inhibitor, pcDNA-ZNF217 and their controls were transfected into HFF-1 cells with lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA) according to the manufacturer's instructions.
2.3. RNA isolation and quantitative real-time PCR (qRT-PCR)
Trizol reagent (Invitrogen) was used to extract total RNA from mice skin granulation tissues and HFF-1 cells according to the manufacturer's protocol. 50 mg of mice skin granulation tissues were collected and cut on ice. Tissue fragments were incubated with 1 mL TRIzol in a 1.5 mL homogenizer at room temperature for 5 min. HFF-1 cells were collected and lysed by TRIzol too. Subsequently, chloroform was added to the lysates at a ratio of 1:5, vigorously shaked for 15 s and maintained at room temperature for 2–3 min. Afterward, the lysates were centrifuged at 12,000 g at 4 °C and 15 min The supernatant (total lysate) was transferred to a new tube, and an equal volume of isopropanol was added. Then the mixture was centrifuged at 12,000 g at 4 °C and for another 15 min. The precipitate was washed with 1 mL 75% ethanol overnight 4 °C, centrifuged and air dried. Extracted RNA was diluted in diethyl pyrocarbonate water (DEPC; Beyotime Biotechnology, Shanghai, China) and stored at −80 °C until use. The RNA was reverse transcribed into cDNA using reverse transcriptase for qPCR (AE341-02, TransGen Biotech, Beijing, China) following the instructions provided by the manufacturer. The mRNA expression was determined by StepOnePlus RT-PCR System (Thermofisher, MA, USA) with a cycling condition of denaturation at 95 °C for 15 s and annealing and extending at 60 °C for 30 s in a total of 40 cycles. Primer sequences used in this study were described below: miR-141-3p forward 5′-GCCGTAACACTGTCTGGTAA-3′, reverse primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCATCT-3’; MALAT1 forward 5′-AAAGCAAGGTCTCCCCACAAG-3′ and reverse 5′-GGTCTGTGCTAGATCAAAAGGCA-3’; ZNF217 forward 5′-GGAAGAAGCCTGTCAGATGC-3′ and reverse 5′-CCTGCACAACTGCCCTTATT-3′. The results were analyzed by the 2-ΔΔCT methods with internal reference of U6 (forward primer 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse primer 5′-CGCTTCACGAATTTGCGTGTCAT-3′) and GAPDH (forward primer 5′-CCAGGTGGTCTCCTCTGA-3′ and reverse primer 5′-GCTGTAGCCAAATCGTTGT-3′). All the qRT-PCR reactions were operated in triplicate.
2.4. Western blotting
Total protein was extracted from mice skin granulation tissues and HFF-1 cells with RIPA lysis buffer (Sigma–Aldrich, MS, USA) supplemented with protease and phosphatase inhibitor cocktail (MCE, NJ, USA). 30 mg of mice skin granulation tissues were collected and cut on ice. Tissue fragments were incubated with 300 μL RIPA lysis buffer in a 1.5 mL homogenizer on ice for 15 min. HFF-1 cells were collected and lysed by RIPA lysis buffer too. Subsequently, the lysates were centrifuged at 12,000 g at 4 °C for 15 min. Then, the total protein concentration in the supernatant was measured by BCA protein assay kit (Kaiji Biotechnology, Nanjing, China). Equivalent amounts of proteins were transferred to a new tube incubate on ice and then 5 × protein loading buffer (Co. Win Biosciences, Beijing, China) was added, heat the eluted samples at 95 °C for 5 min. The supernatant was obtained by centrifuged at 12,000 g. The supernatant was subjected to 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (Millipore, MA, USA) membrane and then blocked with 5% bovine serum albumin (BSA, Sigma–Aldrich). The blotting membrane was incubated with antibodies against Fibronectin (1:1000; Abcam, Cambridge, UK), Collagen I (1:1000; Abcam), Collagen III (1:1000; Abcam), MMP9 (1:1000; Abcam), ZNF217 (1:1000; Abcam), phospho-SMAD2 (1:1000; CST, MA, USA), SMAD2 (1:1000; CST) and GAPDH (1:5000; TransGen Biotech) at 4 °C overnight. The bands of target proteins were showed by horseradish peroxidase -conjugated secondary antibodies and chemiluminescence (ECL, Biosharp, Hefei, China) and the intensity was analyzed by ImageJ v1.35.
2.5. MTT assay
Transfected HFF-1 cells were cultured in 96-well plate at a density of 1 × 104 per well for 2 days, then 20 μL MTT solution (5 mg/mL, M2128, Sigma–Aldrich) were added into each well and incubated for 4 h. Then the formazan was dissolved by the addition of 150 μL DMSO (Sigma–Aldrich) and measured by a microplate reader at 490 nm (Biotek, Winooski, VT).
