Abstract
MicroRNAs (miRNAs) regulate the expression of genes associated with the development of diseases, including type 2 diabetes mellitus (T2DM). However, the use of miRNAs to predict T2DM remission has been poorly studied. Therefore, we aimed to investigate whether circulating miRNAs could be used to predict the probability of T2DM remission in patients with coronary heart disease. We included the newly diagnosed T2DM (n = 190) of the 1,002 patients from the CORDIOPREV study. Seventy-three patients reverted from T2DM after 5 years of dietary intervention with a low-fat or Mediterranean diet. Plasma levels of 56 miRNAs were measured by OpenArray. Generalized linear model, receiver operating characteristic (ROC), Cox regression, and pathway analyses were performed. ROC analysis based on clinical variables showed an area under the curve (AUC) of 0.66. After a linear regression analysis, seven miRNAs were identified as the most important variables in the group’s differentiation. The addition of these miRNAs to clinical variables showed an AUC of 0.79. Cox regression analysis using a T2DM remission score including miRNAs showed that high-score patients have a higher probability of T2DM remission (hazard ratio [HR]low versus high, 4.44). Finally, 26 genes involved in 10 pathways were related to the miRNAs. We have identified miRNAs (hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR-375, and hsa-miR-486) that contribute to the prediction of T2DM remission in patients with coronary heart disease.
Graphical Abstract
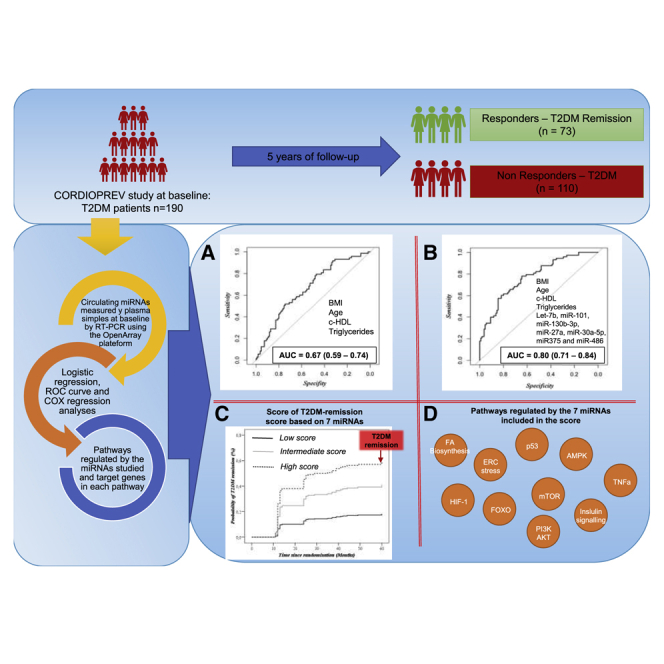
Type 2 diabetes mellitus (T2DM) is one of the most important health problems worldwide, but this disease may revert by the consumption of healthy diets. Our work suggests the use of seven genetic factors (microRNAs) to predict the probability of T2DM remission in patients with coronary heart disease.
Introduction
Type 2 diabetes mellitus (T2DM) is one of the most severe health problems worldwide.1 High blood pressure, dyslipidemia, alcohol intake, and a sedentary lifestyle are risk factors of T2DM development. These factors are directly associated with overweight and obesity characterized by low-grade systemic inflammation and oxidative stress, which damages blood vessels and leads to endothelial dysfunction, thus increasing the risk of heart disease or stroke.2 Patients with acute myocardial infarction (AMI) and T2DM have a higher risk of developing a new cardiovascular event than do those without T2DM.3
With this background, the question arises whether disease remission is possible in patients diagnosed with T2DM. The American Diabetes Association (ADA) has defined the term “remission” as “… achieving glycemia below the diabetic range in the absence of active pharmacologic or surgical therapy.”4 T2DM remission has been associated with the weight loss achieved by bariatric surgery, and the Swedish Obese Subjects (SOS) project showed remission rates of 72.3% after 2 years and 30.4% after 5 years of bariatric surgery.5 Additionally, weight loss has also been achieved through caloric restriction. The Diabetes Remission Clinical Trial (DiRECT), which includes the delivery by a primary care nurse or dietitian of a low-calorie diet replacement (<800 kcal/day), during 12–20 weeks, showed weight loss averaging 15 kg, a decrease in fasting plasma glucose from 148.9 ± 6.8 to 102.2 ± 2.2 mg/dL, and a 1.5-fold decrease in hemoglobin A1c (HbA1c) to 5.9% ± 0.1%.6,7 Finally, intervention with physical exercise has resulted in an average weight loss of 9 kg and a decrease in HbA1c from 6.8 to 6.2.8
T2DM remission involves genetic and epigenetic (DNA methylation and microRNAs [miRNAs]) regulation of several pathways, including those involved in lipid and glucose metabolism.9 Specifically, previous studies have proposed using miRNAs as biomarkers to evaluate the risk of T2DM development.10, 11, 12, 13 Based on this evidence, we aimed to identify whether circulating miRNAs could also predict T2DM remission after 5 years of dietary intervention with a low-fat or Mediterranean (Med) diet in patients with coronary heart disease (CHD).
Results
Baseline characteristics of the study population
After analyzing the baseline characteristics of the patients included in the study, we observed that body weight, body mass index (BMI), waist circumference, HbA1c, glucose, insulin, homeostatic model assessment of insulin resistance (HOMA-IR), and the hepatic insulin resistance index (HIRI) were higher in the non-responders than in responders. In contrast, the insulin sensitivity index (ISI) and disposition index (DI) were lower (Table S1). No differences between diets were observed in the T2DM remission (Table S2). Moreover, the adherence of both diets, that is, low-fat and Mediterranean diets, increased significantly and in a very similar way after a 5-year follow-up period in the two biological groups (responders and non-responders) (Table S3). Despite the baseline differences in body weight between responders and non-responders, these differences were maintained during the dietary intervention and no relevant changes were observed (Table S4).
Receiver operating characteristic curve analysis
Of the 56 miRNAs selected for the study, 9 did not amplify in at least 80% of the samples and 2 (hsa-miR-143 and hsa-miR-144) were used for data normalization, as previously described by our group.10,11
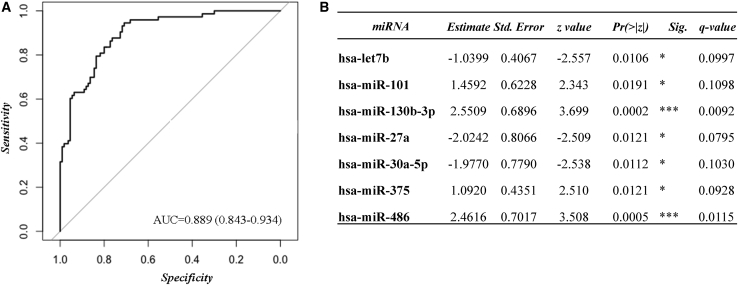
With the remaining 45 miRNAs we carried out a receiver operating characteristic (ROC) curve analysis, getting an area under curve (AUC) of 0.89 (95% confidence interval [CI], 0.84–0.93; sensitivity, 0.85; specificity, 0.77; accuracy, 0.80). Additionally, based on the estimate SE z, or Pr(>|z|), values registered in the summary of the fitting generalized linear models (glm) function, hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR-375, and hsa-miR-486 were identified as the most important miRNAs in the group’s differentiation (responders versus non-responders) (Figure 1; Table S5).
