Summary
Viruses depend on the host metabolic machinery to complete their life cycle in the host cytoplasm. However, the key viral factors initiating the host machinery after the virus enters the cytoplasm remain unclear. Here, we found that compounds packaged in the virions of white spot syndrome virus, such as palmitic amide, could trigger the viral life cycle in the host cytoplasm. Palmitic amide promoted virus infection by enhancing host glycolysis by binding to triosephosphate isomerase to enhance its enzymatic activity. The glycolysis enhancement resulted in lactate accumulation, thereby promoting hypoxia-inducible factor 1 (HIF-1) expression. HIF-1 upregulation further enhanced glycolysis, which in turn promoted virus infection. Therefore, our study presented novel insight into the initiation of the virus life cycle in host cells.
Subject areas: Human Metabolism, Molecular Biology, Virology
Graphical abstract
Highlights
-
•
Palmitic amide packaged in WSSV virions significantly promoted virus infection
-
•
Palmitic amide was released to upregulate HIF-1, leading to the enhanced glycolysis
-
•
Palmitic amide directly bound to TPI and promoted its activity
-
•
The enhanced TPI activity can upregulate glycolysis and the expression of HIF-1
Human Metabolism; Molecular Biology; Virology
Introduction
Viruses are the most abundant life form on earth, inhabiting nearly every ecosystem, including animals, plants, bacteria, and environments. The entry of viruses into host cells is initiated by the recognition of receptors present on the surface of host cells (Sridharan et al., 2017). The receptors on the host cell membrane, the major mediators of virus tropism, interact with the viruses, leading to the occurrence of an apparently programmed series of molecular events to activate the receptors and the downstream pathways (Zhang et al., 2018). The binding of viral spikes to host cell surface receptors not only initiates viral adhesion and the engulfment process necessary for internalization but also can simultaneously initiate direct fusion with the cell membrane (Tsai et al., 2003). After interactions with host cell receptors, viruses enter host cells through either the endosomes or plasma membranes of host cells to undergo infection (Luo et al., 2018). Subsequently, both enveloped and nonenveloped viruses must deliver their genomes into host cells to initiate the virus life cycle in the host cells (Zhang et al., 2018). To move inside host cells, incoming viruses often exploit the cytoskeleton and cellular motor proteins of host cells (Hoffmann et al., 2017). The viruses allow endocytic vesicles to ferry them as passive lumenal cargo, or the penetrated capsid can itself interact with the relevant motors (Fernandez et al., 2019). Then, the viruses replicate within living cells and use the cellular machinery for the synthesis of their genomes and other components. However, it is not clear how the virus manipulates the host metabolic machinery to initiate virus infection in the host cytoplasm.
It is well known that all cells, whether differentiated or not, rely heavily on carbohydrate metabolism to create energy. Carbohydrate metabolism includes glycolysis in the cytoplasm and the tricarboxylic acid (TCA) cycle in mitochondria (Mizock, 1995; Vlashi et al., 2011). Glycolysis is a process whereby glucose is converted anaerobically into pyruvate by a series of intercellular enzymatic reactions to produce adenosine triphosphate (ATP), a high-energy phosphate compound (Noma, 1983; Schulze and Harris, 2012). In the presence of oxygen, cells yield two molecules of pyruvate, which are then converted to acetyl coenzyme A (acetyl CoA), triggering entry into the TCA cycle (Mizock, 1995; Hui et al., 2017). The TCA cycle is highly productive and efficient in terms of energy production (Noma, 1983; Hui et al., 2017). To ensure optimal environments for their replication and spread, viruses have evolved to alter the pathways of glycolysis, as well as glutaminolysis (Chambers et al., 2010). Most viruses examined to date induce aerobic glycolysis, also known as the Warburg effect (Chen et al., 2011). Many viruses can also induce fatty acid synthesis (Heaton and Randall, 2010). These modifications of carbon source utilization by viruses can increase the available energy for virus replication and virion production (Delgado et al., 2010). The expression of hexokinase 2, the first enzyme of glycolysis, is upregulated in dengue virus-infected cells (Fontaine et al., 2015). Pharmacologically, inhibiting the glycolytic pathway dramatically reduces the RNA synthesis of dengue virus and the production of infectious virions, showing the requirement for glycolysis during dengue virus infection (Fontaine et al., 2015). Rhinovirus-infected cells rapidly upregulate glucose uptake in a PI3K-dependent manner, revealing a critical role of glucose mobilization from extracellular and intracellular pools via glycogenolysis for viral replication (Gualdoni et al., 2018). Viruses, as parasitic organisms, completely rely on host metabolic machinery for their survival. However, how viruses trigger host machinery to initiate the virus life cycle in the cytoplasm is unclear.
It is believed that viruses initiate their cellular infection by delivering their genomes, which are packaged in virions, into host cells. Except for viral genomes, however, it remains unknown whether any other molecules can be packaged in virions to manipulate virus infection. To explore whether there were any molecules, such as compounds, to be packaged in virions to initiate virus infection in the host cells, white spot syndrome virus (WSSV) was characterized in this study. WSSV, the type species of Nimaviridae family and Whispovirus genus, contains a circular double-stranded genomic DNA and has a wide range of hosts including shrimp, crab, and crayfish (Zhang et al., 2001; Cui et al., 2015). The results of this study revealed that compounds could be packaged in WSSV virions. The further analysis indicated that palmitic amide packaged in virions promoted virus infection by manipulating glycolysis of host cells.