2.6. Wound healing assay
Wound healing assay was operated as previously reported [25]. Briefly, HFF-1 cells (2 × 106 per well) with or without transfection were cultured in 6-well plate until the cells were grown to confluence. After a pre-incubation with 10 μg/mL mitomycin C (Sigma–Aldrich) for 2 h, the monolayer of the cells was mechanically disrupted with a sterile 10 μL pipette tip. Then, DMEM medium without FCS was added into the plate and photographs were taken at 0 and 24 h post-scratching. The healing area was analyzed by Image Pro Plus 6.0 software and repeated three times.
2.7. Luciferase reporter assay
HFF-1 cells were cultured in 12 well plates at the density of 5 × 105 per well. The luciferase activity assays were carried out 24 h after transfection with a pmirGLO plasmid (Promega, USA) according to the manufacturer's method. The relative luciferase was normalized with renilla luciferase activity.
2.8. Enzyme linked immunosorbent assay (ELISA)
The expression of TGF-β2 of different groups were measured by TGF-β2 ELISA kit (Solarbio, Beijing, China). Cell culture supernatant of HFF-1 cells was collected 48 h after the treatment and the concentration of TGF-β2 was determined by the ELISA kit according to the manufacturer's protocols. Concentrations were recorded as pg/mL. All calibrations and analyses were repeated in triplicate.
2.9. Construction of mouse skin wound model
C57BL/6 mice (18–22 g, male) were got from the Center of Laboratory Animal of the Second Military Medical University (Shanghai, China) and were divided into three groups randomly (5 mice per group): control group, sh-NC group, and sh-MALAT1 group. Full-thickness wounds were created as previously reported [27], and they were housed separately. Lentivirus carrying sh-MALAT1 or sh-NC was mixed with diluted matrigel and administrated at 4 sites around the wound area of each mouse, while matrigel with control virus were added into the wound area of another group of mice. And transparent application was used to cover the surface of wounds. Images of the wounds were captured 0, 3, 7, and 11 days after the surgery. Image Pro Plus software was used to calculate the change in wound area at different time periods. All animal experimental protocols were approved by the ethics committee of Guangzhou Red Cross Hospital (Guangzhou, China).
2.10. Statistical analysis
Three independent biological experiments in triplicate were carried out in all experiments. Results were expressed as mean ± standard deviation (SD). All data were analyzed using Graphpad 6.0 software. Statistical analyses were performed using one-way ANOVA with Dunnett's post-hoc test and student t-test analysis. P < 0.05 indicates that the difference was significant.
3. Results
3.1. Knockdown of MALAT1 suppresses cell proliferation and migration and reduces matrix protein expression in HFF-1 cells
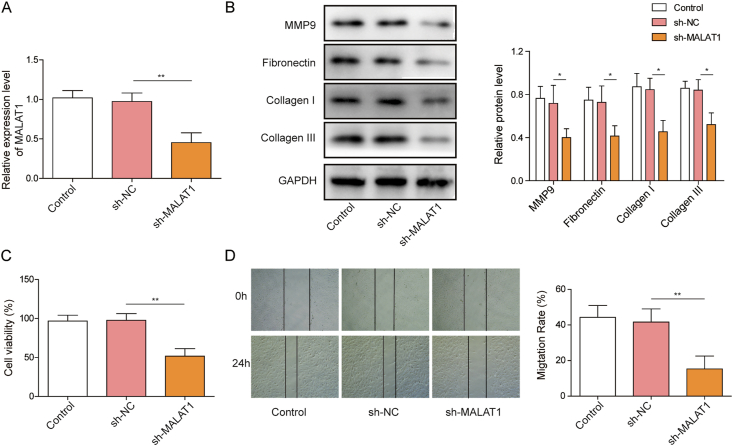
Recent study showed that MALAT1 is a rich noncoding transcript in the nucleus of human primary fibroblasts [12] and the overexpress of MALAT1 increases both wound healing and cellular migration in murine fibroblasts [13]. To investigate whether MALAT1 has similar function in human fibroblast cells (HFF-1), we constructed MALAT1 knockdown plasmid (sh-MALAT1). MALAT1 expression was decreased in HFF-1 cells after transfection with sh-MALAT1 (Fig. 1A), suggesting the plasmid was successfully transfected into the cells. Western blot analysis showed the expression of MMP9, Collagen I/III and Fibronectin were significantly reduced after transfected with sh-MALAT1 (Fig. 1B). MTT assay demonstrated that knockdown of MALAT1 markedly decreased the proliferation of HFF-1 cells compared with control (Fig. 1C). Furthermore, downregulation of MALAT1 significantly suppressed the migration of HFF-1 cells as shown in wound healing assay (Fig. 1D). The above results show that MALAT1 significantly promotes the proliferation, migration and increases the expression of matrix proteins of fibroblasts.
Fig. 1.