Figure 1.
Remission of T2DM assessed by a ROC curve model including 45 miRNAs
(A and B) The ROC analysis was carried out using R software through the glm function (fitting generalized linear models) and pROC libraries, including 45 miRNAs in the model: (A) ROC curve model and (B) summary table with the Pr(>|z|) values of each variable within the model. ∗p < 0.05, ∗∗∗p < 0.01. Multiple comparisons in the large-scale analyses were assessed by false discovery rate (FDR) using the Benjamini and Hochberg method (Q value).
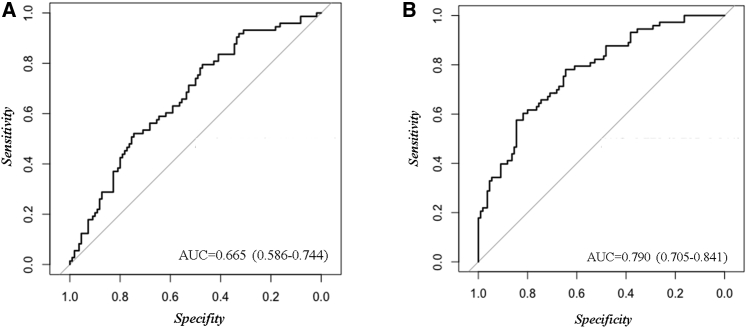
Next, based on the clinical variables used to evaluate the probability of T2DM remission in diabetic patients, we performed a ROC curve analysis, including body mass index, age, high-density lipoprotein cholesterol (HDL-C), and triglycerides (TGs). Our results showed an AUC of 0.66 (95% CI, 0.59–0.74; sensitivity, 0.79; specificity, 0.47; accuracy, 0.60) (Figure 2A). To improve this model, we carried out an ROC analysis based on the seven miRNAs identified above (hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR-375, and hsa-miR-486) added to the clinical variables (BMI, age, HDL-C, and triglycerides). Our results showed an AUC of 0.79 (95% CI, 0.70–0.84; sensitivity, 0.78; specificity, 0.64; accuracy, 0.70) (Figure 2B). A DeLong test between the model based on miRNAs added to the clinical variables and the model based only on the clinical variables showed a Z value = −3.0272 and a p value = 0.0025.
Figure 2.
Remission of T2DM assessed by ROC curve models based on clinical variables and miRNAs
The ROC analysis was carried out using R software through the glm function and pROC libraries. (A) Model based on clinical variables, including body mass index (BMI), age, HDL-C, and triglycerides. (B) Model based on the seven miRNAs (hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR375, and hsa-miR-486) added to clinical variables.
Additionally, to improve the models analyzed so far, we carried out three additional ROC curve analyses based on miRNAs, β cell function indexes, and insulin resistance indexes. According to the AUC of the models, no model was observed to have a higher capacity to differentiate between groups (Table 1).
Table 1.
T2DM remission models performed in our study by ROC curve analysis
| ROC model | AUC | 95% CI | Sensitivity | Specificity | Accuracy | Threshold |
|---|---|---|---|---|---|---|
| 45 miRNAs | 0.889 | 0.843–0.934 | 0.8493 | 0.7727 | 0.8033 | 0.3624 |
| 7 miRNAs | 0.717 | 0.644–0.791 | 0.8767 | 0.4727 | 0.6339 | 0.3313 |
| Clinical variables (age, BMI, HDL-C, triglycerides) | 0.665 | 0.586–0.744 | 0.7945 | 0.4727 | 0.6011 | 0.3484 |
| 7 miRNAs + clinical variables | 0.790 | 0.705–0.841 | 0.7808 | 0.6455 | 0.6995 | 0.3768 |
| Indexes (ISI, HOMA-IR, HIRI, IGI, DI) | 0.716 | 0.638–0.793 | 0.6857 | 0.6606 | 0.6704 | 0.3492 |
| 7 miRNAs + indexes | 0.770 | 0.712–0.849 | 0.6571 | 0.7706 | 0.7263 | 0.4082 |
The models were carried out using R softwar through the glm (fitting generalized linear models) and pROC libraries. The models included clinical variables (BMI, sex, HDL-C, and triglycerides), seven miRNAs (let-7b, miR-101, miR-130b-3p, miR-27a, miR-30a-5p, miR-375, and miR-486), and β cell function and insulin resistance indexes (ISI, HOMA-IR, HIRI, IGI, and DI).
T2DM remission score based on miRNAs
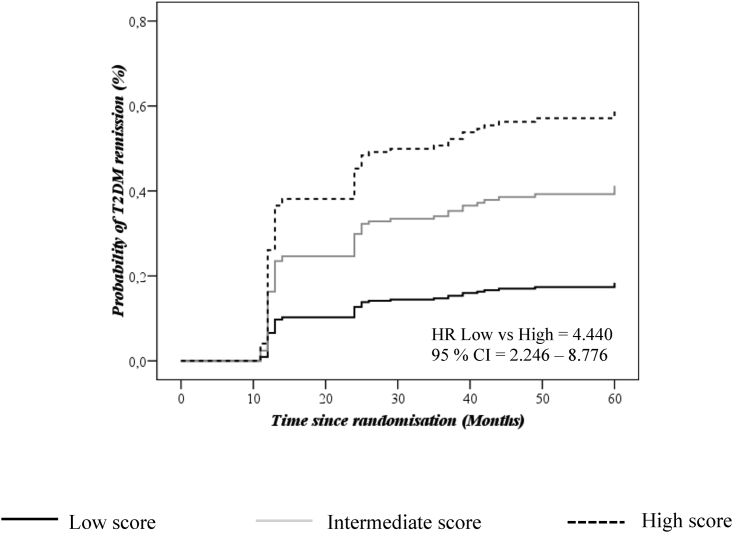
In order to evaluate the probability of remission, a T2DM remission score based on miRNAs was calculated. Next, we classified the population according to tertiles of the score and carried out a Cox regression analysis using as a reference the tertile with a low probability of T2DM remission (tertile 1 = low score). Finally, the hazard ratios (HRs) of the analysis were assessed. Our results showed HRT1 versus T2 of 2.615 (95% CI, 1.284–5.326) and HRT1 versus T3 of 4.440 (95% CI, 2.246–8.776), where T1, T2, and T3 indicate T2DM remission scores (from low to high) (Figure 3).