Results
Metabolites packaged in virions
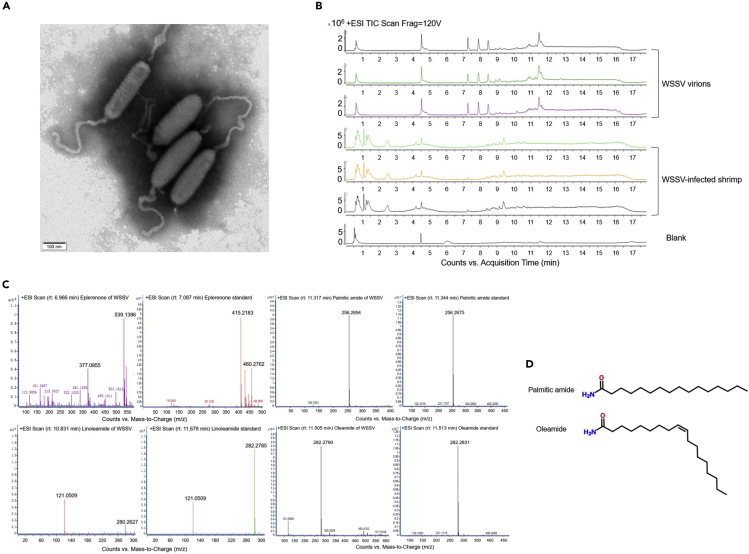
To explore the viral key to initiating host metabolic machinery, molecules packaged in virions, except for the viral genome, were characterized in this study. Virions of WSSV were purified from WSSV-challenged shrimp (Figure 1A). The liquid chromatography-coupled mass spectrometry (LC-MS) data of the metabolites extracted from the purified WSSV virions revealed a total of 1079 unique peaks, 9 of which were enriched in the WSSV virions compared with those in the control (Figure 1B). The nine peaks were identified as palmitic amide, sphinganine, linoleamide, oleamide, clusiacyclol A, myxochelin B, eplerenone, methionyl-leucylphenylalanine, and militarinone A. Therefore, the metabolites packaged in virions were further investigated.
Figure 1.
Metabolites packaged in virions
(A) WSSV virions purified from WSSV-infected shrimp. The purified virions were observed under a transmission electron microscope. Scale bar, 100 nm.
(B) LC-MS profiles of metabolites from the WSSV virions. The images were representatives of three biological repeats. The virus supernatant subjected to metabolite extraction and LC-MS analysis served as “blank”.
(C) Confirmation of compounds packaged in WSSV virions by LC-MS analysis.
(D) Structures of palmitic amide and oleamide.
At present, standards for the 4 compounds (palmitic amide, oleamide, linoleamide, and eplerenone) are available. To confirm the LC-MS data, therefore, the pure standards of palmitic amide, oleamide, linoleamide, and eplerenone were subjected to LC-MS analysis. The LC-MS peaks of palmitic amide and oleamide extracted from the WSSV virions matched those of their corresponding standards (Figure 1C), confirming the LC-MS data. However, the LC-MS peaks of linoleamide and eplerenone extracted from the WSSV virions did not match those of the standards (Figure 1C). These data showed that palmitic amide and oleamide (Figure 1D) were packaged in the WSSV virions.
Effects of the metabolites packaged in virions on virus infection
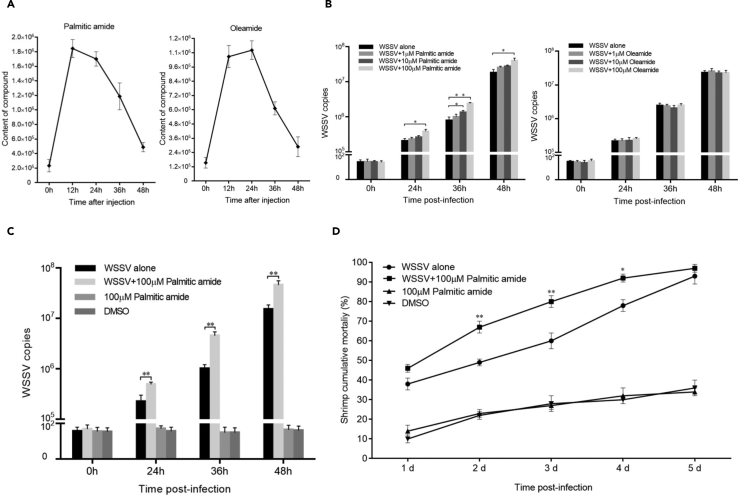
To reveal the effects of palmitic amide and oleamide packaged in virions on virus infection, the compounds were injected into WSSV-infected shrimp, followed by the detection of WSSV infection. The LC-MS results indicated that the content of palmitic amide or oleamide in the shrimp hemocytes reached the highest point at 12 and 24 hr after compound injection, and the level of compounds dropped significantly 48 hr later (Figure 2A), indicating that the injected compound could maintain a high level in shrimp for at least 24 hr. The quantitative real-time polymerase chain reaction (PCR) data revealed that WSSV copies were significantly increased in shrimp treated with palmitic amide in a dose-dependent manner (Figure 2B). The optimal concentration of palmitic amide was 100 μM. However, oleamide had no effect on virus replication (Figure 2B). These results indicated that the palmitic amide packaged in the WSSV virions played a positive role in virus infection.
Figure 2.
Effects of the metabolites packaged in virions on virus infection
(A) Content of compounds in shrimp hemocytes at different times after the injection of compounds. Palmitic amide and oleamide were injected into WSSV-infected shrimp. At different times after the compound injection, the content of compounds in the shrimp hemocytes was examined by LC-MS. The data represented the results of three independent assays.
(B) Influence of palmitic amide and oleamide on WSSV infection. The compounds at various concentrations were injected into the WSSV-infected shrimp. At different times post-infection, the shrimp were subjected to quantitative real-time PCR to quantify the WSSV copies in the shrimp hemocytes. The experiments were biologically repeated for three times (∗p < 0.05; ∗∗p < 0.01).
(C) Impact of constant existence of palmitic amide on WSSV content in shrimp. Shrimp were injected with palmitic amide and/or WSSV. WSSV alone was used as a positive control. Palmitic amide (100 μM) and DMSO (dimethyl sulfoxide) without WSSV were included in the injections as negative controls. For the treatment WSSV+100 μM palmitic amide, shrimp were injected three times to ensure the constant presence of palmitic amide. Firstly, shrimp were injected with 100 μM of palmitic amide. Twenty four hours later, the same shrimp were injected with WSSV and palmitic amide (100 μM). Finally, the shrimp were injected with palmitic amide (100 μM) at 24 hr after infection. At different times post-infection, the WSSV copies were quantified by quantitative real-time PCR (∗∗p < 0.01).