MALAT1 knockdown decreases proliferation, migration and matrix protein expression of fibroblasts (A) MALAT1 level was measured by qRT-PCR in HFF-1 cells transfected with MALAT1 knockdown vector (sh-MALAT1) or the vector contains a non-targeting shRNA sequence (sh-NC) (B) Matrix protein expressions were determined by western blotting in HFF-1 cells (C) The viability of HFF-1 cells was detected by MTT assay after sh-MALAT1 or sh-NC treatment (D) The effect of MALAT1 on cell migration in HFF-1 cells. The data were presented as mean ± SD in triplicate. One-way ANOVA test was performed for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.2. MiR-141-3p inhibits proliferation, migration, and matrix proteins expression of fibroblast and as a direct target of MALAT1
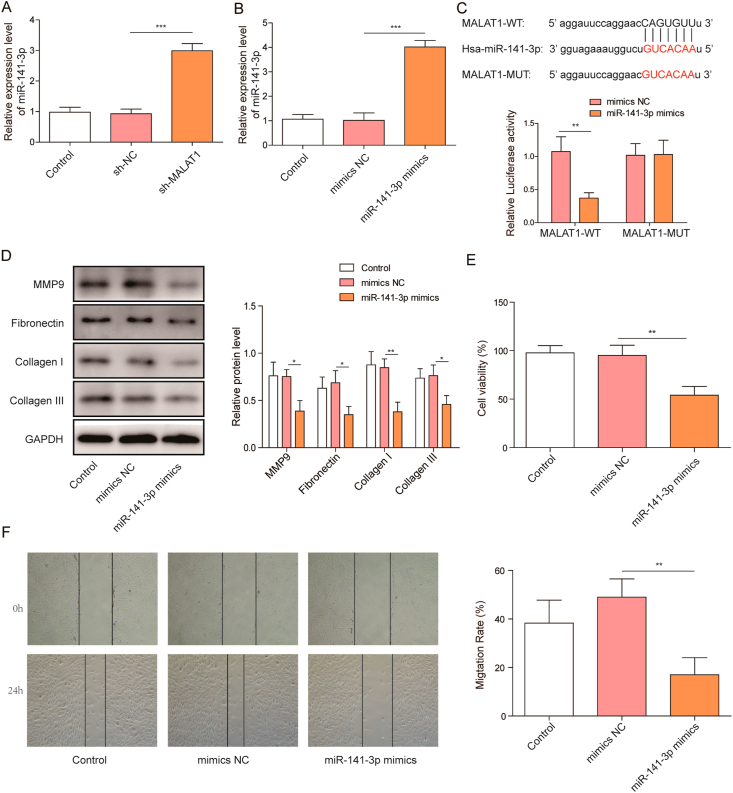
We found that knockdown of MALAT1 increased miR-141-3p level in HFF-1 cells (Fig. 2A). Therefore, bioinformatics prediction was carried out to investigate whether MALAT1 directly regulated miR-141-3p expression. StarBase virtual online screening suggested miR-141-3p might be a potential target of MALAT1 (Fig. 2C). To further investigate whether MALAT1 directly targets miR-141-3p, we constructed luciferase reporters containing MALAT1, which includes wild-type (WT) or mutant (MUT). QRT-PCR analysis indicated that miR-141-3p mimics transfection significantly increased miR-141-3p expression in HFF-1 cells (Fig. 2B). However, overexpression of miR-141-3p significantly reduced the luciferase activity in the MALAT1-WT group without affecting the MALAT1-MUT group in luciferase reporter assay (Fig. 2C), indicating MALAT1 directly targets miR-141-3p. Moreover, the expression of MMP9, Collagen I/III and Fibronectin were significantly decreased after miR-141-3p mimics transfection (Fig. 2D). MTT assay demonstrated that the proliferation of HFF-1 cells was also markedly decreased by the overexpression of miR-141-3p (Fig. 2E). Furthermore, as shown in wound healing assay, miR-141-3p mimics suppressed HFF-1 cell migration significantly (Fig. 2F). As demonstrated above, MALAT1 directly targets miR-141-3p and inhibits its expression level, miR-141-3p overexpression significantly inhibits fibroblast proliferation, migration, and the expression of matrix proteins.
Fig. 2.
MiR-141-3p binds to MALAT1 and inhibits proliferation, migration, and matrix proteins expression of fibroblast (A) QRT-PCR analyzed endogenous miR-141-3p levels in HFF-1 cells treated with sh-MALAT1 or sh-NC (B) Effect of miR-141-3p mimics on the expression of miR-141-3p (C) Interaction of MALAT1 and miR-141-3p was determined by StarBase virtual screening and dual-luciferase reporter gene assay (D) Western blotting was performed to determine the expression of matrix proteins in HFF-1 cells after miR-141-3p mimics treatment (E) Effect of miR-141-3p on HFF-1 cell proliferation (F) Effect of miR-141-3p on HFF-1 cell migration. The data were presented as mean ± SD in triplicate. One-way ANOVA test and student t-test analysis were performed for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P<0.001.
