Abstract
The cell is the unit of life for all organisms, and all cells are certainly not the same. So the technology to generate transcription expression or genomic DNA profiles from single cells is crucial. Since its establishment in 2009, single-cell RNA sequencing (scRNA-seq) has emerged as a major driver of progress in biomedical research. During the last three years, several new single-cell sequencing platforms have emerged. Yet there are only a few systematic comparisons of the advantages and limitations of these commonly used platforms. Here we compare two single-cell sequencing platforms: BD Rhapsody and 10x Genomics Chromium, including their different mechanisms and some scRNA-seq results obtained with them.
Keywords: Single-cell sequencing, DNA, BD rhapsody, 10x genomics chromium, transcriptome sequencing, technology comparison
1. INTRODUCTION
The Cell is the fundamental unit of an organism. When analyzing sequencing data from bulk tissue samples, typically which are composed of millions of cells, it is difficult to resolve cell to cell variations and identify cells that may play an important role in disease progression [1, 2]. The rapidly evolving single-cell sequencing (SCS) technology has become a powerful method to resolve complex tissue compositions and delineate convoluted cellular development [3]. Sequencing the DNA of individual cells can give information about mutations carried by a small population of cells, for example in cancer, while single-cell RNA-seq can investigate transcriptomic cell-to-cell variation, revealing new cell types, and providing insights into developmental processes and transcriptional stochasticity.
These SCS techniques include the detection of DNA mutations, copy-number variants (CNVs), DNA-protein binding, RNA splicing, and the measurement of mRNA expression [4]. More recently, microfluidics platforms and droplet-based methods have enabled massively parallel sequencing of mRNA in large numbers of individual cells [5].
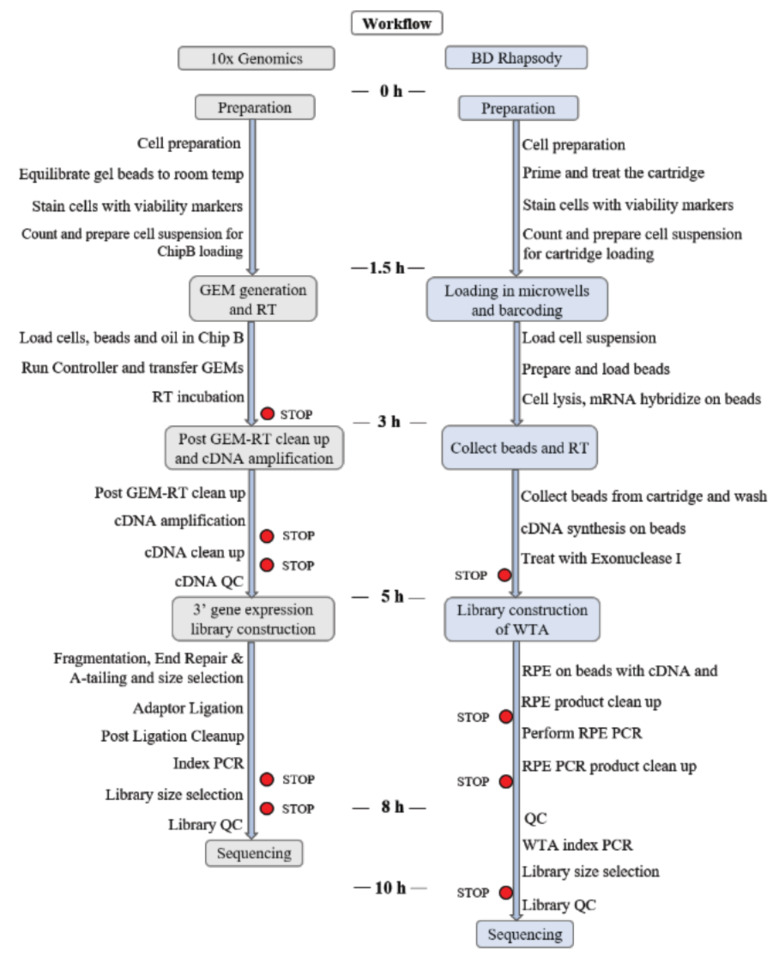
In this review, we compare two of the most successful commercial single-cell sequencing platforms: BD Rhapsody and 10x Genomics Chromium (Fig. 1). We compared them in terms of mechanisms, operations and some scRNA-seq results obtained with them.
