Abstract
Background
Donor-derived, cell-free DNA (dd-cfDNA) level correlates with allograft injury with clinical validity and utility for quiescence and active acute rejection (AR) in kidney transplant recipients. We analyzed trends in dd-cfDNA level immediately preceding and during the coronavirus disease 2019 (COVID-19) pandemic with implemented “shelter in place” and a tele-health strategy with remote home phlebotomy to limit COVID-19 exposure.
Methods
During COVID-19 in the United States (US), we surveyed weekly (January 6, 2020-May 25, 2020) metrics for dd-cfDNA corresponding to both a low risk for active rejection (dd-cfDNA < 0.5%) and cohorts with indeterminate levels of 0.5% to 1.0% and > 1.0%. During the study timeframe, over 11,000 patient samples (67%) from 150 kidney transplantation centers were transitioned from standard facility-based to remote phlebotomy.
Results
The proportion of dd-cfDNA samples, analyzed in 21 weekly aggregated cohorts by risk-stratification category, was unchanged during the COVID-19 escalation in the US. Linearized slopes for numbers of samples corresponding to indeterminate risk for AR cohorts of > 1.0% and 0.5% to 1.0% were -0.31 and -0.12, respectively; indicating that prevalence of these “at risk for AR cohorts” decreased during remote surveillance. Approximately 73% of samples corresponded to low risk of AR (dd-cfDNA < 0.5%), while an additional 15% of samples had dd-cfDNA level ≤ 1.0%.
Conclusion
The combination of remote home phlebotomy including dd-cfDNA analysis and a tele-health program offer a new paradigm that may substantially improve patient compliance and assuage anxiety regarding the state of kidney allograft health during the COVID-19 pandemic. Further prospective multi-center studies with robust outcomes data are warranted.
Keywords: Biomarker, Cell-free nucleic acids, COVID-19, Donor-derived DNA, Kidney transplantation
Introduction
The coronavirus disease 2019 (COVID-19) pandemic has dramatically altered the transplant landscape in all spheres of the process, including both real and perceived risks confronted by transplant recipients seeking outpatient monitoring and allograft surveillance. The ongoing, if incompletely quantified, risk of COVID-19 infection posed to transplant recipients during otherwise innocuous visits for phlebotomy to monitor allograft status has fostered awareness of remote monitoring utilizing at-home phlebotomy as an exposure risk mitigation strategy. Yang et al [1] vividly described the clinical course and outcomes of an initial cohort of 710 critically ill COVID-19 patients hospitalized in Wuhan, China. Subsequently, as the viral pandemic unfolded in the United States (US), the potential impact on solid organ transplant recipients became undeniable. There are more than 220,000 kidney transplant recipients (KTRs) with functioning renal allograft in the US [2]. Those KTRs are at risk of community and nosocomial COVID-19 infection. Akalin et al [3] reported the course of 36 consecutive KTRs infected with COVID-19, finding that 78% required hospitalization, and 28% died of COVID-19 during the study follow-up period. Innovative means of allograft surveillance after kidney transplantation (KT) and implementing tele-health in concert with remote phlebotomy for routine and biomarker laboratory studies offer a new paradigm for monitoring allograft health. This paradigm decreases potential patient exposure to COVID-19 infection and improves recipient adherence to surveillance testing and provider encounters.
Donor-derived, cell-free DNA (dd-cfDNA) measurement as a biomarker after KT is well-established for surveillance and is reimbursed by Centers for Medicare and Medicaid Services based on demonstrated clinical validity and utility [4-6]. Elevation of dd-cfDNA associated with allograft tissue injury is associated with immunologic allograft events such as T-cell-mediated rejection (TCMR) and antibody-mediated rejection (ABMR) [4,7-9]. The pivotal validation trial for dd-cfDNA was the multi-center Circulating Donor-Derived, Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study (NCT02424227). In DART, assessment of dd-cfDNA by targeted next generation sequencing using a 0.21% threshold, which is the median dd-cfDNA of healthy and stable KTRs, the negative predictive value (NPV) for active rejection was 95%, while serum creatinine lacked sensitivity and specificity. Using a threshold of 1.0% dd-cfDNA, the positive predictive value was 61% and 44% for TCMR (Banff criteria TCMR ≥ 1B) [10] and ABMR, respectively [4]. Additional data suggest that dd-cfDNA reflects allograft injury as a continuum rather than at a discrete cut-off, as evidenced by the findings of Stites et al [11] that KTRs with elevated dd-cfDNA (≥ 0.5%) level and a concurrent Banff criteria pathologic diagnosis of borderline or TCMR1A experienced worse clinical outcomes including larger declines in estimated glomerular filtration rate (eGFR) and higher incidences of ABMR and recurrent TCMR than cohorts with dd-cfDNA less than 0.5%.
