Graphical abstract
Keywords: Microbiome, Microbiota, 16S rRNA gene, Human Milk, Microbial barcoding, PCR primers
Abstract
Human milk is the ideal food for infants due to its unique nutritional and immune properties, and more recently human milk has also been recognized as an important source of bacteria for infants. However, a substantial amount of fundamental human milk microbiome information remains unclear, such as the origin, composition and function of the community and its members. There is emerging evidence to suggest that the diversity and composition of the milk microbiome might differ between lactation stages, due to maternal factors and diet, agrarian and urban lifestyles, and geographical location. The evolution of the methods used for studying milk microbiota, transitioning from culture dependent-approaches to include culture-independent approaches, has had an impact on findings and, in particular, primer selection within 16S rRNA gene barcoding studies have led to discrepancies in observed microbiota communities. Here, the current state-of-the-art is reviewed and emerging frontiers essential to improving our understanding of the human milk microbiome are considered.
1. History of milk microbiome
Although many of the nutritional and immune system benefits of human milk have been established [1], [2], [3], [4], the influence of the milk microbiome on health is still in the early days of exploration. A recent review [5], while stressing further evidence is needed, has explored the idea that human milk could represent a reservoir of bacteria which influences infant gut microbiome diversity and aspects of health such as allergy prevention. The existence of the human microbiome is well-established [6] and was studied as far back as Antonie van Leeuwenhoek (1632–1723); however, microbiome study in human milk is very recent due to a long-standing belief that human milk was sterile [7]. In the late ‘60 s, the presence of bacteria in human milk was related to the low levels of personal hygiene and environmental sanitation in women from Guatemala [8] (Table 1). Later, diverse methods were employed in order to make milk ‘bacteriologically safe’, such as heating and freezing [9], [10]. By the late ‘80 s it had been recognized that human milk contained growth-promoting substances postulated to be involved in the development of microbiota [11]. In the early 2000s, interest and study of the microbiome significantly increased and promptly led to important discoveries, including the existence of commensal bacteria in human milk [12]. Bacteria were recognized as components of the natural microbiota rather than contaminants [13] resulting in notable differences among human body sites that included highly diverse communities in skin, less variable communities within the oral cavity compared to other habitats and highly variable gut communities between individuals but with low variability over time [14].
Table 1.
Historical Study of Milk Microbiome.
| Time | Microbiome Concept | Historical Human Milk Microbiome Findings |
|---|---|---|
| ‘60 s | Enterobacteriaceae detected | The presence of Enterobacteriaceae in human breast milk is related to the low levels of personal hygiene and environmental sanitation in women from Guatemala. It is concluded that an interchange of breast milk bacteria between the mother’s breast and the infant’s mouth is possible [8]. |
| ‘70 s | Heat sterilization | Bacteria in milk were detected. Breast milk was heated in order to be “bacteriologically safe” [9]. |
| ‘80 s | Microbial growth promotion suggested | Growth-promoting substances in human milk postulated to be involved in the development of microbiota. Components like lactoferrin and a saccharide containing N-acetyl glucosamine could provide an adequate environment for bacterial growth [11]. |
| ‘90 s | Bacteria considered solely contamination | Presence of bacteria considered to be due to contamination in frozen milk. Contamination levels of human milk compared to pasteurized cow’s milk to develop guidelines for the acceptable microbial quality of human milk [10]. |
| ‘00 s | Commensal bacteria | A study looks for commensal bacteria inhibiting Staphylococcus aureus as published reports at that time had only focused on pathogenic bacteria [12]. |
| Entero-mammary pathway | Bacteria are recognized as components of natural microbiota rather than contaminants. The hypothesis of an Entero-Mammary route is proposed [13]. | |
| Retrograde flow | Description of the “retrograde infant-to-mother transfer” in which microbes are transmitted via the skin and via retrograde-flow of milk into the duct during suckling [16]. | |
| Human Microbiome Project (HMP) | The Human Microbiome Project started in 2007 by the National Institute of Health. The first large-scale effort to characterize the healthy human microbiome across 5 body sites: the digestive tract, mouth, skin, nasal cavity and vagina [120]. | |
| Anaerobic species | Alternative route for microbial transfer due to the presence of the anaerobic genus Bifidobacterium in breast milk samples [39]. Although this led to important new theories about the origin of milk bacteria, Bifidobacterium is now recognised as containing extensive strain variance in O2 tolerance and sensitivity [121], [122]. | |
| Gut and milk link to lymphatic system | Evidence of an internal microbial transfer pathway due to the presence of maternal gut and breast milk bacteria in the lymphatic system is strengthened, although more evidence is necessary due to the characteristics of the study. [27]. | |
| Tissue specific microbiome patterning | Identification of distinct patterns of the microbiome among human body sites [14]. | |
| ‘10 s | Vertical transfer widely accepted | The vertical transfer to the neonate via breastfeeding was strengthened when it was found that the same bacteria is shared by the breast milk, maternal and infant faeces [123]. |
One of the most important advances in microbiome research began in 2007 with the Human Microbiome Project conducted by the National Institutes of Health, the first large-scale effort to characterize the healthy human microbiome [15]. The study included a population of 242 healthy adults and a description of the microbiomes of up to 18 body sites. A total of 5,298 samples were collected within the human airways, skin, oral cavity, gut and vagina. The result was the collection of 11,174 primary biological specimens representing the human microbiome that formed the backdrop for further microbiome research. However, at that time, sites like the mammary glands or human milk were not analysed.
2. Sources of the milk microbiome
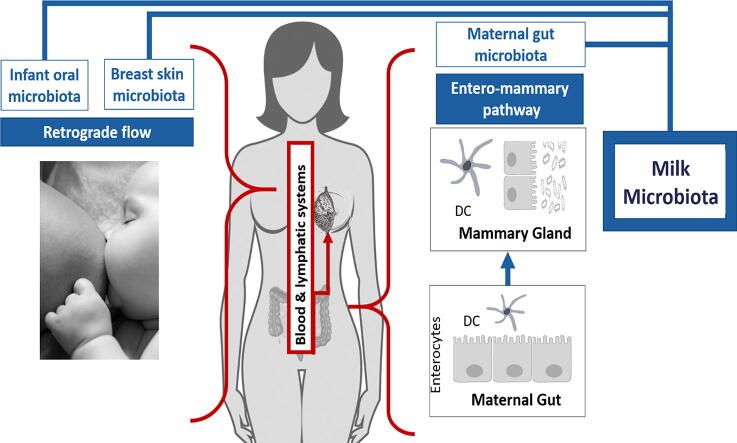
Research has aimed to explain the sources of the milk microbiome (Fig. 1). Two main pathways have been recognized as sources that might potentially contribute to the human milk microbiome: retrograde flow and the entero-mammary pathway.
Fig. 1.
Sources of milk microbiota Two potential sources of bacteria which could contribute to the breast milk microbiome: retrograde flow [16] and the entero-mammary pathway [13], [27], [28], [29]. The retrograde flow pathway suggests that the during suckling, infant oral microbiota and breast skin microbiota can reach the breast milk microbiota. The entero-mammary pathway suggests that dendritic cells in the maternal gut that cross the intestinal epithelium can take up bacteria from the intestinal lumen which are then taken to the mammary gland through the blood and lymphatic systems.
Retrograde flow is an infant-to-mother transfer in which microbes are transmitted via the skin and saliva from the infant’s oral cavity into the duct during suckling [16]. This could explain how bacteria commonly found in the infant oral cavity, such as those within the genera Veillonella, Leptotrichia and Prevotella [17], or bacteria commonly found in the vagina, such as Lactobacillus [18] are sometimes found in human milk. The vaginal bacteria could be acquired by the infant through a vaginal delivery and then transferred to the human milk through retrograde flow. Recent evidence has suggested that the mode of delivery might influence human milk composition [17], [19], [20], [21], [22]. Some studies show higher bacterial diversity and richness in the milk from vaginal deliveries [17], [19], [20], [22]. Human milk from vaginal deliveries has been reported to have higher Bifidobacterium [21], Streptococcus and Haemophilus, and lower abundance of Finegoldia spp., Halomonas spp., Prevotella spp., Pseudomonas spp. and Staphylococcus spp. [22]. However, despite these differences, findings are not consistent as other studies did not report differences [23], [24], [25], [26].
The entero-mammary route has emerged as another potential route for how maternal gut microbiota might enter and enrich the human milk microbiome. The route proposes that maternal gut bacteria are translocated through the intestinal epithelial barrier via dendritic cells which cross the paracellular space of the intestinal epithelium and directly sample bacteria from the intestinal lumen [13], [27], [28], [29]. Circulation of lymph within the mucosal associated lymphoid tissue could then allow the maternal gastrointestinal tract microbiota to reach distant sites such as the lactating mammary gland.
3. Major factors that influence the human milk microbiome
Milk microbiome composition is influenced by diverse factors such as stage of lactation, maternal BMI, diet and use of antibiotics [17], [20], [21], [23], [26], [30], [31], [32], [33], [34], [35]. However, there are other factors that remain unexplored and might have an important influence on human milk composition including age, parity, geographical location, and interactions with the environment.
3.1. Stage of lactation
There is evidence of substantial shifts in microbiota composition at different stages of lactation. Chen et al. [36] found that colostrum and transitional milk share 48.9% of bacterial genera and 42% of bacterial species, so there are common and unique bacteria between the two stages of milk. Cabrera et al. [17] reported initially that the microbiota of colostrum was dominated by Weissella, Leuconostoc, Staphylococcus spp., Streptococcus spp., and Lactococcus spp. Later, milk samples collected 1–6 months post-partum had higher levels of Veillonella, Prevotella, Leptotrichia, Lactobacillus spp, Streptococcus spp., and increasing levels of Bifidobacterium and Enterococcus spp, which the authors speculated was related to the frequent interaction with the infant’s oral microbiota. Similar results were reported by Khodayar et al. [21], in which Bifidobacterium and Enterococcus spp. counts increased throughout the lactation period. Another study reported that colostrum had a higher diversity typical of skin and maternal gut bacteria, and as lactation progressed, mature milk became less diverse but increased in infant oral and skin associated bacteria [37]. While improved technologies will help elucidate the current discrepancies in microbiota across lactation stage, a consistent finding is that the microbiome is dynamic and remodels as lactation progresses [38], [39]. Given this, other factors known to change throughout lactation, such as the nutritional and immunological composition of milk, will need to be explored as modifiers of human milk microbiome.
