Abstract
Monocytes and macrophages provide defense against pathogens and danger signals. These cells respond to stimulation in a fast and stimulus-specific manner by utilizing complex cascaded activation by lineage-determining and signal-dependent transcription factors. The complexity of the functional response is determined by interactions between triggered transcription factors, and depends on the microenvironment and interdependent signaling cascades.
Dysregulation of macrophage phenotypes is a major driver in various diseases, such as atherosclerosis, rheumatoid arthritis and type II diabetes. Furthermore, exposure of macrophage precursor cells, monocytes, to certain stimuli can lead to a hypo-inflammatory tolerized phenotype or a hyper-inflammatory trained phenotype in a macrophage.
In atherosclerosis, macrophages and monocytes are exposed to inflammatory cytokines, oxidized lipids, cholesterol crystals and other factors. All these stimuli induce not only a specific transcriptional response, but also interact extensively, leading to a transcriptional and epigenetic heterogeneity of atherosclerotic plaque macrophages.
Targeting the epigenetic landscape of plaque macrophages can be a powerful tool to modulate pro-atherogenic phenotypes to reduce the rate of plaque formation. In this review we discuss the emerging role of transcription factors and epigenetic remodeling in the context of atherosclerosis and inflammation, and provide a comprehensive overview of epigenetic enzymes and transcription factors shaping macrophage activation.
Introduction
Monocytes are one of the first lines of defense of the body against pathogens or tissue damage. Circulating monocytes comprise three subsets: classical CD14++CD16− monocytes, non-classical CD14+CD16++ and intermediate CD14+CD16+ [1]. After being recruited to danger zones by chemotaxis, monocytes differentiate into macrophages. Together with specialized tissue-resident macrophages that maintain their pool without active contribution of blood monocytes, monocyte-derived macrophages participate in inflammatory process[2]. In response to specifically encountered pathogen- and damage-associated molecular patterns (PAMPs and DAMPs) and host derived signaling molecules, macrophages adopt a plethora of functional phenotypes [3]. The classical macrophage polarization model describes two opposing phenotypic states: a “classical” pro-inflammatory M1 macrophage that is induced by bacterial lipopolysaccharide (LPS) and/or interferon gamma (IFN-ɣ) and an “alternative” anti-inflammatory M2 macrophage that can be triggered by for instance interleukin 4 (IL4). However, recent progress in functional characterization reveals that macrophage phenotypes are not limited to the M1 and M2 extremes but rather represent a continuous spectrum of phenotypes associated with differential cytokine production and functional characteristics [4,5]. This macrophage functional plasticity is tightly regulated by transcriptional reprogramming, which is achieved by modifications of chromatin accessibility and the epigenetic landscape.
In disease, chronic inflammation can lead to aberrant remodeling of macrophage responses and cause a shift in their phenotypes, leading to e.g. an increase of pro-inflammatory macrophages in diabetes or anti-inflammatory macrophages in cancer [6]. In atherosclerosis macrophage dysregulation underlies disease pathogenesis [7]. Exposure of monocytes to i.a. oxidized lipids, cholesterol crystals and inflammatory mediators leads to formation of atypical macrophage activation states combining the characteristics of pro- and anti-inflammatory phenotypes [8].
These changes are associated with transcriptional and epigenetic reprogramming, and are modulated by transcription factors and epigenetic enzymes [9]. Understanding the underlying regulatory mechanisms can help to develop new therapies, by e.g. blocking an unwanted pathway or reprogramming macrophages to a more beneficial phenotype.
In this review we summarize recent efforts in the understanding of transcription factors and epigenetic mechanisms controlling macrophage activation in steady state and in disease with a particular focus on atherosclerosis. We discuss the cooperative action of transcription factors and epigenetic modifiers in the context of inflammation and atherosclerosis and suggest potential ways to ‘re-educate’ macrophages in disease.
Transcriptional regulation
Transcriptional and phenotypic diversity is regulated on multiple interconnected levels through i.a. transcription factor and co-factor binding, the epigenetic landscape, long-range interactions, DNA methylation, RNA editing, and long non-coding RNAs. This multilayered regulation ensures a robust execution of cellular phenotype and function, including macrophage differentiation and activation. Below, we highlight the main mechanisms involved in the transcriptional and epigenetic regulation of macrophage programs.
In the nucleus, DNA is wrapped around histone proteins. This physically hinders its accessibility for transcription factor (TF) binding and the transcriptional machinery [10]. Chromatin accessibility can be modulated by epigenetic modification of histone tails. In particular, the lysine residues of histones H3 and H4 can be chemically modified by e.g. addition of methyl or acetyl groups. For example, activation of regulatory elements is often associated with a gain in histone acetylation. This leads to a more open chromatin configuration and a general increase of TF accessibility and also creates binding sites for ‘reader’ proteins such as Brd4 that recruit additional transcriptional complexes. Di- and tri-methylation of H3K9 (H3K9me2/3), on the other hand, is associated with closed, compacted chromatin and repression of gene transcription through recruitment of proteins such as heterochromatin protein 1 (HP1). Chromatin modification profiling by means of ChIP-sequencing can aid in identification of regulatory elements and transcriptional activity in a genome-wide manner (Box 1). For example, tri-methylation of H3K4 (H3K4me3) is a hallmark of gene promoters and mono-methylation of H3K4 (H3K4me1) is generally associated with distal regulatory elements (enhancers), while acetylation of H3K27 (H3K27ac) near transcription start sites and on enhancers and dimethylation of H3K79 (H3K79me2) or trimethylation of H3K36 across the gene body is associated with actively transcribed genes (Figure 1; Box 2) [11].
Box 1: Common chromatin interrogation methods based on next generation sequencing.
| Method | Description |
|---|---|
| ChIP-seq | DNA fragments bound to a protein of interest, e.g. a histone tail modification or a transcription factor, are captured by immuno precipitation. After sequencing these short reads can be mapped to the genome in order to identify loci bound by the targeted protein. |
| RNA-seq | Total or messenger RNA is extracted from a population of cells, converted into short cDNA fragments, and sequenced. The number of short reads mapped to a gene is subsequently used as a relative measure of gene expression. |
| ATAC-seq | Open chromatin regions are fragmented by transposase and the resulting short DNA fragments are sequenced. A pile-up of mapped reads indicates the chromatin was accessible at these loci. |
Figure 1: Epigenetic landscape of regulatory DNA is set-up by transcription factors.
Chromatin is generally divided into transcription permissive euchromatin and compact inactive heterochromatin. Heterochromatin is often associated with repressive histone modifications such as H3K9me2/3 and H3K27me3. Enhancers and promoters are located in euchromatin and are associated with a distinct epigenetic mark-up (Box 2). Top left: active enhancers can be characterized by the presence of broad H3K4me1 and narrow H3K27ac marks and an ATAC-seq (open chromatin) signal. Top right: active promoters display narrow H3K4me3, H3K27Ac, and ATAC signal. Bottom: transcription factor (TF) binding sites are located within the nucleosome-free regions, characterized by ATAC-seq signal enrichment. In macrophages regulatory elements are associated with binding of lineage determining TFs PU1, CEBPB and AP-1 and can be further activated by binding of signal dependent TFs such as LPS-induced NF-κB or lipid-induced PPAR-ɣ. Activating TFs can recruit histone modifying enzymes (HMEs), such as histone acetyl transferases (HATs) to set up a permissive chromatin landscape. At promoters SDTFs do not regularly bind directly, but the open chromatin allows for the binding of RNA polymerase II, poising the gene for transcription.
Box 2: The Histone Code.
| Selection of histone marks and histone modifying enzymes (HMEs) associated with transcription: | ||||||
| Mark | Function | Location | Writer Family | Eraser Family | Writers | Erasers |
| H3K4me1 | Facultative euchromatin | Enhancers | KMT2 KMT3 KMT7 |
KDM1 KDM5 |
SETD1A MLL1–4 SMYD2 SETD7/9 |
LSD1/2 JMJD9 JARID1B |
| H3K4me3 | Facultative euchromatin | Promoters | KMT2 KMT8 |
KDM5 | SETD1A/B MLL1/2 PRDM9 |
JMJD9 JARID1A-D |
| H3K27ac | Active transcription | Promoters & Enhancers | KAT3 (HAT) | HDAC | EP300 CREBBP |
HDAC3 |
| H3K36me3 | Active transcription | Genebody | KMT3 | KDM4 | SETD2 | JMJD2A-E |
| H3K79me2 | Active transcription | Genebody | KMT4 | Unknown | DOT1L | Unknown |
| Selection of histone marks and HMEs associated with gene silencing: | ||||||
| Mark | Function | Location | Writer Family | Eraser Family | Writers | Erasers |
| H3K9me2 | Facultative heterochromatin | Gene deserts | KMT1 | KDM3 KDM4 |
G9a GLP-1 |
JMJD1A JMJD2D-E |
| H3K9me3 | Constitutive heterochromatin | Intergenic domains promoters enhancers | KMT1 | KDM3 KDM4 |
SUV39H1/2 SETDB1 |
JMJD1B JMJD2A-E |
| H3K27me3 | Facultative heterochromatin | Intergenic domains (bivalent) promoters enhancers | KMT6 | KDM6 | EZH1/2 | UTX UTY JMJD3 |
HME families:
KMT: Lysine methyl transferase
KDM: Lysine demethylase
KAT: Lysine acetyl transferase
HAT: Histone acetyl transferase
HDAC: Histone deacytelase
Histone modifications are written and erased by histone modifying enzymes (HMEs) (Box 2) [12]. HMEs are targeted to specific histones by interactions with TFs and the transcriptional machinery. TFs usually bind to specific DNA motifs that can be located at gene promoters and distally at enhancers. Most TFs require chromatin accessibility to be able to bind to the DNA, while a minority of TFs possess pioneering activity, i.e. are able to bind to their partially exposed motifs within closed chromatin, recruit chromatin remodeling complexes and modify the local chromatin structure leading to a more opened configuration allowing other factors to bind [13].