In this report, we questioned whether lack of direct face-to-face medical care during a “shelter in place” strategy would result in a decline in overall allograft stability within the KTR population, as evidenced by trends in dd-cfDNA level collected and measured during the COVID-19 pandemic.
Methods
At the discretion of individual transplant centers, plasma samples in StreckTM cell-free BCT tubes were obtained either by routine facility-based phlebotomy or by remote home phlebotomy for US living and deceased-donor KTRs. Samples were analyzed for dd-cfDNA (AlloSure®) at a Clinical Laboratory Improvement Amendment (CLIA)-certified laboratory (CareDx; Brisbane, CA, USA) and additionally submitted for routine post-transplant laboratory studies (metabolic profile, complete blood count, tacrolimus trough level, etc.). Informed consent was obtained from patients who were scheduled for home phlebotomy, and appropriate personal protective equipment was provided and utilized by both phlebotomists and patients during all encounters.
Between January 6, 2020 and May 25, 2020, 21 weekly aggregated cohorts of dd-cfDNA samples were analyzed and categorized by week into three clinically-relevant strata: ≥ 1.0%, 0.5% to 1.0%, and < 0.5%. As a reference, in a healthy and stable KT population, approximately 97% of samples show a level less than the threshold of 1.0% dd-cfDNA [8]. The dd-cfDNA-categorized cohorts were analyzed for statistical normality by single-sample Kolmogorov-Smirnov (K-S) test in which P > 