Figure 3.
Probability of disease remission analyzed by a T2DM remission score based on seven miRNAs
The analysis was performed using a Cox regression curve by tertiles of the T2DM remission score and adjusted by BMI, sex, diet, age, HDL-C, triglycerides. and statins. Continuous black line indicates low score (T1), continuous gray line indicates medium score (T2), and black dotted line indicates high score (T3). The analysis was carried out using SPSS (now PASW Statistic for Windows, version 21) (IBM, Chicago, IL, USA).
Pathways regulated by the miRNAs studied and target genes in each pathway
Considering that the miRNAs included in the present study were selected according to their relationship with insulin signaling and β cell function, we analyzed the relationship between the biological processes in which the set of seven miRNAs are involved.
We identified 10 T2DM-related pathways regulated by seven or at least six of the miRNAs studied (hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR-375, and hsa-miR-486) (Table 2).
Table 2.
Pathways regulated by miRNAs included in the T2DM remission score
| KEGG pathways | p value | No. miRNAs |
|---|---|---|
| Protein processing in endoplasmic reticulum | 1.57E−08 | 7 |
| FoxO signaling pathway | 1.93E−07 | 7 |
| p53 Signaling pathway | 3.28E−05 | 7 |
| AMPK signaling pathway | 8.67E−05 | 7 |
| mTOR signaling pathway | 0.000146 | 7 |
| HIF-1 signaling pathway | 0.003033 | 7 |
| PI3K-Akt signaling pathway | 0.007077 | 7 |
| Insulin signaling pathway | 0.012962 | 7 |
| TNF signaling pathway | 0.047134 | 7 |
| Fatty acid biosynthesis | 3.88E−06 | 6 |
The analysis was carried out through the bioinformatics tool DIANA Tools v3. The T2DM-related pathways were selected according to the lower p value and involving all seven miRNAs together or at least six of them.
Additionally, in the analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and TarBase v8 software, a database of experimentally supported miRNA-gene interactions, the results suggested that in the 10 T2DM-related pathways, hsa-let-7b regulated the expression of 16 genes, hsa-miR-101 regulated 87 genes, hsa-miR-130b-3p regulated 44 genes, hsa-miR-27a regulated 131 genes, hsa-miR-30a-5p regulated 80 genes, hsa-miR-375 regulated 50 genes, and hsa-miR-486 regulate 19 genes. In summary, 26 genes were identified as the most significant genes regulated by these seven miRNAs (Table 3; Figures S1–S10). These genes are involved in biological processes related with T2DM, such as apoptosis, cell cycle, ubiquitin-mediated proteolysis, activation of inflammatory cytokines, oxidative stress and DNA repair, inhibition of angiogenesis and metastasis, microtubule organization, lipolysis and lipid biosynthesis, iron metabolism, and glycolysis and gluconeogenesis (Figures S1–S10).
Table 3.
Target genes regulated by the miRNAs included in the T2DM remission score
| Pathway | Genes regulated by miRNAs included in the T2DM remission score |
|---|---|
| Protein processing in endoplasmic reticulum | Sec62, Sec63, Sec23, Sec24, NRF2 |
| FoxO | SET9, CK1, Bim, MnSOD |
| p53 | p21, TSP1 |
| AMPK | cyclin A |
| mTOR | GSK3B, SESN2, CLIP-170 |
| HIF-1 | EDN1, p21, p27, |
| PI3K-AKT | JAK, CCND1, Bim, p21 |
| Insulin signaling | PTP1B, GSK3B |
| TNF | TAK1, CASP3, Edn1, p38 |
| Fatty acid biosynthesis | FASN |
The genes were identified after the pathway analysis through the bioinformatics tool TarBase v8, a database of experimentally supported miRNA-gene interactions. Genes were selected when there was experimental support for a miRNA-gene interaction in T2DM-related pathways and when they were also regulated by the highest number of miRNAs.
Discussion
Our study showed that the circulating plasma levels of 7 out of a selected set of 45 miRNAs measured in 183 diabetic subjects at the baseline of the CORDIOPREV study were associated with the probability of T2DM remission. These seven miRNAs when added to clinical variables (BMI, age, HDL-C, and triglycerides) were able to differentiate between patients continuing as diabetics (non-responders) and those who reverted from T2DM during the 5 years of follow-up (responders), with an AUC in the ROC model of 0.79. Additionally, through a T2DM remission score based on miRNAs, the Cox regression analysis showed that subjects with a high score (T3) have a higher probability of T2DM remission (HRlow versus high, 4.44). Finally, 26 genes regulated by the largest number of miRNAs and involved in biological processes associated with T2DM were identified.
T2DM is one of the leading causes of death globally and is a risk factor for other diseases such as cardiovascular diseases. The etiology of T2DM is complex and is associated with irreversible risk factors such as age, genetics, race, and ethnicity, and reversible factors such as smoking, physical activity, and diet. The common basis of T2DM is overweight and obesity. It has been recognized that the processes underlying T2DM can be reversed and its remission can be achieved.14 T2DM remission is defined as the state in which hyperglycemia decreases to levels below the thresholds for diabetes, according to the ADA. Previous studies based on bariatric surgery, caloric restriction diets (medium, low, and very low of total energy), and physical exercise demonstrated that remission of T2DM is associated with weight loss.