Fig. (1).
Description of the two methods evaluated. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2. OUTLOOK
2.1. Common Mechanism of the Two Methods
These two experiments follow a similar basic strategy [6, 7]. First, individual cells are captured separately and lysed, then reverse transcription is performed to select mRNA (by polyT priming) and to obtain cDNA. Subsequently, the amplified cDNA is used for sequencing library preparation.
These two technologies sample a pool of millions of barcodes to separately index each cell’s transcriptome. They do so by partitioning thousands of cells into nanoliter-scale aqueous compartments, where all generated cDNA share a common cell Barcode. Libraries are generated and sequenced and cell barcodes are used to associate individual reads back to the individual partitions.
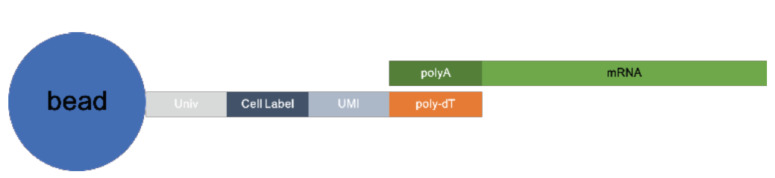
2.1.1. Cell Label, Unique Molecular Index (UMI) and Poly dT Structure of Beads
The oligonucleotides on beads consist of a universal PCR priming site, followed by a combinatorial cell label, a unique molecular index, and an mRNA capture sequence of oligo(deoxythymidine) [oligo(dT)] [8]. All primers on each bead share the same cell label but incorporate a diversity of molecular indices, as shown in Fig. (2).
Fig. (2).
The structure of beads. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
One big challenge in single-cell RNA-seq is the amplification bias (which reduces quantitative accuracy). UMI is designed to detect and quantify unique mRNA transcripts. Such molecular tags have been given various names, such as unique identifiers (UID) [9], unique molecular identifiers (UMI) [10], primer ID [11], etc. Tagging total transcriptome of a single cell has been shown to enable quantitative access of expression level of individual genes in individual cells
[12] and correct the amplification bias [10]. Upon deep sequencing, each UMI is observed multiple times, and the number of original DNA molecules can be determined simply by counting each UMI only once.
2.1.2. Simultaneously Measure Proteins and RNA at a Single-cell Level
Prior to single-cell genomics, defining cell-surface proteins, which are often reliable indicators of cellular activity and function, is critical to understanding the unique characteristics of the various cell types [13]. In 2017, Stoeckius et al. conjugated well-characterized monoclonal antibodies to oligos that were designed to (1) contain a poly-A tail, making it compatible with poly-dT capture RNA-seq-based systems, (2) contain a specific barcode sequence for unique antibody identification [14]. The antibody-oligo complexes are incubated with single-cell suspensions using conditions comparable to staining protocols used in flow cytometry, after which cells are washed to remove unbound antibodies and processed for scRNA-seq.
This kind of antibody is suitable for both platforms. The DNA-barcoded antibodies are incubated with single-cell suspensions under conditions comparable to flow cytometry staining protocols, then the unbound antibody is removed by washing. After cDNA derived from antibody-derived tags (ADTs) is separated from the mRNA-derived cDNA by size selection,the cell-surface-protein library and gene-expression library are prepared independently, and could be sequenced together with different sample indexes.
This technology allows to seamlessly combine cell surface protein expression measurements with immune repertoire and gene expression measurements from the same single cell. Using this to measure both gene and cell surface protein expression in the same cell to identify protein isoforms, proteins can be detected for low abundance transcripts, further increasing the phenotypic specificity. It has plenty of applications as mentioned below.
