Abstract
Despite considerable research progress on SARS-CoV-2, the direct zoonotic origin (intermediate host) of the virus remains ambiguous. The most definitive approach to identify the intermediate host would be the detection of SARS-CoV-2-like coronaviruses in wild animals. However, due to the high number of animal species, it is not feasible to screen all the species in the laboratory. Given that binding to ACE2 proteins is the first step for the coronaviruses to invade host cells, we propose a computational pipeline to identify potential intermediate hosts of SARS-CoV-2 by modeling the binding affinity between the Spike receptor-binding domain (RBD) and host ACE2. Using this pipeline, we systematically examined 285 ACE2 variants from mammals, birds, fish, reptiles, and amphibians, and found that the binding energies calculated for the modeled Spike-RBD/ACE2 complex structures correlated closely with the effectiveness of animal infection as determined by multiple experimental datasets. Built on the optimized binding affinity cutoff, we suggest a set of 96 mammals, including 48 experimentally investigated ones, which are permissive to SARS-CoV-2, with candidates from primates, rodents, and carnivores at the highest risk of infection. Overall, this work not only suggests a limited range of potential intermediate SARS-CoV-2 hosts for further experimental investigation; but more importantly, it proposes a new structure-based approach to general zoonotic origin and susceptibility analyses that are critical for human infectious disease control and wildlife protection.
Keywords: SARS-CoV-2, zoonotic origin, intermediate host, binding affinity, EvoEF2 energy unit
Graphical Abstract

Introduction
Identification of the direct zoonotic origin (intermediate host) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is important for combating the coronavirus disease 2019 (COVID-19) pandemic 1,2. It has become well accepted that SARS-CoV-2 was likely to originate naturally from bats soon after its outbreak, built on the fact that SARS-CoV-2 shares a 96.2% nucleotide sequence identity with the bat coronavirus (CoV) RaTG13 isolated from Rhinolophus affinis 3 and that natural insertions were identified at the S1/S2 cleavage site of the Spike (S) protein of RmYN02-CoV isolated from Rhinolophus malayanus 4. However, it remains unknown how the related CoV was transmitted from bats to humans.
In vitro experiments suggest that RaTG13 also binds to human angiotensin-converting enzyme 2 (hACE2) and can use hACE2 as an entry receptor 5; thus, it could be possible that a progenitor of SARS-CoV-2, e.g. RaTG13 or RaTG13-like CoV, infected humans and evolved during human-to-human transmission 6. However, recent experiments showed that the binding efficiency of RaTG13 to hACE2 is quite low 7, probably due to the lack of critical hACE2-binding residues. Besides, no evidence has shown that RaTG13 can directly infect humans in nature.
It is widely believed that the novel CoV was transmitted from its natural host to humans via some intermediate host, during which a progenitor of SARS-CoV-2 acquired the critical ACE2 binding residues and/or furin cleavage site 6. This point of view is supported in part by the fact that pangolin-CoV isolated from Manis javanica shares almost identical key ACE2-binding residues with SARS-CoV-2 8–11. However, it is controversial whether pangolins are the intermediate host 9,10 or natural host 8,11, or whether they are a host 12,13. Phylogenetic analyses show that some pangolin-CoVs are genetically related to SARS-CoV-2 but do not sufficiently support SARS-CoV-2 emerging directly from these pangolin-CoVs 14. Obtaining related viral sequences from animal sources would be the most definitive approach to identify the zoonotic origin of a virus 6. For instance, the full-length genome sequences of viruses isolated from palm civets and camels are 99.8% and 99.9% identical to human SARS-CoV and MERS-CoV 15,16, respectively, thus consolidating that civets are the intermediate host for SARS-CoV and camels for MERS-CoV. In contrast, RaTG13 shares a genome identity of 96.2% with SARS-CoV-2 3, and pangolin-CoVs only 85-93% 8–11,13, which is not high enough to justify that bats or pangolins are a direct zoonotic host of SARS-CoV-2.
Early studies assumed that the outbreak of SARS-CoV-2 was associated with the Huanan Seafood and Wildlife Market, where one or more animals sold there may be the direct zoonotic source 1,3,8. However, this point of view was challenged by the report that the first case of infection was suggested not to be related to the market 17,18. Therefore, strategies to trace back the origin of SARS-CoV-2 should not be limited to the animals sold in the market, but should also include a wide range of wild animals outside the market. Theoretically, all kinds of animals that may have close contact with humans should be investigated, but this would be extremely laborious as well as time- and money-consuming.
ACE2 recognition by SARS-CoV-2 is an important determinant of viral infectivity and host range 5,19. It has been reported that many animals can be infected by SARS-CoV-2 20–28. In this work, we computationally examined the ACE2 usage of SARS-CoV-2 for 285 vertebrates by modeling the binding energy between the SARS-CoV-2 Spike receptor-binding domain (S-RBD) and host ACE2. The binding data correlate well with the reported experimental studies, perfectly distinguishing the effective ACE2 receptors from the less effective ones. Our results reveal that many mammals could serve as intermediate hosts of SARS-CoV-2. This work presents a fast and reliable computational approach to screen potential animal hosts for further experimental analyses.
Materials and methods
Collection and examination of ACE2 orthologs
A list of ACE2 orthologs from 318 vertebrate species was downloaded from the NCBI website (https://www.ncbi.nlm.nih.gov/gene/59272/ortholog/?scope=7742). Besides these, we also considered the ACE2 orthologs from three mammals that are not included in this list, namely, palm civets, raccoon dogs, and Chinese rufous horseshoe bats, as civets and raccoon dogs were suggested to be intermediate hosts of SARS-CoV 15. Additionally, it was shown that the ACE2 proteins of civets and horseshoe bats can also be utilized by SARS-CoV-2 for viral entry in cell-level experiments 3.
Among the 321 ACE2 orthologs, 30 sequences had one or more amino acids that were either nonstandard or incorrectly parsed, i.e. annotated as ‘X’, and thus these ACE2 orthologs were excluded from the detailed analysis (Supplementary Table S1). Moreover, sequence alignment analysis (see below) showed that the ACE2 proteins of six species had five or more missing S-RBD binding residues (Supplementary Table S2), i.e. Acanthisitta chloris (protein accession ID: XP_009082150.1), Apteryx mantelli mantelli (XP_013805736.1), Salmo salar (XP_014062928.1), Rhinopithecus bieti (XP_017744069.1), Leptonychotes weddellii (XP_030886750.1), and Petromyzon marinus (XP_032835032.1). Subsequent binding analysis (see below) showed that these ACE2 receptors had a much higher binding energy (and thus a lower binding capability) than the others (Supplementary Table S2), partly because of the incomplete binding interface. Therefore, we cannot suggest whether these animals are susceptible to SARS-CoV-2 based on the defective information.
The remaining 285 ACE2 orthologs are summarized in Supplementary Table S3, including 134 mammals (Mammalia), 57 birds (Aves), 69 fish (Actinopterygii (66), Chondrichthyes (2), and Sarcopterygii (1)), 20 reptiles (Reptilia), and five amphibians (Amphibia). The protein ID, scientific classification (Class and Species), and common name are provided for easy retrieval.
Sequence analyses
291 ACE2 sequences, including the six ACE2 proteins with missing S-RBD binding residues in Supplementary Table S2, were subjected to multiple sequence alignment (MSA) analysis using Clustal Omega 29 with default parameters. Pairwise sequence identities between the full-length sequence of hACE2 (accession ID: NP_001358344.1) and the other ACE2 sequences were calculated based on the MSAs. Besides the full-length sequence identities, the sequence identities for the 20 interface residues 30 and five critical S-RBD binding residues 31 were also calculated from the MSAs. The results for these three types of sequence identities are shown in Supplementary Table S4.
Structure modeling
It should be mentioned that in reality, the ACE2 receptors of some animals may not bind to S-RBD. However, to quantitatively compare the capability of different ACE2 receptors to bind to S-RBD, we first constructed initial ACE2/S-RBD complex models through homology modeling, assuming that all the ACE2 receptors could bind to S-RBD, and then computed the binding energies between the two partners.
Pairwise sequence alignments between hACE2 and the other ACE2 orthologs were extracted from the MSAs and trimmed accordingly, as hACE2 was not full-length in the template complex (PDB ID: 6M0J) 30. The trimming should not affect binding analysis because it was shown that the protein-protein interface is unabridged in the experimentally determined hACE2/S-RBD complex structures 5,30–32. We utilized Modeller v9.24 33 to build the initial putative complex models. Each model was first optimized with the variable target function method with conjugate gradients using parameters library_schedule = autosched.slow and max_var_iterations = 300, and then refined using molecular dynamics with simulated annealing (SA) using parameter settings md level = refine.slow. The whole cycle was repeated two times and was not stopped unless the objective function was >1e6 (parameter settings: repeat_optimization = 2 and max_molpdf = 1e6). For each ACE2/S-RBD pair, 100 initial Modeller complex models were constructed.
Binding energy calculation
Before binding energy calculation, each Modeller complex model was first repacked using FASPR 34 to eliminate rotamer outliers and then the interface residue side-chain conformations were thoroughly refined (both for ACE2 and S-RBD) using the EvoEF2 force field in conjunction with a simulated annealing Monte Carlo (SAMC) optimization procedure 35,36, which was also utilized for anti-SARS-CoV-2 peptide design 37. During the side-chain refinement process, both the ACE2 and S-RBD sequences were kept fixed, while the different rotameric side-chain conformations were sampled. Since a stochastic SAMC optimization procedure was used, obtaining the global energy minimum may not always be guaranteed. Therefore, the optimization of the interface residues was performed five times independently to generate five refined low-energy models. Hence, for each ACE2/S-RBD pair, 500 final models were generated and scored using EvoEF2 35. The minimum binding interaction score achieved among all 500 complex models was regarded as the binding energy. The error bar of binding energy was estimated using bootstrapping. Specifically, in each step of bootstrapping, we performed a subsample with replacement for 500 data points from the original dataset of 500 binding scores, and calculated the ensemble statistics (mininum in this case) for the subsampled data. The boostrap steps were repeated for 1000 times to get 1000 minimum binding scores, and the standard deviation of these 1000 minimum values was taken as the length of error bar.