This evidence suggests that it is important for early identification of diabetic patients with a probability of achieving T2DM remission, in order to take personalized, effective, and efficient therapeutic action focused on disease remission. In terms of predicting T2DM remission, a variety of scores and variables have been used to identify subjects with the probability of remission. In a study by Still et al.,15 evaluating 259 clinical variables, four parameters were identified (insulin, age, HbA1c, and type of antidiabetic medication) and the DiaRem score was suggested. This score identified 22% of the partially remitted patients and 78% with total remission. Additionally, previous studies by Lee et al.16 showed, through the application of the ABCD score, which included age, BMI, C-peptide level, and duration of T2DM (years), 25% of patients with complete remission and 23.8% with partial remission were identified. However, all of these variables and scores were designed to predict T2DM remission in the preoperative stages of bariatric surgery and Roux-en-Y gastric bypass to ensure successful surgery and remission. Another recent primary care study in an Egyptian population included 63 patients who gained non-surgical remission from T2DM and 396 patients who served as a control, matched for age, sex, and BMI, all of whom received standards of care according to the updated guidelines. Different variables were analyzed independently by ROC curves, observing that the highest sensitivity was achieved with a short duration of T2DM (85%) and the highest specificity with HDL-C (69%).17
In the field of precision medicine, there are no previous studies that have focused on the use of epigenetic markers such as miRNAs to evaluate the probability of T2DM remission, or which are based on dietary intervention in patients with coronary heart disease. However, recent studies have demonstrated significant alterations in the transcriptome after bariatric surgery. Specifically, in the differential expression of miRNAs, Zhu et al.18 observed in 12 patients with T2DM after 12 months of bariatric surgery the downregulation of miR-29a-3p, miR-122-5p, miR-124-3p, and miR-320a from peripheral blood mononuclear cells. The deregulation of circulating miR-7-5p, let-7f-5p, miR-15b-5p, let-7i-5p, miR-320c, miR-205-5p, and miR-335-5p was also observed in 29 patients with T2DM after 21 days of surgery.19 A meta-analysis identified 13 studies in humans with study periods after surgery of 3, 6, 12, and up to 24 months, in which changes in the expression of miR-93-5p, miR-106b-5p, let-7b-5p, let-7i-5p, miR-16-5p, miR-19b-3p, miR-92a-3p, miR-222-3p, miR-142-3p, miR-140-5p, and miR-155-5p were observed.20
In our study, a dietary intervention study in patients with coronary heart disease, the results showed a ROC curve model based on BMI, age, HDL-C, and triglycerides, with an AUC of 0.66. The addition of let-7b, miR-101, miR-130b-3p, miR-27a, miR-30a-5p, miR-375, and miR-486 to the clinical variables improved the model by approximately 20% (19.6%) (AUC of 0.79), supported by the DeLong test where the p value was statistically significant between the two models (p = 0.0025). In addition, through a T2DM remission score based on these seven miRNAs, we observed that patients with a high score had a higher probability of T2DM remission than did those with a low score (HRlow versus high, 4.44). These results demonstrate that the use of epigenetic markers, such as miRNAs, allows us to assess the probability of T2DM remission during a period of dietary intervention with two healthy diets (low-fat diet and Mediterranean diet) in patients with coronary heart disease.
T2DM remission implies changes at different levels, including decreased liver fat content, the concentration of blood lipid molecules (very-low-density lipoprotein [VLDL], triglycerides, and HDL-C), plasma glucose levels, and increased insulin secretion in the first phase of remission, which suggests recovery of β cell functionality.7 All of these biological processes involve regulating biochemical and metabolic mechanisms such as fatty acid synthesis and metabolism, glucose metabolism, insulin signaling and synthesis, and cell proliferation, among others. Previous studies have shown that after bariatric surgery, the deregulation of miRNAs was associated with biochemical and metabolic mechanisms such as lipid metabolism, insulin secretion, β cell function, and insulin resistance.18, 19, 20 Our results showed that the seven miRNAs included in the T2DM remission score primarily regulate the expression of 26 genes involved in 10 biochemical and metabolic pathways, including insulin, phosphatidylinositol 3-kinase (PI3K)-Akt, AMP-activated protein kinase (AMPK), p53, fatty acid biosynthesis, tumor necrosis factor (TNF), protein processing in the endoplasmic reticulum, FoxO, mTOR, and HIF-1 signaling pathways. Deregulation by miRNAs of these genes leads to changes in biological processes such as activation of inflammatory cytokines, apoptosis, oxidative stress, lipid metabolism, and glycolysis and gluconeogenesis, which are related to T2DM.
The NRF2 and TSP1 genes have been associated with inflammation. The first gene decreases cytokine stress in the β cell, improving insulin synthesis, and the other acts at the level of adipose tissue by regulating pro-inflammatory molecules and insulin resistance.21,22 The NRF2 gene is the central point and primary regulator in conditions of oxidative stress. In this sense, the MnSOD gene actively participates in the elimination of reactive oxygen species and has been associated with an increase in triglyceride levels.23,24 The genes Bim, p21, and CASP3 have shown antiapoptotic activity at the β cell level.25, 26, 27 The GSK3 gene is involved in glycogen synthesis and is a key target for the development of new therapies for T2DM.28 The PTP1B and p38 genes regulate the synthesis and subsequent insulin sensitivity, decreasing fasting glucose levels.29,30 The CCND1 gene activates β cell proliferation and is highly expressed in pancreatic islets.31 Finally, an inverse relationship between FASN levels and lower insulin resistance has been observed in adipose tissue.32 In summary, our in silico analysis suggests that the miRNAs included in the T2DM remission score regulate the expression of genes directly involved in the biological process associated with the disease.
The results of our study open new avenues for the use of miRNAs to evaluate the probability of T2DM remission after an intervention with two healthy diets in patients with coronary heart disease, without differences in the remission rate between diets. Consequently, this should reduce the probability of a new cardiovascular event. Additionally, we unraveled a T2DM remission score based on miRNAs in diabetic patients without relevant weight loss. Nevertheless, our study has limitations. One of them is that we included a set of miRNAs selected based on previous knowledge. Therefore, we did not include other miRNAs that had not previously demonstrated their relationship with the disease. Finally, our study was conducted in a population with a mean age of 60 years with coronary heart disease, which suggests that our results should not be extrapolated to the general population.
In conclusion, we have identified a set of miRNAs (hsa-let-7b, hsa-miR-101, hsa-miR-130b-3p, hsa-miR-27a, hsa-miR-30a-5p, hsa-miR-375, and hsa-miR-486) that have the potential to evaluate the probability of T2DM remission in patients with coronary heart disease.
Materials and methods
Study subjects
This work was conducted within the framework of the CORDIOPREV study. The rationale, methods, and baseline characteristics have been reported by Delgado-Lista et al.33 and provided in ClinicalTrials.gov: NTC00924937. The CORDIOPREV study is an ongoing prospective, randomized, single-blind, controlled dietary intervention trial in 1,002 patients with coronary heart disease, at high cardiovascular risk, aged between 20 and 75 years old, who had their last coronary event more than 6 months before enrolment, and had no severe diseases or a life expectancy of fewer than 5 years. The subjects were randomized into two different dietary models (Mediterranean and low-fat diets) during 8 years. Written consent was obtained from all the subjects before recruitment, and the study protocol and all amendments were approved by the Ethics Committee of Hospital Reina Sofia, all of which follow the Declaration of Helsinki and good clinical practices.