2.1.2.1. High-Throughput Single-Cell Proteogenomic
Several antibodies have been designed for this method, which can be detected together [15-20]. In 2017, Peterson et al. quantified proteins with 82 barcoded antibodies, using the RNA expression and protein sequencing assay. This removes the limitations imposed by the spectral overlap of fluorescent labels [21] or the available number of stable isotopes [22], in flow and mass cytometry.
2.1.2.2. Simultaneous Measurement of Protein and RNA
Using oligonucleotide-labeled antibodies, measurements of cellular surface proteins and transcriptomes can be integrated into a sequencing-based readout of single cells. It enables to simultaneously combine protein with transcriptomic analysis in high-throughput cells in parallel, which affords an enhanced understanding of a diversity of cellular processes.
2.1.2.3. Enhance Cell Type Identification
The use of oligo-conjugated antibodies provides enhanced immunophenotyping as compared to using mRNA analysis alone, especially when analyzing closely related cells. In addition, the antibodies are less prone to dropouts, increasing the resolution of the analysis. This can help the scientific community expedite discovery in biomedical research, moving the frontiers of science forward.
While the surface proteins of individual cells measured by ADTs are also transcriptomically profiled by scRNA-seq, the measurements of these two different molecule species produced from the same genes do not necessarily correlate with each other, presumably because of post-transcriptional and post-translational gene regulation [23]. Therefore, computational integration of single-cell multi-modal profiling data may allow a more accurate characterization of the cells (e.g., cell type identification) [24] and provide new biological insights that may be observable from neither a single data source [25] nor modality [26].
2.1.2.4. Easy Sample Multiplexing
The technology, cell hashing [27], allows robust sample multiplexing, confident multiplet identification, and discrimination of low-quality cells from ambient RNA.
First, cells from distinct samples are labeled with different oligonucleotide-tagged antibodies against ubiquitous cell surface proteins, so that they can subsequently be pooled. By sequencing these tags alongside the cellular transcriptome, it is possible to correlate each cell to its sample of origin and robustly identify doublets originating from multiple samples.
Cell hashing allows multiplexing of many different samples or experimental conditions, and can also be used to optimize existing protocols or experimental designs, testing several conditions in one scRNA-seq run. A further benefit of cell hashtags is that cell hashing can also reduce batch effects.
According to Dr. Smibert, cell hashing also allows super-loading of scRNA-seq instruments to yield four or five times more cells in a single run, driving down the per-experiment cost of single-cell genomic studies.
2.2. VDJ Recombination
Both of the two platforms can detect Single cell VDJ. The Single Cell VDJ offers comprehensive, scalable solutions for measuring immune repertoire information and gene expression from the same cell such as profile full length (5’ UTR to constant region), paired T-cell receptor (TCR), or B-cell immunoglobulin (Ig) transcripts from 100-10,000 individual cells per sample.
3. Difference between the two methods
The BD Rhapsody WTA Amplification kit has allowed the identification of genes interest that can be used for building targeted panels, which 10x Genomics will apply soon.
3.1. Droplet-based vs. Microwell-based
BD Rhapsody is a microwell-based technology, while 10x Genomics is a Droplet-based system.