(D) Shrimp cumulative mortality analysis. The treatments were indicated on the top. The numbers on the horizontal axis represented the days post-infection. Data represented the mean ± standard deviation of triplicate assays (∗p < 0.05; ∗∗p < 0.01).
To explore the influence of the constant presence of palmitic amide in shrimp on virus infection, shrimp were injected with 100 μM palmitic amide first, and twenty four hours later, the same shrimp were injected with WSSV and palmitic amide (100 μM), followed by the injection of palmitic amide (100 μM) at 24 hr after WSSV infection. The results showed that the WSSV content in shrimp was much higher in shrimp treated with WSSV and palmitic amide than in shrimp treated with WSSV alone (Figure 2C). The shrimp mortality analysis yielded similar results (Figure 2D). However, the mortality of palmitic amide-treated shrimp was comparable to that of the negative controls (dimethyl sulfoxide [DMSO]) (Figure 2D), indicating that palmitic amide displayed no cytotoxicity to shrimp. These findings revealed that the metabolite palmitic amide packaged in virions could promote virus infection in vivo.
Activation of the host HIF-1 signaling pathway and glycolysis by palmitic amide
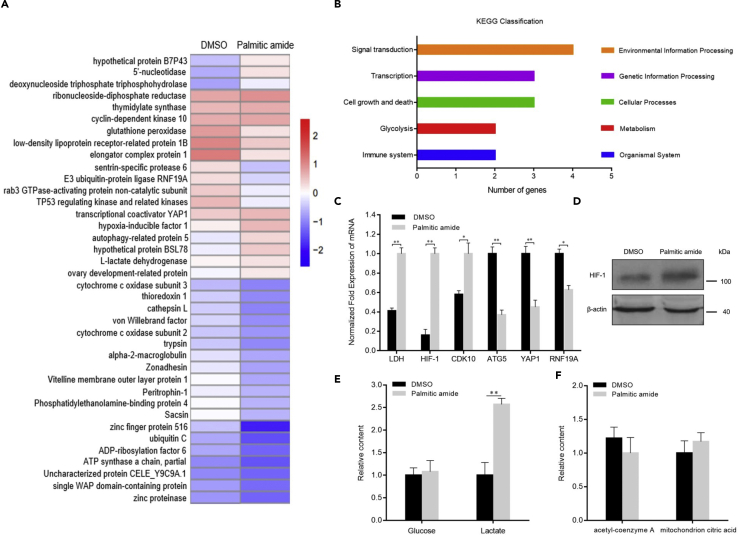
To explore the effects of palmitic amide packaged in virions on host signaling pathways during virus infection, shrimp were injected with palmitic amide, followed by transcriptome analysis of shrimp hemocytes. After removal of repetitive and low-quality reads, a total of 5.04×106 high-quality reads were aligned to the assembled expressed sequence tags of shrimp. In total, 33,323 unigenes were obtained. Cluster analysis revealed that the pattern of mRNA expression levels in shrimp hemocytes was changed in response to palmitic amide treatment. Twelve genes were significantly upregulated and 26 genes were significantly downregulated compared with those in the control (Figure 3A). The upregulated genes included the metabolic activator hypoxia-inducible factor 1 (HIF-1), L-lactate dehydrogenase (LDH), and growth stimulator cyclin-dependent kinase 10 (CDK10), while the downregulated genes contained many genes, such as transcriptional coactivator YAP1, E3 ubiquitin-protein ligase RNF19A, and autophagy-related protein 5 (ATG5). To reveal the functions of these differentially expressed genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted. The results of KEGG analysis revealed that the differentially expressed genes were involved in signal transduction, gene transcription, cell growth and death, glycolysis, and other pathways (Figure 3B). To confirm the transcriptome analysis data, quantitative real-time PCR was conducted. The results showed that the genes encoding LDH, HIF-1, and CDK10 were upregulated, while the genes encoding ATG5, YAP1, and RNF19A were downregulated (Figure 3C), which were consistent with those of transcriptome sequencing, confirming the transcriptome sequencing results. These data indicated that palmitic amide promoted virus proliferation by upregulating the expression of host metabolism-associated genes. As reported, glycolysis of host cells is enhanced by virus infection (Chen et al., 2011; Fontaine et al., 2015), while HIF-1 is a key transcription factor regulating the glycolysis pathway (Yeung et al., 2008). In this context, HIF-1 was further characterized.
Figure 3.
Activation of the host HIF-1 signaling pathway and glycolysis by palmitic amide
(A) Heatmap of differentially expressed genes of shrimp in response to palmitic amide challenge. Shrimp were injected with DMSO or palmitic amide. Twenty-four hours later, the mRNAs of shrimp hemocytes were subjected to sequencing.
(B) KEGG classification of the differentially expressed genes.
(C) Examination of the mRNA level of differentially expressed genes by quantitative real-time PCR. The error bars represent the means ± standard deviations of three independent experiments (∗p < 0.05; ∗∗p < 0.01).
(D) Influence of palmitic amide on the expression of HIF-1. Shrimp were injected with DMSO or palmitic amide. Twenty-four hours later, Western blot analysis was conducted to examine the expression level of HIF-1 in shrimp hemocytes. β-actin was used as a control.
(E) Impact of palmitic amide on glycolysis of shrimp. Shrimp were injected with palmitic amide. As a control, DMSO was included in the injection. At 24 hr after injection, the contents of glucose and lactate in the hemocytes of shrimp were examined. The error bars represented the means ± standard deviations of three independent experiments (∗∗p < 0.01).
(F) Role of palmitic amide in TCA cycle. Shrimp were injected with palmitic amide or DMSO. At 24 hr after injection, the contents of acetyl CoA and mitochondrial citric acid in the hemocytes of shrimp were examined.
To investigate whether palmitic amide promoted virus infection through the HIF-1 signaling pathway, shrimp were injected with palmitic amide (100 μM), and then, the expression level of HIF-1 protein in hemocytes was examined. Western blot results showed that palmitic amide significantly upregulated the HIF-1 protein (Figure 3D), indicating that palmitic amide promoted the expression of HIF-1.