The present study (CORDIOPREV-DIRECT) included all newly diagnosed T2DM patients who had not been receiving glucose-lowering treatment at the beginning of the study (190 out of 1,002 patients). Of these, seven patients were excluded due to their inability to perform the diagnostic test used in this work. T2DM remission was evaluated in the remaining 183 patients during the 5-year follow-up period. Moreover, three participants died during the follow-up period without achieving diabetes remission. The 183 newly diagnosed T2DM patients were classified as responders, that is, patients who reverted from T2DM during the 5 years of dietary intervention without the use of diabetes medication (n = 73); or non-responders, that is, patients who did not achieve diabetes remission at the end of the follow-up period (n = 110) (Table S1). T2DM remission was defined as glycosylated hemoglobin <6.5%, fasting plasma glucose <126 mg/dL, and 2-h plasma glucose after an oral glucose tolerance test (OGTT) <200 mg/dL, for at least 2 consecutive years and without the use of diabetes medication to lower blood glucose levels.34
Diet, dietary assessment, and follow-up visits
The patients were randomized into two different healthy dietary patterns: a Mediterranean diet rich in fat from olive oil, with 35% of calories from fat (22% monounsaturated, 6% polyunsaturated, <10% saturated), and a maximum of 50% carbohydrates; and the low-fat, high-complex carbohydrate diet (LFHCC) recommended by the National Cholesterol Education Program and the American Heart Association, comprising <30% total fat (<10% saturated fat, 12%–14% monounsaturated fatty acid [MUFA] fat, and 6%–8% polyunsaturated fatty acid [PUFA] fat), 15% protein, and a minimum of 55% carbohydrates. Dietitians administered personalized individual interviews at inclusion and every 6 months, and quarterly group education sessions were held with up to 20 participants per session and separate sessions for each group. The general guidelines into the Mediterranean diet group were as follows: abundant use of virgin olive oil for cooking and for dressing salads and other dishes (participants received free extra virgin olive oil and were informed to use the oil as much as they needed in their regular diet), consumption of two or more servings (125 g/serving) per day of vegetables (at least one of them as a salad), three or more servings (125 g/serving) per day of fresh fruit, three or more servings (40 g/serving) per week of legumes, three or more servings (150 g/serving) per week of fish or seafood, three or more servings (25 g/serving) per week of nuts or seeds, white meats instead of red meats or processed meats, and regular preparation of a homemade sauce with tomato, garlic, onion, and spices with olive oil to dress vegetables, pasta, rice, and other dishes. Optionally, for alcohol drinkers, moderate consumption of red wine (seven glasses/week) was also allowed. Recommendations were also given to avoid and limit the consumption of butter, cream, fast foods, sweets, pastries, and sugar-sweetened beverages. The participants randomized to the LFHCC diet received the recommendations according to the American Heart Association and the National Cholesterol Education Program dietary guidelines, which focus on limiting all types of fat (from both animal or vegetable sources) and increasing the intake of complex carbohydrates.
Dietary adherence was assessed using the 14-item Mediterranean Diet Adherence Screener (MEDAS) to measure adherence to the Mediterranean diet. Mediterranean diet adherence was categorized into low (0–5), medium (6–9), and high (10–14) adherence, as previously published.35 A nine-item dietary screener assessing adherence to the low-fat diet guidelines was also administered. This tool was developed and used in the PREDIMED study, and the total score ranged from 0 to 9 points. Low-fat diet adherence was categorized as low (0–3), medium (4–6), and high (7–9) adherence. The dietary adherence from the CORDIOPREV study was previously reported by Quintana-Navarro et al.35 Full study diets, dietary assessment, and follow-up visits have been previously reported.33
Biochemical measurements of metabolic parameters
Venous blood from the participants was collected in tubes containing EDTA after a 12-h overnight fast. Lipid variables, glucose homeostasis variables, and inflammatory variables (hs-PCR [sensibility-Protein C reactive]) were determined as previously reported.11
OGTT, determination of insulin resistance, and secretion by β cell-related indexes
OGTT and the determination of insulin resistance and secretion by β cell-related indexes were previously reported.11,36 In summary, patients underwent a standard Matsuda test at baseline and year-to-year during the follow-up period. After an overnight fast, blood was sampled from a vein before the oral glucose intake (0 min) and again after a 75 g flavored glucose load (75 g dextrose monohydrate in 250 mL water, NUTER-TEC glucosa 50). Blood samples were taken at 30, 60, 90, and 120 min to determine glucose and insulin concentrations.37 The Matsuda ISI, HOMA-IR, insulinogenic index (IGI), DI, and HIRI were calculated as previously reported37, 38, 39, 40, 41 and in our group by Jiménez-Lucena et al.,11 Blanco-Rojo et al.,42 and Roncero-Ramos et al.36
Isolation of circulating miRNAs from plasma samples
Venous blood from the 183 newly diagnosed T2DM patients and who had not been receiving glucose-lowering treatment at the beginning of the study was collected at baseline (day 0 before dietary intervention) in tubes containing EDTA. Since hemolysis can influence the content of miRNAs in plasma samples, and in order to reduce its effect, whole blood was collected in EDTA tubes and subjected to gentle agitation using a rotary tube shaker for six times, immediately after which the samples were kept on ice for no longer than 30 min. Next, the blood samples were centrifuged at 2,000 × g for 10 min at 4°C to separate the plasma from the blood cells. RNA isolation was carried out from plasma samples, as previously described by Jiménez-Lucena et al.11
cDNA synthesis and circulating miRNA levels by real-time PCR
The cDNA synthesis was carried out using the TaqMan miRNA reverse transcription kit (Life Technologies/Thermo Fisher Scientific, Carlsbad, CA, USA) following the manufacturer’s instructions, as previously described in our group by Jiménez-Lucena et al.11
The circulating miRNAs study was carried out on 56 miRNAs, of which our group had previously studied 28 in a population of non-diabetic patients.10,11 The remaining 28 miRNAs were selected based on a bibliographic search according to their association with insulin sensitivity, insulin secretion, inflammation, and growth and proliferation of β cells (Table S6). We measured the levels of miRNAs at the baseline of the CORDIOPREV study with the OpenArray platform (Life Technologies/Thermo Fisher Scientific, Carlsbad, CA, USA) following the manufacturer’s instructions. The relative expression data were analyzed using OpenArray real-time qPCR analysis software (Life Technologies/Thermo Fisher Scientific, Carlsbad, CA, USA), and the normalization method has been previously described.11
Pathways regulated by the miRNAs studied and target genes in each pathway
To study the role of miRNAs in the cellular processes related to T2DM, we performed an analysis with DIANA Tools using miRPath v3 software. DIANA-miRPath is a web server that provides accurate statistics and can accommodate advanced pipelines. miRPath can utilize predicted miRNA targets (in coding sequence [CDS] or 3′ UTR regions) provided by the DIANA-microT-CDS algorithm, or even experimentally validated miRNA interactions derived from Database: DIANA-TarBase.43 The T2DM-related biological processes were selected based on a lower p value and involving all seven miRNAs together or at least six of them.
Additionally, to identify the target genes regulated by the miRNAs studied, we carried out an analysis through the Database: KEGG, listing all of the genes involved in each pathway. Next, through the DIANA Tools bioinformatics tool using TarBase v8 software,44 a database of experimentally supported miRNA-gene interactions, we identified the genes regulated by the seven miRNAs in each pathway associated with the development of T2DM (Figures S1–S10).
