Abstract
Variation and heterogeneity between cells are the basic characteristics of stem cells. Traditional sequencing analysis methods often cover up this difference. Single-cell sequencing technology refers to the technology of high-throughput sequencing analysis of genomes at the single-cell level. It can effectively analyze cell heterogeneity and identify a small number of cell populations. With the continuous progress of cell sorting, nucleic acid extraction and other technologies, single-cell sequencing technology has also made great progress. Encouraging new discoveries have been made in stem cell research, including pluripotent stem cells, tissue-specific stem cells and cancer stem cells. In this review, we discuss the latest progress and future prospects of single-cell sequencing technology in the field of stem cells.
Keywords: Single-cell sequencing, pluripotent stem cell, tissue-specific stem cell, cancer stem cell, RNA sequencing, heterogeneity
1. Introduction
A single cell is the smallest functional unit that makes up an organism. Various expression and regulation of genes are performed in individual cells, and it would be perfect to use single cells for analyses of gene expression. Due to technical reasons, most genome studies use a large number of cells of the same type as raw materials. Analysis and research on the basis of such data are valid, but limitations are obvious. Heterogeneity among cells of the same kind is mercilessly ignored. The products transcribed or expressed by the cell population represent the average level of the cell group, but low molecular weight substances are difficult to detect [1]. Therefore, our conclusions can neither be applied to a single cell, nor can we explain many phenomena; for example, the same drug has different effects when applied to adjacent cells of the same tissue. This biological characteristic of cells is affected by the microenvironment and related to many physiological and pathophysiological processes. At the same time, even in the same individual cell, its gene expression varies in different time phases.
Stem cells are defined as not only being capable of unlimited self-renewal but also having the potential to differentiate into specific types of cells. In general, stem cells can be classified into pluripotent stem cells and tissue-specific stem cells. Among them, pluripotent stem cells can generate cells of all three germ layers (the ectoderm, mesoderm and endoderm), while tissue-specific stem cells play an important role in the development of embryonic tissue and the metabolism of adult tissue. Most tissues are formed by heterogeneous cell masses. The technology of single-cell sequencing provides strong support to describe the biological characteristics of heterogeneous cell populations and its greatest advantage is that it can accurately provide unbiased information of each single cell without being influenced by microenvironment outside factors other than cells [2]. In our view, single-cell sequencing technology has unique advantages for understanding the occurrence and development of stem cells. Stem cells have multiple differentiation potentials. Despite having the same initiation, there are differences in gene expression due to their differentiation into different terminal cells, which may be influenced by intracellular regulators and extracellular signaling molecules. At the same time, during stem cell development, gene expression varies temporally, and these problems are difficult to solve with previous technologies. Single-cell sequencing allows the focus to be on a single cell, either as an individual in a cell population or as a tracer of the same subpopulation at all stages of development. If single-cell sequencing is combined with other professional means, it will be even more fascinating, such as combining scRNA-seq with patch-clamp technology to explore the true meaning of neuropsychiatric diseases. In this review, we briefly discuss the emerging technology of single-cell RNA sequencing (scRNA-seq), its application in the field of stem cells (including pluripotent stem cells, tissue-specific stem cells and cancer stem cells) and its future prospects.
1.1. Single-cell RNA-sequencing Technologies
Since the first scRNA-seq study was reported in 2009, increasing attention has been focused on this new technology [3]. One of the reasons for its popularity is that scRNA-seq can describe RNA molecules in a single cell with high resolution and genome scale [4]. In the past few years, scRNA-seq was more often conducted by specialist research groups. Recently, it is available for common researchers and clinicians to make significant discoveries using this powerful approach.
ScRNA-seq allows the transcriptomes of individual cells to be compared. Therefore, heterogeneity analysis is the core field of its application, and its main purpose is to evaluate transcriptional similarities and differences within cell populations [5]. Evaluation of transcriptional differences is especially suitable for the identification of rare cell populations because of their neglect in previous experimental methods. In addition, scRNA-seq can also provide more important information about characteristics of gene expression, such as gene splicing patterns, expression of single alleles, and identification of co-regulated gene modules by studying gene co-expression patterns at the single-cell level. However, scRNA-seq can provide answers to many scientific questions, but it should be noted that the accuracy of the answers depends on different experimental protocols [6].
