Abstract
Tracking antigen-specific T cell responses over time within individuals is difficult because of lack of knowledge of antigen-specific TCR sequences, limitations in sample size, and assay sensitivities. We hypothesized that analyses of high-throughput sequencing of TCR clonotypes could provide functional readouts of individuals’ immunological histories. Using high-throughput TCR sequencing, we develop a database of TCRβ sequences from large cohorts of mice before (naive) and after smallpox vaccination. We computationally identify 315 vaccine-associated TCR sequences (VATS) that are used to train a diagnostic classifier that distinguishes naive from vaccinated samples in mice up to 9 months post-vaccination with >99% accuracy. We determine that the VATS library contains virus-responsive TCRs by in vitro expansion assays and virus-specific tetramer sorting. These data outline a platform for advancing our capabilities to identify pathogen-specific TCR sequences, which can be used to identify and quantitate low-frequency pathogen-specific TCR sequences in circulation over time with exceptional sensitivity.
Keywords: T cell receptor, vaccinia, monkeypox, DNA sequencing, HLA-A2, biomarkers
Graphical Abstract
Wolf et al. use high-throughput TCR sequencing to survey circulating TCR repertoires of mice before and after Orthopoxvirus infection. Here, they develop a diagnostic assay distinguishing naive and Orthopoxvirus-exposed samples by computational identification of pathogen-associated TCR sequences. The assay is 97% accurate up to 9 months post-exposure.
Introduction
T cell and B cell responses are responsible for generating the adaptive immune response to vaccines and infections. T cells recognize pathogen-specific peptides in the context of the major histocompatibility complex (MHC) through the T cell receptor (TCR). The TCR is a heterodimeric protein composed of TCRα and TCRβ chains. During T cell development, each TCR chain is generated through quasi-random genetic recombination from the germline loci of the variable (V), diversity (D), and joining (J) gene segments (Manfras et al., 1999, Robins et al., 2010). In mice, the tcrb locus has approximately 35 different TCRVβ segments, 2 TCRDβ segments, and 14 TCRJ β segments. During recombination, tcrv, tcrd, and tcrj segments are rearranged together to create and encode complementary determining region 3 (CDR3). CDR3 is the most variable region of the TCR that interacts with foreign peptide. These genetic rearrangement events result in a high degree of diversity in CDR3 of the TCR (Arstila et al., 1999, Cabaniols et al., 2001, Davis and Bjorkman, 1988, Robins et al., 2009).
During an immune response, antigen presentation results in the activation and expansion of T cells with TCR(s) specific to the pathogen (Ishizuka et al., 2009, Venturi et al., 2008b, Venturi et al., 2016). Clonally expanded T cells carry the same unique TCR rearrangement (Manfras et al., 1999). Once the pathogen has been cleared, a subset of T cells with TCRs specific to the pathogen remain as long-lived memory cells. The unique DNA rearrangements have the potential to serve as a stable biomarker, cataloging an individual’s functional T cell memory and immunological history (Emerson and DeWitt, 2017, Estorninho et al., 2013).
On average, ∼107 unique TCRβ chains can be identified from the ∼1012 circulating T cells present in a healthy human adult (Robins et al., 2009). The ability to readily identify identical TCR sequences among multiple individuals (public TCRs) is challenging because an individual has the potential to generate ∼1018 unique TCR recombinants. Nonetheless, in both humans and murine models, there are examples of public T cell responses to infectious disease (such as cytomegalovirus [CMV] and influenza) and in autoimmunity (Elhanati et al., 2014, Emerson and DeWitt, 2017, Li et al., 2012, Lossius et al., 2014, Marrero et al., 2016, Valkenburg et al., 2016, Venturi et al., 2008b). The presence of virus-specific public TCRs may be due partly to preferential use of specific TCR V and J chains in response to conserved hierarchy of epitope recognition (Chen et al., 2000, Hancock et al., 2015, Kim et al., 2013). Public TCR sequences from antigen-experienced T cells should be readily identifiable within the circulating T cell repertoire because of clonal expansion and the formation of memory T cell populations (Emerson and DeWitt, 2017, Heit et al., 2017).
Identifying antigen-specific T cells and tracking an antigen-specific response over time within individuals is a difficult task, especially against emerging pathogens, in which case precise immunogenic epitopes are not well described. Even when antigens are known, the frequencies of antigen-specific T cell populations are notoriously low and can often be difficult to identify (Douillard et al., 1997, Wolf and DiPaolo, 2016, Lim et al., 2000). This is due partly to a lack of knowledge concerning antigen-specific TCR sequences. Another issue is that antigen-specific TCR identification using many traditional immune assays is restricted to the most high-frequency responders (Wolf and DiPaolo, 2016, van der Velden et al., 2014, van der Velden and van Dongen, 2009). However, advancements in next-generation sequencing are allowing researchers to analyze TCR and B cell receptor (BCR) (Ig) repertoires (immunosequencing) with unprecedented depth and sensitivity, identifying 105–107 individual sequences in humans from a very limited volume of whole blood (DeWitt et al., 2015, Faham et al., 2012, Kirsch et al., 2015, Logan et al., 2014, Robins et al., 2009). Recent publications have shown that high-throughput TCR sequencing data are comprehensive enough to be used to accurately predict individual HLA-A and HLA-B alleles in humans and to identify conserved molecular patterns within CDR3 associated with tuberculosis exposure in mice (Emerson and DeWitt, 2017, Thomas et al., 2014). By analyzing the TCRβ repertoires of mice before and after pathogen exposure, we hypothesize that it is possible to computationally identify the comprehensive functional T cell response to a specific pathogen, making it possible to distinguish between exposed and unexposed individuals and to track the functional T cell response over time.
In this study, we sampled the circulating CD4+ and CD8+ T cell populations from cohorts of mice before and after exposure to the smallpox vaccine (ACAM2000) and highly related monkeypox virus (MPXV) Zaire-79, generating extensive TCRβ sequence databases (Shchelkunov et al., 2002). Analyzing the TCR sequence databases, we identified the virus-specific T cell response and identified a library of public vaccine-associated TCRβ sequences (VATS) that were associated with the smallpox-vaccinated but not naive samples. The VATS library was used to train a diagnostic classifier that tracked the virus-specific TCRs circulating in blood over time and correctly distinguished between vaccinated and naive samples with >99% accuracy, including samples from mice up to 9 months post-vaccination. The accuracy of the diagnostic classifier was validated by testing an unrelated cohort of mice infected with the highly related MPXV, distinguishing naive from MPXV-infected mice with 96% accuracy. The VATS library was confirmed to contain vaccine-specific TCRβ sequences by vaccine-specific tetramer sorting of CD8+ T cells and in vitro cultures supplemented with ACAM2000. Tetramer+ VATS were tracked over time in individual mice and were found in mice up to 9 months post-vaccination; this is a reflection of the virus-specific response and shows that from a small sample of blood, the outlined diagnostic approach is capable of determining an individual’s prior pathogen exposure by identifying low-frequency virus-specific TCR sequences.
Results
Vaccination and Infection of AAD Mice with Orthopoxvirus
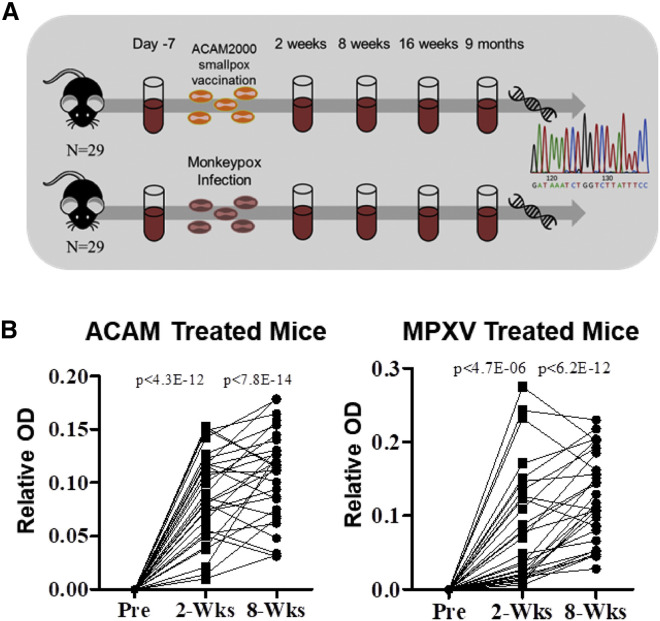
An extensive TCR sequence database was generated from a large cohort of HLA-A2 transgenic (AAD) mice before and up to 9 months after administration of the ACAM2000 smallpox vaccine or infection with MPXV. Fifty-eight AAD HLA-A2 transgenic mice (HLA-A2), pooled from three independent experiments, were entered into the study. The expression of the HLA-A2 transgenic molecule allows a portion of the virus-specific T cell response to be generated in the context of human MHC molecules (Kotturi et al., 2009). A cohort of 29 HLA-A2 mice were vaccinated with the smallpox vaccine, and another cohort of 29 mice were infected with MPXV, and blood samples were collected over time (Figure 1 A). Increased pox-specific antibodies were observed in serum 2 and 8 weeks post-vaccination or infection compared with naive in all mice by ELISA, confirming the generation of an immune response against the vaccine or infection (Figure 1B). Blood samples (∼100 μL) collected before (naive) and 2 weeks, 8 weeks, 16 weeks, and 9 months after exposure had genomic DNA purified for immunosequencing of the TCRβ repertoire to identify all TCRβ clonotypes present in each sample. A unique TCRβ clonotype in this study is defined as a unique combination of a V gene, CDR3 amino acid sequence, and J gene. From the genomic DNA purified from the whole blood, 2.85 × 106 unique TCRβ clonotypes were identified from the TCR repertoires of all naive (n = 32), vaccinated (n = 99), and infected (n = 114) samples (Table 1 ). In brief, after confirming the generation of a pox-specific immune response in all mice, high-throughput sequencing data of TCRβ genes from a large cohort of mice were used to compile a large database of TCR clonotypes in order to computationally identify vaccine- and infection-specific TCRs.