3.2. Maternal BMI and diet
The relationship between BMI and human milk microbiota is not clear. Some studies did not find any significant associations [31], [33] but others have reported that maternal BMI and weight gain during pregnancy do impact the diversity of bacterial community in human milk [17], [40]. Those studies that found an association reported that obesity and excessive weight gain reduced diversity of the milk microbiome [17], [40]. Milk samples of mothers with higher BMI had higher abundance of Lactobacillus in colostrum and higher abundance of Staphylococcus and Akkermansia in mature milk [17] as well as higher Granulicatella [33]. On the other hand, higher BMI has been related to reductions in the genera Bifidobacterium in milk produced at 6 months [17] and Bacteroides [33] and, more broadly, reductions in the phyla Proteobacteria [23] and Firmicutes [35].
Maternal diet, a factor often associated with BMI, could influence microbial composition of human milk directly or as a secondary effect by influencing human milk nutritional content. Some studies have reported that maternal diet influences the nutrient concentration of milk [41], [42], [43] and likely shapes its bacterial community [33]. With regards to macronutrients, maternal intake of saturated fatty acids and monosaturated fatty acids were inversely associated with Corynebacterium, and protein consumption was positively correlated with the relative abundance of Gemella [33]. In relation to micronutrients, a negative correlation was observed between pantothenic acid intake and Streptococcus and between Lactobacillus with thiamin, niacin, vitamin B-6 and chromium and a positive correlation was found between riboflavin and calcium with Veillonella [33]. Since the relationship between diet and gut microbiota has been extensively explored [44], [45], [46], [47], the diet induced alterations of maternal gut microbiota might also influence human milk microbes, and therefore vertical transfer to the infant [48]. Nonetheless, there is limited evidence about the direct relationship of maternal diet on the human milk microbiome, or the interactions between maternal diet, maternal microbiota and infant microbiota. Despite the limited evidence, it is valuable that research suggests that maternal diet can influence human milk microbial composition, as maternal diet is one of the most modifiable factors by which interventions could be explored for modulating the human milk microbiome.
3.3. Use of antibiotics and probiotics
Results regarding the use of antibiotics are contradictory. For instance, in a study of women living in Germany and Austria abundance of lactobacilli or bifidobacteria was lower in women who had received antibiotics during pregnancy or lactation [26]. However, another study reported that the effect of antibiotic exposure on human milk microbiota at 1 month postpartum appeared to be an increase in bacterial richness and diversity [20]. Similarly, it has also been reported that mean bacterial counts in milk produced by women receiving antibiotics were higher than in milk from women taking probiotics [32]. Although probiotics are largely considered to support microbiome diversity, a gut microbiome study found a similar pattern and reported that after the use of antibiotics, the introduction of probiotics actually delayed and impaired mucosal microbiome reconstitution [49]. However, the impact of the consumption of probiotics during pregnancy and lactation on the milk microbiome and infant health following antibiotic use has not been widely explored.
4. Inconsistencies in dominant milk microbiome bacteria persist
Although lactation stage, maternal BMI, diet and use of antibiotics are thought to drastically influence the microbiota present in human milk, a number of taxa are consistently observed across studies. The genera often found include Staphylococcus, Streptococcus, Lactobacillus, Bifidobacterium Pseudomonas, Corynebacteria, Acinetobacter, Sphingomonas, Serratia, Ralstonia, Bradyrhizobiaceae and Cutibacterium [31], [50], [51], [52], [53], [54], [55], [56]. According to the latest systematic review 590 different genera have been detected via sequencing human milk, from which the 10 most frequently found were: Staphylococcus (genera found in 97% of studies; range of relative abundance 5–83%), Streptococcus (95%; < 1 to 74%), Lactobacillus (63%; < 1 to 5%), Pseudomonas (50%; < 1 to 17%), Bifidobacterium (42%; < 1 to 5%), Corynebacterium (42%; < 1 to 6%), Enterococcus (42%; 1%), Acinetobacter (39%; 2 to 4%), Rothia (34%; 1 to 6%) and Cutibacterium (29%; < 1 to 3%) [34].
Some studies have attempted to define a “core” microbiome of human milk [51], [52], but the presence of some bacteria in human milk remains controversial as different studies have considered specific genera as either contaminants or as part of the “core” microbiome. Salter et al. [57] found that laboratory reagents were commonly contaminated with species from genera that are repeatedly reported as present in human milk, such as Acinetobacter, Cutibacterium, Novosphingobium, Pseudomonas, Ralstonia, Sphingomonas and Streptococcus, and which can become spurious signals in samples with low bacterial load during 16S rRNA gene amplification. Jiménez et al. [53] speculated that while species within genera such as Cutibacterium and Streptococcus have been isolated using culture-dependent techniques, species from genera such as Sphingomonas and Pseudomonas (containing many readily culturable species) had not, suggesting these genera could commonly be contamination from laboratory reagents. More recently, similar warnings that reagent contamination can substantially influence low bacterial load human milk microbiome samples were raised within Douglas et al. [58], which provided evidence that species within Pseudomonas are common contaminants and also likely genuinely present in human milk (highlighting the current challenges within the field). Similarly, Acinetobacter, a bacteria commonly found in soil, has been reported previously in human milk [35], [50], [56], [59]. Sakwinska et al. [59] and Urbaniak et al. [56] both detected Acinetobacter in their milk samples where milk collection was done without aseptic cleansing of the breast and also rejection of foremilk. However, Sakwinska et al. [56] found it unlikely that the predominance of Acinetobacter was due to the collection protocol and argued that Acinetobacter might be a specific feature of breastfeeding associated microbiota. Other studies that have reported bacteria commonly associated with soil in their samples correlated it to the maternal diet based in legumes [60] or to the proximity to a soil environment [61].
Care should be taken to clearly report the use of PCR controls in microbiome studies as the presence of species from certain bacterial genera in human milk remains controversial. Because of this, interpretation of observed genera should also include their potential as contamination while not precluding novel discovery within this understudied field. Few studies analysing human milk microbiome have been conducted at species level (Table 2), despite the advantages improved resolution would have by providing more information about functionality and in facilitating biological interpretation. However, research at species level is increasing as 16S rRNA gene barcoding and shotgun metagenomics technologies improve.
Table 2.
Summary of most abundant bacteria, lifestyle or community considered and methods employed in breast milk microbiome studies.
| Most Abundant Taxa | Cohort location | Method | Region | Primera(common label) | Primer sequences (including degeneracy variants) | Template free PCR controlsb | Reference |
|---|---|---|---|---|---|---|---|
| Staphylococcus, Streptococcus, Serratia, Pseudomonas, Corynebacterium, Ralstonia, Cutibacterium/Propionibacterium, Sphingomonas and Bradyrhizobiaceae | USA | 16S rRNA gene sequencing | V1-V2 | 27FC and 338R | 27F: ‘5-AGAGTTTGATCCTGGCTCAG-3′338R: ‘5-TGCTGCCTCCCGTAGGAGT-3′ | Yes | [52] |
| Weisella, Leuconostoc, Staphylococcus, Streptococcus, Lactococcus, Veillonella, Leptotrichia, and Prevotella | Finland | 16S rRNA gene sequencing | V1-V3 | 27F and 533R | N/A | Yes | [17] |
| Cutibacterium acnes, Staphylococcus epidermis, Streptococcus (S. salivarius,S. thermophilus, S.vestibularis, S. mitis, S. pneumoniae, Staphylococcus (S. lugdunensis, . aureus, S. haemolyticus, S. hominis, S. pasteuri, S. wameri) Veillonella (V. atypical, V. dispar, V. parvula), , Rothia mucilaginosa, Propionibacterium granulosum, Bifidobacterium breve, Klebsiella pneumoniae, Escherichia/Shigella, Lactobacillus (L. gasseri, L, brevis), Enterococcus (E. faecalis, E. gallinarum) | Switzerland | 16S rRNA gene sequencing | V5 – V6 | 784F and 1061R | 784F: ‘5-AGGATTAGATACCCTGGTA-3′1061R: ‘5-CRRCACGAGCTGACGAC-3′ | No | [65] |
| Staphylococcus spp., Streptococcus spp., Veillonella spp., Corynebacterium spp., Rothia spp., Enterococcus spp., Lactobacillus spp., Escherichia/Shigella spp., Klebsiella spp. | Switzerland | 16S rRNA gene sequencing | V5 – V6 | 784F and 1061R | 784F: ‘5-AGGATTAGATACCCTGGTA-3′1061R: ‘5-CRRCACGAGCTGACGAC-3′ | No | [123] |
| Eubacterium, Lactobacillus, Acinetobacter, Xanthomonadaceae, Stenotrophomonas | Canada | 16S rRNA gene sequencing | V6 | N/A | Fw: ‘5-CWACGCGARGAACCTTACC-3′Rv: ‘5-ACRACACGAGCTGACGAC-3′ | Yes | [56] |
| Staphylococcus, Streptococcus, Bacteroides, Faecalibacterium, Ruminococcus, Lactobacillus, Cutibacterium/Propionibacterium, Staphylococcus aureus and Staphylococcus epidermis | Spain | Shotgun metagenomic sequencing | – | – | – | – | [53] |