0.050 indicates normal distribution. Linear regression was performed for each dd-cfDNA cohort as number of samples versus time and mean slope of COIVID-19 spread, hospitalizations, and deaths in the US [12]. To assess findings from an epicenter of COVID-19 infection, New York City Metropolitan area was analyzed independently by similar methods.
Results
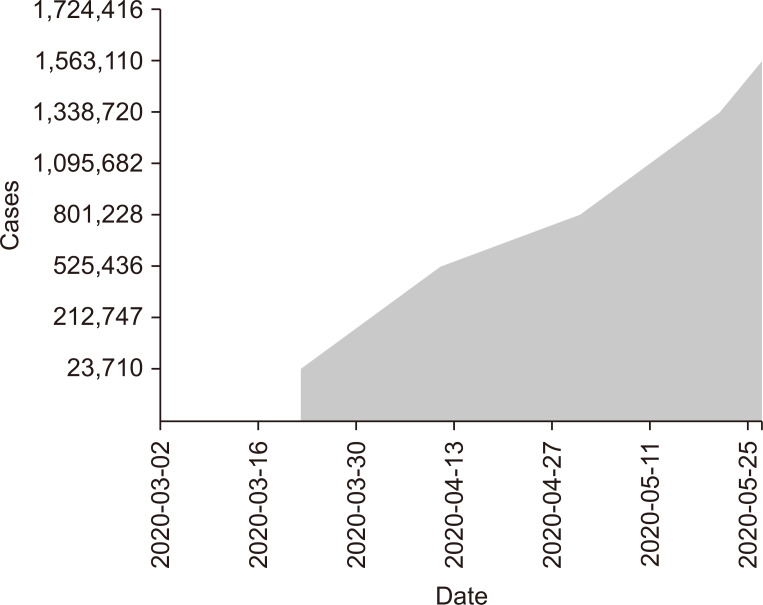
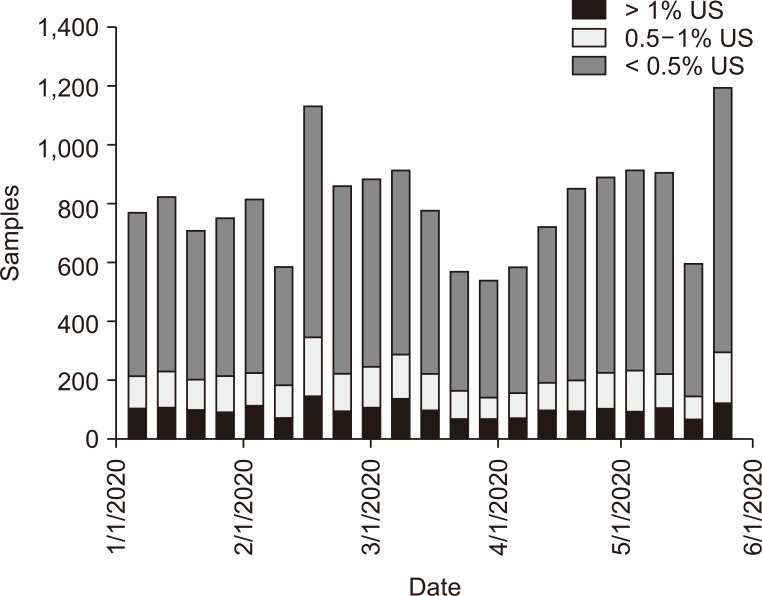
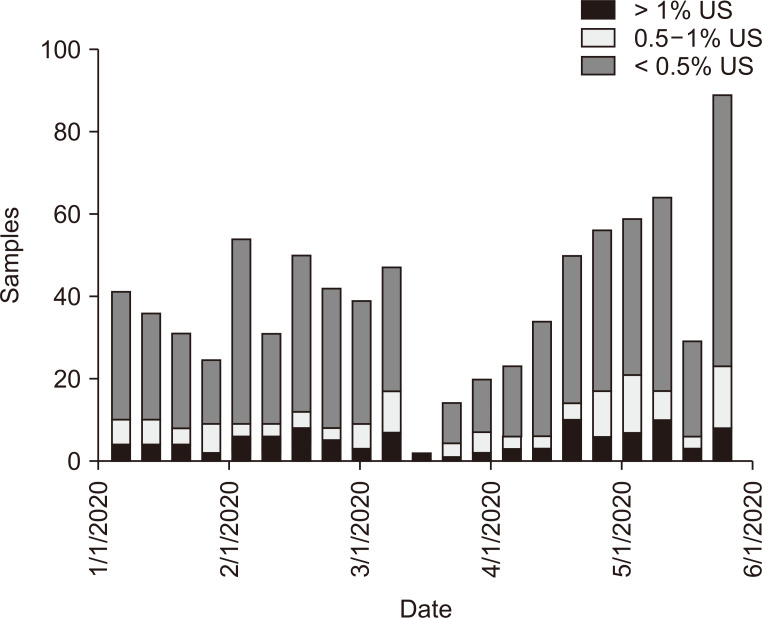
As depicted in Fig. 1, during the collection period, cumulative US cases of COVID-19 exceeded 1.7 million. The highest number of total US submitted dd-cfDNA samples occurred during the weeks of February 10-16, 2020, and May 17-24, 2020 (Fig. 2). The nadir for number of submitted dd-cfDNA samples in the US occurred the week of March 25-31, 2020, while the nadir occurred 2 to 3 weeks earlier in Metropolitan New York City (NYC), on March 7-14, 2020. That nadir in submitted sample volumes occurred prior to implementation of remote phlebotomy in Metropolitan NYC (Fig. 3). Nationally, during the study timeframe, over 11,000 dd-cfDNA samples from 150 KT centers were transitioned from standard to home remote phlebotomy, representing 67% of total samples.
Figure 1. United States (US) coronavirus disease 2019 (COVID-19) infection cases.
Total COVID-19 cases in the US by weekly World Health Organization/Centers for Disease Control and Prevention reports [12].
Figure 2. Donor-derived, cell-free DNA (dd-cfDNA) cohorts in United States (US).