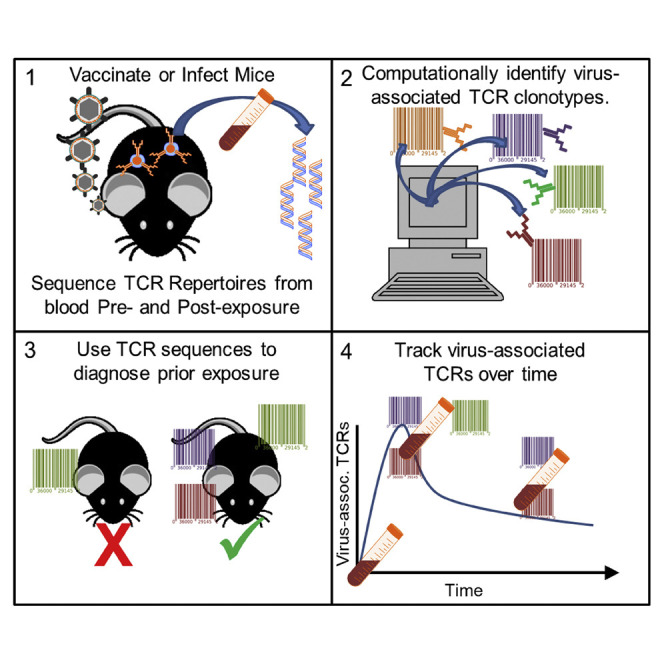
Figure 1.
Overview, Sample Collection, and Immune Responses after Smallpox Vaccination and Monkeypox Infection
(A) Flow diagram depicting the methodology of sample collection, vaccination and infection, DNA extraction, and immunosequencing.
(B) Measurement of pox-specific serum antibodies from HLA-A2 humanized mice before or after ACAM2000 smallpox vaccination (left; n = 29) or monkeypox virus (MPXV) infection (right; n = 29). Serum OD readings from mice post-vaccination or post-infection are displayed as a relative increase in the measured OD compared with their respective pre-treatment serum collection. Significance was determined using a paired t test.
Table 1.
TCRβ Sequence Rearrangements and Clonotypes in Vaccinated and Infected Mice
| Treatment | Time Point | Number of Samples (n) | Number of Unique Clonotypes | Total Number of Rearrangements |
|---|---|---|---|---|
| Naive samples | 32 | 700,000 | 1,469,000 | |
| ACAM2000 vaccinated | 2 week samples | 29 | 391,500 | 700,000 |
| 8 week samples | 29 | 185,000 | 420,000 | |
| 16 week samples | 18 | 222,500 | 397,500 | |
| 9 month samples | 23 | 209,000 | 316,000 | |
| MPXV infected | 2 week samples | 29 | 300,000 | 538,500 |
| 8 week samples | 29 | 177,500 | 326,000 | |
| 16 week samples | 29 | 374,500 | 705,000 | |
| 9 month samples | 27 | 285,000 | 433,000 |
Consolidated data referencing the total number of mice, unique TCRβ sequences (clonotypes), total number of rearranged TCRβ genes sequenced for each time point for naive, ACAM2000 vaccinated, and MPXV infected samples.
Development of the VATS Library
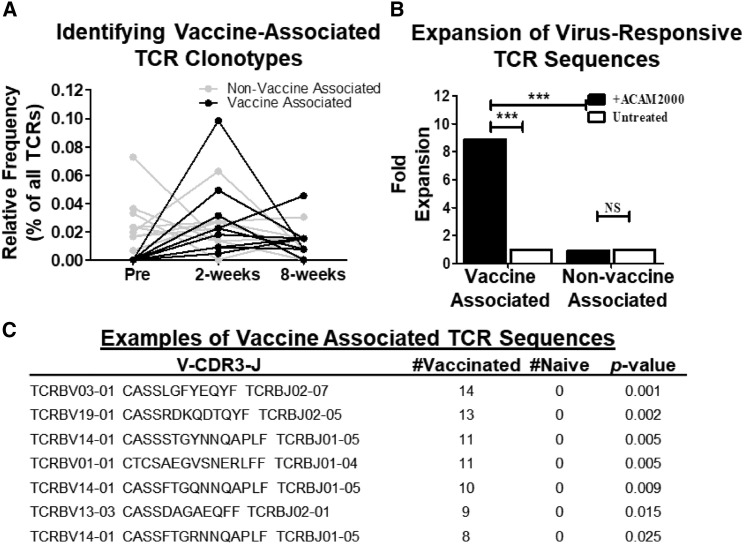
TCR repertoires from whole blood of mice pre- and post-vaccination were analyzed to computationally identify TCR clonotypes present post-vaccination but absent pre-vaccination versus sequences present pre- and post-vaccination. Within individual mice, TCR clonotypes were identified that were expanded in the blood post-vaccination (2 and 8 weeks) and absent prior to vaccination (vaccine-associated) in addition to TCR clonotypes present pre- and post-vaccination (non-vaccine-associated) (Figure 2 A).
Figure 2.
Identifying Vaccine-Associated TCRβ Sequences
(A) Line graph displaying a representative assortment of vaccine-associated (black lines) or non-vaccine-associated (gray lines) T cell receptor β (TCRβ) clonotypes. Each line represents a unique TCRβ clonotype.
(B) Expansion of TCRβ sequences from splenocytes of vaccinated mice cultured with (black bars) or without (white bars) ACAM2000 either found in mice pre-vaccination (non-vaccine-associated) or identified in both 2 and 8 week post-vaccination TCRβ repertoires but absent pre-vaccination (post-vaccine-associated) (n = 29 mice). Significance was calculated using chi-square test with Yates’s correction (∗∗∗p ≤ 0.0001).
(C) Tabular representation of public TCRβ clonotypes enriched in vaccinated versus naive samples. p value calculated using one-tailed Fisher’s exact test.
Next it was determined whether the computationally identified VATS contained TCR clonotypes that functionally expanded in response to smallpox vaccine. Splenocytes of mice from the original cohort 12 weeks after vaccination were cultured with or without the smallpox vaccine for 5 days to induce expansion of vaccine-specific T cells in vitro. Intra-mouse analysis of the TCR repertoires pre- and 2 and 8 weeks post-vaccination were compared with the libraries from splenocytes cultured with or without ACAM2000. It has been previously shown that the smallpox vaccine does not induce bystander activation of CD8+ T cells, which leads us to conclude that TCR sequences from proliferating T cells in this experimental design are virus specific (Miller et al., 2008). The relative abundances of vaccine-associated and non-vaccine-associated TCR clonotypes were compared between vaccine-stimulated and unstimulated cultures. Post-vaccine-associated sequences were significantly expanded (8.9-fold, p < 0.0001) in the vaccine-stimulated versus unstimulated controls; this is significantly greater (p < 0.0001) than the expansion (0.94-fold) measured in non-VATS (Figure 2B). These data show that computational identification of vaccine-associated TCRβ clonotypes enriches for virus-specific TCR sequences.
To distinguish between TCR repertoires from naive and exposed samples, a vaccine-associated public TCRβ library was generated. Pre- and post-vaccination TCRβ sequence libraries were analyzed, computationally detecting the virus-specific T cell response by identifying sequences that were statistically associated with post-vaccination samples. TCRβ sequences from all naive (n = 32) and 2 and 8 week post-vaccination TCRβ repertoires (n = 58) were used for this analysis. Each TCRβ clonotype identified from the 58 post-vaccinated samples (∼576,000) was analyzed using a one-tailed Fisher’s exact test for association with vaccinated libraries compared with naive libraries (Figure 2C). Vaccine-associated TCRβ libraries were designed at various p values to determine a threshold to appropriately filter the virus-associated TCRβ sequences for use in generating the diagnostic assay. Using a heuristic test, comparing the coverage (average number of vaccine-associated sequences present in a single sample) between vaccinated and naive mice, a significance threshold of 0.11 (by one-tailed Fisher’s exact test) was identified as the optimal exclusionary threshold (see STAR Methods). Using this threshold, 315 individual VATS were identified (Table S1).