| Streptococcus, Staphylococcus, and Neisseria | USA | 16S rRNA gene sequencing | V4 | 515F and 806R | 515F: GTGCCAGCMGCCGCGGTAA806R: GGACTACHVGGGTWTCTAAT | No | [30] |
| Staphylococcus, Pseudomonas, Streptococcus and Acinetobacter | Spain | 16S rRNA gene sequencing | V1-V3 | 27F and 533R | 27F: 5′-AGAGTTTGATCMTGGCTCAG-3′) 533R: 5′-GCCTTGCCAGCCCGCTCAGGC-3′ | No | [50] |
| Staphylococcus, Pseudomonas, Enterobacteriaceae,Streptococcus and Lactobacillus | Canada | 16S rRNA gene sequencing | V6 | N/A | Fw: 5′-CWACGCGARGAACCTTACC-3′Rv: 5′- ACRACACGAGCTGACGAC −3′ | Yes | [105] |
| Staphylococcus, Streptococcus, Pseudomonas and Acinetobacter | Spain, Finland, South Africa and China | 16S rRNA gene sequencing | V4 | 515F and 806R | N/A | Yes | [35] |
| Streptococcus Staphylococcus andAcinetobacter | China | 16S rRNA gene sequencing | V4 | 515F and 806R | 515F: 5′-GTGCCAGCMGCCGCGGTAA806R: 5′-GGACTACHVGGGTWTCTAAT-3′ | No | [59] |
| Streptococcus, Pseudomonas, Staphylococcus, Lactobacillus, Propionibacterium,Herbaspirillum, Rothia, Stenotrophomonas, Acinetobacter, Bacteroides, Halomonas, Veillonella, Sphingomonas, Delftia, andCorynebacterium | Taiwan and China | 16S rRNA gene sequencing | V1 – V2 | 27F and 338R | 8F/27F-mod: 5′-AGRGTTTGATYMTGGCTCAG-3′338R: 5′-TGCTGCCTCCCGTAGGAGT-3′ | No | [31] |
| Pseudomonas, Staphylococcus and Streptococcus | Ireland | 16S rRNA gene sequencing | V3 – V4 | Bakt314F-Bakt805R | Bakt314F: 5′-CCTACGGGNGGCWGCAG-3′Bakt805R: 5′-GACTACHVGGGTATCTAATCC-3′ | No | [54] |
| Streptococcaceae, Staphylococcaceae, Gamellaceae, Alicyclobacillaceae, Enterobacteriaceae, Pseudomonadaceae, Moraxellaceae, Xanthomonadaceae, Bradyrhizobiaceae, Caulobacteraceae, Neisseriaceae and Weeksellaceae | USA | 16S rRNA gene sequencing | V4 | 515F and 806R | 515F: GTGCCAGCMGCCGCGGTAA806R: GGACTACHVGGGTWTCTAAT | Yes | [24] |
| Pseudomonas, unclassified Enterobacteriaceae, Enterobacter,unclassified Pseudomonadaceae, Klebsiella, Ralstonia, Acinetobacter andSerratia, Bacillaceae, Staphylococcus, Enterococcus, Bicullus, unclassified Lachnospiraceae, Streptococcus | India | 16S rRNA gene sequencing | V2 – V3 | 101F and 518R | 101F:5′-ACTGGCGGACGGGTGAGTAA 3′518R: 5′-CGTATTACCGCGGCTGCTGG-3′ | No | [99] |
| Streptococcus, Staphylococcus, Veillonella, Corynebacterium, Rhodococcus, Dyella, Lactobacillus, Prevotella, Micrococcus and Hafnia. | Central African Republic | 16S rRNA gene sequencing | V1 – V3 | 27F-YM + 3 and 534R | 27F-YM + 3: 5′-AGMGTTYGATYMTGGCTCAG-3′534R: 5′-ATTACCGCGGCTGCTGGC-3′ | Yes | [92] |
| Streptococcus, Staphylococcus, Ralstonia, Acidovorax, Acinetobacter, Aquabacterium, Massilia, Agrobacterium, Rheinheimera, Veillonella, Vogesella, Nocardioides and Pseudomonas. | Canada | 16S rRNA gene sequencing | V4 | 515F and 806R | 515F: 5′-GTGCCAGCMGCCGCGGTAA-3′806R: 5′-GGACTACHVGGGTWTCTAAT-3′ | Yes | [23] |
| Streptococcus, Staphylococcus, Corynebacterium, Cutibacterium/Propionibacterium, Rhizobium, Lactobacillus, Dyella, Rothia, Kocuria, Veillonella, Bifidobacterium, Acinetobacter, Klebsiella, Gemella, Achromobacter, Escherichia/Shigella, Bacillus, Stenotrophomonas, Enterococcus, Janthinobacterium, Anaerococcus, Acidocella, Enterobacter, Bacteroides, Pseudomonas, Chryseobacterium, Tatumella, Psychrobacter, Clostridium sensu stricto 18. | Ethiopia, Kenya, Ghana, Gambia, Peru, Spain, Sweden and USA | 16S rRNA gene sequencing | V1 – V3 | 27F-YM + 3 and 534R | 27F-YM + 3: 5′-AGMGTTYGATYMTGGCTCAG-3′534R: 5′-ATTACCGCGGCTGCTGGC-3′ | Yes | [89] |
| Staphylococcus, Kaistobacter, Paracoccus, Pseudomonas,Bradyrhizobium, Methylobacterium, Acinetobacter, Cutibacterium/Propionibacterium, Corynebacterium,and Microbacterium; three families Phyllobacteriaceae, Sphingomonadaceae, Gemellaceae | Mexico | 16S rRNA gene sequencing | V3 | 341F and 518R | 341F: CCTACGGGAGGCAGCAG518R: ATTACCGCGGCTGCTGG | Yes | [98] |
Primer labels are often inconsistent in the literature and so exact sequences are provided where reported.
The use of no template PCR controls does not preclude the possibility that any observed sequences were contamination.
“27F” with no degeneracy is sometimes called 8F.
5. Methods shape observed bacterial taxa in human milk
The methods used to study the human milk microbiome continue to advance. The initial studies used bacterial culture techniques followed by phenotyping of isolated strains using morphological and biochemical characteristics. These studies isolated only a limited number of genera, predominantly facultative anaerobes such as members of the Staphylococcus spp., Streptococcus spp., Propionibacterium spp. (now Cutibacterium in the case of C. acnes), Rothia spp., Enterococcus spp. and Lactobacillus spp. [62], [63], [64]. Studies that have used culturing techniques have generally reported that human milk harbours relatively low mean viable bacterial counts often < log 3 cfu/ml [12], [27], [65]. While still a powerful tool for assessing viability of specific bacterial strains, culture-dependent analyses are limited in revealing only taxa capable of surviving sampling procedures and growth under laboratory conditions [25]. This selection can potentially reduce the observed microbiota community in complex habitats to below 1% of the diversity currently estimated by culture-independent approaches [66], although research suggests 50% of bacteria may be a more reasonable rough estimate in the human gut [67]. This difficulty to culture can be due to the lack of specific required nutrients in the culture medium, toxicity of the culture medium, inhibition by other bacteria or a dependence on association with other species (such as present in bacterial consortia or eukaryote host interactions) [68]. Therefore, although culture-based techniques are vital for the study of specific bacteria of clinical importance or functional interest [69], [70], they can be heavily biased and drastically underestimate the diversity when used to assess a microbiome community.
To overcome these challenges, culture-independent metagenomic approaches have been developed in which high-throughput sequencing is harnessed to identify bacteria within microbiome samples using shotgun metagenomics or the 16S ribosomal RNA gene [70]. The major advantage of these approaches is the possibility to detect difficult or yet-to-be-cultured bacteria in addition to improved sample throughput without a requirement of viable cells (allowing the use of frozen samples) [25]. Shotgun metagenomics attempts to sequence DNA directly from DNA fragments derived from all the genomic material present within a microbiome sample and attempts de novo assembly of as many entire genomes or large contiguous sequences as possible in order to infer taxonomic and functional information [70], [71]. As this approach does not target a specific gene for PCR, it does not include the same amplification biases associated to 16S rRNA gene barcoding and is widely considered the gold standard of microbiome research. However, in addition to the high expense currently associated with high depth shotgun metagenomics (which is rapidly dropping [72]), the approach generates large amounts of reads from complex samples which are challenging to assemble de novo [73], [74]. While de novo shotgun metagenomics has yet to become common for the study of the human milk microbiota, recent research has used shotgun metagenomics sequence reads as markers for mapping to reference libraries [53], [75], [76], [77]. While read mapping approaches are still improving in terms of accuracy for quantitative analysis, made difficult by database limitations, very low mapping rates and extensive sequence ambiguity, these rapid improvements in sequencing and bioinformatics methodologies suggest an exciting future for high-resolution identification of metagenomes which includes distinct inventories of genes between human milk microbiome communities. Alternatively, the 16S ribosomal RNA (rRNA) gene has been the most popular approach used for microbial community assessment over recent decades [78], [79], [80], [81], [82]. The value of the 16S rRNA gene as a ‘barcode’ for the identification and phylogenetic classification of bacterial species lies in the very highly conserved function of 16S rRNA leading to regions of hyper-conservation within the gene [83], [84]. These regions of conservation can then be targeted by primers in order to amplify proximal hyper-variable sequence regions (an amplicon) used as a potentially unique barcode of life [85].
In human milk research, 16S rRNA is still the most popular tools for profiling microbiome samples for quantitative comparison of groups or treatments (Table 2). While some limitations of the 16S rRNA gene barcoding approach are generally well recognised in the field, including the impact of low bacterial load in human milk [57] and poor utility for inference of biological functions in the community [70], primer specificity has been less well addressed. Certain primers have, in the past, been considered “universal” to prokaryotes and thought to amplify hypervariable regions from all bacteria. Research including that conducted by Klindworth et al. [85] has now demonstrated that no known primer pair is universal in amplifying 16S rRNA gene regions across all currently known and well-characterised bacterial species, although many have coverage of over 90% of known bacteria. Given that the observed absence or presence of certain bacteria can be determined by primer pair selection and the extensive range of primers used in human milk microbiome research (Table 2), it is perhaps not unsurprising that the “core” human milk microbiome genera or species have been reported as inconsistent in systematic literature reviews [86], [87].