Results
A computational pipeline for ACE2 usage analysis
Since SARS-CoV-2 utilizes ACE2 to invade host cells, ACE2 usage is considered to be an important determinant of infectivity and host range 5,31. To examine the ACE2 usage by SARS-CoV-2, we developed a pipeline to model the binding energy between S-RBD and host ACE2 (Figure 1). We hypothesized that an effective ACE2 receptor should exhibit a low binding energy (or equivalently, a high affinity) while a poor receptor should have a high binding energy. A total of 321 ACE2 orthologs were collected from NCBI, and 285 of them were analyzed in detail after discarding 36 defective sequences (see Methods and Supplementary Tables S1, S2, and S3). Homologous structure models were built by Modeller 33 using the crystal structure of the hACE2/S-RBD complex (PDB ID: 6M0J) 30 as a template. Each initial complex model was then optimized using FASPR 34 and EvoEF2 35 to generate structure ensembles for binding energy calculation (see Methods). The ACE2 that achieved a binding energy below a given cutoff was suggested to be an effective receptor for SARS-CoV-2. During structure modeling and binding energy calculation, the N-glycosylation on ACE2 and S-RBD was ignored because current methods are not well adapted for modeling glycosylated amino acids.
Figure 1. A computational pipeline for ACE2 usage analysis.
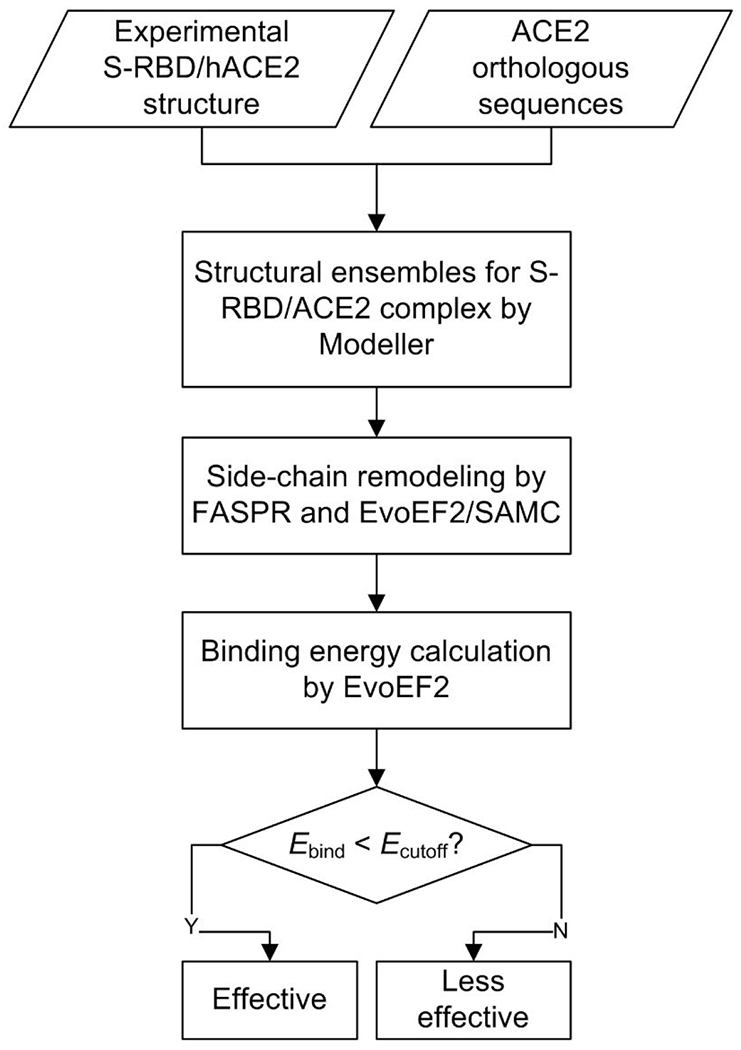
321 ACE2 orthologs were downloaded from NCBI. The crystal structure of the hACE2/S-RBD complex (PDB ID: 6M0J) was used as a template for homology modeling. For each ACE2/S-RBD pair, 100 initial Modeller complex models were constructed and repacked by FASPR, and then five models were generated by EvoEF2/SAMC remodeling for each FASPR model. The binding energy cutoff (Ecutoff) was set to −47 EvoEF2 energy units.
Binding energy assessment and correlation with experimentally determined ACE2 usage information
The length of the 285 ACE2 protein sequences ranged from 431 to 872 amino acids (Supplementary Table S3), where most ACE2 sequences were composed of about 800 amino acids (Supplementary Figure S1a). Five ACE2 sequences were partial (Bison bison bison, Thamnophis sirtalis, Haliaeetus albicilla, Fulmarus glacialis, and Panthera tigris altaica), but there were no missing interface residues according to sequence analysis. The ACE2 orthologs shared a sequence identity of ≥55% with hACE2 (Supplementary Figure S1b), indicating that the ACE2 proteins were conserved. Therefore, reliable structure models could be built by homology modeling. Based on the experimental structure of the hACE2/S-RBD complex, 20 residues (Q24, T27, F28, D30, K31, H34, E35, E37, D38, Y41, Q42, L79, M82, Y83, N330, K353, G354, D355, R357, and R393) were present at the interface of hACE2 within 4 Å of S-RBD 30. Among the ACE2 orthologs, the sequence identities of the 20 interface residues to hACE2 ranged from 30% to 100% (Supplementary Figure S1c), while the sequence identities for the five key interface residues (K31, E35, D38, M82, and K353), which were regarded as important elements to determine host range 31, varied from 0 to 100% (Supplementary Figure S1d).
The binding energy for the 285 ACE2 proteins ranged from −56.21 to −33.30 EvoEF2 energy units (EEU) (Supplementary Table S4), where a lower energy represents a stronger binding affinity, which may correspond to a higher susceptibility to SARS-CoV-2. However, it was unknown how trustable the energy values were, and it was not easy to understand whether or not an ACE2 was suggested to be an effective receptor. To address such issues, we compared the calculated binding energy with the experimental ACE2 usage data. Table 1 summarizes the reported infection cases in nature, and the infection studies in vivo and in vitro. Infections in nature represent those infection cases that take place naturally and have been confirmed by experiments such as quantitative real-time PCR 38. In vivo infection means that the caged experimental animals can be infected by SARS-CoV-2 20,24, while in vitro infection signifies that ACE2-expressing cells (e.g. HeLa cells transiently expressing ACE2) are permissive to SARS-CoV-2 infection 3. Discrepancies may exist between in vivo, in vitro, and natural infections due to different experimental settings. For instance, it was reported that SARS-CoV-2 replicates poorly in dogs and pigs in vivo 20, but it was shown that ACE2 of dogs and pigs could be effectively used for viral entry in vitro 3,39. Moreover, pet dogs were reported to be infected naturally by their owners with COVID-19 23. In this situation, an animal’s ACE2 protein was regarded as an effective receptor to SARS-CoV-2 if any kind of experimental evidence held.
Table 1. The 59 animals whose ACE2 proteins were shown to be effective or less effective for SARS-CoV-2 entry by natural infection and/or experimental studies.
The table is organized by ranking the binding energy from low to high.