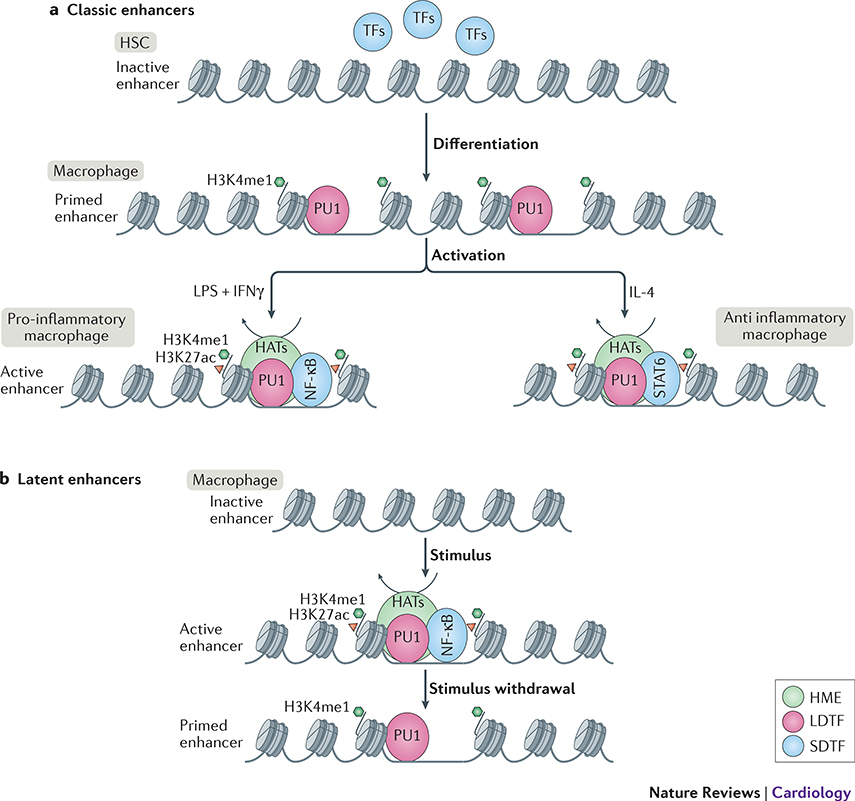
The macrophage epigenetic- and chromatin accessibility landscape is established by differential binding of so-called lineage-determining and signal-dependent transcription factors (LDTFs and SDTFs) [9]. LDTFs serve as master regulators of the cell specific epigenetic landscape. During cell type specification they have pioneering activity, i.e. are able to bind to closed chromatin, set-up a cell-specific regulatory landscape and drive cell-specific transcription programs. For example, combinatorial binding of LDTFs PU1, CEBPB and AP-1 defines macrophage identity, while binding and activity of PU1, E2A and Oct2 is associated with B-cell development [14]. In the case of tissue macrophages, the regulatory landscape can be further diversified by niche-specific LDTFs, such as Runx3 in intestinal macrophages and Mef2c in mouse microglia [15,16](Figure 2). Afterwards, SDTFs such as LPS-induced NF-κB or interferon-induced IRFs can activate a stimulus-specific regulatory program by activating this poised regulatory landscape [9,17]. Besides utilizing the pre-existing regulatory landscape, SDTFs are also able to expand the enhancer repertoire by activating epigenetically unmarked elements termed ‘latent enhancers’ [18]. Upon stimulus removal, these elements remain epigenetically marked and are thereby primed for rapid response to a subsequent activation, acting as cellular memory [18] (Figure2).
Figure 2: Macrophage enhancer selection. Classic enhancers:
are opened by lineage determining transcription factor (LDTF) PU1 during myeloid lineage differentiation to macrophages. Enhancers are primed by deposition of H3K4me1 by histone modifying enzymes (HMEs) from the lysine methyl transferase (KMT) families 2, 3 or 7 (Box 2). Upon activation, signal dependent transcription factor (SDTFs) such as NF-κB for pro-inflammatory M1-like activation or STAT6 for anti-inflammatory M2-like activation, are recruited to the primed enhancers. This in turn leads to recruitment of histone acetyl transferases (HATs) to set up a permissive chromatin landscape and activate the enhancer. Latent enhancers: are unbound by LDTFs such as PU1 in a steady-state macrophage. Upon stimulus, SDTFs such as NF-κB can bind to their partially uncovered motifs and open up the chromatin, leading to PU1, KMT, and HAT recruitment to fully open up the chromatin and activate the enhancer. After removal of the stimulus, the SDTF leaves the chromatin, and activating histone marks are removed by recruitment of histone de-acetylases (HDACs), however H3K4me1 remains unremoved and keeps enhancer in a primed state for subsequent activation. This enhancer memory enables a quicker activation upon the next encounter of the same stimulus.
Enhancers are able to exert their regulatory activity over a large linear distance. This can be achieved via long-range looping of the chromatin to create an interaction between the enhancer and its target promoter, bringing them close together in three-dimensional space. As long-range interactions are associated with specifically primed regulatory elements, human monocytes and macrophages can be readily distinguished based on their long-range interaction landscape [19]. Differentiation of the monocytic cell line THP-1 into macrophages leads to extensive changes in long-range interactions. The majority of interactions are gained in macrophages and these loops are associated with binding of the LDTF AP-1 [20]. Interestingly, while differentiation is associated with interaction landscape remodeling [20], activation of differentiated cells occurs within the pre-established landscape without significant interaction remodeling [21].
DNA methylation is a stable epigenetic mark often found at CpG islands at promoter and enhancer elements. Promoter and enhancer methylation are generally associated with gene repression that can occur through several mechanisms: repelling or recruitment of TFs, nucleosome repositioning, and recruitment of repressive methyl-binding proteins (MBPs) [22]. During differentiation DNA methylation changes ensure a robust silencing of unwanted transcriptional programs, for example lymphoid differentiation is associated with progressive silencing of myeloid lineage transcription factors and their binding sites, ensuring the silencing of this opposing program[23]. Monocyte to macrophage differentiation was also shown to associate with changes in DNA methylation [24–26]. Interestingly, subsequent activation of macrophages leads to very few methylation changes [24,26].
Non-coding RNAs, i.e. transcripts that are generally not translated into protein, play a large role in regulating gene expression on various levels, e.g. by enhancing or blocking transcription or translation, altering the splicing of mRNA, or recruitment of HMEs such as polycomb group complex members [27]. In macrophages, numerous micro RNAs have been shown to influence differentiation and execution of activation programs [28]. Also long non-coding RNAs (lncRNAs) play an important role in regulating macrophage activation. The lncRNA LeXis, for instance, was shown to be an important modulator of the LXR pathway, thus controlling sterol metabolism [29].
Thus, the functional identity and plasticity of macrophages is dynamically controlled by sequential binding of transcription factors, establishing and modifying the epigenetic landscape by recruiting epigenetic enzymes to alter chromatin accessibility, histone modifications, and DNA methylation.. During differentiation, LDTFs setup the ‘macrophage program’ by priming key macrophage related genes, enhancers, and non-coding RNAs. This part of the program is set up and not dynamic in a fully differentiated macrophage. Later, in response to a specific environmental stimulus, SDTFs can bind to a subset of the primed regulatory elements and latent enhancers to execute an activation state specific program, creating a stimulus specific macrophage phenotype. This part of the landscape is dynamic even in a fully differentiated macrophage. Exposure to different stimuli can further expand the enhancer repertoire and prime them for subsequent stimulation. This flexible, yet tightly coordinated regulation makes macrophage phenotype modification an interesting target for intervention in inflammation-related conditions.
Macrophage rewiring in disease
Inflammation is associated with multiple diseases, including diabetes, cancer and atherosclerosis [6]. Macrophages, as modulators of inflammation, play a major role in the development of these inflammation-related conditions and their activation is often affected. In disease, macrophages can shift towards a pro-inflammatory phenotype and lead to an enhanced non-resolving inflammation, as can be seen in atherosclerosis, obesity and diabetes, or to an anti-inflammatory phenotype leading to immune-suppression and a decreased defense such as seen in multiple cancers [6].
Furthermore, during sepsis prolonged or repetitive exposure of monocytes to endotoxins leads to hypo responsiveness of these cells, or “endotoxin tolerance”. In tolerized cells, expression of an endotoxin-induced inflammatory gene program and pro-inflammatory cytokine production is dampened by the first encounter with endotoxin. Moreover, anti-inflammatory cytokines are overexpressed via induction of the TLR4/TRIF/STAT3 pathway, leading to a weaker immune response after a second stimulation [30]. Endotoxin tolerance can in this way result in increased patient mortality after secondary infections by preventing proper mounting of an inflammatory response. Interestingly, in LPS-induced tolerance only some genes are shut-down after repeated stimulation (“tolerized”), while other genes remain active (“responsive”) [25,31]. This gene-specific regulation has been shown to be associated with changes in the chromatin landscape and differential transcription factor binding [25,31]. The non-responding “tolerized” genes fail to accumulate activating acetylation marks on their promoters resulting in a lack of transcriptional activation upon re-exposure [25,31]. This lack of activation was suggested to be caused by direct repression of pro-inflammatory TFs such as IRF and STAT2 and subsequent failure to activate their direct targets, or by repression via tolerance-inducing TFs such as HIF1a [25]. Interestingly, LPS-induced immunological tolerance was shown to be prevented by exposure of monocytes to I-BET, by blocking the initial LPS response, and reversed by β-glucan exposure (at ~60% of tolerized genes), providing a potential future therapeutic approach to combat sepsis [25].
Contrary to sepsis, which is associated with a tolerized, anti-inflammatory macrophage phenotype, autoimmune diseases such as rheumatoid arthritis and systemic lupus erythematosus are linked to a hyper inflammatory macrophage profile [32]. This can in part be explained by the immunological training phenomenon. In training, the primary stimulus will prime specific regulatory elements and open latent enhancers. On the second stimulus, this pre-setup regulatory landscape allows faster and increased transcriptional activation of the stimulus specific gene program, leading to an enhanced inflammatory response. This long-lasting reprogramming is associated with epigenetic and metabolic reprogramming [33,34]. Training can be achieved by exposure of monocytes to e.g. ox-LDL, Candida albicans β-glucan, or tuberculosis vaccine bacillus Calmette-Guerin (BCG) [33,35,36].
The induction of these two opposing programs is regulated via engagement of different pattern recognition receptors (PRRs). For example, engagement of NOD1, NOD2 and dectin-1 receptors leads to formation of trained response, while engagement of TLR receptors with high-dose PRR’s leads to immunological tolerance [37]. On the chromatin level, immunological training and tolerance have been shown to induce long-lasting epigenetic remodeling of different sets of regulatory elements [25,33]. This remodeling was associated with changes in activating histone marks such as H3K27 acetylation and H3K4 tri-methylation [25,33]. Some prominent differences included differential regulation of lipid metabolism and lysosome genes, which are up-regulated by training inducer b-glucan and repressed by tolerance-mediator LPS. This regulation was associated with b-glucan specific induction of EGR2 and MITF transcription factors. The EGR2 and MITF motifs were also strongly enriched in b-glucan specific regulatory elements associated with lipid metabolism genes, strongly suggesting their involvement[25]. Furthermore, transcription factor motif analysis in trained macrophage enhancer elements identified several TFs potentially involved in regulating this innate immune memory, including ATF1, ATF7, and CREB3 [33]. In vivo mouse studies revealed the role of ATF7 in the establishment of trained immunity via association with H3K9 dimethylase G9a, resulting in the repression of a subset of genes involved in the LPS- and β-glucan immune memory programs. Upon stimulation, ATF7 was released from the chromatin, resulting in long-lasting gene activation and providing protection against future re-infection [38].