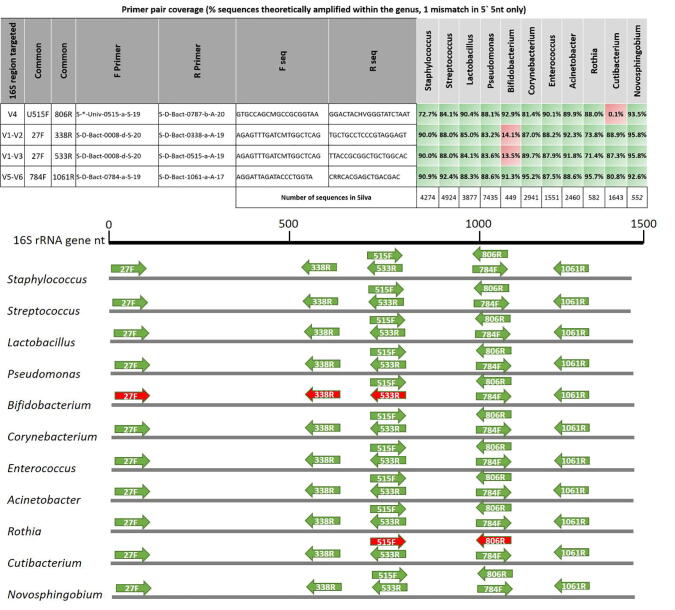
Using the TestPrime tools in the Silva rRNA database [88] and the research by Klindworth et al. [85], it is possible to assess the specific utility of primers commonly used in human milk microbiome research to amplify the putative “core” milk genera in silico. Interestingly, the primers 27F/533R (V1 region targeting), which are often used in human milk microbiome studies [17], [33], [52], [89], [90], have high coverage for amplification of the genus Cutibacterium but not species within the genus Bifidobacterium (with the most common 27F variant, Fig. 2), both of which are considered putative “core” genera [52], [53], [65] (Table 2). This shortfall in the coverage of Bifidobacterium of the commonly used 27F primer was reported by Frank et al. [91], who designed a high degeneracy variant of seven primers (27F YM + 3) with improved coverage in the genera which has been used in human milk microbiome research [92], [89]. Conversely, the other most commonly used primers 515F/806R primer pair (V4 region targeting) will likely amplify species within the genus Bifidobacterium but have very low coverage and do not amplify species from the genera Cutibacterium. To overcome these discrepancies, the 27F YM + 3 high degeneracy variant (within 27F/533R, V1-V3 targeting) or the 784F/1061R primer pair (V5-V6 targeting) should be considered more suitable for human milk research due to high coverage within all of the genera currently considered “core” in human milk (Fig. 2). It is important to recognize that these primers might still fail to amplify yet-to-be-identified species but will allow for the quantitative assessment of relative changes in most species within these important genera when experimentally comparing groups of mothers. While there is a desperate need to increase research into the human milk microbiome using tools such as 16S rRNA gene barcoding, care needs to be taken with all culture-independent techniques to not assume a perfect snapshot of the community, similar to the lessons previously learned with culture-based community assessment [64], [65].
Fig. 2.
16S rRNA gene primers pair coverage of major breast milk microbiome genera Common 16S rRNA gene primer pairs 515F-806R, 27F-338R, 27F-533R and 784F-1061R [34] were tested in silico against all sequences within major breast milk microbiome genera in the SILVA database using the TestPrime tool (set to allow 1 mismatch outside the 3′ first 5 nucleotides; https://www.arb-silva.de/search/testprime/) [34]. Percentage of sequences (coverage) within each genus is reported. High and low coverage primer pairs are illustrated in green and red, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
6. Emerging frontiers in human milk microbiome research
6.1. Overlooked maternal health issues: age and parity
Two overlooked factors that could be impacting the human milk microbiome include maternal age and parity. To date, only one study has analysed the relationship with maternal age, finding no association with human milk microbiota at family level resolution [23]. However, as there are established differences in the development of breast immunity [93], nutritional requirements and human milk nutrient composition [94] throughout a women’s lifetime, these factors could also affect the human milk microbiota and should be assessed at higher taxonomic resolution. Parity has also been reported only in one study finding a sex-dependent association with milk microbiome diversity [23]. This study reported an association of human milk microbiota composition with multiparity in female infants but did not characterize the microbiome. Parity could be a factor influencing milk microbiome if we consider the interactions of milk microbiome such as the retrograde flow from the infant [16]. Assuming retrograde flow, it could be expected that a multiparous mother will have a mammary gland with more diverse microbiota since it has been previously inoculated by the bacteria transferred by her previous infant(s) during earlier pregnancies [95].
6.2. Geographical location
Another factor to consider when identifying dominant bacteria in human milk is geographical location, lifestyle, and community. Multi-country studies confirm that human milk microbiome composition differs between and within countries as well as the presence of unique bacteria exclusively related to some study sites [35], [89], [96].
Some human milk microbiome studies have characterized their study population based on their lifestyle and community, such as rural [92], urban [35], [59], [97], [98], rural and urban [89], [99] or low socioeconomic [30], [100]. These population characteristics have been proposed as factors that might potentially affect the human milk microbiota composition [35], [59], [89], [92], [97]. A recent study by Lackey et al. [89] aimed to elucidate if a “core” microbiome in human milk exists in mothers of different countries. The sample included 413 mothers and their infants from 8 countries: Ethiopia, Gambia, United States, Ghana, Kenya, Peru, Spain and Sweden. In addition, Ethiopia and Gambia populations were subdivided in rural and urban. The researchers used homogenous methodology in the selected countries in order to understand if the differences in milk microbiota previously reported in other studies [17], [23], [52] were due to diverse methodologies, genuine differences among populations, or if they were due to differences in the collection, storage, processing and analysis of milk. This study reported differences in milk microbiota at phyla and genus levels. From the 15 phyla identified, Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes represented 97.7% of the microbiota diversity. At the genus level, there was more variation between countries than within countries. On the other hand, some countries showed unique bacteria. For example, only milk collected in rural Ethiopia contained Acidothermus, Demequina, Flaviflexus and Pediococcus, milk from urban Gambia uniquely contained Chroococcidiopsis and Isoptericolar and milk from urban Ghana uniquely contained Akkermansia and Butyricicoccus. Importantly, none of the developed countries included in the study reported unique bacteria. The authors interpreted these results as the confirmation of the existence of a small “core” microbiome among all countries consisting of Staphylococcus and Streptococcus since they were found in 98.7 and 97.7% of all samples respectively, which aligns to previous conclusions [86]. However, while reporting that a “core” microbiome might exist across different populations, Lackey et al. [89] concluded that geographical location was not the only factor influencing the structure and diversity of microbiota in human milk. They emphasized that the existence of additional factors such as antibiotic use, infant age, parity, infant sex and exclusive breastfeeding status may also play an understudied role in milk microbiome.
The variance observed between countries could be related to disparities between rural and urban populations or socioeconomical status, characteristics previously proposed as factors that might potentially affect the microbiota composition [45], [99]. However, evidence is limited because the majority of published studies characterising microbial communities have been conducted in developed countries and urban settings (Fig. 3). The few studies available in rural communities have reported that human milk composition differs in these populations and suggested this could be due to their agrarian lifestyles’ activities. Meehan et al. [92] compared foragers with horticulturalist women in the Central African Republic, and observed that even though both groups spent considerable time in proximity to each other, milk microbial communities varied significantly within populations and between ethnic groups. Vaidya et al. [99] concluded that human milk microbiota of rural Indian women was more diverse than the milk microbiota of urban women; at phyla level women from rural communities had more species from Firmicutes while human milk from urban women had more Proteobacteria. Lackey et al. [89] noted differences in the milk microbiota of mothers from developed countries and two developing countries (Ethiopia and Gambia), which were also sub-divided into rural and urban populations. Milk from rural-Ethiopia differed from the other populations and was characterized by a relatively high abundance of Rhizobium and Achromobacter; intermediate abundances of Streptococcus and Staphylococcus; and very little Cutibacterium/Propionibacterium, Dyella, and Rothia. However, the methodology used for collection and processing of the milk in Ethiopia differed from other populations, which calls into question that the uniqueness of the Ethiopian results relates to the rural aspect of the community.
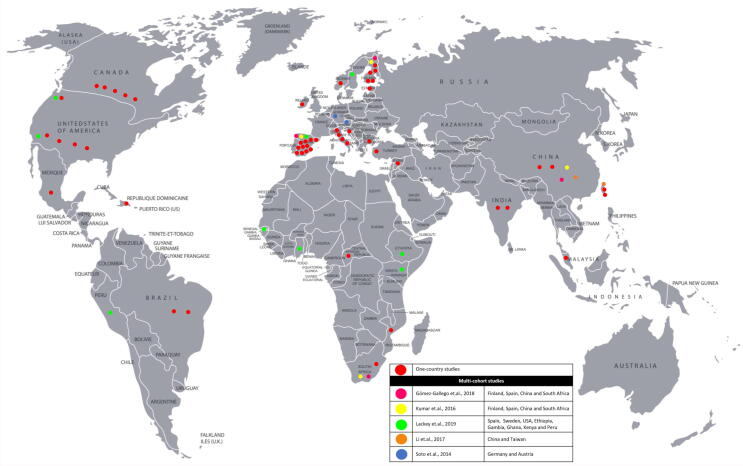
Fig. 3.
Geographical distribution of breast milk microbiome studies Fifty-seven studies into the breast milk microbiome and factors that influence its composition grouped by geographical region. Thirty-two studies were conducted in Europe [12], [17], [26], [35], [38], [50], [53], [54], [60], [76], [89], [97], [103], [19], [20], [21], [22], [63], [64], [65], [124], [125], [126], [127], [128], [129], [130], [131], [132], [133], [134], [135], 11 studies in Asia [31], [35], [36], [59], [62], [97], [99], [104], [108], [136], [137], 11 studies in North America [23], [24], [30], [33], [52], [56], [77], [89], [105], [138], [139], 6 study in Africa [35], [55], [89], [92], [97], [140] and 5 studies in Latin-America [89], [98], [100], [141], [142]. Five multi-country studies included: Germany and Austria [26], China, Finland, South Africa and Spain [35], [97], China and Taiwan, [31], and Ethiopia, Gambia, Ghana, Kenya, Peru, US and Sweden [89]. Studies have most frequently been conducted in Spain (15 studies) [19], [21], [35], [38], [50], [53], [63], [64], [89], [97], [124], [125], [127], [129], [143]
There is only one study that has described its population as a low socioeconomic community. Dave et al. [30] reported an uncommonly high abundance of Streptococcus and therefore a reduced diversity when compared to other populations, suggesting that differences might be related either to ethnicity, socioeconomic status or other factors [17] related to culture including dietary patterns, rituals and customs particular to certain region or geographical location that could affect the interaction of the mother and infant with their environment.