Cohorts of dd-cfDNA plasma number of samples from the US that were categorized as “low risk for rejection” (< 0.5%) and of indeterminate risk with 0.5% to 1.0% and > 1.0% categories during the course of coronavirus disease 2019 (COVID-19). The March-April nadir in total number of samples occurred prior to implementation of home remote phlebotomy.
Figure 3. Donor-derived, cell-free DNA (dd-cfDNA) cohorts in New York City Metropolitan.
Cohorts of dd-cfDNA plasma weekly samples from New York City Metropolitan area categorized as “low risk for rejection” (< 0.5%) and indeterminate risk of rejection with 0.5% to 1.0% and > 1.0% categories, respectively, during the course of coronavirus disease 2019 (COVID-19). The March-April nadir in total number of samples occurred prior to implementation of home remote phlebotomy.
Linearized slope for each dd-cfDNA cohort category by week versus total time are presented in Table 1. There was no statistical difference in slopes for either 0.5% to 1.0% or ≥ 1.0% group compared to the < 0.5% cohort (P < 0.050), while the negative slopes for the indeterminate cohorts that are associated with higher risk for active acute rejection (AR) suggested decreased prevalence over the progressive course of COVID-19. The K-S statistic for each cohort over the 21 weekly-aggregated samples demonstrated normality for each distribution (P > 0.050). Table 2 shows that only the 0.5% to 1% cohort failed normality testing in sub-set analysis for Metropolitan NYC (P = 0.036). The linearized slopes for indeterminate risk of AR dd-cfDNA categories were also negative during progression of the COVID-19 pandemic, representing a declining prevalence for these categories. This implies that the KT population monitored with dd-cfDNA during this COVID-19 timeframe did not experience increased prevalence of indeterminate or higher risk for AR dd-cfDNA level.
Table 1.
United States donor-derived cell-free DNA levels in kidney transplant recipients
| US samples | > 1% | 0.5% to 1% | < 0.5% |
|---|---|---|---|
| No. weeks | 21 | 21 | 21 |
| Mean no. samples | 94.9 (11.9%) | 119.7 (15.0%) | 584.3 (73.1%) |
| SD | 21.4 | 30.0 | 129.2 |
| Regression slope | -0.31 | -0.12 | +0.08 |
| K-S | 0.16 | 0.17 | 0.12 |
United States donor-derived, cell-free DNA weekly grouped samples categorized as cohorts. Linear regression slopes (samples/week) for individual cohort numbers of samples versus time. Normality was assessed with Kolmogorov-Smirnov (K-S) (P < 0.05), and all 3 cohorts were normally distributed.
SD, standard deviation; US, United States.
Table 2.
New York City Metropolitan area donor-derived cell-free DNA levels in kidney transplant recipients
| NYC Metropolitan | > 1% | 0.5% to 1% | < 0.5% |
|---|---|---|---|
| No. weeks | 21 | 21 | 21 |
| Mean no. samples | 4.9 (12.3%) | 5.7 (14.4%) | 29.1 (73.3%) |
| SD | 2.9 | 5.7 | 14.5 |
| Regression Slope | -4.97 | -4.13 | +0.93 |
| K-S | 0.16 | 0.19* | 0.10 |
New York City Metropolitan donor-derived, cell-free DNA samples categorized as cohorts. Linear regression slopes (samples/week) for individual cohort number of samples versus time. Normality was assessed with Kolmogorov-Smirnov (K-S) (P > 0.05), and only the 0.5% to 1% cohort did not show normality, due to small sample size.
SD, standard deviation; NYC, New York City.
*P = 0.036.
Discussion
The vulnerability of KTRs to infection has prompted development of predictive models of likelihood of infection based on novel biomarkers of immune function and allograft integrity [13]. However, in the presence of a novel viral pandemic with unknown biological behavior, prior experience and modeling are of limited value. In the absence of efficacious therapy, prophylaxis, or vaccination, the global transplant community urgently gathered data and developed preliminary guidelines recommending “sheltering in place” as the primary form of prevention of COVID-19 for this vulnerable population [3,14]. This creates a paradox for transplant recipients who must interact with health care providers for routine laboratory studies and maintain follow-up visits to monitor allograft health and rejection but who are vulnerable and advised to “shelter in place” and avoid contact with others. In that setting, remote monitoring combining home phlebotomy and tele-health clinician visits provides a potentially valuable solution. Additionally, remote home phlebotomy with dd-cfDNA surveillance offers a further advantage as a biomarker of potential “allograft tissue injury” that can help diagnose underlying AR [4,5,8,15].