Statistical analysis
The differences between responders and non-responders in the study population’s baseline characteristics were assessed by one-way ANOVA analysis using the SPSS software (now PASW Statistic for Windows, version 21) (IBM, Chicago, IL, USA). p values <0.05 were considered statistically significant. Multiple comparisons in the large-scale analyses were assessed by false discovery rate (FDR) using the Benjamini and Hochberg method (Q value). Before analysis, clinical variables (BMI, age, HDL-C, and triglycerides), β cell function and insulin resistance indexes (ISI, HOMA-IR, HIRI, IGI, and DI), and miRNAs data were transformed (centering and scaling) using the preProcess function from the caret package through the free statistical software R version 3.6.1. This function uses the “method = center” to subtract the mean of the predictor’s data from the predictor values, while “method = scale” divides data by the standard deviation.
ROC curve analysis
We built six models using ROC analysis based on miRNAs, β cell function indexes, insulin resistance indexes, and clinical variables. The statistical analyses were performed with the free statistical software R version 3.6.1, using RStudio version 1.2.5019. The ROC analysis was carried out in two steps, first through the glm function, which is used to fit generalized linear models, and is specified by giving a symbolic description of the linear predictor and a description of the error distribution.45 Next, we performed the ROC curve using the pROC package. pROC is a tool for visualizing, smoothing, and comparing ROC curves. The AUC can be compared with a statistical test based on U statistics or bootstrap anaysis. After the analysis, we evaluated the AUC, sensitivity, specificity, accuracy and threshold values for each model built. The most significant variables were identified by Pr(>|z|) values <0.05, registered in the summary of each glm analysis. Finally, the DeLong test was performed to assess the statistical difference between the ROC curves, and for this, we used the roc.test function of the pROC package using software R. The roc.test function compares the AUC or partial AUC of two correlated (or paired) or uncorrelated (unpaired) ROC curves through the bootstrap and Venkatraman methods with 2,000 bootstrap replicates or permutations.46
T2DM remission score based on miRNAs
In order to evaluate the probability of T2DM remission using seven selected miRNAs, we calculated a T2DM remission score in three steps. First, we performed a glm analysis, including the seven miRNAs and we assessed the z value of each miRNA in the summary analysis (Table S7). Next, we multiplied the z value of each miRNA by the expression value of miRNAs in all patients included in the study. Third, the seven values were then added together to obtain a single value per subject. Finally, the 183 patients included in our study were classified according to the tertiles of the T2DM remission score (T1, low score; T2, intermediate score; T3, high score). Using the SPSS software (now PASW Statistic for Windows, version 21) (IBM, Chicago, IL, USA), we performed a Cox regression analysis adjusting by BMI, sex, diet, age, HDL-C, triglycerides, and statins, and assessing the HR where T1 was the reference.
Acknowledgments
The CORDIOPREV study is supported by the Fundación Patrimonio Comunal Olivarero, Junta de Andalucía (Consejería de Salud, Consejería de Agricultura y Pesca, Consejería de Innovación, Ciencia y Empresa), Diputaciones de Jaén y Córdoba, Centro de Excelencia en Investigación sobre Aceite de Oliva y Salud and Ministerio de Medio Ambiente, Medio Rural y Marino, Gobierno de España; Ministerio de Economía y Competitividad (AGL2012/39615, PIE14/00005, and PIE14/00031 to J.L.-M.; AGL2015-67896-P to J.L.-M. and A.C.); Consejería de Innovación, Ciencia y Empresa, Proyectos de Investigación de Excelencia, Junta de Andalucía (CVI-7450 to J.L.-M.); and by the Fondo Europeo de Desarrollo Regional (FEDER), JPI HDHL-NutriCog (PCIN-2016-084 to J.L.-M.). The CIBEROBN is an initiative of the Instituto de Salud Carlos III, Madrid, Spain. O.A.R.-Z. and A.C. are supported by an ISCIII research contract (Programa Miguel-Servet CP19/00142 and CP14/00114, respectively). J.M.O. is supported by the US Department of Agriculture under agreement no. 8050-51000-098-00D. We would like to express our gratitude to all the subjects who participated in the study, and the Córdoba branch of the Biobank of the Sistema Sanitario Público de Andalucía (Andalusia, Spain) for providing the human biological samples. We would also like to thank the EASP (Escuela Andaluza de Salud Publica), Granada, Spain, which performed the randomization process for this study. Thanks also go to Jose Andres Morales Martinez for providing technical assistance.
Author contributions
O.A.R.-Z., A.C., and J.L.-M. conceived and designed the experiments. J.F.A.-D., G.M.Q.-N., A.L.-A., F.G.-D., J.D.-L., P.P.-M., and J.L.-M. participated in the recruitment and carried out the clinical and nutritional control of the volunteers. O.A.R.-Z., C.V.-D., and Y.K. performed the experiments and collected the data. O.A.R.-Z., C.V.-D., J.F.A.-D., Y.K., J.D.-L., A.C., and J.L.-M. analyzed and interpreted the data. O.A.R.-Z., A.C., and J.L.-M. drafted the manuscript. J.L.-M. conceived and designed the study. O.A.R.-Z., C.V.-D., and Y.K. performed the statistical analysis of data. R.M.L., J.M.O., and J.L.-M. provided critical revision of the paper for the important intellectual content. J.D.-L., P.P.-M., A.C., and J.L.-M. had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All of the authors were involved in writing the paper and gave their final approval to the submitted and published versions.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.11.001.
Contributor Information
Antonio Camargo, Email: antonio.camargo@imibic.org.
Jose Lopez-Miranda, Email: jlopezmir@uco.es.