6.3. Environmental factors
The potential for the environment to modify the human milk microbiome has not been widely researched. Few studies have discussed the source of the bacteria in human milk and have commonly focused on identifying taxa potentially originating from oral, skin or gut habitats [17], [37]. Togo et al. [101] analysed where each species identified in human milk studies (820 species from 242 articles, 38 countries, 11,124 women and 15,489 samples) were originally isolated. The study found that only 40% of breast milk bacteria were first isolated from human tissue (including gut, respiratory tract, oral cavity, urinary tract, skin, vagina, milk) while the other 60% was first observed in association with the environment, plants, animals and food. From this 60%, environment associated bacteria were the most prevalent with 34% [101]. This same study recognized that only one species was initially associated with milk, suggesting extensive interaction between the microbiome of humans and their environment. These observations highlight the potential influence of the environment in shaping the breast milk microbiome, which to-date has most often been perceived as contaminants rather than being normally present in the breast milk [57], [102].
The presence of soil and water associated bacteria has consistently been observed in breast milk studies. These include Acinetobacter [35], [59], [103], [104], [105], Bradyrhizobiaceae [52], Novosphingobium [106], Pseudomonas [23], [24], [52], [53], [59], [104], Ralstonia [23], [33], [35], [52], [99], Sphingobium [106], [107], Sphingomonas [20], [30], [31], [35], [52], [53], [56], [108], Stenotrophomonas [56], and Xanthomonadaceae [30], [56]. Bacteria commonly found in the environment and also found in breast milk samples have the potential to simply be contamination, particularly as contaminants from PCR reagents as suggested by Ruiz et al. [109].. Douglas et al. [58] has provided evidence that, for at least some of these species, their presence in human milk may be genuine.
Soil bacteria observed in breast milk microbiota have been related to the maternal diet, seasonality, the environment and occupation, such as horticulturalists [92] or hunter-gatherers [61], [110], [111], rather than as a product of cross-contamination during the analysis [56], [57], [59]. On the other hand, soil microbes can differ enormously from region to region. There are only a few species that can be found in all soils, while there are numerous rare species that only occur in particular soils or geographical areas [112]. Therefore, generalisations among soil bacteria are difficult to do and further research in milk microbiome that aims to study the source of bacteria should include soil analysis.
Soil is considered to harbour one of the most diverse microbial populations, with several thousand of species often observed in samples. This large microbial diversity in soil results in diverse functional ecology, which includes primary productivity and nutrient cycling such as increased nutrient use efficiency and uptake, which may improve plant resilience and resistance against stressors [61]. For instance, bacteria identified in soil can produce more than 50 antibiotics to protect plants from pathogenic bacteria and rhizobia can associate with plant hosts like alfalfa, soybeans and clover to help provide the plants with nutrients [113]. As soil is part of the habitat of humans providing space for living, recreation and food production [114], it is probable that the soil contributes to the human microbiome due to its close contact as opposed to similar provision of functional diversity. It has been observed that hunter-gatherers have a gut microbiome with a higher species richness than that of humans consuming westernized food or from an urbanized society [61], [99], [110], [111], [115]. While these observations could be related to genetics, diet and their unique environments [116], the findings highlight the potential for largely unexplored links between agricultural practices, soil transmitted parasites and protozoa and human health [117].
6.4. Benefits of environmental bacteria
A milk microbiome enriched in diversity and associated functionality, in contrast to assuming certain observed species are de facto contamination, could be explored as potentially conferring some benefit to mother or infant. For instance, Chan et al. [118] have speculated that Sphingobium yanoikuyae found in nipple aspirate fluid might activate a pathway that could inhibit cancer progression. A human microbiome enriched by soil bacteria could be beneficial through functions well established in soil communities, such as suppression of soil-borne pathogens, exposure to immunoregulation-inducing soil microorganisms, immune tolerance and increase of microbiota diversity [116]. The influence of bacteria associated with soils and water-types have been somewhat explored in the gut microbiome but research in the breast milk microbiome is still scarce. Blum et al. [61] linked the soil microbiome and the human intestinal microbiome as “superorganisms” which, by close contact (soil, faeces and food), can replenish each other as inoculants and provide beneficial microorganisms which could positively impact human health. It is also speculated that urbanization and industrialization of agriculture may have decreased the richness of an overlapping of soil and human microbiota [61]. Additionally, there is evidence that microbes from diverse habitats like soil can colonize the germ-free gut [119] and contribute commensal microbes which enrich gut microbiota diversity, which can reduce inflammatory disease risk, reduce asthma and improve child health [116].
Although the previous studies linked the benefits of environmental bacteria specifically to the gut microbiome, it is plausible that similar relationships exist with the breast milk microbiome. Further research is essential to explore the relationship of breastfeeding practices, such as the introduction of water, beverages and food, as a factor for introducing soil and water bacteria into breast milk microbiome as microbial diversity might influence neonatal gut colonisation, impact the maturation of the immune system, supress pathogenic bacteria such as Staphylococcus aureus and therefore prevent maternal and neonatal infections as well as increase breast milk production.
7. Conclusion
Human milk is the first source of nutrients and immunity that the infant receives, supplying microbes to the newborn infant during a critical period of growth and development. Despite this important role for human development and health, there is scarce evidence of how some factors, such as maternal age and diet, geographical area and environment, might influence milk microbiota composition. Contemporary high-resolution 16S rRNA gene amplicon and shotgun metagenomics are powerful approaches capable of more accurate genus and species-level microbiome assessment. These tools should be used alongside controls for contamination to study the source of environmental bacteria in breast milk and whether they originate from breastfeeding practices, maternal diet or environmental proximity. Additionally, expansion of human milk research to developing countries and rural areas represents low-hanging fruit for important discovery, as the vast majority of available evidence is in developed countries and urban areas. While the study of the breast milk microbiome has faced diverse challenges, there are extensive new strategies and opportunities to advance our understanding and promote future interventions in maternal and infant health.
CRediT authorship contribution statement
Lilian Lopez Leyva: Conceptualization, Writing - original draft. Nicholas J.B. Brereton: Conceptualization, Writing - original draft. Kristine G. Koski: Conceptualization, Writing - original draft.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
LLL was financially supported by a McGill University Graduate Excellence Fellowship and the Cómtie Mexiquense de Ciencia y Tecnología (COMECYT) of Mexico. This research was otherwise funded by the NSERC Discovery Grant #RGPIN-2016-04961.
Contributor Information
Lilian Lopez Leyva, Email: lilian.lopezleyva@mail.mcgill.ca.
Nicholas J.B. Brereton, Email: nicholas.brereton@umontreal.ca.
Kristine G. Koski, Email: kristine.koski@mcgill.ca.
References
- 1.Eriksen K.G., Christensen S.H., Lind M.V., Michaelsen K.F. Human milk composition and infant growth. Curr Opin Clin Nutr Metab Care. 2018;21(3):200–206. doi: 10.1097/MCO.0000000000000466. [DOI] [PubMed] [Google Scholar]
- 2.Mosca F., Giannì M.L. Human milk: composition and health benefits. Pediatr Medica E Chir Med Surg Pediatr. 2017 Jun 28;39(2):155. doi: 10.4081/pmc.2017.155. [DOI] [PubMed] [Google Scholar]
- 3.Triantis V., Bode L., van Neerven R.J.J. Immunological Effects of Human Milk Oligosaccharides. Front Pediatr. 2018;6:190. doi: 10.3389/fped.2018.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Breastfeeding [Internet]. 2020 [cited 2020 May 18]. Available from: https://www.who.int/westernpacific/health-topics/breastfeeding
- 5.van den Elsen LWJ, Garssen J, Burcelin R, Verhasselt V. Shaping the Gut Microbiota by Breastfeeding: The Gateway to Allergy Prevention? Front Pediatr [Internet]. 2019 [cited 2020 Nov 8];7. Available from: https://www.frontiersin.org/articles/10.3389/fped.2019.00047/full [DOI] [PMC free article] [PubMed]
- 6.Savage D.C. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 7.McGuire M.K., McGuire M.A. Got bacteria? The astounding, yet not-so-surprising, microbiome of human milk. Curr Opin Biotechnol. 2017 Apr;44:63–68. doi: 10.1016/j.copbio.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Wyatt R.G., Mata L.J. Bacteria, IN Colostrum, AND Milk, OF GUATEMALAN INDIAN WOMEN. J Trop Pediatr. 1969 Dec 1;15(4):159–162. doi: 10.1093/tropej/15.4.159. [DOI] [PubMed] [Google Scholar]
- 9.Carroll L., Osman M., Davies D.P., Mcneish A.S. Bacteriology of raw breast milk. The Lancet. 1979;314(8153):1186. doi: 10.1016/s0140-6736(79)92409-7. [DOI] [PubMed] [Google Scholar]
- 10.El-Mohandes A.E., Schatz V., Keiser J.F., Jackson B.J. Bacterial contaminants of collected and frozen human milk used in an intensive care nursery. Am J Infect Control. 1993;21(5):226–230. doi: 10.1016/0196-6553(93)90413-x. [DOI] [PubMed] [Google Scholar]
- 11.Hill MJ, Marsh PD. Human Microbial Ecology. CRC Press; 1990. 184 p.
- 12.Heikkilä M.P., Saris P.E.J. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95(3):471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- 13.Martín R, Langa S, Reviriego C, Jiménez E, Marín ML, Olivares M, et al. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol. 2004 Mar;15(3–4):121–7.