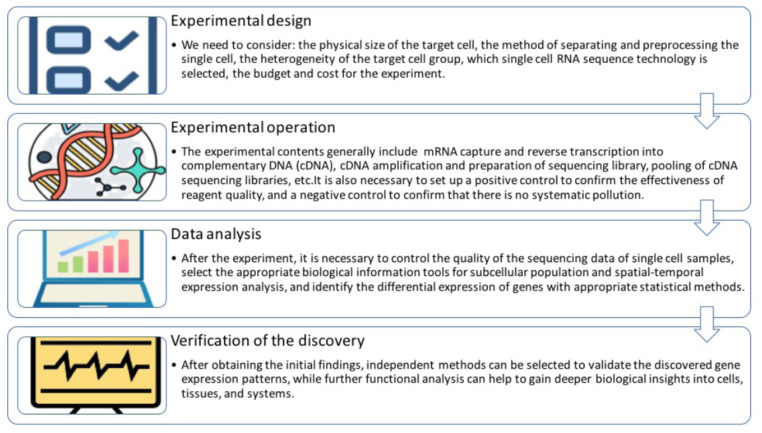
A general workflow for single-cell sequencing includes isolation of single cells, mRNA capture and reverse transcription into complementary DNA (cDNA), cDNA amplification and preparation of sequencing library, pooling of cDNA sequencing libraries, and using bioinformatics and computational methods to interpret robust data biologically [7]. A flow-process diagram of a typical single-cell sequencing project is shown in Fig. (1). Two years after the advent of standard RNA-seq technology using millions of cells, the scRNA-seq technology was first reported in 2009 [8]. Afterwards, many other scRNA-seq methods based on different cell isolation and lysis, RNA capture, cDNA amplification and library establishment strategies have emerged, including Drop-seq [9], SCRB-seq [10], SMART-seq (on Fluidigm C1) [11, 12], SMART-seq V2 [11, 12] and CEL-seq [13]. Ziegenhain et al. made a comparative analysis of these methods [14]. Ziegenhain et al. used the different methods described above to obtain 583 scRNA-seq libraries from mouse embryonic stem cells (mESCs) [14]. In terms of sensitivity, Smart-seq2 is the most sensitive method because it detects the highest number of genes in each cell and the largest number of genes in the total number of transcripts across cells and has the most uniform coverage in transcripts. Smart-seq/C1 was slightly less sensitive to the detection of gene number per cell, with almost the same number of genes detected in the total number of cells across, but with slightly lower coverage. In terms of accuracy, the differences between the above methods in detecting the absolute expression levels across genes within cells are small, and such small differences in accuracy rarely become the decisive factor in method selection. Power is obtained by combining dropout rates and amplification noise. SCRB-seq is the most powerful method when the sequencing depth is 1 million reads and 500,000 reads, and CEL-seq is the most powerful method when the sequencing depth is 250,000 reads. In terms of cost efficiency, the cost of obtaining specific cells, the cost of establishing experimental methods, the cost of reagent consumables required, and the construction of scRNA-seq libraries were considered, Drop-seq is the most cost-effective method for sequencing 254 cells at 250,000 reads, while Smart-seq2 is relatively expensive, mainly using the Tn5 transposase kits from commercial reagent companies, although internally produced transposases could be used if costs were to be controlled. Ziegenhain et al. suggested that Drop-seq be preferred when detecting the transcriptome of large numbers of cells with low sequencing depth, SCRB-seq may be preferred when detecting the transcriptome of fewer cells, and of course, Smart-seq2 may also be considered when using internally produced transposases [14].
Fig. (1).
Flow-process diagram of a typical single-cell sequencing project. Here, we provide a flow-process diagram of a typical single-cell sequencing project. Usually this includes experimental design, experimental operation, data analysis, verification of the discovery. Of course, in a practical project, it often needs to be repeated many times. If there is a mistake in a link, the researcher needs to return to the higher level for inspection and redesign. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
1.2. Pluripotent Stem Cells
At present, scientists still know very little about the development process of embryos. One of the most central questions is when the blastomere begins to change in terms of gene expression and cell fate. Due to the extremely limited number of cells in this process, it is difficult to use traditional experimental methods for research. In the past, scientists generally believed that the earliest time of blastomere differentiation was 8- or 16-cell stage. However, this conjecture lacks molecular evidence. Now scRNA-seq provides unprecedented opportunities to decipher the dynamic changes of gene expression during this process [15-17].
Recently, Biase et al. analyzed individual blastomeres in mouse 2- and 4-cell embryos and found that even in 2-cell embryos, gene expression between blastomeres is different [18]. They further found that the different gene expression characteristics of the two blastomeres in the 2-cell embryo can be transferred to the inner cell mass and trophoblast cells, which indicates that the earliest cell differentiation in embryonic development occurs in the 2-cell stage [18]. Huang et al. conducted a similar study, but concluded that the earliest differentiation of blastomeres was at the 4-cell stage [19].
Embryonic stem cells are recognized as ideal in vitro models for studying the self-renewal ability and differentiation potential of pluripotent stem cells. When it is in appropriate pluripotency supporting conditions, the inner cell mass of the blastocyst form embryonic stem cells, and the derivation of human and mouse embryonic stem cells have been traced by scRNA-seq [16, 20].