| Index | Animal name | Binding energy (EEU) a | ACE2 usage b | Experimental evidence |
|---|---|---|---|---|
| 1 | Sumatran orangutan | −56.21±0.21 | Y | In vitro 39 |
| 2 | Western gorilla | −55.84±0.05 | Y | In vitro 39 |
| 3 | Olive baboon | −55.77±0.05 | Y | In vitro 39 |
| 4 | Silvery gibbon | −55.73±0.37 | Y | In vitro 39 |
| 5 | Crab-eating macaque | −55.38±0.04 | Y | In vitro 39 |
| 6 | Gelada | −55.29±0.09 | Y | In vitro 39 |
| 7 | Rhesus macaque | −55.24±0.53 | Y | In vitro 39, in vivo 27,28 |
| 8 | Human | −55.16±0.10 | Y | Natural 2,40 |
| 9 | Golden snub-nosed monkey | −55.09±0.50 | Y | In vitro 39 |
| 10 | Chimpanzee | −54.97±0.24 | Y | In vitro 39 |
| 11 | Ugandan red colobus | −54.79±0.13 | Y | In vitro 39 |
| 12 | Golden hamster | −53.84±0.05 | Y | In vitro 39, in vivo 25 |
| 13 | Chinese hamster | −53.77±0.03 | Y | In vitro 39 |
| 14 | Steller sea lion | −53.47±0.18 | Y | In vitro 39 |
| 15 | Horse | −52.95±0.41 | Y | In vitro 7,39 |
| 16 | Amur tiger | −52.93±0.32 | Y | Natural 41 |
| 17 | Goat | −52.86±0.06 | Y | In vitro 7,39 |
| 18 | Rabbit | −52.84±0.22 | Y | In vitro 7,39 |
| 19 | Wild yak | −52.83±0.06 | Y | In vitro 39 |
| 20 | Puma | −52.79±0.09 | Y | In vitro 39 |
| 21 | Leopard | −52.74±0.06 | Y | In vitro 39 |
| 22 | Cattle | −52.71±0.02 | Y | In vitro 7,39 |
| 23 | Hawaiian monk seal | −52.56±0.56 | Y | In vitro 39 |
| 24 | Ferret | −52.55±0.63 | Y | In vitro 39, in vivo 20,24; |
| 25 | California sea lion | −52.53±0.80 | Y | In vitro 39 |
| 26 | Water buffalo | −52.45±0.05 | Y | In vitro 39 |
| 27 | Lesser egyptian jerboa | −52.44±0.00 | Y | In vitro 39 |
| 28 | Cat | −52.33±0.20 | Y | In vitro 7,39, in vivo 20, natural 21 |
| 29 | Canada lynx | −52.21±1.16 | Y | In vitro 39 |
| 30 | Giant panda | −52.21±0.25 | Y | In vitro 39 |
| 31 | White-footed mouse | −52.15±0.11 | Y | In vitro 39 |
| 32 | Sheep | −52.09±0.59 | Y | In vitro 7,39 |
| 33 | Beluga whale | −51.98±0.11 | Y | In vitro 39 |
| 34 | Sperm whale | −51.94±0.40 | Y | In vitro 39 |
| 35 | Polar bear | −51.93±0.15 | Y | In vitro 39 |
| 36 | Yangtze finless porpoise | −51.85±0.21 | Y | In vitro 39 |
| 37 | Malayan pangolin | −51.73±0.35 | Y | In vitro 7,39 |
| 38 | Red fox | −51.58±0.18 | Y | In vitro 39 |
| 39 | Dog | −51.38±0.03 | Y | In vitro 39, in vivo 20, natural 23 |
| 40 | Southern white rhinoceros | −51.08±0.05 | Y | In vitro 39 |
| 41 | Pig | −50.74±0.14 | Y | In vitro 3,7 |
| 42 | Arctic ground squirrel | −50.62±0.22 | Y | In vitro 39 |
| 43 | Chinese rufous horseshoe bat | −49.91±0.15 | Y | In vitro 39 |
| 44 | Bactrian camel | −49.88±0.91 | Y | In vitro 7,39 |
| 45 | Killer whale | −49.47±0.03 | Y | In vitro 39 |
| 46 | Long-finned pilot whale | −49.19±0.10 | Y | In vitro 39 |
| 47 | Atlantic bottle-nosed dolphin | −49.05±0.11 | Y | In vitro 39 |
| 48 | Yangtze river dolphin | −49.05±0.02 | Y | In vitro 39 |
| 49 | Masked palm civet | −47.97±0.00 | Y | In vitro 3,7 |
| 50 | Malayan tiger | n.d. | Y | Natural 41 |
| 51 | African lion | n.d. | Y | Natural 41 |
| 52 | Mink | n.d. | Y | Natural 26 |
| 53 | Marmoset | −46.81±0.13 | N | In vitro 39, in vivo 28 |
| 54 | Black-capped squirrel monkey | −46.71±0.07 | N | In vitro 39 |
| 55 | Tufted capuchin | −46.13±0.28 | N | In vitro 39 |
| 56 | Brown rat | −43.14±0.15 | N | In vitro 7,39 |
| 57 | House mouse | −42.62±0.22 | N | In vitro 7,39 |
| 58 | Duck | −42.54±0.82 | N | In vivo 20 |
| 59 | Chicken | −42.07±1.65 | N | In vitro 7, in vivo 20 |
The binding energy was not calculated for Malayan tigers, lions, and minks because their ACE2 proteins were not included in the list of 321 ACE2 orthologs. EEU stands for EvoEF2 energy unit.
Y, effective ACE2 receptors; N, less effective ACE2 receptors. An ACE2 protein is classified as effective if at least one of the three kinds of experimental evidence holds.
The calculated binding energy correlated well with the experimentally determined ACE2 usage data; the ACE2 proteins that can be more effectively used by SARS-CoV-2 achieved a relatively lower binding energy (Table 1). A binding energy cutoff of −47 EEU was able to discriminate the more efficient ACE2 receptors from the less efficient ones (Table 1), with the maximum Matthews correlation coefficient (MCC) of 1.0 (Supplementary Figure S2). Among all the experimental species, apes (Sumatran orangutan, western gorilla, silvery gibbon, and chimpanzee) and Old-World monkeys (olive baboon, crab-eating macaque, gelada, rhesus macaque, golden snub-nosed monkey, and Ugandan red colobus) and humans achieved the lowest binding energy ranging from −56.21 to −54.79 EEU (Table 1). Besides, a few rodents (golden hamster, Chinese hamster, jerboa, white-footed mouse, and Arctic ground squirrel) and carnivores (sea lion, tiger, puma, leopard, seal, ferret, dog, cat, lynx, and bear) also achieved a relatively low binding energy varying from −53.84 to −50.62 EEU (Table 1). Three New-World monkeys (marmoset, black-capped squirrel monkey, and tufted capuchin), rats, mice, ducks, and chickens achieved a higher binding energy score (>−47 EEU), consistent with the reports that these animals are less susceptible to SARS-CoV-2 3,20,39,42.
Binding energy-based intermediate host range prediction
Based on the calculated binding energy and experimental data, we mapped the ACE2 usage effectiveness for each of the 285 species (Figure 2). Fish (including Actinopterygii, Chondrichthyes, and Sarcopterygii), amphibians, reptiles, and birds were predicted to have a relatively high binding energy (>−47 EEU), suggesting the ACE2 proteins of these species may be less permissive to SARS-CoV-2 binding. Mammals showed the broadest binding energy distribution, from −56.21 to −38.67 EEU (Figure 2 and Supplementary Table S4). 97 non-human mammals achieved a binding energy below −47 EEU; that is, besides the experimentally-validated species, another 49 species were also predicted to have an effective ACE2 receptor for SARS-CoV-2 (Figure 2 and Supplementary Table S4). These results suggest that mammals rather than other species are likely to be the main source of SARS-CoV-2 and hence they should be the major focus. This finding is also consistent with previous studies 7,20,39,43–47, but a more quantitative measurement was given here. Our findings also refute isolated reports claiming that non-mammal vertebrates such as reptiles could be the intermediate host 48,49.
Figure 2. Mapping the calculated binding energy to 285 vertebrates.
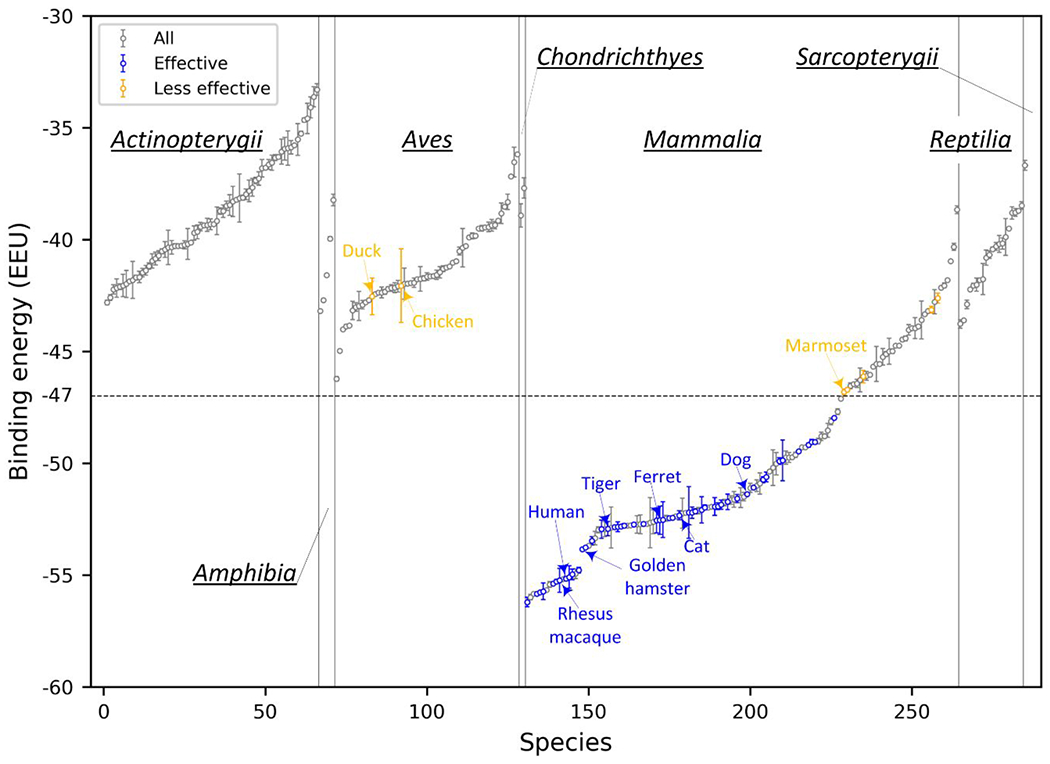
The ACE2 proteins are categorized by their animal Class (Actinopterygii, Amphibia, Aves, Chondrichthyes, Mammalia, Reptilia, and Sarcopterygii) and ranked by the binding energy from low to high in each Class. The ACE2 proteins that are experimentally shown to be effective or less effective to SARS-CoV-2 are shown in blue and orange circles, respectively, while the others are shown in gray circles. Susceptible and insusceptible animals are highlighted in blue and red, respectively. The error bars were estimated via bootstrapping by subsampling with replacement for 500 data points from the original dataset of 500 binding scores for each species; the bootstrap steps were repeated for 1000 times.