To evaluate whether palmitic amide affected the glycolytic reaction of shrimp, shrimp were injected with palmitic amide, followed by the detection of the contents of glucose and lactate in the shrimp hemocytes. The results showed that the lactate content was significantly increased in the palmitic amide-treated shrimp hemocytes compared with that in the controls, but the glucose content was unchanged (Figure 3E), indicating that palmitic amide promoted glycolysis of shrimp. At the same time, examination of the content of acetyl CoA and mitochondrial citric acid showed that palmitic amide had no effect on the host's TCA cycle (Figure 3F). The collective data demonstrated that when the virions entered the hemocytes, palmitic amide packaged in the virions was released to upregulate HIF-1, leading to the enhancement of glycolysis, and thus promoting virus infection.
Role of HIF-1 in virus infection and glycolysis
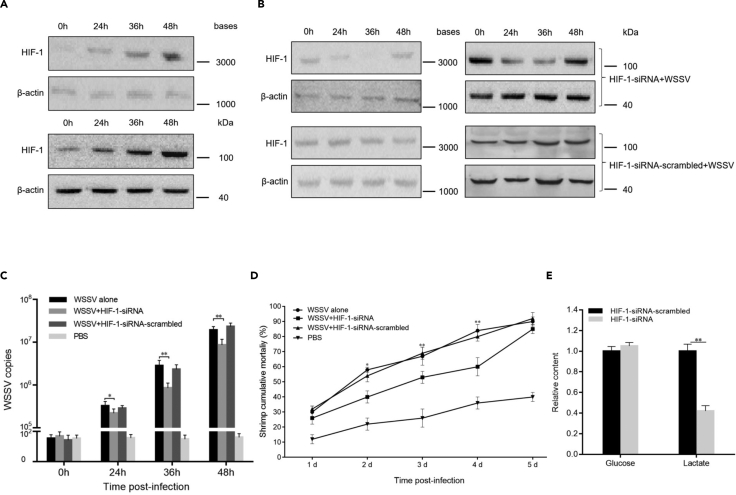
To evaluate the role of shrimp HIF-1 in virus infection, the expression level of HIF-1 was examined in shrimp in response to WSSV challenge. The results indicated that HIF-1 was significantly upregulated in virus-challenged shrimp (Figure 4A), suggesting that HIF-1 was involved in virus infection. To explore the impact of HIF-1 on virus infection, HIF-1 expression was knocked down by sequence-specific siRNA (HIF-1-siRNA) in shrimp (Figure 4B). The results revealed that HIF-1 silencing resulted in significant decreases in WSSV copies (Figure 4C). The mortality of WSSV-infected shrimp with HIF-1-siRNA treatment was also significantly decreased compared with that of the controls (Figure 4D). These data indicated that HIF-1 played a positive role in WSSV infection.
Figure 4.
Role of HIF-1 in virus infection and glycolysis
(A) Upregulation of HIF-1 in shrimp in response to virus infection. Shrimp were challenged with WSSV. At different times post-infection, the expression level of HIF-1 in shrimp hemocytes was examined by Northern blot (up) and Western blot (down). β-actin was used as a control.
(B) Knockdown of HIF-1 by sequence-specific siRNA (HIF-1-siRNA) in WSSV-infected shrimp. Shrimp were co-injected with HIF-1-siRNA and WSSV. As a control, HIF-1-siRNA-scrambled was included in the injection. At different times after injection, the HIF-1 mRNA level and protein level in the hemocytes of shrimp were examined by Northern blot (left) and Western blot (right) separately. β-actin was used as a control.
(C) Influence of HIF-1 silencing on WSSV infection in shrimp. The WSSV content was quantified using quantitative real-time PCR at different times post-infection. WSSV alone, PBS, and HIF-1-scrambled were used as controls (∗p < 0.05; ∗∗p < 0.01).
(D) Effects of HIF-1 silencing on the mortality of WSSV-infected shrimp. The numbers on the horizontal axis indicated the days post-infection (∗p < 0.05; ∗∗p < 0.01).
(E) Role of HIF-1 in glycolysis of shrimp. Shrimp were injected with HIF-1-siRNA. HIF-1-siRNA-scrambled was included in the injection as a control. Thirty-six hours later, the contents of glucose and lactate in the hemocytes of shrimp were examined (∗∗p < 0.01).
To explore whether HIF-1 affected the glycolytic reaction of shrimp, shrimp were injected with HIF-1-siRNA, and then, the contents of glucose and lactate in the hemocytes of shrimp were determined. The results showed that the lactate content was significantly decreased in HIF-1-siRNA-treated shrimp hemocytes compared with that in the controls, while the glucose content was not changed (Figure 4E), indicating that HIF-1 promoted glycolysis in shrimp.
Palmitic amide interacted with the TPI protein and increased the TPI activity
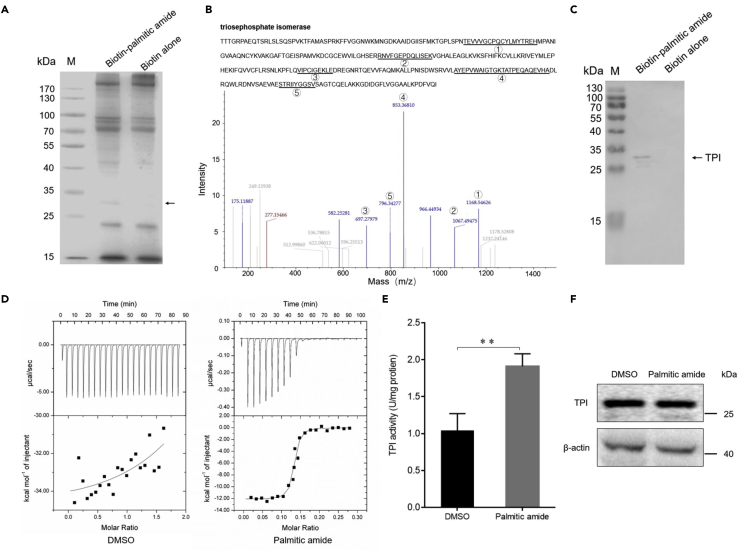
To reveal the role of palmitic amide in the initiation of virus infection in host cells, the host proteins bound to palmitic amide were characterized. The results showed that a protein specifically bound to palmitic amide (Figure 5A). The protein was identified as triosephosphate isomerase (TPI) by mass spectrometry (Figure 5B). Western blot analysis confirmed the mass spectrometric data (Figure 5C). To characterize the direct interaction between palmitic amide and TPI, isothermal titration calorimetry analysis was conducted. The results indicated that the dissociation constant (Kd) of the binding of palmitic amide to the recombinant TPI protein was 4.89 ± 1.23 μM, while the Kd of the binding of DMSO to TPI protein was 0 (Figure 5D). These data showed that palmitic amide was directly bound to the TPI protein.