Mark Van den Hurk et al. combined scRNA-seq with patch-clamp electrophysiological recording and morphological analysis of individual human neurons in vitro on the basis of the accurate analysis of the full transcriptome of single cells [21]. Specific manifestations of neurological or psychiatric disorders often occur only in specific subpopulations (possibly in very small numbers) of cells or at specific stages of development of a certain group of cells, and it is difficult for traditional methods to reveal their essence, while Patch-seq, a new technology, provides a better solution for identifying such unique cell types or specific maturation stages. Patch-seq was able to analyze cytoplasmic RNA and relatively distant RNA in dendrites and axons, and a series of examinations showed that this method could completely collect transcripts from the whole neuron soma and axons (more than 150 μm away from the soma) and adjacent synapses. This has irreplaceable value for the study of physiological or pathological mechanisms that rely on dendrites or neurosynapses for mRNA transport. Patch-seq technology can reveal the association among gene expression profiles, physiological functions and morphology of single cells. So far, Patch-seq has been successfully applied to human neuron culture in vitro [22] and rodent brain slices in vitro [23, 24]. At the same time, thanks to the combination of Patch-seq and optical tools and transneuronal tracing techniques, the structure and function of neuronal circuits and the trajectory of transcriptome projections to specific regions of the brain were elucidated on rodent brain slices [23, 24]. Patch-seq resolves the bias of bulk analysis of cells and tissues. By studying the electrophysiology of a single cell, combined with its morphology and gene expression profile, it is helpful to identify rare or clinically important cell populations and their associated abnormal molecular mechanisms [21].
The retina is very important for light sensing and image processing. There are many kinds of retinal neuronal cells, and the molecular characteristics of these cell subsets were unknown before the application of single-cell sequencing. Macosko et al. reported the first study via Drop-seq to identify 39 different cell clusters [9]. The analysis also confirmed that the amacrine cells are the most diverse types of neurons, which can be divided into three categories: inhibitory (using GABA or glycine as neurotransmitter), excitatory (releasing glutamate) or amacrine cells with unknown neurotransmitter.
Several studies have also applied scRNA-Seq to brain organ-like substances and provided evidence for the existence of progenitor cells and differentiated cells of neurons and mesenchymal mass spectrometry, similar to fetal neocortex [25]. These studies show that brain organ-like substances have the ability to self-organize along the ventral dorsal anterior part and produce mature astrocytes and oligodendrocytes outside the neuron lineage. In some reported examples, these brain organs include retina [26]. Quadrato et al. isolated more than 80,000 individual cells from 31 human brain organoids cultured in vitro and analyzed their gene expression by scRNA-Seq. They found that different groups of cells expressed different characteristic cell markers, and compared with several gene expression datasets in mice and humans, typical genes of glutamatergic, gamma-aminobutyric and dopaminergic neurons were found, thereby identifying miller glial cells, photoreceptors, retinal ganglion, bipolar cells and amacrine cells as well as retinal pigment epithelium of the neuroretina [27].
Projective neurons of retina transmit visual information to midbrain through the optic nerve, and single-cell sequencing technology is also applied to the analysis of retinal projection neurons retinal ganglion cells. Daniszewki et al. identified 3 different cell subpopulations, including progenitor cells and mature retinal ganglion cells, which contain genes related to axonal guidance together with signal interactions, extracellular matrix proteins and down-regulation of cell cycle genes [28]. In a similar study, Langer et al. were able to distinguish between several RGC subtypes along with molecular markers [29]. It can be found that scRNA-Seq plays an important role in deciphering the cellular composition of pluripotent stem cell retinal organ-like cells and retinal cells, discovering new subtype-specific cell markers, and evaluating their similarity and maturity in comparison to their equivalent cell types in the developmental and adult retina.
Human primordial germ cells (hPGCs) are the precursors of mature germ cells. The single-cell transcriptomes of hPGCs are relatively homologous between the migration phase and the gonad phase [30]. Li et al. attempted to clarify the developmental trajectory and heterogeneity of fetal female germ cells [31]. More than 2000 germ cells and their gonadal niche cells were analyzed by scRNA-seq at several developmental stages. The main results of this study include describing different transcriptome characteristics of transcription factor networks at different developmental stages. Single-cell expression profiles show that female embryos contain at least four stages of embryonic germ cells. This work provides a detailed flow chart for the development of human germ cells, providing guidance for the production of female germ cells from human pluripotent stem cells in vitro and also helping to understand germ cell-related diseases.
ScRNA-seq makes it possible to collect a variety of single-cell data from all different types of female and male germ cells at different stages of development and maturity. This will lead to new findings on the development, transformation and fate of germline cells in vivo and in vitro, and the heterogeneity in single-cell expression profiles of human germ cells will clearly define important cell subpopulations [30]. Combining these results, a more comprehensive molecular map of human germline can be established under both normal and pathological conditions.