The binding energy calculation did not consider the impact of possible N-glycosylation of ACE2 and Spike. Although no N-glycosylation site is present at the interface of the hACE2/S-RBD complex 30, some ACE2 variants may have N-glycosylation sites at the interface region, which may prevent their binding to S-RBD due to steric hindrance. Thus, the analysis of interface N-glycosylation may help refine the list of effective ACE2 receptors classified by binding energy. N-glycosylation of asparagine occurs predominantly at the NX(T/S) motif, where X is any amino acid except proline. However, not all N-X-(T/S) sequons are glycosylated, so the motif alone may not be sufficient to discriminate between glycosylated and non-glycosylated asparagines. We tried three predictors, NGlycPred 50, N-GlyDE 51, and NetNGlyc (http://www.cbs.dtu.dk/services/NetNGlyc/), to predict N-glycosylation on hACE2. None of them could accurately predict all the experimentally identified N-glycosylation sites (Supplementary Table S5). All seven NX(T/S) motifs are glycosylated in the experimentally determined structure (PDB ID: 6M17) 52, indicating that ACE2 is highly N-glycosylated. To avoid the omission of potential glycosylation sites, we systematically examined all of the NX(S/T) motifs for the 285 ACE2 proteins and manually checked if any N-glycosylation sites were present at the interface.
64 out of the 285 ACE2 proteins were found to have one or more interface glycosylation sites, including 22 fish, one amphibian, 27 birds, seven mammals, and seven reptiles (Supplementary Table S6). Since many mammals are likely susceptible to SARS-CoV-2 (Figure 2 and Supplementary Table S4), we examined the seven mammals and mapped the putative interface N-glycosylation sites into their structure models. Interestingly, none of the effective ACE2 receptors in Table 1 had an interface N-glycosylation site. The seven mammals were the Eurasian common shrew (Sorex araneus), small Madagascar hedgehog (Echinops telfairi), western European hedgehog (Erinaceus europaeus), aardvark (Orycteropus afer), big brown bat (Eptesicus fuscus), star-nosed mole (Condylura cristata), and greater horseshoe bat (Rhinolophus ferrumequinum), where their binding energies were −40.97, −44.99, −38.67, −48.79, −46.46, −45.56, and −44.47 EEU, respectively. Following the binding energy criterion, aardvark’s ACE2 was predicted to be an effective receptor, but it may be ineffective due to glycosylation. The shrew had two interface glycosylation sites, N23 and N41, which form hydrogen bonds with N487 and Y449, respectively (Figure 3a); the aardvark had only one interface glycosylation site at N38, forming two hydrogen bonds with Y449 and Q498 (Figure 3b). Since these asparagine residues could form direct contact with S-RBD, their glycosylation may hinder the binding of the two proteins (i.e. ACE2 and Spike).
Figure 3. Putative N-glycosylation sites at the interface of two example ACE2/S-RBD complex structures.
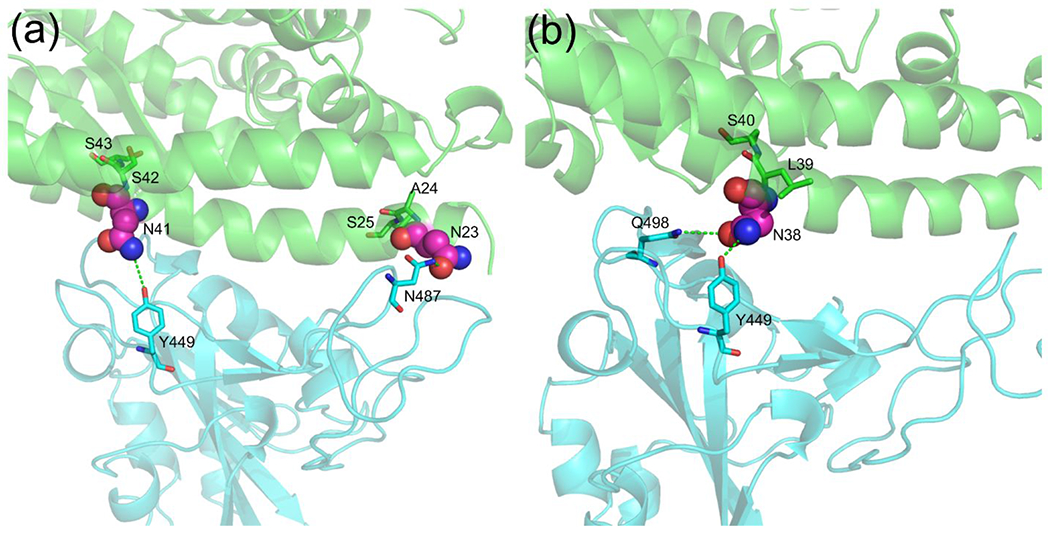
(a) Eurasian common shrew (Sorex araneus); and (b) Aardvark (Orycteropus afer). ACE2 and S-RBD are shown in green and cyan cartoons, respectively. The potential interface N-glycosylation motifs are shown with the asparagine residues highlighted in spheres.
Following the binding energy calculation and interface N-glycosylation site analysis, 96 non-human ACE2 proteins were suggested to be effectively utilized by SARS-CoV-2; half of them have been confirmed by experiments (Table 1) and the other half are summarized in Table 2. Therefore, compared with the original list of 285 animals, our method considerably narrowed the host range. The predicted potential zoonotic animals are distributed widely, including pets, domestic, agricultural, and zoological animals that may have close contact with humans (Tables 1 and 2).
Table 2. 48 other animals were predicted to have an effective ACE2 receptor capable of S-RBD binding.
The table is organized by ranking the binding energy from low to high. The animals in Table 1 were not included in this table. The binding energy cutoff (−47 EEU) was chosen by maximally discriminating the experimentally determined effective ACE2 receptors from the less effective ones.
| Index | Species | Animal name | Binding energy (EEU) |
|---|---|---|---|
| 1 | Pan paniscus | Bonobo | −55.99±0.12 |
| 2 | Nomascus leucogenys | Northern white-cheeked gibbon | −55.84±0.02 |
| 3 | Chlorocebus sabaeus | Green monkey | −55.67±0.04 |
| 4 | Macaca nemestrina | Pig-tailed macaque | −55.42±0.13 |
| 5 | Cercocebus atys | Sooty mangabey | −55.19±0.01 |
| 6 | Mandrillus leucophaeus | Drill | −54.94±0.22 |
| 7 | Nannospalax galili | Northern israeli blind subterranean mole rat | −53.69±0.18 |
| 8 | Propithecus coquereli | Coquerel’s sifaka | −53.35±0.30 |
| 9 | Callorhinus ursinus | Northern fur seal | −52.98±0.14 |
| 10 | Equus przewalskii | Mongolian wild horse | −52.95±0.34 |
| 11 | Acinonyx jubatus | Cheetah | −52.88±0.92 |
| 12 | Heterocephalus glaber | Naked mole-rat | −52.79±0.04 |
| 13 | Bison bison bison | Plains bison | −52.78±0.08 |
| 14 | Mustela erminea | Ermine | −52.74±0.39 |
| 15 | Phoca vitulina | Harbor seal | −52.73±0.42 |
| 16 | Bos indicus x Bos taurus | Hybrid cattle | −52.71±0.02 |
| 17 | Lontra canadensis | Northern american river otter | −52.66±1.12 |
| 18 | Odobenus rosmarus divergens | Walrus | −52.62±0.53 |
| 19 | Bos indicus | Zebu cattle | −52.47±0.13 |
| 20 | Peromyscus maniculatus bairdii | Deer mouse | −52.38±0.07 |
| 21 | Odocoileus virginianus texanus | White-tailed deer | −52.24±0.67 |
| 22 | Fukomys damarensis | Damaraland mole-rat | −52.22±0.02 |
| 23 | Ursus arctos horribilis | Grizzly bear | −52.13±0.14 |
| 24 | Monodon monoceros | Narwhal | −51.98±0.07 |
| 25 | Phocoena sinus | Vaquita | −51.95±0.05 |
| 26 | Microtus ochrogaster | Prairie vole | −51.76±0.14 |
| 27 | Balaenoptera acutorostrata scammoni | North pacific minke whale | −51.65±0.19 |
| 28 | Marmota marmota | Alpine marmot | −51.62±0.35 |
| 29 | Ictidomys tridecemlineatus | Thirteen-lined ground squirrel | −51.54±0.44 |
| 30 | Marmota flaviventris | Yellow-bellied marmot | −51.50±0.19 |
| 31 | Canis lupus dingo | Dingo | −51.11±0.20 |
| 32 | Ochotona princeps | American pika | −51.01±0.26 |
| 33 | Rousettus aegyptiacus | Egyptian rousette | −50.91±0.48 |
| 34 | Lagenorhynchus obliquidens | Pacific white-sided dolphin | −50.37±0.07 |
| 35 | Nyctereutes procyonoides | Raccoon dog | −50.20±0.79 |
| 36 | Equus asinus | Donkey | −50.02±0.50 |
| 37 | Mirounga leonina | Southern elephant seal | −49.75±0.29 |
| 38 | Camelus dromedarius | Arabian camel | −49.74±0.22 |
| 39 | Phyllostomus discolor | Pale spear-nosed bat | −49.73±0.04 |
| 40 | Camelus ferus | Wild bactrian camel | −49.63±0.12 |
| 41 | Pteropus vampyrus | Large flying fox | −49.29±0.00 |
| 42 | Pteropus alecto | Black flying fox | −49.29±0.06 |
| 43 | Dipodomys ordii | Ords kangaroo rat | −49.01±0.13 |
| 44 | Loxodonta africana | African savanna elephant | −48.80±0.19 |
| 45 | Enhydra lutris kenyoni | Sea otter | −48.53±0.31 |
| 46 | Trichechus manatus latirostris | Florida manatee | −48.13±0.16 |
| 47 | Octodon degus | Common degu | −47.70±0.13 |
| 48 | Vicugna pacos | Alpaca | −47.11±0.07 |
Case studies
We then analyzed several ACE2 proteins to show molecular details about why they may or may not be effectively used by SARS-CoV-2 as an entry receptor. The first case is a New-World monkey, marmoset (Callithrix jacchus), which is of extremely low susceptibility to SARS-CoV-2 both in vivo and in vitro 28,39. The marmoset achieved a high binding score of −46.81 EEU. Compared with hACE2, there were four residue substitutions in the marmoset ACE2, i.e. Y41H, Q42E, M82T, and G354Q (Table 3). In hACE2, Y41 could form hydrogen bonds with T500 in the RBD; Q42 could form a hydrogen bond with the carbonyl group of G446 and another hydrogen bond with Y449 where the NE2 atom of Q42 acts as the donor and the OH atom of Y449 as the acceptor (Figure 4a, left). The substitution of Y41 into histidine not only results in a reduced van der Waals packing energy but also disrupts the favorable hydrogen bond with T500; mutation of Q42 into glutamic acid destroys the two hydrogen bonds with G446 and Y449; moreover, the M82T substitution could lead to a reduced packing interaction with F486 due to the smaller side-chain (Figure 4a, right). The loss of three hydrogen bonds and the weakened van der Waals forces result in the poor binding energy. As reported, the double mutant H41Y/E42Q made the variant marmoset receptor more permissive to SARS-CoV-2 infection 39. Besides the New-World monkeys, we found that the ACE2 proteins of four bats (i.e. Eptesicus fuscus, Myotis brandtii, Myotis davidii, and Myotis lucifugus) also have the Y41H/Q42E substitution (Supplementary Table S7); interestingly, they were also predicted to be less effective with a binding score of >−47 EEU.