Figure 5.
Palmitic amide interacted with the TPI protein and increased the TPI activity
(A) Proteins interacted with palmitic amide. Shrimp hemocytes were lysed and then incubated with biotin-labeled palmitic amide. The proteins were analyzed by SDS-PAGE with Coomassie blue staining. Dynabeads alone was used as a control. The arrow indicated the differential protein. M, protein marker.
(B) Identification of the protein bound to palmitic amide by mass spectrometry. The protein was identified to be triosephosphate isomerase. The matched peptides were indicated with underlines and numbers.
(C) Western blot analysis of the proteins interacted with palmitic amide. The arrow indicated the TPI. M, protein marker.
(D) Thermodynamic characterization of the interaction between palmitic amide and TPI protein. The purified recombinant TPI protein (100 μM) and incubated with palmitic amide (1 mM), followed by isothermal titration calorimetry (ITC) analysis. DMSO was used as a control.
(E) Impact of palmitic amide on TPI activity in shrimp. Shrimp were injected with palmitic amide (100 μM). Twenty-four hours later, the activity of TPI of hemocytes was examined. DMSO was used as a control. The error bars denoted the means ± standard deviations of three independent experiments (∗∗p < 0.01).
(F) Influence of palmitic amide on the expression of TPI in shrimp. Shrimp were injected with palmitic amide (100 μM) or DMSO. Twenty-four hours later, the expression level of TPI in the hemocytes of shrimp was examined by Western blot analysis. β-actin was used as a control.
To evaluate the effects of palmitic amide on TPI activity, shrimp were injected with palmitic amide, followed by the detection of TPI enzymatic activity of hemocytes. The results showed that palmitic amide treatment resulted in a significant increase in TPI activity compared with that in the control (Figure 5E), indicating that palmitic amide could enhance TPI activity. To assess whether palmitic amide affected the expression of TPI protein, the hemocytes of shrimp injected with palmitic amide were subjected to Western blot analysis. The results demonstrated that palmitic amide had no effect on the expression of TPI in shrimp compared with that in the controls (Figure 5F). These data indicated that the interaction between palmitic amide and TPI could increase TPI enzymatic activity but did not affect the expression level of TPI.
The influence of TPI on virus infection
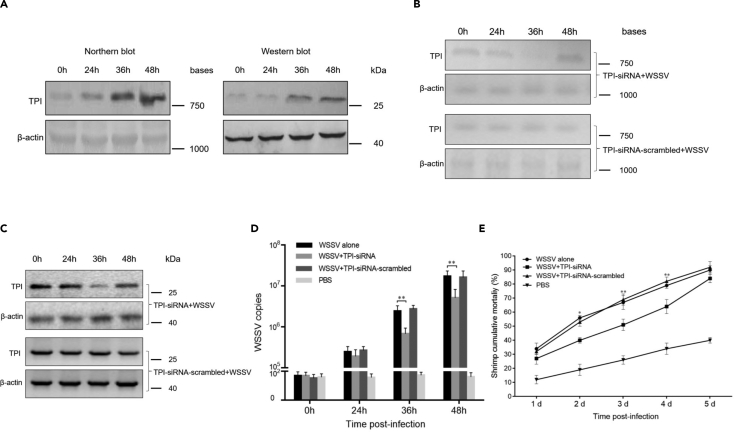
To explore the influence of TPI on virus infection, TPI expression in WSSV-infected shrimp was examined. The results indicated that TPI was upregulated in shrimp in response to virus challenge (Figure 6A). Western blot analysis yielded similar results (Figure 6A). These data demonstrated that TPI played an important role in virus infection. To reveal the role of TPI in virus infection, TPI was knocked down by sequence-specific TPI-siRNA in shrimp. Northern blot and Western blot data showed that TPI was silenced in WSSV-infected shrimp (Figures 6B and 6C). TPI silencing led to a significant decrease in WSSV content in shrimp (Figure 6D). However, TPI-siRNA scrambled had no effect on virus replication (Figure 6D). The mortality of WSSV-infected shrimp treated with TPI-siRNA was significantly decreased compared with that of the controls (Figure 6E). These data indicated that TPI played a positive role in WSSV infection.
Figure 6.
The influence of TPI on virus infection
(A) Upregulation of TPI in shrimp in response to virus infection. Shrimp were challenged with WSSV. At different times post-infection, the expression level of TPI in shrimp hemocytes was examined by Northern blot and Western blot. β-actin was used as a control.
(B) Knockdown of TPI by sequence-specific siRNA (TPI-siRNA) in WSSV-infected shrimp. Shrimp were co-injected with TPI-siRNA and WSSV. As a control, TPI-siRNA-scrambled was included in the injection. At different times after injection, the TPI mRNA level in the hemocytes of shrimp was examined by Northern blot. β-actin was used as a control.
(C) Western blot analysis of TPI protein in siRNA-treated shrimp. β-actin was used as a control.
(D) Influence of TPI silencing on WSSV infection in shrimp. The WSSV content was quantified using quantitative real-time PCR at different times post-infection. WSSV alone and PBS were used as controls. The error bars denoted the means ± standard deviations of three independent experiments (∗∗p < 0.01).
(E) Effects of TPI silencing on the mortality of WSSV-infected shrimp. The numbers on the horizontal axis indicated the post-infection days (∗p < 0.05; ∗∗p < 0.01).