3.3. MiR-141-3p inhibits the activity of the TGF-β2/SMAD2 signaling pathway by targeting ZNF217
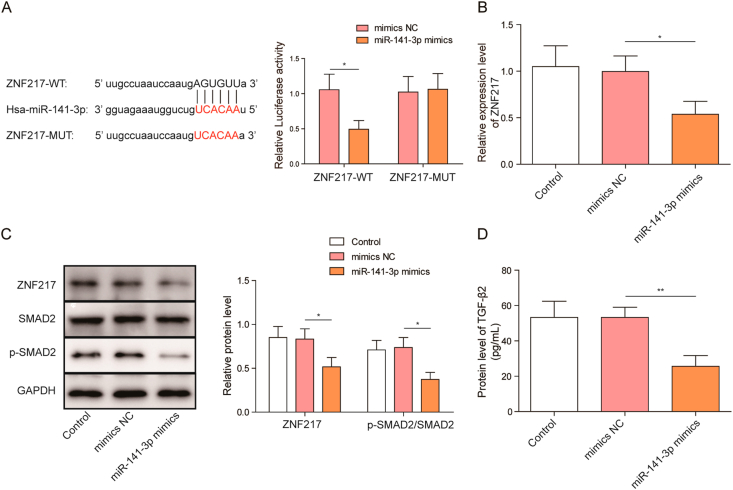
StarBase virtual screening suggested that miR-141-3p might bind to the 3′-UTR of ZNF217 mRNA, and this interaction was further confirmed by luciferase reporter assay (Fig. 3A). Moreover, ZNF217 mRNA expression was significantly inhibited by miR-141-3p mimics (Fig. 3B). In consistent with qRT-PCR experiment, ZNF217 protein level was also significantly decreased by miR-141-3p mimics (Fig. 3C). Previous finding have shown that ZNF217 directly binds to TGF-β gene promoter and promotes TGF-β/SMAD signaling pathway [20,24]. Therefore, we compared the level of SMAD2 phosphorylation and TGF-β2 between control and miR-141-3p overexpressed HFF-1 cells, in order to investigate the role of miR-141-3p on TGF-β/SMAD signaling. SMAD2 phosphorylation (Fig. 3C) and TGF-β2 expression (Fig. 3D) were significantly reduced by miR-141-3p mimics. Taken together, the above data suggests miR-141-3p targets and inhibits ZNF217 expression, thereby suppressing TGF-β2/SMAD2 signaling pathway in fibroblasts.
Fig. 3.
MiR-141-3p regulates TGF-β2/SMAD2 signaling pathway by targeting ZNF217 (A) Interaction of ZNF217 and miR-141-3p was determined by StarBase virtual screening and dual-luciferase reporter gene assay (B) Effect of miR-141-3p mimics on the expression of ZNF217 (C) Effect of miR-141-3p mimics on the expression of ZNF217, SMAD2 and p-SMAD2 (D) Effect of miR-141-3p mimics on the expression of TGF-β2 in HFF-1 cells. The data were presented as mean ± SD in triplicate. One-way ANOVA test and student t-test analysis were performed for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3.4. MALAT1 promotes the activation of ZNF217/TGF-β2/SMAD2 signaling pathway by targeting miR-141-3p
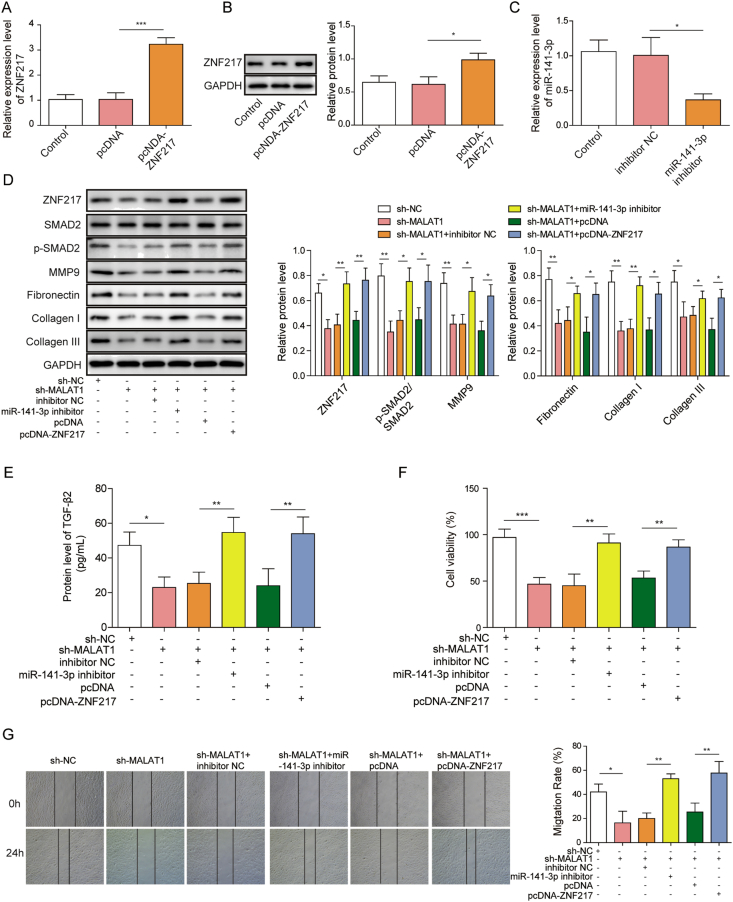
Experiments were carried out to further investigate the mechanism of MALAT1 regulated fibroblast proliferation and migration. Plasmid pcDNA-ZNF217 was used to overexpress ZNF217 (Fig. 4A and B), while miR-141-3p inhibitor were used to down-regulate miR-141-3p, respectively (Fig. 4C). Activation of SMAD2 as well as the expression of matrix proteins were decreased by knockdown of MALAT1 but reversed by both miR-141-3p inhibitor and ZNF217 overexpression (Fig. 4D). ELISA results also showed that the secretion of TGF-β2 was decreased by knockdown of MALAT1 but reversed by both miR-141-3p inhibitor and pcDNA-ZNF217 (Fig. 