Table 3. Comparison of ACE2 interface residues and binding energy for humans, marmosets, pangolins, and turtles.
Five key residues are underlined. Amino acid mutations relative to hACE2 are shown in bold.
| Species | Human (H. sapiens) |
Marmoset (C. jacchus) |
Pangolin (M. javanica) |
Turtle (C. picta) |
Turtle (C. mydas) |
Turtle (P. sinensis) |
|---|---|---|---|---|---|---|
| ACE2 interface residues | Q24 | Q24 | E24 | E24 | E24 | E24 |
| T27 | T27 | T27 | N27 | N27 | N27 | |
| F28 | F28 | F28 | F28 | F28 | F28 | |
| D30 | D30 | E30 | S30 | S30 | S30 | |
| K31 | K31 | K31 | Q31 | Q31 | E31 | |
| H34 | H34 | S34 | V34 | V34 | V34 | |
| E35 | E35 | E35 | R35 | R35 | Q35 | |
| E37 | E37 | E37 | E37 | E37 | E37 | |
| D38 | D38 | E38 | D38 | D38 | D38 | |
| Y41 | H41 | Y41 | Y41 | Y41 | Y41 | |
| Q42 | E42 | Q42 | A42 | A42 | A42 | |
| L79 | L79 | I79 | N79 | N79 | N79 | |
| M82 | T82 | N82 | K82 | K82 | K82 | |
| Y83 | Y83 | Y83 | Y83 | Y83 | Y83 | |
| N330 | N330 | N330 | N330 | N330 | N330 | |
| K353 | K353 | K353 | K353 | K353 | K353 | |
| G354 | Q354 | H354 | K354 | K354 | K354 | |
| D355 | D355 | D355 | D355 | D355 | D355 | |
| R357 | R357 | R357 | R357 | R357 | R357 | |
| R359 | R359 | R359 | R359 | R359 | R359 | |
|
| ||||||
| Binding energy (EEU) | −55.16 | −46.81 | −51.73 | −43.61 | −42.23 | −40.68 |
Figure 4. Comparison of the mutated interface between hACE2/S-RBD and animal-ACE2/S-RBD.
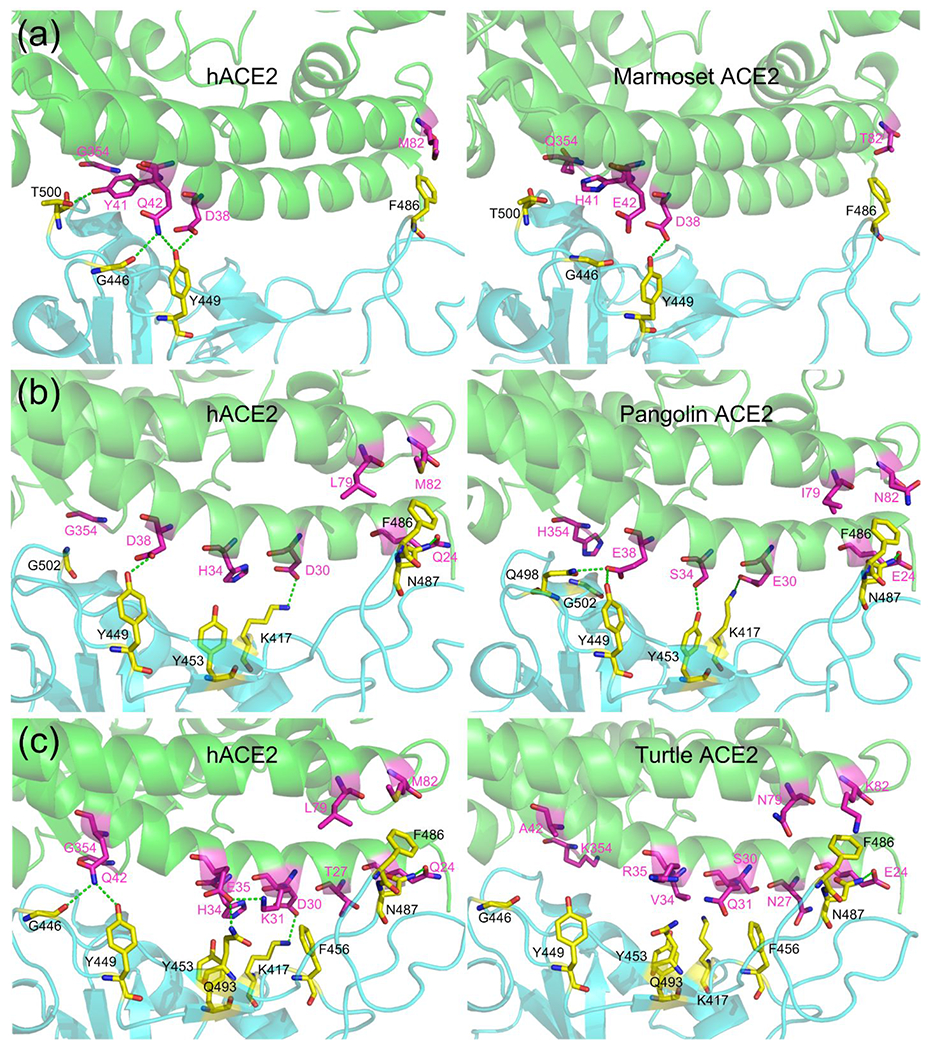
(a) hACE2/S-RBD versus marmoset-ACE2/S-RBD; (b) hACE2/S-RBD versus pangolin-ACE2/S-RBD; and (c) hACE2/S-RBD versus turtle-ACE2/S-RBD. Residues in ACE2 and S-RBD are shown in magenta and yellow, respectively. Hydrogen bonds are shown in green dashed-lines.
The second case is Malayan pangolin (Manis javanica), which has been suggested as a potential intermediate host in a few studies 9,10. Pangolin ACE2 shared an identity of 84.8%, 65%, and 60% with hACE2 for all, interface, and the key residues, respectively (Supplementary Table S4). Although pangolin ACE2 had seven residues mutated compared with hACE2, i.e. Q24E, D30E, H34S, D38E, L79I, M82N, and G354H, it still achieved a relatively low binding energy of −51.73 EEU (Table 3). In the hACE2/S-RBD complex, Q24 forms a hydrogen bond with N487, and D38 forms a hydrogen bond with Y449; D30 forms a salt bridge with K417; L79 and M82 form favorable van der Waals contacts with F486 (Figure 4b, left). In the pangolin-ACE2/S-RBD complex, favorable interactions are also extensively formed. E38 could form two hydrogen bonds with Q498 and Y449; E30 and E24 could form a hydrogen bond with K417 and N487, respectively; S34 could form a hydrogen bond with Y453 though it has a reduced van der Waals interaction due to the small size compared to H34; I79 and N82 could also form favorable packing interactions with F486 (Figure 4b, right). Therefore, although pangolin ACE2 achieved a higher binding score than hACE2, probably due to worse contacting geometries, the extensive favorable interactions demonstrate that pangolin ACE2 can still be an effective receptor to SARS-CoV-2. Thus, the binding analysis and molecular details supported Malayan pangolin as a possible intermediate host.
The third case is turtles (Chrysemys picta, Chelonia mydas, and Pelodiscus sinensis), which have been suggested as a potential intermediate host by Liu et al 49. They argued that turtles have two important residues (Y41 and K353) in their ACE2 that are identical to those in hACE2 and that turtles in the markets were more common than pangolins 49. Although it may be true that Y41 and K353 play an important role in binding S-RBD, it is, however, not a unique feature in the ACE2 of turtles and humans. As shown, many mammals have Y41 and K353 in their ACE2 proteins (Supplementary Table S7). Besides, the first reported case of infection was suggested not to be associated with the market 17. Therefore, their rules for screening intermediate hosts were not persuasive. The ACE2 protein of these turtles has ten amino acid substitutions compared with hACE2 (Table 3). C. picta and C. mydas have identical interface residues in their ACE2 proteins. P. sinensis has two different interface residues (E31 and Q35) compared with C. picta and C. mydas (Table 3). In C. picta, only E24 could form a hydrogen bond with N487 (Figure 4c, right), while the other mutations resulted in a substantial loss of favorable hydrogen bonds and salt bridges compared with those in the hACE2/S-RBD (Figure 4c, left). Expectedly, the three turtles (i.e. C. picta, C. mydas, and P. sinensis) achieved a poor binding score of −43.61, −42.23, and −40.68 EEU, respectively. Therefore, structure modeling did not support turtles as intermediate hosts.