Relationship between TPI, HIF-1, and glycolysis
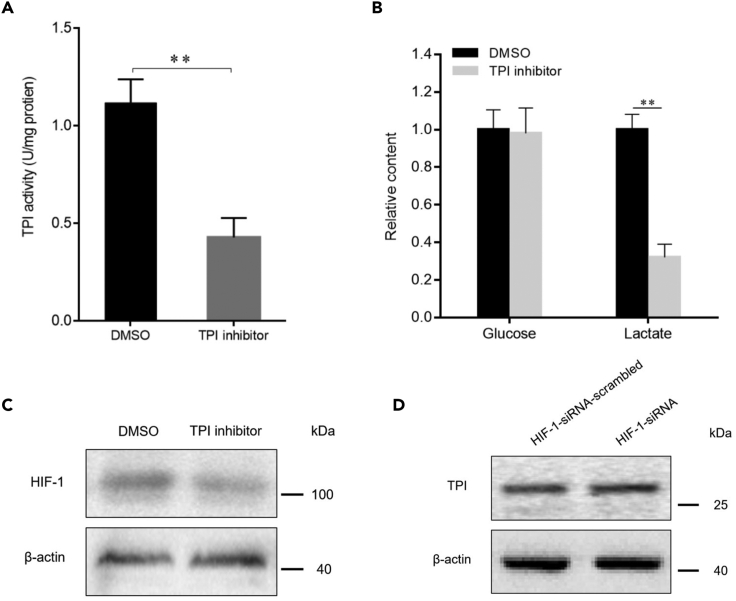
To investigate the relationship between TPI, HIF-1, and glycolysis, shrimp were treated with the TPI enzyme-specific inhibitor (Alvarez et al., 2014), followed by analyses of HIF-1 expression and glycolysis. The results showed that the TPI-specific inhibitor significantly suppressed the enzymatic activity of TPI (Figure 7A). The suppression of TPI activity resulted in a significant decrease in lactate content (Figure 7B), showing that glycolysis was inhibited by a TPI-specific inhibitor. Western blot analysis indicated that the inhibition of TPI enzymatic activity significantly decreased HIF-1 protein levels (Figure 7C). These data demonstrated that the enhanced TPI enzyme activity could upregulate the expression of HIF-1 and promote glycolysis.
Figure 7.
Relationship between TPI, HIF-1, and glycolysis
(A) Influence of TPI inhibitor on TPI activity in shrimp. Shrimp were injected with TPI inhibitor phenazine (100 μM). Twenty-four hours later, the enzymatic activity of TPI in the hemocytes of shrimp was examined. DMSO was used as a control. The error bars denoted the means ± standard deviations of three independent experiments (∗∗p < 0.01).
(B) Impact of the inhibition of TPI activity on glycolysis of shrimp. At 24 hr after the injection of TPI inhibitor, the contents of glucose and lactate in the hemocytes of shrimp were examined. The error bars represented the means ± standard deviations of three independent experiments (∗∗p < 0.01).
(C) Effects of the suppression of TPI activity on the HIF-1 expression in shrimp. At 24 hr after the injection of TPI inhibitor, Western blot analysis was conducted to examine the expression level of HIF-1 protein in shrimp hemocytes. β-actin was used as a control.
(D) Evaluation of the impact of HIF-1 silencing on the expression of TPI in shrimp. Shrimp were injected with HIF-1-siRNA. As a control, HIF-1-siRNA-scrambled was included in the injection. At 36 hr after injection, the expression level of TPI protein in the hemocytes of shrimp was analyzed by Western blot. β-actin was used as a control.
To evaluate the impact of HIF-1 on TPI expression, shrimp were injected with HIF-1-siRNA. Western blot results showed that the TPI protein level in the hemocytes of shrimp treated with HIF-1-siRNA did not change compared with that in the controls (treated with HIF-1-siRNA-scrambled) (Figure 7D), indicating that HIF-1 did not affect the expression of TPI protein.
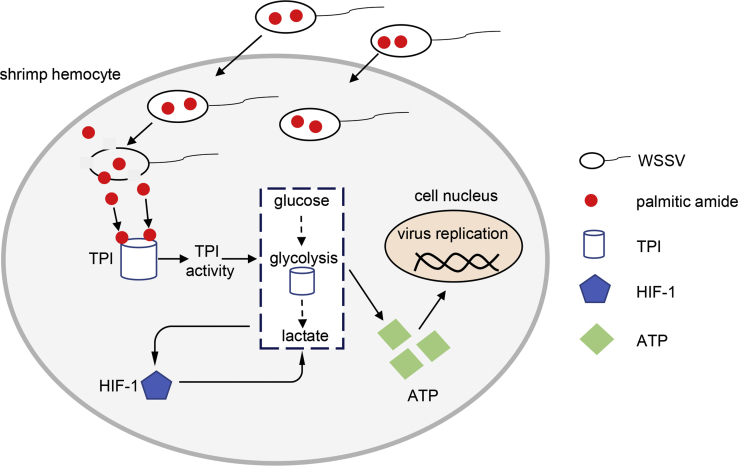
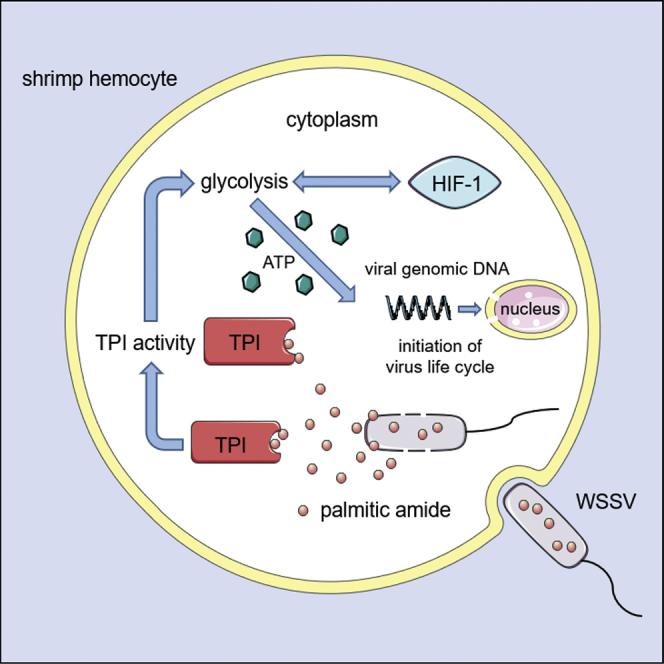
Mechanism of palmitic amide-triggered initiation of virus infection
Collectively, these findings revealed that when the virions invaded the host cells, the virions released packaged palmitic amide (Figure 8). Subsequently, palmitic amide directly bound to host TPI to enhance its enzymatic activity, thus enhancing host glycolysis in the cytoplasm (Figure 8). The enhancement of glycolysis led to the accumulation of lactate in the cytoplasm, which entered the host cells into a hypoxic state, thereby promoting the expression of HIF-1 (Figure 8). The upregulated HIF-1 further promoted glycolysis (Figure 8). Therefore, the enhancement of glycolysis promoted virus infection (Figure 8).