Innate immune memory priming can not only occur at the monocyte level, but already at the level of myeloid precursor cells in the bone marrow [39]. For instance, hypercholesterolemia and western diet were shown to trigger myeloid progenitor reprogramming, resulting in a trained phenotype and enhanced macrophage responses [40,41]. Priming of bone marrow skews differentiation towards the monocytic lineage and is associated with up-regulation of several transcription regulators including the AP-1 family (FOS/JUN). This reprogramming was shown to be associated with chromatin remodeling as detected by ATAC-seq [40]. AP-1 TFs participate in the establishment of a lineage-specific enhancer landscape by interacting with LDTFs, and the recruitment of BAF chromatin remodeling complexes to these enhancers. Simultaneous disruption of three AP-1 TFs (FOS, FOSB, JUNB) was sufficient to decrease nucleosomal remodeling at enhancers as detected by ATAC-seq [42].
Epigenetics of atherosclerosis
Atherosclerosis is a chronic inflammatory disease of the arteries where cholesterol, lipids and cellular debris accumulation in the vessel wall over time can lead to plaque formation, rupture, and thrombosis; this in turn can lead to myocardial infarction or ischemic stroke and is therefore one of the leading causes of death in the western world [43,44]. Major cellular drivers behind atherosclerotic plaque formation are activated macrophages and foam cells [7](Figure 3).
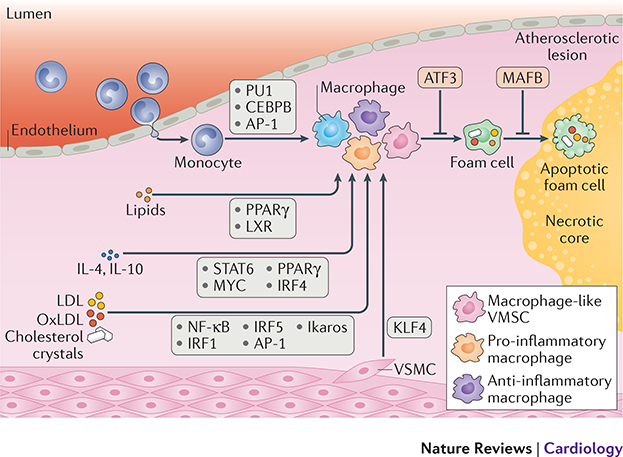
Figure 3: Transcription factors modulate macrophages in atherosclerosis.
Upon recruitment to an atherosclerotic plaque, circulating monocytes are differentiated into macrophages. Lineage determining transcription factors (LDTFs) such as PU1, CEBPB, and AP-1 mark macrophage specific regulatory elements. Smooth muscle cells migrating into the plaque can also differentiate into a macrophage like state by induction of KLF4 TF. Macrophages can subsequently be activated into a plethora of different phenotypes. Lipid accumulation leads to foam cell formation and upregulation of PPAR-ɣ and LXR. Stimulation with anti-inflammatory triggers such as IL4 and IL10 leads to expression of i.a. STAT6, MYC, PPAR-ɣ, and IRF4. Stimulation by pro-inflammatory triggers such as LPS, oxLDL, and cholesterol leads to activation of i.a. NF-κB, IRFs and IKAROS. Foam cell formation can be inhibited by expression of ATF3 while foam cell apoptosis is inhibited by MAFB.
In the atherosclerotic plaque microenvironment, macrophages are exposed to a wide variety of stimuli that impact on their activation state, e.g. oxidized lipids, cytokines and signaling molecules, hypoxia, and necrotic cells [5]. Transcription factors play a major role in setting up the epigenetic landscape of macrophages in atherosclerosis. Here, LDTFs such as CEBPB and PU1 instruct macrophage differentiation. SDTFs such as NF-κB, IRF, and STAT trigger macrophage activation following cytokine stimulation, while nuclear receptors such as PPAR and LXR activate lipid-associated transcriptional programs (Figure 3). A variety of triggers in atherosclerosis and an interplay between TF-induced transcriptional programs leads to functional heterogeneity of macrophages, as evidenced by recent single-cell RNA-seq and mass cytometry (CYTOF) analysis of atherosclerotic lesions in mouse [8,45]. Moreover, the balance of different types of macrophages in the plaque is spatially organized [46] and is considered a major factor determining stability of lesions and clinical consequences [7].
Transcription factors involved in cytokine signaling
Plaque macrophages can gain hyper-inflammatory tissue destroying characteristics [3]. These pro-inflammatory activation states can be induced by cytokines produced by macrophages, such as granulocyte-macrophage colony–stimulating factor (GM-CSF), or T-helper 1 (Th1) cells, such as interferon gamma (IFN-ɣ), or by bacterial products such as lipopolysaccharide (LPS) [47]. In this state, macrophages produce high levels of TNF, IL-6, IL- 1b, IL-12, IL-23, and CCL2, i.a., and propagate Th1 and Th17 responses [48]. On the opposite side of the spectrum, plaque macrophages can obtain anti-inflammatory pro-fibrotic characteristics involved in wound healing and tissue repair by Th2 and Treg cytokines like IL-4, IL10, IL-13, and M-CSF[49]. In this state, macrophages show an increased expression of scavenger and mannose receptors, IL-1 receptor agonist (IL-1RA), IL-10, Fizz1, Ym1, and arginase-1 and will further enhance the Th2 response [50]. Cytokine signaling leading to pro- or anti-inflammatory macrophage activation states is mainly relayed via the NF-κB and JAK/STAT pathways. The pro-inflammatory landscape is regulated by SDTF families such as NF-κB and IRFs, while the anti-inflammatory landscape is set up by transcription factors as STAT6 and HIF-2ɑ [51]. Moreover, LPS induced inflammatory macrophages fine-tune sustained NF-κB binding by co-binding of the pro-inflammatory TF IKAROS in a dose dependent manner [52].
Nuclear factor kappa B (NF-κB) is a family of SDTFs that mediate inflammatory responses. Under basal conditions NF-κB is located in the cytoplasm in an inactive form, but upon stimulation it translocates to the nucleus, and regulates its targets via binding to NF-κB response elements on the DNA [53]. The NF- κB signaling in macrophages has been recently reviewed by Dorrington and Fraser [54]. NF-κB binding to the DNA is dependent on the chromatin environment, and the majority of its binding occurs at pre-accessible chromatin, set-up by LDTFs PU1 and CEBP [14,55]. Upon binding, NF-κB recruits histone acetyltransferase P300, leading to histone acetylation and formation of active chromatin, permissive for transcription [55].
SDTFs belonging to the IRF family also play an important role in setting up macrophage activation programs. While some IRFs, e.g. IRF1, IRF5 and IRF8, are involved in pro-inflammatory activation of macrophages, other IRFs, e.g. IRF3 and IRF4, regulate anti-inflammatory macrophage activation [56]. The majority of Interferon regulatory factors (IRFs) occupy pre-accessible chromatin set-up by e.g. PU1 or indirectly via NF-κB, thus acting as a standard SDTF [57,58]. On the other hand, the inducible binding of Irf8 appears to be independent of a pre-set-up chromatin landscape and suggests a pioneering activity of this factor and an ability to function as an LDTF [57]. Indeed, genome-wide mapping of Irf8 binding sites showed a big overlap with stimulus-induced latent enhancers, thus further suggesting the ability of this TF to bind and mark previously inaccessible loci [57].
IL4-induced alternative macrophage activation is mainly associated with STAT6. This TF regulates the activation of anti-inflammatory program genes, including PPAR-ɣ [59]. Apart from inducing a subset of its target genes, STAT6 is also involved in gene repression through recruitment of HDAC complexes and a decrease in enhancer accessibility. Interestingly, in contrast with gene-activating STAT6 binding sites, STAT6 binding sites associated with a repressive function lack the canonical STAT6 motif suggesting that STAT6 uses either indirect binding or binding to non-canonical motifs to exert its repressive function [60].
Transcription factors involved in lipid signaling and foam cell formation
In an atherosclerotic plaque environment, macrophages can be primed by (oxidized) lipid particles, inducing a ‘foamy’ activation state. Uptake of lipid particles turns macrophages into overloaded foam cells [61] which display an anti-inflammatory phenotype but also acquire a unique transcriptional profile via activation of the LXR-RXR and PPAR-ɣ TF pathways, leading to upregulation of lipid handling proteins such as the ATP-binding cassette (ABC) lipid transporter family [62,63].
Foam cells and other (lipid) activated macrophages occupy both pro- and anti-inflammatory niches in the plaque microenvironment [64]. On the one hand, lipid loading induces anti-inflammatory TFs such as LXR and PPAR-ɣ while on the other hand, lipids can also be recognized by toll-like receptors (TLR), leading to activation of a pro-inflammatory response via the NOD-, LRR- and pyrin domain-containing 3 (NLRP3) pathway [65].
An additional layer of complexity is introduced by a crosstalk between lipid-induced and inflammation-induced TFs in inflammatory signaling. LXR and PPAR-ɣ can inhibit parts of the TLR-induced inflammatory program [66]. Activation of the LXR and PPAR-ɣ pathways inhibits the NF-κB pathway by binding of LXR to co-repressors like nuclear receptor corepressor (N-CoR) and silencing mediator for retinoid and thyroid hormone receptors (SMRT), and to NF-κB target genes by co-binding NF-κB itself [67,68]. Furthermore, IL4-induced STAT6 was shown to induce PPAR-ɣ, facilitate its transcriptional program [59] and expose de novo binding sites for PPAR:RXR heterodimers [69], thus suggesting a complex interaction between TFs during alternative macrophage activation. Interestingly, repeated IL4 stimulation resulted in an activation of additional genes by the PPAR:RXR heterodimer in a ligand-insensitive manner [70].
Several additional macrophage TFs were demonstrated to play an important role in foam cell formation and maintenance, thus suggesting a potential role in atherosclerosis development. For example, loss of transcription factor ATF3 induces foam cell formation and aggravates atherosclerosis in vivo [71]. ATF3 was identified as a regulator of lipid droplet formation in macrophages by repressing the transcription of the Ch25h gene, encoding cholesterol 25-hydroxylase, and thereby repressing 25-hydroxycholesterol formation. Loss of ATF3 resulted in derepression of Ch25h and an increased formation of 25-hydroxycholesterol, which is an LXR ligand, leading to induction of a cellular lipid phenotype and foam cell formation [71]. In agreement with the athero-protective role, ATF3 is induced by HDL and anti-inflammatory HDL action is dependent on ATF3 dependent pro-inflammatory cytokine suppression [72]. Intriguingly, another recent study demonstrated the involvement of TF KLF4 in regulation of the Ch25h gene. Here, KLF4-dependent activation of Ch25h led to LXR activation, repression of the inflammasome and a shift from pro- to anti-inflammatory activation, thus having an atheroprotective effect [73].