Discussion
As the COVID-19 pandemic continues, the direct zoonotic origin (intermediate host) of SARS-CoV-2 remains elusive. Many animals have been reported to be infected by SARS-CoV-2 in nature or the laboratory, suggesting a possibly wide host range for this novel coronavirus. Currently, the number of animals that have been experimentally tested is very small compared to the huge number of animal species. Previous studies suggested that receptor recognition is an important determinant of host range 5,19,31. Therefore, we proposed a computational pipeline for identifying the intermediate hosts of SARS-CoV-2 by modeling the binding affinity between host ACE2 and the viral S-RBD. A recent study showed that, besides ACE2, alternative receptors such as ASGR1 or KREMEN1 may be sufficient to enable entry of SARS-CoV-2 into secretory cells and immune non-resident macrophages 53, suggesting that these receptors may also play a role in SARS-CoV-2 susceptibility. However, in that study it was shown that, as the main receptor, ACE2 has a much higher binding capability for the extracellular domain of S protein than ASGR1 and KREMEN1; furthermore, ACE2 displayed a more significant correlation with virus susceptibility of ciliated cells when compared with the other two receptors 53. At present, little is known about the atomic level protein-protein interactions between S and ASGR1 or KREMEN1, making it difficult to accurately model the binding affinity between the viral proteins and these receptors. Additionally, several in vitro and in vivo studies suggest that the susceptibility of different animals is mainly determined by the ACE2 receptor, as substituting the ACE2 alone is sufficient to enable SARS-CoV-2 infection of otherwise insusceptible cell lines 3,39 or animal models 54,55. Therefore, the potential role of these alternative receptors on host determination was not considered in this work.
The reasonability of ignoring TMPRSS2
It has been shown that SARS-CoV-2 cell entry depends on ACE2 and the serine protease TMPRSS2 56. However, we did not consider the role of TMPRSS2 for host prediction, due to the following reasons. First, TMPRSS2’s role for priming spike may be replaced by some other proteases like cathepsin B and L 57. Second, different from ACE2 which is used as a binding receptor only, TMPRSS2 cleaves Spike through chemical catalysis. Thus, to quantify the impact of TMPRSS2, its catalytic activity for cleavage needs to be predicted; this is, however, an impossible task to achieve at present, as almost all protease cleavage predictors were trained to predict cleavage sites for one known protease of one species 58. Third, TMPRSS2 proteases from different species may be similarly efficient. This is supported in part by the fact that wild-type mice are insusceptible to SARS-CoV-2, while transgenic mice that express hACE2 can be infected 59, suggesting that mouse TMPRSS2 may be sufficiently efficient at cleaving Spike. Besides, a recent study showed that computational modeling failed to distinguish the binding capability of TMPRSS2 from different animals 47. As a result, we believe that it may be reasonable to ignore TMPRSS2 for host prediction.
ACE2 Sequence analysis alone is not accurate enough for host identification
Built on the fact that hACE2 is highly susceptible to SARS-CoV-2, many previous studies only performed sequence analyses and used the sequence identity between animal ACE2 proteins and hACE2 to predict intermediate hosts 44,49,60, as it was believed that the ACE2 proteins that are similar to hACE2 may also be susceptible 6,31. We calculated the MCCs for distinguishing experimentally determined effective ACE2 receptors from the less effective ones listed in Table 1 using sequence identity. The maximum MCCs were 0.51,0.73, and 0.53 with the optimum sequence identity cutoff of 66%-78%, 61%~65%, and ≤60% in terms of all, interface, and key residues, respectively (Supplementary Figure S3), which were much lower than that achieved by the classification via binding energy assessment (Supplementary Figure S2). Four New-World monkeys (Sapajus apella, Aotus nancymaae, Saimiri boliviensis, and Callithrix jacchus) share a relatively high sequence identity of >92%, 80%, and 80% with hACE2 in terms of all, interface, and key residues, respectively (Supplementary Tables S4 and S7). Following the optimum sequence identity cutoffs, the ACE2 proteins of these New-World monkeys were predicted to be very effective receptors. However, in vivo infection studies showed C. jacchus was not susceptible to SARS-CoV-2 28; in vitro experiments also suggested that the ACE2 proteins of S. apella, S. boliviensis, and C. jacchus cannot be used by SARS-CoV-2 39. In contrast, dogs, cats, and ferrets, which have a much lower sequence identity to hACE2 than the new-world monkeys (Supplementary Table S4), can be infected by SARS-CoV-2 in nature and/or in vivo 20–24. These results suggest that an ACE2 protein with a higher sequence identity to hACE2 is not necessarily an effective receptor, whereas those with lower identities are not necessarily poor ones. Therefore, sequence identity between hACE2 and animal ACE2 may not be a good descriptor for host identification.
Binding energy is a better descriptor for host prediction
As indicated by the high MCC achieved (Supplementary Figure S2), structure-based binding energy assessment was more accurate than sequence identity for distinguishing experimentally confirmed species, provided that high-quality structure models were used. Critically, the structure models are highly likely to be very reliable given the high sequence similarity between hACE2 and the ACE2 orthologs and the application of advanced structure modeling tools 33,34. Moreover, we argued that it is critical to model binding energy using structure ensembles rather than a single model. We found that the binding scores that were calculated for different models of the same ACE2/S-RBD complex fluctuated considerably (Supplementary Figure S4). The maximum MCC of the classification by the binding energy derived from the first model was only about 0.63 (Supplementary Figure S5), suggesting a single model was not sufficiently accurate for the classification even if a perfect scoring function was available. To circumvent the randomness of binding energy from a single model, we evaluated a large ensemble of structure models (e.g. 500 models in this work) for each complex and took the lowest binding score as the binding energy for ACE2 usage analysis. With a proper threshold (i.e. −47 EEU), the binding energy calculated in this manner correlated well with experimental data, perfectly distinguishing the experimentally determined effective ACE2 receptors from the less effective ones with a maximum MCC of 1.0 (Supplementary Figure S2). Nevertheless, it should be mentioned that biochemical and biophysical approaches, such as fluorescence resonance energy transfer experiments, are important for verifying the interaction between S and the ACE2 protein (or any other candidate receptors) from different species.
Identification and screening of potential zoonotic origins
The most definitive strategy to identify the direct zoonotic origins of SARS-CoV-2 is to isolate related viruses from animal sources 6. Unlike SARS-CoV and MERS-CoV, whose direct zoonotic origins were identified to be civets 15 and camels 16, respectively, soon after their outbreak, the clue for SARS-CoV-2 remains elusive as the first reported case of infection was suggested not to be associated with the Huanan Seafood and Wildlife Market 17,18. As a result, a large number of animals have to be sampled to isolate viral strains that are highly similar to SARS-CoV-2 (e.g. >99% genome identity); this is a formidable task that would require extensive effort. In this regard, our work presents a fast, yet reliable approach for screening potential animals for further analysis. Our result suggests that many mammals are likely to be potential intermediate hosts of SARS-CoV-2, which is consistent with a few recent studies 39,43,61. Here, the ACE2 proteins of 285 species were assessed because their sequences were of good quality. In reality, there are more animals whose ACE2 proteins have not been sequenced yet. Thus, although 96 mammals in this study were predicted to have an effective ACE2 receptor capable of binding SARS-CoV-2 Spike, it does not necessarily mean that the real intermediate host must be one of them. The list may be further screened by considering the living environment of animals. For instance, some mammals like whales and dolphins live in the water, and therefore the chance for them to transmit bat viruses to humans may be extremely low, considering that bats are terrestrial animals.
Conclusions
The direct zoonotic origin (intermediate host) of SARS-CoV-2 that caused the COVID-19 pandemic remains elusive. In this work, we developed a computational pipeline to facilitate the identification of potential intermediate hosts of SARS-CoV-2 by modeling the binding affinity between the SARS-CoV-2 Spike receptor-binding domain and the ACE2 protein of host animals. The effectiveness of this method was verified by its performance at perfectly distinguishing the experimentally determined effective ACE2 receptors from the less effective ones with a maximum Matthews correlation coefficient (MCC) of 1.0. Although the sequence identity-based descriptors have been widely used for predicting intermediate hosts, our results showed that their performance for discriminating between effective and less effective receptors was much worse than the binding-affinity-based approach proposed here by achieving a maximum MCC of 0.73. Our results reveal that SARS-CoV-2 may have a broad host range and a few mammals, especially some primates, rodents, and carnivores, rather than the non-mammal animals could be potential hosts of SARS-CoV-2. Additionally, as a supplementary to our previous pangolin coronavirus genome assembly study, the detailed structural modeling here also supports pangolins as a possible intermediate host with molecular-level insights. Since these animals are likely to be susceptible to SARS-CoV-2, continuous monitoring of viral circulation in them is very important for disease control and wildlife protection efforts.
Supplementary Material
Table S1. 30 ACE2 proteins that were excluded from analysis due to inaccurate annotations (shown as ‘X’) in their sequences.
Table S2. Six ACE2 proteins with five or more missing interface residues.
Table S3. 285 ACE2 proteins used for detailed analysis in this work.
Table S4. Binding energy and the three kinds of sequence identity for the 285 selected species.
Table S5. Comparison of experimental and predicted N-glycosylation sites on hACE2.