- 14.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knight R., Callewaert C., Marotz C., Hyde E.R., Debelius J.W., McDonald D. The Microbiome and Human Biology. Annu Rev Genomics Hum Genet. 2017;18(1):65–86. doi: 10.1146/annurev-genom-083115-022438. [DOI] [PubMed] [Google Scholar]
- 16.Ramsay D.T., Kent J.C., Owens R.A., Hartmann P.E. Ultrasound Imaging of Milk Ejection in the Breast of Lactating Women. Pediatrics. 2004;113(2):361–367. doi: 10.1542/peds.113.2.361. [DOI] [PubMed] [Google Scholar]
- 17.Cabrera-Rubio R., Collado M.C., Laitinen K., Salminen S., Isolauri E., Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96(3):544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- 18.Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S.K., McCulle S.L. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci. 2011;108(Supplement 1):4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabrera-Rubio R., Mira-Pascual L., Mira A., Collado M.C. Impact of mode of delivery on the milk microbiota composition of healthy women. J Dev Orig Health Dis. 2016;7(1):54–60. doi: 10.1017/S2040174415001397. [DOI] [PubMed] [Google Scholar]
- 20.Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast Milk Microbiota Is Shaped by Mode of Delivery and Intrapartum Antibiotic Exposure. Front Nutr [Internet]. 2019 [cited 2020 Mar 9];6. Available from: https://www.frontiersin.org/articles/10.3389/fnut.2019.00004/full?fbclid=IwAR2O7sMuOX-XlOzkgLCTbEquLBljid0RcKR2lmSYj6MTaNTnKFq6FQfSEVs [DOI] [PMC free article] [PubMed]
- 21.Khodayar-Pardo P., Mira-Pascual L., Collado M.C., Martínez-Costa C. Impact of lactation stage, gestational age and mode of delivery on breast milk microbiota. J Perinatol. 2014 Aug;34(8):599–605. doi: 10.1038/jp.2014.47. [DOI] [PubMed] [Google Scholar]
- 22.Toscano M., De Grandi R., Peroni D.G., Grossi E., Facchin V., Comberiati P. Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 2017;17(1):205. doi: 10.1186/s12866-017-1109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moossavi S., Sepehri S., Robertson B., Bode L., Goruk S., Field C.J. Composition and Variation of the Human Milk Microbiota Are Influenced by Maternal and Early-Life Factors. Cell Host Microbe. 2019 Feb;25(2):324–335.e4. doi: 10.1016/j.chom.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Pannaraj P.S., Li F., Cerini C., Bender J.M., Yang S., Rollie A. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatr. 2017;171(7):647. doi: 10.1001/jamapediatrics.2017.0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakwinska O., Bosco N. Host Microbe Interactions in the Lactating Mammary Gland. Front Microbiol. 2019;13(10):1863. doi: 10.3389/fmicb.2019.01863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soto A., Martín V., Jiménez E., Mader I., Rodríguez J.M., Fernández L. Lactobacilli and Bifidobacteria in Human Breast Milk: Influence of Antibiotherapy and Other Host and Clinical Factors. J Pediatr Gastroenterol Nutr. 2014;59(1):78–88. doi: 10.1097/MPG.0000000000000347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez P.F., Dore J., Leclerc M., Levenez F., Benyacoub J., Serrant P. Bacterial Imprinting of the Neonatal Immune System: Lessons From Maternal Cells? Pediatrics. 2007;119(3):e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 28.Rescigno M., Urbano M., Valzasina B., Francolini M., Rotta G., Bonasio R. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol. 2001;2(4):361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 29.Thum C., Cookson A.L., Otter D.E., McNabb W.C., Hodgkinson A.J., Dyer J. Can Nutritional Modulation of Maternal Intestinal Microbiota Influence the Development of the Infant Gastrointestinal Tract? J Nutr. 2012;142(11):1921–1928. doi: 10.3945/jn.112.166231. [DOI] [PubMed] [Google Scholar]
- 30.Davé V., Street K., Francis S., Bradman A., Riley L., Eskenazi B. Bacterial microbiome of breast milk and child saliva from low-income Mexican-American women and children. Pediatr Res. 2016;79(6):846–854. doi: 10.1038/pr.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S-W, Watanabe K, Hsu C-C, Chao S-H, Yang Z-H, Lin Y-J, et al. Bacterial Composition and Diversity in Breast Milk Samples from Mothers Living in Taiwan and Mainland China. Front Microbiol [Internet]. 2017 May 30 [cited 2019 Sep 16];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5447776/ [DOI] [PMC free article] [PubMed]
- 32.Arroyo R., Martín V., Maldonado A., Jiménez E., Fernández L., Rodríguez J.M. Treatment of Infectious Mastitis during Lactation: Antibiotics versus Oral Administration of Lactobacilli Isolated from Breast Milk. Clin Infect Dis. 2010;50(12):1551–1558. doi: 10.1086/652763. [DOI] [PubMed] [Google Scholar]
- 33.Williams J.E., Carrothers J.M., Lackey K.A., Beatty N.F., York M.A., Brooker S.L. Human Milk Microbial Community Structure Is Relatively Stable and Related to Variations in Macronutrient and Micronutrient Intakes in Healthy Lactating Women. J. Nutr. 2017;147(9):1739–1748. doi: 10.3945/jn.117.248864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmermann P, Curtis N. Breast milk microbiota: A complex microbiome with multiple impacts and conditioning factors. J Infect. 2020;81(1):17–47. doi: 10.1016/j.jinf.2020.01.023. [DOI] [PubMed] [Google Scholar]
- 35.Kumar H, du Toit E, Kulkarni A, Aakko J, Linderborg KM, Zhang Y, et al. Distinct Patterns in Human Milk Microbiota and Fatty Acid Profiles Across Specific Geographic Locations. Front Microbiol [Internet]. 2016 Oct 13 [cited 2019 Sep 4];7. Available from: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01619/full [DOI] [PMC free article] [PubMed]
- 36.Chen P.-W., Lin Y.-L., Huang M.-S. Profiles of commensal and opportunistic bacteria in human milk from healthy donors in Taiwan. J Food Drug Anal. 2018 Oct;26(4):1235–1244. doi: 10.1016/j.jfda.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaTuga M., Stuebe A., Seed P. A Review of the Source and Function of Microbiota in Breast Milk. Semin Reprod Med. 2014 Jan 3;32(01):068–73. doi: 10.1055/s-0033-1361824. [DOI] [PubMed] [Google Scholar]
- 38.Collado M.C., Cernada M., Baüerl C., Vento M., Pérez-Martínez G. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012 Jul 14;3(4):352–365. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gueimonde M., Laitinen K., Salminen S., Isolauri E. Breast Milk: A Source of Bifidobacteria for Infant Gut Development and Maturation? Neonatology. 2007;92(1):64–66. doi: 10.1159/000100088. [DOI] [PubMed] [Google Scholar]
- 40.Collado M.C., Isolauri E., Laitinen K., Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 41.Ballard O., Morrow A.L. Human Milk Composition. Pediatr Clin North Am. 2013;60(1):49–74. doi: 10.1016/j.pcl.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Innis S.M. Impact of maternal diet on human milk composition and neurological development of infants. Am J Clin Nutr. 2014 Mar 1;99(3):734S–741S. doi: 10.3945/ajcn.113.072595. [DOI] [PubMed] [Google Scholar]
- 43.Lönnerdal B. Effects of Maternal Dietary Intake on Human Milk Composition. J Nutr. 1986;116(4):499–513. doi: 10.1093/jn/116.4.499. [DOI] [PubMed] [Google Scholar]
- 44.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014 Jan;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippo C.D., Cavalieri D., Paola M.D., Ramazzotti M., Poullet J.B., Massart S. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ley R.E., Hamady M., Lozupone C., Turnbaugh P.J., Ramey R.R., Bircher J.S. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu G.D., Chen J., Hoffmann C., Bittinger K., Chen Y.-Y., Keilbaugh S.A. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Ryan CA, Boyaval P, Dempsey EM, Ross RP, Stanton C. Maternal Vertical Transmission Affecting Early-life Microbiota Development. Trends Microbiol. 2019 Sep;S0966842X19302082. [DOI] [PubMed]
- 49.Suez J, Zmora N, Zilberman-Schapira G, Mor U, Dori-Bachash M, Bashiardes S, et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell. 2018 06;174(6):1406-1423.e16. [DOI] [PubMed]
- 50.Boix-Amorós A, Collado MC, Mira A. Relationship between Milk Microbiota, Bacterial Load, Macronutrients, and Human Cells during Lactation. Front Microbiol [Internet]. 2016 [cited 2020 Apr 16];7. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2016.00492/full [DOI] [PMC free article] [PubMed]
- 51.Fernández L., Langa S., Martín V., Maldonado A., Jiménez E., Martín R. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol Res. 2013;69(1):1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, et al. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. Zilberstein D, editor. PLoS ONE. 2011 Jun 17;6(6):e21313. [DOI] [PMC free article] [PubMed]
- 53.Jiménez E., de Andrés J., Manrique M., Pareja-Tobes P., Tobes R., Martínez-Blanch J.F. Metagenomic Analysis of Milk of Healthy and Mastitis-Suffering Women. J Hum Lact. 2015;31(3):406–415. doi: 10.1177/0890334415585078. [DOI] [PubMed] [Google Scholar]
- 54.Murphy K, Curley D, O’Callaghan TF, O’Shea C-A, Dempsey EM, O’Toole PW, et al. The Composition of Human Milk and Infant Faecal Microbiota Over the First Three Months of Life: A Pilot Study. Sci Rep [Internet]. 2017 Jan 17 [cited 2019 Nov 21];7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5240090/ [DOI] [PMC free article] [PubMed]
- 55.Ojo-Okunola A, Nicol M, du Toit E. Human Breast Milk Bacteriome in Health and Disease. Nutrients. 2018 Nov 3;10(11). [DOI] [PMC free article] [PubMed]
- 56.Urbaniak C., McMillan A., Angelini M., Gloor G.B., Sumarah M., Burton J.P. Effect of chemotherapy on the microbiota and metabolome of human milk, a case report. Microbiome. 2014;2(1):24. doi: 10.1186/2049-2618-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salter S.J., Cox M.J., Turek E.M., Calus S.T., Cookson W.O., Moffatt M.F. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12(1):87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douglas CA, Ivey KL, Papanicolas LE, Best KP, Muhlhausler BS, Rogers GB. DNA extraction approaches substantially influence the assessment of the human breast milk microbiome. Sci Rep. 2020 Jan 10;10(1):123. [DOI] [PMC free article] [PubMed]