The transcription factor MafB is expressed specifically in the myeloid lineage and is associated with macrophage differentiation. In early atherosclerotic lesions MafB inhibits foam cell apoptosis, thereby promoting plaque development [74]. In advanced plaques MafB depletion causes larger necrotic cores and lower collagen content, potentially destabilizing the plaque [75]. Thus, MafB worsens the lesion in the early stages of plaque development yet stabilizes the plaque in the advance stages of the lesion.
Foam cells are not always macrophage derived
While the classic paradigm of atherosclerotic lesion formation emphasizes foam cell formation from monocyte derived macrophages, a sizable fraction of foam cells is in fact derived from intimal smooth muscle cells (SMCs). As reviewed by Dubland et al. [76], SMCs can under influence of platelet derived growth factor ß (PDGF-ß), lose their contractile phenotype and turn into a more synthetic phenotype, which produces extracellular matrix, and has a restorative, wound healing function that repairs and stabilizes the artery wall [77–79], or in the case of atheroslcerotic lesions, thickens and stabilizes the fibrous cap. As the lesion progresses however, synthetic SMCs are among the first cell types to retain lipoproteins [80]. Further exposure to intra plaque signals such as TGF-ß, oxidized lipids, and cytokines can drive the synthetic SMCs to transdifferentiate into foam cells [81–83]. Interestingly, cholesterol itself was shown to induce transdifferentiation of mouse SMCs and an increase in expression of foam cell markers CD68, Mac-2 and ABCA1[84]. Some reports suggest that as high as 50% of all foam cells in human lesions are of SMC origin [85]. However, this quantification relies on co-staining with SMC- and foam cell specific markers, which does not consider the full phenotype and direction of transdifferentiation and may lead to an overestimation of transdifferentiating cells. However, mouse-based lineage tracing experiments can aid characterization of these in vivo transitions and underlying epigenetic mechanisms. For example, in this way the Kruppel Like Factor 4 (KLF4) TF program was identified as a driver of SMC transdifferentiation[86]. The myeloid LDTF Sp1 (PU1) was shown to bind to the KLF4 promoter in response to PDGF-ß signaling [87]. Like STAT6, KLF4 induces an anti-inflammatory macrophage state via induction of MCP-1 induced protein (encoded by the ZC3H12A gene), which represses NF-κB function and activates the C/EBPb and PPAR-ɣ programs [88]. Although SMC derived foam cells gain macrophage markers such as CD68 and Lgals3, and accumulate cholesterol and lipoproteins, they do not gain phagocytic or efferocytic capabilities, and do therefore not turn into bona fide macrophages [89]. However, simultaneous loss of SMC markers makes it difficult to separate these SMC derived foam cells from their monocyte derived counterparts [90,91]. In monocyte derived macrophages, activation of the KLF4 program leads to an atheroprotective, anti-inflammatory phenotype [92,93]. Epigenetically, expression of KLF4 is regulated by methylation of its promoter by DNMT1. In inflammatory macrophages KLF4 is hypermethylated, leading to suppression of its TF program [94]. While in macrophages KLF4 has an atheroprotective role, KLF4 in SMCs appears to be pro-atherogenic and its loss leads to a decrease in lesion size and an increase in plaque stability. Indeed, in cultured SMCs cholesterol-induced expression of KLF4 led to KLF4-dependent activation of pro-inflammatory cytokines, thus promoting a pro-inflammatory, atherogenic plaque environment [86].
Epigenetic enzymes impacting on plaque macrophages
Transcription factors can modulate macrophage activation via epigenetic remodeling of the chromatin landscape and vice versa. The induction of pro-inflammatory macrophage phenotypes after for instance NF-κB activation is primed by IFN dependent deposition of H3K4me3 at promoters of NF-κB target genes such as IL-6 and TNF [95]. The induction of an anti-inflammatory macrophage phenotype, on the contrary, can be dampened by expression of HDAC3, a histone deacetylase that deactivates key factors needed for IL-4 dependent anti-inflammatory activation [96]. Indeed, macrophage specific KO of Hdac3 in mouse promotes plaque stability and steers the cells into a more wound-healing, fibrotic, anti-inflammatory phenotype [97]. In vivo activity of HDAC3 relies on co-association with nuclear receptor co-repressor complexes N-CoR and SMRT [98]. Thus, similarly to HDAC3, myeloid N-CoR knock-out also leads to an anti-inflammatory macrophage phenotype [99]. Deletion of HDAC9 attenuates atherosclerosis and, similarly to HDAC3, leads to an inhibition of the inflammatory profile in macrophages and an induction of lipid-handling genes and anti-inflammatory activation via upregulation of ABCA1, ABCG1, and PPAR-ɣ [100].
Alternative macrophage activation is also regulated by H3K27me3 via the Jmjd3-Irf4 axis [101]. The repressive histone mark H3K27me3 is deposited by the polycomb repressive complex (PRC) 2 methylase Ezh2 and can be removed by H3K27me3 demethylases KDM6a and KDM6b. KDM6b was shown to be dispensable for establishment of the pro-inflammatory macrophage phenotype in mice, but it was required for a proper anti-inflammatory response via the removal of H3K27me3 from the IRF4 promoter [101]. Indeed, KDM6B expression was shown control a fibrotic, wound healing phenotype in peritoneal macrophages from LDLR KO mice on a high fat diet [102]. Surprisingly, macrophage-specific deletion of Kdm6b resulted in advanced atherosclerosis with an increased collagen content and lesion necrotic core [103]. KDM6B itself was shown to be upregulated by STAT6, induced by IL-4 signaling. KDM6B dependent IRF4 transcription eventually, among other effects, leads to expression of CCL17, an anti-inflammatory macrophage cytokine [104]. Interestingly, KDM6B also modulates IRF4 expression and subsequent CCL17 production after simulation with G-MCSF [105], which normally acts via STAT5 and IRF5 mediated induction of an inflammatory phenotype [106,107].
A recent study by Zhang and colleagues demonstrated that Ezh2 is also involved in modulation of the macrophage phenotype. Ezh2 was shown to impact on H3K27me3 deposition on the promoter of cytokine signaling repressor SOCS3. Depletion of Ezh2 in mouse macrophages led to upregulation of SOCS3, resulting in a decreased activation of Myd88-dependent NF-κB activation and a diminished pro-inflammatory gene expression [108]. Consequently, mice with myeloid Ezh2 deficiency displayed attenuated macrophage activation and reduced development of colitis and experimental autoimmune encephalomyelitis [108], making Ezh2 an attractive target for other inflammatory diseases like atherosclerosis as well.
Somatic mutations in epigenetic enzymes contribute to CVD
Clonal hematopoiesis of indeterminate potential (CHIP) has recently emerged as one of the strongest independent predictors of cardiovascular disease. In CHIP, somatic mutations in hematopoietic precursors lead to clonal expansion of mutation carrying cells in the peripheral blood. Interestingly, these mutations are often found in genes encoding epigenetic enzymes, e.g. DNMT3a and TET2 [109], a DNA methyltransferase and demethylase respectively. Indeed, in vivo studies confirm that TET2 loss aggravates atherosclerosis at least in part through altering the macrophage inflammatory response [109]. While in steady state conditions TET2-deficient macrophages resembled their wild type counterparts, their response to LPS/IFN-ɣ stimulation was associated with an exacerbated upregulation of pro-inflammatory cytokines, including IL1b and IL6 [110]. Unexpectedly, TET2 mediated repression of Il1b appeared to be independent of its demethylase activity. Overexpression of a catalytically inactive TET2 mutant still led to a 90% reduction of Il1b. A similar level of Il1b repression was observed with wild-type TET2, suggesting a methylation-independent mechanism [110]. However, further research is needed to elucidate the mechanism behind TET2-mediated regulation. Interestingly, a recent report identified SIRT1 as an upstream regulator of TET2 and a potential therapeutic target for TET2 mediated myelodysplastic syndrome (MDS). Here, SIRT1 activates TET2 via direct deacetylation, and restores its activity in Tet2 deficient MDS [111]. In line with a potential role of clonal hematopoiesis in cardiovascular disease, SIRT1 is suggested to play a protective role in atherosclerosis [112], which warrants future research of this enzyme in a cardiovascular setting. CRISPR-mediated deletion of DNMT3A led to an increase in inflammatory response upon LPS stimulation with a particular increase in Il6, Ccl5, Cxcl1 and Cxcl2 [113] in the J774 mouse macrophage line. However in mouse peritoneal macrophages, DNMT3A deletion was associated with down-regulation of IFN-α and IFN-β, but pro-inflammatory cytokines TNF and IL6 were unaffected [114], potentially due to the differences in experimental models.
Interplay between signals acting on (opposing) gene programs
In atherosclerotic plaques, macrophages are exposed to a variety of signals, e.g. oxidized lipids, or pro- and anti-inflammatory cytokines, resulting in their complex phenotypes [5]. Recent single-cell RNA-seq analyses of mouse atherosclerotic lesions identified a plethora of activation states associated with plaque macrophages, including pro-inflammatory, resident-like and novel Trem2-high macrophage subsets [8,45,63,115]. Importantly, the identified pro-inflammatory and resident-like anti-inflammatory macrophage subsets did not resemble the classical M1- and M2-polarized macrophages and expressed both pro- and anti-inflammatory macrophage markers simultaneously, highlighting the complexity of inflammatory signaling in an atherosclerotic plaque in vivo [8].