Table S6. Potential N-glycosylation sites in the 285 selected ACE2 proteins.
Table S7. Comparison of the interface residues in the 285 ACE2 proteins.
Figure S1. The distribution of protein length, and sequence identity for all, interface, and the five key residues for 285 ACE2 orthologs.
Figure S2. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by the binding energy calculated from 500 models.
Figure S3. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by sequence identity in terms of all (a), interface (b), and key (c) residues.
Figure S3. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones.
Figure S4. Binding score of 500 models for the hACE2/S-RBD complex.
Figure S5. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by the binding energy calculated from the first model.
Acknowledgements
The work was supported by NIH grants (GM136422, S10OD026825, and AI134678 to Y.Z.; P30ES017885 and U24CA210967 to G.S.O.) and NSF grants (IIS1901191 and DBI2030790 to Y.Z.). The work used the XSEDE clusters62 which is supported by the National Science Foundation (ACI-1548562).
Abbreviations
- ACE2
angiotensin-converting enzyme 2
- COVID-19
coronavirus disease 2019
- EEU
EvoEF2 energy unit
- hACE2
human angiotensin-converting enzyme 2
- MC
Monte Carlo
- MCC
Matthews correlation coefficient
- MSA
multiple sequence alignment
- RBD
receptor-binding domain
- S
Spike
- SA
Simulated Annealing
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
Footnotes
The authors declare no competing financial interest.
References
- (1).Wu F; Zhao S; Yu B; Chen YM; Wang W; Song ZG; Hu Y; Tao ZW; Tian JH; Pei YY; Yuan ML; Zhang YL; Dai FH; Liu Y; Wang QM; Zheng JJ; Xu L; Holmes EC; Zhang YZ, A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhu N; Zhang D; Wang W; Li X; Yang B; Song J; Zhao X; Huang B; Shi W; Lu R; Niu P; Zhan F; Ma X; Wang D; Xu W; Wu G; Gao GF; Tan W, A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med 2020, 382, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zhou P; Yang XL; Wang XG; Hu B; Zhang L; Zhang W; Si HR; Zhu Y; Li B; Huang CL; Chen HD; Chen J; Luo Y; Guo H; Jiang RD; Liu MQ; Chen Y; Shen XR; Wang X; Zheng XS; Zhao K; Chen QJ; Deng F; Liu LL; Yan B; Zhan FX; Wang YY; Xiao GF; Shi ZL, A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Zhou H; Chen X; Hu T; Li J; Song H; Liu Y; Wang P; Liu D; Yang J; Holmes EC; Hughes AC; Bi Y; Shi W, A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein. Curr. Biol 2020, 30, 2196–2203 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Shang J; Ye G; Shi K; Wan Y; Luo C; Aihara H; Geng Q; Auerbach A; Li F, Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Andersen KG; Rambaut A; Lipkin WI; Holmes EC; Garry RF, The proximal origin of SARS-CoV-2. Nat. Med 2020, 26, 450–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Li Y; Wang H; Tang X; Ma D; Du C; Wang Y; Pan H; Zou Q; Zheng J; Xu L; Farzan M; Zhong G, Potential host range of multiple SARS-like coronaviruses and an improved ACE2-Fc variant that is potent against both SARS-CoV-2 and SARS-CoV-1. bioRxiv 2020, 10.1101/2020.04.10.032342. [DOI] [Google Scholar]
- (8).Lam TT; Jia N; Zhang YW; Shum MH; Jiang JF; Zhu HC; Tong YG; Shi YX; Ni XB; Liao YS; Li WJ; Jiang BG; Wei W; Yuan TT; Zheng K; Cui XM; Li J; Pei GQ; Qiang X; Cheung WY; Li LF; Sun FF; Qin S; Huang JC; Leung GM; Holmes EC; Hu YL; Guan Y; Cao WC, Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020, 583, 282–285. [DOI] [PubMed] [Google Scholar]
- (9).Xiao K; Zhai J; Feng Y; Zhou N; Zhang X; Zou JJ; Li N; Guo Y; Li X; Shen X; Zhang Z; Shu F; Huang W; Li Y; Zhang Z; Chen RA; Wu YJ; Peng SM; Huang M; Xie WJ; Cai QH; Hou FH; Chen W; Xiao L; Shen Y, Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 2020, 583, 286–289. [DOI] [PubMed] [Google Scholar]
- (10).Zhang C; Zheng W; Huang X; Bell EW; Zhou X; Zhang Y, Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J. Proteome Res 2020, 19, 1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang T; Wu Q; Zhang Z, Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr. Biol 2020, 30, 1346–1351 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Li X; Zai J; Zhao Q; Nie Q; Li Y; Foley BT; Chaillon A, Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol 2020, 92, 602–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Liu P; Jiang JZ; Wan XF; Hua Y; Li L; Zhou J; Wang X; Hou F; Chen J; Zou J; Chen J, Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020, 16, e1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Zhang X; Chen X; Zhang Z; Roy A; Shen Y, Strategies to trace back the origin of COVID-19. J. Infect 2020, 80, e39–e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Guan Y; Zheng BJ; He YQ; Liu XL; Zhuang ZX; Cheung CL; Luo SW; Li PH; Zhang LJ; Guan YJ; Butt KM; Wong KL; Chan KW; Lim W; Shortridge KF; Yuen KY; Peiris JS; Poon LL, Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [DOI] [PubMed] [Google Scholar]
- (16).Hemida MG; Chu DK; Poon LL; Perera RA; Alhammadi MA; Ng HY; Siu LY; Guan Y; Alnaeem A; Peiris M, MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis 2014, 20, 1231–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Huang C; Wang Y; Li X; Ren L; Zhao J; Hu Y; Zhang L; Fan G; Xu J; Gu X; Cheng Z; Yu T; Xia J; Wei Y; Wu W; Xie X; Yin W; Li H; Liu M; Xiao Y; Gao H; Guo L; Xie J; Wang G; Jiang R; Gao Z; Jin Q; Wang J; Cao B, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cohen J, Wuhan seafood market may not be source of novel virus spreading globally. Science 2020, 10.1126/science.abb0611. [DOI] [Google Scholar]
- (19).Shang J; Wan Y; Luo C; Ye G; Geng Q; Auerbach A; Li F, Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A 2020, 117, 11727–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Shi J; Wen Z; Zhong G; Yang H; Wang C; Huang B; Liu R; He X; Shuai L; Sun Z; Zhao Y; Liu P; Liang L; Cui P; Wang J; Zhang X; Guan Y; Tan W; Wu G; Chen H; Bu Z, Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Halfmann PJ; Hatta M; Chiba S; Maemura T; Fan S; Takeda M; Kinoshita N; Hattori SI; Sakai-Tagawa Y; Iwatsuki-Horimoto K; Imai M; Kawaoka Y, Transmission of SARS-CoV-2 in Domestic Cats. N. Engl. J. Med 2020, 383, 592–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Goumenou M; Spandidos DA; Tsatsakis A, Possibility of transmission through dogs being a contributing factor to the extreme Covid19 outbreak in North Italy. Mol Med Rep 2020, 21, 2293–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sit THC; Brackman CJ; Ip SM; Tam KWS; Law PYT; To EMW; Yu VYT; Sims LD; Tsang DNC; Chu DKW; Perera R; Poon LLM; Peiris M, Infection of dogs with SARS-CoV-2. Nature 2020, 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kim YI; Kim SG; Kim SM; Kim EH; Park SJ; Yu KM; Chang JH; Kim EJ; Lee S; Casel MAB; Um J; Song MS; Jeong HW; Lai VD; Kim Y; Chin BS; Park JS; Chung KH; Foo SS; Poo H; Mo IP; Lee OJ; Webby RJ; Jung JU; Choi YK, Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sia SF; Yan LM; Chin AWH; Fung K; Choy KT; Wong AYL; Kaewpreedee P; Perera R; Poon LLM; Nicholls JM; Peiris M; Yen HL, Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020, 583, 834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Oreshkova N; Molenaar RJ; Vreman S; Harders F; Oude Munnink BB; Hakze-van der Honing RW; Gerhards N; Tolsma P; Bouwstra R; Sikkema RS; Tacken MG; de Rooij MM; Weesendorp E; Engelsma MY; Bruschke CJ; Smit LA; Koopmans M; van der Poel WH; Stegeman A, SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Euro Surveill. 2020, 25, pii=2001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Munster VJ; Feldmann F; Williamson BN; van Doremalen N; Perez-Perez L; Schulz J; Meade-White K; Okumura A; Callison J; Brumbaugh B; Avanzato VA; Rosenke R; Hanley PW; Saturday G; Scott D; Fischer ER; de Wit E, Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lu S; Zhao Y; Yu W; Yang Y; Gao J; Wang J; Kuang D; Yang M; Yang J; Ma C; Xu J; Qian X; Li H; Zhao S; Li J; Wang H; Long H; Zhou J; Luo F; Ding K; Wu D; Zhang Y; Dong Y; Liu Y; Zheng Y; Lin X; Jiao L; Zheng H; Dai Q; Sun Q; Hu Y; Ke C; Liu H; Peng X, Comparison of SARS-CoV-2 infections among 3 species of non-human primates. bioRxiv 2020, 10.1101/2020.04.08.031807. [DOI] [Google Scholar]
- (29).Sievers F; Wilm A; Dineen D; Gibson TJ; Karplus K; Li W; Lopez R; McWilliam H; Remmert M; Söding J, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 2011, 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lan J; Ge J; Yu J; Shan S; Zhou H; Fan S; Zhang Q; Shi X; Wang Q; Zhang L; Wang X, Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [DOI] [PubMed] [Google Scholar]
- (31).Wan Y; Shang J; Graham R; Baric RS; Li F, Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol 2020, 94, e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Wang Q; Zhang Y; Wu L; Niu S; Song C; Zhang Z; Lu G; Qiao C; Hu Y; Yuen KY; Wang Q; Zhou H; Yan J; Qi J, Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell 2020, 181, 894–904 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Webb B; Sali A Protein structure modeling with MODELLER. In Functional Genomics; Springer: 2017, pp 39–54. [DOI] [PubMed] [Google Scholar]