Supplemental information
References
- 1.Ginter E., Simko V. Type 2 diabetes mellitus, pandemic in 21st century. Adv. Exp. Med. Biol. 2012;771:42–50. doi: 10.1007/978-1-4614-5441-0_6. [DOI] [PubMed] [Google Scholar]
- 2.Low Wang C.C., Hess C.N., Hiatt W.R., Goldfine A.B. Clinical update: cardiovascular disease in diabetes mellitus: atherosclerotic cardiovascular disease and heart failure in type 2 diabetes mellitus—mechanisms, management, and clinical considerations. Circulation. 2016;133:2459–2502. doi: 10.1161/CIRCULATIONAHA.116.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martín-Timón I., Sevillano-Collantes C., Segura-Galindo A., Del Cañizo-Gómez F.J. Type 2 diabetes and cardiovascular disease: have all risk factors the same strength? World J. Diabetes. 2014;5:444–470. doi: 10.4239/wjd.v5.i4.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tangelloju S., Little B.B., Esterhay R.J., Brock G., LaJoie A.S. Type 2 diabetes mellitus (T2DM) “remission” in non-bariatric patients 65 years and older. Front. Public Health. 2019;7:82. doi: 10.3389/fpubh.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjöström L., Peltonen M., Jacobson P., Ahlin S., Andersson-Assarsson J., Anveden Å., Bouchard C., Carlsson B., Karason K., Lönroth H. Association of bariatric surgery with long-term remission of type 2 diabetes and with microvascular and macrovascular complications. JAMA. 2014;311:2297–2304. doi: 10.1001/jama.2014.5988. [DOI] [PubMed] [Google Scholar]
- 6.Leslie W.S., Ford I., Sattar N., Hollingsworth K.G., Adamson A., Sniehotta F.F., McCombie L., Brosnahan N., Ross H., Mathers J.C. The Diabetes Remission Clinical Trial (DiRECT): protocol for a cluster randomised trial. BMC Fam. Pract. 2016;17:20. doi: 10.1186/s12875-016-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor R., Al-Mrabeh A., Zhyzhneuskaya S., Peters C., Barnes A.C., Aribisala B.S., Hollingsworth K.G., Mathers J.C., Sattar N., Lean M.E.J. Remission of human type 2 diabetes requires decrease in liver and pancreas fat content but is dependent upon capacity for β cell recovery. Cell Metab. 2018;28:667. doi: 10.1016/j.cmet.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Ades P.A., Savage P.D., Marney A.M., Harvey J., Evans K.A. Remission of recently diagnosed type 2 diabetes mellitus with weight loss and exercise. J. Cardiopulm. Rehabil. Prev. 2015;35:193–197. doi: 10.1097/HCR.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.González-Becerra K., Ramos-Lopez O., Barrón-Cabrera E., Riezu-Boj J.I., Milagro F.I., Martínez-López E., Martínez J.A. Fatty acids, epigenetic mechanisms and chronic diseases: a systematic review. Lipids Health Dis. 2019;18:178. doi: 10.1186/s12944-019-1120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiménez-Lucena R., Camargo A., Alcalá-Diaz J.F., Romero-Baldonado C., Luque R.M., van Ommen B., Delgado-Lista J., Ordovás J.M., Pérez-Martínez P., Rangel-Zúñiga O.A., López-Miranda J. A plasma circulating miRNAs profile predicts type 2 diabetes mellitus and prediabetes: from the CORDIOPREV study. Exp. Mol. Med. 2018;50:1–12. doi: 10.1038/s12276-018-0194-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiménez-Lucena R., Rangel-Zúñiga O.A., Alcalá-Díaz J.F., López-Moreno J., Roncero-Ramos I., Molina-Abril H., Yubero-Serrano E.M., Caballero-Villarraso J., Delgado-Lista J., Castaño J.P. Circulating miRNAs as predictive biomarkers of type 2 diabetes mellitus development in coronary heart disease patients from the CORDIOPREV study. Mol. Ther. Nucleic Acids. 2018;12:146–157. doi: 10.1016/j.omtn.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah R., Murthy V., Pacold M., Danielson K., Tanriverdi K., Larson M.G., Hanspers K., Pico A., Mick E., Reis J. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care. 2017;40:546–553. doi: 10.2337/dc16-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zampetaki A., Kiechl S., Drozdov I., Willeit P., Mayr U., Prokopi M., Mayr A., Weger S., Oberhollenzer F., Bonora E. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 14.Nagi D., Hambling C., Taylor R. Remission of type 2 diabetes: a position statement from the Association of British Clinical Diabetologists (ABCD) and the Primary Care Diabetes Society (PCDS) Br. J. Diabetes. 2019;19:73–76. [Google Scholar]
- 15.Still C.D., Wood G.C., Benotti P., Petrick A.T., Gabrielsen J., Strodel W.E., Ibele A., Seiler J., Irving B.A., Celaya M.P. Preoperative prediction of type 2 diabetes remission after Roux-en-Y gastric bypass surgery: a retrospective cohort study. Lancet Diabetes Endocrinol. 2014;2:38–45. doi: 10.1016/S2213-8587(13)70070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee W.J., Hur K.Y., Lakadawala M., Kasama K., Wong S.K., Chen S.C., Lee Y.C., Ser K.H. Predicting success of metabolic surgery: age, body mass index, C-peptide, and duration score. Surg. Obes. Relat. Dis. 2013;9:379–384. doi: 10.1016/j.soard.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Allam M.M., El-Zawawy H.T. Type 2 diabetes mellitus non-surgical remission: a possible mission. J. Clin. Transl. Endocrinol. 2019;18:100206. doi: 10.1016/j.jcte.2019.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z., Yin J., Li D.C., Mao Z.Q. Role of microRNAs in the treatment of type 2 diabetes mellitus with Roux-en-Y gastric bypass. Braz. J. Med. Biol. Res. 2017;50:e5817. doi: 10.1590/1414-431X20175817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkin S.L., Ramachandran V., Yousri N.A., Benurwar M., Simper S.C., McKinlay R., Adams T.D., Najafi-Shoushtari S.H., Hunt S.C. Changes in blood microRNA expression and early metabolic responsiveness 21 days following bariatric surgery. Front. Endocrinol. (Lausanne) 2019;9:773. doi: 10.3389/fendo.2018.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langi G., Szczerbinski L., Kretowski A. Meta-analysis of differential miRNA expression after bariatric surgery. J. Clin. Med. 2019;8:1220. doi: 10.3390/jcm8081220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sireesh D., Dhamodharan U., Ezhilarasi K., Vijay V., Ramkumar K.M. association of NF-E2 related factor 2 (Nrf2) and inflammatory cytokines in recent onset type 2 diabetes mellitus. Sci. Rep. 2018;8:5126. doi: 10.1038/s41598-018-22913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varma V., Yao-Borengasser A., Bodles A.M., Rasouli N., Phanavanh B., Nolen G.T., Kern E.M., Nagarajan R., Spencer H.J., 3rd, Lee M.J. Thrombospondin-1 is an adipokine associated with obesity, adipose inflammation, and insulin resistance. Diabetes. 2008;57:432–439. doi: 10.2337/db07-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David J.A., Rifkin W.J., Rabbani P.S., Ceradini D.J. The Nrf2/Keap1/ARE pathway and oxidative stress as a therapeutic target in type ii diabetes mellitus. J. Diabetes Res. 2017;2017:4826724. doi: 10.1155/2017/4826724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Yu M., Li M., Zhao R., Zhu Q., Zhou W., Lu M., Lu Y., Zheng T., Jiang J. Polymorphic variations in manganese superoxide dismutase (MnSOD), glutathione peroxidase-1 (GPX1), and catalase (CAT) contribute to elevated plasma triglyceride levels in Chinese patients with type 2 diabetes or diabetic cardiovascular disease. Mol. Cell. Biochem. 2012;363:85–91. doi: 10.1007/s11010-011-1160-3. [DOI] [PubMed] [Google Scholar]