Indeed, several recent studies suggested an extensive epigenetic and transcriptional cross-talk between pro- and anti-inflammatory signaling in macrophages [116,117]. For example, in vitro co-stimulation of bone-marrow derived macrophages with the pro-inflammatory IFN-ɣ and anti-inflammatory IL4 were shown to inhibit parts of opposing programs. IFN-responsive enhancers containing the AP-1 motif were inhibited by IL4, via IL4-dependent inhibition of AP-1 TF family members Cebpb and Junb. On the other hand, IFN-responsive enhancers with minimal IFN response element were not affected by IL4 stimulation. This complex logic allows for a robust antiviral response, while preserving stimulus-dependent plasticity [116]. Furthermore, pre-stimulation of bone-marrow derived macrophages with IL4, followed by pro-inflammatory LPS also resulted in a complex crosstalk between the two signaling pathways with some genes being inhibited to subsequent activation and some genes insensitive to the inhibitory effects. Intriguingly, a small group of genes showed an elevated response to LPS in the pre-treated macrophages, suggesting a cooperative effect of the opposing signaling programs [60]. This inhibitory effect of IL4 pre-treatment was mediated by STAT6 binding and accompanied by a decreased chromatin accessibility, a decrease in transcriptional coactivator P300, and an increase in corepressor HDAC occupancy [60]. Interestingly, the majority of IL4 responsive enhancers were sensitive to the inhibitory effects of IFN, suggesting dominance of the IFN program. The insensitive enhancers were associated with binding of the TF Myc [116]. IFN-ɣ stimulation was also shown to stably repress a subset of anti-inflammatory genes by recruitment of EZH2 and subsequent deposition of the repressive mark H3K27me3 [118], which might explain the dominance of the IFN-induced pro-inflammatory over the anti-inflammatory program [116]. Indeed, targets of the Ezh2-mediated H3K27me3 deposition included IL4-activated PPAR-ɣ, and key glucocorticoid receptor target MERTK [118]. In addition, IFN-ɣ represses anti-inflammatory gene expression program by disassembling, i.e. repressing binding of TFs and chromatin accessibility, at enhancers bound by the anti-inflammatory macrophage TF MAF [119].
Another example of extensive molecular cross-talk between seemingly opposing programs was demonstrated by wound healing enhancer repression by transcription factor Rev-Erb. While some TFs are specifically associated with pro- or anti-inflammatory programs, the Rev-Erb nuclear receptor family was shown to be involved in repression of wound healing regulatory elements irrespective of stimulation type. Accordingly, depletion of Rev-Erb in mouse macrophages led to an enhanced wound-healing capacity. Mechanistically, Rev-Erb was co-occupying NF-κB and AP-1 bound enhancers upon TLR stimulation, and Smad3 enhancers upon TGF-β treatment. In this way, Rev-Erb can integrate these seemingly opposing signals to inhibit wound repair. Exposure of macrophages to the supernatant of skin homogenate, representing a complex skin wound environment, confirmed co-activation and extensive molecular cross-talk between the TLR and TGF-β programs [120].
Apart from mutual suppression between pro- and anti-inflammatory programs, different pro-inflammatory signals also result in complex interactions between TFs [121]. For example, a recent study investigated the molecular mechanism underlying cross-tolerance, i.e. pre-treatment with one inflammatory cytokine, e.g. TNF to decrease the response to a secondary stimulation with another inflammatory mediator, e.g. LPS. TNF stimulation led to tolerance of a subset of pro-inflammatory genes. This TNF induced tolerance was prevented by pre-exposure of macrophages to IFN-ɣ. Pre-exposure of macrophages to IFN primed chromatin for the TNF response, presumably via cooperative action of IRF1 and NF-κB. Interestingly, partial reversal of tolerance by IFN resembled the monocyte profile from SLE patients, where IFN is suggested to play a key role in disease pathogenesis [121].
Thus, extensive molecular crosstalk exists between various inflammatory programs, including those previously considered to be opposing. This crosstalk results in changes in the molecular and functional output of macrophages and should be carefully considered when characterizing these cells in a disease with complex signaling triggers, such as atherosclerosis.
Conclusions
Atherosclerosis is an inflammation driven disease and macrophages play a central role in its modulation. Due to the need to respond in a rapid and specific way these cells adopted a unique permissive epigenetic landscape, established and controlled by sequential binding of LDTFs and SDTFs. These TFs modify the chromatin landscape during differentiation and exposure to stimuli, but also prime elements for future exposure, creating a cellular memory. This allows the cells to mount a faster response upon reactivation. While this strategy allows macrophages to efficiently respond to pathogenic stimuli, this system is also prone to dysregulation. For instance, low-grade systemic inflammation, often associated with the normal ageing process, can lead to changes in macrophage activation states driven by cytokine induced remodeling of the epigenetic landscape of cells.
The flexible yet tight control by transcription factors and epigenetic modifiers make intervention in these pathways in macrophages a promising target for inflammation-driven diseases. In atherosclerosis recent efforts have been directed at understanding the molecular mechanisms driving macrophage activation, with a particular focus on epigenetic enzymes [122]. Indeed, macrophage-specific genetic ablation or pharmacological inhibition of HMEs holds promise for inflammatory disease. However, due to the broad action of epigenetic enzymes, a targeted therapy would be beneficial and possibly even required. Recruiting an HME to the chromatin by modulating specific transcription factors might constrain the broad effects of HME inhibition. Thus, investigating HME recruitment mechanisms and target selection is of particular importance.
In order to develop potential therapeutic strategies to modulate macrophage phenotypes by interfering with HMEs and TFs, we need a better understanding of the molecular mechanisms driving macrophage differentiation and activation. In particular, characterizing the epigenetic landscape can help to pinpoint TFs and upstream signaling pathways regulating cell-type specific gene expression programs. Furthermore, an atherosclerotic plaque in vivo represents a complex environment where different types of signaling cascades act on macrophages in combination. Therefore, studying the effect of signaling crosstalk on gene expression programs and chromatin dynamics can help us understand the in vivo situation better. Additionally, recent efforts towards downscaling cell numbers for chromatin profiling make it feasible to characterize the genome-wide epigenetic landscape of various macrophage populations in an atherosclerotic plaque directly [123].
Genetic loss-of-function phenotypic screens have proven to be a valuable tool for discovering novel regulators of the inflammatory response. Considering macrophage heterogeneity in response to activating stimuli, a combination of loss-of-function screens with single cell read out can aid our understanding of macrophage biology. Several recent studies demonstrated a feasibility of this approach to define key transcription factors in primary immune cells [124,125].
In summary, dysregulation of the tight control over diverse macrophage activation states via transcription factors and epigenetic modulation plays a major role in the pathogenesis of inflammatory diseases such as atherosclerosis and remains a promising avenue of future research.
Key points.
Atherosclerotic plaques represent a complex environment with different activation signals, resulting in intra-plaque macrophage heterogeneity
Plasticity of macrophage phenotype is modulated by an interplay between transcription factors and epigenetic enzymes
Signaling pathways in inflammation have an extensive molecular crosstalk
Modulating transcription factor activity is a promising target for atherosclerosis
Acknowledgements
Our work is supported by The Netherlands Heart Foundation [GENIUS: CVON 2011/B019, GENIUS2: CVON 2017-20 to MW]; The Netherlands Heart Foundation and Spark-Holding BV [2015B002 to MW]; the European Union [ITN-grant EPIMAC to MW]; NIH grants [DK063491, DK091183, HL088093 to CKG] and Fondation Leducq [LEAN Transatlantic Network Grant to MW and CKG].
Glossary
- TF
Transcription Factor. Protein with DNA binding domain that guides the transcriptional machinery to or blocks it from regulatory elements such as promoters and enhancers, thereby facilitating the execution of tailored transcriptional programs.
- LDTF
Lineage Determining TF. Transcription factor with pioneering (i.e. chromatin conformation changing) capabilities that sets up the chromatin for execution of cell type specific gene programs.
- SDTF
Signal Dependent TF. Transcription factor which binds DNA and activates its gene program in reaction to an external stimulus.
- HM
Histone Mark. Chemical group covalently bound to a specific amino acid residue on (the tail of) a histone protein. HMs can change the local chromatin accessibility through electrostatic effects and/or guide the transcription machinery to specific loci in the genome (See Box 1).
- HME
Histone Modifying Enzyme: Reader, writer, or eraser of chemical histone (tail) modifications.
- Heterochromatin
repressive chromatin state. Chromatin in a closed conformation inaccessible for transcription.
- Euchromatin
permissive chromatin state. Chromatin in an open conformation accessible to the transcription machinery.
- Promoter
Regulatory DNA region proximal to a transcription start site. Contains consensus motifs for TFs and the transcriptional machinery.
- Enhancer
Regulatory DNA region distal to a transcription start site. Contains TF binding sites which are able to regulate transcription of target genes, independent of linear distance.
- Long-range interactions
interactions between regulatory DNA occurring over large linear distances. Occur predominantly between enhancers, and between enhancers and promoters. Can have a regulatory role.
- ATAC-seq
assay for transposase accessible chromatin using sequencing. A method for genome-wide identification of open chromatin regions with potential regulatory function.
Footnotes
Competing interests
None
Contributor Information
Tatyana Kuznetsova, Department of Medical Biochemistry, Amsterdam University Medical Centers, location Academic Medical Center, University of Amsterdam.
Koen H.M. Prange, Department of Medical Biochemistry, Amsterdam University Medical Centers, location Academic Medical Center, University of Amsterdam.
Christopher K. Glass, Department of Cellular and Molecular Medicine, Department of Medicine UC San Diego
Menno P.J. De Winther, Department of Medical Biochemistry, Amsterdam University Medical Centers, location Academic Medical Center, University of Amsterdam, Netherlands; Institute for Cardiovascular Prevention (IPEK), Ludwig-Maximilians University, Munich, Germany.