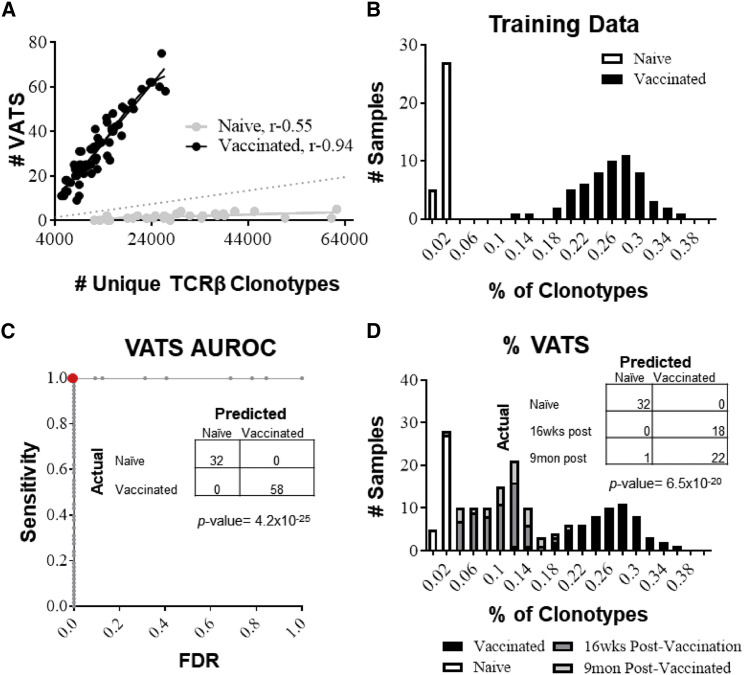
A diagnostic classifier was developed by calculating the number of VATS present relative to the total number of unique TCRβ clonotypes present for each sample. It was observed that the number of VATS present in a sample was significantly correlated with the total number of unique TCRβ clonotypes in both vaccinated and naive samples, indicating that the number of TCRβ clonotypes present directly affects the number of VATS identified (Figure 3 A). To normalize for differences in the number of TCRβ clonotypes identified in various samples, comparisons between naive and vaccinated samples were analyzed in the context of %VATS, the proportion of VATS in an individual sample. This calculation is displayed as a percentage of all unique TCRβ clonotypes:
A binary classification system was constructed to differentiate naive and vaccinated samples on the basis of the normal distribution of %VATS from the TCR repertoires of the naive or vaccinated groups. In this way, the TCR repertoires from the vaccinated and naive samples act as “training data,” teaching the diagnostic classifier to predict the vaccination status of samples (method described in STAR Methods). A comparison of naive and smallpox-vaccinated samples showed a 43-fold increase in the %VATS in vaccinated repertoires (average 0.248 ± 0.047%) compared with naive repertoires (average 0.0057 ± 0.0039%) (Figure 3B). The diagnostic classifier of 315 TCRβ sequences correctly classified 100% (32 of 32) of naive samples and 100% (58 of 58) of the vaccinated samples (Figure 3C). To determine whether the results of the training data were over-fitted from using the complete training dataset to inform the diagnostic classifier, an exhaustive leave-one-out (LOO) analysis was performed. All samples associated with an individual mouse (pre- and post-vaccination) were removed from the training data, and the VATS library was redefined for each individual mouse (average 308 ± 30 clonotypes per library). The same methodology described previously was used to train a diagnostic assay and test the accuracy of the classifier using the sample(s) associated with the mouse left out. Overall, the LOO analysis showed that the diagnostic assay correctly classified 94% of naive samples and 83% of vaccinated samples (Figure S1A). To confirm data from the LOO analysis, TCRβ repertoires from an independent cohort of HLA-A2 mice (n = 20) not used in the construction of the VATS library or training of the diagnostic classifier were analyzed before and after vaccination with the ACAM2000 smallpox vaccine. We show that the diagnostic classifier correctly identified 90% (18 of 20) of naive and 95% (19 of 20) of vaccinated samples, and analysis of %VATS reveals no significant difference between training and cross-validation samples (Figures S1B and S1C). Additionally, a cohort of HLA-A2 mice (n = 15) infected with an unrelated virus (Zika virus) were distinguished from the smallpox-vaccinated mice with 93% accuracy (data not shown). Overall, the training and cross-validation data from the LOO analysis closely resembles the results from the full-training set using the full VATS library of 315 TCRβ sequences.
Figure 3.
Distinguishing Naive from Vaccinated Samples Using the VATS Library
(A) Scatterplot depicting the number of vaccine-associated TCRβ sequences (VATS) present in a sample against the total number of unique TCRβ clonotypes present in vaccinated (black dots) versus naive (gray dots) repertoires. The r value represents the Pearson correlation.
(B) A bar graph displaying the distribution of %VATS in vaccinated (black bars) versus naive (white bars) samples.
(C) Receiver operating characteristic (ROC) graph illustrating the accuracy of the diagnostic classifier at various discrimination thresholds. The graph plots the sensitivity (true positives) against the false discovery rate (FDR; false positives) as the discrimination threshold varies. The area under the ROC curve (AUROC) is a representation of the overall accuracy. The red dot represents the calculated discrimination threshold. The data table within the ROC graph displays the actual identity of the sample(s) (rows) versus how the sample was classified by the diagnostic assay (columns). Thirty-two of 32 naive samples and all 58 of 58 vaccinated samples (p = 4.2 × 10−25) were correctly classified.
(D) Bar graph displaying the distribution of %VATS in mice 16 weeks (dark gray bars) and 9 months (light gray bars) after vaccination compared with naive (white bars) and vaccinated (black bars) training data. The data table displays the actual identity of samples versus the sample’s classification by the diagnostic assay. The assay correctly predicted 18 of 18 (100%) 16 week and 22 of 23 (96%) 9 month post-vaccination samples (p = 6.5 × 10−20).
To test whether the VATS diagnostic classifier was capable of identifying the vaccine-specific T cell response after the primary infection and generation of long-term memory, blood collected 16 weeks and 9 months after vaccination was analyzed. Sixteen week and 9-month post-vaccination samples were collected from the same mice 2 and 8 week samples used for the training data. TCRβ sequences from 16 week and 9 month post-vaccination samples were not used to generate the VATS library or as part of the training data. Enrichment of VATS was observed in the 16 week and 9 month post-vaccinated samples. The VATS library occupied on average 0.091 ± 0.019% and 0.105 ± 0.043% of TCRβ sequences from 16 week and 9 month post-vaccination samples, respectively, compared with naive repertoires (0.0057 ± 0.0039%). Compared with the determination threshold calculated by the diagnostic assay, 100% of 16 week (18 of 18) and 96% of 9 month (22 of 23) post-vaccinated samples were correctly differentiated from naive samples (Figure 3D). These data show that computational assessment of the TCRβ repertoire is capable of tracking the low-frequency virus-specific long-term memory population within the circulating T cell pool.
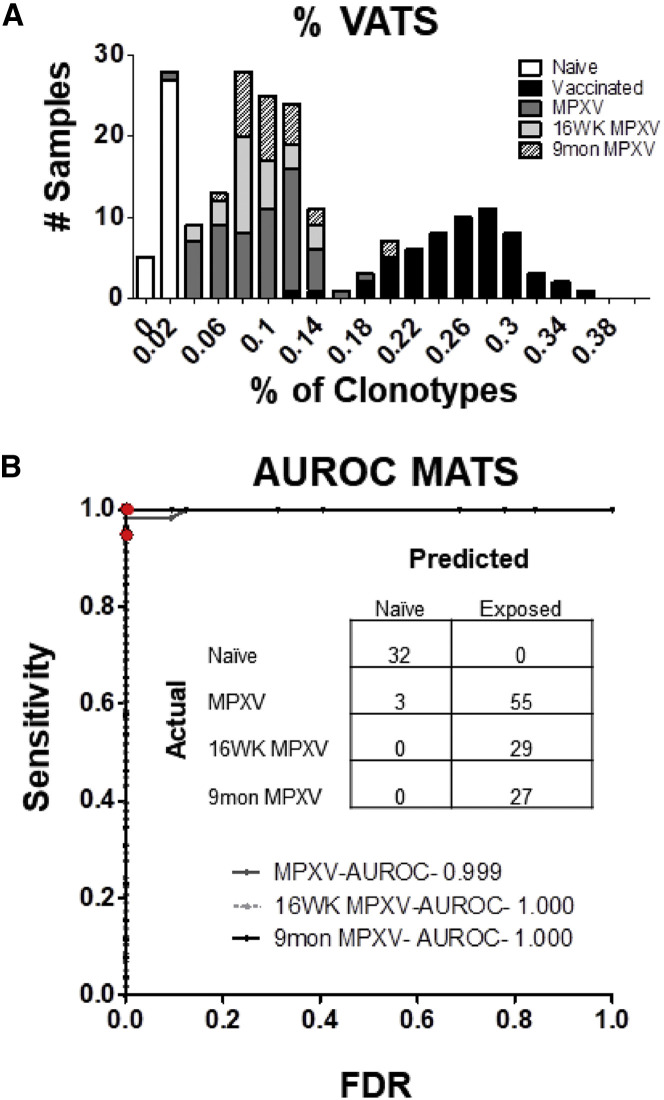
MPXV-Infected Mice Are Distinguished from Naive Samples Using VATS Library
The accuracy of the VATS library and diagnostic classifier was tested using an unrelated cohort of mice infected with a highly related Orthopoxvirus, MPXV. The percentage of sequences in the post-MPXV samples matching sequences from the VATS library was calculated to determine if the vaccine-associated diagnostic assay could distinguish between the naive and MPXV-infected samples. %VATS was significantly increased (p < 6.3 × 10−24) in samples from mice 2 and 8 weeks post-MPXV infection (0.084 ± 0.035%) compared with the 32 naive samples (0.0057 ± 0.0039%). Additionally, %VATS was significantly increased in samples from mice 16 weeks (0.08 ± 0.026%, p 1.7 × 10−15) and 9 months (0.097 ± 0.033%, p 1.5 × 10−13) after infection with MPXV compared with naive samples (Figure 4 A). The VATS library and diagnostic approach developed in the ACAM2000-vaccinated mice correctly distinguished 55 of 58 (95%) 2 and 8 week MPXV-infected mice from naive samples as well as 29 of 29 (100%) and 27 of 27 (100%) samples from mice 16 weeks and 9 months after MPXV infection (Figure 4B). The data show that the diagnostic assay is a robust platform, not only distinguishing ACAM2000-vaccinated mice from naive but also an independent cohort of mice infected with a highly related virus (95% identical) (Shchelkunov et al., 2002).