1.3. Tissue-specific Stem Cells
Tissue-specific stem cells exist in almost all developing or differentiated tissues. They can not only self-renew but also differentiate into various mature tissue cells with specific functions. At present, single-cell sequencing has been used to identify new stem cell subpopulations and reveal the heterogeneity of seemingly homogeneous stem cells [32].
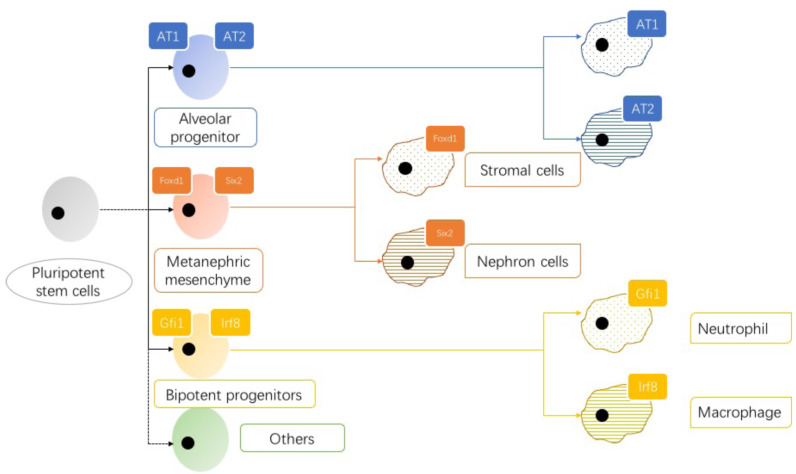
Research on the development of mouse lung epithelium conducted by Treutlein et al. has provided a typical example of how to identify a new stem cell type using scRNA-seq method (Fig. 2) [33]. They used Smart-seq to sequence 198 alveolar epithelial cells from mice to describe the cell type and developmental hierarchy of the developing lung. The alveolar type 1 (AT1) and alveolar type 2 (AT2) cells in the lung are two epithelial cell types, but the characteristics of alveolar progenitor cells are still difficult to determine. Treutlein Desai et al. determined five different cell populations by evaluating 80 epithelial cells from the distal lung regions of mouse embryos, of which four are known cell types and the other is a cell population that has not been reported co-expressing the marker genes of AT1 and AT2. On the principal component analysis chart, this group of cells is located between AT1 and AT2 cell groups, which indicates that this cell group has dual potential to differentiate into AT1 and AT2 cells. This discovery was also confirmed by a series of experiments such as immunostaining, clonal analysis and lineage tracing [34].
Fig. (2).
Schematic diagram of tissue-specific stem cell differentiation with dual differentiation potential. ScRNA-seq can currently identify new tissue-specific stem cell subsets. The oval blue cells are alveolar progenitors that express both AT1 and AT2 cell markers. The oval orange cells co-expressed Foxd1, the marker of stromal-committed cells and Six2, the metanephric mesenchyme of nephron-committed cells, which had dual differentiation potential toward stromal cells and nephron cells. Oval yellow cells are bipotent progenitors that co-express Irf8 and Gfi1, which regulate macrophage and neutrophil specifications, respectively, with the potential to differentiate into macrophage and neutrophil lineages. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
With the application of scRNA-seq technology, increasing stem cells with dual differentiation potential have been found. Brunskill et al. used Smart-seq to sequence 235 cells from mouse kidneys to construct gene expression profiles at different stages of kidney development [35]. They found that Foxd1, the marker of stromal-committed cells and Six2, the marker of nephron-committed cells, were co-expressed in single cells of metanephric mesenchyme using single-cell microarray and Smart-seq. This discovery was later confirmed at the protein level. Different from lung bi-potential progenitor cells, co-expressing metanephric mesenchyme cells do not reflect the overall characteristics of stromal and nephron progenitor cells. However, the co-expressed metanephric mesenchyme cells determine the fate of subsequent cell differentiation, which may differentiate into stromal cells expressing Foxd1 or nephron cells expressing Six2 [35].
By applying Smart-seq to 85 developing olfactory sensory neurons, Hanchate et al. discovered another example of co-expression progenitor cells [36]. In this method, Monocle was used to reconstruct the developmental process map of 85 cells and assign them into four different categories: progenitor cells, precursor cells, and immature and mature neurons. Hanchate et al. found that almost half of immature neurons express more than one receptor, and as cells matured, they showed an increase in the expression of one specific receptor and inhibited other receptor genes, which is also confirmed by single-molecule FISH [36].