Our analyses focused on grouped weekly trends in the US during the initial phases of the COVID-19 pandemic. This analysis did not indicate an increased total number or percentage of dd-cfDNA samples in the indeterminate dd-cfDNA range of 0.5% to 1% or > 1%. Those ranges are categorized as indicating an increased “potential risk of AR” and would typically warrant closer monitoring or allograft biopsy [16]. Linear regression for these cohorts versus time during the 21-week collection period demonstrated negative slopes for these categories. A similar trend analysis for the NYC Metropolitan area did not demonstrate increase in indeterminate “risk of AR” cohorts. The decreased total numbers of samples during the March 2020 weekly cohorts likely reflected the “shelter in place” patient directive, prior to implementation of a remote home phlebotomy solution for dd-cfDNA surveillance. Further, COVID-19 infection in KT patients has been associated with increased risks for acute renal failure and hemodialysis, either by direct viral-mediated processes including collapsing glomerulopathy or indirect acute kidney injury [17,18]. These effects were not expected to impact dd-cfDNA level in our population of stable, outpatient KTRs. Finally, the consistently high proportion of samples throughout this timeframe categorized as “low risk AR” (cfDNA < 0.5%) should offer reassurance of quiescence based on a validated high NPV [4,8].
Limitations to this study relate to the de-identified nature of the data set without associated clinical or pathologic correlations. Nevertheless, the dd-cfDNA cohort weekly distributions during the timeframe of study were comparable to those of previous KT population data [4]. Our observational analysis of KT cohorts based on dd-cfDNA level category did not stratify important data elements such as time since transplantation, pre-transplant panel reactive antibodies, eGFR, race, KDPI, or deceased versus living-donor source. Nevertheless, our analysis of the distribution of dd-cfDNA as cohorts represented a robust spectrum of KT population experience. Future studies are needed to assess patient demographics and individual risk for allograft rejection utilizing this remote surveillance type protocol. There will be a sub-study during final analyses for the Kidney Allograft Outcomes AlloSure® Registry (KOAR) (Clinical Trials.gov; NCT03326076).
Our analysis demonstrates the feasibility of remote home phlebotomy for both routine post-KT lab values and biomarker surveillance with implementation of dd-cfDNA in conjunction with a tele-health platform during the COVID-19 pandemic. During this study timeframe, over 11,000 patients (67%) from 150 KT centers had been transitioned from standard facility-based to home remote phlebotomy. In this observational study of US KT recipients, approximately 73% of samples had a low dd-cfDNA level (dd-cfDNA < 0.5%), indicating a probable underlying low risk for AR. Those results provided reassurance for the “shelter in place” strategy in light of the high NPV of dd-cfDNA for presence of underlying AR [4]. Only 15% of the samples analyzed had dd-cfDNA level of 0.5% to 1%. These results are indicative of an indeterminate risk of underlying AR. Depending on clinical circumstances, these recipients may, based on those dd-cfDNA results, be deemed appropriate for serial monitoring to evaluate for a trend. A dd-cfDNA threshold < 1.0% has been associated with NPV for ACR (Banff criteria ≥ TCMR 1B) and ABMR of 84% and 96%, respectively [4]. Surveillance of longitudinal individual patient trends in dd-cfDNA can offer additional insights for potential risk of AR utilizing the reference change value of 61% for consideration of significance [8]. Therefore, approximately 88% of these samples would not a priori have prompted further evaluation or intervention.
During these challenging times, a surveillance strategy that implements a combination of tele-health with collection of both routine labs and dd-cfDNA level via home phlebotomy may allow avoidance of contagion exposure for a majority of KTRs and serve to assuage patient anxiety regarding the status of allograft health. However, this novel paradigm requires further validation in multi-center prospective clinical trials with robust outcomes data.
Footnotes
Conflicts of interest
All authors have no conflicts of interest to declare.