References
- 1.Woollard KJ, Geissmann F. Monocytes in atherosclerosis: subsets and functions. Nat Rev Cardiol. Nature Publishing Group; 2010. February;7(2):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S, Plüddemann A. Tissue macrophages: heterogeneity and functions. BMC Biology; 2017. June 22;:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ginhoux F, Schultze JL, Murray PJ, Ochando J, Biswas SK. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat Immunol. Nature Publishing Group; 2016. January;17(1):34–40. [DOI] [PubMed] [Google Scholar]
- 4.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, Ganesan H, Nino-Castro A, Mallmann MR, Labzin L, Theis H, Kraut M, Beyer M, Latz E, Freeman TC, Ulas T, Schultze JL. Transcriptome-Based Network Analysis Reveals a Spectrum Model of Human Macrophage Activation. Immunity. Elsevier Inc; 2014. February 20;40(2):274–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colin S, Chinetti-Gbaguidi G, Staels B. Macrophage phenotypes in atherosclerosis. Immunol Rev. John Wiley & Sons, Ltd (10.1111); 2014. November;262(1):153–66. [DOI] [PubMed] [Google Scholar]
- 6.Schultze JL, Schmieder A, Goerdt S. Macrophage activation in human diseases. Semin Immunol. 2015. August;27(4):249–56. [DOI] [PubMed] [Google Scholar]
- 7.Moore KJ, Sheedy FJ, Fisher EA. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol. Nature Publishing Group; 2013. October;13(10):709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochain C, Vafadarnejad E, Arampatzi P, Jaroslav P, Winkels H, Ley K, Wolf D, Saliba A-E, Zernecke A. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res. 2018. March 15;:CIRCRESAHA.117312509–484. [DOI] [PubMed] [Google Scholar]
- 9.Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016. January;17(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Talbert PB, Henikoff S. Histone variants — ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010. March 3;11(4):264–75. [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T Chromatin Modifications and Their Function. Cell. 2007. February;128(4):693–705. [DOI] [PubMed] [Google Scholar]
- 12.Hoeksema MA, de Winther MPJ. Epigenetic Regulation of Monocyte and Macrophage Function. Antioxidants & Redox Signaling. 2016. November 10;25(14):758–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Current Opinion in Genetics & Development. 2016. April;37:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010. May 28;38(4):576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-Resident Macrophage Enhancer Landscapes Are Shaped by the Local Microenvironment. Cell. Elsevier Inc; 2014. December 4;159(6):1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, Stender JD, Chun HB, Garner H, Geissmann F, Glass CK. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014. December 4;159(6):1327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt SV, Krebs W, Ulas T, Xue J, Baßler K, Günther P, Hardt A-L, Schultze H, Sander J, Klee K, Theis H, Kraut M, Beyer M, Schultze JL. The transcriptional regulator network of human inflammatory macrophages is defined by open chromatin. Cell Res. Nature Publishing Group; 2016. February;26(2):151–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013. January 17;152(1–2):157–71. [DOI] [PubMed] [Google Scholar]
- 19.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, Cairns J, Wingett SW, Várnai C, Thiecke MJ, Burden F, Farrow S, Cutler AJ, Rehnström K, Downes K, Grassi L, Kostadima M, Freire-Pritchett P, Wang F, BLUEPRINT Consortium, Stunnenberg HG, Todd JA, Zerbino DR, Stegle O, Ouwehand WH, Frontini M, Wallace C, Spivakov M, Fraser P. Lineage-Specific Genome Architecture Links Enhancers and Non-coding Disease Variants to Target Gene Promoters. Cell. 2016. November 17;167(5):1369–1384.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phanstiel DH, Van Bortle K, Spacek D, Hess GT, Shamim MS, Machol I, Love MI, Aiden EL, Bassik MC, Snyder MP. Static and Dynamic DNA Loops form AP-1-Bound Activation Hubs during Macrophage Development. Molecular Cell. Elsevier Inc; 2017. September 21;67(6):1037–1048.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, Yen C-A, Schmitt AD, Espinoza CA, Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. Nature Publishing Group; 2013. November 14;503(7475):290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schübeler D Function and information content of DNA methylation. Nature. Nature Publishing Group; 2015. January 15;517(7534):321–6. [DOI] [PubMed] [Google Scholar]
- 23.Bock C, Beerman I, Lien W-H, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Molecular Cell. 2012. August 24;47(4):633–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vento-Tormo R, Company C, Rodríguez-Ubreva J, la Rica de L, Urquiza JM, Javierre BM, Sabarinathan R, Luque A, Esteller M, Aran JM, Álvarez-Errico D, Ballestar E. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. BioMed Central; 2016. January 13;17(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novakovic B, Habibi E, Wang S-Y, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen-Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JHA, Logie C, Stunnenberg HG. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016. November 17;167(5):1354–1368.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dekkers KF, Neele AE, Jukema JW, Heijmans BT, de Winther MPJ. Human monocyte-to-macrophage differentiation involves highly localized gain and loss of DNA methylation at transcription factor binding sites. Epigenetics & Chromatin. BioMed Central; 2019. June 6;12(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, RIKEN Genome Exploration Research Group, Genome Science Group (Genome Network Project Core Group), FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. American Association for the Advancement of Science; 2005. September 2;309(5740):1564–6. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Jiang T, Li M-Q, Zheng X-L, Zhao G-J. Transcriptional Regulation of Macrophages Polarization by MicroRNAs. Front Immunol. 2018. May 28;9:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.PhD ZZ, MS DS, PhD TSM. Long Noncoding RNAs in Atherosclerosis. Journal of the American College of Cardiology. Elsevier; 2018. November 6;72(19):2380–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009. October;30(10):475–87. [DOI] [PubMed] [Google Scholar]
- 31.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. Nature Publishing Group; 2007. June 21;447(7147):972–8. [DOI] [PubMed] [Google Scholar]
- 32.Arts RJW, Joosten LAB, Netea MG. The Potential Role of Trained Immunity in Autoimmune and Autoinflammatory Disorders. Front Immunol. Frontiers; 2018;9:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeed S, Quintin J, Kerstens HHD, Rao NA, Aghajanirefah A, Matarese F, Cheng S-C, Ratter J, Berentsen K, van der Ent MA, Sharifi N, Janssen-Megens EM, Huurne Ter M, Mandoli A, van Schaik T, Ng A, Burden F, Downes K, Frontini M, Kumar V, Giamarellos-Bourboulis EJ, Ouwehand WH, van der Meer JWM, Joosten LAB, Wijmenga C, Martens JHA, Xavier RJ, Logie C, Netea MG, Stunnenberg HG. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. American Association for the Advancement of Science; 2014. September 26;345(6204):1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JHA, Rao NA, Aghajanirefah A, Manjeri GR, Li Y, Ifrim DC, Arts RJW, van der Veer BMJW, Deen PMT, Logie C, O’Neill LA, Willems P, van de Veerdonk FL, van der Meer JWM, Ng A, Joosten LAB, Wijmenga C, Stunnenberg HG, Xavier RJ, Netea MG. mTOR- and HIF-1 -mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014. September 25;345(6204):1250684–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bekkering S, Quintin J, Joosten LAB, van der Meer JWM, Netea MG, Riksen NP. Oxidized low-density lipoprotein induces long-term proinflammatory cytokine production and foam cell formation via epigenetic reprogramming of monocytes. Arteriosclerosis, Thrombosis, and Vascular Biology. American Heart Association, Inc; 2014. August;34(8):1731–8. [DOI] [PubMed] [Google Scholar]
- 36.Arts RJW, Moorlag SJCFM, Novakovic B, Li Y, Wang S-Y, Oosting M, Kumar V, Xavier RJ, Wijmenga C, Joosten LAB, Reusken CBEM, Benn CS, Aaby P, Koopmans MP, Stunnenberg HG, van Crevel R, Netea MG. BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe. 2018. January 10;23(1):89–100.e5. [DOI] [PubMed] [Google Scholar]
- 37.Ifrim DC, Quintin J, Joosten LAB, Jacobs C, Jansen T, Jacobs L, Gow NAR, Williams DL, van der Meer JWM, Netea MG. Trained immunity or tolerance: opposing functional programs induced in human monocytes after engagement of various pattern recognition receptors. Papasian CJ, editor. Clin Vaccine Immunol. American Society for Microbiology; 2014. April;21(4):534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida K, Maekawa T, Zhu Y, Renard-Guillet C, Chatton B, Inoue K, Uchiyama T, Ishibashi K-I, Yamada T, Ohno N, Shirahige K, Okada-Hatakeyama M, Ishii S. The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immunological memory. Nature Publishing Group. Nature Publishing Group; 2015. October;16(10):1034–43. [DOI] [PubMed] [Google Scholar]
- 39.Mitroulis I, Ruppova K, Wang B, Chen L-S, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, Chatzigeorgiou A, Schlitzer A, Beyer M, Joosten LAB, Isermann B, Lesche M, Petzold A, Simons K, Henry I, Dahl A, Schultze JL, Ben Wielockx, Zamboni N, Mirtschink P, Coskun Ü, Hajishengallis G, Netea MG, Chavakis T. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity . Cell. Elsevier Inc; 2018. January 11;172(1–2):147–155.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Christ A, Günther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, Scholz CJ, Oosting M, Haendler K, Baßler K, Klee K, Schulte-Schrepping J, Ulas T, Moorlag SJCFM, Kumar V, Park MH, Joosten LAB, Groh LA, Riksen NP, Espevik T, Schlitzer A, Li Y, Fitzgerald ML, Netea MG, Schultze JL, Latz E. Western Diet Triggers NLRP3-Dependent Innate Immune Reprogramming. Cell. Elsevier Inc; 2018. January 11;172(1–2):162–168.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seijkens T, Hoeksema MA, Beckers L, Smeets E, Meiler S, Levels J, Tjwa M, de Winther MPJ, Lutgens E. Hypercholesterolemia-induced priming of hematopoietic stem and progenitor cells aggravates atherosclerosis. FASEB J. Federation of American Societies for Experimental Biology Bethesda, MD, USA; 2014. May;28(5):2202–13. [DOI] [PubMed] [Google Scholar]
- 42.Vierbuchen T, Ling E, Cowley CJ, Couch CH, Wang X, Harmin DA, Roberts CWM, Greenberg ME. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Molecular Cell. 2017. December 21;68(6):1067–1082.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ Res. 2016. February 19;118(4):535–46. [DOI] [PubMed] [Google Scholar]