Figure 4.
VATS Diagnostic Classification of MPXV-Infected Mice
(A) Bar graph displaying the distribution of VATS of mice 2 and 8 weeks (dark gray bars; n = 58), 16 weeks (light gray bars; n = 29), and 9 months (shaded bars; 27 of 27) post-MPXV infection compared with the vaccinated (black bars) or naive (white bars) training data.
(B) ROC curve representing the overall accuracy of the diagnostic classifier to distinguish between naive samples and samples from mice infected with MPXV at the various time points post-infection. Overall, 55 of 58 (95%) of samples 2 and 8 weeks post-infection and 100% of samples 16 weeks (29 of 29) and 9 months (27 of 27) post-infection were correctly differentiated from naive samples.
Cross-Validation of Diagnostic Classification System through Identification of MPXV-Associated TCRβ Sequences
To determine whether the platform used to generate the VATS could be replicated independent of the ACAM2000 analysis, the same protocol was used with the TCRβ sequences identified in the MPXV-infected mice to generate a separate library of MPXV-associated TCR sequences (MATS). A total of 120 MATS were identified (Table S2). Using the same diagnostic approach implemented with the VATS library, a diagnostic classifier using the MATS library was generated. The proportion of a samples unique TCRβ clonotypes occupied by MATS (%MATS) was calculated and used to distinguish between naive and infected or vaccinated samples. Compared with naive samples (0.0009 ± 0.0018%), there were significant increases in the %MATS of MPXV-infected samples (0.114 ± 0.037%, 126.7-fold increase) and ACAM2000-vaccinated samples (0.036% ± 0.02%, 40-fold increase) (Figure S2A). The diagnostic assay using the MATS library correctly classified 97% of the naive samples (31 of 32), 100% of the MPXV-infected samples (58 of 58), and 97% of the ACAM2000 smallpox-vaccinated (56 of 58) samples (Figure S2B). With these data, the methodology for producing a diagnostic assay capable of distinguishing between naive and MPXV- or vaccine-exposed samples with a high degree of accuracy was replicated using a large independent cohort of mice. This confirms that construction of pathogen-associated TCR libraries as diagnostic assays can be used as a highly robust, accurate, and reproducible methodology for the identification and tracking of vaccine- and pathogen-specific T cell populations.
The VATS Library Contains Virus-Specific TCR Sequences
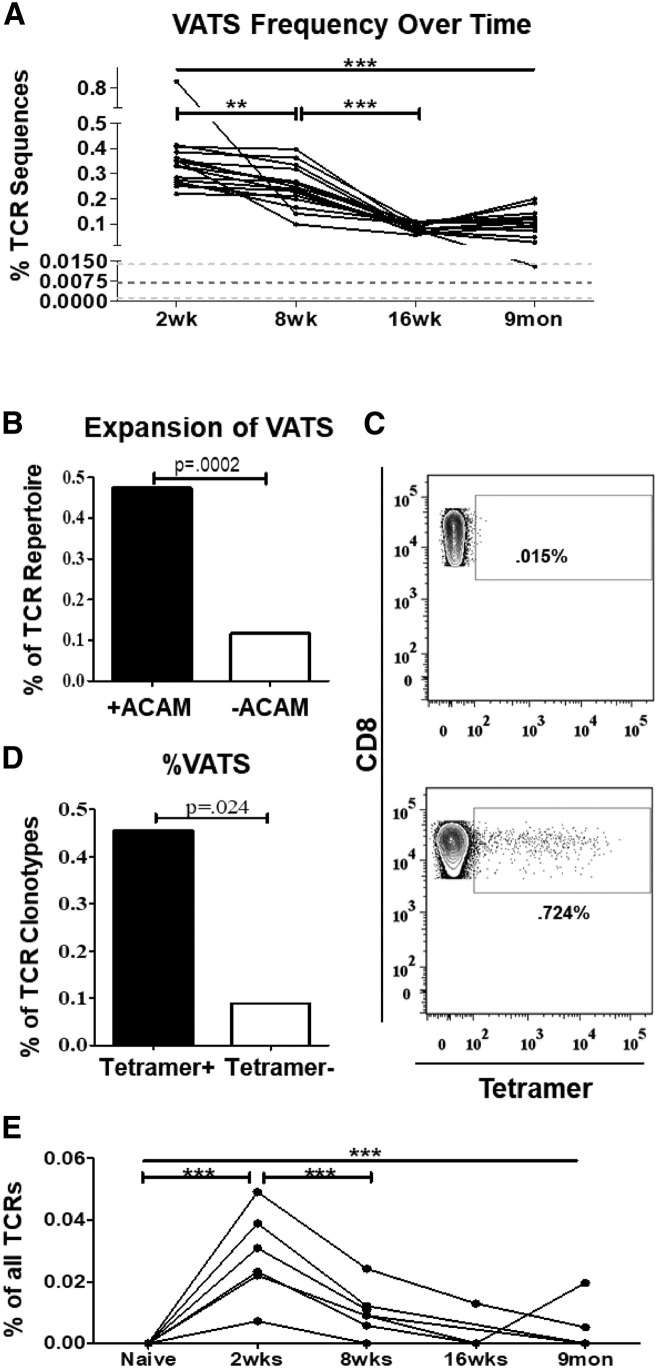
The diagnostic assay has shown the ability to monitor and track the presence of sequences from the VATS library over time in vaccinated or infected mice. Using sequence analyses, the relative frequencies of TCR sequences from the VATS library within the circulating T cell repertoire was determined. The frequencies of VATS sequences significantly decrease in mice over time from 2 weeks (0.35 ± 0.13%) through 9 months (0.11 ± 0.05%) after exposure (Figure 5 A). The decrease in frequency of VATS sequences over time is consistent with that of an antigen-specific response, displaying virus-specific T cells at higher frequencies early after vaccination (2 weeks) before declining in frequency and remaining as long-lived memory T cells (8 weeks, 16 weeks, and 9 months) (Figure 5E). Previously in Figure 2, it was shown that in vitro stimulation of splenocytes from vaccinated mice with the ACAM2000 smallpox vaccine resulted in preferential expansion of TCR clonotypes associated with post-vaccinated samples. To determine if the VATS library contained virus-specific TCRs, splenocytes from vaccinated mice were cultured in vitro with or without ACAM2000 for 5 days to induce expansion of vaccine-specific T cells. The TCR repertoire from T cells cultured with ACAM2000 was analyzed for the expansion of sequences included in the VATS library compared with the untreated control. The frequencies of VATS were 4-fold higher after culture with ACAM2000, representing ∼0.47% of all T cells sequenced compared with 0.12% in unstimulated controls (Figure 5B). These data confirm that TCRs recognizing smallpox vaccine antigens are present in the VATS library.
Figure 5.
The VATS Library Is Representative of the Functional Virus-Specific Response
(A) The frequency of VATS in mice vaccinated with the ACAM2000 smallpox vaccine over time. Each line represents the summed frequency of VATS in a single mouse from 2 weeks to 9 months post-vaccination. Dark dotted line represents the mean frequency of VATS in naive TCR repertoires ± SD (light dotted lines). Significance (p < 0.0001) was determined using one-way ANOVA testing with Bonferroni’s multiple comparison test.
(B) Graphical representation of the expansion of VATS after in vitro culture with (black bar) or without (white bar) ACAM2000. Represented as the summed frequency (percentage of all TCRβ sequences) of VATS. Significance was calculated using chi-square test with Yates’s correction.
(C) Representative flow plots displaying irrelevant tetramer (top) and nine pooled HLA-A2 tetramers loaded with vaccinia-specific peptides (bottom) binding to CD8+ T cells.
(D) Bar graph displaying the proportion of tetramer− or tetramer+ sequences that were included in the VATS library. p value calculated using a two-tailed Fisher’s exact test.
(E) Summed frequency of tetramer+ VATS in ACAM2000-vaccinated mice over time. Each line represents the frequency of the tetramer+ VATS in an individual mouse from prior to vaccination through 9 months post-vaccination. Significance (p < 0.0001) was determined using one-way ANOVA testing with Bonferroni’s multiple-comparison test.