Authors’ contributions
David J. Ross and Steven R. Potter participated in the data collection and wrote the initial draft of manuscript. David J. Ross, Steven R. Potter, Randall Hinojosa, Dan O’Brien, and Cliff D. Miles participated in the study design. Statistical analysis performed by David J. Ross, Steven R. Potter, and Dan O’Brien. David J. Ross, Steven R. Potter, Randall Hinojosa, and Cliff D. Miles participated in the conception, analysis, and interpretation of data. All participated in manuscript review, edits and approval of final manuscript.
References
- 1.Yang X, Yu Y, Xu J, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart A, Smith JM, Skeans MA, et al. OPTN/SRTR 2018 annual data report: kidney. Am J Transplant. 2020;20 Suppl s1:20–130. doi: 10.1111/ajt.15672. [DOI] [PubMed] [Google Scholar]
- 3.Akalin E, Azzi Y, Bartash R, et al. Covid-19 and kidney transplantation. N Engl J Med. 2020;382:2475–2477. doi: 10.1056/NEJMc2011117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloom RD, Bromberg JS, Poggio ED, et al. Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators, author. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–2232. doi: 10.1681/ASN.2016091034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gielis EM, Ledeganck KJ, Dendooven A, et al. The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant. 2020;35:714–721. doi: 10.1093/ndt/gfz091. [DOI] [PubMed] [Google Scholar]
- 6.Altuğ Y, Liang N, Ram R, et al. Analytical validation of a single-nucleotide polymorphism-based donor-derived cell-free DNA assay for detecting rejection in kidney transplant patients. Transplantation. 2019;103:2657–2665. doi: 10.1097/TP.0000000000002665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grskovic M, Hiller DJ, Eubank LA, et al. Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn. 2016;18:890–902. doi: 10.1016/j.jmoldx.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Bromberg JS, Brennan DC, Poggio E, et al. Biological variation of donor-derived cell-free DNA in renal transplant recipients: clinical implications. J Appl Lab Med. 2017;2:309–321. doi: 10.1373/jalm.2016.022731. [DOI] [PubMed] [Google Scholar]
- 9.Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: a prospective multicenter study. Am J Transplant. 2019;19:2889–2899. doi: 10.1111/ajt.15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loupy A, Haas M, Solez K, et al. The Banff 2015 kidney meeting report: current challenges in rejection classification and prospects for adopting molecular pathology. Am J Transplant. 2017;17:28–41. doi: 10.1111/ajt.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020;20:2491–2498. doi: 10.1111/ajt.15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention, author. [cited 2020 Jun 1];COVID-19 infection prevalence U.S. [Internet] c2020 Available from: https://covid.cdc.gov/covid-data-tracker/#trends_dailytrendscases .
- 13.Fernández-Ruiz M, López-Medrano F, Aguado JM. Predictive tools to determine risk of infection in kidney transplant recipients. Expert Rev Anti Infect Ther. 2020;18:423–441. doi: 10.1080/14787210.2020.1733976. [DOI] [PubMed] [Google Scholar]
- 14.Samavat S, Nafar M, Firozan A, et al. COVID-19 rapid guideline in kidney transplant recipients. Iran J Kidney Dis. 2020;14:231–234. [PubMed] [Google Scholar]
- 15.Jordan SC, Bunnapradist S, Bromberg JS, et al. Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct. 2018;4:e379. doi: 10.1097/TXD.0000000000000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oellerich M, Christenson RH, Beck J, et al. Donor-derived cell-free DNA testing in solid organ transplantation: a value proposition. J Appl Lab Med. 2020;5:993–1004. doi: 10.1093/jalm/jfaa062. [DOI] [PubMed] [Google Scholar]
- 17.Durvasula R, Wellington T, McNamara E, Watnick S. COVID-19 and kidney failure in the acute care setting: our experience from Seattle. Am J Kidney Dis. 2020;76:4–6. doi: 10.1053/j.ajkd.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasr SH, Kopp JB. COVID-19-associated collapsing glomerulopathy: an emerging entity. Kidney Int Rep. 2020;5:759–761. doi: 10.1016/j.ekir.2020.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]