- 44.GBD 2016 Causes of Death Collaborators. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017. September 16;390(10100):1151–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh H, Kobiyama K, Hamers A, Cochain C, Vafadarnejad E, Saliba A-E, Zernecke A, Pramod A, Ghosh A, Anto Michel N, Hoppe N, Hilgendorf I, Zirlik A, Hedrick C, Ley K, Wolf D. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ Res. American Heart Association, Inc; 2018. March 15;:CIRCRESAHA.117312513–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stöger JL, Gijbels MJJ, van der Velden S, Manca M, van der Loos CM, Biessen EAL, Daemen MJAP, Lutgens E, de Winther MPJ. Distribution of macrophage polarization markers in human atherosclerosis. ATH. Elsevier Ltd; 2012. December 1;225(2):461–8. [DOI] [PubMed] [Google Scholar]
- 47.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008. January 1;13:453–61. [DOI] [PubMed] [Google Scholar]
- 48.Benoit M, Desnues B, Mege JL. Macrophage Polarization in Bacterial Infections. J Immunol. 2008. September 3;181(6):3733–9. [DOI] [PubMed] [Google Scholar]
- 49.Gordon S, Martinez FO. Alternative Activation of Macrophages: Mechanism and Functions. Immunity. Elsevier Inc; 2010. May 28;32(5):593–604. [DOI] [PubMed] [Google Scholar]
- 50.Müller U, Stenzel W, Köhler G, Werner C, Polte T, Hansen G, Schütze N, Straubinger RK, Blessing M, McKenzie ANJ, Brombacher F, Alber G. IL-13 Induces Disease-Promoting Type 2 Cytokines, Alternatively Activated Macrophages and Allergic Inflammation during Pulmonary Infection of Mice with Cryptococcus neoformans. J Immunol. 2007. October 2;179(8):5367–77. [DOI] [PubMed] [Google Scholar]
- 51.Tugal D, Liao X, Jain MK. Transcriptional Control of Macrophage Polarization. Arteriosclerosis, Thrombosis, and Vascular Biology. 2013. June;33(6):1135–44. [DOI] [PubMed] [Google Scholar]
- 52.Dual Roles for Ikaros in Regulation of Macrophage Chromatin State and Inflammatory Gene Expression. 2018. June 16;:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Nature Publishing Group. The Author(s); 2017. July 14;2:1–9. [Google Scholar]
- 54.Dorrington MG, Fraser IDC. NF-κB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front Immunol. Frontiers; 2019;10:705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei C-L, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010. March 26;32(3):317–28. [DOI] [PubMed] [Google Scholar]
- 56.Günthner R, Anders H-J. Interferon-regulatory factors determine macrophage phenotype polarization. Mediators of Inflammation. Hindawi; 2013;2013(11):731023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mancino A, Termanini A, Barozzi I, Ghisletti S, Ostuni R, Prosperini E, Ozato K, Natoli G. A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes Dev Cold Spring Harbor Lab; 2015. February 15;29(4):394–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saliba DG, Heger A, Eames HL, Oikonomopoulos S, Teixeira A, Blazek K, Androulidaki A, Wong D, Goh FG, Weiss M, Byrne A, Pasparakis M, Ragoussis J, Udalova IA. IRF5:RelA Interaction Targets Inflammatory Genes in Macrophages. Cell Rep. The Authors; 2014. September 11;8(5):1308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szanto A, Balint BL, Nagy ZS, Barta E, Dezso B, Pap A, Szeles L, Poliska S, Oros M, Evans RM, Barak Y, Schwabe J, Nagy L. STAT6 Transcription Factor Is a Facilitator of the Nuclear Receptor PPAR&gamma-Regulated Gene Expression in Macrophages and Dendritic Cells. Immunity Elsevier Inc; 2010. November 24;33(5):699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta-Monroy I, Simandi Z, Steiner L, Nagy B Jr, Poliska S, Banko C, Bacso Z, Schulman IG, Sauer S, Deleuze J-F, Allen JE, Benko S, Nagy L. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity Elsevier Inc; 2018. January 16;48(1):75–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chistiakov DA, Melnichenko AA, Myasoedova VA, Grechko AV, Orekhov AN. Mechanisms of foam cell formation in atherosclerosis. J Mol Med. Journal of Molecular Medicine; 2017. October 26;95(11):1–13. [DOI] [PubMed] [Google Scholar]
- 62.Tangirala RK, Bischoff ED, Joseph SB, Wagner BL, Walczak R, Laffitte BA, Daige CL, Thomas D, Heyman RA, Mangelsdorf DJ, Wang X, Lusis AJ, Tontonoz P, Schulman IG. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc Natl Acad Sci USA. National Academy of Sciences; 2002. September 3;99(18):11896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim K, Shim D, Lee JS, Zaitsev K, Williams JW, Kim K-W, Jang M-Y, Seok Jang H, Yun TJ, Lee SH, Yoon WK, Prat A, Seidah NG, Choi J, Lee S-P, Yoon S-H, Nam JW, Seong JK, Oh GT, Randolph GJ, Artyomov MN, Cheong C, Choi J-H. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ Res. Lippincott Williams & Wilkins Hagerstown, MD; 2018. October 26;123(10):1127–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wolfs IMJ, Donners MMPC, de Winther MPJ. Differentiation factors and cytokines in the atherosclerotic plaque micro-environment as a trigger for macrophage polarisation. Thromb Haemost. Schattauer Publishers; 2011. November;106(5):763–71. [DOI] [PubMed] [Google Scholar]
- 65.Tall AR, Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol. Nature Publishing Group; 2015. February 1;15(2):104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005. September 9;122(5):707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghisletti S, Huang W, Ogawa S, Pascual G, Lin M-E, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Molecular Cell. 2007. January 12;25(1):57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes & Development. 2009. March 15;23(6):681–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Daniel B, Nagy G, Horvath A, Czimmerer Z, Cuaranta-Monroy I, Poliska S, Hays TT, Sauer S, Francois-Deleuze J, Nagy L. The IL-4/STAT6/PPARγ signaling axis is driving the expansion of the RXR heterodimer cistrome, providing complex ligand responsiveness in macrophages. Nucleic Acids Res. 2018. May 18;46(9):4425–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Cuaranta-Monroy I, Poliska S, Tzerpos P, Kolostyak Z, Hays TT, Patsalos A, Houtman R, Sauer S, Francois-Deleuze J, Rastinejad F, Balint BL, Sweeney HL, Nagy L. The Nuclear Receptor PPAR&gamma Controls Progressive Macrophage Polarization as a Ligand-Insensitive Epigenomic Ratchet of Transcriptional Memory. Immunity. Elsevier Inc; 2018. October 16;49(4):615–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gold ES, Ramsey SA, Sartain MJ, Selinummi J, Podolsky I, Rodriguez DJ, Moritz RL, Aderem A. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J Exp Med. Rockefeller University Press; 2012. April 9;209(4):807–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Nardo D, Labzin LI, Kono H, Seki R, Schmidt SV, Beyer M, Xu D, Zimmer S, Lahrmann C, Schildberg FA, Vogelhuber J, Kraut M, Ulas T, Kerksiek A, Krebs W, Bode N, Grebe A, Fitzgerald ML, Hernandez NJ, Williams BRG, Knolle P, Kneilling M, Röcken M, Lütjohann D, Wright SD, Schultze JL, Latz E. High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nature Publishing Group. Nature Publishing Group; 2014. February;15(2):152–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li Z, Martin M, Zhang J, Huang H-Y, Bai L, Zhang J, Kang J, He M, Li J, Maurya MR, Gupta S, Zhou G, Sangwung P, Xu Y-J, Lei T, Huang H-D, Jain M, Jain MK, Subramaniam S, Shyy JY-J. Krüppel-Like Factor 4 Regulation of Cholesterol-25-Hydroxylase and Liver X Receptor Mitigates Atherosclerosis Susceptibility. Circulation. 2017. October 3;136(14):1315–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamada M, Nakamura M, Tran MTN, Moriguchi T, Hong C, Ohsumi T, Dinh TTH, Kusakabe M, Hattori M, Katsumata T, Arai S, Nakashima K, Kudo T, Kuroda E, Wu C-H, Kao P-H, Sakai M, Shimano H, Miyazaki T, Tontonoz P, Takahashi S. MafB promotes atherosclerosis by inhibiting foam-cell apoptosis. Nature Communications. Nature Publishing Group; 2014;5(1):3147. [DOI] [PubMed] [Google Scholar]
- 75.Hasegawa H, Watanabe T, Kato S, Toshima T, Yokoyama M, Aida Y, Nishiwaki M, Kadowaki S, Narumi T, Honda Y, Otaki Y, Honda S, Shunsuke N, Funayama A, Nishiyama S, Takahashi H, Arimoto T, Shishido T, Miyamoto T, Abe S, Shibata Y, Kubota I. The role of macrophage transcription factor MafB in atherosclerotic plaque stability. ATH. Elsevier Ltd; 2016. July 1;250(C):133–43. [DOI] [PubMed] [Google Scholar]
- 76.Dubland JA, Francis GA. So Much Cholesterol: the unrecognized importance of smooth muscle cells in atherosclerotic foam cell formation. Curr Opin Lipidol. 2016. April;27(2):155–61. [DOI] [PubMed] [Google Scholar]
- 77.Holycross BJ, Blank RS, Thompson MM, Peach MJ, Owens GK. Platelet-derived growth factor-BB-induced suppression of smooth muscle cell differentiation. Circ Res. 1992. December;71(6):1525–32. [DOI] [PubMed] [Google Scholar]
- 78.Barrett TB, Benditt EP. Platelet-derived growth factor gene expression in human atherosclerotic plaques and normal artery wall. Proc Natl Acad Sci USA. National Academy of Sciences; 1988. April;85(8):2810–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Owens GK, Kumar MS, Wamhoff BR. Molecular Regulation of Vascular Smooth Muscle Cell Differentiation in Development and Disease. Physiol Rev. 2004. July;84(3):767–801. [DOI] [PubMed] [Google Scholar]
- 80.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology. 1995. May;15(5):551–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma S, Yang D, De Li, Tang B, Yang Y. Oleic acid induces smooth muscle foam cell formation and enhances atherosclerotic lesion development via CD36. Lipids in Health and Disease. BioMed Central Ltd; 2011. April 12;10(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Costales P, Fuentes-Prior P, Castellano J, Revuelta-Lopez E, Corral-Rodríguez MÁ, Nasarre L, Badimon L, Llorente-Cortes V. K Domain CR9 of Low Density Lipoprotein (LDL) Receptor-related Protein 1 (LRP1) Is Critical for Aggregated LDL-induced Foam Cell Formation from Human Vascular Smooth Muscle Cells. J Biol Chem. 2015. June 11;290(24):14852–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rivera J, Walduck AK, Thomas SR, Glaros EN, Hooker EU, Guida E, Sobey CG, Drummond GR. Accumulation of serum lipids by vascular smooth muscle cells involves a macropinocytosis-like uptake pathway and is associated with the downregulation of the ATP-binding cassette transporter A1. Naunyn-Schmiedeberg’s Arch Pharmacol. 2013. August 30;386(12):1081–93. [DOI] [PubMed] [Google Scholar]
- 84.Rong JX, Shapiro M, Trogan E, Fisher EA. Transdifferentiation of mouse aortic smooth muscle cells to a macrophage-like state after cholesterol loading. Proc Natl Acad Sci USA. National Academy of Sciences; 2003. November 11;100(23):13531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation. 2014. April 15;129(15):1551–9. [DOI] [PubMed] [Google Scholar]