TCR sequences identified in the VATS library were examined for sequences specific for known HLA-A2 epitopes previously identified in VACV-immune humans and HLA-A2 transgenic mice (Gilchuk et al., 2013). HLA-A2 tetramers loaded with nine different vaccinia peptides were used to identify and isolate HLA-A2-restricted vaccinia-specific T cells (Table 2 ). The vaccinia peptides loaded onto HLA-A2 tetramers had been previously shown by Gilchuk et al. (2013) to elicit strong CD8+ T cells responses in HLA-A2 transgenic mice. Mice approximately 6 months post-vaccination were boosted with ACAM2000; after 4 days, splenocytes were isolated, and tetramer-binding CD8+ T cells were isolated using fluorescence-activated cell sorting (FACS) with pooled tetramers (Figure 5C). TCRβ sequences from tetramer sorted cells were compared with the VATS library to determine if the VATS library contained HLA-A2 vaccine-specific sequences. Overall, tetramer+ sorted T cells were enriched for VATS sequences, representing 0.46% of all TCRβ clonotypes (3 of 658) compared with 0.09% of clonotypes (51 of 56,772) in the tetramer- T cell population (Figure 5D). The frequencies of the HLA-A2 tetramer+ VATS were tracked over time in vaccinated mice from before vaccination through 9 months after exposure. Significant increases were observed in the frequencies of the tetramer+ VATS 2 weeks after vaccination before decreasing in subsequent time points (Figure 5E). This is a clear representation of the antigen-specific response to the vaccine, from the expansion of virus-specific T cells during primary exposure to the induction of the low-frequency memory T cells that remain in circulation. Despite being present at low frequencies in the circulation (<1:50,000), by identifying the VATS library, it is possible to identify the virus-specific sequences from a very small volume of blood. Overall, these data confirm the computational identification of the virus-specific response and that virus-specific TCRβ sequences can be readily identified even at very low frequencies within the circulating memory T cell population over long periods of time.
Table 2.
HLA-A2.1 Vaccinia-Specific Peptides
| Peptides | ORF |
|---|---|
| RLYDYFTRV | I1L211–219 |
| ILDDNLYKV | G5R18–26 |
| ALDEKLFLI | A23R273–281 |
| GLFDFVNFV | A46R142–150 |
| KVDDTFYYV | C7L74–82 |
| RVYEALYYV | D12L251–259 |
| KIDDMIEEV | C9L602–610 |
| SLSNLDFRL | F11L340–348 |
| ILSDENYLL | A6L171–179 |
Previously published HLA-A2 restricted peptides and corresponding open reading frames.
Discussion
In this study, high-throughput TCR repertoire analyses from a large cohort of mice (n = 58) were used to identify and track TCR sequences responding to either the ACAM2000 smallpox vaccine or infection with MPXV. In total, >2.8 × 106 unique TCRβ clonotypes were analyzed from 245 individual blood samples collected before and after exposure. Data from mice administered the ACAM2000 smallpox vaccine were used to identify a library of 315 VATS. The VATS library acted as a diagnostic classifier, differentiating between naive and vaccinated or infected mice on the basis of the presence or absence of the public TCRβ sequences. The VATS library correctly identified samples from mice vaccinated with the smallpox vaccine and infected with MPXV from 2 weeks up to 9 months post-vaccination or infection. Overall, the diagnostic classifier was capable of distinguishing between vaccinated or infected samples and naive repertoires with >95% accuracy, which was replicated with MPXV-infected mice and the generation of the MATS library.
It was confirmed that the VATS library represented the public vaccine-specific T cell population by comparing the VATS library with TCRs expanded after in vitro culture with ACAM2000 and from vaccinia-specific HLA-A2.1 tetramer sorted T cells. The overlap between the TCR repertoires identified by in vitro expansion or tetramer sorting and the VATS library was limited. This was expected given the large number of immune-recognized Orthopoxvirus epitopes, various MHC molecules in mice, and that the majority of antigen-specific TCR clonotypes are private (specific to the individual mouse). This was previously shown in the human TCR repertoire, analyzing HLA-A2-restricted CD8+ T cells recognizing Epstein-Barr virus- and CMV-specific epitopes (Venturi et al., 2008a). Although only a small number of VATS sequences were found in the tetramer+ TCRβ repertoire, those sequences could be readily tracked and identified in mice up to 9 months after vaccination. Thus, this approach allowed the development of a tool to follow the virus-specific response over sequential time points in an aging population of mice. The frequencies at which the tetramer+ VATS were found in the circulation by TCR sequencing were as low as 1:50,000, meaning that using the VATS library to probe the TCRβ repertoire can be more sensitive than other technologies such as tetramer staining, intracellular cytokine staining, or allele-specific oligonucleotide PCR (Campana, 2010, Faham et al., 2012, Wolf and DiPaolo, 2016, van der Velden et al., 2014, van der Velden and van Dongen, 2009).
The low-frequency virus-specific memory response previously could only be readily measured through immune assays screening for serum antibodies against vaccinia and other pox viruses, which have historically been shown to be a very powerful and accurate tool for determining an individual’s prior exposure to Orthopoxvirus (Frey et al., 2003, Newman et al., 2003, Yin et al., 2013). However, we have shown that immunosequencing of the TCRβ chain can differentiate between naive and vaccinated or infected individuals with approximately the same level of accuracy (Hammarlund et al., 2005). Considering the nature of the two methods, there are some key differences between immunosequencing of the TCR repertoire and serum antibody profiling. A major difference is that immunosequencing relies on genomic DNA or cDNA. In areas of the world where resources are limited, the molecular stability and shelf life of DNA compared with serum and antibodies offers significant benefit when collecting and transporting samples. Additionally, once the appropriate pathogen-associated TCR sequence libraries are developed, an individual’s TCR repertoire could easily be used to determine prior exposure to a host of different pathogens with virtually no increase in effort, whereas testing serum for antibodies would require multiple tests for subsequent infectious agents (Emerson and DeWitt, 2017). Although the initial effort of collecting and analyzing large training cohorts for each target pathogen may be substantial, as sequencing data from subsequent studies become available, the resources required to produce large datasets becomes less (Emerson and DeWitt, 2017). Immunosequencing data, when published, are archived in public databases. This allows researchers to use previously published TCR repertoire data to increase diagnostic power while lowering the resources required to achieve large sample sizes (DeWitt et al., 2016).
Using this technology, it may be possible to develop panels of TCR sequence libraries capable of determining individuals’ prior exposure to multiple pathogens simultaneously. The ability to discern an individual’s immunological history has significant clinical benefits. Additionally, it is known that multiple viruses are able to undergo rapid mutation to evade immune detection. Analysis of the TCR repertoire could potentially be used in an attempt to track viral variants. However, we recognize that there are significant challenges involved in recapitulating this approach in human populations. Compared with genetically identical mice, humans display significant diversity in their HLA haplotypes, including rare HLAs, and TCR repertoires will likely limit the detection of public TCRs. Additionally, human populations are under constant exposure to different commensals, pathogens, and environmental stimuli, which can make identifying TCR sequences recognizing specific pathogens significantly more difficult. However, by acquiring larger blood volumes (5 mL in human versus 100 μL in mouse) and performing sequencing at greater depths, generating TCR repertoires of hundreds of thousands of clonotypes per sample, it is possible to perform similar studies in humans. This has been shown to be possible using populations of CMV+ and CMV− human populations (Emerson and DeWitt, 2017). Future studies are focused on adapting the current methodology to determine prior pathogen-specific exposure in human populations.
In summary, we have demonstrated that immunosequencing is a powerful and highly versatile tool for analyzing the TCR repertoire. In this study, an extensive database of TCRβ sequences was generated from the circulating TCR repertoires from a large cohort of mice (n = 58) and used to identify and track the vaccine-specific T cell response over time. This allowed a comprehensive analysis of Orthopoxvirus-associated TCR sequences and shows that analyses of TCRβ repertoires can be used to determine individuals’ prior exposure to ACAM2000 or MPXV with a high degree of accuracy and is capable of tracking the virus-specific populations present at ultra-low frequencies long after primary exposure resolved.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse IgG-Peroxidase | Sigma-Aldrich | RRID: AB_258426 |
| anti-mouse CD4 PerCP-efluor710 (Clone GK1.5) | ebioscience | RRID: AB_11149869 |
| anti-mouse CD8 BV450 (Clone 53-6.7) | BD Horizon | RRID: AB_1645281 |
| anti-mouse CD19 BV605 (clone ID3) | BD Horizon | RRID: AB_2732057 |
| Bacterial and Virus Strains | ||
| ACAM2000 smallpox vaccine | CDC | NA |
| MPXV ZAI-79 | Stabenow et. al. (2010) | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| HLA-A2.1 Tetramer-PE | Gilchuk et. al. (2013) | NA |
| ACK Lysis Buffer | Lonza | www.lonza.com |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# N4637 |
| RPMI-1640 | Fisher | Cat# MT15040CV |
| Critical Commercial Assays | ||
| QIAGEN Blood and Tissue Kit | QIAGEN | Cat # 69506 |
| ImmunoSeq | Adaptive Biotechnologies | RRID:SCR_014709 |
| Deposited Data | ||
| High-Throughput TCRβ data | This paper. Mendeley Data | https://doi.org/10.17632/cf92gt44zf.1 |
| Experimental Models: Cell Lines | ||
| BSC-1 cells | ATCC | RRID:CVCL_0607 |
| Experimental Models: Organisms/Strains | ||
| Mouse: AAD C57BL/6J | The Jackson Laboratory | RRID:IMSR_JAX:004191 |
| Software and Algorithms | ||
| FlowJo 7.5 | TreeStar Inc. | RRID:SCR_008520 |
| GraphPad Prism 5.0 | GraphPad Software, Inc. | RRID:SCR_002798 |
| ImmunoSeq Analyzer | Adaptive Biotechnologies | RRID:SCR_014709 |
| Other | ||
| BD FACSAria II cell sorter | BD Biosciences | NA |
| ImmunoSeq System | Illumina | NA |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Richard DiPaolo (richard.dipaolo@health.slu.edu).