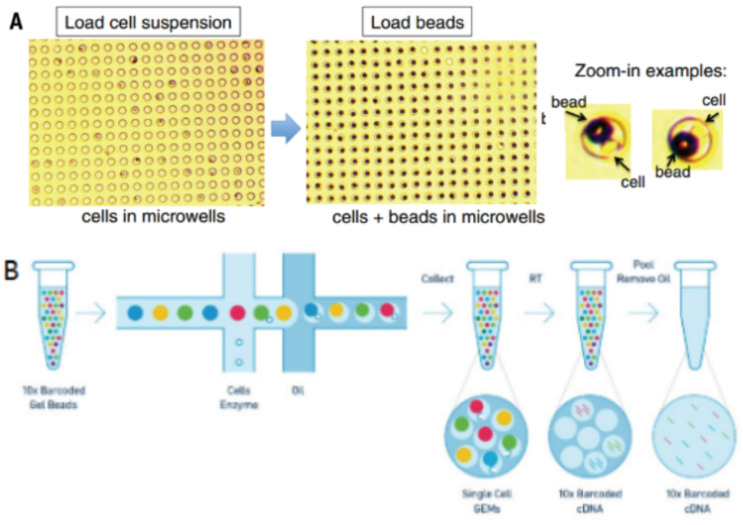
Droplets nanoliter-scale aqueous compartments are formed by precisely combining aqueous and oil flows in a microfluidic device [28, 29]. 10x Genomics system, as shown in Fig. (3), does so by partitioning thousands of cells into nanoliter-scale Gel Bead-In-Emulsions (GEMs), where all generated cDNA from one cell share a common cell barcode [30]. During reverse transcription, the cell barcodes are labeled, then the tiny reaction chambers are broken and the pooled fractions are recovered.
Fig. (3A, B).
Single-cell barcoding process. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
BD Rhapsody is a well-based system like Microwell-seq (a well-based inDrop system) [5, 29], as shown in Fig. (3). Firstly, individual cells are randomly deposited into an array of picoliter wells under the effect of gravity. Then, a combinatorial library of beads bearing cell barcodes and UMI is then loaded onto the microwell array to saturation, so that most wells become filled. The dimensions of the beads and wells are optimized to prevent double occupancy of beads. After cell lysis, mRNAs hybridize to beads, which are pooled for reverse transcription, amplification and sequencing.
3.2. Gel Emulsion Microbeads vs. Magnetic Beads
The two technologies sample a pool of about a million cell barcodes to separately index each cell’s transcriptome. 10x does so by partitioning thousands of cells into nanoliter-scale Gel Beads-in-emulsion (GEMs), while BD by using magnetic microspheres in a microfluidic device [6, 30].
Gel emulsion microbeads are prepared by the emulsion-gelation method with chemical reagents such as polymeric surfactants and dispersants, which are often used in the drug delivery systems [1, 31]. In this case, they deliver oligonucleotides consisting of a universal PCR priming site, UMI, cell barcode and poly-dT. The beads are stable under one condition, while they are swelled by the action of some chemical reagents and the oligonucleotides meet the lysed cells. The RT react and the cDNAs are mixed.
Magnetic beads have been designed for life science applications, such as immunoprecipitation, cell isolation, RNA/DNA extraction, and protein purification. Magnetic particles can be used to isolate specific target molecules and protein complexes. In BD Rhapsody, magnetic beads bind to mRNA from single-cell lysate. Then the beads are mixed, and the RT react in one tube.
3.3. Random Primer (No Tagmentation) vs. Template Switch Oligo (TSO) Full-length cDNA
Different from the cDNA synthesis approach of 10x Genomics, which primes cDNA second-strand synthesis using TSO after first-strand synthesis and complete double-strand cDNA synthesis with a continuous two-step PCR procedure, BD synthesizes cDNA in two steps with two PCR procedures.
10x Genomics obtains full-length cDNA by using TSO, while BD gets fragment cDNA by random primer [32]. Like 10x Genomics, each BD bead captures single-cell mRNAs by primers with oligo(deoxythymidine)[oligo(dT)]. Then BD Beads with captured mRNAs in individual microwells are retrieved by the magnet and synthesize first-strand cDNA in a microcentrifuge tube. After first-strand synthesis, random priming and extension (RPE) are performed on BD beads with cDNA.
Accurate quantification of individual transcripts requires an efficient approach to convert mRNA molecules into full-length cDNA. However, conventional cDNA construction methods usually result in an underrepresentation of the 5’ ends of cDNA. The TSO hybridizes to untemplated C nucleotides added by the reverse transcriptase during reverse transcription [33, 34]. The TSO adds a common 5' sequence to full-length cDNA that is used for downstream cDNA amplification. The TSO makes it possible to efficiently amplify the entire full-length transcript pool in a completely sequence-independent manner.