4E). Knockdown of MALAT1 significantly inhibited fibroblast proliferation and migration, while miR-141-3p inhibitor as well as ZNF217 overexpression reversed these phenomenon, respectively (Fig. 4F and G), which suggested that MALAT1 promotes the activation of TGF-β2/SMAD2 signaling pathway via targeting miR-141-3p, thus facilitates fibroblast proliferation, migration and matrix protein expression.
Fig. 4.
MALAT1 promotes fibroblast proliferation, migration, and matrix protein expression by activating ZNF217/TGF-β2/SMAD2 signal pathway (A) ZNF217 expression after transfection of pcDNA-ZNF217 and the empty plasmids in HFF-1 cells (B) ZNF217 protein levels were tested by western blotting (C) Effect of miR-141-3p inhibitor on the expression of miR-141-3p in HFF-1 (D) Fibroblast ZNF217, matrix protein and SMAD2 phosphorylation levels were detected by western blotting after indicated treatments (E) Expression levels of TGF-β2 were determined by ELISA in HFF-1 cells after indicated treatments (F) Proliferation of HFF-1 cells was subjected to MTT assays after indicated treatments (G) Cell migration of HFF-1 cells after indicated treatments. The data were presented as mean ± SD in triplicate. One-way ANOVA test was performed for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
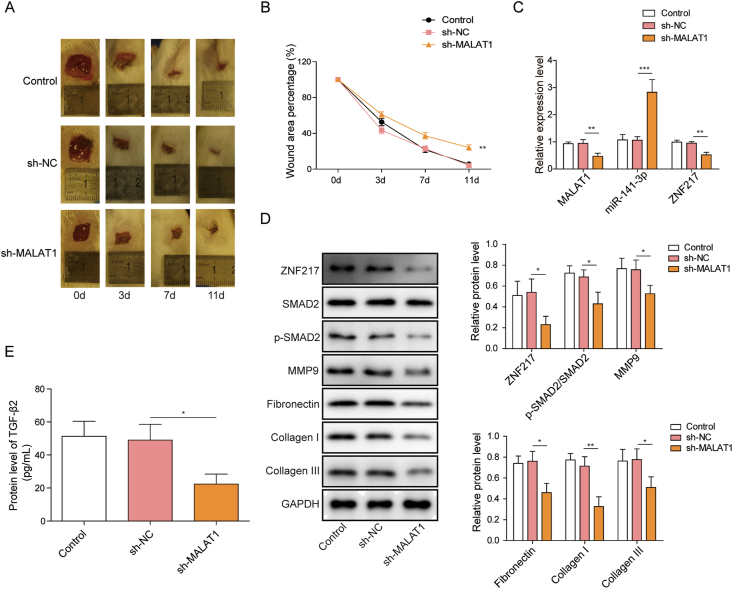
3.5. Knockdown of MALAT1 inhibits the wound healing of mice through ZNF217/TGF-β2/SMAD2 pathway via miR-141-3p
The findings so far have shown that MALAT1 promotes fibroblast proliferation and migration via miR-141-3p mediated ZNF217/TGF-β/SMAD2 signaling pathway. To further evaluate whether MALAT1 affects wound healing via ZNF217/TGF-β/SMAD2 signaling pathway, we conducted full-thickness wound model in C57BL/6 mice. Compared with sh-NC group, knockdown of MALAT1 significantly suppressed the wound healing process in vivo (Fig. 5A and B). Furthermore, the MALAT1 expression level was significantly reduced and miR-141-3p expression level was elevated in granulation tissue transfected with sh-MALAT1 (Fig. 5C). The mRNA (Fig. 5C) and protein levels (Fig. 5D) of ZNF217 were markedly decreased by sh-MALAT1. SMAD2 activation (Fig. 5D) and TGF-β2 level (Fig. 5E) in granulation tissue were significantly inhibited by sh-MALAT1. Last, the expression of matrix proteins were also decreased by knockdown of MALAT1 (Fig. 5D). These data suggest that knockdown of MALAT1 reduces the rate of wound healing by inhibiting ZNF217/TGF-β2/Smad 2 signal axis.