However, the discovery of cells with dual differentiation potential also brings a series of questions worthy of our consideration. First, what is the regulation of the differentiation direction of tissue-specific stem cells. Hopx, which is a transcriptional repressor, because of inhibiting the expression of some genes, usually only expressed in terminal cells, but its expression was also currently detected in tissue-specific stem cells, so it cannot be used as a starting event for cell differentiation direction selection. In fact, in the existing scRNA-seq sequence libraries, no inducing differentiation factor specific to each stem cell has been found. Many authors have also speculated that this transcriptional repressor, which determines the direction of cell differentiation, may not be detectable by existing scRNA-seq because of its extremely low expression, or that microRNAs that determine the line of differentiation is not described by current scRNA-seq. Of course, we have reason to speculate that some post-transcriptional events may become decisive events in the direction of final cell differentiation [33-37].
Second, whether this is an accidental or an inevitable phenomenon is how common it is for cells with dual (or possibly more) differentiation potential. As mentioned above, post-renal mesenchymal cells co-expressing Foxd1 and Six2 do not reflect the overall characteristics of interstitial cells and renal progenitor cells. Next, we need to sequence more cells at earlier time points during cell development to help answer our questions. As more and more cells with dual differentiation potential are discovered, we need to better understand when dual states are performed, and of the molecular basis for their activation, permission and resolution, in order to address our most interesting topic: what is the purpose of cells to maintain this dual (multiple) differentiation potentials. We have some speculation about this, that whether it allows tissue-specific stem cells to perform certain functions during development that are subsequently assigned to more specialized and mature cell types or whether it allows tissue-specific stem cells to undergo activation by extracellular signaling molecules and differentiate into cell types that are more suitable for the time to perform corresponding functions, such as tissue self-repair [37].
1.4. Cancer Stem Cells
Cancer tissues commonly contain sub-population of cells with definite phenotype and functional heterogeneity. According to the cancer stem cell (CSC) hypothesis, CSCs are a kind of highly malignant cell sub-population, and considered responsible for many important biological characteristics of tumors such as their self-renewal, tumor-initiating, drug-resistance, disease recurrence and metastasis [32]. However, there is still a dispute as to whether the current viewpoint on cancer stem cells is applicable to all tumor types. ScRNA-seq can detect extremely trace nucleic acid sequences so it is possible to help identify these cells and, more generally, it may provide new insights for the heterogeneity in complex tumors and new ideas for the occurrence and development of tumors. ScRNA-Seq analyses of different types of cancer stem cells were summarized in Table 1.
Table 1.
Single cell sequencing analyses of different types of cancer stem cells.
| Study | Cell Type(s) | Primary | Cells | Method | Summary |
|---|---|---|---|---|---|
| Patel et al. (2014) | Glioblastoma | Primary | 672 | SMART-Seq | Patel et al. determined continuous stem cell-related expression status in each tumor by examining a group of “stem cell” genes, reflecting the complex stem cell status in primary tumors. |
| Yang et al. (2017) | Bladder cancer | Primary | 59 | SMART-Seq | Yang et al. determined the co-mutation of ARID1A, GPRC5A and MLL2, significantly enhance the self-renewal and tumor initiation ability of bladder cancer non-stem cells, which made them have the stability of stem cells. |
| Navin et al. (2011) | Breast cancer | Primary & metastasis | 100 | single nucleus sequencing (SNS) | Navin et al. described three different subclones that may represent sequential cloning amplification, and the copy number aberrations analysis also showed that clonal amplification led to the formation and metastasis of primary tumors respectively. |
| Wang et al. (2014) | Breast cancer | Primary | >200 | Nuc-Seq | Wang et al. determined that the tumors had highly similar copy number aberrations, indicating the existence of monoclonal cell populations. |
| Gao et al. (2016) | Breast cancer | Primary | 1,000 | SNS | Gao et al. determined a model established by single cell copy number aberrations provides important enlightenment for tumor evolution, diagnosis and treatment of breast cancer. |
| Karaayvaz et al. (2018) | Breast cancer (triple-negative) | Primary | >1,500 | SMART-seq V2 | Karaayvaz et al. described the degree of diversity of TNBC cells with regard to their expression of normal breast tissue, established TNBC characteristics, and determined the cell sub-populations shared among tumors. |
Patel et al. sequenced 672 single cells from five glioblastoma samples [38]. Each tumor tissue showed a high degree of intratumoral cellular heterogeneity in many aspects, such as copy number changes, cell cycle, hypoxia and immune response. Patel et al. determined continuous stem cell-related expression status in each tumor by examining a group of “stem cell” genes, reflecting the complex stem cell status in primary tumors [38].