Figure 8.
Mechanism of palmitic amide-triggered initiation of virus infection
Schematic diagram of the underlying mechanism of palmitic amide packaged in virions during virus infection.
Discussion
Viruses initiate their life cycle and replicate within living cells by using the cellular machinery for the synthesis of their genomes and other components (Zhang et al., 2018). To gain access to host cells, viruses have evolved a variety of elegant mechanisms to utilize host energy. Following successful penetration into host cells, the virus particles need to reach an appropriate site in the cell for genome replication (Luo et al., 2018). The viral nucleocapsids are routed to the perinuclear area via microtubule-mediated transport. During this process, a dynein motor powers the movement of virus particles (Chou, 2007). For viruses that replicate in the nucleus, the viral genome needs to enter the nucleus via a nuclear pore (Smith and Helenius, 2004). Multiple distinct strategies are utilized, largely depending on the viral genome size (Schrad et al., 2020). Viral capsids of viruses with smaller genomes enter the nucleus, while for viruses with large genomes, the docking of nucleocapsids to a nuclear pore complex causes a partial disruption of the capsid or induces a minimal change in the viral capsid, allowing the transit of the DNA genome into the nucleus (Lakadamyali et al., 2003; Xie et al., 2019). At present, it is accepted that when a virus enters the cytoplasm of its host cell, the viral genome is released for gene expression and viral genome replication. However, there exists a gap between “after the virus enters the cytoplasm” and “before the virus initiates replication of the viral genome”. It remains unclear how the virus initiates its life cycle in the host cytoplasm. Our findings revealed that compounds packaged in virions, such as palmitic amide, could initiate the virus life cycle by enhancing host glycolysis to provide energy for virus proliferation in the host cytoplasm. In this context, our study demonstrated novel insights into the initiation of the virus life cycle in host cells.
Glycolysis and the TCA cycle represent the two main metabolic pathways used to support the energetic needs of cells, which are the basis for all cellular activities (Pavlova and Thompson, 2016; Sanchez et al., 2017). As reported, eukaryotic cells under normal growth conditions predominantly utilize glucose for oxidative phosphorylation in the mitochondria via the TCA cycle to generate the bulk of ATP (Janiszewska et al., 2012; Wang et al., 2019; Lebleu et al., 2014). In this study, however, the findings indicated that viruses initiate their life cycle by utilizing glycolysis but not the TCA cycle to support the energetic needs of virus proliferation in the cytoplasm of host cells. As well known, glycolysis is conducted in the cytoplasm, while the TCA cycle occurs in mitochondria. Therefore, it was easy for the compounds packaged in virions to utilize glycolysis in the cytoplasm without the entrance into mitochondria to trigger the TCA cycle. When the virus entered host cells, palmitic amide packaged in the virions was released to promote host glycolysis, thus initiating the virus life cycle. Although the production of lactic acid results in less ATP per molecule of glucose, it has been proposed that increased glycolysis and decreased oxidative phosphorylation may serve to increase the rate of ATP production without producing reactive oxygen species (Pinho and Reis, 2015; Courtnay et al., 2015), showing that energy is the first requirement for the initiation of virus infection in host cells. The large requirement of energy for virus restricted the energy supply for host cells, which might lead to host mortality. The compounds packaged in virions might be one of the important factors causing host mortality. Our study demonstrated that viruses could induce distinct alterations in host cellular metabolism to provide the energy and molecular building blocks needed for successful viral replication. In the initiation of the virus life cycle in host cells, the compounds packaged in virions played essential roles in manipulating the host's machinery for energy supply and could be the targets for therapy and diagnosis of virus-caused diseases. This issue merits to be explored in the future.
Limitations of the study
For different viruses, the compounds packaged in virions may be different from each other. However, our study was limited to WSSV. It is unclear whether compounds packaged by other viruses interfere with host metabolism and affect viral infection.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Xiaobo Zhang (zxb0812@zju.edu.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Original samples and data are available upon request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was financially supported by the National Key Research and Development Program of China (2018YFD0900504) and China Ocean Mineral Resources R & D Association (DY135-B-04).
Author contributions
X.Z. designed the experiments. S.Z. and X.F. conducted experiments and analyzed the data. S.Z. and X.Z. interpreted the results. S.Z. and X.Z. wrote the manuscript.
Declaration of interests
All authors declare no competing interests.
Published: January 22, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101915.