- 59.Sakwinska O, Moine D, Delley M, Combremont S, Rezzonico E, Descombes P, et al. Microbiota in Breast Milk of Chinese Lactating Mothers. Riedel CU, editor. PLOS ONE. 2016 Aug 16;11(8):e0160856. [DOI] [PMC free article] [PubMed]
- 60.Drago L., Toscano M., De Grandi R., Grossi E., Padovani E.M., Peroni D.G. Microbiota network and mathematic microbe mutualism in colostrum and mature milk collected in two different geographic areas: Italy versus Burundi. ISME J. 2017;11(4):875–884. doi: 10.1038/ismej.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blum WEH, Zechmeister-Boltenstern S, Keiblinger KM. Does Soil Contribute to the Human Gut Microbiome? Microorganisms. 2019 Aug 23;7(9). [DOI] [PMC free article] [PubMed]
- 62.Albesharat R., Ehrmann M.A., Korakli M., Yazaji S., Vogel R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst Appl Microbiol. 2011;34(2):148–155. doi: 10.1016/j.syapm.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Jiménez E., Delgado S., Maldonado A., Arroyo R., Albújar M., García N. Staphylococcus epidermidis: A differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 2008;10(8):143. doi: 10.1186/1471-2180-8-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martín R., Langa S., Reviriego C., Jimínez E., Marín M.L., Xaus J. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143(6):754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 65.Jost T., Lacroix C., Braegger C., Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110(7):1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 66.Handelsman J. Metagenomics: Application of Genomics to Uncultured Microorganisms. Microbiol Mol Biol Rev. 2004;68(4):669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lagkouvardos I, Overmann J, Clavel T. Cultured microbes represent a substantial fraction of the human and mouse gut microbiota. Gut Microbes. 2017 03;8(5):493–503. [DOI] [PMC free article] [PubMed]
- 68.Wade W. Unculturable bacteria—the uncharacterized organisms that cause oral infections. J R Soc. Med. 2002;95(2):81–83. doi: 10.1258/jrsm.95.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarangi A.N., Goel A., Aggarwal R. Methods for Studying Gut Microbiota: A Primer for Physicians. J Clin Exp Hepatol. 2019;9(1):62–73. doi: 10.1016/j.jceh.2018.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A. A new genomic blueprint of the human gut microbiota. Nature. 2019 Apr;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Nimwegen K.J.M., van Soest R.A., Veltman J.A., Nelen M.R., van der Wilt G.J., Vissers L.E.L.M. Is the $1000 Genome as Near as We Think? A Cost Analysis of Next-Generation Sequencing. Clin Chem. 2016 Nov 1;62(11):1458–1464. doi: 10.1373/clinchem.2016.258632. [DOI] [PubMed] [Google Scholar]
- 73.Sczyrba A., Hofmann P., Belmann P., Koslicki D., Janssen S., Dröge J. Critical Assessment of Metagenome Interpretation—a benchmark of metagenomics software. Nat Methods. 2017;14(11):1063–1071. doi: 10.1038/nmeth.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vollmers J, Wiegand S, Kaster A-K. Comparing and Evaluating Metagenome Assembly Tools from a Microbiologist’s Perspective - Not Only Size Matters! PLOS ONE. 2017 ene;12(1):e0169662. [DOI] [PMC free article] [PubMed]
- 75.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems [Internet]. 2017 Feb 28 [cited 2020 Aug 29];2(1). Available from: https://msystems.asm.org/content/2/1/e00164-16 [DOI] [PMC free article] [PubMed]
- 76.Pärnänen K., Karkman A., Hultman J., Lyra C., Bengtsson-Palme J., Larsson D.G.J. Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat Commun. 2018;9(1):3891. doi: 10.1038/s41467-018-06393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward T.L., Hosid S., Ioshikhes I., Altosaar I. Human milk metagenome: a functional capacity analysis. BMC Microbiol. 2013;25(13):116. doi: 10.1186/1471-2180-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fox G.E., Pechman K.R., Woese C.R. Comparative Cataloging of 16S Ribosomal Ribonucleic Acid: Molecular Approach to Procaryotic Systematics. Int J Syst Evol Microbiol. 1977;27(1):44–57. [Google Scholar]
- 80.Muyzer G., de Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pace NR, Stahl DA, Lane DJ, Olsen GJ. The Analysis of Natural Microbial Populations by Ribosomal RNA Sequences. In: Marshall KC, editor. Advances in Microbial Ecology [Internet]. Boston, MA: Springer US; 1986 [cited 2020 Aug 30]. p. 1–55. (Advances in Microbial Ecology). Available from: https://doi.org/10.1007/978-1-4757-0611-6_1
- 82.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Martinez-Porchas M, Villalpando-Canchola E, Ortiz Suarez LE, Vargas-Albores F. How conserved are the conserved 16S-rRNA regions? PeerJ [Internet]. 2017 Feb 28 [cited 2020 Aug 29];5. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5333541/ [DOI] [PMC free article] [PubMed]
- 84.Woese C.R., Gutell R., Gupta R., Noller H.F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012;41(1):e1. [DOI] [PMC free article] [PubMed]
- 86.Fitzstevens J.L., Smith K.C., Hagadorn J.I., Caimano M.J., Matson A.P., Brownell E.A. Systematic Review of the Human Milk Microbiota. Nutr Clin Pract. 2017;32(3):354–364. doi: 10.1177/0884533616670150. [DOI] [PubMed] [Google Scholar]
- 87.Jeurink P v., van Bergenhenegouwen J, Jiménez E, Knippels L m. j., Fernández L, Garssen J, et al. Human milk: a source of more life than we imagine. Benef Microbes. 2013 Mar 1;4(1):17–30. [DOI] [PubMed]
- 88.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013 Jan;41(Database issue):D590–6. [DOI] [PMC free article] [PubMed]
- 89.Lackey K.A., Williams J.E., Meehan C.L., Zachek J.A., Benda E.D., Price W.J. What’s Normal? Microbiomes in Human Milk and Infant Feces Are Related to Each Other but Vary Geographically: The INSPIRE Study. Front Nutr. 2019;17(6):45. doi: 10.3389/fnut.2019.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mediano P., Fernández L., Jiménez E., Arroyo R., Espinosa-Martos I., Rodríguez J.M. Microbial Diversity in Milk of Women With Mastitis: Potential Role of Coagulase-Negative Staphylococci, Viridans Group Streptococci, and Corynebacteria. J Hum Lact Off J Int Lact Consult Assoc. 2017 May;33(2):309–318. doi: 10.1177/0890334417692968. [DOI] [PubMed] [Google Scholar]
- 91.Frank J.A., Reich C.I., Sharma S., Weisbaum J.S., Wilson B.A., Olsen G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. 2008;74(8):2461–2470. doi: 10.1128/AEM.02272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Meehan C.L., Lackey K.A., Hagen E.H., Williams J.E., Roulette J., Helfrecht C. Social networks, cooperative breeding, and the human milk microbiome. Am J Hum Biol. 2018;30(4) doi: 10.1002/ajhb.23131. [DOI] [PubMed] [Google Scholar]
- 93.Bartlett J.A., Schleifer S.J., Demetrikopoulos M.K., Delaney B.R., Shiflett S.C., Keller S.E. Immune Function in Healthy Adolescents. Clin Diagn Lab Immunol. 1998 Jan;5(1):105–113. doi: 10.1128/cdli.5.1.105-113.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dror DK, Allen LH. Overview of Nutrients in Human Milk. Adv Nutr. 2018 May 1;9(suppl_1):278S-294S. [DOI] [PMC free article] [PubMed]
- 95.Nasioudis D., Forney L.J., Schneider G.M., Gliniewicz K., France M., Boester A. Influence of Pregnancy History on the Vaginal Microbiome of Pregnant Women in their First Trimester. Sci Rep. 2017;7(1):10201. doi: 10.1038/s41598-017-09857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gómez-Gallego C, Morales JM, Monleón D, du Toit E, Kumar H, Linderborg KM, et al. Human Breast Milk NMR Metabolomic Profile across Specific Geographical Locations and Its Association with the Milk Microbiota. Nutrients [Internet]. 2018 Sep 21 [cited 2019 Sep 19];10(10). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6213536/ [DOI] [PMC free article] [PubMed]
- 97.Gomez-Gallego C., Garcia-Mantrana I., Salminen S., Collado M.C. The human milk microbiome and factors influencing its composition and activity. Semin Fetal Neonatal Med. 2016;21(6):400–405. doi: 10.1016/j.siny.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 98.Corona-Cervantes K., García-González I., Villalobos-Flores L.E., Hernández-Quiroz F., Piña-Escobedo A., Hoyo-Vadillo C. Human milk microbiota associated with early colonization of the neonatal gut in Mexican newborns. PeerJ. 2020;22(8) doi: 10.7717/peerj.9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vaidya Y.H., Patel S.H., Patel R.J., Pandit R.J., Joshi C.G., Kunjadia A.P. Human milk microbiome in urban and rural populations of India. Meta Gene. 2017;13:13–22. [Google Scholar]
- 100.Carvalho-Ramos I.I., Duarte R.T.D., Brandt K.G., Martinez M.B., Taddei C.R. Breastfeeding increases microbial community resilience. J Pediatr (Rio J). 2018;94(3):258–267. doi: 10.1016/j.jped.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 101.Togo A., Dufour J.-C., Lagier J.-C., Dubourg G., Raoult D., Million M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. 2019;14:623–641. doi: 10.2217/fmb-2018-0317. [DOI] [PubMed] [Google Scholar]
- 102.Browne PD, Aparicio M, Alba C, Hechler C, Beijers R, Rodríguez JM, et al. Human Milk Microbiome and Maternal Postnatal Psychosocial Distress. Front Microbiol [Internet]. 2019 Oct 22 [cited 2020 Apr 15];10. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6817470/ [DOI] [PMC free article] [PubMed]
- 103.Drell T., Štšepetova J., Simm J., Rull K., Aleksejeva A., Antson A. The Influence of Different Maternal Microbial Communities on the Development of Infant Gut and Oral Microbiota. Sci Rep. 2017;7(1):9940. doi: 10.1038/s41598-017-09278-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel S.H., Vaidya Y.H., Patel R.J., Pandit R.J., Joshi C.G., Kunjadiya A.P. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. 2017;7(1):7804. doi: 10.1038/s41598-017-08451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016 Dec;4(1):1. [DOI] [PMC free article] [PubMed]
- 106.Jiménez E, Espinosa I, Arroyo R, Fernández L, Rodríguez JM. Book of abstracts, 3rd TNO Beneficial Microbes Conferences, 26-28 March 2012. Noordwijkerhout, the Netherlands; 2012. 76 p.