Bladder cancer is one of the most common malignant tumors of the urinary system, but the genetic basis and origin of human bladder cancer stem cells are still unknown. Yang et al. performed scRNA-seq on 59 cells from three bladder cancer specimens, including bladder cancer stem cells, bladder cancer non-stem cells, bladder epithelial stem cells and bladder epithelial non-stem cells [39]. Specifically, bladder cancer stem cells showed clonal homogeneity, and it was proved by phylogenetic analysis that they were derived from bladder epithelial stem cells or bladder epithelial non-stem cells. In addition, 21 key modification genes have been described in bladder cancer stem cells, of which 6 genes have never been reported (ETS1, GPRC5A, MKL1, PAWR, PITX2 and RGS9BP). More importantly, the co-mutation of ARID1A, GPRC5A and MLL2, significantly enhanced the self-renewal and tumor initiation ability of bladder cancer non-stem cells, which made them have the stability of stem cells [39]. These findings provide the possibility for new targeted therapy for bladder cancers.
Breast cancer is the most common malignant tumor among women in the world, showing amazing genetic and phenotypic diversity. According to the histological characteristics of primary tumors, breast cancer can be divided into 18 subtypes [40]. If only divided into major categories, it is impossible to really identify the specific cell types in each kind of breast cancer, and accurate treatment can not be achieved. Navin et al. published the first study on the application of Single-Nucleus-Sequencing (SNS, a kind of single-cell sequencing) in breast cancer in 2011 [41]. They sequenced 100 single cells in a polygenomic triple-negative (ER/PR/Her2) breast cancer and 100 single cells in a monogenomic triple-negative breast cancer and its liver metastasis cells through a low coverage single-cell sequencing method and described three different subclones that may represent sequential cloning amplification. The copy number aberrations analysis also showed that clonal amplification led to the formation and metastasis of primary tumors, respectively. Wang et al. sequenced 50 monocytes of an estrogen receptor-positive (ER+/PR+/Her2) breast cancer [42]. The results showed that the tumors had highly similar copy number aberrations, indicating the existence of monoclonal cell populations. These findings indicate that tumors develop through early chromosome rearrangement, followed by stable clonal expansion. Gao et al. further confirmed this finding by sequencing 1000 single cells of triple-negative breast cancers from 12 patients [43]. This model established by single-cell copy number aberrations provides important enlightenment for tumor evolution, diagnosis and treatment of breast cancer.
Triple-negative breast cancer (TNBC) is an invasive subtype characterized by extensive inter-tumoral and intra-tumoral heterogeneity [44]. Karaayvaz et al. sequenced more than 1500 single cells from 6 primary TNBC specimens [44]. They described the degree of diversity of TNBC cells with regard to their expression of normal breast tissue, established TNBC characteristics, and determined the cell sub-populations shared among tumors. The characteristics of these cells led to adverse results, that is, intra-tumoral heterogeneity is the core of the clinical behavior of the disease [44]. Clinically, the failure to achieve a complete pathological response after preoperative chemotherapy is related to the high recurrence rate, which is consistent with the results of scRNA-seq, that is, a small proportion of refractory cell sub-populations determine the prognosis of patients. The most common tumor-derived signal in all cells was basal-like 1 proliferation signal, which corresponds to these proliferating luminal progenitor-like cells to a large extent. Clustering of Gene Expression Profiles has identified a number of cell subpopulations, which were functionally characterized by activation of glycosphingolipid metabolism and related innate immunity pathways, and glycosphingolipid pathway signals themselves were important predictors of prognosis in TNBC patients. Karaayvaz et al. also provided many potential attractive therapeutic targets for treating TNBC, like S1PR1, highly expressed in tumors [44].
2. Future perspectives
In the field of pluripotent stem cells, at present, scRNA-seq has explained many unsolved mysteries in the early stage of the embryo, such as the blastomere changing gene expression and cell fate at the 4-cell stage, etc., which allows people to observe the origin of life and the formation of vital organs from the cell level, which will greatly help to enhance the understanding of tissue growth and development processes, providing the basis for tissue regeneration and other emerging areas. In the field of tissue-specific stem cells, scRNA-seq seems to be a “detection radar” for people. It can find a minority group of unknown (stem) cells from the cell level, which provides a theoretical basis for the previously unexplained phenomenon. It also provides researchers with some selectors for cell differentiation, triggers specific molecular switches, and can move towards specific mature cell differentiation, giving a new way for people to understand the tissue microenvironment and the occurrence and development of some diseases. In the field of tumor stem cells, scRNA-seq technology has become a hot topic in recent years. Tumor stem cells seem to be a kind of cells with the highest degree of “malignancy”. Decoding them will undoubtedly help to have a more comprehensive understanding of the disease. At present, it has been found that some gene mutations will make tumor tissue grow rapidly and give tumor cells the properties of self-renewal and being immortal, and these mutated genes will become effective targets for future cancer treatment. The research on tumor stem cells can control the disease from the root.