Experimental Model and Subject Details
Mice
HLA-A2.1 AAD C57BL/6 male and female mice were purchased from Jackson Laboratories and maintained under specific pathogen conditions (Gilchuk et al., 2013). The HLA-A2.1 AAD mice express a transgenic HLA-A2.1 chimeric molecule containing the human β-2 microglobulin and HLA-A2.1 α1 and α2 domains with a mouse α3 and transmembrane domain (AAD HLA-A2). Mice entered the study at approximately 6-8 weeks of age. Male mice weighed between 18 and 23 g, female mice weighed between 16-21 g. Mice were housed in groups of 3 to 5 mice per cage. All animal work has been conducted in accordance with the Guide for Care and Use of Laboratory Animals of the National Institute of Health with approval from the Saint Louis University Institutional Animal Care and Use Committee.
Virus
The ACAM2000 (Acambis, Inc.) smallpox vaccine is a live virus derived from the original Dryvax (Wyeth Laboratories, Inc.). MPXV is a member of the Orthopoxvirus family and is 95% genetically identical to the smallpox vaccine. Vaccination with the smallpox vaccine confers protection against MPXV infection (Handley et al., 2009, Parker and Buller, 2013). MPXV Zaire 79 was obtained from the Saint Louis University School of Medicine, department of molecular microbiology and immunology Biosafety Level 3/Select Agent program. ACAM2000 smallpox vaccine was a gift from the center for disease control (Atlanta, GA). Both MPXV and ACAM2000 smallpox vaccine titers and infectivity were estimated plaque forming assay (Handley et al., 2009, Parker et al., 2014). 1x105 BSC-1 cells were plated in DMEM+ 10%FCS in 24-well plates at 0.5mL final volume and cultured for 24 hours at 37°C and 5% CO2. Stock viral suspensions were serially diluted 1:10 in DMEM+ 1%FCS, and 300uL of supernatant from BSC-1 cells was removed. 100uL of diluted virus solutions were added to BSC-1 cells in triplicate, gently swirled, and incubated for 1 hour at 37°C and 5% CO2. Overlay media (1% carboxyl methyl cellulose in DMEM with 5% FCS) was warmed to 37°C and 1mL added to each well and cultures were returned to the incubator. Cultures were maintained for 3-4 days at 37°C until plaques were visible. Plaques were visualized and virus inactivated with 200uL 0.3% crystal violet/10% formalin solution for 1 hour. All liquid was aspirated and plates were washed with water, inverted, and dried. Plaques were counted for each viral dilution, the average plaque count is divided by product of the dilution and volume of virus overlay.
Primary T cell cultures
Spleens from male and female mice previously vaccinated with the ACAM2000 smallpox vaccine were mechanically disrupted to form single cell suspensions. Cells were filtered through a 40um nylon mesh cell strainer and washed with complete RPMI supplemented with 10% fetal bovine serum (cRPMI10F, 1% Penicillin/Streptomycin (Sigma-Aldrich, P0871, 10,000U/10mg per mL), 1% L-glutamine (Sigma-Aldrich, G7513, 200mM), 1% non-essential amino acids (Sigma-Aldrich, M7145, 100x), 1% HEPES (Sigma-Aldrich, H3537), 1% sodium pyruvate (Sigma-Aldrich, S8636, 100mM)). Red blood cells were lysed by incubation with 5mL 1x ACK (ammonium-chloride-potassium) lysing buffer for 5 minutes at 37°C and 5% CO2. Cells were washed with cRPMI10F and cell count determined. Splenocytes were cultured in cRPMI10F at a concentration of 1x106/mL at 37°C and 5% CO2.
Method Details
Vaccination/Infection of HLA-A2 Mice
At 6-8 weeks of age blood mice were anesthetized by intraperitoneal injection with 0.01mL/g body weight Ketamine (6mg/mL)/Xylazine (0.5mg/mL) cocktail and intranasally administered sub-lethal doses of either the ACAM2000 smallpox vaccine (∼5x104 PFU) or MPXV Zaire-79 strain (0.5x104 PFU) in 25uL total volume (12.5uL per naris) (Moutaftsi et al., 2009, Parker and Buller, 2013, Parker et al., 2014, Stabenow et al., 2010). Male and female mice were entered into each treatment group with approximate equality by cage. All mice recovered from the viral challenge. Blood samples were collected via the submandibular cheek bleed 1 week prior to vaccination or infection and at 2 weeks, 8 weeks, 16 weeks, and 9 months post-vaccination or post-infection.
Anti-pox Serum ELISA
Serum from HLA-A2 mice was collected by aspiration from whole blood prior to vaccination or infection and at 2-weeks and 8-weeks after viral exposure by centrifugation at 11,000 g for 5min. Samples were tested for vaccinia-specific serum antibody by neutralizing antibody ELISA(Frey et al., 2002). 96-well maxSorp plates (ThermoFisher 44-2404-21) were coated with crude extract from lysed BSC-1 cells infected with live ACAM2000 smallpox vaccine. Plates were washed 3x with PBS and wells were coated with ∼5x104 PFU live ACAM2000 diluted in carbonate coating buffer (0.1M Na2CO3 0.1M NaHCO3 pH 9.3) in 100ul overnight at 4°C. Wells were washed and blocked with blocking buffer containing 5% BSA in PBS for 30min at RT. Plates were washed 3x with PBS and serum samples diluted 1:20 in blocking buffer were added (in duplicate) to wells and incubated for 2 hours at RT. Plate was washed 3x with PBS, then 100uL anti-mouse HRP-conjugated antibody (Sigma A8924) was diluted 1:2500 in blocking buffer was added and allowed to incubate for 1h. Plates were washed 3x with PBS and 75ul of True Blue peroxidase substrate (KPL, 71-00-65) was added to each well and incubated for 15min in the dark at RT. 75ul of 1N HCl was added to each well and OD measured at 450nm.
Sample preparation and DNA Sequencing
Genomic DNA was extracted and purified using the QIAGEN Blood and Tissue Kit (item # 69506). Genomic DNA was amplified using multiplexed primers targeting all V and J gene segments by (Carlson et al., 2013). tcrb CDR3 regions were amplified and sequenced using ImmunoSEQ (Adaptive Biotechnologies). Synthetic templates mimicking natural V(D)J rearrangements were used to measure and correct potential amplification bias (Carlson et al., 2013, Wolf et al., 2016). CDR3 segments were annotated according to the International ImMunoGeneTics (IMGT) collaboration (Yousfi Monod et al., 2004), identifying V, D, and J genes contributing to each rearrangement.
T cell Expansion Assay
Cells were maintained in cRPMI10F media alone or supplemented with 0.2MOI live ACAM2000 smallpox vaccine and allowed to incubate at 37°C for 5 days. T cell blasting and proliferation were observed prior to DNA extraction for Immunosequencing. Cultured cells were pelleted by centrifugation at 1500rpm for 10 minutes and supernatant aspirated. Pelleted cells were re-suspended in 200ul PBS prior to DNA extraction.
Flow Cytometry and HLA-A2 Tetramer Sorting
PE-conjugated HLA-A2.1 chimeric tetramers (HLA-A2 tetramers) loaded with vaccinia-derived peptides (Gilchuk et al., 2013) were a kind gift from Dr. Sebastian Joyce (Vanderbilt). Pooled splenocytes from previously vaccinated mice were stained with T cell and B cell lineage markers CD4 (ebioscience, PerCP-efluor710,clone GK1.5), CD8 (BD Horizon, BV450, clone 53-6.7), and CD19 (BD Horizon, BV605, clone ID3) for 20min at 4°C. Cells were washed 2x in PBS+2% FBS, then cells were incubated with vaccinia-peptide loaded HLA-A2 tetramers (25ug/mL, 2uL per 1x106 splenocytes) for 1h at RT. TCR-epitope-tetramer binding CD4-CD19-CD8+ T cell populations were purified by FACS into tetramer- and TCR-epitope-tetramer binding tetramer+ populations. Tetramer sorted T cells were centrifuged at 1500rpm for 10 minutes. Supernatant was aspirated and cells re-suspended in 200ul PBS prior to DNA extraction.
Quantification and Statistical Analysis
VATS and MATS Library Development
Alignment of shared and non-shared TCRβ sequences was completed using ImmunoSEQ software provided by Adaptive Biotechnologies. Alignments of all 2-week and 8-week post-ACAM2000 smallpox vaccination TCR repertoires were used to identify public TCR sequences. The list of public TCR sequences was compared to alignments of all TCRβ repertoires from naive samples in order to perform an association analysis to identify a set of TCRβ sequences that had significantly increased incidence among vaccinated but not naive TCRβ repertoires. For the association analysis, we performed a one-tailed Fisher’s Exact test on all sequences, comparing the number of naive and vaccinated samples each TCRβ sequence was present in.