- 25.Tomita T. Apoptosis in pancreatic β-islet cells in type 2 diabetes. Bosn. J. Basic Med. Sci. 2016;16:162–179. doi: 10.17305/bjbms.2016.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nogueira T.C., Paula F.M., Villate O., Colli M.L., Moura R.F., Cunha D.A., Marselli L., Marchetti P., Cnop M., Julier C., Eizirik D.L. GLIS3, a susceptibility gene for type 1 and type 2 diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9:e1003532. doi: 10.1371/journal.pgen.1003532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay V.S.Y., Devaraj S., Koh T., Ke G., Crasta K.C., Ali Y. Increased double strand breaks in diabetic β-cells with a p21 response that limits apoptosis. Sci. Rep. 2019;9:19341. doi: 10.1038/s41598-019-54554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao C., Hölscher C., Liu Y., Li L. GSK3: a key target for the development of novel treatments for type 2 diabetes mellitus and Alzheimer disease. Rev. Neurosci. 2011;23:1–11. doi: 10.1515/rns.2011.061. [DOI] [PubMed] [Google Scholar]
- 29.Bento J.L., Palmer N.D., Mychaleckyj J.C., Lange L.A., Langefeld C.D., Rich S.S., Freedman B.I., Bowden D.W. Association of protein tyrosine phosphatase 1B gene polymorphisms with type 2 diabetes. Diabetes. 2004;53:3007–3012. doi: 10.2337/diabetes.53.11.3007. [DOI] [PubMed] [Google Scholar]
- 30.Carlson C.J., Koterski S., Sciotti R.J., Poccard G.B., Rondinone C.M. Enhanced basal activation of mitogen-activated protein kinases in adipocytes from type 2 diabetes: potential role of p38 in the downregulation of GLUT4 expression. Diabetes. 2003;52:634–641. doi: 10.2337/diabetes.52.3.634. [DOI] [PubMed] [Google Scholar]
- 31.Taneera J., Fadista J., Ahlqvist E., Zhang M., Wierup N., Renström E., Groop L. Expression profiling of cell cycle genes in human pancreatic islets with and without type 2 diabetes. Mol. Cell. Endocrinol. 2013;375:35–42. doi: 10.1016/j.mce.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Mayas M.D., Ortega F.J., Macías-González M., Bernal R., Gómez-Huelgas R., Fernández-Real J.M., Tinahones F.J. Inverse relation between FASN expression in human adipose tissue and the insulin resistance level. Nutr. Metab. (Lond.) 2010;7:3. doi: 10.1186/1743-7075-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delgado-Lista J., Perez-Martinez P., Garcia-Rios A., Alcala-Diaz J.F., Perez-Caballero A.I., Gomez-Delgado F., Fuentes F., Quintana-Navarro G., Lopez-Segura F., Ortiz-Morales A.M. CORonary Diet Intervention with Olive oil and cardiovascular PREVention study (the CORDIOPREV study): rationale, methods, and baseline characteristics: a clinical trial comparing the efficacy of a Mediterranean diet rich in olive oil versus a low-fat diet on cardiovascular disease in coronary patients. Am. Heart J. 2016;177:42–50. doi: 10.1016/j.ahj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buse J.B., Caprio S., Cefalu W.T., Ceriello A., Del Prato S., Inzucchi S.E., McLaughlin S., Phillips G.L., 2nd, Robertson R.P., Rubino F. How do we define cure of diabetes? Diabetes Care. 2009;32:2133–2135. doi: 10.2337/dc09-9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quintana-Navarro G.M., Alcala-Diaz J.F., Lopez-Moreno J., Perez-Corral I., Leon-Acuña A., Torres-Peña J.D., Rangel-Zuñiga O.A., Arenas de Larriva A.P., Corina A., Camargo A. Long-term dietary adherence and changes in dietary intake in coronary patients after intervention with a Mediterranean diet or a low-fat diet: the CORDIOPREV randomized trial. Eur. J. Nutr. 2020;59:2099–2110. doi: 10.1007/s00394-019-02059-5. [DOI] [PubMed] [Google Scholar]
- 36.Roncero-Ramos I., Jimenez-Lucena R., Alcala-Diaz J.F., Vals-Delgado C., Arenas-Larriva A.P., Rangel-Zuñiga O.A., Leon-Acuña A., Malagon M.M., Delgado-Lista J., Perez-Martinez P. Alpha cell function interacts with diet to modulate prediabetes and type 2 diabetes. J. Nutr. Biochem. 2018;62:247–256. doi: 10.1016/j.jnutbio.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Matsuda M., DeFronzo R.A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 38.Abdul-Ghani M.A., Matsuda M., Balas B., DeFronzo R.A. Muscle and liver insulin resistance indexes derived from the oral glucose tolerance test. Diabetes Care. 2007;30:89–94. doi: 10.2337/dc06-1519. [DOI] [PubMed] [Google Scholar]
- 39.Hanson R.L., Pratley R.E., Bogardus C., Narayan K.M., Roumain J.M., Imperatore G., Fagot-Campagna A., Pettitt D.J., Bennett P.H., Knowler W.C. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am. J. Epidemiol. 2000;151:190–198. doi: 10.1093/oxfordjournals.aje.a010187. [DOI] [PubMed] [Google Scholar]
- 40.Song Y., Manson J.E., Tinker L., Howard B.V., Kuller L.H., Nathan L., Rifai N., Liu S. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women’s Health Initiative Observational Study. Diabetes Care. 2007;30:1747–1752. doi: 10.2337/dc07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang W., Fu Q., Zhang Q., Sun M., Gao Y., Liu X., Qian L., Shan S., Yang T. The association between serum uric acid and residual β -cell function in type 2 diabetes. J. Diabetes Res. 2014;2014:709691. doi: 10.1155/2014/709691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanco-Rojo R., Alcala-Diaz J.F., Wopereis S., Perez-Martinez P., Quintana-Navarro G.M., Marin C., Ordovas J.M., van Ommen B., Perez-Jimenez F., Delgado-Lista J., Lopez-Miranda J. The insulin resistance phenotype (muscle or liver) interacts with the type of diet to determine changes in disposition index after 2 years of intervention: the CORDIOPREV-DIAB randomised clinical trial. Diabetologia. 2016;59:67–76. doi: 10.1007/s00125-015-3776-4. [DOI] [PubMed] [Google Scholar]
- 43.Vlachos I.S., Zagganas K., Paraskevopoulou M.D., Georgakilas G., Karagkouni D., Vergoulis T., Dalamagas T., Hatzigeorgiou A.G. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karagkouni D., Paraskevopoulou M.D., Chatzopoulos S., Vlachos I.S., Tastsoglou S., Kanellos I., Papadimitriou D., Kavakiotis I., Maniou S., Skoufos G. DIANA-TarBase v8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46(D1):D239–D245. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marschener I. glm2: fitting generalized linear models with convergence problems. R J. 2011;3:12–15. [Google Scholar]
- 46.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.