Of course, as a new technology, scRNA-seq still has many shortcomings. There are still some problems in single-cell preparation, genome-wide amplification, sequencing and data analysis, such as low coverage, bias and error, also depending on different sequencing platforms and methods. In the future, we need better single-cell preparation methods, better whole genome amplification patterns, better sequencing and data analysis algorithms. In the hot field of cancer stem cells, some fundamental biological problems remain unsolved, such as which genes are the real driving force of tumor occurrence and development; whether these driving factors will change their roles in the process of tumor occurrence; what are the differences and connections between different tumor stem cells within a single tumor tissue; and which cell or group of cells are the origins of the tumor. With the development of technology platforms and the decrease in single sequencing costs, these problems will be reasonably explained, and scRNA-seq technology will also serve the cause of human health.
Conclusion
The technology of scRNA-seq was just started a decade ago, and now it has achieved amazing results. Remarkable progress has been made in many aspects, such as the construction of cell maps of whole tissues or organs, redefinition of cell types, investigation of new marker genes and cell sub-populations; delineation of cell differentiation and development trajectories; identification of tumor molecular markers, exploration of tumor heterogeneity and tumor micro-environment characteristics, and mechanism study of diseases or drug effects. In the field of stem cells, scRNA-seq has made a great contribution to the exploration of the origin of life, and also plays a role in tissue repair and tumor heterogeneity research. Of course, it also has some disadvantages, such as high price, complicated sample processing and operation procedures, some technical noise and amplification errors. It is believed that with the improvement of cell sorting technology and the continuous improvement of whole genome and whole transcriptome amplification technology, the sensitivity and accuracy of single-cell analysis will be further improved, and this technology will be widely applied in various fields.
Acknowledgements
Declared none.
List of abbreviations
- scRNA-seq
single-cell RNA sequencing
- mRNA
messenger RNA
- cDNA
complementary DNA
- SCRB-seq
Single-Cell RNA Barcoding and Sequencing
- SMART-seq
Switching Mechanism At 5’ End of the RNA Transcript and Sequencing
- CEL-seq
Cell Expression by Linear Amplification and Sequencing
Consent for Publication
Not applicable.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0107501 to R.R), National Natural Science Foundation of China (81770747 and 81970646 to R.R, 81400688 to Y.Z).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Navin N., Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3(5):31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Junker J.P., van Oudenaarden A. Every cell is special: genome-wide studies add a new dimension to single-cell biology. Cell. 2014;157(1):8–11. doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picelli S., Faridani O.R., Björklund Å.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9(1):171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 5.Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33(2):155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 6.Spiro A., Shapiro E. Accuracy of answers to cell lineage questions depends on single-cell genomics data quality and quantity. PLOS Comput. Biol. 2016;12(6):e1004983. doi: 10.1371/journal.pcbi.1004983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haque A., Engel J., Teichmann S.A., Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9(1):75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6(5):377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 9.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., Trombetta J.J., Weitz D.A., Sanes J.R., Shalek A.K., Regev A., McCarroll S.A. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soumillon M., Cacchiarelli D., Semrau S., van Oudenaarden A., Mikkelsen T.S. Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv. 2014;•••:003236 [Google Scholar]
- 11.Ramsköld D., Luo S., Wang Y-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C., Schroth G.P., Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30(8):777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10(11):1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 13.Hashimshony T., Wagner F., Sher N., Yanai I. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2012;2(3):666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65(4):631–643. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Tang F., Barbacioru C., Nordman E., Bao S., Lee C., Wang X., Tuch B.B., Heard E., Lao K., Surani M.A. Deterministic and stochastic allele specific gene expression in single mouse blastomeres. PLoS One. 2011;6(6):e21208. doi: 10.1371/journal.pone.0021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., Tang F. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20(9):1131–1139. doi: 10.1038/nsmb.2660. [DOI] [PubMed] [Google Scholar]