To determine an optimal p value threshold for identifying VATS, we applied a heuristic test that selected the optimal p value threshold based on the “coverage” provided by the library for both vaccinated (Cv) and naive samples (Cn). “Coverage” is defined as the summation of the number of samples containing each VATS divided by the number of samples. In the equations below, xi denotes the number of vaccinated samples a single TCRβ is identified in (yi denotes naive samples) and nv represents the number of samples in the training data (nn represents naive samples).
The ratio of Cv to Cn is determined for each p value. Additionally, the Cv and Cn of each p value (rounded to the nearest whole integer), are applied to a one-tailed Fisher’s Exact test against the total number of sequences in the prospective library to determine if there is sufficient coverage to distinguish vaccinated from naive samples (p < 0.05).The p value with the largest Cv:Cn ratio and offers significant coverage to distinguish vaccinated from naive samples was chosen.
Classification of vaccinated or MPXV exposed versus naive samples
To distinguish between vaccinated and naive samples, the proportion of VATS present in a sample was compared against the normal distribution of the naive and vaccinated training data. Normal distribution for our purposes is used to measure the distance a sample is from the mean. The normal distributions for the naive and vaccinated populations in our training data were calculated based on a function of the difference between a single sample value (x) and the mean of a set of data (μ) over the standard deviation of that set of data (σ). The greater the value, the greater association that sample has with the training group. By comparing a sample against the normal distribution of vaccinated and naive training groups, we can determine which group a sample is more statistically associated with.
The Leave-one-out (LOO) analysis was completed as previously described (Emerson and DeWitt, 2017). Briefly, all samples associated with a single mouse were removed from the training data and the VATS library was re-derived using the remaining training cohort. The % VATS was calculated for all samples and used to train the diagnostic classifier.
Statistical analyses
Alignment of shared and non-shared TCRs was completed using ImmunoSeq software provided by Adaptive Biotechnologies. Graphical analyses were created using GraphPad Prism 5.0. 1-way ANOVA and Bonferroni’s multiple comparison test was accomplished using GraphPad Prism 5.0. Pearson correlation was calculated using GraphPad Prism 5.0.
Data and Software Availability
TCR Sequencing Repertoires: https://doi.org/10.17632/cf92gt44zf.1
Acknowledgments
We would like to thank Sherri Koehm and Joy Eslick of the Saint Louis University School of Medicine flow cytometry core for their efforts in purifying tetramer-bound T cells, Dr. Amelia Pinto and James Brian of Saint Louis University for their assistance with handling MPXV, and the Department of Comparative Medicine at the Saint Louis University School of Medicine for assisting with mouse care after infection and vaccination. Finally, we thank Dr. Bruce Budowle for critiques and peer review of the manuscript. Funding was provided through a research contract from the Federal Bureau of Investigation to R.J.D., and S.J. was supported by a U.S. Department of Veterans Affairs (VA) merit award (BX001444).
Author Contributions
K.W. jointly conceived the study with R.M.B., R.J.D., and J.M., performed all experiments, and prepared the initial draft of the manuscript. T.H. and K.W. performed data analyses. T.H. assisted with manuscript preparation. A.R., C.S., and T.-H.A. assisted in the design of the diagnostic classifier and LOO analysis. R.M.B. was instrumental in managing access to select agents (MPXV). P.G., A.K., and S.J. constructed and provided HLA-A2 tetramers bound with vaccinia-specific peptide and assisted with manuscript preparation.
Declaration of Interests
T.H. has full-time employment and equity ownership at Adaptive Biotechnologies Corporation. There are no patents, products in development, or marketed products to declare. This does not alter the authors’ adherence to all Cell Reports policies on sharing data and materials.
Published: November 27, 2018
Footnotes
Supplemental Information includes two figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.009.
Supplemental Information
References
- Arstila T.P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286:958–961. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- Cabaniols J.P., Fazilleau N., Casrouge A., Kourilsky P., Kanellopoulos J.M. Most alpha/beta T cell receptor diversity is due to terminal deoxynucleotidyl transferase. J. Exp. Med. 2001;194:1385–1390. doi: 10.1084/jem.194.9.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campana D. Minimal residual disease in acute lymphoblastic leukemia. Hematology Am. Soc. Hematol. Educ. Program. 2010;2010:7–12. doi: 10.1182/asheducation-2010.1.7. [DOI] [PubMed] [Google Scholar]
- Carlson C.S., Emerson R.O., Sherwood A.M., Desmarais C., Chung M.W., Parsons J.M., Steen M.S., LaMadrid-Herrmannsfeldt M.A., Williamson D.W., Livingston R.J., et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat. Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
- Chen W., Antón L.C., Bennink J.R., Yewdell J.W. Dissecting the multifactorial causes of immunodominance in class I-restricted T cell responses to viruses. Immunity. 2000;12:83–93. doi: 10.1016/s1074-7613(00)80161-2. [DOI] [PubMed] [Google Scholar]
- Davis M.M., Bjorkman P.J. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- DeWitt W.S., Emerson R.O., Lindau P., Vignali M., Snyder T.M., Desmarais C., Sanders C., Utsugi H., Warren E.H., McElrath J., et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J. Virol. 2015;89:4517–4526. doi: 10.1128/JVI.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt W.S., Lindau P., Snyder T.M., Sherwood A.M., Vignali M., Carlson C.S., Greenberg P.D., Duerkopp N., Emerson R.O., Robins H.S. A public database of memory and naive B-Cell receptor sequences. PLoS ONE. 2016;11:e0160853. doi: 10.1371/journal.pone.0160853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douillard P., Josien R., Pannetier C., Menoret S., Kourilsky P., Soulillou J.P., Cuturi M.C. TCR V beta repertoire in LEW.1W heart allografts acutely rejected by LEW.1A recipients is restricted and comprises a public response. Transplant. Proc. 1997;29:1054. doi: 10.1016/s0041-1345(96)00637-9. [DOI] [PubMed] [Google Scholar]
- Elhanati Y., Murugan A., Callan C.G., Jr., Mora T., Walczak A.M. Quantifying selection in immune receptor repertoires. Proc. Natl. Acad. Sci. U S A. 2014;111:9875–9880. doi: 10.1073/pnas.1409572111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson R.O., DeWitt W.S. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat. Genet. 2017;49:659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- Estorninho M., Gibson V.B., Kronenberg-Versteeg D., Liu Y.F., Ni C., Cerosaletti K., Peakman M. A novel approach to tracking antigen-experienced CD4 T cells into functional compartments via tandem deep and shallow TCR clonotyping. J. Immunol. 2013;191:5430–5440. doi: 10.4049/jimmunol.1300622. [DOI] [PubMed] [Google Scholar]
- Faham M., Zheng J., Moorhead M., Carlton V.E., Stow P., Coustan-Smith E., Pui C.H., Campana D. Deep-sequencing approach for minimal residual disease detection in acute lymphoblastic leukemia. Blood. 2012;120:5173–5180. doi: 10.1182/blood-2012-07-444042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S.E., Newman F.K., Cruz J., Shelton W.B., Tennant J.M., Polach T., Rothman A.L., Kennedy J.S., Wolff M., Belshe R.B., Ennis F.A. Dose-related effects of smallpox vaccine. N. Engl. J. Med. 2002;346:1275–1280. doi: 10.1056/NEJMoa013431. [DOI] [PubMed] [Google Scholar]
- Frey S.E., Newman F.K., Yan L., Lottenbach K.R., Belshe R.B. Response to smallpox vaccine in persons immunized in the distant past. JAMA. 2003;289:3295–3299. doi: 10.1001/jama.289.24.3295. [DOI] [PubMed] [Google Scholar]
- Gilchuk P., Spencer C.T., Conant S.B., Hill T., Gray J.J., Niu X., Zheng M., Erickson J.J., Boyd K.L., McAfee K.J., et al. Discovering naturally processed antigenic determinants that confer protective T cell immunity. J. Clin. Invest. 2013;123:1976–1987. doi: 10.1172/JCI67388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund E., Lewis M.W., Carter S.V., Amanna I., Hansen S.G., Strelow L.I., Wong S.W., Yoshihara P., Hanifin J.M., Slifka M.K. Multiple diagnostic techniques identify previously vaccinated individuals with protective immunity against monkeypox. Nat. Med. 2005;11:1005–1011. doi: 10.1038/nm1273. [DOI] [PubMed] [Google Scholar]
- Hancock G., Yang H., Yorke E., Wainwright E., Bourne V., Frisbee A., Payne T.L., Berrong M., Ferrari G., Chopera D., et al. Identification of effective subdominant anti-HIV-1 CD8+ T cells within entire post-infection and post-vaccination immune responses. PLoS Pathog. 2015;11:e1004658. doi: 10.1371/journal.ppat.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley L., Buller R.M., Frey S.E., Bellone C., Parker S. The new ACAM2000 vaccine and other therapies to control orthopoxvirus outbreaks and bioterror attacks. Expert Rev. Vaccines. 2009;8:841–850. doi: 10.1586/erv.09.55. [DOI] [PubMed] [Google Scholar]