- 86.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC, Isakson B, Randolph GJ, Owens GK. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015. May 18;21(6):628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deaton RA, Gan Q, Owens GK. Sp1-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. American Journal of Physiology-Heart and Circulatory Physiology. 2009. April;296(4):H1027–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kapoor N, Niu J, Saad Y, Kumar S, Sirakova T, Becerra E, Li X, Kolattukudy PE. Transcription Factors STAT6 and KLF4 Implement Macrophage Polarization via the Dual Catalytic Powers of MCPIP. J Immunol. 2015. June 5;194(12):6011–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vengrenyuk Y, Nishi H, Long X, Ouimet M, Savji N, Martinez FO, Cassella CP, Moore KJ, Ramsey SA, Miano JM, Fisher EA. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015. March;35(3):535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014. September 12;115(7):662–7. [DOI] [PubMed] [Google Scholar]
- 91.Huff MW, Pickering JG. Can a vascular smooth muscle-derived foam-cell really change its spots? Arteriosclerosis, Thrombosis, and Vascular Biology. 2015. March;35(3):492–5. [DOI] [PubMed] [Google Scholar]
- 92.Li B, Sheng Z, Liu C, Qian L, Wu Y, Wu Y, Ma G, Yao Y. Kallistatin Inhibits Atherosclerotic Inflammation by Regulating Macrophage Polarization. Hum Gene Ther. Mary Ann Liebert, Inc., publishers 140 Huguenot Street, 3rd Floor New Rochelle, NY 10801 USA; 2019. March;30(3):339–51. [DOI] [PubMed] [Google Scholar]
- 93.Chen W, Li X, Guo S, Song N, Wang J, Jia L, Zhu A. Tanshinone IIA harmonizes the crosstalk of autophagy and polarization in macrophages via miR-375/KLF4 pathway to attenuate atherosclerosis. Int Immunopharmacol. 2019. May;70:486–97. [DOI] [PubMed] [Google Scholar]
- 94.Tang R-Z, Zhu J-J, Yang F-F, Zhang Y-P, Xie S-A, Liu Y-F, Yao W-J, Pang W, Han L-L, Kong W, Wang Y-X, Zhang T, Zhou J. DNA methyltransferase 1 and Krüppel-like factor 4 axis regulates macrophage inflammation and atherosclerosis. J Mol Cell Cardiol. 2019. March;128:11–24. [DOI] [PubMed] [Google Scholar]
- 95.Ivashkiv LB. Epigenetic regulation of macrophage polarization and function. Trends Immunol. 2013. May;34(5):216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mullican SE, Gaddis CA, Alenghat T, Nair MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D, Lazar MA. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes Dev. Cold Spring Harbor Lab; 2011. December 1;25(23):2480–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoeksema MA, Gijbels MJ, Van den Bossche J, van der Velden S, Sijm A, Neele AE, Seijkens T, Stöger JL, Meiler S, Boshuizen MC, Dallinga-Thie GM, Levels JH, Boon L, Mullican SE, Spann NJ, Cleutjens JP, Glass CK, Lazar MA, de Vries CJ, Biessen EA, Daemen MJ, Lutgens E, de Winther MP. Targeting macrophage Histone deacetylase 3 stabilizes atherosclerotic lesions. EMBO Molecular Medicine EMBO Press; 2014. September;6(9):1124–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.You S-H, Lim H-W, Sun Z, Broache M, Won K-J, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. Nature Publishing Group; 2013. January 6;20(2):182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li P, Spann NJ, Kaikkonen MU, Lu M, Oh DY, Fox JN, Bandyopadhyay G, Talukdar S, Xu J, Lagakos WS, Patsouris D, Armando A, Quehenberger O, Dennis EA, Watkins SM, Auwerx J, Glass CK, Olefsky JM. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell. 2013. September 26;155(1):200–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cao Q, Rong S, Repa JJ, St Clair R, Parks JS, Mishra N. Histone deacetylase 9 represses cholesterol efflux and alternatively activated macrophages in atherosclerosis development. Arteriosclerosis, Thrombosis, and Vascular Biology. Lippincott Williams & Wilkins Hagerstown, MD; 2014. September;34(9):1871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol Nature Publishing Group; 2010. August 22;11(10):936–44. [DOI] [PubMed] [Google Scholar]
- 102.Neele AE, Prange KH, Hoeksema MA, van der Velden S, Lucas T, Dimmeler S, Lutgens E, Van den Bossche J, de Winther MP. Macrophage Kdm6b controls the pro-fibrotic transcriptome signature of foam cells. Epigenomics. 2017. April;9(4):383–91. [DOI] [PubMed] [Google Scholar]
- 103.Neele AE, Gijbels MJJ, van der Velden S, Hoeksema MA, Boshuizen MCS, Prange K, Chen H-J, Van den Bossche J, van Roomen CPPA, Shami A, Levels JHM, Kroon J, Lucas T, Dimmeler S, Lutgens E, de Winther MPJ. Myeloid Kdm6b deficiency results in advanced atherosclerosis. Atherosclerosis. 2018. August;275:156–65. [DOI] [PubMed] [Google Scholar]
- 104.Hsu AT, Lupancu TJ, Lee M-C, Fleetwood AJ, Cook AD, Hamilton JA, Achuthan A. Epigenetic and transcriptional regulation of IL4-induced CCL17 production in human monocytes and murine macrophages. J Biol Chem. 2018. July 20;293(29):11415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Achuthan A, Cook AD, Lee M-C, Saleh R, Khiew H-W, Chang MWN, Louis C, Fleetwood AJ, Lacey DC, Christensen AD, Frye AT, Lam PY, Kusano H, Nomura K, Steiner N, Förster I, Nutt SL, Olshansky M, Turner SJ, Hamilton JA. Granulocyte macrophage colony-stimulating factor induces CCL17 production via IRF4 to mediate inflammation. J Clin Invest. 2016. September 1;126(9):3453–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lehtonen A, Matikainen S, Miettinen M, Julkunen I. Granulocyte-macrophage colony-stimulating factor (GM-CSF)-induced STAT5 activation and target-gene expression during human monocyte/macrophage differentiation. J Leukoc Biol. 2002. March;71(3):511–9. [PubMed] [Google Scholar]
- 107.Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, Hussell T, Feldmann M, Udalova IA. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nature Publishing Group. Nature Publishing Group; 2011. January 16;12(3):231–8. [DOI] [PubMed] [Google Scholar]
- 108.Zhang X, Wang Y, Yuan J, Li N, Pei S, Xu J, Luo X, Mao C, Liu J, Yu T, Gan S, Zheng Q, Liang Y, Guo W, Qiu J, Constantin G, Jin J, Qin J, Xiao Y. Macrophage/microglial Ezh2 facilitates autoimmune inflammation through inhibition of Socs3. J Exp Med. Rockefeller University Press; 2018. May 7;215(5):1365–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, Baber U, Mehran R, Fuster V, Danesh J, Frossard P, Saleheen D, Melander O, Sukhova GK, Neuberg D, Libby P, Kathiresan S, Ebert BL. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N Engl J Med. 2017. July 13;377(2):111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu C-L, Sano S, Muralidharan S, Rius C, Vuong J, Jacob S, Muralidhar V, Robertson AAB, Cooper MA, Andrés V, Hirschi KK, Martin KA, Walsh K. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017. February 24;355(6327):842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, Du J, Wang H, Wu X, Zhang L, Yu X, Lin A, McDonald T, Zhao D, Wu H, Hua W-K, Bin Zhang, Feng L, Tohyama K, Bhatia R, Oberdoerffer P, Chung YJ, Aplan PD, Boultwood J, Pellagatti A, Khaled S, Kortylewski M, Pichiorri F, Kuo Y-H, Carlesso N, Marcucci G, Jin H, Li L. SIRT1 Activation Disrupts Maintenance of Myelodysplastic Syndrome Stem and Progenitor Cells by Restoring TET2 Function. Stem Cell. Elsevier Inc; 2018. September 6;23(3):355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stein S, Matter CM. Protective roles of SIRT1 in atherosclerosis. Cell Cycle. 2014. October 31;10(4):640–7. [DOI] [PubMed] [Google Scholar]
- 113.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-Mediated Gene Editing to Assess the Roles of Tet2 and Dnmt3a in Clonal Hematopoiesis and Cardiovascular Disease. Circ Res. 2018. July 20;123(3):335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li X, Zhang Q, Ding Y, Liu Y, Zhao D, Zhao K, Shen Q, Liu X, Zhu X, Li N, Cheng Z, Fan G, Wang Q, Cao X. Methyltransferase Dnmt3a upregulates HDAC9 to deacetylate the kinase TBK1 for activation of antiviral innate immunity. Nat Immunol. 2016. May 30;17(7):806–15. [DOI] [PubMed] [Google Scholar]
- 115.Lin J-D, Nishi H, Poles J, Niu X, Mccauley C, Rahman K, Brown EJ, Yeung ST, Vozhilla N, Weinstock A, Ramsey SA, Fisher EA, Loke P. Single-cell analysis of fate-mapped macrophages reveals heterogeneity, including stem-like properties, during atherosclerosis progression and regression. JCI Insight. 2019. February 21;4(4):e21–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Piccolo V, Curina A, Genua M, Ghisletti S, Simonatto M, Sabò A, Amati B, Ostuni R, Natoli G. Opposing macrophage polarization programs show extensive epigenomic and transcriptional cross-talk. Nat Immunol. 2017. May;18(5):530–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Czimmerer Z, Daniel B, Horvath A, Rückerl D, Nagy G, Kiss M, Peloquin M, Budai MM, Cuaranta-Monroy I, Simandi Z, Steiner L, Nagy B Jr, Poliska S, Banko C, Bacso Z, Schulman IG, Sauer S, Deleuze J-F, Allen JE, Benko S, Nagy L. The Transcription Factor STAT6 Mediates Direct Repression of Inflammatory Enhancers and Limits Activation of Alternatively Polarized Macrophages. Immunity. Elsevier Inc; 2018. January 16;48(1):75–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiao Y, Kang K, Giannopoulou E, Fang C, Ivashkiv LB. IFN-γ Induces Histone 3 Lysine 27 Trimethylation in a Small Subset of Promoters to Stably Silence Gene Expression in Human Macrophages. Cell Rep. 2016. September 20;16(12):3121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kang K, Park SH, Chen J, Qiao Y, Giannopoulou E, Berg K, Hanidu A, Li J, Nabozny G, Kang K, Park-Min K-H, Ivashkiv LB. Interferon-&gamma Represses M2 Gene Expression in Human Macrophages by Disassembling Enhancers Bound by the Transcription Factor MAF. Immunity. Elsevier Inc; 2017. August 15;47(2):235–250.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Eichenfield DZ, Troutman TD, Link VM, Lam MT, Cho H, Gosselin D, Spann NJ, Lesch HP, Tao J, Muto J, Gallo RL, Evans RM, Glass CK. Tissue damage drives co-localization of NF-κB, Smad3, and Nrf2 to direct Rev-erb sensitive wound repair in mouse macrophages. Elife. eLife Sciences Publications Limited; 2016. July 27;5:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park SH, Kang K, Giannopoulou E, Qiao Y, Kang K, Kim G, Park-Min K-H, Ivashkiv LB. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nature Publishing Group. Nature Publishing Group; 2017. October;18(10):1104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neele AE, Van den Bossche J, Hoeksema MA, de Winther MPJ. Epigenetic pathways in macrophages emerge as novel targets in atherosclerosis. Eur J Pharmacol. 2015. September 15;763(Pt A):79–89. [DOI] [PubMed] [Google Scholar]
- 123.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nature Communications. Springer US; 2019. April 18;:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, Shalem O, Satija R, Raychowdhury R, Mertins P, Carr SA, Zhang F, Hacohen N, Regev A. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. Elsevier Inc; 2015. July 30;162(3):675–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell. Elsevier Inc; 2016. December 15;167(7):1853–1857.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]