However, in the 3' assay of 10x Genomics, amplified full-length cDNAs are sheared. Only 3’ end of cDNA, adapter and sample indices are incorporated into finished libraries, which are compatible with next-generation short-read sequencing. So that the TSO and random primers show no difference in the final data.
3.4. Costs
BD and 10x possess little difference in terms of costs. According to our suppliers, single-cell transcriptome kit of BD is a little cheaper.
When the antibody panel is used for the protein expression, actually there are two libraries, one for single-cell transcriptomes and the other for protein expression. So more reads and more reagents are needed to downstream which consequently impacts the costs.
4. Results
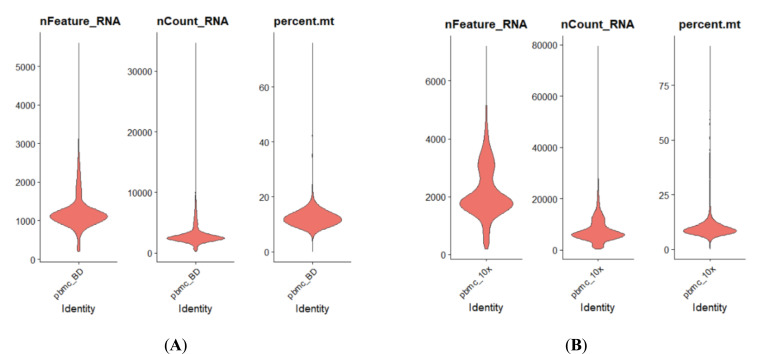
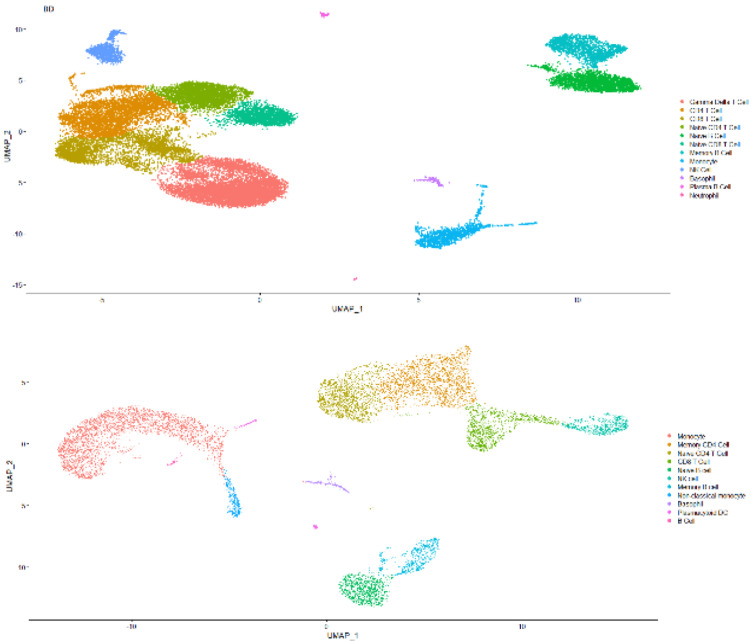
The single-cell transcriptome data are downloaded from 10x Genomics website or given by the Technical Support of BD Company. They are both demo human PBMC data, and the average number of reads is >50,000 per cell. Some results are shown in Figs. (4 and 5).
As shown in Fig. (4), the capability of BD Rhapsody capturing RNA species ranges from 500 to 2500 per cell
Fig. (4A, B).
Comparison of basic parameters for human PBMC scRNASeq results acquired by either BD Rhapsody (Left Panel) or 10x 3’ RNASeq V3 (Right Panel). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
(A) (B)
concluding remarks
In this review, we systematically compared two high throughput single-cell sequencing platforms. Different scenarios and the applicability of the two techniques are shown in Table (1).