- Heit A., Schmitz F., Gerdts S., Flach B., Moore M.S., Perkins J.A. Vaccination establishes clonal relatives of germinal center T cells in the blood of humans. J. Exp. Med. 2017;214:2139–2152. doi: 10.1084/jem.20161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka J., Grebe K., Shenderov E., Peters B., Chen Q., Peng Y., Wang L., Dong T., Pasquetto V., Oseroff C., et al. Quantitating T cell cross-reactivity for unrelated peptide antigens. J. Immunol. 2009;183:4337–4345. doi: 10.4049/jimmunol.0901607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Yewdell J.W., Sette A., Peters B. Positional bias of MHC class I restricted T-cell epitopes in viral antigens is likely due to a bias in conservation. PLoS Comput. Biol. 2013;9:e1002884. doi: 10.1371/journal.pcbi.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch I., Vignali M., Robins H. T-cell receptor profiling in cancer. Mol. Oncol. 2015;9:2063–2070. doi: 10.1016/j.molonc.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotturi M.F., Assarsson E., Peters B., Grey H., Oseroff C., Pasquetto V., Sette A. Of mice and humans: how good are HLA transgenic mice as a model of human immune responses? Immunome Res. 2009;5:3. doi: 10.1186/1745-7580-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ye C., Ji G., Han J. Determinants of public T cell responses. Cell Res. 2012;22:33–42. doi: 10.1038/cr.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A., Trautmann L., Peyrat M.A., Couedel C., Davodeau F., Romagné F., Kourilsky P., Bonneville M. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J. Immunol. 2000;165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- Logan A.C., Vashi N., Faham M., Carlton V., Kong K., Buno I., Zheng J., Moorhead M., Klinger M., Zhang B., et al. Immunoglobulin and T cell receptor gene high-throughput sequencing quantifies minimal residual disease in acute lymphoblastic leukemia and predicts post-transplantation relapse and survival. Biol. Blood Marrow Transplant. 2014;20:1307–1313. doi: 10.1016/j.bbmt.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossius A., Johansen J.N., Vartdal F., Robins H., Jūratė Šaltytė B., Holmøy T., Olweus J. High-throughput sequencing of TCR repertoires in multiple sclerosis reveals intrathecal enrichment of EBV-reactive CD8+ T cells. Eur. J. Immunol. 2014;44:3439–3452. doi: 10.1002/eji.201444662. [DOI] [PubMed] [Google Scholar]
- Manfras B.J., Terjung D., Boehm B.O. Non-productive human TCR beta chain genes represent V-D-J diversity before selection upon function: insight into biased usage of TCRBD and TCRBJ genes and diversity of CDR3 region length. Hum. Immunol. 1999;60:1090–1100. doi: 10.1016/s0198-8859(99)00099-3. [DOI] [PubMed] [Google Scholar]
- Marrero I., Aguilera C., Hamm D.E., Quinn A., Kumar V. High-throughput sequencing reveals restricted TCR Vβ usage and public TCRβ clonotypes among pancreatic lymph node memory CD4(+) T cells and their involvement in autoimmune diabetes. Mol. Immunol. 2016;74:82–95. doi: 10.1016/j.molimm.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., van der Most R.G., Akondy R.S., Glidewell J.T., Albott S., Masopust D., Murali-Krishna K., Mahar P.L., Edupuganti S., Lalor S., et al. Human effector and memory CD8+ T cell responses to smallpox and yellow fever vaccines. Immunity. 2008;28:710–722. doi: 10.1016/j.immuni.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Moutaftsi M., Salek-Ardakani S., Croft M., Peters B., Sidney J., Grey H., Sette A. Correlates of protection efficacy induced by vaccinia virus-specific CD8+ T-cell epitopes in the murine intranasal challenge model. Eur. J. Immunol. 2009;39:717–722. doi: 10.1002/eji.200838815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman F.K., Frey S.E., Blevins T.P., Mandava M., Bonifacio A., Jr., Yan L., Belshe R.B. Improved assay to detect neutralizing antibody following vaccination with diluted or undiluted vaccinia (Dryvax) vaccine. J. Clin. Microbiol. 2003;41:3154–3157. doi: 10.1128/JCM.41.7.3154-3157.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S., Buller R.M. A review of experimental and natural infections of animals with monkeypox virus between 1958 and 2012. Future Virol. 2013;8:129–157. doi: 10.2217/fvl.12.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker S., Crump R., Foster S., Hartzler H., Hembrador E., Lanier E.R., Painter G., Schriewer J., Trost L.C., Buller R.M. Co-administration of the broad-spectrum antiviral, brincidofovir (CMX001), with smallpox vaccine does not compromise vaccine protection in mice challenged with ectromelia virus. Antiviral Res. 2014;111:42–52. doi: 10.1016/j.antiviral.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H.S., Campregher P.V., Srivastava S.K., Wacher A., Turtle C.J., Kahsai O., Riddell S.R., Warren E.H., Carlson C.S. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H.S., Srivastava S.K., Campregher P.V., Turtle C.J., Andriesen J., Riddell S.R., Carlson C.S., Warren E.H. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci. Transl. Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S.N., Totmenin A.V., Safronov P.F., Mikheev M.V., Gutorov V.V., Ryazankina O.I., Petrov N.A., Babkin I.V., Uvarova E.A., Sandakhchiev L.S., et al. Analysis of the monkeypox virus genome. Virology. 2002;297:172–194. doi: 10.1006/viro.2002.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabenow J., Buller R.M., Schriewer J., West C., Sagartz J.E., Parker S. A mouse model of lethal infection for evaluating prophylactics and therapeutics against Monkeypox virus. J. Virol. 2010;84:3909–3920. doi: 10.1128/JVI.02012-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N., Best K., Cinelli M., Reich-Zeliger S., Gal H., Shifrut E., Madi A., Friedman N., Shawe-Taylor J., Chain B. Tracking global changes induced in the CD4 T-cell receptor repertoire by immunization with a complex antigen using short stretches of CDR3 protein sequence. Bioinformatics. 2014;30:3181–3188. doi: 10.1093/bioinformatics/btu523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg S.A., Josephs T.M., Clemens E.B., Grant E.J., Nguyen T.H., Wang G.C., Price D.A., Miller A., Tong S.Y., Thomas P.G., et al. Molecular basis for universal HLA-A∗0201-restricted CD8+ T-cell immunity against influenza viruses. Proc. Natl. Acad. Sci. U S A. 2016;113:4440–4445. doi: 10.1073/pnas.1603106113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden V.H., van Dongen J.J. MRD detection in acute lymphoblastic leukemia patients using Ig/TCR gene rearrangements as targets for real-time quantitative PCR. Methods Mol. Biol. 2009;538:115–150. doi: 10.1007/978-1-59745-418-6_7. [DOI] [PubMed] [Google Scholar]
- van der Velden V.H., Noordijk R., Brussee M., Hoogeveen P.G., Homburg C., de Haas V., van der Schoot C.E., van Dongen J.J. Minimal residual disease diagnostics in acute lymphoblastic leukaemia: impact of primer characteristics and size of junctional regions. Br. J. Haematol. 2014;164:451–453. doi: 10.1111/bjh.12621. [DOI] [PubMed] [Google Scholar]
- Venturi V., Chin H.Y., Asher T.E., Ladell K., Scheinberg P., Bornstein E., van Bockel D., Kelleher A.D., Douek D.C., Price D.A., Davenport M.P. TCR beta-chain sharing in human CD8+ T cell responses to cytomegalovirus and EBV. J. Immunol. 2008;181:7853–7862. doi: 10.4049/jimmunol.181.11.7853. [DOI] [PubMed] [Google Scholar]
- Venturi V., Price D.A., Douek D.C., Davenport M.P. The molecular basis for public T-cell responses? Nat. Rev. Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- Venturi V., Nzingha K., Amos T.G. The neonatal CD8+ T cell repertoire rapidly diversifies during persistent viral infection. J. Immunol. 2016;196:1604–1616. doi: 10.4049/jimmunol.1501867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K., DiPaolo R. Immunosequencing: accelerating discovery in immunology and medicine. Curr. Trends Immunol. 2016;17:85–93. [Google Scholar]
- Wolf K.J., Emerson R.O., Pingel J., Buller R.M., DiPaolo R.J. Conventional and regulatory CD4+ T cells that share identical TCRs are derived from common clones. PLoS ONE. 2016;11:e0153705. doi: 10.1371/journal.pone.0153705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Calvo-Calle J.M., Cruz J., Newman F.K., Frey S.E., Ennis F.A., Stern L.J. CD4+ T cells provide intermolecular help to generate robust antibody responses in vaccinia virus-vaccinated humans. J. Immunol. 2013;190:6023–6033. doi: 10.4049/jimmunol.1202523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousfi Monod M., Giudicelli V., Chaume D., Lefranc M.P. IMGT/JunctionAnalysis: the first tool for the analysis of the immunoglobulin and T cell receptor complex V-J and V-D-J JUNCTIONs. Bioinformatics. 2004;20(Suppl 1):i379–i385. doi: 10.1093/bioinformatics/bth945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCR Sequencing Repertoires: https://doi.org/10.17632/cf92gt44zf.1