Supplemental information
References
- Alvarez G., Martinez J., Aguirrelopez B., Cabrera N., Perezdiaz L., de Gomez-Puyou M.T., GómezPuyou A., Pérez-Montfort R., Garat B., Merlino A. New chemotypes as Trypanosoma cruzi triosephosphate isomerase inhibitors: a deeper insight into the mechanism of inhibition. J. Enzyme Inhib. Med. Chem. 2014;29:198–204. doi: 10.3109/14756366.2013.765415. [DOI] [PubMed] [Google Scholar]
- Chambers J.W., Maguire T.G., Alwine J.C. Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 2010;84:1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.T., Aoki T., Huang Y.T., Hirono I., Chen T.C., Huang J.Y., Chang G.D., Lo C.F., Wang H.C. White Spot Syndrome Virus induces metabolic changes resembling the Warburg effect in shrimp hemocytes in the early stage of infection. J. Virol. 2011;85:12919–12928. doi: 10.1128/JVI.05385-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T. Stochastic entry of enveloped viruses: fusion versus endocytosis. Biophys. J. 2007;93:1116–1123. doi: 10.1529/biophysj.107.106708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Huang T., Zhang X. RNA editing of microRNA prevents RNA-induced silencing complex recognition of target mRNA. Open Biol. 2015;5:150126. doi: 10.1098/rsob.150126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Courtnay R., Ngo D.C., Malik N., Ververis K., Tortorella S.M., Karagiannis T.C. Cancer metabolism and the Warburg effect: the role of HIF-1 and PI3K. Mol. Biol. Rep. 2015;42:841–851. doi: 10.1007/s11033-015-3858-x. [DOI] [PubMed] [Google Scholar]
- Delgado T., Carroll P.A., Punjabi A.S., Margineantu D., Hockenbery D.M., Lagunoff M. Induction of the Warburg effect by Kaposi's sarcoma herpesvirus is required for the maintenance of latently infected endothelial cells. Proc. Natl. Acad. Sci. U S A. 2010;107:10696–10701. doi: 10.1073/pnas.1004882107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J., Machado A.K., Lyonnais S., Chamontin C., Gartner K., Leger T., Henriquet C., Garcia C., Portilho D.M., Pugnière M. Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat. Microbiol. 2019;4:1840–1850. doi: 10.1038/s41564-019-0575-6. [DOI] [PubMed] [Google Scholar]
- Fontaine K.A., Sanchez E.L., Camarda R., Lagunoff M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015;89:2358–2366. doi: 10.1128/JVI.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdoni G.A., Mayer K.A., Kapsch A., Kreuzberg K., Puck A., Kienzl P., Oberndorfer F., Frühwirth K., Winkler S., Blaas D. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc. Natl. Acad. Sci. U S A. 2018;115:158–165. doi: 10.1073/pnas.1800525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann H.H., Schneider W.M., Blomen V.A., Scull M.A., Hovnanian A., Brummelkamp T.R., Rice C.M. Diverse viruses require the calcium transporter SPCA1 for maturation and spread. Cell Host Microbe. 2017;22:460–470. doi: 10.1016/j.chom.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui S., Ghergurovich J.M., Morscher R.J., Jang C., Teng X., Lu W., Esparza L.A., Reya T., Zhan L., Guo J.Y. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiszewska M., Suva M.L., Riggi N., Houtkooper R.H., Auwerx J., Clement-Schatlo V., Radovanovic I., Rheinbay E., Provero P., Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Gene Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Yang W., Gao G. SUN1 regulates HIV-1 nuclear import in a manner dependent on the interaction between the viral capsid and cellular cyclophilin A. J. Virol. 2018;92 doi: 10.1128/JVI.00229-18. e00229–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakadamyali M., Rust M.J., Babcock H.P., Zhuang X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U S A. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebleu V.S., Oconnell J.T., Herrera K.N., Wikman H., Pantel K., Haigis M.C., de Carvalho F.M., Damascena A., Chinen L.T.D., Rocha R.M. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:992–1003. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizock B.A. Alterations in carbohydrate metabolism during stress: a review of the literature. Am. J. Med. 1995;98:75–84. doi: 10.1016/S0002-9343(99)80083-7. [DOI] [PubMed] [Google Scholar]
- Noma A. ATP-regulated K+ channels in cardiac muscle. Nature. 1983;305:147–148. doi: 10.1038/305147a0. [DOI] [PubMed] [Google Scholar]
- Pinho S.S., Reis C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan H., Ragan K.B., Guo H., Gilley R.P., Landsteiner V.J., Kaiser W.J., Upton J.W. Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 2017;18:1429–1441. doi: 10.15252/embr.201743947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez E.L., Pulliam T., Dimaio T.A., Thalhofer A.B., Delgado T., Lagunoff M. Glycolysis, glutaminolysis, and fatty acid synthesis are required for distinct stages of kaposi's sarcoma-associated herpesvirus lytic replication. J. Virol. 2017;91 doi: 10.1128/JVI.02237-16. e02237–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.E., Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Schrad J.R., Abrahão J.S., Cortines J.R., Parent K.N. Structural and proteomic characterization of the initiation of giant virus infection. Cell. 2020;181:1046–1061. doi: 10.1016/j.cell.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai B., Gilbert J.M., Stehle T., Lencer W.I., Benjamin T.L., Rapoport T.A. Gangliosides are receptors for murine polyoma virus and SV40. EMBO J. 2003;22:4346–4355. doi: 10.1093/emboj/cdg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E., Lagadec C., Vergnes L., Matsutani T., Masui K., Poulou M., Popescua R., Donnaa L.D., Eversa P., Dekmeziana C. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. U S A. 2011;108:16062–16067. doi: 10.1073/pnas.1106704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yip L.Y., Lee J.H., Wu Z., Chew H.Y., Chong P.K., Teo C.C., Ang H.Y.K., Peh K.L.E., Yuan J. Methionine is a metabolic dependency of tumor-initiating cells. Nat. Med. 2019;25:825–837. doi: 10.1038/s41591-019-0423-5. [DOI] [PubMed] [Google Scholar]
- Xie X., Zou J., Zhang X., Zhou Y., Routh A., Kang C., Popov V.L., Chen X., Wang Q.Y., Dong H., Shi P.Y. Dengue NS2A protein orchestrates virus assembly. Cell Host Microbe. 2019;26:606–622. doi: 10.1016/j.chom.2019.09.015. [DOI] [PubMed] [Google Scholar]
- Yeung S.J., Pan J., Lee M. Roles of p53, Myc and HIF-1 in regulating glycolysis - the seventh hallmark of cancer. Cell. Mol. Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Kim A.S., Fox J.M., Nair S., Basore K., Klimstra W.B., Rimkunas R., Fong R.H., Lin H., Poddar S. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature. 2018;557:570–574. doi: 10.1038/s41586-018-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xu X., Hew C.L. The structure and function of a gene encoding a basic peptide from prawn white spot syndrome virus. Virus Res. 2001;79:137–144. doi: 10.1016/s0168-1702(01)00340-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original samples and data are available upon request.