Table 1.
Comparison between 10x genomics and BD rhapsody.
| Items | 10x Genomics | BD Rhapsody |
|---|---|---|
| Library Costs | $ 1600 | $ 1400 |
| Time Costs | ~8 hours | ~10 hours |
| Reproducibility | Pretty good | good |
| Cell viability demand | >80% | >50% |
| Cells loading | 500-25000 | 2000-40000 |
| Cells recovery | 100-16000 | 100-25000 |
| Safe stop points | 5 | 4 |
| Single cell isolation | Microfluidics | Nanowells |
| Cell Surface Protein | yes | yes |
| Single Cell VDJ | yes | yes |
| Single Cell ATAC | yes | no |
| RNA capture beads | Gel beads | Magnetic beads |
| Second strand cDNA synthesis | TSO | RPE |
| Full-length cDNA | yes | no |
| Advantage | High throughput Easy operation High cell recovery |
Monitor cells viability with imaging system Fewer doublets |
| Limitations | Fresh samples needed Detect only 10% mRNA |
Fresh samples needed More hands work More cells input One sample per assay |
(median: over 1000), while 10x 3’ V3 ranges from 1000 to nearly 4000 per cell (median: around 2000) with each cell capturing around 2500 RNA molecules or 5000, respectively. The median mitochondrial genes percentile for BD methods is a little bit more than 10x, while 10x delivers the range of mitochondrial genes percentile from 5 to 15. This result indicates that due to different capture mechanisms, the BD system could tolerate less cell viability, while 10x scRNASeq clearly delivers a better RNA capture rate. Therefore, the BD system might be better for a less accessible patient sample. As for cell type capture capability, there is not much difference as indicated in Fig. (4). Both methods could capture lymphocytes, myeloid cells and granulocytes well.
Also there are low-throughput methods for scRNA-seq, which sort a cell into a well of a multi-well plate, such as Smart-seq2 [35] and CEL-Seq2 [36]. But these methods analyze only tens to hundreds of single-cell transcriptomes at a time.
When starting the single-cell analysis experiment, several points need to be checked. Such as, if only one rare subset needs to be examined, low-throughput methods can be selected to load in pre-purified cells. Or more cells can be loaded in high throughput platforms, which will give more information on all loaded cell subsets. Of course, the cost should be considered. Low-throughput methods can be a better choice when the cells are fewer than one hundred, otherwise, high throughput platforms perform well. Also when dealing with fewer cells, sample multiplexing technology enables the use of high throughput platforms [27, 37].
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) is an efficient method to probe genome-wide open chromatin sites, using the Tn5 transposase to tag them with sequencing adapters [38]. 10x Genomics also profiles the ATAC at single-cell level (scATAC-seq) [39], but the BD Rhapsody has not incorporated this technology yet. scATAC-seq maps hundreds of single cells in aggregate of closely resembling accessibility profiles from tens of millions of cells and provides insights into cell-to-cell variation, offering a similar power of resolution and generates additional information regarding gene regulatory processes.
Fig. (5A, B).
Comparison of captured cell type for human PBMC scRNASeq results acquired by either BD Rhapsody (Up Panel) or 10x 3’ RNASeq V3 (Down Panel). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Acknowledgements
Declared none.
Consent for Publication
Not applicable.