Fig. 5.
Knockdown of MALAT1 inhibits the wound healing of mice by inhibiting ZNF217/TGF-β2/SMAD2 signal pathway (A) Images of the mouse skin wounds in sh-NC and sh-MALAT1 treated groups 3, 7, and 11 days after surgery (B) The comparison of the percentage of wound area remaining in wounds transfection with sh-NC and sh-MALAT1 (C) MALAT1, ZNF217 and miR-141-3p levels in granulation tissues after indicated treatments (D) The protein levels of ZNF217, matrix proteins and SMAD2 phosphorylation were determined by western blotting after indicated treatments (E) Expression levels of TGF-β2 in granulation tissues were determined by ELISA in HFF-1 cells after indicated treatments. The data were presented as mean ± SD in triplicate. One-way ANOVA test was performed for statistical analysis. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
4. Discussion
Chronic wound affects millions of people around the world and is one of the major challenge to public health [3]. Fibroblasts play an important role in wound healing process [4]. MALAT1 overexpression in fibroblasts [12] accelerates wound healing by promoting fibroblasts migration [13,14]. However, the mechanism of how MALAT1 affect wound healing is not fully understood. Here we show that MALAT1 promotes wound healing process by targeting miR-141-3p/ZNF217 axis.
Previous studies have found that MALAT1 contributes cancer cell migration such as bladder cancer [30], lung cancer [31], colorectal cancer cells [32]. In recent years, MALAT1 was found to be engaged in wound healing process. MALAT1 containing exosome promotes human fibroblast migration [14]. Here we show that MALAT1 promotes human fibroblast cell (HFF-1) proliferation and migration. Fibroblasts express matrix metalloproteinases (MMPs) which degrade the fibrin clot and generate large amount of extracellular matrix (ECM) components, such as Fibronectin and collagen, to fill the wound site [33]. In present study, knockdown of MALAT1 reduced MMP9, Fibronectin and collagen I/III expression in HFF-1 cells, and inhibited the speed of wound healing in mouse skin wound model, suggesting MALAT1 facilitates wound healing process.
Despite that MALAT1 is engaged in wound healing, the mechanism of how MALAT1 affect wound healing needs further investigation. MiR-141-3p inhibites proliferation and migration of cancer cells [18,19,34] and fibroblasts [23]. Recent studies found that miR-141-3p targeted to the 3′-UTR of Zinc-finger E-box binding homeobox 1 (ZEB1) and inhibited epithelial cell migration [20,35]. We first demonstrated that MALAT1 directly inhibited miR-141-3p expression. Moreover, miR-141-3p regulated fibroblasts proliferation, migration as well as ECM protein expression, and might have an inhibitory effect on wound healing process.
Prevailing theory posits that ZNF217, acts as a transcription activator, promotes TGF-β expression via directly binding to the promoter of TGF-β [24]. TGF-β signaling accelerates the wound healing process by activating phosphorylation of SMAD family proteins [27,28]. Except that ZNF217 increases migration and proliferation of many cancer cells, such as colon, breast and ovarian cancer cells [[36], [37], [38]], recent study also finds that ZNF217 positively regulates wound healing process [26]. Since TGF-β/SMAD signaling enhances the synthesis of matrix proteins and accelerates wound healing process both in vitro and in vivo [24,25,29], it's not surprising that overexpression of ZNF217 promoted TGF-β2 and matrix protein expression, as well as proliferation and migration of fibroblast cells with knockdown MALAT1. However, no previous evidence demonstrates the connections of MALAT1, miR-141-3p and ZNF217. Here we showed that miR-141-3p inhibitor and ZNF217 overexpression increased fibroblast proliferation and migration which were inhibited by MALAT1 knockdown. Our findings suggested that miR-141-3p directly targeted ZNF217 mRNA and inhibited ZNF217 expression. Thus, MALAT1 may promote ZNF217/TGF-β/SMAD axis via regulating miR-141-3p expression.
In this study, we show that MALAT1 up-regulates ZNF217 by targeting miR-141-3p, enhances TGF-β2/SMAD2 signaling pathway and increases the expression of matrix protein in fibroblasts, thus promotes wound healing process. This study provides novel insights into the understanding of the role of MALAT1 in wound healing, and may provide potential target for the treatment of abnormal wound healing.