- 107.Mueller N.T., Bakacs E., Combellick J., Grigoryan Z., Dominguez-Bello M.G. The infant microbiome development: mom matters. Trends Mol Med. 2015 Feb;21(2):109–117. doi: 10.1016/j.molmed.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding M., Qi C., Yang Z., Jiang S., Bi Y., Lai J. Geographical location specific composition of cultured microbiota and Lactobacillus occurrence in human breast milk in China. Food Funct. 2019;10(2):554–564. doi: 10.1039/c8fo02182a. [DOI] [PubMed] [Google Scholar]
- 109.Ruiz L., García-Carral C., Rodriguez J.M. Unfolding the Human Milk Microbiome Landscape in the Omics Era. Front Microbiol. 2019;25(10):1378. doi: 10.3389/fmicb.2019.01378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clemente J.C., Pehrsson E.C., Blaser M.J., Sandhu K., Gao Z., Wang B. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wall D.H., Nielsen U.N., Six J. Soil biodiversity and human health. Nature. 2015;528(7580):69–76. doi: 10.1038/nature15744. [DOI] [PubMed] [Google Scholar]
- 113.Brereton N.J.B., Gonzalez E., Desjardins D., Labrecque M., Pitre F.E. Co-cropping with three phytoremediation crops influences rhizosphere microbiome community in contaminated soil. Sci Total Environ. 2020;1(711) doi: 10.1016/j.scitotenv.2019.135067. [DOI] [PubMed] [Google Scholar]
- 114.Brevik EC, Slaughter L, Singh BR, Steffan JJ, Collier D, Barnhart P, et al. Soil and Human Health: Current Status and Future Needs. Air Soil Water Res. 2020 Jan 1;13:1178622120934441.
- 115.Schnorr S.L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5(1):3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tasnim N, Abulizi N, Pither J, Hart MM, Gibson DL. Linking the Gut Microbial Ecosystem with the Environment: Does Gut Health Depend on Where We Live? Front Microbiol [Internet]. 2017 Oct 6 [cited 2020 Jun 16];8. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5635058/ [DOI] [PMC free article] [PubMed]
- 117.Scott M, Koski K. Soil-Transmitted Helminths. In: Does Nutrition Make a Difference? In: Nutrition and Infectious Diseases: Shifting the Clinical Paradigm. Boston: Springer; In press.
- 118.Chan A.A., Bashir M., Rivas M.N., Duvall K., Sieling P.A., Pieber T.R. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci Rep. 2016;6(1):28061. doi: 10.1038/srep28061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Seedorf H., Griffin N.W., Ridaura V.K., Reyes A., Cheng J., Rey F.E. Bacteria from Diverse Habitats Colonize and Compete in the Mouse Gut. Cell. 2014;159(2):253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, et al. The NIH Human Microbiome Project. Genome Res. 2009 Dec;19(12):2317–23. [DOI] [PMC free article] [PubMed]
- 121.Shimamura S., Abe F., Ishibashi N., Miyakawa H., Yaeshima T., Araya T. Relationship between oxygen sensitivity and oxygen metabolism of Bifidobacterium species. J Dairy Sci. 1992;75(12):3296–3306. doi: 10.3168/jds.S0022-0302(92)78105-3. [DOI] [PubMed] [Google Scholar]
- 122.Simpson P.J., Stanton C., Fitzgerald G.F., Ross R.P. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J Appl Microbiol. 2005;99(3):493–501. doi: 10.1111/j.1365-2672.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 123.Jost T., Lacroix C., Braegger C.P., Rochat F., Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding: Mother-neonate bacterial transfer. Environ Microbiol. 2014 Sep;16(9):2891–2904. doi: 10.1111/1462-2920.12238. [DOI] [PubMed] [Google Scholar]
- 124.Martín R., Heilig H.G.H.J., ErwinG Zoetendal, Jiménez E., Fernández L., Smidt H. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Res Microbiol. 2007 Jan 1;158(1):31–37. doi: 10.1016/j.resmic.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 125.Collado M.C., Delgado S., Maldonado A., Rodríguez J.M. Assessment of the bacterial diversity of breast milk of healthy women by quantitative real-time PCR. Lett Appl Microbiol. 2009;48(5):523–528. doi: 10.1111/j.1472-765X.2009.02567.x. [DOI] [PubMed] [Google Scholar]
- 126.Tuzun F., Kumral A., Duman N., Ozkan H. Breast milk jaundice: effect of bacteria present in breast milk and infant feces. J Pediatr Gastroenterol Nutr. 2013;56(3):328–332. doi: 10.1097/MPG.0b013e31827a964b. [DOI] [PubMed] [Google Scholar]
- 127.Olivares M., Albrecht S., De Palma G., Ferrer M.D., Castillejo G., Schols H.A. Human milk composition differs in healthy mothers and mothers with celiac disease. Eur J Nutr. 2015;54(1):119–128. doi: 10.1007/s00394-014-0692-1. [DOI] [PubMed] [Google Scholar]
- 128.Boix-Amorós A, Puente-Sánchez F, du Toit E, Linderborg KM, Zhang Y, Yang B, et al. Mycobiome Profiles in Breast Milk from Healthy Women Depend on Mode of Delivery, Geographic Location, and Interaction with Bacteria. Appl Environ Microbiol. 2019 01;85(9). [DOI] [PMC free article] [PubMed]
- 129.Moles L, Escribano E, de Andrés J, Montes MT, Rodríguez JM, Jiménez E, et al. Administration of Bifidobacterium breve PS12929 and Lactobacillus salivarius PS12934, Two Strains Isolated from Human Milk, to Very Low and Extremely Low Birth Weight Preterm Infants: A Pilot Study [Internet]. Vol. 2015, Journal of Immunology Research. Hindawi; 2015 [cited 2020 Aug 30]. p. e538171. Available from: https://www.hindawi.com/journals/jir/2015/538171/ [DOI] [PMC free article] [PubMed]
- 130.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, et al. The Bacterial Ecosystem of Mother’s Milk and Infant’s Mouth and Gut. Front Microbiol [Internet]. 2017 [cited 2020 Aug 30];8. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01214/full [DOI] [PMC free article] [PubMed]
- 131.Biagi E, Aceti A, Quercia S, Beghetti I, Rampelli S, Turroni S, et al. Microbial Community Dynamics in Mother’s Milk and Infant’s Mouth and Gut in Moderately Preterm Infants. Front Microbiol [Internet]. 2018 [cited 2020 Aug 30];9. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02512/full [DOI] [PMC free article] [PubMed]
- 132.Simpson M.R., Avershina E., Storrø O., Johnsen R., Rudi K., Øien T. Breastfeeding-associated microbiota in human milk following supplementation with Lactobacillus rhamnosus GG, Lactobacillus acidophilus La-5, and Bifidobacterium animalis ssp. lactis Bb-12. J Dairy Sci. 2018;101(2):889–899. doi: 10.3168/jds.2017-13411. [DOI] [PubMed] [Google Scholar]
- 133.Tuominen H., Rautava S., Collado M.C., Syrjänen S., Rautava J. HPV infection and bacterial microbiota in breast milk and infant oral mucosa. PLoS ONE. 2018;13(11) doi: 10.1371/journal.pone.0207016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aakko J., Kumar H., Rautava S., Wise A., Autran C., Bode L. Human milk oligosaccharide categories define the microbiota composition in human colostrum. Benef Microbes. 2017;8(4):563–567. doi: 10.3920/BM2016.0185. [DOI] [PubMed] [Google Scholar]
- 135.Obermajer T, Lipoglavšek L, Tompa G, Treven P, Lorbeg PM, Matijašić BB, et al. Colostrum of Healthy Slovenian Mothers: Microbiota Composition and Bacteriocin Gene Prevalence. PLOS ONE. 2015 abr;10(4):e0123324. [DOI] [PMC free article] [PubMed]
- 136.Huang M.-S., Cheng C.-C., Tseng S.-Y., Lin Y.-L., Lo H., Chen P.-W. Most commensally bacterial strains in human milk of healthy mothers display multiple antibiotic resistance. MicrobiologyOpen. 2019;8(1) doi: 10.1002/mbo3.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dahaban N.M., Romli M.F., Roslan N.R., Kong S.S.-S., Cheah F.-C. Bacteria in Expressed Breastmilk from Mothers of Premature Infants and Maternal Hygienic Status. Breastfeed Med. 2013;8(4):422–423. doi: 10.1089/bfm.2012.0109. [DOI] [PubMed] [Google Scholar]
- 138.Moossavi S., Atakora F., Miliku K., Sepehri S., Robertson B., Duan Q.L. Integrated Analysis of Human Milk Microbiota With Oligosaccharides and Fatty Acids in the CHILD Cohort. Front Nutr. 2019 May;16(6):58. doi: 10.3389/fnut.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cacho NT, Harrison NA, Parker LA, Padgett KA, Lemas DJ, Marcial GE, et al. Personalization of the Microbiota of Donor Human Milk with Mother’s Own Milk. Front Microbiol [Internet]. 2017 [cited 2020 Mar 24];8. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2017.01470/full [DOI] [PMC free article] [PubMed]
- 140.González R, Mandomando I, Fumadó V, Sacoor C, Macete E, Alonso PL, et al. Breast Milk and Gut Microbiota in African Mothers and Infants from an Area of High HIV Prevalence. PLoS ONE [Internet]. 2013 Nov 26 [cited 2019 Sep 19];8(11). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3841168/ [DOI] [PMC free article] [PubMed]
- 141.Damaceno Q.S., Souza J.P., Nicoli J.R., Paula R.L., Assis G.B., Figueiredo H.C. Evaluation of Potential Probiotics Isolated from Human Milk and Colostrum. Probiotics Antimicrob Proteins. 2017;9(4):371–379. doi: 10.1007/s12602-017-9270-1. [DOI] [PubMed] [Google Scholar]
- 142.Bender JM, Li F, Martelly S, Byrt E, Rouzier V, Leo M, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med. 2016 Jul 27;8(349):349ra100-349ra100. [DOI] [PMC free article] [PubMed]
- 143.Boix-Amorós A, Martinez-Costa C, Querol A, Collado MC, Mira A. Multiple Approaches Detect the Presence of Fungi in Human Breastmilk Samples from Healthy Mothers. Sci Rep [Internet]. 2017 Oct 12 [cited 2020 Aug 30];7. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5638952/ [DOI] [PMC free article] [PubMed]