Funding
The study was supported by the Shanghai Municipal Commission of health and family planning (20184Y0147).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015;58(4):610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Tabula M.C., Tabula Muris Consortium Overall coordination; Logistical coordination; Organ collection and processing; Library preparation and sequencing; Computational data analysis; Cell type annotation; Writing group; Supplemental text writing group; Principal investigators. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727):367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Navin N.E. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58(4):598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haque A., Engel J., Teichmann S.A., Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudapati H., Dey M., Ozbolat I. A comprehensive review on droplet-based bioprinting: Past, present and future. Biomaterials. 2016;102:20–42. doi: 10.1016/j.biomaterials.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Fan H.C., Fu G.K., Fodor S.P.A. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science. 2015;347(6222):1258367. doi: 10.1126/science.1258367. [DOI] [PubMed] [Google Scholar]
- 7.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., Gregory M.T., Shuga J., Montesclaros L., Underwood J.G., Masquelier D.A., Nishimura S.Y., Schnall-Levin M., Wyatt P.W., Hindson C.M., Bharadwaj R., Wong A., Ness K.D., Beppu L.W., Deeg H.J., McFarland C., Loeb K.R., Valente W.J., Ericson N.G., Stevens E.A., Radich J.P., Mikkelsen T.S., Hindson B.J., Bielas J.H. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8(1):14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA. 2011;108(23):9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kivioja T., Vähärautio A., Karlsson K., Bonke M., Enge M., Linnarsson S., Taipale J. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods. 2011;9(1):72–74. doi: 10.1038/nmeth.1778. [DOI] [PubMed] [Google Scholar]
- 11.Liang R.H., Mo T., Dong W., Lee G.Q., Swenson L.C., McCloskey R.M., Woods C.K., Brumme C.J., Ho C.K.Y., Schinkel J., Joy J.B., Harrigan P.R., Poon A.F. Theoretical and experimental assessment of degenerate primer tagging in ultra-deep applications of next-generation sequencing. Nucleic Acids Res. 2014;42(12):e98–e98. doi: 10.1093/nar/gku355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam S., Zeisel A., Joost S., La Manno G., Zajac P., Kasper M., Lönnerberg P., Linnarsson S. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods. 2014;11(2):163–166. doi: 10.1038/nmeth.2772. [DOI] [PubMed] [Google Scholar]
- 13.Pontén F., Gry M., Fagerberg L., Lundberg E., Asplund A., Berglund L., Oksvold P., Björling E., Hober S., Kampf C., Navani S., Nilsson P., Ottosson J., Persson A., Wernérus H., Wester K., Uhlén M. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009;5:337. doi: 10.1038/msb.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14(9):865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Large-scale simultaneous measurement of epitopes and transcriptomes in single cells. bioRxiv. 2017:113068. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang X., Zhang S., Peng Q., Ling L., Shi H., Liu Y., Cheng L., Xu L., Cheng L., Chakrabarti L.A. Sustained IFN-I stimulation impairs MAIT cell responses to bacteria by inducing IL-10 during chronic HIV-1 infection. 2020. [DOI] [PMC free article] [PubMed]
- 17.Mair F., Erickson J.R., Voillet V., Simoni Y., Bi T., Tyznik A.J., Martin J., Gottardo R., Newell E.W., Prlic M. A targeted multi-omic analysis approach measures protein expression and low-abundance transcripts on the single-cell level. Cell Rep. 2020;31(1):107499. doi: 10.1016/j.celrep.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson V.M., Zhang K.X., Kumar N., Wong J., Li L., Wilson D.C., Moore R., McClanahan T.K., Sadekova S., Klappenbach J.A. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 2017;35(10):936–939. doi: 10.1038/nbt.3973. [DOI] [PubMed] [Google Scholar]