Authors’ contributions
Guarantor of integrity of the entire study: Zun-Hong Liang; study concepts: Zhi Zhang
Study design: Zun-Hong Liang; definition of intellectual content: Yun-Chuan Pan; literature research: Shi-Shuai Lin
Experimental studies: Zun-Hong Liang; data analysis: Zhi-Yang Qiu; statistical analysis: Zhi-Yang Qiu;
Manuscript preparation: Zun-Hong Liang;
Manuscript editing: Zun-Hong Liang;
Manuscript review: Zhi Zhang.
Declaration of competing interest
The authors have no conflict of interest to disclose.
Acknowledgements
This work was supported by the Wang Zhengguo Trauma Medicine Development Fund Project of Shanghai (No. 2017KJB-SS-002) and the High Level Talent Project in Natural Science of Hainan Province (No. 2019RC369).
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Gurtner G.C., Werner S., Barrandon Y., Longaker M.T. Wound repair and regeneration. Nature. 2008;453(7193):314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 2.Jones R.E., Foster D.S., Longaker M.T. Management of chronic wounds-2018. J Am Med Assoc. 2018;320(14):1481–1482. doi: 10.1001/jama.2018.12426. [DOI] [PubMed] [Google Scholar]
- 3.Frykberg R.G., Banks J. Challenges in the treatment of chronic wounds. Adv Wound Care. 2015;4(9):560–582. doi: 10.1089/wound.2015.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bainbridge P. Wound healing and the role of fibroblasts. J Wound Care. 2013;22(8):407–408. doi: 10.12968/jowc.2013.22.8.407. 410-12. [DOI] [PubMed] [Google Scholar]
- 5.Tao S.C., Rui B.Y., Wang Q.Y., Zhou D., Zhang Y., Guo S.C. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018;25(1):241–255. doi: 10.1080/10717544.2018.1425774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya A.P., Pastar I., Stojadinovic O., Lazovic S., Davis S.C., Gil J. Topical mevastatin promotes wound healing by inhibiting the transcription factor c-Myc via the glucocorticoid receptor and the long non-coding RNA Gas5. J Biol Chem. 2018;293(4):1439–1449. doi: 10.1074/jbc.M117.811240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herter E.K., Xu Landén N. Non-coding RNAs: new players in skin wound healing. Adv Wound Care. 2017;6(3):93–107. doi: 10.1089/wound.2016.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutschner T., Hämmerle M., Eißmann M., Hsu J., Kim Y., Hung G. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Canc Res. 2013;73(3):1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutschner T., Hämmerle M., Diederichs S. MALAT1—a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91(7):791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 10.Li L., Chen H., Gao Y., Wang Y.W., Zhang G.Q., Pan S.H. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Canc Therapeut. 2016;15(9):2232–2243. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 11.Kim J., Piao H.L., Kim B.J., Yao F., Han Z., Wang Y. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50(12):1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hutchinson J.N., Ensminger A.W., Clemson C.M., Lynch C.R., Lawrence J.B., Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8(1):39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt L.H., Spieker T., Humberg J., Marra A., Hillejan L., Berdel W.E. D78. Pathways and models OF lung oncogenesis. American Thoracic Society; 2012. MALAT-1 NcRNA enhances cellular migration and wound healing. A6369-A6369. [Google Scholar]
- 14.Cooper D.R., Wang C., Patel R., Trujillo A., Patel N.A., Prather J. Human adipose-derived stem cell conditioned media and exosomes containing malat1 promote human dermal fibroblast migration and ischemic wound healing. Adv Wound Care. 2018;7(9):299–308. doi: 10.1089/wound.2017.0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S., Zhang L., Song J., Wang Z., Huang X., Guo Z. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J Cell Physiol. 2019;234(3):2997–3006. doi: 10.1002/jcp.27117. [DOI] [PubMed] [Google Scholar]
- 16.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat Rev Canc. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 17.Du Y., Xu Y., Ding L., Yao H., Yu H., Zhou T. Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol. 2009;44(6):556–561. doi: 10.1007/s00535-009-0037-7. [DOI] [PubMed] [Google Scholar]
- 18.Liang Z., Li X., Liu S., Li C., Wang X., Xing J. MiR-141-3p inhibits cell proliferation, migration and invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys Res Commun. 2019;514(3):699–705. doi: 10.1016/j.bbrc.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Li W., Cui Y., Wang D., Wang Y., Wang L. MiR-141-3p functions as a tumor suppressor through directly targeting ZFR in non-small cell lung cancer. Biochem Biophys Res Commun. 2019;509(3):647–656. doi: 10.1016/j.bbrc.2018.12.089. [DOI] [PubMed] [Google Scholar]