- 17.Xue Z., Huang K., Cai C., Cai L., Jiang C.Y., Feng Y., Liu Z., Zeng Q., Cheng L., Sun Y.E., Liu J.Y., Horvath S., Fan G. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature. 2013;500(7464):593–597. doi: 10.1038/nature12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biase F.H., Cao X., Zhong S. Cell fate inclination within 2-cell and 4-cell mouse embryos revealed by single-cell RNA sequencing. Genome Res. 2014;24(11):1787–1796. doi: 10.1101/gr.177725.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang W., Cao X., Biase F.H., Yu P., Zhong S. Time-variant clustering model for understanding cell fate decisions. Proc. Natl. Acad. Sci. USA. 2014;111(44):E4797–E4806. doi: 10.1073/pnas.1407388111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010;6(5):468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van den Hurk M., Erwin J.A., Yeo G.W., Gage F.H., Bardy C. Corrigendum: Patch-Seq protocol to analyze the electrophysiology, morphology and transcriptome of whole single neurons derived from human pluripotent stem cells. Front. Mol. Neurosci. 2019;12:150. doi: 10.3389/fnmol.2019.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bardy C., van den Hurk M., Kakaradov B., Erwin J.A., Jaeger B.N., Hernandez R.V., Eames T., Paucar A.A., Gorris M., Marchand C., Jappelli R., Barron J., Bryant A.K., Kellogg M., Lasken R.S., Rutten B.P., Steinbusch H.W., Yeo G.W., Gage F.H. Predicting the functional states of human iPSC-derived neurons with single-cell RNA-seq and electrophysiology. Mol. Psychiatry. 2016;21(11):1573–1588. doi: 10.1038/mp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadwell C.R., Palasantza A., Jiang X., Berens P., Deng Q., Yilmaz M., Reimer J., Shen S., Bethge M., Tolias K.F., Sandberg R., Tolias A.S. Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq. Nat. Biotechnol. 2016;34(2):199–203. doi: 10.1038/nbt.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuzik J., Zeisel A., Máté Z., Calvigioni D., Yanagawa Y., Szabó G., Linnarsson S., Harkany T. Integration of electrophysiological recordings with single-cell RNA-seq data identifies neuronal subtypes. Nat. Biotechnol. 2016;34(2):175–183. doi: 10.1038/nbt.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camp J.G., Badsha F., Florio M., Kanton S., Gerber T., Wilsch-Bräuninger M., Lewitus E., Sykes A., Hevers W., Lancaster M., Knoblich J.A., Lachmann R., Pääbo S., Huttner W.B., Treutlein B. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA. 2015;112(51):15672–15677. doi: 10.1073/pnas.1520760112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36(10):1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Yang M. S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; Boyden, E.S.; Lichtman, J.W.; Williams, Z.M.; McCarroll, S.A.; Arlotta, P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545(7652):48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniszewski M., Senabouth A., Nguyen Q.H., Crombie D.E., Lukowski S.W., Kulkarni T., Sluch V.M., Jabbari J.S., Chamling X., Zack D.J., Pébay A., Powell J.E., Hewitt A.W. Single cell RNA sequencing of stem cell-derived retinal ganglion cells. Sci. Data. 2018;5:180013. doi: 10.1038/sdata.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langer K.B., Ohlemacher S.K., Phillips M.J., Fligor C.M., Jiang P., Gamm D.M., Meyer J.S. Retinal ganglion cell diversity and subtype specification from human pluripotent stem cells. Stem Cell Reports. 2018;10(4):1282–1293. doi: 10.1016/j.stemcr.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad S., Azizi H., Skutella T. Single-cell expression profiling and proteomics of primordial germ cells, spermatogonial stem cells, adult germ stem cells, and oocytes.Stem Cells: Biology and Engineering. Springer; 2017. pp. 77–87. [DOI] [PubMed] [Google Scholar]
- 31.Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20(6):858–873. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Wen L., Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016;17(1):71. doi: 10.1186/s13059-016-0941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509(7500):371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desai T.J., Brownfield D.G., Krasnow M.A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507(7491):190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunskill E.W., Park J-S., Chung E., Chen F., Magella B., Potter S.S. Single cell dissection of early kidney development: multilineage priming. Development. 2014;141(15):3093–3101. doi: 10.1242/dev.110601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanchate N.K., Kondoh K., Lu Z., Kuang D., Ye X., Qiu X., Pachter L., Trapnell C., Buck L.B. Single-cell transcriptomics reveals receptor transformations during olfactory neurogenesis. Science. 2015;350(6265):1251–1255. doi: 10.1126/science.aad2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar P., Tan Y., Cahan P. Understanding development and stem cells using single cell-based analyses of gene expression. Development. 2017;144(1):17–32. doi: 10.1242/dev.133058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., Louis D.N., Rozenblatt-Rosen O., Suvà M.L., Regev A., Bernstein B.E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z., Li C., Fan Z., Liu H., Zhang X., Cai Z., Xu L., Luo J., Huang Y., He L., Liu C., Wu S. Single-cell sequencing reveals variants in ARID1A, GPRC5A and MLL2 driving self-renewal of human bladder cancer stem cells. Eur. Urol. 2017;71(1):8–12. doi: 10.1016/j.eururo.2016.06.025. [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Adhav R., Xu X. Current progresses of single cell DNA sequencing in breast cancer research. Int. J. Biol. Sci. 2017;13(8):949–960. doi: 10.7150/ijbs.19627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W.R., Hicks J., Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Waters J., Leung M.L., Unruh A., Roh W., Shi X., Chen K., Scheet P., Vattathil S., Liang H., Multani A., Zhang H., Zhao R., Michor F., Meric-Bernstam F., Navin N.E. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao R., Davis A., McDonald T.O., Sei E., Shi X., Wang Y., Tsai P-C., Casasent A., Waters J., Zhang H., Meric-Bernstam F., Michor F., Navin N.E. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat. Genet. 2016;48(10):1119–1130. doi: 10.1038/ng.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., Ellisen L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9(1):3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]