- 19.Granja J.M., Klemm S., McGinnis L.M., Kathiria A.S., Mezger A., Corces M.R., Parks B., Gars E., Liedtke M., Zheng G.X.Y., Chang H.Y., Majeti R., Greenleaf W.J. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 2019;37(12):1458–1465. doi: 10.1038/s41587-019-0332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotliarov Y., Sparks R., Martins A.J., Mulè M.P., Lu Y., Goswami M., Kardava L., Banchereau R., Pascual V., Biancotto A., Chen J., Schwartzberg P.L., Bansal N., Liu C.C., Cheung F., Moir S., Tsang J.S. Broad immune activation underlies shared set point signatures for vaccine responsiveness in healthy individuals and disease activity in patients with lupus. Nat. Med. 2020;26(4):618–629. doi: 10.1038/s41591-020-0769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perfetto S.P., Chattopadhyay P.K., Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 2004;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 22.Bendall S.C., Simonds E.F., Qiu P., Amir A.D., Krutzik P.O., Finck R., Bruggner R.V., Melamed R., Trejo A., Ornatsky O.I., Balderas R.S., Plevritis S.K., Sachs K., Pe’er D., Tanner S.D., Nolan G.P. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.See P., Lum J., Chen J., Ginhoux F. A single-cell sequencing guide for immunologists. Front. Immunol. 2018;9:2425. doi: 10.3389/fimmu.2018.02425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33(2):155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 25.Lin Y., Ghazanfar S., Wang K.Y.X., Gagnon-Bartsch J.A., Lo K.K., Su X., Han Z-G., Ormerod J.T., Speed T.P., Yang P., Yang J.Y.H. scMerge leverages factor analysis, stable expression, and pseudoreplication to merge multiple single-cell RNA-seq datasets. Proc. Natl. Acad. Sci. USA. 2019;116(20):9775–9784. doi: 10.1073/pnas.1820006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji Y., Qi D., Li L., Su H., Li X., Luo Y., Sun B., Zhang F., Lin B., Liu T., Lu Y. Multiplexed profiling of single-cell extracellular vesicles secretion. Proc. Natl. Acad. Sci. USA. 2019;116(13):5979–5984. doi: 10.1073/pnas.1814348116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M., III, Smibert P., Satija R. Cell hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19(1):224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorsen T., Roberts R.W., Arnold F.H., Quake S.R. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett. 2001;86(18):4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 29.Umbanhowar P.B., Prasad V., Weitz D.A. Monodisperse emulsion generation via drop break off in a coflowing stream. Langmuir. 2000;16(2):347–351. doi: 10.1021/la990101e. [DOI] [Google Scholar]
- 30.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J. Microwell-seq of single cells. Nat. Commun. 2017;8(1):14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smriti M.J.P. Sumeet Dwivedi: Formulation and evaluation of floating microbeads of ciprofloxacin HCl by emulsion gelation method. Int. J. Pharm Life Sci. 2013;4(8):2876–2884. [Google Scholar]
- 32.Xiang C.C., Chen M., Ma L., Phan Q.N., Inman J.M., Kozhich O.A., Brownstein M.J. A new strategy to amplify degraded RNA from small tissue samples for microarray studies. Nucleic Acids Res. 2003;31(9):e53–e53. doi: 10.1093/nar/gng053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo G.X., Taylor J. Template switching by reverse transcriptase during DNA synthesis. J. Virol. 1990;64(9):4321–4328. doi: 10.1128/JVI.64.9.4321-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y.Y., Machleder E.M., Chenchik A., Li R., Siebert P.D. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30(4):892–897. doi: 10.2144/01304pf02. [DOI] [PubMed] [Google Scholar]
- 35.Ramsköld D., Luo S., Wang Y-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C., Schroth G.P., Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., Gennert D., Li S., Livak K.J., Rozenblatt-Rosen O., Dor Y., Regev A., Yanai I. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77–77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGinnis C.S., Patterson D.M., Winkler J., Conrad D.N., Hein M.Y., Srivastava V., Hu J.L., Murrow L.M., Weissman J.S., Werb Z., Chow E.D., Gartner Z.J. MULTI-seq: sample multiplexing for single-cell RNA sequencing using lipid-tagged indices. Nat. Methods. 2019;16(7):619–626. doi: 10.1038/s41592-019-0433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10(12):1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satpathy A.T., Granja J.M., Yost K.E., Qi Y., Meschi F., McDermott G.P., Olsen B.N., Mumbach M.R., Pierce S.E., Corces M.R., Shah P., Bell J.C., Jhutty D., Nemec C.M., Wang J., Wang L., Yin Y., Giresi P.G., Chang A.L.S., Zheng G.X.Y., Greenleaf W.J., Chang H.Y. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 2019;37(8):925–936. doi: 10.1038/s41587-019-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]