- 20.Qian W., Cai X., Qian Q., Peng W., Yu J., Zhang X. lncRNA ZEB1-AS1 promotes pulmonary fibrosis through ZEB1-mediated epithelial–mesenchymal transition by competitively binding miR-141-3p. Cell Death Dis. 2019;10(2):1–12. doi: 10.1038/s41419-019-1339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta 1. Biochem Biophys Res Commun. 2017;487(4):769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 22.Quan B., Zhang H., Xue R. miR-141 alleviates LPS-induced inflammation injury in WI-38 fibroblasts by up-regulation of NOX2. Life Sci. 2019;216:271–278. doi: 10.1016/j.lfs.2018.11.056. [DOI] [PubMed] [Google Scholar]
- 23.Feng J., Xue S., Pang Q., Rang Z., Cui F. miR-141-3p inhibits fibroblast proliferation and migration by targeting GAB1 in keloids. Biochem Biophys Res Commun. 2017;490(2):302–308. doi: 10.1016/j.bbrc.2017.06.040. [DOI] [PubMed] [Google Scholar]
- 24.Cohen P.A., Donini C.F., Nguyen N.T., Lincet H., Vendrell J.A. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015;6(39):41566. doi: 10.18632/oncotarget.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bran G.M., Goessler U.R., Schardt C., Hormann K., Riedel F., Sadick H. Effect of the abrogation of TGF-β1 by antisense oligonucleotides on the expression of TGF-β-isoforms and their receptors I and II in isolated fibroblasts from keloid scars. Int J Mol Med. 2010;25(6):915–921. doi: 10.3892/ijmm_00000422. [DOI] [PubMed] [Google Scholar]
- 26.Zhu H.Y., Bai W.D., Li C., Zheng Z., Guan H., Liu J.Q. Knockdown of lncRNA-ATB suppresses autocrine secretion of TGF-β2 by targeting ZNF217 via miR-200c in keloid fibroblasts. Sci Rep. 2016;6(1):1–9. doi: 10.1038/srep24728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim Y.I., Kim K.S., Ahn H.J., Kang I.H., Shin M.K. Reduced matrix metalloproteinase and collagen transcription mediated by the TGF-beta/Smad pathway in passaged normal human dermal fibroblasts. J Cosmet Dermatol. 2020;19(5):1211–1218. doi: 10.1111/jocd.13114. [DOI] [PubMed] [Google Scholar]
- 28.Palumbo-Zerr K., Zerr P., Distler A., Fliehr J., Mancuso R., Huang J. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat Med. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 29.Bran G.M., Sommer U.J., Goessler U.R., Hoermann K., Riedel F., Sadick H. TGF-β1 antisense impacts the SMAD signalling system in fibroblasts from keloid scars. Anticancer Res. 2010;30(9):3459–3463. [PubMed] [Google Scholar]
- 30.Ying L., Chen Q., Wang Y., Zhou Z., Huang Y., Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt L.H., Spieker T., Koschmieder S., Schaffers S., Humberg J., Jungen D. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 32.Yang M.H., Hu Z.Y., Xu C., Xie L.Y., Wang X.Y., Chen S.Y. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta (BBA) - Mol Basis Dis. 2015;1852(1):166–174. doi: 10.1016/j.bbadis.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li B., Wang J.H.-C. Fibroblasts and myofibroblasts in wound healing: force generation and measurement. J Tissue Viability. 2011;20(4):108–120. doi: 10.1016/j.jtv.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S., Wa Q., Pan J., Peng X., Ren D., Huang Y. Downregulation of miR-141-3p promotes bone metastasis via activating NF-κB signaling in prostate cancer. J Exp Clin Canc Res. 2017;36(1):173. doi: 10.1186/s13046-017-0645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L., Xu M., Xu J., Wu K., Fang Q., Liang Y. ELF3 promotes epithelial–mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 2018;9(3):1–14. doi: 10.1038/s41419-018-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z.C., Zheng L.Q., Pan L.J., Guo J.X., Yang G.S. ZNF217 is overexpressed and enhances cell migration and invasion in colorectal carcinoma. Asian Pac J Cancer Prev APJCP. 2015;16(6):2459–2463. doi: 10.7314/apjcp.2015.16.6.2459. [DOI] [PubMed] [Google Scholar]
- 37.Bai W.D., Ye X.M., Zhang M.Y., Zhu H.Y., Xi W.J., Huang X. MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. Int J Canc. 2014;135(6):1356–1368. doi: 10.1002/ijc.28782. [DOI] [PubMed] [Google Scholar]
- 38.Rahman M.T., Nakayama K., Rahman M., Katagiri H., Katagiri A., Ishibashi T. Gene amplification of ZNF217 located at chr20q13. 2 is associated with lymph node metastasis in ovarian clear cell carcinoma. Anticancer Res. 2012;32(8):3091–3095. [PubMed] [Google Scholar]