- (34).Huang X; Pearce R; Zhang Y, FASPR: an open-source tool for fast and accurate protein side-chain packing. Bioinformatics 2020, 36, 3758–3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Huang X; Pearce R; Zhang Y, EvoEF2: accurate and fast energy function for computational protein design. Bioinformatics 2020, 36, 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Huang X; Pearce R; Zhang Y, Toward the Accuracy and Speed of Protein Side-Chain Packing: A Systematic Study on Rotamer Libraries. J. Chem. Inf. Model 2020, 60, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Huang X; Pearce R; Zhang Y, De novo design of protein peptides to block association of the SARS-CoV-2 spike protein with human ACE2. Aging 2020, 12, 11263–11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Corman VM; Landt O; Kaiser M; Molenkamp R; Meijer A; Chu DKW; Bleicker T; Brunink S; Schneider J; Schmidt ML; Mulders D; Haagmans BL; van der Veer B; van den Brink S; Wijsman L; Goderski G; Romette JL; Ellis J; Zambon M; Peiris M; Goossens H; Reusken C; Koopmans MPG; Drosten C, Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Liu Y; Hu G; Wang Y; Zhao X; Ji F; Ren W; Gong M; Ju X; Li C; Hong J; Zhu Y; Cai X; Wu J; Lan X; Xie Y; Wang X; Yuan Z; Zhang R; Ding Q, Functional and Genetic Analysis of Viral Receptor ACE2 Orthologs Reveals Broad Potential Host Range of SARS-CoV-2. bioRxiv 2020, 10.1101/2020.04.22.046565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Holshue ML; DeBolt C; Lindquist S; Lofy KH; Wiesman J; Bruce H; Spitters C; Ericson K; Wilkerson S; Tural A; Diaz G; Cohn A; Fox L; Patel A; Gerber SI; Kim L; Tong S; Lu X; Lindstrom S; Pallansch MA; Weldon WC; Biggs HM; Uyeki TM; Pillai SK, First Case of 2019 Novel Coronavirus in the United States. N. Engl. J. Med 2020, 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Bartlett SL; Diel DG; Wang L; Zee S; Laverack M; Martins M; Caserta LC; Killian ML; Terio K; Olmstead C; Delaney MA; Stokol T; Ivančić M; Jenkins-Moore M; Ingerman K; Teegan T; McCann C; Thomas P; McAloose D; Sykes JM; Calle PP, SARS-CoV-2 Infection And Longitudinal Fecal Screening In Malayan Tigers (Panthera tigris jacksoni), Amur Tigers (Panthera tigris altaica), And African Lions (Panthera leo krugeri) At The Bronx Zoo, New York, USA. bioRxiv 2020, 10.1101/2020.08.14.250928. [DOI] [PubMed] [Google Scholar]
- (42).Zhao X; Chen D; Szabla R; Zheng M; Li G; Du P; Zheng S; Li X; Song C; Li R ; Guo J-T; Junop M; Zeng H; Lin H, Broad and differential animal ACE2 receptor usage by SARS-CoV-2. J. Virol 2020, 10.1128/JVI.00940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Damas J; Hughes GM; Keough KC; Painter CA; Persky NS; Corbo M; Hiller M; Koepfli K-P; Pfenning AR; Zhao H; Genereux DP; Swofford R; Pollard KS; Ryder OA; Nweeia MT; Lindblad-Toh K; Teeling EC; Karlsson EK; Lewin HA, Broad Host Range of SARS-CoV-2 Predicted by Comparative and Structural Analysis of ACE2 in Vertebrates. bioRxiv 2020, 10.1101/2020.04.16.045302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Luan J; Lu Y; Jin X; Zhang L, Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun 2020, 526, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Qiu Y; Zhao YB; Wang Q; Li JY; Zhou ZJ; Liao CH; Ge XY, Predicting the angiotensin converting enzyme 2 (ACE2) utilizing capability as the receptor of SARS-CoV-2. Microbes Infect 2020, 22, 221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Zhai X; Sun J; Yan Z; Zhang J; Zhao J; Zhao Z; Gao Q; He WT; Veit M; Su S, Comparison of Severe Acute Respiratory Syndrome Coronavirus 2 Spike Protein Binding to ACE2 Receptors from Human, Pets, Farm Animals, and Putative Intermediate Hosts. J. Virol 2020, 94, e00831–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lam SD; Bordin N; Waman VP; Scholes HM; Ashford P; Sen N; van Dorp L; Rauer C; Dawson NL; Pang CSM; Abbasian M; Sillitoe I; Edwards SJL; Fraternali F; Lees JG; Santini JM; Orengo CA, SARS-CoV-2 spike protein predicted to form stable complexes with host receptor protein orthologues from mammals, but not fish, birds or reptiles. bioRxiv 2020, 10.1101/2020.05.01.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Ji W; Wang W; Zhao X; Zai J; Li X, Cross-species transmission of the newly identified coronavirus 2019-nCoV. J. Med. Virol 2020, 92, 433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Liu Z; Xiao X; Wei X; Li J; Yang J; Tan H; Zhu J; Zhang Q; Wu J; Liu L, Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol 2020, 92, 595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Chuang G-Y; Boyington JC; Joyce MG; Zhu J; Nabel GJ; Kwong PD; Georgiev I, Computational prediction of N-linked glycosylation incorporating structural properties and patterns. Bioinformatics 2012, 28, 2249–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Pitti T; Chen C-T; Lin H-N; Choong W-K; Hsu W-L; Sung T-Y, N-GlyDE: a two-stage N-linked glycosylation site prediction incorporating gapped dipeptides and pattern-based encoding. Sci. Rep 2019, 9, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Yan R; Zhang Y; Li Y; Xia L; Guo Y; Zhou Q, Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Gu Y; Cao J; Zhang X; Gao H; Wang Y; Wang J; Zhang J; Shen G; Jiang X; Yang J; Zheng X; Xu J; Zhang CC; Lan F; Qu D; Zhao Y; Xu G; Xie Y; Luo M; Lu Z, Interaction network of SARS-CoV-2 with host receptome through spike protein. bioRxiv 2020, 10.1101/2020.09.09.287508. [DOI] [Google Scholar]
- (54).Dinnon KH; Leist SR; Schäfer A; Edwards CE; Martinez DR; Montgomery SA; West A; Yount BL; Hou YJ; Adams LE, A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586, 560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Goldman-Israelow B; Song E; Mao T; Lu P; Meir A; Liu F; Alfajaro MM; Wei J; Dong H; Homer R, Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med 2020, 217, e20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hoffmann M; Kleine-Weber H; Schroeder S; Kruger N; Herrler T; Erichsen S; Schiergens TS; Herrler G; Wu NH; Nitsche A; Muller MA; Drosten C; Pohlmann S, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Simmons G; Gosalia DN; Rennekamp AJ; Reeves JD; Diamond SL; Bates P, Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U.S.A 2005, 102, 11876–11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Fan YX; Zhang Y; Shen HB, LabCaS: labeling calpain substrate cleavage sites from amino acid sequence using conditional random fields. Proteins 2013, 81, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Bao L; Deng W; Huang B; Gao H; Liu J; Ren L; Wei Q; Yu P; Xu Y; Qi F; Qu Y; Li F; Lv Q; Wang W; Xue J; Gong S; Liu M; Wang G; Wang S; Song Z; Zhao L; Liu P; Zhao L; Ye F; Wang H; Zhou W; Zhu N; Zhen W; Yu H; Zhang X; Guo L; Chen L; Wang C; Wang Y; Wang X; Xiao Y; Sun Q; Liu H; Zhu F; Ma C; Yan L; Yang M; Han J; Xu W; Tan W; Peng X; Jin Q; Wu G; Qin C, The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [DOI] [PubMed] [Google Scholar]
- (60).Luan J; Jin X; Lu Y; Zhang L, SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol 2020, 10.1002/jmv.25817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Li F, Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol 2015, 89, 1954–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Towns J; Cockerill T; Dahan M; Foster I; Gaither K; Grimshaw A; Hazlewood V; Lathrop S; Lifka D; Peterson GD, XSEDE: accelerating scientific discovery. Comput. Sci. Eng 2014, 16, 62–74. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. 30 ACE2 proteins that were excluded from analysis due to inaccurate annotations (shown as ‘X’) in their sequences.
Table S2. Six ACE2 proteins with five or more missing interface residues.
Table S3. 285 ACE2 proteins used for detailed analysis in this work.
Table S4. Binding energy and the three kinds of sequence identity for the 285 selected species.
Table S5. Comparison of experimental and predicted N-glycosylation sites on hACE2.
Table S6. Potential N-glycosylation sites in the 285 selected ACE2 proteins.
Table S7. Comparison of the interface residues in the 285 ACE2 proteins.
Figure S1. The distribution of protein length, and sequence identity for all, interface, and the five key residues for 285 ACE2 orthologs.
Figure S2. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by the binding energy calculated from 500 models.
Figure S3. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by sequence identity in terms of all (a), interface (b), and key (c) residues.
Figure S3. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones.
Figure S4. Binding score of 500 models for the hACE2/S-RBD complex.
Figure S5. Matthews correlation coefficient for classifying experimentally determined effective ACE2 receptors from the less effective ones by the binding energy calculated from the first model.


